Abstract
Nonalcoholic fatty liver disease (NAFLD) is associated with an increased ratio of classically activated M1 macrophages/Kupffer cells to alternatively activated M2 macrophages, which plays an imperative role in the development and progression of NAFLD. However, little is known about the precise mechanism behind macrophage polarization shift. Here, we provide evidence regarding the relationship between the polarization shift in Kupffer cells and autophagy resulting from lipid exposure. High-fat and high-fructose diet supplementation for 10 weeks significantly increased the abundance of Kupffer cells with an M1-predominant phenotype in mice. Interestingly, at the molecular level, we also observed a concomitant increase in expression of DNA methyltransferases DNMT1 and reduced autophagy in the NAFLD mice. We also observed hypermethylation at the promotor regions of autophagy genes (LC3B, ATG-5, and ATG-7). Furthermore, the pharmacological inhibition of DNMT1 by using DNA hypomethylating agents (azacitidine and zebularine) restored Kupffer cell autophagy, M1/M2 polarization, and therefore prevented the progression of NAFLD. We report the presence of a link between epigenetic regulation of autophagy gene and macrophage polarization switch. We provide the evidence that epigenetic modulators restore the lipid-induced imbalance in macrophage polarization, therefore preventing the development and progression of NAFLD.
Keywords: macrophage polarization, autophagy, DNA hypomethylating agents, epigenetic reprogramming, NAFLD
NAFLD is the hepatic manifestation of a metabolic syndrome characterized by chronic systemic low-grade inflammation and insulin resistance (1, 2, 3). An increasing body of evidence documented that Kupffer cells play a significant role in the pathophysiology of NAFLD (4). They play a vital role in controlling the function and phenotype of nearby parenchymal and nonparenchymal cells (5, 6, 7). However, the extent and stage of tissue inflammation is evaluated by determining macrophage polarization status, i.e., levels of proinflammatory M1 and anti-inflammatory M2 macrophages. Several investigations have recently reported that lipid buildup in the adipose tissue during obesity promoted the M1 polarization shift in infiltrated macrophages, while the M2 phenotype was predominant in lean adipose tissue (8, 9). Similarly, in the liver, Kupffer cells become activated and promote NAFLD development (10, 11). However, the dynamic change in Kupffer cell polarization in the development of NAFLD is still elusive.
In this last decade, much due emphasis has been given to how autophagy contributes to the pathology of various disorders. Autophagy provides the cell with the necessary lysosomal clearance of intracellular lipids in hepatocytes and may contribute to developing hepatic steatosis. Recent studies have begun to decipher the role of macrophage autophagy in regulating inflammation and immune responses (12, 13). Several studies have documented that genetic obesity or diet-induced obesity reduces the autophagy activity in the liver (9, 14). An increasing volume of literature documented that p-ULK/AMPK and SIRT-1 axis is essential for regulating the pathways for glucose and hepatic lipid metabolism as well as dysregulated hepatic autophagy in NAFLD condition (15, 16, 17). However, the mechanism for the decline in hepatic resident macrophage autophagy activity with the NAFLD progression is still obscure. The most prevalent epigenetic change is DNA methylation, which mainly occurs at the CpG dinucleotide of cytosines. CpGs are frequently abundant in the promotor and first exon/5′ untranslated region of genes. Hypomethylation is prevalent at the promoter favoring the euchromatin form and providing transcriptional activity to the respective genes. In contrast, DNA hypermethylation hinders the binding of methylation-specific DNA-binding elements and/or interaction with histone modifications and corepressors that modify DNA accessibility, resulting in gene silencing (18, 19, 20, 21). However, whether DNA methylation affects the NAFLD-induced polarization switch and inflammation in Kupffer cells is unknown.
The present study was undertaken to evaluate the hypothesis that the epigenetic modulation of hepatic resident macrophage autophagy and their polarization to inflammatory phenotype is anchored by DNA methylation. We observed that ablation of DNMT-1 inhibits DNA methylation, suppressing macrophage inflammation and correcting the impaired macrophage autophagy in Western diet–induced NAFLD. As a result, our finding offers a unique explanation for decreasing autophagy activity with the NAFLD progression. Furthermore, our current study deciphers the role of DNA methyltransferase 1 inhibitors in improving macrophage autophagy and limits their polarization into proinflammatory phenotype.
Results
Western diet–induced NAFLD impairs hepatic resident macrophage autophagy
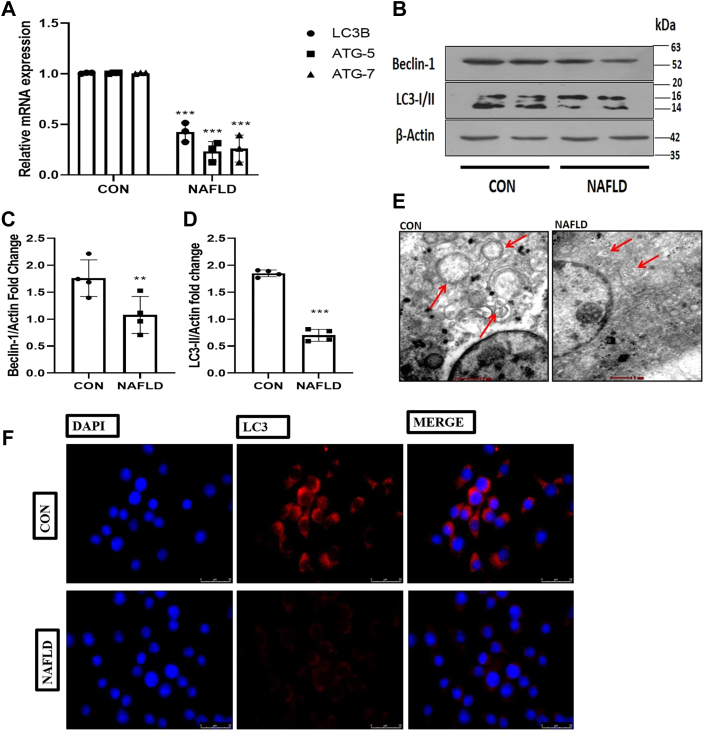
The effect of Western diet–induced NAFLD on macrophage autophagic function was investigated to assess the potential implication of altered macrophage autophagy as a mechanism for the hyperactive inflammatory response in NAFLD. We began by supplementing high-fat and high-fructose diet (HF-HFR) for 10 weeks to induced NAFLD model (Fig. S1). After that we isolated and characterized the hepatic macrophages from control and NAFLD mice (Figs. S2 and S3) and measured their autophagy. To determine their autophagic function, first we determined the expression of autophagy genes (LC3B, ATG-7, and ATG-5) in the macrophages derived from control and NAFLD mice. We have found that the gene expression levels of crucial autophagic genes (LC3B, ATG5, and ATG7) were significantly (p < 0.001; p < 0.001; p < 0.001) decreased in the hepatic resident macrophages as compared with the normal control (Fig. 1A). Next, we checked the protein expression of autophagic proteins (LC3I/II and Beclin) and found that the levels of autophagic proteins were significantly (p < 0.001; p < 0.001) downregulated in the hepatic macrophage derived from the NAFLD mice (Fig. 1, B–D). From the above findings we speculate that NAFLD may cause a malfunction in autophagosomes production, resulting in decreased macrophage autophagy. To determine whether the Western diet impaired the autophagosomes formation, we fixed the hepatic macrophages, and the sections of macrophages were subjected to transmission electron microscopy analysis. Images taken from the sections of isolated hepatic macrophages derived from the NAFLD mice showed fewer autophagosomes than the normal control (Fig. 1E). In order to make our claim more robust, we next performed immunofluorescence microscopy to detect autophagic marker LC3 in the isolated hepatic macrophages to confirm the impairment of macrophage autophagy in NAFLD mice. Macrophages from NAFLD and control mice were fixed, and LC3 was detected using the primary antibody and a secondary fluorescent antibody. The fluorescent intensity of LC3 was significantly reduced in macrophages derived from the NAFLD mice compared with the normal control (Fig. 1F). Taken together, these results indicate that the expression and activity of essential autophagy proteins were diminished in macrophages isolated from NAFLD mice.
Figure 1.
Western diet–induced NAFLD impairs hepatic resident macrophage autophagy.A, 10 weeks of high-fat diet and high-fructose supplementation decreases the mRNA expression of key autophagic genes (LC3B, ATG5, and ATG7) in macrophages derived from NAFLD mice. B, cells were lysed and subjected to immunoblotting against LC3 I/II and Beclin1. C, bar graphs represent the densitometric ratio of Beclin1 to the corresponding β-actin. D, bar graph represents the densitometric ratio of LC3-II to the corresponding β-actin. E, transmission electron microscopy images of sections showing autophagy in control and NAFLD Kupffer cells (red arrows indicate the autophagosomes formation) (the scale bar represents 2 μm in CON and 1 μm in NAFLD). F, images indicating higher immunofluorescence of LC3 protein in control compared with NAFLD macrophage-derived cells (the scale bar represents 25 μm). Data are represented as mean ±SD; ∗∗p < 0.01 and ∗∗∗p < 0.001 versus Control group. Error bars (wherever applicable in the figure) represent standard deviations from 3 to 4 independent experiments. NAFLD, nonalcoholic fatty liver disease.
