Graphical abstract
Keywords: Pneumonia identification, Chest X-ray imaging, Transfer ensemble learning, Transformer encoder (TE), Self-attention network, Explainable artificial intelligence (XAI)
Highlights
-
•
A new hybrid explainable AI pneumonia identification framework is proposed based on the ensemble Transformer Strategy.
-
•
The multi-head attention mechanism is used to address the relationships of the distant pixels among the chest X-ray images.
-
•
A comprehensive and robust binary and multi-class pneumonia classification study is conducted using two Chest X-ray datasets.
-
•
The XAI framework improves the performance by 2.05% (binary) and 1.3% (multiclass) against the individual ensemble scenarios.
Abstract
Introduction
Pneumonia is a microorganism infection that causes chronic inflammation of the human lung cells. Chest X-ray imaging is the most well-known screening approach used for detecting pneumonia in the early stages. While chest-Xray images are mostly blurry with low illumination, a strong feature extraction approach is required for promising identification performance.
Objectives
A new hybrid explainable deep learning framework is proposed for accurate pneumonia disease identification using chest X-ray images.
Methods
The proposed hybrid workflow is developed by fusing the capabilities of both ensemble convolutional networks and the Transformer Encoder mechanism. The ensemble learning backbone is used to extract strong features from the raw input X-ray images in two different scenarios: ensemble A (i.e., DenseNet201, VGG16, and GoogleNet) and ensemble B (i.e., DenseNet201, InceptionResNetV2, and Xception). Whereas, the Transformer Encoder is built based on the self-attention mechanism with multilayer perceptron (MLP) for accurate disease identification. The visual explainable saliency maps are derived to emphasize the crucial predicted regions on the input X-ray images. The end-to-end training process of the proposed deep learning models over all scenarios is performed for binary and multi-class classification scenarios.
Results
The proposed hybrid deep learning model recorded 99.21% classification performance in terms of overall accuracy and F1-score for the binary classification task, while it achieved 98.19% accuracy and 97.29% F1-score for multi-classification task. For the ensemble binary identification scenario, ensemble A recorded 97.22% accuracy and 97.14% F1-score, while ensemble B achieved 96.44% for both accuracy and F1-score. For the ensemble multiclass identification scenario, ensemble A recorded 97.2% accuracy and 95.8% F1-score, while ensemble B recorded 96.4% accuracy and 94.9% F1-score.
Conclusion
The proposed hybrid deep learning framework could provide promising and encouraging explainable identification performance comparing with the individual, ensemble models, or even the latest AI models in the literature. The code is available here: https://github.com/chiagoziemchima/Pneumonia_Identificaton.
Introduction
The human lung is seen as one of the vital organs of the human body; hence lung diseases are hazardous for humans. Pneumonia is a lung disease that is caused by an acute respiratory infection. There are several methods for classifying Pneumonia; Infectious and non-infectious. Pneumonia is classified as infectious according to the etiologic agents such as bacteria, viruses, mycoplasmas, chlamydial pneumonia and so on. Non-infectious pneumonia is seen as the body's immune caused by chemical, physical, or radiation pneumonia. Pneumonia is further classified as either ventilator-associated Pneumonia (VAP), community-acquired pneumonia (CAP), or hospital-acquired pneumonia (HAP). Because of the broad spectrum of diseases, HAP is more resistant to medicines and easier to proliferate, making the treatment difficult [1], [2]. Pneumonia can be caused by various causes including age, malnutrition, alcohol consumption, and smoking. Pneumonia may affect people of all ages although it is more dangerous in two-year-old and younger newborns as well as persons aged 65 and older due to their weakened immune systems (https://www.nhlbi.nih.gov/health/pneumonia accessed on 16 May 2022). In Western nations, pneumonia is the primary cause of infectious disease-related death. Pneumonia is a condition that may be controlled if detected and treated early. Pneumonia kills about 800,000 children under the age of five every year with over 2,200 dying every day. Pneumonia affects almost 1,400 children per 100,000 children [3]. However, because clinical, biochemical, and imaging symptoms are not always specific, diagnosing Pneumonia in an emergency context can be difficult.
X-ray scans of the human body have long been used to detect sensitive regions such as the head, teeth, chest and bones. For numerous years, health experts have used this technology to analyze and see fractures or irregularities in the bodily organ pattern. X-rays are effective diagnostic instruments for detecting neurotic changes despite their quasi features and cost. Pneumonia is diagnosed using X-ray images of the chest. Even for professional radiologists, diagnosing Pneumonia from chest X-ray images is challenging. Pneumonia's appearance on X-ray images is often ambiguous and it can be mistaken for other diseases and act as symptoms of other typical disorders. These discrepancies have resulted in significant subjective judgments and differences in pneumonia diagnosis among radiologists [4]. To assist radiologists in diagnosing Pneumonia from chest X-ray images, computerized assistance technologies are required. Artificial intelligence has proven to be more successful in vision tasks, especially its sub-field; Machine learning [5] and deep learning [6], [7] which have been utilized in a variety of disciplines in recent times to aid specialists in the early diagnosis of diseases such as lung disease prediction, diabetic retinopathy, sleep disorders [8], [9], stress [10], [11], anxiety [12], and brain tumor detection among others. These models are based on X-ray images and detect pneumonia disease thanks to their success. Although identifying pneumonia from chest X-ray images remains a challenging task, there is a dire need to develop highly accurate and efficient automated diagnostic methods for pneumonia identification to facilitate early-stage detection, thereby reducing the mortality rate yearly.
Deep learning has risen to prominence being one of the most promising technologies in previous research due to its ability to handle massive quantities of data [1], [13]. The most widely recognized Deep learning approach is the Convolutional Neural Network (CNN), which has made an outstanding performance in image processing, voice recognition and pattern recognition. Their operation is an end-to-end method where they make predictions from the extracted useful and relevant features of the input images. CNN approaches are preferred over the conventional approach due to their automatic feature extraction from the input image, thus performing much better, making it more popular among researchers for image classification. According to the findings of the prior studies, utilizing deep learning algorithms to identify Pneumonia on chest X-rays can relieve the load on radiologists; however, because numerous researchers use different deep learning approaches, it's uncertain which model is superior. Secondly, most current deep learning algorithms for pneumonia identification rely on a single CNN model and the applicability of the ensemble learning method in this classification problem has yet to be investigated. Finally, most studies focus on binary categorizing pneumonia vs normal with just a few capturing multi-class classifications due to the scarcity of biological data. On the other hand, the CNN model only analyzes the correlation between spatially neighboring pixels in the receptive area determined by the filter size [14]. As a result, it is difficult to identify relationships with distant pixels. Recent attempts have been made to use attention mechanisms to tackle this difficulty. Attention is a method of locating and concentrating on the most informative portion of the data.
As earlier stated, Pneumonia impacts a large number of people, adolescents and children, notably in rural and undeveloped nations defined by adverse outcomes such as overpopulation, poor sanitary conditions, hunger as well as a lack of sufficient medical services. Pneumonia must be diagnosed as soon as possible in order to be cured. The most prevalent method of diagnosis is the evaluation of X-ray images. However, it is dependent on the interpretive abilities of the physician and is usually not accepted by other physicians. To detect the condition, a timely and accurate model with generalizing capacity is necessary. Since Traditional approaches suffer from the extraction of relevant input image features, current researchers prefer the use of deep learning models (CNN) that extract relevant and informative features of the input data automatically to perform much better. However, the CNN model only analyzes the correlation between spatially neighboring pixels in the receptive area determined by the filter size. As a result, it is difficult to identify relationships with distant pixels. Recent attempts have been made to use attention mechanisms to tackle such difficulty. Also, for result generalization, most researchers applied CNN focused on single or individual models, thus ensemble model approaches have not been well investigated. On the other hand, image enhancement techniques (e.g., adaptive histogram equalization, contrast stretching, denoising, etc.) have been employed by several researchers leading a good classification performance of the deep learning models. Using the enhancement techniques, the performance could be slightly improved compared to the raw image performance [14]. Furthermore, few authors have employed the attention mechanism which has proven to have more balanced and accurate image feature extraction techniques than the convention deep learning models for lung disease identification from chest X-ray images, however, no focus has been given to the recent multi-head self-attention which have proven to be more promising compared to single head attention mechanism in computer vision tasks.
Having taken note of the drawbacks, this paper proposed an end-to-end binary and multiclass deep learning framework for pneumonia identification from chest X-ray images using Transformer Encoder: Self-Attention Network and Multilayer Perceptron. The proposed framework could directly handle the input raw images without any prior enhancement on the chest X-ray images as recently done by several researchers for performance gain to mitigate these limitations [15]. First, this study is established to tackle the issue of feature extraction from the input raw images by using the latest transformer encoder techniques against the conventional CNN models. Indeed, the Transformer is a self-attention-based architecture that emerged as the preferred paradigm in today’s visual challenges [15], [16], [17]. The adoption of transformer architecture enabled substantial parallelization and translation quality optimization. On the other hand, the CNN-based model worked on a fixed-sized window and struggled to capture connections at low resolution in both spatial and temporal domains [15]. Furthermore, because the filter weights in CNN’s stay constant after training, the process cannot adjust interactively to changes in input. Objects are processed as sequences in Vision Transformers (VisTransf) and class labels are inferred, enabling algorithms to understand image hierarchy autonomously [14]. Second, this study overlooked the strategy of employing image enhancement techniques on the chest X-ray images as done by other researchers for visual improvement as our proposed model process the input images as a series of patches, with each patch squashed into a single feature vector by combining the layers of all pixels in a patch and then exponentially expanding it to the appropriate input dimension by so doing, attend to the features patch by patch yielding better performance. This paper first examined the performance of six pre-trained deep learning models (i.e. EfficientNetB7, DensetNet201, VGG16, InceptionResNetV2, Xception, and GoogleNet network) via a transfer learning approach on the chest X-ray images. The ensemble learning of the pre-trained models served as feature extractors to the transformer encoder network in two scenarios: ensemble A (i.e., DenseNet201, VGG16, and GoogleNet) and ensemble B (i.e., DenseNet201, InceptionResNetV2, and Xception). We propose an automated technology that can distinguish between chest diseases like pneumonia and healthy people to aid medical diagnosis even when professional radiologists are unavailable. Furthermore, the proposed method was compared to other baseline models and recently published researches to provide a point of comparison for our findings. The contributions of this paper are summarized as follows;
-
•
Explainability-driven, medically explainable visuals that emphasize the crucial regions relevant to the model's prediction of the input image are proposed based on the Transformer Encoder and ensemble learning for detecting the pneumonia disease using chest X-ray images in early stages.
-
•
Ensemble deep learning-based feature extraction framework that is significantly discriminative in identifying pneumonia and COVID-19, as a backbone of the proposed hybrid framework is investigated in two binary and multiclass classification scenarios.
-
•
A comprehensive and robust binary and multi-class pneumonia classification study is conducted using two different public chest X-ray datasets: Mendeley data [12] and Chest X-ray [18].
-
•
Investigating the classification performance of the several pre-trained deep learning models in individual and ensemble forms.
-
•
Based on the detailed comprehensive experimental evaluation of the proposed deep learning scenarios, a robust deep learning framework compared with the state-of-the-art techniques is recommended for the early stage of pneumonia disease.
-
•
Lastly, the proposed model shows its ability to improve the ensemble model identification accuracy, thereby serving as a robust performance enhancement framework of ensemble models for disease detection using chest X-ray images.
The remainder of this paper is outlined as follows; Section “Introduction” focuses on the introductory part and the related works of this research. Section “Related work” describes the related works in detail while Section “Materials and methods” describes the materials, the proposed method, and evaluation metrics. Section “Results and discussion” introduces the experimental setup and results, the proposed method's discussion, application, and limitation. Section “Conclusion” presents the conclusion and future works.
Related work
Deep learning models
Chest X-ray Pneumonia detection has been an identified problem by researchers for a while [19], [20]. Researchers in addressing this issue have identified several approaches. The well-established approach is the conventional approach. However, this approach has problems that make it not acceptable in the medical field. Such problems include the scarcity of publicly available training datasets, the extraction of pneumonia features etc., which are time-wasting and inaccurate, resulting in false-positive results. Recent advancements in the research field have seen applying the deep learning approach in disease identification and predicting Pneumonia inclusive. Deep learning is a computational intelligence-based machine learning algorithm that simulates deep visual representations in information stored through various input and output layers with advanced systems or other methods [21]. Deep learning has risen to prominence being one of the most promising technologies in previous research due to its ability to handle massive quantities of data [22]. The most widely recognized Deep learning approach is the Convolutional Neural Network (CNN), which has made an outstanding performance in image processing, voice recognition and pattern recognition. Their operation (Classification) [23] is an end-to-end method where they make predictions from the extracted useful and relevant features of the input images. CNN approaches are preferred over the conventional approach due to their automatic feature extraction from the input image, thus performing much better, making it more popular among researchers for image classification.
