Abstract
The Phytophthora Genome Initiative (PGI) is a distributed collaboration to study the genome and evolution of a particularly destructive group of plant pathogenic oomycete, with the goal of understanding the mechanisms of infection and resistance. NCGR provides informatics support for the collaboration as well as a centralized data repository. In the pilot phase of the project, several investigators prepared Phytophthora infestans and Phytophthora sojae EST and Phytophthora sojae BAC libraries and sent them to another laboratory for sequencing. Data from sequencing reactions were transferred to NCGR for analysis and curation. An analysis pipeline transforms raw data by performing simple analyses (i.e., vector removal and similarity searching) that are stored and can be retrieved by investigators using a web browser. Here we describe the database and access tools, provide an overview of the data therein and outline future plans. This resource has provided a unique opportunity for the distributed, collaborative study of a genus from which relatively little sequence data are available. Results may lead to insight into how better to control these pathogens. The homepage of PGI can be accessed at http:www.ncgr.org/pgi , with database access through the database access hyperlink.
INTRODUCTION
NCGR is a not-for-profit organization located in Santa Fe, New Mexico, dedicated to furthering the progress of biological research through novel bioinformatic and database solutions. NCGR is the home of the Genome Sequence Database (GSDB), Gepasi biochemical kinetics simulator, the R-Genes plant disease resistance database, and the Phytophthora Genome Initiative database (PGI), as well as a range of data visualization and analysis tools. The home page URL is http://www.ncgr.org
The PGI is a distributed sequencing initiative whose goal is better to understand the mechanisms of infection and resistance between host-pathogen pairs, specifically as they relate to the genus Phytophthora and their plant hosts. Members of the initiative providing data include Brett Tyler’s laboratory at University of California at Davis, Francine Gover’s laboratory at Wageningen Agricultural University in The Netherlands, Don Nuss’ laboratory at the Center for Agricultural Biotechnology at the University of Maryland Biotechnology Institute and Mark Gijzen’s laboratory at the Southern Crop Protection and Food Research Center at Agriculture Canada (http://www. ncgr.org/pgi/aboutpgi.html ). As part of its agricultural genomics initiative, NCGR facilitates PGI by providing analysis services and database access utilities. Sequence data are integrated into a shared resource and data and analysis results are easily viewed via the WWW (http://www.ncgr.org/pgi ).
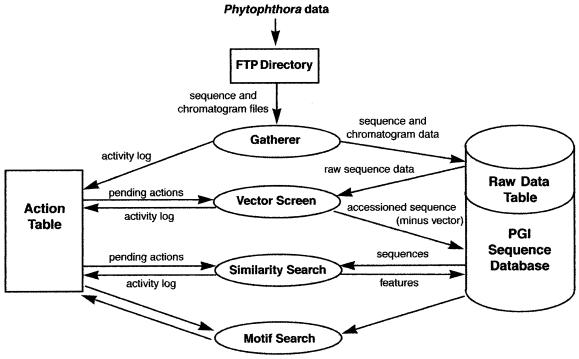
Within the oomycete genus Phytophthora are about sixty species of destructive plant pathogens that cause rots of roots, stems, leaves and fruits in a large range of plant hosts (1). Given the economic importance of this group of pathogens and the questionable utility of comparisons with model pathosystems, it is clear that in-depth molecular studies of Phytophthora pathogenicity will be critical to its eventual control (2). The PGI database contains public sequence and annotation data from Phytophthora infestans, which causes late blight of potato, and Phytophthora sojae, which affects soybeans (Glycine max), as well as from sequences correlated with the compatible interaction of P.sojae and soybean. Specifically, the sequences have been derived from the Phytophthora zoospore and mycelial morphotypes as well as from infected host tissue. The data derive from both expressed sequence tag (EST) and genomic BAC sequencing projects. The annotation is a result of a series of automated analysis processes which form the core of the PGI analysis pipeline (Fig. 1; http://www. ncgr.org/graphics/pgi/pgiflow1.jpg ).
Figure 1.
