Abstract
A high-throughput genotyping platform with customized flexibility, high genotyping accuracy, and low cost is critical for marker-assisted selection and genetic mapping in soybean. Three assay panels were selected from the SoySNP50K, 40K, 20K, and 10K arrays, containing 41,541, 20,748, and 9670 SNP markers, respectively, for genotyping by target sequencing (GBTS). Fifteen representative accessions were used to assess the accuracy and consistency of the SNP alleles identified by the SNP panels and sequencing platform. The SNP alleles were 99.87% identical between technical replicates and 98.86% identical between the 40K SNP GBTS panel and 10× resequencing analysis. The GBTS method was also accurate in the sense that the genotypic dataset of the 15 representative accessions correctly revealed the pedigree of the accessions, and the biparental progeny datasets correctly constructed the linkage maps of the SNPs. The 10K panel was also used to genotype two parent-derived populations and analyze QTLs controlling 100-seed weight, resulting in the identification of the stable associated genetic locus Locus_OSW_06 on chromosome 06. The markers flanking the QTL explained 7.05% and 9.83% of the phenotypic variation, respectively. Compared with GBS and DNA chips, the 40K, 20K, and 10K panels reduced costs by 5.07% and 58.28%, 21.44% and 65.48%, and 35.74% and 71.76%, respectively. Low-cost genotyping panels could facilitate soybean germplasm assessment, genetic linkage map construction, QTL identification, and genomic selection.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-023-01372-6.
Keywords: Soybean breeding, Marker-assisted selection (MAS), SNP marker panel, Genotyping by target sequencing (GBTS), Germplasm evaluation, QTL identification
Introduction
Increasing crop productivity to meet demands is critical in the face of a rapidly growing global population (Ray et al. 2013; Arabzai et al. 2021). Breeding new varieties with high yields is often considered a solution. Traditional selection–based breeding mainly relies on visual observation of plant traits and performance in the field, which is a time-consuming and labor-intensive task (Guo et al. 2019; Yang et al. 2021). In recent decades, marker-assisted selection (MAS) has been developed for crop improvement (Cuo et al. 2017). Molecular markers have been shown to play a key role in accelerating the “Green Revolution” (Liu et al. 2020). For instance, markers associated with the sd1 (semi-dwarf) gene have been widely used to reduce plant height and improve lodging resistance and yield in rice (Srivastava et al. 2019), while markers associated with a well-known dwarfing gene in wheat, Rht (reduced height), have been used for the selective breeding of commercial wheat varieties (Grover et al. 2018; Miedaner et al. 2018).
Many types of molecular markers, such as amplified fragment length polymorphisms (AFLPs), restriction fragment length polymorphisms (RFLPs), random amplified polymorphism DNA (RAPD), simple sequence repeats (SSRs), insertions/deletions (indels), and single nucleotide polymorphism (SNP) markers, have been developed and applied in crop breeding (Chen et al. 2021). Due to their abundance in the genome, high degree of polymorphism, and ease of genotyping, SNPs have become the most commonly used markers (Song et al. 2013; Lee et al. 2015; Chen et al. 2021; Sun et al. 2022). In addition, advances in next-generation sequencing technologies have enabled the detection of a large number of high-quality SNP markers in plant populations.
SNP assays have become a key technology in soybean genetic research, and a variety of soybean SNP chips/assays have been developed. For example, the SoySNP50K assay, an Illumina Infinium BeadChip containing 52,041 SNPs, was the first to be developed (Song et al. 2013). The SoySNP180K assay, with 170,223 SNP markers (Lee et al. 2015); the SoySNP355K assay, with 355,595 SNP markers (Wang et al. 2016); the BARCSoySNP6K assay, with 6000 SNP markers (Song et al. 2020); the SoySNP200K assay, containing 158,959 SNP markers (Sun et al. 2022); and the GenoBaits Soy40K assay, with 40,334 SNPs/indels (Liu et al. 2022), have been developed and used for soybean population evaluation and breeding.
Bead chip and genome-by-sequencing (GBS) genotyping platforms have been widely applied in soybean breeding programs. GBS is commonly used for genotyping, but SNPs are not fixed among populations, which is a major limiting factor for MAS application (Guo et al. 2019). Bead chip assays are easy to use, and the SNPs in the assay are fixed. The SoySNP50K assay is one of the most widely used bead chip assays in soybean research. For example, the SoySNP50K assay has been used to genotype thousands of soybean germplasms, including 18,480 domesticated soybean and 1168 wild soybean accessions in the USDA Soybean Germplasm Collection (Song et al. 2015), and the dataset is widely used by soybean researchers. However, access to the SoySNP50K assay outside the USA is limited due to high costs and a lack of genotyping equipment in other countries. Genotyping by target sequencing (GBTS), as a targeted sequence capture strategy, has been used to develop SNP marker panels in maize and cucumber breeding (Guo et al. 2019). However, there have been very few reports on the application of GBTS in soybean. Therefore, our objectives are to develop and evaluate SNP marker panels, including 40K, 20K, and 10K panels, from the SoySNP50K assay (Song et al. 2013) and apply the assays to QTL mapping. This study will provide an alternative approach to accurately genotype soybean populations at low cost. In addition, because the SNPs from the SoySNP50K assays are highly polymorphic and the SoySNP50K dataset from the USDA Soybean Germplasm Collection is publicly available from SoyBase (https://www.soybase.org/snps/), the SNP panels developed in this study will enable the comparison of new germplasm with US accessions.
