Abstract
Tomato spotted wilt virus (TSWV) poses a serious threat to tomato (Solanum lycopersicum) production. In this study, tomato inbred line YNAU335 was developed without the Sw-5 locus, which confers resistance or immunity to TSWV (absence of infection). Genetic analysis demonstrated that immunity to TSWV was controlled by a dominant nuclear gene. The candidate genes were mapped into a 20-kb region in the terminal of the long arm of chromosome 9 using bulk segregant analysis and linkage analysis. In this candidate region, a chalcone synthase–encoding gene (SlCHS3) was identified as a strong candidate gene for TSWV resistance. Silencing SlCHS3 reduced flavonoid synthesis, and SlCHS3 overexpression increased flavonoid content. The increase in flavonoids improved TSWV resistance in tomato. These findings indicate that SlCHS3 is indeed involved in the regulation of flavonoid synthesis and plays a significant role in TSWV resistance of YNAU335. This could provide new insights and lay the foundation for analyzing TSWV resistance mechanisms.
Supplementary information
The online version contains supplementary material available at 10.1007/s11032-022-01325-5.
Keywords: Tomato spotted wilt virus, Fine mapping, Resistance gene, Gene function, Chalcone synthase, Flavonoids
Introduction
Tomato spotted wilt virus (TSWV) is an important virus in the Tospovirus genus of the Bunyaviridae family (Lopez et al. 2011; Peiro et al. 2014). The virus is vectored by several species of thrips, most importantly the western flower thrip (Boonham et al. 2002). It has an extensive host range of more than 800 plant species in 82 families (Soler et al. 2003; Chung et al. 2018). The virus has spread to most countries in the world and causes serious losses to the production of crops, such as tomatoes (Hoffmann et al. 2001; Boonham et al. 2002; Lopez et al. 2011).
Tomato germplasm resources with TSWV resistance are mostly wild species, such as Peruvian (Solanum peruvianum Mill.) and Chilean (S. chilense Mill.) tomatoes (Stevens et al. 1994; Gordillo and Stevens, 2008). Multiple TSWV-resistant genes, namely Sw-1a, Sw-1b, Sw-2, Sw-3, Sw-4, Sw-5a, Sw-5b, Sw-6, and Sw-7, are derived from common tomato (Solanum lycopersicum L.) or wild tomato plants (Finlay et al., 1952; Finlay et al., 1953; Stevens et al., 1992; Canady et al., 2001; Dockter et al., 2009; Lee et al., 2015). Among these, Sw-5 and Sw-7 have been widely used in resistance breeding (Stevens et al. 1991; Boiteux and Giordano, 1993). Multiple molecular markers such as RAPDs, RFLPs, and SCARs, have been developed for Sw-5 (Stevens et al. 1995; Chague et al. 1996; Dianese et al. 2010). A series of technologies including marker linkage analysis (Stevens et al. 1995), YAC library construction (Brommonschenkel and Tanksley, 1997), and homology cloning (Folkertsma et al. 1999) revealed two CC-(NB-ARC)-LRR genes, Sw5-a and Sw5-b, but only Sw5-b increased resistance to TSWV (Spassova et al. 2001). TSWV resistance–breaking isolates have emerged in different countries after using resistant cultivars carrying Sw-5 including the T992 isolate in Italy (Ciuffo et al. 2005) and the Pujol1TL3 isolate in Spain (Debreczeni et al. 2015). Mutations in the TSWV genome, for example, a substitution of C to Y at position 118 or T to N at position 120 in the TSWV movement protein have been found to inhibit resistance induced by Sw-5 (Hoffmann et al. 2001; Lopez et al. 2011). Sw-7 has been reported to exhibit field resistance against TSWV, but the molecular mechanism of Sw-7 remains unknown (Ramesh et al., 2017).
Flavonoids are important secondary metabolites in plants that play significant roles in the regulation of plant growth and development (Peer et al. 2004; Wu et al. 2018). Flavonoids are induced by stress such as ultraviolet light, free radicals, and pathogens, thereby increasing resistance to these stressors in plants (Yamasaki et al. 1997; Ryan et al. 2002; Silva et al. 2015; Wu et al. 2018). Previous research has shown that the transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum (Wang et al. 2020). VqWRKY31 can directly or indirectly bind to the promoters of structural genes in the flavonoid synthesis pathway and promote flavonoid accumulation; it contributes to strong resistance to powdery mildew in grapes (Yin et al. 2022). Chalcone synthase (CHS) is a key enzyme in the plant flavonoid synthesis pathway (Nagamatsu et al. 2007; Wang et al. 2010). Based on the whole tomato genome sequence, eight CHS genes have been identified, with protein sequence lengths varying from 160 to 438 distributed on chromosomes 1, 5, 6, 9, and 12 (Ruan et al. 2013). The CHS genes are induced by a variety of pathogens including viruses and fungi, thus further increasing the synthesis of flavonoids, and potentially enhancing resistance to these pathogens and contributing to the strong resistance of plants (Gutha et al. 2010; Samac et al. 2011). Moreover, the expression levels of CHS genes are induced by abiotic stress, such as exposure to UV light, mechanical damage, and salt (Christie and Jenkins, 1996; Dehghan et al. 2014).