Despite the outstanding performance of CNN models, the problem of overfitting and spatial information loss induced by normal convolution operation leading to poor result generalization persists. To tackle this issue of the CNN models, Liang et al. [24] proposed residual blocks and dilated convolution layers in their network backbone achieving a recall rate of 96.7 % and F1-Score of 92.7 %. On the other hand, the works of [25], [26] claimed that the Transfer learning (the use of features learned from a substantial trained dataset) accelerated the model training time and overcame the overfitting. They used the Kaggle competition dataset to train their proposed model and achieve their experiments. The dataset is divided into three groups: training, validation, and testing. The transfer learning approach is widely accepted by researchers in solving training dataset limitations, especially in the medical field classification task. This uses previous knowledge gained from a large trained dataset for a new task of limited training data samples via finetuning method. The research work presented in [25], [26], [27], [28], [29] employed the Transformer technique for pneumonia disease classification. In [29], Rahman et al. proposed transfer deep learning model for pneumonia detection using chest X-ray images. They used the pre-trained deep learning models of AlexNet, ResNet18, DenseNet201, and SqueezeNet to classify the chest images into bacterial, viral, and normal classes using 5,247 images. They designed their experiments with three classification schemes normal vs pneumonia, bacterial vs viral pneumonia, and combined schema with three classes of normal, bacterial, and viral pneumonia. They achieved the overall classification accuracy of 98 %, 95 %, and 93.3 % over their three schemes, respectively. Similarly, Ayan et al. [4] fine-tuned two conventional CNN architectures (i.e., VGG16 and Xception) via Transfer Learning for pneumonia disease classification. The authors of [30] pointed out a different approach to tackle the issue of training dataset samples. They suggested using the data augmentation method to increase the training sample. They use the data augmentation functions such as random rotation and random horizontal and vertical translation to enhance the representation ability of their CNN model, which resulted in an outstanding performance. Elshennawy et al. [30] evaluated the performance of four deep learning models, of which two of the evaluated models were pre-trained models (MobileNetV2 and ResNet152V2), from scratch CNN model and an LSTM model. All models were evaluated with varying parameters using the conventional classification evaluation metrics. Wang et al. [31] stressed the necessity of early identification of pneumonia illness. They employed transfer learning and model adaption approaches to predict the disease using the VGG-16 and Xception models, achieving 87 % and 82 % detection accuracy for the VGG-16 model and Xception model, respectively. Talo et al. [32] employed the ResNet152 model to identify pneumonia disease using the transfer learning technique. It recognized 97.4 % set without any preprocessing or feature extraction. In DICOM format, O'Quinn, Haddad, and Moore in [33] attempted to detect the presence of Pneumonia. AlexNet obtained a recognition success rate of 76 % using the transfer learning technique. Another study by Varshni et al. [34] examined the identification of pneumonia using several models based on a convolutional neural network (CNN), which they utilized for extracting features via transfer learning and different classifiers as predictors. Their findings demonstrate that pre-trained CNN models combined with supervised classifier models might help assess chest X-ray images, particularly for detecting Pneumonia. The authors also reported that utilizing DenseNet-169 for extracting features and Support Vector Machines (SVM) as the predictor yielded the best performance. In contrast to works based on transfer learning, such as Stephen et al. [34] used data augmentation to develop a trained CNN for pneumonia detection. The model's efficiency was evaluated with various image dimensions with the maximum performance being 93.73 % for a 200 × 200 RGB image. Urey et al. [35] pre-processed the chest X-ray data in three distinct ways before applying three different networks to it. During the feature extraction phase, they employed the CNN model to extract the feature maps of the pre-processed chest X-ray images using image contrast and image unpacking. They identified the chest X-ray images into three classes which were normal, bacterial pneumonia and viral pneumonia achieving an overall accuracy of 79 %. Hammoudi et al. [36] employed several pre-trained deep learning models to identify the chest X-ray diseases: ResNet50, ResNet34, VGG-19, DenseNet169 and Inception ResNetV2, and RNN. They compared the results among all deep learning models but they did not introduce the Transformer for more improvement of the disease classification performance. Sirazitdinov et al. [37] used two pre-trained deep learning models (i.e., RetinaNet and Mask R-CNN) to diagnose lung pneumonia using Chest X-ray images. The best classification accuracy was achieved with 79.3 %. Liang and Zheng [24] provided a transfer learning approach for detecting kid Pneumonia with a recall rate of 96.7 % and an F1-score of 92.7 % in identifying kid Pneumonia. The Author also employed the standard CNN and VGG16 deep learning models, recording 90.5 %, 89.1 %, 96.7 %, and 92.7 % for accuracy, precision, recall, and F1-score for the CNN model, respectively. On the other hand, the accuracy, precision, recall and F1-score score of the VGG16 were recorded to be 74.2 %, 72.3 % and 82.2 %, respectively. Guangzhou Women's and Children's Medical Center was used by Chouhan et al. [38] via transfer learning algorithm recording an accuracy of 96.4 %. Siddiqi [39] proposed a sequential 18-layer CNN to detect pneumonia disease from chest X-ray images. The overall accuracy of 93.75 % was achieved and compared with the accuracy of the CapsNet recorded by 82.50 % [39]. Yadav and Jadhav [40] suggested a CNN model that obtained 84.18 % accuracy, 78.33 % recall, 94.05 % precision and 85.66 % F1-score classification performance. E3CC and VGG16 + CapsNet were used by Asnaoui, Chawki and Idri in [41] with E3CC achieving an accuracy rate of 81.54 % and VGG16 + CapsNet achieving an accuracy rate of 88.30 %. Mittal et al. [42] attained 85.26 % accuracy, 94 % recall and 89 % F1-score, respectively, while Jain et al. in [26] earned 95.62 % accuracy, 95 % recall and 96 % precision respectively, for pneumonia detection from chest X-ray images.
Unfortunately, X-ray-based pneumonia diagnosis remains a monumental challenge even for trained and experienced clinicians as X-ray images have identical area information for other illnesses, such as lung cancer. As a result, diagnosing Pneumonia using traditional methods is time-consuming and energy-intensive, and it is hard to use a uniform methodology to determine whether a patient has Pneumonia. Many researchers have worked on further improving CNN's performance and demonstrated considerable increases over time. On the other hand, the CNN model only analyzes the correlation between spatially neighboring pixels in the receptive area determined by the filter size. As a result, it is difficult to identify relationships with distant pixels. Recent attempts have been made to use attention mechanisms to tackle this difficulty. Attention is a method of locating and concentrating on the most informative portion of the data.
Attention mechanism
The current state-of-the-art advancement in computer vision tasks is the attention mechanism [43], [44], [45]. Due to the ability of the attention mechanism to focus more on the valuable features while discarding redundant features, it has superseded the conventional CNNs models, which focus on the whole features of the input image hence paying attention to redundant and essential information, thereby leading to false-negative results. Sequel to the attention mechanism's ability to boost interpretation and optimize the classifier's performance without increasing the computational cost of the model, it is applied to replace the current CNN framework. The study of Ref. [46] presented SENet to extract specific image channel weights, demonstrating that decreasing superfluous information may boost the classification strength of a deep learning model. SENet significantly decreased prediction error while incurring only a little computation complexity. Zhang et al. [47] suggested a unique Shuffle Attention mechanism to tackle the computation complexity. They subjected the input features to both channel and spatial attention, achieving a classification improvement on the ImageNet-1 k with more than 1.34 % accuracy. This approach performs much better in the medical field because it captures tiny lesions' information. ECA-Net [47] raised the issue of SENet's rising model complexity. ECA-Net eliminates the dimensionality reduction procedure in the fully connected layer via cross-channel connections using 1D Convolutions. As a result, they can boost speed while lowering model complexity. The study in Ref. [48] employed a two-stage network for Pneumonia diagnosing. The U-Net architecture was used for the lungs RIO segmentation with the SE-ResNet34 as the network backbone. Ref [49] saw the abnormal-aware attention mechanism to transform the input image low-level features into high-level features based on the feature relevance, yielding a framework that dynamically gave one attentiveness score for every densely linked layer. In addition, they used a unique angular contrastive loss to minimize intra-class loss and increase inter-class loss. In WCE images, their approach attained an accuracy of 89.4 %. As a result, embedding spatial and channel attention in deep learning models to detect Pneumonia.
Vision transformer (VT)
Transformers [15], [17] are attention mechanism-based deep neural network architectures initially designed for natural language processing (NLP) tasks. After obtaining state-of-the-art performance in NLP, it motivated the vision community to investigate its usefulness in vision challenges to take advantage of its capacity to represent long-range dependence inside an image [50]. The Vision Transformer (ViT) is among the successful efforts to deploy Transformer explicitly on images, obtaining good results in image classification tests compared to state-of-the-art CNN [51]. Despite its better performance, it has a simple modular architecture allowing several extensive applicability in various jobs with minimum alteration. Chen et al. [52] bintroduced an image recognition transformer, one of the promising multi-task frameworks for various computer vision problems, partitioning ViT into the common core and undertaking heads and tails. This implies that the Transformer has enough profound architectural body potential to be distributed among required tasks. Meanwhile, they utilized an encoder-decoder architecture and the utility of the multi-task ViT concept was not examined alongside the decentralized learning approaches. ViT was reportedly and effectively used for the diagnosing and symptomatic prognosis of COVID-19, demonstrating state-of-the-art efficiency [53]. To eliminate the over-fitting issue caused by limited training samples, the total architecture is divided into two phases: (1) the pre-trained backbone structure is used to categorize common deep features, and (2) the extracted deep features then exploited by the Transformer-based architecture for a high-level diagnosis and symptomatic prognosis of the COVID-19 chest X-ray images. Even with a few training samples, the model achieved a steady result generalization and state-of-the-art performance in a range of external test sets from various institutions.
Materials and methods
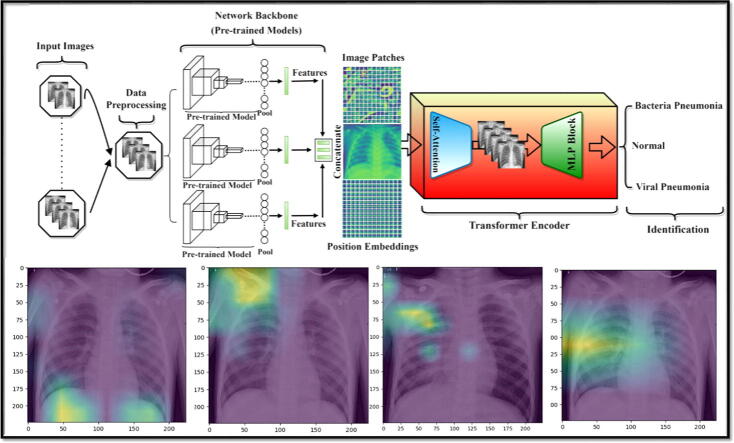
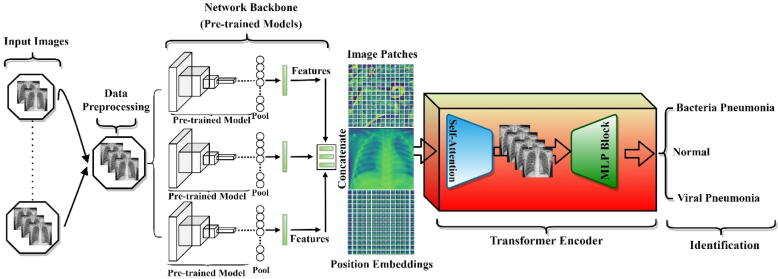
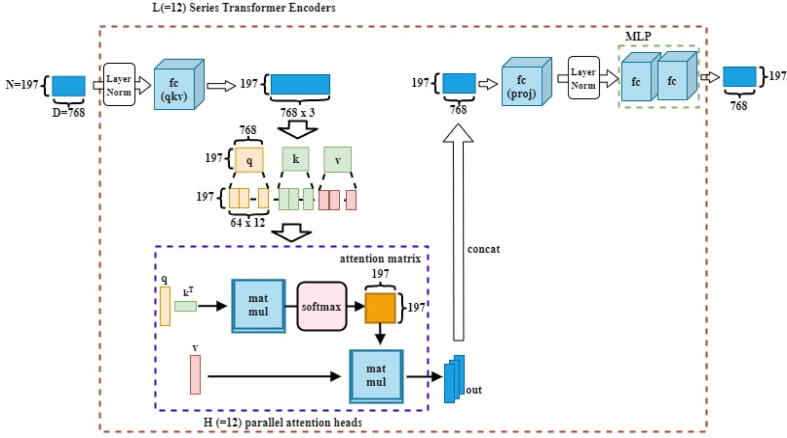
This section presents the proposed hybrid deep learning architecture in detail as shown in Fig. 1. This model is inspired to combine both deep convolutional networks for strong deep feature extraction as well as transformer encoder with self-attention and MLP mechanisms in an end-to-end manner for accurate identification performance. As it is known, the deep learning CNN models could analyze the spatial correlation among the neighboring pixels in the receptive area determined by the convolutional filter size ignoring the directional relationships with the distance among these pixels [15], [17]. To solve this, transformers based on the attention deep learning mechanism have recently been presented and proven to be more powerful and robust to consider both spatial pixel correlation with their distance relations for more accurate visual recognition tasks.
Fig. 1.
The abstract organizational structure of the proposed hybrid deep learning framework for pneumonia identification from chest X-ray images. The pre-trained ensemble deep learning models serve as deep feature extractors, while the transformer encoder based on the self-attention mechanism and perceptron multilayer (MLP Block) is used for pneumonia accurate identification.
The proposed hybrid deep learning framework has the following overall processing steps. First, the data preprocessing techniques were performed to prepare and clean the data including resizing, rotation, cropping, normalization and data splitting into training-validation and testing sets. Second, deep learning features are extracted using the capability of the latest ensemble deep convolutional network structures. To select the best backbone network of the proposed hybrid framework, a comprehensive experimental analysis for six deep learning pre-trained models were performed. Finally, the ensemble concatenated deep features of the models are passed through the proposed transformer encoder where the self-attention network distinguishes the different symptoms in the fed images. The block of multilayer perceptron (MLP) is further applied to enhance the results of the self-attention network in false symptom detection in the fed dataset. We further discuss the implementation steps of our proposed model as follows;
-
•
Step 1: Collection of multiple chest X-ray images from various benchmark databases, data preprocessing, and splitting.
