Abstract
Peanut is an essential food and oilseed crop. One of the most critical factors contributing to the low yield and destruction of peanut plant growth is leaf disease attack, which will directly reduce the yield and quality of peanut plants. The existing works have shortcomings such as strong subjectivity and insufficient generalization ability. So, we proposed a new deep learning model for peanut leaf disease identification. The proposed model is a combination of an improved X-ception, a parts-activated feature fusion module, and two attention-augmented branches. We obtained an accuracy of 99.69%, which was 9.67%–23.34% higher than those of Inception-V4, ResNet 34, and MobileNet-V3. Besides, supplementary experiments were performed to confirm the generality of the proposed model. The proposed model was applied to cucumber, apple, rice, corn, and wheat leaf disease identification, and yielded an average accuracy of 99.61%. The experimental results demonstrate that the proposed model can identify different crop leaf diseases, proving its feasibility and generalization. The proposed model has a positive significance for exploring other crop diseases’ detection.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11032-023-01370-8.
Keywords: Crop diseases, Peanut leaf, Deep learning, Generalization
Introduction
Crop development is irreversibly affected by agricultural diseases and pests (Fedele et al. 2022). Accurate crop disease type identification and degree determination are critical to selecting pesticides, which are critical to winning the opportunity of plant protection operation and accurate treatment and have become an important goal of intelligent field production control (Che'Ya et al. 2022).
Peanut is an economically significant crop with a vast planting range. Its production and quality are crucial to economic growth (Tang et al. 2022). The occurrence of diseases has a serious impact on yield and quality. Consequently, quick identification and management of peanut disorders are crucial.
Because of the complex field environment, peanut leaves are easily infected by pathogens. The pathogen can spread rapidly through natural factors, destroying green tissues and chlorophyll in leaves, resulting in decreased peanut yield and quality (Rathod et al. 2020). Peanut leaf disease identification methods are mainly judged by the professional knowledge and experience of relevant experts and farmers (Mandloi et al. 2022). These methods are highly subjective and inefficient, and it is easy to miss the best time for disease warning and treatment.
Crop leaf disease recognition is one of the most basic and significant activities in agricultural production, and it is also the core research content in the field of image processing. With the development of computer vision (Wang et al. 2022) and artificial intelligence (Khan et al. 2022), there are two classes of methods for crop disease identification: traditional machine learning methods and prevalent deep learning algorithms.
Traditional machine learning image processing approaches rely heavily on color, shape, texture, and other data to establish feature vectors, which are subsequently classified using support vector machines (SVM) (Wang et al. 2022) and neural networks (Korchagin et al. 2021). Although the model trained by using the fixed feature extraction method can achieve a certain classification effect (Shen et al. 2022), it generally needs to segment diseased spots or diseased leaves, which increases the workload in the early stages. Moreover, the feature extraction method is designed for different disease combinations each time, which lacks robustness and makes it difficult to distinguish similar diseases.
Traditional machine learning methods have high accuracy, but they rely heavily on lesion segmentation and manual design features, and the preprocessing method is not universal, limited to small data sets, and time-consuming (Cravero et al. 2022). For instance, Astani et al. (2022) presented an image processing technique based on a gray-level occurrence matrix and SVM for the automatic detection and treatment of leaf diseases in tomato crops. Its average accuracy was 92.5% for four diseases. A new molecular method based on loop-mediated isothermal amplification was published by Gomez-Gutierrez and Goodwin (2022) for the identification of wheat pathogens. In order to identify crop diseases, Ali et al. (2022) introduced a method called feature fusion and principle component analysis (PCA), which had a 98.2% accuracy rate. Its average accuracy improved by 1.69% over ResNet50. Rahman et al. (2022) developed a gray-level co-occurrence matrix and an SVM-based image processing technique for the autonomous detection and management of tomato leaf diseases. Its average accuracy for four different tomato leaf diseases was 92.5%. Kumar and Kannan (2022) created an adaptive SVM classifier for detecting diseases in rice plants such as bacterial leaf blight, brown spots, and leaf smut. It detected and classified rice leaf diseases with an accuracy of up to 98.8%. Most methods use SVM and PCA to recognize crop leaf disease images. These methods not only rely heavily on feature extraction but also require the design of different identification models for different crop diseases, which lack universal applicability.
