Abstract
The expression patterns of the NRT2 genes have been well described; however, the role of OsNRT2.4 in root growth is not well known. In this study, we thus aimed at investigating the role of high-affinity NO3− transport OsNRT2.4 in root growth modulation. Through the amiRNA-mediated gene silencing technique, we successfully obtained osnrt2.4 knockdown lines to study the role of OsNRT2.4 on root growth under low nitrate conditions. We performed real-time PCR analysis to investigate the relative gene expression level in root and shoot, soluble metabolites, and measurement of root system. Knockdown of OsNRT2.4 decreased rice growth. The comparison with wild-type (WT) plants showed that (i) knockdown of OsNRT2.4 inhibited root formation under low NO3− supply; (ii) we demonstrated that the mutant lines had significantly increased NO3− uptake than WT plants when grown in different nitrate supplies; (iii) osnrt2.4 knockdown lines showed an alteration in nitrogen metabolism, and this affected the root growth; and (iv) the downregulation of OsNRT2.4 enhanced the expression of gene response of low external NO3− concentrations. Herein we provide new insights in OsNRT2.4 functions. Our data demonstrated that OsNRT2.4 plays a role in root growth, nitrogen metabolic pathway and probably have functions in nitrate transport from root to shoot under low nitrate availability in rice.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-021-01273-6.
Keywords: Root morphology, Nitrogen, OsNRT2.4, Oryza sativa
Introduction
Rice (Oryza sativa L.) is one of the most consumed and produced cereals around the world, representing the staple food of more than half the world’s population (FAO 2017). In the diet of the Brazilian population, rice provides 20% of the energy and 15% of the total protein needed (Lam-Sánchez et al. 1994). Sustainable Development Goals (SDGs) 2 and 12 predict that by 2030 it will be necessary to double agricultural productivity (Sustainable Development Goals 2015), making it crucial to adopt new techniques in rice cultivation to increase its production in a sustainable way.
The roots are important anchorage organs, as well water and nutrients absorption, all of which are important aspects to growth and yield crops. The ability to modulate the spatial arrangement of the root system is an essential adaptive function to adjust to environmental variations and nutrient bioavailability in the soil (López-Bucio et al. 2003), enabling crops such as rice overcome yield limitations at low environmental cost by reducing chemical fertilizer use, such as nitrogen fertilizers (Jiao et al. 2016).
Nitrogen is an essential nutrient with high metabolic demand, and their availability in nitrate (NO3−) forms acts as a signal which trigger molecular mechanism responsible for plant growth and development (Crawford 1995; López-Bucio et al. 2003; Miller et al. 2007; Krapp et al. 2014). Several genes have been identified and characterized as important regulators of root development, increasing crop production in a sustainable way (Zhang and Forde 2000; Forde and Lorenzo 2001; Nacry et al. 2013). However, fewer NO3− transporter genes with a functional role in root development were characterized in crop models (Krapp et al. 2014; Kiba and Krapp 2016; Fan et al. 2017).
In higher plants, four gene families have been shown to function as nitrate transporter (Nacry et al. 2013; Krapp et al. 2014), and OsNRT2.4 (Nitrate Transporter 2 family) was the focus in this study. The NRT2 gene family are high-affinity nitrate transporters (HATS—High-Affinity Transport System), and in the rice genome (Oryza sativa L.) have previously been identified at least four putative NRT2 genes (OsNRT2.1, OsNRT2.2, OsNRT2.3, OsNRT2.4) and two putative NAR2 genes (OsNAR2.1 and OsNAR2.2) (Araki and Hasegawa 2006; Cai et al. 2008).
Overexpression of OsNRT2.1 affected positively total root length under 0.5 mM NO3−, and increased 15NO3− influx rate under low nitrate condition (Naz et al. 2019). In addition, osnar2.1 knockdown lines decreased the OsNRT2.1, OsNRT2.2, and OsNRT2.3a expression levels in the roots and affected the growth and total N concentration in rice plants (Yan et al. 2011a, 2011b). Diverse studies suggest the function of OsNRT2.1 and OsNAR2.1 in NO3− uptake and their contribution in lateral root initiation (Feng et al. 2011; Huang et al. 2015). OsNRT2.4 functional role in crop plants is still lacking. Current studies have highlighted the expression of OsNRT2.4 in the base of lateral root primordia, in the hull and vascular tissue of the anther (Feng et al. 2011; Wei et al. 2018).
The use of genetic engineering remains a powerful tool to study the largely unknow mechanism underlying rice root development by strategy for modification target genes enabling an improvement in the rice yield and quality (Yan et al. 2011a, 2011b; Baldrich and San Segundo 2016). Artificial microRNA (amiRNA) is one of a powerful tool in the study of gene functions in plants and is a strategy for sequence-specific cleavage of any target transcript, and provides a highly specific approach for gene silencing in plants, enabling to be useful for biotechnology approaches for the breeding (Ossowski et al. 2008; Warthmann et al. 2008).
In this study, we thus aimed at investigating the role of high-affinity NO3− transport OsNRT2.4 in NO3−-regulated and the modulation of root growth, and we observed shoot growth was similar in osnrt2.4 knockdown lines compared to wild-type plants.
Materials and methods
Plasmid construction and rice transformation
Artificial microRNA (amiRNA) to OsNRT2.4 was designed by Web MicroRNA Designer platform (WMD) (http://wmd3.weigelworld.org/) based on the Max Planck Institute for Developmental Biology. Each primary amiRNA was constructed replacing the bases of the natural osa-MIR528 and was cloned by PCR into the pNW55 vector (Warthmann et al. 2008). The amiRNA sequences and amplification primers are shown in Online Resource 1.
To the construction of plasmid were following the gene cloning step. The natural MIR528/MIR528* sequences were replaced by artificial amiRNA/amiRNA* sequences desired, and then the DNA fragments were fused and transferred into the multiple cloning site of the binary vector IRS154 to obtain plant expression vectors. The IRS154 containing the amiRNA was transferred into DH5α Escherichia coli using electroporation protocol (Sambrook and Russel 2001), and the vector was extracted using the PureYield™ Plasmid Miniprep System (Promega) according to the manufacturer’s instructions.
Finally, amiRNA plant expression vector was introduced into Agrobacterium tumefaciens strain LBA4404 by freeze-thaw method (Holsters et al. 1978) and then rice plants variety Nipponbare were transformed into wild-type according to the method described by Toki et al. (2006) and Sahoo et al. (2011). The transformed T2 lineages were selected by hygromycin (50 μg.L−1), based on low levels of OsNRT2.4 transcripts (real-time PCR analyses) and following the root morphology analysis. Thus, we selected two knockdown lines (r1 and r2) to show lower root growth without negative effect on the shoots.
Plant material and growth conditions
The Japonica rice variety Nipponbare was used as the wild-type (WT), and all rice materials were grown in a control growth chamber, under a light intensity of around 318–330 μmol m−2 s−1, a 14/10 h light/dark photoperiod (light starts at 06.00 h and dark starts at 20.00 h), 70–75% relative humidity, and a 28 °C day/night temperature, in the Department of Soil Sciences of the Federal Rural University of Rio de Janeiro (UFRRJ).
After the transformation process, we observed that knockdown line seeds did not germinate adequately. Then, we stablished a protocol to break seed dormancy based on the “Rules for Seed Analysis” (RAS 2009) in the following way. The seeds were preheated at 45°C for 2 h in a forced ventilation oven following a disinfection in a solution containing sodium hypochlorite 2% (v/v) for 15 min, washed five times with deionized water and placed in pots covered by deionized water overnight in the growth chamber. Subsequently, the seeds covered by deionized water were preheated a second time at 45°C for 2 h and placed in 3 L pots covered by deionized water to induce germination.