Materials and methods
Plant materials and field trials
A total of 15 representative soybean accessions, including five wild soybeans (PI407222, ZYD03247, ZYD02878, PI507600, and ZYD04569) and ten cultivars (HJ117, JD12, Hobbit, JD17, Suinong14, Qihuang34, Zhonghuang42, Xudou16, Zhonghuang13, and Zheng196), were used to develop and evaluate the 40K, 20K, and 10K SNP marker panels for GBTS technology (Table 2, Table 3, Fig. 1c and Fig. S3).
Table 2.
Consistency of SNP alleles in repeated sequencing of 15 soybean accessions based on the 40K panel
| Soybean accession | Number of SNPs | Consistency rate | ||
|---|---|---|---|---|
| Consistent | Inconsistent | Not available | ||
| PI407222 | 41209 | 21 | 311 | 99.95% |
| PI507600 | 41035 | 14 | 492 | 99.97% |
| ZYD02878 | 41217 | 23 | 301 | 99.94% |
| ZYD03247 | 40952 | 162 | 427 | 99.61% |
| ZYD04569 | 41054 | 297 | 190 | 99.28% |
| HJ117 | 40766 | 88 | 687 | 99.78% |
| Hobbit | 41293 | 7 | 241 | 99.98% |
| JD12 | 38888 | 43 | 2610 | 99.89% |
| JD17 | 39361 | 25 | 2155 | 99.94% |
| Qihuang34 | 41405 | 12 | 124 | 99.97% |
| Suinong14 | 40666 | 13 | 862 | 99.97% |
| Xudou16 | 41206 | 19 | 316 | 99.95% |
| Zheng196 | 41281 | 20 | 240 | 99.95% |
| Zhonghuang13 | 40756 | 55 | 730 | 99.87% |
| Zhonghuang42 | 41148 | 16 | 377 | 99.96% |
Table 3.
Consistency of SNP alleles between the GBTS data with 40K markers and resequencing data with a sequence depth of 10× among 15 soybean accessions
| Soybean accession | Number of loci | Consistency rate | ||
|---|---|---|---|---|
| Consistent | Inconsistent | Not available | ||
| PI407222 | 40837 | 296 | 408 | 99.28% |
| PI507600 | 40744 | 239 | 558 | 99.42% |
| ZYD02878 | 39499 | 1802 | 240 | 95.64% |
| ZYD03247 | 40919 | 286 | 336 | 99.31% |
| ZYD04569 | 39978 | 1031 | 532 | 97.49% |
| HJ117 | 41183 | 241 | 117 | 99.42% |
| Hobbit | 41314 | 126 | 101 | 99.70% |
| JD12 | 40932 | 535 | 74 | 98.71% |
| JD17 | 40767 | 697 | 77 | 98.32% |
| Qihuang34 | 41238 | 236 | 67 | 99.43% |
| Suinong14 | 40941 | 160 | 440 | 99.61% |
| Xudou16 | 41247 | 189 | 105 | 99.54% |
| Zheng196 | 41223 | 214 | 104 | 99.48% |
| Zhonghuang13 | 40484 | 834 | 223 | 97.98% |
| Zhonghuang42 | 41291 | 204 | 46 | 99.51% |
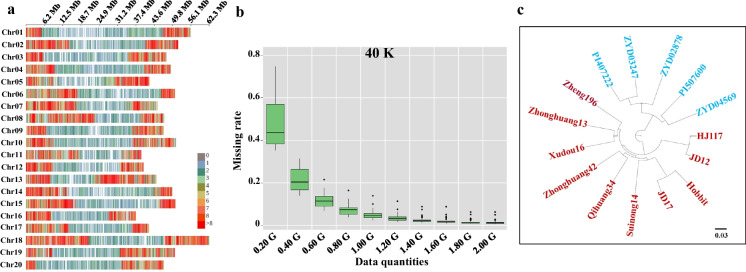
Fig. 1.
Distribution of SNP markers on chromosomes, Data missing rates and application for the 40K SNP marker panel in soybean. a, the distribution of SNP markers on chromosomes for the 40K SNP marker panel, the marker density is indicated by different bar colors, and each bar represents a 1 Mb window size; b, the data missing rates for the 40K marker panel, the X axis represents the number of reads, and the Y axis is the missing rate; c, the phylogenetic trees constructed for the 15 representative soybean accessions using 40K SNP marker panel, scale indicates that 3 substitutions occurred in 100 nucleotides
In addition, two RIL populations, QH3417 and XH1617, consisting of 171 and 183 lines, respectively, were created from crosses between Qihuang34 × HJ117 and Xudou16 × HJ117 with the single-seed descent (SSD) method and were used to construct genetic linkage maps and assess the quality of the SNP panels through collinearity analysis. The genotypic dataset was also used to analyze QTLs for 100-seed weight in soybean. Two RIL populations and their parents were grown in Shijiazhuang, Hebei, China (located at 114.8 E longitude, 38.0 N latitude) in June 2019. A field experiment with a randomized complete block design and two replicates was conducted. Each line and its parents were planted in a single row, 2.0 m long and 0.5 m wide, with a distance of 0.1 m between plants.
Plant sampling and genetic analysis
After harvest, the seeds were dried at room temperature, and the 100-seed weight for each line of QH3417 and XH1617 was measured in 4 replicates by electronic scales. The broad-sense heritability (h2b) of 100-seed weight was estimated based on the formula h2b = VG/(VG + VE), where VG is the genetic variance among RILs and VE is the error variance (Cui et al. 2020).
