Abstract
Using imbalanced historical yield data to predict performance and select new lines is an arduous breeding task. Genome-wide association studies (GWAS) and high throughput genotyping based on sequencing techniques can increase prediction accuracy. An association mapping panel of 227 Texas elite (TXE) wheat breeding lines was used for GWAS and a training population to develop prediction models for grain yield selection. An imbalanced set of yield data collected from 102 environments (year-by-location) over 10 years, through testing yield in 40–66 lines each year at 6–14 locations with 38–41 lines repeated in the test in any two consecutive years, was used. Based on correlations among data from different environments within two adjacent years and heritability estimated in each environment, yield data from 87 environments were selected and assigned to two correlation-based groups. The yield best linear unbiased estimation (BLUE) from each group, along with reaction to greenbug and Hessian fly in each line, was used for GWAS to reveal genomic regions associated with yield and insect resistance. A total of 74 genomic regions were associated with grain yield and two of them were commonly detected in both correlation-based groups. Greenbug resistance in TXE lines was mainly controlled by Gb3 on chromosome 7DL in addition to two novel regions on 3DL and 6DS, and Hessian fly resistance was conferred by the region on 1AS. Genomic prediction models developed in two correlation-based groups were validated using a set of 105 new advanced breeding lines and the model from correlation-based group G2 was more reliable for prediction. This research not only identified genomic regions associated with yield and insect resistance but also established the method of using historical imbalanced breeding data to develop a genomic prediction model for crop improvement.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-022-01287-8.
Keywords: Genetic correlation, Wheat breeding, Imbalanced data, Insect resistance, Genomic prediction accuracy
Introduction
The complex nature of grain yield makes it difficult for precise selection of lines with high yield potential. Accumulation of favorite allele combinations for yield would lead to lines with improved yields. For identifying favorite alleles, genome-wide association studies (GWAS) (Atwell et al. 2010; Rafalski 2010) have showed advantages over traditional QTL mapping using bi-parental mapping populations. GWAS uses the natural collection of germplasm lines such as landraces, varieties, and breeding lines as mapping panels, and detects historical recombination events and linkage disequilibrium (LD) to identify the non-random association between allele loci and traits (Flint-Garcia et al. 2003). GWAS can identify multiple alleles simultaneously since a wider range of germplasms in a panel would contain more diverse genetic composition (Zhu et al. 2008; Myles et al. 2009; Atwell et al. 2010).
Increasing marker coverage in the genome will enhance the power of GWAS for allele identification. Single nucleotide polymorphisms (SNPs), the variations on a single nucleotide at the specific position, are the most abundant and widely distributed genome markers (Agarwal et al. 2008). With the advances in DNA sequencing technology, the genotype-by-sequencing (GBS) (Elshire 2011) and later double-digested restriction-site associated DNA sequencing (ddRADseq) (Baird et al. 2008; Peterson et al. 2012) are robust approaches of identifying SNPs that are randomly distributed throughout the whole genome and thus are suitable for investigating genome-wide genetic variations. Particularly, by aligning SNP flanking DNA sequences to assembled whole genome sequences of hexaploid wheat (IWGSC 2014; Zimin et al. 2017), tetraploid wheat (Avni et al. 2017), and Aegilops tauschii (Jia et al. 2013b), chromosomal location of each SNP can be precisely located to accurately track favorite alleles.
Previously, QTL analysis was widely used to identify genomic regions associated with the traits and then develop molecular markers for marker-assisted selection (MAS). However, most important traits such as grain yield, yield components, and end-use quality are all highly polygenic with each locus contributing only a very small proportion of total phenotypic variance (Simmonds et al. 2014; Tyagi et al. 2014; Jia et al. 2013a; Collard and Mackill 2008), which leads to weak stability and low repeatability in those QTLs and thus limits the application of MAS for accumulating desirable genes in crop improvement. Genomic selection (GS) utilizes a large set of markers covering a whole genome to detect all possible alleles within the LD and their effects on the trait, and to estimate the genomic breeding value of each line to conduct selection in breeding (Meuwissen et al. 2001; Bernardo and Yu 2007; Bhat et al. 2016; Rutkoski et al. 2016; Daetwyler et al. 2008; Sun et al. 2019; Tsai et al. 2020). Therefore, using GWAS to identify favorite alleles and form the training population to develop prediction models for conducting GS will be an efficient way of accumulating desirable alleles for improving yield and other polygenic traits.
To conduct GWAS and GS, the composition of training populations and precision of phenotyping are two additional critical factors affecting prediction accuracy (He et al. 2016; Michel et al. 2017; Marulanda et al. 2015). Using advanced lines from the same breeding program as a training population has showed a positive effect on prediction accuracy (Endelman et al. 2014; Daetwyler et al. 2008). It was demonstrated that a training population including lines from the same family, half sibs, and more distant lines could be efficient for a GS scheme (Verges and Van Sanford 2020). Using historical advanced breeding lines developed at different periods together with germplasm lines from the same breeding program may also be a good strategy for association mapping and genomic prediction studies, simply because all those lines could represent all genetic sources in the program with historical recombination maintained to ensure mapping resolution, particularly when the training population is keeping updated as the new germplasm lines were introduced into the program.
Another advantage of using advanced breeding lines in GWAS and genomic prediction is that the lines have been evaluated in many years under different environmental conditions and have phenotypic data already available. This would be especially helpful for traits such as grain yield that requires many resources for phenotyping (Verges and Van Sanford 2020). However, the most significant difficulty in using phenotypic data of the historical breeding lines for GWAS is that the lines are evaluated at different time and that the data were typically imbalanced. Research using imbalanced data in historical breeding lines for GWAS and genomic prediction have been seldom reported. The possibility of using imbalanced data for GS was explored by clustering analysis of data through pre-defined mega-environments based on climatic patterns, farming systems, water regimes, and the incidence of biotic and abiotic stress, but this strategy appears ineffective for genomic selection (Dawson et al. 2013). Therefore, it is necessary to identify an appropriate way that can directly utilize the imbalanced historical data for GWAS and genomic prediction.
During the past few decades, the Texas A&M AgriLife Research and Extension Center at Amarillo, TX, has released several drought tolerant cultivars such as ‘TAM 105’, ‘TAM 107’, ‘TAM 111’, and ‘TAM 112’ (Porter et al. 1980, 1987; Lazar et al. 2004; Rudd et al. 2014). Among them, TAM 111 and TAM 112 were two broadly planted cultivars in the Great Plains hard red winter wheat regions since 2010 based on planted acreages (NASS, 2011–2013 http://www.nass.usda.gov). They have been the core parents in many HWW breeding programs due to their drought tolerance and wide adaptability that have been confirmed using RNA-seq (Chu et al. 2021). A newer release ‘TAM 114’ has superior bread-making quality and drought tolerance (Rudd et al. 2018). A grain and forage dual purpose awnless wheat ‘TAM 204’ has higher yield, drought tolerance, and a good level of resistance to insects such as greenbug, Hessian fly, and wheat curl mite (Rudd et al. 2019). These widely adapted winter wheat cultivars have been used as germplasm lines in wheat breeding programs in the USA and many other countries. To localize their genes conferring the superior traits will greatly improve selection efficiency for improving yield, end-use quality, and tolerance to biotic and abiotic stresses (Liu et al. 2016a; Yang et al. 2020, 2019; Yu et al. 2021).
In this research, we used a set of 227 elite breeding lines (including the aforementioned released cultivars) developed by Texas A&M AgriLife Research wheat breeding programs in the last 10 years as the mapping panel for conducting GWAS to identify favorite alleles and as the training population to build a genomic prediction model for selecting grain yield. By combining correlation analysis with genetic heritability estimation using the imbalanced yield data collected from diverse environments, we successfully developed a data management strategy of using the imbalanced historical yield data for conducting GWAS and building genomic prediction models. In addition, the genomic prediction model was further validated using a set of newly developed advanced breeding lines from Texas wheat breeding programs.
Materials and methods
Plant materials
The set of 227 Texas elite (TXE, F9) breeding lines were developed by the two Texas A&M AgriLife Research wheat breeding programs located at Amarillo and College Station, TX, during 2009–2018, which included 13 released TAM cultivars. Briefly, the lines were originally selected at the F6 generation according to their performance in the observation yield trials conducted at two locations followed by evaluation preliminary yield trials in six locations and advanced yield trials in ten locations at the F7 and F8 generations, respectively. The state-wide yield TXE trials were conducted at 16 locations across Texas with three replicates per location. The superior TXE lines were further evaluated for traits of yield, end-use quality, biotic and abiotic tolerance, and agronomic traits either toward the new cultivar release or as the germplasms to enter the new breeding cycles. Therefore, the set of TXE lines represented the major gene sources in Texas wheat breeding programs and were appropriate materials for building genomic prediction models to improve selection efficiency. In addition, a set of 105 lines entered into the advanced yield trials was evaluated at ten locations with two replications per location, and their yield data were used to validate the genomic prediction model developed from the TXE collection.
