Abstract
Sprouty-related EVH-1 domain-containing (SPRED) proteins are a family of proteins that negatively regulate the RAS-Mitogen-Activated Protein Kinase (MAPK) pathway, which is involved in the regulation of the mitogenic response and cell proliferation. However, the mechanism by which these proteins affect RAS-MAPK signaling has not been elucidated. Patients with mutations in SPRED give rise to unique disease phenotypes; thus, we hypothesized that distinct interactions across SPRED proteins may account for alternative nodes of regulation. To characterize the SPRED interactome and evaluate how members of the SPRED family function through unique binding partners, we performed affinity purification mass spectrometry. We identified 90-kDa ribosomal S6 kinase 2 (RSK2) as a specific interactor of SPRED2 but not SPRED1 or SPRED3. We identified that the N-terminal kinase domain of RSK2 mediates the interaction between amino acids 123 to 201 of SPRED2. Using X-ray crystallography, we determined the structure of the SPRED2-RSK2 complex and identified the SPRED2 motif, F145A, as critical for interaction. We found that the formation of this interaction is regulated by MAPK signaling events. We also find that this interaction between SPRED2 and RSK2 has functional consequences, whereby the knockdown of SPRED2 resulted in increased phosphorylation of RSK substrates, YB1 and CREB. Furthermore, SPRED2 knockdown hindered phospho-RSK membrane and nuclear subcellular localization. We report that disruption of the SPRED2–RSK complex has effects on RAS-MAPK signaling dynamics. Our analysis reveals that members of the SPRED family have unique protein binding partners and describes the molecular and functional determinants of SPRED2–RSK2 complex dynamics.
Keywords: SPRED2, RSK2, neurofibromin, RASopathy, RAS signaling, kinase
Mitogenic signaling mediated by Receptor Tyrosine Kinases (RTKs) activates the RAS-Mitogen-Activated Protein Kinase (MAPK) pathway to regulate various cellular processes such as proliferation and differentiation (1, 2). Hyperactivation of this pathway is known to drive many diseases, including cancer, and is also the underlying cause of a group of developmental disorders collectively termed RASopathies (3, 4). Given the importance of this pathway for cellular homeostasis, these mitogenic signals are stringently regulated at different levels of the pathway by various negative feedback and feedforward loops that coordinate signaling events following RTK activation (2). Sprouty-related protein with EVH-1 domain (SPRED) are a family of proteins that negatively regulate the RAS-MAPK pathway (5). This family consists of SPRED1, SPRED2, and SPRED3, each containing an N- terminal Ena/Vasp Homology (EVH-1) domain, a c-KIT binding domain (KBD), a C-terminal sprouty related (SPR) domain, and two uncharacterized domains (Fig. 1A) (5, 6). Although SPREDs do not exhibit any enzymatic activity, they can regulate RAS–MAPK signaling, mainly through a direct interaction between their EVH-1 domain binding to the RAS-GTPase Activating Protein (GAP), neurofibromin. This interaction localizes neurofibromin to the plasma membrane, which catalyzes RAS GTPase activity and inactivates the cascade (7, 8, 9). Germline loss-of-function variants in SPRED1 cause Legius syndrome, a RASopathy characterized by varying developmental abnormalities such as café-au-lait skin spots and behavioral traits (10). Although SPRED2 or SPRED3 pathogenic variants are not found in individuals with Legius syndrome, a recent study has observed SPRED2 loss-of-function variants in individuals with Noonan syndrome, another RASopathy caused by hyperactivation of the RAS-MAPK pathway (11). Furthermore, Spred1 knockout (KO) mice exhibit a phenotype that resembles Legius syndrome characterized by shortened faces and learning disabilities (12, 13). In contrast, Spred2 KO mice show discrepancies in their reported phenotypes; while initial studies did not describe any significant phenotypes in these mice, another group has recently proposed that these mice exhibit a Noonan syndrome-like phenotype (11, 14). Despite these inconsistencies, there is a clear phenotypic difference between Spred1 and Spred2 null mice, suggesting unique functions for these genes that have yet to be identified (15). We reasoned that unique protein interactions across SPRED proteins may account for their specific phenotypes and functions. Here, we used a systematic approach to characterize the interactome of SPRED proteins and identify unique protein binding partners that may contribute to their distinct roles. Using affinity purification mass spectrometry, we identified an interaction between SPRED2, but not SPRED1 or SPRED3, and serine/threonine kinase ribosomal protein s6 kinase alpha-2 (RSK2). We further characterized the biochemical and structural properties of this interaction and describe a potential mechanism of the SPRED2-RSK2 complex in regulating the RAS-MAPK signaling pathway.
Figure 1.
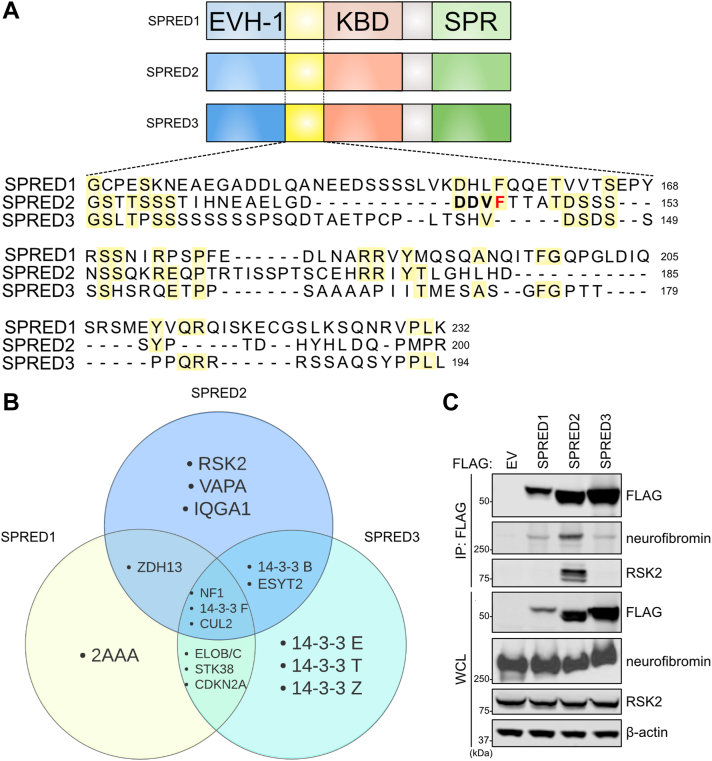
A proteomic screen reveals common and unique interactors of SPRED proteins.A, Schematic of SPRED1-3 domain structure and alignment of the uncharacterized unique domains found between EVH-1 and KBD. B, Venn diagram of SPRED protein binding partners identified from affinity purification mass spectrometry screen. C, Immunoblot of indicated proteins from a co-immunoprecipitation of FLAG-tagged SPRED proteins expressed in HEK293T cells. 2AAA, Serine/threonine-protein phosphatase 2A regulatory subunit A alpha isoform; B, beta; CDKN2A, Cyclin-dependent kinase inhibitor 2A; CUL2, Cullin-2 14-3-3 protein; ELOB/C, Elongin-B/C; EV, Empty vector; EVH-1, Ena/Vasp Homology; F, eta; IP, immunoprecipitation; IQGA1, Ras GTPase-activating-like protein; KBD, KIT binding domain; NF1, neurofibromin; RSK2, ribosomal protein s6 kinase alpha-2; SPR, sprouty related domain; SPRED, Sprouty-related EVH-1 domain-containing; STK38, Serine/threonine kinase 38; T, theta; VAPA, VAMP Associated Protein A; WCL, whole cell lysate; Z, zeta; ZDH13, Palmitoyltransferase ZDHHC13.
Results
A proteomic screen reveals common and unique interactors of SPRED proteins
To identify potential binding partners of the SPRED family, we performed affinity purification-mass spectrometry of FLAG-tagged SPRED proteins expressed in cultured cells. Neurofibromin was a top common interactor across all SPRED proteins, as previously reported (Fig. 1B) (7). In addition, we identified RSK2 and 14-3-3 proteins as unique interactors of SPRED2 and SPRED3, respectively (Fig. S1A). The results from our proteomics screening were validated by immunoprecipitation of the individual SPRED proteins from cell extracts and immunoblotting of endogenous proteins (Figs. 1C and S1B). 14-3-3 proteins are a family of proteins that bind to phospho-serine and phospho-threonine recognition motifs to function as molecular scaffolds, regulate protein function, or alter protein localization (16). RSK2 is a serine/threonine kinase that acts as a downstream effector of the RAS-MAPK pathway to regulate cell cycle progression, transcription, and apoptosis (17). Given the connection with the RAS-MAPK pathway, we focused on the SPRED2–RSK2 interaction for further characterization.