Autophagy genes were hypermethylated in hepatic resident macrophages derived from NAFLD mice
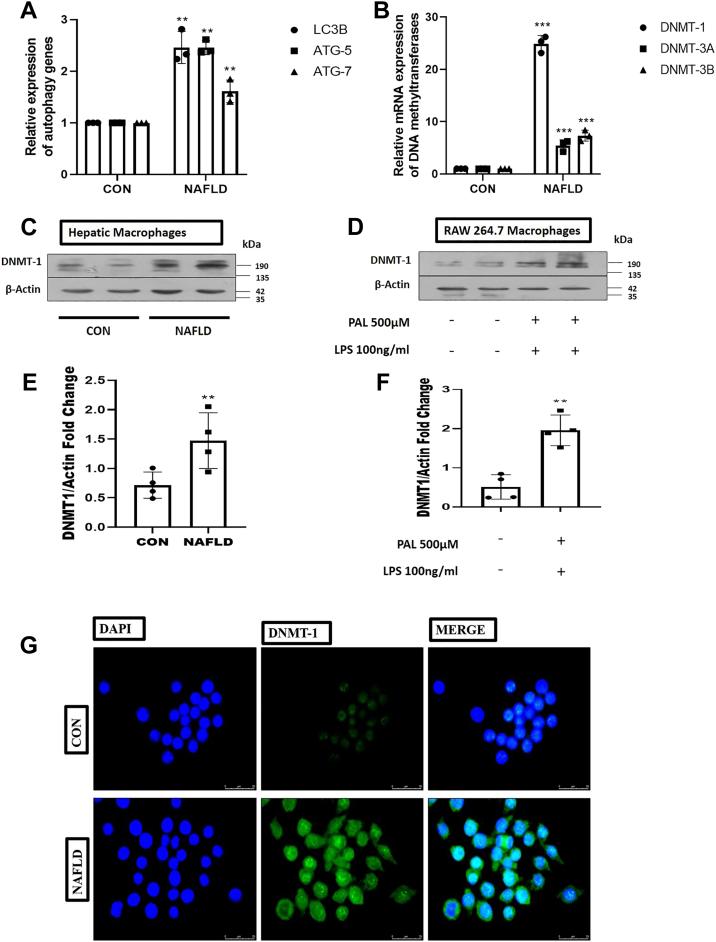
Although mounting research, including ours, has proven a significant association between the reduction of autophagy activity and the progression of NAFLD, the underlying mechanism is still unknown. To explore the possible mechanism of reduced autophagy gene expression in NAFLD, we have examined the methylation status of the promoter regions of essential autophagy genes by bisulfite-specific quantitative PCR using BSP-specific primers. Our data revealed that autophagy genes LC3B, ATG5, and ATG7 were hypermethylated in the hepatic macrophage derived from the NAFLD mice (Fig. 2A). Furthermore, we questioned which DNMT expression was escalated in the macrophages that were responsible for the hypermethylation of autophagy genes. To explore this, we examined the gene expression of different DNA methylating enzymes (DNMT1, DNMT3A, and DNMTB) in macrophages isolated from control and NAFLD mice. Among these three DNA methylating enzymes, the gene expression of DNMT1 was significantly higher in macrophages derived from NAFLD mice (Fig. 2B). Furthermore, Western blotting was used to confirm DNMT1 expression in macrophages derived from NAFLD mice and in RAW 264.7 cell lines treated with palmitate and lipopolysaccharide (LPS). Interestingly, we observed that the protein expression of DNMT-1 was significantly higher (p < 0.001) in macrophages from NAFLD mice and in RAW 264.7 cell lines treated with palmitate and LPS (Fig. 2, C–F). In order to support our findings, we fixed hepatic macrophages and incubated the cells with a primary DNMT-1 antibody followed by the secondary fluorescent antibody. Using confocal microscopy, we measured the fluorescence level of DNMT-1 and observed that the fluorescence intensity of DNMT-1 was greater in macrophages obtained from NAFLD animals as compared with the normal control mice (Fig. 2G).
Figure 2.
Autophagy genes were hypermethylated in hepatic macrophages derived from NAFLD mice.A, BSP-specific quantitative PCR analysis of levels of expression of autophagic genes (LC3B, ATG5, and ATG7). B, mRNA expression of DNA methylating enzymes (DNMT1, DNMT3A, and DNMT3B) in macrophage derived from control and NAFLD mice. C, immunoblotting of DNMT1 in primary macrophages derived from control and NAFLD mice. D, RAW 264.7 macrophage cells treated with or without palmitate (500 μM) and LPS (100 ng/ml) followed by Western blot against DNMT1. E, bar graphs represent the densitometric ratio of DNMT1 to the corresponding β-actin in primary macrophages derived from the control and NAFLD mice. F, bar graphs represent the densitometric ratio of DNMT1 to the corresponding β-actin in palmitate (500 μM) and LPS (100 ng/ml)-treated RAW 264.7 macrophage cells. G, immunofluorescence microscopy shows an increase in the fluorescence intensity of DNMT1 in primary macrophages derived from NAFLD as compared with control mice (the scale bar represents 25 μm). Data are represented as mean ±SD; ∗∗p < 0.01 and ∗∗∗p < 0.001 versus Control group. Error bars (wherever applicable in the figure) represent standard deviations from 3 to 4 independent experiments. LPS, lipopolysaccharide; NAFLD, nonalcoholic fatty liver disease.
Knockdown of DNMT1 modulates autophagy and prevents macrophage inflammation as well as polarization in palmitate and LPS-treated RAW 264.7 macrophages
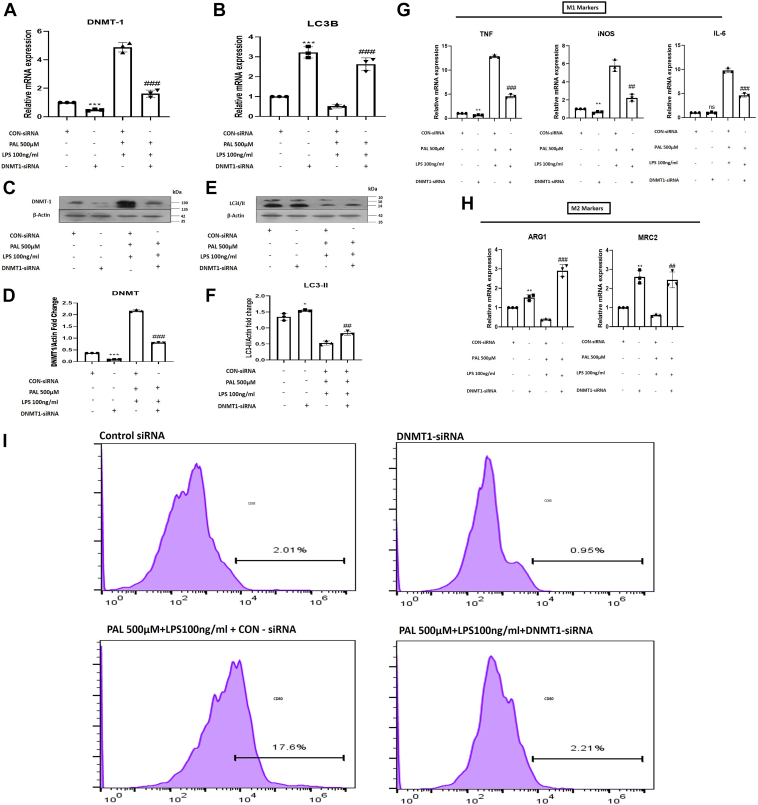
To determine whether the selective inhibition of DNMT1 induces autophagy and influences macrophage polarization, we used DNMT1-siRNA, which was transfected in palmitate 500 μM and LPS 100 ng/ml treated RAW264.7 macrophage cells. Coexposure of palmitate and LPS for 48 h robustly increased the protein and gene expression of DNMT1 and simultaneously decreased the autophagy marker (LC3-B) in RAW 264.7 cells. Moreover, we also evaluated the effect of coexposure of palmitate and LPS on the M1/M2 macrophage polarization marker and observed that palmitate and LPS treatment favored polarization to the M1 (inflammatory) phenotype of macrophages. To gain further insight into the impact of DNMT1 activation on LC3B expression and macrophage polarization, palmitate and LPS-treated Raw 264.7 cells were transfected with DNMT1-siRNA. We found that DNMT1-siRNA transfection markedly inhibited DNMT1 hyperactivity and concomitantly led to the upregulation of LC3B (Fig. 3, A–F).