With the excellent performance of deep learning technologies in the field of image recognition (Chakraborty et al. 2022), convolutional neural networks (CNNs) have a strong independent driving ability in feature extraction (Tugrul et al. 2022). They have good adaptability to image deformations such as displacement, scaling, and distortion (Dhaka et al. 2021). In crop disease and pest detection, deep learning algorithms have been proven to perform better than traditional machine learning methods. For instance, Qi et al. (2021) used a traditional machine learning method to ensemble the output of a deep learning model for identifying diseases in peanut leaves. Its accuracy was as high as 97.59% on the test data set, compared to Inception-V3, which increased by 7.5%. Yang et al. (2021) built a novel method based on improved VGG-16 for peanut variety identification and classification, the average accuracy was 96.7% on 12 varieties of peanuts, which was 8.9% higher than that of the traditional VGG-16. Yuan et al. (2022) created a point-centered convolutional neural network combined with embedded feature selection for moldy peanut identification and achieved an accuracy of 97.98%. Bi et al. (2020) designed a novel fingerprint modeling and profiling process based on several machine learning models, including convolutional neural networks for peanut oil flavor prediction, its accuracy was 93%. Although deep learning algorithms have been applied to feature extraction and crop leaf disease recognition, most of the above test samples are simple, the disease area is single, and the disease category is small. A large number of training samples are required to learn the ability of feature extraction for complex multiple disease categories and regional images.
To solve the above problems, we deploy a novel deep learning identification framework for peanut leaf diseases. Compared with the outstanding Inception-V4, ResNet34, and MobileNet-V3, the proposed model outperforms their classification performances.
The remaining paper is organized as follows: the “Proposed methods” section mainly introduces the proposed method for peanut leaf disease identification. The “Experimental results and discussion” section mainly reports the experimental results and analysis. Finally, the “Conclusions and future work” section presents the conclusions and future directions of research.
Proposed methods
X-ception network
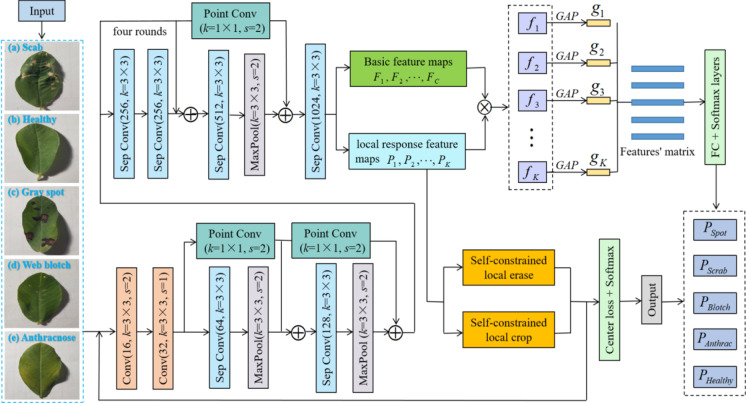
To fully extract the important features of peanut leaf disease images and reduce the computational parameters of the network. The backbone feature extraction network was X-ception (Sutaji and Yildiz 2022). The improved X-ception was based on Inception-V3 (Kaur and Kaur 2022). It replaces the convolution operation in Inception-V3 with the depthwise separable convolution module. A 1 × 1 convolution with cross-channel correlation is used in the module. There is an independent spatial convolution in the output channel of each convolution to map spatial correlation.
There is an input, eight modules, fully connected (FC) layers, the softmax layer, and the output in the improved X-ception. Every module mainly includes the rectified linear unit (ReLu) and depthwise separable convolution (Sep Conv). The depthwise separable convolution is mainly composed of depthwise convolution kernel 3 × 3 and pointwise convolution kernel 1 × 1. Peanut leaf disease images are the input, and the global average value is aggregated through the middle layer for four cycles. Finally, it is output through the FC layer and the output layer. Hence, all convolutions and separable convolutions are batch normalized, and the depth multiplier of the separated convolution layer is 1.
Depthwise convolution can be expressed as follows:
| 1 |
where K and L are the width and the height of the convolutional kernels, respectively. Wk,l means the value of the position (k, l) in the convolution kernel, and represents the value of the position (i + k, j + l) in the input feature maps.
The point-wise convolution can be computed as follows:
| 2 |
where Wn is n-th 1 × 1 convolutional kernel, yi,j,m is the value of position (i, j) in the m-th input feature map, and M is the number of input feature maps.