PGI analysis pipeline. Raw sequencing data are processed by a set of steps to add incoming data to the database, screen for vector contamination, perform similarity searches, and find instances of characterized amino acid motifs. Each step records its activity in a separate table, called the Action table, and prepares for the next step.
DATABASE OVERVIEW
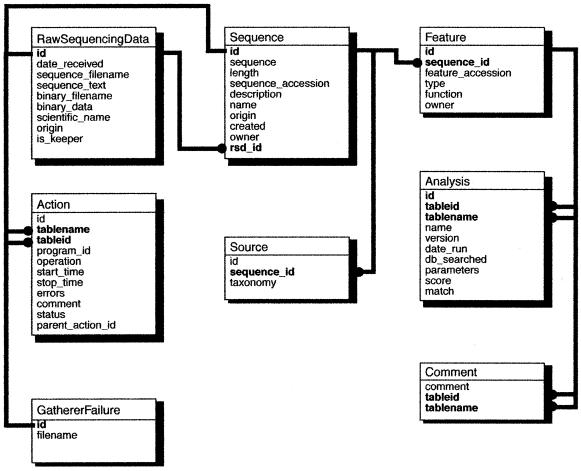
The PGI database was implemented using the relational model. The schema (Fig. 2) is a modified descendant of the GSDB (3). A total of eight tables provide storage for sequence data, metadata and functional annotation. Descriptions of the schema can be viewed at (http://www.ncgr.org/pgi/user_docs/pipeschm.html ). A subset of the schema provides an opportunity to automate the analysis workflow and is used by the analysis pipeline (Fig. 1), described below. Inputs to the database are raw sequence and chromatogram trace files, accessed using the WWW as a convenient mechanism for data exchange (4). Outputs are reports that contain formatted query results. Support of each is described below.
Figure 2.
PGI schema. Eight tables store sequence data, metadata and functional annotation using the relational model.
AUTOMATED ANALYSIS PIPELINE
The automated analysis pipeline was developed expressly to meet the needs of the PGI collaborators, and not as a general-purpose public or commercial software package. Details are provided here to give users a full understanding of the processing steps and resultant annotation and therefore the types of data available from the PGI database.
Raw data
A ‘Gatherer’ program examines incoming files for obvious errors, such as improper file headers or contents that do not appear to be sequencing or chromatogram data, before adding the data to the database.
Vector screening
The vector screening algorithm employs several steps to remove any vector sequence which contaminates the insert sequence. Similarities to known vector are identified using the BLASTN algorithm, version 2.0.3. (5). Matching regions are sorted and considered one at a time. The working 5′ and 3′ ends of the raw sequence are reset to exclude a region if two criteria are met. The expect value for the match must be below a specified expect value (the default is 0.001), and the distance to the near end of the match must be less than a given interval (the default is 25 nt) from the current end of the sequence.
After excluding vector sequence, restriction sites used in cloning are considered. If an apparent end of the insert sequence occurs near the cloning restriction site, the insert end is reset to the cleavage point of the restriction site. The insert sequence is then saved to the database. If the sequence is entirely contaminated by vector or there are chimeric vector regions within the insert sequence the sequences are flagged for further manual review and are not accessioned.
Similarity searching
To search for similarity to other sequences, an interface to BLAST was developed, which adds database connectivity and parses BLAST output into data objects. Each sequence is queried against current versions of NCBI’s ‘nr’ non-redundant amino acid reference library (ftp://ftp.ncbi.nlm.nih.gov/blast.db ) and a non-redundant set of nucleotide sequences compiled from the GSDB at NCGR.
Similarity regions which result from BLAST analysis are added to the database as features linked to the query sequence. The BLAST output is also stored intact in the Tracker database. In most cases, the default BLAST parameters are used, and no similarity regions with expect values above 0.01 are stored.
Motif searching
The PGI pipeline uses the PROSITE dictionary to define ‘motifs’. Translated sequences with known motifs are summarized in tabular form with hyperlinks to corresponding descriptions.
