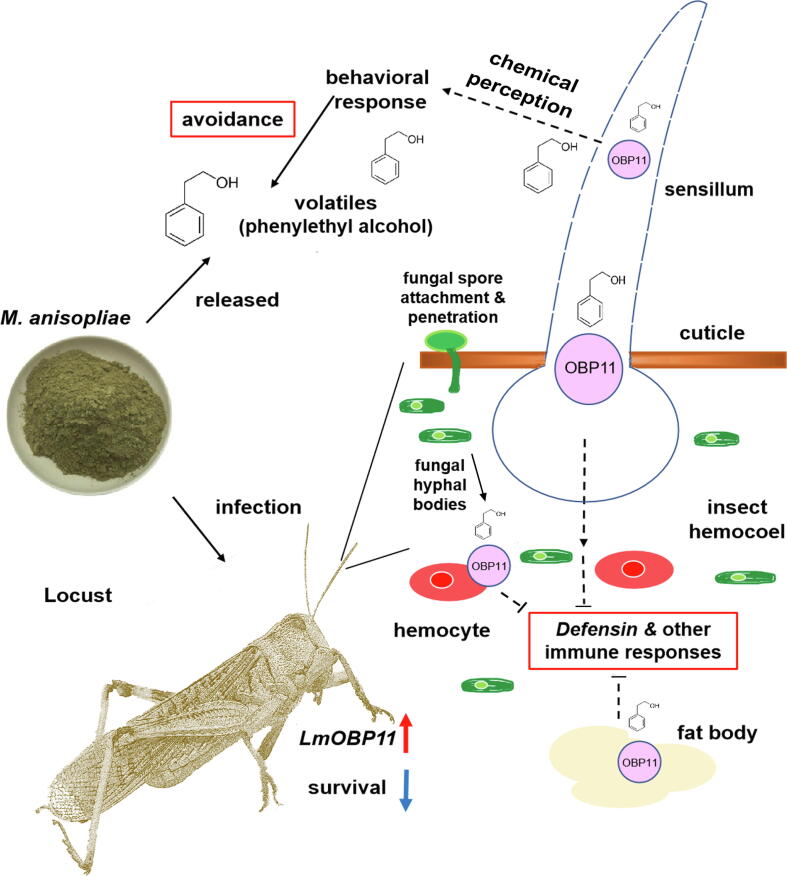
Graphical abstract
Keywords: Insect innate immunity, Entomopathogenic fungi, Locust, Antimicrobial peptides, Olfactory-mediated behaviors, Volatile organic compounds (VOCs)
Highlights
-
•
M. anisopliae phenylethyl alcohol exerts an avoidance effect to locusts.
-
•
M. anisopliae infection elevates the expression of LmOBP11 in locusts.
-
•
Treatment with phenylethyl alcohol upregulates LmOBP11 in locusts.
-
•
Locust LmOBP11 is involved in the detection of fungal phenylethyl alcohol.
-
•
LmOBP11 negatively regulates the locust immune system to benefit fungal infection.
Abstract
Introduction
Odorant-binding proteins (OBPs) are a class of small molecular weight soluble proteins that exist as expanded gene families in all insects, acting as ligand carriers mediating olfaction and other physiological processes. During fungal infection, a subset of insect OBPs were shown to be differentially expressed.
Objectives
We tested whether the altered expression of insect OBPs during pathogenic infection plays a role in behavioral or immune interactions between insect hosts and their pathogens.
Methods
A wide range of techniques including RNAi-directed knockdown, heterologous protein expression, electrophysiological/behavioral analyses, transcriptomics, gut microbiome analyses, coupled with tandem mass spectrometry ion monitoring, were used to characterize the function of a locust OBP in host behavioral and immune responses.
Results
The entomopathogenic fungus Metarhizium anisopliae produces the volatile compound phenylethyl alcohol (PEA) that causes behavioral avoidance in locusts. This is mediated by the locust odorant binding protein 11 (LmOBP11). Expression of LmOBP11 is induced by M. anisopliae infection and PEA treatment. LmOBP11 participates in insect detection of the fungal-produced PEA and avoidance of PEA-contaminated food, but the upregulation of LmOBP11 upon M. anisopliae infection negatively affects the insect immune responses to ultimately benefit successful mycosis by the pathogen. RNAi knockdown of LmOBP11 increases the production of antimicrobial peptides and enhances locust resistance to M. anisopliae infection, while reducing host antennal electrophysiological responses to PEA and locust avoidance of PEA treated food. Also, transcriptomic and gut microbiome analyses reveal microbiome dysbiosis and changes in host genes involved in behavior and immunity. These results are consistent with the elevated expression of LmOBP11 leading to enhanced volatile detection and suppression of immune responses.
Conclusion
These findings suggest a crosstalk between olfaction and immunity, indicating manipulation of host OBPs as a novel target exploited by fungal pathogens to alter immune activation and thus promote the successful infection of the host.
Introduction
Insects in their natural environments are constantly challenged by versatile pathogens. Effective strategies of insects to respond to the invading microbes are essential for successful insect survival and propagation. Physical insect barriers are composed by the cuticle (exoskeleton), that expresses antimicrobial compounds (epidermis), which form the first line to defense against the direct entry of microbial pathogens [1]. The insect innate immune system consists of cellular immunity and humoral immunity [2], [3], which compose a quasi-adaptive physiological defense strategy that lacks antibody production and other aspects of adaptive immunity. A number of insect species including mole crickets [4], ladybirds [5], Anopheles gambiae mosquitoes [6] and Coptotermes lacteus milk termites [7], have evolved strategies to recognize and respond to volatiles from such pathogens, such as the fungal entomopathogens Beauveria bassiana and Metarhizium anisopliae, which assist the insect hosts to engage in avoidance behaviors.
Metarhizium anisopliae, is a potent natural pathogen of insects. This fungus is able to first attach to the surface of the host exoskeleton and then penetrate the insect cuticle using a combination of mechanical pressure and enzymatic activities, which mostly consist of cuticle degrading enzymes [8], [9]. Once the cuticle has been breached, the fungus can proliferate within the insect hemocoel by nutrition exploitation and evading host innate immune processes through releasing siRNA, enzymes and secondary metabolites with toxic properties [10], and ultimately work its way out to sporulate on the host cadaver [11]. Although avoidance of contacting with virulence pathogens is probably a more cost-effective insect strategy than regulating innate immune responses, the exact relationship between behavioral and physiological immune mechanisms is not well understood.
The volatile organic compounds (VOCs) released by insect hosts are suggested to vary depending on the virulence of the infecting microbial strain [12], and the repellant effect of certain VOCs has been confirmed previously, i.e., 1–3-cyclohepten-1-one, 1,3-dimethoxy-benzene [13], naphthalene, 1-octen-3-ol [14], and phenylethyl alcohol [15]. Apart from the behavioral effects, some of the repellent VOCs, such as 1-octen-3-ol [16] and phenylethyl alcohol [17], also exhibit toxic effect against insects. This indicates a potential link between pathogen detection and immune capacity.
Olfaction has been linked to fungal avoidance in a number of insects [18], [19]; however, no specific olfactory protein(s) have been implicated in mediating perception of fungal cells and/or volatiles. Chemosensory proteins (CSPs) and odorant-binding proteins (OBPs) are small molecular weight ligand carrier proteins originally considered to act to transport odorants to receptors within sensory cells and modulate the insect behavioral response [20], [21], [22]. The function of CSPs and OBPs in VOCs recognition has been widely confirmed [23], [24], [25], [26]. However, increasing evidence shows that some CSPs and OBPs function in alternate ways to regulate other physiological processes beyond olfaction by binding to exogenous or endogenous metabolites [27], [28]. For instance, a mosquito saliva-secreted odorant binding protein is suggested to possess anti-inflammatory characteristics by binding host biogenic amine [29]. In addition, some olfactory proteins have been shown to be expressed in insect immune tissues, including hemocytes and fat bodies, beyond olfactory tissues [30]. During the various stages of M. anisopliae infection of the migratory locust Locusta migratoria, many elements of insect olfactory pathways including CSPs, OBPs, and odorant and ionotropic receptors (ORs/IRs) were shown to be differentially expressed [31], [32]. Further proteomic analyses validated these alterations of olfactory pathway components at the protein level in the antennae of M. anisopliae infected locusts [33]. Changes in the expression of olfactory genes were shown to occur as early as four hours after infection which coincides with conidial attachment to the cuticle surface, a time point at which the fungal spores have not germinated yet, let alone begun the process of penetrating the cuticle [32]. These data imply the importance of olfactory proteins in mediating insect behavioral and immune reactions.
Herein, we show that M. anisopliae produces a volatile compound (phenylethyl alcohol, PEA) that causes a behavioral avoidance effect in locusts. Also, the expression of locust odorant binding protein 11 (LmOBP11) can be induced by M. anisopliae infection and PEA treatment. Further, RNAi mediated suppression of LmOBP11 expression decreases insect chemical perception to the M. anisopliae produced volatile compound PEA, including reduced avoidance to PEA contaminated food, and reduced antennal electrophysiological responses to PEA. However, suppression of LmOBP11 expression surprisingly enhances host immune responses during M. anisopliae infection, which increases insect resistance to fungal infection by activating the expression of Toll-pathway related genes and associated effectors. These results suggest that elevated levels of LmOBP11 observed during M. anisopliae infection may benefit the host by increasing chemoperception of the pathogen but comes with a trade-off that benefits the pathogen by inhibiting host innate immune responses. These data provide a novel link between olfaction and innate immunity and represent a typical example of the subversion of a host olfactory protein for increasing successful mycosis by the microbial pathogen.
Materials and Methods
Insects and fungal strain
Insects (Locusta migratoria; NCBI: txid229990) were maintained in cages at 30 ± 3 °C with 90 % relative humidity, a photoperiod of 14:10 h light:dark, and supplied with fresh wheat shoots, wheat bran and water daily. Synchronized fifth instar locusts were used in all experiments. M. anisopliae CQMa421, stored at the Genetic Engineering Center of Chongqing University, was cultured on 1/4-strength Sabouraud dextrose agar (1/4-SDA) for 15 days at 28 °C. Conidia were harvested from plates via suspension in paraffin oil for topically inoculation or Tween 80 (0.05 %) for hemocoel injection, and mycelial debris were removed by filtration through sterile lens paper. Spore concentrations were calculated using a Neubauer hemocytometer and adjusted as described (typically to 1 × 108 spores/mL).
Tissue dissection and gene expression analyses
Locust tissues including cuticle, gut, head, thoracic muscles and reproductive glands, wings, fat body, Malpighian tubules and central nervous system, and hemocytes were dissected from 5, 10, and 20 fifth instars, respectively, for each experimental replicate. The samples were immediately frozen in liquid nitrogen and stored at −80 °C until used. Hemolymph was isolated by puncturing the arthrodial membrane at the base of hind legs with a sterile needle. Subsequently, the liquid was added into a tube with the same volume of ice-cold anticoagulant solution (26 mM citric acid, 30 mM sodium citrate, 60 mM NaCl, 100 mM d-glucose and 10 mM EDTA) to prevent coagulation. Total RNA was extracted using the Trizol reagent (Takara, Japan) according to the manufacturer’s protocols. TURB DNase (Thermofisher, USA) was used to remove the DNA contamination from the RNA samples in terms of manufacture’s instruction. cDNA libraries were prepared using a high-capacity cDNA Reverse transcription kit (Thermofisher, USA).
Primers for qRT-PCR (Supplemental Table S6) were designed using the Beacon Designer Software (Palo Alto, CA, USA). Actin was used as an internal reference gene as previously described [34]. qRT-PCR experiments were performed in accordance with the Minimum Information Required for Publication of Quantitative Real-Time PCR Experiments guidelines [35]. Premix Ex TaqTM II (Tli RNaseH Plus) Kit (Takara, Shiga, Japan) was used as the qRT-PCR reagent. Reaction conditions were set as follows: 95 °C for 3 min, 40 cycles of 95 °C for 15 s and 60 °C for 30 s, following by a dissociation protocol in the iCycler iQ real-time PCR detection system (Bio-Rad, Hercules, CA, USA). Each treatment contained three technical replicates and conducted with three separate biological triplicates. Data were analyzed based on the 2−ΔΔCT [ΔΔCt = ΔCt (test) - ΔCt (calibrator)] method [36]. qRT-PCR quality control additional data, including amplification efficiencies (%) of the qRT-PCR reactions, and correlation coefficients (R2) is supplemented in the SI information (Supplemental Table S7).
