Abstract
Genome-wide association studies (GWAS) are effectively applied to detect the marker trait associations (MTAs) using whole genome-wide variants for complex quantitative traits in different crop species. GWAS has been applied in wheat for different quality, biotic and abiotic stresses, and agronomic and yield-related traits. Predictions for marker-trait associations are controlled with the development of better statistical models taking population structure and familial relatedness into account. In this review, we have provided a detailed overview of the importance of association mapping, population design, high-throughput genotyping and phenotyping platforms, advancements in statistical models and multiple threshold comparisons, and recent GWA studies conducted in wheat. The information about MTAs utilized for gene characterization and adopted in breeding programs is also provided. In the literature that we surveyed, as many as 86,122 wheat lines have been studied under various GWA studies reporting 46,940 loci. However, further utilization of these is largely limited. The future breakthroughs in area of genomic selection, multi-omics-based approaches, machine, and deep learning models in wheat breeding after exploring the complex genetic structure with the GWAS are also discussed. This is a most comprehensive study of a large number of reports on wheat GWAS and gives a comparison and timeline of technological developments in this area. This will be useful to new researchers or groups who wish to invest in GWAS.
Keywords: Genome-wide association studies, Genomic selection, High-throughput phenotyping, Machine and deep learning, Wheat
Introduction
Wheat is a crop having great historical significance as it marks the turning point of human civilization 10,000 years ago with its domestication. It is grown worldwide and ranks third after maize and rice in global production (Shiferaw et al. 2013). Numerous efforts have resulted in the improvement of wheat genetic maps since the last 3 decades, beginning from restriction fragment length polymorphism (RFLP) to the exon capture analysis (Botstein et al. 1980; Saintenac et al. 2011). The development of molecular markers, since the 1980s, has been based on advanced statistical models, and high-speed computer software which aids in the detection of genomic regions associated with both simple and complex traits in crops. Linkage mapping involves the mapping of quantitative trait loci (QTLs) at a specific/particular location over the genome using a bi-parental population. It is a regression analysis that unravel an association between a genomic locus and variation in the phenotypic data collected from the population (Lander and Botstein 1989; Xie et al. 1993). The important factors affecting linkage mapping include the molecular markers density on genetic maps, quality of phenotypic data, and size of the mapping population. The biggest issue with linkage mapping involves low genetic resolution.
Recently, association or linkage disequilibrium (LD) mapping utilizing genome-wide markers is being adopted in wheat because of its two main advantages: (i) association mapping does not require the cost and time associated with the population development and (ii) GWAS provides high mapping resolution as it efficiently uses the multiple historical crossover events occurred in the diverse association panel used. GWAS detects the association between the particular genotype and trait of interest using conserved LD present in the selected panel of accessions (Myles et al. 2009). It is being adopted at a rapid pace by the plant geneticists/breeders because of the reduction in the genotyping cost, which was a major bottleneck previously. In wheat, the development of next-generation sequencing (NGS) tools such as genotyping by sequencing (GBS) and different SNP arrays provides a plethora of information for conducting whole genome-wide analysis at a very low and affordable price (Tibbs Cortes et al. 2021; Sandhu et al. 2021e).
There has been rapid advancement in the GWAS statistical algorithms which ranged from single to multi-locus models for detecting the real association with complete avoidance of false positives and false negatives (Yu et al. 2006; Huang et al. 2018). Population structure and familial relatedness/kinship are the main causes for the spurious associations, and these associations are avoided in the modified GWAS models with the inclusion of population structure and kinship matrix components (Price et al. 2006; Vanraden 2008). Since the first association mapping in wheat, there was a rapid adoption of GWAS for dissecting the genetic architecture of various important traits (Breseghello and Sorrells 2006). Since, majority of QTLs identified through GWAS have minor effect and are population specific, and difficulties in estimation of exact QTLs’ effect create several challenges for adoption of this technique. However, still, hundreds of GWA studies have been conducted in wheat since the last 15 years for different traits (Tables 3, 4, 5 and 6).
Table 3.
Recent GWAS in wheat and characterization of candidate genes conducted over the last decade (2010–2020) for biotic stress traits
| Traits | Population size | Total identified MTAs | Putative or candidate gene IDs/QTLs/ Significant markers associated with the trait |
GWAS model used | Significance threshold | References |
|---|---|---|---|---|---|---|
| Black point resistance | 101 accessions | 23 | LOC109758065, LOC109744537, TaMATE1B | MLM | -log10(P) > 3.0 | (Li et al. 2020b) |
| Black point resistance | 166 cultivars | 221 | PPO-18, IWA5463, AX-111518195 | MLM and FarmCPU | -log10(P) > 3 | (Liu et al. 2017a) |
| Barley yellow dwarf virus resistance | 335 accessions | 36 | TraesCS2B01G037300, TraesCS2B01G038300, TraesCS2B01G038200, TraesCS6A01G368200 | GLM and MLM | -log10(P) > 3 | (Choudhury et al. 2019) |
| Common bunt resistance | 125 accessions | 15 | TraesCS2A01G440100.1, TraesCS2A01G438200.1, TraesCS2B01G627300.1 | FarmCPU | log10(P) > 4.05 | (Bhatta et al. 2018b) |
| Common bunt resistance | 318 accessions | 123 | TraesCS1B01G116700.2, TraesCS1B01G121600.2, TraesCS1B01G123200.2, TraesCS1B01G130000.1 | MLM | FDR < 0.05 | (Mourad et al. 2018b) |
| Multiple disease resistance | 81 cultivars | 94 | QTs.ksu-1AS, Tsn1 | MLM | P value < 5 × 10−5 | (Perez-Lara et al. 2017) |
| Dwarf bunt resistance | 292 accessions | 4 | DB-6D2,DB-6D1 | MLM | P value < 0.05 | (Gordon et al. 2020) |
| Eyespot resistance | 868 accessions | 92 | Pch1, Pch2, IWB8331, IWB73709, IWB472981, IWB47160 | CMLM | P value < 0.001 | (Lewien et al. 2018) |
| Fungal pathogens resistance | 173 accessions | 74 | Lr01, Lr05, SNG04, SNG05, SNL01, Sr01, Sr06 | MLM | P value < 0.001 | (Jighly et al. 2016) |
| Fusarium crown rot | 358 accessions | 104 | TraesCS5D01G138700.1, TraesCS5D01G142400.1, TraesCS5D01G143300.1 | MLM | P value < 0.001 | (Jin et al. 2020) |
| Fusarium crown rot resistance | 234 cultivars | 286 | AX-111106634, AX-94534539, AX-11170401, AX-109474774 | MLM | P value < 0.0001 | (Yang et al. 2019) |
| Fusarium head blight resistance | 171 accessions | 88 | TraesCS4A02G341700, TraesCS4A02G507700LC | MLM | -log10(P) > 3 | (Hu et al. 2020) |
| Fusarium head blight resistance | 240 accessions | 5 | TraesCS1A01G015500, TraesCS1A01G015600, TraesCS1A01G015700, TraesCS1A01G015800 | MLM | -log10(P) > 3.0 | (Zhu et al. 2020) |
| Fusarium head blight resistance | 354 accessions | 10 | S4B_577008759 | FarmCPU | FDR < 0.1 | (Larkin et al. 2020) |
| Fusarium head blight and Septoria tritici blotch resistance | 1023 | 9 | wsnp_BE444720A_TA_2_1 | MLM | Bonf. P value < 0.05 | (Herter et al. 2019) |
| Fusarium head blight resistance | 439 accessions | 6 | S1A_477852878, S1A_477852881 | GLM + Structure. | FDR < 0.1 | (Liu et al. 2019b) |
| Fusarium head blight resistance | 256 cultivars | 16 | M11423,M9432, M6959 | CMLM | P value < 0.001 | (Tessmann and Van Sanford 2018) |
| Fusarium head blight resistance | 238 accessions | 19 | M1563, M6959, M5744, M13020, M5748 | CMLM | LOD score >3 | (Tessmann et al. 2019) |
| Fusarium head blight resistance | 273 accessions | 10 | IWGSC_CSS_3B_scaff_10676713_7175, IWGSC_CSS_7DS_scaff_3876750_2023 | CMLM | FDR < 0.01 | (Arruda et al. 2016) |
| Karnal bunt resistance | 179 accessions | 15 | TraesCS4D02G352200,TraesCS4D02G350300 | MLM | P value < 0.001 | (Singh et al. 2020) |
| Karnal bunt resistance | 119 accessions | 8 | IWA3835, IWA2087 | MLMM | Bonf. P value < 0.05 | (Emebiri et al. 2019) |
| Karnal bunt resistance | 339 accessions | 18 | TraesCS2B02G496800, TraesCS2B01G535000, ,TraesCS5B01G506000 | MLM | P value < 0.001 | (Gupta et al. 2019b) |
| Leaf and stripe rust resistance | 294 accessions | 165 | Yrswp-3B, 3B_t2 | MLM | P value < 0.001 | (Liu et al. 2019a) |
| Leaf rust resistance | 331 accessions | 32 | LrA2K | MLM | FDR < 0.1 | (Sapkota et al. 2019) |
| Leaf rust, stripe rust and tan spot resistance | 646 accessions | 36 | Traes_1DS_3CC12E215, Traes_1DS_3C6EAAFFD, Traes_1AS_BF353B963, Traes_1AS_F098402B4 | MLM | FDR < 0.2 | (Juliana et al. 2018) |
| Leaf rust resistance | 295 accessions | 52 | qNV.Lr-2B.3, qNV.Lr-3A.3, and qNV.Lr-7B.2 | MLM | -log10(P) > 3.5 | (Riaz et al. 2018) |
| Leaf and stem rust resistance | 2111 accessions | 82 | IWA4151, IWA3812, IWA7699, IWA5391IWA7191 | MLM | FDR < 0.05 | (Elbasyoni et al. 2017) |
| Leaf rust resistance | 496 accessions | 88 | Lr.locus-2A2, Lr.locus-2A4, Lr.locus-3A2, Lr.locus-5A1, Lr.locus-6B | MLM | P value < 0.01 | (Aoun et al. 2016) |
| Leaf rust resistance | 338 accessions | 333 | QTLs 6B_3, 6B_4, 5D_1, 2B_2, 4A_1,,1A_3, 2B_2 | GLM and MLM | P value < 0.001 | (Gao et al. 2016) |
| Seedling leaf rust resistance | 1596 accessions | 14 | QLr.stars-1BC1, QLr.stars-2DS1, QLr.stars-1BS1 | MLM | P value < 0.01 | (Li et al. 2016) |
| Leaf spot | 528 landraces | 48 | wsnp_Ex_c10596_17293363, wsnp_CAP11_rep_c4157_1965583, wsnp_Ex_c5998_10513766 | GLM | P value < 0.001 | (Kang et al. 2020) |
| Powdery mildew resistance | 97 varieties | 33 | BS00100185_51, Excalibur_rep_c115510_314, Kukri_c8835_112, BS00094095_51 | MLM | FDR < 0.001 | (Tsai et al. 2020) |
| Powdery mildew resistance | 1292 accessions | 7 | QPm.stars-2BL1, QPm.stars-2BL2 | MLM | FDR < 0.01 | (Li et al. 2019b) |
| Powdery mildew resistance | 97 accessions | 262 | TraesCS1B02G264000, TraesCS2B02G536100, TraesCS2B02G536200 | CMLM | FDR < 0.05 | (Mohler and Stadlmeier 2019) |
| Rust resistance | 2300 accessions | 493 | QRYr1A.1, Yr29, Sr33, Sr45, Lr3 | MLM | P value < 0.001 | (Joukhadar et al. 2020) |
| Rust resistance | 483 accessions | 481 | QYr.ramp-1B.4, QLr.ramp-1B.3, QSr.ramp-1A.2, QYr.ramp-2D.1,,QSr.ramp-2B.6 | CMLM | FDR < 0.001 | (Kumar et al. 2020) |
| Rust resistance | 190 accessions | 15 | IWB11553, IWB14375, IWB39306, IWB12320 | CMLM | FDR <0.10 | (Muleta et al. 2017b) |
| Rust resistance | 676 accessions | 30 [5(Lr), 14 (Yr), and 11 (Sr)] | IWA3295, IWA5474, IWB23955, IWB34703, IWA7440, IWB44883 | MLM | -log10(P) > 4.438 | (Pasam et al. 2017) |
| Stem rust resistance | 158 accessions | 84 | wsnp_Ra_c5346_9501281, Tdurum_contig75595_586 | MLM | P value < 0.05 | (Leonova et al. 2020) |
| Stem rust resistance | 270 accessions | 32 | TraesCS2D01G108000.1, TraesCS2D01G104700.1 | MLM | Bonf. P value < 0.05 | (Mourad et al. 2018a) |
| Stem rust resistance | 2152 accessions | 47 | Sr2, Sr6, Sr7a, Sr8a, Sr9h | MLMM | P value < 0.05 | (Gao et al. 2017) |
| Stem rust resistance | 277 accessions | 12 | RPP13, MDAR6, PP2C, AG4, DSDS1, IPT9, RGA3 | MLM | FDR < 0.05 | (Yu et al. 2017) |
| Stem rust resistance | 250 breeding lines | 247 | IWB11987, IWB65634, IWB12193, IWB52694, IWB5070, IWB4830 | MLM | P value < 0.001 | (Bajgain et al. 2015) |
| Stem rust resistance | 232 breeding lines | 12 | Sr2, Lr34 | MLM | FDR < 0.05 | (Yu et al. 2012) |
| Stem rust resistance | 276 accessions | 15 | wPt7763, wPt9822, wPt664017 | GLM and MLM | FDR < 0.05 | (Yu et al. 2011) |
| Stripe rust resistance | 616 accessions | 34 | Yr5, Yr15, Yr17, Yr18, Yr27, QYrSW.wgp-1A, QYrSW.wgp-2B.2, QYrSW.wgp-4A.2, QYrSW.wgp-4B | BLINK | -log10(P) > 4.61 | (Liu et al. 2020b) |
| Stripe rust resistance | 213 accessions | 11 | QYr.sicau-2BS, QYr.sicau-5AL, QYr.sicau-5DL, QYr.sicau-7BL, QYr.sicau-7DS | MLM | P value < 0.0001 | (Li et al. 2020a) |
| Stripe rust resistance | 410 exotic germplasm | 35 | Yr25, Yr33, Yr47, Yr51, Yr56, Yr57 | MLM | P value < 2.90×10-4 | (Zhou et al. 2020) |
| Stripe rust resistance | 120 accessions | 182 | E3 ubiquitin-protein ligase KEG | MLM | -log10(P) > 3 | (Yang et al. 2020a) |
| Stripe rust resistance | 188 accessions | 21 | QYr.sicau-3B.2,QYr.sicau-5B.3,QYr.sicau-1B.1,QYr.sicau-4A | MLM | -log10(P) > 3.8 | (Cheng et al. 2019) |
| Stripe rust resistance | 1794 accessions | 24 | TraesCS1A01G015900, TraesCS1B01G046300, TraesCS2B01G501500 | MLM | P value < 0.01 | (Elbasyoni et al. 2019) |
| Stripe rust resistance | 152 accessions | 51 | QDS.sicau-2A, QIT.sicau-4B, QDS.sicau-4B.2 | MLM | -log10(P) > 3 | (Long et al. 2019) |
| Stripe rust resistance | 244 accessions | 19 | TraesCS5A02G079700, TraesCS5A02G364700, TraesCS5A02G365300, TraesCS5A02G365600 | MLM | P value < 0.001 | (Ye et al. 2019) |
| Stripe rust resistance | 237 elite lines | 13 | QYr.tsw-1A.1, QYr.tsw-1A.2, QYr.tsw-1B.1, QYr.tsw-1D | MLM | P value < 0.001 | (Godoy et al. 2018b) |
| Stripe rust resistance | 465 accessions | 20 | IWB60567, IWB24342, IWB46564, IWB70554 | MLM | FDR < 0.1 | (Liu et al. 2018c) |
| Stripe rust resistance | 232 accessions | 82 | Yrdurum-1BS.1, Yrdurum-7BL, QYrdurum-3BL, QYrdurum-4BS, QYrdurum-5BL | MLM | P value < 0.005 | (Liu et al. 2017b) |
| Stripe rust resistance | 176 accessions | 81 | YrTtd-6AS.1, YrTtd-6AL.1, YrTtd-7AS, YrTtd-7AL.2, YrTtd-1BS, YrTtd-2BL.1 | MLM | P value < 0.001 | (Liu et al. 2017c) |
| Stripe rust resistance | 182 accessions | 154 | YrEDWL-1ASYrEDWL-1BS.1, YrEDWL-1BS.2, YrEDWL-3AS, YrEDWL-4BL, YrEDWL-5BL | MLM | P value < 0.005 | (Liu et al. 2017d) |
| Stripe rust resistance | 959 accessions | 70 | IWA1191, IWA5861, IWA3621, IWA1040, IWA2194, IWA2145, IWA7815 | CMLM | P value < 0.01 | (Muleta et al. 2017a) |
| Stripe rust resistance | 1175 accessions | 127 | IWA3215, IWA5915, IWA2526, IWA5824 | MLM | P value < 0.01 | (Bulli et al. 2016) |
| Stripe rust resistance | 875 accessions | 97 | QYr.ucw-1B, QYr.ucw-1D, QYr.ucw-2A.2, QYr.ucw-2A.3, QYr.ucw-3B.2 | CMLM | P value < 0.0001 | (Maccaferri et al. 2015) |
| Stripe rust resistance | 402 accessions | 47 | Qyr.wpg-1B.1, Qyr.wpg-1B.2, Qyr.wpg-1B.3, Qyr.wpg-1D.1, Qyr.wpg-2A.3, Qyr.wpg-2B.2 | MLM | P value < 0.01 | (Naruoka et al. 2015) |
| Stripe rust resistance | 181 accessions | 65 | TaADF7, TaLSD1 | MLM | FDR < 0.05 | (Zegeye et al. 2014) |
| Stripe rust resistance | 410 accessions | 292 | TraesCS2B01G512900, TraesCS2B01G513000, and TraesCS2B01G513100 | MLM | P-value < 2.90 × 10-4 | (Wu et al. 2020a) |
| Stripe rust resistance | 857 accessions | 20 | QYrww.wgp.1D-3, QYrww.wgp.2B-2, QYrww.wgp.2B-3, QYrww.wgp.2B-4, QYrww.wgp.3A, QYrww.wgp.5A, QYrww.wgp.5B, QYrww.wgp.5D | MLM | P value < 0.001 | (Mu et al. 2020) |
| Stripe rust resistance | 140 accessions | 12 | QYrcl.sicau-1B.3, QYrcl.sicau-4A.3, QYrcl.sicau-6A.2, QYrcl.sicau-7B.2, QYrcl.sicau-2D.1 | MLM | P value < 0.001 | (Yao et al. 2020) |
| Stripe rust resistance | 120 accessions | 16 | BS00067586_51, BS00086365_51, wsnp_Ex_c965_1846161, TA002369-0369 | MLM and FarmCPU | -log10(P) > 4.69 | (Cheng et al. 2020) |
| Yellow rust resistance | 319 varieties | 47 | TraesCS2B01G486100, TraesCS2B01G486200, TraesCS2B01G486300, TraesCS2B01G486400 | CMLM | -log10(P) > 4.5 | (Gardiner et al. 2020) |
| Yellow rust resistance | 419 accessions | 14 | TraesCS2A02G047700.1, TraesCS2A02G029900, TraesCS2A02G030800 | MLM | P value <10−6 | (Ledesma-Ramírez et al. 2019) |
| Septoria nodorum blotch resistance | 232 accessions | 47 | QSnl08.daw-5B,QSnl07.daw-5B, QSnl07.daw-1B | MLM | -log10(P) > 5.61 | (Francki et al. 2020) |
| Septoria nodorum blotch resistance | 295 accessions | 15 | Snn1, Snn3, Snn2c,Snn5 | MLM | -log10(P) > 3.5 | (Phan et al. 2018) |
| Septoria tritici blotch resistance | 371 varieties | 44 | TraesCS1A01G323600, TraesCS6D01G365100 | MLM | -log10(P) > 3 | (Muqaddasi et al. 2019b) |
| Septoria tritici blotch resistance | 175 accessions | 10 | TraesCS1B01G390100, TraesCS1B01G390500, TraesCS1B01G390200 | GLM | FDR < 0.05 | (Odilbekov et al. 2019) |
| Septoria tritici blotch resistance | 164 accessions | 4 | TRIAE_CS42_5DL_TGACv1_433779_AA1421900, TRIAE_CS42_1BL_TGACv1_030884_AA0102990 | GLM | P value < 0.001 | (Vagndorf et al. 2017) |
| Snow mold tolerance | 458 accessions | 100 | IWB36501, IWB3779, IWB65663 | FarmCPU | FDR < 0.05 | (Lozada et al. 2019) |
| Spot blotch resistance | 141 accessions | 70 | TraesCS5A01G402800, TraesCS5B01G128000, TraesCS2B01G018200 | MLM | FDR < 0.05 | (Tomar et al. 2020) |
| Spot blotch resistance | 294 accessions | 13 | QSb.sdsu-2D.1, QSb.sdsu-3A.1, QSb.sdsu-4A.1, QSb.sdsu-4B.1 | MLM | P value < 0.001 | (Halder et al. 2019) |
| Tan spot resistance | 474 accessions | 93 | S5B_54681021,S1A_2988049, S3B_472110858 | MLM | FDR < 0.05 | (Galagedara et al. 2020) |
|
Tan spot, Stagonospora nodorum blotch, and fusarium head blight resistance |
118 accessions | 30 | Q.Ts1.sdsu-4BS, Q.Ts1.sdsu-5BS,Q.Ts5.sdsu-1BL ,Q.Ts5.sdsu-2DL | MLM | -log10(P) > 3 | (Halder et al. 2019) |
| Tan spot resistance | 170 lines | 33 | GLM | P value < 0.001 | (Singh et al. 2016) | |
| Yellow spot resistance | 295 accessions | 89 | QTL qNV.YS-5B, qNV.YS-2B.1 | CMLM | -log10(P) > 3.0 | (Dinglasan et al. 2019) |
| Cereal cyst nematode resistance | 161 accessions | 11 | DMAP-1 | MLM | FDR < 0.01 | (Pariyar et al. 2016) |
| Multiple pest resistances | 408 accessions | 55 | IWA860, IWA8314, IWA6803, IWA4653, IWA3207 | MLM | P value < 0.001 | (Ando et al. 2018) |
| Multiple biotic stresses | 125 accessions | 124 | TraesCS4D01G096900, TraesCS7A01G517300, TraesCS4B01G007000 | FarmCPU | P value <9.18×10−5 | (Bhatta et al. 2019) |
| Pest resistances | 134 accessions | 26 | wPt-9032, wPt-731493, wPt-666174 | MLM | FDR < 0.01 | (Joukhadar et al. 2013) |
Table 4.
Recent GWAS in wheat and characterization of candidate genes conducted over the last decade (2010-2020) for abiotic stress traits
| Traits | Population size | Total identified MTAs | Putative or candidate gene IDs/QTLs/ Significant markers associated with the trait |
GWAS model used | Significance threshold | References |
|---|---|---|---|---|---|---|
| Aluminum toxicity tolerance | 860 | 70 | CMLM | Bonf. P value < 0.01 | (Froese et al. 2016) | |
| Selenium sensitivity | 480 accessions | 10 | Excalibur_c47452_183, GENE-3324_338, BobWhite_c4838_58 | CMLM | Bonf. P value < 0.05 | (Downie et al. 2018) |
| Drought traits | 277 accessions | 295 | TraesCS6A02G124100, TraesCS6D02G114400 | GLM, MLM, and FarmCPU | P value < 1.88 × 10−5 | (Li et al. 2019c) |
| Drought tolerance and biomass allocation | 100 accessions | 75 | TraesCS2D02G462600, TraesCS2D02G514100, TraesCS2D02G370400, TraesCS1B02G340800 | CMLM | FDR < 0.05 | (Deshmukh et al. 2014) |
| Drought and heat stress tolerance | 315 accessions | 472 | QGWt.adh-3A, QGWp.adr-2D, QGWt.ara-7A.1, QGWt.ara-6D.2, QGWt.ara-6B.6 | CMLM | FDR < 0.05 | (Schmidt et al. 2020) |
| Drought response of wheat | 111 accessions | 263 | SDP6 | MLM | FDR < 0.01 | (Tarawneh et al. 2019) |
| Grain yield and related traits under drought stress | 123 accessions | 90 | TraesCS7A01G158200.1, TraesCS3D01G002700, TraesCS3A01G343700 | FarmCPU | P value <9.99×0-5 | (Bhatta et al. 2018a) |
| Drought tolerance traits | 108 accessions | 28 | MLM | P value < 0.01 | (Muhu-Din Ahmed et al. 2020) | |
| Cold tolerance | 543 accessions | 76 | TraesCS7B01G466300, TraesCS5B01G351200 | MLM | -log10(P) > 4.05 | (Zhao et al. 2020b) |
| Heat-responsive physiological traits | 236 accessions | 500 | TraesCS5B01G325000, TraesCS6B01G063500, TraesCS2B01G496300, TraesCS5B01G436700 | FarmCPU | P value < 9.99 × 10− 4 | (Pradhan et al. 2019) |
| Yield and related traits under heat and drought stress | 192 accessions | 487 | GENE-1752_162, RFL_Contig2471_119, IACX203, IACX5767, Kukri_rep_c68068_95 | MLM | -log10(P) > 3.96 | (Qaseem et al. 2019) |
| Seedling heat tolerance | 200 accessions | 15 | QLCCOT.nri-1B, QLNOT.nri-2A, QLNHR.nri-2A.2 | MLM | P value <0.001 | (Maulana et al. 2018) |
| Salt tolerance | 307 accessions | 117 | QSt.nwafu-1A, QSt.nwafu-3B, QSt.nwafu-6B | MLM | P value < 1.0×10-5 | (Yu et al. 2020) |
| Salinity tolerance | 227 varieties | 24 | Wmc120, barc151, gwm274, barc318, gwm275 | MLM | P value < 0.01 | (Liu et al. 2018d) |
| Salt stress tolerance | 150 cultivars | 187 |
Traes_1BS_D68F0- BED6.1.mrna1-E4(ZIP-7), Traes_2AL_A2CBDB5F7.1.mrna1-E2(KeFc), AtABC8 |
MMLM | P value < 0.01 | (Oyiga et al. 2018) |
| Herbicide resistance | 697 accessions | 329 | gHR-5B, gHR-7D, gHR-1B | MLM | P value < 0.001 | (Shi et al. 2020) |
Table 5.