We use the adaptive moment estimation (Adam) optimization algorithm to update adaptive parameters. The optimal solution and minimum loss value are achieved by updating parameters. The expression of the update parameter is as follows:
| 3 |
where θ is the parameter to be updated, represents the learning rate. and are the average value of the gradient at the first time and the variance at the second time, respectively.
Using the depthwise separable convolution (e.g., a point wise convolution kernel of 1 × 1 and a depth convolution kernel of 3 × 3) to train the suggested network can reduce the calculation of parameters and the characteristic dimension. It not only plays the role of stabilizing the deep features of the network but also provides feasibility for the following self-constraints.
Parts-activated feature fusion
The difference between different peanut leaf disease categories is often reflected in each local area of the leaf. If the networks can extract local area features, the classification performance will be improved. Based on the analysis, we propose the parts-activated feature fusion strategy. We use the convolution operation to generate parts-activated feature maps of different areas. When the basic feature map is , the parts-activated map can be defined as follows:
| 4 |
where is the k-th feature map, it mainly extracts position information and weights. K means the channels of the parts-activated map.
First, we multiply the basic feature map F and the parts-activated feature map PK of the k-th part by the element bit and obtain the fusion feature fk. Then, the global average pooling (GAP) adaptive fusion feature maps get the corresponding parts-activated features . Finally, we obtain the feature matrix by contacting these features. The process can be described as follows:
| 5 |
where is the multiplication operation.
The fusion module of the feature maps and the parts-activated maps can calculate the features of different peanut disease locations, and the global average pooling can obtain the global features. The fusion operation captures the details of different feature channels and locations and provides stronger features than standard linear models.
Self-constrained attention-augmented branches
Most of the existing multi-branch networks obtain better object features by cropping images. Because parts-activated maps can reflect the locations of local attention features, we crop the regions of the parts-activated maps and excavate the detailed features on the original image. The operation can enhance the object’s regions. However, the simple enhancement operation will make the network tend to cyclically train on the same part of the objects, which may cause the network to be insensitive to secondary features and over-fitting. We use the image erasure enhancement method to force the network to learn the features of other regions. The general random image erasure strategy may erase the irrelevant background regions, but the quality and effect of the image enhancement are poor. Therefore, we design the local erasure branches. It can prevent the network from focusing excessively on one local characteristic while disregarding others, and it can enhance the network’s capacity for generalization.
The attention-augmented basis on the local parts-activated maps is the threshold regions. The regions are too small or too large for cropping and erasing. It reduces the attention-augmented effect. Hence, we propose a self-constrained strategy in the attention-augmented branches and constrain the regions for cropping and erasing. We train the regions, extract more effective spatial features, and obtain more accurate local enhancement features. Combined with the above analysis, we propose two self-constrained attention-augmented branches: the attention local cropping branch and the attention local erasing branch.
We use Pk to guide the training process of the attention-augmented branches in the parts-activated feature maps . Pk is normalized, and it can be calculated as follows:
| 6 |
where Pk(u,v) is the activation value in the u-th row and the v-th column and represents the basis of judgment for cropping and erasing.
On the local attention-cropped branches, when is less than the cropping threshold , is 1. If not, is 0. Then, a group of bounding box is used to cover the regions of and enlarge it to the original images to show more detail.
On the local attention-erased branches, when is less than the cropping threshold , is 1. If not, is 0. If the value of the corresponding pixel regions is higher than the threshold in the input image, it will be set to 0. Then, we obtain the erasure images and solve the problem that multiple feature maps of the network focus on the same part.
Combined to the features of different network layers, the self-constrained mechanism makes the network models gradually focus on the local regions. The proposed self-constrained technique is capable of filtering out more precise local information and rejecting as much sample noise as possible. For samples with large internal changes, it can reduce intra-class differences and improve the performance of cropping and erasing branches.
Classifier
The loss function plays an important role in the classification performance of convolutional neural networks. The current activation function is Softmax. It can be formulated as follows:
| 7 |
where m is the sample quantity, yi represents the real category label of the i-th image, xi means the feature vector of the i-th image before putting the fully connected layer. W is the last fully connected layer, b is the offset value, and s is the quantity of the category.
The Softmax reflects inter-class differences but fails to reflect intra-class similarities well. Therefore, we use the center regularization loss function to reflect intra-class similarities well. The center loss can be defined as follows:
| 8 |
where K is the quantity of the category, gk(Ii) means the k-th parts-activated features of the i-th image, represents the feature center of the k-th parts-activated features in the category yi of the i-th image Ii.