Grain yield data analysis
The TXE trials conducted during 2009–2018 were conducted in three replications at 16 locations that represented four typical wheat growing regions (High Plains, Rolling Plains, Blacklands, and South Texas) in Texas (Fig. S1), and grain yield data were collected from 6 to 14 locations each year (Table 1) due to the abandoned harvest in some locations experienced serious damage caused by severe weather. Totally, yield data were from 102 environments defined as year-by-location combinations (Table 1). However, TXE yield data were typically imbalanced with 40 to 66 lines evaluated each year and 38 to 41 lines were also tested in the following year (Table 1). Three cultivars, ‘TAM 112’, ‘TAM 401’, and ‘TAM W-101’ were used as controls across all years.
Table 1.
List of datasets in two correlation-based groups formed by correlation analysis and broad-sense heritability estimation using yield data of 227 Texas elite (TXE) breeding lines collected during 2009–2018
| Year | Yield dataset | Dataset grouped through correlation | Dataset group adjusted by heritability test | ||||
|---|---|---|---|---|---|---|---|
| Correlation-group 1 | Correlation-group 2 | Non-correlated | Correlation-group 1 | Correlation-group 2 | Abandoned | ||
| 2009 | 9 | 5 | 3 | 1 | 7 | 2 | 0 |
| 2010 | 14 | 6 | 5 | 3 | 7 | 6 | 1 |
| 2011 | 10 | 4 | 4 | 2 | 6 | 4 | 0 |
| 2012 | 14 | 3 | 8 | 3 | 3 | 9 | 2 |
| 2013 | 9 | 5 | 3 | 1 | 2 | 6 | 1 |
| 2014 | 10 | 2 | 7 | 1 | 3 | 7 | 0 |
| 2015 | 6 | 1 | 4 | 1 | 3 | 3 | 0 |
| 2016 | 9 | 6 | 3 | 0 | 6 | 1 | 2 |
| 2017 | 11 | 3 | 5 | 3 | 3 | 5 | 3 |
| 2018 | 10 | 1 | 6 | 3 | 1 | 3 | 6 |
| Total | 102 | 36 | 48 | 18 | 41 | 46 | 15 |
To manage the imbalanced yield data for using in GWAS and then developing genomic prediction model for grain yield in Texas wheat breeding programs, genetic correlations coefficients among yield data of common TXE lines in different environments were calculated through R package META-R (Alvarado et al. 2020), and the significantly positive correlation indicated that those common TXE lines showed similar trends of reacting to growing condition under those environments. Yield data collected from all environments were then grouped according to their correlations. For example, if data of common lines in datasets A and B were correlated and data of common lines in datasets B and C were correlated, the three datasets A, B, and C will be kept in one correlated group though no common lines between A and C for correlation calculation. The best linear unbiased prediction (BLUP) in each correlation group was calculated using the mixed model y = Xβ + Zu + ε using the R package lme4 (Bates et al. 2015) with genotype was set as the only random effect, where y represents the vector of observations, β and u mean fixed and random effects, respectively, X and Z are matrices of observations related to fixed and random effects, respectively, and ε is the residual of the model. Since genotype was the only random effect in this model, variation from random effect will be the genetic variance (VG) and thus can be used to estimate the heritability (H2) of grain yield in each correlation group using the formula H2 = VG/(VG + Ve), where Ve is the residual variance. If a dataset from one environment was included in heritability estimation and lead to an increase in yield heritability of one correlation-based group, the dataset was kept in that correlation-based group. Such a heritability estimation was conducted for each environment, including the datasets that were non-correlated in correlation analysis in the previous step, to finally determine if the dataset should be kept in the corresponding correlation-based group.
Once correlation-based groups of yield data were finalized through heritability estimation, the R package lme4 and the mixed model y = Xβ + Zu + ε were used again to calculate the best linear unbiased estimation (BLUE) of each line in each correlation-based group with genotype was set as the fixed and environment set as the random effects. The yield BLUEs calculated in each group were used for GWAS and developing genomic prediction models.
Evaluation resistance to greenbug and Hessian fly
Growth chamber and greenhouse experiments were conducted to evaluate resistance to greenbug (Schizaphis graminum Rondani) and Hessian fly (Mayetiola destructor Say) in the TXE lines. Briefly, wheat plants grown in one-gallon pots were maintained in 60 × 60 × 60 cm cages (MegaView Science Co., Ltd., Taichung, Taiwan) equipped with insect proof mesh. Greenbug biotype E and Hessian fly biotype GP colonies were established and maintained on caged wheat plants for approximately 6 weeks prior to the evaluation experiments and were used as the source for the subsequent assays.
Greenbug infestation was done according to Weng and Lazar (2002). Cultivar ‘TAM 105’ or TAM 111 was the susceptible control while ‘TAM 110’ was the resistant control. Sixteen lines and two controls were grown in a 30 × 50 cm flat with 20 seeds per line. At the three-leaf stage, about 500 greenbugs were scattered over each test flat and the flats were then kept in a growth chamber at 22 °C with a day length of 8 h. The plant was scored as either resistant (normal healthy) or susceptible (chlorotic leaf and necrotic stem lesions) 10–14 days after infestation. Percentage of resistant plants in each line was recorded for GWAS.
Hessian fly infestation was conducted as described in Chen et al. (2009). Wheat accessions ‘Carol’ (H3), ‘Cardwell’ (H6), and ‘Molly’ (H13) were used as the resistant checks, and ‘Danby’ as the susceptible control. Twenty lines and four checks with 25 seeds per line were planted in one plastic flat (56 × 36 cm) in a greenhouse at 18 ± 3 °C with day length as 14 h. When the first leaf was fully expanded and the second leaf started emerging, about 200 newly mated female flies were released to each flat covered with a cheesecloth tent (540 × 120 × 40 cm). Resistance rating was conducted 3 weeks later and the stunted plants having bloated live larvae at stem base were considered as susceptible (S), and the normally healthy plants with small dead larvae or tiny live larvae between leaf sheaths as resistant (R). Percentage of resistant plants per line was calculated for GWAS.
SNP genotyping and marker data management
Whole genomic DNA was extracted from leaf samples using CTAB (cetyl trimethylammonium bromide) method (Stewart and Via 1993) with slight modification (Liu et al. 2013). SNP genotyping was done through ddRADSeq procedure (Peterson et al. 2012). Briefly, genomic DNA was co-digested with two restriction enzymes PstI (CTGCAG) and MspI (CCGG) and barcoded adapters were then ligated to DNA segments of each individual sample. Adapter oligos were synthesized from Integrated DNA Technologies (IDT), Inc. (Coralville, IA), and were mixed in equimolar amounts (30 µM of top and bottom oligos). After denaturing at 95 °C for 10 s, oligos were cooled to 12 °C at a rate of 0.1 °C/s. P5-Index adapters were made through annealing the top and bottom oligos (top oligo (5’–3’): AAT GAT ACG GCG ACC ACC GAG ATC TAC ACX XXX XXX XTC TTT CCC T; bottom oligo (5’–3’): /5Phos/AXX XXX XXX GTG TAG ATC TCG GTG GTC GCC GTA TCA TT, where XXXXXXXX represents 8-base i5 index sequences). The P5-PstI-Bridge adapters were made by annealing top (Pster_T, 5’ to 3’): /5Phos/ACA CGA CGC TCT TCC GAT CTT GCA and bottom (Pster_B, 5’ to 3’): AGA TCG GAA GAG CGT CGT GTA GGG AAA G oligos. P7-MluCI adapter was made by annealing top (P7-MluCI_T, 5’ to 3’): AAT TAG ATC GGA AGA GCA CAC GTC TGA ACT CCA GTC AC and bottom (P7-MluCI_B, 5’ to 3’): GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T.
The ddRADSeq libraries were constructed using 96-plex plate with a single random blank well used for quality control and were then sequenced through an Illumina HiSeq 2000 at the Genomics & Bioinformatics Services of Texas A&M AgriLife Research at College Station, TX (Yang et al. 2020), and SNP calls were made using the reference-based Stacks Pipeline (Catchen et al. 2013) using IWGSC v1.0 as the reference genome (IWGSC 2014), which obtained over 247,000 raw SNP data with missing rate below 50%. Considering all TXE lines were at F9 generation or later that have a very low level of heterozygosity and thus the homozygous SNP readings should be more reliable, which is also approved by comparison of SNP readings of few control lines with 2–4 replications included for DNA sequencing and SNP calling (data not shown). Majority of heterozygous SNP readings were more likely due to technique error during sequencing according to their extra high heterozygosity rate. Therefore, heterozygous marker data from TXEs thus were all converted as the missing data, and all SNP data with > 30% missing rate or MAF < 5% were removed using the computer package Tassel v5.0 (http://www.maizegenetics.net/) (Bradbury et al. 2007), which retained over 75,000 SNPs with a higher level of reliability. Genotype imputation with accuracy of 98% was then conducted using computer program Beagle (v5.0) (Browning and Browning 2007) and achieved data missing rate to less than 10%. Imputed data were filtered again by removing SNPs with MAF less than 5% and obtained the final set of 70,525 SNPs were used for GWAS.