-
•
Step 2: Proper backbone model selection for deep feature extraction from various state-of-the-art pre-trained deep learning models. A comprehensive experimental study is performed on six deep learning pre-trained models: DenseNet201, Xception, VGG16, GoogleNet, GoogleNet, InceptionResNetV2 and EfficientNetB7. For this purpose, the Chest X-ray dataset is used for the Multi-classification scenario.
-
•
Step 3: Concatenating the selected pre-trained models for richer features and accurate results generalization. Experiments were carried out with only the ensemble models to record their performance. The implemented ensemble models are of two scenarios: Ensemble A is the concatenation of DenseNet201, VGG16 and GoogleNet architecture, while Ensemble B is the concatenation of DenseNet201, InceptionResNetV2 and Xception network architecture.
-
•
Step 4: Employ the proposed fine-tuned Transformer Encoder (TE) based on the ensemble model features as the network backbone.
-
•
Step 5: Identification and classification stage which is the last stage of the proposed hybrid XAI model. The learned features are passed into the classification layer for the final result prediction. The main components of the proposed model architecture are explained in the following sections.
Fig. 1 shows the consecutive processing steps of the proposed hybrid deep learning framework: data preprocessing, deep feature extraction via the backbone pre-trained ensemble scenario, and the feature enhancement using the Transformer Encoder with and the self-attention and MLP mechanisms for the final classification purpose.
Dataset
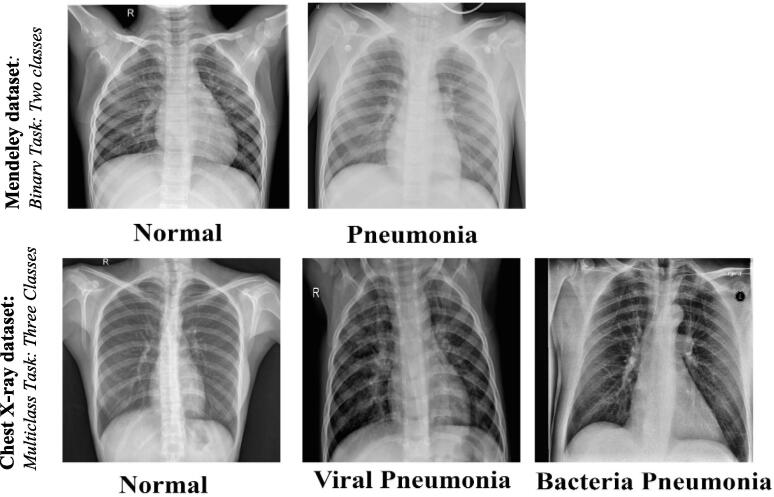
The proposed deep learning models were trained, validated, and assessed using two different chest X-ray datasets: Mendeley [54] and Chest X-ray [31] datasets. The Mendeley dataset represents the binary classification scenario with two classes of Normal Vs. Pneumonia, while the Chest X-ray dataset reflects the multi-classification scenario via three different respiratory disease classes: Normal, Bacteria Pneumonia, and Viral Pneumonia. Fig. 2 depicts the pictorial representations of some Chest X-ray images from both datasets.
Fig. 2.
Some samples of the deployed chest X-ray datasets. The first row depicts some images from the Mendeley dataset, while the second row depicts some images from the chest X-ray dataset.
Mendeley dataset
This dataset was collected for kid patients over the age of one to five years old at the Guangzhou Women and Children's medical center in Guangzhou, China. All images in this dataset were in the format of the joint photographic experts’ group (.jpeg) but with different spatial resolutions. Despite this dataset having three different classes of Normal, Bacteria Pneumonia, and Viral Pneumonia, both Bacteria and Viral Pneumonia classes were merged to form the binary classification dataset set (i.e., Normal vs Pneumonia). The number of normal images for training, validation, and testing were collected and randomly split to be 1341, 8, and 234, respectively. Whereas, the number of pneumonia images after combining was 3875 for training, 8 for validation, and 390 for testing. To enlarge the number of images in a balanced manner, more additional images are collected, for the respective classes, from another dataset source, called COVID-19 Radiography Dataset which is publically available [55]. The final dataset distribution is reported in Table 1.
Table 1.
Chest X-ray dataset distribution over binary and multiclass classification scenarios. Training and validation images are combined for fine-tuning the deep learning models.
Chest X-ray dataset
The Chest X-ray-15 k dataset is collected from eleven different sources by Badawi et al [18]. This dataset has a balanced number of Chest X-ray images in terms of training/validation and testing with 3,500 and 1,500 images, respectively. It has three different chest X-ray classes: Normal, Bacteria Pneumonia, and Viral Pneumonia. The data distribution per class is summarized in Table 1. All images in this category are in the portable network graphics (.png) but with different spatial resolutions.
Preprocessing
The chest X-ray images employed in this study have varied width and height values, thus they were resized to 224 × 224 pixels before the training process. The reshape size of 224 × 224 pixels was selected to allow us to do some data augmentation. Each deep learning model could internally resize the input images to fit its structure. Since deep learning models require a massive quantity of data to increase their performance, data augmentation is one solution for dealing with sparse data and enlarging the number of images in the training sets. The images in the training set were rescaled (i.e., image magnification or reduction) using the ratio of 1.0/255, Zoom range of 0.2, rotation range equal to 1, and horizontal flip. The rotation range specifies the span under which the images were spontaneously rotated throughout training. The zoom range dynamically zooms the images to a ratio of 0.2 percent as well as the images were eventually flipped horizontally. While noting the significance of recent data augmentation techniques used by the researchers such as denoising and histogram equalization and their contribution to the improvement of model results, this study focused more on the real-time implementation strategy where models are applied directly to the whole input chest X-ray images. Also, this paper is much concerned with the ability of the attention mechanism to detect the affected pixel of the chest X-ray images.
Pre-trained models
The selected pre-trained backbone deep learning models are DenseNet201 [56], Xception [57], VGG16 [4], GoogleNet [58], InceptionResNetV2, and EfficientNetB7 [59] are discussed as follows;
-
•
DenseNet201[56]: This model ensures information flow across layers in the network by connecting each layer to every other layer in a feed-forward approach (with equal feature-map size). It concatenates (.) the previous layer's output with the output of the future layer. The transition layers are 1 × 1 convolution, followed by 2 × 2 average pooling. The global pooling is used at the end of the last dense block before the SoftMax is applied.
-
•
VGG16[4]: VGG16 has 16 layers. After the preprocessing step, the extracted values are passed into a stacked Convolutional layer with 3 × 3 receptive-field filters, a fixed stride of 1. After that, spatial pooling is done by five max-pooling of several convolutional layers. The max-pooling layer of a 2 × 2 filter is performed with a stride of 2. At the end of the last convolution, two fully connected layers (FC) and SoftMax (for the output) are added to complete the architecture.
-
•
GoogleNet[58]: This architecture was built to overcome the problem of overfitting while going deeper with the network layer. The idea was basically of having multiple filters that can work on the same level, thus making the network broader instead of deeper. A typical GoogleNet architecture has 22 layers and 27 max-pooling layers with nine linearly stacked Inception modules. The global average pooling is at the end of the inception module.
-
•
Xception[57]: This is a stack of the depth-wise separable convolution layers linearly with residual connections. It consists of 36 convolutional layers structured into 14 modules with linear residual connections around each, serving as the feature extraction backbone of the network.
-
•
InceptionResNetV2[60]: This is an improvement of InceptionResNetv1 as the network schema is that of InceptionResNetv1, and the stem is that of InceptionV4. Each module has a shortcut connection at the left. It combines inception architecture with residual connections to improve classification accuracy. For the residual links to be effective, the inception module convolutional operation must have the same input and output; thus, 1 × 1 convolution must be used after the original convolution to match the depth sizes. The addition of the residual connections replaced the pooling operations.
-
•
EfficientNetB7[59]: EfficientNetB7 is a non-linear and non-recurrent neural networks search that balances network depth, breadth, and resolution to maximize accuracy and FLOPS. The architecture employs seven inverted residual blocks, each with its parameters. Squeeze and excitation blocks as well as swish activation, are used in these blocks.
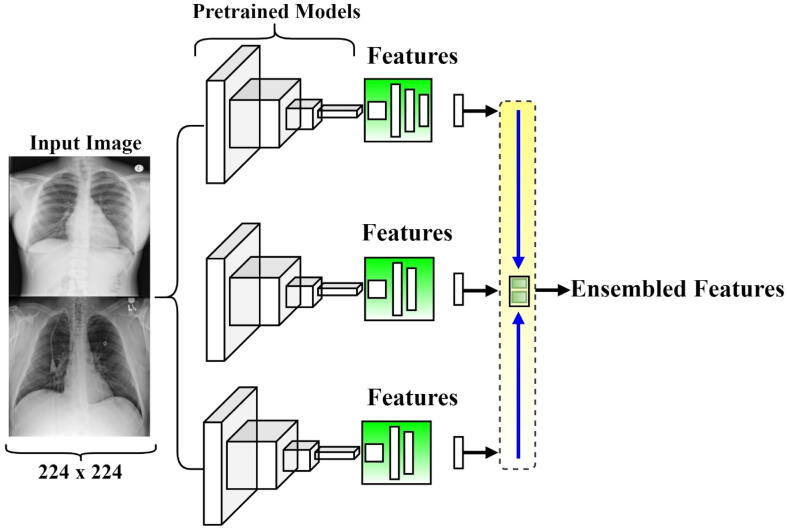
Multi-model ensemble deep learning
Indeed, deep learning networks are non-linear models and they could provide great flexibility to the scarcity amount of training datasets [42]. They are very sensitive to the training data details since they are fine-tuned via random algorithms and produce some variation in the weight sets every training time. This pushes the neural networks to achieve different predictions at a time giving the neural network a high variance. To reduce such variance of deep neural networks, the ensemble learning technique is recently used to learn multiple deep learning models instead of a single one [47]. Then, the final prediction result is achieved by combining the predictions of these multiple models. Indeed, ensemble learning allows the fusing of different models' decisions, thus allowing more detailed salient image features and capturing more useful information from the different classifiers hence yielding more robust classification results. In the literature, we found that the majority of cutting-edge deep learning techniques for pneumonia identification rely on a single convolutional network. Ensemble learning perspective has received little attention in the context of the pneumonia identification and classification task. In this paper, ensemble deep learning models were investigated and considered for designing the backbone network. This paper defines its ensemble method as the concatenation of different pre-trained deep learning models for rich feature extraction and enhancement of performance results as shown in Fig. 3. The ensemble method varies on the choice of researchers. We experimented with two ensemble scenarios as explained below,
-
•
Ensemble A is the concatenation of deep learning architectures of DenseNet201, VGG16, and GoogleNet. The first top layer which is the classification layer of each model is removed, while the deep features were extracted from the last block convolution layer of each model. The DenseNet architecture outputted (None, 7, 7, 1920) after the classification layer was removed, while the VGG16 and GoogleNet architecture output are formed to be (None, 7, 7, 512) and (None, 5, 5, 2048), respectively. Looking at the 3 models, the GoogleNet architecture had a different output hence there is a need for uniformization of all the output features. The GoogleNet architecture was zero-padded before the features were concatenated.
-
•
Ensemble B is the deep learning concatenation of DenseNet201, InceptionResNetV2, and Xception convolutional networks. Same as ensemble A, the classification layer of the three models were removed and the outputted features from the last convolutional block/layer were concatenated. The output of the deep learning models of DenseNet, InceptionResNetV2, and Xception network is formed to be (None, 7, 7, 2048), (None, 7, 7, 1920), and (None, 5, 5, 1536), respectively. Just the GoogleNet in Ensemble A, we zero-padded the InceptionResNetV2 architecture for uniformity of the outputted features before concatenating them.
Fig. 3.
Diagrammatical illustration of the proposed Ensemble architecture.
Transformer encoder (fine-tuned vision transformer)
A transformer is a deep learning model that uses the self-attention mechanism to weigh the importance of each element of the input data differently. It is presented in an encoder-decoder manner. In this paper, we adopt and fine-tune the vision transformer (ViT) via encoder approach for early pneumonia disease detection from chest X-ray images. The proposed transformer encoder consists of a self-attention network, a multi-linear perceptron block, and a classification layer. Two output results are yielded from a given input image by the ViT encoder; the total input token embedding vector (i.e., image and class tokens), all layers and head attention weights. The number of tokens makes up the dimension of the embedding tensor. Since we are interested in only the embedding vector of the class token to be used as the image feature vector for classification, we extracted it at index 0 before passing it through the classifier to yield a given input of length equivalent to the total number of classes. The fine-tuned model components are described in detail as follows,
Self-attention network
The self-attention mechanism could compute a representation of a single input sequence by linking distinct places of the same series [15]. The self-attention network and MLP block represent the encoded structure where the normalization layer with residual connections is used in each block intermediately. The attention function is the mapping to an output of a set of keys, value pairs, and a query [16]. The weights allocated to each value are determined by the query compatibility function with the relevant key, whereas the weighted sum of the values results in the output. Considering an input with dimension of queries and keys and dimension , the dot product of all queries with keys are computed by dividing each with , while using SoftMax to ascertain the weights on the value pairs. The attention matrix contains the set of queries (Q), the keys (K), and values (V), which are used to compute the attention function simultaneously. The attention (Q, K, V) is computed as follows,
| (1) |
Multi-headed attention allows the model to simultaneously attend to input from several representation subspaces at various locations. Fig. 4 elaborates on the computation done by the multi-head self-attention in the encoder.
| (2) |
Fig. 4.