Figure 3.
DNMT1 silencing restores autophagy and macrophage polarization in RAW 264.7 macrophages.A, mRNA expression of DNMT1 in control-siRNA, DNMT1-siRNA, palmitate (500 μM)+LPS (100 ng/ml)+Control-siRNA, and palmitate (500 μM)+LPS (100 ng/ml)+DNMT1-siRNA-treated RAW 264.7 macrophages. B, mRNA expression of LC3B in Control-siRNA, DNMT1-siRNA, palmitate+LPS+Control-siRNA, and palmitate (500 μM)+LPS (100 ng/ml)+DNMT1-siRNA-treated RAW 264.7 macrophages. C, immunoblotting of DNMT1 in different groups indicated in the figure. D, bar graphs represent the densitometric ratio of DNMT1 to the corresponding β-actin. E, immunoblotting of LC3-II in different groups indicated in the figure. F, bar graphs represent the densitometric ratio of LC3-II to the corresponding β-actin. G, quantitative RT-PCR analysis of M1 markers (iNOS, TNF, and IL6) in different groups as indicated in the figure. H, quantitative RT-PCR analysis of M2 markers (ARG1 and MRC2) in different groups as indicated in the figure. I, flow cytometric analysis showed higher percentage of CD86+ (M1 macrophage marker) RAW 264.7 cells in palmitate+LPS+Control-siRNA-treated cells and decreased in palmitate (500 μM)+LPS (100 ng/ml) +DNMT1-siRNA-treated RAW 264.7 macrophages. Data are represented as mean ±SD, ∗p < 0.05, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus Control-siRNA group; ##p < 0.01, and ###p < 0.001 versus Palmitate and LPS-treated group. Error bars (wherever applicable in the figure) represent standard deviations from 3 to 4 independent experiments.
Further confirmation regarding the direct effect of DNMT1 on macrophage polarization and inflammation status was afforded by transfecting RAW 264.7 cells with DNMT1-siRNA in the absence and presence of coexposure of palmitate and LPS followed by the analysis of CD80 levels through flow cytometry and M1 and M2 markers gene expression. We noticed suppression of gene expression of M1 markers (TNF, iNOS, and IL6) along with the upregulation of M2 markers (MRC2 and ARG1) expression in DNMT1 siRNA-transfected palmitate and LPS-treated macrophages (Fig. 3, G and H). Furthermore, our flowcytometry data revealed that selective knockdown of DNMT1 by using DNMT1 siRNA in palmitate and LPS-treated macrophages abolished the CD80 levels (Fig. 3I). Our findings testify that ablation of DNMT1 suppresses macrophage inflammation by regulating macrophage autophagy gene expression.
Inhibiting DNMT1 restores autophagy in palmitate and LPS-treated RAW 264.7 macrophages through the p-ULK/AMPK and SIRT-1 pathway
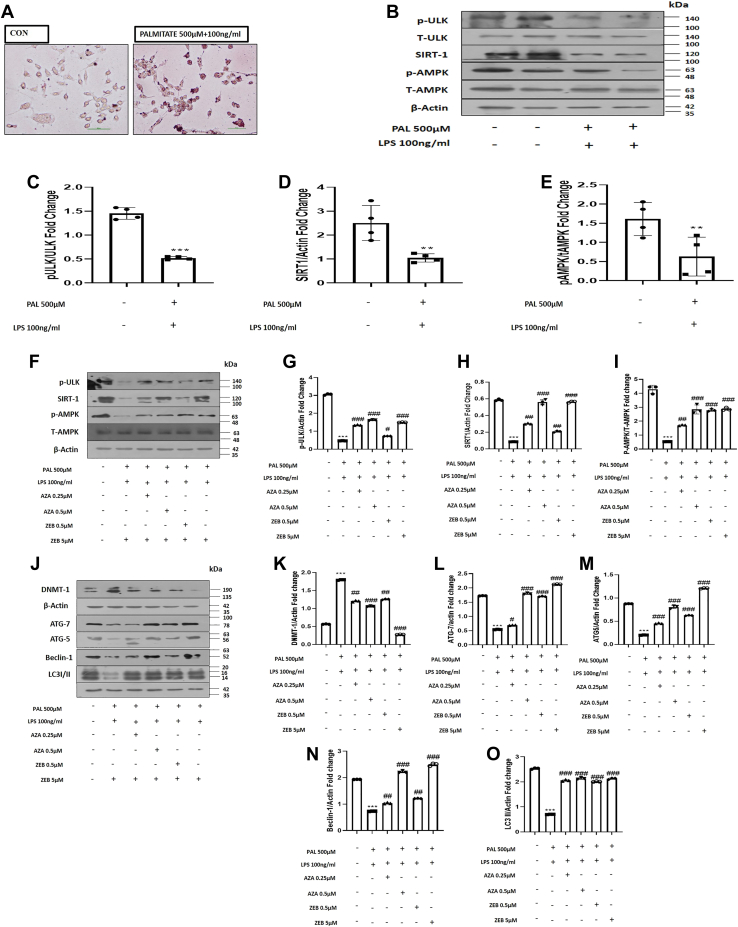
A deep insight into the molecular mechanism by which DNA methylation may contribute to the downregulation of macrophage autophagy is required to reveal the underlying mechanism. RAW 264.7 macrophages were cultured and treated with palmitate 500 μM and LPS 100 ng/ml for further mechanistic confirmation to induce in vitro NAFLD condition. We performed Oil Red O staining and observed that RAW 264.7 cells treated with palmitate and LPS accumulated more fat, indicated by the prominent red staining in palmitate and LPS-treated cells, confirming the establishment of a successful in vitro NAFLD model (Fig. 4A). Furthermore, an increasing volume of the literature suggests that the p-ULK/AMPK and SIRT-1 axis plays a critical role in controlling lipid metabolism pathways and dysregulated hepatic autophagy in NAFLD conditions (15). Therefore, evaluating the expression of p-ULK, p-AMPK, and SIRT-1 protein expression is necessary. Our results have demonstrated that palmitate and LPS-treated cells exhibit low expression of p-ULK (p < 0.001), p-AMPK (p < 0.01), and SIRT-1 (p < 0.01) (Fig. 4, B–E). 5-Azacitidine (Aza) and zebularine (Zeb) are nucleoside-based DNMT inhibitors extensively utilized to investigate the role of DNA methylation in cancer and other illnesses. The in vitro studies for both the drugs, i.e., Zeb and Aza, were conducted using the doses as per the previous studies, and at these selected doses, both agents showed minimal cytotoxicity in the in vitro models (22, 23). We found a dose-dependent increase in the expression of pULK, p-AMPK, and SIRT1 protein expression following the treatment of palmitate and LPS-treated cells with Aza (0.25 μM and 0.5 μM) and Zeb (0.5 μM and 5 μM). We also measured the impact of DNA hypomethylating agents on the protein expression of autophagy-related genes. DNA hypomethylating agents increased autophagic gene expression (ATG7, ATG-5, Beclin-1, and LC3-II), pointing toward restored autophagy in treated groups. On the other hand, DNA hypomethylating agents treatment also leads to the rapid dose-dependent depletion of DNMT1 protein expression in palmitate and LPS-treated RAW 264.7 cells (Fig. 4, F–O). Interestingly, our findings suggest that DNMT1 regulates the expression of autophagy-related genes, pointing toward the role of the pULK, p-AMPK, and SIRT1 axis acting as a downstream signaling for modulating autophagy pathways.
Figure 4.