DNA extraction
At the first fully expanded trifoliate leaf stage (V2 stage), ten leaves were collected from each soybean accession and from each line of each of the two RIL populations to extract genomic DNA using the cetyltrimethylammonium bromide (CTAB) method (Saghai-Maroof et al. 1984). The quality and concentration of DNA were examined by agarose gel electrophoresis and a NanoDrop spectrophotometer.
Selection of SNP loci for development of the marker panels
A total of 45,000 SNPs, including 42,509 SNPs in the SoySNP50K dataset, were selected from the SoySNP50K assay (Song et al. 2013). A set of probes, each with 110 nt and a GC content greater than 30%, were designed using GenoBaits Probe Designer (Guo et al. 2019) to target each SNP flanking region. The specificity of all probes from each SNP region in the reference genome (Schmutz et al. 2010) was assessed. Each SNP was captured by 2 cross-covered probes. The probe set was synthesized by a semiconductor-based in situ synthesis process. All the SNPs were ranked by the average missing rate per locus and the average sequencing depth according to the 15 accessions. The lowest-ranking 3459 SNPs were removed. Finally, 41,541 SNPs were selected and included in the 40K SNP marker panel. Two additional marker panels, 20K and 10K, were generated from the 40K SNP marker panel by sequencing at low depths.
DNA library construction and probe hybridization
DNA library construction and probe hybridization were performed according to Guo et al. (2019). Briefly, the genomic DNA was fragmented and end repaired. After adding the A-tail and adapter ligation, the ligation products with 200–300 bp sizes were indexed with different barcode sequences. The resulting DNA library was eluted with Tris-HCl at pH=8.0.
For probe hybridization, 500 ng library DNA, 5 μL GenoBaits Block I and 2 μL GenoBaits Block II were mixed in the tube and vacuum-dried. The dry powder and hybridization buffer were mixed and incubated at 65°C for 2 h in a PCR instrument for library hybridization. Then, 100 μL of GenoBaits DNA Probe Beads were added to the reaction for target capture. Library amplification was carried out by PCR with probe beads, which were washed at room temperature. The probe beads with post-PCR products were washed with 75% ethanol. Finally, the DNA library was checked with a Qubit 2.0 Fluorometer (Thermo Fisher Scientific, CA); the portion that passed quality control was loaded onto the flow cell and sequenced with PE150 on the MGISEQ-2000 platform (MGI, Shenzhen, China).
SNP identification and genetic linkage map construction
Sequence quality was assessed using FastQC (www.bioinformatics.babraham.ac.uk/project), and the reads were mapped onto the reference genome using BWA (biobwa.sourceforge.net) with default parameters. SNP identification was performed using GATK (software.broadinstitute.org/gatk). The genotypic datasets of the 40K, 20K, and 10K panels were obtained using a Perl script written by Mol Breeding.
The SNPs in the 10K genotypic files of the two RIL populations QH3417 and XH1617 were eliminated if alleles in the parent were identical or missing, the missing rate was > 20.00% among RILs, or segregation was distorted at P value < 0.05 in Chi-square tests. Genetic linkage maps for the two RIL populations of QH3417 and XH1617 were constructed as described by Yang et al. (2019). A total of 2055 SNP markers in QH3417 and 2111 in XH1617 were used to construct the genetic linkage maps using Join Map 4.1 (Van Ooijen 2006).
Phylogenetic tree construction and collinearity analysis
Phylogenetic trees and the collinearity of the physical and genetic positions of the markers were analyzed to evaluate the quality of the 40K, 20K, and 10K SNP marker panels developed by GBTS technology. Phylogenetic trees were constructed by FigTree v1.4.3 software (Gadissa et al. 2021). The collinearity of SNP positions between the physical map (PM, measured in megabases/Mb) of the Wm82.a2.v1 reference genome and the genetic maps (GMs, measured in centimorgans/cM) in the RIL populations QH3417 and XH1617 was calculated and displayed using an R program. The coordinate scales of the physical map (PM, Mb) and the genetic map (GM, cM) of each SNP marker in the RIL populations were calculated as follows:
where Mr means the rth (r=1, 2, …, n) SNP marker; the maximum values of n for the RIL populations QH3417 and XH1617 are 2055 and 2111, respectively. A statistical program to detect the collinearity between the PM and the CM of each RIL population was written in the form of R scripts, and the results were displayed in R.
QTL identification
The average 100-seed weight of the replicates of each line in the two RIL populations was used for QTL detection. The analysis was performed using the composite interval mapping (CIM) method in Windows QTL Cartographer software 2.5 (Wang 2007). A permutation test with 500 iterations was conducted to obtain a proper log of the odds (LOD) score or threshold to declare QTLs (Doerge and Churchil 1996). Model 6 (standard model) was chosen with a control marker and window size of 5 and 10 cM, respectively. The walk speed of precision selection was 1 cM.
Results
Development of high-quality SNP marker panels
Based on the analysis of 15 soybean accessions, three high-quality SNP marker panels, namely, 40K, 20K, and 10K, were developed for GBTS. The 40K, 20K, and 10K marker panels consisted of 41,541, 20,748, and 9670 SNP markers, respectively, well distributed along the entire genome (Table S1). The average numbers of SNPs were 2077, 1037, and 483 per chromosome for the 40K, 20K, and 10K marker panels, respectively. The number of SNPs per chromosome ranged from 1623 (chromosome 20) to 3189 (chromosome 18) for the 40K SNP marker panel, 752 (chromosome 11) to 1576 (chromosome 18) for the 20K SNP marker panel, and 348 (chromosome 11) to 623 (chromosome 18) for the 10K SNP marker panel (Table 1).