Visualization of a Multi-head self-attention Network and MLP blocks.
The parameter matrices are the projections and .
Multilayer perceptron layer (MLP)
The multilayer perceptron (MLP) is referred to as a feed-forward neural network model with some dense and dropout layers [15]. In this study, the MLP is designed using two non-linear layers of Gaussian error linear units (GELU). The MLP blocks receive identical stacked layer blocks and structures. For instance, let be the token features with its length of sequence denoted and dimension denoted . Defining each block mathematically;
| (3) |
| (4) |
| (5) |
| (6) |
| (7) |
is an activation function, and denote the dimension of the channel’s linear projections and denotes the identity mapping. From Eq. (3), the spatial interaction is captured by the layer denoted as where the individual tokens are computed separately without any token interactions. Eqs. (4), (5) explain in detail the individual layers class token, learnable imbedding positioning and the patch embedding before being stacked as shown in Eq. (3). Eq. (7) depicts the final output of the Encoder as formulated to be,
| (8) |
where ⊙ reflects the dot product or the element-wise multiplication.
Classification layer
The classification head is implemented with one hidden layer during pre-training (Eq. (5)) and a single linear layer (Eq. (6)) during fine-tuning by the MLP. We use the SoftMax layer after the MLP block to accurately detect a sample. The SoftMax layer's primary function converts the encoding layer's output information into a likelihood interval [0,1]. In this work, we consider pneumonia identification for binary and multi-classification scenarios. After that, the input samples are sent to the encoding network, whose outputs are then transferred into the likelihood interval [0,n] via the SoftMax layer, as seen below:
| (9) |
where the weight matrix and the bias term are denoted as and , respectively. The categorical smooth loss function is used to calculate the loss between the ground truth and the detected labels. This loss function is the addition of smoothing of the labels function to the cross-entropy loss function as presented here,
| (10) |
Despite the fact that VisTransf architectures demand a greater number of training samples than CNN architectures, the most common strategy is to utilize a pre-trained network and afterward fine-tune it on a smaller undertaking sample which was done in this paper. The following are the benefits of our fine-tuned architecture over CNN design: (1) The proposed hybrid architecture integrates more spatial information than ResNet (CNN) at lower layers, resulting in statistically distinct features. (2) In the implemented model, skip connections are significantly more prominent than in ResNet (CNN), with significant effects on efficiency and representation comparability. (3) ResNet needed lower layers to calculate representations that were equivalent to a limited number of the implemented lower layers. (4) Using bigger pre-training samples, our models create much better intermediary representations.
Experimental setup
The proposed model is trained in an end-to-end manner. The training parameters used in this experiment include a learning rate of 0.0001 with a reduced learning rate by 0.2 factors, epsilon of 0.001, and patience of 10 were utilized. Early stopping strategy of es_callback with the patience of 10 was considered as well. An es_callback is a component that may execute operations at multiple phases of learning at different batch intervals, different epoch intervals, etc. For hyper-parameter optimization, Adam optimizer with clip value of 0.2 and epoch of 100 was utilized. During the selection of the pre-trained models, we used an epoch of 50, while other parameters remain constant as in the main experiment. In the encoder part, a patch size of 2, a drop rate of 0.01 for all layers, and 8 heads were used. Meanwhile, embed_dim of 64 (i.e., embed_dim indicates the dimension by which high-dimensional vectors are converted to low-dimensional vector without any loss during conversion), num_mlp of 256 (i.e., this indicate the number of Multi-linear perceptrons), a window size of 2 and global average pooling (GAP) for the shift size were utilized. The comparison among all deep learning models, used in this study, in terms of the final output shape and number of the trainable parameters were summarized in Table 2. The ensemble deep features were extracted via ensemble learning scenario and passed as an input to the transformer encoder for the final prediction purpose. We fine-tuned the vision transformer to suit our identification task where the transformer received an input size of (12, 12, 4480) and (12, 12, 5504) in terms of Ensemble A and B, respectively. Since different deep learning models are used for this study, the ReLU activation function is used in the internal convolutional layers, while the the softmax is used for the output layer as a prediction regression to find the final class probabilitieis.
Table 2.
The parameters of the proposed deep learning models in terms of the final output shape and number of the trainable and non-trainable parameters.
| Models | Last Convolution Layer Output | Trainable Parameter | Non-Trainable Parameter | Total Parameter |
|---|---|---|---|---|
| DenseNet201 | None, 7, 7, 1920 | 1,106,179 | 18,321,984 | 19,428,163 |
| VGG16 | None, 7, 7, 512 | 598,403 | 14,714,688 | 15,313,091 |
| GoogleNet | None, 5, 5, 2048 | 524,547 | 21,802,784 | 22,327,331 |
| EfficientNetB7 | None, 7, 7, 2560 | 1,474,819 | 64,097,680 | 65,572,499 |
| InceptResNetV2 | None, 5, 5, 1536 | 393,475 | 54,336,736 | 54,730,211 |
| Xception | None, 7, 7, 2048 | 1,179,907 | 20,861,480 | 22,041,387 |
| Ensemble A | None, 7, 7, 4460 | 286,979 | 54,839,456 | 55,126,435 |
| Ensemble B | None, 7, 7, 5504 | 352,515 | 93,520,200 | 93,872,715 |
| Proposed hybrid model with Ensemble A backbone | – | 1,290,579 | 54,839,632 | 56,130,211 |
| Proposed hybrid model with Ensemble B backbone | – | 1,552,723 | 93,520,376 | 95,073,099 |
Experimental environment
All experiments were performed on PC with the following hardware specifications: Intel(R) Core (TM) i9-10850 K CPU @ 3.60 GHz, 64.0 GB RAM, and an NVIDIA Geforce RTX-3080 Ti 10 GB graphical processing unit (GPU). This paper utilized the open-source library of Keras and TensorFlow for the implementation purpose.
Evaluation metrics
Various evaluation matrices were used to evaluate the performance of the proposed hybrid deep learning framework including Accuracy (ACC), Precision (PRE), Specificity (SPE), Sensitivity (SEN), F1-score, and area under the receiver operating characteristic (ROC) curve [61]. The overall classification accuracy (ACC) was defined as,
| (11) |
where denotes the marked positive and were correctly predicted as positive (True Positive, TP), denotes observed negative and was correctly predicted as negative (True Negative, TN), and denote the positive and negative predictions, respectively. Precision (PRE) can also be referred to as the percentage of the predicted positive value and is defined as,
| (12) |
where denotes the marked negative but was predicted positive (False Negative, FN). Specificity is the percentage of the classification of marked negative and was correctly predicted to be negative and is defined as,
| (13) |
The percentage of correctly classified and positively marked was referred to as sensitivity (SEN) and is mathematically represented by,
| (14) |
The precision and sensitivity harmonic mean was referred to as the mathematically represented as,
| (15) |
TP, TN, FP, and FN were driven based on the confusion matrices. The AUC measures a classifier's performance, while the probability curve is gotten from plotting at different threshold settings, the FP rate is referred to as the receiver operating characteristic (ROC) curve. The AUC indicates how well the model distinguishes between Pneumonia and non-Pneumonia instances. A higher AUC means better identification performance. The AUC equals one implies a perfect classification performance, whereas AUC = 0.5 suggests a classifier randomizing class observation [1]. To determine the area under the ROC curve, AUC is calculated using the trapezoidal integration process. Moreover, the precision-recall (PR) curve is also used to check the average precision (AP) evaluation performance.
Results and discussion
The experimental results of the proposed deep learning models were presented and explained in this section. First, the parameter sensitivity analysis of the proposed models was investigated and the research findings were reported accordingly. The classification performance of the deep learning models in terms of individual and combined models were presented and discussed. In addition, the explainable visual heat maps of the attention mechanism were performed in an ablation study scenario.
Parameter sensitivity analysis of the proposed hybrid deep learning method
Before going straight into the implementation stage of the proposed hybrid deep learning model, the parameter sensitivity analysis was carried out to select the optimal number of Transformer heads and the best configuration of the ensemble deep learning feature extractors. During this analysis, the number of epochs and learning rate were kept constant at 50 and 0.0001, respectively. Due to the computation cost and time effort, this study had been experimented with using transformer heads of 2, 4, and 8 with different ensemble deep learning options. For each transformer head, three different ensemble models were used with 1, 2, and 3 deep learning models. The combination number of the ensemble deep learning models was randomly selected without any priority from the six deep learning models: DenseNet201, VGG16, GoogleNet, InceptionResNetV2, Xception, and EfficientNetB7. Table 3 shows the classification evaluation results via the multiclass classification scenario using overall identification accuracy (ACC), precision (PRE), and F-1 score. The best classification performance of the proposed model was achieved while using three ensemble feature extractor models with 8 heads of the self-attention network. Although with the minor difference of using four heads with three ensemble models, the eight heads with three ensemble feature extractors were used to perform all the experiments in this study.
Table 3.
Parameter sensitivity analysis of the proposed hybrid deep learning model in terms of the number of transformer heads with three ensemble deep learning options.
| No. of Pre-trained Ensemble Models | Transformer Head | ACC | PRE | F1-score |
|---|---|---|---|---|
| One Model | 2 | 0.9684 | 0.9612 | 0.9602 |
| Two Models | 0.9695 | 0.9711 | 0.9608 | |
| Three Models | 0.9771 | 0.9721 | 0.9729 | |
| One Model | 4 | 0.9696 | 0.9623 | 0.9609 |
| Two Models | 0.9770 | 0.9801 | 0.9711 | |
| Three Models | 0.9820 | 0.9756 | 0.9744 | |
| One Model | 8 | 0.9699 | 0.9701 | 0.9703 |
| Two Models | 0.9799 | 0.9800 | 0.9708 | |
| Three Models | 0.9898 | 0.9806 | 0.9801 |
Classification results
This section presents the classification results of the various approaches implemented in this paper. The model's classification performances were evaluated over three scenarios: (1) individual pre-trained transfer deep learning models, (2) ensemble deep learning models: Ensemble A and Ensemble B, and (3) the proposed hybrid deep learning model based on the ensemble transformer encoder mechanism. All experiments in this study, for all different scenarios, were carried out using the multiclass classification task via the Chest X-ray dataset. This is because the performance with a multiclass scenario could be more reliable and show the capability of the models to deal with multiple classes at once.
Individual Pre-trained transfer deep learning models
Six state-of-the-art deep learning models were selected and investigated for the backbone ensemble network: DenseNet201 [56], Xception [57], VGG16 [4], GoogleNet [58], InceptionResNetV2 [60], and EfficientNetB7 [59]. The individual classification performances of each model were recorded in Table 4. Same training settings and system setup were used for these models. The DenseNet201 shows superiority in the classification result among the other pre-trained models in terms of accuracy, sensitivity, specificity, precision, F-1 score and AUC. Among all the pre-trained models, efficientNetB7 had the lowest classification performance. Thus, the efficientNetB7 was excluded during the formation of our proposed ensemble deep learning scenarios. To further evaluate the classification performance of the transfer learning models, we assessed with the PR and ROC curves as shown in Table 5. The normal class received the lowest AUC and AP scores among other pneumonia classes. For the Bacteria Pneumonia class, the DenseNet201 and VGG16 achieved the highest identification accuracy with 97.01 % and 97.38 %, respectively. The best AP score was achieved by DenseNet201 for viral pneumonia class with 95.78 % accuracy, while it was 94.74 % and 85.15 % for bacteria pneumonia and normal classes, respectively.
Table 4.
Classification evaluation performance of the individual pre-trained deep learning models.
| Pre-trained Models | ACC | SEN | SPE | PRE | F1-score | AUC |
|---|---|---|---|---|---|---|
| DenseNet201 | 0.9695 | 0.9542 | 0.9771 | 0.9551 | 0.9544 | 0.9657 |
| VGG16 | 0.9585 | 0.9378 | 0.9689 | 0.9392 | 0.9378 | 0.9533 |
| GoogleNet | 0.9446 | 0.9169 | 0.9584 | 0.9214 | 0.9178 | 0.9377 |
| EfficientNetB7 | 0.9276 | 0.8913 | 0.9457 | 0.9214 | 0.8930 | 0.9185 |
| InceptResNetV2 | 0.9444 | 0.9167 | 0.9583 | 0.9197 | 0.9169 | 0.9375 |
| Xception | 0.9579 | 0.9369 | 0.9684 | 0.9397 | 0.9374 | 0.9527 |
Table 5.
The individual Pre-trained deep learning model classification performance in terms of the ROC and PR evaluation curves.
| Deep Learning Model | Receiver Operating Characteristic (ROC) curve |
||||
|---|---|---|---|---|---|
| Macro-Average Area | Micro-Average Area | Bacteria Pneumonia Area | Normal Class Area | Viral Pneumonia Class Area | |
| DenseNet201 | 0.97 | 0.97 | 0.9701 | 0.9548 | 0.9723 |
| VGG16 | 0.95 | 0.95 | 0.9738 | 0.9363 | 0.9498 |
| Xception | 0.95 | 0.95 | 0.9627 | 0.9432 | 0.9522 |
| GoogleNet | 0.94 | 0.94 | 0.9438 | 0.9231 | 0.9518 |
| InceptResNetV2 | 0.94 | 0.94 | 0.9681 | 0.9258 | 0.9487 |
| EfficientNetB7 | 0.92 | 0.92 | 0.9383 | 0.9023 | 0.9248 |
| Deep Learning Model | Precision-Recall (PR) Curve |
|||
|---|---|---|---|---|
| Micro-Average Precision-Recall | Bacteria Pneumonia Class AP | Normal Class AP | Viral Pneumonia Class AP | |
| DenseNet201 | 0.9322 | 0.9474 | 0.8915 | 0.9578 |
| VGG16 | 0.9051 | 0.9238 | 0.8639 | 0.92765 |
| Xception | 0.9016 | 0.9302 | 0.8524 | 0.9224 |
| GoogleNet | 0.8761 | 0.8901 | 0.8164 | 0.9219 |
| InceptResNetV2 | 0.8751 | 0.8864 | 0.8202 | 0.9188 |
| EfficientNetB7 | 0.8399 | 0.8734 | 0.7614 | 0.8868 |
Ensemble deep learning classification results
Based on the classification performance of the former individual deep learning models, two ensemble learning scenarios were designed and carried out using the selected best five performing pre-trained models: Ensemble A (i.e., concatenation of the DenseNet201, VGG16, and GoogleNet) and Ensemble B (i.e., concatenation of the DenseNet201, InceptionReseNetv4, and Xception). The classification performance of each ensemble deep learning model is reported in Table 6. Meanwhile, the classification evaluation results in terms of ROC and PR curves were presented in Table 7 and Table 8 for binary and multiclass classification scenarios, respectively.
