Abstract
The EcoGene database provides a set of gene and protein sequences derived from the genome sequence of Escherichia coli K-12. EcoGene is a source of re-annotated sequences for the SWISS-PROT and Colibri databases. EcoGene is used for genetic and physical map compilations in collaboration with the Coli Genetic Stock Center. The EcoGene12 release includes 4293 genes. EcoGene12 differs from the GenBank annotation of the complete genome sequence in several ways, including (i) the revision of 706 predicted or confirmed gene start sites, (ii) the correction or hypothetical reconstruction of 61 frame-shifts caused by either sequence error or mutation, (iii) the reconstruction of 14 protein sequences interrupted by the insertion of IS elements, and (iv) pre-dictions that 92 genes are partially deleted gene fragments. A literature survey identified 717 proteins whose N-terminal amino acids have been verified by sequencing. 12 446 cross-references to 6835 literature citations and abstracts are provided. EcoGene is accessible at a new website: http://bmb.med.miami. edu/EcoGene/EcoWeb . Users can search and retrieve individual EcoGene GenePages or they can download large datasets for incorporation into database management systems, facilitating various genome-scale computational and functional analyses.
INTRODUCTION
EcoGene originated as a collection of Escherichia coli K-12 gene and protein sequences derived from EcoSeq, a set of DNA sequence contigs assembled in the pre-genomic era from hundreds of individual E.coli GenBank sequences (1–3). New genes were identified in unannotated regions of the EcoSeq contigs using both protein sequence similarity searches and the prediction of protein coding potential using GeneMark (4). These two methods were also used to review and revise the starts of coding region intervals, as well as to identify potential frameshift errors. Deciding which of several possible start codons to choose was often aided by the assessment of potential ribosome binding sites (RBSs), including a relative ranking of RBSs based on their estimated individual contributions to the overall information content in a collection of E.coli RBSs (5).
EcoSeq was superseded by the complete genome sequence of the standard E.coli laboratory strain MG1655, although the EcoSeq7 set of DNA sequence melds, a mosaic containing GenBank DNA sequences from a variety of E.coli K-12 strains, was helpful to the genome sequencing project (6). The pre-genome era EcoGene revisions and the systematic open reading frame (ORF) nomenclature are reflected in the annotation of the complete genome sequence since they were present in SWISS-PROT release 34, the protein database used for similarity searches of the complete genome sequence (6). The EcoGene database has been reconstituted using the complete genome sequence. All the previous methods of gene annotation review and revision were applied systematically to the complete genome sequence and several new methods were adopted (unpublished results). EcoGene is an alternative representation of the gene and protein features of the E.coli genome sequence and is under constant review and revision. Many computational and functional research projects utilize the gene annotations of the E.coli genome sequence. EcoGene provides an alternative view of the E.coli genome sequence gene and protein annotation and should be useful for the design of E.coli research and database projects.
EcoGene COLLABORATORS
In collaboration with A. Bairoch, all EcoGene protein sequence revisions become part of the SWISS-PROT database, with cross-references to EcoGene EG accession numbers (7). SWISS-PROT has adopted the use of EcoGene’s systematic nomenclature devised for E.coli ORFs of unknown function, the y-gene nomenclature (8). Functionally characterized E.coli genes often have synonymous gene names. The choice of the primary gene name used in EcoGene is decided in consultation with Mary Berlyn of the Coli Genetic Stock Center. This is part of a genome map collaboration to produce well-coordinated E.coli genetic maps. (1,8,9). The EcoGene model has also been applied to Salmonella typhimurium to create StyGene in collaboration with K. Sanderson of the Salmonella Genetic Stock Center (10). The systematic y-gene nomenclature for uncharacterized genes has been extended to include Salmonella genes. The EcoGene data populates the WWW-based Colibri database management, graphic display and query software developed by A. Danchin and colleagues at the Institut Pasteur, bioweb.pasteur.fr/GenoList/Colibri (11). Other databases can utilize the EcoGene accession numbers, as has been done within the EcoCyc database (12), to facilitate E.coli database linkages using WWW hyperlinks.
