Abstract
Breeding crop varieties with high yield and ideal plant architecture is a desirable goal of agricultural science. The success of “Green Revolution” in cereal crops provides opportunities to incorporate phytohormones in crop breeding. Auxin is a critical phytohormone to determine nearly all the aspects of plant development. Despite the current knowledge regarding auxin biosynthesis, auxin transport and auxin signaling have been well characterized in model Arabidopsis (Arabidopsis thaliana) plants, how auxin regulates crop architecture is far from being understood, and the introduction of auxin biology in crop breeding stays in the theoretical stage. Here, we give an overview on molecular mechanisms of auxin biology in Arabidopsis, and mainly summarize auxin contributions for crop plant development. Furthermore, we propose potential opportunities to integrate auxin biology in soybean (Glycine max) breeding.
Keywords: Auxin, Crop, Soybean, Breeding
Introduction
To meet human demand for food, the “Green Revolution” in agriculture has saved more than a billion people from starvation in the 1960s. The Green Revolution refers to the modification of cereal crop architecture, like rice (Oryza sativa) and wheat (Triticum aestivum), from tall to short and compact plants which are suitable for high-density planting with high-yielding unit area (Peng et al. 1999; Silverstone and Sun 2000; Khush 2001; Boss and Thomas 2002; Sasaki et al. 2002; Spielmeyer et al. 2002; Evenson and Gollin 2003). The semi-dwarf varieties generated from the Green Revolution are lodging resistance which respond to fertilizer inputs properly with an increased yield. Consequently, semi-dwarf plants become an ideal shoot architecture during rice and wheat breeding (Hedden 2003). Semi-dwarf plants in Green Revolution are mostly realized by reduced gibberellin (GA) synthesis or signaling (review in Liu et al. 2021b; Gao and Chu 2020). DELLA proteins act as negative regulators of GA signaling pathway, and GA-induced DELLA degradation is a central regulatory event for GA-mediated plant development (Sun 2011). Generation of semi-dwarf plants is derived from the reduced GA response caused by DELLA accumulation (Liu et al. 2021b). Apparently, modification of GA pathways becomes an excellent example, providing opportunities to introduce phytohormones in crop breeding.
Among all phytohormones, auxin is the first phytohormone to be discovered, which influences a wide variety of plant developmental processes throughout the plant life span (Benjamins and Scheres 2008). The natural auxin in plants is indole-3-acetic acid (IAA). Naphthalene acetic acid (NAA), indole butryic acid (IBA), and 2,4-dichloro phenoxyacetic acid (2,4-D) are synthetic auxin products, performing similar functions as the endogenous auxin to promote plant rooting during vegetative propagation, promote the fruit development of parthenocarpy, prevent premature dropping, and prevent sprouting and killing weeds. Dynamic regulation of auxin concentration and maintenance of auxin gradient at the level of individual cells determinate cell fate and cell growth plasticity (Leyser 2002). Appropriate auxin gradient is established by coordination of auxin synthesis, auxin transport, and auxin signaling. In Arabidopsis, a large number of auxin regulators have been identified, and the molecular mechanisms of auxin regulation on plant development are relatively well characterized among all phytohormones. However, auxin regulation on crop development is far from being understood, and the introduction of auxin pathways in crop breeding stays in the theoretical stage. In this review, we give an overview on recent progress of auxin biology in Arabidopsis, and summarize the current knowledge on auxin regulations in cereal crop development. Meanwhile, we discuss the opportunities to integrate available auxin biology in soybean breeding.
Overview on regulatory mechanisms of auxin in model plant system
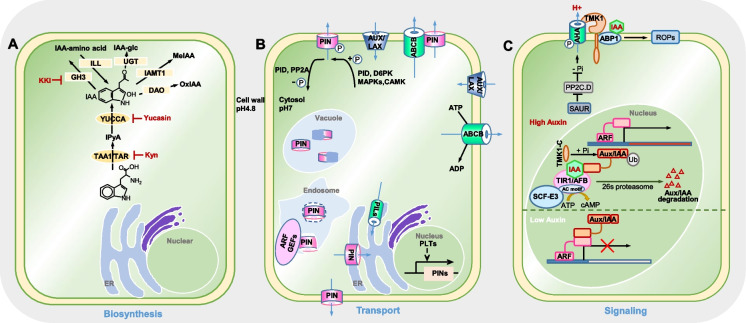
Auxin molecules tend to be concentrated in meristems and young plant tissues, which are at the forefront of growth. The optimal auxin distribution within a tissue depends on local auxin biosynthesis and cell-to-cell polar auxin transport. Auxin precisely determines cell fate, by specifying auxin signal in individual cells and converting auxin perception into a diversity of cell responses on a transcriptional, post-transcriptional, or post-translational level. In this way, plant cells coordinate their size, division, and differentiation based on auxin concentration and auxin gradient. Here, we briefly introduce the recent progress of auxin regulations on biosynthesis, metabolism, transport, and signaling level (Fig. 1) to provide the basic knowledge for crop development.
Fig. 1.
Overview of molecular mechanisms of auxin biosynthesis, auxin transport, and auxin biosynthesis in Arabidopsis plants. The coordination of auxin biosynthesis, transport, and signaling enables the plants to initiate new organs and rapidly adapt to environmental stresses. A Auxin is synthesized by cytosol-located TAA-YUCCA enzymes; then, auxin inactivation is carried out by IAA conjugation (UGT-mediated IAA-sugar conjugates, GH3 and ILL-mediated IAA-amino acid conjugates), IAMT1-mediated IAA oxidation, and DAO-mediated IAA methylation. KKI, Yucasin, and Kyn act as specific inhibitors at each step. B Auxin is polarly transported by influx AUX/LAX transporters, efflux PIN, and ABCB transporters; PIN proteins confer polar distribution on the plasma membrane via the endosomal trafficking between membrane and endosomes, and part of PIN proteins are degraded in vacuole; PIN activity can be modulated by phosphorylation through the kinases of PID/WAG, D6PK, CAMK, and MAPK cascades; PP2A acts antagonistically with PID on PIN polar targeting, mediating PIN dephosphorylation. C Auxin signaling is perceived by TIR1/AFBs-mediated nucleus signaling and TMK1-ABP1-mediated cell surface signaling; in the nucleus, TIR1/AFBs serve as auxin receptors, mediating Aux/IAA degradation via E3 ligase-mediated ubiquitination; TIR1/AFB adenylate cyclase activity is also required for rapid auxin response by promotion of cAMP production; on the cell surface, TMK1, ABP1, AHA, and ROP proteins interact with each other to establish membrane-resident auxin perception events. Created with PowerPoint2019
Auxin biosynthesis and metabolism
Trp is the precursor of multiple auxin biosynthesis pathways. Currently, the best defined pathway of auxin biosynthesis is the tryptophan aminotransferase of Arabidopsis (TAA)/YUCCA route (Won et al. 2011), in which the TAA/tryptophan aminotransferase related (TAR) catalyzes the conversion of Trp to indole-3-pyruvic acid (IPyA) (Stepanova et al. 2008; Tao et al. 2008) and YUCCA flavin-dependent monooxygenases catalyze oxidative decarboxylation of IPyA to be IAA (Zhao et al. 2001; Zhao 2010, 2012). Through chemical genetic screening, small molecules, l-kynurenine (Kyn) and 5-(4-chlorophenyl)-4H-1,2,4–triazole-3–thiol (yucasin), were successively identified as inhibitors that individually inhibit TAA1/TAR (He et al. 2011) and YUCCA activity (Nishimura et al. 2014). Currently, Kyn or yucasin treatment has been widely used to mimic the deficiency of TAA1/TAR or YUCCA.
Auxin is present in active and inactivate forms. The majority of auxin is amino acid-linked auxins, glycosylated conjugates, and the inactive methyl ester form. Only less than 5% of auxins exist in free form (Percival and Bandurski 1976). Maintenance of appropriate auxin level by auxin homeostasis between inactive (storage) and active (free) form is vital for plant developmental plasticity. IAA inactivation is carried out by IAA conjugation (IAA-sugar conjugates, IAA-amino acid conjugates), IAA oxidation, and IAA methylation (review in Korasick et al. 2013). GRETCHEN HAGEN 3 (GH3) enzymes encode IAA-amido synthetases, which conjugate excess free IAA to IAA-amino acid conjugates, such as IAA–aspartate (IAA-Asp) and IAA–glutamate (IAA-Glu) (Staswick et al. 2005). Kakeimide (KKI) was identified as a specific inhibitor that rapidly (about 10 min) suppresses GH3 activity (Fukui et al. 2022). IAA-Asp and IAA-Glu are storage forms of IAA, which can be reverted to IAA by IAA-Leu-Resistant1 (ILR1)/ILR1-like (ILL) amidohydrolases (Bartel and Fink 1995; Davies et al. 1999). Ester-linked IAA conjugates with glucose (IAA-glc) are another pathway to regulate IAA homeostasis by UDP-glucosyltransferase (UGT) (Jackson et al. 2001; Grubb et al. 2004; Tognetti et al. 2010). Besides auxin conjugation and glycosylation, IAA can be methylated or oxidized. IAA carboxyl methyltransferase1 (IAMT1) is responsible for converting activate IAA to methyl-IAA ester (MeIAA) (Qin et al. 2005), and Dioxygenase for Auxin Oxidation 1 (DAO1) converts free IAA to inactive auxin form of 2-oxoindole-3-acetic acid (OxIAA) (Porco et al. 2016; Zhao et al. 2013b; Zhang et al. 2016). Interestingly, DAO1 mainly results in the steady-state–level changes between OxIAA and IAA conjugates, whereas free IAA level is not significantly influenced (Zhang et al. 2016; Porco et al. 2016). Hence, auxin homeostasis and catabolism became important features to control endogenous free auxin level.
Auxin transport
Auxin molecules are mainly synthesized in young tissues with a high activity of cell division. The natural auxin, IAA, is a weak acid (pKa = 4.85), which is not able to cross the PM from the neutral cytoplasm (pH = 7.0) (Zazimalova et al. 2010). Therefore, cell-to-cell movement of auxin requires an export system. Auxin moves from distal to proximal cells in a polar manner by specific efflux and influx transporters, contributing to the formation of auxin gradient. Local auxin gradient is vital for plant organogenesis and subsequent development (Benková et al. 2003). It has been well studied that polar auxin transport is achieved via the efflux PIN-FROMED (PIN) auxin transporters (Petrasek et al. 2006; Glanc et al. 2018), influx AUXIN-RESISTANT1/LIKE AUX1 (AUX1/LAX) transporters, and some of the B subclass of ATP-BINDING CASSETTE (ABCB) efflux transporters (Geisler et al. 2005; Geisler 2021).
PIN exporters are plant-specific transmembrane proteins which determinate polar auxin transport in a cell-to-cell manner (review in Bennett 2015; Adamowski and Friml 2015; Feraru and Friml 2008). Expression pattern associated with cellular polarity of PIN proteins controls the direction of auxin flow during diverse aspects of organ development (Muller et al. 1998; Friml et al. 2002; Benková et al. 2003; Okada et al. 1991; Weijers et al. 2005). Current plant developmental modules support that PIN-dependent auxin transport is required for nearly all the processes of polar organ growth (Han et al. 2021; Adamowski and Friml 2015). Auxin-induced PLETHORA (PLT) transcription factors display a similar graded distribution as the auxin gradient (Aida et al. 2004). The tissue-specific PLT gradients are sufficient to determine stem cell identity, cell expansion, and cell differentiation (Galinha et al. 2007). Auxin fine-tunes PLT expression, meanwhile PLTs regulate PIN functions on the transcriptional level (Mahonen et al. 2014; Galinha et al. 2007). Endocytic trafficking and kinase-mediated phosphorylation control cellular PIN distribution and polar targeting. PM-associated PIN proteins are dynamically endocytosed into endosomal compartments via clathrin-dependent endocytosis (Dhonukshe et al. 2007). Internalized PIN proteins either target the vacuole for degradation or be exocytosed to PM, via ADP-RIBOSYLATION FACTOR GUANINE-NUCLEOTIDE EXCHANGE FACTORS (ARF-GEFs) (Kleine-Vehn et al. 2008; Geldner et al. 2003). The endosomal trafficking mainly affects distribution of PIN proteins on PM or within cytosol (review in Adamowski and Friml 2015). Recent studies have revealed that phosphorylation and dephosphorylation are essential for PIN activation and polarization (review in Lanassa Bassukas et al. 2022). PIN polarity is under the control of phosphorylation events by PINOID AGC kinase (PID), D6 PROTEIN kinase (D6PK), MITOGEN-ACTIVATED PROTEIN kinase cascades (MAPKs), and Ca2 + /CALMODULIN-DEPENDENT PROTEIN kinase (CAMK) (review in Lanassa Bassukas et al. 2022; Barbosa et al. 2018). PROTEIN PHOSPHATASE 2A (PP2A) acts antagonistically with PINOID on PIN polar targeting, mediating PIN dephosphorylation (Michniewicz et al. 2007). Besides the canonical PIN proteins, non-canonical PIN proteins with dual localization at both plasma membrane (PM) and endoplasmic reticulum (ER), as well as ER-resident PIN-LIKES (PILS), modulate intracellular auxin transport and auxin homeostasis (Ding et al. 2012; Mravec et al. 2009; Barbez et al. 2012).
Auxin efflux also requires ATP-binding cassette superfamily B (ABCB) proteins, which interact with PIN proteins (Geisler et al. 2005). ABCB19 co-localizes and stabilizes PIN1 in membrane lipid microdomains to enhance PIN1 transport activity (Titapiwatanakun et al. 2009). Computational modelling further supports the synergistic interactions of ABCB and PIN proteins for directional auxin flow (Mellor et al. 2022). Besides the auxin export system, the auxin uptake is also an active process which requires proton motive force. AUX1/LAX proteins which encode amino acid/auxin permease (AAAP) family of proton symport permeases serve as an auxin influx transporter to establish auxin influx pathway in different organs (review in Swarup and Peret 2012).
The coordination of influx and efflux auxin transportation is tightly associated with an appropriate auxin gradient, and thereby plants are able to initiate new organs, display tropisms, and rapidly adapt to environmental stresses.
Auxin signaling
Local biosynthesis and polar transport specify auxin concentration and distribution within plant tissues. Upon auxin perception in single cells, the cell surface and nucleus-allocated auxin signaling cascades trigger specific auxin response on a transcriptional or post-transcriptional level. Nuclear auxin signaling comprises three protein families: the F-box TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX PROTEIN (TIR1/AFB) auxin co-receptors, the Auxin/INDOLE-3-ACETIC ACID (Aux/IAA) transcriptional repressors, and the AUXIN RESPONSE FACTOR (ARF) transcription factors. TIR1/AFB auxin receptors are F-box subunits of SCF E3 ubiquitin ligase complexes, which mediate Aux/IAA degradation via ubiquitination. In the presence of auxin, auxin binds to TIR1/AFB receptors; auxin promotes the interaction between Aux/IAA proteins and SCF TIR1/AFB complex, resulting in the proteasome-mediated Aux/IAA degradation, by SCF E3 ubiquitin ligase (Chapman and Estelle 2009; Leyser 2006). Aux/IAAs are auxin signaling repressor, forming interactive pairs with ARFs. Degradation of Aux/IAAs releases ARFs to active transcriptional regulation of ARF-targeted downstream genes (review in Cance et al. 2022). Besides the E3 ligase activity, a recent study identified that adenylate cyclase (AC) activity is an additional function of TIR1/AFB receptors. AC activity of TIR1/AFB is controlled by the C-terminal AC motif, which is enhanced by auxin. TIR1/AFB AC activity is required for rapid auxin response by promotion of cAMP production, independent of E3 ligase-mediated ubiquitination activity (Qi et al. 2022).
Auxin perception and signal transduction not only happen in the nucleus, but auxin molecules have to also firstly cross PM. Hence, another auxin signaling pathway must happen on the cell surface to mediate rapid auxin response. Two decades ago, ABP1 protein was purified from maize (Zea mays L.) coleoptiles by several independent laboratories (Löbler and Klämbt 1985a; Shimomura et al. 1986; Hesse et al. 1989; Tillmann et al. 1989; Lobler and Klambt 1985), and be shown to bind auxin with high affinity in the acidic environment (Löbler and Klämbt 1985b, 1985a; Jones and Venis 1989). A large number of physiological and developmental studies demonstrated important roles of ABP1 in various auxin-dependent processes (Jones et al. 1998; Chen et al. 2001a, 2001b; Bauly et al. 2000). Despite eminent research efforts, CRISPR-CAS9-based ABP1-knockout genetic materials bring debates and it is puzzling whether ABP1 acts as a dedicated receptor for auxin on cell surface (Gao et al. 2015).