Table 1.
Number of SNPs per chromosome for the three soybean assay panels
| Chr a | Number of SNPs | ||
|---|---|---|---|
| 40K | 20k | 10K | |
| Chr01 | 1733 | 1044 | 536 |
| Chr02 | 2425 | 1160 | 504 |
| Chr03 | 1732 | 905 | 437 |
| Chr04 | 2000 | 1093 | 550 |
| Chr05 | 2058 | 908 | 399 |
| Chr06 | 2013 | 1020 | 519 |
| Chr07 | 2217 | 1095 | 468 |
| Chr08 | 2587 | 1118 | 508 |
| Chr09 | 1796 | 1011 | 504 |
| Chr10 | 2150 | 1106 | 544 |
| Chr11 | 1664 | 752 | 348 |
| Chr12 | 1694 | 866 | 390 |
| Chr13 | 2601 | 1172 | 502 |
| Chr14 | 1888 | 982 | 476 |
| Chr15 | 2333 | 1129 | 514 |
| Chr16 | 1646 | 803 | 383 |
| Chr17 | 1944 | 911 | 453 |
| Chr18 | 3189 | 1576 | 623 |
| Chr19 | 2248 | 1137 | 524 |
| Chr20 | 1623 | 960 | 488 |
| Total | 41541 | 20748 | 9670 |
aChr, Chromosome. The same as below
The SNP density per 1 Mb was higher in euchromatic regions than in the centromeres of the Wm82.a2.v1 assembly (Fig. 1a and Fig. S1) because recombination events in the heterochromatic regions were infrequent, and the source panel, SoySNP50K, was purposely selected to reduce the number of SNPs in this region (Song et al. 2013).
The missing rate (Fig. 1b and Fig. S2) decreased as the number of sequencing reads in each panel increased. A total of 1.2 Gbp, 0.6 Gbp, and 0.4 Gbp sequencing reads were required to reduce the missing rate to less than 0.02 for 40K, 20K, and 10K, respectively (Fig. 1b and Fig. S2). Sequencing reads were reduced by 50.00% and 66.67% in the 20K and 10K SNP marker panels, respectively, compared to the 40K panel.
Consistency of SNPs from different sequencing technologies among 15 soybean accessions
To examine the reliability and consistency of SNP alleles from GBTS, the 15 representative soybean accessions were sequenced twice with a 40K SNP GBTS panel, and the SNP alleles from these replicates were compared (Table 2). The concordance of the SNP genotypes from repeat target sequencing averaged 99.87% and ranged from 99.28% in ZYD04569 with 41054 SNPs to 99.98% in Hobbit with 41293 SNPs. The concordance of SNP alleles of the 15 soybean accessions between 10× resequencing data and GBTS data averaged 98.86% and ranged from 95.64% for ZYD02878 with 39499 SNPs to 99.70% for Hobbit with 41314 SNPs (Table 3). The results suggested that the 40K SNP marker panel was highly reliable and consistent.
Phylogenetic relationships revealed by different marker panels
Phylogenetic trees were constructed for the 15 representative soybean accessions using 40K, 20K and 10K SNP marker panels to further evaluate the quality of the SNP marker panels. As shown in Fig. 1c and Fig. S3, identical phylogenetic relationships among the soybean accessions were observed based on the 40K, 20K, and 10K SNP markers. The wild soybean accessions were in one cluster, and the cultivated soybean accessions were in another cluster. Although all three GBTS panels could be used for germplasm evaluation, the 10K panel might be a better choice in terms of minimizing costs.
High-density genetic linkage map construction using the 10K SNP marker panel
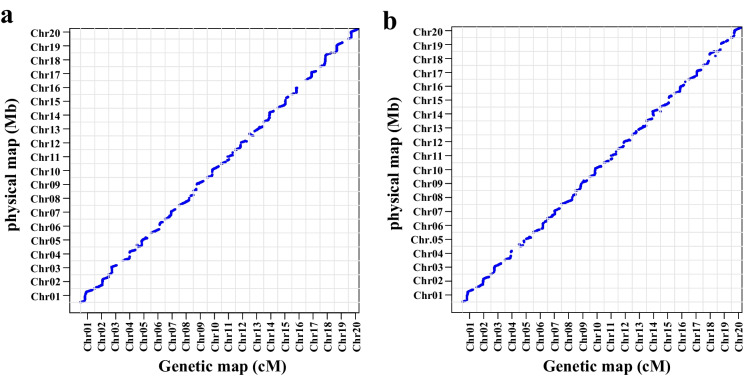
To evaluate the application of these marker panels from GBTS in genetic linkage map construction in soybean. Two RIL populations, derived from Qihuang34 × HJ117 (QH3417) and Xudou16 × HJ117 (XH1617), were sequenced using the 10K SNP marker panel. The linkage map length of 2055 SNP markers obtained in QH3417 was 4490.83 cM, and the average interval between adjacent SNP markers was 2.19 cM. Among the 20 chromosomes, chromosome 18 had the largest number of SNP markers (164), with a genetic length of 238.58 cM, and chromosome 16 had the smallest number of SNP markers (57), with a genetic length of 108.70 cM (Table S2). The linkage map of XH1617 consisted of 2111 SNP markers (Table S3). The total linkage map length and average genetic distance between two adjacent SNP markers were 4798.81 cM and 2.27 cM, respectively. The largest and smallest numbers of SNP markers were 134 on chromosome 18 and 71 on chromosome 04 and chromosome 12, respectively. A relatively high degree of collinearity between the SNP positions in the reference genome and the genetic maps was observed in these two RIL populations, as there was a distinct diagonal direction to the relationship between physical length and genetic distance in the plot of each population (Fig. 2).