Table 6.
Ensemble learning classification evaluation performance for ensemble A and ensemble B with binary and multi-class identification scenarios. These results were recorded as an average for all respiratory classes.
| Models | Accuracy (ACC) | Sensitivity (SEN) | Specificity (SPE) | Precision (PRE) | F1-score | AUC |
|---|---|---|---|---|---|---|
| Binary Classification Results | ||||||
| Ensemble A | 0.9723 | 0.9722 | 0.9722 | 0.9724 | 0.9722 | 0.9722 |
| Ensemble B | 0.9644 | 0.9644 | 0.9644 | 0.9651 | 0.9644 | 0.9644 |
| Multi-class Classification Results | ||||||
| Ensemble A | 0.9720 | 0.9580 | 0.9790 | 0.9583 | 0.9580 | 0.9686 |
| Ensemble B | 0.9643 | 0.9464 | 0.9732 | 0.9493 | 0.9468 | 0.9598 |
Table 7.
Ensemble learning evaluation performance of the binary identification scenario using the Mendeley dataset. These results were recorded as per class of the respiratory classes.
| Ensemble Learning Scenario | Receiver Operating Characteristic (ROC) curve |
||||
|---|---|---|---|---|---|
| Macro-Average Area | Micro-Average Area | Bacteria Pneumonia Class Area | Normal Class Area | Viral Pneumonia Area | |
| Ensemble A | 0.9722 | 0.9722 | 0.9722 | 0.9714 | 0.9722 |
| Ensemble B | 0.9644 | 0.9644 | 0.9644 | 0.944 | 0.9644 |
| Ensemble Learning Scenario | Precision-Recall (PR) Curve |
|||
|---|---|---|---|---|
| Micro-Average Precision-Recall | Bacteria Pneumonia Class AP | Normal Class AP | Viral Pneumonia AP | |
| Ensemble A | 0.96 | 0.9488 | 0.9488 | 0.96 |
| Ensemble B | 0.95 | 0.9301 | 0.9301 | 0.95 |
Table 8.
Ensemble learning evaluation performance of the multiclass identification scenario using the Chest X-ray dataset. These results were recorded as per class of the respiratory classes.
| Ensemble Learning Scenario | Receiver Operating Characteristic (ROC) curve |
||||
|---|---|---|---|---|---|
| Macro-Average Area | Micro-Average Area | Bacteria Pneumonia Class Area | Normal Class Area | Viral Pneumonia Area | |
| Ensemble A | 0.97 | 0.97 | 0.9771 | 0.9541 | 0.9741 |
| Ensemble B | 0.96 | 0.96 | 0.9745 | 0.9517 | 0.9533 |
| Ensemble Learning Scenario | Precision-Recall (PR) Curve |
|||
|---|---|---|---|---|
| Micro-Average Precision-Recall | Bacteria Pneumonia Class AP | Normal Class AP | Viral Pneumonia AP | |
| Ensemble A | 0.93 | 0.9461 | 0.9077 | 0.9574 |
| Ensemble B | 0.91 | 0.9538 | 0.8687 | 0.9460 |
For both binary and multiclass identification performance, Ensemble A achieved always better and more accurate results than Ensemble B in terms of all quantitative evaluation results as well as the ROC and PR evaluation curves as reported in Table 6, Table 7, Table 8. For the binary classification class performance, Ensemble A achieved higher and better AUC of the ROC against Ensemble B in terms of macro and micro average areas with more than 97 %. Similarly, the average precision was better achieved in the Ensemble A scenario with more than 94 % for all classes. For the multiclass identification scenario, the best macro and micro AUC areas of the ROC curves were recorded to be 97 %, while the micro average precision-recall rate of the PR curve was 93 %.
The proposed hybrid deep learning model classification results
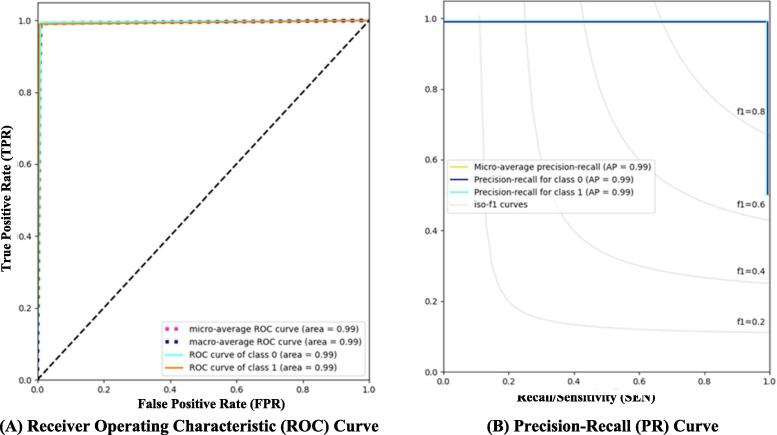
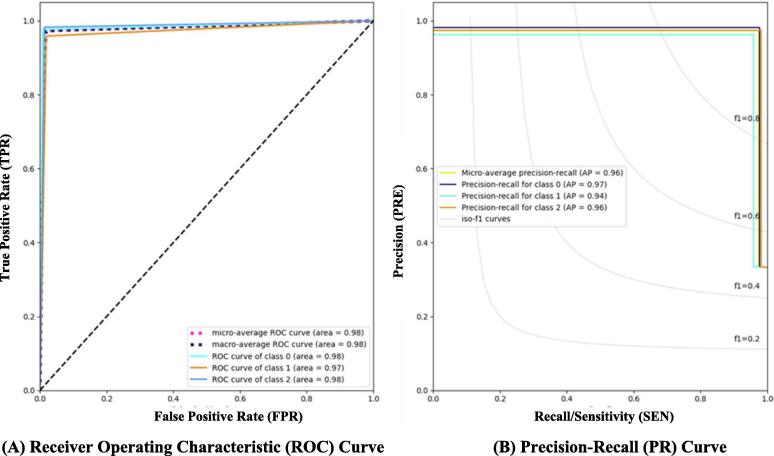
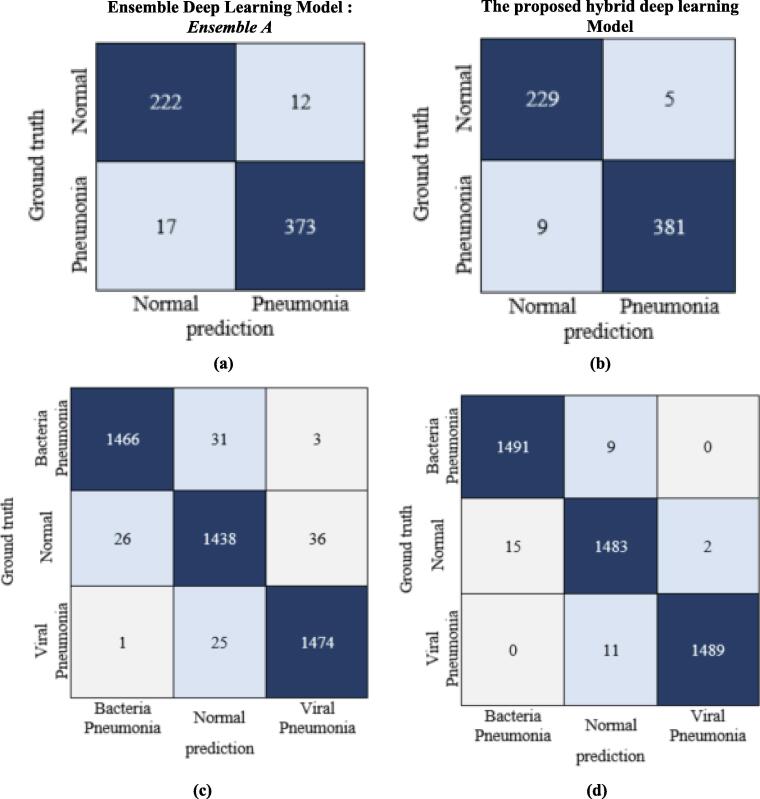
Based on the recommended ensemble learning investigation results, the Ensemble A model with DenseNet201, VGG16, and GoogleNet achieved better classification performance than Ensemble B. As presented in Table 9, the proposed hybrid deep learning model (i.e., a hybrid Ensemble Transformer Encoder) recorded much better classification performance with overall accuracies of 99.21 % and 98.19 % for binary and multiclass scenarios, respectively. This means the proposed hybrid transformer based on ensemble A improves the overall classification performance by 2.05 % and 1.3 % compared with the individual ensemble A scenario for binary and multiclass identification tasks, respectively. Meanwhile, the qualitative evaluation results in terms of ROC and PR curves depicted similar findings as shown in Fig. 5. For the binary classification scenario, the macro and micro-average ROC, as well as the micro-average PR, were equally recorded to be 99.21 %. A comprehensive experimental study with several different scenarios was performed to prove the capability and reliability of the proposed hybrid deep learning framework. For the multiclass classification scenario, the bacteria pneumonia samples were predicted more correctly than the other two classes in terms of the ROC and the precision-recall curves as depicted in Fig. 6. The bacteria pneumonia class had an AUC of 98.42 % with the viral pneumonia class, while the AP was more significant than the viral pneumonia class with an average of 96.0 %. The bacteria pneumonia had an AP of 97.96 %, while viral pneumonia recorded 97.85 %. In the case of normal class, the evaluation results were achieved with light lower AUC and even PR rates with 97 % and 96.06 %, respectively. This was due to the random deep learning process for fine-tuning the trainable parameters as it is commonly known. To measure the number of accurately and mistakenly identified samples, the confusion matrix or contingency table was used for binary and multiclass scenarios as shown in Fig. 7. It is shown that the ensemble strategy alone could not accurately identify the pneumonia respiratory diseases where the number of misclassified cases was larger compared with the proposed hybrid deep learning model. The improving rate of the misclassification identification cases was almost twice improved for both binary and multiclass scenarios. For the binary classification, 12 normal samples were wrongly identified with the ensemble A model, while the misclassification cases decreased to 5 samples with the capability of the proposed hybrid deep learning model. Similarly, 17 pneumonia samples were misclassified with the ensemble learning strategy compared with only 9 cases of the proposed hybrid model as shown in Fig. 7 (a) and (b). For the multiclass identification scenario via the proposed hybrid model, the same findings were concluded where the number of misclassified bacteria pneumonia was decreased from 31 and 3 to 9 and 0 as a normal and viral pneumonia case, respectively. Fig. 7 (c) and (d) shows an example of confusion matrices of the proposed hybrid models in the case of multiclass identification task vis Chest X-ray dataset. In a summary, in terms of all quantitative and qualitative evaluation results, the proposed hybrid deep learning framework performed well compared with the ensemble models individually. This means besides the extracted deep features by the ensemble backbone network, the transformer encoder contributed well to improving the identification performance in general. Such findings were concluded in the literature as mentioned in Section “Related work”.
Table 9.
Identification evaluation performance of the proposed hybrid deep learning framework (i.e., Transformer Encoder based on the ensemble backbone feature extractor). These experimental findings were performed in terms of binary and multiclass recognition scenarios using Mendeley and Chest X-ray datasets, respectively. These results were recorded as an average and per class of the respiratory classes.
| Models | ACC | SEN | SPE | PRE | F1-score | AUC | |
|---|---|---|---|---|---|---|---|
| (1) Binary Classification Scenario | Hybrid TE with Ensemble A backbone | 0.9920 | 0.9919 | 0.9920 | 0.9921 | 0.9921 | 0.9918 |
| Hybrid TE with Ensemble B backbone | 0.9828 | 0.9828 | 0.9828 | 0.9828 | 0.9828 | 0.9828 | |
|
Models |
Receiver Operating Characteristic (ROC) curve |
||||||
|
Macro-Average Area |
Micro-Average Area |
Normal Area |
Pneumonia Area |
||||
| Hybrid TE with Ensemble B backbone | 0.9921 | 0.9921 | 0.9921 | 0.9921 | |||
| Hybrid TE with Ensemble A backbone | 0.9828 | 0.9828 | 0.9828 | 0.9828 | |||
|
Models |
Precision-Recall (PR) Curve |
||||||
|
Micro-Average Precision-Recall |
Normal AP |
Pneumonia AP |
|||||
| Hybrid TE with Ensemble A backbone | 0.9921 | 0.9920 | 0.9921 | ||||
| Hybrid TE with Ensemble B backbone | 0.9828 | 0.9828 | 0.9828 | ||||
| (2) Multiclass Classification Scenario | Hybrid TE with Ensemble A backbone | 0.9819 | 0.9729 | 0.9864 | 0.9729 | 0.9729 | 0.9810 |
| Hybrid TE with Ensemble B backbone | 0.9784 | 0.9676 | 0.9838 | 0.9680 | 0.9676 | 0.9757 | |
|
Models |
Receiver Operating Characteristic (ROC) curve |
||||||
|
Macro-Average Area |
Micro-Average Area |
Bacteria Pneumonia Area |
Normal Area |
Viral Pneumonia Area |
|||
| Hybrid TE with Ensemble A backbone | 0.98 | 0.98 | 0.9842 | 0.9700 | 0.9848 | ||
| Hybrid TE with Ensemble B backbone | 0.98 | 0.98 | 0.9868 | 0.9681 | 0.9720 | ||
|
Models |
Precision-Recall (PR) Curve |
||||||
|
Micro-Average Precision-Recall |
Bacteria Pneumonia AP |
Normal AP |
Viral Pneumonia AP |
||||
| Hybrid TE with Ensemble A backbone | 0.96 | 0.9796 | 0.9606 | 0.9785 | |||
| Hybrid TE with Ensemble B backbone | 0.95 | 0.9802 | 0.9541 | 0.9691 | |||
Fig. 5.