Double-stranded RNA construction and survival analysis
Sequence alignment analysis of a transcript previously identified in a transcriptomic study of locust gene expression in response to M. anisopliae infection indicated that Unigene66453 was identical to LmOBP11 as annotated in the L. migratoria genome [32]. The open reading frame (ORF) of LmOBP11 was amplified according to our transcriptomic dataset and the amplified fragment was subcloned into the pUC19-T vector for sequencing. A target sequence for dsRNA mediated gene expression knockdown of LmOBP11 (Supplemental Table S6) was designed according to the specificity and RNAi efficiency analysis. The MEGAscript high yield transcription kit (Ambion, Austin, TX, USA) was used to synthesize the dsRNA targeting LmOBP11, LmOBP2 and LmOBP4 as well as one designed towards GFP (control), according to the manufacturer’s protocol. GFP was used as a negative control as GFP dsRNA shows no side effects [34] and it has been broadly used in locusts [37], [38]. Briefly, dsRNA template was prepared by PCR and purified before dsRNA synthesis. dsRNA was synthesized by mixing plasmid template and the in vitro transcription reaction in a RNase free tube. The dsRNA product was precipitated with LiCl solution. The DNA template in the reaction product was digested by DNase treatment followed by phenol/chloroform purification to remove any extra components. The concentration of the final dsRNA product was determined using a NanoVue Plus spectrophotometer (GE Healthcare Life Sciences, Little Chalfont, UK). For RNAi assay treatments, 5 μL of the dsRNA solution (300 ng/μL) was injected into the hemocoel of each locust through the abdomen. For co-treatment of locusts with (LmOBP11 or GFP) dsRNA and M. anisopliae infection, dsRNA was injected initially and 3 days later M. anisopliae conidia were topically inoculated. Hemocytes and hyphal body numbers were qualified by collecting the hemolymph at three days post M. anisopliae inoculation, as described in detail below. For testing the efficiency of the dsRNA in target gene knockdown, locust antennae were dissected from five randomly selected locusts at three- and seven-days post dsRNA injection. Expression of LmOBP11 and other genes as noted (e.g., defensin) was examined in corresponding tissues of various treatments at three days post M. anisopliae inoculation. For the synergy effect of PEA in fungal infection, PEA was mixed with 5 μL M. anisopliae conidia suspension with the final concentration of 0.0001 mol/L and injected into locusts 72 h after dsRNA treatment. Antennae, cuticle, hemocyte, and fat body of locust topically treated with PEA or ethanol (control) were collected to identify the expression of LmOBP11 and (or) defensin in response to PEA treatment. The number of surviving insects was measured twice daily after fungal inoculation. All experiments consisted of three replicates (n = 20), and the entire experiment was performed with at least three independent batches of insects and fungal conidia.
Volatile organic compound analyses of M. anisopliae spores and determination of PEA concentrations in locust hemolymph
To detect VOCs produced by M. anisopliae, conidia cultured on 1/4 SDAY medium were collected and transferred into 20 mL SPME vials with 6 replications in total. The SPME (headspace solid phase microextraction) syringe with a 100 μm PA (polyacrylate) fiber (CTC Analytics AG, Zwingen, Switzerland) was used for the extraction of volatiles emitted by the fungus [39]. The new fiber was conditioned at the GC inlet according to the manufacturer’s instructions before use. Samples were preheated in the incubator at 30 °C for 30 min, and then the fiber was inserted into the vessel and exposed to the headspace gas phase. After 4 h exposure to the headspace, the fiber was immediately introduced to the GC–MS for thermal desorption and analysis. Volatile organic compounds were detected by GC–MS analysis with a GCMS-TQ8040 (Shimadzu Corporation, Kyoto, Japan), equipped with a WAX-HT capillary column (30 m × 0.25 mm × 0.25 μm) (Shimadzu Corporation, Kyoto, Japan). The PA fiber was desorbed at 240 °C in the injection port for 5 min in the spitless mode and helium was used as carrier gas. The front inlet purge flow velocity was 3 mL/min, and the gas flow rate through the column was 1 mL/min. The oven temperature ramp was programmed as follows: 40 °C for 5 min, followed by a temperature gradient of 5 °C/min up to 240 °C, with a 5 min hold at the end. The transfer line and ion source temperatures were 240 °C and 230 °C, respectively. Mass spectroscopy (MS) analyses were performed in full-spectrum scanning mode (m/z 20–400) and in Selected Ion Monitoring (SIM) mode at 70 eV electron energy, with a solvent delay of 5 min. Volatile compounds were identified by comparing their mass spectra with the standard spectral NIST14 library. The PEA analysis in locust hemolymph was analyzed similarly by GC–MS but without SPME treatment, unless hemolymph samples were collected on three days post M. anisopliae inoculation as previous described.
Protein expression and quantitative fluorescence-based ligand-binding assay
LmOBP11 protein was expressed using a prokaryotic protein expression system, as previous described [40]. Briefly, the ORF of LmOBP11 (468 bp), after removal of its corresponding predicted signal peptide sequence, was cloned into the pET32a vector (Novagen, Darmstadt, Germany) and transformed into the Escherichia coli BL21 strain. Individual positive colonies grown on LB-agar plates (100 μg/mL, ampicillin) were selected and cultured in 5 mL of LB liquid medium (100 μg/mL, ampicillin) overnight at 37 °C with shaking at 220 rpm. The cell culture was inoculated in 500 mL LB/ampicillin liquid medium until OD (600 nm) reached 0.6 to 0.8. Then, 0.5 mM IPTG was added into the culture to induce protein expression at 11°C with overnight shaking at 220 rpm/min. Cell pellet was collected by centrifugation at 3,000 rpm and resuspended in HEPES buffer (10 mM HEPES, 100 mM NaCl, pH 7.5). After sonication and centrifugation at 14,000 rpm and 4 °C for 30 min, the supernatant and pellet were collected separately. SDS-polyacrylamide gel electrophoresis (SDS-PAGE) confirmed that the recombinant protein was soluble. The solution was applied to His-trap affinity columns (Cobalt Chelating Resin, G-Biosciences, USA). Bound protein was eluted with HEPES buffer containing gradually increased imidazole from 50 mM to 500 mM. After electrophoretic analysis. The fractions with the recombinant protein were pooled and dialyzed three times against 3 L of HEPES buffer at 4 °C, overnight. His-tag antibody (Thermofisher, USA) was used to perform western blot to further confirm the size of the recombinant protein.
GC grade authentic standards were purchased, including phenylethyl alcohol (PEA; >98.0 %, TCI chemicals, China), n-hexadecanoic acid (95 %, Kaiwei Chemical, China), 2,4-di-tert-butylphenol (99 %, Solarbio, China), 2-ethoxy-ethanol (99 %, Aladdin, China), 2-hexyldecanoic acid (98 %, Macklin, China), and 1,2-ethanediol monoacetate (60 %, Macklin, China). N-phenyl-1-naphthylamine (1-NPN, 98 %, Sigma-Aldrich) was used as a probe for the fluorescent binding assay, as previously described [22]. A Hitachi F-2000 fluorescence spectrophotometer was used for the binding assay, with an excitation wavelength of 337 nm and an emission wavelength of 350–500 nm. The binding constant of the purified protein and 1-NPN were analyzed using the change in the fluorescent value from the machine when the 1-NPN concentration was gradually increased from 2 to 24 μM in a cuvette with a set protein concentration (2.0 μM; in 10 mM HEPES and 100 mM NaCl buffer pH 7.5). The competitive binding of the chemical ligand was measured by gradually increasing the chemicals into the mixture with 2 μM 1-NPN and 2 μM protein or 2 μM 1-NPN alone to determine chemical background levels. Fluorescence values were recorded in the presence of protein, 1-NPN, and differing concentrations of competing ligands. Final fluorescence values were calculated using the corrected fluorescence values through subtracting the chemical backgrounds.. The dissociation constants (Kd) of LmOBP11 and 1-NPN were calculated by Scatchard plots through the GraphPad Prism 8 Software (GraphPad, La Jolla, CA, USA). The dissociation constants of competitor ligands (Kapp) to 1-NPN were calculated according to the IC50 values using the following equation: Kapp = [IC50]/(1 + [1 − NPN]/K1 − NPN).
Electroantennography (EAG) response and behavioral analysis
EAG response (mV) was analyzed by an EAG system (Syntech, the Netherlands) through insertion of an excised locust antenna into the EAG glass capillary tube containing two silver electrodes (recording electrode and reference electrode). The amplified electrophysiological signals were recorded with a Syntech PC-based signal processing system (Kirchzarten, Germany). In the dose–response experiment, five doses of PEA (10 µL of 10−4, 10−3, 10−2, 0.1 and 1 M, respectively, used as the standard stimulus) were prepared in ethanol (concomitantly used as control), and each dose was tested five times on an antenna at 1-min intervals between stimulations. LmOBP11 dsRNA-injected and GFP dsRNA-injected locusts were tested respectively for all doses. In each group of locusts, five antennae were analyzed, and three batches of treated locusts were tested entirely.
The avoidance behavior of locusts to PEA was analyzed by mixing 1 mL of PEA solution (0.01 M, in ethanol) or 1 mL of ethanol with 1.5 g of wheat bran. A total of 15 locusts/technical replicate were placed in metal cages with PEA or ethanol control and wheat bran on random opposite corners of the cage. Food consumption was measured 12 h after feeding. Three replicates with triplicate locusts were analyzed entirely.
Hemocyte quantification and measurement of phenoloxidase (PO) activity
Hemolymph samples were collected from locusts via puncture of the leg segmacoria and collection in anticoagulant buffer as described above. An aliquot of 10 μL hemolymph was used to count hemocyte numbers using a hemocytometer under a microscope, with three regions examined for each sample. The remaining hemolymph was used for measurement of PO activity after centrifugation (4 °C, 20,000 rpm, 5 min) to collect supernatant. PO activity was measured as previously described with minor modification [41]. Briefly, 30 μL of supernatant were mixed with 210 μL of 10 mM sodium phosphate buffer (pH 6.0). Aliquots (160 μL) of the mixture were used for PO activity assay and 30 μL of the mixture was used for protein concentration assay. Protein concentrations were determined using the Bradford method (Bradford Protein Assay Kit, Solarbio). For PO activity measurement, 480 μL of 10 mM l-Dopa were added to the 160 μL hemolymph aliquot and incubated at 25 °C for 20 min after which the initial absorbance at 492 nm was measured, and the absorbance values were recorded every 5 min for 1 h. The PO activity was calculated according to the slope of the absorbance values divided by the corresponding protein concentration (U/mg). All experiments were performed with three independent batches of locusts.
Transcriptomic analysis
For transcriptomics analyses, four treatments were examined: (1) GFP dsRNA injected locusts, (2) LmOBP11 dsRNA injected locusts, (3) GFP dsRNA injected + M. anisopliae infected locusts, and (4) LmOBP11 dsRNA + M. anisopliae infected locusts. dsRNA treatment (injection of 1.5 μg dsRNA) was performed first, 72 h after which locusts (where indicated) were infected (5 μL of 108 conidia/ml topically on the locust abdomen). For all treatments, locusts were collected three days post-inoculation (six days post-treatment if no infection) with three replications. The fat body, antennae, and cuticle of five locusts per treatment were dissected into locust physiological saline solution (147 mM NaCl, 10 mM KCl, 4 mM CaCl2, 3 mM NaOH, 10 mM HEPES, pH 7.2–7.4). Samples were homogenized for RNA extraction using the Trizol reagent (100 mg/mL, Invitrogen, USA) according to the manufacturer’s instructions. RNA samples were treated with RNase-free DNase I (Takara, Shiga, Japan) for 1 h at 37 °C to remove any residual genomic DNA. The quality of RNA samples was determined by agarose gel electrophoresis and NanoVue Plus (GE) analyses. Samples were sequenced via a commercial company (Lianchuan, Hangzhou, China) using a next generation sequencing platform (Illumina NovaseqTM 6000). Briefly, the poly-T oligo-attached magnetic beads (Invitrogen) were used to enrich for polyadenylated RNA and for library preparation in accordance with the protocol for the mRNASeq sample preparation kit (Illumina, San Diego, USA), and then the paired-end sequencing was performed on an IlluminaHiseq4000 at the (LC Sciences, USA) following the vendor’s recommended protocol. Differentially expressed genes between two treatments were analyzed by DESeq R package (1.10.1), which provides statistical routines based on a model of negative binomial distribution. The resulting P values were adjusted using the Benjamini and Hochberg’s approach to control the false discovery rate [42]. Genes with an adjusted p-value < 0.05 found were assigned as differential expression.