EcoGene12 TABLES AND EcoWeb
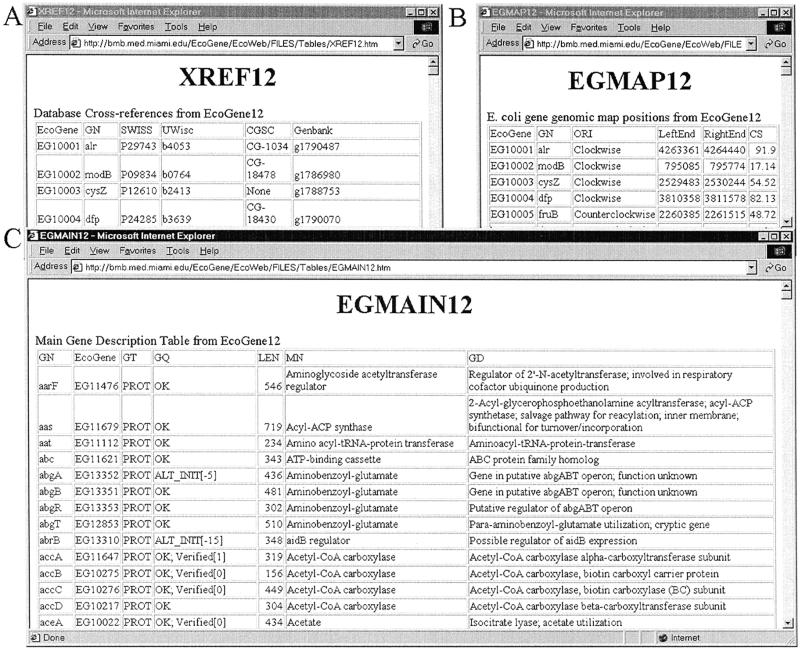
Tables of EcoGene12 data are available from a new website, http://bmb.med.miami.edu/EcoGene/EcoWeb . Figure 1 shows excerpts from three of these database tables. The EcoGene tables utilize the EG accession numbers as key fields. XREF12 (Fig. 1A) cross-references gene and protein accession numbers from the EcoGene, SWISS-PROT, GenBank and the Coli Genetic Stock Center databases, as well as the E.coli genome project ‘b’ numbers.
Figure 1.
Three tables of EcoGene data. (A) XREF12 contains cross-references of EcoGene EG accession numbers (EcoGene) and gene names (GN) to E.coli gene and protein record accession numbers in the SWISS-PROT, Coli Genetic Stock Center (CGSC) and GenBank databases, as well as to the University of Wisconsin ‘b’ numbers. (B) EGMAP12 contains genomic sequence and map locations for E.coli genes. EcoGene, EG accession numbers; GN, gene name; ORI, orientation of transcription; LeftEnd, the counterclockwise end of a gene; RightEnd, the clockwise end of a gene; CS, the centisome (= % = minute) map position of a gene, derived by dividing the LeftEnd basepair position by the length of the genome sequence (4 639 221 bp). (C) EGMAIN12 contains descriptive information about E.coli genes: the primary gene name (GN), the gene name mnemonic (MN), the gene description (GD), the gene type (GT; PROT or RNA), the gene product sequence length (LEN) and gene quality (GQ) fields.
Figure 1B shows some of EGMAP12, which has the genomic map positions, in both base pairs and centisomes, and genomic orientations of E.coli genes. Other EcoMap tables with the genomic basepair coordinates indicating the locations of IS elements (ISMAP12), DNA sequence-derived restriction sites (RSMAP12), the Kohara lambda clone library (KOHMAP12), and GenBank records AE000111–AE000510 (BGBMAP12), individual records derived from the complete genomic sequence record GenBank U00096, are also available. These tables were used to create EcoMap12 (see below).
Figure 1C depicts a portion of EGMAIN12 with primary gene name, the gene name mnemonic, gene description, gene type (protein or RNA), gene product sequence length (number of amino acids or RNA nucleotides), and gene quality fields. Gene quality field indicates problems or changes with regards to the ability to conceptually translate the gene intervals in GenBank U00096. Gene quality options are described in Table 1.