Evaluation results of the proposed hybrid deep learning model in terms of ROC and PR curves for the binary classification scenario. Class 0 and 1 reflect the normal and pneumonia classes from Mendeley Dataset, respectively.
Fig. 6.
Evaluation results of the proposed hybrid deep learning model in terms of ROC and PR curves for the multiclass identification scenario. Class 0, 1, and 2 reflect the bacteria pneumonia, normal, and viral pneumonia classes from Chest X-ray Dataset, respectively.
Fig. 7.
The identification evaluation performance of the binary and multicalssification scenarios in terms of confusion matrices. (a), and (b) represent the binary classification confusion matrices of the Ensemble A model and the proposed hybrid deep learning model, respectively. Whereas, the (c) and (d) depict the multiclassifcation confusion matrices of the Ensemble A model and the proposed hybrid deep learning model, respectively.
Visualization of the transformer encoder implementation steps
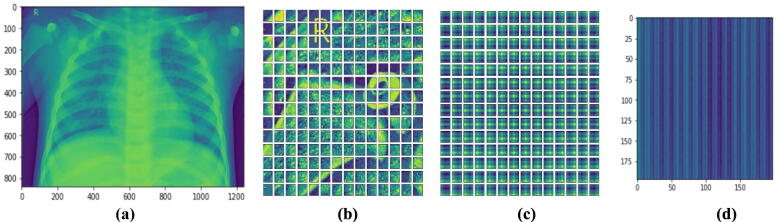
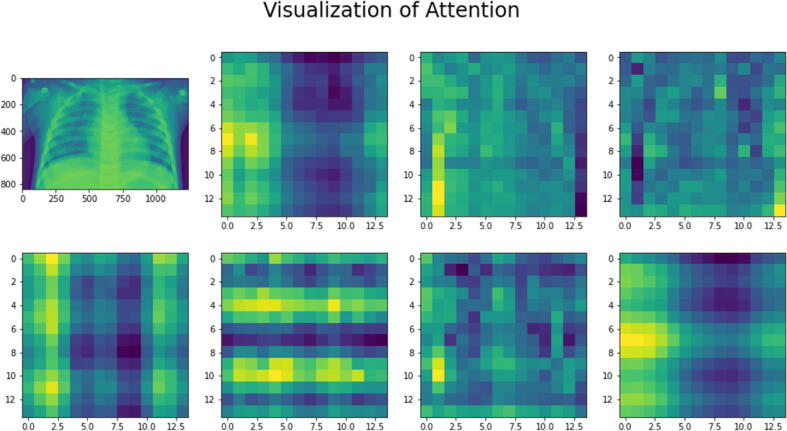
To implement and evaluate the TE, as shown in Fig. 8 (b), the input chest X-ray image was equally split into multiple non-overlapping fixed-size patches and projected after flattening to a feature space where the encoder could process them to provide the final prediction score [17]. By combining the pixel layers in a patch and then immensely extending it to the suitable input dimension, each patch was squeezed into a vector representation. Fig. 8 (c) demonstrates the model understanding to encrypt distance within the input image in the comparability of position linear embeddings. The relatively close patches have much more position similar embeddings. The reason for the patches and the learnable embeddings was to treat each patch separately for accurate feature extraction and identification. The positional embedding helps the model to know where each patch was at the initial input during the output. The patches were first converted using 2D learnable convolutions to further analyze the impact of the patch and embedding combinations. Fig. 8 (d) validates the envisaged approach efficacy in improving the prospective patches enabling the model to efficiently and successfully concentrate on these areas and determine abnormalities. Finally, Fig. 8(d) shows how the Self-attention heads enable the transformer encoder to generalize across the input frame, even within the minimum layers. Accordingly, the total distance in the input images across which relevant data is assimilated is comparable to the receptive scale factor in CNNs and is highly recognized in our model due to our network backbone, which is an ensemble A, and thus we observed continuously small attention scales in the small layers. Indeed, implementing the transformer encoder model without a backbone ensemble network, i.e. generating features from scratch, causes the attention heads to focus on the majority of the image in the lowest layers. Then, it demonstrates that the model's potential to consolidate information globally was really useful for more accurate identification results. Furthermore, as the network depth increases, so does the attention proximity. We discover that the model focuses on visual features that are semantic information significant for classification as depicted in Fig. 9.
Fig. 8.
The visualization steps of the proposed transformer encoder model: (a) depicts the input chest X-ray image, (b) illustrates the divided input image into equal size non-overlapping patches, (c) shows learnable position embeddings of the input image patches, and (d) demonstrates the corresponding attention matrix.
Fig. 9.
The transformer Encoder visualization based on the attention mechanism via the input chest X-ray image.
Ablation studies of the proposed hybrid Model: Explainable Artificial intelligence (XAI)
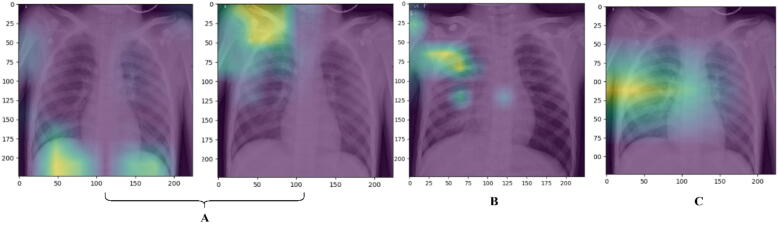
Fig. 10 shows the explainable deep learning results in terms of heat maps (i.e., saliency maps). Such qualitative heat maps were achieved using the recent attention mechanism to depict the most important disease-related geographic localization on the chest X-ray images that the AI model was paying attention to [62]. Indeed, the visual heat maps are powerful for visualizing the areas that the neural network focuses on for classification and also it could explain, in some sense, the internal working of the black-box deep learning models [63]. This is especially significant considering that AI functions in a high-dimensional environment. These heat maps allow for repetitions and render AI readable and understandable in terms of clinical results. In this study, the heat map generation was done using the Grad-CAM which is a method that uses the gradient of a subject idea to “conveys information” to neural network models. The subject (i.e., derived feature maps) was sent into the last convolutional layer, which generates a fine localization map that highlights the significant locations via the classification feature maps. In this study, the attention mechanism helps our model highlights the useful features in the Chest X-ray images that will guide the model’s prediction capability. Fig. 10 illustrates the derived explainable heat maps from different implemented models for the same input chest X-ray image: pre-trained deep learning models (i.e., DenseNet201and VGG16), the ensemble A model, and the proposed hybrid deep learning model (i.e., Transformer Encoder based on the ensemble convolutional networks). As shown in Fig. 10, more robust and powerful visualization explainable results were achieved using the proposed hybrid model which determined the disease localization well. Using ensemble strategy alone was not enough to produce more powerful and feasible classification results. Table 10 shows the quantitative ablation study of the proposed deep learning models over all scenarios: individual pre-train deep learning models, ensemble strategy, and the proposed hybrid framework (i.e., ensemble backbone and Transformer Encoder). To achieve the desired results for this study, the Mendeley data with a binary classification scenario was used. We set the performance threshold limit of 90 % to investigate the contribution of each model starting from the individual pre-trained models to the proposed fine-tuned hybrid model.
Fig. 10.
Visual explainable heat maps (i.e., saliency maps) of the chest X-ray pneumonia image: (A) Depicts the heat maps of the pre-train DenseNet201and VGG16 models, (B) shows the heat map of the ensemble A deep learning model, and (C) illustrates the saliency map for the proposed hybrid deep learning framework.
Table 10.
Quantitative ablation study of the proposed deep learning models to illustrate the contribution rate of each scenario: (1) individual pre-trained transfer deep learning models, (2) ensemble deep learning models, and (3) the proposed hybrid deep learning model based on the ensemble transformer encoder.
| Model | ACC | SEN | SPE | PRE | F1-score | AUC |
|---|---|---|---|---|---|---|
| DenseNet201 | 0.695 | 0.542 | 0.771 | 0.551 | 0.455 | 0.657 |
| VGG16 | 0.585 | 0.378 | 0.689 | 0.392 | 0.378 | 0.533 |
| GoogleNet | 0.446 | 0.169 | 0.584 | 0.214 | 0.178 | 0.377 |
| EfficientNetB7 | 0.276 | – | 0.457 | 0.214 | – | 0.185 |
| InceptResNetV2 | 0.444 | 0.167 | 0.583 | 0.197 | 0.169 | 0.375 |
| Xception | 0.579 | 0.369 | 0.684 | 0.397 | 0.374 | 0.527 |
| Ensemble A | 0.723 | 0.722 | 0.722 | 0.724 | 0.722 | 0.722 |
| Ensemble B | 0.644 | 0.644 | 0.644 | 0.651 | 0.644 | 0.644 |
| Proposed hybrid TE with Ensemble A backbone | 0.921 | 0.921 | 0.9212 | 0.9212 | 0.921 | 0.921 |
| Proposed hybrid TE with Ensemble B backbone | 0.828 | 0.828 | 0.828 | 0.828 | 0.828 | 0.828 |
Validation and statistics of false positive (FP) on unseen data using the CheXpert dataset
To validate the capability of the proposed hybrid deep learning model on unseen data, we used the CheXpert Dataset [64]. The CheXpert dataset is a huge medical chest X-ray dataset that is used for computerized chest X-ray diagnosis competitions with confidence labels annotated by radiologists. It consists of 224,316 chest radiographs (i.e., chest X-ray images) and accompanying imaging records from 65,240 patients. It was gathered and collected across both outpatient and inpatient departments from the Stanford Hospital in “COUNTRY” between October 2002 and July 2017. Every study was categorized as + ve (i.e., positive case), -ve (i.e., negative or healthy case), or Nil based on the existence of 14 variables (blank = unmentioned, 0 = -ve, −1 = uncertain, and 1=+ve). During this experiment, we preprocessed the dataset into a binary classification set: Normal vs pneumonia. The pneumonia and pneumothorax are/were combined to represent the anomaly class (i.e., pneumonia class 1), while the rest of the images are/were gathered to reflect the normal class 0). From the preprocessed dataset, we randomly selected 1,500 each and performed our experiments for ensemble strategy against the proposed hybrid model. The classification evaluation results in terms of accuracy, sensitivity, specificity, precision, F1-score, and AUC were reported in Table 11.
Table 11.
Validation and statistics false positive (FP) result using the unseen chest X-ray dataset: CheXpert Dataset. This is to validate the reliability and feasibility of the proposed hybrid deep learning model based on the new and unseen dataset.
| Models | ACC | SEN | SPE | PRE | F1-score | AUC |
|---|---|---|---|---|---|---|
| Ensemble A | 0.4973 | 0.4973 | 0.4973 | 0.4464 | 0.4973 | 0.4973 |
| Ensemble B | 0.4857 | 0.4857 | 0.4857 | 0.4857 | 0.4857 | 0.4857 |
| Proposed Model With Ensemble A backbone | 0.5053 | 0.5053 | 0.5053 | 0.5053 | 0.4088 | 0.5053 |
| Proposed Model with Ensemble B backbone | 0.5013 | 0.5013 | 0.5013 | 0.5013 | 0.5013 | 0.4991 |
Comparison with the latest state-of-the-art deep learning models
To check the availability of the proposed hybrid deep learning mode, we compared its performance with the latest state-of-the-art deep learning models for binary and multiclass identification scenarios. For this comparison, we focused only on the published findings in the literature starting 2019 up to date. The comparison evaluation results were summarized in Table 12, Table 13 for binary and multiclass scenarios, respectively.
Table 12.