In the set of 105 advanced breeding lines from yield trial in 2018, SNP genotyping and marker data management was done using the similar methods as indicated for TXE collections. A total of 384,648 SNPs was called and imputed in the 105 advanced lines with marker data missing rate less than 10%. Among those SNPs, 37,975 were the common set between TXE and advanced breeding lines and were extracted for validating the accuracy of genomic prediction model developed from TXE by comparing the predicted with observed yield in those 105 advanced lines.
Population structure analysis
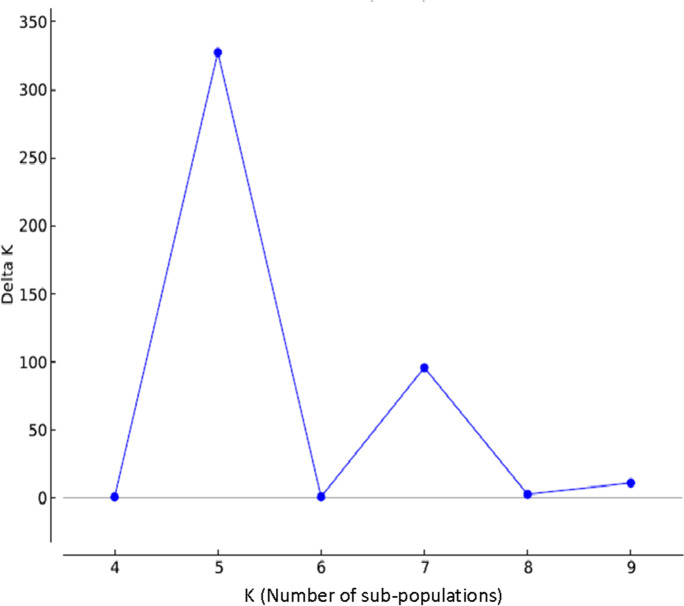
From the raw 247,000 SNPs in TXE with a data missing rate of less than 50%, a set of 8,401 SNPs with data missing rate less than 18% and heterozygosity less than 5% was considered the most reliable markers for analyzing population structure in TXE lines. The computer program Structure v2.3.4 (https://web.stanford.edu/group/pritchardlab/structure.html) (Falush et al. 2003; Pritchard et al. 2000) was used with the number of presumable sub-populations (K) set from three to ten with iteration number equal to ten based on preliminary structure scan in the TXE collection. For simulation running under each K, length of burn-in period was set to 10,000 and number of MCMC replicates was set to 100,000 with the model of admixture and correlated allelic frequency was used. The number of sub-populations was then determined using delta K (ΔK) method described in Evanno et al. (2005) through the online tool Structure Harvester (http://taylor0.biology.ucla.edu/structureHarvester/). Meanwhile, phylogenetic tree using 70,525 imputed SNP data through UPGMA (unweighted pair group method with arithmetic mean) hierarchical clustering method was also carried out through Tassel v5.0 (Bradbury et al. 2007) and the clade tree was drawn using the online tool Interactive Tree Of Life (iTOL v5) (Letunic and Bork 2019). The phylogenetic tree was used to verify the results obtained from Structure v2.3.4.
Genome-wide association studies
GWAS was carried out using the set of 70,525 imputed SNPs through Tassel v5.0 (Bradbury et al. 2007). Principal component analysis (PCA) was conducted with the number of sub-populations determined by Structure v2.3.4 to generate the Q-matrix that incorporated as the covariate in association analysis, and the fixed and random effects mixed model (MLM) (Liu et al. 2016b; Yu et al. 2006; Zhang et al. 2010) was used for association mapping with the K-matrix showing the relationship of all individuals that was used to account for effects due to kinship. For detecting genomic regions associated with grain yield, the yield BLUE of each line calculated using the R package lme4 in correlation-based groups was used as the trait data and GWAS was separately conducted in each correlation-based group. Bonferroni adjustment using R package simpleM (http://simplem.sourceforge.net/) (Gao et al. 2010) determined the significant threshold of − log10(P) = 4.0 for grain yield and − log10(P) = 6.0 for insect resistance.
SNP allele frequency change during new TXE line development
According to the time of line development, the 227 TXE lines were divided into three groups using a 3-year interval with the first group including 92 lines developed during 2009–2011 (namely the old group), the second group containing 67 lines from 2012 to 2014, and the third group had 68 lines from 2015 to 2017 (namely the newly developed group). Therefore, comparing allele frequency between the first and the third groups would have a good indication of allele drifting due to breeding selection in Texas wheat breeding programs. Allele frequency change was investigated focusing on the major allele genotype of SNPs in the TXE collection. The frequency of each SNP major allele was respectively calculated in the first and third groups and then to find the difference between the two frequencies.
Genomic prediction model development
For developing genomic prediction models, 7,573 SNP markers from all chromosomes with at least one million bases (Mb) apart were selected for estimating the mean effect of each marker. The R package rrBLUP (ridge regression best linear unbiased prediction) (Endelman 2011) was used for developing genomic prediction model. The mixed model y = µ + Xβ + ε was used with y as the vector of phenotypic means, µ as the overall mean, X as the marker matrix, β as the vector of marker effects, and ε as the vector of residual effects. The genomic estimated breeding values (GEBVs) of each line were calculated by adding the grand mean to the product of genotypic matrix and the vector of mean effect of each marker. The prediction accuracy was measured by the correlation between the predicted and observed yield BLUEs. Genomic prediction models were developed separately in each of the correlation-based groups, and the prediction accuracy was estimated at three times using 60%, 70%, and 80% of TXE lines as training sets and 40%, 30%, and 20% as testing sets, accordingly. For each training/testing set, prediction accuracy was obtained based on a calculation using 500 repeated runs.
To validate the prediction models developed in each correlation-based group, all TXE lines were used as the training set and 105 advanced breeding lines were used as the testing set. The common SNPs between TXEs and advanced breeding lines with at least 1-Mb apart on each chromosome were selected. Marker effects were estimated using BLUEs of each correlation-based group through rrBLUP mixed model y = µ + Xβ + ε. The GEBVs of each advanced breeding line were calculated by adding the grand mean to the product of genotypic matrix and the vector of mean effect of each marker. Prediction accuracy was then measured through the correlation between predicted and observed yield in 105 advanced breeding lines.
Results
Grain yield in two correlation-based groups
Based on correlations among yield of overlapped lines in different environments, yield data from eighteen environments were not correlated with any of the remaining 84 environments (Tables S1 and S2). Data from those 84 environments were divided into two correlation-based groups with groups G1 containing 36 and G2 including 48 environments (Table 1). After heritability estimation, data from five non-correlated environments were added into the group G1 but data from two environments were dropped from the group G2. Therefore, group G1 contained data of 41 environments and group G2 carried data from 46 environments for further analysis, and data from 15 non-correlated environments were abandoned (Table S3). Interestingly, the majority of dataset in correlation-based group G1 included environments from the High Plains and Rolling Plains that normally have low level of rainfall and represented the drought-prone areas of wheat acreage in Texas, whereas the majority of data in group G2 contained environments from Blacklands and South Texas that usually received relatively a higher level of rainfall and represented the wet growing conditions of Texas (Table S3).
Of the two correlation-based groups that respectively included data collected from 41 and 46 environments, the best linear unbiased estimation (BLUE) of each line was calculated through R package lme4 and the skewed yield distributions were observed in both groups (Fig. 1). In correlation-based group G1, the grain yield of all lines ranged from 2,000 to 4,500 kg/ha, but 201 (88.5%) lines have grain yield in the range of 3,250–4,000 kg/ha. Whereas in the correlation-based group G2, grain yield of each line was in the range of 2,250–4,750 kg/ha but with more lines distributed in a broader range (Fig. 1, Table S11). The grain yield in two datasets was not correlated and Bartlett’s test also indicated heterogeneous variances in the two datasets. Since the correlation-based groups G1 and G2 were corresponding to dry and wet growing condition in north and south Texas, respectively, the broader trait variation in group G2 corresponding to the less stressful growing conditions in south Texas may more likely explain the genetic variance.
Fig. 1.
Distribution of the best linear unbiased estimations (BLUEs) for grain yield in the two correlation-based groups G1 and G2. BLUEs were calculated using the R package lme4 (Bates et al. 2015)
SNP genotyping and population structure in TXE collection
Of the 70,525 imputed SNPs with data missing rate less than 10%, the B-genome chromosomes carried the most (37,773) SNPs, followed by the chromosomes in the A-genome (23,782) and the D-genome (8,970) (Table S4), indicating the relatively lower level of genetic diversity in the D-genome. Particularly, only 731 and 708 SNPs were identified on chromosomes 4D and 5D, respectively, suggesting that the two chromosomes in TXE lines have the least variations.
Population structure analysis revealed five sub-populations in the TXE collection (Fig. 2) and the population was mainly admixed with all released cultivars (Fig. S2) spread into different sub-populations, which indicated that the TXE collection covered primary gene sources in Texas wheat breeding programs. Phylogenetic tree developed using 70,525 imputed SNPs also suggested the similar population structure (Fig. 3).
Fig. 2.