Fig. 2.
Collinearity analyses of SNP positions in the physical maps vs. genetic maps of QH3417 and XH1617. a, Collinearity analysis in the population of QH3417; b, Collinearity analysis in the population of XH1617. The abscissa and ordinate represent the physical position on each chromosome and the genetic length of each linkage group, respectively
Genetic analysis and QTL identification for the 100-seed weight trait in soybean
The 100-seed weight of three representative plants from each line of QH3417 and XH1617 was determined and used for QTL identification. As shown in Table S4, the 100-seed weight was between 16.60 g and 31.75 g in QH3417 and 11.45 g and 28.05 g in XH1617. Genetic analysis suggested that the distributions of the 100-seed weight approximated a normal distribution, and the skewness and kurtosis were 0.16 and 0.05 in QH3417, respectively, and −0.19 and −0.09 in XH1617, respectively. The broad-sense heritability (h2b) for the two RILs was 0.96 and 0.92, demonstrating that the variation in 100-seed weight caused by experimental error was small.
QTL analysis in the two RIL populations identified a common stable locus on chromosome 06, Locus_OSW_06 (Table 4). In QH3417, qOSW-34 was mapped to the 6,541,348–6,611,603 bp intervals between the markers Gm06_6611603_A_G and Gm06_6541348_G_T with an LOD value of 4.85, which explained 9.83% of the phenotypic variation. In XH1617, qOSW-16 was detected in the interval of 6187779-6370390 bp between the markers of Gm06_6187779_T_C-Gm06_6370390_G_A with an LOD value of 4.09, explaining 7.05% of the phenotypic variation.
Table 4.
Putative QTLs detected for 100-seed weight in the QH3417 and XH1617 populations
| Locus | QTL a | Chr | Position | Marker or interval b | LOD c | PVE(%) d | Add e |
|---|---|---|---|---|---|---|---|
| Locus_OSW_06 | qOSW-34 | 06 |
6541348- 6611603 |
Gm06_6611603_A_G- Gm06_6541348_G_T |
4.85 | 9.83 | -0.95 |
| qOSW-16 | 06 |
6187779- 6370390 |
Gm06_6187779_T_C- Gm06_6370390_G_A |
4.09 | 7.05 | -0.81 |
aqOSW-34 and qOSW-16, the QTLs detected for 100-seed weight in the QH3417 and XH1617 populations, respectively
bMarker or interval, markers or support intervals on the linkage map in which the LOD is the largest
cLOD, logarithm of odds
dPVE (%), percentage of phenotypic variance explained by the QTL
eAdd, additive effects, negative values represent increasing effects of the QTLs derived from HJ117
Cost of GBTS compared with GBS and DNA chips
The cost in soybean breeding programs, especially the high genotyping cost, is the major constraint for breeders. The costs of GBTS, GBS, and DNA chips were analyzed (Table 5). According to the Mol Breeding Company, the costs for DNA extraction, library construction, probe hybridization, and labor were $0.44, $2.94, $2.21, and $1.47 per sample, respectively, for each panel of GBTS; however, the costs for sequencing, bioinformatics analysis and equipment depreciation varied depending on the panels. Compared to the costs of the 40K marker panel, the costs of the 20K and 10K panels were reduced by 33.33% and 66.67% for sequencing, 60.81% and 79.73% for data analysis, and 29.93% and 59.86% for equipment depreciation, respectively. The total genotyping costs of GBTS for 40K, 20K, 10K, GBS with a sequence depth of 2× and DNA chips containing 50K SNP markers were $13.68, $11.32, $9.26, $14.46, and $32.79 per sample, respectively. Compared to GBS and DNA chips, the costs for 40K, 20K, and 10K were reduced by 5.07% and 58.28%, 21.44% and 65.48%, and 35.74% and 71.76%, respectively. GBTS has a cost advantage over GBS and DNA chips, especially when the 10K marker panel is used.
Table 5.
Genotyping cost (US$ per sample) for different sequencing technologies
| Procedure | GBTS a | GBS b | DNA chips c | ||
|---|---|---|---|---|---|
| 40K | 20K | 10K | |||
| DNA extraction | 0.44 | 0.44 | 0.44 | 0.44 | 0.44 |
| Library construction | 2.94 | 2.94 | 2.94 | 5.88 | 29.41 |
| Probe hybridization | 2.21 | 2.21 | 2.21 | 0.00 | |
| Sequencing | 4.41 | 2.94 | 1.47 | 4.41 | |
| Bioinformatics analysis | 0.74 | 0.29 | 0.15 | 0.74 | |
| Labor | 1.47 | 1.47 | 1.47 | 1.47 | 1.47 |
| Depreciation cost | 1.47 | 1.03 | 0.59 | 1.47 | 1.47 |
| Total | 13.68 | 11.32 | 9.26 | 14.41 | 32.79 |
aGBTS, genotyping by target sequencing
bGBS, genotyping by sequencing, where the sequence depth is 2×
cDNA chips, chips containing 50K SNP markers with a genotyping cost of $32.79
Discussion
In recent decades, sequencing technologies, such as first-generation DNA sequencing, next-generation DNA sequencing, and Affymetrix GeneChips, have been used for genotyping and application of MAS in soybean breeding (Song et al. 2013; Lee et al. 2015; Heather and Chain 2016; Wang et al. 2016; Song et al. 2020; Sun et al. 2022). However, whole-genome resequencing of all genotypes is not cost-effective and is sometimes unnecessary, while gene chips require expensive equipment. Breeders frequently need high-efficiency, low-cost, and flexible genotyping platforms for breeding (Rasheed et al. 2017). The new sequencing technology of GBTS has the advantages of customized flexibility, high sequencing accuracy, and low sequencing cost and has been widely used in maize breeding (Guo et al. 2019).
