Abstract
REBASE is a comprehensive database of information about restriction enzymes and related proteins. It contains published and unpublished references, recognition and cleavage sites, isoschizomers, commercial availability, methylation sensitivity, crystal and sequence data. DNA methyltransferases, homing endonucleases, nicking enzymes, specificity subunits and control proteins are also included. Most recently, putative DNA methyltransferases and restriction enzymes, as predicted from analysis of genomic sequences, are also listed. The data is distributed via Email, ftp (ftp.neb.com), and the Web (http://rebase.neb.com ).
INTRODUCTION
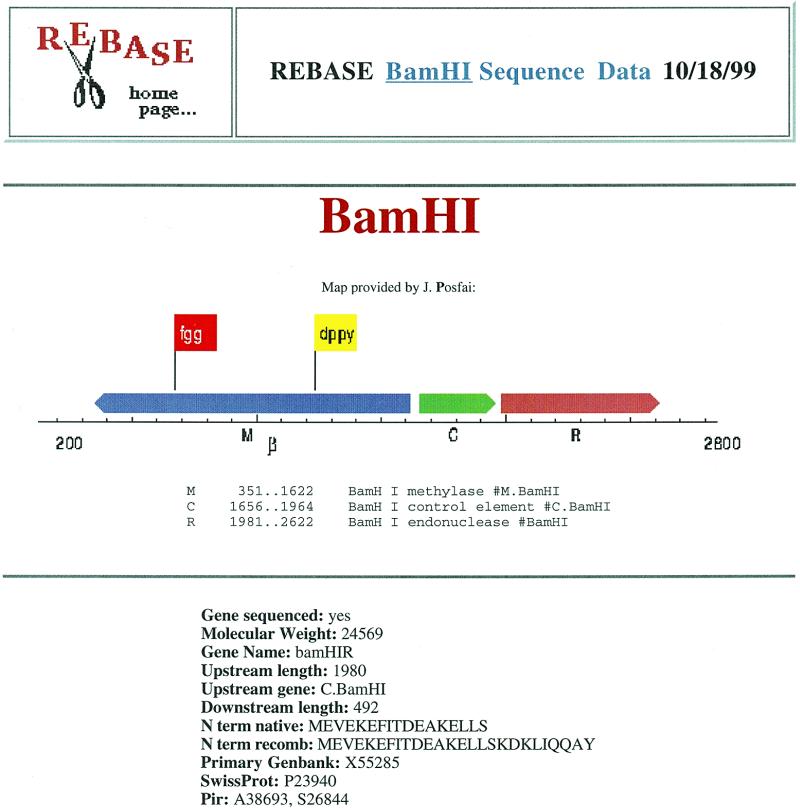
REBASE has undergone much growth since the 1999 NAR Database Issue (1). We have more data, more kinds of data and more ways to access the data. In addition to restriction enzymes, methylases and homing endonucleases, REBASE now includes information about other types of related enzymes: nicking enzymes, specificity subunits of the Type I enzymes, control proteins and methyl-directed restriction enzymes. With the explosion of bacterial and archaeal genome sequences that are appearing in GenBank we now include putative enzymes predicted from these sequences. Such enzymes are given names that resemble those of normal restriction enzymes [using the conventions of Smith and Nathans (2)], but with the suffix ‘P’ added to indicate their unproven status. The REBASE web site provides details of whatever information is known about each enzyme, such as commercial availability, sequence data, crystal structures, cleavage sites, recognition sequences, isoschizomers, growth temperatures and methylation sensitivity. During this last year we have increased our focus on the genes that encode restriction systems. Among other features, we now present schematics showing the organization of restriction system genes (Fig. 1). Further analysis of these gene sequences will appear in the near future.
Figure 1.
The genetic organization of the BamHI restriction–modification system (4). The red and yellow flags above the M.BamHI methyltransferase gene indicate the locations of the two sequence motifs characteristic of amino-methyltransferases.
There are currently 3154 restriction enzymes in REBASE. 139 well-characterized restriction enzymes have been added since the last review (1). These include four new Type II specificities shown in Table 1. Of the 3100 Type II restriction enzymes, 506 are commercially available (189 distinct specificities of the 225 total specificities known). In addition, 20 DNA methyltransferases and seven homing endonucleases are commercially available. We currently have 5604 references in REBASE (journal and book publications, patents and unpublished observations). These are complete with abstracts when available. References are provided for every enzyme.