In this study, the inbred line YNAU335 was not infected after inoculation with the TSWV isolate YNAU2015. Immunity was controlled by the dominant nuclear gene SlCHS3, which was successfully located at the terminal of the long arm of chromosome 9. Its presence resulted in altered expression patterns in different resistant inbred lines and affected tomato resistance to TSWV.
Materials and methods
Tomato materials and TSWV isolate
The tomato materials used in this experiment were YNAU335, No. 5, and 96172I, and S. peruvianum LA2823, LA3858, PI128657. YNAU335, No. 5, and 96172I were developed from local tomato varieties collected from Yuanmou County, Yunnan Province, China. S. peruvianum LA2823, LA3858, and PI128657 were donated from the Institute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences. TSWV isolate YNAU2015 was identified and preserved in our laboratory.
To demonstrate that the TSWV resistance locus of YNAU335 was different from that of Sw-5, the co-dominant SCAR marker “Sw-5–2” (Dianese et al., 2010) linked to Sw-5 was employed to phenotype YNAU335. Genomic DNA was extracted from the leaves of tomato plants using a DNA Secure Plant Kit (BioTeke) and used as a template for PCR amplification. The primer pSw-5–2 for Sw-5 is shown in Table S1.
TSWV inoculation
The TSWV mechanical inoculation method used was based on that of Sundaraj et al. (2014) with minor changes. TSWV was stored on tobacco (Nicotiana tabacum L.) and spread by mechanical inoculation, as follows. Diseased tobacco leaves were ground in 0.1 M phosphate buffer at a ratio of 1:10 (wt/vol). The buffer (pH = 7.0) contained 0.2% sodium sulfite, 0.01 M mercaptoethanol, 0.01 g/mL emery, and 0.01 g/mL Celite 545. The whole grinding process was performed in an ice bath, and the grinding solution was used as the viral homogenate. The tomato leaves were gently wiped with emery, and the homogenate was applied to the leaves. For the non-inoculated control group, phosphate buffer alone was applied. After inoculation, all seedlings were placed in an incubator for 3 weeks. For inoculation with TSWV using thrips, healthy tomato seedlings were transferred to the Vegetable Cognitive Center of Yunnan Agricultural University, where a population of TSWV-harboring thrips has existed for many years.
Bulk segregant analysis, genome resequencing, and genetic map construction
Two DNA pools consisting of a TSWV-resistant pool and a TSWV- sensitive pool from the F2 population were used for bulk segregant analysis (BSA) (Lv et al. 2019). For each pool, genomic DNA was extracted from the leaves of 20 individual plants from the F2 population. The parental YNAU335 and No. 5 inbred lines and segregated populations of the F2 generation were established for genome resequencing to detect single nucleotide polymorphisms (SNPs) and InDels between the parents and F2 generation. The SNP index and delta SNP were used to identify candidate chromosomal regions related to the TSWV-resistant gene (Win et al. 2017). The polymorphic InDel markers were developed in this region to identify the genotypes of F2 individuals in expanded F2 populations and to construct a genetic map (Chi et al. 2010). A total of 460 recessive F2 individuals were used for the linkage analysis. The primers are shown in Table S1.
Generation of transgenic lines
To generate virus-induced gene silencing (VIGS) transgenic plants, the gene segments of Solyc09g091500, Solyc09g091510, and Solyc09g091520 were cloned and inserted independently into the pTRV2 plasmid. The primers p500-V, pSlCHS3-V, and p520-V for the three candidate genes are shown in Table S1. The positive Agrobacterium tumefaciens strain GV3101 containing pTRV2, pTRV2-PDS, and pTRV2-target gene segments were each co-injected with positive GV3101 containing pTRV1 into cotyledons of the YNAU335 inbred line (Sheng et al. 2015).
To generate overexpressing transgenic No. 5 plants, full-length SlCHS3 cDNA was amplified using the specific primer pSlCHS3-O (Table S1). The PCR product was fused to the binary plant transformation vector pBI121. The pBI121 plasmid and pBI121-35S-SlCHS3 fusion plasmid were introduced into GV3101. Positive GV3101 was transformed into No. 5 cotyledons (Sheng et al. 2015). The primers used in this study are shown in Table S1.
DAS-ELISA and reverse transcription and real-time quantitative PCR
Inoculated tomato plants were used to detect TSWV accumulation using DAS-ELISA and real-time quantitative PCR (RT-qPCR) employing the primer pN-Q for the TSWV nucleoprotein (N) gene. A 100-μL tomato leaf extract was used for detection using the TSWV DAS-ELISA kit from Agdia (Elkhart, IN, USA) according to the manufacturer’s instructions. Healthy tomato leaves were used as controls. The chromogenic reaction took place for 30 min in the dark, after which the optical density (OD) readings were recorded at 415 nm in iMark Microplate Reader (Bio-RAD, Hercules, CA, USA). If the sample OD415/control OD415 ≥ 2, the sample was considered positive, while if the sample OD415/control OD415 < 2, the sample was considered negative (Canady et al. 2001). Each analysis was performed in biological and technical triplicate.