As two core SNP capture technologies in GBTS, GenoPlex (based on multiplexing PCR) and GenoBaits (based on sequence capture in solution) can assay many types of molecular markers, including SSRs, SNPs, and indels (Xu et al. 2020; Guo et al. 2021). Compared to DNA chips/assays, GBTS is more flexible in meeting the requirement of various numbers of markers by controlling the sequencing depth (Guo et al. 2019; Xu et al. 2020). In this study, three high-quality SNP marker panels were developed for GBTS (Table S1) and could be utilized for soybean germplasm evaluation, genetic mapping, marker-assisted selection, etc. In addition, the relatively high cost of genotyping accessions is the main constraint on the application of MAS in soybean breeding (Guo et al. 2019); GBTS provides an alternative approach to genotype lines at relatively low cost, according to our cost comparison.
Recently, one GBTS liquid chip, the GenoBaits Soy40K assay, was developed based on the whole genome sequences of approximately 2900 soybean accessions (Liu et al. 2022). The 40K, 20K, and 10K SNP marker panels were developed based on the SoySNP50K assay (Song et al. 2015), which will greatly facilitate the comparison of the genotypes to the 18,480 domesticated soybean and 1168 wild soybean accessions in the USDA Soybean Germplasm Collection, which were genotyped with SoySNP50K chips (Fig. 1a and Fig. S1).
Molecular markers have been widely used for soybean germplasm evaluation (Song et al. 2013; Lee et al. 2015; Wang et al. 2016; Song et al. 2020; Sun et al. 2022), genetic linkage map construction (Song et al. 2016), and QTL identification (Song et al. 2020; Yang et al. 2021). SNPs have become the mainstream marker type due to their advantages of high abundance and ease of high-throughput genotyping, which could greatly improve the efficiency of soybean genetic studies and breeding programs (Chen et al. 2021). However, the quality of the SNP marker is the key factor in the success of MAS application and genetic studies (Ben-Ari and Lavi 2012; Chen et al. 2021). In this study, the 40K, 20K and 10K marker panels were all from the well-tested SoySNP50K assay panel reported by Song et al. (2013). The density of the SNPs in these three marker panels was higher in the euchromatic regions than in the centromeres (Fig. 1a and Fig. S1). The SNP alleles were consistent between resequencing data and the 40K marker panel developed by GBTS among 15 soybean accessions (Tables and Table 3), suggesting that the SNP markers in these three marker panels were reliable and of high quality.
The results from phylogenetic tree analysis of the 15 tested soybean accessions showed that the phylogenetic relationships were consistent based on the markers in the three panels. Interestingly, HJ117 was grouped with JD12, JD17 with Hobbit, and Zhonghuang42 with Qihuang34, which was consistent with their pedigrees. HJ117 is the progeny of JD12; JD17 is the progeny of Hobbit; and Zhonghuang42 and Qihuang34 share the same parent, Youchu4. Additionally, Zhonghuang13 and Zheng196 share the same parents, Juxuan23 and 5905 (Zhang et al. 2014).
A relatively high degree of collinearity of SNP positions between the reference genome and genetic map constructed by the 10K SNP marker panel was observed in the two RIL populations. As expected, one stable QTL associated with soybean 100-seed weight, namely, Locus_OSW_06, was identified on chromosome 06 with a LOD value of 4.09–4.85, explaining 7.05–9.83% of the phenotypic variation. This QTL was reported in a previous study by Pathan et al. (2013). This result suggested that the linkage map constructed by GBTS technology is appropriate for QTL identification.
The SoySNP50K assay is one of the most widely used bead chip assays in the soybean community and has accelerated soybean breeding progress. However, some developing countries have not been able to take advantage of the SoySNP50K assay due to the high costs of purchasing genotyping equipment and carrying out the sequencing process. The 40K, 20K, and 10K SNP marker panels in this study are low cost and have high accuracy, which may solve the problem more effectively in some developing countries. In addition, the 40K, 20K, and 10K panels contained fewer markers than the SoySNP50K assay. However, the evaluation of too many markers will increase the cost and generate redundant information (Song et al. 2020). Marker panels with fewer SNPs could also have certain advantages if they are widely used and commercialized for soybean population evaluation and breeding. For instance, the BARCSoySNP6K assay, with 6000 SNP markers, was carefully chosen from the SoySNP50K assay and was found to be an excellent tool for QTL detection, genomic selection and genetic relationship assessment (Song et al. 2020). In this study, all three marker panels were used for germplasm evaluation, and the 10K marker panel can used for linkage map construction and QTL identification in soybean. Overall, the application of these three marker panels was developed by new sequencing technology for GBTS and is reasonable and beneficial for soybean breeding programs.
Conclusion
This study provided an alternative approach for genotyping soybean lines at low cost and with high accuracy. Three SNP marker panels, 40K, 20K, and 10K, were selected from the SoySNP50K assay and were developed by GBTS. The marker panels could be used for soybean germplasm evaluation, linkage map construction, and QTL identification. Most importantly, the resulting genotypic data of soybean accessions from these three marker panels could be compared to the SoySNP50K dataset of the USDA Soybean Germplasm Collection.