Table 1. New Type II restriction enzyme specificities.
| Enzymea | Recognition sequenceb | References |
|---|---|---|
| Hpy99I | CGWCG | Xu,Q., Morgan,R., Blaser,M. (unpublished) |
| MjaIVc | GTNNAC | Morgan,R., Posfai,J., Patti,J., Roberts,R.J. (unpublished; 3) |
| Sse232I | CG↓CCGGCG | Nomura,Y., Ishizaki,I., Oshima,K., Kato,I. (Japanese Patent, JP10127279) |
| UbaPI | CGAACG | Padegimiene,E., Petrusyte,M., Capskaja,L., Kiuduliene,L., Butkus,V., Janulaitis,A. (unpublished) |
aThe endonucleases are named in accordance with the proposal of Smith and Nathans (2).
bCleavage sites are indicated as ↓ when cleavage is at the identical position on both strands. W = A or T; N = any base.
cThree isoschizomers have also been found in strains of Helicobacter pylori: Hpy8I (Vitkute,J., Trinkunaite,L., Kiuduliene,E., Butkus,V., Janulaitis,A., unpublished); HpyBII (5); Hpy166II (Xu,Q., Morgan,R., Blaser,M., unpublished).
REBASE has its own dedicated web server (http://rebase. neb.com ). Many new features are available. Searches are now launched directly from the homepage. Users select the search category via menu: enzyme name, recognition sequence, author, journal, citation, GenBank number, microorganism, etc. The search results are tabular and are provided with links leading to further details.
The new REBASE Lists icon leads to tables of specialized information: crystal data, cloned/sequenced enzymes, N-terminal protein sequence data, Newest Enzymes, Type IIs enzymes, enzymes grouped by type (Type I, Type II, Type III, methylases, homing, nicking,...), etc. Additionally, the REBASE Lists page offers new ways to navigate through enzyme homepages using the acronyms. There is also a table of all prototypes and neoschizomers, with an optional sort order by either enzyme name or recognition sequence.
The NEWS page now includes a link to back issues of previous editions of the NEWS. The References icon allows users to choose to view recently added references. The supplier pages now include distributors and buffer data for the commercially available enzymes. There are now online forms available to submit new references, new enzymes and sequence data for inclusion into REBASE or to suggest corrections.
The REBASE Files icon brings up the growing list of currently available monthly data formats. Click on any of the numbered choices for their descriptions or to download these files. GeneRunner, EMBOSS, DNAssist and DNATools users will be glad to find these software packages have now been added to the list. Users who prefer retrieving REBASE data via anonymous ftp may continue to do so at ftp://ftp.neb.com (cd/pub/rebase). We also continue to maintain a monthly Emailing list. Send a request to macelis@neb.com to join.
Acknowledgments
ACKNOWLEDGEMENTS
Special thanks are due to the many individuals who have so kindly contributed their unpublished results for inclusion in this compilation and to the REBASE users who continue to steer our efforts with their helpful comments. We are especially grateful to Janos Posfai for developing the software that enables the display of the sequence organization of RM genes. This database is supported by the National Library of Medicine (LM04971).
REFERENCES
- 1.Roberts R.J. and Macelis,D. (1999) Nucleic Acids Res., 27, 312–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith H.O. and Nathans,D.J. (1973) J. Mol. Biol., 81, 419–423. [DOI] [PubMed] [Google Scholar]
- 3.Bult C.J., White,O., Olsen,G.J., Zhou,L., Fleischmann,R.D.,Sutton, G.G., Blake,J.A., FitzGerald,L.M., Clayton,R.A., Gocayne,J.D. et al. (1996) Science, 273, 1058–1073. [DOI] [PubMed] [Google Scholar]
- 4.Brooks J.E., Nathan,P.D., Landry,D., Sznyter,L.A., Waite-Rees,P., Ives,C.L., Moran,L.S., Slatko,B.E. and Benner,J.S. (1991) Nucleic Acids Res., 19, 841–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ivic A., Jakeman,K.J., Penn,C.W. and Brown,N.L. (1999) FEMS Microbiol. Lett., 179, 175–180. [DOI] [PubMed] [Google Scholar]