DATA ACCESS
At this time PGI sequences are not submitted to GSDB or GenBank. Ownership of the sequences themselves remains with the originating laboratory and are not the property of NCGR. Any decision to submit these sequences to GenBank lies solely with the owners. However, the full set of Phytophthora nucleotide sequences are available to run a BLAST search against. Detailed instructions on how one may utilize this resource are available at http://www.ncgr.org/research/pgi/howto-blast-pgi.html . In addition PGI sequence information is freely available through browser cut-and-paste operations and it will soon be possible to apply to these sequences the more advanced similarity search algorithms available on NCGR’s TimeLogic server, such as HMM, FRAME, available at http://seqsim.ncgr.org (for details see 6). A complete and current set of sequences contained in the PGI database will be distributed upon e-mail request to pgi-admin@ncgr.org
The database, accessed through CGI-based forms via the Web, is organized by data source, and contains primary data as well as annotated analysis results generated automatically by the PGI analysis pipeline. Each source laboratory is focusing on a particular sequence type (i.e. genomic versus EST) and species (P.sojae versus P.infestans). Currently the data can be sorted by date or clone name to facilitate access, or the analysis results can be searched directly. Search criteria include keyword searches of BLAST results (such as ‘elicitin’) and dynamic nucleotide sequence pattern recognition.
Once a clone has been selected from a sorted list (http://www.ncgr.org/cgi-bin/pgi/database_front.pl ), information is summarized in the Sequence Data window. Information includes full text of the sequence including highlighted known (in red) and putative (in blue) vector, as well as flanking restriction sites from the sequencing library construction process (underlined) and the base composition of the sequence. A Java-based chromatogram trace file viewer can also be launched to examine the raw data, which can be downloaded if desired. From this window one can also get to all of the results from the automated analysis. This includes detailed reports on the vector screening process including alignments, results from BLAST2 blastn (non-redundant nucleotide database derived from GSDB) and blastx (non-redundant amino acid database obtained from GenBank). Detailed results are also available for automated amino acid motif searching against the PROSITE dictionary. In the case of similarity searches, high-scoring hits (parameters defined by PGI participants) are hyperlinked to GSDB Maestro for immediate sequence retrieval, and PROSITE results are hyperlinked to descriptive references.
Keyword searches of BLAST results, based upon specific query sets or the entire set of Phytophthora clones, search the description lines of matching target sequences for the keyword. The results are presented in tabular form as matched sets between query and target sequences with full text descriptions and hyperlinks to the corresponding GSDB or GenBank entry. Nucleotide sequence pattern matching can be performed to discover the frequency of short nucleotide sequence patterns such as restriction sites or simple sequence repeats in either the entire set or a subset of the clones. Tabular results list all clones containing the match and the associated frequencies of the patterns as well as hyperlinks back to the sequence data view.
RESULTS
The PGI database contains sequences from several different sources. For P.infestans, about 1400 ESTs have been obtained from mycelia cultures to date, and for P.sojae, a total of 3125 ESTs have so far been obtained from mycelial cultures, zoospores and a compatible interaction with soybean (G.max), 2 days after infection. A total of 325 genomic DNA sequences from a BAC library near an avirulence locus have also been sequenced.
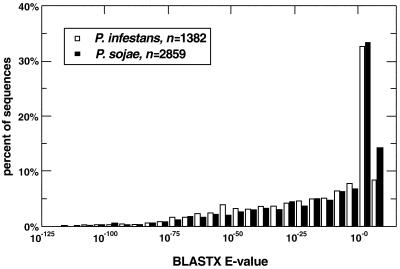
Figure 3 represents a histogram of best BLASTX E-values for each Phytophthora sequence. Also shown are 111 P.infestans and 385 P.sojae ESTs that yielded no hit against the nr target, as the group with E-values >1. Totaled, the portion of sequences that could not be well characterized by this search (E-value exponent >–5) was 28% for P.infestans (mycelia) and 34% for P.sojae (all libraries).
Figure 3.
Frequency distribution of best BLASTX E-value exponents per sequence. More negative exponents indicate a greater degree of similarity. Translated sequences with an E-value >1 show no similarity to amino acid sequences in the target library.