Based on previous studies that highlighted OsNRT2.4 expression in rice was much higher in roots supplied with NO3− when compared with NH4+ solution (Feng et al. 2011; Wei et al. 2018), we performed an experiment at different N concentrations, N starvation (without N), and 0.1 mM NO3− (low N), to determine the role of OsNRT2.4 transporter in nitrate uptake under low nitrogen conditions.
The experimental design was completely randomized with four replicates. The seedlings were deprived of N for 1 week from germination, to reduce endogenous N reserves, and supplied with nutrient solution containing a modified ¼ total ionic strength (IS) Hoagland solution (Hoagland and Arnon 1950) containing 1 mM NH4NO3 (pH 5.8.) for 3 days (d). At 10 days after germination (DAG), the seedlings were transferred into 0.7 L pots and the solution was replaced by the nutrient solution containing a modified ½ IS Hoagland solution with 0.1 mM NO3− (pH 5.5). At 13 DAG, the plants were divided into two groups: Hoagland solution without nitrogen (N starvation) or continued 0.1 mM NO3− treatment. Before harvest at 17 DAG, the pots received the same Hoagland solution containing 0.1mM NO3− and the plants were harvested after 04.00 h (04) (light start). The plants were harvested separately in root and shoot, frozen in liquid nitrogen, and stored in a −80 °C freezer for RNA extraction. For soluble metabolites, 0.5 g root and shoot fresh tissue were stored in 80% ethanol.
RNA preparation and real-time PCR analysis
The total RNA was extracted according to Gao et al. (2001) and quantified using Nanodrop (Thermo Scientific). The quality was verified by A260/230 e A260/280 proportion, with the ratio between 1,9 e 2,1 and then using agarose (1%) gel electrophoresis stained with gel red was verified the RNA integrity. One microgram of total RNA sample was DNase-treated using DNAse I Amplification Grade (Sigma-Aldrich) and cDNA synthesis was performed cDNA with High-Capacity RNA-to-cDNA Kit (Thermo Scientific) according to the manufacturer’s instructions.
The real-time PCR reactions were performed in a “StepOne Plus Real-Time PCR System” (Applied Biosystems) using the HOT FIREPol® EvaGreen® qPCR Mix Plus (ROX) Kit (Solis Biodyne) according to the manufacturer’s instructions. Each reaction was performed in the following way: 15 min at 95 °C and 40 cycles of amplification at 95°C for 15 s and 60°C for 1 min and 72°C for 30 s. Specificity of the primer sequences was analyzed using NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi) as well as experimentally at the end of the PCR reactions through the melting curve. Gene expression data were analyzed by the comparative CT method (2−∆∆CT) (Livak and Schmittgen 2001), using the OsUBC (Ubiquitin–conjugating enzyme–E2) and OseEF-1α (1α elongation factor) as housekeeping genes (Jain et al. 2006). All primers are listed in Online Resource 2.
Soluble metabolites
Shoot and roots were analyzed in an experiment identical to the previously described in the “Plant material and growth conditions” section. Samples of 0.5 g were homogenized 80% ethanol, separated with chloroform and water according to the method described by Fernandes (1983). The extract was used for the determination of soluble sugars (Yemm and Willis 1954), free amino-N (Yemm and Cocking 1955), nitrate (Miranda et al. 2001), and ammonium (Felker 1977).
Measurement of root system
The root morphology of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2) was performed using four biological replicates for each genotype and harvested at 13 days after germination (DAG) at 04.00 h (04) (light start) grown in a hydroponic system, Hoagland solution (Hoagland and Arnon 1950) containing 0.1 mM NO3−. The roots were stored in 50% alcohol and scanned subsequently.
The rice root system was scanned at 600 dpi with an Epson Expression 10000XL scanner, and the images were analyzed using the WinRhizo Arabdopsis scanner–based image analysis system (Regent Instruments, Montreal, QC, Canada). Total root length (mm), total number of tips, total root surface area (mm2), total root volume (mm3), and total average diameter (mm) were measured.
Data analysis
The data were analyzed using the R software. Subsequently, the data were analyzed using the analysis of variance: test of data normality (Shapiro-Wilk test), homocedasticity of variances (Bartlett’s test), and t-test, where treatment means were compared using Dunnet’s test at 5% probability (p< 0.05). The effects of the variables were verified by F test (5% probability). Asterisk (*) indicates a significant difference between WT and the knockdown lines.
Results
Knockdown of OsNRT2.4 inhibited root growth in response to nitrogen availability
To validate OsNRT2.4 expression pattern, we performed a real-time PCR analysis in rice (cv. Nipponbare), and we demonstrate that OsNRT2.4 was not very abundant in the two osnrt2.4 knockdown lines (r1 and r2) roots grown in 0.1 mM nitrate solution when compared with WT (Fig. 1).
Fig. 1.
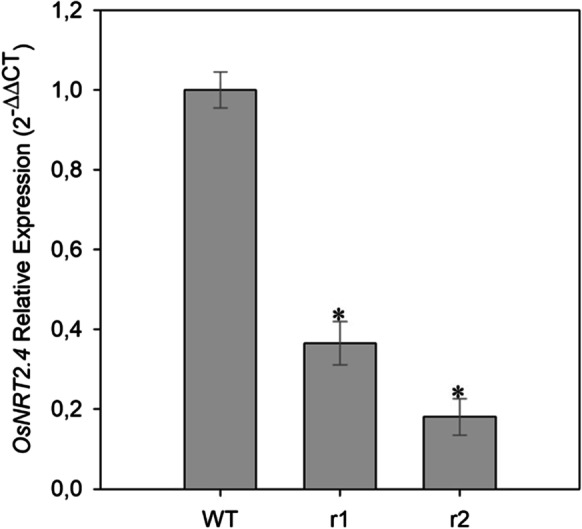
Expression level of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2) in root rice. Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution containing 0.1mM N-NO3− for 4 days. Total RNA was isolated from roots at 04.00 h (04) (light start). OsNRT2.4 expression levels were normalized to the expression of the OsUBC and OseEF-1α reference genes. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
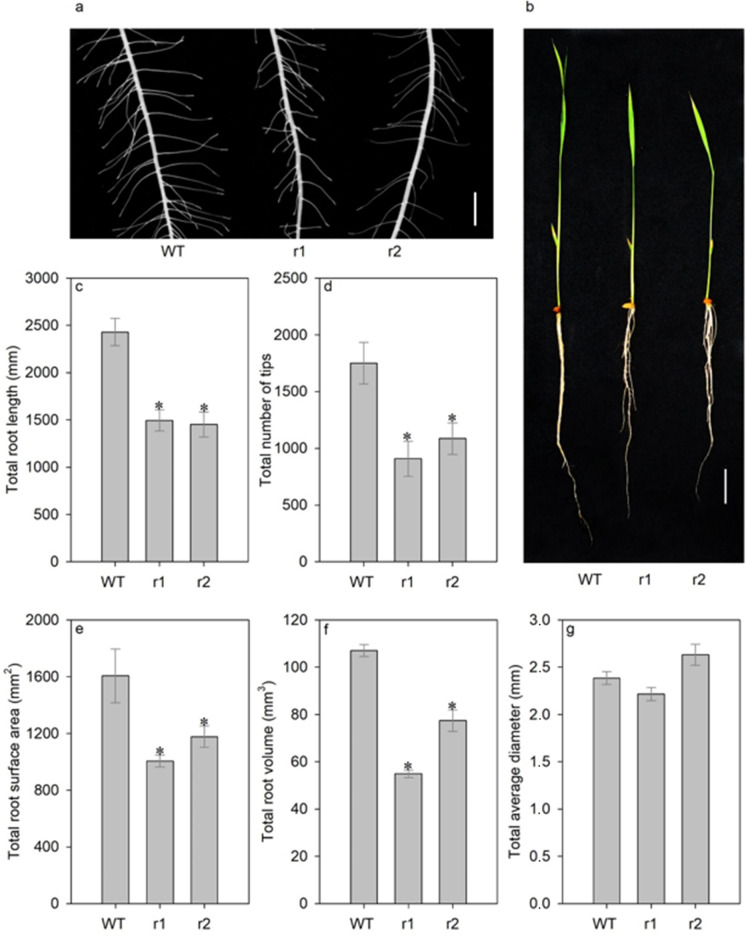
Knockdown of OsNRT2.4 decreased root growth. Total root length was 38% shorter in osnrt2.4 knockdown lines (r1 and r2) than in WT plants (Fig. 2b, c). The difference in total number of tips between the mutant lines was 38% of those in WT plants (Fig. 2a, d). Reductions in total root surface area were 26% of those in WT plants (Fig. 2e) and were largely attributable to reduced total number of tips (Fig. 2d). In comparison with WT plants, total root volume decreased 27% in mutant lines (Fig. 2f). However, total root average diameter was similar between genotypes (Fig. 2g).