Recent GWAS in wheat and characterization of candidate genes conducted over the last decade (2010–2020) for agronomic traits
| Traits | Population size | Total identified MTAs | Putative or candidate gene IDs/QTLs/ Significant markers associated with the trait |
GWAS model used | Significance threshold | References |
|---|---|---|---|---|---|---|
| Agronomic traits | 184 accessions | 83 | Ppd-A gene, Ppd-B1, QHD.td.ipbb_5A.2, QHD.td.ipbb_5B.1, QPH.td.ipbb_4B.1 | MLM | P value <1.96E-4 | (Anuarbek et al. 2020) |
| Agronomic traits | 197 accessions | 1118 |
AX-109824320, AX-108740226, AX-110928491, AX-110044748 |
MLM | P value < 0.0001 | (Chen et al. 2020b) |
| Agronomic traits | 768 accessions | 807 | TraesCS4B02G049100, TraesCS4B02G049300, TraesCS5A02G377600 | CMLM | P value < 0.00001 | (Pang et al. 2020) |
| Agronomic traits | 298 varieties | 313 | GLM and MLM | -log10(P) > 3 | (Rahimi et al. 2019) | |
| Agronomic traits | 404 accessions | 146 | TraesCS1A02G329900, TraesCS5A02G554200, TraesCS3A02G368200 | CMLM | -log10(P) > 4 | (Sheoran et al. 2019) |
| Agronomic traits | 237 accessions | 226 | IWA4057 (5B), IWB73713.1, IWB27650 | FarmCPU | P value < 0.001 | (Godoy et al. 2018a) |
| Agronomic traits | 723 landraces | 149 | PM, Ppd-A1 gene, MFT-3B-1, TaAP2-D, CKX2.3 | GLM and MLM | -log10(P) > 4.72 | (Liu et al. 2017e) |
| Agronomic and physiological traits evaluated in a range of heat prone environments | 188 accessions | 245 | wPt-741686, wPt-2822, wPt-664276, wPt-1038, wPt-742925 | MLM | FDR < 0.05 | (Ogbonnaya et al. 2017) |
| Agronomic traits | 163 cultivars | 1769 | BS00021705_51,wsnp_Ex_c32624_41252144, BS00010573_51, IACX9238, IAAV416 , Kukri_rep_c68594_530 | MLM | P value < 0.001 | (Sun et al. 2017) |
| Anther length | 305 accessions | 17 | wsnp_Ex_c53364_56625806, Excalibur_c13242_1178, RAC875_c25375_236 | MLM | P value < 0.0001 | (Song et al. 2018) |
| Anther extrusion | 207 accessions | 23 | Traes_3DL_C1FD5EB05, Traes_3DL_4A818FD98, Traes_7AL_3ACEF0277 | MLM | -log10(P) > 3.0 | (Muqaddasi et al. 2017) |
| Anther extrusion | 514 varieties | 51 | Rht-D1a, Traes_2AL_5BA7E2623.1 | MLM | -log10(P) > 3 | (Muqaddasi et al. 2017) |
| Coleoptile length | 707 accessions | 29 | TraesCS6B01G282600, TraesCS6B01G282500, TraesCS6B01G531200LC | CMLM | Bonf. P value < 0.05. | (Ma et al. 2020) |
| Coleoptile length | 298 accessions | 46 | TraesCS2A02G025800, TraesCS2A02G033900, TraesCS2B02G009100, TraesCS2B02G010100 | MLM | −log10(P) > 3 | (Sidhu et al. 2020) |
| Coleoptile length | 893 accessions | 13 | Rht2, Rht1, QCL.stars-4DC1, QCL.stars-4BS1 | MLM | FDR < 0.05 | (Li et al. 2017a) |
| Flag leaf traits | 163 cultivars | 618 | TaFLL-5B1 | MLM | P value <1.07e−3 | (Yan et al. 2020) |
| Grain weight and grain number | 287 elite lines | 92 | 2B (96 cM), 3B (99 cM), 5A (98 cM), and 6A (77–85 cM) | GLM and MLM | P value < 0.001 | (Sukumaran et al. 2018a) |
| Grain weight | 94 accessions | 37 | wPt-6965, Xgwm299, rPt-1806 | MLM | P value < 0.01 | (Zhang et al. 2013) |
| Kernel Traits | 319 accessions | 24 | TraesCS4B01G018900, TraesCS3B01G409500, TraesCS5B01G330500, TraesCS7A01G533900 | FarmCPU, MLM, and MLMM | LOD ≥ 3 | (Muhammad et al. 2020) |
| Kernel size | 163 cultivars | 8239 | wsnp_JD_C26552_21868492, wsnp_Ex_C2426_4542393, Excalibur_C1845_4911 | MLM | P value < 0.001 | (Yan et al. 2019) |
| Kernel number per spike | 264 accessions | 62 | BS00022896_51-2ATT , BobWhite_c10539_201-2DAA | MLM | -log10(P) > 4.30 | (Shi et al. 2017) |
| Kernel weight-related traits | 205 accessions | 271 | Ku_c9210_10, BS00023893_51 | MLM | P value < 0.001 | (Chen et al. 2016) |
| Rachis nodes per rachis | 220 accessions | 12(haplotypes) | TraesCS7A01G481600 | MLM | -log10(P) > 4 | (Voss-Fels et al. 2019) |
| Total spikelet number per spike | 518 varieties | 43 | TaAPO-A1 | MLM | -log10(P) > 10 | (Muqaddasi et al. 2019a) |
| Wheat spike related traits | 192 accessions | 184 | TraesCS7B01G456300, TraesCS2B01G592100, TraesCS2A01G439500 | CMLM | –log10(P) > 3 | (Liu et al. 2018b) |
| Thousand grain weight | 4302 accessions | 15 haplotype blocks | TaCwi, TaSus1, TaGW2, TaGS5, TaTGW6 | MLM | P value < 0.05 | (Sehgal et al. 2019) |
| Thousand grain weight | 358 lines | 86 | BobWhite_c10402_140, wsnp_JD_c2623_3541255, BS00076190_51, TaGW-6A | MLM | -log10(P) > 4.82 | (Zanke et al. 2015) |
| Grain yield and related traits | 166 accessions | 3106 | MLM | -log10(P) >1 | (Ali et al. 2020) | |
| Yield related traits | 384 accessions | 209 | AX-111600193, AX-109860828, AX-108838800, AX-110982403 | MLM | FDR < 0.05. | (Li et al. 2020c) |
| Tiller number | 92 accessions | 24 | IWB44377 and IWB39005 | FarmCPU | FDR < 0.05 | (Bilgrami et al. 2020) |
| Grain yield and stability | 4302 accessions | 73 haplotype blocks and 125 SNPs | HB17.1, HB8.26, HB20.29,HB20.33, HB20.38 | MLM | P value < 0.001 | (Sehgal et al. 2020) |
| Yield, quality and disease-related traits | 1325 breeding lines | 23 | SNP15,SNP16,SNP17,SNP18 | MLM | P value < 0.05 | (Tsai et al. 2020) |
| Yield and its contributing traits under different water regimes | 320 accessions | 46 | TraesCS3B02G123600, TraesCS4A02G389900, TraesCS5B02G193100 | FarmCPU | -log10(P) > 6 | (Gahlaut et al. 2019) |
| Grain yield | 568 accessions | 33 | Ppd-D1, Rht-D1, QPh.aww-6A, QYld.aww-6B | CMLM | P value < 0.01 | (Garcia et al. 2019) |
| Grain yield | 166 varieties | 120 | IWB75191, AX_111183518, AX_111819405, IWB17930 | MLM | -log10(P) > 4 | (Li et al. 2019a) |
| Yield components | 102 cultivars | 97 | Ppd-D1 gene, TaGW2-A1, WPCL1 | SUPER | P value < 0.05 | (Luján Basile et al. 2019) |
| Yield related traits | 322 accessions | 9 | TaGA2ox-A1, TaAPO1, TaMFT, TaB2 | MLM and FarmCPU | P value < 0.05 | (Ward et al. 2019) |
| Yield related traits | 215 accessions | 117 | wsnp_Ku_c99567_87349060-5BCC,wsnp_Ex_c1630_3105100-5BAA | MLM | -log10(P) > 3.51 | (Guo et al. 2018a) |
| Yield and quality related traits | 192 lines | 148 | TaUBP24, Traes_2AS_DFDA79E58, Traes_7BL_10754D81F | CMLM | -log10(P) > 3 | (Liu et al. 2018a) |
| Grain yield components | 233 accessions | 18 | QKns.mgb-4A, QKns.mgb-2A, QKns.mgb-3B, QTkw.mgb-3B, QTkw.mgb-6A.1 | MLM | -log10(P) > 3 | (Mangini et al. 2018) |
| Yield related traits | 66 accessions | 803 | QPh-3A, QFss-2D, QTkw-5B | GLM and MLM | P value < 0.01. | (Ma et al. 2018a) |
| Yield and component traits | 208 accessions | 392 | MLM | -log10(P) > 3 | (Sukumaran et al. 2018b) | |
| Yield and related traits | 372 varieties | 345 | Rht-B1, TaGW2-6B | MLM | FDR < 0.05 | (Schulthess et al. 2017) |
| Grain yield and related traits | 105 varieties | 24 | Td99211, Excalibur_c14451_1313, Kukri_c19251_579, wsnp_Ex_rep_c68762_67626384 | MLM | P value < 0.001 | (Wang et al. 2017) |
| Yield and yield components | 287 accessions | 565 | wpt3457, wpt0286, wpt0419, wpt6531, wpt5072 | MLM | P value < 0.001 | (Edae et al. 2014) |
| Heading and flowering | 163 cultivars | 306 | Vrn-B1, Vrn-D1, Ppd-D1, RAC875_c41145_189 | MLM | P value < 0.05 | (Zhang et al. 2018) |
| Arbuscular mycorrhizal colonization | 108 varieties | 7 | QTamf-1A, QTamf-2B, QTamf-6A.1 | MLM, CMLM and SUPER | FDR < 0.05 | (De Vita et al. 2018) |
| Response to mycorrhizae | 94 accessions | 187 | QTL_BMmyc25_4A, QTL_BMc_4A, QTL_STI_BMmyc_4A | CMLM | -log10(P) > 4.25 | (Lehnert et al. 2018) |
| Mycorrhizal symbiosis | 94 accessions | 30 | QTL_myc_3A, QTL_myc_4A, QTL_myc_7A1, QTL_myc_7A2, QTL_myc_7A3 | CMLM | P value < 0.001 | (Lehnert et al. 2017) |
| Nitrogen-use efficiency | 56 accessions | 2 | Excalibur c8879293,Exc5594 2818 | CMLM | LOD score >3 | (Hitz et al. 2017) |
| Nitrogen use efficiency | 93 cultivars | 183 | MLM | P value < 0.05 | (Monostori et al. 2017) | |
| Nitrogen use efficiency components | 214 accessions | 1010 | GSe, Rbcs, NADH-Gogat | MLM | −log10(P) > 3 | (Cormier et al. 2014) |
| Potassium use efficiency | 150 accessions | 534 | TraesCS1A02G288500, TraesCS2B02G201400, TraesCS2B02G201500 | MLM | P value < 0.000001 | (Bin Safdar et al. 2020) |
| Pre-harvest sprouting tolerance | 192 varieties | 39 | EX06323-G, IA1142, WS5431 | MLM | P value < 0.0001 | (Zhu et al. 2019) |
| Tolerance to preharvest sprouting and low falling numbers | 469 accessions | 43 | QPHS.wsu-1A.2, QPHS.wsu-1B.2, QPHS.wsu-2D, QPHS.wsu-1D, QPHS.wsu-5A.1 | FarmCPU | Bonf. P value < 0.05 | (Martinez et al. 2018) |
| Pre-harvest sprouting resistance | 717 accessions | 10 | Tamyb10-A1, Tamyb10-D1 | CMLM | -log10 (P) > 6.55 | (Zhou et al. 2017) |
| Pre-harvest sprouting | 185 accessions | 60 | Tamyb10-A1, Qphs.hwwgr-3AS, Qphs.hwwgr-4A | GLM and MLM | P value < 0.0001 | (Lin et al. 2016) |
| Plant growth traits during the stem elongation phase | 210 accessions | 18 | BobWhite_c11808_975, RAC875_rep_c118305_446, RAC875_c57371_238 | MLM | -log10(P) > 4 | (Guo et al. 2018b) |
| Root traits | 196 accessions | 1105 | TraesCS5A02G022100, TraesCS5A02G022300, TraesCS4A02G484800, TraesCS4A02G493900 | MLM | -log10(P) ≥ 3.5 | (Xu et al.2020) |
| Root architecture | 393 accessions | 7 | TraesCS6A01G381700, TraesCS6A01G392400, TraesCS6A01G396400 | MLM | Bonf. P value < 0.05 | (Alahmad et al. 2019) |
| Root traits | 201 accessions | 63 | TraesCS5B01G487600, TraesCS1A01G430800, TraesCS4B01G360900 | MLM | -log10 (P) > 3.5 | (Beyer et al. 2019) |
| Root and shoot traits | 323 accessions | 93 | NRT2 | GLM and MLM | -log10(P) > 4 | (Li et al. 2019d) |
| Root architecture | 160 accessions | 233 | TraesCS2A01G250600, TraesCS2A01G541700, TraesCS1A01G363600 | MLM | -log10(P) > 3 | (Roselló et al. 2019) |
| Root length | 91 accessions | 5 | wPt6278,wPt1159, wPt0021,wPt4487 | CMLM | FDR < 0.005 | (Ayalew et al. 2018) |
| Spikelet sterility and yield-related traits | 710 accessions | 118 | TraesCS1B01G144500, TraesCS1B01G145500, TraesCS2A01G108900 | MLM | FDR < 0.01 | (Alqudah et al. 2020) |
| Spike fertility | 236 accessions | 109 | TraesCS3B01G526100, TraesCS6B01G199000, TraesCS7D01G002400 | FarmCPU | -log10(P) > 4.00 | (Pradhan et al. 2019) |
| Apical and basal spikelet fertility | 212 lines | 112 | w s n p _ E x _ c 3 1 7 9 9 _ 4 0 5 4 5 3 7 6, wsnp_BF293620A_Ta_2_3 | MLM | P value < 3.06 × 10− 4 | (Shi et al. 2018) |
| Floret fertility | 210 accessions | 218 | TaVrs 1 gene, TaSUSIBA2 gene, TaSus1-7A, TaBRI1,TaCO4, TaCO6, eps2, TaCEN, TaFT2, TaHXK9, TaD3 | MLM | P value < 1.30 × 10-4 | (Guo et al. 2017) |
| Seed dormancy | 166 accessions | 153 | Qgr.cas-1AS, Qgr.cas-5AL.2 | MLM | -log10(P) > 3 | (Ma et al. 2020) |
| Seedling emergence and tiller number | 205 accessions | 31 | Ra_c14761_1348-T, Excalibur_c11045_236-A, BobWhite_c8436_391-T | GLM | Bonf. P value < 0.05 | (Chen et al. 2017) |
Table 6.
Recent GWAS in wheat and characterization of candidate genes conducted over the last decade (2010–2020) for end-use quality traits
| Traits | Population size | Total identified MTAs | Putative or candidate gene IDs/QTLs/ Significant markers associated with the trait |
GWAS model used | Significance threshold | References |
|---|---|---|---|---|---|---|
| Baking and milling traits | 270 accessions | 84 | TraesCS1B01G12950, TraesCS7A01G01360 | MLM | -log10(P) > 4.0 | (Gaire et al. 2019) |
| Milling and baking quality | 4095 lines | 52 | Glu-D1 | MLM | P value < 0.001 | (Battenfield et al. 2018) |
| β-Glucan content | 230 accessions | 9 | WSs2A, Bamy1, Wxl1, 1-FEH | GLM and MLM | -log10(P) > 3 | (Marcotuli et al. 2016) |
| Biomass accumulation and radiation use efficiency | 150 accessions | 94 | CPT1, RAN1 | MLM | -log10(P) > 3 | (Molero et al. 2019) |
| Culm cellulose content | 288 accessions | 9 | TRIAE_CS42_5AL_TGACv1_376159_AA1232950S, TRIUR3_05395 | FarmCPU | FDR < 0.001 | (Smiley et al. 2009) |
| Carotenoid biosynthetic and catabolic genes | 233 accessions | 32 |
PSY1, PSY2, BCH1, CRTISO, LYCE,CYP97A3 |
MLM | −log10(p) ≥ 3 | (Colasuonno et al. 2017) |
| Flavonoid decoration pathway of wheat kernels | 182 accessions | 1098 | TraesCS4B01G371700, TraesCS4D01G365800, TraesCS1A01G347100, TraesCS2D01G043500 | MLM | Bonf. P value < 6.83 × 105 | (Chen et al. 2020a) |
| Essential minerals accumulation | 246 accessions | 222 | M15616, M802, M14494, M3205, M10371 | GLM and MLM | P value < 0.001 | (Kumar et al. 2018) |
| Zinc, iron, copper, manganese and phosphorus in grain and rachis | 330 accessions | 279 | TraesCS1A01G373100, TraesCS1A01G374800, TraesCS1A01G375300 | MLMM | P value < 2×10−4 | (Cu et al. 2020) |
| Calcium accumulation in grains | 353 accessions | 485 | Traes_5AL_F49663738, Traes_5AL_320913F7A.1, Traes_5AL_19637DE03 | MLM | -log10(P)> 3.0 | (Alomari et al. 2017) |
| Zinc accumulation | 207 varieties | 125 | TraesCS3D02G078500, TraesCS4A02G428500 | MLM | P value < 1.0 × 10-4 | (Zhou et al. 2020) |
| Grain zinc concentration | 369 accessions | 40 | mRNA_2.1, mRNA_3.1, mRNA_10.1, mRNA_32.1 | MLM | -log10(P) > 3 | (Alomari et al. 2018b) |
| Grain zinc concentration | 330 lines | 39 | TraesCS2A01G072500 | MLM | P value < 0.0001 | (Velu et al. 2018) |
| Increased zinc and decreased cadmium concentration in grain | 286 accessions | 29 | Traes_5AL_9C03C779C.1, Traes_5AL_60CB3D93A.1 | MLM | P value < 0.01 | (Guttieri et al. 2015) |
| Iron accumulation | 369 varieties | 178 |
TraesCS2A01G565900, TraesCS2A01G566000 |
MLM | -log10(P) > 3 | (Alomari et al. 2018a) |
| Micronutrients accumulation | 123 accessions | 92 | TraesCS2A01G519900,TraesCS2A01G520000,TraesCS6B01G117700 | FarmCPU | P value <1.4026×10-6 | (Bhatta et al. 2018b) |
| Grain copper content | 243 accessions | 489 | GCC_Hap_2A1, GCC_Hap_3B1,GCC_Hap_5A1 | MLM | P value < 0.05 | (Zhao et al. 2020a) |
| Vitamins B1 and B2 | 166 accessions |
17(B1) 7(B2) |
IWB43809, IWB69903, IWB23595 | MLM | P-value < 0.001 | (Li et al. 2018) |
| Feruloyl arabinoxylan content | 256 varieties | 116 | RAC875_c6916_860, BS00062894_51, wsnp_Ku_c7458_12842353 | MLM | P value < 0.001 | (Zhan et al. 2019) |
| Polyphenol oxidase activity | 166 cultivars | 465 | Ppo-A1, Ppo-A2, Ppo-B2, Ppo-D2, Pinb-D1 | MLM | FDR < 0.05 | (Zhai et al. 2020) |
| Starch and its components | 205 accessions | 47 |
TRIAE_CS42_2AL_TGACv1_093900_AA0288950, TRIAE_CS42_2AL_TGACv1_ 094920_AA0304860 |
MLM | P value < 0.01 | (Chen et al. 2019b) |
| Starch granule size | 166 accessions | 15 | IWB34623, IWB22624, IWA4574, IWA3693, | MLM | P value < 0.001 | (Li et al. 2017b) |
| Grain protein content | 161 accessions | 145 | TraesCS1D01G029200.2, TraesCS2A01G328100.1, TraesCS2A01G322500.1, TraesCS2B01G003000 | GLM and MLM | Bonf. P value < 0.05 | (Wang et al. 2004) |
| Grain protein content and protein deviation | 240 accessions | 81 | AlaAT, NADH-GOGAT , ASN1, NIR, NR, NRT2, GS1, GS2, GSr | GLM and MLM | P value < 0.001 | (Nigro et al. 2019) |
| Grain protein concentration | 2111 wheat accessions | 46 | IWA3169, IWA3501, IWA7937, IWA6649 | MLM | P value < 0.001 | (Elbasyoni et al. 2018) |
| Water-soluble carbohydrate content | 166 accessions | 5916 | TraesCS6B01G421500, TraesCS7D01G521400, TraesCS2A02G505000 | MLM and FarmCPU | -log10(P) > 3 | (Fu et al. 2020) |
| Water soluble carbohydrate | 1314 accessions | 26 | CMLM | FDR < 0.1 | (Dong et al. 2016) | |
| Water soluble carbohydrates | 166 accessions | 52 | TaSST-D1, SDP6, Hgsnat, CBL7, PPR-repeat, RPD8L3 | MLM | P value < 0.001 | (Dong et al. 2016) |
| Quality traits | 163 cultivars | 846 | TaRPP13L1 | MLM | P value < 0.001 | (Chen et al. 2019a) |
| Quality traits | 190 accessions | 35 | wsnp_Ex_c9362_15546626, Tdurum_contig91519_224, GENE-2712_130,Ku_c18920_414,BS00009777_51 | CMLM, SUPER, and FarmCPU | P value < 9.01 × 10−6 | (Malik et al. 2019) |
| Quality traits | 267 accessions | 40 | TraesCS1B02G330000, TraesCS1D02G041200, TraesCS5B02G431100, | MLM | LOD > 3 | (Yang et al. 2020c) |
| Quality traits | 96 accessions | 72 | TraesCS1B02G440200, TraesCS1B02G434000, TraesCS3B02G367600, TraesCS3A02G359700 | MLM | -log10(P) > 3 | (Muhu-Din Ahmed et al. 2020) |
| Grain and semolina quality | 1184 lines | 320 | ABA2, AAO3, Glu-3, Gli-1 | MLM | P value < 0.01 | (Fiedler et al. 2017) |
| End use quality traits | 469 accessions | 105 | IWB77391, IWB18115, IWB4446, IWB30179, IWB51337 | FarmCPU | Bonf. P value < 0.05 | (Jernigan et al. 2018) |
| Flour color related traits | 166 accessions | 100 | AX_109030196, IWB35120, IWB31766 | MLM | FDR < 0.05 | (Zhai et al. 2018) |
| Grain texture | 92 varieties | 121 | Pin-D1k, TraesCS5B02G010800, TraesCS7B02G055300, TraesCS5B02G010300 | MLM | -log10(P) > 3 | (Kiseleva et al. 2020) |
| Kernel weight and length | 324 accessions | 122 | TraesCS2B01G034100, TraesCS2D01G020800, TraesCS3B01G582000 | MLM | -log10 (P) > 3.5 | (Daba et al. 2018) |
| Straw fodder quality | 287 accessions | 17 | wsnp_Ex_rep_c68058_66805898, Excalibur_c49875_479 | MLM | P value < 0.001 | (Joshi et al. 2019) |
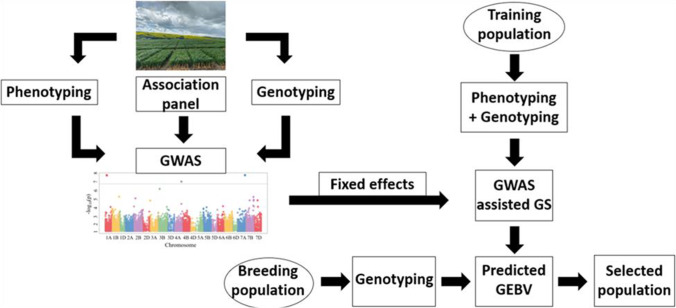
We have provided detailed information about experimental designs adopted for performing GWA studies, and their comparison is made with conventional linkage mapping and nested association mapping (NAM) population designs. Furthermore, various next-generation sequencing platforms, high-throughput phenotyping (HTP), and statistical models are discussed for explaining the whole GWAS analysis pipeline to a novice with previously conducted studies. We tried to cover most of the GWA studies being conducted in the wheat, and critical analysis was performed to detect whether results from these studies were used by the wheat breeding programs for marker-assisted selection (MAS). The genetic architecture of most of the agronomic traits deciphered from various GWAS is also provided. In the end, we made a transition for the future prospectus of genomic selection, OMICS approaches, and machine and deep learning studies after exploration of most of the economically important traits with association studies.