Recent work on PM-localized transmembrane kinase (TMK) defines a novel cell surface-resident auxin perception pathway. TMK1 interacts with ABP1 to stimulate the activity of PM-associated Rho-like guanosine triphosphatases GTPase (ROPs), shaping leaf pavement cell interdigitation (Xu et al. 2014). The interaction between ABP1 and TMK at the cell surface is dependent on ABP1-mediated auxin sensing. Upon auxin perception, TMK1 is also able to interact with and phosphorylate PM-associated H+-ATPases, thereby promoting cell-wall acidification and rapid cell expansion (Lin et al. 2021). Meanwhile, small Auxin Up RNA (SAUR) proteins can also stimulate H+-ATPase proton pumping activity by inhibiting activity of PP2C.D phosphatases, thereby preventing the dephosphorylation of H+-ATPase to stimulate cell expansion (Spartz et al. 2014; Ren et al. 2018; Wong et al. 2021). Strikingly, TMK1 functions not only on the cell surface, but also interacts with nuclear-allocated auxin signaling. Auxin promotes the C-terminal cleavage of TMK1. The cytosolic and nucleus-translocated C terminus of TMK1 specifically interacts with and phosphorylates the nuclear auxin core components, Aux/IAA proteins (IAA32 and IAA34), thereby regulating ARF-dependent nuclear auxin signaling (Cao et al. 2019). Therefore, cell surface-resident auxin signaling collaborates with nuclear auxin signaling via TMK1-Aux/IAA complex, enabling specific auxin response under different concentrations of cellular auxin and leading to complex developmental outcomes (Cao et al. 2019). In comparison to global auxin-triggered phospho-response of abp1 and tmk1 loss-of-function mutants, TMK1 and ABP1 indeed mediate a large number of overlapping rapid phosphor-responses (Friml et al. 2022). Therefore, ABP1 and TMK1 serve as the auxin co-receptors to perceive cell surface auxin signaling, mediating rapid auxin response on the level of protein phosphorylation.
Auxin contribution on crops
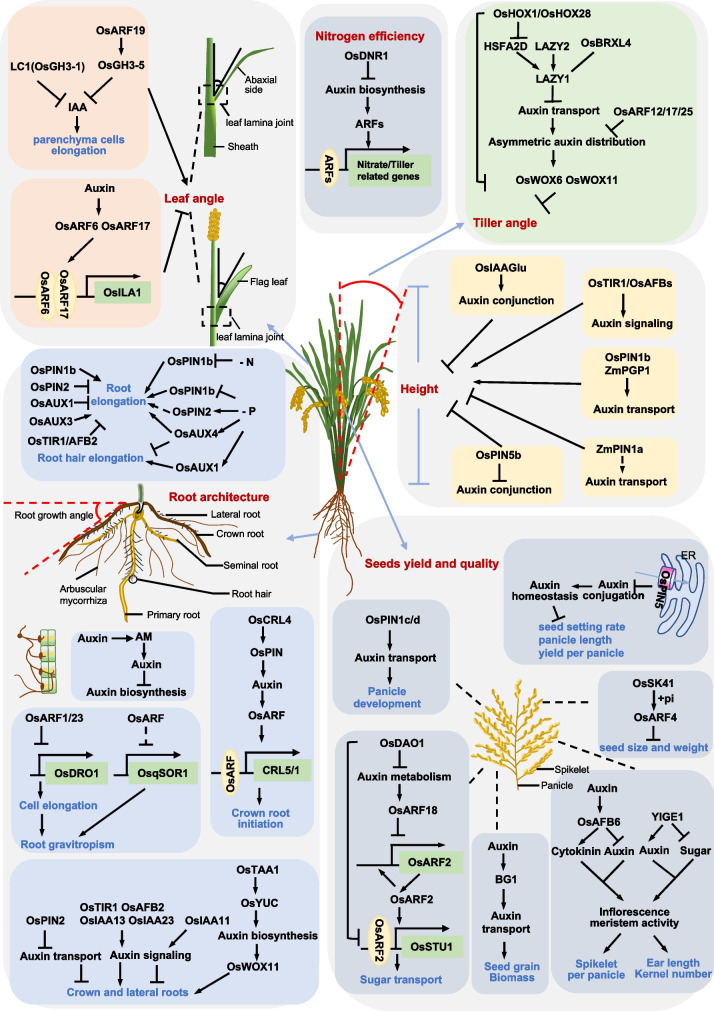
The major objective of plant science was to apply our knowledge to improve agronomic traits. Yield promotion in cereal crops, such as rice, is realized through the introduction of semidwarf varieties which reduce GA synthesis and response (review in Gao and Chu 2020; Liu et al. 2021b). The resultant semi-dwarf plant not only prevents lodging, but also facilitates fertilizer inputs to improve grain yield. Based on the experience of rice breeding, shaping an ideal shoot architecture in cereal crops is proposed, including semi-dwarf height, being suitable for high-density planting and high yielding (review in Gao and Chu 2020). Besides ideal shoot architecture, shaping desirable crop plants with an ideal root architecture which has great potential to increase resource-use efficiency is also highlighted in recent years (Panda et al. 2021). Auxin functionality in shoot and root plasticity makes a potential application value of auxin in crop breeding. We here summarize the recent studies of auxin biosynthesis, auxin transport, and auxin signaling in crop development (mainly in rice, maize, and soybean) (Fig. 2) and discuss the possible application of auxin for crop breeding.
Fig. 2.
Regulatory network of auxin biology for rice plant development, regarding auxin contributions to shoot height, leaf petiole angle, tiller angle, seed development, root architecture, and nutrient absorption. Arrows indicate promotion or activation; truncated connectors indicate repression or negative effects; dashed arrows indicate the regulatory mechanisms remain unclear. Created with PowerPoint2019
Shoot height
Semi-dwarf traits have been widely introduced into cereal crop breeding, including reduced plant height, reduced tiller angle, and reduced leaf angle. Crop breeding interest in short plant height is mainly derived from their ability of lodging resistance. In rice, indole-3-acetic acid glucosyltransferase (OsIAAGLU), which catalyzes the conjugation of free IAA with glucose to generate IAA-glc, is involved in the regulation of plant height. Overexpression of OsIAAGLU that decreases free auxin level significantly reduces plant height (Yu et al. 2019). On the cellular level, loss of auxin receptors OsTIR1, OsAFB2, OsAFB3, OsAFB4, and OsAFB5 in rice, which disrupts auxin perception, also significantly reduces plant height (Guo et al. 2021). Apparently, maintenance of shoot height requires sufficient auxin level. On a tissue level, specific auxin distribution and auxin gradient determine plastic organ formation, which is under the control of auxin transporters. In rice, mutation of the PIN orthologs OsPIN1b which disrupts polar auxin transport decreases shoot height (Wang et al. 2022a), whereas overexpression of OsPIN5b elevates free auxin level but suppresses conjugated auxin level in shoot, thereby resulting in the reduction of plant height (Lu et al. 2015). In maize, two classes of mutants have a dramatic reduction in shoot height, named the dwarf mutants and the brachytic mutants. Among them, brachytic mutant2 (br2) shows compact lower stalk internodes, which is caused by loss of P-glycoprotein (PGP, also named ABCB) function and impaired polar auxin transport (Multani et al. 2003). In Arabidopsis, loss of PGP function is associated with multiple phenotypes in roots (Terasaka et al. 2005; Geisler et al. 2005), whereas the disruption of ZmPGP1 (Br2) in maize has different consequences that cause a specific shortening of lower stalk internodes but not in other tissues. Gene duplication and the specific expression pattern of ZmPGP1 in elongating internodes provide an opportunity to apply Zmpgp1 mutant to shape a dwarfish shoot height during maize breeding (Multani et al. 2003; Xing et al. 2015). To explore natural variation and domestication selection of ZmPGP1 gene, sequence polymorphism of ZmPGP1 in 349 inbred lines, 68 landraces, and 32 teosintes of maize varieties identified several SNP variants and indel regions in ZmPGP1 which have great potential to associate with maize shoot architecture (Li et al. 2019a). Strikingly, overexpression of ZmPIN1a in maize displays consistent shoot phenotype as ZmPGP1 deficiency, resulting in reduced internode length and shorter height (Li et al. 2018a). Although these two types of auxin efflux transporters, PGP and PIN, act synergistically to direct polar auxin flow in Arabidopsis (review in Mravec et al. 2008), we could not simply imitate the conserved mechanism from Arabidopsis to crop plants. In maize, the reduction of internode is caused by the increased acropetal auxin transport from shoot to root in ZmPIN1a overexpressors but decreased auxin basipetal transport from lower to upper internodes in Zmpgp1 mutant (Li et al. 2018a; Zhang et al. 2018a).
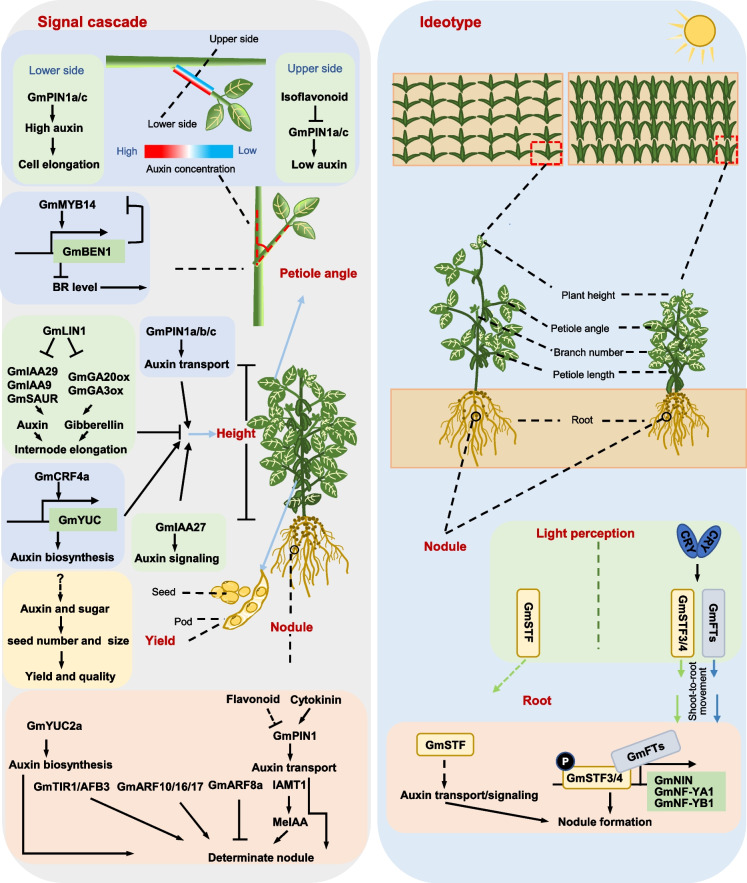
Soybean plants have two growth habits: indeterminate and determinate. The determinate type completes vegetative growth once the plant starts flowering, whereas the indeterminate type has continuous vegetative growth during the reproductive phase (Tian et al. 2010). Thus, the different growth pattern of soybean cultivars results in the variation of shoot height and yield. Shoot height is a complex quantitative trait, which is influenced by a variety of phytohormones, including GA, auxin, and cytokinin (Wang et al. 2017). Diverse auxin biosynthesis metabolites accumulate differently in the apical shoot, elongation zone, or mature zone of soybean stem (Jiang et al. 2020). Apparently, there is a correlation between auxin metabolism and soybean stem elongation. Consistent with the finding in rice, GA plays a vital role in soybean plant height. A number of genes involved in GA biosynthesis or signaling pathways have been reported to regulate soybean height, such as GmDW1 (Dwarf Mutant 1) (Li et al. 2018b), GmAP1s (Apetala1) (Chen et al. 2020), and GmLHY (Late elongated hypocotyl) (Cheng et al. 2019), ABI3/VP1 gene GmRAV (Xue et al. 2022). It is well known that auxin promotes GA biosynthesis (Wolbang and Ross 2001; Ross et al. 2000, 2001). Thus, it is highly possible that auxin interacts with GA to influence shoot height in soybean. A mutant of the Glycine max long internode 1 (Gmlin1), which encodes a homologue of the Arabidopsis long hypocotyl 2 (HY2) gene, exhibits extremely taller and longer internode than the soybean wild-type plant (Zhang et al. 2022b). Auxin-responsive genes GmIAA29, GmIAA9, and GmSAUR (Small Auxin Up RNA) and GA biosynthesis genes GmGA20ox and GmGA3ox are all upregulated in Gmlin1 mutant, implying auxin interplays with GA to promote internode elongation (Zhang et al. 2022b). Knockout of auxin transporters GmPIN1a, GmPIN1b, and GmPIN1c in soybean displays a semi-dwarf phenotype, indicating an involvement of auxin transport in shoot height regulation (Zhang et al. 2022a). Jiang et al. (2018) identified 11 major quantitative trait loci (QTLs) associated with the first pod height from 147 recombinant soybean inbred lines (Jiang et al. 2018). Among this QTL interval, 8 candidate genes including an auxin response factor 9 (ARF9), a SAUR family gene, and an AUX influx permease are components of auxin signaling or auxin transport (Jiang et al. 2018). A dwarf soybean mutant named dmbn was identified from ethyl methanesulfonate (EMS)-mutagenized soybean seeds, and the dwarf phenotype might be associated with an auxin signaling regulator, Aux/IAA protein GmIAA27 (Su et al. 2022). Furthermore, the cytokinin response factors 4a (GmCRF4a) promotes cell elongation and shoot height in soybean, as GmCRF4 is required for GmYUC expression which controls auxin biosynthesis (Xu et al. 2022). All these studies imply auxin is associated with soybean plant height (Fig. 3).
Fig. 3.
Recent progress of auxin biology for soybean plant development, regarding auxin contributions to shoot height, leaf petiole angle, and nodule development. The ideal soybean model with compact shoot architecture, high photosynthesis efficiency, and high nitrogen-fixation ability is proposed in the right panel. Arrows indicate promotion or activation; truncated connectors indicate repression or negative effects; dashed arrows indicate the regulatory mechanisms remain unclear. Created with PowerPoint2019
Despite the relationship between auxin and shoot height having been studied, the underlying mechanisms are largely unknown. With the help of fundamental research in model plant Arabidopsis, auxin contribution in shoot height can be partially interpreted by the acid growth model. The decades-old auxin-acid growth model has put forward a promotional effect of auxin on cell expansion/elongation through its effect on cell wall acidification (Du et al. 2020). As mentioned above, TMK1-dependent auxin signaling stimulates rapid phosphorylation of PM H+-ATPase, and the active H+-ATPase (also named as AHA) causes apoplastic acidification to increase cell wall extensibility (Lin et al. 2021). A type 2C protein phosphatases PP2C.D dephosphorylates AHA to decrease H+ pump activity (Spartz et al. 2014; Ren et al. 2018). SAURs inhibit de-phosphorylation activity of PP2C.D, conferring high AHA activity (Spartz et al. 2014; Ren et al. 2018; Wong et al. 2021). Besides the cell surface auxin signaling, TIR1/AFB-mediated nuclear auxin signaling also mediates auxin acid growth (Fendrych et al. 2018). Therefore, manipulation of TMK1-PP2C.D-SAUR-AHA and TIR1/AFB-AUX/IAA-ARF auxin signaling cascades can be considered new strategies to optimize crop shoot height.
Leaf and tiller angle
In recent years, improvement of photosynthesis efficiency becomes a major goal for scientists to increase crop yielding, as canopy photosynthesis determines carbohydrate accumulation which is tightly correlated with biomass growth and crop production (Song et al. 2016). Leaves at the top of a canopy are usually light saturated, while leaves at the bottom layers are light limited. Canopy growth pattern not only influences photosynthesis efficiency, but also determines the planting population in a unit area.