Fig. 2.
Root morphology of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system containing 0.1mM N-NO3− and harvested at 13 days after germination (DAG) at 04.00 h (04) (light start). (a) Morphology of lateral roots; (b) morphology of rice plants (bar = 5 cm); (c) total root length (mm); (d) total number of tips; (e) total root surface area (mm2); (f) total root volume (mm3); (g) total average diameter (mm). Root system was analyzed using the WinRhizo Arabdopsis software. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
Effects of osnrt2.4 knockdown on rice growth and metabolite contents in response to nitrogen availability
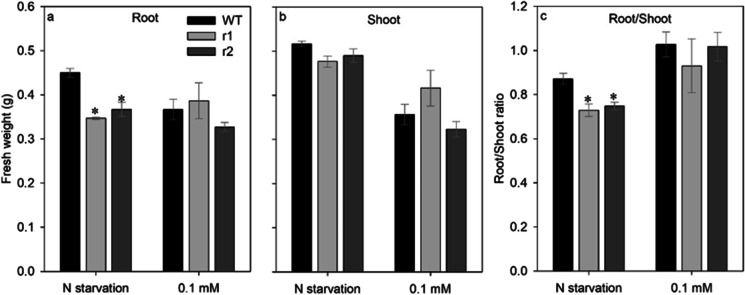
Knockdown of OsNRT2.4 decreased rice growth and altered metabolite contents in the different plant tissues under different nitrate treatments. In comparison with WT, osnrt2.4 knockdown lines (r1 and r2) showed lower fresh weight in root when cultivated with N starvation (Fig. 3a), but not when cultivated with 0.1 mM N-NO3−. The root/shoot ratio was lower in mutant lines when cultivated with N starvation (Fig. 3c), but not when cultivated with 0.1 mM N-NO3−. The vegetative growth of osnrt2.4 knockdown lines showed a visible difference from WT plants when supplied with 0.1 mM (Fig. 2b). However, when the shoot fresh weight was analyzed, there was no significant difference in both treatments (Fig. 3b). Our data suggest that the knockdown of OsNRT2.4 does not affect biomass production in shoot, demonstrating that OsNRT2.4 plays a key role in root growth.
Fig. 3.
Biomass weight in response to different nitrate treatments in root and shoot of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution without nitrogen for 4 days (starvation) or with 0.1mM N-NO3− for 4 days (low nitrogen) and harvested at 04.00 h (04) (light start). (a) Root biomass; (b) shoot biomass; (c) root:shoot (R/S) ratio. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
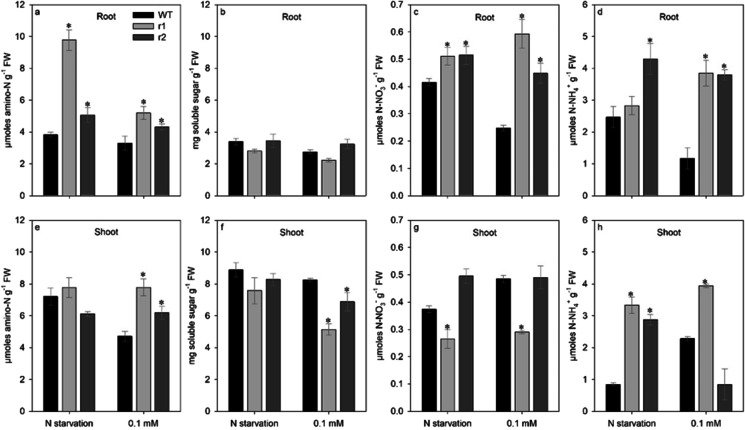
In comparison with WT, osnrt2.4 knockdown lines (r1 and r2) showed higher amino-N content in root growth with both N starvation and low (0.1 mM) NO3− supply (Fig. 4a). However, when analyzed in shoot, amino-N content was increased in mutant lines only when grown in 0.1 mM N-NO3− (Fig. 4e). There was no significant difference in shoot amino-N content between the osnrt2.4 knockdown lines (r1 and r2) and WT when grown in N starvation (Fig. 4e). We also analyzed soluble sugar, and there was no significant difference in root soluble sugar content between the osnrt2.4 knockdown lines (r1 and r2) and WT when grown in both N starvation and low (0.1 mM) NO3− supply (Fig. 4b). In shoot, the soluble sugar content decreased only in mutant lines when cultivated with 0.1 mM N-NO3−, but not when cultivated with N starvation (Fig. 4f)
Fig. 4.
Soluble metabolites in response to different nitrate treatments in root and shoot of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution without nitrogen for 4 days (starvation) or with 0.1mM N-NO3− for 4 days (low nitrogen) and harvested at 04.00 h (04) (light start). amino-N, soluble sugar, N-NO3−, and NH4+ in roots (a, b, c, and d) and shoots (e, f, g, and h). The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
Nitrate content (N-NO3−) in root was significantly higher in osnrt2.4 knockdown lines (r1 and r2) than in WT plants when grown in both N starvation and low (0.1 mM) NO3− supply (Fig. 4c). In contrast, shoot nitrate content decreased in osnrt2.4 knockdown lines (r1 and r2) in comparison with WT, but the significant difference was only in one line, r1, to both treatments (Fig. 4g). Compared to WT plants, the ammonium content (N-NH4+) in root and shoot was significantly higher in osnrt2.4 knockdown lines (r1 and r2) growth with both N starvation and low (0.1 mM) NO3− supply (Fig. 4d, h), but the significant difference was only in one line, r2 for N starvation (root) and r1 for low supply (shoot).