Gene expression profiles of Solyc09g091500, SlCHS3, and Solyc09g091520 in transgenic tomatoes were estimated by RT-qPCR using the Eppendorf Mastercycler ep Realplex real-time PCR system, and tomato housekeeping ribosomal protein L2 (RPL2) was used as the reference gene (Lovdal et al., 2009). Total RNA was extracted using a Quick RNA Isolation Kit (HuaYueYang Biotech Co., Ltd., Beijing, China) and then treated with RNase-free DNase I (TAKARA, Japan) to remove genomic DNA. Total RNA (2 μg) was reverse transcribed into first-strand cDNA using the M-MLV Reverse Transcriptase Kit (TAKARA, Japan), according to the manufacturer’s protocol, and oligo(dT) primer and random primers were used in the reverse transcription reactions. The cDNA samples were diluted fivefold and used as a template for RT-qPCR. Forty PCR cycles were performed according to the following temperature scheme: 95 °C for 15 s, 60 °C for 15 s, and 72 °C for 20 s. The cycle threshold (Ct) values were read from the quantification curves. Each analysis was performed in biological and technical triplicate using the 2−ΔΔCt method (Zhao et al. 2013). The sequence of Solyc09g091510 was amplified using the PCR method. The primers pN-Q, pRPL2-Q, p500-Q, pSlCHS3-Q, p520-Q, and p510-cds for N, RPL2, Solyc09g091500, SlCHS3, Solyc09g091520, and Solyc09g091510, respectively, are shown in Table S1.
Flavonoid content measurement
The total flavonoid content was measured using an aluminum chloride colorimetric method (Djeridane et al. 2006). In brief, 3.0 g of tomato leaf was weighed, and flavonoids were extracted using an ultrasonic method. Rutin was used as the standard substance to generate a standard curve (Rasha 2017). Flavonoid extracts were stained using aluminum chloride, and the absorptions were measured at 415 nm. The total flavonoid content was calculated using the following formula:
C (mg·g−1) = C1 × V/m,
where C represents the total flavonoid content; C1 represents the concentration of the sample extract; V represents the volume of the extract; and m represents the weight of the fresh sample.
Results
Phenotyping identification of tomato materials
The “Sw-5–2” primer pair was used to fingerprint the TSWV susceptible and resistant tomato lines in our experiment. The Sw-5-derived amplicon of 574 bp was observed in S. peruvianum LA3858, LA2823, and PI128657 (Fig. S1A, lanes 1–3). However, YNAU335, No. 5, and 96172I displayed only a smaller amplicon of 464 bp (Fig. S1A, lanes 4–6). A sequence comparison of “Sw-5–2” PCR amplicons from the six tomato varieties is shown in Fig.S2. Mechanical inoculation with TSWV (YNAU2015 isolate) was performed in LA3858 and YNAU335 to evaluate resistance to TSWV using a double antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) and RT-qPCR. Both LA3858 and YNAU335 showed no TSWV symptoms after being inoculated with phosphate buffer (Fig. S1B). After being inoculated with TSWV, LA3858 showed typical TSWV symptoms and was considered positive by ELISA with an OD415 ratio of 3.1. YNAU335 displayed no TSWV symptoms and a Ct value which was considered negative with an OD415 ratio of ~ 1 (Fig. S1C). The YNAU335 inbred line without the Sw-5 locus had immunity to TSWV.
Resistance identification and genetic control
TSWV inoculation was performed using a mechanical method in the laboratory and by thrips in the field. The resistance levels of YNAU335 and No. 5 inbred lines, as well as their F1 generation, were determined. Both YNAU335 and the F1 generation had no necrotic lesions in the inoculation zones. In contrast to the Ct values of the TSWV N gene in infected No. 5, the N gene was not detected in the TSWV-inoculated plants of the YNAU335 line or in the F1 generation of the YANAU355 or No. 5 lines. The OD415 ratios of the TSWV-inoculated YNAU335 and F1 plants were ~ 1, while the OD415 ratios of TSWV-inoculated No. 5 were ~ 3 (Fig. 1). The F2 and BC1 population was constructed with the resistant and susceptible parental inbred lines of YNAU335 and No. 5. The segregation ratios of the F2 and BC1 generations between the YANAU355 and No. 5 lines were 1:3 and 1:1, respectively (Table 1).
Fig. 1.
Identification of resistance in tomato materials infected with the TSWV isolate YNAU2015. Tomato materials were inoculated with phosphate buffer (A) and TSWV using a mechanical method (B) and thrips (C). The numbers in the first and second brackets indicate the Ct values of the N gene using RT-qPCR and the ratios of the sample OD415 to the control OD415 using ELISA, which are shown as the means of the three biological replicates ± standard deviations (SDs), “-”: not detected
Table 1.