The smaller the value of LCenter is, the smaller the feature difference of the same category image in the same part is. The proposed total function can be expressed as follows:
| 9 |
where is the linear combination coefficient of two loss functions.
To intuitively show the proposed peanut leaf disease identification framework, we present it in Fig. 1.
Fig. 1.
Proposed overall identification framework
The proposed frameworks for peanut leaf disease identification based on deep learning mainly include the improved backbone feature extraction network X-ception, the parts-activated feature fusion (PFF) module, the self-constrained attention-augmented branches (SAB), and the center regularization loss function. The improved X-ception is used to extract the shallow features of images of peanut leaf disease. The parts-activated feature fusion not only preserves the overall features of the original image but also increases the feature weight of each local region. The network pays more attention to the local features of peanut leaf diseases. Attention-augmented branches highlight the distinguishing features through a training method similar to confrontation, which can strengthen the ability to capture local features. In addition, we use the center regularization loss function to correct the attention region in weakly supervised learning.
Experimental results and discussion
Experimental setup
Experimental hardware was the operating system, Windows 10 (64-bit). The central processing unit (CPU) was configured with an Intel Core i7-9700. The graphical processing unit (GPU) was an NVIDIA GeForce GTX 1080. The dedicated GPU memory was 8 GB. The software was managed by Anaconda 3, and the programming language was Python 3.7. We used Spyder 3.3.3 as the Python interpreter and used the popular deep learning framework Keras 2.2.4 on the TensorFlow-gpu 1.14.0 backend on our personal computer.
In order to determine the network parameters of the proposed model, we used different learning rates, iteration times, and batch sizes to analyze and compare different parameter settings and corresponding results. Table 1 lists the comparison of different parameter settings.
Table 1.
Comparison of different parameter settings
| Parameter names | Learning rate | Iteration times | Batch sizes | Test loss | Test accuracy |
|---|---|---|---|---|---|
| Parameter settings and testing results | 0.001 | 500 | 16 | 0.5658 | 0.8135 |
| 0.002 | 1000 | 32 | 0.3946 | 0.8651 | |
| 0.001 | 1500 | 64 | 0.2753 | 0.8927 | |
| 0.001 | 2000 | 32 | 0.0647 | 0.9154 | |
| 0.002 | 2500 | 64 | 0.1991 | 0.9358 | |
| 0.001 | 3000 | 32 | 0.0489 | 0.9969 | |
| 0.001 | 3500 | 16 | 0.0892 | 0.9586 |
As can be seen in Table 1, different network parameter settings have different testing results. When the iteration time is 3000, the testing loss and accuracy are better than those of other iteration times. When the iteration time is 3000 and the learning rate is 0.001, the testing loss is lower than that of other iteration times and learning rates. When the iteration time is 3000 and the batch size is 32, the testing accuracy is higher than that of other iteration times and batch sizes. Therefore, the learning rate, iteration time, and batch size are 0.001, 3000, and 32, respectively.
Construction of the dataset
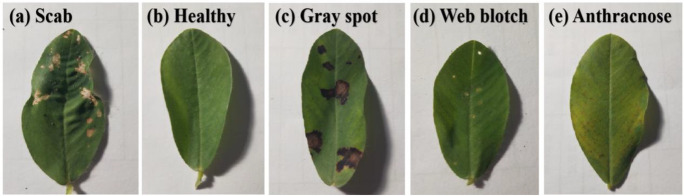
In the natural environment, we collected experimental samples from the peanut planting base in Henan Province, China. We took the peanut leaves with the mobile phone camera (Vivo X20A, China) in a laboratory environment. The image was 3024 × 4032 pixel. The format is JPEG, and each image is a 24-bit color bitmap. Here, each sample we collected contains only one disease type. There are a total of 9420 samples, mainly including the healthy and four kinds of diseases, which are scab, gray spot, web blotch, and anthracnose. Five samples are presented in Fig. 2.
Fig. 2.
Five samples
We obtained scabby peanut leaves from the moderate degree and middle period. The gray spots on the peanut leaves date from the late period and are of moderate severity. The blotchy peanut leaves are from the early period and light degree. The peanut leaves with anthracnose are from the early period and light degree. In our experiment, we have not collected peanut leaf samples at other stages and degrees of disease. In addition to the disease period and disease degree of the four peanut leaf diseases mentioned above, we have not collected peanut leaf samples for any other disease period or disease degree.