Population structure analysis using Structure v2.3.4 (Evanno et al. 2005; Pritchard et al. 2000) based on the most reliable set of 8,401 SNPs in 227 TXE lines. Five sub-populations were contained in the collection. Number of sub-populations was determined using delta K method through Structure Harvester (Earl and vonHoldt 2012)
Fig. 3.
Phylogenetic tree developed using 70,525 imputed SNPs in 227 TXE lines. The released cultivars were indicated in the corresponding clusters. Phylogenetic tree was produced using Tassel v5.0 (Bradbury et al. 2007) through UPGMA (unweighted pair group method with arithmetic mean) hierarchical clustering method and was drawn using the Interactive Tree Of Life (iTOL v5) (Letunic and Bork 2019)
Genome-wide association studies of grain yield in TXE lines
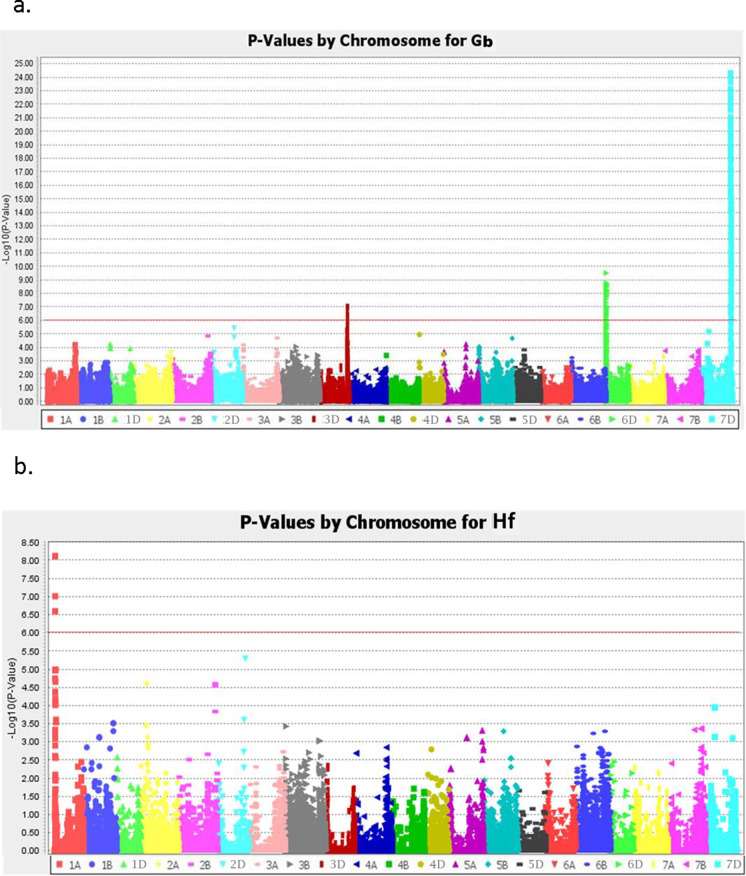
Association analysis identified 74 genomic regions in two correlation-based groups associated with grain yield with significant level above threshold of − log10(P) = 4.0 (Fig. 4, Tables S5 and S6). Of those associations, two regions were commonly detected in both groups and located on chromosome 1D at 229.6 and 345.4 Mb with − log10(P) scores of 4.4 and 4.8 and each contributing around 10% of yield variation. The favorite alleles at two regions had the additive effects of increasing yield by 330.5 and 325.0 kg/ha in group G1 and 406.7 and 343.5 kg/ha in group G2, respectively. In addition, 17 genomic regions identified only in correlation-based group G1 and 55 regions only in group G2 were associated with grain yield. In group G1, those genomic regions were located on nine chromosomes, namely, 2A, 3A, 3B, 5A, 5D, 6B, 6D, 7B, and 7D. The − log10(P) scores ranged from 4.0 to 5.8 and explained 7 to 14% of yield variations with favorite alleles having the potential of increasing yield 220.9–816.3 kg/ha. There were ten genomic regions with each explaining 10% or more of phenotypic variations located on chromosome 2A at 308.1 Mb, 3A at 12.8 Mb, 3B at 245.3 Mb, 5A at 24.1 Mb, 6B at 32.8 Mb and 682.6 Mb, 6D at 454.1 Mb, and 7B at 627.2, 637.8, and 676.8 Mb. In group G2, the significant genomic regions were detected from all chromosomes with the significance varying from − log10(P) = 4.0 to 8.1 and each accounting for 7 to 16% of trait variations with the favorite allele having the additive effects of increasing yield 258.0–516.1 kg/ha. The most significant association was located at 14.7 Mb on chromosome 7D and explained 16% of the phenotypic variations with the favorite allele having the potential of increasing yield by 516.1 kg/ha (Tables S5 and S6). A total of 16 regions on seven chromosomes each explained over 10% of trait variations in group G2 (Table S5 and Fig. 4).
Fig. 4.
GWAS in TXE collections using Tassel v5.0 identified genomic regions significantly associated with grain yield in correlation-based groups G1 and G2. Critical threshold was set at − log10(P) = 4.0
Genome-wide association studies of greenbug and Hessian fly resistance in TXE lines
For reaction to greenbug, 173 and 42 TXE lines were susceptible and resistant, respectively, and twelve lines showed partial resistance (Table S7). GWAS indicated that three genomic regions on chromosome 3DL (565.0 Mb), 6DS (7.2 Mb), and 7DL (597.9 Mb) were associated with greenbug resistance in TXE lines (Fig. 5a and Tables S8 and S9). A region on chromosome 7DL showed the largest effect and explained 48.7% of the trait variation in TXE lines, and the regions on 3DL and 6DS explained 10.7% and 15.3% of phenotypic variation, respectively. For reaction to Hessian fly, data were obtained from 219 TXE lines with 166 susceptible, 18 resistant, and 35 partially resistant (Table S7). Only the genomic region on 1AS at 7.8 Mb was significantly associated with the resistance and explained 17.0% of trait variation in TXE lines (Fig. 5b and Tables S8 and S9).
Fig. 5.
GWAS in TXE collections using Tassel v5.0 identified genomic regions significantly associated with greenbug resistance (a) and Hessian fly resistance (b). Critical threshold was set at − log10(P) = 6.0
SNP allele drift in TXE lines due to breeding selection
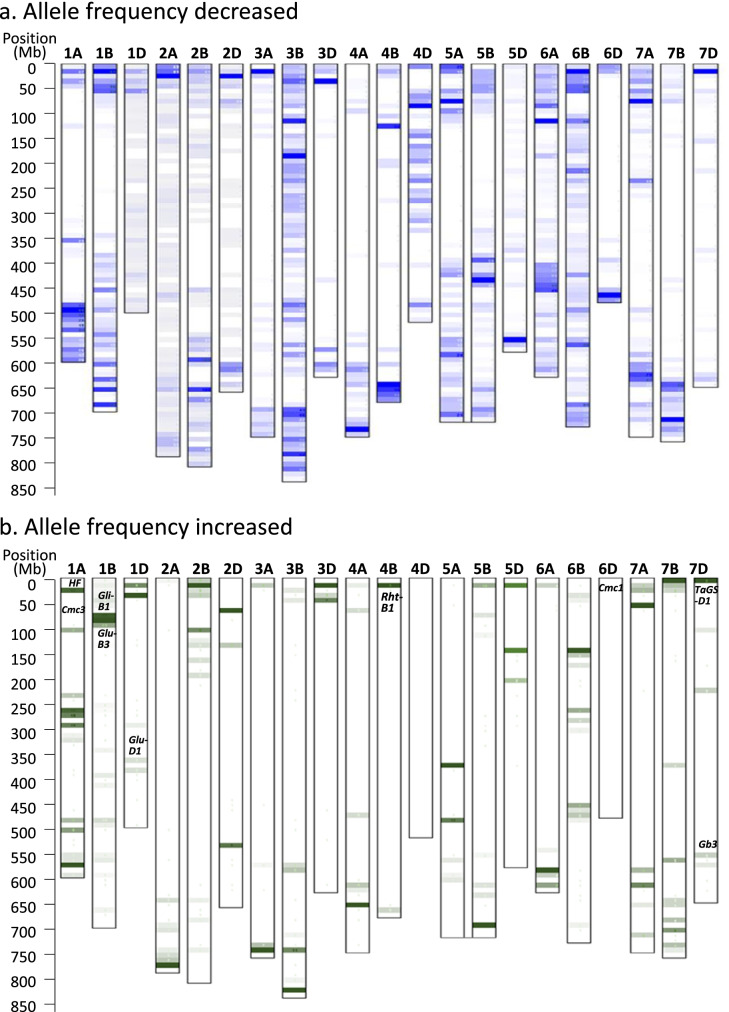
By comparing frequency of major alleles in 70,525 SNPs between TXE groups of 2009–2011 (old) and 2015–2017 (new), allele frequencies of 10,034 SNPs decreased. Meanwhile, allele frequency in a different set of 974 SNPs each increased over 20% in the new TXE group. Of the SNPs with allele frequency decreasing in the newly developed TXEs, the allele genotype of 2,000 SNPs that have been the major alleles in TXE group of 2009–2011 was changed to minor allele. Whereas in SNPs with allele frequency increasing in the newly developed TXEs, 300 SNPs changed the allele status from minor in the group of 2009–2011 to major in the group of 2015–2017 (Table S10). Comparing the SNPs that had significant associations in increasing yield, several were located in the vicinity of the SNPs that had allele status changing from minor to major in new TXEs, such as the ones on 2A at 101.9 Mb, 3B at 245.3 and 738.3 Mb, 6D at 454.1 Mb, 7A at 620.3 Mb, 7D at 621.2 Mb, and 7B at 654.1, 711.9, and 741.0 Mb. Each of these alleles showed the potential of increasing yield by 281.8–611.3 kg/ha (Table S5). Among the 74 SNPs significantly associated with yield in groups G1 or G2, there were trends that the newer lines or cultivars had more favorite alleles for increasing yield (Table S6). Based on the pseudomolecule physical position indicated in the reference wheat genome sequence (IWGSC 2014), markers with allele frequency changing over 20% were mostly located at the distal sides of the chromosomes (Fig. 6) and agreed with the higher rate of recombination observed at the distal regions of the chromosomes.