Evaluation performance comparison with the latest state-of-the-art deep learning models for the binary classification scenario.
| Ref. | Year | AI Model Architecture | ACC (%) | PRE (%) | SEN (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Ayan et al. [4] | 2019 | VGG16 Xception |
87 82 |
– – |
82 85 |
– |
| Chouhan et al. [38] | 2020 | CNN | 96 | 96 | 95 | – |
| Yadav et al. [40] | 2019 | CapsNet | 83 | – | – | – |
| Liang et al. [24] | 2020 | CNN VGG16 |
91 74 |
89 72 |
97 95 |
93 82 |
| Asnaoui et al. [41] | 2020 | CNN | 84 | 94 | 78 | 86 |
| Mittal et al. [42] | 2020 | E3CC VGG16 + CapsNet |
82 88 |
– | – | – |
| Jain et al. [26] | 2020 | CNN | 85 | – | 94 | 89 |
| ERDEM et al. [65] | 2020 | CNN | 87 | 86 | 97 | 92 |
| Darici et al. [66] | 2020 | CNN Ensemble |
95 95 |
95 94 |
95 95 |
95 95 |
| Talo et al. [32] | 2019 | ResNet152 | 97 | – | – | – |
| O’Quinn et al. [33] | 2019 | AlexNet | 76 | – | – | – |
| Stephen et al. [67] | 2019 | CNN | 93 | – | – | – |
| Urey et al. [35] | 2019 | ResNet | 78 | – | – | – |
| Khalid et al. [41] | 2020 | CNN VGG16 VGG19 InceptionV3 Xception DenseNet201 MobileNetV2 InceptionResNetV2 ResNet50 |
84 86 86 95 83 94 96 96 97 |
94 88 80 94 96 99 98 99 98 |
– – – – – – – – – |
86 86 85 95 85 94 96 96 97 |
| Mohammad et al. [68] | 2021 | ResNet50 Compound Scaled ResNet50 |
97 98 |
97 98 |
98 98 |
98 98 |
| Chomsin et al. [69] | 2021 | UBNetV1 | 95 | 86 | 97 | 91 |
| Shazia et al. [70] | 2021 | VGG16 VGG19 DenseNet121 InceptionResNetV2 InceptionV3 ResNet50 Xception |
99.09 99.10 99.18 98.21 98.96 99.12 98.34 |
99.28 99.14 99.14 98.79 99.17 99.12 98.83 |
99.09 99.18 99.18 98.21 98.96 99.12 98.34 |
99.14 99.12 99.19 98.38 99.02 99.15 98.49 |
| Juan et al. [71] | 2020 | Xception CNN | 97.3 | 84.3 | 99.2 | 91.2 |
| Rohit et al. [72] | 2021 | Ensemble | 98.81 | 98.82 | 98.80 | 98.79 |
| The proposed Hybrid Deep Learning Model | 2022 | Transformer Encoder | 99.21 | 99.21 | 99.21 | 99.21 |
Table 13.
Evaluation performance comparison with the latest state-of-the-art deep learning models for the multiclass identification scenario.
| Reference | Year | AI Model Architecture | ACC (%) | PRE (%) | SEN (%) | F1-Score (%) |
|---|---|---|---|---|---|---|
| Darici et al. [66] | 2020 | CNN Ensemble |
78 75 |
80 77 |
78 75 |
78 75 |
| Al-antari et al. [61] | 2021 | End-to-end CAD-based YOLO | 97.40 | – | 85.15 | 84.81 |
| Hammoudi et al. [36] | 2020 | VGG19 ResNet + RNN1 ResNet + RNN2 DenseNet169 |
83 78 80 96 |
– – – – |
– – – – |
– – – – |
| Chuhan et al. [38] | 2021 | UBNetV1 UBNetV2 |
88 88 |
89 89 |
86 85 |
86 86 |
| Ibrahim et al. [27] | 2020 | AlexNet | 97.40 | – | – | – |
| The proposed Hybrid Deep Learning Model | 2022 | Transformer Encoder | 98.19 | 97.29 | 97.29 | 97.29 |
From Table 12, transfer learning techniques were seen as the primary approach for detecting pneumonia from normal using the chest X-ray images. The models built from the scratch achieved lower performance compared with the recent transfer learning methods which also affect the employed dataset. Analyzing the transfer learning approaches, the authors of Ref. [21] employed the AlexNet architecture via transfer learning and achieved a binary classification of 72 %. In [22], several deep learning models were implemented for pneumonia detection feature extraction. Among the implemented models the DenseNet169, DenseNet121, Xception, VGG16, VGG19 and ResNet50 were the best. The conventional models were also experimented with for such X-ray pneumonia classification tasks as K-Nearest Neighbor (KNN), Naïve Bayes (NB), Support Vector Machine (SVM), and Random Forest (RF). The authors concluded that DenseNet-169 followed by optimal SVM RBF kernel hyper-parameter values beat all other experimented models. The authors of Ref. [24] stated that the several deep learning customized models had demonstrated encouraging classification results as they all outperformed the pneumonia prediction results by more than 84 % accuracy. They further reported that the InceptionResNetV2 model achieves better results. Judging from experimental results using the same dataset [29], the authors employed several deep learning pre-trained, models, via transfer learning for the task of pneumonia disease identification where the DenseNet121 achieved the best result among the used networks, whereas the InceptionResNetV2 had the lowest performance score. In contrast, the authors of Ref [46] employed the same pre-trained deep learning models via transfer learning for the same task of early pneumonia detection where the Resnet50 achieved the best classification accuracy, while the Xception had the lowest results. Both authors argued that shallow models perform poorly compared to deeper models. Thus, we used the pre-trained models as the network backbone for this experimental study, Table 8 indicated that only a few authors have considered ensemble models [42], which was the second step of the proposed model. Ensemble models give room to understand the task better and yield better results. Since this paper architecture summarizes the identified research, the results of the proposed hybrid model presented its superiority over all other deep learning models listed in the literature by recording 99.21 % in term of evaluation metrics. From the literature, only a few works were seen for multi-class detection as indicated in Table 13. The employed models were basically trained from scratch networks compared to the binary classification, thus meaning that deeper networks are preferred for multi-class tasks and the single models achieved less performance. The proposed architecture attains the best result using the employed evaluation metrics where it achieved an overall accuracy and F1-soce of 98.19 % and 97.29 %, respectively. In general, the improvement of our proposed model is achieved in the range of 0.01 %–24.19 % for the binary classification and in the range of 0.79 %–20.19 % for multi-classification performance in terms of accuracy, precision, sensitivity and F1-Score.
Applications and limitations
This paper presents the early identification of pneumonia from chest X-ray images using the transformer encoder strategy: self-attention network and multilinear perceptron block. This approach would provide more accurate detection and classification accuracy of various lung diseases such as COVID-19 detection, heart diseases, oral cancer, skin cancer, breast cancer, microscopic images for various medical applications. As much as the proposed method outperforms the state-of-the-art AI models, it still has some limitations. The exact affected pneumonia area of the chest X-ray image was not accurately enough indicated. Also, the chest X-ray dataset depicts just a single series for a patient, which supports the argument that a limited dataset (a patient single chest X-ray series) cannot determine if a patient developed or will develop a radiographic finding as the disease progresses.
Future works
Future research will investigate the ability to use the proposed hybrid deep learning for different medical imaging modalities. Furthermore, we would experiment with an automatic parameter-tuning approach and apply the suggested algorithm to numerous medical image collections to conduct a statistical study of its performance. In addition, detecting and classifying X-ray images of pneumonia and lung cancer has become a significant difficulty in recent years, and the proposed technique could be addressed for such issue. The textual explainable besides the visual heatmaps could be more useful once the appropriate annotated text dataset becomes available.
Conclusion
This paper explores the ability of early pneumonia identification from chest X-ray images using the capability of backbone ensemble deep learning as well as Transformer Encoder: Multi-Head Self-Attention Network and MLP Block for better, accurate, and generalized identification performance. Thus, preventing the acute inflammation of the lung cells, which has become one of the commonest among kids under the age of 5, accounting for 15 % of all mortality in underdeveloped nations each year. The backbone architecture of the proposed hybrid framework was an ensemble of pre-trained deep learning models for more successful feature extraction. An end-to-end training using binary and multi-class datasets was carried out using accuracy, F-1 score, sensitivity, specificity, precision, confusion matrix, ROC and PR curves for the model evaluation. We recorded the model's performance developed in three stages, (1) individual pre-trained transfer learning, (2) ensemble deep learning, and (3) the proposed hybrid XAI model based on the ensemble transformer encoder. The proposed hybrid deep learning model recorded 99.21% classification performance in terms of overall accuracy and F1-score in the case of binary classification, while it achieved 98.19% accuracy and 97.29% F1-score for multiclass identification performance. The proposed model has better binary and multi-class classification results than the state-of-the-art deep learning methods based on the employed evaluation metrics, thus showing its superiority and efficiency in the early identification of pneumonia from chest X-ray images. Moreover, the proposed hybrid XAI model shows its capability to provide better and more accurate explainable identification results in terms of heat maps or saliency maps.
Compliance with ethics requirements
All procedures followed were in accordance with the ethical standards research because we have used public available Chest X-ray datasets: Mendeley and Chest X-ray datasets.
CRediT authorship contribution statement
Chiagoziem C. Ukwuoma: Data curation, Methodology. Zhiguang Qin: Conceptualization, Supervision. Md Belal Bin Heyat: Validation, Resources. Faijan Akhtar: Data curation, Writing – original draft. Olusola Bamisile: Writing – original draft, Validation. Abdullah Y. Muad: Formal analysis, Validation. Daniel Addo: Software, Visualization. Mugahed A. Al-antari: Supervision, Writing – review & editing, Funding acquisition.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
This research was supported by the National Natural Science Foundation of China (62027827). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. RS-2022-00166402).
Footnotes
Peer review under responsibility of Cairo University.
Contributor Information
Zhiguang Qin, Email: qinzg@uestc.edu.cn.
Mugahed A. Al-antari, Email: en.mualshz@sejong.ac.kr.
References
- 1.Al-antari M.A., Al-masni M.A., Choi M.T., Han S.M., Kim T.S. A fully integrated computer-aided diagnosis system for digital X-ray mammograms via deep learning detection, segmentation, and classification. Int J Med Inform. 2018;117(May):44–54. doi: 10.1016/j.ijmedinf.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Al-antari M.A., Han S.M., Kim T.S. Evaluation of deep learning detection and classification towards computer-aided diagnosis of breast lesions in digital X-ray mammograms. Comput Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105584. [DOI] [PubMed] [Google Scholar]
- 3.UNICEF. A child dies of pneumonia every 39 seconds; 2018. [Online]. Available from: https://data.unicef.org/topic/child-health/pneumonia.
- 4.Ayan E, Ünver HM. Diagnosis of pneumonia from chest X-ray images using deep learning; 2019. doi: 10.1109/EBBT.2019.8741582.
- 5.Akhtar F, Bin Heyat MB, Li JP, Patel PK, Rishipal Guragai B. Role of machine learning in human stress: a review. In: 2020 17th international computer conference on wavelet active media technology and information processing, ICCWAMTIP 2020; Dec. 2020. p. 170–4. doi: 10.1109/ICCWAMTIP51612.2020.9317396.
- 6.Ukwuoma C.C., Zhiguang Q., Bin Heyat M.B., Ali L., Almaspoor Z., Monday H.N., et al. Recent advancements in fruit detection and classification using deep learning techniques. Math Prob Eng. 2022;2022:1–29. [Google Scholar]
- 7.Guragai B, Alshorman O, Masadeh M, Bin Heyat MB. A survey on deep learning classification algorithms for motor imagery. In: Proceedings of the international conference on microelectronics, ICM, Dec. 2020, vol. 2020-December, p. 1–4. doi: 10.1109/ICM50269.2020.9331503.
- 8.Heyat M.B.B., Akhtar F., Khan M.H., Ullah N., Gul I., Khan H., et al. Detection, treatment planning, and genetic predisposition of bruxism: a systematic mapping process and network visualization technique. CNSNDDT. 2021;20(8):755–775. doi: 10.2174/1871527319666201110124954. [DOI] [PubMed] [Google Scholar]
- 9.Bin Heyat MB et al. A novel hybrid machine learning classification for the detection of bruxism patients using physiological signals. Appl. Sci. 2020;10(21):1–16. doi: 10.3390/app10217410.
- 10.AlShorman O., Masadeh M., Heyat M.B.B., Akhtar F., Almahasneh H., Ashraf G.M., et al. Frontal lobe real-time EEG analysis using machine learning techniques for mental stress detection. J Integr Neurosci. 2022;21(1):020. doi: 10.31083/j.jin2101020. [DOI] [PubMed] [Google Scholar]
- 11.Bin Heyat MB et al. Wearable flexible electronics based cardiac electrode for researcher mental stress detection system using machine learning models on single lead electrocardiogram signal. Biosensors 2022;12(6):427. doi: 10.3390/bios12060427. [DOI] [PMC free article] [PubMed]
- 12.Teelhawod BN et al. Machine learning in E-health: a comprehensive survey of anxiety. In: 2021 International conference on data analytics for business and industry, ICDABI 2021; Oct. 2021. p. 167–12. doi: 10.1109/ICDABI53623.2021.9655966.
- 13.Al-masni M.A., Al-antari M.A., Park J.-M., Gi G., Kim T.-Y., Rivera P., et al. Simultaneous detection and classification of breast masses in digital mammograms via a deep learning YOLO-based CAD system. Comput Methods Programs Biomed. 2018;157:85–94. doi: 10.1016/j.cmpb.2018.01.017. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, Lyu S, Wang X, Zhao Q. TPH-YOLOv5: improved YOLOv5 based on transformer prediction head for object detection on drone-captured scenarios. In: Proc IEEE Int Conf Comput Vis, vol. 2021-Octob; 2021. p. 2778–88. doi: 10.1109/ICCVW54120.2021.00312.
- 15.Vaswani A et al. Attention is all you need attention is all you need. Adv Neural Inf Process Syst 2017:5999–6009.
- 16.Alayrac J.B., et al. Self-supervised multimodal versatile networks. Adv Neural Inf Process Syst. 2020:25–37. [Google Scholar]
- 17.Shamshad F et al. Transformers in medical imaging: a survey. arXiv preprint arXiv:2201.09873; 2022. p. 1–41.