Canopy architecture consists of leaf size and leaf growth angle in dicot, but includes tiller number and tiller angle in monocot. More upright-growing leaves increase light interception across the abaxial and adaxial leaf surface, which has been shown to improve photosynthesis rate (Pendleton et al. 1968). According to the difference in leaf angle, maize plants are divided into three types, namely compact type, intermediate type, and flat type (Donald 1968). There is a correlation between leaf angle and canopy photosynthesis, as well as yield: very upright-growing leaves is beneficial for the middle leaf to receive light, thereby improving the global photosynthesis efficiency and production (Stewart et al. 2003). Exogenous application of auxin makes increased leaf angle in maize (Ji et al. 2022). During leaf angle formation in rice, auxin promotes the elongation of parenchyma cells in leaf lamina joint. Reduction of auxin level by overexpressed IAA–amido Synthetase, Rice LEAF INCLINATION1 (LC1), in lamina joint increases flag leaf angle (Zhao et al. 2013a), suggesting auxin level is negatively correlated with leaf angle. Through genetic screening, mutant of Increased Leaf Angle1 (ILA1), a Raf-like mitogen-activated protein kinase kinase kinase (MAPKKK), exhibits a larger leaf angle due to the reduced mechanical strength of lamina joints (Ning et al. 2011). The crucial nuclear auxin signaling components, OsARF6, OsARF17, and OsARF19, show high abundance in the lamina joint (Zhang et al. 2015; Huang et al. 2021a). OsARF19 binds to the promoter of OsGH3-5, which converts free IAA to IAA conjugates. Overexpression of OsARF19 or OsGH3-5 both decreases free IAA level at lamina joints, resulting in an increase of adaxial cell division and an enlarged leaf angle (Zhang et al. 2015). Moreover, OsARF6 and OsARF17 directly bind to the ILA1 promoter and activate ILA1 expression (Huang et al. 2021a). Consequently, osarf6 osarf17 double mutant shows an exaggerated enlarged leaf angle (Huang et al. 2021a). The opposite phenotype of OsARF6, OsARF17, and OsARF19 genetic materials indicates a complicated regulation of auxin signaling for lamina joint development in rice.
Besides leaf angle, tiller angle is another core constituent of canopy architecture in monocot to influence light perception and planting density. Significant progress in rice has defined that tiller angle formation is associated with shoot gravitropism (review in Wang et al. 2022b). Most knowledge regarding the molecular basis of gravitropism has been mainly obtained from the studies in Arabidopsis. Gravitropism is caused by the asymmetric auxin distribution in the upper and lower flanks of root tip or in the abaxial and adaxial cells of shoot upon gravity stimulation (review in Strohm et al. 2012). PIN efflux transporters and AUX/LAX influx transporters serve as core regulators to control auxin gradient and asymmetric auxin distribution (review in Swarup and Peret 2012). Current understandings on tiller angle regulation are mostly related to auxin. LAZY1 controls the rice tiller angle by negatively regulating polar auxin transport (Zhu et al. 2020; Li et al. 2007). LAZY2 controls shoot gravitropism and tiller angle by acting upstream of LAZY1 (Huang et al. 2021b). Downregulation of auxin signaling by disrupting OsARF12, OsARF17, and OsARF25 promotes tiller angle (Li et al. 2020b), indicating that auxin signaling negatively correlates with tiller angle. PIN-controlled asymmetric auxin distribution happens on PM, whereas all the core regulators that control tiller angle in rice are mostly nuclear-localized transcriptional factors. Questions remain whether LAZY1 and other tiller angle-related transcriptional factors directly regulate PM-associated PIN cascade. Interestingly, LAZY1 displays dual localization in both PM and nuclear. LAZY1-interacting protein, named Brevis Radix Like 4 (OsBRXL4), interacts with LAZY1 at the PM, and their interaction determines nuclear localization and functionality of LAZY1 (Li et al. 2019b). PROTEIN KINASE ASSOCIATED WITH BRX (PAX) is able to recruit AtBRX to the PM, in turn inhibiting PIN-dependent auxin efflux at a lower auxin level in Arabidopsis root (Marhava et al. 2020). A regulatory network of LAZY1-BRX-PIN probably happens for tiller angle formation in rice. To further understand the functional relationship between shoot gravitropism and till angle, Zhang et al. (2018b) performed a large-scale transcriptome analysis to identify the regulatory network in upper and lower flanks of rice tiller upon gravi-stimulation. Transcriptional factor HEAT STRESS TRANSCRIPTION FACTOR 2D (HSFA2D) acts as positive regulator upstream of LAZY1, and WUSCHEL RELATED HOMEOBOX6 (WOX6) and WOX11 are expressed asymmetrically in tiller flanks in response to gravi-stimulation (Zhang et al. 2018b). OsHOX1 and OsHOX28 are further found to restrain lateral auxin transport and then repress the expression of WOX6 and WOX11 in the lower side of shoot (Hu et al. 2020). OsHOX1 and OsHOX28 proteins are also able to bind to the promoter region of HSFA2D and suppress the expression of HSFA2D and LAZY1 (Hu et al. 2020).
Compared with the knowledge in rice, soybean study is far behind. Soybean is a typical light-sensitive plant, displaying rapid growing of petioles under shade compared with normal light irradiation (Yang et al. 2018). Reduced red–to–far-red light intensity ratio promotes the elongation of petiole length, probably caused by the decreased IAA-to-GA content ratio of soybean petiole (Yang et al. 2018). In soybean, an appropriate leaf angle is not simply determined by a general change of auxin content. Auxin displays asymmetric accumulation in leaf petiole base, with more auxin in the lower side than in the upper side, corresponding with the consistent asymmetric pattern of GmPIN1a/c abundance. The asymmetric GmPIN1 expression and auxin accumulation in petiole base result in asymmetric cell expansion to create a desirable petiole curvature in soybean (Zhang et al. 2022a). Accumulating evidences have proved the interactive regulation of phytohormones for leaf angle formation, such as the interaction between brassinosteroid (BR) and auxin (review in Luo et al. 2016; Li et al. 2020a). High BR content in the lamina joint promotes leaf petiole angle (Tong and Chu 2018). GmMYB14 transcriptional factor directly binds to the promoter of BRASSINOSTEROID-INSENSITIVE 1 ENHANCED 1 (BEN1), in turn negatively regulating BR level. The reduction of BR content in overexpressed GmMYB14 soybean plants causes a smaller petiole angle and compacter plant architecture than wild type (Chen et al. 2021). Very limited mechanisms have been studied for soybean petiole angle formation. Currently, the theory of GmPIN-dependent asymmetric auxin distribution in leaf petiole base can be a reference to study petiole angle formation and to search for an optimal approach for high-density planting in soybean (Fig. 3). With the help of time-series phenotypic data collected by an unmanned aircraft system from 1303 soybean varieties, 35 QTL regions are identified to associate with canopy coverage (Li et al. 2022a), which offers a promising opportunity for soybean breeding with canopy architecture.
Seed quality and yield
During crop breeding, improving yield and seed quality are the most important challenges among all breeding issues. Seed number per pod is determined by ovule number which has been well studied in Arabidopsis (review in Cucinotta et al. 2020). Phytohormone communication, including the crosstalk among auxin, cytokinin, BR, and GA, promotes ovule initiation (Cucinotta et al. 2020). Among them, PIN transporters-driven auxin flow creates an auxin maximum in the founder cells of ovule primordia to direct ovule initiation (Hu et al. 2022; Yu et al. 2020). Hence, it is easy to understand that deficiency of auxin biosynthesis or auxin transport causes defects of ovule initiation and the decrease of seed number. For instance, Ospin1c Ospin1d double mutants exhibit severe defects of panicle development in rice (Liu et al. 2022); overexpression of OsPIN5b decreases seed setting rate, panicle length, and yield per panicle (Lu et al. 2015). However, promotion of seed number is not simply realized by elevation of auxin level. Overexpression of OsAFB6 in rice greatly elevates spikelet number per panicle and grain yield per plant by increasing cytokinin but decreasing auxin level (He et al. 2018). Therefore, it is not a good idea to improve crop yield directly by increasing auxin transport or auxin biosynthesis. A dominant rice mutant big grain1 (Bg1-D) shows obvious bigger seed sizes. BG1 gene is specially and rapidly induced by IAA, and overexpression of BG1 significantly enhances the basipetal IAA transport and IAA level (Liu et al. 2015b). Consequently, BG1-overexpressing plants improved biomass and grain yield in rice (Liu et al. 2015b), illuminating a new strategy to apply auxin in crop yielding.
Seed size and weight are important traits for crop, which are influenced by auxin and communications with other phytohormones. The GSK3/SHAGGY-Like Kinase 41 (OsSK41, also called OsGSK5) negatively regulates grain size. Loss of function of OsSK41 increases grain length and weight. OsSK41 interacts and phosphorylates OsARF4; therefore, deficiency of OsARF4 also enhances rice grain size and weight (Hu et al. 2018). Auxin and BR share a large number of co-regulated downstream genes, synergistically affecting various physiological events. For instance, the small organ size 1 (SMOS1) and SMOS2 are individually involved in auxin and BR signaling (Aya et al. 2014; Hirano et al. 2016). Knockout of SMOS1 or SMOS2 results in a decreased size of various organs in rice. SMOS1 is positively regulated by OsARF1-mediated auxin signaling (Aya et al. 2014), and SMOS2 is negatively regulated by BRASSINOSTEROID INSENSITIVE 2 (BIN2)-mediated BR signaling (Hirano et al. 2016). The coordination of SMOS1 and SMOS2 might be a crosstalk point of auxin and BR signaling for seed size regulation.
Grain filling quality is determined by carbohydrate distribution that is derived from leaf photosynthesis. Sucrose is the major carbohydrate, transporting from source to sink to supply for grain filling. Exogenous IAA application enhances the initiation of grain filling by transcriptional upregulation of sucrose transporters and increase of sucrose level (Deng et al. 2021). In maize, overexpression of YIGE1 enlarges inflorescence meristem size, increases ear length, and promotes kernel number, thus enhancing grain yield. YIGE1-overexpressors accumulate more IAA but decrease sugar content, implying a feedback regulation between auxin and sugar for seed development (Luo et al. 2022). Sucrose transport is mainly controlled by two types of sucrose transporters: Sugar Will Eventually be Exported Transporters (SWEETs) and sucrose transporters (SUTs/SUCs) (Kuhn and Grof 2010; Baker et al. 2012). Deficiency of sugar transporters blocks sugar transportation, in turn elevating sugar in the source leaves but depriving of sugar in the sink tissues (seeds) (Julius et al. 2017). Hence, maintenance of appropriate source-to-sink sugar transport is crucial to control seed quality. In barley (Hordeum vulgare), mutation of auxin biosynthesis HvYUCCA4 gene fails to establish the carbon metabolism pathway during grain development, thereby affecting starch production for pollen maturation (Amanda et al. 2022). Knockdown of tomato (Solanum lycopersicum) SlARF4 enhances sugar accumulation in fruit, implying a connection between sugar partitioning and auxin signaling (Sagar et al. 2013). Zhao et al. (2022c) recently found that rice dao mutant, which failed to convert IAA into OxIAA, had defects in sugar partition between source (leaf) to sink (spikelet) tissue. dao mutant upregulates auxin signaling component OsARF18 but downregulates OsARF2. OsARF2 directly binds to the Sugar-Responsive Elements (SuREs) of the OsSUT1 promoter, enabling the activation of sugar transport. Strikingly, the cis-element motif of SuREs (GTCTC) shows high-sequence similarity with auxin-responsive elements (AuxREs: TGTCTC), implying a synergistic or antagonistic regulation between auxin and sugar on the transcriptional level. OsARF18 acts as a repressor of OsARF2-OsSUT1 complex, and feedback regulates sugar transport through auxin signaling (Zhao et al. 2022c). Hence, auxin signaling cascade is also involved in carbohydrate partitioning, providing an important implication for raising seed quality.
Improving the ratio of four seeds per pod is a long-term demand for ideal soybean plant breeding (Liu et al. 2020). In the past years, efforts have been made to search for master regulators that control seed number per pod in soybean. So far, the Ln gene has been cloned as a key factor to control seed set by regulating the ovule number per pistil. Accordingly, ln mutants display a higher percentage of four seeds per pod (Jeong et al. 2011, 2012). However, limited advance is achieved on seed number control in the recent 10 years. To explore the possible regulatory network, Liu et al. (2021a) compared the differences of auxin and cytokinin pathways which individually determine ovule initiation and cell division events in soybean and cowpea, in trying to understand the underlying mechanism by which cowpea has much more seeds per pod than soybean. In Arabidopsis, PIN1-dependent auxin transport determines ovule initiation by establishing auxin maximum in the apex of the ovule primordium (Benková et al. 2003). Interestingly, gene abundance and phosphorylation level of PIN1 in soybean are less than in cowpea, possible leading to the lower efficiency of auxin transport in soybean (Liu et al. 2021a). Thus, elevation of auxin concentration in ovule primordium by blocking auxin efflux becomes a possible idea to raise soybean seed set. Within our expectation, application of auxin efflux inhibitor, methylchlorflurenol (MCF), on soybean foliage during pod initiation elevates pod number by 40% upregulation (Noodén and Noodén 1985). However, the global seed yield per plant did not rise after MCF treatment because the seeds were smaller in the treated plants than normal (Noodén and Noodén 1985). Consistent with the finding in rice, the carbohydrate source, sucrose flux, across seed coat is crucial to fulfill soybean seed filling quality. SWEET transporters GmSWEET10a and GmSWEET10b, responsible for sucrose and hexose transport, serve as master regulators to determine seed size, oil, and protein content. GmSWEET10a and GmSWEET10b direct sucrose flow to the developing embryos at the rapid-seed-growth stage, in turn enabling a high seed-growth rate, larger seed, and higher oil content (Wang et al. 2020). Therefore, improvement of soybean yielding requires integration of multiple signaling instead of sole change of auxin transport. A combination of auxin and sugar transport pathways could be an ideal approach to breed new soybean varieties with high quality and high yield.
Root architecture
Root elongation is an important adaptive strategy for crops responding to nitrogen and phosphate deficiency (Sun et al. 2018; Li et al. 2015). Under phosphate deficiency, root hair number and root elongation are essential for phosphate absorption. Root elongation relies on both cell division and cell elongation in the root apical meristem (RAM) and elongation zones, which requires appropriate auxin concentration (Fu et al. 2021; Jiang and Feldman 2003; Di Mambro et al. 2017). Auxin gradient mediated by polar auxin transport has been well known to regulate root elongation. In rice, loss of function or gain of function of PINs, AUX/LAX, and ABAC auxin transporter disrupted auxin gradient in RAM, thereby affecting root elongation. For instance, Ospin1b mutant decreases seminal root length (Sun et al. 2018), overexpressed OsPIN2 inhibits root elongation (Sun et al. 2019), Osaux1 mutant had longer primary roots but Osaux3 had shorter primary roots (Yu et al. 2015; Wang et al. 2019a), ZmPIN1a overexpressors increase lateral root density but decrease lateral root elongation (Li et al. 2018a), and Zmpgp1 mutant had shortened roots (Zhang et al. 2018a). Apparently, deficiency of auxin transporters all affects root elongation, owing to the disruption of auxin gradient in root meristem. Distinct with the shorter primary roots of auxin transporter mutants, deficiency of auxin signaling components in Ostir1 afb2 single and multiple mutants all had longer primary roots than WT (Guo et al. 2021). Compared with the dwarfed roots of tir1 afbs multiple mutants in Arabidopsis (Prigge et al. 2020), a longer root length in Ostir1 and Osafbs mutants implies a possibility that auxin is overproduced and had an inhibitory effect on root growth in rice (Prigge et al. 2020). Besides the endogenous auxin regulation, exogenous nutrient supply integrating auxin can reconstruct root architecture. Low nitrate and low phosphate induce root elongation but decrease auxin transport from shoots to roots (Sun et al. 2014). The promotion of root length by nitrate and phosphate starvation was less effective in Ospin1b mutant, since OsPIN1b transcript is suppressed under low nitrate and low phosphate conditions (Sun et al. 2018). In contrast to the decreased level of OsPIN1b, OsPIN2 expression is enhanced upon phosphate deprivation (Sun et al. 2019). Low phosphate suppresses auxin transport in shoot-to-root direction by OsPIN1b but promotes auxin transport in root-to-shoot direction by OsPIN2, implying that the global auxin concentration should be reduced in RAM during phosphate starvation (Sun et al. 2018, 2019). However, more auxin is deposited in root tip under low-phosphate conditions (Sun et al. 2019), indicating that not only PIN transporters are involved in low-phosphate regulation. OsAUX1 transports the accumulated auxin from the root apex to the root differentiation zone, facilitating phosphate acquisition in rice (Giri et al. 2018). Besides OsAUX1, OsAUX4 also participates in phosphate absorption during root growth (Ye et al. 2021). It has been well known that available phosphate has a significant effect on root hair elongation via auxin regulation. OsAUX1 promotes root hair growth during phosphate starvation, facilitating phosphate absorption via root hairs (Giri et al. 2018).