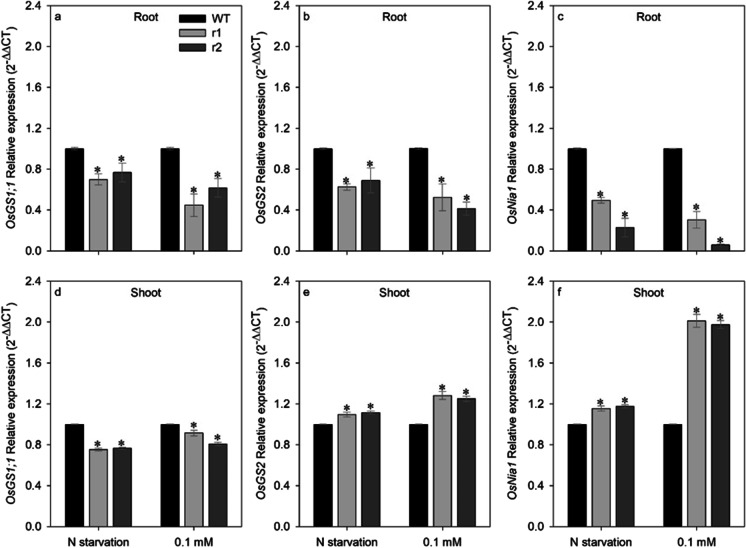
Knockdown of OsNRT2.4 alters expression levels of nitrogen metabolism–related genes
The root and shoot expression of glutamine synthetase1;1 (OsGS1;1) were significantly lower in both knockdown lines relative to the WT growth with both N starvation and low (0.1 mM) NO3− supply (Fig. 5a, d). In root, the expression of glutamine synthetase2 (OsGS2) decreased in mutant lines when cultivated with both N starvation and low (0.1 mM) NO3− supply (Fig. 5b), but in contrast, shoot expression of OsGS2 was significantly increased in both knockdown lines relative to the WT growth with both treatment (Fig. 5e). With both N starvation and low (0.1 mM) NO3− supply, the root expression of nitrate reductase (OsNia1) in mutant lines was lower than the WT (Fig. 5c). However, knockdown of OsNRT2.4 increased the shoot expression of OsNia1 in both mutant lines in comparison with WT growth with both N starvation and low (0.1 mM) NO3− supply (Fig. 5f).
Fig. 5.
Comparison of root and shoot expression pattern of nitrogen metabolism genes in wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution without nitrogen for 4 days (starvation) or with 0.1mM N-NO3− for 4 days (low nitrogen). Total RNA was isolated from roots at 04.00h (04) (light start). OsGS1;1, OsGS2, and OsNIA1 relative expression in roots (a, b, and c) and shoots (d, e, and f). The expression levels were normalized to the expression of the OsUBC and OseEF-1α reference genes. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett's test (p<0.05)
Expression patterns of OsNRT2.1 and OsNAR2.1 genes in response to different nitrate treatment in the wild-type and osnrt2.4 knockdown lines
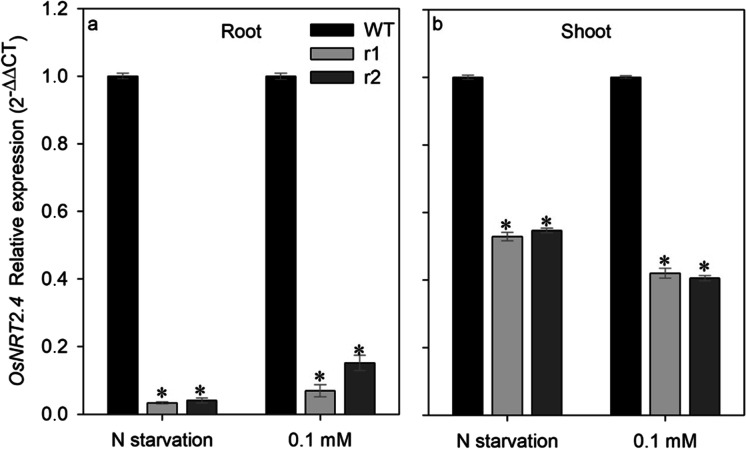
In the different plant tissues under different nitrate treatments, the expression of OsNRT2.4 in mutant lines was smaller than in WT plants (Fig. 6a, b). Note that even with a significant reduction in both tissues in mutant lines, knockdown of the expression of OsNRT2.4 was much smaller in root than in shoot in both N starvation and low (0.1 mM) NO3− supply.
Fig. 6.
OsNRT2.4 expression pattern in response to different nitrate treatment in the root and shoot of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution without nitrogen for 4 days (starvation) or with 0.1mM N-NO3− for 4 days (low nitrogen). Total RNA was isolated from roots at 04.00h (04) (light start). (a) OsNRT2.4 relative expression in root and (b) OsNRT2.4 relative expression in shoot. The expression levels were normalized to the expression of the OsUBC and OseEF-1α reference genes. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
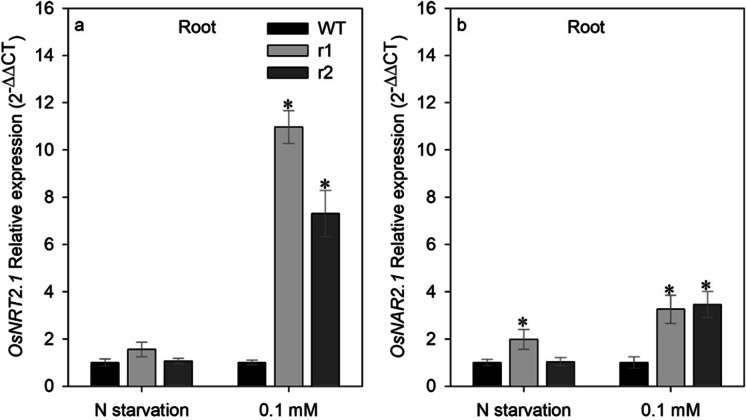
Previous studies (Araki and Hasegawa 2006; Feng et al. 2011) had demonstrated that OsNRT2.1 and OsNAR2.1 have a slight expression in shoot but an elevated expression in roots, suggesting that both transporters are a root-specifics genes. Therefore, we decided to analyze expression patterns of OsNRT2.1 and OsNAR2.1 only in roots, and we noted that knockdown of OsNRT2.4 resulted in the increase in the root expression of OsNRT2.1 and OsNAR2.1 genes in mutant lines. There was no significant difference in root expression of OsNRT2.1 between the osnrt2.4 knockdown lines (r1 and r2) and WT when grown in N starvation (Fig. 7a). However, knockdown of OsNRT2.4 increased the root expression of OsNRT2.1 in both mutant lines in comparison with WT when growth with low (0.1 mM) NO3− supply (Fig. 7a). The expression of OsNAR2.1 was higher in both treatments, but the significant difference in N starvation treatment was only in r1 line.
Fig. 7.
OsNRT2.1 and OsNAR2.1 expression pattern in response to different nitrate treatments in the roots of wild-type (WT) and osnrt2.4 knockdown lines (r1 and r2). Rice seedlings were grown in a hydroponic system. Thirteen-day-old seedlings were transferred into nutrient solution without nitrogen for 4 days (starvation) or with 0.1mM N-NO3− for 4 days (low nitrogen). Total RNA was isolated from roots at 04.00h (04) (light start). (a) OsNRT2.1 relative expression and (b) OsNAR2.1 relative expression. The expression levels were normalized to the expression of the OsUBC and OseEF-1α reference genes. The experiment was performed using four biological replicates for each genotype. Bars indicate the standard error of the mean (n =16). Asterisk (*) indicates statistically significant differences between WT and two mutant lines by the Dunnett’s test (p<0.05)
Discussion
Nutrient availability in the soil modulates root growth and development (Kiba and Krapp 2016) and is known that the plants have diverse phenotypic responses for adapting to nitrogen (N) fluctuation (Huang et al. 2015) as developing a deeper root system that acquired more N (Koevoets et al. 2016). Understanding the molecular mechanism of NO3−-regulated root growth is crucial for improved N uptake. Under low NO3− concentration, Huang et al. (2015) cultivated OsNAR2.1 knockdown rice plants and the mutant lines exhibited a decreased lateral root formation. We investigated knockdown of OsNRT2.4 in rice root growth and compared with WT plants, knockdown of OsNRT2.4 inhibited root growth under low NO3− concentration (0.1 mM) (Fig. 2).