Gut microbial community analysis
Locusts were treated under identical conditions (four different treatments) as described for the transcriptomic analyses above, and then their midguts were collected by dissection under a stereoscope. Bacterial and fungal DNA from each sample were extracted using separate protocols and overall DNA quality was verified by NanoDrop spectrophotometer. The V3-V4 region of the bacterial 16S rRNA gene was amplified using the primer pair (Forward primer, 5′-ACTCCTACGGGAGGCAGCA-3′, reverse primer, 5′-GGACTACHVGGGTWTCTAAT-3′) combined with adapter sequences and barcode sequences, whereas the ITS1 region of fungal 18S rRNA was amplified with the primer pairs (5′-CTTGGTCATTTAGAGGAAGTAA-3′, 5′-TGCGTTCTTCATCGATGC-3′) combined with adapter sequences and barcode sequences. PCR products were purified and sequenced using the Illumina Hiseq 2500 platform (2 × 250 paired ends, Biomarker Technologies Corporation, Beijing, China). Three replications (five midguts/replication) were performed to analyze the gut bacterial and fungi species, respectively.
Statistical analysis
SPSS (SPSS Inc., Chicago, IL) was used to perform significance tests. The half lethal time (LT50) for survival curves were estimated by a probit analysis with the same software. PO activity and gene expression results across various treatments were analyzed by one-way ANOVA with the Post Hoc analysis of Bonferroni (equal variance) or Dunnett T (unequal variance). The significances of relative expression of LmOBP11 in each set of treatment, food consumption, hemocyte and hyphal body numbers in the hemolymph were analyzed by Student t test. p < 0.05 and p < 0.01 were considered as significant and highly significant, respectively. Unless otherwise stated, all experiments included three technical replicates, and each experiment was replicated three times.
Results
LmOBP11 expression is activated during fungal infection and RNAi mediated knockdown of LmOBP11 increases resistance to fungal infection
LmOBP11 was identified as a differentially expressed gene in M. anisopliae infected locusts as compared to controls in previous transcriptomic studies (Supplemental Fig. S1A) [31], [32]. More detailed tissue expression pattern analyses revealed that LmOBP11 was highly expressed in the antennae, followed by the sexual gland, hemocytes, head, central nervous system, gut, Malpighian tubules, muscle, and wing, with low levels seen in the cuticle and fat body tissues (Supplemental Fig. S1B). A time course of LmOBP11 expression in response to M. anisopliae infection further indicated increased expression in the antennae, cuticle, and fat body ∼72 h post-infection (p < 0.05), with only a marginal increase seen in hemocytes (Supplemental Fig. S1C).
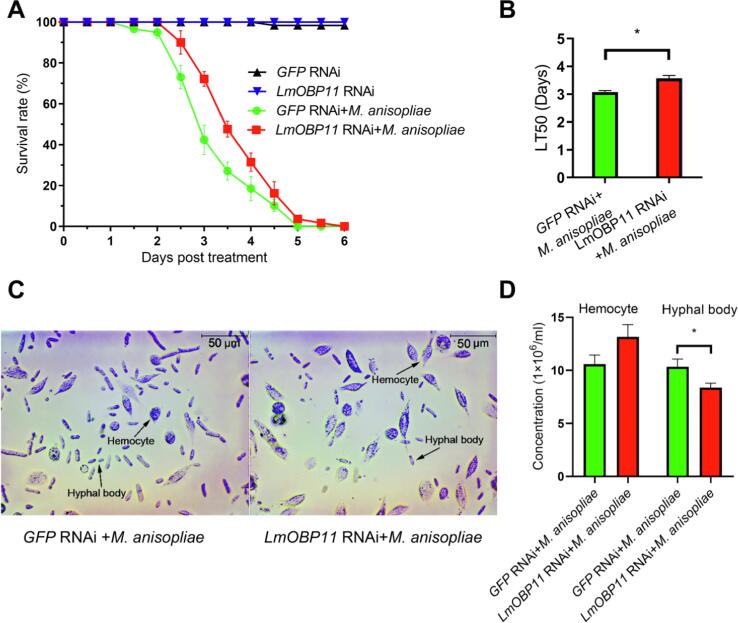
In order to probe the function of LmOBP11 in response to M. anisopliae infection, constructs for RNAi LmOBP11 gene knockdown were synthesized and injected into locusts. RNAi efficiency analysis showed that LmOBP11 expression in dsRNA injected locusts was reduced by ∼90 % 72 h after the injection, and that reduced expression was maintained for at least 7 d after injection (Supplemental Fig. S2). Surprisingly, insect bioassays revealed an increase in host resistance to fungal infection following LmOBP11 RNAi knockdown (Fig. 1A). The mean lethal time to kill 50 % of host locusts (LT50) increased from 3.08 ± 0.10 d for wild type locusts to 3.57 ± 0.17 d for locusts treated with LmOBP11 dsRNA (Fig. 1B, p < 0.05), with no changes seen using a control dsRNA (GFP, LmOBP2 and LmOBP4) (Supplemental Fig. S3) or in response to the LmOBP11 dsRNA alone in the absence of infection. Measurements of fungal hyphal body production and host hemocyte numbers indicated decreased fungal proliferation (from 10.4 ± 0.7 × 106 to 8.4 ± 0.4 × 106 hyphal bodies/mL of hemolymph, p < 0.05) when locusts were treated with M. anisopliae + LmOBP11 dsRNA as compared to M. anisopliae alone, and with a slight but not statistically significant (p = 0.091) increase in hemocyte numbers in M. anisopliae + LmOBP11 dsRNA treated locusts as compared to M. anisopliae + GFP controls (Fig. 1C & D).
Fig. 1.
Survival rate (A, B) and immune responses analysis (C, D) of LmOBP11-silenced and GFP control locusts to fungal infection. The Student's t-test was used to compare the means between treatment pairs, assuming they have equal variances. Asterisks indicate the level of statistical significance; *p < 0.05.
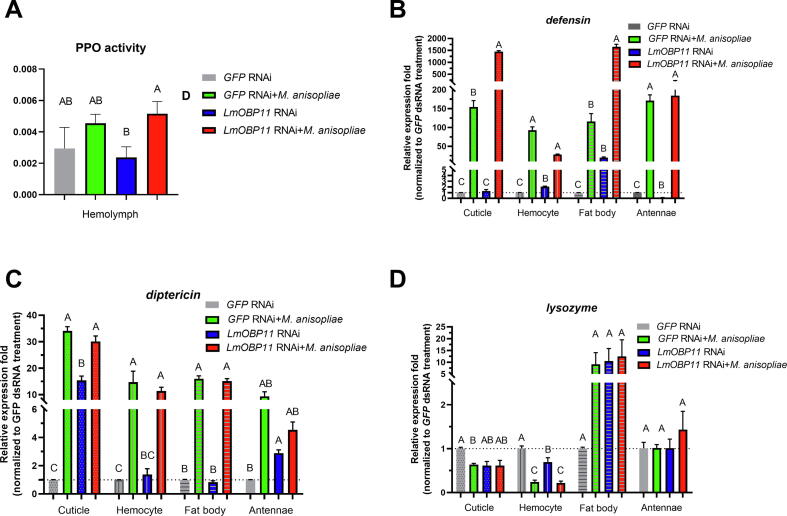
Host prophenoloxidase (PO) activity was similar between LmOBP11 dsRNA and control (GFP RNAi treated) locusts with infection by M. anisopliae resulting in similar levels of PO activation independent of LmOBP11 knockdown (Fig. 2A). The expression of three host antimicrobial defense responses, including the Toll pathway regulated AMPs Defensin and Diptericin, and the antimicrobial enzyme, lysozyme, were examined in four different host tissues including cuticle, fat body, hemocytes, and antennae and normalized to expression levels seen in GFP dsRNA treated samples. These results revealed that knockdown of LmOBP11 increased Defensin expression in the locust cuticle, fat body, and hemocyte tissues, with fold changes of 1.3, 20.3, and 2.0, respectively. However, Defensin expression decreased in antennae with a fold change of 0.15 as compared to controls (Fig. 2B). Significantly higher (p < 0.01) levels of Defensin expression were seen in locusts that were subjected to M. anisopliae infection + LmOBP11 dsRNA as compared to M. anisopliae + control GFP dsRNA and LmOBP11 dsRNA alone in both cuticle and fat body tissues, whereas equivalent (between treatments) Defensin expression was seen in hemocytes and antenna (Fig. 2B). With respect to Diptericin expression, treatment with LmOBP11 dsRNA activated its expression in both cuticle (∼15.4-fold) and antennae (2.9-fold); however, similar levels of Diptericin expression were seen in hemocytes and fat body. In addition, similar levels of Diptericin expression were observed in locusts treated with M. anisopliae + LmOBP11 dsRNA and M. anisopliae treatments alone in all examined tissues (Fig. 2C). Lysozyme gene expression was upregulated in the fat body (∼11.7-fold) but downregulated in hemocytes (0.69-fold) in locusts treated with LmOBP11 dsRNA (Fig. 2D). Similar levels of lysozyme expression were seen in locusts treated with M. anisopliae + LmOBP11 dsRNA and M. anisopliae alone treatments in all examined tissues.
Fig. 2.
Immune effector, PPO (A), Defensin (B), Diptericin (C) and Lysozyme (D) analysis of LmOBP11-silenced and GFP control locusts to fungal infection by q-RT-PCR. Means followed by the same letter are not significantly different at p < 0.05 according to one-way ANOVA with the Post Hoc analysis of Bonferroni (equal variance) or with the Post Hoc analysis of Dunnett's test (unequal variance).