Experimental populations for association mapping
Association or LD mapping is a powerful tool for dissecting the genetic architecture of a trait with the help of phenotypic and genotypic information derived from a set of diverse panels (Kaur et al. 2021). It has been used for deciphering the genetic architecture of most of the complex quantitative traits in wheat (Edae et al. 2014; Arora et al. 2017) due to the availability of high-throughput genotyping and phenotyping platforms along with larger population size. Association mapping has high mapping resolution; this can be attributed to historical recombination events and greater allele richness. The population panel/sets frequently used in association mapping involve lines or accessions having vast genetic diversity. Linkage disequilibrium (LD) in these accessions is used for associating the marker with the QTLs (Nordborg and Weigel 2008).
The association results provided by LD-based mapping not only depends on the linkage between different genomic regions but also on the population structure and relatedness among the lines in the population (Korte and Ashley 2013). The association panel used for association mapping should have genetic diversity with negligible population structure, which otherwise might result in spurious associations or subgrouping. Family-based association mapping was proposed to overcome this obstacle. However, efforts were also made for combining traditional linkage mapping and association mapping for conducting the joint linkage association mapping, which can combine the advantages of both mating designs with avoidance of their pitfalls, but this technique did not get importance in wheat (Würschum et al. 2012). One biggest issue with these populations includes already fixed and rare alleles in the sub-populations which results in missing the identification of rare alleles even after involving a large population size.
However, multi-parent population designs such as multi-parental advanced generation intercross (MAGIC) and nested association mapping (NAM) populations can address these issues for identifying rare variants while retaining the higher mapping resolution at the same time (Beyer et al. 2008; Sandhu et al. 2021e). To our knowledge, in wheat, eight sets of the MAGIC population are available and have been used for genetic characterization of various traits, viz. grain protein content, disease resistance, and grain yield (Mackay et al. 2014; Delhaize et al. 2015). MAGIC populations are developed by several generations of inter-mating cycles among the multiple founder parents. The multiple founder parents maintain the relatively high allelic diversity depending upon the number of parents used compared to the bi-parental mapping population. Several generations of inter-crossing in MAGIC populations create opportunities for the number of recombination events and ultimately results in high resolution in the mapping of MTAs. Mackay et al. (2014) developed the MAGIC population in wheat using eight diverse founder parents for studying the genetic architecture of awns in the UK and European wheats. Similarly, Delhaize et al. (2015) developed the two MAGIC populations of wheat using four and eight founder parents, respectively for studying the genetics of rhizosheath size.
Nested-association mapping (NAM) population is a novel genetic approach for mapping the complex traits which combine the advantages of both association and linkage mapping (Sandhu et al. 2021e). NAM population involves crossing the diverse founder parents with a common cultivar and the resulting F1 are selfed for creating the recombinant inbred lines (Sandhu et al. 2021b). In this way, NAM populations have high allelic variation because of the diverse founder parents and high mapping resolution due to the creation of recombinant inbred lines (Song et al. 2017). For the first time, a NAM population was developed in maize, where 25 lines were crossed with one single parent B73, followed by selfing for creation of a population of 5000 recombinant inbred lines (200 per cross) (Yu et al. 2008). Till now, a couple of studies have reported the use of the NAM population for mapping the complex traits in wheat (Ren et al. 2018; Kidane et al. 2019). Jordan et al. (2018) utilized the NAM population of 2100 RILs derived from 26 founder parents, for genetic mapping and studying the recombination events in different regions of the chromosome to identify recombination hotspots. Linked top cross populations in wheat are another source and can have even more parents than NAM populations with a higher possibility of allele discovery than any other population.
Recent advances in genotyping technologies
The extent of LD provides the estimation of the marker density needed for GWAS in any crop. Linkage disequilibrium (LD) measure or D refers to the difference between the observed and expected gametic frequencies of haplotypes under linkage equilibrium (Cortes et al. 2021). Besides D, several other measures of LD (D', r2, R, D2, D*, Q*, F', X(2), and δ) have been developed to quantify LD in various bi-allelic and multi-allelic situations (Devlin and Risch 1995; Gupta et al. 2005). The detailed description and formulae of LD quantification along with sensitivity, merits, comparison, suitable statistical tests, and calculation methodology for these different LD measures have been extensively described in the literature (Gupta et al. 2005). A low level of LD in crop accessions implies that a higher number of markers will be required for the sufficient coverage of the genome that is crucial for finding the MTAs and/or QTLs for the concerning traits and vice versa. In wheat, the extent of LD patterns have been analyzed extensively (Maccaferri et al. 2005; Chao et al. 2010). Analysis of LD patterns using simple sequence repeats (SSRs) markers in hexaploid wheat revealed significant LD extension to 5 cM (Breseghello and Sorrells 2006) or 10 cM (Chao et al. 2007), whereas tetraploid wheat populations maintained around 50% of their initial LD value at distances up to 20 cM (Maccaferri et al. 2005). Moreover, different genomes of wheat have shown different extent of significant LD; the comparatively higher extent of LD has been observed in the D genome than A and B genomes (Chao et al. 2010). Theoretically, the extent of LD can be increased by selecting a set of closely related accessions or genotypes, and then only fewer markers can be sufficient for detecting the associations. In contrast, accessions having high genetic diversity are employed for GWAS; therefore, availability of a large number of markers, sufficiently covering the whole genome, becomes essential. Initially, SSR markers were used for association analysis in wheat (Prasad et al. 1999; Chen et al. 2014). The number of SSR markers used in these studies was very less which did not cover the genome sufficiently. Hence, numerous causal genomic regions might have gone unnoticed. Later, various advances in genotyping techniques enabled successful GWA studies capable of identifying maximum genetic variation in diverse accessions. To overcome the limitations associated with SSR markers, a high-throughput genotyping system, i.e. diversity array technology (DArT), was developed which allowed the rapid and cost-effective genome-wide genotyping in wheat (Crossa et al. 2007).
DArT is a microarray-based genotyping technique that is independent of sequence information. Since the first study was published in 2007, several studies have reported the successful use of DArT markers for GWAS in wheat for many complex traits (Joukhadar et al. 2013; Jighly et al. 2016; Kalia et al. 2018). The number of DArT markers used in various studies in wheat ranged from 242 (Crossa et al. 2007) to 1728 markers which covered a total genetic distance of 2,851.7 cM at an average distance of 1.7 cM (Joukhadar et al. 2013). Although these DArT markers were quite abundant than SSRs, several researchers found some chromosomes to be poorly covered (Crossa et al. 2007; Langer et al. 2014). The arrival of next-generation sequencing technologies allowed the development of an advanced genotyping technology/platform, i.e. GBS (Elshire et al. 2011), which provided access to a comparatively large number of single nucleotide polymorphism (SNP) markers in a cost-effective manner. GBS-based genotyping has been widely used in wheat for GWAS (Arruda et al. 2016; Liu et al. 2018a; Pradhan et al. 2019). The number of GBS-SNPs used in GWA studies in wheat ranged from 17,937 (Kumar et al. 2018) to 327,609 SNPs (Pang et al. 2020).
Owing to low read coverage, sometimes GBS shows genotyping errors and poor ability to sufficiently detect the true homozygotes. Moreover, its performance is highly affected by the quality of the reference genome. Wheat has a large genome size (1C = 16Gb) comprising three homoeologous genomes which contain more than 85% of repetitive DNA sequences and significant gaps (only 14.1Gb of the total have been accurately assigned and ordered so far). Therefore, the relative incidence of genotyping errors may get increase further as the paralog sequences might be treated as the same reads (Appels et al. 2018; Rahimi et al. 2019). Moreover, the Chinese Spring (whose genome is generally used as a reference genome for SNP calling) is derived from landrace which is known to have wide genetic variation compared to modern/advanced cultivars, resulting in low coverage of SNP markers shown by GBS, therefore restricting the utilization of GBS in modern wheat cultivars. It is also frequently troubled by a high amount of missing data that can potentially reduce the accuracy of any GWA study. One approach to deal with this missing data is imputation and this has widely been applied in many studies in many crops including wheat (Arruda et al. 2016; Liu et al. 2018a; Pradhan et al. 2019). It can increase the number of variants that are used for GWAS by relying on linkage information obtained from common haplotypes. Furthermore, the GBS-SNPs imputed based on the reference genome of ‘W7984’ have shown the highest imputation accuracy (Alipour et al. 2019).
Later, a comparatively cheaper and easier but efficient genotyping technique, i.e. DArTseq™, was also developed by combining DArT with next-generation sequencing platforms, which provides a relatively large number of markers to build more dense linkage maps cost-effectively. These highly dense linkage maps have been widely used for various GWAS in wheat (Dinglasan et al. 2019; Alahmad et al. 2019; Long et al. 2019). In some studies, to get more marker density, both DArT and DArTseq markers have also been used together (Ledesma-Ramírez et al. 2019).
SNP array/platform is another genotyping platform that has various features such as high marker density, low cost, high-throughput, high commercialization, and flexibility which are highly recommended for GWAS in wheat. Unlike NGS and PCR-based markers, these SNP arrays are flexible with respect to customization of sample and data point number, which donates to its high-density scanning and comparatively higher and robust call rates. To date, a number of high-density SNP genotyping arrays have been developed and employed for various GWA studies in wheat, for instance, the Illumina Wheat 9K iSelect SNP array (Cavanagh et al. 2013; Guo et al. 2018a), the Wheat 15K SNP array (Boeven et al. 2016; Qaseem et al. 2019), the Wheat Breeders’ 35K Axiom array developed from 820K SNP array (Sheoran et al. 2019; Kumar et al. 2020), the Wheat 55K SNP array developed from 660K array (Ye et al. 2019; Jin et al. 2020), the Illumina Wheat 90K iSelect SNP genotyping array (Dhakal et al. 2018; Mellers et al. 2020), TaBW280K (Rimbert et al. 2018), and the Axiom Wheat 660K SNP array (Yang et al. 2019). These arrays have been very promising for detecting extensive variation in secondary and tertiary gene pools in addition to the primary gene pool of wheat. For covering maximum genetic variation present in the large germplasm collections, different combinations of SNP arrays have also been utilized in wheat such as the combination of 9K and 90K SNP arrays (Lewien et al. 2018), 35K and 90K SNP arrays (Muqaddasi et al. 2017), and 90K and 660K SNP arrays (Liu et al. 2017e). However, a more recent study showed that the Wheat 660K SNP array could be used as a substitute for other SNP arrays for a great range of possible applications including GWAS, as it comprises the highest percentage of genome-specific SNPs with precise physical positions (Sun et al. 2020). The timeline of advancements in genotyping of whole-genome variants employed for GWAS in wheat is diagrammatically represented in Fig. 1.
Fig. 1.
Timeline of advancements in genotyping of whole-genome variants employed for GWAS in wheat
The Wheat 660K SNP array, developed by the Chinese Academy of Agricultural Sciences, is based on tetraploid and hexaploid wheat, Aegilops tauschii, and emmer wheat and has the advantages of being genome-specific, efficient, high-density, cost-effective, and with a wide range of possible applications, as well as adding numerous markers to the D genome (Sun et al. 2020). However, the choice of array largely depends upon the aim of the researcher, available resources, and the kind of population used for genetic dissection of the trait(s). The use of SNP arrays has allowed access to an unprecedented number of markers for genomic studies; however, there are drawbacks in using these technologies (Lachance and Tishkoff 2013; Elbasyoni et al. 2018; Chu et al. 2020). Inherent ascertainment owing to smaller population size is one of the major drawbacks in using SNP arrays for genotyping (Lachance and Tishkoff 2013). Since the SNP probes on arrays are static, sub-population-specific variants or rare variants are generally not assayed. This causes bias in population genetics studies including GWAS and does not permit the identification of rare functional variants controlling traits in question. By allowing access to all potential polymorphisms in the population of interest and not restricting the analysis to discrete markers on an array, a method of detecting markers directly from sequence data may reduce ascertainment bias on an experiment-by-experiment basis. Access to all possible polymorphisms can improve the resolution of genetic mapping and GWA studies. However, the confidence in sequence-based prediction of genotypes is confounded by the uncertain alignment of short reads in the genome of polyploids such as wheat. To overcome these challenges, a method of sequence-based genotyping has been proposed (Clevenger et al. 2018) which instead of applying a filter to individual sites collects observed haplotypes from sequence reads and contrasts those haplotypes between genotypes/accessions to identify available polymorphic markers in polyploids. Haplotype-based genotyping should be broadly applicable in wheat and other allopolyploids.
On the other hand, in most techniques of association analysis, SNP markers are evaluated individually for their association with the trait of interest, which can be problematic for complex traits regulated by several genetic loci (Gupta et al. 2014; Zhai et al. 2018). Furthermore, due to the bi-allelic nature of SNPs, a single model may be unable to describe true allelic diversity available in the population (Lu et al. 2011). Therefore, for better description of the genetic architecture of complex traits, researchers recommend testing numerous SNP markers, either with a multi-locus model that evaluates adjacent markers simultaneously or with haplotype blocks where closely linked markers are converted to a solitary multi-locus haplotype block (Da 2015). The use of haplotype blocks minimizes the cost and time spent on GWAS since it eliminates the need to study every individual SNP. Several GWA studies have demonstrated the importance of this approach in the identification of QTLs associated with different traits in wheat (Liu et al. 2020a). Furthermore, deep analysis for the identified causative loci by GWAS, e.g., haplotype-based analysis, is considered a key for genomics-assisted crop breeding. Using “wheat AND GWAS” as the keywords, we screened papers published on GWAS in wheat in the PubMed database (https://pubmed.ncbi.nlm.nih.gov/). A total of 552 research articles were published since December 31, 2020, which reported the successful application of GWAS for dissecting various complex traits in wheat. This analysis shows that wheat geneticists and/or breeders are rapidly utilizing and realizing the importance of GWAS for genetic dissection of complex traits and the number of papers on GWAS in wheat is expanding rapidly with the maximum number of papers being reported in the year 2020 (Fig. 2).
Fig. 2.
The number of publications related to GWAS in wheat published from 01/01/2009 to 31/12/2020. Source: PubMed (keywords “wheat AND GWAS” were used to search the number of publications in PubMed
Alleviating the phenomics bottleneck: high-throughput phenotyping
Associating genotype to phenotype for complex traits on a genome level requires an association panel having diverse accessions or mapping populations along with high density of molecular markers. Plant phenotyping refers to the assessment and measurement of observable characteristics of the plants in the field or under controlled conditions. The capability to collect accurate phenotypic data in the field and/or greenhouse conditions is a major bottleneck for precise genetic dissection of complex traits. The recent advancements in genotyping technologies have already provided almost limitless access to high-density molecular markers; therefore, it becomes an urgent need to shift plant science research from genomics to phenomics. Several efforts have already been made in the recent past for replacing low throughput and invasive phenotyping methods with high-throughput, rapid, and non-invasive phenotyping technologies (Mir et al. 2019). Table 1 includes several such imaging techniques developed in the last decade which have revolutionized crop phenomics.
Table 1.
Various high throughput phenotyping techniques or platforms presently used for phenotyping different traits in various crops including wheat
| Sr. No. | Phenotyping techniques/platforms | Purpose | Working efficiency [construction cost] | Reference |
|---|---|---|---|---|
| 1. | Phenovator | For phenotyping hundreds of samples for growth, photosynthesis, and spectral reflectance | Can phenotype >1400 crop plants multiple times per day | (Flood et al. 2016) |
| 2. | GROWSCREEN FLUORO | For simultaneous phenotyping of chlorophyll fluorescence, and leaf growth | Can phenotype roughly 60 plants per hour | (Jansen et al. 2009) |
| 3. | LEAF-E | For measuring leaf growth | - | (Voorend et al. 2014) |
| 4. | Zeppelin NT aircraft | For field-based high throughput phenotyping | The experimental field of 0.4 ha can be imaged from the air within 10 seconds | (Liebisch et al. 2015) |
| 5. | TRiP (Tracking Rhythms in Plants) | For computing circadian period by leaf movement | 118 plants/camera | (Greenham et al. 2015) |
| 6. | Push Carts | For recording data on crop development and growth including responses to drought, heat, and other abiotic stresses | Can record data with average speed of approximately 0.36 m/s | (White and Conley 2013) |
| 7. | BreedVision | For measuring various complex traits such as lodging, tiller density, and biomass yield | Can record data on >2000 plots (4 × 1m) per day | (Busemeyer et al. 2013) |
| 8. | Phenocart | For field-based high throughput phenotyping | At an average walking speed (of ~1 to 2 m/s), it can generate ~100 data points for CT and NDVI per plot (2 × 0.8 m) [~US$12,000] | (Crain et al. 2016) |
| 9. | PHENOPSIS | For dissecting the plant responses to drought stress | Can phenotype more than 500 plants per 1.5 h | (Granier et al. 2006) |
| 10. | Heliaphen | For phenotyping leaf or whole plant morphology | Can hold up to 1,300 plants | (Gosseau et al. 2019) |
| 11. | MVS-Pheno | For measuring shoot architecture | One sample per 60–120 s [$7560 ] | (Wu et al. 2020b) |
| 12. | GPhenoVision | For phenotyping different morphological traits | At a constant walking speed of 1 m/s, it can store up to 346 MB/s data | (Kim et al. 2011) |
| 13. | Field Scanalyzer (LemnaTec, GmbH, Aachen, German) | For monitoring the crop development under field conditions | Can take images of 30–90 plots/h (field view of ~0.5 m width and 0.5 m depth) | (Sadeghi-Tehran et al. 2017) |
| 14. | Plant Accelerator® (LemnaTec, GmbH, Aachen, German) | For assessing the tolerance to various stresses such as salinity and nitrogen/water deficiency in crops and for phenotyping different morphological traits | Can assess >1500 plants in smart houses | (Neilson et al. 2015) |
Hyperspectral imaging was used in wheat to determine spectral changes during salt stress (Moghimi et al. 2018). Moreover, a “hyperspectral absorption-reflectance-transmittance imaging (HyperART)” system was employed for the non-invasive quantification of different leaf traits (Bergsträsser et al. 2015). Various “unmanned aerial vehicles” (UAVs) having different sensors attached to them were utilized effectively to measure different traits in various crops including wheat (Yang et al. 2020b).
These non-invasive high-throughput phenotyping platforms involve the utilization of various sophisticated technologies such as (a) fluorescent spectroscopy to evaluate photosynthetic rates; (b) infrared imagery and thermography to examine transpiration/temperature profiles; (c) light detection and ranging (LIDAR) to measure development/growth rates; (d) 3-dimensional reconstruction to measure plant structure and growth rate; (e) canopy spectral reflectance for monitoring dynamic complex quantitative traits; (f) magnetic resonance imaging (MRI) and positron emission tomography (PET) to measure leaf/root physiology, growth/development patterns, photosynthetic assimilate translocation properties, and water relations; (g) digital RGB (red, green, and blue colour) imaging for recording data on several characteristics of shoots, roots, leaves, and seeds; and (h) nuclear magnetic resonance (NMR) for monitoring the sucrose allocation and the structure of tissues (Yang et al. 2020b).
Although the potential of these high-throughput phenotyping technologies have already been and being continuously demonstrated for various applications in wheat (Crain et al. 2018; Sandhu et al. 2021d), somehow these technologies have not been fully explored for GWA studies in wheat. Only a few papers have been published so far which utilized phenotypic data recorded via high-throughput phenotyping platforms for dissecting the different complex traits such as normalized difference vegetation index (NDVI) (Condorelli et al. 2018), lodging (Singh et al. 2019), and transpiration efficiency (Gehan and Kellogg 2017) in wheat. For the first time in wheat, a study reported the increased ability of aerial platforms, viz. UAVs over ground-based phenotyping platforms to identify the QTLs by GWAS for NDVI under terminal drought stress conditions (Condorelli et al. 2018). Recently in 2019, one more study provided a proof-of-concept application of UAS-based phenotyping of a complex phenological trait, i.e. lodging for describing the genetic architecture of lodging tolerance in wheat through GWAS. Phenotypic data recorded on transpiration efficiency (TE) via a high-throughput lysimeter platform was also successfully used for identifying the QTLs associated with TE in wheat (Fletcher et al. 2019).
Several state-of-the-art phenomics centers have been established to increase the visibility and impact of plant phenotyping in crops including wheat. Moreover, an association, known as International Plant Phenotyping Network (IPPN) (https://www.plant-phenotyping.org/) has also been established to disseminate information about high-throughput phenotyping. This network has mainly six national partners/centers: (i) the Austrian Plant Phenotyping Network (https://www.appn.at/), (ii) Australian Plant Phenomics Facility (https://www.plantphenomics.org.au/), (iii) China Plant Phenotyping Network, (iv) German Plant Phenotyping Network (https://dppn.plant-phenotyping-network.de/), (v) Phen-Italy (http://www.phen-italy.it/index.php), and (vi) the PHENOME-The French plant phenomic network (https://www6.dijon.inrae.fr/umragroecologie_eng/Research-Programs/Investissement-Avenir/PHENOME). Other major centers are the Julich Plant Phenotyping Centre (https://www.fz-juelich.de/ibg/ibg-2/EN/_organisation/JPPC/JPPC_node.html) in Germany and High-Resolution Plant Phenomics Centre located in Canberra at CSIRO Plant Industry, whereas Nanaji Deshmukh Plant Phenomics Centre (developed by Saveer Biotech Limited) at ICAR-IARI, high-throughput plant phenomics facility at the ICAR-Indian Institute of Horticultural Research (IIHR), high-throughput automated phenotyping platform at ICRISAT (https://www.icrisat.org/researchfacilities/), and phenomics facility (http://www.niam.res.in/Phenomics-facility) at ICAR-National Institute of Abiotic Stress Management (developed by LemnaTech, Germany) are the major high-throughput phenotyping centres in India.
These centres use platforms designed mainly for phenotyping under artificial/controlled conditions; however, efforts are being made to create relevant technologies and tools for use under field conditions at both industrial and experimental scales. Moreover, the establishment of high-throughput phenotyping systems is time-consuming and costly and needs in-depth knowledge of computational and engineering sciences to maintain functionality and flexibility. The implementation of such systems may only be justified at big research centres and companies as the unit cost depends on throughput. Several private companies like ‘LemnaTec’, ‘PhenoSpex’, ‘Phenokey’, ‘Photon System Instruments’, ‘We Provide Solutions’, ‘WIWAM’, and ‘Saveer Biotech Limited’ offer large-scale, custom, high-throughput phenotyping platforms for both controlled and field environments (Gehan and Kellogg 2017).
One of the biggest problems associated with high-throughput phenotyping platforms is the handling of large volume, velocity, and variety of data. This might be one of the possible reasons also why high-throughput phenotyping technologies have not been fully explored in wheat for genetic studies. To overcome these challenges associated with the analysis and interpretation of enormous datasets, machine learning (ML) and deep learning (DL) algorithms can be employed (Ma et al. 2018b; Sandhu et al. 2021a). These ML and DL algorithms are multidisciplinary approaches that provide more efficient, accurate, and faster data analytics by utilizing the concepts from statistics, probability theories, decision theories, and optimization (González-Camacho et al. 2018). Application of these machine learning and deep learning algorithms/methods in the prediction of phenotypes holds big promise, and therefore, these methods are likely to be integral tools for future breeding programs (Shah et al. 2019).