Lateral and adventitious roots increase the volume of soil reached by the root, whose number and distribution are major determinants of root architecture. In rice, the crown root plays a vital role in generating lateral root. Auxin signaling is required for crown and lateral root development, as the deficiencies of auxin signaling components such as Ostir1, Osafb2, Osiaa23, and Osiaa13 mutants all suppress crown and lateral root growth (Guo et al. 2021; Yamauchi et al. 2019; Kitomi et al. 2012), and gain of function of OsIAA11 blocks lateral root initiation (Zhu et al. 2012; Kitomi et al. 2012). In particular in the Osiaa23 mutant, lateral and crown root primordium are absent and the quiescent center (QC) identity is lost in root meristem, indicating the central role of OsIAA23 in specifying crown and lateral root initiation (Jun et al. 2011). OsIAA23 is specifically expressed in the root QC; therefore, OsIAA23-mediated auxin signaling determines the maintenance of postembryonic root stem cells (Jun et al. 2011). Through mutant screening, the rice crown rootless1 (crl1) mutant is identified which is incapable of producing any crown root primordia and the crl5 mutant has very few crown roots (Inukai et al. 2005; Kitomi et al. 2011). Auxin activates Crl1 and Crl5 expression, and OsARF1 binds to the promoter region of these two genes to initiate crown roots (Inukai et al. 2005; Kitomi et al. 2011). Thus, Crl1 and Crl5 function as mediators to connect auxin signaling with crown and lateral root development. Few crown roots of crl4 mutants are caused by the mutation of a guanine nucleotide exchange factor for ADP-ribosylation factor (OsGNOM1) gene, which is responsible for vesicle trafficking of PIN transporters (Liu et al. 2009). Correspondingly, overexpressed OsPIN2 disrupts basipetal auxin transport in root epidermis and reduces lateral root density (Sun et al. 2019). Besides auxin signaling and transport, auxin synthesis by the TAA/YUC pathway is sufficient for crown root initiation. In rice, the Ostaa1 mutant has few crown roots and a lack of lateral roots (Yoshikawa et al. 2014). Interestingly, the crown rootless phenotype of Ostaa1 mutant was partially rescued by overexpression of WUSCHEL RELATED HOMEOBOX11 (OsWOX11), which is expressed in emerging crown roots and in cell division regions of the root meristem (Zhao et al. 2009). OsWOX11 abundance is stimulated by OsYUC overexpressors but repressed in Ostaa1 mutants (Zhang et al. 2018c). Accordingly, overexpression of OsYUCs induces ectopic adventitious root and shortened seminal roots (Zhang et al. 2018c). Apparently, OsYUC, auxin, and OsWOX11 establish a fine-tuning module for crown root development (Zhang et al. 2018c). Altogether, despite crown root behaving as a new root organ in rice, auxin regulation for root development is generally conserved in dicot Arabidopsis and monocot rice plants.
Crop plant growth in the field is comprehensively affected by a general root architecture; thus, it is not possible to consider root growth by a single factor of primary root or lateral root. The root system architecture (RAS) becomes a new criterion in crop planting, since RAS is responsible for nutrient and water absorption which interacts with soil microbial communities. Optimal RAS determines the utilization efficiency of nutrient fertilizer and water, to determine crop adaptation, yield, and biomass. RAS has been proposed as the key to the second Green Revolution (Bender et al. 2016). However, there is no explicit quality index of the RAS ideotype for crop breeding due to a complex soil environment (Uga 2021; Lynch 2019). Evidences from a certain amount of studies highlight the central role of auxin in orchestrating RAS; thus, design-oriented root breeding can be improved by genetic manipulation of auxin pathways. Actually, to adapt to different abiotic stress conditions, such as drought, hypoxic, nitrogen deficiency, or phosphorus deficiency, roots display very different structures which are reflected by the depth of primary roots and the angle and direction of lateral roots (Uga 2021). The steeper root growth angle (RGA), which is formed in response to gravity, enhances rice resistance to drought condition by accessing deep-soil water (Uga et al. 2015). At the same time, a deeper root system was beneficial for acquisition of nitrate and other nutrients that are more abundant in deeper soil layers (Wasson et al. 2012). In rice, DEEPER ROOTING 1 (DRO1) is a major QTL, which controls root growth angle and improves rice grains under drought condition (Uga et al. 2013). DRO1 is involved in cell elongation in the root tip that causes asymmetric root growth and downward bending of root tip in response to gravity. DRO1 is negatively regulated by auxin signaling regulator OsARF1/OsARF23, which binds to the promoter of DRO1. Therefore, overexpression of DRO1 confers greater gravitropic curvature to grow roots to deeper soil (Uga et al. 2013). In contrast, in response to hypoxic stress, rice develops soil-surface root (shallow roots) to access oxygen from air by reducing gravity response (Kitomi et al. 2020; Uga 2021). QTL for SOIL SURFACE ROOTING1 (qSOR1), a homolog of DOR1, regulates rice yields in high-salt conditions. Roots of qsor1 mutant develop particular structures on the soil surface, which enable plants to avoid hypoxic stress. The expression of qSOR1 is negatively regulated by auxin signaling, which may be caused by the presence of auxin response element in qSOR1 promoter (Kitomi et al. 2020). DULL NITROGEN RESPONSE1 (DNR1) is a major QTL associated with nitrogen uptake, which encodes an aminotransferase that catalyzes the conversion of indole-3-pyruvate to L-Trp, thus antagonizing auxin biosynthesis. A 520-bp segment in the promoter of DNR1 is absent in indica varieties but present in japonica varieties, which coincides with the generally lower DNR1 abundance in indica than in japonica. In indica, lower protein abundances of DNR1 promote auxin biosynthesis, and consequently stimulate OsARF-mediated activation of nitrate metabolic pathways. As a result, DNR1indica allele improves rice yielding with reduced nitrate requirement (Zhang et al. 2021). Apparently, auxin biology has been incorporated in optimizing crop root architecture and has great potential for the new Green Revolution.
Root symbiosis
In recent years, the goal of crop breeding has gradually moved from high yield demand to sustainable crop developing with high quality and safety. To solve the pollution problem from high amounts of fertilizer in soil, utilization of plant growth–promoting microbes could be a wise choice. The beneficial microbes associated with roots facilitate plant growth by providing them with resources like nitrogen or phosphorus (Perez-Montano et al. 2014). Thanks to the development of “omics” approaches, the molecular mechanisms of plant–microbe interaction are gradually uncovered. The promotional effect of beneficial microbes for plant growth can be partially interpreted by auxin production from microbes.
Arbuscular mycorrhiza (AM) is a major root symbiosis in rice to provide phosphate and other nutrients (review in Huang et al. 2022). AM colonization promotes auxin accumulation in soybean, maize, Medicago (Medicago truncatula), and tomato (Shaul-Keinan et al. 2002; Kaldorf and Ludwig-Müller 2000; Meixner et al. 2005). In tomato, exogenous auxin treatment or auxin overproduction by loss of SlGH3.4 function increases arbuscule abundance, and the elevated auxin response was mainly present within arbuscule-containing cells (Chen et al. 2022). AM colonization stimulates auxin response and auxin level in host roots, meanwhile the overaccumulated auxin-level feedback constrains auxin biosynthesis in host plants (Chen et al. 2022). Inhibition of auxin transport by NPA inhibitor greatly suppresses the mycorrhization level (Chen et al. 2022). Hence, auxin is positively correlated with AM colonization, as shown by the fact that high auxin level/signaling promotes but auxin deficiency suppresses AM symbiosis (Hanlon and Coenen 2011; Etemadi et al. 2014; Guillotin et al. 2017; Chen et al. 2022). Despite a certain amount of genetic evidence in support of auxin required for AM colonization, the underlying mechanism remains to be explored.
Legume plants can form symbiosis not only with AM, but also with soil bacteria (rhizobia) to generate nodules. Nodules convert atmospheric nitrogen into ammonia to overcome nitrogen shortage in the host plant. Nodules are mainly divided into two types, determinate and indeterminate nodules, relying on the maintenance duration of nodule meristems (Sprent and James 2007). Soybean and lotus (Lotus japonicus) produce determinate nodules whose meristem is lost soon after nodule formation, but the Medicago nodule is an indeterminate type which possesses a persistent apical meristem (Popp and Ott 2011). Auxin response in root hair occurs rapidly in response to rhizobia infection (Breakspear et al. 2014; Nadzieja et al. 2018). An auxin maximum is established by collaborating local auxin biosynthesis, auxin transport, and auxin signaling pathways during nodule organogenesis. Local auxin biosynthesis is activated below the site of rhizobia inoculation in indeterminate nodules (Schiessl et al. 2019). In soybean, the expression of auxin biosynthesis gene GmYUC2a reaches the peak at the very early stage before the emergence of nodule primordium (Wang et al. 2019b). Besides local auxin biosynthesis, auxin methyl ester form (MeIAA) is elevated at the early stage of nodule initiation. Silence of IAA carboxyl methyltransferase 1 (IAMT1), which converts free IAA into MeIAA, inhibits nodule development in the root cortex layer (Goto et al. 2022). PIN-dependent auxin transport is a determinant to establish auxin maximum for organ initiation, including nodule initiation. Loss of GmPIN1 function in Gmpin1abc triple mutant generates clusters of abnormal dividing cells and defective nodule primordia, thus reducing nodule number (Gao et al. 2021). Once auxin response is defected, nodule initiation and development is also severely influenced, as seen by the decreased nodule number in deficiency of auxin signaling components in GmTIR1/AFB3-knockdown lines (Cai et al. 2017) and GmARF8a-overexpression lines (Wang et al. 2015).
Not only the sole auxin regulation, but auxin also interacts with cytokinin and flavonoid to coordinate nodule initiation. In Medicago, cytokinin perception mutant cytokinin response1 (cre1) fails to initiate nodules. Meanwhile, polar auxin transport, auxin biosynthesis, and auxin signaling pathways are all defective in cre1 mutant. Surprisingly, nodulation defects of cre1 mutant can be rescued by application of auxin transport inhibitors (Ng et al. 2015). Deficiency of flavonoid production by silencing chalcone synthase (CHS) promotes auxin transport but fails to initiate nodules (Wasson et al. 2006). In lotus, the dominant snf2 mutation, which constitutively activates cytokinin signaling pathway, causes spontaneous nodule formation in the absence of rhizobia. Auxin is overaccumulated in snf2 nodules, indicating that cytokinin positively regulates auxin accumulation during nodule development (Suzaki et al. 2012). Apparently, an unknown causal relationship exists among auxin, cytokinin, and flavonoid for nodule initiation. During soybean nodule primordium formation, flavonoids induce lateral distribution of GmPIN1 and cytokinin re-orientates GmPIN1 cellular localization (Gao et al. 2021). The coordination of flavonoids and cytokinin on GmPIN1 establishes an appropriate auxin gradient at nodule primordium (Gao et al. 2021). ARF10/16/17 transcription factors, targeted by microRNA160 (miR160), are key signaling components that determine the balance of auxin and cytokinin during soybean nodule development (Nizampatnam et al. 2015). miR160 overexpression causes the silence of ARF10/16/17 and the reduction of nodules, which is partially restored by exogenous auxin application (Nizampatnam et al. 2015). Interestingly, nodule and lateral root, as lateral root organs, share overlapping developmental program at their initiation stage, which are both dependent on a local auxin biosynthesis and cytokinin. The core regulator NODULE INCEPTION (NIN) stimulates LOB-DOMAIN PROTEIN 16 (LBD16), in turn activating YUC-dependent auxin biosynthesis for nodule initiation in Medicago (Schiessl et al. 2019). Therefore, cytokinin signaling positively regulates auxin accumulation for nodulation, but whether flavonoids have a general inhibitory effect on auxin transport during nodule organogenesis is still unknown. Recent studies on Cryptochrome 1 (CRY1)-dependent light signaling highlight the promotional effect of light signaling in shoots for soybean root nodulation. The mobile transcriptional factors TGACG-motif binding factor 3/4 (GmSTF3/4) and FLOWERING LOCUS T (GmFTs), which move from shoots to roots, activate nodulation events including upregulation of GmNIN, nuclear factor Y (GmNF-YA1 and NF-YB1) and GmENOD40 transcripts (Wang et al. 2021; Li et al. 2022b). Soybean GmSTFs are orthologs of Arabidopsis HY5, which has been well characterized as a central regulator of light signaling, directly or indirectly controlling auxin transport and auxin signaling on a transcriptional level (Cluis et al. 2004; Sibout et al. 2006; van Gelderen et al. 2018). Thus, auxin regulation possibly also contributes to GmSTF-dependent soybean nodulation. Taken together, auxin plays a critical role for determinate and indeterminate nodule development (Fig. 3). How we can apply the current molecular mechanisms in legume plant breeding, and how we can integrate the belowground root symbiosis with aboveground yield deserve further investigation.
Perspective and opportunities for soybean breeding
The studies in Arabidopsis are important for understanding the molecular mechanisms of auxin regulations on organ development, and the added information from other crop systems is indispensable to learn the possible applications of auxin in agricultural system. More and more QTLs associated with important traits are related to auxin (Zhao et al. 2021, 2022b, 2022a; Bettembourg et al. 2017; Guo et al. 2022; Liu et al. 2015a; Wang et al. 2016; Ping et al. 2022), implying auxin engineering has tremendous potential for manipulation of crop architecture in breeding. According to the Green Revolution concept in rice, Liu et al. (2020) proposed a potential strategy to achieve ideal soybean plant, with desirable soybean traits of appropriate plant height, more internodes but short internode length, few branches, high podding rate, high ratio of four seed per pod, and small leaf petiole angle. However, it is impossible to simply transform the experience from rice and maize in soybean breeding.
Firstly, compared with the compact shoot architecture for high-density planting in rice and maize, it is hard to quantitatively define the ideal soybean shoot architecture since the soybean traits, such as leaf angle, leaf size, podding rate, internode length, and branch number, show intensive feedback regulation on each other. Overexpression or knockout of certain auxin genes in soybean may affect the global auxin responses in the whole plant level. The CRISPR-based tissue-specific knockout system (CRISPR-TSKO) (Decaestecker et al. 2019), which conditionally silences genes in a tissue-specific manner, becomes a good approach to integrate auxin engineering for desirable soybean traits.
Secondly, soybean breeding requires a global consideration of above- and underground tissues with energy consumption and nutrient partitioning (Fig. 3). The soybean nodule system has been considered in most cropping systems; thus, the ideal root architecture and nodule-nitrogen fixation ability should be included in the ideal soybean model. The ideal soybean plant should display a compact shoot architecture with high photosynthesis efficiency, as well as appropriate root architecture with high ability of nitrogen fixation and nutrient utilization.
Thirdly, compared with model plant Arabidopsis, two whole-genome duplication results in high redundancy of gene function in soybean. Gene redundancy offers a good opportunity of auxin manipulation to generate mild auxin-related aberrant soybean plants although it makes troubles for investigation of gene function. By knocking out a single gene or quantitative interference of redundant genes according to the specific tissue expression manner, auxin engineering becomes a possibility to breed an ideal soybean plant.
Author contribution
Linfang Li and Xu Chen carried out the literature review, designed the figures, and wrote the manuscript. All the authors read and approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation-Outstanding Youth Foundation (32222009) and Major Program of Natural Science Foundation in Fujian Province (2021J02011) to Xu Chen.