The low nutrient availability induces morphological changes in root growth (López-Bucio et al. 2003) and can trigger an impact on root system by altering the root diameter, angle, length, and number of roots (Forde and Lorenzo 2001). Despite the innumerous papers reporting nitrate effects on root growth, the mechanisms underlying the NO3− effects on primary root elongation are still controversial because even when stimulated primary root growth, an inhibitory effect has been observed according to nitrate concentration medium (Trevisan et al. 2014). Arabidopsis plants under limited N supply may alter lateral root elongation (Gruber et al. 2013). In maize (Zea mays L.), Ravazzolo et al. (2020) observed that nitrate positively influences the primary root length after 7 days of growth at a nitrate concentration of 1 mM.
In this study, knockdown of OsNRT2.4 did not affect root diameter (Fig. 2g), and instead decreased total root length, number of tips, root surface area, and root volume. Despite the similar root diameter between mutant lines and WT, according to Forde and Lorenzo (2001), root diameter generally shows less plasticity than stem diameter to the environmental conditions. Length and root surface area are major factors of water and nutrient uptake rates because they determine the total volume of soil explored by the roots; in addition, the number of tips indicates lateral root initiation and all these parameters have a great impact on root growth and development (Forde and Lorenzo 2001; López-Bucio et al. 2003; Nacry et al. 2013). Our data suggest that OsNRT2.4 had a key function in coordinating root formation at low external NO3− concentration.
Changes in the nutritional status of the plants trigger morphological responses different in the overall root system, and primary root growth and lateral root formation are particularly sensitive to changes in the external N availability and internal N status of the plants (López-Bucio et al. 2003; Nacry et al. 2013; Kiba and Krapp 2016). The plant growth rate, measured as the biomass of root and shoot during the different nitrate treatments, showed that N starvation treatment (plants without N for 4 days) affected only root growth of osnrt2.4 knockdown lines (r1 and r2) and when cultivated with low N concentration (0.1 mM N-NO3−) showed no significant difference (Fig. 3). In a recent report, Wei et al. (2018) observed inhibition of root and shoot growth when the osnrt2.4 knockout lines were cultivated in low (0.25 mM) and high (2.5 mM) NO3− supply that resulted in lower dry weight and less total N accumulation. The mechanisms underlying the nitrate effects are still controversial. Our results suggest differences in the nitrate feedback signaling in rice, possibly resulting from the fact that changes in the external and internal N status are the predominant in affecting root biomass, and also additional factors must play a role.
Nitrate (NO3−) is a major N source for most higher plants and acts as a signaling molecule (Miller et al. 2007; Bloom 2015) and might be stored in the vacuole or reduced to nitrite in the cytosol by nitrate reductase (NR) enzyme and, then, nitrite is transported into plastids where occurs its reduction to ammonium by nitrite reductase (NiR) enzyme and subsequently incorporated into amino acid (Fernandes and Rossiello 1995; Tegeder and Masclaux-Daubresse 2018). In addition, plants can reduce nitrate to ammonium (process named N assimilation) in both tissues root and shoot. We investigated nitrogen metabolism in rice plants, and we observed nitrate content (N-NO3−) in root was significantly higher in osnrt2.4 knockdown lines (r1 and r2) than in WT plants when grown in both N starvation and low (0.1 mM) NO3− supply (Fig. 4c).
A previous study by Tang et al. (2012) in rice showed general pattern with a decrease of nitrate accumulation in shoots and increase in roots. The two osnrt2.3a knockdown lines clearly accumulated much higher nitrate in their roots at low nitrate solution (0.5 mM) than in the shoots. By contrast, osnar2.1 knockdown mutants decreased total N concentration in both root and shoot treated with 0.2 mM NO3− (Yan et al. 2011a, 2011b). Besides, Huang et al. (2015) when cultivating osnar2.1 knockdown lines have observed that the mutants had reduced N accumulations in the shoots and the roots under 0.2 mM nitrate supply. In another study, total N uptake in osnrt2.4 knockout lines was significantly lower than WT in root and shoot in low (0.25 mM) and high (2.5 mM) NO3− supply (Wei et al. 2018). Thus, we hypothesize that the higher root NO3− content indicates an increase in nitrate uptake and it was remained in sufficient amounts to attend metabolic demands. Intriguingly, shoot nitrate content was significantly lower in mutant lines when compared to WT suggesting that OsNRT2.4 alters nitrate translocation from the roots to shoots in both N starvation and low (0.1 mM) NO3− supply.
NH4+ uptake and assimilation are strictly regulated because excess ammonium is toxic to plant cells and may trigger morphology and physiology mechanisms damaging to the plant (Tegeder and Masclaux-Daubresse 2018). Generally, higher ammonium concentration is a result of amino acid catabolism and photorespiratory recycling (Fernandes and Rossiello 1995). This is why the increased NH4+ contents in the knockdown lines were also shown in the root and shoot in both treatments (Fig. 4d, h)
The accumulation of large amount of amino-N in mutant lines in roots, in both treatments, and shoot only in 0.1 mM NO3− supply (Fig. 4a, e) agree with the observations reported by Fernandes (1991) and Souza et al. (1999) that when plants are in a medium with low N concentration, occurs increases amino-N because of higher nitrate rate uptake, and this demands a greater metabolic energy. We observed that there were negative relationships between amino-N and free sugars (Fig. 4a, b, e, f). The assimilation of NH4+ during amino acid biosynthesis requires carbon skeletons (Fernandes and Rossiello 1995). Thus, in agreement with Souza et al. (1999), the decrease of soluble sugar observed in this work is due to the mobilization of carbon skeletons required to incorporate N uptake.
We investigated expression patterns of genes involved in nitrogen metabolic pathway, as NR (nitrate reductase), GS (glutamine synthetase), and NRT (nitrate transporter). Knockdown of OsNRT2.4 altered the relative gene expression level in the root and shoot under different nitrogen levels (Fig. 5). Nitrate reductase is a critical step for nitrogen assimilation and is positively regulated by nitrate, light, and carbohydrates and repressed by ammonium, glutamine, and darkness (Crawford 1995; Yanagisawa 2014). Huarancca Reyes et al. (2018) investigated the ability of Arabidopsis thaliana to assimilate N in the presence of NO3− trough NR activity. The NR activity under control condition with 30 mM NO3− and 100 mM glucose was lower in comparison with that under treated condition with 0.3 mM NO3− and 200 mM glucose reaching the highest level. When compared with WT, osnrt2.3a knockdown lines accumulated more root nitrate and increased OsNia1 expression under 0.5 mM NO3−, even though the shoot nitrate concentration was decreased (Tang et al. 2012).
It is well established that GS1 is usually localized in cytosol whereas GS2 is found in chloroplasts/plastids which also indicated its restriction to green photosynthetic tissues (Tabuchi et al. 2005; Tegeder and Masclaux-Daubresse 2018). Most previous studies showed higher levels of GS1 in root while GS2 is predominant in leaves (Tabuchi et al. 2005; Prinsi and Espen 2015; Zhang et al. 2017). Bao et al. (2014) showed that under 0 mM N and 0.1 mM N condition, GS1;1-overexpressing rice plants significantly increased in roots the expression levels of GS1;1, and GS2 was increased only in shoot. In maize (Zea mays), the three mutants, gln1-3, gln1-4, and gln1-3/gln1-4, have demonstrated a high level of GS1-1 transcripts in the root cortex, thus confirming that they are likely to be involved in Gln synthesis for vegetative growth (Martin et al. 2006).