Genetic ratios of YNAU335 and No. 5 inbred lines resistant to TSWV
| Samples | Numbers of immune plants | Numbers of susceptible plants | Ratios (immune:susceptible) | χ2 |
|---|---|---|---|---|
| P1: YNAU335 | 67 | 0 | —— | —— |
| P2: No. 5 | 0 | 84 | —— | —— |
| F2: P1♂ × P2♀ | 512 | 165 | 3:1 | 0.58 |
| RF2: P1♀ × P2♂ | 431 | 139 | 3:1 | 1.02 |
| BCP2: (P1 × P2) ♂ × P2♀ | 121 | 116 | 1:1 | 2.03 |
| RBCP2: (P1 × P2) ♀ × P2♂ | 140 | 131 | 1:1 | 0.01 |
Fine mapping of TSWV-resistant genes
We established parental inbred lines of YNAU335 and No. 5, as well as susceptible and resistant selections from the F2 generation. BSA genome resequencing was used to map the candidate chromosomal region, which was located at the terminal of the long arm of chromosome 9. Nine InDel markers in this region were used to construct a genetic map using the screening results obtained using polymorphic InDel markers in the 304 recessive individuals from the F2 segregating population, and the recombinant numbers were identified with the markers. Eventually, the candidate region was reduced to a physical interval of about 20 kb which was between the InDel 2 and InDel 3 markers (Fig. 2). Three candidate genes were identified between the InDel2 and InDel3 markers according to the tomato reference genome: Solyc09g091500, Solyc09g091510 (SlCHS3), and Solyc09g091520 (Table 2).
Fig. 2.
Fine mapping of the TSWV-resistant candidate genes
Table 2.
Annotation of the disease-resistance candidate genes
| Gene ID | Chromosome | Function description |
|---|---|---|
| Solyc09g091500 | ch09 | U6 snRNA-associated Sm-like protein LSm5 IPR006649 Like-Sm ribonucleoprotein, eukaryotic and archaea-type, core |
| Solyc09g091510 | ch09 | Chalcone synthase IPR011141 Polyketide synthase, type III |
| Solyc09g091520 | ch09 | 60S acidic ribosomal protein P0IPR001790 Ribosomal protein L10 |
Functional verification of TSWV-resistant candidate genes
The VIGS system was used to preliminarily verify the functions of the three candidate genes: Solyc09g091500, Solyc09g091510, and Solyc09g091520. The YNAU335 inbred line was susceptible to TSWV after silencing Solyc09g091510 (Fig. 3). However, the resistance of YNAU335 to TSWV did not change after silencing Solyc09g091500 or Solyc09g091520 (Fig. 3). Therefore, Solyc09g091510 appeared to be the disease resistance gene in YNAU335, which encoded CHS and was named SlCHS3. A SlCHS3-overexpression transgenic system in transgenic No. 5 lines showed increased resistance to TSWV(Fig. 4). SlCHS3 was amplified using the PCR method, the sequence alignment of this gene revealed one vital SNP mutation (C to T) among the TSWV susceptible and resistant tomato lines (Fig. S3A), and one SNP caused a non-synonymous mutation in YNAU335 (Fig. S3B), further suggesting SlCHS3 as a strong candidate gene involved in the TSWV resistance in tomato (Fig. S3).
Fig. 3.
Functional identification of the SlCHS3 gene using a transgenic silencing system. Identification of transgenic plants using a phenotypic analysis of phytoene desaturase (PDS) silencing and RT-qPCR. WT-1 and WT-2 represent empty vector-transformed plants, which were assigned a value of 1. The error bar on each column represents the SD of three biological replicates (Student’s t-test, *P < 0.05, **P < 0.01) (A). The resistance performances of transgenic plants after inoculation with TSWV using a mechanical method (VIGS-1, -3, and -4) (B) and thrips (VIGS-8, -9, and -17) (C). The ratios of the sample OD415 to the control OD415 using ELISA, which are shown as the means of three biological replicates ± standard deviations (SDs)
Fig. 4.
Functional identification of the SlCHS3 gene using a transgenic overexpression system. SlCHS3 gene expression levels in transgenic lines were assessed by RT-qPCR. WT-1 and WT-2 represent empty vector-transformed plants, which were assigned a value of 1. The error bar on each column represents the SD of three biological replicates (Student’s t-test, **P < 0.01) (A). The resistance performances of transgenic lines after inoculation with TSWV using a mechanical method (L-1, -4, and -6) (B) and thrips (L-9, -10, and -12) (C).The ratios of the sample OD415 to the control OD415 using ELISA, which are shown as the means of three biological replicates ± SDs
Impact of TSWV on flavonoid content in different tomato materials
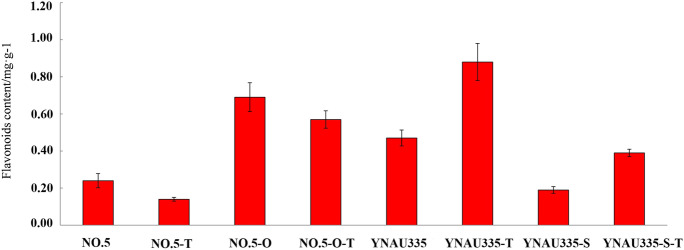
The flavonoid content in SlCHS3-overexpressing No. 5 inbred lines was higher than that of the wild type, and the flavonoid content in silenced YNAU335 inbred lines was lower than that of the wild type. TSWV decreased the flavonoid content in the wild type and SlCHS3-overexpressing No. 5 inbred lines and increased the flavonoid content in the wild type and SlCHS3-silenced YNAU335 inbred lines (Fig. 5).
Fig. 5.