Data availability
All data generated or analyzed during this study are included in this published article.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
This article is part of the Topical Collection on Soybean Functional Genomics
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Adamowski M, Friml J. PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell. 2015;27:20–32. doi: 10.1105/tpc.114.134874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aida M, Beis D, Heidstra R, Willemsen V, Blilou I, Galinha C, Nussaume L, Noh YS, Amasino R, Scheres B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell. 2004;119:109–120. doi: 10.1016/j.cell.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Amanda D, Frey FP, Neumann U, Przybyl M, Simura J, Zhang Y, Chen Z, Gallavotti A, Fernie AR, Ljung K, Acosta IF. Auxin boosts energy generation pathways to fuel pollen maturation in barley. Curr Biol. 2022;32:1798–1811. doi: 10.1016/j.cub.2022.02.073. [DOI] [PubMed] [Google Scholar]
- Aya K, Hobo T, Sato-Izawa K, Ueguchi-Tanaka M, Kitano H, Matsuoka M. A novel AP2-type transcription factor, SMALL ORGAN SIZE1, controls organ size downstream of an auxin signaling pathway. Plant Cell Physiol. 2014;55:897–912. doi: 10.1093/pcp/pcu023. [DOI] [PubMed] [Google Scholar]
- Baker RF, Leach KA, Braun DM. SWEET as sugar: new sucrose effluxers in plants. Mol Plant. 2012;5:766–768. doi: 10.1093/mp/sss054. [DOI] [PubMed] [Google Scholar]
- Barbez E, Kubes M, Rolcik J, Beziat C, Pencik A, Wang B, Rosquete MR, Zhu J, Dobrev PI, Lee Y, Zazimalova E, Petrasek J, Geisler M, Friml J, Kleine-Vehn J. A novel putative auxin carrier family regulates intracellular auxin homeostasis in plants. Nature. 2012;485:119–122. doi: 10.1038/nature11001. [DOI] [PubMed] [Google Scholar]
- Barbosa ICR, Hammes UZ, Schwechheimer C. Activation and polarity control of PIN-FORMED auxin transporters by phosphorylation. Trends Plant Sci. 2018;23:523–538. doi: 10.1016/j.tplants.2018.03.009. [DOI] [PubMed] [Google Scholar]
- Bartel B, Fink GR. ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science. 1995;268:1745–1748. doi: 10.1126/science.7792599. [DOI] [PubMed] [Google Scholar]
- Bauly JM, Sealy IM, Macdonald H, Brearley J, Dröge S, Hillmer S, Robinson DG, Venis MA, Blatt MR, Lazarus CM, Napier RM. Overexpression of auxin-binding protein enhances the sensitivity of guard cells to auxin. Plant Physiol. 2000;124:1229–1238. doi: 10.1104/pp.124.3.1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender SF, Wagg C, van der Heijden MGA. An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol Evol. 2016;31:440–452. doi: 10.1016/j.tree.2016.02.016. [DOI] [PubMed] [Google Scholar]
- Benjamins R, Scheres B. Auxin: the looping star in plant development. Annu Rev Plant Biol. 2008;59:443–465. doi: 10.1146/annurev.arplant.58.032806.103805. [DOI] [PubMed] [Google Scholar]
- Benková E, Michniewicz M, Sauer M, Teichmann T, Seifertová D, Jürgens G, Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- Bennett T. PIN proteins and the evolution of plant development. Trends Plant Sci. 2015;20:498–507. doi: 10.1016/j.tplants.2015.05.005. [DOI] [PubMed] [Google Scholar]
- Bettembourg M, Dardou A, Audebert A, Thomas E, Frouin J, Guiderdoni E, Ahmadi N, Perin C, Dievart A, Courtois B. Genome-wide association mapping for root cone angle in rice. Rice (n y) 2017;10:45. doi: 10.1186/s12284-017-0184-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boss PK, Thomas MR. Association of dwarfism and floral induction with a grape ‘green revolution’ mutation. Nature. 2002;416:847–850. doi: 10.1038/416847a. [DOI] [PubMed] [Google Scholar]
- Breakspear A, Liu C, Roy S, Stacey N, Rogers C, Trick M, Morieri G, Mysore KS, Wen J, Oldroyd GE, Downie JA, Murray JD. The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell. 2014;26:4680–4701. doi: 10.1105/tpc.114.133496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai Z, Wang Y, Zhu L, Tian Y, Chen L, Sun Z, Ullah I, Li X. GmTIR1/GmAFB3-based auxin perception regulated by miR393 modulates soybean nodulation. New Phytol. 2017;215:672–686. doi: 10.1111/nph.14632. [DOI] [PubMed] [Google Scholar]
- Cance C, Martin-Arevalillo R, Boubekeur K, Dumas R. Auxin response factors are keys to the many auxin doors. New Phytol. 2022;235:402–419. doi: 10.1111/nph.18159. [DOI] [PubMed] [Google Scholar]
- Cao M, Chen R, Li P, Yu Y, Zheng R, Ge D, Zheng W, Wang X, Gu Y, Gelova Z, Friml J, Zhang H, Liu R, He J, Xu T. TMK1-mediated auxin signalling regulates differential growth of the apical hook. Nature. 2019;568:240–243. doi: 10.1038/s41586-019-1069-7. [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M. Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet. 2009;43:265–285. doi: 10.1146/annurev-genet-102108-134148. [DOI] [PubMed] [Google Scholar]
- Chen JG, Shimomura S, Sitbon F, Sandberg G, Jones AM. The role of auxin-binding protein 1 in the expansion of tobacco leaf cells. Plant J. 2001;28:607–617. doi: 10.1046/j.1365-313x.2001.01152.x. [DOI] [PubMed] [Google Scholar]
- Chen JG, Ullah H, Young JC, Sussman MR, Jones AM. ABP1 is required for organized cell elongation and division in Arabidopsis embryogenesis. Genes Dev. 2001;15:902–911. doi: 10.1101/gad.866201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Nan H, Kong L, Yue L, Yang H, Zhao Q, Fang C, Li H, Cheng Q, Lu S, Kong F, Liu B, Dong L. Soybean AP1 homologs control flowering time and plant height. J Integr Plant Biol. 2020;62:1868–1879. doi: 10.1111/jipb.12988. [DOI] [PubMed] [Google Scholar]
- Chen L, Yang H, Fang Y, Guo W, Chen H, Zhang X, Dai W, Chen S, Hao Q, Yuan S, Zhang C, Huang Y, Shan Z, Yang Z, Qiu D, Liu X, Tran LP, Zhou X, Cao D. Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol J. 2021;19:702–716. doi: 10.1111/pbi.13496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Chen J, Liao D, Ye H, Li C, Luo Z, Yan A, Zhao Q, Xie K, Li Y, Wang D, Chen J, Chen A, Xu G. Auxin-mediated regulation of arbuscular mycorrhizal symbiosis: a role of SlGH3.4 in tomato. Plant Cell Environ. 2022;45:955–968. doi: 10.1111/pce.14210. [DOI] [PubMed] [Google Scholar]
- Cheng Q, Dong L, Su T, Li T, Gan Z, Nan H, Lu S, Fang C, Kong L, Li H, Hou Z, Kou K, Tang Y, Lin X, Zhao X, Chen L, Liu B, Kong F. CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 2019;19:562. doi: 10.1186/s12870-019-2145-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J. 2004;38:332–347. doi: 10.1111/j.1365-313X.2004.02052.x. [DOI] [PubMed] [Google Scholar]
- Cucinotta M, Di Marzo M, Guazzotti A, de Folter S, Kater MM, Colombo L. Gynoecium size and ovule number are interconnected traits that impact seed yield. J Exp Bot. 2020;71:2479–2489. doi: 10.1093/jxb/eraa050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies RT, Goetz DH, Lasswell J, Anderson MN, Bartel B. IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell. 1999;11:365–376. doi: 10.1105/tpc.11.3.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decaestecker W, Buono RA, Pfeiffer ML, Vangheluwe N, Jourquin J, Karimi M, Van Isterdael G, Beeckman T, Nowack MK, Jacobs TB. CRISPR-TSKO: a technique for efficient mutagenesis in specific cell types, tissues, or organs in Arabidopsis. Plant Cell. 2019;31:2868–2887. doi: 10.1105/tpc.19.00454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng Y, Yu Y, Hu Y, Ma L, Lin Y, Wu Y, Wang Z, Wang Z, Bai J, Ding Y, Chen L. Auxin-mediated regulation of dorsal vascular cell development may be responsible for sucrose phloem unloading in large panicle rice. Front Plant Sci. 2021;12:630997. doi: 10.3389/fpls.2021.630997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P, Aniento F, Hwang I, Robinson DG, Mravec J, Stierhof YD, Friml J. Clathrin-mediated constitutive endocytosis of PIN auxin efflux carriers in Arabidopsis. Curr Biol. 2007;17:520–527. doi: 10.1016/j.cub.2007.01.052. [DOI] [PubMed] [Google Scholar]
- Di Mambro R, De Ruvo M, Pacifici E, Salvi E, Sozzani R, Benfey PN, Busch W, Novak O, Ljung K, Di Paola L, Maree AFM, Costantino P, Grieneisen VA, Sabatini S. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc Natl Acad Sci U S A. 2017;114:E7641–E7649. doi: 10.1073/pnas.1705833114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Wang B, Moreno I, Duplakova N, Simon S, Carraro N, Reemmer J, Pencik A, Chen X, Tejos R, Skupa P, Pollmann S, Mravec J, Petrasek J, Zazimalova E, Honys D, Rolcik J, Murphy A, Orellana A, Geisler M, Friml J. ER-localized auxin transporter PIN8 regulates auxin homeostasis and male gametophyte development in Arabidopsis. Nat Commun. 2012;3:941. doi: 10.1038/ncomms1941. [DOI] [PubMed] [Google Scholar]
- Donald CM. The breeding of crop ideotypes. Euphytica. 1968;17:385–403. doi: 10.1007/BF00056241. [DOI] [Google Scholar]
- Du M, Spalding EP, Gray WM. Rapid auxin-mediated cell expansion. Annu Rev Plant Biol. 2020;71:379–402. doi: 10.1146/annurev-arplant-073019-025907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etemadi M, Gutjahr C, Couzigou JM, Zouine M, Lauressergues D, Timmers A, Audran C, Bouzayen M, Becard G, Combier JP. Auxin perception is required for arbuscule development in arbuscular mycorrhizal symbiosis. Plant Physiol. 2014;166:281–292. doi: 10.1104/pp.114.246595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evenson RE, Gollin D. Assessing the impact of the green revolution, 1960 to 2000. Science. 2003;300:758–762. doi: 10.1126/science.1078710. [DOI] [PubMed] [Google Scholar]
- Fendrych M, Akhmanova M, Merrin J, Glanc M, Hagihara S, Takahashi K, Uchida N, Torii KU, Friml J. Rapid and reversible root growth inhibition by TIR1 auxin signalling. Nat Plants. 2018;4:453–459. doi: 10.1038/s41477-018-0190-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Friml J. PIN polar targeting. Plant Physiol. 2008;147:1553–1559. doi: 10.1104/pp.108.121756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Wiśniewska J, Benková E, Mendgen K, Palme K. Lateral relocation of auxin efflux regulator PIN3 mediates tropism in Arabidopsis. Nature. 2002;415:806–809. doi: 10.1038/415806a. [DOI] [PubMed] [Google Scholar]
- Friml J, Gallei M, Gelova Z, Johnson A, Mazur E, Monzer A, Rodriguez L, Roosjen M, Verstraeten I, Zivanovic BD, Zou M, Fiedler L, Giannini C, Grones P, Hrtyan M, Kaufmann WA, Kuhn A, Narasimhan M, Randuch M, Rydza N, Takahashi K, Tan S, Teplova A, Kinoshita T, Weijers D, Rakusova H. ABP1-TMK auxin perception for global phosphorylation and auxin canalization. Nature. 2022;609:575–581. doi: 10.1038/s41586-022-05187-x. [DOI] [PubMed] [Google Scholar]
- Fu J, Zhang X, Liu J, Gao X, Bai J, Hao Y, Cui H. A mechanism coordinating root elongation, endodermal differentiation, redox homeostasis and stress response. Plant J. 2021;107:1029–1039. doi: 10.1111/tpj.15361. [DOI] [PubMed] [Google Scholar]
- Fukui K, Arai K, Tanaka Y, Aoi Y, Kukshal V, Jez JM, Kubes MF, Napier R, Zhao Y, Kasahara H, Hayashi KI. Chemical inhibition of the auxin inactivation pathway uncovers the roles of metabolic turnover in auxin homeostasis. Proc Natl Acad Sci U S A. 2022;119:e2206869119. doi: 10.1073/pnas.2206869119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galinha C, Hofhuis H, Luijten M, Willemsen V, Blilou I, Heidstra R, Scheres B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature. 2007;449:1053–1057. doi: 10.1038/nature06206. [DOI] [PubMed] [Google Scholar]
- Gao S, Chu C. Gibberellin metabolism and signaling: targets for improving agronomic performance of crops. Plant Cell Physiol. 2020;61:1902–1911. doi: 10.1093/pcp/pcaa104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci U S A. 2015;112:2275–2280. doi: 10.1073/pnas.1500365112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z, Chen Z, Cui Y, Ke M, Xu H, Xu Q, Chen J, Li Y, Huang L, Zhao H, Huang D, Mai S, Xu T, Liu X, Li S, Guan Y, Yang W, Friml J, Petrasek J, Zhang J, Chen X. GmPIN-dependent polar auxin transport is involved in soybean nodule development. Plant Cell. 2021;33:2981–3003. doi: 10.1093/plcell/koab183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler MM. A retro-perspective on auxin transport. Front Plant Sci. 2021;12:756968. doi: 10.3389/fpls.2021.756968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisler M, Blakeslee JJ, Bouchard R, Lee OR, Vincenzetti V, Bandyopadhyay A, Titapiwatanakun B, Peer WA, Bailly A, Richards EL, Ejendal KF, Smith AP, Baroux C, Grossniklaus U, Muller A, Hrycyna CA, Dudler R, Murphy AS, Martinoia E. Cellular efflux of auxin catalyzed by the Arabidopsis MDR/PGP transporter AtPGP1. Plant J. 2005;44:179–194. doi: 10.1111/j.1365-313X.2005.02519.x. [DOI] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell. 2003;112:219–230. doi: 10.1016/s0092-8674(03)00003-5. [DOI] [PubMed] [Google Scholar]
- Giri J, Bhosale R, Huang G, Pandey BK, Parker H, Zappala S, Yang J, Dievart A, Bureau C, Ljung K, Price A, Rose T, Larrieu A, Mairhofer S, Sturrock CJ, White P, Dupuy L, Hawkesford M, Perin C, Liang W, Peret B, Hodgman CT, Lynch J, Wissuwa M, Zhang D, Pridmore T, Mooney SJ, Guiderdoni E, Swarup R, Bennett MJ. Rice auxin influx carrier OsAUX1 facilitates root hair elongation in response to low external phosphate. Nat Commun. 2018;9:1408. doi: 10.1038/s41467-018-03850-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glanc M, Fendrych M, Friml J. Mechanistic framework for cell-intrinsic re-establishment of PIN2 polarity after cell division. Nat Plants. 2018;4:1082–1088. doi: 10.1038/s41477-018-0318-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto T, Soyano T, Liu M, Mori T, Kawaguchi M. Auxin methylation by IAMT1, duplicated in the legume lineage, promotes root nodule development in Lotus japonicus. Proc Natl Acad Sci U S A. 2022;119:e2116549119. doi: 10.1073/pnas.2116549119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grubb CD, Zipp BJ, Ludwig-Muller J, Masuno MN, Molinski TF, Abel S. Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 2004;40:893–908. doi: 10.1111/j.1365-313X.2004.02261.x. [DOI] [PubMed] [Google Scholar]
- Guillotin B, Etemadi M, Audran C, Bouzayen M, Becard G, Combier JP. Sl-IAA27 regulates strigolactone biosynthesis and mycorrhization in tomato (var. MicroTom) New Phytol. 2017;213:1124–1132. doi: 10.1111/nph.14246. [DOI] [PubMed] [Google Scholar]
- Guo F, Huang Y, Qi P, Lian G, Hu X, Han N, Wang J, Zhu M, Qian Q, Bian H. Functional analysis of auxin receptor OsTIR1/OsAFB family members in rice grain yield, tillering, plant height, root system, germination, and auxinic herbicide resistance. New Phytol. 2021;229:2676–2692. doi: 10.1111/nph.17061. [DOI] [PubMed] [Google Scholar]