Identification of a unique SPRED2 domain that mediates RSK2 interaction
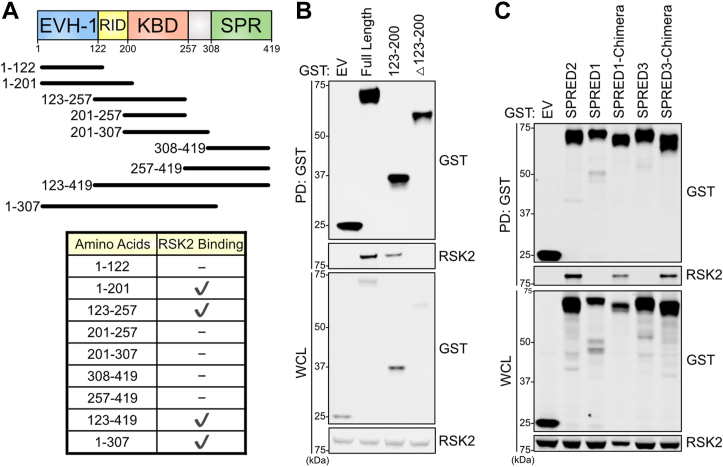
To evaluate the motifs that mediate the interaction between SPRED2 and RSK2, we performed binding assays using a series of SPRED2 truncation mutants (Figs. 2A and S2A). We identified amino acids 123 to 200 as necessary and sufficient for interaction with RSK2 (Fig. 2B). These amino acids encompass a domain of SPRED2 that does not exhibit sequence homology with other family members (Fig. 1A). To further confirm the role of this domain in RSK2 interaction, we generated chimeric proteins in which the SPRED2 amino acids 123 to 200 were replaced with the corresponding amino acids in SPRED1 and SPRED3 and tested binding to RSK2. Using these proteins, we found RSK2 binding to SPRED1 and SPRED3 chimeras (Fig. 2C). We refer to this motif as the RSK interacting domain (RID); however, it is unknown whether SPRED2 may have additional protein interactors in this region (Fig. 2A).
Figure 2.
SPRED2 interacts with RSK2.A, Truncation mutations of SPRED2 expressed in HEK293Ts and immunoblotted to assess for binding to endogenous RSK2. B and C, Immunoblots of indicated proteins from a pulldown of GST -EV, -SPRED2, -SPRED2 mutants, -SPRED1, or -SPRED3 expressed in HEK293T cells. EV, Empty vector; EVH-1, Ena/Vasp Homology; IP, immunoprecipitation; KBD, KIT binding domain; RID, RSK interacting domain; RSK2, ribosomal S6 kinase 2; SPR, sprouty related domain; SPRED, Sprouty-related EVH-1 domain-containing; SPRED1-Chimera/SPRED3-Chimera, SPRED1/3 construct replaced with corresponding SPRED2-RID amino acids; WCL, whole cell lysate.
RSK kinases phosphorylate SPRED2
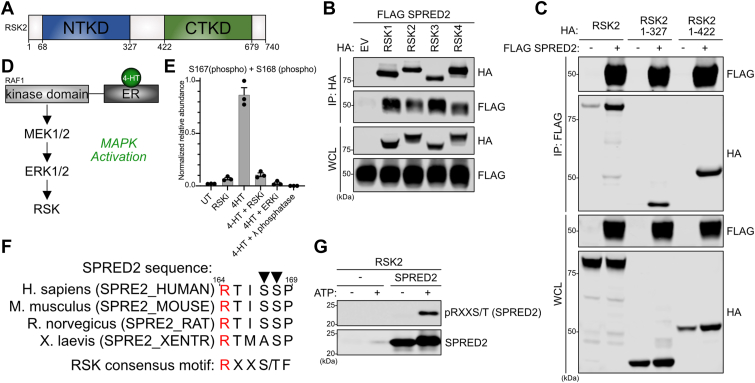
RSK kinases contain an N-terminal kinase domain (NTKD) and a C-terminal kinase domain (CTKD) that are activated through a cascade of phosphorylation events mediated by upstream ERK1/2 and PDK1 kinases (Fig. 3A) (17). The RSK family consists of RSK1-4, and they have greater than 90% and 80% homology in their NTKD and CTKD domains, respectively (18). Using co-immunoprecipitation, we found that SPRED2 could interact with all four RSK family members (Fig. 3B). To identify the specific RSK2 domains that mediate interaction with SPRED2, we generated truncations of RSK2 and found that SPRED2 interacted with the RSK2 NTKD (Fig. 3C). While RSK proteins have two kinase domains, only the NTKD is responsible for phosphorylating substrates (19). Hence, we speculated that SPRED2 might be a substrate of RSK kinases. RSK kinases phosphorylate substrates containing a consensus site that follows the motif RXX(S/T), where R is an Arginine, X is any amino acid, and (S/T) are phosphorylatable Serine or Threonine (20). We identified a putative RSK substrate consensus site (RXXS/T) within the SPRED2 protein sequence within amino acids 123 to 200, suggesting that SPRED2 may be phosphorylated by RSK2 at S167 and S168 (20). To evaluate the phosphorylation events on SPRED2 that may be mediated by RSK, we used HEK293 cells stably expressing the kinase domain of RAF1 (which is constitutively active) fused to the estrogen receptor (ER), henceforth referred as HR1 cells (Fig. 3D) (21, 22). When HR1 cells are treated with 4-hydroxytamoxifen (4-HT), the ligand binds ER, allowing for RAF1-mediated activation of canonical MAPK pathway, including downstream RSK kinases. Using this inducible model, we transiently transfected FLAG-tagged SPRED2 and assessed phosphorylation events through mass spectrometry when cells were treated with 4-HT to activate MAPK, or co-treated with an ERK1/2 inhibitor, pan-RSK inhibitor, or lambda phosphatase to remove all phosphorylation events. We identified two phosphorylation sites on SPRED2, S167 and S168, that were abrogated when cells were treated with a pan-RSK inhibitor (Figs. 3E and S3A). In support of our hypothesis, these sites reside on the putative RSK phosphorylation motif, RXXS/T, and are evolutionarily conserved in SPRED2 protein sequences (Fig. 3F). To validate these findings in vitro, we performed a kinase assay with recombinant RSK2 kinase domain and SPRED2 (104–297) proteins and assessed SPRED2 phosphorylation using a phospho-specific antibody that detects the RXX(S/T) motif. We detected ATP-dependent phosphorylation of SPRED2 (104–297) at RXXS/T sites, suggesting RSK can directly phosphorylate SPRED2 and likely mediates the RSK-dependent phosphorylation of SPRED2 we observed in cells. (Fig. 3G). Furthermore, we found that mutating the S167/S168 phosphorylation sites to alanine, aspartic acid, or glutamic acid had no effect on SPRED2 binding to RSK2 or neurofibromin (Fig. S4B)
Figure 3.
SPRED2 interacts with RSK family and is phosphorylatable.A, Schematic of RSK2 domain architecture. B and C, Immunoblots of indicated proteins from a co-immunoprecipitation of HA-RSK(1–4) or -RSK mutants expressed in HEK 293T cells. D, Schematic of HR1 cell system; ER (estrogen receptor). E, Analysis of phosphorylation events on FLAG-SPRED2-expressing HR1 cells treated with 200 nM 4-HT for 2 h with 4-HT (4-hydroxytamoxifen) and the indicated kinase inhibitors or λ phosphatase (lambda phosphatase). Relative qualification of the peptide abundance in three biological replicates based on extracted MS1 chromatographic peaks. Bars represent mean normalized relative abundance, and error bars show the corresponding standard deviation. F, Analysis of AGC Kinase phosphorylation motif RXX(S/T). R1 highlights in red, and phosphorylatable S is indicated by the black triangle. Alignment was performed with Uniprot using SPRED2 protein sequences from humans (Homo sapiens), mouse (Mus musculus), rat (Rattus norvegicus), and frog (Xenopus laevis). G, SPRED2 (104–297) recombinant protein was subjected to in vitro kinase assay and immunoblotted for phosphorylation by RSK2 with pRXXS/T antibody. CTKD, C-terminal kinase domain; EV, Empty vector; IP, immunoprecipitation; NTKD, N-terminal kinase domain; R, arginine; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing; S/T, phosphorylatable serine or threonine; WCL, whole cell lysate; X, any amino acid.