LmOBP11 binds to phenylethyl alcohol (PEA), and LmOBP11 knockdown results in loss of PEA perception and locust ability to avoid PEA contaminated food
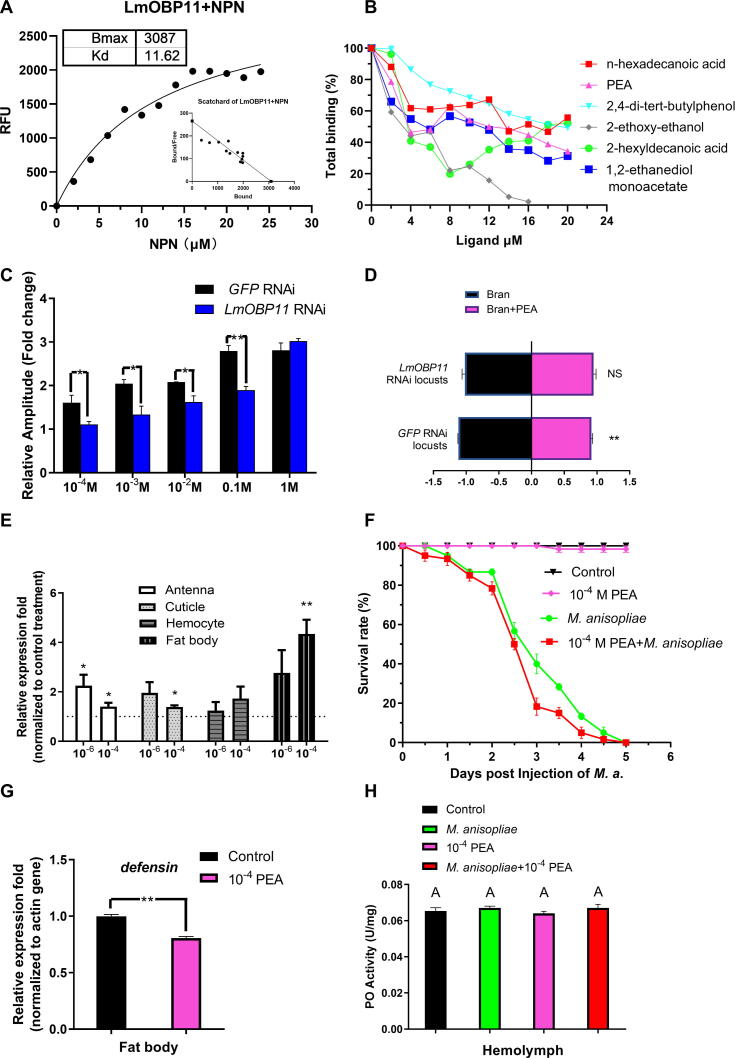
A set of VOCs produced by M. anisopliae was detected by HS-SPME-GC–MS. A total of 14 M. anisopliae derived volatile compounds not found in the control blank were identified (Supplemental Table S1), and included five different alcohols (2-ethoxy-ethanol, 11-hexadecyn-1-ol, phenylethyl alcohol, 3-(1-methylbutoxy)-2-butanol and 4-nonanol), four esters (1,2-ethanediol monoacetate, sulfurous acid, nonyl 2-pentyl ester, formic acid, 2,4-dimethylpent-3-yl ester and tetrahydrofurfuryl acrylate), three carboxylic acids (acetic acid, 2-hexyldecanoic acid and n-hexadecanoic acid), as well as 2,5-dimethyl-4-hydroxy-3-hexanone and tetrahydrofurfuryl acrylate. We therefore tested whether LmOBP11 could bind PEA and five more compounds, n-hexadecanoic acid, 2,4-di-tert-butylphenol, 2-ethoxy-ethanol, 2-hexyldecanoic acid and 1,2-ethanediol monoacetate. LmOBP11 was purified after expression of the recombinant protein using an E. coli heterologous expression system (Supplemental Fig. S4), and the binding constant of purified LmOBP11 to the fluorophore N-phenyl 1-napthylamine (1-NPN) was determined (Kd = 11.62 µM) in order to use this substrate in (competition/displacement) reporter assays as has been extensively used for OBP ligand characterization [43] (Fig. 3A). The 1-NPN competition assay was then used to determine the binding affinities of purified LmOBP11 to PEA, n-hexadecanoic acid, 2,4-di-tert-butylphenol, 2-ethoxy-ethanol, 2-hexyldecanoic acid and 1,2-ethanediol monoacetate (Fig. 3B). These data revealed an observed binding affinity of 9.37 µM for PEA, and binding affinities of 15.4, 20.6, 3.6, 10.8, and 7.23 µM for n-hexadecanoic acid, 2,4-di-tert-butylphenol, 2-ethoxy-ethanol, 2-hexyldecanoic acid and 1,2-ethanediol monoacetate, respectively (Fig. 3B). Among them, PEA was shown to act as a repellant/avoidance molecule in insects and this avoidance effect also confirmed in locusts (Fig. 3D). However, this avoidance behavior was suppressed in LmOBP11 dsRNA treated locusts (Fig. 3D). In order to examine antennal responses, electroantennography was used to determine the average antennal output in response to PEA in control and LmOBP11 dsRNA treated locust antennae. These data showed a clear decrease in response (65.3–78.0 %) to PEA in LmOBP11 dsRNA treated locusts over a concentration range from 10−4 to 0.1 M (p < 0.01), which was only overcome at 1 M of the odorant (Fig. 3C, Fig. S6).
Fig. 3.
LmOBP11 binds to phenylethyl alcohol (PEA), and LmOBP11 knockdown results in loss of PEA avoidance behavior. (A) OBP binding to N-phenyl 1-napthylamine (1-NPN) and (B) binding affinities to volatile produced by M. anisopliae, namely phenylethyl alcohol, and another five volatiles, n-hexadecanoic acid, 2,4-di-tert-butylphenol, 2-ethoxy-ethanol, 2-hexyldecanoic acid and 1,2-ethanediol monoacetate, (C) Electroantennography of average antennal output in response to PEA, (D) aversion behavior of LmOBP11 dsRNA treated locusts to phenylethyl alcohol. (E) Relative expression analysis of LmOBP11 under various concentrations of PEA (mol/L). (F) Survival rates of locusts in response to PEA and M. anisopliae separately and collectively. (G) Expression of defensin in the fat body, and (H) PO activity in the hemolymph following treatment with 10−4 mol/L PEA and fungus infection. The Student's t-test was used to compare the means between treatment pairs, assuming they have equal variances. Asterisks indicate the level of statistical significance; *p < 0.05, *p < 0.01. Means followed by the same letter are not significantly different at p < 0.05 according to one-way ANOVA with the Post Hoc analysis of Bonferroni (equal variance).
PEA is known to be produced by M. anisopliae [44]; however, in order to determine whether PEA concentrations increased during M. anisopliae infection, insect hemolymph was isolated three days post-treatment and analyzed for PEA levels using mass spectroscopy. These data showed a 2–3-fold increase in PEA concentrations (p < 0.05) after M. anisopliae infection and after M. anisopliae + LmOBP11 dsRNA treatments, whereas LmOBP11 dsRNA treatment alone did not result in a significant change in host PEA levels (p = 0.72, Supplemental Fig. S5). Analyses of LmOBP11 expression in response to injection of 10–4 mol/L PEA (normalized to no PEA control) revealed increased expression of LmOBP11 within 24 h to levels similar to that seen after M. anisopliae infection three days post inoculation (Fig. 3E). Furthermore, co-treatment of M. anisopliae + PEA slightly increased locust mortality. The LT50 decreased from 2.81 days for M. anisopliae alone to 2.45 days for M. anisopliae + PEA (p = 0.06), with PEA alone having no effect on insect mortality (Fig. 3F). In addition, treatment of locusts with PEA decreased Defensin expression (p < 0.01), although no significant effects were seen in terms of PO activity (Fig. 3G & H).
Transcriptomic analyses indicate that LmOBP11 knockdown alters global expression of innate immune and chemical perception pathways
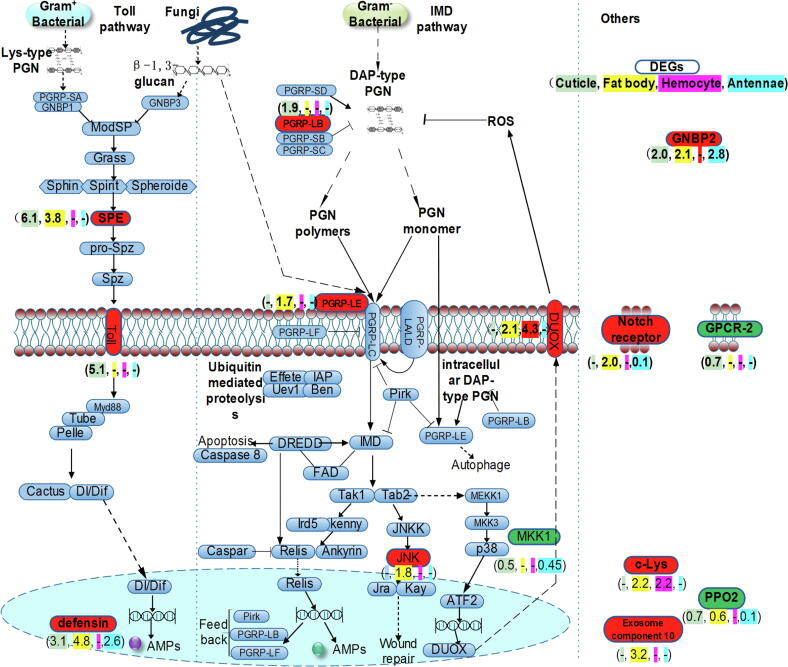
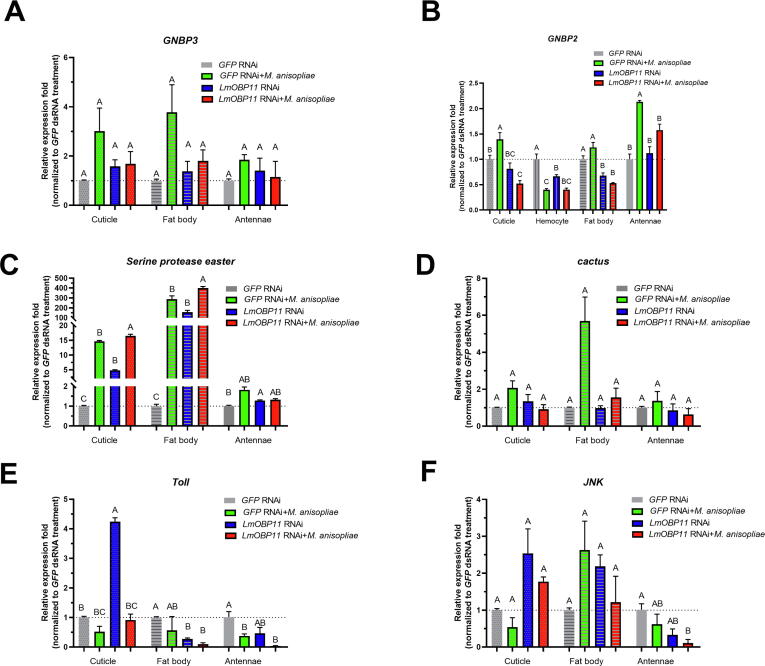
We next sought to analyze global gene expression changes related to LmOBP11 knockdown that affected innate immunity and chemical perception. A comparative transcriptomic analysis with four locust tissues including (a) antennae, (b) cuticle, (c) hemocytes, and (d) the fat body, along with four treatments: (1) control, GFP RNAi, (2) LmOBP11 RNAi, (3) GFP RNAi + M. anisopliae infection, and (4) LmOBP11 RNAi + M. anisopliae infection, three days post-treatment was performed. Pairwise comparisons between the various treatments to controls across all four tissues and four treatment groups were performed (Supplemental Fig. S7 & Supplemental Table S2). In order to determine the effects of LmOBP11 knockdown on M. anisopliae infection, differentially expressed transcripts in the locust cuticle, fat body, hemocytes and antennae samples comparing M. anisopliae infection to M. anisopliae infection + LmOBP11 dsRNA were analyzed for immune pathway components (Fig. 4). A general pattern of increased expression of select immune pathway genes was found in locusts treated with LmOBP11 targeting dsRNA. In total, 8, 10, 3, and 5 immune related DEGs (comparing M. anisopliae infection to M. anisopliae infection + LmOBP11 dsRNA) were identified in cuticle, fat body, hemocytes, and antennal tissues with fold change over 1.5 and below 0.5. Within the Toll pathway SPE (MH393893.1, Serine protease easter or Spätzle-processing enzyme) was upregulated in the cuticle and fat body, Toll was upregulated in the cuticle, and Defensin was upregulated in the cuticle, fat body and antennae in LmOBP11 RNAi + M. anisopliae co-treated locusts as compared with M. anisopliae infection alone. Within the IMD immune pathway, PGRP-LB, PGRP-LE, Duox, and JNK were upregulated, whereas the expression of MKK14 in the cuticle and antennae decreased. Expression of GNBP2, c-Lys, and exosome component 10 in the fat body also increased, whereas expression of GPCR-2 and prophenoloxidase 2 (PPO2) in the cuticle decreased. The Notch receptor 9 gene was upregulated in the fat body but downregulated in the antennae of locusts co-treated with M. anisopliae + LmOBP11 dsRNA as compared to locusts infected with M. anisopliae alone.
Fig. 4.
Differential expressed immune related genes in locust cuticle, fat body, hemocytes and antennae under various treatments. Genes highlighted with red and green are significantly up- (>1.5-fold) and down-regulated (<0.5) in locusts treated with LmOBP11 RNAi + M. anisopliae when compared with locusts treated with GFP RNAi + M. anisopliae treatment. Numbers in the brackets highlighted with cyan, yellow, purple, and turquoise represent the differential expression fold in cuticle, fat body, hemocytes and antennae for locusts treated with LmOBP11 RNAi + M. anisopliae compared with GFP RNAi + M. anisopliae infection treated locusts.