According to a ratio of 3:1:1, we randomly divided the 9420 samples into a training set, a validation set, and a testing set. Table 2 shows the results of the experimental specific division.
Table 2.
The division of each category
| Categories | Train set | Validate set | Test set | Total number |
|---|---|---|---|---|
| Scab | 1051 | 350 | 351 | 1752 |
| Healthy | 1009 | 336 | 336 | 1681 |
| Gray spot | 1163 | 387 | 387 | 1937 |
| Web blotch | 1310 | 437 | 436 | 2183 |
| Anthracnose | 1121 | 373 | 373 | 1867 |
| Total number | 5654 | 1883 | 1883 | 9420 |
Experimental results
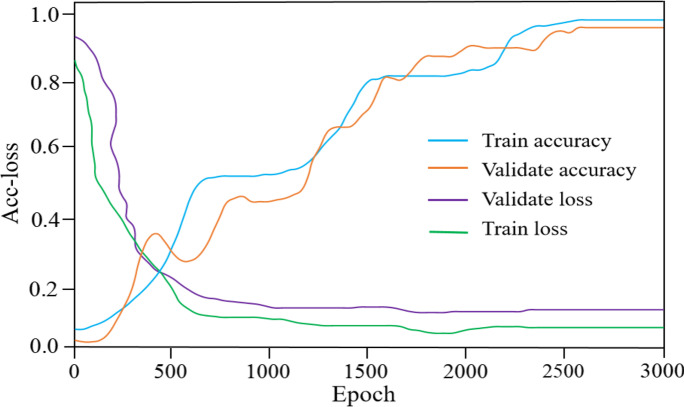
We trained the proposed model on the training set and the validation set. The results of the accuracy and loss (acc-loss) values are revealed in Fig. 3.
Fig. 3.
Acc-loss curves for training and validating
Figure 3 contains some significant observations. When the epoch is between 0 and 2400, the training and validation losses exhibit irregular fluctuations. After 2550 iterations, the training and validation accuracy continue to improve until they tend toward a stable trend and there is no longer a significant difference. The training and validation losses become increasingly small as the number of iterations increases. As a result, the suggested model can better learn the features of peanut leaves.
By comparing the original image with the heat map, it can be seen that the model accurately locates some key areas in the original image, such as the diseased area on the diseased leaves. To verify the feature extraction ability of the proposed model, we visualize the heat maps of the test samples. The visual results of the proposed model are shown in Fig. 4.
Fig. 4.
Heat maps of different layers with the proposed model
From Fig. 4, we can see that the proposed model can extract key information from images. The activated area covers the diseased area comprehensively. It indicates that the proposed model can give an accurate recognition of peanut leaf diseases according to the active regions in the thermodynamic diagram. The model can focus on some key information related to recognition in images and has strong feature extraction abilities.
To verify the performance of the proposed model, we used specificity, precision, sensitivity, the f1-score, and accuracy to evaluate it. The evaluation metrics can be expressed as follows:
| 10 |
| 11 |
| 12 |
| 13 |
| 14 |
where TP and TN are true positives (TP) and true negatives (TN), respectively. FP and FN are false positives (FP) and false negatives (FN), respectively. In Eqs. (12) and (13), the sensitivity is the same concept as the recall, and the recall can be directly used to refer to the sensitivity. Therefore, we used the sensitivity calculation formula instead of the recall to compute the f1-score value.
Due to the fact that the proposed model utilized numerous enhanced techniques based on an enhanced X-ception, a parts-activated feature fusion (PFF), and the self-constrained attention-augmented branches (SAB). To verify the effectiveness of the improved strategies, we conducted ablation experiments and used some popular evaluation metrics, such as specificity (Spe), precision (Pre), sensitivity (Sen), the f1-score (F1-s), and accuracy (Acc) to explore the performance of each module.
In addition, to validate the effectivity of the proposed method, we compared it with other state-of-the-art algorithms, such as ResNet34, Inception-V4, and MobileNet-V3. The results are presented in Table 3. The network parameters of the outstanding models ResNet34, Inception-V4, and MobileNet-V3 are the same. Their learning rate, iteration time, and batch size are 0.005, 5000, and 128, respectively. The network parameter settings of ResNet34, Inception-V4, and MobileNet-V3 are different from those of the combination of an improved X-ception, PFF, and SAB. The experimental environment is the same.