Table 1. EcoGene gene quality types.
| Quality type | No.a | Descriptionb |
|---|---|---|
| OK | 3482 | Unchanged from the gene interval in U00096, version M54 |
| FRM | 42 | A hypothetical frameshift reconstruction |
| STP | 22 | An in-frame stop codon is present |
| CORR | 19 | An experimental sequence correction of a frameshift mutation |
| ALT_INIT[number] | 706 | An alternative start codon is used (number of codons added or removed relative to GenBank U00096) |
| VERIFIED[number] | 717 | N-terminal protein sequence is known {number of amino acid residues, if any, that are removed during processing) |
| PART | 92 | Partial gene, a segment of the gene appears to be deleted |
| MUTANT | 6 | Strain MG1655 is known to have frameshift or in-frame stop codon mutations in these genes |
| EXCEPT | 5 | The gene has a translational exception, i.e., a programmed frameshift, a selenocysteine-encoding UGA codon, or an AUU start codon |
aThe number of genes of this type in EcoGene12.
bComplete gene quality descriptions are available at http://bmb.med.miami.edu/EcoGene/EcoWeb/Translation_Status.htm
Various other EcoGene12 tables are available, including SYN12, a table of gene name synonyms, and EGREF12, a literature citation table with 12 446 cross-references linking EG numbers to 6835 different MEDLINE unique identifiers. In addition, a web page listing the publications and GenBank records from several research laboratories that participated in the systematic sequencing of the E.coli K-12 genome can be found at http://bmb.med.miami.edu/EcoGene/EcoWeb/GenSeqRef.htm
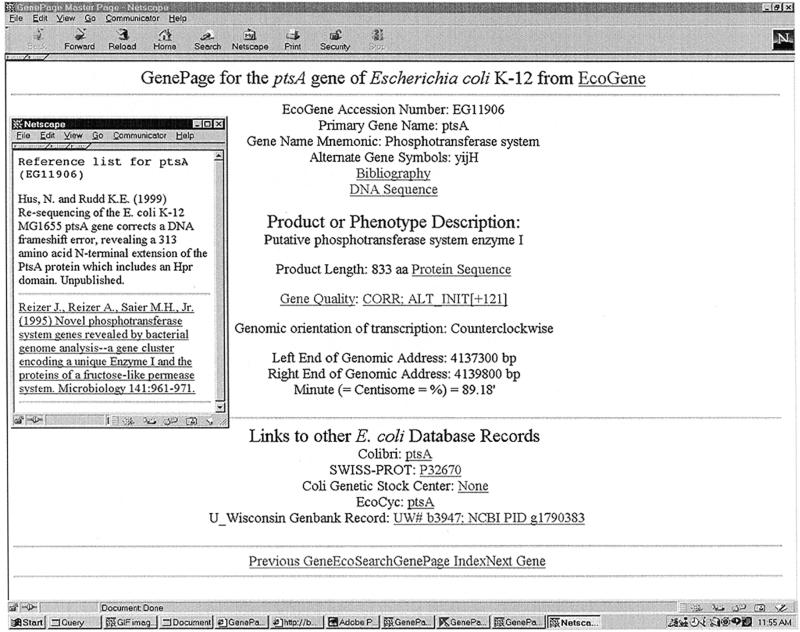
Database reports for each individual E.coli gene are derived from the EcoGene tables and are available as web pages, called GenePages. EcoWeb GenePages can be accessed by browsing an alphabetical index of all genes or by using an indexed search engine. A sample GenePage is shown in Figure 2. The GenePages include hyperlinks to the corresponding gene records in Colibri, SWISS-PROT, EcoCyc and the Coli Genetic Stock Center databases. These databases contain additional information about E.coli genes and proteins.
Figure 2.
The EcoWeb GenePage for the E.coli ptsA gene. The GenePage contains descriptive and genomic position information, hyperlinks to DNA and protein sequences and hyperlinks to gene records in other databases. The Gene Quality information indicates that a frameshift sequencing error has been corrected and that the N-terminus of the protein has been extended by 313 amino acids. The inset shows the Bibliography page for ptsA.