Structural analysis of SPRED2–RSK2 complex
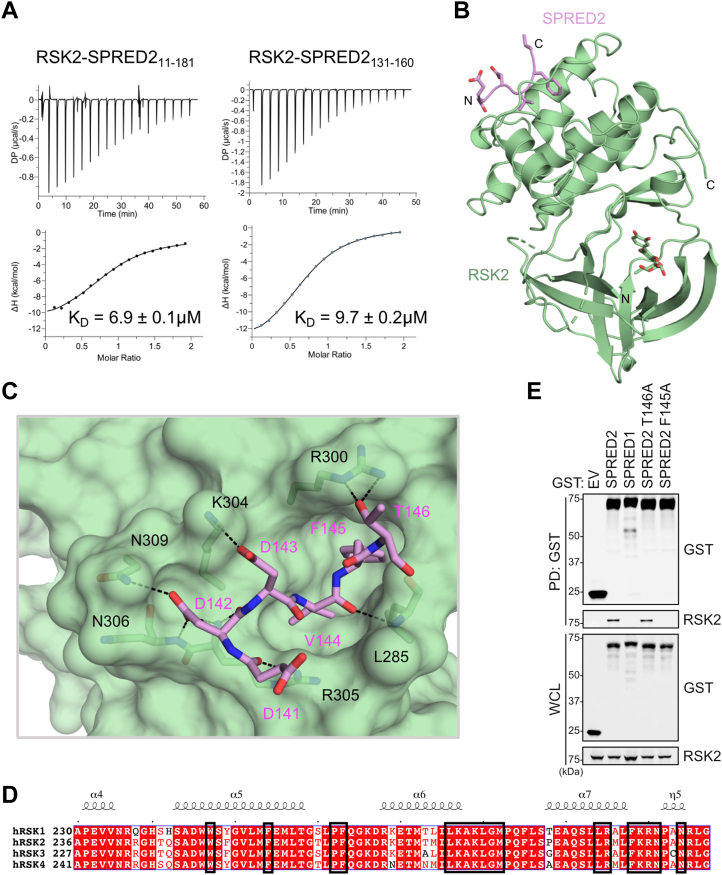
To further characterize the SPRED2–RSK2 interaction, we used isothermal titration calorimetry (ITC) to measure the affinity of the interaction. We observed that the SPRED2 (104–181) protein bound the NTKD of RSK2 (44–367) with an affinity of approximately 9 μM (Fig. S4A). We synthesized additional peptides of various lengths and found the SPRED2 (140–151) and (138–153) peptides bound to the NTKD of RSK2 although with significantly weaker affinities of 36 and 20 μM, respectively (Fig. S4A). A slightly longer peptide, SPRED2 (131–160), bound the NTKD of RSK2 with similar thermodynamics and affinity to the SPRED2 (104–181) (Fig. 4A). The binding of SPRED2 (11–181), which includes the entirety of the EVH-1 domain, is also bound with identical thermodynamics and affinity as SPRED2 (131–160) (Fig. 4A). This further suggests the complete SPRED2 RID is found within residues 131 to 160 (Fig. 2B). We used X-ray crystallography to solve the structure of the complex of the NTKD of RSK2 (46–346) in a complex with SPRED2 RID (131–160). To obtain crystals of the complex, we used the pan-kinase inhibitor, quercitrin, to rigidify and stabilize the active site to achieve a final resolution of 1.8 Å. Despite using a 30-mer peptide, we only observed electron density for six residues of SPRED2 (141DDDVFT146) distal to the active site (Fig. 4B). These residues are bound within a cleft of RSK2, formed by two helices and a loop (residues 281–309), and residues 252, 259, 267, and 268. The SPRED2 hydrophobic residues, V144 and F145, were inserted within a hydrophobic pocket formed from the RSK2 residues W252, F259, F268, L285, G286, M287, F303, and R305. The acidic residues of RSK2 formed several hydrogen bonds to basic residues that line the RSK2 cleft (Fig. S4B). Specifically, SPRED2-D142 formed three hydrogen bonds to RSK2-R305, -N306, and -N307, whereas SPRED2-D143 forms a single hydrogen bond to RSK2-K304 (Fig. 4C). Two hydrogen bonds formed between SPRED2-T146 and RSK-R300 (Fig. 4C). Three additional hydrogen bonds formed between the main chain of SPRED2 (D141, D142, and V144) to RSK2 (Fig. 4C). In total, nine hydrogen bonds formed at the interface and 940 Å2 of RSK2 surface area was buried upon complex formation. The RSK2 binding pocket identified from our structural analysis is conserved across the RSK family, supporting our findings that SPRED2 can interact with the entire RSK family (Figs. 3B and 4D). Given that we only observed six residues of SPRED2 that interact with RSK2, we hypothesized that mutations at these sites would be critical for RSK2 binding. Accordingly, we found that the SPRED2 F145A mutant failed to pulldown RSK2, whereas the SPRED2 T146A mutant retained binding to RSK2 (Fig. 4E).
Figure 4.
StructuralanalysisoftheSPRED2-RSK2interaction.A, Isothermal titration calorimetry of RSK2 (44–367) with SPRED2 (11–181) (left) or SPRED2 (131–160) (right). Dissociation constant (KD) is shown as a mean ± SD. B, The 1.8 Å crystal structure of RSK2 (46–346) (green) in complex with SPRED2 (131–160) (pink) shown as a cartoon. The pan-RSK ligand (quercitrin) bound to the active site is shown as green sticks. C, SPRED2 peptide (shown as pink sticks) bound to the surface of RSK2 (green). Hydrogen bonds are shown as black dashes. D, Sequence alignment of all four human members of the RSK family. Residues that SPRED2 contact on RSK2 are shown in black boxes. Conserved and non-conserved residues are shown in red and white, respectively. E, Immunoblot of indicated proteins from a pulldown of GST- EV, -SPRED2, or -SPRED2 mutants expressed in HEK293T cells. EV, Empty vector; PD, pull down; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing; WCL, whole cell lysate.
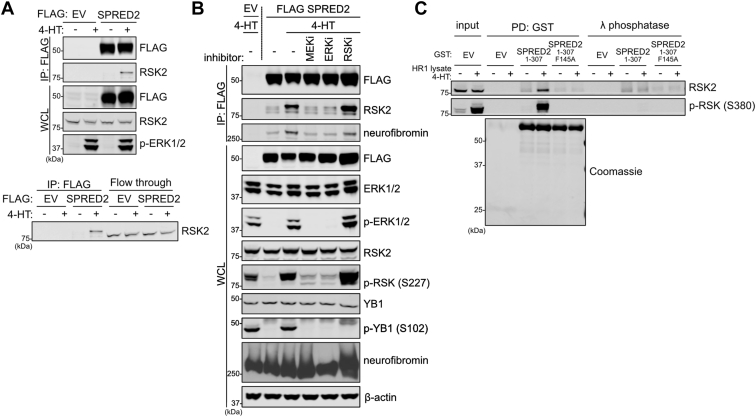
The MAPK pathway regulates SPRED2–RSK interaction
RSK activity is regulated by phosphorylation events that relieve auto-inhibition (17). Since ERK1/2 phosphorylation is a critical step for RSK activation, we assessed whether stimulation of the MAPK pathway could influence the SPRED2–RSK2 interaction. We transiently transfected FLAG-tagged SPRED2 into our inducible HR1 cells and observed that MAPK pathway activation strongly enhanced the interaction between SPRED2 and RSK1/RSK2 (Figs. 5A and S5A). Additionally, we observed a significant band shift (corresponding to an increased apparent molecular weight in SDS-PAGE) in the species of RSK2 bound with SPRED2 compared to the flow-through of unbound RSK2. In co-immunoprecipitation experiments, we also found that SPRED2 interacted with the phosphorylated form of RSK2 (Fig. S5B). Together, these data suggest that post-translational modifications on RSK are required to enhance this interaction. To test if phosphorylation of RSK2 by the upstream MAPK pathway would affect interaction with SPRED2, we treated cells with various MAPK inhibitors. Treatment with MEK1/2 or ERK1/2 inhibitors resulted in disruption of the SPRED2–RSK2 complex, whereas this interaction was unaffected by treatment with an RSK inhibitor (Fig. 5B). Additionally, treating lysates with lambda phosphatase to indiscriminately remove all phosphorylation events present in these lysates abolished the SPRED2–RSK2 interaction (Fig. 5C and S5C). Altogether, these data support a model in which MAPK signaling regulates the dynamics of the SPRED2-RSK complex.
Figure 5.
SPRED2–RSK interaction is regulated by MAPK activation.A, Immunoblot of indicated proteins from a co-immunoprecipitation of FLAG-SPRED2-expressing HR1 cells treated with 200 nM 4-HT for 2 h. Co-immunoprecipitate and flow through were immunoblotted for endogenous RSK2. B, FLAG-SPRED2-expressing HR1 cells were treated with 4-HT and the indicated kinase inhibitors. FLAG-immunoprecipitates and non-binding lysates (flow through) were probed for interacting proteins and phosphorylated proteins respectively. C, Immunoblot of indicated proteins from a pulldown of GST -EV, -SPRED2 and -SPRED2 F415A purified from bacteria, were probed for endogenous RSK2 binding following incubation with lysate from HR1 cells. EV, Empty vector; PD, pull down; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing; WCL, whole cell lysate.
SPRED2 regulates the phosphorylation of RSK substrates and phospho-RSK membrane localization
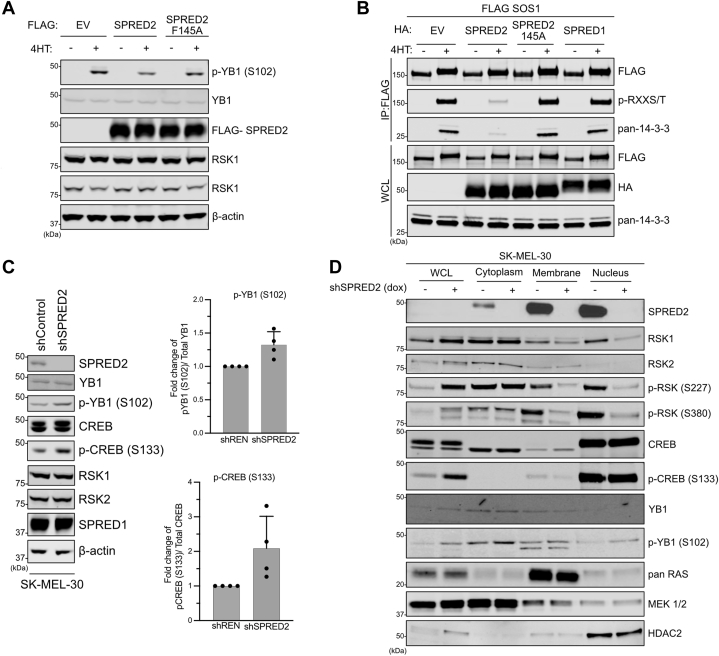
A recent study has shown that RSK kinases bind to proteins containing a DDVF motif and that protein–protein interactions influence the phosphorylation of RSK substrates (23). Our biochemical and structural analyses also identified such a motif in SPRED2, and this prompted us to test whether the association with SPRED2 alters the phosphorylation of known RSK substrates. We overexpressed SPRED2 in cells and observed a decrease in the phosphorylation of the canonical RSK substrate, YB1 at S102 (Fig. 6A) (24). Similarly, we observed a decrease in RSK-mediated phosphorylation of Son of sevenless homolog 1 (SOS1) when SPRED2 but not when the RSK-binding deficient SPRED2 F145A mutant was overexpressed (Fig. 6B) (25). To further test these findings in an endogenous setting, we knocked down SPRED2 in several cell lines (Figs. 6C and S6A). SPRED2 knockdown resulted in an increase in phosphorylated CREB (S133) and phosphorylated YB1 (S102), further supporting a model whereby SPRED2 regulates RSK substrate availability. SPRED proteins are known to promote the membrane localization of neurofibromin, and thus we hypothesized that SPRED2 may also regulate RSK localization. To test this, we performed subcellular fractionation in cells with SPRED2 knockdown. We observed a reduction in the phosphorylation of RSK2 at S386 (S380 in RSK1), the phosphorylation site that recruits PDK1, and S227 (S221 in RSK1), the site in the active loop of the NTKD that is phosphorylated by PDK1, at the plasma membrane and nucleus in these cells (Figs. 6D and S6A) (18). Together, these data indicate that SPRED2 regulates the phosphorylation of RSK substrates, possibly by modulating its subcellular localization.