Flavonoids contents in different tomato materials. No. 5 and YNAU335 represent the No. 5 and YNAU335 inbred lines under normal growth conditions, respectively. No. 5-T and YNAU335-T represent the No. 5 and YNAU335 inbred lines inoculated with TSWV, respectively. No. 5-O represents the No. 5 inbred line overexpressing SlCHS3. No. 5-O-T represents the SlCHS3-overexpressing No. 5 inbred lines inoculated with TSWV. YNAU335-S represents the SlCHS3-silenced YNAU335 inbred line. YNAU335-S-T represents the -silenced YNAU335 inbred line inoculated with TSWV. The error bar on each column represents the SD of three biological replicates
Discussion
TSWV-resistant tomato resources are concentrated in wild species, and interspecific hybridization challenges the genetic-based improvement of these crop resources (Stevens et al. 1994; Gordillo and Stevens 2008). To date, eight TSWV-resistant genes have been discovered: Sw-1a, Sw-1b, Sw-2, Sw-3, Sw-4, Sw-5, Sw-6, and Sw-7 (Finlay 1953; Stevens et al. 1991; Boiteux and Giordano 1993; Rosello et al. 1998; Dockter et al. 2009). Of these, Sw-5 is the only verified gene that has been applied to tomato production (Spassova et al. 2001). However, TSWV isolates that break Sw-5 resistance have been observed (Lopez et al. 2011). In this study, we produced tomato inbred line YNAU335 without the Sw-5 locus that is immune to TSWV. YNAU335 is an indeterminate growth inbred line with an average fruit weight of 335 g. It is normally hybridized with other tomato inbred lines, and its immunity to TSWV is controlled by a dominant gene. We successfully mapped this resistance gene in the YNAU335 inbred line and verified its functions.
The TSWV resistance of YNAU335 is regulated by the quality gene SlCHS3, which was identified using BSA genome sequencing, genetic mapping, and functional identification. SlCHS3 encodes CHS, which is the first key enzyme that regulates flavonoid synthesis (Nagamatsu et al. 2007; Wang et al. 2010). The CHS gene can be induced by a variety of pathogens (Gutha et al. 2010; Samac et al. 2011). Flavonoids regulated by CHS also play important roles in pathogen resistance (Nagamatsu et al. 2007; Wang et al. 2010). In summary, SlCHS3 is the TSWV-resistant quality gene in the YNAU335 inbred line.
Previous reports have shown that gene expression levels in secondary metabolite biosynthesis are highly downregulated upon TSWV infection in tomato plants (Catoni et al., 2009). Salicylic acid, mechanical damage, and salt can induce flavonoid synthesis in plants, and flavonoid content can regulate resistance ability (Christie et al., 1996; Dehghan et al., 2014). In addition, stress, such as ultraviolet light, free radicals, and pathogenic bacteria, can induce flavonoid synthesis to regulate plant resistance (Wu et al., 2018; Yamasaki et al., 1997; Ryan et al., 2002; Franco et al., 2015). The transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in the sorghum (Wang et al. 2020). VqWRKY31 can regulate the flavonoid synthesis pathway and promote flavonoid accumulation; it contributes to the strong resistance of grapes to powdery mildew (Yin et al. 2022). The enriched flavonoid content may improve the defense response and increase the nutritional values of sorghum grain/bran (Hsu et al. 2009). In this study, SlCHS3 expression influenced flavonoid content. The reason for this may be that the exon, intron, and promoter sequences of SlCHS3 have mutations between the TSWV-resistant and TSWV-susceptible inbred lines. Therefore, we hypothesized that the differential expression levels of SlCHS3 influence resistance in tomato.
Conclusions
We used BSA and linkage analysis to map the TSWV-resistant gene into a 20-kb region on chromosome 9, and there were three candidate genes in the region. Based on the results of gene silencing and overexpression, SlCHS3 was considered a strong candidate gene. SlCHS3 silencing reduced flavonoid synthesis, and SlCHS3 overexpression increased flavonoid content. The increased flavonoid content improved TSWV resistance in tomato. We speculated that the gene expression level controls flavonoid synthesis and influences TSWV resistance in tomato. Our study provides new insights and lays the foundation for analyzing TSWV-resistant mechanisms.
Supplementary information
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable
Author contribution
Wrote first draft: JL and KZ. Designed experimental work: JL, MD, SJ, HZ, and KZ. Investigation: ZL, ZW, and JL. Provided experimental materials: JL, ZY, and YL. Analyzed data: JL, KZ, and JX. Wrote original manuscript: JL and KZ. Wrote and edit review: JL and KZ. Visualization: JL. Supervised the whole work: KZ. Project administration: KZ. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (32160715, 31660576, 31760583), the Joint Project of Basic Agricultural Research in Yunnan Province (2018FG001-004), the General Project of Yunnan Science and Technology plan (2016FB064), and Research and Integrated Applications of Key Technology in Standardized Production of Facility Vegetables (202102AE090005).