GENE AND PROTEIN SEQUENCES
EcoGene gene sequences start with the first base of the start codon and stop with the last base of the stop codon. EcoGene protein sequences are the primary translation products of the gene sequence. The protein sequences derived from EcoGene, collectively referred to as EcoProt, share the use of the EG number as an identifier. Individual FASTA format DNA sequence files and protein sequence files are named using the EG number followed by the extensions ‘.seq’ and ‘.aas’, respectively. EcoGene and EcoProt sequences can be downloaded as complete sets from EcoWeb as either a single sequence library file format or as a UNIX tarfile containing a directory of individual sequence files. EcoWeb pages of each individual DNA and protein sequence are also linked to the individual GenePages.
The individual sequence files may differ from the complete genome sequence in GenBank U00096 if a sequence correction or hypothetical reconstruction of a frameshift has been implemented. The complete genome sequence file of GenBank U00096 has not been altered in EcoGene, thus preserving the genomic base pair coordinates of genes, which are often referred to in publications and other databases. Thus, the genomic position intervals of apparently frameshifted genes are not multiples of three basepairs and one must look to the individual EcoGene sequence files for the EcoGene version of those genes. The number of EcoGene entries that have been modified in various ways are given in Table 1.
EcoMap12 IN ADOBE ACROBAT PDF FORMAT
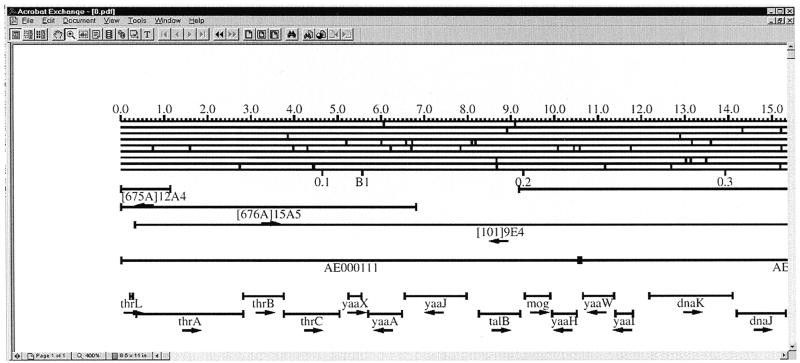
The WWW version of EcoGene12 is accompanied by an updated version of the EcoMap physical and gene map graphic representation. The format is identical to the published version of EcoMap10 (8), except that the EcoMap12 Postscript files have been converted to Adobe Acrobat PDF format. The EcoGene12 and EcoMap12 datasets are converted into Postscript files using the PrintMap C program (3). These Postscript files are computer programs that implement the Plasmid Description Language Postscript definitions created by Craig Werner (3). Figure 3 depicts a portion of EcoMap12 in PDF format viewed at 400% magnification using Adobe Acrobat.
Figure 3.
A portion of the EcoMap12 Adobe Acrobat PDF format genome map file. The format is identical to that from EcoMap10 in edition 10 of the E.coli linkage map (8). DNA sequence derived restriction sites (top line to bottom line) are BamHI, HindIII, EcoRI, EcoRV, BglI, KpnI, PtsI and PvuII. Also depicted are kilobase coordinates, centisomes, Kohara clone map positions, GenBank MG1655 genome sequence record alignments, gene positions and gene orientations.
The periodic release of updated versions of EcoMap and EcoGene tables, GenePage reports and map figures through EcoWeb represents a transition from the publication of new versions of EcoMap in a print journal (8) to electronic publication in digital formats. Electronic publication has some advantages over journal publication for both the author and the research community, including the ability to provide updated versions frequently, keeping the map information as current as possible. CD-ROM versions or hard copy printouts of EcoMap12 can be made available upon request to anyone without reliable Internet access.
PUBLIC USE OF ECOGENE DATASETS
Permission to use the EcoGene and EcoMap datasets for genome analysis, for incorporation into existing databases, or for the development of new genome database management systems is not required. The use of EcoGene12 and EcoMap12 as data sources should be prominently noted and referenced in any database publication, implementation or distribution. GenBank U00096 and Blattner et al., 1997 (6) also should be cited as primary data sources if any EcoGene DNA sequence-based datasets are utilized. Any extensive utilization of the gene description, mnemonic, gene name synonym or the literature citations from EcoGene tables should be accompanied by an acknowledgment of Mary Berlyn of the Coli Genetic Stock Center (9) as an original source for those datasets.