Inhibiting DNA methylation in vitro restores autophagy.A, Oil red O staining confirmed increased oil globules in palmitate (500 μM) and LPS (100 ng/ml)-treated compared with control RAW 264.7 macrophages (the scale bar represents 200 pixels). B, RAW 264.7 cells with or without palmitate and LPS treatment are subjected to Western blotting against p-ULK, T-ULK, SIRT-1, p-AMPK, and T-AMPK, and β-actin. C, bar graphs represent the densitometric ratio of p-ULK to T-ULK. D, bar graphs represent the densitometric ratio of SIRT-1 to the corresponding β-actin. E, bar graphs represent the densitometric ratio of pAMPK to T-AMPK. F, RAW 264.7 cells pretreated with palmitate and LPS treated with Zebularine (0.5 μM and 5 μM) and Azacytidine (0.25 μM and 0.5 μM) where subjected to Western blotting against protein expression of SIRT1, p-ULK, p-AMPK, and T-AMPK. G, bar graphs represent the densitometric ratio of p-ULK to the corresponding β-actin. H, bar graphs represent the densitometric ratio of SIRT-1 to the corresponding β-actin. I, bar graphs represent the densitometric ratio of pAMPK to T-AMPK. J, RAW 264.7 cells pretreated with palmitate and LPS treated with Zebularine (0.5 μM and 5 μM) and Azacytidine (0.25 μM and 0.5 μM) were subjected to Western blotting against DNMT1 and autophagic proteins (ATG-7, ATG-5, Beclin1, and LC3I/II). K, bar graphs represent the densitometric ratio of DNMT1 to the corresponding β-actin. L, bar graphs represent the densitometric ratio of ATG-7 to the corresponding β-actin. M, bar graphs represent the densitometric ratio of ATG5 to the corresponding β-actin. N, bar graphs represent the densitometric ratio of Beclin-1 to the corresponding β-actin. O, bar graphs represent the densitometric ratio of LC3-II to the corresponding β-actin. Data are represented as mean ±SD, ∗∗p < 0.01, and ∗∗∗p < 0.001 versus Control group; #p < 0.05, ##p < 0.01, and ###p < 0.001 versus Palmitate and LPS-treated group. Error bars (wherever applicable in the figure) represent standard deviations from 3 to 4 independent experiments.
Treatment of mice with DNA hypomethylating agents for 8 weeks prevents the progression of Western diet–induced NAFLD
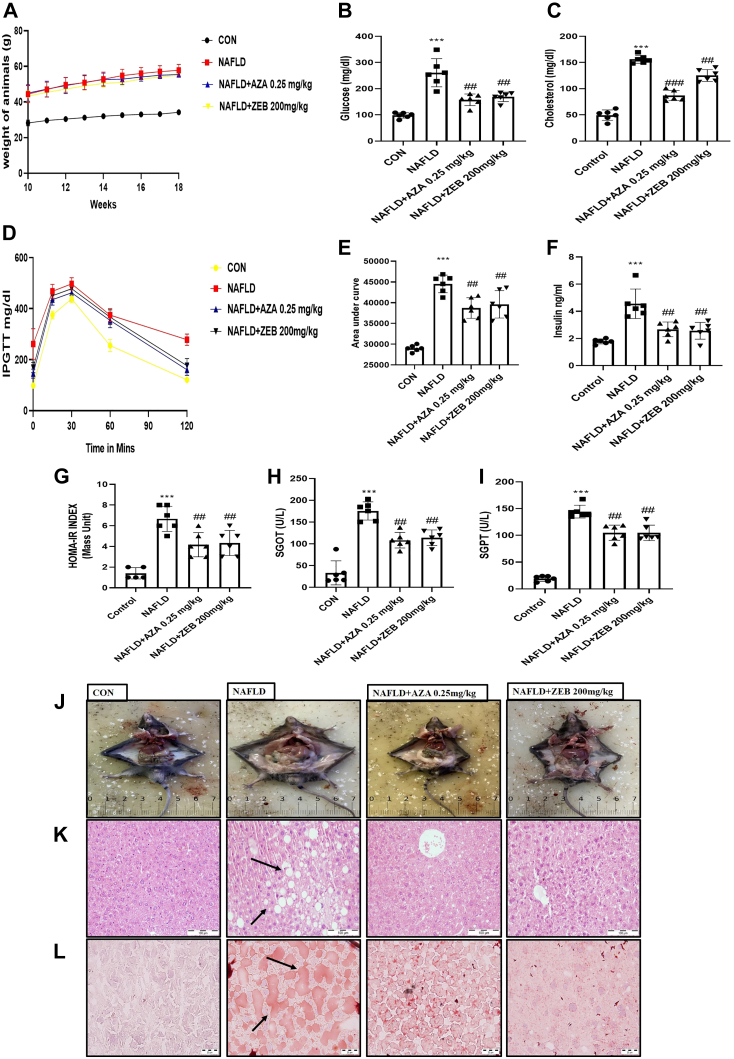
To inspect the therapeutic potential of DNA hypomethylating agents in rescuing Western diet–induced hepatic steatosis, we administered Aza (0.25 mg/kg 3 days per week; i.p.) and Zeb (200 mg/kg daily; i.p.) for eight consecutive weeks. Zeb and Aza were deemed safe at the doses used in animal studies derived from previous research (22, 23). After 8 weeks of treatment, animals were sacrificed, and various biochemical parameters were obtained. The body weight of the treated animals remained constant (Fig. 5A). The plasma glucose level in the NAFLD control group was substantially higher than in the normal control group, whereas the plasma glucose level in the Aza and Zeb-treated groups considerably reduced (p < 0.01; p < 0.01) after 8 weeks of continuous treatment (Fig. 5B). In addition, the treatment positively impacted the lipid profile of animals. The plasma total cholesterol levels were significantly decreased (p < 0.001; p < 0.01) in the Aza and Zeb-treated group compared with the NAFLD control group (Fig. 5C). intraperitoneal glucose tolerance test results demonstrated that Aza and Zeb treatment enhanced insulin sensitivity. All experimental treatments lowered plasma glucose concentrations at various time points and normalized plasma glucose concentrations at the 120-min time point (Fig. 5D). Analysis of the area under the curve was used to corroborate the significance level (p < 0.01; p < 0.01) (Fig. 5E). In addition, ELISA kits were used to validate enhanced insulin sensitivity and measure fasting insulin concentrations. Compared with the NAFLD control group, the treatment group experienced a substantial decrease (p < 0.01; p < 0.01) in plasma insulin levels (Fig. 5F). Furthermore, the homeostatic model assessment for insulin resistance level in the NAFLD control group was considerably more significant than in the normal control group. Compared with the NAFLD control group, homeostatic model assessment for insulin resistance levels in the treatment groups decreased significantly (p < 0.01; p < 0.01) (Fig. 5G). Moreover, increased levels of serum glutamic oxaloacetic transaminase and serum glutamic pyruvic transaminase were observed in the NAFLD control group as compared with the normal control animals, with a significant reduction (p < 0.01; p < 0.01) observed in the treatment groups (Fig. 5, H and I). In HF-HFR diet-induced NAFLD mice, liver histological alterations were evaluated to confirm the positive effect of Aza and Zeb on the etiology of hepatic steatosis. Hepatic steatosis and fat accumulation in the hepatic tissue slices were quantified using hematoxylin and eosin and Oil Red O staining. HF-HFR diet supplementation continuously for 18 weeks had various detrimental effects, validated by observation of multiple structural aberrations in the liver of NAFLD mice. Unfavorable traits such as hepatic vacuolations with steatohepatitis, hepatic sinusoidal dilatation, and fat buildup in the livers help characterize diseased liver. The frequency and extent of hepatic vacuolations and Oil and Red O stain were reduced in the treated groups, indicating an improvement in liver histology (Fig. 5, J–L).
Figure 5.
DNA hypomethylating agents treatment for 8 weeks improved metabolic parameters and histopathological alteration.A, body weight of animals showed no significant change after 8 weeks of Aza (0.25 mg/kg) and Zeb (200 mg/kg) treatment. B, significant reduction in fasting blood glucose after 8 weeks of treatment. C, reduction in cholesterol was achieved by 8 weeks of Zeb and Aza treatment. D, improvement in insulin sensitivity of treatment groups and improved glucose tolerance as determined by intraperitoneal glucose (2 g/kg body weight) tolerance test. E, decreased area under the curve. F, decreased insulin concentration estimated using ELISA. G, reduced HOMA-IR index. Aza (0.25 mg/kg) and Zeb (200 mg/kg) treatment improved liver function in NAFLD mice estimated using (H) serum glutamic oxaloacetic transaminase (SGOT) and (I) serum glutamic pyruvic transaminase (SGPT). J, phenotypic changes occur in control and NAFLD mice after 8 weeks of Aza and Zeb treatment. K, hepatic sections stained with H&E showing hepatic lipid accumulation and sinusoidal dilation (indicated by black arrows) in NAFLD group with improvement in treatment groups (the scale bar represents 100 μm). L, hepatic sections stained with Oil Red O showing fat accumulation inside the liver with NAFLD with improvement in different treatment groups (black arrows indicate the fat accumulation inside the liver) (the scale bar represents 20 μm). Data are represented as mean ±SD, ∗∗∗p < 0.001 versus Control group; ##p < 0.01 and ###p < 0.001 versus NAFLD group. Error bars (wherever applicable in the figure) represent standard deviations from six independent experiments. HOMA-IR, homeostatic model assessment for insulin resistance; NAFLD, nonalcoholic fatty liver disease.