In our study, osnrt2.4 knockdown lines had an increased nitrate accumulation in roots in detrimental of shoots despite NO3− supply (Fig. 4c, g), but OsGS1;1, OsGS2, and OsNia1 expression levels were lower in root when compared with shoot (Fig. 5). Our data to this point suggested that higher metabolites accumulated in root such as amino-N decreased nitrogen metabolism-related gene expression. Furthermore, we hypothesize that after N uptake, a high amount of nitrate was accumulated in the root vacuole as an N reserve and might result in slower root growth (Fig. 3a), while cytoplasmatic NO3− residual was assimilated in amino acid into plastids decreasing NO3− content of the cytoplasm and negatively affected OsNia1 and OsGS1;1 expression. This idea may be supported by studies Fernandes and Rossiello (1995), Souza et al. (1999), and Santos et al. (2009). The expression pattern of glutamine synthetase 2 is well characterized and known to be involved in NO3− reduction during NH4+ assimilation into plastids (Cren and Hirel 1999; Bernard and Habash 2009). The decreased OsGS2 in roots, even though the root amino-N was higher, suggests its restriction to green photosynthetic tissues. Thus, our results suggest that knockdown of OsNRT2.4 is likely to be involved in nitrogen metabolic pathway and affect the plant growth.
Nitrate availability can determine numerous changes for it acts as signal for several physiological, morphological, and metabolic processes (Zhang and Forde 2000; Nacry et al. 2013), such as in coordinating the expression of nitrate-related genes (Miller et al. 2007; Krapp et al. 2014). In the high-affinity NRT2 family, OsNRT2.1 and OsNAR2.1 are well characterized and known to be expressed abundantly in primary and lateral roots and to play a key role in NO3− uptake under low nitrate availability (Feng et al. 2011). However, the role of OsNRT2.4 in root growth is not well known. In this study, we demonstrated that in rice OsNRT2.4 is highly expressed in shoot, with a low level in roots (Fig. 6), altering root growth (Fig. 2) and nitrogen metabolism (Figs. 4 and 5).
Intriguingly, the expression levels of OsNRT2.1 and OsNAR2.1 in the roots of both osnrt2.4 knockdown lines were markedly increased when compared with WT plants under low (0.1 mM) NO3− treatment. Yan et al. (2011a, 2011b) reported the expression of OsNRT2.1, OsNRT2.2, and OsNRT2.3a were affected by OsNAR2.1 knockdown, and OsNRT2.3b and OsNRT2.4 showed not significant difference between lines and WT plants. In addition, Wei et al. (2018) confirmed in osnrt2.4 knockout mutants, OsNRT2.4 did not require the associate protein OsNAR2.1 in NO3− uptake when expressed in oocytes. These results confirm OsNRT2.1 and OsNAR2.1 mainly act in nitrate uptake under N starvation and low (0.1mM) NO3− supply conditions.
The expression patterns of the Arabidopsis NRT2 genes have been well described (Okamoto et al. 2003) and indicate that AtNRT2.4 and AtNRT2.5 are responsible for nitrate uptake from the soil, and AtNRT2.1 plays a role in apoplastic nitrate absorption (Lezhneva et al. 2014; Kiba and Krapp 2016). Through phylogenic analysis of NRT2 proteins (Araki and Hasegawa 2006; Cai et al. 2008), OsNRT2.4 is more closely related to Arabidopsis AtNRT2.7 proteins. According to Chopin et al. (2007), AtNRT2.7 plays a specific role in nitrate loading into the seeds vacuole and might be useful in nutrient reserve to young seedlings after germination. On the other hand, OsNRT2.4 is expressed mainly in the base of the lateral root primordia and is a transporter localized to the plasma membrane (Wei et al. 2018). Our data suggested that OsNRT2.4 had a key function in coordinating NO3− transport from the roots to shoots (Fig. 4c, g).
A summary of OsNRT2.4 role plays on root growth
The novel regulatory role involving root growth, nitrogen metabolic pathway, and nitrate transport function from root-to-shoot herein provides new insights in OsNRT2.4 functions that modulates root growth as well regulates nitrogen metabolism under low N availability. Here, we showcased that efficient amiRNA-mediated gene silencing in plants has been successfully used to downregulate the expression of OsNRT2.4.
Knowledge on root phenotyping, and how to enable the acquisition of water and nutrients in the soil, is critical for future food security (Tracy et al. 2020). Developing crops with roots able to uptake more efficiently nutrients is critical for sustainable agriculture given less than 50% of nitrogen, like nitrate-based fertilizer that is one of the most factor environmental degradation, is taken up by cereal crops (Zhang et al. 2020).
In this study, the downregulation of OsNRT2.4 improved nitrate uptake and enhanced the expression of gene response of low external NO3− concentrations. This effect on root growth suggests the roles in NO3− transport from roots to shoot.
Supplementary Information
(PDF 112 kb)
Acknowledgements
We would like to thank Sonia Regina de Souza (in memoriam) for their substantial assistance and useful discussions on this research project. We thank the undergraduate students from the scientific initiation program for their help with the experiments, and the funding agencies Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) (Finance Code 001) and the National Council for Scientific and Technological Development (CNPq), and the Graduate Program in Agronomy - Soil Science (PPGA-CS) at Federal Rural University of Rio de Janeiro (UFRRJ). We thank the anonymous reviewers for their rigorous work to improve this paper.
Author contribution
This article is a joint effort by several authors. A.F.F.S. designed the experiments, obtained funding, performed the data analysis, and drafted the manuscript with contributions of authors. L.N.A. and R.P.R. performed part of the experiment, measurements and provided assistance for data analysis. M.S.F. and L.A.S. contributed to research plan and experiment support. S.R.S (in memoriam) supervised all experiments and together with C.A.B conceived the research plan and project design. The authors read and approved the final manuscript.
Funding
This research was funded in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior – Brasil (CAPES) (Finance Code 001) and the National Council for Scientific and Technological Development (CNPq).
Availability of data and material
Not applicable.
Code availability
WinRhizo Arabdopsis software (2012b).