The expression of a set of immune related genes from the differential immune pathways including the Toll and IMD was further examined by qRT-PCR in GFP RNAi control, LmOBP11 RNAi, GFP RNAi + M. anisopliae infected locusts and LmOBP11 RNAi + M. anisopliae infected locusts, and relative expression level divided to GFP RNAi control locusts were analyzed. These included Gram-positive bacteria-binding protein 3 (GNBP3, Toll pathway), cactus (Toll pathway), Toll (Toll pathway) and c-Jun N-terminal kinase (JNK, IMD pathway). No significant differences in GNBP3 expression were observed for any of the treatments in fat body, cuticle, or antenna similar as the transcriptomic data (Fig. 5A). GNBP2 expression showed no change in cuticle, fat body and antennae, but a slight decrease in hemocytes in LmOBP11 dsRNA treated locusts was observed (i.e., during infection, Fig. 5B). Serine protease easter levels increased in the cuticle, fat body and antennae of LmOBP11 dsRNA treated locusts, and significantly increased in M. anisopliae + LmOBP11 RNAi treated insects as compared to the cuticle and fat body of M. anisopliae + GFP RNAi controls (Fig. 5C). Cactus expression was not significantly different in all examined samples irrespective of the treatments (Fig. 3D), and Toll expression significantly increased (p < 0.05) in the cuticle but decreased in fat body tissues of LmOBP11 dsRNA treated locusts but no significant difference was noticed between M. anisopliae + GFP RNAi controls and M. anisopliae + LmOBP11 RNAi treated insects (Fig. 5E). JNK expression in host cuticle, fat body and antennae tissue were not significantly different in LmOBP11 dsRNA treated locusts (Fig. 5F). Among them, the Toll pathway components, the expression of the Serine protease easter showed a similar tendency as defensin in cuticle and fat body tissues.
Fig. 5.
Differentially expressed Immune pathway related genes, GNBP3 (A), GNBP2 (B), Serine protease easter (C), Cactus (D), Toll (E) and JNK (F) analysis of LmOBP11-silenced and GFP control locusts to fungal infection. Mean values followed with different uppercase letters are significantly different (p < 0 0.05), using one-way ANOVA with the Post Hoc analysis of Bonferroni (equal variance) or with the Post Hoc analysis of Dunnett's test (unequal variance).
An analysis of olfactory pathway components, including OBPs, CSPs, odorant receptors (ORs) and ionotropic receptors (IRs) was performed with respect to four different comparisons, (i) M. anisopliae infection + GFP RNAi/GFP RNAi locusts, (ii) LmOBP11 RNAi/GFP RNAi locusts, (iii) (M. anisopliae infection + LmOBP11 RNAi)/(M. anisopliae infection + GFP RNAi), and (iv) (M. anisopliae infection + LmOBP11 RNAi)/(LmOBP11 RNAi), across the four locust tissues (antennae, cuticle, hemocytes, and fat body) (Supplemental Fig. S9). As expected, expression (FPKM) of LmOBP11 decreased from ∼9.7 to 0 in antennae of locusts injected with LmOBP11 dsRNA when compared with GFP control locusts. Among the OBPs, LmOBP7 showed the highest homology with LmOBP11, but no significant change in its expression was seen in any of the tissues (Supplemental Fig. S2). However, significant changes in the expression of olfactory pathway proteins were seen in the different comparisons. Infection by M. anisopliae resulted in a general downregulation of a range of OBPs, CSPs, ORs and IRs in most tissues with the exception of the fat body, which had increased expression of five CSPs (CSPs 4, 6, 21, 46, and I-1) and one IR (OR8). Increased expression in the antennae of ORs 8, 15, 42, and 47 as well as IR12, and in the cuticle of ORs 8, 11, and 87, and of OR11 in hemocytes were also noted. In locusts injected with LmOBP11 dsRNA, there were a number of olfactory genes with decreased expression levels as compared to GFP dsRNA control. These genes also showed similar patterns of upregulated olfactory pathway genes in antennae and fat body (but not in cuticle or hemocytes) as M. anisopliae infection with a number of differences; (i) in the antennae, LmORs40 and 55 were also upregulated (in addition to the ones listed for M. anisopliae infection above), and (ii) in the fat body, expression of LmCSP6 and I-1, were unaffected. Comparisons between (LmOBP11 RNAi + M. anisopliae infection)/(M. anisopliae infection) revealed a different pattern of effect on olfactory pathway genes. In the antennae, expression of a different set (as compared to (LmOBP11 RNAi + M. anisopliae infected)/(M. anisopliae infection)) of ORs were downregulated, whereas LmOR4, 40, 55, 85, and 96 showed increased expression. In addition, LmOBP8 and LmIR28 also showed decreased expression in the antennae. In the same comparison, only minor changes were noted for olfactory pathway components in the cuticle or fat body; however, LmOBP14 and LmIR25a, and to a lesser extent LmCSP2 all showed decreased expression (again comparing (LmOBP11 RNAi + M. anisopliae infection)/(M. anisopliae infection) samples). In addition, in the hemocytes, LmCSP3, I-1, and LmOR2 were all downregulated. In the final comparison between (LmOBP11 RNAi + M. anisopliae infection) and (LmOBP11 RNAi) treatments, the general downregulation of olfactory pathway components in the antennae was similar to M. anisopliae infected/control, although none of the upregulated genes in the latter were found in the former, in which instead, expression of LmOR95 and LmIR28 were elevated. In the cuticle and fat body tissues, expression of olfactory genes was similar between (LmOBP11 RNAi + M. anisopliae infection)/(LmOBP11 RNAi) and (GFP RNAi + M. anisopliae infected)/(GFP RNAi) samples, although in the case of the fat body, expression levels were not as pronounced as in the former comparison. Relative expression analyses in hemocytes were also found to be similar, with the exception of LmIR25a which showed opposite trends when comparing (LmOBP11 RNAi + M. anisopliae infection)/(LmOBP11 RNAi) to (GFP RNAi + M. anisopliae infected)/(GFP RNAi) samples.
In terms of identification of behavior-related GO terms: DEGs corresponding to synaptic target inhibition, presynaptic membrane, presynaptic active zone, dopamine biosynthesis process, detection of mechanical stimulus involved in sensory perception, and catecholamine metabolic processes, were found to be significantly enriched in the antennae of LmOBP11 dsRNA treated locusts as compared to the antennae of GFP control locusts (Supplemental Fig. S8). Similarly, DEGs corresponding to synaptic target inhibition, response to ethanol, post-mating behavior, multicellular organism reproduction, motor neuron axon guidance, and circadian rhythm were significantly enriched in the antennae of GFP RNAi + M. anisopliae infection treated locusts (no LmOBP11 dsRNA treatment) as compared to antennae of GFP control locusts (Supplemental Fig. S8). The set of significantly enriched overlapping DEGs found in the antennae of these two comparisons included 28 transcripts (Supplemental Table S3). Among them, 15 transcripts showed differential expression tendencies in locust antennal samples when comparing LmOBP11 dsRNA treatment to GFP dsRNA controls and M. anisopliae infected locusts as compared to control individuals. Most of these transcripts were involved in metabolism related functions, and included acyl-CoA synthetase family members, fructose-1,6-bisphosphatase, acetaldehyde dehydrogenase class 2, beta-galactosidase-like, and peroxidase, which are potentially implicated in the chemical perception and behavioral responses during fungal infection. An overview of a model derived from these results is given in Fig. 6.
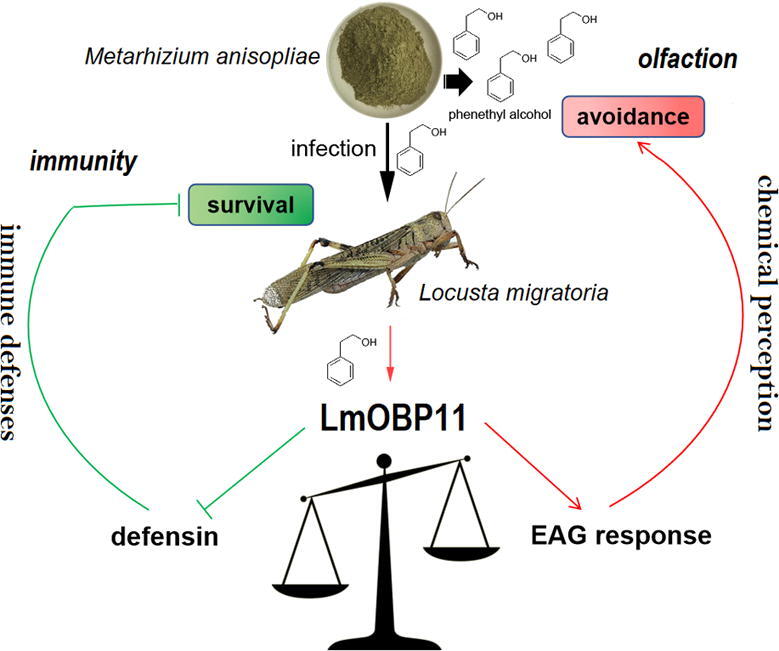
Fig. 6.
Model diagram of the trade-off between innate immunity and chemical perception of locust to M. anisopliae infection mediated by LmOBP11.
Knockdown of LmOBP11 affects gut microbiome dysbiosis caused by M. anisopliae infection
Changes in the gut microbiome have been reported as one aspect of the infection of host insects by entomopathogenic fungi (see the Discussion section). We therefore sought to examine the effect of LmOBP11 gene expression knockdown on the gut microbiome of M. anisopliae infected locusts. As expected, infection by M. anisopliae + GFP RNAi resulted in a ∼ 140-fold increase in M. robertsii (which show high homology with M. anisopliae) OTUs as compared to GFP RNAi locusts, however, co-treatment with LmOBP11 dsRNA decreased the overall M. robertsii fold increase to ∼38 (p < 0.01) (Supplemental Fig. S10 and Supplemental Table S4). As previously reported, infection by M. anisopliae leads to increased presence of opportunistic bacterial pathogens, including Pseudarcicella sp., and Gluconobacter sp., however co-treatment with LmOBP11 dsRNA suppressed this increase (p < 0.01) (Supplemental Fig. S11 and Supplemental Table S5). Conversely, OTUs corresponding to Thauera sp (implicated in aromatic compound degradation), were increased ∼194-fold in locusts cotreated with M. anisopliae + LmOBP11 dsRNA, but only ∼4.4-fold in M. anisopliae + GFP dsRNA, as compared to GFP control locusts (p < 0.01) (Supplemental Fig. S11 and Supplemental Table S5).
Discussion
Pathogen recognition is a central aspect of the innate immune response to eliminate invading microbes. Microbial sensing can occur at the level of chemoreception of exogenous cues (i.e., volatiles) via olfaction [45], [46], [47], [48], or detection of pathogen molecules once the invading microbe has gained access into the organism [3], [49]. With respect to the latter, although lacking antibody production and other aspects of adaptive immunity, insects and other arthropods have evolved sophisticated innate immune systems that can be considered as quasi-adaptive [2], [50]. These responses include induction of hemocyte proliferation, secretion of antimicrobial compounds, and activation of immune processes (e.g., prophenoloxidase, melanization) via pathogen recognition mechanisms (e.g., Toll, Imd) that target invading microbes [2], [3], [51], [52]. With respect to olfaction, M. anisopliae produces PEA that possesses repellent properties against termites [44], but no insect olfactory proteins have been shown to participate in pathogen avoidance, apart from the characterization of a mutation of the general odorant receptor co-receptor (orco1, required for the activity of most if not all ORs) in Drosophila that reduces grooming behavior in response to entomopathogenic fungi [53]. Changes in expression levels of olfactory proteins, including OBPs, have been reported in locusts through transcriptomics and antennal proteomics and in fire ants through gene expression analysis in response to infection with entomopathogenic fungi [32], [33]; however, the impact(s) and/or role(s) of these changes in mediating and/or affecting infection remains unknown. Here, we show that the expression of one OBP, LmOBP11, is elevated in locusts infected by the insect fungal pathogen, M. anisopliae [32]. Our initial hypothesis was that enhanced LmOBP11 expression is a host response which is potentially linked to detection, avoidance, and/or defense response aimed towards mitigating infection. In support of this, our data show that LmOBP11 can bind to the fungal produced volatile, PEA. Exposure to PEA results in the induction of LmOBP11 expression and wild type locusts show avoidance behavior towards PEA contaminated food. This avoidance behavior was diminished after LmOBP11 gene knockdown, further indicating the LmOBP11 acts as a mechanism by which pathogen volatiles can be detected, resulting in avoidance behaviors. This information suggests that LmOBP11 upregulation benefits the locust via enhanced sensitization to fungal volatiles. It is noted that the slight increase in fluorescence observed in chemical competitive binding assay has been reported previously [54], [55], [56], [57], [58]. The 6xHis tag (His-tag) might affect the competitive binding assay by interfering with the binding cavity. Nevertheless, the good binding ability of LmOBP11 to PEA and other chemicals indicates that the binding cavity is not affected. Some chemicals can form micelles by encapsulating the fluorescent probe or form complexes with proteins [58].