Mixed models and significance thresholds for GWAS in wheat
Several GWAS models are available, which range from simple to increasingly complex for associating phenotypic variation with the particular genotype configuration in wheat (Huang et al. 2018). Traditionally, linear models such as ANOVA, t-tests, and linear regression were used for studying MTAs, but these models usually resulted in several spurious associations because of the ignorance of population structure and familial relatedness (Price et al. 2006; Yu et al. 2006). The wide geographical distribution of wheat parents in the association panel results in a strong population structure and it is important to use GWAS models which reduce the false associations due to the population structure. However, analysis using structure and principal component analysis (PCA) packages accounts for the population structure generated with the help of molecular information. The inclusion of these structure parameters as a covariate in the GWAS model controls the false positives (Pritchard et al. 2000). General linear models (GLM) perform the association using a single marker at a time with the inclusion of population structure as a fixed effect in the model and can be represented as:
where Y is the trait of interest, SNP represents the matrix of genotypic information, Q is population structure obtained using structure or PCA, and e is residual error. The results from structure and PCA are usually similar, but PCA is more often utilized because of less computational cost and resources required for its generation (Wang et al. 2009; Wu et al. 2011); initially, GLM was most frequently used for GWAS analysis in wheat, but later it was realized that GLM results in various false-positive associations because of ignorance of relatedness among the populations and hence, it was then replaced by recent mixed models (Segura et al. 2012). GLM only accounts for the population structure, completely ignoring the relatedness among the individuals in the population. Yu et al. (2006) developed the unified mixed model approach with inclusion the family relatedness as a random effect in the GLM, resulting in the creation of a mixed linear model (MLM) for GWAS, which can be represented as:
All the terms of this equation are described above. At the same time, kinship denotes the random components of the model, demonstrating the relationship between individuals in the population obtained using pedigree or genotypic information. This model complements the previously developed models that only account for either population structure or familial relatedness, thus resulting in the creation of the powerful GWAS model (Abecasis et al. 2000). Presently, this is the most often utilized GWAS model. More than 50% of the GWAS conducted in wheat were performed with this model and it is also evident from the information on models provided in Table 2.
Table 2.
A brief comparison of different GWAS models
| GLM | MLM | CMLM | MLMM | SUPER | FarmCPU | BLINK | |
|---|---|---|---|---|---|---|---|
| Year of release | 2004 | 2006 | 2010 | 2012 | 2014 | 2016 | 2019 |
| Single or multi-locus | Single locus | Single locus | Single locus | Multi-locus | Multi-locus | Multi-locus | Multi-locus |
| Population structure | Ignored | Included | Included | Included | Included | Included | Included |
| Kinship | No | Yes | Yes | Yes | Yes | Yes | No |
| Number of SNPs in kinship | Not applicable | All markers | Among associated groups | Among pseudo QTNs | Among pseudo QTNs | Among pseudo QTNs | Not applicable |
| False Positives | Present | Controlled | Controlled | Controlled | Present | Controlled | Controlled |
| False negatives | Absent | Present | Present | Present | Absent | Absent | Absent |
| LD criteria | No | No | No | No | No | No | Yes |
| Model effect | Fixed | Mixed | Mixed | Mixed | Mixed | Mixed | Fixed |
| Time or computational burden | Quick | Large | Large | Large | Large | Medium | Quick |
| Use | ++ | +++++ | ++ | +++ | ++ | ++++ | ++ * |
| References | (Price et al. 2006) | (Yu et al. 2006) | (Zhang et al. 2010) | (Segura et al.2012) | (Wang et al. 2014a) | (Liu et al. 2016) | (Huang et al. 2018) |
*(less, as it is new)
MLM was shown to be superior regarding control of false positives in the simulation models, but this model suffers from substantial computational cost. Computational time varies in MLM as mpn3 where m is the number of markers, p is the number of iterations required to solve the model, and n is the number of individuals in the random effect model. The computational time increases with the cube of individuals in the random component of the MLM. Zhang et al. (2010) developed the compressed mixed linear model (CMLM) that reduces the computational time of the MLM by the grouping of a number of individuals in the random effect model. This model reduces the computational time compared to MLM by retaining the same or higher statistical power than MLM. CMLM can be represented as
Here, kinship is obtained among the groups using the maximum likelihood method. If all the individuals are classified into one group, it is equivalent to GLM, while if all the individuals are in separate groups, this will result in MLM. In this regard, CMLM is intermediate to MLM and GLM. CMLM gained its popularity in wheat due to the grouping of lines from the same breeding programs or regions into one pool, to account for the relatedness for controlling false positives. Several studies utilized CMLM for association analysis and reported its computational superiority over the MLM in wheat (Arruda et al. 2016).
The first mixed linear model was published in 2006 for GWAS analysis, and since then, many MLMs have been proposed to account for population structure and family relatedness for controlling the false positives (Breseghello and Sorrells 2006). However, all these MLM were single-locus models, studying a single association at a time, but the majority of traits in wheat are controlled by a large number of QTLs which show that these models fail to mimic the true genetic architecture of the traits (Segura et al., 2012; Liu et al. 2016). This required the use of multi-locus GWAS models in wheat to reduce the false negatives produced by single-locus GWAS models because of overfitting in the models. This overfitting happens because single-locus models explain the variation individually for each marker, which completely ignores the other significant marker and interactions between markers, resulting in missing some real associations. Segura et al. (2012) developed the multi-locus mixed model (MLMM), which studies multiple associations using stepwise regression and heritability as criteria for forward inclusion and backward elimination of markers in the model. This model can be represented as:
Quantitative trait nucleotides (QTNs) are added in the model using the heritability estimate for stopping further inclusion. Once forward inclusion is done, backward elimination is performed, where each added QTN is removed individually, to identify the exact number of QTNs which are controlling the variation in the trait, using heritability estimate.
MLMM uses all the SNP marker information for extracting the kinship matrix among the individuals. The settlement of MLM under the progressive exclusive relationship (SUPER) model was developed, which used significant QTNs for extracting the kinship matrix. This model produces higher statistical power and is also computationally efficient than MLMM (Wang et al. 2014a). Both MLMM and SUPER models incorporate the significant QTNs to remove the confounding problem between the testing markers and kinship. To altogether remove the confounding issue in the analysis, a fixed and random model circulating probability unification (FarmCPU) was developed, which divides the model into a fixed and random effect model (Liu et al. 2016). The fixed-effect model tests a single marker at a time, while the random effect model utilizes the multiple associated markers for obtaining kinship as a covariate in the model, and this also controls the false positives in the model. This model is superior compared to previous multi-locus models, having high statistical power and less computational time (Liu et al. 2016). This model is represented as:
This is a fixed component of the model where each QTN is tested individually at a time. The random effect component of the model is represented as:
Kinship in this model is obtained using multiple associated markers from the fixed-effect model, thus controlling the false positives. Several studies using the FarmCPU model for association analysis have been reported in wheat (Bhatta et al. 2018b). FarmCPU has been reported to be superior for GWAS analysis because of its computational advantage owing to the separation of fixed and random effects. This is particularly important for wheat, owing to its hexaploid and complex genome nature.
Malik et al. (2019) compared the performances for three GWAS models, namely, SUPER, CMLM, and FarmCPU for association analysis of yield and straw quality traits in wheat. The superiority of FarmCPU was reported for MTAs for plant height, yield, lodging, Septoria tritici blotch, and harvest index. Q-Q plots and P-value inflations were compared to conclude that FarmCPU performed superior for all the traits analyzed in this study and validated that FarmCPU should be used for future analysis in wheat. Similar results were obtained by Ward et al. (2019) during the comparison of MLM and FarmCPU for GWAS for yield and yield-related components in wheat. They showed that MLM results in a large number of false negatives, as MLM only identified nine significant MTAs while FarmCPU identified 74 significant MTAs. These results were also validated using Q-Q plots from association analysis.
FarmCPU model assumes that QTNs are randomly distributed across the genome, thus eliminating the LD along the genome. Furthermore, FarmCPU has a random effect model that has associated computational cost with bigger data sets. Bayesian information and LD iteratively nested keyway (BLINK) is the most recent GWAS model which removes the problem available in the FarmCPU (Huang et al. 2018). BLINK uses Bayesian information criteria for replacing the random effect component of the FarmCPU with the fixed effect model. Furthermore, LD information is used for the inclusion of a single marker at a time in the model and eliminating the confounding problem (Huang et al. 2018). This model is reported superior for analysis in wheat, but till now, there are only a few studies available that have reported the use of this model due to its recent release (Liu et al. 2020b). A comparison of different GWAS models is presented in Table 2.
False positives and false negatives occur not only by GWAS models, but they can also arise because of over-conservative or less stringent threshold, suggesting that identification of significant threshold is crucial in wheat (Dudbridge and Gusnanto 2008; Pe’er et al. 2008). The commonly utilized significant thresholds in wheat are Bonferroni correction, false discovery rate (FDR), and positive false discovery rate (PFDR) (Benjamini and Hochberg 1995). Bonferroni correction of 0.05 is a strict significant threshold (obtained using P-value/number of markers), and this causes a number of false negatives because of the over-conservative nature of this threshold, as it does not consider that markers on the same chromosome could be independent (Hayes 2013). MTAs identified with Bonferroni correction are highly significant and provide high confidence for incorporating particular MTAs in a breeding program. A very few studies usually report the significant MTAs with Bonferroni correction as evident from Table 3. FDR and PFDR are somewhat less stringent threshold criteria and are often used for reporting the significant associations (Tables 3, 4, 5, and 6). FDR is calculated from the expected portion of MTAs that are in fact the false positives (Hayes 2013). There is a high need for deciding the strict threshold for controlling the false positive associations in wheat. Permutation testing was proposed for solving the multiple testing problem in humans to select a significant threshold by analyzing the large number of simulated data sets generated from the real data set by randomly shuffling the population (Churchill and Doerge 1994). This led to the selection of a P-value < 5 x 10−8 as a strict cutoff for reporting significant MTAs in humans, and this kind of cutoff is needed for association studies in crops including wheat, in spite of freedom to the researchers to report their own subjective threshold P-value.
GWAS and characterization of candidate genes
Over the years, GWA studies have been successfully conducted for better defining the relative role of genes in various crops and further assisted in exploring the genetic basis of natural selection and population differences among the individuals of a population, developing into a briefly verified and mature method today. GWAS has been extensively used to investigate various biological and physiological traits in the wheat crop during the last decade (Tables 3, 4, 5, and 6). In the literature that we surveyed, 86,122 wheat lines have been studied under various GWA studies reporting 46,940 loci. However, further utilization of these is largely limited. Nevertheless, this huge information source can be further utilized for identifying meta-QTLs through meta-GWAS. Meta-analysis of QTLs identified through interval mapping has regularly been conducted in wheat (Saini et al. 2021a, 2021b, 2021c). The traits considered under evaluation include cold tolerance (Zhao et al. 2020b), seed dormancy (Zuo et al. 2019), coleoptile length (Ma et al. 2020), spike fertility (Pradhan et al. 2019), agro-morphological traits (Sheoran et al. 2019), kernel weight and length (Daba et al. 2018), end-use quality traits (Jernigan et al. 2018), anther extrusion (Muqaddasi et al. 2017), root traits (Beyer et al. 2019), disease resistance (Bhatta et al. 2018b), micro-nutrients in grain (Cu et al. 2020), and multiple pest resistances (Ando et al. 2018) (see Tables 3, 4, 5, 6). Bar graphs have been provided to represent the number of GWA studies conducted over the last decade, and further, the recorded studies have been divided into four major categories to compile the data for this study. Maximum number of GWA studies have been conducted for agronomic and yield traits, followed by biotic stress resistance, quality traits, and abiotic stress tolerance in wheat over the last decade (Figs. 3, 4, and 5).
Fig. 3.
Histogram showing the number of GWAS over the last decade under different categories
Fig. 4.
Histogram showing the number of GWAS over the last decade for various a biotic stresses and b abiotic stresses. Trait name is given as reported in published reports
Fig. 5.
Histogram showing the number of GWAS over the last decade for various a agronomic and b quality traits. Trait name is given as reported in published reports
While the number of studies for four major categories has been represented here in the form of histograms, a comprehensive table has also been provided to summarize all these GWAS conducted in wheat during the last decade, with information related to the population size for a particular study, the number of QTLs identified, putative genes, and GWAS models used for analysis in the study. Only high confidence putative genes or associated markers having a significant PVE (Phenotypic Variation Explained) or R2 value for a particular trait have been documented in this review (Tables 3, 4, 5, 6).
Multiple disease resistance for leaf rust (Puccinia triticina), stripe rust (Puccinia striiformis var. tritici), common bunt (Tilletia tritici), and tan spot (Pyrenophora tritici-repentis) was phenotyped in 81 accessions where 94 MTAs were identified on seven chromosomes for the studied traits. Identified major effect genomic regions were found to be coinciding with previously identified genes like Tsn1 gene (Perez-Lara et al. 2017). Resistance to powdery mildew was phenotyped in 97 accessions, and 262 significant loci were identified in these accessions. Based on GWAS and the linkage map-based QTL analysis, two large effect QTLs with dynamic gene action were identified on chromosome 1BL and 2BL for adult plant resistance to powdery mildew which may be used in breeding programs; some candidate genes were also identified and annotated like TraesCS1B02G264000 (Mohler and Stadlmeier 2019).
In a recent study, a total of 319 varieties were phenotyped for the resistance to stripe rust, and 47 significant loci were found significantly associated with the trait. Moreover, using the gene enrichment with mapping-by-sequencing and the homozygosity haplotyping algorithm, 589 high confidence genes were detected, and out of these 589 genes, 10 genes (e.g. TraesCS2B01G486100, TraesCS2B01G486200) were annotated for diseases resistance which had homology to a previously characterized Yr7 candidate gene (Gardiner et al. 2020). Similarly, fusarium crown rot resistance was phenotyped in 358 accessions, and 104 loci were found to be significantly associated with it. A novel significant region was detected on chromosome 5DL; qRt-PCR was used to validate the involvement of candidate genes in providing resistance to the fusarium crown rot disease. Validated candidate genes, namely, TraesCS5D01G138700.1 and TraesCS5D01G142400.1, encode the proteins belonging to the widely known disease resistance protein (TIR-NBS-LRR class) family (Jin et al. 2020).
The combination of advanced techniques with GWAS provides precision targeting of the candidates for the trait of interest. Conversely, advancements in breeding methodologies based on GWAS also provide a key role in developing resistant lines; efficient methods such as genomic selections can be used to assist in it. A similar study was associated with Septoria tritici blotch (STB) resistance; a total of 371 accessions were phenotyped, and 44 loci were found to be significantly associated with STB resistance. Putative candidate genes TraesCS1A01G323600 and TraesCS6D01G365100 were also identified and annotated. Along with candidate gene identification, the potential of using the results of GWAS in genomic prediction was also assessed in order to highlight the potential of combined use of GWAS and genomic selection in STB resistance (Muqaddasi et al. 2019a).
Multiple GWAS models have also been used for conducting association analysis in wheat. For instance, a study was conducted for Barley yellow dwarf (BYD) virus resistance where 335 accessions were phenotyped and both MLM and GLM models of GWAS were employed which resulted in the identification of 36 loci significantly associated with the target trait. The candidate genes, namely, TraesCS2B01G037300 and TraesCS2B01G038300 were defined which may be useful to breeders in breeding programs to achieve the stable resistance to BYD virus (Choudhury et al. 2019). Along with disease resistance, traits like quality of the wheat grain and its processed products were also well studied by using GWAS. A comprehensive study of baking and milling traits which include flour yield, softness equivalent, flour protein, and four solvent was conducted in 270 accessions and 84 loci were found to be significantly associated; except one, all the identified associations were novel. Moreover, two putative genes, viz. TraesCS1B01G12950 and TraesCS7A01G01360 were also identified (Gaire et al. 2019). Likewise, STB resistance, more than one GWAS analysis model, namely GLM and MLM, were used to study grain protein content. Grain protein content was phenotyped in 161 accessions, and 145 loci were found to be significantly associated with it. Furthermore, two large effect QTLs on chromosome arms 2B and 7B and underlying putative genes namely TraesCS1D01G029200.2 and TraesCS2A01G328100.1 were identified.
Abiotic stresses pose a great threat to the crops and can cause a huge loss (Kaur et al. 2021). In order to have a better insight into the genetics of traits associated with abiotic stresses, several GWA studies have been conducted in wheat. Drought, nutrient toxicity, extreme temperatures, and salinity are some of the major abiotic stresses that can significantly impede the normal development of plants. Among these abiotic stresses, the effect of drought on various traits like yield and other related traits has been extensively studied by GWAS. Li et al. (2019a) conducted GWAS using 277 accessions and identified 295 significantly associated loci using three different models, namely, GLM, MLM, and FarmCPU. Candidate genes including TraesCS6A02G124100 and TraesCS6D02G114400 were also identified (Li et al. 2019a, 2019b, 2019c, 2019d). Similarly, the effect of drought stress on grain yield was studied by Bhatta et al. (2018a) using the FarmCPU model of GWAS, the trait was phenotyped among 123 accessions, and 90 loci were found to be significantly associated. Candidate genes TraesCS7A01G158200.1 and TraesCS3D01G002700 were also identified. As evident from the histogram (Fig. 4), nine GWA studies have been conducted for drought tolerance alone, followed by heat stress (3), salt tolerance (3), and drought and heat together (2), while many other abiotic stresses have been studied at least once (Fig. 4).
Quality traits including the micronutrients like vitamins and minerals have been the topics of keen interest with respect to market pricing and consumer preference; in this regard, GWAS was performed for evaluating the variation for vitamins B1 and B2 content in wheat, which were phenotyped among 166 cultivars. A total of 24 loci were declared significant (17 loci for Vitamin B1 and 7 loci for Vitamin B2) in this study. IWB43809, IWB69903, and IWB23595 were identified as putative markers which can be of interest to the breeders. However, the candidate genes remain unidentified as little is known about biosynthetic pathways of Vitamins B1 and B2 in plants (Li et al. 2018). Similarly, copper content in wheat grains was phenotyped using 243 accessions, and 489 loci were found to be significantly associated with the trait. Furthermore, haplotype analysis revealed three important genetic loci, GCC_Hap_2A1, GCC_Hap_3B1, and GCC_Hap_5A1 associated with grain copper content. Linkage mapping identified four QTLs on chromosomes 1D, 6A, 6B, and 7D, associated with copper content in wheat grains. Two of the significant SNPs, detected on chromosome 1D via GWAS, were mapped within the interval of one QTL (QGCC.hau-1D), implying that this locus has an important role in regulating copper content in wheat grains (Zhao et al. 2020a). Fig. 5 represents the number of GWA studies considered or covered under a particular quality trait over the last decade.
Agronomic traits such as plant architecture, root structure, and most importantly yield affecting traits have also been studied by GWAS. For example, a GWA study for twelve agronomic traits phenotyped in 768 accessions under multiple environments resulted in the identification of a total of 807 loci significantly associated with the traits under study. A total of 9 environmentally stable QTLs were identified which can be of great use in breeding programs. Candidate genes TraesCS4B02G049100 and TraesCS1B02G415500 were reported for QTLs associated with spike seed setting and grain size, respectively (Pang et al. 2020). For root traits, six related traits were phenotyped in 196 accessions, and 1,105 loci were found to be significantly associated with the traits under study. Three candidate genes TraesCS5A02G022300, TraesCS4A02G484800, and TraesCS4A02G493900 were also reported; the proteins of these genes were found to be associated with carbon metabolism, nitrogen metabolism, signal induction, stress responses, and DNA synthesis (Xu et al. 2020). Similarly, yield and its contributing traits were phenotyped in 320 accessions of a highly diverse wheat association mapping panel, and 46 loci were found to be significantly associated with five traits. Candidate genes TraesCS3B02G123600 and TraesCS4A02G389900 were also reported (Gahlaut et al. 2019). Generally, a single locus GWAS model is used to identify significant MTAs, but for complex traits like nutrient use efficiency which are being controlled by multiple loci, a more stringent model is required. Hence, multi-locus models were developed, as they can detect potential MTAs using lower significance criteria. Such a study was conducted for potassium use efficiency which was phenotyped in 150 accessions. In this study, both single and multi-locus GWAS models were used from which a total of 534 loci were found to be significantly associated with the traits in question. Candidate genes TraesCS1A02G288500 and TraesCS2B02G201400 were reported (Bin Safdar et al. 2020). Similarly, spikelet sterility was phenotyped in 710 accessions, and 118 loci were found to be significant using GWAS. Candidate genes TraesCS1B01G144500 and TraesCS1B01G145500 were reported (Alqudah et al. 2020). Fig. 4 represents the number of GWA studies for each agronomic trait over the last decade. Grain yield remains the most extensively studied trait under GWAS of various agronomic traits of wheat.
Applications in breeding
Recent advancements in molecular genetics have made it possible to use molecular technologies in breeding programs and to develop diverse molecular breeding strategies for efficient and effective crop improvement. One of such tools is GWAS, which has been extensively used to search for genomic regions associated with various traits. These identified genomic regions, then, can be used to develop breeder-friendly markers for use in the breeding programs. Many traits, including phenology, height, and resistance to rusts in wheat, are affected by some key genes (such as Ppd, Vrn, Yr, Lr, Sr, and Rht) with major effects. These genes can reduce the sensitivity for other minor QTLs (or hinder the detection of minor effect QTLs) since the different alleles/QTLs can only be analyzed accurately in the respective group of lines. Actually, the estimation of the total number of QTLs depends on the distribution of QTL effects. If the overall distribution of the effects is delineated by an exponential distribution, the distribution of identified QTL effects becomes a truncated exponential distribution after incorporating the Beavis effect. This must not be confused with the original Beavis experiment where all simulated QTLs are supposed to have an equal genetic effect. According to the Beavis experiment, when only 100 progeny are evaluated, the average estimates of phenotypic variances associated with correctly identified QTL are greatly overestimated, slightly overestimated when 500 progeny are evaluated, and fairly close to the actual magnitude when 1000 progeny are evaluated (Beavis et al. 1994; Beavis 2019). The statistical power of detecting a minor QTL is as low as 3% when the sample size is modest, say 100, and the predicted effects are frequently inflated 10-fold. This phenomenon has since been termed the Beavis effect and has formed the basis of a number of subsequent analyses (Beavis et al. 1994; Beavis 2019). Furthermore, minor alleles/genes can also be detected for any trait that has been measured in response to the major genes (at least background genes such as Vrn, Ppd, and Rht) or using the wheat genotypes having null alleles for these major genes and using a genotyping technology which facilitate the selection of most-informative SNPs (by adding or removing targeted loci) in a custom-designed fashion.
Various modifications of GWAS like (a) eGWAS (uses data from gene expression profiling) which can be very useful for identification and annotation of candidate genes involved in the metabolic pathways (Luo 2015), (b) PWAS (proteome wide association study) which can be used to link proteome abundance variation and phenotypic variation (Brandes et al. 2020), (c) mGWAS (metabolic GWAS) which is used to define the relationship between genetic factors and the metabolome of a tissue or the complete plant (Luo 2015), and (d) TWAS (transcriptome wide association study) which conducts expression mapping by creating functionally relevant maps that correspond to genes and their expression have broadened the application of GWAS in genetic studies from gene to the molecule level (metabolites) (Wainberg et al. 2019). Another concept, PheWAS (phenome wide association studies) applies a contrasting phenotype to genotype approach for assessing the sequence polymorphisms across diverse phenotypes, thereby complementing the data from GWAS (Denny et al. 2010). The markers derived from GWAS can be involved in genomic selection/genomic prediction models as fixed effects for enhancing the prediction accuracy (e.g. for grain yield and yield-related traits in wheat) (Odilbekov et al. 2019). Despite all these applications and advantages of GWAS over conventional breeding, the true potential of GWAS still awaits full exploitation in wheat breeding because there are many gaps between genomic studies and breeding (Samantara et al. 2021). One of the gaps is that the breeders who hold molecular biology training still fall short in the handling of genomic data. More user-friendly software systems are required to fill this gap. Similarly, various modifications of GWAS like eGWAS, PWAS, and TWAS are still relatively new concepts to the breeders and thus require a wider adoption. Moreover, the effectiveness of molecular breeding for highly complex traits like yield and related traits based on GWAS data needs to be further improved. The complex traits are controlled by multiple alleles, thus the conventional GWAS or the single-locus GWAS cannot be used to search for the associated MTAs, as the marker selection criteria like FDR implemented in single-locus GWAS make the criteria stringent for the detection of multiple MTAs. Hence, multi-locus GWAS models with higher sensitivity are required in order to detect MTAs of such complex traits using a less stringent criterion. Another gap arises, as the QTL×QTL interactions and QTL×E interactions of complex traits are not completely described in genetic studies. The knowledge of these interactions is very important for the better understanding of complex quantitative traits and effectively using GWAS for them (Samantara et al. 2021).