Supplementary Information
Tables S1–S4 (XLSX 2831 kb)
Figures S1–S3 (PPTX 444 kb)
Author contribution
Q.J.S., L.Y., C.Y.Y., and M.C.Z. designed the experiments and critically revised the manuscript. Q.Y., J.N.Z., L.C., and L.Y. analyzed the data. Y.Q. wrote the manuscript. L.Y. and X.L.S. carried out the experiments. All authors have read and approved the manuscript.
Funding
This research was supported by the National Key R & D Project (2021YFD1201602), Natural Science Foundation of Hebei Province (C2020301020), National Natural Science Foundation of China (31871652, 32072092), Modern Agricultural Science and Technology Innovation Project of Hebei Province (2019-4-3), and China Agriculture Research System of MOF and MARA (CARS-04-PS06).
Data availability
The datasets generated and/or analyzed during the current study are available from Long Yan on reasonable request.
Declarations
Ethics approval and consent to participate
No applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Qing Yang, Jianan Zhang, and Xiaolei Shi contributed equally to this study.
Contributor Information
Chunyan Yang, Email: chyyang66@163.com.
Qijian Song, Email: Qijian.Song@usda.gov.
Long Yan, Email: dragonyan1979@163.com.
References
- Arabzai MG, Gul H. Application techniques of molecular marker and achievement of marker assisted selection (MAS) in three major crops rice, wheat and maize. Int J Res Appl Sci Biotechnol. 2021;8:82–93. doi: 10.31033/ijrasb.8.1.10. [DOI] [Google Scholar]
- Ben-Ari G, Lavi U. Marker-assisted selection in plant breeding in plant biotechnology and agriculture. ArieAltman, Cambridge: Academic Press, England; 2012. pp. 163–184. [Google Scholar]
- Cao YC, Li SG, He XH, Chang FG, Kong JJ, Gai JY, Zhao TJ. Mapping QTLs for plant height and flowering time in a Chinese summer planting soybean RIL population. Euphytica. 2017;213:1–13. doi: 10.1007/s10681-016-1834-8. [DOI] [Google Scholar]
- Chen ZJ, Tang DG, Ni JX, Li P, Wang L, Zhou JH, Li CY, Lan H, Li LJ, Liu J. Development of genic KASP SNP markers from RNA-Seq data for map-based cloning and marker-assisted selection in maize. BMC Plant Biol. 2021;21:1–11. doi: 10.1186/s12870-021-02932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui BF, Chen L, Yang YQ, Liao H. Genetic analysis and map-based delimitation of a major locus qSS3 for seed size in soybean. Plant Breed. 2020;00:1–13. [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadissa F, Abebe M, Bekele T. Agro-morphological traits-based genetic diversity assessment in Ethiopian barley (Hordeum vulgare L.) landrace collections from Bale highlands, Southeast Ethiopia. Agric Food Secur. 2021;10:1–14. doi: 10.1186/s40066-021-00335-4. [DOI] [Google Scholar]
- Grover G, Sharma A, Gill HS, Srivastava P, Bains NS. Rht8 gene as an alternate dwarfing gene in elite Indian spring wheat cultivars. PLoS One. 2018;13:1–11. doi: 10.1371/journal.pone.0199330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo ZF, Wang HG, Tao JJ, Ren YH, Xu C, Wu KS, Zou C, Zhang JN, Xu YB. Development of multiple SNP marker panels affordable to breeders through genotyping by target sequencing (GBTS) in maize. Mol Breed. 2019;39:1–12. doi: 10.1007/s11032-019-0940-4. [DOI] [Google Scholar]
- Guo ZF, Yang QN, Huang FF, Zheng HJ, Sang ZQ, Xu YF, Zhang C, Wu KH, Tao JJ, Prasanna BM, Olsen MS MS, Wang YB, Zhang JN, Xu YB. Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Communications. 2021;100230:1–15. doi: 10.1016/j.xplc.2021.100230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heather JM, Chain B. The sequence of sequencers: the history of sequencing DNA. Genomics. 2016;107:1–8. doi: 10.1016/j.ygeno.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YG, Jeong N, Kim JH, Lee K, Kim KH, Pirani A, Ha BK, Kang ST, Park BS, Moon JK, Kim N, Soon-Chun Jeong SC. Development, validation and genetic analysis of a large soybean SNP genotyping array. Plant J. 2015;81:625–636. doi: 10.1111/tpj.12755. [DOI] [PubMed] [Google Scholar]
- Liu SL, Zhang M, Feng F, Tian ZX. Toward a “Green Revolution” for soybean. Mol Plant. 2020;13:688–697. doi: 10.1016/j.molp.2020.03.002. [DOI] [PubMed] [Google Scholar]
- Liu YC, Liu SL, Zhang ZF, Ni LB, Chen XM, Ge YX, Zhou GA, Tian ZX. GenoBaits Soy40K: a highly flexible and low-cost SNP array for soybean studies. Sci China Life Sci. 2022;65:1–4. doi: 10.1007/s11427-022-2130-8. [DOI] [PubMed] [Google Scholar]
- Miedaner T, Herter CP, Ebmeyer E, Kollers S, Korzun V. Use of non-adapted quantitative trait loci for increasing Fusarium head blight resistance for breeding semi-dwarf wheat. Plant Breed. 2018;138:140–147. doi: 10.1111/pbr.12683. [DOI] [Google Scholar]