Table 3.
Results of ablation experiments on our dataset
| Algorithms | Spe (%) | Pre (%) | Sen (%) | F1-s (%) | Acc (%) |
|---|---|---|---|---|---|
| ResNet34 | 75.64 | 76.17 | 75.62 | 75.89 | 76.35 |
| Inception-V4 | 81.73 | 81.79 | 80.95 | 81.37 | 82.74 |
| MobileNet-V3 | 89.85 | 90.69 | 88.93 | 89.80 | 90.02 |
| Improved X-ception | 75.68 | 78.72 | 78.63 | 78.67 | 78.76 |
| Improved X-ception + PFF | 80.72 | 78.96 | 78.91 | 78.93 | 78.98 |
| Improved X-ception + SAB | 92.59 | 93.45 | 91.84 | 92.64 | 91.67 |
| Improved X-ception + PFF + SAB | 98.93 | 99.72 | 98.87 | 99.29 | 99.69 |
As can be seen in Table 3, the improved X-ception + PFF algorithm is superior to ResNet. It illustrates that adding the PFF module can promote the performance. The improved X-ception + SAB model is better than Inception-V4, MobileNet-V3, and the improved X-ception + PFF model. It illustrates that adding the SAB module also can promote the performance and is superior to adding the PFF module. The improved X-ception + PFF + SAB model is best in the seven algorithms. It illustrates that both PFF and SAB can improve the score. Relatively speaking, SAB increases a higher score than PFF. In summary, the current testing results demonstrate that the integrated deep-learning models, viz., an improved X-ception, a parts-activated feature fusion (PFF) module, and a self-constrained attention-augmented branch (SAB) module, can perform accurate peanut leaf disease identification.
Comparing Table 3 to the single network model, it can be seen that the method based on model fusion has some advantages and compensates for the limitations of the single network model. In particular, the proposed model outperforms the single X-ception on the accuracy. To a certain extent, the fusion model has better stability and generalization ability than the single network structure. While the results indicate that the three improvements proposed do not conflict with each other, In addition, the proposed model outperforms outstanding deep-learning networks MobileNet-V3, ResNet34, and Inception-V4. The proposed research results will hopefully serve as useful feedback for improving the detection of peanut leaf diseases.
To verify the performance of the suggested model on other datasets, we used specificity, precision, sensitivity, the f1-score, and accuracy to estimate it. The results are presented in Table 4.
Table 4.
Comparisons of different datasets
| Data sets, number, and class | Spe (%) | Pre (%) | Sen (%) | F1-s (%) | Acc (%) |
|---|---|---|---|---|---|
| Cucumber Plant Diseases Dataset, 695, 2-class | 99.16 | 98.85 | 98.54 | 98.69 | 99.47 |
| Corn Leaf Diseases (NLB), 4115, 2-class | 98.97 | 98.92 | 98.89 | 98.90 | 99.83 |
| Plant Pathology Apple Dataset, 3171, 4-class | 98.71 | 98.49 | 97.62 | 98.05 | 99.51 |
| LWDCD 2020 Wheat Dataset, 4500, 4-class | 98.68 | 99.46 | 98.83 | 99.14 | 99.65 |
| Rice Diseases Image Dataset, 5447, 4-class | 99.03 | 98.99 | 98.95 | 98.97 | 99.52 |
| Our dataset, 9420, 5-class | 98.93 | 99.72 | 98.87 | 99.29 | 99.69 |
On the other five datasets, as shown in Table 4, the suggested method performs better. The average across five evaluation metrics is 97%. According to the findings of recent testing, it is proved that the suggested system offers strong portability and generalizability.
We collected recently published literature on the identification of crop disease images. The results are elucidated in Table 5.
Table 5.
Comparisons of the proposed model with recent literature
Table 5 shows that our test accuracy is higher than the other four models. Math and Dharwadkar (2022) and Anari (2022) have comparable classification performances that outperform Rimal et al. (2022) and Liu et al. (2022). Taken as a whole, the proposed strategy for identifying peanut leaf disease is promising.