EcoGene12 and EcoMap12, like all existing genome datasets, contain different types of unrecognized errors. No guarantees with regard to data integrity, consistency or accuracy are stated or implied. All changes in the sequence data not specifically noted as experimentally derived corrections obtained from the biomedical literature or personal communications should be assumed to be hypothetical reconstructions. These reconstructions are not based on experimental evidence other than computer analysis-based predictions. Feedback from users is welcome in order to improve the quality of the EcoGene datasets in future releases. Provisions can be made to include unpublished results and the comments of EcoGene users as part of the GenePage bibliographies.
Acknowledgments
ACKNOWLEDGEMENTS
Special thanks are given to Mary Berlyn, Antoine Danchin, Amos Bairoch and their associates for years of compilation, analysis and revision of E.coli datasets. K.E.R. appreciates their continuing encouragement and collaboration. EcoGene12 would not be possible without the diligent work of Fred Blattner and the members of his laboratory to produce the complete genome sequence of E.coli K-12 MG1655. K.E.R. is pleased to acknowledge many helpful discussions and collaborative research with Nir Hus, Rick Mitchell, Mark DelCampo, Gerry Bouffard, Yuhong Zuo, Amos Bairoch, Anne-Lise Veuthey, Ivan Moszer, Tom Schneider, Ryan Shultzaberger, John Clark, Webb Miller, Karl Sirotkin, Russell Bishop, Mark Emmett, Arun Malhotra and Gabrielle Redfern. EcoGene12 was made possible by a Markey Foundation grant to the Department of Biochemistry and Molecular Biology, University of Miami School of Medicine.
REFERENCES
- 1.Berlyn M.B., Low,K.B. and Rudd,K.E. (1996) In Neidhardt,F.C., Curtiss,R., Ingraham,J.L., Lin,E.C.C., Low,K.B., Magasanik,B., Reznikoff,W.S., Riley,M., Schaechter,M. and Umbarger,H.E. (eds), Escherichia coli and Salmonella: Cellular and Molecular Biology. ASM Press, Washington, DC, Vol. 2, pp. 1715–1902.
- 2.Rudd K.E. (1993) ASM News, 59, 335–341. [Google Scholar]
- 3.Rudd K.E., Miller,W., Werner,C., Ostell,J., Tolstoshev,C. and Satterfield,S.G. (1991) Nucleic Acids Res., 19, 637–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borodovsky M., Koonin,E.V. and Rudd,K.E. (1994) Trends Biochem. Sci., 19, 309–313. [DOI] [PubMed] [Google Scholar]
- 5.Rudd K.E. and Schneider,T.D. (1992) In Miller,J. (ed.), A Short Course in Bacterial Genetics; a Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. Cold Spring Harbor Press, Cold Spring Harbor, NY, pp. 17.19–17.45.
- 6.Blattner F.R., Plunkett,G., Bloch,C.A., Perna,N.T., Burland,V., Riley,M., Collado-Vides,J., Glasner,J.D., Rode,C.K., Mayhew,G.F., Gregor,J., Davis,N.W., Kirkpatrick,H.A., Goeden,M.A., Rose,D.J., Mau,B. and Shao,Y. (1997) Science, 277, 1453–1462. [DOI] [PubMed] [Google Scholar]
- 7.Bairoch A. and Apweiler,R. (1999) Nucleic Acids Res., 27, 49–54. Updated article in this issue: Nucleic Acids Res. (2000), 28, 45–48.9847139 [Google Scholar]
- 8.Rudd K.E. (1998) Microbiol. Mol. Biol. Rev., 62, 985–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berlyn M.K.B. (1998) Microbiol. Mol. Biol. Rev., 62, 814–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanderson K.E., Hessel,A. and Rudd,K.E. (1995) Microbiol. Rev., 59, 241–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Medigue C., Viari,A., Henaut,A. and Danchin,A. (1993) Microbiol. Rev., 57, 623–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karp P.D., Riley,M., Paley,S.M., Pellegrini-Toole,A. and Krummenacker,M. (1999) Nucleic Acids Res., 27, 55–58. Updated article in this issue: Nucleic Acids Res. (2000), 28, 56–59.9847140 [Google Scholar]