Being an allopolyploid, genetic redundancy is no new concept to wheat. A plethora of traits like seed dormancy (Abe et al. 2019) and broad-spectrum resistance to diseases like powdery mildew (Wang et al. 2014b) can be identified in wheat where genetic analysis has been very difficult because of the presence of multiple homeoalleles, as completely recessive mutant does not exist in natural population to understand the functioning and effects of underlying alleles (homeoalleles). To solve this issue and for the improvement or better understanding of polyploid crops, simultaneous editing of multiple homeoalleles of a trait is required. The CRISPR-Cas9 system and its predecessors ZFN (zinc finger nucleases) and TALEN (transcription activator–like effector nuclease) are powerful tools for genome editing which can be used to precisely edit multiple QTLs simultaneously and to generate novel alleles, providing rapid genetic enhancements (Abe et al. 2019). The results of significant MTAs from GWAS can be put into candidate gene identification approaches to find putative genes. The CRISPR-Cas9 or TALEN can then be used to generate genome-edited organisms in order to validate the function of associated putative genes or they can be directly used in the editing of candidate genes if the data is already available. So far, CRISPR-Cas9 and TALEN have been successfully used in wheat for editing traits like male sterility (Okada et al. 2019), powdery mildew resistance (Wang et al. 2014b), and quality traits like gluten content of grains (Jouanin et al. 2020). Hence, genome editing tools enlighten the path to the era of ‘GWAS-plus’ in wheat, a concept given in rice (Wang et al. 2020).
In wheat, several recent studies have also demonstrated the power of association mapping in identifying and characterizing the candidate genes that control the target traits (Li et al. 2019a; Wang et al. 2019; Sandhu et al. 2021e). For instance, using wheat 90 K SNP assay, an association mapping was performed for grain length and thousand-grain weight leading to the identification of numerous significant SNPs located on chromosome 7B. Furthermore, haplotype analysis of these significant SNPs on 7B generated the block containing the predicted TaGW8-B1 gene, which was then cloned by sequencing in bread wheat. Analysis of agronomic traits revealed that genotypes with TaGW8-B1a allele possessed significantly more grain number per spike, wider grain length, higher thousand-grain weight, longer grain length, and more spikelets per spike than the genotypes with TaGW8-B1b (Yan et al. 2019). Another GWA study conducted in wheat using a 90K genotyping assay for the six quality-related traits in Chinese wheat cultivars in eight environments over 4 years led to the identification of a total of 846 significant SNPs, involving 103 multi-environment significant SNPs detected in more than four environments (Chen et al. 2019b). Furthermore, it was discovered that some important genes, including some known functional genes and annotated unknown functional genes, were linked to the six quality traits. TaRPP13L1 was found to be associated with flour colour among the annotated unknown functional genes. Wheat cultivars or lines with the TaRPP13L1-B1a allele showed considerably higher flour redness and lower yellowness than those with TaRPP13L1-B1b in the Chinese wheat natural population and the bi-parental population. This study provided valuable information for further dissection of the genetic basis of flour colour and also provided potential genes or genetic loci for marker-assisted selection to improve the process of breeding quality wheat (Chen et al. 2019b). The aforementioned successful examples of genes discovered using GWAS give strong evidence that GWAS can be utilized as a part of a rapid gene-cloning strategy.
Researchers may have been misled by early optimism regarding QTL deployment in populations using marker-assisted selection. Because favourable alleles frequently have population-specific effects, QTL found in one population may not have the same amount of effect in other populations. This could be the result of epistatic interactions between the QTL and the total genetic background, resulting in low penetrance and varying degrees of expression (Gaire et al. 2020). After interrogating simultaneously, a large number of QTLs/alleles in natural populations through GWAS, bi-parental populations can be used to validate a subset of the detected QTLs. This validation step is required to choose a parental line confirmed to have the favourable allele that can be used as a donor in the marker-assisted breeding program. For instance, a GWA study identified several QTLs associated with grain yield, yield components, and plant water status in wheat. Fourteen of these QTLs detected as significant in at least three environments in the GWAS were further validated using a panel of eight bi-parental mapping populations (Zhang et al. 2018). QTL identified and validated in this study provided beneficial information for the improvement of wheat under full and limited irrigation.
Moreover, GWAS results can also be validated using meta-QTL analysis. For instance, a GWAS analysis identified a total of 13 and 11 significant MTAs for fertile tiller number and total tiller number, respectively, in Iranian wheat under varying water regimes (Bilgrami et al. 2020). Then, a meta-analysis was conducted using 30 previously published independent studies, which led to the identification of 30 meta-QTL regions on 11 wheat chromosomes, that validated at least 5 significant MTAs (identified through GWAS) associated with the trait in question (Bilgrami et al. 2020). GWAS offers the opportunity to identify genes that contribute to naturally occurring variation in complex quantitative traits. However, GWAS relies largely on the statistical association, so functional validation is necessary to make strong claims about gene function. The genes identified through GWAS can be validated using different strategies including transgenesis, gene silencing, gene, and genome editing (Curtin et al. 2017).
GWAS can be considered an exploratory analysis for the right selection of true segregating genotypes/accessions that may be used as parents in the bi-parental mapping population, as well as for further genetic and molecular validation of the associations (Alqudah et al. 2020). GWAS can also be utilized to get insights into breeding-program variation (the genetic variation in the natural population used to develop improved breeding material) or MAS (where candidates are screened for target markers, their phenotypes are predicted based on allelic states, and then selections are made based on these predictions) because the association mapping population can be considered as a source of favourable alleles that are not or rarely present in the bi-parental populations. QTLs/MTAs identified through GWAS can be followed by MAS if a significant proportion of trait genetic variation is explained by the associated markers. Initial limitation of retrieving large number of loci based on hundreds of identified SNPs through simple MAS was practically not possible and required re-genotyping making it difficult cost-wise; however, platforms now have come up for multiplexed SNP identification which may now lead to practical utilization of information generated through GWAS or GS. For instance, the AgriSeq targeted GBS can target and uniformly amplify the hundreds to thousand of markers in a single PCR reaction utilizing a highly efficient multiplexed PCR chemistry (Gujjula et al. 2019). A targeted sequence-based, scalable, and flexible multiplexed genotyping technology known as KeyGene SNPSelect technology was also proposed which facilitates the selection of most informative SNPs (by adding or removing loci), permitting cost efficient yet highly informative genotyping in a custom-made fashion (Hogers et al. 2018). Most recently in 2020, a method known as SNP-seq was developed which combines the advantages of multiplex PCR amplification and high-throughput sequencing. This is flexible both in number of SNPs and samples targeted, yields high accuracy, particularly when genotyping genome wide perfect SNPs with high polymorphism and conserved flanking sequences, and is also cost-effective (Zhang et al. 2020a). Under significant epistasis, interacting loci distributed across the genome alter the outcome of a major single-locus QTL. The epistatic background influence limits the usefulness of QTLs in other populations (Korte et al. 2012; Bocianowski 2013). The QTL and the interacting loci act as a package within the specific genetic background of the discovery population (Bocianowski 2013); in these scenarios, special statistical techniques may be required to identify and minimize background epistasis effects. Xavier et al. (2015) (Bocianowski 2013) advocated simultaneously assessing marker effects in different populations to eliminate variations in QTL phasing, genetic background, and effect sizes from one population to another (Bocianowski 2013). Most recently in 2021, Malosetti et al. combined a QTL discovery method employing pre-breeding populations that used intensive phenotypic selection for the target trait across several plant generations with accelerated generation turnover (i.e. ‘speed breeding’) to allow the cycling of multiple plant generations each year. They demonstrated that QTL detection using breeding populations under selection for the target trait can detect QTLs associated with the trait in question and that the frequency of the favourable alleles gets increased as a response to selection, thereby validating the QTLs identified. This is a useful opportunistic approach that may provide QTL information that is more readily transferred to breeding applications (Malosetti et al. 2020). They also envisaged great potential for integrating speed breeding with GWAS, accelerating the rate of crop improvement.
GWAS-assisted genomic selection
Conventionally plant breeders used to rely on the use of phenotypic information for selections of desirable plants in the field. With the development of high throughput genotyping tools, the selection process got complemented with the use of MAS. The MAS allows the rapid selection of superior genotypes by identifying QTLs having a major effect on the trait. Still, it fails in most of the complex quantitative traits in crop plants, which are usually controlled by a large number of small-effect QTLs (Xu and Crouch 2008). Furthermore, these small effect QTLs are highly affected by environmental conditions, different genetic backgrounds, and QTL by environment interactions (Bernardo 2016). Even linkage and association mapping have not been able to properly account for such small effect QTLs. Moreover, MAS is used to introduce a single gene at a time and thus increasing the time required for variety release especially in wheat, which has a large number of contributing genes due to its hexaploid nature. Hence, genome-wide prediction (GP) or genomic selection (GS) came in handy, which uses the whole genome-wide marker information for predicting the breeding value of the plant, known as genomic estimated breeding values (GEBVs), and these GEBVs further assist in making selection (Meuwissen et al. 2001).
Originally proposed by Meuwissen et al. (2001) in animal breeding, GS could be considered an upgraded version of MAS, where all the markers are used to calculate the GEBVs of the plant. It is believed that each QTLs is in LD with at least one of the molecular markers, and this marker accounts for all the genetic variances for that QTLs in the GS models for predicting GEBVs (Lorenz et al. 2011). Genomic selection requires a training population that is genotyped and phenotyped, and the generated information is then used for defining the GS model. This model calculates the effect of all molecular markers using phenotypic data from the training population in the GS model (Rutkoski et al. 2011). Once the GS model is trained, it is used to predict the GEBVs of the breeding/testing population, which is only genotyped. Plant breeders can choose the parents for inter-mating, generate segregating population, genotype the population, make the selection based on these GEBVs, and develop cultivars without further testing, thus accelerating the breeding cycle and ultimately increasing the genetic gain per unit time (Bernardo 2016). Genomic selection is being applied in breeding programs for the selection of parents for crossing, selection of top-performing lines in the breeding trials, and prediction of multi-environmental trials breeding values, and assists in the maintenance of high performing lines in the program (Sandhu et al. 2021b, a).
Several factors affect the GS prediction accuracies, namely, the heritability of the trait, relatedness between training and testing population, sample size, cross-validation scenario, marker density, and GS model used (Lorenz et al. 2011; Sandhu et al. 2021c). Some of the traits in wheat are controlled by large effect QTLs, and hence, the inclusion of those QTLs’ effects in the GS model may provide an excellent opportunity (Fig. 6). Several studies have shown that incorporation of GWAS results as a fixed effect in GS models resulted in an increase in prediction accuracy for quantitative traits (Boichard et al. 2012; Bernardo 2014). Bernardo (2014) showed in a simulation study that the inclusion of a QTL as a fixed effect in GS model which explains more than 10% of the genetic variance resulted in a significant increase in model performance. GWAS-assisted GS has several benefits as it does not require the additional data and results in the increase of prediction accuracy, and furthermore, this is easily accessible to plant breeders without the need for considering the underlying genetic architecture of the trait (Spindel et al. 2016), and the structure of a population can be accounted by using PCA as a fixed effect in the GS models. In a recent study from CIMMYT, authors showed that the inclusion of GWAS loci as a fixed effect in the GS model results in a 9 to10% increase in prediction accuracy for grain yield in spring wheat (Sehgal et al. 2020). Similarly, Odilbekov et al. (2019) demonstrated the ability of GWAS-assisted GS for predicting resistance to Septoria tritici blotch in winter wheat. They showed that prediction accuracy increased from 47 to 62% with the inclusion of all significant QTLs in the GS model. The GWAS-assisted GS has demonstrated significant results for traits controlled by a smaller number of QTLs; however, for grain yield, significant improvement has not been observed.
Fig. 6.
Steps are depicted for GWAS assisted genomic selection. The results from GWA studies (association population) are used as fixed effects in the genomic selection pipeline. Genomic selection models are trained on previous data sets and significant QTLs are included as fixed effects during the prediction of breeding values. Concept for development of this figure is taken from (Crossa et al. 2017; Sehgal et al. 2020b; Sandhu et al. 2021c)
Mixed models used in GS take only the additive genetic effects into account completely ignoring the dominance, epistatic, and environmental variances (Crossa et al. 2019). With the rapid adoption of machine learning (ML) and deep learning (DL) approaches in other disciplines, there is also a need for these highly efficient approaches for conducting GS in wheat breeding. ML and DL models are flexible in regard to modelling the large and small effect QTLs in the GS model, and hence, these models have completely overcome the need for separately including the GWAS-assisted fixed effects in the GS models. ML and DL models have shown their superiority for predicting grain length (Ma et al. 2018b), grain yield (Sandhu et al. 2021c), and rust resistance (González-Camacho et al. 2018) in wheat. These models remove the assumptions of traditional GS models during training of the models due to the use of nonlinear activation functions (Bellot et al. 2018; Sandhu et al. 2021c). Commonly used models are random forest, reproducing kernel Hilbert space, support vector machine, multilayer perceptron, convolutional neural network, and recurrent neural network. This review opens up the avenue where we can shift from GWAS to the GS using ML and DL models for making the best selection, and thus increasing the genetic gain in crop plants for complex quantitative traits.
Transcriptome-wide association studies (TWAS) and probabilistic TWAS (PTWAS)
GWAS is performed in humans, animals, and plants to associate the various traits to genomic loci (MacArthur et al. 2017). Most GWAS loci lie in the intronic region of the genome; therefore, information about casual genes for the gene-trait association is largely lacking. GWAS has failed to determine the exact causal genes that have a major effect on the trait variant and causal genomic loci that drive the association (Gallagher and Chen-Plotkin 2018). This limitation has led to the development of new methods to prioritize causal genes at GWAS loci. Transcriptome-wide association study (TWAS) is one such method, which uses gene expression data to determine gene-complex trait association and prioritizes likely causal genes at GWAS loci (Gamazon et al. 2015). Transcriptome-wide association study follows a three-step procedure: firstly, it uses expression panels to train the simulation models for expression prediction from genotype; secondly, these models are used to predict an individual’s expression in the GWAS cohort; and the final step involves the estimation of a statistical association between predicted gene expression and phenotypic traits. Transcriptome-wide association study could be performed with individual data and summary of GWAS data using PrediXcan (Gamazon et al. 2015) and Fusion (Gusev et al. 2016) or S-PrediXcan (Barbeira et al. 2018), respectively.
However, it is also found that TWAS makes false prioritization with expression panels from non-related tissues. TWAS’s Fusion platform performs better in prioritizing genes at loci than two simple baselines, i.e. random per locus ranking and expression ranking (Wainberg et al. 2019). But TWAS is challenged by two factors, i.e. tissue biasness and co-regulation. Tissue biases can be reduced by using mechanistically most related tissue. If tissue is too small to get a sufficient sample size, then other related tissues can be taken to increase the sample size. Co-regulation can be addressed by using TWAS fine mapping. But TWAS fine mapping is more challenging to perform than GWAS fine mapping. Therefore, there is a need for more computational methods along with TWAS to make it perform better in gene prioritization at GWAS loci. The other two limitations of TWAS are that (i) it does not validate the causal implications of association and (ii) it lacks estimation of the causal effect of gene-trait associations (Zhang et al. 2020b). Probabilistic transcriptome genome-wide association studies (PTWAS) address these limitations of TWAS analysis by testing relationships between causal genes and complex traits and allow validation of causal implication and estimation of the causal effect.
Probabilistic transcriptome genome-wide association studies use instrumental variance analysis and probabilistic eQTLs annotations to estimate the causal relationship between causal gene expression and phenotypic traits (Zhang et al. 2020a, 2020b). It is more powerful than other existing methods as it provides causal implications and estimates tissue-specific genes to trait effect using multi-tissue eQTL data for analysis. The phenome-wide association study (PheWAS) is a high-throughput tool that determines the association between the genotypic variation and phenotype of the organism to get a better understanding of the effect of genotype. GWAS determines genotype-phenotype association by linking a number of genotypic variants like SNPs to a phenotypic trait or disease, whereas PheWAS studies the link of genotypic variation to a number of phenotypic traits. GWAS focuses on the study of a single target phenotype over a number of genotypes (maybe up to 500,000 SNPs) and PheWAS studies of single target genotype to a number of phenotypes (up to 1,000). Phenome-wide association study was recently used in the field of medicines to identify the association of genetic loci with many diseases. However, the application of PheWAS in the field of plant science is not explored yet.
Conclusion and future perspectives
With the arrival of high-throughput next-generation sequencing technologies and the development of various efficient statistical models, GWAS has become a method of choice for the genetic dissection of complex quantitative traits in many crops including wheat. The information generated in various GWA studies reporting 46,940 loci is apparently for great use in breeding and may form the base of meta-GWAS analysis, while actual utilization of these is not apparent and we are yet to see them being transferred from publications to actual varieties. Using GWAS, the genetic architecture of several different agronomic, physiological, and quality traits has been widely investigated and thousands of MTAs or causal SNPs have been revealed for these studied traits in wheat (Tables 3, 4, 5, and 6). These identified causal SNPs or MTAs have largely allowed the identification of candidate genes for different complex traits (Tables 3, 4, 5, and 6). The use of these identified significant MTAs as fixed effects in the genomic prediction models has also resulted in the increased prediction accuracy of GS for various traits in wheat (Sehgal et al. 2020) which indicates that these highly significant and robust genomic regions identified via GWAS can largely improve the utility of GS in future wheat breeding programs.
Almost 14 years have passed since the first paper of association study in wheat was published (Breseghello and Sorrells 2006), but still, GWAS faces some challenges which need to be addressed carefully to exploit this important approach. These challenges or limitations include false discovery rate (FDR), ‘large p small n problem’, markers with rare genetic variants and rare alleles, family-wise error rate (FWER), and reproducibility of identified loci. These issues have been discussed elsewhere and solutions have also been sought to manage these issues/concerns (Gupta et al. 2019a). Moreover, epistatic interactions and G × E interactions have largely been ignored in wheat GWAS, although these genetic interactions have been demonstrated to be important for complex quantitative traits (Sehgal et al. 2017). Improved statistical models/methods and the experimental designs for dissecting these genetic interactions need to be explored in the future. Furthermore, for a detailed understanding of the underlying molecular mechanisms of genotype-phenotype relationships, causative genes along with other causative sequence variants need to be identified. Also, we do not have enough knowledge of the potential effects of sequence variants on untranslated regions (UTRs) and promoter regions. Integrated use of multi-omics data can also help in getting insights into these molecular mechanisms. Various modifications of GWAS like eGWAS, PWAS, and TWAS have emerged but these are still relatively new concepts to the wheat breeders/geneticists. A new method, meta-GWAS, has recently emerged that can enable more robust and significant genomic regions associated with the target traits. Nevertheless, a few meta-GWA studies have been used within a wheat breeding program to reveal associated genomic regions and directly implement genomics-assisted breeding. Moreover, with the increasing interest in the ML and DL techniques, the analysis of multi-dimensional data will become much easier soon (Sandhu et al. 2021a; c). With these advancements, it will be possible to develop the networks that might be involved in the expression of target phenotypes of the complex traits. We believe that these efforts will greatly facilitate molecular breeding in wheat.
Availability of data and materials
Not applicable.
Author contribution
DKS, YC, JS, KSS, and AK: Writing-original draft, conceptualized the idea and collaborate; SB and PS: Writing-review and editing; PS: Supervised and monitored the study.
Declarations
Ethics approval and consent to participate
The study was conducted following all ethical guidelines.
Consent for publication
On behalf of all authors, corresponding author provides the consent for publication.
Competing interest
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
All authors contributed equally to this work.