- Pathan SM, Vuong T, Clark K, Lee JD, Shannon JG, Roberts CA, Ellersieck MR, Burton JW, Cregan PB, Hyten DL, Nguyen HT, Sleper DA. Genetic mapping and confirmation of quantitative trait loci for seed protein and oil contents and seed weight in soybean. Crop Sci. 2013;53:765–774. doi: 10.2135/cropsci2012.03.0153. [DOI] [Google Scholar]
- Rasheed A, Hao YF, Xia XC, Khan A, Xu YB, Varshney RK, He ZH. Crop breeding chips and genotyping platforms: progress, challenges and perspectives. Mol Plant. 2017;10:1047–1064. doi: 10.1016/j.molp.2017.06.008. [DOI] [PubMed] [Google Scholar]
- Ray DK, Mueller ND, West PC, Foley JA. Yield trends are insufficient to double global crop production by 2050. PLoS One. 2013;8:e66428. doi: 10.1371/journal.pone.0066428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saghai-Maroof MA, Soliman KM, Jorgensen RA, Allard RW. Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. P Natl Acad Sci USA. 1984;81:8014–8018. doi: 10.1073/pnas.81.24.8014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J, Cannon SB, Schlueter J, Ma JX, Mitros T, Nelson W, Hyten DL, Song QJ, Thelen JJ, Cheng JL, Xu D, Hellsten U, May GD, Yu Y, Sakurai T, Umezawa T, Bhattacharyya MK, Sandhu D, Valliyodan B, Lindquist E, Peto M, Grant D, Shu SQ, Goodstein D, Barry K, Futrell-Griggs M, Abernathy B, Du JC, Tian ZX, Zhu LC, Gill N, Joshi T, Libault M, Sethuraman A, Zhang XC, Shinozaki K, Nguyen HT, Wing RA, Cregan P, Specht J, Grimwood J, Rokhsar D, Stacey G, Shoemaker RC, Jackson SA. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–183. doi: 10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- Song QJ, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS One. 2013;8:e54985. doi: 10.1371/journal.pone.0054985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Q, Hyten DL, Jia G, Quigley CV, Fickus EW, Nelson RL, Cregan PB. Fingerprinting soybean germplasm and its utility in genomic research. G3-Genes Genom Genet. 2015;5:1999–2006. doi: 10.1534/g3.115.019000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QJ, Jenkins J, Gf J, Hyten DL, Pantalone V, Jackson SA, Schmutz J, Cregan PB. Construction of high resolution genetic linkage maps to improve the soybean genome sequence assembly Glyma1.01. BMC Genomics. 2016;17:1–11. doi: 10.1186/s12864-015-2344-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song QJ, Yan L, Quigley C, Fickus E, Wei H, Chen LF, Dong F, Araya S, Liu JL, Hyten D, Pantalone V, Nelson RL. Soybean BARCSoySNP6K: an assay for soybean genetics and breeding research. Plant J. 2020;104:800–811. doi: 10.1111/tpj.14960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava DS, Shamim M, Mishra A, Yadav P, Kumar D, Pandey P, Khan NA, Singh KN. Introgression of semi-dwarf gene in Kalanamak rice using marker-assisted selection breeding. Curr Sci India. 2019;116:597–603. doi: 10.18520/cs/v116/i4/597-603. [DOI] [Google Scholar]
- Sun RJ, Sun BC, Tian Y, Su SS, Zhang Y, Zhang WH, Wang JS, Yu P, Guo BF, Li HH, Li YF, Gao HW, Gu YZ, Yu LL, Ma YS, Su EH, Li Q, Hu XG, Zhang Q, Guo RQ, Chai S, Feng L, Wang J, Hong HL, Xu JY, Yao XD, Wen J, Liu JQ, Li YH, Qiu LJ. Dissection of the practical soybean breeding pipeline by developing ZDX1, a high-throughput functional array. Theor Appl Genet. 2022;135:1413–1427. doi: 10.1007/s00122-022-04043-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Ooijen JW. JoinMap® 4.0: software for the calculation of genetic linkage maps in experimental populations. Wageningen: Kyazma BV; 2006. [Google Scholar]
- Wang S. Windows QTL Cartographer 2.5. (Software); 2007. [Google Scholar]
- Wang J, Chu SS, Zhang H, Zhu Y, Cheng H, Yu DY. Development and application of a novel genome-wide SNP array reveals domestication history in soybean. Sci Rep-UK. 2016;6:1–10. doi: 10.1038/srep20728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu YB, Yang QN, Zheng HJ, Xu YF, Sang ZQ, Guo ZF, Peng H, Zhang C, Lan HF, Wang YB, Wu KS, Tao JJ, Zhang JN. Genotyping by target sequencing (GBTS) and its applications. Sci Agric Sin. 2020;53:2983–3004. [Google Scholar]
- Yang Q, Yang YQ, Xu RN, Lv HY, Liao H. Genetic analysis and mapping of QTLs for soybean biological nitrogen fixation traits under varied field conditions. Front Plant Sci. 2019;10:1–11. doi: 10.3389/fpls.2019.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q, Lin GM, Lv HY, Wang CH, Yang YQ, Liao H. Environmental and genetic regulation of plant height in soybean. BMC Plant Biol. 2021;21:1–15. doi: 10.1186/s12870-021-02836-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang MC, Zhang L, Liu XY. Soybean improved germplasm in Huanghuaihai region. China: Beijing; 2014. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1–S4 (XLSX 2831 kb)
Figures S1–S3 (PPTX 444 kb)
Data Availability Statement
The datasets generated and/or analyzed during the current study are available from Long Yan on reasonable request.