- Guo N, Wang Y, Chen W, Tang S, An R, Wei X, Hu S, Tang S, Shao G, Jiao G, Xie L, Wang L, Sheng Z, Hu P. Fine mapping and target gene identification of qSE4, a QTL for stigma exsertion rate in rice (Oryza sativa L.) Front Plant Sci. 2022;13:959859. doi: 10.3389/fpls.2022.959859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han H, Adamowski M, Qi L, Alotaibi SS, Friml J. PIN-mediated polar auxin transport regulations in plant tropic responses. New Phytol. 2021;232:510–522. doi: 10.1111/nph.17617. [DOI] [PubMed] [Google Scholar]
- Hanlon MT, Coenen C. Genetic evidence for auxin involvement in arbuscular mycorrhiza initiation. New Phytol. 2011;189:701–709. doi: 10.1111/j.1469-8137.2010.03567.x. [DOI] [PubMed] [Google Scholar]
- He W, Brumos J, Li H, Ji Y, Ke M, Gong X, Zeng Q, Li W, Zhang X, An F, Wen X, Li P, Chu J, Sun X, Yan C, Yan N, Xie DY, Raikhel N, Yang Z, Stepanova AN, Alonso JM, Guo H. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Yang L, Hu W, Zhang J, Xing Y. Overexpression of an auxin receptor OsAFB6 significantly enhanced grain yield by increasing cytokinin and decreasing auxin concentrations in rice panicle. Sci Rep. 2018;8:14051. doi: 10.1038/s41598-018-32450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/s0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- Hesse T, Feldwisch J, Balshüsemann D, Bauw G, Puype M, Vandekerckhove J, Löbler M, Klämbt D, Schell J, Palme K. Molecular cloning and structural analysis of a gene from Zea mays (L.) coding for a putative receptor for the plant hormone auxin. Embo j. 1989;8:2453–2461. doi: 10.1002/j.1460-2075.1989.tb08380.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano K, Yoshida H, Aya K, Kawamura M, Hayashi M, Hobo T, Sato-Izawa K, Kitano H, Ueguchi-Tanaka M, Matsuoka M. SMALL ORGAN SIZE 1 and SMALL ORGAN SIZE 2/DWARF AND LOW-TILLERING Form a Complex to Integrate Auxin and Brassinosteroid Signaling in Rice. Mol Plant. 2017;10:590–604. doi: 10.1016/j.molp.2016.12.013. [DOI] [PubMed] [Google Scholar]
- Hu Z, Lu SJ, Wang MJ, He H, Sun L, Wang H, Liu XH, Jiang L, Sun JL, Xin X, Kong W, Chu C, Xue HW, Yang J, Luo X, Liu JX. A novel QTL qTGW3 encodes the GSK3/SHAGGY-Like Kinase OsGSK5/OsSK41 that interacts with OsARF4 to negatively regulate grain size and weight in rice. Mol Plant. 2018;11:736–749. doi: 10.1016/j.molp.2018.03.005. [DOI] [PubMed] [Google Scholar]
- Hu Y, Li S, Fan X, Song S, Zhou X, Weng X, Xiao J, Li X, Xiong L, You A, Xing Y. OsHOX1 and OsHOX28 redundantly shape rice tiller angle by reducing HSFA2D expression and auxin content. Plant Physiol. 2020;184:1424–1437. doi: 10.1104/pp.20.00536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu LQ, Chang JH, Yu SX, Jiang YT, Li RH, Zheng JX, Zhang YJ, Xue HW, Lin WH. PIN3 positively regulates the late initiation of ovule primordia in Arabidopsis thaliana. PLoS Genet. 2022;18:e1010077. doi: 10.1371/journal.pgen.1010077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang G, Hu H, van de Meene A, Zhang J, Dong L, Zheng S, Zhang F, Betts NS, Liang W, Bennett MJ, Persson S, Zhang D. AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell. 2021;33:3120–3133. doi: 10.1093/plcell/koab175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L, Wang W, Zhang N, Cai Y, Liang Y, Meng X, Yuan Y, Li J, Wu D, Wang Y. LAZY2 controls rice tiller angle through regulating starch biosynthesis in gravity-sensing cells. New Phytol. 2021;231:1073–1087. doi: 10.1111/nph.17426. [DOI] [PubMed] [Google Scholar]
- Huang R, Li Z, Shen X, Choi J, Cao Y (2022) The Perspective of Arbuscular Mycorrhizal Symbiosis in Rice Domestication and Breeding. Int J Mol Sci 23. 10.3390/ijms232012383 [DOI] [PMC free article] [PubMed]
- Inukai Y, Sakamoto T, Ueguchi-Tanaka M, Shibata Y, Gomi K, Umemura I, Hasegawa Y, Ashikari M, Kitano H, Matsuoka M. Crown rootless1, which is essential for crown root formation in rice, is a target of an AUXIN RESPONSE FACTOR in auxin signaling. Plant Cell. 2005;17:1387–1396. doi: 10.1105/tpc.105.030981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson RG, Lim EK, Li Y, Kowalczyk M, Sandberg G, Hoggett J, Ashford DA, Bowles DJ. Identification and biochemical characterization of an Arabidopsis indole-3-acetic acid glucosyltransferase. J Biol Chem. 2001;276:4350–4356. doi: 10.1074/jbc.M006185200. [DOI] [PubMed] [Google Scholar]
- Jeong N, Moon JK, Kim HS, Kim CG, Jeong SC. Fine genetic mapping of the genomic region controlling leaflet shape and number of seeds per pod in the soybean. Theor Appl Genet. 2011;122:865–874. doi: 10.1007/s00122-010-1492-5. [DOI] [PubMed] [Google Scholar]
- Jeong N, Suh SJ, Kim MH, Lee S, Moon JK, Kim HS, Jeong SC. Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell. 2012;24:4807–4818. doi: 10.1105/tpc.112.104968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji X, Gao Q, Chen F, Bai M, Zhuang Z, Peng Y (2022) Mutant lpa1 Analysis of ZmLPA1 Gene Regulates Maize Leaf-Angle Development through the Auxin Pathway. Int J Mol Sci 23. 10.3390/ijms23094886 [DOI] [PMC free article] [PubMed]
- Jiang K, Feldman LJ. Root Meristem Establishment and Maintenance: The Role of Auxin. J Plant Growth Regul. 2003;21:432–440. doi: 10.1007/s00344-002-0037-9. [DOI] [Google Scholar]
- Jiang H, Li Y, Qin H, Li Y, Qi H, Li C, Wang N, Li R, Zhao Y, Huang S, Yu J, Wang X, Zhu R, Liu C, Hu Z, Qi Z, Xin D, Wu X, Chen Q. Identification of Major QTLs Associated With First Pod Height and Candidate Gene Mining in Soybean. Front Plant Sci. 2018;9:1280. doi: 10.3389/fpls.2018.01280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z-f, Liu D-d, Wang T-q, Liang X-l, Cui Y-h, Liu Z-h, Li W-b. Concentration difference of auxin involved in stem development in soybean. J Integr Agric. 2020;19:953–964. doi: 10.1016/s2095-3119(19)62676-6. [DOI] [Google Scholar]
- Jones AM, Venis MA. Photoaffinity labeling of indole-3-acetic acid-binding proteins in maize. Proc Natl Acad Sci U S A. 1989;86:6153–6156. doi: 10.1073/pnas.86.16.6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones AM, Im KH, Savka MA, Wu MJ, DeWitt NG, Shillito R, Binns AN. Auxin-dependent cell expansion mediated by overexpressed auxin-binding protein 1. Science. 1998;282:1114–1117. doi: 10.1126/science.282.5391.1114. [DOI] [PubMed] [Google Scholar]
- Julius BT, Leach KA, Tran TM, Mertz RA, Braun DM. Sugar Transporters in Plants: New Insights and Discoveries. Plant Cell Physiol. 2017;58:1442–1460. doi: 10.1093/pcp/pcx090. [DOI] [PubMed] [Google Scholar]
- Jun N, Gaohang W, Zhenxing Z, Huanhuan Z, Yunrong W, Ping W. OsIAA23-mediated auxin signaling defines postembryonic maintenance of QC in rice. Plant J. 2011;68:433–442. doi: 10.1111/j.1365-313X.2011.04698.x. [DOI] [PubMed] [Google Scholar]
- Kaldorf M, Ludwig-Müller J. AM fungi might affect the root morphology of maize by increasing indole-3-butyric acid biosynthesis. Physiol Plant. 2000;109:58–67. doi: 10.1034/j.1399-3054.2000.100109.x. [DOI] [Google Scholar]
- Khush GS. Green revolution: the way forward. Nat Rev Genet. 2001;2:815–822. doi: 10.1038/35093585. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Ito H, Hobo T, Aya K, Kitano H, Inukai Y. The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 2011;67:472–484. doi: 10.1111/j.1365-313X.2011.04610.x. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Inahashi H, Takehisa H, Sato Y, Inukai Y. OsIAA13-mediated auxin signaling is involved in lateral root initiation in rice. Plant Sci. 2012;190:116–122. doi: 10.1016/j.plantsci.2012.04.005. [DOI] [PubMed] [Google Scholar]
- Kitomi Y, Hanzawa E, Kuya N, Inoue H, Hara N, Kawai S, Kanno N, Endo M, Sugimoto K, Yamazaki T, Sakamoto S, Sentoku N, Wu J, Kanno H, Mitsuda N, Toriyama K, Sato T, Uga Y. Root angle modifications by the DRO1 homolog improve rice yields in saline paddy fields. Proc Natl Acad Sci U S A. 2020;117:21242–21250. doi: 10.1073/pnas.2005911117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J. Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci U S A. 2008;105:17812–17817. doi: 10.1073/pnas.0808073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korasick DA, Enders TA, Strader LC. Auxin biosynthesis and storage forms. J Exp Bot. 2013;64:2541–2555. doi: 10.1093/jxb/ert080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhn C, Grof CP. Sucrose transporters of higher plants. Curr Opin Plant Biol. 2010;13:288–298. doi: 10.1016/j.pbi.2010.02.001. [DOI] [PubMed] [Google Scholar]
- LanassaBassukas AE, Xiao Y, Schwechheimer C. Phosphorylation control of PIN auxin transporters. Curr Opin Plant Biol. 2022;65:102146. doi: 10.1016/j.pbi.2021.102146. [DOI] [PubMed] [Google Scholar]
- Leyser O. Molecular genetics of auxin signaling. Annu Rev Plant Biol. 2002;53:377–398. doi: 10.1146/annurev.arplant.53.100301.135227. [DOI] [PubMed] [Google Scholar]
- Leyser O. Dynamic integration of auxin transport and signalling. Curr Biol. 2006;16:R424–433. doi: 10.1016/j.cub.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Li P, Wang Y, Qian Q, Fu Z, Wang M, Zeng D, Li B, Wang X, Li J. LAZY1 controls rice shoot gravitropism through regulating polar auxin transport. Cell Res. 2007;17:402–410. doi: 10.1038/cr.2007.38. [DOI] [PubMed] [Google Scholar]
- Li P, Chen F, Cai H, Liu J, Pan Q, Liu Z, Gu R, Mi G, Zhang F, Yuan L. A genetic relationship between nitrogen use efficiency and seedling root traits in maize as revealed by QTL analysis. J Exp Bot. 2015;66:3175–3188. doi: 10.1093/jxb/erv127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Zhang X, Zhao Y, Li Y, Zhang G, Peng Z, Zhang J. Enhancing auxin accumulation in maize root tips improves root growth and dwarfs plant height. Plant Biotechnol J. 2018;16:86–99. doi: 10.1111/pbi.12751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li ZF, Guo Y, Ou L, Hong H, Wang J, Liu ZX, Guo B, Zhang L, Qiu L. Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor Appl Genet. 2018;131:1001–1016. doi: 10.1007/s00122-017-3044-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Liang Y, Yuan Y, Wang L, Meng X, Xiong G, Zhou J, Cai Y, Han N, Hua L, Liu G, Li J, Wang Y. OsBRXL4 Regulates Shoot Gravitropism and Rice Tiller Angle through Affecting LAZY1 Nuclear Localization. Mol Plant. 2019;12:1143–1156. doi: 10.1016/j.molp.2019.05.014. [DOI] [PubMed] [Google Scholar]
- Li Y, Li J, Chen Z, Wei Y, Qi Y, Wu C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Bai D, Tian Y, Li YH, Zhao C, Wang Q, Guo S, Gu Y, Luan X, Wang R, Yang J, Hawkesford MJ, Schnable JC, Jin X, Qiu LJ. Time Series Canopy Phenotyping Enables the Identification of Genetic Variants Controlling Dynamic Phenotypes in Soybean. J Integr Plant Biol. 2022 doi: 10.1111/jipb.13380. [DOI] [PubMed] [Google Scholar]
- Li X, Zhou H, Cheng L, Ma N, Cui B, Wang W, Zhong Y, Liao H. Shoot-to-root translocated GmNN1/FT2a triggers nodulation and regulates soybean nitrogen nutrition. PLoS Biol. 2022;20:e3001739. doi: 10.1371/journal.pbio.3001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li P, Wei J, Wang H, Fang Y, Yin S, Xu Y, Liu J, Yang Z, Xu C (2019a) Natural Variation and Domestication Selection of ZmPGP1 Affects Plant Architecture and Yield-Related Traits in Maize. Genes (Basel) 10. 10.3390/genes10090664 [DOI] [PMC free article] [PubMed]
- Li X, Wu P, Lu Y, Guo S, Zhong Z, Shen R, Xie Q (2020a) Synergistic Interaction of Phytohormones in Determining Leaf Angle in Crops. Int J Mol Sci 21. 10.3390/ijms21145052 [DOI] [PMC free article] [PubMed]
- Lin W, Zhou X, Tang W, Takahashi K, Pan X, Dai J, Ren H, Zhu X, Pan S, Zheng H, Gray WM, Xu T, Kinoshita T, Yang Z. TMK-based cell-surface auxin signalling activates cell-wall acidification. Nature. 2021;599:278–282. doi: 10.1038/s41586-021-03976-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Wang J, Wang L, Wang X, Xue Y, Wu P, Shou H. Adventitious root formation in rice requires OsGNOM1 and is mediated by the OsPINs family. Cell Res. 2009;19:1110–1119. doi: 10.1038/cr.2009.70. [DOI] [PubMed] [Google Scholar]
- Liu J, Hua W, Hu Z, Yang H, Zhang L, Li R, Deng L, Sun X, Wang X, Wang H. Natural variation in ARF18 gene simultaneously affects seed weight and silique length in polyploid rapeseed. Proc Natl Acad Sci U S A. 2015;112:E5123–5132. doi: 10.1073/pnas.1502160112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tong H, Xiao Y, Che R, Xu F, Hu B, Liang C, Chu J, Li J, Chu C. Activation of Big Grain1 significantly improves grain size by regulating auxin transport in rice. Proc Natl Acad Sci U S A. 2015;112:11102–11107. doi: 10.1073/pnas.1512748112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Zhang M, Feng F, Tian Z. Toward a "Green Revolution" for Soybean. Mol Plant. 2020;13:688–697. doi: 10.1016/j.molp.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Liu LM, Zhang HQ, Cheng K, Zhang YM. Integrated Bioinformatics Analyses of PIN1, CKX, and Yield-Related Genes Reveals the Molecular Mechanisms for the Difference of Seed Number Per Pod Between Soybean and Cowpea. Front Plant Sci. 2021;12:749902. doi: 10.3389/fpls.2021.749902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Wu K, Harberd NP, Fu X. Green Revolution DELLAs: From translational reinitiation to future sustainable agriculture. Mol Plant. 2021;14:547–549. doi: 10.1016/j.molp.2021.03.015. [DOI] [PubMed] [Google Scholar]
- Liu J, Shi X, Chang Z, Ding Y, Ding C. Auxin Efflux Transporters OsPIN1c and OsPIN1d Function Redundantly in Regulating Rice (Oryza sativa L.) Panicle Development. Plant Cell Physiol. 2022;63:305–316. doi: 10.1093/pcp/pcab172. [DOI] [PubMed] [Google Scholar]
- Lobler M, Klambt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). II. Localization of a putative auxin receptor. J Biol Chem. 1985;260:9854–9859. doi: 10.1016/S0021-9258(17)39315-8. [DOI] [PubMed] [Google Scholar]
- Löbler M, Klämbt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). I. Purification by immunological methods and characterization. J Biol Chem. 1985;260:9848–9853. doi: 10.1016/S0021-9258(17)39314-6. [DOI] [PubMed] [Google Scholar]
- Löbler M, Klämbt D. Auxin-binding protein from coleoptile membranes of corn (Zea mays L.). II. Localization of a putative auxin receptor. J Biol Chem. 1985;260:9854–9859. doi: 10.1016/s0021-9258(17)39315-8. [DOI] [PubMed] [Google Scholar]
- Lu G, Coneva V, Casaretto JA, Ying S, Mahmood K, Liu F, Nambara E, Bi YM, Rothstein SJ. OsPIN5b modulates rice (Oryza sativa) plant architecture and yield by changing auxin homeostasis, transport and distribution. Plant J. 2015;83:913–925. doi: 10.1111/tpj.12939. [DOI] [PubMed] [Google Scholar]
- Luo X, Zheng J, Huang R, Huang Y, Wang H, Jiang L, Fang X. Phytohormones signaling and crosstalk regulating leaf angle in rice. Plant Cell Rep. 2016;35:2423–2433. doi: 10.1007/s00299-016-2052-5. [DOI] [PubMed] [Google Scholar]
- Luo Y, Zhang M, Liu Y, Liu J, Li W, Chen G, Peng Y, Jin M, Wei W, Jian L, Yan J, Fernie AR, Yan J. Genetic variation in YIGE1 contributes to ear length and grain yield in maize. New Phytol. 2022;234:513–526. doi: 10.1111/nph.17882. [DOI] [PubMed] [Google Scholar]