Data availability
All of the whole-genome resequencing data used in the BSA analysis is available from the NCBI Short Read Archive (SRA, BioProject ID: PRJNA751989), and the raw data are freely available at http://www.ncbi.nlm.nih.gov/bioproject/751989. The other supporting data are included as additional files.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Boiteux LS, Giordano LB. Genetic basis of resistance against two Tospovirus species in tomato (Lycopersicon esculentum) Euphytica. 1993;71:151–154. doi: 10.1007/BF00023478. [DOI] [Google Scholar]
- Boonham N, Smith P, Walsh K, Tame J, Morris J, Spence N, Bennison J, Barker I. The detection of Tomato spotted wilt virus (TSWV) in individual thrips using real time fluorescent RT-PCR (TaqMan) J Virol Methods. 2002;101:37–48. doi: 10.1016/S0166-0934(01)00418-9. [DOI] [PubMed] [Google Scholar]
- Brommonschenkel SH, Tanksley SD. Map-based cloning of the tomato genomic region that spans the Sw-5 Tospovirus resistance gene in tomato. Mol Gen Genet. 1997;256:121–126. doi: 10.1007/s004380050553. [DOI] [PubMed] [Google Scholar]
- Canady MA, Stevens MR, Barineau MS, Scott JW. Tomato spotted wilt virus (TSWV) resistance in tomato derived from Lycopersicon chilense Dun. LA 1938. Euphytica. 2001;117:19–25. doi: 10.1023/A:1004089504051. [DOI] [Google Scholar]
- Catoni M, Miozzi L, Fiorilli V, Lanfranco L, Accotto GP. Comparative analysis of expression profiles in shoots and roots of tomato systemically infected by tomato spotted wilt virus reveals organ-specific transcriptional responses. Mol Plant Microbe Interactions. 2009;22:1504–1513. doi: 10.1094/MPMI-22-12-1504. [DOI] [PubMed] [Google Scholar]
- Chague V, Mercier JC, Guenard M, de Courcel A, Vedel F. Identification and mapping on chromosome 9 of RAPD markers linked to Sw-5 in tomato by bulked segregant analysis. Theor Appl Genet. 1996;92:1045–1051. doi: 10.1007/BF00224047. [DOI] [PubMed] [Google Scholar]
- Chi XF, Zhou XS, Shu QY. Fine mapping of a Xantha mutation in rice (Oryza sativa L.) Euphytica. 2010;172:215–220. doi: 10.1007/s10681-009-0048-8. [DOI] [Google Scholar]
- Christie JM, Jenkins GI. Distinct UV-B and UV-A/Blue light signal transduction pathways induce chalcone synthase gene expression in Arabidopsis cells. Plant Cell. 1996;8:1555–1567. doi: 10.1105/tpc.8.9.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BN, Lee JH, Kang BC, Koh SW, Joa JH, Choi KS, Ahn JJ. HR-mediated defense response is overcome at high temperatures in Capsicum Species. Plant Pathol J. 2018;34:71–77. doi: 10.5423/PPJ.NT.06.2017.0120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciuffo M, Finetti-Sialer MM, Gallitelli D, Turina M. First report in Italy of a resistance-breaking strain of tomato spotted wilt virus infecting tomato cultivars carrying the Sw5 resistance gene. Plant Pathol. 2005;54:564. doi: 10.1111/j.1365-3059.2005.01203.x. [DOI] [Google Scholar]
- Debreczeni DE, Lopez C, Aramburu J, Daros JA, Soler S, Galipienso L, Falk BW, Rubio L. Complete sequence of three different biotypes of tomato spotted wilt virus (wild type, tomato Sw-5 resistance-breaking and pepper Tsw resistance-breaking) from Spain. Arch Virol. 2015;160:2117–2123. doi: 10.1007/s00705-015-2453-8. [DOI] [PubMed] [Google Scholar]
- Dehghan S, Sadeghi M, Anne PA, Fischer R, Lakes-Harlan R, Kavousi HR, Vilcinskas A, Rahnamaeian M. Differential inductions of phenylalanine ammonia-lyase and chalcone synthase during wounding, salicylic acid treatment, and salinity stress in safflower. Carthamus Tinctorius Biosci Rep. 2014;34:273–282. doi: 10.1042/BSR20140026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dianese EC, de Fonseca MEN, Goldbach R, Kormelink R, Inoue-Nagata AK, Resende RO, Boiteux LS. Development of a locus-specific, co-dominant SCAR marker for assisted-selection of the Sw-5 (Tospovirus resistance) gene cluster in a wide range of tomato accessions. Mol Breeding. 2010;25:133–142. doi: 10.1007/s11032-009-9313-8. [DOI] [Google Scholar]
- Djeridane A, Yousfi M, Nadjemi B, Boutassouna D, Stocker P, Vidal N. Antioxidant activity of some algerian medicinal plants extracts containing phenolic compounds. Food Chem. 2006;97:654–660. doi: 10.1016/j.foodchem.2005.04.028. [DOI] [Google Scholar]
- Dockter KG, O'Neil DS, Price DL, Scott JW, Stevens MR (2009) Molecular mapping of the tomato spotted wilt virus resistance gene "Sw-7" in tomato. ASHS Conference