DNMT-1 inhibition modulates hepatic macrophage autophagy in NAFLD mice
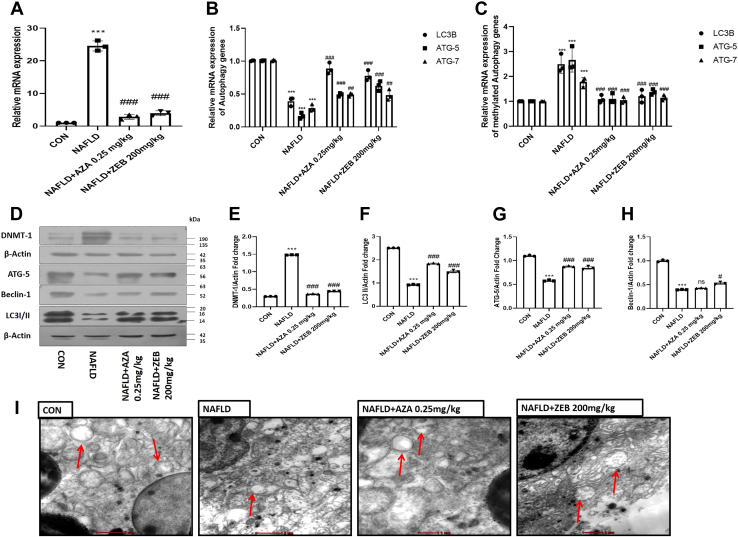
To investigate whether the in vivo administration of DNA hypomethylating agents modulates macrophage autophagy, we have administered DNA hypomethylating agents for eight consecutive weeks. After 8 weeks of treatment, we checked the mRNA and protein expression of DNMT1 and autophagic proteins. We found that the gene expression of DNMT1 was significantly downregulated (p < 0.001; p < 0.001) by Aza and Zeb in hepatic macrophages derived from the treated NAFLD mice (Fig. 6A). Interestingly, to check the methylation status of the promotor of autophagic genes after the DNA hypomethylating treatment, we performed bisulfite-specific quantitative PCR. We found that the expression of methylated autophagy genes was significantly downregulated in the Aza and Zeb-treated group as compared with the NAFLD group (Fig. 6B). On contrary, treatment with DNA hypomethylating agents significantly increased ([p < 0.001; p < 0.001], [p < 0.05; p < 0.001], and [p < 0.01; p < 0.05]) the gene expression of the key autophagic genes (LC3B, ATG5, and ATG7) (Fig. 6C). Similarly, the protein expression of DNMT1 was also downregulated (p < 0.001; p < 0.001) in treated groups (Fig. 6, D and E). Furthermore, the protein expression of vital autophagic proteins (LC3B, ATG-5, and Beclin1) were significantly increased ([p < 0.001; p < 0.001], [p < 0.001; p < 0.001], [p < 0.01; p < 0.05]) in NAFLD treated mice (Fig. 6, D–H). To make our claim more robust, further, we fixed the hepatic macrophages and the sections of macrophages were subjected to transmission electron microscopy analysis. Images taken from the sections of isolated hepatic macrophages revealed Aza and Zeb-treated mice showed an increase in the number of autophagosomes (Fig. 6I). Taken together, our finding demonstrated that in vivo treatment of NAFLD mice by DNA hypomethylating agents restores the diminished autophagy activity in macrophages derived from the NAFLD treated mice.
Figure 6.
DNA hypomethylating agents modulate hepatic macrophage autophagy in NAFLD mice.A, mRNA expression of DNMT1 in the macrophage derived from Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated mice. B, mRNA expression of LC3B, ATG5, and ATG7 in different groups indicated in the figure. C, mRNA expression of methylated LC3B, ATG5, and ATG7 in different groups indicated in the figure. D, Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated macrophages derived from NAFLD and treated mice were subjected to Western blotting against DNMT1, β-actin, ATG5, Beclin-1, LC3I/II, β-actin. E, bar graphs represent the densitometric ratio of DNMT1 to the corresponding β-actin. F, bar graphs represent the densitometric ratio of LC3I/II to the corresponding β-actin. G, bar graphs represent the densitometric ratio of ATG5 to the corresponding β-actin. H, bar graphs represent the densitometric ratio of Beclin-1 to the corresponding β-actin. I, transmission electron microscopy images of macrophage derived from Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated mice indicating upregulation of autophagy (red arrows indicate the autophagosomes formation) compared with NAFLD mice (the scale bar represents 2 μm in CON and NAFLD and 1 μm in Aza [0.25 mg/kg] and Zeb [200 mg/kg]). Data are represented as mean ±SD, ∗∗∗p < 0.001 versus Control group #p < 0.05, ##p < 0.01, and ###p < 0.001 versus NAFLD group. Error bars (wherever applicable in the figure) represent standard deviations from 3 to 6 independent experiments.
Treatment of mice with DNA hypomethylating agents switches macrophages polarity toward M2 phenotype
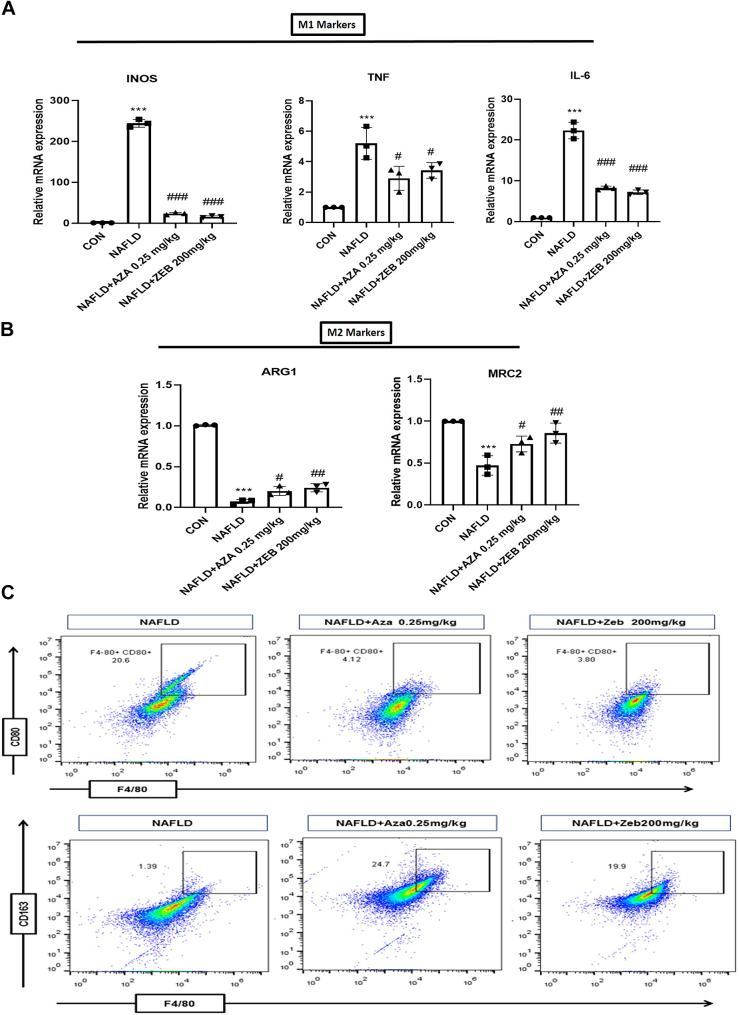
To examine whether the administration of DNA hypomethylating agents influences macrophage polarization, we started by checking the gene expression of M1 markers (iNOS, TNF, and IL6) and M2 markers (ARG1 and MRC2). Treatment with Aza and Zeb significantly downregulates the gene expression of M1 markers (iNOS, TNF, and IL6) and simultaneously upregulates the gene expression of M2 markers (ARG1 and MRC2) in the macrophages derived from the NAFLD treated mice as compared with the NAFLD control mice (Fig. 7, A and B). Furthermore, to strengthen our claims, flow cytometry was used to study the effect of DNA hypomethylating agents on macrophage polarity. Our findings revealed that the macrophages derived from the Aza and Zeb-treated animals showed reduced CD80+ M1 inflammatory phenotype and increased CD163+ M2 (Fig. 7C). These findings demonstrated that in vivo treatment of NAFLD mice by DNA hypomethylating agents skewed macrophages to their anti-inflammatory M2 phenotype.
Figure 7.
DNA hypomethylating agents regulate alternative activation of M2 macrophage polarization in NAFLD mice.A, decreased mRNA expression of M1 markers (iNOS, TNF-α, and IL-6) in macrophage derived from Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated mice. B, increased mRNA expression of M2 markers (ARG and MRC2) in macrophage derived from Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated mice. C, flow cytometric analysis indicated higher percentage of F4/80+CD80+ (M1 macrophage marker) cells from macrophage derived from NAFLD mice and higher percentage of F4/80+CD163+ (M2 macrophage marker) cells from macrophage derived from Aza (0.25 mg/kg) and Zeb (200 mg/kg)-treated mice. Data are represented as mean ± SD; ∗∗∗p < 0.001 versus Control group; #p < 0.05, ##p < 0.01 and ###p < 0.001 versus NAFLD group. Error bars (wherever applicable in the figure) represent standard deviations from three independent experiments.