- Lynch JP. Root phenotypes for improved nutrient capture: an underexploited opportunity for global agriculture. New Phytol. 2019;223:548–564. doi: 10.1111/nph.15738. [DOI] [PubMed] [Google Scholar]
- Mahonen AP, Ten Tusscher K, Siligato R, Smetana O, Diaz-Trivino S, Salojarvi J, Wachsman G, Prasad K, Heidstra R, Scheres B. PLETHORA gradient formation mechanism separates auxin responses. Nature. 2014;515:125–129. doi: 10.1038/nature13663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhava P, Aliaga Fandino AC, Koh SWH, Jelinkova A, Kolb M, Janacek DP, Breda AS, Cattaneo P, Hammes UZ, Petrasek J, Hardtke CS. Plasma Membrane Domain Patterning and Self-Reinforcing Polarity in Arabidopsis. Dev Cell. 2020;52(223–235):e225. doi: 10.1016/j.devcel.2019.11.015. [DOI] [PubMed] [Google Scholar]
- Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H. Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta. 2005;222:709–715. doi: 10.1007/s00425-005-0003-4. [DOI] [PubMed] [Google Scholar]
- Mellor NL, Voss U, Ware A, Janes G, Barrack D, Bishopp A, Bennett MJ, Geisler M, Wells DM, Band LR. Systems approaches reveal that ABCB and PIN proteins mediate co-dependent auxin efflux. Plant Cell. 2022;34:2309–2327. doi: 10.1093/plcell/koac086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michniewicz M, Zago MK, Abas L, Weijers D, Schweighofer A, Meskiene I, Heisler MG, Ohno C, Zhang J, Huang F, Schwab R, Weigel D, Meyerowitz EM, Luschnig C, Offringa R, Friml J. Antagonistic regulation of PIN phosphorylation by PP2A and PINOID directs auxin flux. Cell. 2007;130:1044–1056. doi: 10.1016/j.cell.2007.07.033. [DOI] [PubMed] [Google Scholar]
- Mravec J, Kubes M, Bielach A, Gaykova V, Petrasek J, Skupa P, Chand S, Benkova E, Zazimalova E, Friml J. Interaction of PIN and PGP transport mechanisms in auxin distribution-dependent development. Development. 2008;135:3345–3354. doi: 10.1242/dev.021071. [DOI] [PubMed] [Google Scholar]
- Mravec J, Skůpa P, Bailly A, Hoyerová K, Krecek P, Bielach A, Petrásek J, Zhang J, Gaykova V, Stierhof YD, Dobrev PI, Schwarzerová K, Rolcík J, Seifertová D, Luschnig C, Benková E, Zazímalová E, Geisler M, Friml J. Subcellular homeostasis of phytohormone auxin is mediated by the ER-localized PIN5 transporter. Nature. 2009;459:1136–1140. doi: 10.1038/nature08066. [DOI] [PubMed] [Google Scholar]
- Muller A, Guan C, Galweiler L, Tanzler P, Huijser P, Marchant A, Parry G, Bennett M, Wisman E, Palme K. AtPIN2 defines a locus of Arabidopsis for root gravitropism control. EMBO J. 1998;17:6903–6911. doi: 10.1093/emboj/17.23.6903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Multani DS, Briggs SP, Chamberlin MA, Blakeslee JJ, Murphy AS, Johal GS. Loss of an MDR transporter in compact stalks of maize br2 and sorghum dw3 mutants. Science. 2003;302:81–84. doi: 10.1126/science.1086072. [DOI] [PubMed] [Google Scholar]
- Nadzieja M, Kelly S, Stougaard J, Reid D. Epidermal auxin biosynthesis facilitates rhizobial infection in Lotus japonicus. Plant J. 2018;95:101–111. doi: 10.1111/tpj.13934. [DOI] [PubMed] [Google Scholar]
- Ng JL, Hassan S, Truong TT, Hocart CH, Laffont C, Frugier F, Mathesius U. Flavonoids and Auxin Transport Inhibitors Rescue Symbiotic Nodulation in the Medicago truncatula Cytokinin Perception Mutant cre1. Plant Cell. 2015;27:2210–2226. doi: 10.1105/tpc.15.00231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J, Zhang B, Wang N, Zhou Y, Xiong L. Increased leaf angle1, a Raf-like MAPKKK that interacts with a nuclear protein family, regulates mechanical tissue formation in the Lamina joint of rice. Plant Cell. 2011;23:4334–4347. doi: 10.1105/tpc.111.093419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura T, Hayashi K, Suzuki H, Gyohda A, Takaoka C, Sakaguchi Y, Matsumoto S, Kasahara H, Sakai T, Kato J, Kamiya Y, Koshiba T. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- Nizampatnam NR, Schreier SJ, Damodaran S, Adhikari S, Subramanian S. microRNA160 dictates stage-specific auxin and cytokinin sensitivities and directs soybean nodule development. Plant J. 2015;84:140–153. doi: 10.1111/tpj.12965. [DOI] [PubMed] [Google Scholar]
- Noodén LD, Noodén SM. Effects of morphactin and other auxin transport inhibitors on soybean senescence and pod development. Plant Physiol. 1985;78:263–266. doi: 10.1104/pp.78.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K, Ueda J, Komaki MK, Bell CJ, Shimura Y. Requirement of the Auxin Polar Transport System in Early Stages of Arabidopsis Floral Bud Formation. Plant Cell. 1991;3:677–684. doi: 10.1105/tpc.3.7.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panda S, Majhi PK, Anandan A, Mahender A, Veludandi S, Bastia D, Guttala SB, Singh SK, Saha S, Ali J (2021) Proofing Direct-Seeded Rice with Better Root Plasticity and Architecture. Int J Mol Sci 22. 10.3390/ijms22116058 [DOI] [PMC free article] [PubMed]
- Pendleton JW, Smith GE, Winter SR, Johnston TJ. Field investigations of the relationships of leaf Angle in Corn (Zea mays L.) to grain yield and apparent photosynthesis 1. Agron J. 1968;60:422–424. doi: 10.2134/agronj1968.00021962006000040027x. [DOI] [Google Scholar]
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KM, Flintham JE, Beales J, Fish LJ, Worland AJ, Pelica F, Sudhakar D, Christou P, Snape JW, Gale MD, Harberd NP. 'Green revolution' genes encode mutant gibberellin response modulators. Nature. 1999;400:256–261. doi: 10.1038/22307. [DOI] [PubMed] [Google Scholar]
- Percival FW, Bandurski RS. Esters of indole-3-acetic Acid from Avena seeds. Plant Physiol. 1976;58:60–67. doi: 10.1104/pp.58.1.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Montano F, Jimenez-Guerrero I, Del Cerro P, Baena-Ropero I, Lopez-Baena FJ, Ollero FJ, Bellogin R, Lloret J, Espuny R. The symbiotic biofilm of Sinorhizobium fredii SMH12, necessary for successful colonization and symbiosis of Glycine max cv Osumi, is regulated by Quorum Sensing systems and inducing flavonoids via NodD1. PLoS One. 2014;9:e105901. doi: 10.1371/journal.pone.0105901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrasek J, Mravec J, Bouchard R, Blakeslee JJ, Abas M, Seifertova D, Wisniewska J, Tadele Z, Kubes M, Covanova M, Dhonukshe P, Skupa P, Benkova E, Perry L, Krecek P, Lee OR, Fink GR, Geisler M, Murphy AS, Luschnig C, Zazimalova E, Friml J. PIN proteins perform a rate-limiting function in cellular auxin efflux. Science. 2006;312:914–918. doi: 10.1126/science.1123542. [DOI] [PubMed] [Google Scholar]
- Ping X, Ye Q, Yan M, Zeng J, Yan X, Li H, Li J, Liu L. Integrated genetic mapping and transcriptome analysis reveal the BnaA03.IAA7 protein regulates plant architecture and gibberellin signaling in Brassica napus L. Theor Appl Genet. 2022;135:3497–3510. doi: 10.1007/s00122-022-04196-8. [DOI] [PubMed] [Google Scholar]
- Popp C, Ott T. Regulation of signal transduction and bacterial infection during root nodule symbiosis. Curr Opin Plant Biol. 2011;14:458–467. doi: 10.1016/j.pbi.2011.03.016. [DOI] [PubMed] [Google Scholar]
- Porco S, Pencik A, Rashed A, Voss U, Casanova-Saez R, Bishopp A, Golebiowska A, Bhosale R, Swarup R, Swarup K, Penakova P, Novak O, Staswick P, Hedden P, Phillips AL, Vissenberg K, Bennett MJ, Ljung K. Dioxygenase-encoding AtDAO1 gene controls IAA oxidation and homeostasis in Arabidopsis. Proc Natl Acad Sci U S A. 2016;113:11016–11021. doi: 10.1073/pnas.1604375113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prigge MJ, Platre M, Kadakia N, Zhang Y, Greenham K, Szutu W, Pandey BK, Bhosale RA, Bennett MJ, Busch W, Estelle M (2020) Genetic analysis of the Arabidopsis TIR1/AFB auxin receptors reveals both overlapping and specialized functions. Elife 9. 10.7554/eLife.54740 [DOI] [PMC free article] [PubMed]
- Qi L, Kwiatkowski M, Chen H, Hoermayer L, Sinclair S, Zou M, Del Genio CI, Kubes MF, Napier R, Jaworski K, Friml J. Adenylate cyclase activity of TIR1/AFB auxin receptors in plants. Nature. 2022;611:133–138. doi: 10.1038/s41586-022-05369-7. [DOI] [PubMed] [Google Scholar]
- Qin G, Gu H, Zhao Y, Ma Z, Shi G, Yang Y, Pichersky E, Chen H, Liu M, Chen Z, Qu LJ. An indole-3-acetic acid carboxyl methyltransferase regulates Arabidopsis leaf development. Plant Cell. 2005;17:2693–2704. doi: 10.1105/tpc.105.034959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren H, Park MY, Spartz AK, Wong JH, Gray WM. A subset of plasma membrane-localized PP2C.D phosphatases negatively regulate SAUR-mediated cell expansion in Arabidopsis. PLoS Genet. 2018;14:e1007455. doi: 10.1371/journal.pgen.1007455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JJ, O'Neill DP, Smith JJ, Kerckhoffs LH, Elliott RC. Evidence that auxin promotes gibberellin A1 biosynthesis in pea. Plant J. 2000;21:547–552. doi: 10.1046/j.1365-313x.2000.00702.x. [DOI] [PubMed] [Google Scholar]
- Ross JJ, O'Neill DP, Wolbang CM, Symons GM, Reid JB. Auxin-Gibberellin Interactions and Their Role in Plant Growth. J Plant Growth Regul. 2001;20:336–353. doi: 10.1007/s003440010034. [DOI] [PubMed] [Google Scholar]
- Sagar M, Chervin C, Mila I, Hao Y, Roustan JP, Benichou M, Gibon Y, Biais B, Maury P, Latche A, Pech JC, Bouzayen M, Zouine M. SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 2013;161:1362–1374. doi: 10.1104/pp.113.213843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki A, Ashikari M, Ueguchi-Tanaka M, Itoh H, Nishimura A, Swapan D, Ishiyama K, Saito T, Kobayashi M, Khush GS, Kitano H, Matsuoka M. Green revolution: a mutant gibberellin-synthesis gene in rice. Nature. 2002;416:701–702. doi: 10.1038/416701a. [DOI] [PubMed] [Google Scholar]
- Schiessl K, Lilley JLS, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD, Mysore KS, Wen J, Ahnert S, Grieneisen VA, Oldroyd GED. NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Curr Biol. 2019;29(3657–3668):e3655. doi: 10.1016/j.cub.2019.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaul-Keinan O, Gadkar V, Ginzberg I, Grunzweig JM, Chet I, Elad Y, Wininger S, Belausov E, Eshed Y, Atzmon N, Ben-Tal Y, Kapulnik Y. Hormone concentrations in tobacco roots change during arbuscular mycorrhizal colonization with Glomus intraradices. New Phytol. 2002;154:501–507. doi: 10.1046/j.1469-8137.2002.00388.x. [DOI] [PubMed] [Google Scholar]
- Shimomura S, Sotobayashi T, Futai M, Fukui T. Purification and properties of an auxin-binding protein from maize shoot membranes. J Biochem. 1986;99:1513–1524. doi: 10.1093/oxfordjournals.jbchem.a135621. [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet. 2006;2:e202. doi: 10.1371/journal.pgen.0020202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverstone AL, Sun T. Gibberellins and the Green Revolution. Trends Plant Sci. 2000;5:1–2. doi: 10.1016/s1360-1385(99)01516-2. [DOI] [PubMed] [Google Scholar]
- Song Q, Chu C, Parry MA, Zhu XG. Genetics-based dynamic systems model of canopy photosynthesis: the key to improve light and resource use efficiencies for crops. Food Energy Secur. 2016;5:18–25. doi: 10.1002/fes3.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM. SAUR inhibition of PP2C-D phosphatases activates plasma Membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell. 2014;26:2129–2142. doi: 10.1105/tpc.114.126037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielmeyer W, Ellis MH, Chandler PM. Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc Natl Acad Sci U S A. 2002;99:9043–9048. doi: 10.1073/pnas.132266399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprent JI, James EK. Legume evolution: where do nodules and mycorrhizas fit in? Plant Physiol. 2007;144:575–581. doi: 10.1104/pp.107.096156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W. Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell. 2005;17:616–627. doi: 10.1105/tpc.104.026690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- Stewart DW, Costa C, Dwyer LM, Smith DL, Hamilton RI, Ma BL. Canopy structure, light interception, and photosynthesis in maize. Agron J. 2003;95:1465–1474. doi: 10.2134/agronj2003.1465. [DOI] [Google Scholar]
- Strohm AK, Baldwin KL, Masson PH. Multiple roles for membrane-associated protein trafficking and signaling in gravitropism. Front Plant Sci. 2012;3:274. doi: 10.3389/fpls.2012.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su B, Wu H, Guo Y, Gao H, Wei Z, Zhao Y, Qiu L (2022) GmIAA27 encodes an AUX/IAA protein involved in dwarfing and multi-branching in soybean. Int J Mol Sci 23. 10.3390/ijms23158643 [DOI] [PMC free article] [PubMed]
- Sun TP. The molecular mechanism and evolution of the GA-GID1-DELLA signaling module in plants. Curr Biol. 2011;21:R338–345. doi: 10.1016/j.cub.2011.02.036. [DOI] [PubMed] [Google Scholar]
- Sun H, Tao J, Liu S, Huang S, Chen S, Xie X, Yoneyama K, Zhang Y, Xu G. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J Exp Bot. 2014;65:6735–6746. doi: 10.1093/jxb/eru029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Tao J, Bi Y, Hou M, Lou J, Chen X, Zhang X, Luo L, Xie X, Yoneyama K, Zhao Q, Xu G, Zhang Y. OsPIN1b is involved in rice seminal root elongation by regulating root apical meristem activity in response to low nitrogen and phosphate. Sci Rep. 2018;8:13014. doi: 10.1038/s41598-018-29784-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Guo X, Xu F, Wu D, Zhang X, Lou M, Luo F, Xu G, Zhang Y (2019) Overexpression of OsPIN2 regulates root growth and formation in response to phosphate deficiency in rice. Int J Mol Sci 20. 10.3390/ijms20205144 [DOI] [PMC free article] [PubMed]
- Suzaki T, Yano K, Ito M, Umehara Y, Suganuma N, Kawaguchi M. Positive and negative regulation of cortical cell division during root nodule development in Lotus japonicus is accompanied by auxin response. Development. 2012;139:3997–4006. doi: 10.1242/dev.084079. [DOI] [PubMed] [Google Scholar]
- Swarup R, Peret B. AUX/LAX family of auxin influx carriers-an overview. Front Plant Sci. 2012;3:225. doi: 10.3389/fpls.2012.00225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, Cheng Y, Lim J, Zhao Y, Ballare CL, Sandberg G, Noel JP, Chory J. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell. 2008;133:164–176. doi: 10.1016/j.cell.2008.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terasaka K, Blakeslee JJ, Titapiwatanakun B, Peer WA, Bandyopadhyay A, Makam SN, Lee OR, Richards EL, Murphy AS, Sato F, Yazaki K. PGP4, an ATP binding cassette P-glycoprotein, catalyzes auxin transport in Arabidopsis thaliana roots. Plant Cell. 2005;17:2922–2939. doi: 10.1105/tpc.105.035816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Z, Wang X, Lee R, Li Y, Specht JE, Nelson RL, McClean PE, Qiu L, Ma J. Artificial selection for determinate growth habit in soybean. Proc Natl Acad Sci U S A. 2010;107:8563–8568. doi: 10.1073/pnas.1000088107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmann U, Viola G, Kayser B, Siemeister G, Hesse T, Palme K, Lobler M, Klambt D. cDNA clones of the auxin-binding protein from corn coleoptiles (Zea mays L.): isolation and characterization by immunological methods. EMBO J. 1989;8:2463–2467. doi: 10.1002/j.1460-2075.1989.tb08381.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titapiwatanakun B, Blakeslee JJ, Bandyopadhyay A, Yang H, Mravec J, Sauer M, Cheng Y, Adamec J, Nagashima A, Geisler M, Sakai T, Friml J, Peer WA, Murphy AS. ABCB19/PGP19 stabilises PIN1 in membrane microdomains in Arabidopsis. Plant J. 2009;57:27–44. doi: 10.1111/j.1365-313X.2008.03668.x. [DOI] [PubMed] [Google Scholar]