- Finlay KW. Inheritance of spotted wilt resistance in the tomato. II. Five genes controlling spotted wilt resistance in four tomato types. Aust J Biol Sci. 1953;6:153–163. doi: 10.1071/BI9530153. [DOI] [PubMed] [Google Scholar]
- Folkertsma RT, Spassova M, Prins M, Stevens MR, Hille J, Goldbach RW. Construction of a bacterial artificial chromosome (BAC) library of Lycopersicon esculentum cv. Stevens and its application to physically map the Sw-5 locus. Mol Breed. 1999;5:197–207. doi: 10.1023/A:1009650424891. [DOI] [Google Scholar]
- Franco DM, Silva EM, Saldanha LL, Adachi SA, Schley TR, Rodrigues TM, Dokkedal AL, Nogueira FT, de Almeida LFR. Flavonoids modify root growth and modulate expression of SHORT-ROOT and HD-ZIP III. J Plant Physiol. 2015;188:89–95. doi: 10.1016/j.jplph.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Gordillo LF, Stevens MR. Screening two Lycopersicon peruvianum collections for resistance to tomato spotted wilt virus. Plant Dis. 2008;92:694–704. doi: 10.1094/PDIS-92-5-0694. [DOI] [PubMed] [Google Scholar]
- Gutha LR, Casassa LF, Harbertson JF, Naidu RA. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010;10:187. doi: 10.1186/1471-2229-10-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann K, Qiu WP, Moyer M. Overcoming host and pathogen-mediated resistance in tomato and tobacco maps to the M RNA of Tomato spotted wilt virus. Mol Plant Microbe In. 2001;14:242–249. doi: 10.1094/MPMI.2001.14.2.242. [DOI] [PubMed] [Google Scholar]
- Hsu YL, Liang HL, Hung CH, et al. Syringetin, a flavonoid derivative in grape and wine, induces human osteoblast differentiation through bone morphogenetic protein-2/extracellular signal-regulated kinase 1/2 pathway. Mol Nutr Food Res. 2009;53:1452–1461. doi: 10.1002/mnfr.200800483. [DOI] [PubMed] [Google Scholar]
- Lopez C, Aramburu J, Galipienso L, Soler S, Nuez F, Rubio L. Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of tomato spotted wilt virus. J Gen Virol. 2011;92:210–215. doi: 10.1099/vir.0.026708-0. [DOI] [PubMed] [Google Scholar]
- Løvdal T, Lillo C. Reference gene selection for quantitative real-time PCR normalization in tomato subjected to nitrogen, cold, and light stress. Anal Biochem. 2009;387:238–242. doi: 10.1016/j.ab.2009.01.024. [DOI] [PubMed] [Google Scholar]
- Lv JH, Liu YH, Liu ZB, Wang J, Ma YQ, Zhang ZQ, Yang S, Ou LJ, Chen WC, Zou XX. Mapping and identifying candidate genes involved in the novel fasciculate inflorescence in pepper (Capsicum annuum L.) Mol Breed. 2019;39:148. doi: 10.1007/s11032-019-1050-z. [DOI] [Google Scholar]
- Nagamatsu A, Masuta C, Senda M, Matsuura H, Kasai A, Hong JS, Kitamura K, Abe J, Kanazawa A. Functional analysis of soybean genes involved in flavonoid biosynthesis by virus-induced gene silencing. Plant Biotechnol J. 2007;5:778–790. doi: 10.1111/j.1467-7652.2007.00288.x. [DOI] [PubMed] [Google Scholar]
- Peer WA, Bandyopadhyay A, Blakeslee JJ, Makam SN, Chen RJ, Masson PH, Murphya AS. Variation in expression and protein localization of the PIN family of auxin efflux facilitator proteins in flavonoid mutants with altered auxin transport in Arabidopsis thaliana. Plant Cell. 2004;16:1898–1911. doi: 10.1105/tpc.021501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiro A, Canizares MC, Rubio L, Lopez C, Moriones E, Aramburu J, Sanchez-Navarro J. The movement protein (NSm) of Tomato spotted wilt virus is the avirulence determinant in the tomato Sw-5 gene-based resistance. Mol Plant Pathol. 2014;15:802–813. doi: 10.1111/mpp.12142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasha E. Quantitative estimation of rutin in rue (Ruta graveolens L.) cultivated in Iraq with the evaluation of its antioxidant activity. Asian J Pharm Clin Res. 2017;10:353–355. [Google Scholar]
- Rosello S, Diez MJ, Nuez F. Genetics of tomato spotted wilt virus resistance coming from Lycopersicon peruvianum. Eur J Plant Pathol. 1998;104:499–509. doi: 10.1023/A:1008622128504. [DOI] [Google Scholar]
- Ruan MY, Wan HJ, Ye QJ, Wang RQ, Yao ZP, Yu K, Yuan W, Liu YF, Yang YJ. Identification and bioinformatics analysis of chalcone synthase genes in tomato. Molecular Plant Breeding. 2013;11:379–384. [Google Scholar]
- Ryan K, Swinny E, Markham K, Winefiled C. Flavonoid gene expression and UV photoprotection in transgenic and mutant Petunia leaves. Phytochemistry. 2002;59:23–32. doi: 10.1016/S0031-9422(01)00404-6. [DOI] [PubMed] [Google Scholar]