Figure 6.
SPRED2 regulates the phosphorylation of RSK substrates and subcellular localization.A, Immunoblot of indicated proteins from HR1 cells expressing FLAG-tagged EV, SPRED2, or SPRED2 F145A. B, Immunoblot from a co-immunoprecipitation of HR1 cells co-expressing FLAG-SOS1 and HA-tagged EV, SPRED2, SPRED2 F145A, or SPRED1. C, Immunoblots showing SPRED2 stably knocked down in SK-MEL-30 cells. Relative qualification from three biological replicates based on densitometry of the pCREB and pYB1 in three biological replicates. Bars represent mean normalized relative abundance and error bars show the corresponding standard deviation. Knockdown was induced in cells with doxycycline (1 μg/ml) treatment. D, Immunoblot of subcellular fractionation from SK-MEL-30 cells with SPRED2 knocked down. EV, Empty vector; PD, pull down; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing; WCL, whole cell lysate.
RSK binding promotes SPRED2 complex formation with neurofibromin and negative feedback of MAPK signaling
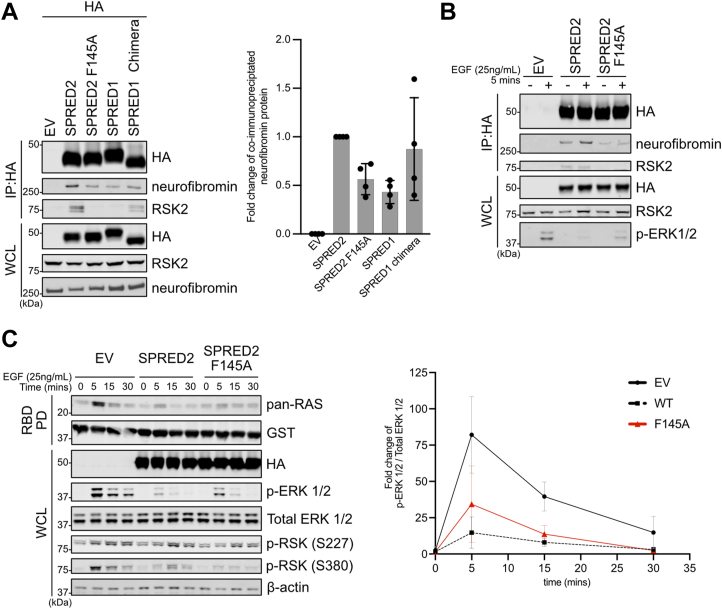
Using the HR1 cell system, we observed that MAPK activation also promoted interaction between SPRED2 and neurofibromin, in a similar manner as SPRED2–RSK2 (Fig. 5C). These findings led us to hypothesize that RSK2 promotes SPRED2 complex formation with neurofibromin. To test this, we stably expressed SPRED2 or the RSK2 binding deficient mutant F145A in HEK293T cells and tested SPRED2 interaction with endogenous neurofibromin (Fig. 7A). We noted a decrease in neurofibromin binding with the SPRED2 F145A mutant compared to WT. Furthermore, we observed that inserting the RID from SPRED2 into SPRED1 (SPRED1 chimera), which confers RSK binding capacity to SPRED1, also enhanced binding to neurofibromin compared to SPRED1. These differences in binding were observed both in normal serum media and epidermal growth factor (EGF) stimulated conditions, further suggesting that RSK2 promotes SPRED2-neurofibromin complex formation (Figs. 7B and S7A). Since neurofibromin is a negative regulator of the RAS-MAPK pathway, we hypothesized that this deficiency in SPRED2 binding has functional consequences on MAPK signaling. To test this, we performed an EGF time course stimulation and observed that cells expressing SPRED2 F145A had higher RAS activation (as assessed by RAS GTP loading) and elevated ERK1/2 phosphorylation, compared to cells expressing SPRED2 WT (Fig. 7C). Given the importance of SPREDs in neurofibromin membrane localization, we hypothesized that this difference in MAPK activation may be due to the inability of SPRED2 F145A to localize neurofibromin to the plasma membrane. However, when performing subcellular fractionation, we observed no statistically significant difference in neurofibromin membrane localization between SPRED2 F145A mutant and SPRED2 expressing cells (Fig. S7B).
Figure 7.
RSK2 negative feedback of MAPK signaling through regulation of SPRED binding to neurofibromin.A–C, Immunoblots of HEK293T cells with stable expression of HA-tagged EV, SPRED2, SPRED2 F145A, SPRED1, or SPRED1 chimera probing against endogenous neurofibromin and RSK2. Relative qualification based on densitometry of neurofibromin in four biological replicates. Bars represent mean fold change, and error bars show the corresponding standard deviation. C, Cells were serum starved for 15-18 h and then stimulated for the indicated time with 25 ng/ml of EGF. Relative qualification based on densitometry of the pERK1/2 from three biological replicates. Bars represent mean fold change, and error bars show the corresponding standard deviation. Lysate was incubated on RBD-GST beads and immunoblotted against endogenous pan-RAS. EV, Empty vector; PD, pull down; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing; WCL, whole cell lysate.
Discussion
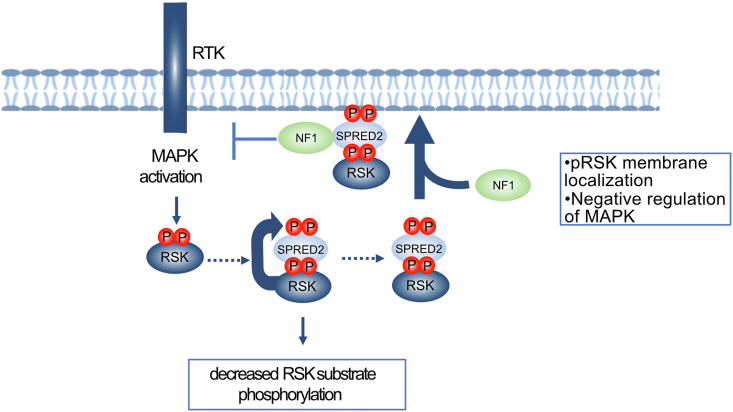
In this work, we have conducted a systematic analysis of the SPRED interactome and identified common and unique interactors among the SPRED family that suggests unique functions of these proteins. Here, we focused on the biochemical properties of the interaction between SPRED2 and RSK2 and demonstrate that this interaction is implicated in regulating RSK and the negative feedback of MAPK signaling. While kinase–substrate interactions are typically transient (26, 27), we report a strong molecular interaction between RSK2 and SPRED2 and characterize SPRED2 as an RSK substrate (26, 27). Recent reports have identified several viral and bacterial proteins that hijack RSK proteins through a conserved DDVF motif (23, 28). A crystal structure of the Kaposi’s sarcoma-associated herpesvirus protein (ORF45) bound to RSK2 shows that the DDVFT motif of ORF45 binds and interacts with RSK2 similarly as SPRED2 (Fig. S4C). However, unlike SPRED2, ORF45 uses additional ∼45 residues to interact with RSK2 forming a complex with an affinity of ∼1 nM, approximately 8000-fold tighter than the RSK2–SPRED2 interaction that we have described (Fig. S4D). This interaction allows ORF45 to hijack the cell to promote pathogen replication (23). In addition to regulating the ability of RSK to phosphorylate substrates, our findings demonstrate that SPRED2 plays an important role in modulating RSK's subcellular localization. Localization to the membrane is a step of RSK activation and previous work has shown that the proximity of RSK1 to RAS depends on SPRED2 (29, 30). This is consistent with our findings and supports our model that SPRED2 regulates phosphorylated RSK at the membrane, presumably to facilitate negative feedback of MAPK signaling (Fig. 8A).
Figure 8.
A RSK–SPRED2 complex regulates the phosphorylation of RSK substrates and MAPK signaling. Formation of the SPRED2-RSK complex is regulated by MAPK activation. SPRED2 binding perturbs phosphorylation of other RSK substrates and leads to SPRED2 phosphorylation. SPRED2 binding promotes pRSK membrane translocation. RSK binding enhances NF1 interaction with SPRED2, leading to negative feedback on MAPK signaling. MAPK, Mitogen-Activated Protein Kinase; RSK2, ribosomal S6 kinase 2; SPRED, Sprouty-related EVH-1 domain-containing.