Odorant binding proteins represent a diverse protein family initially considered as carrier proteins shuttling odorants to odorant receptors in insect sensilla. More recently, their functions beyond olfaction have been recognized although significant aspects of their functioning remain obscure. Deletion of all the major antennal OBPs in Drosophila has been reported to result in little change in the magnitude of olfactory responses, although select OBPs appeared linked to behavioral responses involving odor aversion and ovipositing [59]. Some OBPs, even when abundant in antennal tissues, appear to be involved in responses to humidity (rather than an odorant per se) [60]. As OBP expression can be found in tissues beyond the olfactory system, their functions in processes ranging from wound healing to development have been reported [29]. Regarding cellular immunity, RNAi mediated knockdown of a Drosophila OBP (OBP28a) was upregulated during gut bacterial colonization and resulted in reduced cellular immunity via a lozenge transcription factor [61]. Indirectly, odorant receptors (ORs) have been hypothesized to participate in the regulation of cellular immunity by affecting the synthesis of gamma-aminobutyric acid (GABA) [62]. Regarding humoral immunity, sensory neuron membrane proteins (SNMPs) have been speculated to regulate Toll receptors [63]. RNAi mediated knockdown of a mosquito hormone-binding protein mPBP in Aedes aegypti resulted in the differential expression of antimicrobial peptides (AMPs) and affected hemocyte proliferation [64]. Changes in expression levels of olfactory component proteins, including OBPs, have been reported in locusts (transcriptomic and antennal proteome) and fire ants (gene expression) in response to infection by entomopathogenic fungi [32], [33], however, the impact(s) and/or role(s) of these changes in mediating and/or affecting infection remains unknown. Unexpectedly, knockdown of locust LmOBP11 expression during infection also resulted in increased resistance of the locust to the invading pathogen. The mechanism for this counter-intuitive result appears to be that the elevated levels of LmOBP11 dampen immune responses including signaling pathway components, pattern recognition molecules, and the production of Toll output antimicrobial peptides including diptericin and defensin. Immune dampening was further reflected by the increased gut microbiome dysbiosis that was noted in comparison between locusts treated with LmOBP11 dsRNA + M. anisopliae as compared to locusts infected by M. anisopliae alone.
Previous research has demonstrated the effect of entomopathogenic fungi on insect gut microbial load and revealed strategies that gut microbes have acquired to combat fungal pathogens [65]. For example, drastic changes have been found in the structure of the gut microbial community in the brown planthopper Nilaparvata lugens upon topical application with M. anisopliae, a process that was linked to the decreased gut immune response [66]. Similarly, microbial diversity in the gut of the termite Odontotermes formosanus was reduced upon challenge with the insect pathogenic fungus Metarhizium robertsii [67]. The current findings implicate OBPs as a potential component that bridges insect gut microbiota composition and host resistance to microbial pathogens. In particular, our findings imply that innate immune capacity in L. migratoria locusts can be modified not only by the level of expression of certain OBPs, but also by disrupting the gut microbiota homeostasis. Alternatively, the current results suggest a direct or indirect connection between OBP function and gut dysbiosis that in turn can affect immune signaling activity and host antifungal defense. Future efforts will concentrate on identifying the microbial species-specific modifications in the gut microbiome of locusts infected with different fungal pathogens and understanding whether the regulatory role of OBPs is restricted to locusts only or is also found in other insect pests or vectors of infectious disease.
With respect to the specific immune responses affected by LmOBP11, little to no changes were seen in either hemocyte numbers or PO activity in the locust hemolymph following LmOBP11 knockdown, suggesting that LmOBP11 is more involved in the negative regulation of humoral-mediated immunity pathways. The recognition of fungal pathogen-associated molecular pattern (PAMP) molecules (e.g., beta-glucan) relies on pathogen recognition receptors (PRRs), such as GNBPs and then allows for modulation of serine protease activity, ultimately resulting in pro-Spaetzle hydrolysis, activation of downstream regulators in the Toll pathway, and AMP production [3]. Our data show that Serine protease easter, encoding an extracellular serine protease that processes the pro-Spätzle protein to generate the Drosophila Toll ligand Spätzle [68], was significantly upregulated in locusts co-treated with LmOBP11 dsRNA and M. anisopliae infection. This finding provides indirect evidence that LmOBP11 is a negative regulator controling immune-protease related functions. Thus, during wild type locust infection, elevated levels of LmOBP11 may act to suppress the Toll-mediated immune cascade leading to AMP production. Decreased production of AMPs coupled to other potential aspects of immune suppression results in a decreased ability of the locust to mount a successful challenge to the fungus, thus allowing for proliferation of the fungus within the hemocoel, a key aspect for the fungus to complete its lifecycle.
As PEA levels in the hemolymph were shown to increase during M. anisopliae infection, this can potentially provide an in situ within host signal for increasing LmOBP11 expression, which in turn would suppress immune responses to benefit the pathogen. Thus, our data suggest a trade-off in LmOBP11 functioning, with the hypothesis that LmOBP11 acts as a sensor in antennae mediating avoidance, but functions as a negative regulator of immune activation in hemocytes and the fat body. Entomopathogenic fungi have evolved to evade host defenses and a number of mechanisms employed by these fungi have been reported [10], [69], [70]. These include the production of (i) host antimicrobial detoxifying enzymes, (ii) proteins that mask the fungal cell during growth within the insect hemocoel [71], (iii) secondary metabolites that target specific host processes including immunity [72], and (iv) manipulation of host hormone levels to facilitate infection [73]. In addition, entomopathogenic fungi have been shown to release siRNAs that can subsequently inhibit the expression of immune genes during the attachment and/or penetration stage [74]. PEA can be synthesized by the catalysis of alcohol dehydrogenase (ADH) in both fungi and bacteria [75], [76]. Consistent with the fungus actively subverting host processes via PEA production, it has been previously noted that knockdown of alcohol dehydrogenase (ADH) results in reduced Metarhizium virulence, and that this gene is highly expressed during the later stage of growth within the insect host and conidiation stage [77]. Our data show that although external PEA may act as an avoidance signal, internal production of PEA by the fungus, may be adaptive for the pathogen by acting on LmOBP11. From the pathogen perspective, M. anisopliae infection-mediated expression of LmOBP11 acts to subvert (repress) host regulation of immune responses to the fungal pathogen. For the insect, PEA-induced LmOBP11 expression, may lead to increased sensitization, which benefit the insects by reducing the chance of repeated contact with pathogen considering that exposure to low doses of fungi in the environment sometimes is not lethal. However, this consequence is counteracted due to the observation that increased LmOBP11 expression suppresses important immune responses. This trade-off effect of LmOBP11 in behavior sensation and innate immune regulation could lead to a selection pressure on insect populations to modulate increased sensitization behaviors toward pathogen-produced chemicals that can enhance survival with the antagonistic effects of compromised innate immunity.
Conclusion
Our study suggests that certain pathogens may have evolved mechanisms for exploiting host olfactory processes to facilitate and enable infection. By inducing the expression of a host protein that acts to suppress immune responses, the pathogen gains time and a critical threshold in order to successfully infect the host. The finding that elevated levels of olfactory proteins can overstimulate olfactory responses, dampen immunity, and disturb the gut microbiome, highlights the importance of both behavioral responses and the consequences of pathways that can then undermine immunity and other critical physiological processes. These data expand the functional role(s) of OBPs to the mediation of both detection and protection against microbial pathogens, the former via antennal chemoperception and the latter via modulation of insect immunity, providing a novel, previously undescribed, link between olfaction and innate immunity. This link represents a superb example that natural selection facilitates insects with stronger behavior and immunity to avoid exposure to virulence pathogens.
Author contributions
Wei Zhang: Conceptualization, Project administration, Investigation, Methodology, Data Curation, Formal analysis, Data interpretation, Funding acquisition, Resources, Software, Validation, Writing - original draft, Writing - review & editing. Mushan Xie: Investigation, Methodology, Formal Analysis, Visualization, Software, Data Analysis. Ioannis Eleftherianos: Methodology, Data interpretation, Writing - original draft, Writing - review & editing. Amr Mohamed: Methodology, Data interpretation, Writing - original draft, Writing - review & editing. Yueqing Cao: Methodology, Validation, Data Analysis, Writing - review & editing. Baoan Song: Methodology, Resources, Supervision, Validation, Data Analysis. Lian-Sheng Zang: Methodology, Resources, Supervision, Validation, Data Analysis. Chen Jia: Investigation, Methodology, Data Analysis. Jing Bian: Investigation, Methodology, Data Analysis. Nemat O. Keyhani: Conceptualization, Supervision, Resources, Funding acquisition, Data Curation, Methodology, Investigation, Validation, Writing - original draft, Writing - review & editing. Yuxian Xia: Conceptualization, Supervision, Resources, Funding acquisition, Data Curation, Methodology, Investigation, Validation, Writing - original draft, Writing - review & editing.
Compliance with ethics requirements
-
•
For studies on the insect species which has been used in our research works all the institutional and national guidelines for the care and use of laboratory animals were followed.
-
•
This article does not contain any studies with human subjects.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgments
We thank Prof. Zhengbo He (Chongqing Normal University, Chongqing, China) for the assistance with the insect EAG response analysis and Prof. David Stanley (USDA ARS, Missouri, USA) for his critical reading and editing of the manuscript.
Funding
This work was supported by funds from the National Natural Science Foundation of China (No. 32001961, 32160666), the Guizhou Province Science and Technology Support Project ([2002] General 239) and the Program of Introducing Talents to Chinese Universities (111 Program, D20023) to WZ, the University of Florida Institute of Food and Agricultural Sciences and USDA NIFA grant 2019-05150 to NOK, the Natural Science Foundation of China (No. 31540089, 32111530121) and National Key R&D Program of China (2017YFD0201208) to YX, and The Research Support Fund Program of the Faculty of Science, Cairo University to AM.
Biographies

Wei Zhang, PhD, is a Professor in the State Key Laboratory Breeding Base of Green Pesticide and Agricultural Bioengineering, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China. His research focuses on the innate, behavioral, and ecological immunity in insect hosts against entomopathogenic fungi and parasitic wasps as well as the interactions between insects and pesticides. He is an editor at different peer-reviewed international journals, including Frontiers in Physiology and Frontiers in Microbiology.

Mushan Xie is a PhD candidate at the Life Sciences College of Chongqing University, Shapingba District, Chongqing, China. She published two peer-reviewed papers in the fungal genetics.

Ioannis Eleftherianos, PhD, FRES. Dr. Ioannis Eleftherianos’ current research involves Drosophila immune responses to nematode parasites and their mutualistic bacteria as well as Drosophila antiviral immunity. His lab also investigates host innate immune signaling and function in relation to microbiome composition and diversity. He is currently a Professor of Biology in the Department of Biological Sciences and Institute for Biomedical Sciences at George Washington University in Washington DC (USA).

Amr Mohamed (PhD, FRES) is an Associate Professor of Insect Biochemistry and Molecular Sciences at Cairo University, Egypt and a Research Associate at the American Museum of Natural History, USA. His research focus on insect molecular science and immunology. Dr Mohamed is an amateur myrmecologist with expertise in insect systematics and invasion biology. He has been involved in international collaborations with entomologists from China, Czech, Ecuador, France, Germany, Italy, India, Japan, and USA. He is an editor at international journals like Front. Physiol., J. Insect Sci., Int. J. Trop. Insect Sci., and an editorial board member of J. Nat. Hist.

Yueqing Cao, PhD, is the Professor and doctoral mentor of the Life Sciences College of Chongqing University, Chongqing, China. Her research interests include the regulation mechanisms of fungal conidia production, conidial stress resistance, interactions between entomopathogenic fungi and both insect and plant hosts. She has published more than 40 papers in mycoinsecticides.