A recent study, to which two of the authors of this article contributed (7), details the results of an analysis of 1000 ESTs from P.infestans. Of particular interest to the plant pathology community was the identification of several novel members of the elicitin family of proteins. Elicitins may prove to be the first step in triggering the signal transduction cascades in plant disease resistance mechanisms to microbial infection. Elicitins, as their name implies, have been shown to elicit, or induce, the hypersensitive response and condition avirulence in plant hosts, and are thought to be sterol carrier proteins. In addition, a majority of the ESTs were classified into functional categories. Interested users are invited to browse the site for the category of most interest to them. Similar investigations of the P.sojae EST data are underway.
FUTURE PLANS
The international PGI is being used as a development/testing/implementation ground for a distributed genomics project. Because sequence data are consistent across all organisms, the general analytic strategy remains fairly constant across genome initiatives and is a model example of the deployment of web-accessible solutions for information management (8). The system we have described is essentially reusable for other distributed genome collaborations as needed. The intent is to provide to our collaborators mechanisms that significantly reduce their involvement in repetitive tasks and provide a first-pass filter for the data output so that it is easy to focus on the sequences of most interest for the biologist to pursue for further characterization. Such an approach was recently described as ‘framework annotation’ (9) and is consistent with the philosophy of NCGR to provide both first-pass annotation as well as tools for subsequent expert annotation.
In the near future we plan to evolve the system to allow collaborators to interact with the analysis tools in real time, by changing specific input parameters and customizing their reports. Additional changes will include support for multiple sequence alignments and to facilitate the process of assembling contiguous regions of a genome from an ordered set of sequenced fragments. Gene prediction algorithms will be added. Tools for the integrated comparison of genetic and physical maps are also being developed.
The PGI database is one of many independent, thematic databases as evidenced by this volume. Such databases are of great value to researchers in specialized domains. The data gain value when they can be considered in an aggregate view. Thus, we embrace standard representations for the exchange of sequence data and annotation.
Acknowledgments
ACKNOWLEDGEMENTS
The authors are deeply indebted to Mark Gijzen, Francine Govers, Sophien Kamoun, Don Nuss and Brett Tyler for their involvement in and support of the Phytophthora Genome Initiative. The USDA/ARS provided partial financial support for sequencing of P.infestans ESTs.
REFERENCES
- 1.Erwin D.C. and Ribeiro,O.K. (1996) Phytophthora Diseases Worldwide. APS Press, St. Paul, MN.
- 2.Waugh M.E., Waugh,K.O., Snead,Q.L. and Liddell,C.M. (1996) Phytopathology Suppl., 86, Abstract. [Google Scholar]
- 3.Harger C., Skupski,M., Allen,E., Clark,C., Crowley,D.,Dickenson,E., Easley,D., Espinosa-Lujan,A., Farmer,A., Fields,C. et al. (1997) Nucleic Acids Res., 25, 18–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lewis S. (1994) In Adams,M.D., Fields,C. and Venter,J.C. (eds), Automated DNA Sequencing and Analysis. Academic Press, San Diego, CA, pp. 329–338.
- 5.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Skupski M., Booker,M., Farmer,A., Harpold,M., Huang,W., Inman,J., Kiphart,D., Kodira,C., Root,S., Schilkey,F. et al. (1999) Nucleic Acids Res., 27, 35–38. Updated article in this issue: Nucleic Acids Res. (2000), 28, 31–32.9847136 [Google Scholar]
- 7.Kamoun S., Hraber,P., Sobral,B.W.S., Nuss,D. and Govers,F. (1999) Fungal Genet. Biol., in press. [DOI] [PubMed] [Google Scholar]
- 8.Rouzé P., Pavy,N. and Rombauts,S. (1999) Curr. Opin. Plant Biol., 2, 90–95. [DOI] [PubMed] [Google Scholar]
- 9.Bailey L.C. Jr, Fischer,S., Schug,J., Crabtree,J., Gibson,M. and Overton,G.C. (1998) Genome Res., 8, 234–250. [DOI] [PubMed] [Google Scholar]