Discussion
The present study was undertaken to decipher the role of epigenetic alterations (DNA methylation) in regulating Western diet–induced Kupffer cells phenotypic transition (macrophage polarization) and inflammation. An emerging volume of literature has revealed that long-term supplementation of a high-fat diet induces M1-predominant polarization of adipose tissue macrophages, contributing to the development of obesity and insulin resistance (22). We have also characterized the M1 and M2 markers expression profiles in different Kupffer cells population derived from control and NAFLD mice based on the surface expression of CD86+ and CD163+ through flow cytometry. Our flow cytometry data show that macrophages derived from NAFLD mice show higher expression levels of F4/80+CD86+ (M1 marker) than those derived from the control mice. In addition, hepatic macrophages derived from mice with NAFLD exhibit elevated expression of iNOS, TNF, and IL6 (M1 markers) and decreased expression of ARG1 and MRC2 genes (M2 markers). Recent studies have identified an immune control system in which autophagy modulates innate immunity by influencing the polarization of M1 and M2 macrophages (15). However, it remains unclear how obesity leads to anomalous hepatic macrophage activation.
Our study was designed to know how autophagy dysregulation causes aberrant macrophage polarization in NAFLD. We provide evidence that DNA methylation plays an important role in macrophage polarization and inflammatory response by modulating autophagy. This was further supported by an increase in the gene as well as protein expression of DNMT-1 in these mice. We show that the promotor region of autophagy genes (LC3B, ATG5, and ATG7) was hypermethylated in HF-HFR-fed NAFLD mice. These results are of profound clinical significance, not only from a mechanistic standpoint but also for crucial explanation of the pathophysiology of the disease. Deciphering the downstream pathway of reduced macrophage autophagy, RAW 264.7 macrophages treated with palmitate and LPS mimicking in vitro NAFLD conditions show downregulation of p-ULK, AMPK, SIRT1 and autophagic mediators LC3B, ATG-5, ATG-7, Beclin1 protein expression. Several reports indicate that p-ULK/AMPK and SIRT-1 play a crucial role in controlling glucose, lipid metabolism, and autophagy in NAFLD (15).
The interactions between genes and the environment cause most complex disorders, such as diabetes and obesity. One of the underlying mechanisms is that environmental factors, such as nutrition, can influence gene expression and reprogram the epigenome (24, 25). Recent reports also support our results that dietary saturated fatty acid and proinflammatory cytokine (TNF-α) levels were increased in obesity, leading to the significant upregulation of the DNMT1 expression in macrophages (22). In view of altered DNMT-1 protein expression, we studied the role of potent demethylating agents Aza and Zeb in in vitro and in vivo NAFLD models. Treatment with DNA hypomethylating agents for 8 weeks ameliorated different metabolic biochemical parameters and increased insulin sensitivity, preventing the progression and development of NAFLD. In addition, the liver function test and liver histopathological alteration were also ameliorated by DNA hypomethylating agents. We found that Aza and Zeb treatment resulted in rapid depletion of DNMT1, similar to the previous reports (23, 26). Interestingly, treatment with DNA hypomethylating agents significantly increased the protein expression of autophagic mediators LC3B, ATG-5, ATG-7, and Beclin-1 and also upregulated the protein expression of pULK, AMPK, and SIRT1 in in vitro NAFLD conditions. Taken together, our findings suggest that DNMT1 is responsible for the hypermethylation of promotors of autophagic genes and dysregulation of the p-ULK/AMPK and SIRT1 axis. Furthermore, the in vitro knockdown of DNMT1 provided much more conclusive evidence regarding the role of DNMT1 in regulating the expression of autophagy genes and thereby altering the inflammatory status. The silencing of DNMT1 using DNMT1-siRNA in palmitate and LPS-treated Raw 264.7 cells increased LC3B expression, thereby ameliorating macrophage autophagy, accompanied by a reduction in M1 markers (TNF, iNOS, and IL6) and an increase in M2 markers (MRC2 and ARG1) expression. The DNMT1 silencing bolstered our strong claim concerning the interaction between DNMT1, autophagy, and macrophage polarization. Furthermore, the pharmacological inhibition of DNMT1 specifically reprogrammed the hepatic macrophages by increasing the abundance of the M2 marker (F4/80+CD163+) as well as the upregulation of ARG1 and MRC2 genes. On the other hand, the abundance of M1 markers (F4/80+CD80+) and gene expression of iNOS, TNF-α, and IL-6 also got downregulated by treatment of DNA hypomethylating agents in NAFLD mice. Our findings lay the groundwork for developing epigenetic-based therapeutics for the prevention and treatment of NAFLD and other metabolic disorders.
Conclusion
The present study concluded that DNMT1 regulates macrophage polarization and inflammation, which is manifested by decreased hepatic macrophage autophagy through DNA methylation. The pharmacological inhibition of DNMT1 leads to decreased DNA methylation at the promotor of autophagic genes, promotes M2 polarization, decreases inflammation, and prevents NAFLD progression.
Experimental procedures
Animals
For this particular study, male C57BL6/J mice (18–20 g) of 4 to 5 weeks were used. Its procurement was done from IISER Mohali. Throughout the study, the ambient environmental temperature 22 °C ± 2 deg. C, humidity 50 ± 10%, and circadian rhythmicity of 12 h light 12 h dark cycle with food and water ad libitum were adequately maintained. According to their grouping, animals were fed regular rodent diet, i.e., normal pellet diet or high-fat diet (60% kcal of fat) (Research Diet # D12492) and high-fructose diet (30% w/v) for 18 weeks. The Institutional Animal Ethics Committee authorized the study protocol (approval number IAEC20/10).
Experimental design and animal treatment
Male C57BL6/J mice aged 4 to 5 weeks and weighing 18 to 20 g were kept on a combination diet of high fat and high fructose for 10 weeks, whereas control mice were kept on normal pellet diet (3.8 kcal/g) (Normal Control). Alteration in the metabolic parameters such as an increase in plasma insulin, glucose, triglycerides, and circulating lipids indicates successful NAFLD induction. It was followed by the randomization of animals into four groups, which were administered treatment for 8 weeks:
Group 1: Normal Control; received 0.9% w/v normal saline (CON); n = 6,
Group 2: NAFLD Disease Control; received a combination diet of high fat and high fructose (HF-HFR); n = 6,
Group 3: NAFLD mice administered Aza (0.25 mg/kg i.p., NAFLD + Aza); n = 6.
Group 4: NAFLD mice administered Zeb (200 mg mg/kg i.p., NAFLD + Zeb); n = 6.
The dose of Zeb (TCI # Z0022) and Aza (Sigma # A2385) used for animal studies was taken from previous studies, and at these doses both agents were considered safe (22, 23).
Cell lines, culture condition, and methyl transferase inhibitor treatment
Raw 264.7 cells (ATCC cat#TIB-71) were cultured in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum and 1% penicillin–streptomycin, followed by 48-h treatment with 500 μM palmitate and 100 ng/ml LPS in accordance with the pre-established methodology (27). After this the cells were treated with or without Aza at the dose of (0.25 μM and 0.5 μM) and Zeb (0.5 μM and 5 μM) for 48 h. Bovine serum albumin was used as vehicle control. RAW macrophage with passage no. 8-12 was used for experiments.
DNMT1-siRNA transfection
For the transfection of DNMT-siRNA (Silencer predesigned siRNA DNMT1 #AM16708) and the respective control (siRNA #AM 4611), Lipofectamine 3000 Transfection kit (Invitrogen #L3000-015) was used according to manufacturer protocol. Briefly, RAW264.7 macrophages were seeded (0.1 × 106 cells/well) in a six-well plate in an antibiotic-free complete growth medium for 24 h before transfection. For each well, 100 pmol of DNMT1-siRNA/Control-siRNA and lipofectamine 3000 were added separately into OptiMEM serum-free medium (Thermo Scientific). Then, both these solutions were mixed and incubated for 5 min. The transfection mixture was then added to the cells containing medium and incubated for 48 h. After 48 h of transfection, cells were washed and samples were prepared for analyzing gene and protein expression.
Isolation of hepatic macrophages
Liver macrophage extraction was done following a previously established protocol (28). After severing the inferior vena cava, a 20-G catheter was inserted through the mouse portal vein. PBS was infused into the liver, followed by digestion using type IV collagenase. Single cells were filtered using a 100-mesh sieve cell strainer and fractionated using 25% and 50% Percoll (Sigma-Aldrich # 17-5445-02). The intercushion fraction was then washed and affixed to six-well plate containing medium. Kupffer cells attached to the plate, and the nonadherent fraction was removed by subsequent washing with Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum.