References
- Abe F, Haque E, Hisano H, et al. Genome-edited triple-recessive mutation alters seed dormancy in wheat. Cell Rep. 2019;28:1362–1369.e4. doi: 10.1016/j.celrep.2019.06.090. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cardon LR, Cookson WOC. A general test of association for quantitative traits in nuclear families. Am J Hum Genet. 2000;66:279–292. doi: 10.1086/302698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alahmad S, El Hassouni K, Bassi FM, et al. A major root architecture QTL responding to water limitation in durum wheat. Front Plant Sci. 2019;10:436. doi: 10.3389/fpls.2019.00436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali M, Zhang Y, Rasheed A, et al. Genomic prediction for grain yield and yield-related traits in Chinese winter wheat. Int J Mol Sci. 2020;21:1342. doi: 10.3390/ijms21041342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alipour H, Bai G, Zhang G, et al. Imputation accuracy of wheat genotyping-by-sequencing (GBS) data using barley and wheat genome references. PLoS One. 2019;14:e0208614. doi: 10.1371/journal.pone.0208614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alomari DZ, Eggert K, von Wirén N, et al. Genome-wide association study of calcium accumulation in grains of European wheat cultivars. Front Plant Sci. 2017;8:1797. doi: 10.3389/fpls.2017.01797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alomari D, Eggert K, von Wirén N, et al. Whole-genome association mapping and genomic prediction for iron concentration in wheat grains. Int J Mol Sci. 2018;20:76. doi: 10.3390/ijms20010076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alomari DZ, Eggert K, von Wirén N, et al. Identifying Candidate Genes for Enhancing Grain Zn Concentration in Wheat. Front Plant Sci. 2018;9:1313. doi: 10.3389/fpls.2018.01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alqudah AM, Haile JK, Alomari DZ, et al. Genome-wide and SNP network analyses reveal genetic control of spikelet sterility and yield-related traits in wheat. Sci Rep. 2020;10:1–12. doi: 10.1038/s41598-020-59004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Rynearson S, Muleta KT, et al (2018) Genome-wide associations for multiple pest resistances in a Northwestern United States elite spring wheat panel. PLoS One 13. 10.1371/journal.pone.0191305 [DOI] [PMC free article] [PubMed]
- Anuarbek S, Abugalieva S, Pecchioni N, et al. Quantitative trait loci for agronomic traits in tetraploid wheat for enhancing grain yield in Kazakhstan environments. PLoS One. 2020;15:e0234863. doi: 10.1371/journal.pone.0234863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoun M, Breiland M, Kathryn Turner M, et al (2016) Genome-wide association mapping of leaf rust response in a durum wheat worldwide germplasm collection. Plant Genome 9:plantgenome2016.01.0008. 10.3835/plantgenome2016.01.0008 [DOI] [PubMed]
- Appels R, Eversole K, Feuillet C, et al (2018) Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (80-) 361. 10.1126/science.aar7191 [DOI] [PubMed]
- Arora S, Singh N, Kaur S, et al. Genome-wide association study of grain architecture in wild wheat aegilops tauschii. Front Plant Sci. 2017;8:1–13. doi: 10.3389/fpls.2017.00886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arruda MP, Brown P, Brown-Guedira G, et al (2016) Genome-wide association mapping of fusarium head blight resistance in wheat using genotyping-by-sequencing. Plant Genome 9:plantgenome2015.04.0028. 10.3835/plantgenome2015.04.0028 [DOI] [PubMed]
- Ayalew H, Liu H, Börner A, et al. Genome-Wide Association Mapping of Major Root Length QTLs Under PEG Induced Water Stress in Wheat. Front Plant Sci. 2018;9:1759. doi: 10.3389/fpls.2018.01759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajgain P, Rouse MN, Bulli P, et al. Association mapping of North American spring wheat breeding germplasm reveals loci conferring resistance to Ug99 and other African stem rust races. BMC Plant Biol. 2015;15:1–19. doi: 10.1186/s12870-015-0628-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbeira AN, Dickinson SP, Bonazzola R, et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat Commun. 2018;9:1–20. doi: 10.1038/s41467-018-03621-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battenfield SD, Sheridan JL, Silva LDCE, et al. Breeding-assisted genomics: Applying meta-GWAS for milling and baking quality in CIMMYT wheat breeding program. PLoS One. 2018;13:e0204757. doi: 10.1371/journal.pone.0204757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beavis WD (2019) QTL analyses: power, precision, and accuracy. Mol Dissection Complex Trait 145–162. 10.1201/9780429117770-10
- Beavis WD, Smith OS, Grant D, Fincher R. Identification of quantitative trait loci using a small sample of topcrossed and F4 progeny from maize. Crop Sci. 1994;34:882–896. doi: 10.2135/CROPSCI1994.0011183X003400040010X. [DOI] [Google Scholar]
- Bellot P, de los Campos G, Pérez-Enciso M. Can deep learning improve genomic prediction of complex human traits? Genetics. 2018;210:809–819. doi: 10.1534/genetics.118.301298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x. [DOI] [Google Scholar]
- Bergsträsser S, Fanourakis D, Schmittgen S, et al. HyperART: Non-invasive quantification of leaf traits using hyperspectral absorption-reflectance-transmittance imaging. Plant Methods. 2015;11:1–17. doi: 10.1186/s13007-015-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo R. Genomewide Selection when Major Genes Are Known. Crop Sci. 2014;54:68–75. doi: 10.2135/cropsci2013.05.0315. [DOI] [Google Scholar]
- Bernardo R. Bandwagons I, too, have known. Theor Appl Genet. 2016;129:2323–2332. doi: 10.1007/s00122-016-2772-5. [DOI] [PubMed] [Google Scholar]
- Beyer P, Morell M, Mackay I, Powell W. From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr Opin Plant Biol. 2008;11:215–221. doi: 10.1016/j.pbi.2008.01.002. [DOI] [PubMed] [Google Scholar]
- Beyer S, Daba S, Tyagi P, et al. Loci and candidate genes controlling root traits in wheat seedlings—a wheat root GWAS. Funct Integr Genomics. 2019;19:91–107. doi: 10.1007/s10142-018-0630-z. [DOI] [PubMed] [Google Scholar]
- Bhatta M, Morgounov A, Belamkar V, Baenziger P. Genome-wide association study reveals novel genomic regions for grain yield and yield-related traits in drought-stressed synthetic hexaploid wheat. Int J Mol Sci. 2018;19:3011. doi: 10.3390/ijms19103011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta M, Stephen Baenziger P, Waters BM, et al. Genome-wide association study reveals novel genomic regions associated with 10 grain minerals in synthetic hexaploid wheat. Int J Mol Sci. 2018;19:1–18. doi: 10.3390/ijms19103237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatta M, Morgounov A, Belamkar V, et al. Genome-wide association study for multiple biotic stress resistance in synthetic hexaploid wheat. Int J Mol Sci. 2019;20:3667. doi: 10.3390/ijms20153667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilgrami SS, Ramandi HD, Shariati V, et al. Detection of genomic regions associated with tiller number in Iranian bread wheat under different water regimes using genome-wide association study. Sci Rep. 2020;10:14034. doi: 10.1038/s41598-020-69442-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bin Safdar L, Andleeb T, Latif S, et al. Genome-Wide Association Study and QTL Meta-Analysis Identified Novel Genomic Loci Controlling Potassium Use Efficiency and Agronomic Traits in Bread Wheat. Front Plant Sci. 2020;11:70. doi: 10.3389/fpls.2020.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bocianowski J. Epistasis interaction of QTL effects as a genetic parameter influencing estimation of the genetic additive effect. Genet Mol Biol. 2013;36:093–100. doi: 10.1590/S1415-47572013000100013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeven PHG, Longin CFH, Leiser WL, et al. Genetic architecture of male floral traits required for hybrid wheat breeding. Theor Appl Genet. 2016;129:2343–2357. doi: 10.1007/s00122-016-2771-6. [DOI] [PubMed] [Google Scholar]
- Boichard D, Guillaume F, Baur A, et al. Genomic selection in French dairy cattle. Anim Prod Sci. 2012;52:115. doi: 10.1071/AN11119. [DOI] [Google Scholar]
- Botstein D, White RL, Skolnick M, Davis RW. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet. 1980;32:314–331. [PMC free article] [PubMed] [Google Scholar]
- Brandes N, Linial N, Linial M. PWAS: Proteome-wide association study - linking genes and phenotypes by functional variation in proteins. Genome Biol. 2020;21:1–22. doi: 10.1186/s13059-020-02089-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172:1165–1177. 10.1534/genetics.105.044586 [DOI] [PMC free article] [PubMed]
- Bulli P, Zhang J, Chao S, et al. Genetic architecture of resistance to stripe rust in a global winter wheat germplasm collection. G3 Genes, Genomes, Genet. 2016;6:2237–2253. doi: 10.1534/g3.116.028407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busemeyer L, Mentrup D, Möller K, et al. BreedVision — A Multi-Sensor Platform for Non-Destructive Field-Based Phenotyping in Plant Breeding. Sensors. 2013;13:2830–2847. doi: 10.3390/s130302830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh CR, Chao S, Wang S, et al. Genome-wide comparative diversity uncovers multiple targets of selection for improvement in hexaploid wheat landraces and cultivars. Proc Natl Acad Sci U S A. 2013;110:8057–8062. doi: 10.1073/pnas.1217133110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao S, Zhang W, Dubcovsky J, Sorrells M. Evaluation of genetic diversity and genome-wide linkage disequilibrium among U.S. wheat (Triticum aestivum L.) germplasm representing different market classes. Crop Sci. 2007;47:1018–1030. doi: 10.2135/cropsci2006.06.0434. [DOI] [Google Scholar]
- Chao S, Dubcovsky J, Dvorak J, et al (2010) Population- and genome-specific patterns of linkage disequilibrium and SNP variation in spring and winter wheat (Triticum aestivum L.). BMC Genomics 11:. 10.1186/1471-2164-11-727 [DOI] [PMC free article] [PubMed]
- Chen H, Xie W, He H, et al. A high-density SNP genotyping array for rice biology and molecular breeding. Mol Plant. 2014;7:541–553. doi: 10.1093/mp/sst135. [DOI] [PubMed] [Google Scholar]
- Chen G, Zhang H, Deng Z, et al. Genome-wide association study for kernel weight-related traits using SNPs in a Chinese winter wheat population. Euphytica. 2016;212:173–185. doi: 10.1007/s10681-016-1750-y. [DOI] [Google Scholar]
- Chen GF, Wu RG, Li DM, et al. Genomewide association study for seeding emergence and tiller number using SNP markers in an elite winter wheat population. J Genet. 2017;96:177–186. doi: 10.1007/s12041-016-0731-1. [DOI] [PubMed] [Google Scholar]
- Chen J, Zhang F, Zhao C, et al. Genome-wide association study of six quality traits reveals the association of the TaRPP13L1 gene with flour colour in Chinese bread wheat. Plant Biotechnol J. 2019;17:2106–2122. doi: 10.1111/pbi.13126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Fang W, Ji M, et al. Genome-wide association study of total starch and its components in common wheat. Euphytica. 2019;215:1–13. doi: 10.1007/s10681-019-2517-z. [DOI] [Google Scholar]
- Chen J, Hu X, Shi T, et al. Metabolite-based genome-wide association study enables dissection of the flavonoid decoration pathway of wheat kernels. Plant Biotechnol J. 2020;18:1722–1735. doi: 10.1111/pbi.13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Cheng X, Yu K, et al. Genome-wide association study of differences in 14 agronomic traits under low- and high-density planting models based on the 660k SNP array for common wheat. Plant Breed. 2020;139:272–283. doi: 10.1111/pbr.12774. [DOI] [Google Scholar]
- Cheng Y, Li J, Yao F, et al. Dissection of loci conferring resistance to stripe rust in Chinese wheat landraces from the middle and lower reaches of the Yangtze River via genome-wide association study. Plant Sci. 2019;287:110204. doi: 10.1016/j.plantsci.2019.110204. [DOI] [PubMed] [Google Scholar]
- Cheng B, Gao X, Cao N, et al. Genome-wide association analysis of stripe rust resistance loci in wheat accessions from southwestern China. J Appl Genet. 2020;61:37–50. doi: 10.1007/s13353-019-00533-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhury S, Larkin P, Xu R, et al. Genome wide association study reveals novel QTL for barley yellow dwarf virus resistance in wheat. BMC Genomics. 2019;20:891. doi: 10.1186/s12864-019-6249-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu J, Zhao Y, Beier S, et al. Suitability of single-nucleotide polymorphism arrays versus genotyping-by-sequencing for Genebank genomics in wheat. Front Plant Sci. 2020;0:42. doi: 10.3389/FPLS.2020.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138: [DOI] [PMC free article] [PubMed]
- Clevenger JP, Korani W, Ozias-Akins P, Jackson S. Haplotype-based genotyping in polyploids. Front Plant Sci. 2018;0:564. doi: 10.3389/FPLS.2018.00564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colasuonno P, Lozito ML, Marcotuli I, et al. The carotenoid biosynthetic and catabolic genes in wheat and their association with yellow pigments. BMC Genomics. 2017;18:1–18. doi: 10.1186/s12864-016-3395-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli GE, Maccaferri M, Newcomb M, et al. Comparative aerial and ground based high throughput phenotyping for the genetic dissection of NDVI as a proxy for drought adaptive traits in durum wheat. Front Plant Sci. 2018;9:893. doi: 10.3389/fpls.2018.00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cormier F, Le Gouis J, Dubreuil P, et al. A genome-wide identification of chromosomal regions determining nitrogen use efficiency components in wheat (Triticum aestivum L.) Theor Appl Genet. 2014;127:2679–2693. doi: 10.1007/s00122-014-2407-7. [DOI] [PubMed] [Google Scholar]
- Cortés LAG, Austerlitz F, de Cara MÁR (2021) A method to estimate effective population size from linkage disequilibrium when generations overlap. bioRxiv. 10.1101/2021.02.17.431658
- Crain JL, Wei Y, Barker J, et al. Development and deployment of a portable field phenotyping platform. Crop Sci. 2016;56:965–975. doi: 10.2135/cropsci2015.05.0290. [DOI] [Google Scholar]
- Crain J, Mondal S, Rutkoski J, et al. Combining high-throughput phenotyping and genomic information to increase prediction and selection accuracy in wheat breeding. Plant Genome. 2018;11:1–14. doi: 10.3835/plantgenome2017.05.0043. [DOI] [PubMed] [Google Scholar]
- Crossa J, Burgueño J, Dreisigacker S, et al. Association analysis of historical bread wheat germplasm using additive genetic covariance of relatives and population structure. Genetics. 2007;177:1889–1913. doi: 10.1534/genetics.107.078659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crossa J, Pérez-Rodríguez P, Cuevas J, et al. Genomic Selection in Plant Breeding: Methods, Models, and Perspectives. Trends Plant Sci. 2017;22:961–975. doi: 10.1016/j.tplants.2017.08.011. [DOI] [PubMed] [Google Scholar]
- Crossa J, Martini JWR, Gianola D, et al. Deep kernel and deep learning for genome-based prediction of single traits in multienvironment breeding trials. Front Genet. 2019;10:1–13. doi: 10.3389/fgene.2019.01168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cu ST, Guild G, Nicolson A, et al. Genetic dissection of zinc, iron, copper, manganese and phosphorus in wheat (Triticum aestivum L.) grain and rachis at two developmental stages. Plant Sci. 2020;291:110338. doi: 10.1016/j.plantsci.2019.110338. [DOI] [PubMed] [Google Scholar]
- Curtin SJ, Tiffin P, Guhlin J, et al. Validating genome-wide association candidates controlling quantitative variation in nodulation. Plant Physiol. 2017;173:921–931. doi: 10.1104/PP.16.01923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Y. Multi-allelic haplotype model based on genetic partition for genomic prediction and variance component estimation using SNP markers. BMC Genet. 2015;161(16):1–12. doi: 10.1186/S12863-015-0301-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba SD, Tyagi P, Brown-Guedira G, Mohammadi M. Genome-wide association studies to identify loci and candidate genes controlling kernel weight and length in a historical United States wheat population. Front Plant Sci. 2018;9:1045. doi: 10.3389/fpls.2018.01045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Vita P, Avio L, Sbrana C, et al. Genetic markers associated to arbuscular mycorrhizal colonization in durum wheat. Sci Rep. 2018;8:1–12. doi: 10.1038/s41598-018-29020-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize E, Rathjen TM, Cavanagh CR. The genetics of rhizosheath size in a multiparent mapping population of wheat. J Exp Bot. 2015;66:4527–4536. doi: 10.1093/jxb/erv223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny JC, Ritchie MD, Basford MA, et al. PheWAS: demonstrating the feasibility of a phenome-wide scan to discover gene-disease associations. Bioinformatics. 2010;26:1205–1210. doi: 10.1093/bioinformatics/btq126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deshmukh R, Sonah H, Patil G, et al. Integrating omic approaches for abiotic stress tolerance in soybean. Front Plant Sci. 2014;5:244. doi: 10.3389/fpls.2014.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devlin B, Risch N. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 1995;29:311–322. doi: 10.1006/geno.1995.9003. [DOI] [PubMed] [Google Scholar]
- Dhakal S, Tan CT, Anderson V, et al. Mapping and KASP marker development for wheat curl mite resistance in “TAM 112” wheat using linkage and association analysis. Mol Breed. 2018;38:1–13. doi: 10.1007/s11032-018-0879-x. [DOI] [Google Scholar]
- Dinglasan EG, Singh D, Shankar M, et al. Discovering new alleles for yellow spot resistance in the Vavilov wheat collection. Theor Appl Genet. 2019;132:149–162. doi: 10.1007/s00122-018-3204-5. [DOI] [PubMed] [Google Scholar]
- Dong Y, Liu J, Zhang Y, et al. Genome-wide association of stem water soluble carbohydrates in bread wheat. PLoS One. 2016;11:e0164293. doi: 10.1371/journal.pone.0164293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie RC, Bouvet L, Furuki E, et al. Assessing European wheat sensitivities to Parastagonospora nodorum necrotrophic effectors and fine-mapping the Snn3-B1 locus conferring sensitivity to the effector SnTox3. Front Plant Sci. 2018;9:881. doi: 10.3389/fpls.2018.00881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 2008;32:227–234. doi: 10.1002/gepi.20297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edae EA, Byrne PF, Haley SD, et al. Genome-wide association mapping of yield and yield components of spring wheat under contrasting moisture regimes. Theor Appl Genet. 2014;127:791–807. doi: 10.1007/s00122-013-2257-8. [DOI] [PubMed] [Google Scholar]
- Elbasyoni I, El-Orabey W, Baenziger P, Eskridge K. Association mapping for leaf and stem rust resistance using worldwide spring wheat collection. Asian J Biol. 2017;4:1–25. doi: 10.9734/ajob/2017/38120. [DOI] [Google Scholar]
- Elbasyoni I, Morsy S, Ramamurthy R, Nassar A. Identification of Genomic Regions Contributing to Protein Accumulation in Wheat under Well-Watered and Water Deficit Growth Conditions. Plants. 2018;7:56. doi: 10.3390/plants7030056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elbasyoni IS, El-Orabey WM, Morsy S, et al. Evaluation of a global spring wheat panel for stripe rust: Resistance loci validation and novel resources identification. PLoS One. 2019;14:e0222755. doi: 10.1371/journal.pone.0222755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, et al. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One. 2011;6:1–10. doi: 10.1371/journal.pone.0019379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emebiri L, Singh S, Tan M-K, et al. Unravelling the complex genetics of karnal bunt (Tilletia indica) resistance in common wheat (Triticum aestivum) by genetic linkage and genome-wide association analyses. G3 Genes|Genomes|Genetics. 2019;9:1437–1447. doi: 10.1534/g3.119.400103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiedler JD, Salsman E, Liu Y et al (2017) Genome-wide association and prediction of grain and semolina quality traits in durum wheat breeding populations. Plant Genome 10. 10.3835/plantgenome2017.05.0038 [DOI] [PubMed]
- Fletcher A, Kelly A, Christopher J, et al (2019) Using high-throughput phenotyping and genome wide association study ( GWAS ) techniques to identify molecular markers for transpiration efficiency in wheat. 16:25–29
- Flood PJ, Kruijer W, Schnabel SK, et al. Phenomics for photosynthesis, growth and reflectance in Arabidopsis thaliana reveals circadian and long-term fluctuations in heritability. Plant Methods. 2016;12:14. doi: 10.1186/s13007-016-0113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francki MG, Walker E, McMullan CJ, Morris WG. Multi-location evaluation of global wheat lines reveal multiple QTL for adult plant resistance to Septoria nodorum blotch (SNB) detected in specific environments and in response to different isolates. Front Plant Sci. 2020;11:771. doi: 10.3389/fpls.2020.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Froese PS, Murray TD, Carter AH. Quantitative Cephalosporium stripe disease resistance mapped in the wheat genome. Crop Sci. 2016;56:1586–1601. doi: 10.2135/cropsci2015.09.0568. [DOI] [Google Scholar]
- Fu L, Wu J, Yang S, et al. Genome-wide association analysis of stem water-soluble carbohydrate content in bread wheat. Theor Appl Genet. 2020;133:2897–2914. doi: 10.1007/s00122-020-03640-x. [DOI] [PubMed] [Google Scholar]
- Gahlaut V, Jaiswal V, Singh S, et al. Multi-locus genome wide association mapping for yield and its contributing traits in hexaploid wheat under different water regimes. Sci Rep. 2019;9:1–15. doi: 10.1038/s41598-019-55520-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaire R, Huang M, Sneller C, et al. Association analysis of baking and milling quality traits in an elite soft red winter wheat population. Crop Sci. 2019;59:1085–1094. doi: 10.2135/cropsci2018.12.0751. [DOI] [Google Scholar]
- Gaire R, Ohm H, Brown-Guedira G, Mohammadi M (2020) Identification of regions under selection and loci controlling agronomic traits in a soft red winter wheat population. Plant Genome 13. 10.1002/tpg2.20031 [DOI] [PubMed]
- Galagedara N, Liu Y, Fiedler J, et al. Genome-wide association mapping of tan spot resistance in a worldwide collection of durum wheat. Theor Appl Genet. 2020;133:2227–2237. doi: 10.1007/s00122-020-03593-1. [DOI] [PubMed] [Google Scholar]
- Gallagher MD, Chen-Plotkin AS. The Post-GWAS Era: From Association to Function. Am J Hum Genet. 2018;102:717–730. doi: 10.1016/j.ajhg.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamazon ER, Wheeler HE, Shah KP, et al. A gene-based association method for mapping traits using reference transcriptome data. Nat Genet. 2015;47:1091–1098. doi: 10.1038/ng.3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Turner MK, Chao S, et al. Genome wide association study of seedling and adult plant leaf rust resistance in elite spring wheat breeding lines. PLoS One. 2016;11:e0148671. doi: 10.1371/journal.pone.0148671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao L, Rouse MN, Mihalyov PD, et al. Genetic characterization of stem rust resistance in a global spring wheat germplasm collection. Crop Sci. 2017;57:2575–2589. doi: 10.2135/cropsci2017.03.0159. [DOI] [Google Scholar]
- Garcia M, Eckermann P, Haefele S, et al. Genome-wide association mapping of grain yield in a diverse collection of spring wheat (Triticum aestivum L.) evaluated in southern Australia. PLoS One. 2019;14:e0211730. doi: 10.1371/journal.pone.0211730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner L-J, Bansept-Basler P, El-Soda M, et al. A framework for gene mapping in wheat demonstrated using the Yr7 yellow rust resistance gene. PLoS One. 2020;15:e0231157. doi: 10.1371/journal.pone.0231157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehan MA, Kellogg EA. High-throughput phenotyping. Am J Bot. 2017;104:505–508. doi: 10.3732/ajb.1700044. [DOI] [PubMed] [Google Scholar]
- Godoy J, Gizaw S, Chao S, et al. Genome-wide association study of agronomic traits in a spring-planted North American elite hard red spring wheat panel. Crop Sci. 2018;58:1838–1852. doi: 10.2135/cropsci2017.07.0423. [DOI] [Google Scholar]
- Godoy JG, Rynearson S, Chen X, Pumphrey M. Genome-wide association mapping of loci for resistance to stripe rust in North American elite spring wheat germplasm. Phytopathology®. 2018;108:234–245. doi: 10.1094/PHYTO-06-17-0195-R. [DOI] [PubMed] [Google Scholar]
- González-Camacho JM, Ornella L, Pérez-Rodríguez P, et al. Applications of Machine Learning Methods to Genomic Selection in Breeding Wheat for Rust Resistance. Plant Genome. 2018;11:170104. doi: 10.3835/plantgenome2017.11.0104. [DOI] [PubMed] [Google Scholar]
- Gordon T, Wang R, Hole D, et al. Genetic characterization and genome-wide association mapping for dwarf bunt resistance in bread wheat accessions from the USDA National Small Grains Collection. Theor Appl Genet. 2020;133:1069–1080. doi: 10.1007/s00122-020-03532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosseau F, Blanchet N, Varès D, et al. Heliaphen, an outdoor high-throughput phenotyping platform for genetic studies and crop modeling. Front Plant Sci. 2019;9:1908. doi: 10.3389/fpls.2018.01908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granier C, Aguirrezabal L, Chenu K, et al. PHENOPSIS, an automated platform for reproducible phenotyping of plant responses to soil water deficit in Arabidopsis thaliana permitted the identification of an accession with low sensitivity to soil water deficit. New Phytol. 2006;169:623–635. doi: 10.1111/j.1469-8137.2005.01609.x. [DOI] [PubMed] [Google Scholar]
- Greenham K, Lou P, Remsen SE, et al. TRiP: Tracking Rhythms in Plants, an automated leaf movement analysis program for circadian period estimation. Plant Methods. 2015;11:1–11. doi: 10.1186/s13007-015-0075-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujjula KR, Baselgia L, Wall J, et al (2019) The hallmark of AgriSeq TM technology: highly reproducible genotype calls and identification of novel genotypes. In Plant and Animal Genome XXVII Conference (January 12-16, 2019). PAG
- Guo Z, Chen D, Alqudah AM, et al. Genome-wide association analyses of 54 traits identified multiple loci for the determination of floret fertility in wheat. New Phytol. 2017;214:257–270. doi: 10.1111/nph.14342. [DOI] [PubMed] [Google Scholar]
- Guo J, Shi W, Zhang Z, et al. Association of yield-related traits in founder genotypes and derivatives of common wheat (Triticum aestivum L.) BMC Plant Biol. 2018;18:1–15. doi: 10.1186/s12870-018-1234-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Liu G, Röder MS, et al. Genome-wide association analyses of plant growth traits during the stem elongation phase in wheat. Plant Biotechnol J. 2018;16:2042–2052. doi: 10.1111/pbi.12937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta PK, Rustgi S, Kulwal PL. Linkage disequilibrium and association studies in higher plants: Present status and future prospects. Plant Mol Biol. 2005;57:461–485. doi: 10.1007/s11103-005-0257-z. [DOI] [PubMed] [Google Scholar]