Conclusions and future work
In this study, we gathered five distinct peanut leaf disease samples to use in developing, validating, and evaluating the suggested model. First, images of peanut disease are transferred to an improved X-ception network to obtain basic features. Second, we use the attention-augmented mechanism to perform self-constrained parts-cropping and self-constrained parts-erasing in the parts-activated fusion regions. Finally, we utilize the center regularization loss function to reflect intra-class similarities and promote the more accurate detection of the disease regions. Experimental results demonstrate that the proposed method is superior to the outstanding CNNs, such as ResNet 34, Inception-V4, and MobileNet-V3. The classification accuracy is 99.69% on the testing set.
However, the suggested model was only suitable for identifying the healthy peanut leaves, the scabby peanut leaves of the middle period and moderate degree, the gray spot peanut leaves of the late period and moderate degree, the blotchy peanut leaves of the early period and light degree, and the peanut leaves with anthracnose from the early period and light degree. It is not able to accurately judge the degree of other diseases. The recognition effect of other disease periods is not ideal. It cannot identify multiple diseases on a leaf. Additionally, we do not get enough samples of disease periods and degrees, and the disease categories are not comprehensive.
In the future, we will continue to collect more samples of peanut disease leaves and expand the number and types of crop disease databases. In addition, aiming at the sharp geometric edge of the blade target, further optimization and improvement of the proposed algorithm will be used to eliminate its limitations and improve the accuracy of feature extraction. In this way, the application scope and robustness of automatic detection algorithms can be increased for crop diseases and pests.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
The authors would like to thank the university colleagues for their valuable feedback and worthwhile discussions.
Author contribution
XL and BC studied conception and design. XL, WZ, and FZ collected experimental data. LX, BC, SN, and FZ analyzed experimental results. LX and BC prepared draft manuscript. XL, BC, and WZ wrote the main manuscript text. All authors reviewed the manuscript and approved the final version.
Data availability
Not applicable.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Laixiang Xu, Email: xulaixiang@hainanu.edu.cn.
Bingxu Cao, Email: caobxlctcedu@163.com.
Shiyuan Ning, Email: yuancetgc@yeah.net.
Wenbo Zhang, Email: zhangwenbo@hainanu.edu.cn.
Fengjie Zhao, Email: pediatricszhao@yeah.net.
References
- Ali S, Hassan M, Kim J, Farid M (2022) FF-PCA-LDA: intelligent feature fusion based PCA-LDA classification system for plant leaf diseases. Appl Sci-Basel 12(7). 10.3390/app12073514
- Anari M (2022) A hybrid model for leaf diseases classification based on the modified deep transfer learning and ensemble approach for agricultural AIoT-based monitoring. Comput Intell Neurosci. 10.1155/2022/6504616 [DOI] [PMC free article] [PubMed]
- Astani M, Hasheminejad M, Vaghefi M (2022) A diverse ensemble classifier for tomato disease recognition. Comput Electron Agric 198. 10.1016/j.compag.2022.107054.s
- Bi K, Zhang D, Qiu T (2020) GC-MS fingerprints profiling using machine learning models for food flavor prediction. Processes 8(1). 10.3390/pr8010023
- Chakraborty SK, Chandel NS, Jat D, Tiwari MK, Rajwade YA, Subeesh A (2022) Deep learning approaches and interventions for futuristic engineering in agriculture. Neural Comput Appl 34(23). 10.1007/s00521-022-07744-x
- Che'Ya N, Mohidem N, Roslin N (2022) Mobile computing for pest and disease management using spectral signature analysis: a review. Agronomy-Basel 12(4). 10.3390/agronomy12040967
- Cravero A, Pardo S, Sepulveda S, Munoz L. Challenges to use machine learning in agricultural big data: a systematic literature review. Agronomy-Basel. 2022;12(3):10. doi: 10.3390/agronomy12030748. [DOI] [Google Scholar]
- Dhaka V, Meena S, Rani G (2021) A survey of deep convolutional neural networks applied for prediction of plant leaf diseases. Sensors 21(14). 10.3390/s21144749 [DOI] [PMC free article] [PubMed]