- 18.Badawi A., Elgazzar K. Detecting coronavirus from chest X-rays using transfer learning. Covid. 2021;1(1):403–415. doi: 10.3390/covid1010034. [DOI] [Google Scholar]
- 19.Albahli S., Rauf H.T., Algosaibi A., Balas V.E. AI-driven deep CNN approach for multilabel pathology classification using chest X-Rays. PeerJ Comput Sci. 2021;7:1–17. doi: 10.7717/peerj-cs.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albahli S., Rauf H.T., Arif M., Nafis M.T., Algosaibi A. Identification of thoracic diseases by exploiting deep neural networks. Comput Mater Contin. 2020;66(3):3139–3149. doi: 10.32604/cmc.2021.014134. [DOI] [Google Scholar]
- 21.Woźniak M., Połap D. Bio-inspired methods modeled for respiratory disease detection from medical images. Swarm Evol Comput. 2018;41:69–96. doi: 10.1016/j.swevo.2018.01.008. [DOI] [Google Scholar]
- 22.Ukwuoma CC et al. Holistic attention on pooling based cascaded partial decoder for real- time salient object detection; 2021. doi: 10.1109/PRAI53619.2021.9551094.
- 23.Rajinikanth V, Kadry S, Taniar D, Damasevicius R, Rauf HT. Breast-cancer detection using thermal images with marine-predators-algorithm selected features. In: Proc 2021 IEEE 7th Int Conf Bio Signals, Images Instrumentation, ICBSII 2021; 2021. p. 1–6. doi: 10.1109/ICBSII51839.2021.9445166.
- 24.Liang G., Zheng L. A transfer learning method with deep residual network for pediatric pneumonia diagnosis. Comput Methods Programs Biomed. 2020;187 doi: 10.1016/j.cmpb.2019.06.023. [DOI] [PubMed] [Google Scholar]
- 25.Lu C., Zhu R., Yu F., Jiang X., Liu Z., Dong L., et al. Gear rotational speed sensor based on FeCoSiB/Pb(Zr, Ti)O3 magnetoelectric composite. Meas J Int Meas Confed. 2021;168:108409. [Google Scholar]
- 26.Jain R., Nagrath P., Kataria G., Sirish Kaushik V., Jude Hemanth D. Pneumonia detection in chest X-ray images using convolutional neural networks and transfer learning. Measurement. 2020;165:108046. [Google Scholar]
- 27.Ibrahim A.U., Ozsoz M., Serte S., Al-Turjman F., Yakoi P.S. Pneumonia classification using deep learning from chest X-ray images during COVID-19. Cognit Comput. 2021 doi: 10.1007/s12559-020-09787-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mirmohammadi S., Yazdani J., Etemadinejad S., Asgarinejad H. A cross-sectional study on work-related musculoskeletal disorders and associated risk factors among hospital health cares. Proc Manuf. 2015;3:4528–4534. doi: 10.1016/j.promfg.2015.07.468. [DOI] [Google Scholar]
- 29.Rahman T., Chowdhury M.E.H., Khandakar A. Applied sciences transfer learning with deep convolutional neural network (CNN) for pneumonia detection using. MDPI J Appl Sci. 2020;3233:1–17. [Google Scholar]
- 30.Elshennawy NM, Ibrahim DM. Deep-pneumonia framework using deep learning models based on chest X-ray images. Diagnostics 2020;10(9). doi: 10.3390/diagnostics10090649. [DOI] [PMC free article] [PubMed]
- 31.Wang X, Peng Y, Lu L, Lu Z, Bagheri M, Summers RM. ChestX-ray8: hospital-scale chest X-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. In: Proc - 30th IEEE Conf Comput Vis Pattern Recognition, CVPR 2017, vol. 2017-January; 2017. p. 3462–71. doi: 10.1109/CVPR.2017.369.
- 32.Talo M. Pneumonia detection from radiography images using convolutional neural networks. In: 27th signal processing and communications applications conference, SIU 2019; Apr. 2019. p. 1–4. doi: 10.1109/SIU.2019.8806614.
- 33.O’Quinn W, Haddad RJ, MooreDL. Pneumonia radiograph diagnosis utilizing deep learning network; 2019. doi: 10.1109/ICEICT.2019.8846438.
- 34.Varshni D, Thakral K, Agarwal L, Nijhawan R, Mittal A. Pneumonia detection using CNN based feature extraction. In: Proceedings of 2019 3rd IEEE international conference on electrical, computer and communication technologies, ICECCT 2019; Feb. 2019. p. 1–7, doi: 10.1109/ICECCT.2019.8869364.
- 35.Urey DY, Saul CJ, Taktakoglu CD. Early diagnosis of pneumonia with deep learning. 2019;1904:00937. [Google Scholar]
- 36.Hammoudi K., Benhabiles H., Melkemi M., Dornaika F., Arganda-Carreras I., Collard D., et al. Deep learning on chest X-ray images to detect and evaluate pneumonia cases at the era of COVID-19. J Med Syst. 2021;45(7) doi: 10.1007/s10916-021-01745-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jaiswal R.K. Position-based routing protocol using Kalman filter as a prediction module for vehicular ad hoc networks. Comput Electr Eng. 2020;83 doi: 10.1016/j.compeleceng.2020.106599. [DOI] [Google Scholar]
- 38.Chouhan V., Singh S.K., Khamparia A., Gupta D., Tiwari P., Moreira C., et al. A novel transfer learning based approach for pneumonia detection in chest X-ray images. Appl Sci. 2020;10(2):559. [Google Scholar]
- 39.Siddiqi R. ACM international conference proceeding series. 2019. Automated pneumonia diagnosis using a customized sequential convolutional neural network; pp. 64–70. [DOI] [Google Scholar]
- 40.Yadav S.S., Jadhav S.M. Deep convolutional neural network based medical image classification for disease diagnosis. J Big Data. 2019;6(1):1–18. [Google Scholar]
- 41.El Asnaoui K, Chawki Y, Idri A. Automated methods for detection and classification pneumonia based on X-ray images using deep learning; 2021.
- 42.Mittal A et al. Detecting pneumonia using convolutions and dynamic capsule routing for chest X-ray images. Sensors (Switzerland) 2020;20(4). doi: 10.3390/s20041068. [DOI] [PMC free article] [PubMed]
- 43.Li L., Xu M., Liu H., Li Y., Wang X., Jiang L., et al. A large-scale database and a CNN model for attention-based glaucoma detection. IEEE Trans Med Imaging. 2020;39(2):413–424. doi: 10.1109/TMI.2019.2927226. [DOI] [PubMed] [Google Scholar]
- 44.Fu J., Li W., Du J., Huang Y. A multiscale residual pyramid attention network for medical image fusion. Biomed Signal Process Control. 2021;66 doi: 10.1016/j.bspc.2021.102488. [DOI] [Google Scholar]
- 45.Gu R., Wang G., Song T., Huang R., Aertsen M., Deprest J., et al. CA-Net: comprehensive attention convolutional neural networks for explainable medical image segmentation. IEEE Trans Med Imaging. 2021;40(2):699–711. doi: 10.1109/TMI.2020.3035253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hu J., Shen L., Albanie S., Sun G., Wu E. Squeeze-and-excitation networks. IEEE Trans Pattern Anal Mach Intell. 2020;42(8):2011–2023. doi: 10.1109/TPAMI.2019.2913372. [DOI] [PubMed] [Google Scholar]
- 47.Zhang QL, Bin Yang Y. ECA-Net: Efficient channel attention for deep convolutional neural networks. In: ICASSP, IEEE Int Conf Acoust Speech Signal Process - Proc, vol. 2021-June; 2021. p. 2235–9. doi: 10.1109/ICASSP39728.2021.9414568.
- 48.Li B, Kang G, Cheng K, Zhang N. Attention-guided convolutional neural network for detecting pneumonia on chest X-rays. In: Proc Annu Int Conf IEEE Eng Med Biol Soc, EMBS; 2019. p. 4851–4. doi: 10.1109/EMBC.2019.8857277. [DOI] [PubMed]
- 49.Guo X., Yuan Y. Triple ANet: Adaptive abnormal-aware attention network for WCE image classification. Lect Notes Comput Sci (including Subser Lect Notes Artif Intell Lect Notes Bioinformatics) 2019;11764:293–301. doi: 10.1007/978-3-030-32239-7_33. [DOI] [Google Scholar]
- 50.Khan S., Naseer M., Hayat M., Zamir S.W., Khan F.S., Shah M. Transformers in vision: a survey. ACM Comput Surv. 2022 doi: 10.1145/3505244. [DOI] [Google Scholar]
- 51.Dosovitskiy A et al. An image is worth 16×16 words: transformers for image recognition at scale. arXiv preprint arXiv:2010.11929; 2020.
- 52.Chen H., et al. Proc IEEE Comput Soc Conf Comput Vis Pattern Recognit. 2021. Pre-trained image processing transformer; pp. 12294–12305. [DOI] [Google Scholar]
- 53.Park S., Kim G., Oh Y., Seo J.B., Lee S.M., Kim J.H., et al. Multi-task vision transformer using low-level chest X-ray feature corpus for COVID-19 diagnosis and severity quantification. Med Image Anal. 2022;75 doi: 10.1016/j.media.2021.102299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kermany D.S., Goldbaum M., Cai W., Valentim C.C.S., Liang H., Baxter S.L., et al. Identifying medical diagnoses and treatable diseases by image-based deep learning. Cell. 2018;172(5):1122–1131.e9. doi: 10.1016/j.cell.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 55.Chowdhury M.E.H., Rahman T., Khandakar A., Mazhar R., Kadir M.A., Mahbub Z.B., et al. Can AI help in screening viral and COVID-19 pneumonia? IEEE Access. 2020;8:132665–132676. [Google Scholar]
- 56.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: Proceedings of the IEEE computer society conference on computer vision and pattern recognition, vol. 2016-Decem; 2016. p. 770–8. doi: 10.1109/CVPR.2016.90.
- 57.Chollet F. Xception: deep learning with depthwise separable convolutions. In: Proceedings - 30th IEEE conference on computer vision and pattern recognition, CVPR 2017, vol. 2017-January; 2017. p. 1800–7. doi: 10.1109/CVPR.2017.195.
- 58.Szegedy C et al. Going deeper with convolutions (GoogleLeNet). J Chem Technol Biotechnol 2016;91(8).
- 59.Tan M, Le QV. EfficientNet: Rethinking model scaling for convolutional neural networks. In: 36th International conference on machine learning, ICML 2019, vol. 2019-June; 2019. p. 10691–700.
- 60.Szegedy C, Ioffe S, Vanhoucke V, Alemi A. Inception-v4, inception-ResNet and the impact of residual connections on learning. In: Proc AAAI Conf Artif Intell, Vol. 31, no. 1; Feb. 2017. doi: 10.1609/aaai.v31i1.11231.
- 61.Al-antari M.A., Hua C.H., Bang J., Lee S. Fast deep learning computer-aided diagnosis of COVID-19 based on digital chest X-ray images. Appl Intell. 2021;51(5):2890–2907. doi: 10.1007/s10489-020-02076-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus COVID-19 detection from X-rays. Comput Methods Programs Biomed. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kremers R. Artificial intelligence. Lev Des. AK Peters/CRC Press. 2009:341–368. doi: 10.1201/b10933-22.
- 64.Irvin J et al. CheXpert: a large chest radiograph dataset with uncertainty labels and expert comparison. In: 33rd AAAI Conf Artif. Intell AAAI 2019, 31st Innov Appl Artif Intell Conf IAAI 2019 9th AAAI Symp Educ Adv Artif Intell, EAAI 2019; 2019. p. 590–7. doi: 10.1609/aaai.v33i01.3301590.
- 65.Erdem E, Aydin T. Detection of pneumonia with a novel CNN-based approach. Sak Univ J Comput Inf Sci 2021;4(1):26–34. doi: 10.35377/saucis.04.01.787030.
- 66.Paquin F., Rivnay J., Salleo A., Stingelin N., Silva C. Pneumonia detection and classification using deep learning on chest X-Ray images Muazzez. J Mater Chem C. 2015;3(4):10715–10722. doi: 10.1039/b000000x. [DOI] [Google Scholar]
- 67.Stephen O., Sain M., Maduh U.J., Jeong D.U. An efficient deep learning approach to pneumonia classification in healthcare. J Healthc Eng. 2019;2019:1–7. doi: 10.1155/2019/4180949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hashmi M.F., Katiyar S., Hashmi A.W., Keskar A.G. Pneumonia detection in chest X-ray images using compound scaled deep learning model. Automatika. 2021;62(3–4):397–406. doi: 10.1080/00051144.2021.1973297. [DOI] [Google Scholar]
- 69.Widodo C.S., Naba A., Mahasin M.M., Yueniwati Y., Putranto T.A., Patra P.I. UBNet: deep learning-based approach for automatic X-ray image detection of pneumonia and COVID-19 patients. J X-ray Sci Technol. 2022;30(1):57–71. doi: 10.3233/XST-211005. [DOI] [PubMed] [Google Scholar]
- 70.Shazia A., Xuan T.Z., Chuah J.H., Usman J., Qian P., Lai K.W. A comparative study of multiple neural network for detection of COVID-19 on chest X-ray. EURASIP J Adv Signal Process. 2021;2021(1):1–16. doi: 10.1186/s13634-021-00755-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Luján-García JE, Yáñez-Márquez C, Villuendas-Rey Y, Camacho-Nieto O. A transfer learning method for pneumonia classification and visualization. Appl Sci 2020;10(8). doi: 10.3390/APP10082908.
- 72.Kundu R, Das R, Geem ZW, Han GT, Sarkar R. Pneumonia detection in chest X-ray images using an ensemble of deep learning models. PLoS One 2021;16(9). doi: 10.1371/journal.pone.0256630. [DOI] [PMC free article] [PubMed]