Evaluation of metabolic profile
After an overnight fast, blood was collected via the retro-orbital route, and plasma and serum were separated by centrifuging the blood samples at 4 °C, 2500g for 10 min. Next, biochemical parameters such as serum glutamic pyruvic transaminase and serum glutamic oxaloacetic transaminase were estimated using serum samples, glucose and total cholesterol were estimated using plasma samples, and hepatic triglyceride levels were also estimated in accordance with the manufacturer's assay kit protocol (Accurex, India). In addition, an Insulin ELISA kit was used to determine insulin levels in the mice (Crystal Chem).
Intraperitoneal glucose tolerance test
On completion of 18 weeks, followed by an overnight fast, animals were administered a glucose load of 2 g/kg intraperitoneally. Plasma glucose was measured at several time points, including 0 min (right before glucose load injection) and 15, 30, 60, and 120 min. Then, using GraphPad Prism 8 software, the total area under curves was determined from a plot of blood glucose concentration versus time.
Histopathological studies
Liver tissue samples were collected from mice and placed in the neutral formalin (10%) for overnight fixation at 4 °C. After the fixation, liver tissue was embedded in OCT (optimal cutting temperature compound, Sigma) and frozen at −60 °C followed by cryosections using cryotome (Leica CM 1860, Leica Biosystem). Subsequently, 5-μm sections of the processed tissue were taken on the gelatin-coated glass slides. After this the sections were eventually stained using H&E and Oil Red O, mounted with glycerin for histopathological examination under the microscope (Olympus BX51 microscope).
RNA extraction and quantitative RT-PCR
Total RNA was extracted from hepatic macrophages using the Triazole technique, and subsequently, cDNA was synthesized using a verso cDNA synthesis kit (Thermo Fisher) following the manufacturer's instructions. QuantiTect SYBR Green PCR Kit (Qiagen) and oligonucleotides specific for each gene (Tables 1 and 2) were used to determine the relative expression of Macrophage polarization genes, autophagy genes, and DNA methylating enzymes. The 18s level was amplified using particular oligonucleotides and was used for normalization. ΔΔCt equations were used to analyze the result.
Table 1.
Sequence of bisulfite-specific primers of autophagy genes
| S.No | Gene promoter | BSP forward primer | BSP reverse primer |
|---|---|---|---|
| 1. | LC3B | YGATTTYGATTTAGGAGTAAATAT | CTATCTACRTAAATCRAACRC |
| 2. | ATG5 | TTTATYGAGTAAATGAATGTGGTT | CTCRAAACRAATTTACCRAT |
| 3. | ATG7 | TTTTGTTAGTTYGAGATTAGATTG | CAATAACRAAACRAAACTAAAAC |
| 4. | β-Actin | CATCACCAACTGGGACGACA | ATACATGGCAGGCACGTTGA |
Table 2.
Primer sequence of various genes
| S.No | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| 1. | DNMT1 | CGGCTCAAAGACTTGGAAAG | TAGCCAGGTAGCCTTCCTCA |
| 2. | DNMT3a | GAGCCGCCTGAAGCCC | TGCCGTGGTCTTTGTAAGCA |
| 3. | DNMT3b | GCCAGACCTTGGAAACCTCA | GCTGGCACCCTCTTCTTCAT |
| 4. | ATG5 | TGCATCAAGTTCAGCTCTTCCT | ATCCAGAGCTGCTTGTGGTC |
| 5. | ATG7 | CGGAAGTTGAGCGGCGA | AGGAAAGCAGTGTGGAGTTG |
| 6. | Beclin1 | GCCTCTGAAACTGGACACGA | TAGCCTCTTCCTCCTGGGTC |
| 7. | LC3B | ACAAGGGAAGTGATCGTCGC | TCGCTCTATAATCACTGGGATCT |
| 8. | INOS2 | ACCTTGGTGAAGGGACTGAG | ACTCCGTGGAGTGAACAAGAC |
| 9. | TNF-α | CTACCTTGTTGCCTCCTCTTT | GAGCAGAGGTTCAGTGATGTAG |
| 10. | IL-6 | CCCCAATTTCCAATGCTCTCC | CGCACTAGGTTTGCCGAGTA |
| 11. | Arg1 | CTCCAAGCCAAAGTCCTTAGAG | AGGAGCTGTCATTAGGGACATC |
| 12. | MRC2 | TACAGCTCCACGCTATGGATT | CACTCTCCCAGTTGAGGTACT |
| 13. | IL-1β | TGCCACCTTTTGACAGTGATG | TGATGTGCTGCTGCGAGATT |
| 14. | 18s | GTTCAGCCACCCGAGATTGA | CCCATCACGAATGGGGTTCA |
Western blotting
Evaluation of protein expression was conducted by Western blot analysis, as reported (29). Protein samples were initially resolved on 8 to 14% SDS-PAGE, transferred to PVDF, followed by incubation with primary antibodies usually used at dilution 1:1000. DNMT1 (CST #5032S); AMPK (CST #2532S); β-Actin (Santa Cruz Biotechnology; #sc-47 778); SIRT1 (CST #9475S); LC3A/B, ATG-5, ATG-7, and Beclin-1 (Autophagy Sampler Kit, Cell Signalling Technology; #sc-4445T) and p-AMPKalpha (Thr172) (CST #2531S); p-ULK (CST #688T); T-ULK (CST #8054T). The immunoblots were then incubated with a secondary antibody (Promega). The blot visualization was done using the enhanced chemiluminescence substrate. Finally, densitometry analysis was used to quantify immunoblots using ImageJ software, NIH.
Fluorescence confocal microscopy
Macrophages derived from the control and NAFLD and treated mice were seeded at the density of 1 × 105 in 24-well plates and incubated overnight. Following this, cells were fixed with 4% paraformaldehyde for 25 min at room temperature, after that cell were permeabilized with cold methanol for 10 s and then incubated with a primary LC3, CD86, and DNMT1 antibody overnight at 4 °C. After that, cells were treated with secondary antibody Goat anti-Mouse IgG Highly Cross-Adsorbed Alexa Fluor Plus 488 and Goat Anti-Rabbit IgG Highly Cross-Adsorbed Alexa Fluor Plus 647 by Invitrogen. Finally, cells were then stained for 15 min with the fluorescent DNA dye DAPI at 1 mg/ml. In addition, we have performed the secondary antibody control staining as an internal standard control in macrophages derived from the NAFLD and control mice in order to show that labeling is specific to the primary antibody (Fig. S4, A and B). Images were captured by an inverted fluorescent microscope (Leica DMi8).
Flow cytometry
The cell pellets of macrophages derived from the control, NAFLD, and treated mice were resuspended in cell staining buffer (PBS containing 0.2% fetal bovine serum and 0.09% NaNO3) and blocked with TruStain FcX (Fcγ blocker, mouse anti-CD16/32 antibody, BioLegend) for 15 min at 4 °C. Cells were then stained with fluorochrome-labeled primary antibodies against CD80 (anti-mouse), CD163 (anti-mouse), and F4/80 (anti-mouse) (BioLegend) for 1 h on ice. Cells were then washed twice with chilled PBS, resuspended in cell staining buffer and analyzed in a Flow cytometer (BD Accuri C6+, BD Biosciences) using FlowJo v10.6.1 software.
Sample preparation for the detection of autophagosomes through transmission electron microscopy
The previously mentioned technique was used to prepare the samples (30). Briefly, the cultured hepatic macrophages were scraped and centrifuged at 4000g at 4 °C for 5 min. The pellet was washed with phosphate buffer solution and fixed with 2.5% phosphate-buffered glutaraldehyde for 1 h and then postfixed in 2% osmium tetroxide. The cells were dehydrated in ethanol followed by propylene oxide and then embedded in resin. Sections were taken by the glass knife, using 4% uranyl acetate. These thin sections were placed on the copper grid and autophagosomes inside the cells were examined by using transmission electron microscopy (Model TF-20), FEI, at NIPER, S.A.S Nagar, Punjab, India.
Statistical analysis
All data analyses were performed using GraphPad Prism software (v.8.0). Data were represented as mean ± SD. Statistical comparison between two groups was performed using Student's t test. One-way ANOVA testing was used for the comparisons among multiple groups. p < 0.05 was considered as statistically significant.
Data availability
All the data described in this study are contained within the article.
Supporting information
This article contains supporting information.
Conflict of interest
The authors declare that they have no conflicts of interest with the contents of this article.