- Fedele G, Brischetto C, Rossi V (2022) A systematic map of the research on disease modelling for agricultural crops worldwide. Plant-Basel 11(6). 10.3390/plants11060724 [DOI] [PMC free article] [PubMed]
- Gomez-Gutierrez S, Goodwin S (2022) Loop-mediated isothermal amplification for detection of plant pathogens in wheat (Triticum aestivum). Front Plant Sci 13. 10.3389/fpls.2022.857673 [DOI] [PMC free article] [PubMed]
- Kaur J, Kaur P (2022) UNIConv: An enhanced U-Net based InceptionV3 convolutional model for DR semantic segmentation in retinal fundus images. Concurrency and Computation Practice and Experience.10.1002/cpe.7138
- Khan MHU, Wang SD, Wang J, Ahmar S, Saeed S, Khan SU, Xu XG, Chen HY, Bhat JA, Feng XZ (2022) Applications of Artificial Intelligence in Climate-Resilient Smart-Crop Breeding. Int J Mol Sci 23(19). 10.3390/ijms231911156 [DOI] [PMC free article] [PubMed]
- Korchagin S, Gataullin S, Osipov A (2021) Development of an optimal algorithm for detecting damaged and diseased potato tubers moving along a conveyor belt using computer vision systems. Agronomy-Basel 11(10). 10.3390/agronomy11101980
- Kumar K, Kannan E. Detection of rice plant disease using AdaBoostSVM classifier. Agron J. 2022;114(4):2213–2229. doi: 10.1002/agj2.21070. [DOI] [Google Scholar]
- Liu Y, Zhang X, Gao Y. Improved CNN method for crop pest identification based on transfer learning. Comput Intell Neurosci. 2022 doi: 10.1155/2022/9709648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandloi S, Tripathi M, Tiwari S (2022) Genetic diversity analysis among late leaf spot and rust resistant and susceptible germplasm in groundnut (Arachis hypogea L.). Israel J Plant Sci 69(3–4):163–171. 10.1163/22238980-bja10058
- Math R, Dharwadkar N. Early detection and identification of grape diseases using convolutional neural networks. J Plant Dis Prot. 2022;129(3):521–532. doi: 10.1007/s41348-022-00589-5. [DOI] [Google Scholar]
- Qi H, Liang Y, Ding Q (2021) Automatic identification of peanut-leaf diseases based on stack. Appl Sci-Basel 11(4). 10.3390/app11041950
- Rahman S, Alam F, Ahmad N. Image processing based system for the detection, identification and treatment of tomato leaf diseases. Multimed Tools Appl. 2022 doi: 10.1007/s11042-022-13715-0. [DOI] [Google Scholar]
- Rathod V, Hamid R, Tomar R (2020) Comparative RNA-Seq profiling of a resistant and susceptible peanut (Arachis hypogaea) genotypes in response to leaf rust infection caused by Puccinia arachidis. Biotech 10(6). 10.1007/s13205-020-02270-w [DOI] [PMC free article] [PubMed]
- Rimal K, Shah K, Jha A. Advanced multi-class deep learning convolution neural network approach for insect pest classification using TensorFlow. Int J Environ Sci Technol. 2022 doi: 10.1007/s13762-022-04277-7. [DOI] [Google Scholar]
- Shen FZ, Deng HC, Yu LJ, Cai FH (2022) Open-source mobile multispectral imaging system and its applications in biological sample sensing. Spectrochim Acta Part A-Mol Biomol Spectrosc 280. 10.1016/j.saa.2022.121504 [DOI] [PubMed]
- Sutaji D, Yildiz O (2022) LEMOXINET: Lite ensemble MobileNetV2 and Xception models to predict plant disease. Ecol Inform 70. 10.1016/j.ecoinf.2022.101698
- Tang Y, Qiu X, Hu C (2022) Breeding of a new variety of peanut with high-oleic-acid content and high-yield by marker-assisted backcrossing. Mol Breed 42(7). 10.1007/s11032-022-01313-9 [DOI] [PMC free article] [PubMed]
- Tugrul B, Elfatimi E, Eryigit R (2022) Convolutional neural networks in detection of plant leaf diseases: a review. Agriculture-Basel 12(8). 10.3390/agriculture2081192
- Wang TC, Shen FZ, Deng HC, Cai FH, Chen SF. Smartphone imaging spectrometer for egg/meat freshness monitoring. Anal Methods. 2022;14(5):508–517. doi: 10.1039/d1ay01726h. [DOI] [PubMed] [Google Scholar]
- Yang H, Ni J, Gao J (2021) A novel method for peanut variety identification and classification by Improved VGG16. Sci Rep 11(1). 10.1038/s41598-021-95240-y [DOI] [PMC free article] [PubMed]
- Yuan D, Jiang J, Gong Z (2022) Moldy peanuts identification based on hyperspectral images and Point-centered convolutional neural network combined with embedded feature selection. Comput Electron Agric 197. 10.1016/j.compag.2022.106963
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.