- Tognetti VB, Van Aken O, Morreel K, Vandenbroucke K, van de Cotte B, De Clercq I, Chiwocha S, Fenske R, Prinsen E, Boerjan W, Genty B, Stubbs KA, Inze D, Van Breusegem F. Perturbation of indole-3-butyric acid homeostasis by the UDP-glucosyltransferase UGT74E2 modulates Arabidopsis architecture and water stress tolerance. Plant Cell. 2010;22:2660–2679. doi: 10.1105/tpc.109.071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong H, Chu C. Functional specificities of brassinosteroid and potential utilization for crop improvement. Trends Plant Sci. 2018;23:1016–1028. doi: 10.1016/j.tplants.2018.08.007. [DOI] [PubMed] [Google Scholar]
- Uga Y. Challenges to design-oriented breeding of root system architecture adapted to climate change. Breed Sci. 2021;71:3–12. doi: 10.1270/jsbbs.20118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, Inoue H, Takehisa H, Motoyama R, Nagamura Y, Wu J, Matsumoto T, Takai T, Okuno K, Yano M. Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet. 2013;45:1097–1102. doi: 10.1038/ng.2725. [DOI] [PubMed] [Google Scholar]
- Uga Y, Kitomi Y, Ishikawa S, Yano M. Genetic improvement for root growth angle to enhance crop production. Breed Sci. 2015;65:111–119. doi: 10.1270/jsbbs.65.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gelderen K, Kang C, Pierik R. Light signaling, root development, and plasticity. Plant Physiol. 2018;176:1049–1060. doi: 10.1104/pp.17.01079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li K, Chen L, Zou Y, Liu H, Tian Y, Li D, Wang R, Zhao F, Ferguson BJ, Gresshoff PM, Li X. MicroRNA167-directed regulation of the auxin response factors GmARF8a and GmARF8b is required for soybean nodulation and lateral root development. Plant Physiol. 2015;168:984–999. doi: 10.1104/pp.15.00265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Cheng H, Wang W, Liu J, Hao M, Mei D, Zhou R, Fu L, Hu Q. Identification of BnaYUCCA6 as a candidate gene for branch angle in Brassica napus by QTL-seq. Sci Rep. 2016;6:38493. doi: 10.1038/srep38493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao J, Lu W, Deng D. Gibberellin in plant height control: old player, new story. Plant Cell Rep. 2017;36:391–398. doi: 10.1007/s00299-017-2104-5. [DOI] [PubMed] [Google Scholar]
- Wang M, Qiao J, Yu C, Chen H, Sun C, Huang L, Li C, Geisler M, Qian Q, Jiang A, Qi Y. The auxin influx carrier, OsAUX3, regulates rice root development and responses to aluminium stress. Plant Cell Environ. 2019;42:1125–1138. doi: 10.1111/pce.13478. [DOI] [PubMed] [Google Scholar]
- Wang Y, Yang W, Zuo Y, Zhu L, Hastwell AH, Chen L, Tian Y, Su C, Ferguson BJ, Li X. GmYUC2a mediates auxin biosynthesis during root development and nodulation in soybean. J Exp Bot. 2019;70:3165–3176. doi: 10.1093/jxb/erz144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liu S, Wang J, Yokosho K, Zhou B, Yu YC, Liu Z, Frommer WB, Ma JF, Chen LQ, Guan Y, Shou H, Tian Z. Simultaneous changes in seed size, oil content and protein content driven by selection of SWEET homologues during soybean domestication. Natl Sci Rev. 2020;7:1776–1786. doi: 10.1093/nsr/nwaa110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Guo J, Peng Y, Lyu X, Liu B, Sun S, Wang X. Light-induced mobile factors from shoots regulate rhizobium-triggered soybean root nodulation. Science. 2021;374:65–71. doi: 10.1126/science.abh2890. [DOI] [PubMed] [Google Scholar]
- Wang W, Gao H, Liang Y, Li J, Wang Y. Molecular basis underlying rice tiller angle: current progress and future perspectives. Mol Plant. 2022;15:125–137. doi: 10.1016/j.molp.2021.12.002. [DOI] [PubMed] [Google Scholar]
- Wang H, Ouyang Q, Yang C, Zhang Z, Hou D, Liu H, Xu H (2022a) Mutation of OsPIN1b by CRISPR/Cas9 reveals a role for auxin transport in modulating rice architecture and root gravitropism. Int J Mol Sci 23. 10.3390/ijms23168965 [DOI] [PMC free article] [PubMed]
- Wasson AP, Pellerone FI, Mathesius U. Silencing the flavonoid pathway in Medicago truncatula inhibits root nodule formation and prevents auxin transport regulation by rhizobia. Plant Cell. 2006;18:1617–1629. doi: 10.1105/tpc.105.038232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasson AP, Richards RA, Chatrath R, Misra SC, Prasad SV, Rebetzke GJ, Kirkegaard JA, Christopher J, Watt M. Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J Exp Bot. 2012;63:3485–3498. doi: 10.1093/jxb/ers111. [DOI] [PubMed] [Google Scholar]
- Weijers D, Sauer M, Meurette O, Friml J, Ljung K, Sandberg G, Hooykaas P, Offringa R. Maintenance of embryonic auxin distribution for apical-basal patterning by PIN-FORMED-dependent auxin transport in Arabidopsis. Plant Cell. 2005;17:2517–2526. doi: 10.1105/tpc.105.034637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolbang CM, Ross JJ. Auxin promotes gibberellin biosynthesis in decapitated tobacco plants. Planta. 2001;214:153–157. doi: 10.1007/s004250100663. [DOI] [PubMed] [Google Scholar]
- Won C, Shen X, Mashiguchi K, Zheng Z, Dai X, Cheng Y, Kasahara H, Kamiya Y, Chory J, Zhao Y. Conversion of tryptophan to indole-3-acetic acid by TRYPTOPHAN AMINOTRANSFERASES OF ARABIDOPSIS and YUCCAs in Arabidopsis. Proc Natl Acad Sci U S A. 2011;108:18518–18523. doi: 10.1073/pnas.1108436108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JH, Klejchova M, Snipes SA, Nagpal P, Bak G, Wang B, Dunlap S, Park MY, Kunkel EN, Trinidad B, Reed JW, Blatt MR, Gray WM. SAUR proteins and PP2C.D phosphatases regulate H+-ATPases and K+ channels to control stomatal movements. Plant Physiol. 2021;185:256–273. doi: 10.1093/plphys/kiaa023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing A, Gao Y, Ye L, Zhang W, Cai L, Ching A, Llaca V, Johnson B, Liu L, Yang X, Kang D, Yan J, Li J. A rare SNP mutation in Brachytic2 moderately reduces plant height and increases yield potential in maize. J Exp Bot. 2015;66:3791–3802. doi: 10.1093/jxb/erv182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Dai N, Chen J, Nagawa S, Cao M, Li H, Zhou Z, Chen X, De Rycke R, Rakusová H, Wang W, Jones AM, Friml J, Patterson SE, Bleecker AB, Yang Z. Cell surface ABP1-TMK auxin-sensing complex activates ROP GTPase signaling. Science. 2014;343:1025–1028. doi: 10.1126/science.1245125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wang R, Kong K, Begum N, Almakas A, Liu J, Li H, Liu B, Zhao T, Zhao T. An APETALA2/ethylene responsive factor transcription factor GmCRF4a regulates plant height and auxin biosynthesis in soybean. Front Plant Sci. 2022;13:983650. doi: 10.3389/fpls.2022.983650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y, Zhang Y, Shan J, Ji Y, Zhang X, Li W, Li D, Zhao L (2022) Growth repressor GmRAV binds to the GmGA3ox promoter to negatively regulate plant height development in soybean. Int J Mol Sci 23. 10.3390/ijms23031721 [DOI] [PMC free article] [PubMed]
- Yamauchi T, Tanaka A, Inahashi H, Nishizawa NK, Tsutsumi N, Inukai Y, Nakazono M. Fine control of aerenchyma and lateral root development through AUX/IAA- and ARF-dependent auxin signaling. Proc Natl Acad Sci U S A. 2019;116:20770–20775. doi: 10.1073/pnas.1907181116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Fan Y, Wu X, Cheng Y, Liu Q, Feng L, Chen J, Wang Z, Wang X, Yong T, Liu W, Liu J, Du J, Shu K, Yang W. Auxin-to-gibberellin ratio as a signal for light intensity and quality in regulating soybean growth and matter partitioning. Front Plant Sci. 2018;9:56. doi: 10.3389/fpls.2018.00056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye R, Wu Y, Gao Z, Chen H, Jia L, Li D, Li X, Qian Q, Qi Y. Primary root and root hair development regulation by OsAUX4 and its participation in the phosphate starvation response. J Integr Plant Biol. 2021;63:1555–1567. doi: 10.1111/jipb.13142. [DOI] [PubMed] [Google Scholar]
- Yoshikawa T, Ito M, Sumikura T, Nakayama A, Nishimura T, Kitano H, Yamaguchi I, Koshiba T, Hibara K, Nagato Y, Itoh J. The rice FISH BONE gene encodes a tryptophan aminotransferase, which affects pleiotropic auxin-related processes. Plant J. 2014;78:927–936. doi: 10.1111/tpj.12517. [DOI] [PubMed] [Google Scholar]
- Yu C, Sun C, Shen C, Wang S, Liu F, Liu Y, Chen Y, Li C, Qian Q, Aryal B, Geisler M, de Jiang A, Qi Y. The auxin transporter, OsAUX1, is involved in primary root and root hair elongation and in Cd stress responses in rice (Oryza sativa L.) Plant J. 2015;83:818–830. doi: 10.1111/tpj.12929. [DOI] [PubMed] [Google Scholar]
- Yu XL, Wang HY, Leung DWM, He ZD, Zhang JJ, Peng XX, Liu EE. Overexpression of OsIAAGLU reveals a role for IAA-glucose conjugation in modulating rice plant architecture. Plant Cell Rep. 2019;38:731–739. doi: 10.1007/s00299-019-02402-4. [DOI] [PubMed] [Google Scholar]
- Yu SX, Zhou LW, Hu LQ, Jiang YT, Zhang YJ, Feng SL, Jiao Y, Xu L, Lin WH (2020) Asynchrony of ovule primordia initiation in Arabidopsis. Development 147. 10.1242/dev.196618 [DOI] [PMC free article] [PubMed]
- Zazimalova E, Murphy AS, Yang H, Hoyerova K, Hosek P. Auxin transporters–why so many? Cold Spring Harb Perspect Biol. 2010;2:a001552. doi: 10.1101/cshperspect.a001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Wang S, Xu Y, Yu C, Shen C, Qian Q, Geisler M, de Jiang A, Qi Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015;38:638–654. doi: 10.1111/pce.12397. [DOI] [PubMed] [Google Scholar]
- Zhang J, Lin JE, Harris C, Campos Mastrotti Pereira F, Wu F, Blakeslee JJ, Peer WA. DAO1 catalyzes temporal and tissue-specific oxidative inactivation of auxin in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2016;113:11010–11015. doi: 10.1073/pnas.1604769113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Lu X, Li C, Zhang B, Zhang C, Zhang XS, Ding Z. Auxin efflux carrier ZmPGP1 mediates root growth inhibition under aluminum stress. Plant Physiol. 2018;177:819–832. doi: 10.1104/pp.17.01379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang N, Yu H, Yu H, Cai Y, Huang L, Xu C, Xiong G, Meng X, Wang J, Chen H, Liu G, Jing Y, Yuan Y, Liang Y, Li S, Smith SM, Li J, Wang Y. A core regulatory pathway controlling rice tiller angle mediated by the LAZY1-dependent asymmetric distribution of auxin. Plant Cell. 2018;30:1461–1475. doi: 10.1105/tpc.18.00063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T, Li R, Xing J, Yan L, Wang R, Zhao Y. The YUCCA-Auxin-WOX11 module controls crown root development in rice. Front Plant Sci. 2018;9:523. doi: 10.3389/fpls.2018.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Zhu L, Shen C, Ji Z, Zhang H, Zhang T, Li Y, Yu J, Yang N, He Y, Tian Y, Wu K, Wu J, Harberd NP, Zhao Y, Fu X, Wang S, Li S. Natural allelic variation in a modulator of auxin homeostasis improves grain yield and nitrogen use efficiency in rice. Plant Cell. 2021;33:566–580. doi: 10.1093/plcell/koaa037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Gao L, Ke M, Gao Z, Tu T, Huang L, Chen J, Guan Y, Huang X, Chen X. GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean. J Integr Plant Biol. 2022;64:1325–1338. doi: 10.1111/jipb.13269. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Yang S, Wang Q, Yu H, Zhao B, Wu T, Tang K, Ma J, Yang X, Feng X. Soybean GmHY2a encodes a phytochromobilin synthase that regulates internode length and flowering time. J Exp Bot. 2022;73:6646–6662. doi: 10.1093/jxb/erac318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol. 2010;61:49–64. doi: 10.1146/annurev-arplant-042809-112308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant. 2012;5:334–338. doi: 10.1093/mp/ssr104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Christensen SK, Fankhauser C, Cashman JR, Cohen JD, Weigel D, Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- Zhao Y, Hu Y, Dai M, Huang L, Zhou DX. The WUSCHEL-related homeobox gene WOX11 is required to activate shoot-borne crown root development in rice. Plant Cell. 2009;21:736–748. doi: 10.1105/tpc.108.061655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao SQ, Xiang JJ, Xue HW. Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol Plant. 2013;6:174–187. doi: 10.1093/mp/sss064. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, Guo X, Gai Y, Wu C, Zhai H, Wang H, Wan J. A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell. 2013;27:113–122. doi: 10.1016/j.devcel.2013.09.005. [DOI] [PubMed] [Google Scholar]
- Zhao J, Yang B, Li W, Sun S, Peng L, Feng D, Li L, Di H, He Y, Wang Z. A genome-wide association study reveals that the glucosyltransferase OsIAGLU regulates root growth in rice. J Exp Bot. 2021;72:1119–1134. doi: 10.1093/jxb/eraa512. [DOI] [PubMed] [Google Scholar]
- Zhao Z, Wang C, Yu X, Tian Y, Wang W, Zhang Y, Bai W, Yang N, Zhang T, Zheng H, Wang Q, Lu J, Lei D, He X, Chen K, Gao J, Liu X, Liu S, Jiang L, Wang H, Wan J. Auxin regulates source-sink carbohydrate partitioning and reproductive organ development in rice. Proc Natl Acad Sci U S A. 2022;119:e2121671119. doi: 10.1073/pnas.2121671119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao DD, Jang YH, Farooq M, Park JR, Kim EG, Du XX, Jan R, Kim KH, Lee SI, Lee GS, Kim KM (2022a) Identification of a major QTL and validation of related genes for tiller angle in rice based on QTL analysis. Int J Mol Sci 23. 10.3390/ijms23095192 [DOI] [PMC free article] [PubMed]
- Zhao DD, Park JR, Jang YH, Kim EG, Du XX, Farooq M, Yun BJ, Kim KM (2022b) Identification of one major QTL and a novel gene OsIAA17q5 associated with tiller number in rice using QTL analysis. Plants (Basel) 11. 10.3390/plants11040538 [DOI] [PMC free article] [PubMed]
- Zhu ZX, Liu Y, Liu SJ, Mao CZ, Wu YR, Wu P. A gain-of-function mutation in OsIAA11 affects lateral root development in rice. Mol Plant. 2012;5:154–161. doi: 10.1093/mp/ssr074. [DOI] [PubMed] [Google Scholar]
- Zhu M, Hu Y, Tong A, Yan B, Lv Y, Wang S, Ma W, Cui Z, Wang X. LAZY1 controls tiller angle and shoot gravitropism by regulating the expression of auxin transporters and signaling factors in rice. Plant Cell Physiol. 2020;61:2111–2125. doi: 10.1093/pcp/pcaa131. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.