Abstract
Background
Adamantinoma craniopharyngioma (ACP) is a non-malignant tumour of unknown pathogenesis that frequently occurs in children and has malignant potential. The main treatment options are currently surgical resection and radiotherapy. These treatments can lead to serious complications that greatly affect the overall survival and quality of life of patients. It is therefore important to use bioinformatics to explore the mechanisms of ACP development and progression and to identify new molecules.
Methods
Sequencing data of ACP was downloaded from the comprehensive gene expression database for differentially expressed gene identification and visualized by Gene Ontology, Kyoto Gene, and gene set enrichment analyses (GSEAs). Weighted correlation network analysis was used to identify the genes most strongly associated with ACP. GSE94349 was used as the training set and five diagnostic markers were screened using machine learning algorithms to assess diagnostic accuracy using receiver operating characteristic (ROC) curves, while GSE68015 was used as the validation set for verification.
Results
Type I cytoskeletal 15 (KRT15), Follicular dendritic cell secreted peptide (FDCSP), Rho-related GTP-binding protein RhoC (RHOC), Modulates negatively TGFB1 signaling in keratinocytes (CD109), and type II cytoskeletal 6A (KRT6A) (area under their receiver operating characteristic curves is 1 for both the training and validation sets), Nomograms constructed using these five markers can predict progression of ACP patients. Whereas ACP tissues with activated T-cell surface glycoprotein CD4, Gamma delta T cells, eosinophils and regulatory T cells were expressed at higher levels than in normal tissues, which may contribute to the pathogenesis of ACP. According to the analysis of the CellMiner database (Tumor cell and drug related database tools), high CD109 levels showed significant drug sensitivity to Dexrazoxane, which has the potential to be a therapeutic agent for ACP.
Conclusions
Our findings extend understandings of the molecular immune mechanisms of ACP and suggest possible biomarkers for the targeted and precise treatment of ACP.
Keywords: Adamantinomatous craniopharyngioma (ACP), weighted correlation network (WGANA), machine learning, immune infiltration, therapeutic
Highlight box.
Key findings
• CP is prone to local recurrence, and the disease is prone to lead to neuroendocrine disorders. At present, the treatment of CP is currently based on surgical excision, and the diagnosis of CP relies on imaging. We sought to find new diagnostic and therapeutic methods for CP.
What is known and what is new?
• The CP main treatment methods are surgical resection and radiotherapy.
• Models constructed for KRT15, FDCSP, RHOC, CD109, and KRT6A could serve as new diagnostic tools for CP.
What is the implication, and what should change now?
• RHOC high expression showed significant drug sensitivity to LDK-378 (Ceritinib), oxaliplatin, Palbociclib, and CUDC-305 (Hsp90 Inhibitor Debio 0932). This provides new ideas for the direction we should take in targeting pc drug therapy.
Introduction
Craniopharyngioma (CP) is a low-grade aggressive intracranial tumor (1). There are 2 histological subtypes of CP (i.e., adamantine tumor and papilloma), both of which occur as embryonal brain tumors in the saddle and para-saddle regions, are usually benign central nervous system tumors, and account for 2–3% of intracranial tumors (2). adamantinoma craniopharyngioma (ACP) can develop at any age and shows a bimodal distribution in age. In terms of the histopathology, the adamantine type often presents in children aged 5–14 years, and the papillary subtype predominantly presents in middle-aged and elderly people aged 50–74 years (2).
The ACP may progress from the pterygoid saddle upwards to the third ventricle, affecting the hypothalamic-pituitary and visual nerve pathway area. Patients may also develop symptoms due to the mass effect and infiltration of the surrounding tissues. Due to the close proximity of important neurovascular structures, such as the pituitary, hypothalamus, optic nerve, and internal carotid artery, endocrine disorders and abnormalities in hypothalamic function and visual disturbances are often observed before and after ACP, resulting in reduced survival and a poor quality of life for ACP patients (1,3-5).
The current tools of choice for the diagnosis of ACP are computed tomography (CT) and magnetic resonance imaging (MRI). MRI shows the relationship between the tumor and the surrounding neurovascular anatomy. On the MRI of the ACP, the lesion is heterogeneously enhanced, with both solid and cystic portions. Both the solid portion and the cystic wall undergo inhomogeneous enhancement (1).
The majority of patients choose hospital treatments for non-specific manifestations of increased intracranial pressure (e.g., nausea and headache), visual impairment (loss of vision and the visual field) (62–84%), and endocrine defects (52–87%) (6). Additionally, many patients experience growth disturbances and significant weight gain before diagnosis (3). Thus, this disease can be physically and psychologically devastating for patients.
In terms of the typical imaging features of ACP, about 90% of tumors are predominantly cystic; 90% of tumors have more or less visible calcifications; and about 90% of tumors are contrast visible in the cyst wall (2). On MRI, the solid portion and the cyst wall may show various T1 signals. On T2-weighted images, tumors usually show both hypo- and hyper-enhancing manifestations (7,8). Calcification in tumors is usually not possible with MRI. The ideal sequence for identifying calcification is a T2- or susceptibility-weighted sequence, but both sequences are affected by the amount of air in the sinus. Thus, CT presentation of peritumoral calcification is currently the gold standard for the diagnosis of craniopharyngioma, and there is no diagnostic modality that is specific for CPs, which is an area that requires further research. Bioinformatics analysis may enable the screening of a biomarker to specifically diagnose ACP and predict the progression of the disease in patients.