- Gupta PK, Kulwal PL, Jaiswal V (2014) Association mapping in crop plants: opportunities and challenges. In: Advances in Genetics. Academic Press Inc., pp 109–147 [DOI] [PubMed]
- Gupta PK, Kulwal PL, Jaiswal V (2019a) Association mapping in plants in the post-GWAS genomics era. In: Advances in Genetics. Academic Press Inc., pp 75–154 [DOI] [PubMed]
- Gupta V, He X, Kumar N, et al. Genome wide association study of karnal bunt resistance in a wheat germplasm collection from Afghanistan. Int J Mol Sci. 2019;20:3124. doi: 10.3390/ijms20133124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48:245–252. doi: 10.1038/ng.3506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttieri MJ, Stephen Baenziger P, Frels K, et al. Prospects for selecting wheat with increased zinc and decreased cadmium concentration in grain. Crop Sci. 2015;55:1712–1728. doi: 10.2135/cropsci2014.08.0559. [DOI] [Google Scholar]
- Halder J, Zhang J, Ali S, et al. Mining and genomic characterization of resistance to tan spot, Stagonospora nodorum blotch (SNB), and Fusarium head blight in Watkins core collection of wheat landraces. BMC Plant Biol. 2019;19:1–15. doi: 10.1186/s12870-019-2093-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes B. Overview of statistical methods for genome-wide association studies (GWAS) Totowa, NJ: Humana Press; 2013. pp. 149–169. [DOI] [PubMed] [Google Scholar]
- Herter CP, Ebmeyer E, Kollers S, et al. Accuracy of within- and among-family genomic prediction for Fusarium head blight and Septoria tritici blotch in winter wheat. Theor Appl Genet. 2019;132:1121–1135. doi: 10.1007/s00122-018-3264-6. [DOI] [PubMed] [Google Scholar]
- Hitz K, Clark AJ, Van Sanford DA. Identifying nitrogen-use efficient soft red winter wheat lines in high and low nitrogen environments. F Crop Res. 2017;200:1–9. doi: 10.1016/j.fcr.2016.10.001. [DOI] [Google Scholar]
- Hogers RC, de Ruiter M, Huvenaars KH, et al. SNPSelect: A scalable and flexible targeted sequence-based genotyping solution. PLoS One. 2018;13:e0205577. doi: 10.1371/journal.pone.0205577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu W, Gao D, Wu H, et al. Genome-wide association mapping revealed syntenic loci QFhb-4AL and QFhb-5DL for Fusarium head blight resistance in common wheat (Triticum aestivum L.) BMC Plant Biol. 2020;20:29. doi: 10.1186/s12870-019-2177-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Liu X, Zhou Y, et al. BLINK: A package for the next level of genome-wide association studies with both individuals and markers in the millions. Gigascience. 2018;8:1–12. doi: 10.1093/gigascience/giy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen M, Gilmer F, Biskup B, et al. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via GROWSCREEN FLUORO allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct Plant Biol. 2009;36:902. doi: 10.1071/FP09095. [DOI] [PubMed] [Google Scholar]
- Jernigan KL, Godoy JV, Huang M, et al. Genetic dissection of end-use quality traits in adapted soft white winter wheat. Front Plant Sci. 2018;9:1–15. doi: 10.3389/fpls.2018.00271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jighly A, Alagu M, Makdis F, et al. Genomic regions conferring resistance to multiple fungal pathogens in synthetic hexaploid wheat. Mol Breed. 2016;36:127. doi: 10.1007/s11032-016-0541-4. [DOI] [Google Scholar]
- Jin J, Duan S, Qi Y, et al. Identification of a novel genomic region associated with resistance to Fusarium crown rot in wheat. Theor Appl Genet. 2020;133:2063–2073. doi: 10.1007/s00122-020-03577-1. [DOI] [PubMed] [Google Scholar]
- Jordan KW, Wang S, He F, Chao S, et al. The genetic architecture of genome-wide recombination rate variation in allopolyploid wheat revealed by nested association mapping. Plant J. 2018;95:1039–1054. doi: 10.1111/tpj.14009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AK, Kumar U, Mishra VK, et al. Variations in straw fodder quality and grain–straw relationships in a mapping population of 287 diverse spring wheat lines. F Crop Res. 2019;243:107627. doi: 10.1016/j.fcr.2019.107627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouanin A, Gilissen LJWJ, Schaart JG, et al. CRISPR/Cas9 gene editing of gluten in wheat to reduce gluten content and exposure—reviewing methods to screen for coeliac safety. Front Nutr. 2020;7:51. doi: 10.3389/fnut.2020.00051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukhadar R, El-Bouhssini M, Jighly A, Ogbonnaya FC. Genome-wide association mapping for five major pest resistances in wheat. Mol Breed. 2013;32:943–960. doi: 10.1007/s11032-013-9924-y. [DOI] [Google Scholar]
- Joukhadar R, Hollaway G, Shi F, et al. Genome-wide association reveals a complex architecture for rust resistance in 2300 worldwide bread wheat accessions screened under various Australian conditions. Theor Appl Genet. 2020;133:2695–2712. doi: 10.1007/s00122-020-03626-9. [DOI] [PubMed] [Google Scholar]
- Juliana P, Singh RP, Singh PK, et al. Genome-wide association mapping for resistance to leaf rust, stripe rust and tan spot in wheat reveals potential candidate genes. Theor Appl Genet. 2018;131:1405–1422. doi: 10.1007/s00122-018-3086-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalia B, Bockus WW, Singh S, et al. Mapping of quantitative trait loci for resistance to race 1 of Pyrenophora tritici-repentis in synthetic hexaploid wheat. Plant Breed. 2018;137:313–319. doi: 10.1111/pbr.12586. [DOI] [Google Scholar]
- Kang Y, Barry K, Cao F, Zhou M. Genome-wide association mapping for adult resistance to powdery mildew in common wheat. Mol Biol Rep. 2020;47:1241–1256. doi: 10.1007/s11033-019-05225-4. [DOI] [PubMed] [Google Scholar]
- Kaur B, Sandhu KS, Kamal R, et al (2021) Omics for the improvement of abiotic, biotic and agronomic traits in major cereals: applications, challenges, and prospects. Plants [DOI] [PMC free article] [PubMed]
- Kidane YG, Gesesse CA, Hailemariam BN, et al. A large nested association mapping population for breeding and quantitative trait locus mapping in Ethiopian durum wheat. Plant Biotechnol J. 2019;17:1380–1393. doi: 10.1111/pbi.13062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SW, Kim HJ, Kim JH, et al. A rapid, simple method for the genetic discrimination of intact Arabidopsis thaliana mutant seeds using metabolic profiling by direct analysis in real-time mass spectrometry. Plant Methods. 2011;71(7):1–10. doi: 10.1186/1746-4811-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva AA, Leonova IN, Pshenichnikova TA, Salina EA. Dissection of novel candidate genes for grain texture in Russian wheat varieties. Plant Mol Biol. 2020;104:219–233. doi: 10.1007/s11103-020-01025-8. [DOI] [PubMed] [Google Scholar]
- Korte A, Ashley F. The advantages and limitations of trait analysis with GWAS : a review. Plant Methods. 2013;9:29. doi: 10.1186/1746-4811-9-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korte A, Vilhjálmsson BJ, Segura V, et al. A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet. 2012;44:1066–1071. doi: 10.1038/ng.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar J, Saripalli G, Gahlaut V, et al. Genetics of Fe, Zn, β-carotene, GPC and yield traits in bread wheat (Triticum aestivum L.) using multi-locus and multi-traits GWAS. Euphytica. 2018;214:1–17. doi: 10.1007/s10681-018-2284-2. [DOI] [Google Scholar]
- Kumar D, Kumar A, Chhokar V, et al. Genome-wide association studies in diverse spring wheat panel for stripe, stem, and leaf rust resistance. Front Plant Sci. 2020;11:748. doi: 10.3389/fpls.2020.00748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance J, Tishkoff SA. SNP ascertainment bias in population genetic analyses: Why it is important, and how to correct it. BioEssays. 2013;35:780–786. doi: 10.1002/BIES.201300014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121: [DOI] [PMC free article] [PubMed]
- Langer SM, Longin CFH, Würschum T (2014) Flowering time control in European winter wheat. Front Plant Sci 5:537. 10.3389/fpls.2014.00537 [DOI] [PMC free article] [PubMed]
- Larkin DL, Holder AL, Mason RE, et al. Genome-wide analysis and prediction of Fusarium head blight resistance in soft red winter wheat. Crop Sci. 2020;60:2882–2900. doi: 10.1002/csc2.20273. [DOI] [Google Scholar]
- Ledesma-Ramírez L, Solís-Moya E, Iturriaga G, et al. GWAS to Identify Genetic Loci for Resistance to Yellow Rust in Wheat Pre-Breeding Lines Derived From Diverse Exotic Crosses. Front Plant Sci. 2019;10:1390. doi: 10.3389/fpls.2019.01390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehnert H, Serfling A, Enders M, et al. Genetics of mycorrhizal symbiosis in winter wheat ( Triticum aestivum ) New Phytol. 2017;215:779–791. doi: 10.1111/nph.14595. [DOI] [PubMed] [Google Scholar]
- Lehnert H, Serfling A, Friedt W, Ordon F. Genome-wide association studies reveal genomic regions associated with the response of wheat (Triticum aestivum L.) to mycorrhizae under drought stress conditions. Front Plant Sci. 2018;871:1728. doi: 10.3389/fpls.2018.01728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonova IN, Skolotneva ES, Orlova EA, et al. Detection of genomic regions associated with resistance to stem rust in Russian spring wheat varieties and breeding germplasm. Int J Mol Sci. 2020;21:4706. doi: 10.3390/ijms21134706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewien MJ, Murray TD, Jernigan KL, et al. Genome-wide association mapping for eyespot disease in US Pacific Northwest winter wheat. PLoS One. 2018;13:1–19. doi: 10.1371/journal.pone.0194698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xu X, Bai G, et al (2016) Genome-wide association mapping reveals novel QTL for seedling leaf rust resistance in a worldwide collection of winter wheat. Plant Genome 9:plantgenome2016.06.0051. 10.3835/plantgenome2016.06.0051 [DOI] [PubMed]
- Li G, Bai G, Carver BF, et al. Genome-wide association study reveals genetic architecture of coleoptile length in wheat. Theor Appl Genet. 2017;130:391–401. doi: 10.1007/s00122-016-2820-1. [DOI] [PubMed] [Google Scholar]
- Li J, Rasheed A, Guo Q, et al. Genome-wide association mapping of starch granule size distribution in common wheat. J Cereal Sci. 2017;77:211–218. doi: 10.1016/j.jcs.2017.08.016. [DOI] [Google Scholar]
- Li J, Liu J, Wen W, et al. Genome-wide association mapping of vitamins B1 and B2 in common wheat. Crop J. 2018;6:263–270. doi: 10.1016/j.cj.2017.08.002. [DOI] [Google Scholar]
- Li F, Wen W, Liu J, et al. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019;19:168. doi: 10.1186/s12870-019-1781-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Xu X, Tan C, et al. Identification of powdery mildew resistance loci in wheat by integrating genome-wide association study (GWAS) and linkage mapping. Crop J. 2019;7:294–306. doi: 10.1016/j.cj.2019.01.005. [DOI] [Google Scholar]
- Li L, Mao X, Wang J, et al. Genetic dissection of drought and heat-responsive agronomic traits in wheat. Plant Cell Environ. 2019;42:2540–2553. doi: 10.1111/pce.13577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Peng Z, Mao X, et al. Genome-wide association study reveals genomic regions controlling root and shoot traits at late growth stages in wheat. Ann Bot. 2019;124:993–1006. doi: 10.1093/aob/mcz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Jiang Y, Yao F, et al. Genome-wide association study reveals the genetic architecture of stripe rust resistance at the adult plant stage in Chinese endemic wheat. Front Plant Sci. 2020;11:625. doi: 10.3389/fpls.2020.00625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Niu H, Xu K, et al. GWAS for resistance against black point caused by Bipolaris sorokiniana in wheat. J Cereal Sci. 2020;91:102859. doi: 10.1016/j.jcs.2019.102859. [DOI] [Google Scholar]
- Li X, Xu X, Liu W, et al. Dissection of superior alleles for yield-related traits and their distribution in important cultivars of wheat by association mapping. Front Plant Sci. 2020;11:14034. doi: 10.3389/fpls.2020.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liebisch F, Kirchgessner N, Schneider D, et al. Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods. 2015;11:9. doi: 10.1186/s13007-015-0048-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Zhang D, Liu S, et al. Genome-wide association analysis on pre-harvest sprouting resistance and grain color in U.S. winter wheat. BMC Genomics. 2016;17:1–16. doi: 10.1186/s12864-016-3148-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, et al. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet. 2016;12:e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, He Z, Rasheed A, et al. Genome-wide association mapping of black point reaction in common wheat (Triticum aestivum L.) BMC Plant Biol. 2017;17:220. doi: 10.1186/s12870-017-1167-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Maccaferri M, Bulli P, et al. Genome-wide association mapping for seedling and field resistance to Puccinia striiformis f. sp. tritici in elite durum wheat. Theor Appl Genet. 2017;130:649–667. doi: 10.1007/s00122-016-2841-9. [DOI] [PubMed] [Google Scholar]
- Liu W, Maccaferri M, Chen X, et al. Genome-wide association mapping reveals a rich genetic architecture of stripe rust resistance loci in emmer wheat (Triticum turgidum ssp. dicoccum) Theor Appl Genet. 2017;130:2249–2270. doi: 10.1007/s00122-017-2957-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Maccaferri M, Rynearson S, et al. Novel sources of stripe rust resistance identified by genome-wide association mapping in Ethiopian durum wheat (Triticum turgidum ssp. durum) Front Plant Sci. 2017;8:774. doi: 10.3389/fpls.2017.00774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Lin Y, Gao S, et al. A genome-wide association study of 23 agronomic traits in Chinese wheat landraces. Plant J. 2017;91:861–873. doi: 10.1111/tpj.13614. [DOI] [PubMed] [Google Scholar]
- Liu J, Feng B, Xu Z, et al. A genome-wide association study of wheat yield and quality-related traits in southwest China. Mol Breed. 2018;38:1–11. doi: 10.1007/s11032-017-0759-9. [DOI] [Google Scholar]
- Liu J, Xu Z, Fan X, et al. A genome-wide association study of wheat spike related traits in China. Front Plant Sci. 2018;871:1584. doi: 10.3389/fpls.2018.01584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Naruoka Y, Miller K, et al. Characterizing and validating stripe rust resistance loci in US Pacific Northwest winter wheat accessions (Triticum aestivum L.) by genome-wide association and linkage mapping. Plant Genome. 2018;11:170087. doi: 10.3835/plantgenome2017.10.0087. [DOI] [PubMed] [Google Scholar]
- Liu Y, Liu Y, Zhang Q, et al. Genome-wide association analysis of quantitative trait loci for salinity-tolerance related morphological indices in bread wheat. Euphytica. 2018;214:1–11. doi: 10.1007/s10681-018-2265-5. [DOI] [Google Scholar]
- Liu W, Kolmer J, Rynearson S, et al. Identifying loci conferring resistance to leaf and stripe rusts in a spring wheat population (Triticum aestivum) via genome-wide association mapping. Phytopathology®. 2019;109:1932–1940. doi: 10.1094/PHYTO-04-19-0143-R. [DOI] [PubMed] [Google Scholar]
- Liu Y, Salsman E, Fiedler JD, et al. Genetic mapping and prediction analysis of FHB resistance in a hard red spring wheat breeding population. Front Plant Sci. 2019;10:1007. doi: 10.3389/fpls.2019.01007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Jiang Y, Zhao Y, et al. Haplotype-based genome-wide association increases the predictability of leaf rust (Puccinia triticina) resistance in wheat. J Exp Bot. 2020;71:6958–6968. doi: 10.1093/JXB/ERAA387. [DOI] [PubMed] [Google Scholar]
- Liu L, Wang M, Zhang Z, et al. Identification of stripe rust resistance loci in U.S. spring wheat cultivars and breeding lines using genome-wide association mapping and Yr gene markers. Plant Dis. 2020;104:2181–2192. doi: 10.1094/PDIS-11-19-2402-RE. [DOI] [PubMed] [Google Scholar]
- Long L, Yao F, Yu C et al (2019) Genome-wide association study for adult-plant resistance to stripe rust in Chinese Wheat landraces (Triticum aestivum L.) From the Yellow and Huai River Valleys. Front Plant Sci 10(596). 10.3389/fpls.2019.00596 [DOI] [PMC free article] [PubMed]
- Lorenz AJ, Chao S, Asoro FG, et al (2011) Genomic selection in plant breeding : knowledge and prospects, 1st edn. Elsevier Inc.
- Lozada D, Godoy J V, Murray TD, et al (2019) Genetic dissection of snow mold tolerance in US Pacific Northwest winter wheat through genome-wide association study and genomic selection. 10:1–15. 10.3389/fpls.2019.01337 [DOI] [PMC free article] [PubMed]
- Lu Y, Shah T, Hao Z, et al. Comparative SNP and haplotype analysis reveals a higher genetic diversity and rapider LD decay in tropical than temperate germplasm in maize. PLoS One. 2011;6:e24861. doi: 10.1371/JOURNAL.PONE.0024861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luján Basile SM, Ramírez IA, Crescente JM, et al. Haplotype block analysis of an Argentinean hexaploid wheat collection and GWAS for yield components and adaptation. BMC Plant Biol. 2019;19:1–16. doi: 10.1186/s12870-019-2015-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. Metabolite-based genome-wide association studies in plants. Curr Opin Plant Biol. 2015;24:31–38. doi: 10.1016/j.pbi.2015.01.006. [DOI] [PubMed] [Google Scholar]
- Ma F, Xu Y, Ma Z, et al. Genome-wide association and validation of key loci for yield-related traits in wheat founder parent Xiaoyan 6. Mol Breed. 2018;38:1–15. doi: 10.1007/s11032-018-0837-7. [DOI] [Google Scholar]
- Ma W, Qiu Z, Song J, et al. A deep convolutional neural network approach for predicting phenotypes from genotypes. Planta. 2018;248:1307–1318. doi: 10.1007/s00425-018-2976-9. [DOI] [PubMed] [Google Scholar]
- Ma J, Lin Y, Tang S, et al. A genome-wide association study of coleoptile length in different Chinese wheat landraces. Front Plant Sci. 2020;11:677. doi: 10.3389/fpls.2020.00677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog) Nucleic Acids Res. 2017;45:D896–D901. doi: 10.1093/nar/gkw1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maccaferri M, Sanguineti MC, Noli E, Tuberosa R. Population structure and long-range linkage disequilibrium in a durum wheat elite collection. Mol Breed. 2005;15:271–290. doi: 10.1007/s11032-004-7012-z. [DOI] [Google Scholar]
- Maccaferri M, Zhang J, Bulli P, et al. A genome-wide association study of resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a worldwide collection of hexaploid spring wheat (Triticum aestivum L.) G3 Genes, Genomes, Genet. 2015;5:449–465. doi: 10.1534/g3.114.014563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay IJ, Bansept-Basler P, Bentley AR, et al. An eight-parent multiparent advanced generation inter-cross population for winter-sown wheat: Creation, properties, and validation. G3 Genes, Genomes, Genet. 2014;4:1603–1610. doi: 10.1534/g3.114.012963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik PL, Janss L, Nielsen LK, et al. Breeding for dual-purpose wheat varieties using marker–trait associations for biomass yield and quality traits. Theor Appl Genet. 2019;132:3375–3398. doi: 10.1007/s00122-019-03431-z. [DOI] [PubMed] [Google Scholar]
- Malosetti M, Zwep LB, Forrest K, et al. Lessons from a GWA study of a wheat pre-breeding program: pyramiding resistance alleles to Fusarium crown rot. Theor Appl Genet. 2020;1:3. doi: 10.1007/s00122-020-03740-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangini G, Gadaleta A, Colasuonno P, et al. Genetic dissection of the relationships between grain yield components by genome-wide association mapping in a collection of tetraploid wheats. PLoS One. 2018;13:e0190162. doi: 10.1371/journal.pone.0190162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcotuli I, Houston K, Schwerdt JG, et al. Genetic diversity and genome wide association study of β-glucan content in tetraploid wheat grains. PLoS One. 2016;11:e0152590. doi: 10.1371/journal.pone.0152590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez SA, Godoy J, Huang M, et al. Genome-wide association mapping for tolerance to preharvest sprouting and low falling numbers in wheat. Front Plant Sci. 2018;9:141. doi: 10.3389/fpls.2018.00141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maulana F, Ayalew H, Anderson JD, et al. Genome-wide association mapping of seedling heat tolerance in winter wheat. Front Plant Sci. 2018;9:1272. doi: 10.3389/fpls.2018.01272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellers G, Aguilera JG, Bird N, et al. Genetic characterization of a wheat association mapping panel relevant to Brazilian breeding using a high-density single nucleotide polymorphism array. G3 Genes|Genomes|Genetics. 2020;10:2229–2239. doi: 10.1534/g3.120.401234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuwissen THE, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157:1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir RR, Reynolds M, Pinto F, et al. High-throughput phenotyping for crop improvement in the genomics era. Plant Sci. 2019;282:60–72. doi: 10.1016/j.plantsci.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Moghimi A, Yang C, Miller ME, et al. A novel approach to assess salt stress tolerance in wheat using hyperspectral imaging. Front Plant Sci. 2018;9:1182. doi: 10.3389/fpls.2018.01182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohler V, Stadlmeier M. Dynamic QTL for adult plant resistance to powdery mildew in common wheat (Triticum aestivum L.) J Appl Genet. 2019;60:291–300. doi: 10.1007/s13353-019-00518-7. [DOI] [PubMed] [Google Scholar]
- Molero G, Joynson R, Pinera-Chavez FJ, et al. Elucidating the genetic basis of biomass accumulation and radiation use efficiency in spring wheat and its role in yield potential. Plant Biotechnol J. 2019;17:1276–1288. doi: 10.1111/pbi.13052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monostori I, Szira F, Tondelli A, et al. Genome-wide association study and genetic diversity analysis on nitrogen use efficiency in a Central European winter wheat (Triticum aestivum L.) collection. PLoS One. 2017;12:e0189265. doi: 10.1371/journal.pone.0189265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad AMI, Sallam A, Belamkar V, et al. Genome-wide association study for identification and validation of novel SNP markers for Sr6 stem rust resistance gene in bread wheat. Front Plant Sci. 2018;9:380. doi: 10.3389/fpls.2018.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourad AMI, Sallam A, Belamkar V, et al. Genetic architecture of common bunt resistance in winter wheat using genome-wide association study. BMC Plant Biol. 2018;18:1–14. doi: 10.1186/s12870-018-1435-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu J, Liu L, Liu Y, et al. Genome-wide association study and gene specific markers identified 51 genes or QTL for resistance to stripe rust in U.S. winter wheat cultivars and breeding lines. Front Plant Sci. 2020;11:998. doi: 10.3389/fpls.2020.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhammad A, Hu W, Li Z, et al. Appraising the genetic architecture of kernel traits in hexaploid wheat using GWAS. Int J Mol Sci. 2020;21:5649. doi: 10.3390/ijms21165649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muhu-Din Ahmed HG, Sajjad M, Zeng Y, et al. Genome-wide association mapping through 90K SNP array for quality and yield attributes in bread wheat against water-deficit conditions. Agriculture. 2020;10:392. doi: 10.3390/agriculture10090392. [DOI] [Google Scholar]
- Muleta KT, Bulli P, Rynearson S, et al. Loci associated with resistance to stripe rust (Puccinia striiformis f. sp. tritici) in a core collection of spring wheat (Triticum aestivum) PLoS One. 2017;12:e0179087. doi: 10.1371/journal.pone.0179087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muleta KT, Rouse MN, Rynearson S, et al. Characterization of molecular diversity and genome-wide mapping of loci associated with resistance to stripe rust and stem rust in Ethiopian bread wheat accessions. BMC Plant Biol. 2017;17:134. doi: 10.1186/s12870-017-1082-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqaddasi QH, Brassac J, Börner A, et al. Genetic architecture of anther extrusion in spring and winter wheat. Front Plant Sci. 2017;8:754. doi: 10.3389/fpls.2017.00754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqaddasi QH, Brassac J, Koppolu R, et al. TaAPO-A1, an ortholog of rice ABERRANT PANICLE ORGANIZATION 1, is associated with total spikelet number per spike in elite European hexaploid winter wheat (Triticum aestivum L.) varieties. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-50331-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muqaddasi QH, Zhao Y, Rodemann B, et al. Genome-wide association mapping and prediction of adult stage Septoria tritici blotch infection in european winter wheat via high-density marker arrays. Plant Genome. 2019;12:180029. doi: 10.3835/plantgenome2018.05.0029. [DOI] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, et al. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21:2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naruoka Y, Garland-Campbell KA, Carter AH. Genome-wide association mapping for stripe rust (Puccinia striiformis F. sp. tritici) in US Pacific Northwest winter wheat (Triticum aestivum L.) Theor Appl Genet. 2015;128:1083–1101. doi: 10.1007/s00122-015-2492-2. [DOI] [PubMed] [Google Scholar]
- Neilson EH, Edwards AM, Blomstedt CK, et al. Utilization of a high-throughput shoot imaging system to examine the dynamic phenotypic responses of a C4 cereal crop plant to nitrogen and water deficiency over time. J Exp Bot. 2015;66:1817–1832. doi: 10.1093/jxb/eru526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro D, Gadaleta A, Mangini G, et al. Candidate genes and genome-wide association study of grain protein content and protein deviation in durum wheat. Planta. 2019;249:1157–1175. doi: 10.1007/s00425-018-03075-1. [DOI] [PubMed] [Google Scholar]
- Nordborg M, Weigel D. Next-generation genetics in plants. Nature. 2008;456:720–723. doi: 10.1038/nature07629. [DOI] [PubMed] [Google Scholar]
- Odilbekov F, Armoniené R, Koc A, et al. GWAS-assisted genomic prediction to predict resistance to Septoria tritici blotch in nordic winter wheat at seedling stage. Front Genet. 2019;10:1224. doi: 10.3389/fgene.2019.01224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogbonnaya FC, Rasheed A, Okechukwu EC, et al. Genome-wide association study for agronomic and physiological traits in spring wheat evaluated in a range of heat prone environments. Theor Appl Genet. 2017;130:1819–1835. doi: 10.1007/s00122-017-2927-z. [DOI] [PubMed] [Google Scholar]
- Okada A, Arndell T, Borisjuk N, et al (2019) CRISPR/Cas9-mediated knockout of Ms1 enables the rapid generation of male-sterile hexaploid wheat lines for use in hybrid seed production. Plant Biotechnol J 17:1905–1913. 10.1111/pbi.13106 [DOI] [PMC free article] [PubMed]