Additionally, we speculate that the SPRED2–RSK complex provides an extra layer of regulation for MAPK signaling. Our finding shows that SPRED2–RSK complex formation is regulated by MAPK activation. Upon MAPK stimulation, active ERK1/2 phosphorylates RSK2 at T369, S365, and T577 (T363, S359, and T573 in RSK1) (17). This activates the RSK-CTKD and results in auto-phosphorylation at S386 (31). PDK1 binds at the S386 (S380 in RSK1) phosphorylation site, and then phosphorylates S227 (S221 in RSK1) on the RSK NTKD, allowing RSK to activate and phosphorylate substrates (31, 32). We suspect that phosphorylation by ERK1/2 relieves the autoinhibition of RSK and causes a conformational change to promote SPRED2 binding. Our study highlights the crucial role of the interaction between SPRED2 and RSK in the negative regulation of MAPK signaling. Disruption of this interaction leads to hindered interaction with neurofibromin, which in turn results in elevated MAPK signaling (Fig. 8A). This finding is consistent with previous reports that RSK2 plays a role in the negative feedback of RAS-MAPK signaling (33, 34). By elucidating the molecular determinants of the SPRED2-RSK interaction, our study provides a mechanism for negative feedback. Moreover, our proposed model suggests a dual layer of regulation to ensure negative feedback of MAPK signaling, wherein SPRED2 binds and localizes phosphorylated RSK at the membrane, leading to alterations in RSK substrate phosphorylation. Although our studies did not reveal any significant quantitative effects on neurofibromin membrane localization between SPRED2 WT and SPRED2 F145A expressing cells, this plausible mechanism requires further investigation using a more sensitive methodology in future studies (Fig. S7B).
Phosphorylation of substrates by RSK2 is important during development; thus, germline loss-of-function variants in RPS6KA3, the gene encoding RSK2, cause Coffin-Lowry syndrome, a disorder characterized by growth and developmental delay as well as other skeletal anomalies (35, 36, 37, 38). The recent identification of SPRED2 loss-of-function variants in individuals with Noonan syndrome, which also has phenotypic skeletal features, suggests a genetic link between SPRED2 and RSK2 with implications for proper skeletal development (11, 39).
An additional interaction we identified was between SPRED3 and 14-3-3 proteins. While pathogenic mutations in SPRED3 have not been reported, both SPRED3 and 14-3-3 proteins are highly expressed in the brain, suggesting an important neuronal function of SPRED3 (6, 40). Further analysis of this interaction may shed light on a function of SPRED3 that cooperates with 14-3-3 proteins in the context of neuronal development.
In all, our screening results show ways in which SPRED proteins evolved to have unique protein binding partners that, in turn, delegate their functions and alter how they can regulate signaling.
Experimental procedures
Cells and culture conditions
HEK293T, A375, CHL-1, and H441 cells were obtained from the American Type Culture Collection (ATCC), and SK-MEL-30 cells were obtained from DSMZ (Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures, Braunschweig, Germany). HEK293 ΔRaf-1:ER∗ (HR1) cells were kindly provided by Simon Cook (Babraham Institute, Cambridge, UK). HEK293T, A375, CHL1, and HR1 cells were cultured in Dulbecco’s modified Eagle’s medium supplemented with 10% Fetal Bovine Serum (FBS) and Penicillin-Streptomycin. HR1 cells were supplemented with 1 mg/ml G418. SK-MEL-30 were cultured in Roswell Park Memorial Institute (RPMI) 1640 media supplemented with 10% FB) and Penicillin-Streptomycin. Cell lines were regularly tested with MycoAlert PLUS Mycoplasma Detection Kit (Lonza).
Reagents, antibodies, and immunoblotting
Antibodies against Neurofibromin 1 (#14623; 1:1000), RSK2 (5528; 1:1000), HA-Tag (#3724; 1:1000), Phospho-RSK2 (Ser227) (#3556; 1:1000), Phospho-p90RSK (Ser380) (#11989; 1:1000), Phospho-Akt Substrate (RXXS∗/T∗) (#9614; 1:1000), Phospho-p44/42 MAPK (Erk1/2) (Thr202/Tyr204)(#4370; 1:1000), p44/42 MAPK (Erk1/2) (#4696; 1:1000), CREB (#4820; 1:1000), Phospho-CREB (Ser133) (#9198; 1:1000), Phospho-YB1 (Ser102) (#2900; 1:1000), MEK1/2 (# 8727; 1:1000), HDAC2 (#57156, 1:1000), and RAS (#67648, 1:1000) were all purchased from Cell Signaling Technology. Antibodies against GST (#sc-138; 1:1000), pan 14-3-3 (#sc-629; 1:1000), YB1 (#sc101198, 1:1000) were purchased from Santa Cruz Biotechnology. ANTI-FLAG (# F7425) and β-Actin (A5441) were purchased from Sigma-Aldrich, and THE DYKDDDDK (#A00187; 1:1000) was purchased from GenScript. Antibodies against Neurofibromin 1 (#MABE1820, 1:1000) were purchased from Millipore and pan RAS (#108602; 1:1000) was purchased from Abcam. A customized rabbit polyclonal antibody against Spred2_122 to 212aa was generated with GenScript. Lysates were prepared with RIPA Buffer (150 mM NaCl, 1% NP-40, 0.5% Deoxycholate, 0.1% SDS, 50 mM Tris-HCl 7.5) and normalized to load 20 to 30 μg of protein. Immunoblots were performed with Bis-Tris NuPAGE gels (Life Technologies) and transferred to nitrocellulose membranes. Membranes were blocked at room temperature for 1 h in 5% skimmed milk in TBS-T buffer. Primary antibodies were diluted in 3% BSA and incubated at 4 °C overnight. Immunoblots were developed by washing with TBS-T three times and then incubating with secondary antibodies; DyLight680 (611-144-002; 1:10,000) or DyLight800 (610-145-002; 1:10,000) (Rockland), and scanning with LI-COR Odyssey. To detect SPRED2, HRP-conjugated secondary antibodies were used and developed using Immobilon Forte Western HRP Substrate (# WBLUF05000) and HyBlot CL Autoradiography Film (#XC6A2). Fractionation was performed using Subcellular Protein Fractionation Kit (# 7884) from Thermo Fisher Scientific following the manufacturers protocol.
Plasmids, cloning, and transfection
SPRED1-3, RSK1-4, and SOS1 entry clones were obtained from the RAS Pathway 2.0 Clone Collection (Addgene #1000000070). SPRED2 plasmids used for mammalian expression were generated by mutagenesis in pDONR255-SPRED2 template: (1–122, 1–201, 123–257, 201–257, 201–307, 308–419, 257–419, 123–419, 1–307, △123–200, 123–200, SPRED2 F145A, SPRED2 1–307 F145A). All 14-3-3 plasmids were obtained from Addgene. SPRED2 △KBD, SPRED1 chimera, and SPRED3 chimera were generated as a gene fragment with GenScript. RSK2 plasmids used for mammalian expression were generated by mutagenesis in pDONR255-RSK2 template: (RSK2 1–327, 1–422). All mutagenesis primer sequences are available upon request. All entry clones were generated using BP reactions into DONR255 vectors. All expression vectors were generated with LR Gateway cloning containing pDEST302 or pDEST663, EF1α promoter, and either N-terminal 3xFLAG or N-terminal HA tag. All plasmid transfections were performed with jetOPTIMUS DNA Transfection Reagent (# 101000006) following the manufacturers protocol.
Genes for protein expression were generated from DNA constructs initially synthesized as Gateway Entry clones (ATUM). Constructs consisted of E. coli gene optimized fragments containing an upstream tobacco etch virus (TEV) protease site (ENLYFQ/G) followed by fragments of the coding sequence of human SPRED2 or human RSK2 (RPS6KA3) as defined below. Entry clones were transferred to E. coli destination vectors containing aminoterminal His6 (pDest-527, Addgene #11518) or His6-MBP (pDest-566, Addgene #11517) fusions by Gateway LR recombination (Thermo Scientific, Waltham, MA). The shorter RSK2 clone (R765-X33–527) contained an additional non-native tripeptide sequence (Met-Ala-Gly) at the N-terminus of the final construct to improve protein production and crystallization. Constructs generated were: His6-MBP-tev-Hs.SPRED2(11–181); His6-MBP-tev-Hs.SPRED2(104–181); His6-tev-Hs.RSK2(44–367; His6-tev-GMAG-Hs.RSK2(46–346).
Lentiviral transduction
Constructs for stable lentiviral expression were generated as described above with pDEST663 vector. HEK293T cells were co-transfected with the construct of interest, pδ8.91, and pMD2.G in a 3:2:1 ratio, respectively. 48 h post-transfection, the supernatant was collected, passaged through a 0.45-μm filter, and transduced into cells with polybrene. Cells were cultured with the appropriate selection media to select for expressing cells.
To generate knock-down cell lines, miR-E constructs were designed to target SPRED2 as previously reported (41). Lentiviral particles were generated using these miR-E constructs (LT3GEPIR), as described earlier, and transduced into cells. Cells were kept in culture with puromycin (0.5 μg/ml) and doxycycline (1 μg/ml) was used to induce knockdown.