Fig. 6.
Diagram of allele frequency change among sets of TXE lines developed during 2009–2011 (old TXE, 92 lines) and 2015–2017 (newly developed TXE, 68 lines). a Allele frequency decreased in newly developed TXEs. b Allele frequency increased in newly developed TXEs with some major genes indicted in the corresponding position according to previous research (Liu et al. 2014; Dhakal et al. 2018; Zhang et al. 2014). Physical position of SNPs was determined according to pseudomolecule position in wheat reference genome v1.0 (IWGSC 2014). Darker regions indicated that more markers have frequency changed
Genomic prediction in TXE lines and model validation using advanced breeding lines
Genomic prediction models were tested in three situations that randomly picked 60%, 70%, and 80% of TXE lines as training populations to predict the remaining TXE lines. After 500 independent runs in each situation, prediction accuracy using yield data from correlation-based group G2 is higher than using data from correlation-based group G1 (Table 2). Average prediction accuracies in group G1 varied from 0.42 to 0.44 but that increased from 0.68 to 0.70 in group G2 as the size of training population increased. The lowest range of the prediction accuracies was 0.14–0.19 in group G1 and 0.49–0.52 in group G2 and the maximum prediction accuracies were 0.61–0.75 in group G1 and 0.81–0.89 in group G2. This indicated that yield data in correlation-based group G2 were more reliable for genomic prediction.
Table 2.
Prediction accuracy using different portions of TXE lines as training and testing sets in two correlation-based groups G1 and G2
| Percentage ration of training:testing | Correlation-based group G1 | Correlation-based group G2 | ||||
|---|---|---|---|---|---|---|
| Average | Minimum | Maximum | Average | Minimum | Maximum | |
| 60%:40% | 0.42 ± 0.07 | 0.19 | 0.61 | 0.66 ± 0.04 | 0.52 | 0.81 |
| 70%:30% | 0.45 ± 0.08 | 0.14 | 0.65 | 0.68 ± 0.04 | 0.52 | 0.82 |
| 80%:20% | 0.46 ± 0.10 | 0.18 | 0.75 | 0.69 ± 0.06 | 0.49 | 0.89 |
To validate the prediction models developed in two correlation-based groups, yield data in a set of 105 advanced breeding lines were collected in 2018 from four environments including rain-fed and irrigated location in Bushland, TX, irrigated location in Etter, TX, and the rain-fed location in McGregor, TX. All TXE lines were used as the training set and 105 advanced breeding lines were used as the testing set. From the common 37,975 SNPs between TXE and advanced breeding lines, a total of 5,542 SNPs that were at least 1-Mb apart on each chromosome were selected for genomic prediction, and the prediction accuracies using the models developed from the correlation-group G2 ranged from 0.12 to 0.29, but none of the predictions based on the models from correlation-group G1 was correlated with the observed yield (Table 3). This is consistent with previous results of models from correlation-group G2 which were more reliable for genomic prediction when using TXE lines as both training and testing sets.
Table 3.
Prediction validation using yield data obtained from a set of advanced breeding lines in 2019 based on genomic prediction models developed in two correlation-based groups G1 and G2
| Model | Environmenta | |||
|---|---|---|---|---|
| BD | BI | EI | MCG | |
| Correlation-based group G1 | − 0.06 | − 0.03 | 0.01 | − 0.06 |
| Correlation-based group G2 | 0.29 | 0.12 | 0.14 | 0.23 |
aBD and BI mean rain-fed and irrigated land in location at Bushland, TX, respectively. EI means irrigated land in location at Etter, TX. MCG means rain-fed land location in McGregor, TX
Discussion
Wheat grain yield is a very complex trait and is affected by numerous genes involved in many different biological processes affecting plant development, photosynthesis, carbon mobilization, grain filling, and maturity. The effect of each gene is very limited and varied under different environments. Testing grain yield in breeding lines thus demands major efforts and breeding resources since it needs to be done in many locations under multiple years with replications included. Using historical yield data in the past or current breeding lines or cultivars to conduct genome-wide association analysis and genomic prediction will provide a cost-effective way of identifying beneficial genes for increasing yield and cumulating favorite alleles for crop improvement. However, imbalanced historical data obtained during breeding are hard to use since each environmental condition is unique and cannot be repeated. Interactions between genotypes and environments vary at different times and locations, which greatly increased difficulties of identifying favorite alleles. In this study, we developed a strategy of using correlations among yield data of overlapped lines evaluated at different times and locations to group different environments that have showed interactions with similar magnitudes. The grouping is further tested through heritability estimation. The best linear unbiased estimation (BLUE) calculated in each correlation-based group was then used for GWAS and developing genomic prediction models, which were further validated through a set of advanced breeding lines. This research thus developed a new strategy of using the imbalanced historical data for detecting beneficial genes and conducting genomic prediction in breeding.
There are numerous QTLs for yield and yield components identified from bread wheat trials worldwide. Chromosome regions with significant SNPs associated with yield from GWAS in this study were almost coincided to the position of several QTLs identified from previous research from bread wheat trials conducted in the US Great Plains or other regions (Table S5). Yield-associated genomic regions in this research at 71.4 Mb on 2A, 21.8 Mb on 2D, 12.8 Mb on 3A, 42.6 Mb on 3B, 682.6 Mb on 6B, 627.2 Mb and 654.1 Mb on 7B, and 592.2 Mb on 7D were very close to the QTLs at 79.8 Mb on 2A, 15.7 Mb on 2D, 9.6 Mb on 3A, 48.6 Mb on 3B, 673.8 Mb on 6B, 617.0 Mb and 647.8 Mb on 7B, and 591.2 Mb on 7D that were associated with yield and yield components identified from a bi-parental mapping population derived from the cross between two popular cultivars TAM 111 and TAM 112 (Yang et al. 2020). Both cultivars and their derivatives have been the core parents in both Texas whet breeding programs. Particularly, the region around 591.2 Mb on 7D is harboring gene Gb3 conferring greenbug resistance (Liu et al. 2014). Breeders found that the majority of the TAM 112 derivatives had a decent yield in dry environments as Gb3 was kept (J Rudd, personal communication, 2020).
Yield-associated regions at 603.0 Mb on chromosome 3D and 25.4 Mb on 7B were very close to the QTLs associated with spikes per square meter at 603.8 Mb on 3D linked to XIWA6485 and kernel per spike at 22.6–24.7 Mb on 7B linked to XIWB71684 with favorite alleles from ‘TAM 111’ (Assanga et al. 2017). The yield-associated region at 603.0 Mb on 3D was very close to a QTL at 603.4 Mb on 3D of ‘ND 705’ that was associated with spikes per square meter and linked to XIWB17317 (Kumar et al. 2019).
Yield-associated regions at 532.8 Mb on 1A and at 711.5 Mb on 7B from this study were physically close to a flour yield QTL at 533.4 Mb on 1A and a grain volume weight QTL at 709.6 Mb on 7B detected in a recombinant inbred mapping population derived from cross between TAM 111 and TAM 112 population (Yang et al. 2020; Dhakal et al. 2021a). The yield-associated region at 531.3 Mb on 2D from this study was also identified in the mapping population derived from TAM 112/TAM 111 and associated with thousand kernel weight and kernel diameter (Yang et al. 2020; Dhakal et al. 2021b). Since cultivars TAM 111 and TAM 112 were core parents used in the Texas A&M AgriLife Research wheat breeding programs, it is very possible that these favorite alleles were carried through generations due to selections.
The yield-associated region at 16.2 Mb on 7D was close to gene TaGS3-D1 located in region 6.5–6.8 Mb on 7D affecting wheat kernel weight and length (Rasheed et al. 2016; Zhang et al. 2014). Two QTLs at 32.8 Mb and 47.5 Mb on 6B associated with thousand kernel weight (Zou et al. 2017) coincided with the two yield-associated regions at 32.8 and 47.5 Mb on 6B in this study. The regions at 709.2 Mb on 3A, 625.7 Mb on 5A, and 633.9 Mb on 6B from this study were very close to gene TaTGW6-A1 at 711.1 Mb on 3A, a QTL at 619.5 Mb on 5A for thousand kernel weight, and a QTL at 631.8 Mb on 6B for grain volume weight (Juliana et al. 2019). Therefore, the consistence between findings of this research and those from previous reports further proved that the strategy we developed for using imbalanced historical breeding data is effective for identifying important beneficial genes.