- Oyiga BC, Sharma RC, Baum M, et al. Allelic variations and differential expressions detected at quantitative trait loci for salt stress tolerance in wheat. Plant Cell Environ. 2018;41:919–935. doi: 10.1111/pce.12898. [DOI] [PubMed] [Google Scholar]
- Pang Y, Liu C, Wang D, et al. High-resolution genome-wide association study identifies genomic regions and candidate genes for important agronomic traits in wheat. Mol Plant. 2020;13:1311–1327. doi: 10.1016/j.molp.2020.07.008. [DOI] [PubMed] [Google Scholar]
- Pariyar SR, Dababat AA, Sannemann W, et al. Genome-wide association study in wheat identifies resistance to the cereal cyst nematode Heterodera Filipjevi. Phytopathology. 2016;106:1128–1138. doi: 10.1094/PHYTO-02-16-0054-FI. [DOI] [PubMed] [Google Scholar]
- Pasam RK, Bansal U, Daetwyler HD, et al. Detection and validation of genomic regions associated with resistance to rust diseases in a worldwide hexaploid wheat landrace collection using BayesR and mixed linear model approaches. Theor Appl Genet. 2017;130:777–793. doi: 10.1007/s00122-016-2851-7. [DOI] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Perez-Lara E, Semagn K, Tran VA, et al. Population structure and genomewide association analysis of resistance to disease and insensitivity to Ptr toxins in Canadian spring wheat using 90k SNP array. Crop Sci. 2017;57:1522–1539. doi: 10.2135/cropsci2016.10.0859. [DOI] [Google Scholar]
- Phan HTT, Rybak K, Bertazzoni S, et al. Novel sources of resistance to Septoria nodorum blotch in the Vavilov wheat collection identified by genome-wide association studies. Theor Appl Genet. 2018;131:1223–1238. doi: 10.1007/s00122-018-3073-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradhan S, Babar MA, Robbins K, et al. Understanding the genetic basis of spike fertility to improve grain number, harvest index, and grain yield in wheat under high temperature stress environments. Front Plant Sci. 2019;10:1481. doi: 10.3389/fpls.2019.01481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad M, Varshney RK, Kumar A, et al. A microsatellite marker associated with a QTL for grain protein content on chromosome arm 2DL of bread wheat. Theor Appl Genet. 1999;99:341–345. doi: 10.1007/s001220051242. [DOI] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, et al (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 10.1038/ng1847 [DOI] [PubMed]
- Pritchard JK, Stephens M, Rosenberg NA, Donnelly P. Association mapping in structured populations. Am J Hum Genet. 2000;67:170–181. doi: 10.1086/302959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaseem MF, Qureshi R, Shaheen H, Shafqat N. Genome-wide association analyses for yield and yield-related traits in bread wheat (Triticum aestivum L.) under pre-anthesis combined heat and drought stress in field conditions. PLoS One. 2019;14:e0213407. doi: 10.1371/journal.pone.0213407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimi Y, Bihamta MR, Taleei A, et al. Genome-wide association study of agronomic traits in bread wheat reveals novel putative alleles for future breeding programs. BMC Plant Biol. 2019;19:541. doi: 10.1186/s12870-019-2165-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren D, Fang X, Jiang P, et al. Genetic architecture of nitrogen-deficiency tolerance in wheat seedlings based on a nested association mapping (NAM) population. Front Plant Sci. 2018;9:845. doi: 10.3389/fpls.2018.00845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riaz A, Athiyannan N, Periyannan SK, et al. Unlocking new alleles for leaf rust resistance in the Vavilov wheat collection. Theor Appl Genet. 2018;131:127–144. doi: 10.1007/s00122-017-2990-5. [DOI] [PubMed] [Google Scholar]
- Rimbert H, Darrier B, Navarro J, et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS One. 2018;13:1–19. doi: 10.1371/journal.pone.0186329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roselló M, Royo C, Sanchez-Garcia M, Soriano JM. Genetic dissection of the seminal root system architecture in mediterranean durum wheat landraces by genome-wide association study. Agronomy. 2019;9:364. doi: 10.3390/agronomy9070364. [DOI] [Google Scholar]
- Rutkoski JE, Heffner EL, Sorrells ME. Genomic selection for durable stem rust resistance in wheat. Euphytica. 2011;179:161–173. doi: 10.1007/s10681-010-0301-1. [DOI] [Google Scholar]
- Sadeghi-Tehran P, Sabermanesh K, Virlet N, Hawkesford MJ. Automated method to determine two critical growth stages of wheat: heading and flowering. Front Plant Sci. 2017;8:252. doi: 10.3389/fpls.2017.00252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini DK, Chahal A, Pal N, Srivastava P, Gupta PK (2021a) Meta-analysis reveals consensus genomic regions associated with multiple disease resistance in wheat (Triticum Aestivum L.). Research Square 29 September 2021. 10.21203/rs.3.rs-773587/v1 [DOI] [PMC free article] [PubMed]
- Saini DK, Chopra Y, Pal N, et al (2021b) Meta-QTLs, ortho-MQTLs and candidate genes for nitrogen use efficiency and root system architecture in bread wheat (Triticum aestivum L.). Physiol Mol Biol Plants. 10.1007/s12298-021-01085-0 [DOI] [PMC free article] [PubMed]
- Saini DK, Srivastava P, Pal N, Gupta PK (2021c) Meta-QTLs, Ortho-MetaQTLs and Candidate Genes for Grain yield and Associated Traits in Wheat (Triticum aestivum L.). Research Square 27 April 2021. 10.21203/rs.3.rs-430452/v1 [DOI] [PubMed]
- Saintenac C, Jiang D, Akhunov ED. Targeted analysis of nucleotide and copy number variation by exon capture in allotetraploid wheat genome. Genome Biol. 2011;12:R88. doi: 10.1186/gb-2011-12-9-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samantara K, Shiv A, de Sousa LL, et al. A comprehensive review on epigenetic mechanisms and application of epigenetic modifications for crop improvement. Environ Exp Bot. 2021;188:104479. doi: 10.1016/j.envexpbot.2021.104479. [DOI] [Google Scholar]
- Sandhu K, Patil SS, Pumphrey M, Carter A (2021a) Multitrait machine- and deep-learning models for genomic selection using spectral information in a wheat breeding program. Plant Genome e20119. 10.1002/TPG2.20119 [DOI] [PubMed]
- Sandhu KS, Aoun M, Morris CF, Carter AH. Genomic selection for end-use quality and processing traits in soft white winter wheat breeding program with machine and deep learning models. Biol. 2021;10:689–689. doi: 10.3390/BIOLOGY10070689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Lozada DN, Zhang Z, et al. Deep learning for predicting complex traits in spring wheat breeding program. Front Plant Sci. 2021;11:613325. doi: 10.3389/fpls.2020.613325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Mihalyov PD, Lewien MJ, et al. Combining genomic and phenomic information for predicting grain protein content and grain yield in spring Wheat. Front Plant Sci. 2021;12:170. doi: 10.3389/fpls.2021.613300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu KS, Mihalyov PD, Lewien MJ, et al (2021e) Genome-wide association studies and genomic selection for grain protein content stability in a nested association mapping population of spring wheat. bioRxiv.04.15.440064. 10.1101/2021.04.15.440064
- Sapkota S, Hao Y, Johnson J, et al. Genome-wide association study of a worldwide collection of wheat genotypes reveals novel quantitative trait loci for leaf rust resistance. Plant Genome. 2019;12:190033. doi: 10.3835/plantgenome2019.05.0033. [DOI] [PubMed] [Google Scholar]
- Schmidt J, Tricker PJ, Eckermann P, et al. Novel Alleles for Combined Drought and Heat Stress Tolerance in Wheat. Front Plant Sci. 2020;10:1800. doi: 10.3389/fpls.2019.01800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess AW, Reif JC, Ling J, et al. The roles of pleiotropy and close linkage as revealed by association mapping of yield and correlated traits of wheat (Triticum aestivum L.) J Exp Bot. 2017;68:4089–4101. doi: 10.1093/jxb/erx214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura V, Vilhjálmsson BJ, Platt A, et al. An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet. 2012;44:825–830. doi: 10.1038/ng.2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D, Autrique E, Singh R, et al. Identification of genomic regions for grain yield and yield stability and their epistatic interactions. Sci Rep. 2017;7:1–12. doi: 10.1038/srep41578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D, Mondal S, Guzman C, et al. Validation of candidate gene-based markers and identification of novel loci for thousand-grain weight in spring bread wheat. Front Plant Sci. 2019;10:1189. doi: 10.3389/fpls.2019.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sehgal D, Rosyara U, Mondal S, et al. Incorporating genome-wide association mapping results into genomic prediction models for grain yield and yield stability in CIMMYT spring bread wheat. Front Plant Sci. 2020;11:197. doi: 10.3389/fpls.2020.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Angel Y, Houborg R, et al. A random forest machine learning approach for the retrieval of leaf chlorophyll content in wheat. Remote Sens. 2019;11:920. doi: 10.3390/rs11080920. [DOI] [Google Scholar]
- Sheoran S, Jaiswal S, Kumar D, et al. Uncovering genomic regions associated with 36 agro-morphological traits in Indian spring wheat using GWAS. Front Plant Sci. 2019;10:527. doi: 10.3389/fpls.2019.00527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Hao C, Zhang Y, et al. A combined association mapping and linkage analysis of kernel number per spike in common wheat (Triticum aestivum L.) Front Plant Sci. 2017;8:1412. doi: 10.3389/fpls.2017.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W, Yue L, Cheng J, et al. A genome-wide associate study reveals favorable alleles conferring apical and basal spikelet fertility in wheat (Triticum aestivum L.) Mol Breed. 2018;38:1–12. doi: 10.1007/s11032-018-0906-y. [DOI] [Google Scholar]
- Shi C, Zheng Y, Geng J, et al. Identification of herbicide resistance loci using a genome-wide association study and linkage mapping in Chinese common wheat. Crop J. 2020;8:666–675. doi: 10.1016/j.cj.2020.02.004. [DOI] [Google Scholar]
- Shiferaw B, Smale M, Braun HJ, et al. Crops that feed the world 10. Past successes and future challenges to the role played by wheat in global food security. Food Secur. 2013;5:291–317. doi: 10.1007/s12571-013-0263-y. [DOI] [Google Scholar]
- Sidhu JS, Singh D, Gill HS, et al. Genome-Wide Association Study Uncovers Novel Genomic Regions Associated With Coleoptile Length in Hard Winter Wheat. Front Genet. 2020;10:1. doi: 10.3389/fgene.2019.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh PK, Crossa J, Duveiller E, et al. Association mapping for resistance to tan spot induced by Pyrenophora tritici-repentis race 1 in CIMMYTs historical bread wheat set. Euphytica. 2016;207:515–525. doi: 10.1007/s10681-015-1528-7. [DOI] [Google Scholar]
- Singh D, Wang X, Kumar U, et al. High-throughput phenotyping enabled genetic dissection of crop lodging in wheat. Front Plant Sci. 2019;10:394. doi: 10.3389/fpls.2019.00394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Sehgal D, Kumar S, et al. GWAS revealed a novel resistance locus on chromosome 4D for the quarantine disease Karnal bunt in diverse wheat pre-breeding germplasm. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-62711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smiley RW, Backhouse D, Lucas P, Paulitz TC (2009) Diseases which challenge global wheat production-root, crown, and culm rots. Wheat Sci Trade 125–153. 10.1002/9780813818832.ch6
- Song Q, Yan L, Quigley C, et al (2017) Genetic Characterization of the Soybean Nested Association Mapping Population. Plant Genome 10. 10.3835/plantgenome2016.10.0109 [DOI] [PubMed]
- Song X, Feng J, Cui Z, et al. Genome-wide association study for anther length in some elite bread wheat germplasm. Czech J Genet Plant Breed. 2018;54:109–114. doi: 10.17221/70/2017-CJGPB. [DOI] [Google Scholar]
- Spindel JE, Begum H, Akdemir D, et al. Genome-wide prediction models that incorporate de novo GWAS are a powerful new tool for tropical rice improvement. Heredity (Edinb) 2016;116:395–408. doi: 10.1038/hdy.2015.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukumaran S, Lopes M, Dreisigacker S, Reynolds M. Genetic analysis of multi-environmental spring wheat trials identifies genomic regions for locus-specific trade-offs for grain weight and grain number. Theor Appl Genet. 2018;131:985–998. doi: 10.1007/s00122-017-3037-7. [DOI] [PubMed] [Google Scholar]
- Sukumaran S, Reynolds MP, Sansaloni C. Genome-wide association analyses identify QTL hotspots for yield and component traits in durum wheat grown under yield potential, drought, and heat stress environments. Front Plant Sci. 2018;9:81. doi: 10.3389/fpls.2018.00081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Zhang F, Yan X, et al. Genome-wide association study for 13 agronomic traits reveals distribution of superior alleles in bread wheat from the Yellow and Huai Valley of China. Plant Biotechnol J. 2017;15:953–969. doi: 10.1111/pbi.12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Dong Z, Zhao L, et al. The Wheat 660K SNP array demonstrates great potential for marker-assisted selection in polyploid wheat. Plant Biotechnol J. 2020;18:1354–1360. doi: 10.1111/pbi.13361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarawneh RA, Szira F, Monostori I, et al. Genetic analysis of drought response of wheat following either chemical desiccation or the use of a rain-out shelter. J Appl Genet. 2019;60:137–146. doi: 10.1007/s13353-019-00494-y. [DOI] [PubMed] [Google Scholar]
- Tessmann E, Van Sanford D (2018) GWAS for Fusarium head blight related traits in winter wheat (Triticum Aestivum L.) in an artificially warmed treatment. Agronomy 8(68). 10.3390/agronomy8050068
- Tessmann EW, Dong Y, Van Sanford DA. GWAS for Fusarium head blight traits in a soft red winter wheat mapping panel. Crop Sci. 2019;59:1823–1837. doi: 10.2135/cropsci2018.08.0492. [DOI] [Google Scholar]
- Tibbs Cortes L, Zhang Z, Yu J (2021) Status and prospects of genome-wide association studies in plants. Plant Genome e20077 [DOI] [PubMed]
- Tomar V, Singh R, Poland J, et al (2020) Genome-wide association study and Genomic Prediction of spot blotch disease in wheat (Triticum aestivum L.) using genotyping by sequencing. 1–47. 10.21203/rs.2.22818/v1
- Tsai HY, Janss LL, Andersen JR, et al. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci Rep. 2020;10:1–15. doi: 10.1038/s41598-020-60203-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagndorf N, Nielsen NH, Edriss V, et al. Genomewide association study reveals novel quantitative trait loci associated with resistance towards Septoria tritici blotch in North European winter wheat. Plant Breed. 2017;136:474–482. doi: 10.1111/pbr.12490. [DOI] [Google Scholar]
- Vanraden PM. Efficient Methods to Compute Genomic Predictions. J Dairy Sci. 2008;91:4414–4423. doi: 10.3168/jds.2007-0980. [DOI] [PubMed] [Google Scholar]
- Velu G, Singh RP, Crespo-Herrera L, et al. Genetic dissection of grain zinc concentration in spring wheat for mainstreaming biofortification in CIMMYT wheat breeding. Sci Rep. 2018;8:1–10. doi: 10.1038/s41598-018-31951-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voorend W, Lootens P, Nelissen H, et al. LEAF-E: A tool to analyze grass leaf growth using function fitting. Plant Methods. 2014;10:1–13. doi: 10.1186/1746-4811-10-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voss-Fels KP, Keeble-Gagnère G, Hickey LT, et al. High-resolution mapping of rachis nodes per rachis, a critical determinant of grain yield components in wheat. Theor Appl Genet. 2019;132:2707–2719. doi: 10.1007/s00122-019-03383-4. [DOI] [PubMed] [Google Scholar]
- Wainberg M, Sinnott-Armstrong N, Mancuso N, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZJ, Wang JH, Liu LY, et al. Prediction of grain protein content in winter wheat (Triticum aestivum L.) using plant pigment ratio (PPR) F Crop Res. 2004;90:311–321. doi: 10.1016/j.fcr.2004.04.004. [DOI] [Google Scholar]
- Wang D, Sun Y, Stang P, et al. Comparison of methods for correcting population stratification in a genome-wide association study of rheumatoid arthritis: principal-component analysis versus multidimensional scaling. BMC Proc. 2009;3:109. doi: 10.1186/1753-6561-3-s7-s109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Tian F, Pan Y, et al. A SUPER powerful method for genome wide association study. PLoS One. 2014;9:e107684. doi: 10.1371/journal.pone.0107684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Wong D, Forrest K, et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12:787–796. doi: 10.1111/pbi.12183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Cheng X, Shan Q, et al. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32:947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- Wang S-X, Zhu Y-L, Zhang D-X, et al. Genome-wide association study for grain yield and related traits in elite wheat varieties and advanced lines using SNP markers. PLoS One. 2017;12:e0188662. doi: 10.1371/journal.pone.0188662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R, Gordon T, Hole D, et al (2019) Identification and assessment of two major QTLs for dwarf bunt resistance in winter wheat line ‘IDO835.’ Theor Appl Genet 132:2755–2766. 10.1007/s00122-019-03385-2 [DOI] [PubMed]
- Wang Q, Tang J, Han B, Huang X. Advances in genome-wide association studies of complex traits in rice. Theor Appl Genet. 2020;133:1415–1425. doi: 10.1007/s00122-019-03473-3. [DOI] [PubMed] [Google Scholar]
- Ward BP, Brown-Guedira G, Tyagi P, et al. Multienvironment and multitrait genomic selection models in unbalanced early-generation wheat yield trials. Crop Sci. 2019;59:491–507. doi: 10.2135/cropsci2018.03.0189. [DOI] [Google Scholar]
- White JW, Conley MM. A flexible, low-cost cart for proximal sensing. Crop Sci. 2013;53:1646–1649. doi: 10.2135/cropsci2013.01.0054. [DOI] [Google Scholar]
- Wu C, DeWan A, Hoh J, Wang Z. A comparison of association methods correcting for population stratification in case-control studies. Ann Hum Genet. 2011;75:418–427. doi: 10.1111/j.1469-1809.2010.00639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu J, Wang X, Chen N, et al. Association analysis identifies new loci for resistance to Chinese Yr26 -virulent races of the stripe rust pathogen in a diverse panel of wheat germplasm. Plant Dis. 2020;104:1751–1762. doi: 10.1094/PDIS-12-19-2663-RE. [DOI] [PubMed] [Google Scholar]
- Wu S, Wen W, Wang Y, et al (2020b) MVS-pheno: a portable and low-cost phenotyping platform for maize shoots using multiview stereo 3D reconstruction. Plant Phenomics 1–17. 10.34133/2020/1848437 [DOI] [PMC free article] [PubMed]
- Würschum T, Liu W, Gowda M, et al. Comparison of biometrical models for joint linkage association mapping. Heredity (Edinb) 2012;108:332–340. doi: 10.1038/hdy.2011.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xavier A, Xu S, Muir WM, Rainey KM. NAM: association studies in multiple populations. Bioinformatics. 2015;31:3862–3864. doi: 10.1093/BIOINFORMATICS/BTV448. [DOI] [PubMed] [Google Scholar]
- Xie DX, Devos KM, Moore G, Gale MD. RFLP-based genetic maps of the homoeologous group 5 chromosomes of bread wheat (Triticum aestivum L.) Theor Appl Genet. 1993;87:70–74. doi: 10.1007/BF00223747. [DOI] [PubMed] [Google Scholar]
- Xu Y, Crouch JH. Marker-assisted selection in plant breeding: From publications to practice. Crop Sci. 2008;48:391–407. doi: 10.2135/cropsci2007.04.0191. [DOI] [Google Scholar]
- Xu F, Chen S, Yang X, et al (2020) Genome-wide association study on root traits under different cultivation patterns in wheat. 10.21203/rs.3.rs-27846/v1
- Yan X, Zhao L, Ren Y, et al. Genome-wide association study revealed that the TaGW8 gene was associated with kernel size in Chinese bread wheat. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-38570-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X, Zhao L, Ren Y, et al. Identification of genetic loci and a candidate gene related to flag leaf traits in common wheat by genome-wide association study and linkage mapping. Mol Breed. 2020;40:1–15. doi: 10.1007/s11032-020-01135-7. [DOI] [Google Scholar]
- Yang X, Pan Y, Singh PK, et al. Investigation and genome-wide association study for Fusarium crown rot resistance in Chinese common wheat. BMC Plant Biol. 2019;19:153. doi: 10.1186/s12870-019-1758-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Liu J, Guo Y, et al. Genome-wide association mapping of adult-plant resistance to stripe rust in common wheat (Triticum aestivum ) Plant Dis. 2020;104:2174–2180. doi: 10.1094/PDIS-10-19-2116-RE. [DOI] [PubMed] [Google Scholar]
- Yang W, Feng H, Zhang X, et al. Crop phenomics and high-throughput phenotyping: past decades, current challenges, and future perspectives. Mol Plant. 2020;13:187–214. doi: 10.1016/j.molp.2020.01.008. [DOI] [PubMed] [Google Scholar]
- Yang Y, Chai Y, Zhang X, et al. Multi-locus GWAS of quality traits in bread wheat: mining more candidate genes and possible regulatory network. Front Plant Sci. 2020;11:1091. doi: 10.3389/fpls.2020.01091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao F, Long L, Wang Y, et al. Population structure and genetic basis of the stripe rust resistance of 140 Chinese wheat landraces revealed by a genome-wide association study. Plant Sci. 2020;301:110688. doi: 10.1016/j.plantsci.2020.110688. [DOI] [PubMed] [Google Scholar]
- Ye X, Li J, Cheng Y, et al. Genome-wide association study reveals new loci for yield-related traits in Sichuan wheat germplasm under stripe rust stress. BMC Genomics. 2019;20:1–17. doi: 10.1186/s12864-019-6005-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J, Pressoir G, Briggs WH, et al. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38:203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Yu J, Holland JB, Mcmullen MD, Buckler ES. Genetic design and statistical power of nested association mapping in maize. Genetics. 2008;178:539–551. doi: 10.1534/genetics.107.074245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu LX, Lorenz A, Rutkoski J, et al. Association mapping and gene-gene interaction for stem rust resistance in CIMMYT spring wheat germplasm. Theor Appl Genet. 2011;123:1257–1268. doi: 10.1007/s00122-011-1664-y. [DOI] [PubMed] [Google Scholar]
- Yu LX, Morgounov A, Wanyera R, et al. Identification of Ug99 stem rust resistance loci in winter wheat germplasm using genome-wide association analysis. Theor Appl Genet. 2012;125:749–758. doi: 10.1007/s00122-012-1867-x. [DOI] [PubMed] [Google Scholar]
- Yu L-X, Chao S, Singh RP, Sorrells ME. Identification and validation of single nucleotide polymorphic markers linked to Ug99 stem rust resistance in spring wheat. PLoS One. 2017;12:e0171963. doi: 10.1371/journal.pone.0171963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu S, Wu J, Wang M, et al. Haplotype variations in QTL for salt tolerance in Chinese wheat accessions identified by marker-based and pedigree-based kinship analyses. Crop J. 2020;8:1011–1024. doi: 10.1016/j.cj.2020.03.007. [DOI] [Google Scholar]
- Zanke CD, Ling J, Plieske J, et al. Analysis of main effect QTL for thousand grain weight in European winter wheat (Triticum aestivum L.) by genome-wide association mapping. Front. Plant Sci. 2015;6:644. doi: 10.3389/fpls.2015.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegeye H, Rasheed A, Makdis F, et al. Genome-wide association mapping for seedling and adult plant resistance to stripe rust in synthetic hexaploid wheat. PLoS One. 2014;9:1–18. doi: 10.1371/journal.pone.0105593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S, Liu J, Xu D, et al. A genome-wide association study reveals a rich genetic architecture of flour color-related traits in bread wheat. Front Plant Sci. 2018;9:1136. doi: 10.3389/fpls.2018.01136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai S, He Z, Wen W, et al. Genetic architecture of polyphenol oxidase activity in wheat flour by genome-wide association study. Crop Sci. 2020;60:1281–1293. doi: 10.1002/csc2.20038. [DOI] [Google Scholar]
- Zhan S, Ren Y, Liu J, et al. Genome-wide association study of feruloyl arabinoxylan content in common wheat grain. J Cereal Sci. 2019;89:102787. doi: 10.1016/j.jcs.2019.06.001. [DOI] [Google Scholar]
- Zhang Z, Ersoz E, Lai C, et al. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42:355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang K, Wang J, Zhang L, et al. Association analysis of genomic loci important for grain weight control in elite common wheat varieties cultivated with variable water and fertiliser supply. PLoS One. 2013;8:e57853. doi: 10.1371/journal.pone.0057853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Chen J, Yan Y, et al. Genome-wide association study of heading and flowering dates and construction of its prediction equation in Chinese common wheat. Theor Appl Genet. 2018;131:2271–2285. doi: 10.1007/s00122-018-3181-8. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yang J, Zhang L, Luo J, Zhao H, Zhang J, Wen C. A new SNP genotyping technology Target SNP-seq and its application in genetic analysis of cucumber varieties. Sci Rep. 2020;10:1–11. doi: 10.1038/s41598-020-62518-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Quick C, Yu K, et al. PTWAS: investigating tissue-relevant causal molecular mechanisms of complex traits using probabilistic TWAS analysis. Genome Biol. 2020;21:232. doi: 10.1186/s13059-020-02026-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Pan Y, Dong Z, et al. Investigation and genome-wide association study of grain copper content in Chinese common wheat. J Cereal Sci. 2020;95:102991. doi: 10.1016/j.jcs.2020.102991. [DOI] [Google Scholar]
- Zhao Y, Li J, Zhao R, et al. Genome-wide association study reveals the genetic basis of cold tolerance in wheat. Mol Breed. 2020;40:1–13. doi: 10.1007/s11032-020-01115-x. [DOI] [Google Scholar]
- Zhou Y, Tang H, Cheng M-P, et al. Genome-wide association study for pre-harvest sprouting resistance in a large germplasm collection of Chinese wheat landraces. Front Plant Sci. 2017;08:401. doi: 10.3389/fpls.2017.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Shi X, Zhao G, et al. Identification of novel genomic regions and superior alleles associated with Zn accumulation in wheat using a genome-wide association analysis method. Int J Mol Sci. 2020;21:1928. doi: 10.3390/ijms21061928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y, Wang S, Wei W, et al. Genome-wide association study of pre-harvest sprouting tolerance using a 90K SNP array in common wheat (Triticum aestivum L.) Theor Appl Genet. 2019;132:2947–2963. doi: 10.1007/s00122-019-03398-x. [DOI] [PubMed] [Google Scholar]
- Zhu Z, Chen L, Zhang W, et al. Genome-wide association analysis of Fusarium head blight resistance in Chinese elite wheat lines. Front Plant Sci. 2020;11:206. doi: 10.3389/fpls.2020.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuo J, Lin CT, Cao H, et al. Genome-wide association study and quantitative trait loci mapping of seed dormancy in common wheat (Triticum aestivum L.) Planta. 2019;250:187–198. doi: 10.1007/s00425-019-03164-9. [DOI] [PubMed] [Google Scholar]