Bacterial protein expression and purification
Proteins were expressed as described for the Dynamite expression protocol (42). Proteins were purified essentially as previously outlined for KRAS (1–169) with some exceptions. Specifically, the Hs.SPRED2(104–181), Hs.RSK2(44–367), and GMAG-Hs.RSK2(46–346) purifications included 300 mM NaCl and 10% glycerol (w/v) in all buffers, MgCl2 was omitted, and the final buffer was 20 mM HEPES, pH 7.4, 300 mM NaCl, 1 mM TCEP, and 10% glycerol (w/v) (43). Final buffer for Hs.SPRED2 (11–181) was 20 mM HEPES, pH 7.4, 150 mM NaCl, and 1 mM TCEP. Recombinant GST-tagged proteins -EV, -SPRED2 1 to 307, and -SPRED2 1 to 307 F145A were generated by gateway cloning into pGEX-6. Plasmids were then transformed into BL21 (DE3) cells. Cultures were inoculated with starter cultures and grown at 37 °C for 3 to 4 h. Following, expression was induced with 0.2 mM IPTG for 14 to 16 h at 18 °C. Cells were lysed in 50 mM Tris HCl (pH 8.0), 300 mM NaCl, 5% glycerol, 1 mM DTT by sonication. Lysate was put over Glutathione Sepharose 4B beads (Cytiva Life Sciences), washed, and stored in 50% glycerol at −20 °C.
Immunoprecipitation and GST pulldown assays
Cells were lysed in buffer containing 150 mM NaCl, 1% IGEPAL CA-630, 10% Glycerol, 50 mM Tris-HCl 7.5 with Phosphatase Inhibitor Cocktail 1(P2850; 1:100), Phosphatase Inhibitor Cocktail 2 (P5726; 1:100), and Protease Inhibitor Cocktail (mammalian cell extracts) (P8340; 1:100) from Sigma-Aldrich. Lysates were rotated for 1 h at 4 °C with Glutathione Sepharose 4B beads (Cytiva Life Sciences), ANTI-FLAG M2 Affinity Gel (Sigma), or Pierce Anti-HA Agarose (ThermoFisher Scientific). Beads were washed three times with lysis buffer and resuspended with LDS sample buffer. For immunoprecipitation from HR1 cells, cells were treated with 200 nM 4-HT for 2 h and with 100 nM trametinib (MEKi), 1 μM SCH772984 (ERKi), or 10 μM LJH685 (RSKi) where indicated.
To measure RAS.GTP levels, cells were serum starved for 15 to 18 h and stimulated with 25 ng/ml of Epidermal Growth Factor (EGF). Lysates rotated for 1 h at 4 °C with RBD-GST beads.
Kinase assays
Recombinant RSK2 was purchased from SignalChem (R17-10G) and SPRED2 was made as described in the bacterial protein expression and purification section. Reactions contained 100 ng of RSK2, 1 μg of SPRED2, and 1 mM of ATP (Sigma- Aldrich; #A7699). Reactions were brought up to 25 μl with Kinase Assay Buffer (5 mM MOPS, pH7.2, 2.5 mM β-glycerol-phosphate, 10 mM MgCl2, 1 mM EGTA, 0.4 mM EDTA, 50 ng/ml BSA) and were incubated at 30 °C and were stopped after 15 min with LDS sample buffer.
Mass spectrometry
To evaluate bindings partners, 5 μg of 3x-FLAG-SPRED1-3 was transiently transfected into 106 HEK293T cells. Cells were lysed as described above and incubated with magnetic flag beads.
To evaluate phosphorylation events on SPRED2, 5 μg of 3x-FLAG-SPRED2 was transiently transfected into 5 × 106 HR1 cells. Cells were treated with 4-HT to activate MAPK and co-treated with 1 μM SCH772984 (ERKi) or 10 μM LJH685 (RSKi) for 2 h.
Cells were lysed as described above. Conditions with lambda phosphatase were treated post-lysis, following manufacturers’ guidelines (New England BioLabs; P0753S). The lysates were then incubated with magnetic flag beads.
The beads were washed with ice-cold 20 mM Tris-HCl (pH 8.0), 2 mM CaCl2 buffer, and frozen prior to trypsin digest. On-bead trypsin digests were performed as previously described (44). Digested peptides were desalted with ZipTip u-C18 pipette tips (Millipore), vacuum dried, and reconstituted in 15 μl of 0.1% formic acid. 2.5 uL of each sample were injected on Acquity UPLC M-Class system (Waters) and analyzed by LC-MS/MS on Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher Scientific) equipped with 15 cm silica-C18 EasySpray column (Thermo Fisher Scientific). The column was operated at 45 °C, and reversed-phase chromatography was performed using a conventional two-buffer system (Buffer A - 0.1% formic acid in water; Buffer B - 0.1% formic acid in acetonitrile) at 400 nl/min flow rate, as follows: Load at 2% B for 20 min; 2 to 60% B gradient over 60 min; brief wash at 80% B; equilibration at 2% B. The mass spectrometer was operated in DDA (Full-MS/ddMS2) mode with one survey scan (375–1500 m/z, R = 120,000, AGC target of 4e5) followed by HCD MS2 scans up to 3 s limit (AGC target of 5e4, max IT 100 ms, R = 30,000, isolation window 1.6 m/z, NCE 30%, stepped collision 5%, and 30 s dynamic exclusion).
Acquired MS data were analyzed as previously described (45). Briefly, spectra were searched using Protein Prospector (version 6.2.4) against the human SwissProt database downloaded on 01/18/2021 and the corresponding decoy database of randomly shuffled peptide sequences (46, 47, 48). Default “ESI-Q-high-res” parameters with trypsin as the protease were used. Up to two missed cleavages were allowed. Carbamidomethyl-C was a constant modification, with default variable modifications plus phosphorylation at STY. Up to three modifications per peptide were considered. Mass tolerance was 5 ppm for precursor ions and 15 ppm for fragment ions. The false discovery rate was set for <1% for peptides, and at least three unique peptides per protein were required. Protein Prospector search results were exported as BiblioSpec spectral library for downstream analysis in Skyline (v21) to quantify peptide and protein abundances (49). Statistical analysis of peptide and protein abundances in different biological conditions was carried out using MSstats package integrated in Skyline (50).
Peptides
SPRED2 (140–151) (GDDDVFTTATDS), SPRED2 (138–153) (ELGDDDVFTTATDSSS) and SPRED2 (131–160) (STIHNEAELGDDDVFTTATDSSSNSSQKRE) peptides were synthesized by Genscript with the N-terminal residues acetylated.
Isothermal titration calorimetry measurements
Proteins were extensively dialyzed against 50 mM HEPES, 500 mM NaCl, 3 mM β-mercaptoethanol, 5 mM ETDA, pH 8.0. Duplicate ITC measurements were performed on a MicroCal PEAQ-ITC instrument (Malvern Panalytical). An ITC experiment typically consisted of 40 to 70 μM of RSK2 (44–367) in the cell with 400 to 700 μM of either purified SPRED2 (11–181) and SPRED2 (104–181), or SPRED2 (131–160), SPRED2 (138–153) or SPRED2 (140–151) peptides in the syringe. All measurements were carried out at 25 °C, with a stirring speed of 750 rpm and 19 injections of 2.2 μl injected at 210 s intervals. Data analysis was performed using a “one set of sites” model using the MicroCal PEAQ-ITC analysis software (v1.41, Malvern Panalytical).
Crystallization and data collection
RSK2 (46–346) (1.1 ml of 138 μM) was mixed with a 1.1-fold excess of quercitrin (20 mM stock dissolved in ethylene glycol, Selleck Chemicals S3824) and incubated on ice for 15 min. Protein–ligand complex was dialysis against 50 mM Tris, 600 mM NaCl, 5 mM EDTA, 5 mM β-mercaptoethanol, pH 8.0 for 16 h at 4 °C. No precipitation had occurred, and the volume had increased to 1.35 ml. Quercitrin interferes with UV absorbance, so the concentration was estimated based on the volume change (final concentration ∼110 μM). The SPRED2 (131–160) peptide was added at an equimolar ratio from a stock of 1.24 mM stock dissolved in dialysis buffer. The RSK2-SPRED2 complex was incubated for 1 h on ice before crystallization screening was carried out at 20 °C using the sitting-drop vapor diffusion method by mixing the complex with an equal volume of reservoir solution (200 nl:200 nl). A single crystal grew over the course of 5 days in condition B12 of the PEGRx screen (30% w/v PEG 2K MME, 0.1 M Tris, pH 8.0, Hampton Research). The crystal was cryoprotected with 15% v/v glycerol. Using a single crystal, a 1.8 Å dataset was collected on beamline 24-ID-C at the Advance Photon Source (Argonne).
Structure determination and analysis
The dataset was indexed and integrated with XDS before scaling, truncating, and converting to structure factors with Aimless from the CCP4 suite (51, 52). The Matthews coefficient suggested a single copy of RSK2 (46–346) in the asymmetric unit. Molecular replacement was carried out with MOLREP using RSK2 (PDB 4GUE) as a search model (52). Refinement was initially carried out by Refmac5 and manual building in COOT (53, 54). After a few rounds of refinement, an inspection of the difference (Fo-Fc) map showed positive density for the six residues in the 30-mer SPRED2 peptide, suggesting the remaining residues are disordered inside the crystal. Final refinement was carried out in Phenix.Refine with occupancy refinement for the SPRED2 peptide (final occupancy for the SPRED2 peptide was 0.89 (55). Figures were generated with PyMOL (54). All crystallographic and structural analysis software were supported and provided by the SBGrid Consortium (56). Crystallographic data collection and refinement statistics are shown in Table 1.