The region at 597.9 Mb on 7DL showed significant association with greenbug resistance and is corresponding to Gb3, the gene known to be carried by germplasms used for developing TXE lines and present in cultivars such as TAM 110, TAM 112, ‘TAM 115’, and TAM 204 (Lazar et al. 1997; Rudd et al. 2014, 2019; Weng and Lazar 2002; Liu et al. 2014). The other two regions on 3DL and 6DS with minor effects on greenbug resistance might be novel genes since no greenbug resistance have been reported from these two genomic regions. Hessian fly resistance was associated with a region on 1AS (7.8 Mb) in this study; the position coincided with a Hessian fly resistance QTL on 1AS in ‘Duster’ (PI 644,016) (Li et al. 2015; Edwards et al. 2012). It is likely that Hessian fly resistance in TXE lines is derived from Duster since many TXE lines had this cultivar in their pedigree. It thus indicated that the additional resistance sources need to be included in the future Texas wheat breeding to maintain the long-lasting resistance to Hessian fly.
From this study, eight chromosome regions significantly associated with yield, along with several major genes in TXE lines such as wheat curl mite resistance genes CmcTAM112 and Cmc3 (Dhakal et al. 2018; Dhakal et al. 2017; Zhao et al. 2021; Gaurav et al. 2021), seed storage protein subunit genes Gli-B1 and Glu-D1, dwarf gene Rht-B1, and grain weight and length gene TaGS-D1 (Zhang et al. 2014; Liu et al. 2014). These mostly coincided with regions in which allele frequencies were greatly increased in the newly developed lines (Fig. 6b; Table S10), which may be a good indication of accumulating favorite alleles during selection. Particularly, the recently released cultivars fall into different sub-populations (Figs. 3 and S2) showing improved yield, disease and/or insect resistance, drought tolerance, and enhanced baking and milling quality attributes. As aforementioned, cultivars TAM 111 (Lazar et al. 2004) and TAM 112 (Rudd et al. 2014) were used as parental lines in new releases due to their high yield and superior drought tolerance in addition to the greenbug and wheat curl mite resistance carried in TAM 112. For example, cultivar TAM 114 derived from crosses of using TAM 111 as parents showed excellent baking and milling quality, and intermediate resistance to Hessian fly (Rudd et al. 2018), and cultivar TAM 204 selected from the crosses involving TAM 112 showed a good level of resistance to greenbug, Hessian fly, and wheat curl mite in addition to the high grain yield (Rudd et al. 2019). The newly released TAM 115 is also a selection from crosses involving TAM 112 and showed high yield, good drought tolerance, and resistance to greenbug and wheat curl mite (Rudd et al., not published). Therefore, further research focusing on the regions where allele frequency greatly increased in those newly developed TXE lines may provide an efficient way of revealing beneficial alleles for wheat improvement.
Of the two correlation-based groups G1 and G2 developed through historical yield data of TXE lines, results from GWAS and genomic prediction both indicated that yield data from group G2 may be more reliable. This is supported by the fact that group G2 contained environments either in north Texas under irrigated conditions or from south Texas with relatively higher level of rainfall and thus had better growing conditions. On the other hand, the group G1 included mainly dryland environments with severer drought stress, which greatly limited the expression of yield potential in each line and led to a much narrow yield variation (Fig. 1). Similarly, GWAS detected fewer genomic regions significantly associated with yield in group G1 than in G2 (Table S5), and genomic prediction models in two groups using 60–80% of lines as training set also pointed to lower prediction accuracy in group G1 (0.14–0.75) than in G2 (0.49–0.89) (Table 2). Validation of genomic prediction models through a set of advanced breeding lines also indicated that predictions made using yield data of group G2 showed stronger correlation with the observed data (Table 3). Therefore, the strategy of combining correlation and heritability estimates to group data from different environments used in this study also provided a way of selecting appropriate data from diverse environments for genomic predicting in breeding.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
Great thanks to Dr. Shuhao Yu in Texas A&M AgriLife Research at Amarillo for providing critical review to this manuscript.
Author contribution
C. Chu conducted data analysis and prepared the manuscript; S. Wang conducted SNP calls and imputation of GBS genotyping; J.C. Rudd, A.M.H. Ibrahim, and Q. Xue designed yield trial experiments and helped on yield data collection and manuscript editing; R.N. Devkota, J.A. Baker, S. Baker, B. Simoneaux, G. Opena, X. Liu, K. Hui, and K.E. Jessup involved in yield data collection or DNA sample preparation; H. Dong and Z.S. Zhang provided helps in data analyses and reviewed the manuscript; M-S. Chen evaluated Hessian fly resistance and reviewed the manuscript; R. Metz and C.D. Johnson conducted GBS library preparation and sequencing; and S. Liu designed the project and helped on data analyses and manuscript preparation.
Funding
This research was supported partially by Texas A&M AgriLife Research, Texas Wheat Producers Board, U.S. Department of Agriculture, Agricultural Research Service, and National Research Initiative Competitive Grants 2017–67007-25939, 2019–67013-29172, and 2021–67013-33940.
Declarations
Ethics approval
The experiments comply with the ethical standards in the country in which they were performed.
Conflict of interest
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Chenggen Chu, Email: chenggen.chu@usda.gov.
Shuyu Liu, Email: Shuyu.Liu@ag.tamu.edu.
References
- Agarwal M, Shrivastava N, Padh H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008;27(4):617–631. doi: 10.1007/s00299-008-0507-z. [DOI] [PubMed] [Google Scholar]
- Alvarado G, Rodríguez FM, Pacheco A, Burgueño J, Crossa J, Vargas M, Pérez-Rodríguez P, Lopez-Cruz MA. META-R: a software to analyze data from multi-environment plant breeding trials. The Crop Journal. 2020;8(5):745–756. doi: 10.1016/j.cj.2020.03.010. [DOI] [Google Scholar]
- Assanga SO, Fuentealba M, Zhang G, Tan C, Dhakal S, Rudd JC, Ibrahim AMH, Xue Q, Haley S, Chen J, Chao S, Baker J, Jessup K, Liu S. Mapping of quantitative trait loci for grain yield and its components in a US popular winter wheat TAM 111 using 90K SNPs. PLoS ONE. 2017;12(12):e0189669. doi: 10.1371/journal.pone.0189669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT. Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature. 2010;465(7298):627–631. doi: 10.1038/nature08800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avni R, Nave M, Barad O, Baruch K, Twardziok SO, Gundlach H, Hale I, Mascher M, Spannagl M, Wiebe K. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science. 2017;357(6346):93–97. doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PloS One. 2008;3(10):e3376. doi: 10.1371/journal.pone.0003376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S. Fitting linear mixed-effects models using lme4. J Stat Softw. 2015;67:48. doi: 10.18637/jss.v067.i01. [DOI] [Google Scholar]
- Bernardo R, Yu J. Prospects for genomewide selection for quantitative traits in maize. Crop Sci. 2007;47(3):1082–1090. doi: 10.2135/cropsci2006.11.0690. [DOI] [Google Scholar]
- Bhat JA, Ali S, Salgotra RK, Mir ZA, Dutta S, Jadon V, Tyagi A, Mushtaq M, Jain N, Singh PK. Genomic selection in the era of next generation sequencing for complex traits in plant breeding. Front Genet. 2016;7:221. doi: 10.3389/fgene.2016.00221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23(19):2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Human Genetics. 2007;81(5):1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA. Stacks: an analysis tool set for population genomics. Mol Ecol. 2013;22(11):3124–3140. doi: 10.1111/mec.12354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M-S, Echegaray E, Whitworth RJ, Wang H, Sloderbeck PE, Knutson A, Giles KL, Royer TA. Virulence analysis of hessian fly populations from Texas, Oklahoma, and Kansas. J Econ Entomol. 2009;102(2):774–780. doi: 10.1603/029.102.0239. [DOI] [PubMed] [Google Scholar]