Acknowledgments
We are thankful to Archana from the Department of Pharmacology and Toxicology, NIPER S.A.S Nagar for providing assistance in preparing cryogenic sections of liver tissue and Dr Durba Pal Department of BME, Indian Institute of Technology Ropar for extending the facilities required for the present investigation. The authors are grateful to the National Institute of Pharmaceutical Education and Research, S.A.S Nagar and Department of Pharmaceuticals, Ministry of Chemical and Fertilizer, Government of India grant number (NPLC-KBT-(2020-2021)) for providing the necessary resources and funding to complete this study.
Author contributions
R. P. and K. T. conceptualization; R. P., S. W. K., S. S., V. K., Debarun Patra, and Durba Pal formal analysis; R. P. investigation; R. P., S. W. K., S. S., V. K., and Debarun Patra data curation; R. P. writing – original draft; S. W. K., S. S., V. K., Debarun Patra, Durba Pal, and K. T. writing – review & editing; K. T. supervision.
Funding and additional information
R. P. expresses his gratitude to the Indian Council of Medical Research (ICMR) for the award of the Research Associate Fellowship Award no. (3/1/2(14)/OBS/2022-NCD-II).
Reviewed by members of the JBC Editorial Board. Edited by Qi-Qun Tang
Supporting information
References
- 1.Machado M.V., Diehl A.M. Pathogenesis of nonalcoholic steatohepatitis. Gastroenterology. 2016;150:1769–1777. doi: 10.1053/j.gastro.2016.02.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.McNelis J.C., Olefsky J.M. Macrophages, immunity, and metabolic disease. Immunity. 2014;41:36–48. doi: 10.1016/j.immuni.2014.05.010. [DOI] [PubMed] [Google Scholar]
- 3.Tilg H., Moschen A.R. Insulin resistance, inflammation, and non-alcoholic fatty liver disease. Trends Endocrinol. Metab. 2008;19:371–379. doi: 10.1016/j.tem.2008.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Baffy G. Kupffer cells in non-alcoholic fatty liver disease: the emerging view. J. Hepatol. 2009;51:212–223. doi: 10.1016/j.jhep.2009.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouwens L., Baekeland M., De Zanger R., Wisse E. Quantitation, tissue distribution and proliferation kinetics of Kupffer cells in normal rat liver. Hepatology. 1986;6:718–722. doi: 10.1002/hep.1840060430. [DOI] [PubMed] [Google Scholar]
- 6.Dixon L.J., Barnes M., Tang H., Pritchard M.T., Nagy L.E. Kupffer cells in the liver. Compr. Physiol. 2013;3:785–797. doi: 10.1002/cphy.c120026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao B., Jeong W.I., Tian Z. Liver: an organ with predominant innate immunity. Hepatology. 2008;47:729–736. doi: 10.1002/hep.22034. [DOI] [PubMed] [Google Scholar]
- 8.Lumeng C.N., Bodzin J.L., Saltiel A.R. Obesity induces a phenotypic switch in adipose tissue macrophage polarization. J. Clin. Invest. 2007;117:175–184. doi: 10.1172/JCI29881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Morris D.L., Singer K., Lumeng C.N. Adipose tissue macrophages: phenotypic plasticity and diversity in lean and obese states. Curr. Opin. Clin. Nutr. Metab. Care. 2011;14:341–346. doi: 10.1097/MCO.0b013e328347970b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang W., Metlakunta A., Dedousis N., Zhang P., Sipula I., Dube J., et al. Depletion of liver Kupffer cells prevents the development of diet-induced hepatic steatosis and insulin resistance. Diabetes. 2010;59:347–357. doi: 10.2337/db09-0016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Odegaard J., Ricardo-Gonzalez R., Eagle A., Vats D., Morel C., Goforth M., et al. Alternative M2 activation of Kupffer cells by PPARδ ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Czaja M.J. Function of autophagy in nonalcoholic fatty liver disease. Dig. Dis. Sci. 2016;61:1304–1313. doi: 10.1007/s10620-015-4025-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martinez-Lopez N., Singh R. Autophagy and lipid droplets in the liver. Annu. Rev. Nutr. 2015;35:215–237. doi: 10.1146/annurev-nutr-071813-105336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang L., Li P., Fu S., Calay E.S., Hotamisligil G.S. Defective hepatic autophagy in obesity promotes ER stress and causes insulin resistance. Cell Metab. 2010;11:467–478. doi: 10.1016/j.cmet.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma A., Anand S.K., Singh N., Dwarkanath A., Dwivedi U.N., Kakkar P. Berbamine induced activation of the SIRT1/LKB1/AMPK signaling axis attenuates the development of hepatic steatosis in high-fat diet-induced NAFLD rats. Food Funct. 2021;12:892–909. doi: 10.1039/d0fo02501a. [DOI] [PubMed] [Google Scholar]
- 16.Pant R., Sharma N., Kabeer S.W., Sharma S., Tikoo K. Selenium-enriched probiotic alleviates western diet-induced non-alcoholic fatty liver disease in rats via modulation of autophagy through AMPK/SIRT-1 pathway. Biol. Trace Elem. Res. 2023;201:1344–1357. doi: 10.1007/s12011-022-03247-x. [DOI] [PubMed] [Google Scholar]
- 17.Navik U., Sheth V.G., Sharma N., Tikoo K. L-Methionine supplementation attenuates high-fat fructose diet-induced non-alcoholic steatohepatitis by modulating lipid metabolism, fibrosis, and inflammation in rats. Food Funct. 2022;13:4941–4953. doi: 10.1039/d1fo03403k. [DOI] [PubMed] [Google Scholar]
- 18.Bäckdahl L., Bushell A., Beck S. Inflammatory signalling as mediator of epigenetic modulation in tissue-specific chronic inflammation. Int. J. Biochem. Cell Biol. 2009;41:176–184. doi: 10.1016/j.biocel.2008.08.023. [DOI] [PubMed] [Google Scholar]
- 19.Luczak M.W., Jagodzinski P.P. The role of DNA methylation in cancer development. Folia Histochem. Cytobiol. 2006;44:143–154. [PubMed] [Google Scholar]
- 20.Maunakea A.K., Chepelev I., Zhao K. Epigenome mapping in normal and disease states. Circ. Res. 2010;107:327–339. doi: 10.1161/CIRCRESAHA.110.222463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suzuki M.M., Bird A. DNA methylation landscapes: provocative insights from epigenomics. Nat. Rev. Genet. 2008;9:465–476. doi: 10.1038/nrg2341. [DOI] [PubMed] [Google Scholar]
- 22.Wang X., Cao Q., Yu L., Shi H., Xue B. Epigenetic regulation of macrophage polarization and inflammation by DNA methylation in obesity. JCI Insight. 2016;1 doi: 10.1172/jci.insight.87748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sass P., Sosnowski P., Podolak-Popinigis J., Górnikiewicz B., Kamińska J., Deptuła M., et al. Epigenetic inhibitor zebularine activates ear pinna wound closure in the mouse. EBioMedicine. 2019;46:317–329. doi: 10.1016/j.ebiom.2019.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Edwards T.M., Myers J.P. Environmental exposures and gene regulation in disease etiology. Environ. Health Perspect. 2007;115:1264–1270. doi: 10.1289/ehp.9951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Skinner M.K., Manikkam M., Guerrero-Bosagna C. Epigenetic transgenerational actions of environmental factors in disease etiology. Trends Endocrinol. Metab. 2010;21:214–222. doi: 10.1016/j.tem.2009.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Christman J.K. 5-Azacytidine and 5-aza-2′-deoxycytidine as inhibitors of DNA methylation: mechanistic studies and their implications for cancer therapy. J. Oncogene. 2002;21:5483–5495. doi: 10.1038/sj.onc.1205699. [DOI] [PubMed] [Google Scholar]
- 27.Schilling J.D., Machkovech H.M., He L., Sidhu R., Fujiwara H., Weber K., et al. Palmitate and lipopolysaccharide trigger synergistic ceramide production in primary macrophages. J. Biol. Chem. 2013;288:2923–2932. doi: 10.1074/jbc.M112.419978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aparicio-Vergara M., Tencerova M., Morgantini C., Barreby E., Aouadi M. Alpha-1 Antitrypsin Deficiency: Methods and Protocols. Springer; New York, NY: 2017. Isolation of Kupffer cells and hepatocytes from a single mouse liver; pp. 161–171. [DOI] [PubMed] [Google Scholar]
- 29.Luo W., Xu Q., Wang Q., Wu H., Hua J. Effect of modulation of PPAR-gamma activity on Kupffer cells M1/M2 polarization in the development of non-alcoholic fatty liver disease. Sci. Rep. 2017;7 doi: 10.1038/srep44612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Graham L., Orenstein J.M. Processing tissue and cells for transmission electron microscopy in diagnostic pathology and research. Nat. Protoc. 2007;2:2439–2450. doi: 10.1038/nprot.2007.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All the data described in this study are contained within the article.