Baoan Song, PhD, is an Academician at the Chinese Academy of Engineering, full professor and president of Guizhou University, China. His primary areas of investigation include organic chemistry, stereochemistry, moiety, chemical synthesis and catalysis. He designed and synthesized a series of green pesticides with superior anti-viral, anti-bacterial or anti-fungal activities that have been mass produced with large-scale applications. He has published more than 200 academic papers, more than 36 patents. He obtained 3 second prizes and 1 third prize for National Prize for Progress in Science and Technology (China), in addition to 4 first prizes and 7 second prizes for provincial scientific awards (China).

Liansheng Zang, PhD, Lian-Sheng Zang, PhD, is a Professor of Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China. He focuses on studying on biological control with natural enemies, insect ecology, and IPM.

Chen Jia is a graduate student of the State Key Laboratory Breeding Base of Green Pesticide and Agricultural Bioengineering, Key Laboratory of Green Pesticide and Agricultural Bioengineering, Ministry of Education, Guizhou University, Guiyang, China.

Jing Bian is a PhD candidate of College of Environmental and Ecology, Chongqing University, China.

Nemat O. Keyhani, PhD, is a Professor in the Department of Microbiology and Cell Science at the University of Florida, Gainesville, FL, USA. His research involves fungal biology and genetics. Much of his career has been focused on the biological intricacies of fungal-insect interactions including entomopathogenic fungi and insect fungal mutualists. Recently, Keyhani’s research interests have broadened to include insect communication and chemical detection. Understanding how insect perceive and respond to chemicals in their environment and how they communicate to produce cooperative or self-organized behaviors is integral to insect behavior and can have practical application in identifying insect attractants and/or repellants.

Yuxian Xia, PhD, is a professor in College of Life Sciences and director of Genetic Engineering Research Center of Chongqing University, expert of Plant Protection Panel in the Ministry of Agriculture of China, and the recipient of special allowance of State Council of China. He has presided more than 10 national scientific research and industrialization projects in fungal insecticides, published over 120 papers in understanding insect and their fungal pathogens. He developed a fungal-based locusticide, a broad-spectrum fungal pesticide for rice pest insects and many other pests. For large scale application, he established mass production systems for mycoinsecticides.
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2022.08.013.
Contributor Information
Wei Zhang, Email: wzhang9@gzu.edu.cn.
Nemat O. Keyhani, Email: keyhani@ufl.edu.
Yuxian Xia, Email: yuxianxia@cqu.edu.cn.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
The sequencing files reported in this paper have been deposited as raw reads in the GenBank SRA database (accession number: SRR16556343, SRR16556342, SRR16556335, SRR16556334, SRR16556333, SRR16556332, SRR16556331, SRR16556330, SRR16556329, SRR16556328, SRR16556341, SRR16556340, SRR16556339, SRR16556338, SRR16556337 and SRR16556336).
References
- 1.Ortiz-Urquiza A., Keyhani N. Action on the surface: entomopathogenic fungi versus the insect cuticle. Insects. 2013;4(3):357–374. doi: 10.3390/insects4030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eleftherianos I., Heryanto C., Bassal T., Zhang W., Tettamanti G., Mohamed A. Haemocyte-mediated immunity in insects: cells, processes and associated components in the fight against pathogens and parasites. Immunology. 2021;164(3):401–432. doi: 10.1111/imm.13390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W., Tettamanti G., Bassal T., Heryanto C., Eleftherianos I., Mohamed A. Regulators and signalling in insect antimicrobial innate immunity: functional molecules and cellular pathways. Cell Signall. 2021;83 doi: 10.1016/j.cellsig.2021.110003. [DOI] [PubMed] [Google Scholar]
- 4.Thompson S.R., Brandenburg R.L. Tunneling responses of mole crickets (Orthoptera: Gryllotalpidae) to the entomopathogenic fungus, Beauveria bassiana. Environ Entomol. 2005;34(1):140–147. [Google Scholar]
- 5.Ormond E.L., Thomas A.P., Pell J.K., Freeman S.N., Roy H.E. Avoidance of a generalist entomopathogenic fungus by the ladybird, Coccinella septempunctata. FEMS Microbiol Ecol. 2011;77(2):229–237. doi: 10.1111/j.1574-6941.2011.01100.x. [DOI] [PubMed] [Google Scholar]
- 6.Scholte E.-J., Knols B.G., Takken W. A study on avoidance and repellency of the African malaria vector Anopheles gambiae upon exposure to the entomopathogenic fungus Metarhizium anisopliae. Proc Neth Entomol Soc Meet. 2005;16(4):131–138. [Google Scholar]
- 7.Staples J.A., Milner R.J. A laboratory evaluation of the repellency of Metarhizium anisopliae conidia to Coptotermes lacteus (Isoptera: Rhinotermitidae) Sociobiology. 2000;36(1):133–148. [Google Scholar]
- 8.Wang C., Leger R.J.S. The Metarhizium anisopliae perilipin homolog MPL1 regulates lipid metabolism, appressorial turgor pressure, and virulence. J Biol Chem. 2007;282(29):21110–21115. doi: 10.1074/jbc.M609592200. [DOI] [PubMed] [Google Scholar]
- 9.Fang W., Leng B., Xiao Y., Jin K., Ma J., Fan Y., et al. Cloning of Beauveria bassiana chitinase gene Bbchit1 and its application to improve fungal strain virulence. Appl Environ Microbiol. 2005;71(1):363–370. doi: 10.1128/AEM.71.1.363-370.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang C., Wang S. Insect pathogenic fungi: genomics, molecular interactions, and genetic improvements. Annu Rev Entomol. 2017;62:73–90. doi: 10.1146/annurev-ento-031616-035509. [DOI] [PubMed] [Google Scholar]
- 11.Feng P., Shang Y., Cen K., Wang C. Fungal biosynthesis of the bibenzoquinone oosporein to evade insect immunity. Proc Natl Acad Sci U S A. 2015;112(36):11365–11370. doi: 10.1073/pnas.1503200112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hussain A., Tian M.Y., He Y.R., Bland J.M., Gu W.X. Behavioral and electrophysiological responses of Coptotermes formosanus Shiraki towards entomopathogenic fungal volatiles. Biol Control. 2010;55(3):166–173. [Google Scholar]
- 13.Lozano-Soria A., Picciotti U., Lopez-Moya F., Lopez-Cepero J., Porcelli F., Lopez-Llorca L.V. Volatile organic compounds from entomopathogenic and nematophagous fungi, repel banana black weevil (Cosmopolites sordidus) Insects. 2020;11(8):509. doi: 10.3390/insects11080509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boucias D.G., Lietze V.U., Teal P. In: Biocommunication of fungi. Witzany G., editor. Springer; Dordrecht: 2012. Chemical signals that mediate insect-fungal interactions; pp. 305–336. [Google Scholar]
- 15.Wang L.X., Ren L.L., Liu X.B., Shi J., Wang J.Z., Luo Y.Q. Effects of endophytic fungi in Mongolian pine on the selection behavior of woodwasp (Sirex noctilio) and the growth of its fungal symbiont. Pest Manage Sci. 2019;75(2):492–505. doi: 10.1002/ps.5146. [DOI] [PubMed] [Google Scholar]
- 16.Inamdar A.A., Masurekar P., Bennett J.W. Neurotoxicity of fungal volatile organic compounds in Drosophila melanogaster. Toxicol Sci. 2010;117(2):418–426. doi: 10.1093/toxsci/kfq222. [DOI] [PubMed] [Google Scholar]
- 17.Mackled M.I., EL-Hefny M., Bin-Jumah M., Wahba T.F., Allam A.A. Assessment of the toxicity of natural oils from Mentha piperita, Pinus roxburghii, and Rosa spp. against three stored product insects. Processes. 2019;7(11):861. [Google Scholar]
- 18.Kecskeméti S., Szelényi M.O., Erdei A.L., Geösel A., Fail J., Molnár B.P. Fungal volatiles as olfactory cues for female fungus gnat, Lycoriella ingenua in the avoidance of mycelia colonized compost. J Chem Ecol. 2020;46(10):917–926. doi: 10.1007/s10886-020-01210-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu L., Zhao X.Y., Tang Q.B., Lei C.L., Huang Q.Y. The mechanisms of social immunity against fungal infections in eusocial insects. Toxins. 2019;11(5):244. doi: 10.3390/toxins11050244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelosi P., Zhou J.J., Ban L.P., Calvello M. Soluble proteins in insect chemical communication. Cell Mol Life Sci. 2006;63(14):1658–1676. doi: 10.1007/s00018-005-5607-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jia X.J., Wang H.X., Yan Z.G., Zhang M.Z., Wei C.H., Qin X.C., et al. Antennal transcriptome and differential expression of olfactory genes in the yellow peach moth, Conogethes punctiferalis (Lepidoptera: Crambidae) Sci Rep. 2016;6(1):29067. doi: 10.1038/srep29067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.D'Onofrio C., Zaremska V., Zhu J., Knoll W., Pelosi P. Ligand-binding assays with OBPs and CSPs. Methods Enzymol. 2020;642:229–258. doi: 10.1016/bs.mie.2020.05.006. [DOI] [PubMed] [Google Scholar]
- 23.Pevsner J., Hou V., Snowman A.M., Snyder S.H. Odorant-binding protein. Characterization of ligand binding. J Biol Chem. 1990;265(11):6118–6125. [PubMed] [Google Scholar]
- 24.Campanacci V., Lartigue A., Hällberg B.M., Jones T.A., Giudici-Orticoni M.T., Tegoni M., et al. Moth chemosensory protein exhibits drastic conformational changes and cooperativity on ligand binding. Proc Natl Acad Sci U S A. 2003;100(9):5069–5074. doi: 10.1073/pnas.0836654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scieuzo C., Nardiello M., Farina D., Scala A., Cammack J.A., Tomberlin J.K., et al. Hermetia illucens (L.) (Diptera: Stratiomyidae) odorant binding proteins and their interactions with selected volatile organic compounds: an in silico approach. Insects. 2021;12(9):814. doi: 10.3390/insects12090814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nardiello M., Scieuzo C., Salvia R., Farina D., Franco A., Cammack J.A., et al. Odorant binding proteins from Hermetia illucens: potential sensing elements for detecting volatile aldehydes involved in early stages of organic decomposition. Nanotechnology. 2022;33(20) doi: 10.1088/1361-6528/ac51ab. [DOI] [PubMed] [Google Scholar]
- 27.Zhu J., Iovinella I., Dani F.R., Liu Y.L., Huang L.Q., Liu Y., et al. Conserved chemosensory proteins in the proboscis and eyes of Lepidoptera. Int J Biol Sci. 2016;12(11):1394–1404. doi: 10.7150/ijbs.16517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh S., Tyagi C., Rather I.A., Sabir J.S., Hassan M., Singh A., et al. Molecular modeling of chemosensory protein 3 from Spodoptera litura and its binding property with plant defensive metabolites. Int J Mol Sci. 2020;21(11):4073. doi: 10.3390/ijms21114073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Calvo E., Mans B.J., Ribeiro J.M., Andersen J.F. Multifunctionality and mechanism of ligand binding in a mosquito antiinflammatory protein. Proc Natl Acad Sci U S A. 2009;106(10):3728–3733. doi: 10.1073/pnas.0813190106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas T., De T.D., Sharma P., Lata S., Saraswat P., Pandey K.C., et al. Hemocytome: deep sequencing analysis of mosquito blood cells in Indian malarial vector Anopheles stephensi. Gene. 2016;585(2):177–190. doi: 10.1016/j.gene.2016.02.031. [DOI] [PubMed] [Google Scholar]
- 31.Zhang W., Chen J., Keyhani N.O., Jin K., Wei Q., Xia Y. Central nervous system responses of the Oriental migratory, Locusta migratoria manilensis, to fungal Infection. Sci Rep. 2017;7(1):10340. doi: 10.1038/s41598-017-10622-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang W., Zheng X., Chen J., Keyhani N.O., Cai K., Xia Y. Spatial and temporal transcriptomic analyses reveal locust initiation of immune responses to Metarhizium acridum at the pre-penetration stage. Dev Comp Immunol. 2020;104 doi: 10.1016/j.dci.2019.103524. [DOI] [PubMed] [Google Scholar]