- Chu C, Wang S, Paetzold L, Wang Z, Hui K, Rudd JC, Xue Q, Ibrahim AMH, Metz R, Johnson CD, Rush CM, Liu S. RNA-seq analysis reveals different drought tolerance mechanisms in two broadly adapted wheat cultivars ‘TAM 111’ and ‘TAM 112’. Sci Rep. 2021;11(1):4301. doi: 10.1038/s41598-021-83372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collard BCY, Mackill DJ. Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Philosophical Transactions of the Royal Society b: Biological Sciences. 2008;363(1491):557–572. doi: 10.1098/rstb.2007.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daetwyler HD, Villanueva B, Woolliams JA. Accuracy of predicting the genetic risk of disease using a genome-wide approach. PloS One. 2008;3(10):e3395. doi: 10.1371/journal.pone.0003395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson JC, Endelman JB, Heslot N, Crossa J, Poland J, Dreisigacker S, Manès Y, Sorrells ME, Jannink J-L. The use of unbalanced historical data for genomic selection in an international wheat breeding program. Field Crop Res. 2013;154:12–22. doi: 10.1016/j.fcr.2013.07.020. [DOI] [Google Scholar]
- Dhakal S, Liu X, Chu C, Yang Y, Rudd JC, Ibrahim AM, Xue Q, Devkota RN, Baker JA, Baker SA. Genome-wide QTL mapping of yield and agronomic traits in two widely adapted winter wheat cultivars from multiple mega-environments. PeerJ. 2021;9:e12350. doi: 10.7717/peerj.12350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhakal S, Liu X, Girard A, Chu C, Yang Y, Wang S, Xue Q, Rudd JC, Ibrahim AMH, Awika JM, Jessup KE, Baker JA, Garza L, Devkota RN, Baker S, Johnson CD, Metz RP, Liu S. Genetic dissection of end-use quality traits in two widely-adapted wheat cultivars ‘TAM 111’ and ‘TAM 112’. Crop Sci. 2021;61(3):1944–1959. doi: 10.1002/csc2.20415. [DOI] [Google Scholar]
- Dhakal S, Tan C-T, Anderson V, Yu H, Fuentealba MP, Rudd JC, Haley SD, Xue Q, Ibrahim AMH, Garza L, Devkota RN, Liu S. Mapping and KASP marker development for wheat curl mite resistance in “TAM 112” wheat using linkage and association analysis. Mol Breed. 2018;38(10):119. doi: 10.1007/s11032-018-0879-x. [DOI] [Google Scholar]
- Earl DA, vonHoldt BM. STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour. 2012;4(2):359–361. doi: 10.1007/s12686-011-9548-7. [DOI] [Google Scholar]
- Edwards JT, Hunger RM, Smith EL, Horn GW, Chen M-S, Yan L, Bai G, Bowden RL, Klatt AR, Rayas-Duarte P, Osburn RD, Giles KL, Kolmer JA, Jin Y, Porter DR, Seabourn BW, Bayles MB, Carver BF. ‘Duster’ wheat: a durable, dual-purpose cultivar adapted to the Southern Great Plains of the USA. J Plant Reg. 2012;6(1):37–48. doi: 10.3198/jpr2011.04.0195crc. [DOI] [Google Scholar]
- Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLOS One 6:e19379 [DOI] [PMC free article] [PubMed]
- Endelman JB. Ridge regression and other kernels for genomic selection with R package rrBLUP. The Plant Genome. 2011;4(3):250–255. doi: 10.3835/plantgenome2011.08.0024. [DOI] [Google Scholar]
- Endelman JB, Atlin GN, Beyene Y, Semagn K, Zhang X, Sorrells ME, Jannink J-L. Optimal design of preliminary yield trials with genome-wide markers. Crop Sci. 2014;54(1):48–59. doi: 10.2135/cropsci2013.03.0154. [DOI] [Google Scholar]
- Evanno G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14(8):2611–2620. doi: 10.1111/j.1365-294X.2005.02553.x. [DOI] [PubMed] [Google Scholar]
- Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164 [DOI] [PMC free article] [PubMed]
- Flint-Garcia SA, Thornsberry JM, Buckler ES., IV Structure of linkage disequilibrium in plants. Annu Rev Plant Biol. 2003;54(1):357–374. doi: 10.1146/annurev.arplant.54.031902.134907. [DOI] [PubMed] [Google Scholar]
- Gao X, Becker LC, Becker DM, Starmer JD, Province MA. Avoiding the high Bonferroni penalty in genome-wide association studies. Genet Epidemiol. 2010;34(1):100–105. doi: 10.1002/gepi.20430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaurav K, Arora S, Silva P, Sánchez-Martín J, Horsnell R, Gao L, Brar GS, Widrig V, John Raupp W, Singh N (2021) Population genomic analysis of Aegilops tauschii identifies targets for bread wheat improvement. Nature biotechnology:1–10 [DOI] [PMC free article] [PubMed]
- He S, Schulthess AW, Mirdita V, Zhao Y, Korzun V, Bothe R, Ebmeyer E, Reif JC, Jiang Y. Genomic selection in a commercial winter wheat population. Theor Appl Genet. 2016;129(3):641–651. doi: 10.1007/s00122-015-2655-1. [DOI] [PubMed] [Google Scholar]
- IWGSC (2014) A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Science 345. 10.1126/science.1251788 [DOI] [PubMed]
- Jia H, Wan H, Yang S, Zhang Z, Kong Z, Xue S, Zhang L, Ma Z. Genetic dissection of yield-related traits in a recombinant inbred line population created using a key breeding parent in China’s wheat breeding. Theor Appl Genet. 2013;126(8):2123–2139. doi: 10.1007/s00122-013-2123-8. [DOI] [PubMed] [Google Scholar]
- Jia J, Zhao S, Kong X, Li Y, Guangyao Zhao WH, Appels R, Pfeifer M, Tao Y, Zhang X, Jing R, Zhang C, Ma Y, Gao L, Gao C, Spannagl M, Mayer KFX, Li D, Pan S, Zheng F, Qun Hu, Xia X, Li J, Liang Q, Chen J, et al. Aegilops tauschii draft genome sequence reveals a gene repertoire for wheat adaptation. Nature. 2013 doi: 10.1038/nature12028. [DOI] [PubMed] [Google Scholar]
- Juliana P, Poland J, Huerta-Espino J, Shrestha S, Crossa J, Crespo-Herrera L, Toledo FH, Govindan V, Mondal S, Kumar U, Bhavani S, Singh PK, Randhawa MS, He X, Guzman C, Dreisigacker S, Rouse MN, Jin Y, Pérez-Rodríguez P, Montesinos-López OA, Singh D, Mokhlesur Rahman M, Marza F, Singh RP. Improving grain yield, stress resilience and quality of bread wheat using large-scale genomics. Nat Genet. 2019;51(10):1530–1539. doi: 10.1038/s41588-019-0496-6. [DOI] [PubMed] [Google Scholar]
- Kumar A, Mantovani EE, Simsek S, Jain S, Elias EM, Mergoum M. Genome wide genetic dissection of wheat quality and yield related traits and their relationship with grain shape and size traits in an elite × non-adapted bread wheat cross. PLoS ONE. 2019;14(9):e0221826. doi: 10.1371/journal.pone.0221826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar MD, Worrall WD, Peterson GL, Porter Lwrnat KB, Marshall Mem DS, Nelson LR. Registration of ‘TAM 110’ wheat. Crop Science. 1997;37(6):2. doi: 10.2135/cropsci1997.0011183X003700060055x. [DOI] [Google Scholar]
- Lazar MD, Worrall WD, Peterson GL, Fritz AK, Marshall D, Nelson LR, Rooney LW. Registration of ‘TAM 111’ wheat. Crop Sci. 2004;44(1):355–356. doi: 10.2135/cropsci2004.3550. [DOI] [Google Scholar]
- Letunic I, Bork P. Interactive Tree Of Life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. 2019;47(W1):W256–W259. doi: 10.1093/nar/gkz239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Wang Y, Chen M-S, Edae E, Poland J, Akhunov E, Chao S, Bai G, Carver BF, Yan L. Precisely mapping a major gene conferring resistance to Hessian fly in bread wheat using genotyping-by-sequencing. BMC Genomics. 2015;16(1):108. doi: 10.1186/s12864-015-1297-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Assanga SO, Dhakal S, Gu X, Tan C-T, Yang Y, Rudd J, Hays D, Ibrahim A, Xue Q, Chao S, Devkota R, Shachter C, Huggins T, Mohammed S, Fuentealba MP. Validation of chromosomal locations of 90K array single nucleotide polymorphisms in US wheat. Crop Sci. 2016;56(1):10. doi: 10.2135/cropsci2015.03.0194. [DOI] [Google Scholar]
- Liu S, Griffey C, Hall M, McKendry A, Chen J, Brooks W, Brown-Guedira G, Sanford D, Schmale D. Molecular characterization of field resistance to Fusarium head blight in two US soft red winter wheat cultivars. Theor Appl Genet. 2013;126(10):2485–2498. doi: 10.1007/s00122-013-2149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Rudd JC, Bai G, Haley SD, Ibrahim AMH, Xue Q, Hays DB, Graybosch RA, Devkota RN, St. Amand P. Molecular markers linked to important genes in hard winter wheat. Crop Sci. 2014;54(4):1304–1321. doi: 10.2135/cropsci2013.08.0564. [DOI] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z. Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genetics. 2016;12(2):e1005767. doi: 10.1371/journal.pgen.1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marulanda JJ, Melchinger AE, Würschum T. Genomic selection in biparental populations: assessment of parameters for optimum estimation set design. Plant Breeding. 2015;134(6):623–630. doi: 10.1111/pbr.12317. [DOI] [Google Scholar]