Declarations
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Araki R, Hasegawa H. Expression of rice (Oryza sativa L.) genes involved in high-affinity nitrate transport during the period of nitrate induction. Breed Sci. 2006;56:295–302. doi: 10.1270/jsbbs.56.295. [DOI] [Google Scholar]
- Baldrich P, San Segundo B. MicroRNAs in rice innate immunity. Rice. 2016;9:6. doi: 10.1186/s12284-016-0078-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao A, Zhao Z, Ding G, Shi L, Xu F, Cai H. Accumulated expression level of cytosolic Glutamine Synthetase 1 gene (OsGS1;1 or OsGS1;2) alter plant development and the carbon-nitrogen metabolic status in rice. PLoS ONE. 2014;9:e95581. doi: 10.1371/journal.pone.0095581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard SM, Habash DZ. The importance of cytosolic glutamine synthetase in nitrogen assimilation and recycling. New Phytol. 2009;182:608–620. doi: 10.1111/j.1469-8137.2009.02823.x. [DOI] [PubMed] [Google Scholar]
- Bloom AJ. The increasing importance of distinguishing among plant nitrogen sources. Curr Opin Plant Biol. 2015;25:10–16. doi: 10.1016/j.pbi.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Cai C, Wang J-Y, Zhu Y-G, Shen Q-R, Li B, Tong Y-P, Li Z-S. Gene structure and expression of the high-affinity nitrate transport system in rice roots. J Integr Plant Biol. 2008;50:443–451. doi: 10.1111/j.1744-7909.2008.00642.x. [DOI] [PubMed] [Google Scholar]
- Chopin F, Orsel M, Dorbe M-F, Chardon F, Truong H-N, Miller AJ, Krapp A, Daniel-Vedele F. The arabidopsis ATNRT2.7 nitrate transporter controls nitrate content in seeds. Plant Cell. 2007;19:1590–1602. doi: 10.1105/tpc.107.050542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford NM. Nitrate: nutrient and signal for plant growth. Plant Cell. 1995;7:859–868. doi: 10.1105/tpc.7.7.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cren M, Hirel B. Glutamine synthetase in higher plants: regulation of gene and protein expression from the organ to the cell. Plant Cell Physiol. 1999;40:1187–1193. doi: 10.1093/oxfordjournals.pcp.a029506. [DOI] [Google Scholar]
- Fan X, Naz M, Fan X, Xuan W, Miller AJ, Xu G. Plant nitrate transporters: from gene function to application. J Exp Bot. 2017;68:2463–2475. doi: 10.1093/jxb/erx011. [DOI] [PubMed] [Google Scholar]
- FAO (2017) Food and Agriculture Organization of the United Nations. FAOSTAT database: agriculture production. Available via http://www.fao.org/faostat/en/#home.
- Felker P. Micro determination of nitrogen in seed protein extracts with the salicylate-dichloroisocyanurate color reaction. Anal Chem. 1977;49:1080. doi: 10.1021/ac50015a053. [DOI] [Google Scholar]
- Feng H, Yan M, Fan X, Li B, Shen Q, Miller AJ, Xu G. Spatial expression and regulation of rice high-affinity nitrate transporters by nitrogen and carbon status. J Exp Bot. 2011;62:2319–2332. doi: 10.1093/jxb/erq403. [DOI] [PubMed] [Google Scholar]
- Fernandes MS (1983) N-carriers, light and temperature influences on the free amino acid pool composition of rice plants. Turrialba 33:297-301. Available via http://orton.catie.ac.cr/repdoc/A0782e/A0782e03.html
- Fernandes MS. Effects of environmental stress on the relationship of free amino-N to fresh weight of rice plants. J Plant Nutr. 1991;14:1151–1164. doi: 10.1080/01904169109364274. [DOI] [Google Scholar]
- Fernandes MS, Rossiello ROP. Mineral nitrogen in plant physiology and plant nutrition. Crit Rev Plant Sci. 1995;14:111–148. doi: 10.1080/07352689509701924. [DOI] [Google Scholar]
- Forde B, Lorenzo H. The nutritional control of root development. Plant Soil. 2001;232:51–68. doi: 10.1023/A:1010329902165. [DOI] [Google Scholar]
- Gao J, Liu J, Li B, Li Z. Isolation and purification of functional total RNA from blue-grained wheat endosperm tissues containing high levels of starches and flavonoids. Plant Mol Biol Rep. 2001;19:185–186. doi: 10.1007/BF02772163. [DOI] [Google Scholar]
- Gruber BD, Giehl RFH, Friedel S, von Wirén N. Plasticity of the Arabidopsis root system under nutrient deficiencies. Plant Physiol. 2013;163:161–179. doi: 10.1104/pp.113.218453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Available via CAB Direct. https://www.cabdirect.org/cabdirect/abstract/19500302257
- Holsters M, de Waele D, Depicker A, Messens E, Montagu M, Schell J. Transfection and transformation of Agrobacterium tumefaciens. Molec Gen Genet. 1978;163:181–187. doi: 10.1007/BF00267408. [DOI] [PubMed] [Google Scholar]
- Huang S, Chen S, Liang Z, Zhang C, Yan M, Chen J, Xu G, Fan X, Zhang Y (2015) Knockdown of the partner protein OsNAR2.1 for high-affinity nitrate transport represses lateral root formation in a nitrate-dependent manner. Sci Rep 5:18192. 10.1038/srep18192 [DOI] [PMC free article] [PubMed]
- Huarancca Reyes T, Scartazza A, Pompeiano A, Ciurli A, Lu Y, Guglielminetti L, Yamaguchi J. Nitrate reductase modulation in response to changes in C/N balance and nitrogen source in arabidopsis. Plant Cell Physiol. 2018;59:1248–1254. doi: 10.1093/pcp/pcy065. [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Jiao X, Lyu Y, Wu X, Li H, Cheng L, Zhang C, Yuan L, Jiang R, Jiang B, Rengel Z, Zhang F, Davies W, Shen J. Grain production versus resource and environmental costs: towards increasing sustainability of nutrient use in China. J Exp Bot. 2016;67:4935–4949. doi: 10.1093/jxb/erw282. [DOI] [PubMed] [Google Scholar]
- Kiba T, Krapp A. Plant nitrogen acquisition under low availability: regulation of uptake and root architecture. Plant Cell Physiol. 2016;57:707–714. doi: 10.1093/pcp/pcw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koevoets IT, Venema JH, Elzenga JTM, Testerink C. Roots withstanding their environment: exploiting root system architecture responses to abiotic stress to improve crop tolerance. Front Plant Sci. 2016;7:1335–1354. doi: 10.3389/fpls.2016.01335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapp A, David LC, Chardin C, Girin T, Marmagne A, Leprince A-S, Chaillou S, Ferrario-Méry S, Meyer C, Daniel-Vedele F. Nitrate transport and signalling in Arabidopsis. J Exp Bot. 2014;65:789–798. doi: 10.1093/jxb/eru001. [DOI] [PubMed] [Google Scholar]
- Lam-Sánchez A, Santos JE, Takamura K, Treptow RMO, Oliveira JED (1994) Estudos Nutricionais com arroz (Oryza sativa, L.). Alim Nutr 5:37-48. Available via https://agris.fao.org/agris-search/search.do?recordID=DJ2012066293
- Lezhneva L, Kiba T, Feria-Bourrellier A-B, Lafouge F, Boutet-Mercey S, Zoufan P, Sakakibara H, Daniel-Vedele F, Krapp A (2014). The Arabidopsis nitrate transporter NRT2.5 plays a role in nitrate acquisition and remobilization in nitrogen-starved plants. Plant J 80:230-241. 10.1111/tpj.12626 [DOI] [PubMed]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- López-Bucio J, Cruz-Ramírez A, Herrera-Estrella L. The role of nutrient availability in regulating root architecture. Curr Opin Plant Biol. 2003;6:280–287. doi: 10.1016/S1369-5266(03)00035-9. [DOI] [PubMed] [Google Scholar]
- Martin A, Lee J, Kichey T, Gerentes D, Zivy M, Tatout C, Dubois F, Balliau T, Valot B, Davanture M, Tercé-Laforgue T, Quilleré I, Coque M, Gallais A, Gonzalez-Moro M-B, Bethencourt L, Habash DZ, Lea PJ, Charcosset A, Perez P, Murigneux A, Sakakibara H, Edwards KJ, Hirel B. Two cytosolic glutamine synthetase isoforms of maize are specifically involved in the control of grain production. Plant Cell. 2006;18:3252–3274. doi: 10.1105/tpc.106.042689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller AJ, Fan X, Orsel M, Smith SJ, Wells DM. Nitrate transport and signalling. J Exp Bot. 2007;58:2297–2306. doi: 10.1093/jxb/erm066. [DOI] [PubMed] [Google Scholar]
- Miranda KM, Espey MG, Wink DA. A rapid, simple spectrophotometric method for simultaneous detection of nitrate and nitrite. Nitric Oxide. 2001;5:62–71. doi: 10.1006/niox.2000.0319. [DOI] [PubMed] [Google Scholar]
- Nacry P, Bouguyon E, Gojon A. Nitrogen acquisition by roots: physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil. 2013;370:1–29. doi: 10.1007/s11104-013-1645-9. [DOI] [Google Scholar]
- Naz M, Luo B, Guo X, Li B, Chen J, Fan X (2019) Overexpression of nitrate transporter OsNRT2.1 enhances nitrate-dependent root elongation. Genes 10:290. 10.3390/genes10040290 [DOI] [PMC free article] [PubMed]
- Okamoto M, Vidmar JJ, Glass ADM (2003) Regulation of NRT1 and NRT2 gene families of Arabidopsis thaliana: responses to nitrate provision. Plant Cell Physiol 44:304-317. 10.1093/pcp/pcg036 [DOI] [PubMed]
- Ossowski S, Schwab R, Weigel D. Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J. 2008;53:674–690. doi: 10.1111/j.1365-313X.2007.03328.x. [DOI] [PubMed] [Google Scholar]
- Prinsi B, Espen L. Mineral nitrogen sources differently affect root glutamine synthetase isoforms and amino acid balance among organs in maize. BMC Plant Biol. 2015;15:96. doi: 10.1186/s12870-015-0482-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RAS (2009) Regras para análise de sementes. Ministério da Agricultura, Pecuária e Abastecimento. 1st edn. Mapa/ACS, p 399. Available via MAPA. https://www.gov.br/agricultura/pt-br/assuntos/insumos-agropecuarios/arquivos-publicacoes-insumos/2946_regras_analise__sementes.pdf.