Table 1.
Crystallographic data collection and refinement statistics
| RSK246–346-SPRED2131-160 | |
|---|---|
| Data collection | |
| Space group | I 1 2 1 |
| Cell dimensions | |
| a, b, c (Å) | 83.44, 41.10, 99.26 |
| α, β, γ (°) | 90.0, 113.89, 90.0 |
| Resolution (Å) | 49.39–1.80 (1.84–1.80) |
| Rmerge | 0.059 (0.771) |
| Rpim | 0.057 (0.752) |
| I/σI | 8.8 (1.4) |
| Completeness (%) | 97.7 (98.8) |
| Redundancy | 3.3 (3.3) |
| CC(1/2) | 0.99 (0.57) |
| Refinement | |
| Resolution (Å) | 49.39–1.80 |
| No. reflections | 93,088 |
| Rwork/Rfree | 18.3/22.5 |
| No. atoms | |
| Protein | 2343 |
| Ligand/ion | 32 |
| Water | 139 |
| B-factors | |
| RSK2 | 38.9 |
| SPRED2 | 62.7 |
| Ligand/ion | 31.2 |
| Water | 40.3 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.007 |
| Bond angles (°) | 0.903 |
Data availability
The atomic coordinates and structure factors of the RSK2-SPRED2 complex have been deposited in the Protein Data Bank and are available under the accession number 8EQ5. The authors confirm that the data supporting the findings of this study are available within the article and its supporting information.
Supporting information
This article contains supporting information.
Conflict of interest
P. C is a founder and advisory board of Venthera, Inc. The other authors declare no competing interests. F. M. is a consultant for the following companies: Amgen; Daiichi Ltd, Frontier Medicines, Exuma Biotech, Ideaya Biosciences, Kura Oncology, Leidos Biomedical Research, Inc, PellePharm, Pfizer Inc, PMV Pharma and Quanta Therapeutics. F. M. is a consultant and co-founder for the following companies (with ownership interest including stock options): BridgeBio; DNAtrix Inc; Olema Pharmaceuticals, Inc; and Quartz. F. M. is the scientific director of the National Cancer Institute (NCI) RAS Initiative at the Frederick National Laboratory for Cancer Research/Leidos Biomedical Research, Inc. F. M. has been a recipient of research grants from Daiichi Sankyo, Gilead Sciences and has a current grant from Boehringer-Ingelheim.
Acknowledgments
The authors wish to acknowledge Dominic Esposito, Peter Frank, Simon Messing, Matt Drew, Gulcin Gulten, Ashley Mitchell, Nitya Ramakrishnan, Matt Smith, Kelly Snead, Troy Taylor, and Vanessa Wall from the Protein Expression Laboratory at the Frederick National Laboratory for cloning, protein expression, and protein purification support. We thank the UCSF Mass Spectrometry Facility and A. L. Burlingame for providing MS instrumentation support for this. We also thank members of the McCormick lab for their support and input.
Author contributions
J. L., P. C., and F. M. conceptualization; J. L., D. A. B., A. U., J. L. M., M. C.-D., and D. K. S. investigation; D. A. B., A. U., and D. K. S. formal analysis; J. L. writing - original draft; D. A. B., M. J. S., A. U., P. C., D. K. S., and F. M. writing- reviewing and editing; M. J. S., P. C. methodology; M. J. S. resources; F. M. supervision.
Funding and additional information
This work was supported by the NCI 1R35CA197709–01 and DOD US Army Med. Res. Acq. Activity W81XWH2010129 to F. M. P. C.’s work is supported by NCI (K99CA245122) and DOD CDMRP NFRP (W81XWH-20-1-0391). J. L. and M. C.-D are supported by UCSF's IMSD program, funded by the National Institute of General Medical Sciences (5R25GM056847). J. L. is also a UCSF discovery fellow. D. S. and D. A. B. are supported by the NCI (HHSN261200800001E).
Reviewed by members of the JBC Editorial Board. Edited by Donita Brady
Supporting information
References
- 1.Roovers K., Assoian R.K. Integrating the MAP kinase signal into the G1 phase cell cycle machinery. Bioessays. 2000;22:818–826. doi: 10.1002/1521-1878(200009)22:9<818::AID-BIES7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 2.Neben C.L., Lo M., Jura N., Klein O.D. Feedback regulation of RTK signaling in development. Dev. Biol. 2017;447:71–89. doi: 10.1016/j.ydbio.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simanshu D.K., Nissley D.V., McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castel P., Rauen K.A., McCormick F. The duality of human oncoproteins: drivers of cancer and congenital disorders. Nat. Rev. Cancer. 2020;20:383–397. doi: 10.1038/s41568-020-0256-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wakioka T., Sasaki A., Kato R., Shouda T., Matsumoto A., Miyoshi K., et al. Spred is a sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- 6.Kato R., Nonami A., Taketomi T., Wakioka T., Kuroiwa A., Matsuda Y., et al. Molecular cloning of mammalian Spred-3 which suppresses tyrosine kinase-mediated Erk activation. Biochem. Bioph Res. Commun. 2003;302:767–772. doi: 10.1016/s0006-291x(03)00259-6. [DOI] [PubMed] [Google Scholar]
- 7.Stowe I.B., Mercado E.L., Stowe T.R., Bell E.L., Oses-Prieto J.A., Hernández H., et al. A shared molecular mechanism underlies the human rasopathies legius syndrome and neurofibromatosis-1. Genes Dev. 2012;26:1421–1426. doi: 10.1101/gad.190876.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan W., Markegard E., Dharmaiah S., Urisman A., Drew M., Esposito D., et al. Structural insights into the SPRED1-neurofibromin-KRAS complex and disruption of SPRED1-neurofibromin interaction by Oncogenic EGFR. Cell Rep. 2020;32 doi: 10.1016/j.celrep.2020.107909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dunzendorfer-Matt T., Mercado E.L., Maly K., McCormick F., Scheffzek K. The neurofibromin recruitment factor Spred1 binds to the GAP related domain without affecting Ras inactivation. Proc. Natl. Acad. Sci. U. S. A. 2016;113:7497–7502. doi: 10.1073/pnas.1607298113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brems H., Chmara M., Sahbatou M., Denayer E., Taniguchi K., Kato R., et al. Germline loss-of-function mutations in SPRED1 cause a neurofibromatosis 1–like phenotype. Nat. Genet. 2007;39:1120–1126. doi: 10.1038/ng2113. [DOI] [PubMed] [Google Scholar]
- 11.Motta M., Fasano G., Gredy S., Brinkmann J., Bonnard A.A., Simsek-Kiper P.O., et al. SPRED2 loss-of-function causes a recessive Noonan syndrome-like phenotype. Am. J. Hum. Genet. 2021;108:2112–2129. doi: 10.1016/j.ajhg.2021.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inoue H., Kato R., Fukuyama S., Nonami A., Taniguchi K., Matsumoto K., et al. Spred-1 negatively regulates allergen-induced airway eosinophilia and hyperresponsiveness. J. Exp. Med. 2005;201:73–82. doi: 10.1084/jem.20040616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Denayer E., Ahmed T., Brems H., Woerden G.V., Borgesius N.Z., Callaerts-Vegh Z., et al. Spred1 is required for synaptic plasticity and hippocampus-dependent learning. J. Neurosci. 2008;28:14443–14449. doi: 10.1523/JNEUROSCI.4698-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nobuhisa I., Kato R., Inoue H., Takizawa M., Okita K., Yoshimura A., et al. Spred-2 suppresses Aorta-Gonad-Mesonephros Hematopoiesis by inhibiting MAP kinase activation. J. Exp. Med. 2004;199:737–742. doi: 10.1084/jem.20030830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lorenzo C., McCormick F. SPRED proteins and their roles in signal transduction, development, and malignancy. Genes Dev. 2020;34:1410–1421. doi: 10.1101/gad.341222.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Freeman A.K., Morrison D.K. 14-3-3 Proteins: diverse functions in cell proliferation and cancer progression. Semin. Cell Dev. Biol. 2011;22:681–687. doi: 10.1016/j.semcdb.2011.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anjum R., Blenis J. The RSK family of kinases: emerging roles in cellular signalling. Nat. Rev. Mol. Cell Biol. 2008;9:747–758. doi: 10.1038/nrm2509. [DOI] [PubMed] [Google Scholar]
- 18.Carriere A., Ray H., Blenis J., Roux P.P. The RSK factors of activating the Ras/MAPK signaling cascade. Front. Biosci. 2008;13:4258–4275. doi: 10.2741/3003. [DOI] [PubMed] [Google Scholar]
- 19.Chrestensen C.A., Sturgill T.W. Characterization of the p90 ribosomal S6 kinase 2 Carboxyl-terminal domain as a protein kinase. J. Biol. Chem. 2002;277:27733–27741. doi: 10.1074/jbc.M202663200. [DOI] [PubMed] [Google Scholar]
- 20.Pearce L.R., Komander D., Alessi D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010;11:9–22. doi: 10.1038/nrm2822. [DOI] [PubMed] [Google Scholar]
- 21.Samuels M.L., Weber M.J., Bishop J.M., McMahon M. Conditional transformation of cells and rapid activation of the mitogen-activated protein kinase cascade by an estradiol-dependent human raf-1 protein kinase. Mol. Cell Biol. 1993;13:6241–6252. doi: 10.1128/mcb.13.10.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boughan P.K., Argent R.H., Body-Malapel M., Park J.-H., Ewings K.E., Bowie A.G., et al. Nucleotide-binding oligomerization domain-1 and epidermal growth factor receptor critical regulators of β-defensins during helicobacter pylori infection. J. Biol. Chem. 2006;281:11637–11648. doi: 10.1074/jbc.M510275200. [DOI] [PubMed] [Google Scholar]