- Samac DA, Penuela S, Schnurr JA, Hunt EN, Foster-Hartnett D, Vandenbosch KA, Gantt JS. Expression of coordinately regulated defence response genes and analysis of their role in disease resistance in Medicago truncatula. Mol Plant Pathol. 2011;12:786–798. doi: 10.1111/j.1364-3703.2011.00712.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng S, Kang XP, Xing XJ, Xu XY, Cheng J, Zheng SW, Xing GM. Agrobacterium-mediated transformation of tomato (Lycopersicon esculentum L. cv. Hezuo 908) with improved efficiency. Biotechnol Biotec Eq. 2015;29:861–868. doi: 10.1080/13102818.2015.1056753. [DOI] [Google Scholar]
- Silva EM, Saldanha LL, Adachi SA, Schley TR, Rodrigues TM, Dokkedal AL, Nogueira FTS, de Almeida LFR. Flavonoids modify root growth and modulate expression of SHORT-ROOT and HD-ZIP III. J Plant Physiol. 2015;188:89–95. doi: 10.1016/j.jplph.2015.09.009. [DOI] [PubMed] [Google Scholar]
- Soler SJ, Cebolla-Cornejo J, Nuez F. Control of disease induced by tospoviruses in tomato: an update of the genetic approach. Phytopathol Mediterr. 2003;42:207–219. [Google Scholar]
- Spassova M, Prins TW, Folkertsma RT, Klein-Lankhorst RM, Hille J, Goldbac RW. The tomato gene Sw-5 is a member of the coiled coil, nucleotide binding, leucine-rich repeat class of plant resistance genes and confers resistance to TSWV in tobacco. Mol Breed. 2001;7:151–161. doi: 10.1023/A:1011363119763. [DOI] [Google Scholar]
- Stevens MR, Scott SJ, Gererrich RC. Inheritance of a gene for resistance to tomato spotted wilt virus (TSWV) from Lycopersicon peruvianum Mill. Euphytica. 1991;59:9–17. doi: 10.1007/BF00025356. [DOI] [Google Scholar]
- Stevens MR, Scott SJ, Gererrich RC. Evaluation of seven Lycopersicon species for resistance to tomato spotted wilt virus (TSWV) Euphytica. 1994;80:79–84. doi: 10.1007/BF00039301. [DOI] [Google Scholar]
- Stevens MR, Lamb EM, Rhoads DD. Mapping the Sw-5 locus for tomato spotted wilt virus resistance in tomatoes using RAPD and RFLP analyses. Theor Appl Genet. 1995;90:451–456. doi: 10.1007/BF00221989. [DOI] [PubMed] [Google Scholar]
- Sundaraj S, Srinivasan R, Culbreath AK, Riley DG, Pappu HR. Host plant resistance against tomato spotted wilt virus in peanut (Arachis hypogaea) and its impact on susceptibility to the virus, virus population genetics, and vector feeding behavior and survival. Phytopathology. 2014;104:202–210. doi: 10.1094/PHYTO-04-13-0107-R. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li J, Xia R. Expression of chalcone synthase and chalcone isomerase genes and accumulation of corresponding flavonoids during fruit maturation of Guoqing No. 4 satsuma mandarin (Citrus unshiu Marcow) Sci Hortic. 2010;125:110–116. doi: 10.1016/j.scienta.2010.02.001. [DOI] [Google Scholar]
- Wang L, Lui ACW, Lam PY, et al. Transgenic expression of flavanone 3-hydroxylase redirects flavonoid biosynthesis and alleviates anthracnose susceptibility in sorghum. Plant Biotechnol J. 2020;18:2170–2172. doi: 10.1111/pbi.13397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Win KT, Vegas J, Zhang CY, Song K, Lee S. QTL mapping for downy mildew resistance in cucumber via bulked segregant analysis using next-generation sequencing and conventional methods. Theor Appl Genet. 2017;130:199–211. doi: 10.1007/s00122-016-2806-z. [DOI] [PubMed] [Google Scholar]
- Wu Q, Li PC, Zhang HJ, Feng CY, Li SS, Yin DD, Tian J, Xu WZ, Wang LS. Relationship between the flavonoid composition and flower colour variation in Victoria. Plant Biol. 2018;20:674–681. doi: 10.1111/plb.12835. [DOI] [PubMed] [Google Scholar]
- Yamasaki H, Sakihama Y, Ikehara N. Flavonoid-peroxidase reaction as a detoxification mechanism of plant cells against H2O2. Plant Physiol. 1997;11:1405–1412. doi: 10.1104/pp.115.4.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin WC, Wang XH, Liu H, Wang Y, Nocker SV, Tu MX, Fang JH, Guo JQ, Li Z, Wang XP. Overexpression of VqWRKY31 enhances powdery mildew resistance in grapevine by promoting salicylic acid signaling and specific metabolite synthesis. Hortic Res 2022. 2022;9:uhab 064. doi: 10.1093/hr/uhab064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao K, Shen XJ, Yuan YZ, Liu Y, Liao X, Wang Q, Liu LL, Li F, Li TH. Isolation and characterization of dehydration-responsive element-binding factor 2C (MsDREB2C) from Malus sieversii Roem. Plant Cell Physiol. 2013;54:1415–1430. doi: 10.1093/pcp/pct087. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All of the whole-genome resequencing data used in the BSA analysis is available from the NCBI Short Read Archive (SRA, BioProject ID: PRJNA751989), and the raw data are freely available at http://www.ncbi.nlm.nih.gov/bioproject/751989. The other supporting data are included as additional files.