- Ravazzolo L, Trevisan S, Forestan C, Varotto S, Sut S, Dall’Acqua S, Malagoli M, Quaggiotti S. Nitrate and ammonium affect the overall maize response to nitrogen availability by triggering specific and common transcriptional signatures in roots. Int J Mol Sci. 2020;21:686. doi: 10.3390/ijms21020686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo KK, Tripathi AK, Pareek A, Sopory SK, Pareek SLS. An improved protocol for efficient transformation and regeneration of diverse indica rice cultivars. Plant Methods. 2011;7:49–59. doi: 10.1186/1746-4811-7-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russel DW. Molecular cloning: a laboratory manual. 3. New York: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Santos LA, Bucher CA, Souza SR, Fernandes MS. Effects of nitrogen stress on proton-pumping and nitrogen metabolism in rice. J Plant Nutr. 2009;32:549–564. doi: 10.1080/01904160802714953. [DOI] [Google Scholar]
- Souza SR, Stark EMLM, Fernandes MS, Magalhaes JR. Effects of supplemental nitrogen on nitrogen-assimilation enzymes, free amino nitrogen, soluble sugars, and crude protein of rice. Comm Soil Sci Plant Anal. 1999;30:711–724. doi: 10.1080/00103629909370240. [DOI] [Google Scholar]
- Sustainable Development Goals – SDG (2015). Available via https://sdgs.un.org/goals.
- Tabuchi M, Sugiyama K, Ishiyama K, Inoue ST, Takahashi H, Yamaya T. Severe reduction in growth rate and grain filling of rice mutants lacking OsGS1;1, a cytosolic glutamine synthetase 1;1. Plant J. 2005;42:641–651. doi: 10.1111/j.1365-313X.2005.02406.x. [DOI] [PubMed] [Google Scholar]
- Tang Z, Fan X, Li Q, Feng H, Miller AJ, Shen Q, Xu G. Knockdown of a rice stelar nitrate transporter alters long-distance translocation but not root influx. Plant Physiol. 2012;160:2052–2063. doi: 10.1104/pp.112.204461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tegeder M, Masclaux-Daubresse C. Source and sink mechanisms of nitrogen transport and use. New Phytol. 2018;217:35–53. doi: 10.1111/nph.14876. [DOI] [PubMed] [Google Scholar]
- Toki S, Hara N, Ono K, Onodera H, Tagiri A, Oka S, Tanaka H. Early infection of scutellum tissue with Agrobacterium allows high-speed transformation of rice. Plant J. 2006;47:969–976. doi: 10.1111/j.1365-313X.2006.02836.x. [DOI] [PubMed] [Google Scholar]
- Tracy SR, Nagel KA, Postma JA, Fassbender H, Wasson A, Watt M. Crop improvement from phenotyping roots: highlights reveal expanding opportunities. Trends Plant Sci. 2020;25:105–118. doi: 10.1016/j.tplants.2019.10.015. [DOI] [PubMed] [Google Scholar]
- Trevisan S, Manoli A, Quaggiotti S. NO signaling is a key component of the root growth response to nitrate in Zea mays L. Plant Signal Behav. 2014;9(6):e28290. doi: 10.4161/psb.28290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warthmann N, Chen H, Ossowski S, Weigel D, Herve P. Highly Specific gene silencing by artificial miRNAs in rice. Plos One. 2008;3:e1829. doi: 10.1371/journal.pone.0001829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei J, Zheng Y, Feng H, Qu H, Fan X, Yamaji N, Ma JF, Xu G. OsNRT2.4 encodes a dual-affinity nitrate transporter and functions in nitrate-regulated root growth and nitrate distribution in rice. J Exp Bot. 2018;69:1095–1107. doi: 10.1093/jxb/erx486. [DOI] [PubMed] [Google Scholar]
- Yan H, Deng X, Cao Y, Huang J, Ma L, Zhao B. A novel approach for the construction of plant amiRNA expression vectors. J Biotechnol. 2011;151:9–14. doi: 10.1016/j.jbiotec.2010.10.078. [DOI] [PubMed] [Google Scholar]
- Yan M, Fan X, Feng H, Miller AJ, Shen Q, Xu G. Rice OsNAR2.1 interacts with OsNRT2.1, OsNRT2.2 and OsNRT2.3a nitrate transporters to provide uptake over high and low concentration ranges. Plant Cell Environ. 2011;34:1360–1372. doi: 10.1111/j.1365-3040.2011.02335.x. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. Transcription factors involved in controlling the expression of nitrate reductase genes in higher plants. Plant Sci. 2014;229:167–171. doi: 10.1016/j.plantsci.2014.09.006. [DOI] [PubMed] [Google Scholar]
- Yemm EW, Cocking EC. The determination of amino-acid with ninhydrin. Analyst. 1955;80:209–213. doi: 10.1039/AN9558000209. [DOI] [Google Scholar]
- Yemm EW, Willis AJ. The estimation of carbohydrates in plant extracts by anthrone. Biochem J. 1954;57:508–514. doi: 10.1042/bj0570508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Forde BG. Regulation of Arabidopsis root development by nitrate availability. J Exp Bot. 2000;51:51–59. doi: 10.1093/jexbot/51.342.51. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Xiong S, Wei Y, Meng X, Wang X, Ma X (2017) The role of glutamine synthetase isozymes in enhancing nitrogen use efficiency of N-efficient winter wheat. Sci Rep. 10.1038/s41598-017-01071-1 [DOI] [PMC free article] [PubMed]
- Zhang Z, Gao S, Chu C. Improvement of nutrient use efficiency in rice: current toolbox and future perspectives. Theor Appl Genet. 2020;133:1365–1384. doi: 10.1007/s00122-019-03527-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(PDF 112 kb)
Data Availability Statement
Not applicable.
WinRhizo Arabdopsis software (2012b).