- 23.Sorgeloos F., Peeters M., Hayashi Y., Borghese F., Capelli N., Drappier M., et al. A case of convergent evolution: several viral and bacterial pathogens hijack RSK kinases through a common linear motif. Proc. Natl. Acad. Sci. U. S. A. 2022;119 doi: 10.1073/pnas.2114647119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cesare D.D., Jacquot S., Hanauer A., Sassone-Corsi P. Rsk-2 activity is necessary for epidermal growth factor-induced phosphorylation of CREB protein and transcription of c-fos gene. Proc. Natl. Acad. Sci. U. S. A. 1998;95:12202–12207. doi: 10.1073/pnas.95.21.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Saha M., Carriere A., Cheerathodi M., Zhang X., Lavoie G., Rush J., et al. RSK phosphorylates SOS1 creating 14-3-3-docking sites and negatively regulating MAPK activation. Biochem. J. 2012;447:159–166. doi: 10.1042/BJ20120938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waas W.F., Dalby K.N. Transient protein-protein interactions and a random-ordered kinetic mechanism for the phosphorylation of a transcription factor by extracellular-regulated protein kinase 2. J. Biol. Chem. 2002;277:12532–12540. doi: 10.1074/jbc.M110523200. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Cole P.A. Catalytic mechanisms and regulation of protein kinases. Methods Enzymol. 2014;548:1–21. doi: 10.1016/B978-0-12-397918-6.00001-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alexa A., Sok P., Gross F., Albert K., Kobori E., Póti Á.L., et al. A non-catalytic herpesviral protein reconfigures ERK-RSK signaling by targeting kinase docking systems in the host. Nat. Commun. 2022;13:472. doi: 10.1038/s41467-022-28109-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Richards S.A., Dreisbach V.C., Murphy L.O., Blenis J. Characterization of regulatory events associated with membrane targeting of p90 ribosomal S6 kinase 1. Mol. Cell. Biol. 2001;21:7470–7480. doi: 10.1128/MCB.21.21.7470-7480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng D.K., Oni T.E., Park Y., Thalappillil J.S., Ting H.-C., Alagesan B., et al. Oncogenic KRAS engages an RSK1/NF1 complex in pancreatic cancer. Biorxiv. 2020 doi: 10.1101/2020.09.14.295394. [preprint] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vik T.A., Ryder J.W. Identification of serine 380 as the Major site of Autophosphorylation of Xenopus pp90rsk. Biochem. Biophys. Res. Commun. 1997;235:398–402. doi: 10.1006/bbrc.1997.6794. [DOI] [PubMed] [Google Scholar]
- 32.Dalby K.N., Morrice N., Caudwell F.B., Avruch J., Cohen P. Identification of regulatory phosphorylation sites in mitogen-activated protein kinase (MAPK)-activated protein kinase-1a/p90 rsk that are inducible by MAPK. J. Biol. Chem. 1998;273:1496–1505. doi: 10.1074/jbc.273.3.1496. [DOI] [PubMed] [Google Scholar]
- 33.Cheng D.K., Oni T.E., Thalappillil J.S., Park Y., Ting H.-C., Alagesan B., et al. Oncogenic KRAS engages an RSK1/NF1 pathway to inhibit wild-type RAS signaling in pancreatic cancer. Proc. Natl. Acad. Sci. U. S. A. 2021;118 doi: 10.1073/pnas.2016904118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nadratowska-Wesolowska B., Haugsten E.M., Zakrzewska M., Jakimowicz P., Zhen Y., Pajdzik D., et al. RSK2 regulates endocytosis of FGF receptor 1 by phosphorylation on serine 789. Oncogene. 2014;33:4823–4836. doi: 10.1038/onc.2013.425. [DOI] [PubMed] [Google Scholar]
- 35.Trivier E., Cesare D.D., Jacquot S., Pannetier S., Zackai E., Young I., et al. Mutations in the kinase Rsk-2 associated with coffin-lowry syndrome. Nature. 1996;384:567–570. doi: 10.1038/384567a0. [DOI] [PubMed] [Google Scholar]
- 36.Lv Y., Zhu L., Zheng J., Wu D., Shao J. Growth concerns in coffin–lowry syndrome: a case report and literature review. Front. Pediatr. 2019;6:430. doi: 10.3389/fped.2018.00430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harum K.H., Alemi L., Johnston M.V. Cognitive impairment in coffin-lowry syndrome correlates with reduced RSK2 activation. Neurology. 2001;56:207–214. doi: 10.1212/wnl.56.2.207. [DOI] [PubMed] [Google Scholar]
- 38.Hanauer A., Young I.D. Coffin-lowry syndrome: clinical and molecular features. J. Med. Genet. 2002;39:705–713. doi: 10.1136/jmg.39.10.705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rauen K.A. The RASopathies. Annu. Rev. Genomics Hum. Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cornell B., Toyo-oka K. 14-3-3 proteins in brain development: neurogenesis, neuronal Migration and Neuromorphogenesis. Front. Mol. Neurosci. 2017;10:318. doi: 10.3389/fnmol.2017.00318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fellmann C., Hoffmann T., Sridhar V., Hopfgartner B., Muhar M., Roth M., et al. An optimized microRNA Backbone for effective single-copy RNAi. Cell Rep. 2013;5:1704–1713. doi: 10.1016/j.celrep.2013.11.020. [DOI] [PubMed] [Google Scholar]
- 42.Taylor T., Denson J.-P., Esposito D. Optimizing expression and solubility of proteins in E. coli using modified media and Induction parameters. Methods Mol. Biol. 2017;1586:65–82. doi: 10.1007/978-1-4939-6887-9_5. [DOI] [PubMed] [Google Scholar]
- 43.Kopra K., Vuorinen E., Abreu-Blanco M., Wang Q., Eskonen V., Gillette W., et al. Homogeneous dual-Parametric-Coupled assay for simultaneous nucleotide exchange and KRAS/RAF-RBD interaction monitoring. Anal. Chem. 2020;92:4971–4979. doi: 10.1021/acs.analchem.9b05126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Castel P., Cheng A., Cuevas-Navarro A., Everman D.B., Papageorge A.G., Simanshu D.K., et al. RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science. 2019;363:1226–1230. doi: 10.1126/science.aav1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cuevas-Navarro A., Van R., Cheng A., Urisman A., Castel P., McCormick F. The RAS GTPase RIT1 compromises mitotic fidelity through spindle assembly checkpoint suppression. Curr. Biol. 2021;31:3915–3924.e9. doi: 10.1016/j.cub.2021.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Baker P.R., Trinidad J.C., Chalkley R.J. Modification site localization scoring integrated into a search Engine. Mol. Cell. Proteomics. 2011;10 doi: 10.1074/mcp.M111.008078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chalkley R.J., Baker P.R. Use of a glycosylation site database to improve glycopeptide identification from complex mixtures. Anal. Bioanal. Chem. 2017;409:571–577. doi: 10.1007/s00216-016-9981-2. [DOI] [PubMed] [Google Scholar]
- 48.Bateman A., Martin M.-J., Orchard S., Magrane M., Alpi E., Bely B., et al. UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47:D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pino L.K., Searle B.C., Bollinger J.G., Nunn B., MacLean B., MacCoss M.J. The Skyline ecosystem: informatics for quantitative mass spectrometry proteomics. Mass Spectrom. Rev. 2020;39:229–244. doi: 10.1002/mas.21540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Choi M., Chang C.-Y., Clough T., Broudy D., Killeen T., MacLean B., et al. MSstats: an R package for statistical analysis of quantitative mass spectrometry-based proteomic experiments. Bioinformatics. 2014;30:2524–2526. doi: 10.1093/bioinformatics/btu305. [DOI] [PubMed] [Google Scholar]
- 51.Kabsch W. Xds. Acta Crystallogr. D Biol. Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Winn M.D., Ballard C.C., Cowtan K.D., Dodson E.J., Emsley P., Evans P.R., et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. D Biol. Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Murshudov G.N., Skubák P., Lebedev A.A., Pannu N.S., Steiner R.A., Nicholls R.A., et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Emsley P., Lohkamp B., Scott W.G., Cowtan K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liebschner D., Afonine P.V., Baker M.L., Bunkóczi G., Chen V.B., Croll T.I., et al. Macromolecular structure determination using X-rays, neutrons and electrons: recent developments in phenix. Acta Crystallogr. D Struct. Biol. 2019;75:861–877. doi: 10.1107/S2059798319011471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Morin A., Eisenbraun B., Key J., Sanschagrin P.C., Timony M.A., Ottaviano M., et al. Collaboration gets the most out of software. Elife. 2013;2 doi: 10.7554/eLife.01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates and structure factors of the RSK2-SPRED2 complex have been deposited in the Protein Data Bank and are available under the accession number 8EQ5. The authors confirm that the data supporting the findings of this study are available within the article and its supporting information.