- Meuwissen TH, Hayes BJ, Goddard ME. Prediction of total genetic value using genome-wide dense marker maps. Genetics. 2001;157(4):1819–1829. doi: 10.1093/genetics/157.4.1819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel S, Ametz C, Gungor H, Akgöl B, Epure D, Grausgruber H, Löschenberger F, Buerstmayr H. Genomic assisted selection for enhancing line breeding: merging genomic and phenotypic selection in winter wheat breeding programs with preliminary yield trials. Theor Appl Genet. 2017;130(2):363–376. doi: 10.1007/s00122-016-2818-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES. Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell. 2009;21(8):2194–2202. doi: 10.1105/tpc.109.068437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson BK, Weber JN, Kay EH, Fisher HS, Hoekstra HE. Double digest RADseq: an inexpensive method for de novo SNP discovery and genotyping in model and non-model species. PloS One. 2012;7(5):e37135. doi: 10.1371/journal.pone.0037135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter KB, Gilmore EC, Tuleen NA (1980) Registration of Tam 105 wheat1 (reg. no. 624). Crop Sci 20(1):114–114. 10.2135/cropsci1980.0011183X002000010034x
- Porter KB, Worrall WD, Gardenhire JH, Gilmore EC, McDaniel ME, Tuleen NA. Registration of ‘TAM 107’ wheat. Crop Sci. 1987;27(4):818–819. doi: 10.2135/cropsci1987.0011183X002700040050x. [DOI] [Google Scholar]
- Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155(2):945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafalski JA. Association genetics in crop improvement. Curr Opin Plant Biol. 2010;13(2):174–180. doi: 10.1016/j.pbi.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Rasheed A, Wen W, Gao F, Zhai S, Jin H, Liu J, Guo Q, Zhang Y, Dreisigacker S, Xia X, He Z (2016) Development and validation of KASP assays for genes underpinning key economic traits in bread wheat. Theor Appl Genet 129(10):18 [DOI] [PubMed]
- Rudd JC, Devkota RN, Baker JA, Peterson GL, Lazar MD, Bean B, Worrall D, Baughman T, Marshall D, Sutton R, Rooney LW, Nelson LR, Fritz AK, Weng Y, Morgan GD, Seabourn BW. ‘TAM 112’ wheat, resistant to greenbug and wheat curl mite and adapted to the dryland production system in the Southern High Plains. Journal of Plant Registrations. 2014;8(3):291–297. doi: 10.3198/jpr2014.03.0016crc. [DOI] [Google Scholar]
- Rudd JC, Devkota RN, Ibrahim AM, Baker JA, Baker S, Lazar MD, Sutton R, Simoneaux B, Opena G, Rooney LW, Awika JM, Liu S, Xue Q, Bean B, Duncan RW, Seabourn BW, Bowden RL, Jin Y, Chen M-S, Graybosch RA. ‘TAM 114’ wheat, excellent bread-making quality hard red winter wheat cultivar adapted to the Southern High Plains. Journal of Plant Registrations. 2018;12(3):367–372. doi: 10.3198/jpr2017.11.0081crc. [DOI] [Google Scholar]
- Rudd JC, Devkota RN, Ibrahim AM, Baker JA, Baker S, Sutton R, Simoneaux B, Opena G, Hathcoat D, Awika JM, Nelson LR, Liu S, Xue Q, Bean B, Neely CB, Duncan RW, Seabourn BW, Bowden RL, Jin Y, Chen M-S, Graybosch RA. ‘TAM 204’ wheat, adapted to grazing, grain, and graze-out production systems in the Southern High Plains. Journal of Plant Registrations. 2019;13(3):6. doi: 10.3198/jpr2018.12.0080crc. [DOI] [Google Scholar]
- Rutkoski J, Poland J, Mondal S, Autrique E, Pérez LG, Crossa J, Reynolds M, Singh R (2016) Canopy temperature and vegetation indices from high-throughput phenotyping improve accuracy of pedigree and genomic selection for grain yield in wheat. G3: Genes, Genomes, Genetics 6 (9):2799–2808 [DOI] [PMC free article] [PubMed]
- Simmonds J, Scott P, Leverington-Waite M, Turner AS, Brinton J, Korzun V, Snape J, Uauy C. Identification and independent validation of a stable yield and thousand grain weight QTL on chromosome 6A of hexaploid wheat (Triticum aestivum L.) BMC plant biology. 2014;14(1):1–13. doi: 10.1186/s12870-014-0191-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Via LE. A rapid CTAB DNA isolation technique useful for RAPD fingerprinting and other PCR applications. Biotechniques. 1993;14(5):748–751. [PubMed] [Google Scholar]
- Sun J, Poland JA, Mondal S, Crossa J, Juliana P, Singh RP, Rutkoski JE, Jannink J-L, Crespo-Herrera L, Velu G. High-throughput phenotyping platforms enhance genomic selection for wheat grain yield across populations and cycles in early stage. Theor Appl Genet. 2019;132(6):1705–1720. doi: 10.1007/s00122-019-03309-0. [DOI] [PubMed] [Google Scholar]
- Tsai H-Y, Janss LL, Andersen JR, Orabi J, Jensen JD, Jahoor A, Jensen J. Genomic prediction and GWAS of yield, quality and disease-related traits in spring barley and winter wheat. Sci Rep. 2020;10(1):1–15. doi: 10.1038/s41598-020-63862-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyagi S, Mir R, Kaur H, Chhuneja P, Ramesh B, Balyan H, Gupta P. Marker-assisted pyramiding of eight QTLs/genes for seven different traits in common wheat (Triticum aestivum L.) Molecular Breeding. 2014;34(1):167–175. doi: 10.1007/s11032-014-0027-1. [DOI] [Google Scholar]
- Verges VL, Van Sanford DA. Genomic selection at preliminary yield trial stage: training population design to predict untested lines. Agronomy. 2020;10(1):60. doi: 10.3390/agronomy10010060. [DOI] [Google Scholar]
- Weng Y, Lazar M. Amplified fragment length polymorphism-and simple sequence repeat-based molecular tagging and mapping of greenbug resistance gene Gb3 in wheat. Plant Breeding. 2002;121(3):218–223. doi: 10.1046/j.1439-0523.2002.00693.x. [DOI] [Google Scholar]
- Yang Y, Basnet BR, Ibrahim AMH, Rudd JC, Chen X, Bowden RL, Xue Q, Wang S, Johnson CD, Metz R, Mason RE, Hays DB, Liu S. Developing KASP markers on a major stripe rust resistance QTL in a popular wheat TAM 111 using 90K array and genotyping-by-sequencing SNPs. Crop Sci. 2019;59(1):165–175. doi: 10.2135/cropsci2018.05.0349. [DOI] [Google Scholar]
- Yang Y, Dhakal S, Chu C, Wang S, Xue Q, Rudd JC, Ibrahim AMH, Jessup K, Baker J, Fuentealba MP, Devkota R, Baker S, Johnson CD, Metz R, Liu S (2020) Genome wide identification of QTL associated with yield and yield components in two popular wheat cultivars TAM 111 and TAM 112. PLOS ONE 15 (12) 10.1371/journal.pone.0237293 [DOI] [PMC free article] [PubMed]
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, Doebley JF, McMullen MD, Gaut BS, Nielsen DM, Holland JB. A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet. 2006;38(2):203–208. doi: 10.1038/ng1702. [DOI] [PubMed] [Google Scholar]
- Yu S, Assanga SO, Awika JM, Ibrahim AM, Rudd JC, Xue Q, Guttieri MJ, Zhang G, Baker JA, Jessup KE. Genetic mapping of quantitative trait loci for end-use quality and grain minerals in hard red winter wheat. Agronomy. 2021;11(12):2519. doi: 10.3390/agronomy11122519. [DOI] [Google Scholar]
- Zhang Y, Liu J, Xia X, He Z (2014) TaGS-D1, an ortholog of rice OsGS3, is associated with grain weight and grain length in common wheat. Mol Breed 34(3):1097–1107. 10.1007/s11032-014-0102-7
- Zhang Z, Ersoz E, Lai C-Q, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES. Mixed linear model approach adapted for genome-wide association studies. Nat Genet. 2010;42(4):355–360. doi: 10.1038/ng.546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Liu S, Abdelsalam NR, Carver BF, Bai G. Characterization of wheat curl mite resistance gene Cmc4 in OK05312. Theor Appl Genet. 2021;134(4):993–1005. doi: 10.1007/s00122-020-03737-3. [DOI] [PubMed] [Google Scholar]
- Zhu C, Gore M, Buckler ES, Yu J. Status and prospects of association mapping in plants. The Plant Genome. 2008;1(1):5–20. doi: 10.3835/plantgenome2008.02.0089. [DOI] [Google Scholar]
- Zimin AV, Puiu D, Hall R, Kingan S, Clavijo BJ, Salzberg SL (2017) The first near-complete assembly of the hexaploid bread wheat genome. Triticum aestivum. Gigascience 6(11):gix097 [DOI] [PMC free article] [PubMed]
- Zou J, Semagn K, Iqbal M, N’Diaye A, Chen H, Asif M, Navabi A, Perez-Lara E, Pozniak C, Yang R-C, Randhawa H, Spaner D. Mapping QTLs controlling agronomic traits in the ‘Attila’ × ‘CDC Go’ spring wheat population under organic management using 90K SNP array. Crop Sci. 2017;57(1):365–377. doi: 10.2135/cropsci2016.06.0459. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.