- 33.Zheng R., Xia Y., Keyhani N.O. Differential responses of the antennal proteome of male and female migratory locusts to infection by a fungal pathogen. J Proteomics. 2021;232 doi: 10.1016/j.jprot.2020.104050. [DOI] [PubMed] [Google Scholar]
- 34.Guo W., Wang X., Ma Z., Xue L., Han J., Yu D., et al. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLoS Genet. 2011;7(2) doi: 10.1371/journal.pgen.1001291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M., et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55(4):611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 36.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 37.Guo X., Ma Z., Kang L. Two dopamine receptors play different roles in phase change of the migratory locust. Front Behav Neurosci. 2015;9:80. doi: 10.3389/fnbeh.2015.00080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu R., Wu Z., Wang X., Yang P., Yu D., Zhao C., et al. Metabolomic analysis reveals that carnitines are key regulatory metabolites in phase transition of the locusts. Proc Natl Acad Sci U S A. 2012;109(9):3259–3263. doi: 10.1073/pnas.1119155109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bojke A., Tkaczuk C., Bauer M., Kamysz W., Gołębiowski M. Application of HS-SPME-GC-MS for the analysis of aldehydes produced by different insect species and their antifungal activity. J Microbiol Meth. 2020;169 doi: 10.1016/j.mimet.2020.105835. [DOI] [PubMed] [Google Scholar]
- 40.Calvello M., Guerra N., Brandazza A., D'ambrosio C., Scaloni A., Dani F.R., et al. Soluble proteins of chemical communication in the social wasp Polistes dominulus. Cell Mol Life Sci. 2003;60(9):1933–1943. doi: 10.1007/s00018-003-3186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gillespie J.P., Burnett C., Charnley A.K. The immune response of the desert locust Schistocerca gregaria during mycosis of the entomopathogenic fungus, Metarhizium anisopliae var acridum. J Insect Physiol. 2000;46(4):429–437. doi: 10.1016/s0022-1910(99)00128-6. [DOI] [PubMed] [Google Scholar]
- 42.Benjamini Y., Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann Stat. 2001;29(4):1165–1188. [Google Scholar]
- 43.Zhou J.J., Zhang G.A., Huang W., Birkett M.A., Field L.M., Pickett J.A., et al. Revisiting the odorant-binding protein LUSH of Drosophila melanogaster: evidence for odour recognition and discrimination. FEBS Lett. 2004;558(1–3):23–26. doi: 10.1016/S0014-5793(03)01521-7. [DOI] [PubMed] [Google Scholar]
- 44.Mburu D.M., Ndung'u M.W., Maniania N.K., Hassanali A. Comparison of volatile blends and gene sequences of two isolates of Metarhizium anisopliae of different virulence and repellency toward the termite Macrotermes michaelseni. J Exp Biol. 2011;214(Pt 6):956–962. doi: 10.1242/jeb.050419. [DOI] [PubMed] [Google Scholar]
- 45.Stensmyr M.C., Dweck H.K., Farhan A., Ibba I., Strutz A., Mukunda L., et al. A conserved dedicated olfactory circuit for detecting harmful microbes in Drosophila. Cell. 2012;151(6):1345–1357. doi: 10.1016/j.cell.2012.09.046. [DOI] [PubMed] [Google Scholar]
- 46.Cho B., Spratford C.M., Yoon S., Cha N., Banerjee U., Shim J. Systemic control of immune cell development by integrated carbon dioxide and hypoxia chemosensation in Drosophila. Nat Commun. 2018;9:2679. doi: 10.1038/s41467-018-04990-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Moisan K., Cordovez V., van de Zande E.M., Raaijmakers J.M., Dicke M., Lucas-Barbosa D. Volatiles of pathogenic and non-pathogenic soil-borne fungi affect plant development and resistance to insects. Oecologia. 2019;190(3):589–604. doi: 10.1007/s00442-019-04433-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Montanari M., Royet J. Impact of microorganisms and parasites on neuronally controlled drosophila behaviours. Cells. 2021;10(9):2350. doi: 10.3390/cells10092350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liegeois S., Ferrandon D. Sensing microbial infections in the Drosophila melanogaster genetic model organism. Immunogenetics. 2022;74(1):35–62. doi: 10.1007/s00251-021-01239-0. [DOI] [PubMed] [Google Scholar]
- 50.Schmid-Hempel P. Function and mechanisms in defence strategies. Curr Opin Insect Sci. 2022;49:31–36. doi: 10.1016/j.cois.2021.10.005. [DOI] [PubMed] [Google Scholar]
- 51.Manniello M.D., Moretta A., Salvia R., Scieuzo C., Lucchetti D., Vogel H., et al. Insect antimicrobial peptides: potential weapons to counteract the antibiotic resistance. Cell Mol Life Sci. 2021;78(9):4259–4282. doi: 10.1007/s00018-021-03784-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wei G., Lai Y., Wang G., Chen H., Li F., Wang S. Insect pathogenic fungus interacts with the gut microbiota to accelerate mosquito mortality. Proc Natl Acad Sci U S A. 2017;114(23):5994–5999. doi: 10.1073/pnas.1703546114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yanagawa A., Chabaud M.A., Imai T., Marion-Poll F. Olfactory cues play a significant role in removing fungus from the body surface of Drosophila melanogaster. J Invertebr Pathol. 2018;151:144–150. doi: 10.1016/j.jip.2017.11.011. [DOI] [PubMed] [Google Scholar]
- 54.Zhang X.Q., Yan Q., Li L.L., Xu J.W., Mang D., Wang X.L., et al. Different binding properties of two general-odorant binding proteins in Athetis lepigone with sex pheromones, host plant volatiles and insecticides. Pestic Biochem Physiol. 2020;164:173–182. doi: 10.1016/j.pestbp.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 55.Wang B., Dong W., Li H., D’Onofrio C., Bai P., Chen R., et al. Molecular basis of (E)-β-farnesene-mediated aphid location in the predator Eupeodes corollae. Curr Biol. 2022;32(5):951–962. doi: 10.1016/j.cub.2021.12.054. [DOI] [PubMed] [Google Scholar]
- 56.Sun Y.F., De Biasio F., Qiao H.L., Iovinella I., Yang S.X., Ling Y., et al. Two odorant-binding proteins mediate the behavioural response of aphids to the alarm pheromone (E)-ß-farnesene and structural analogues. PLoS ONE. 2012;7(3) doi: 10.1371/journal.pone.0032759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pelosi P., Zhu J., Knoll W. From radioactive ligands to biosensors: binding methods with olfactory proteins. Appl Microbiol Biot. 2018;102(19):8213–8227. doi: 10.1007/s00253-018-9253-5. [DOI] [PubMed] [Google Scholar]
- 58.Majorek K.A., Kuhn M.L., Chruszcz M., Chruszcz M., Anderson W.F., Minor W. Double trouble-buffer selection and His-tag presence may be responsible for nonreproducibility of biomedical experiments. Protein Sci. 2014;23(10):1359–1368. doi: 10.1002/pro.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Larter N.K., Sun J.S., Carlson J.R. Organization and function of Drosophila odorant binding proteins. 2016;eLife. 5 doi: 10.7554/eLife.20242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sun J.S., Larter N.K., Chahda J.S., Rioux D., Gumaste A., Carlson J.R. Humidity response depends on the small soluble protein Obp59a in Drosophila. eLife. 2018;7 doi: 10.7554/eLife.39249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Benoit J.B., Vigneron A., Broderick N.A., Wu Y., Sun J.S., Carlson J.R., et al. Symbiont-induced odorant binding proteins mediate insect host hematopoiesis. 2017;eLife 6 doi: 10.7554/eLife.19535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shim J., Mukherjee T., Mondal B.C., Liu T., Young G.C., Wijewarnasuriya D.P., et al. Olfactory control of blood progenitor maintenance. Cell. 2013;155(5):1141–1153. doi: 10.1016/j.cell.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Benton R., Vannice K.S., Vosshall L.B. An essential role for a CD36-related receptor in pheromone detection in Drosophila. Nature. 2007;450(7167):289. doi: 10.1038/nature06328. [DOI] [PubMed] [Google Scholar]
- 64.Kim I.H., Castillo J.C., Aryan A., Martin-Martin I., Nouzova M., Noriega F.G., et al. A mosquito juvenile hormone binding protein (mJHBP) regulates the activation of innate immune defenses and hemocyte development. PLoS Pathog. 2020;16(1) doi: 10.1371/journal.ppat.1008288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Boucias D.G., Zhou Y., Huang S., Keyhani N.O. Microbiota in insect fungal pathology. Appl Microbiol Biotechnol. 2018;102(14):5873–5888. doi: 10.1007/s00253-018-9089-z. [DOI] [PubMed] [Google Scholar]
- 66.Wang Z., Cheng Y., Wang Y., Yu X. Topical fungal infection induces shifts in the gut microbiota structure of brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae) Insects. 2022;13(6):528. doi: 10.3390/insects13060528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wu C.Y., Meng J., Merchant A., Zhang Y.X., Li M.W., Zhou X.G., et al. Microbial response to fungal infection in a fungus-growing termite, Odontotermes formosanus (Shiraki) Front Microbiol. 2021;12 doi: 10.3389/fmicb.2021.723508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Jang I.H., Chosa N., Kim S.H., Nam H.J., Lemaitre B., Ochiai M., et al. A Spätzle-processing enzyme required for toll signaling activation in Drosophila innate immunity. Dev Cell. 2006;10(1):45–55. doi: 10.1016/j.devcel.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 69.Butt T.M., Coates C.J., Dubovskiy I.M., Ratcliffe N.A. Entomopathogenic fungi: new insights into host-pathogen interactions. Adv Genet. 2016;94:307–364. doi: 10.1016/bs.adgen.2016.01.006. [DOI] [PubMed] [Google Scholar]
- 70.Qu S., Wang S. Interaction of entomopathogenic fungi with the host immune system. Dev Comp Immunol. 2018;83:96–103. doi: 10.1016/j.dci.2018.01.010. [DOI] [PubMed] [Google Scholar]
- 71.Wang C., St Leger R.J. A collagenous protective coat enables Metarhizium anisopliae to evade insect immune responses. Proc Natl Acad Sci U S A. 2006;103(17):6647–6652. doi: 10.1073/pnas.0601951103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang B., Kang Q., Lu Y., Bai L., Wang C. Unveiling the biosynthetic puzzle of destruxins in Metarhizium species. Proc Natl Acad Sci U S A. 2012;109(4):1287–1292. doi: 10.1073/pnas.1115983109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pedrini N., Ortiz-Urquiza A., Huarte-Bonnet C., Fan Y., Juárez M.P., Keyhani N.O. Tenebrionid secretions and a fungal benzoquinone oxidoreductase form competing components of an arms race between a host and pathogen. Proc Natl Acad Sci U S A. 2015;112(28):E3651–E3660. doi: 10.1073/pnas.1504552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cui C., Wang Y., Liu J., Zhao J., Sun P., Wang S. A fungal pathogen deploys a small silencing RNA that attenuates mosquito immunity and facilitates infection. Nat Commun. 2019;10(1):4298. doi: 10.1038/s41467-019-12323-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Homola P., Kurák T., Illeová V., Polakovič M. Cultivation of Pichia capsulata as a whole-cell biocatalyst with NADH-dependent alcohol dehydrogenase activity for R-1-phenylethanol production. Food Bioprod Process. 2015;96:126–132. [Google Scholar]
- 76.Hummel W. Reduction of acetophenone to R(+)-phenylethanol by a new alcohol dehydrogenase from Lactobacillus kefir. Appl Microbiol Biotechnol. 1990;34(1):15–19. [Google Scholar]
- 77.Callejas-Negrete O.A., Torres-Guzmán J.C., Padilla-Guerrero I.E., Esquivel-Naranjo U., Padilla-Ballesteros M.F., García-Tapia A., et al. The Adh1 gene of the fungus Metarhizium anisopliae is expressed during insect colonization and required for full virulence. Microbiol Res. 2015;172:57–67. doi: 10.1016/j.micres.2014.11.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing files reported in this paper have been deposited as raw reads in the GenBank SRA database (accession number: SRR16556343, SRR16556342, SRR16556335, SRR16556334, SRR16556333, SRR16556332, SRR16556331, SRR16556330, SRR16556329, SRR16556328, SRR16556341, SRR16556340, SRR16556339, SRR16556338, SRR16556337 and SRR16556336).