To date, the main treatment option for ACP has been surgery. The clinical aim is to remove the tumor as completely as possible without leaving sequelae, such as neuroendocrine disorders. However, as the tumor is located in the suprasellar or parasellar regions, it is highly susceptible to postoperative complications (due to damage to the hypothalamic-pituitary and optic nerve pathways). These complications reduce the quality of life of patients after surgery and increase the morbidity and mortality of patients suffering from cardiovascular disease (9). Consequently, the clinical treatment options often include maximal and safe surgical resection and thus also increase the risk of long-term recurrence and morbidity, and patients often have to undergo follow-up combination therapy, such as radiation and chemotherapy after surgery. However, these treatments often result in patients being accompanied by a variety of complications (10).
Proton-beam therapy has recently been found to be useful in the treatment of ACP, but due to its high costs and the fact that the clinical dose is unclear due to tissue heterogeneity, it requires further study (11,12). Chemotherapy is now extensively used as an active treatment for a large variety of cancers. Chemotherapy can be used preoperatively to make tumor tissue smaller and facilitate surgical removal, or it can be used to treat recurrent tumors to destroy residual tumor cells. However, chemotherapy is not the most efficacious treatment modality for ACP, and many studies have shown that better results can be achieved by direct intracranial injection of the chemotherapeutic agents bleomycin and interferon-α (13,14).
Immunotherapy is known to have a huge effect in the treatment of many tumors, and the role of immunotherapy in the development of primary and secondary ACP has been closely examined. Many potential targets for immunotherapy of ACP are being investigated, but no suitable targeted therapeutic agent has yet found its way into clinical trials for the treatment of adult ACP (15). Thus, a marketed immunotherapeutic target for ACP needs to be identified to provide a novel and feasible option for the postoperative treatment of ACP.
In this study, the GSE94349 data set was downloaded for ACP-EDGS (differentially expressed genes). A weighted correlation network analysis (WGCNA) least absolute shrinkage and selection operator (LASSO) models, and 4 machine-learning algorithms were used to screen the core diagnostic markers. The final marker genes were then validated using the GSE68015 data set. Next, the correlations between the immune infiltrating cells and biomarkers were analyzed. The CellMiner database (Tumor cell and drug related database tools) was then used to predict the drug sensitivity of the identified diagnostic biomarkers.
Our systematic analysis identified ACP diagnostic genes and their molecular immune mechanisms and therapeutic agents. This study also explored these diagnostic marker targets and their corresponding potentially therapeutic small molecule drugs. Our findings may provide novel ideas for the subsequent immunotherapeutic targeting of drugs for ACP. This article is presented in accordance with the TRIPOD reporting checklist (available at https://tp.amegroups.com/article/view/10.21037/tp-23-152/rc).
Methods
Data download
The ACP expression profile GSE68015 data set (comprising 15 ACP sample tissues and 16 control brain sample tissues) and the GSE94349 data set (comprising 9 ACP sample tissues and 17 control brain sample tissues) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/) using “craniopharyngioma” as the keyword and selecting “Homo sapiens” as the species. The platform data were downloaded from GPL 570 (HG-U133_Plus_2) Affymetrix Human Genome U133+2.0 Array. The raw data from the dataset was extracted and gene names were transformed using Perl software. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Identification of ACP-EDG
The data from the GSE94349 data set were used in the Limma (16) package to screen the 9 ACP sample tissues and 17 control brain sample tissues for differentially expressed genes (DEGs) in ACP. The following screening criteria were set: |log fold change (FC)| ≥1, adjusted P value <0.05. The results were also presented in volcano plots and heatmaps using the ggplot2 package.
Functional and pathway enrichment analyses
The Gene Ontology (GO) and Kyoto Gene and Genome Encyclopedia (KEGG) enrichment analyses were performed on the ACP-EDG data using R packages, such as clusterProfiler, org.Hs.eg.db, and enrichplot (17-19). The GO analyses included the annotation of the molecular functions (MFs), biological processes (BPs), and cellular components (CCs). A false discovery rate <1 and q value <0.05 indicated significant enrichment. The enrichment results were visualized using the ggplot2 package.
PPI network construction and identification of central genes
The interaction information for the proteins with a minimum required interaction score of >0.9 was obtained by importing the ACPs-EDG data into the search tool of the STRING database (20) (https://string-db.org/). The CytoNCA plug-in in the Cytoscape software (21) was used to visualize the network and identify the key active ingredients and key candidate targets.
Weighted co-expression network construction
The co-expression networks for the ACP disease module were constructed using the “WGCNA” package in R (22). Genes were aligned using Pearson correlation matrices. Outliers were detected by clustering the samples, and dissimilarity was calculated for the module signature genes, and the modules were then merged. The module-trait relationships between the modular trait genes and clinical traits were also analyzed, and the most important trait modules and DEGs were intersected to find the common trait genes for the clinical traits and DEGs.
Model building and validation of machine-learning methods
The common feature genes of the identified clinical traits and DEGs were validated using R software, and the most relevant genes were found by a LASSO regression crossover. Algorithms, such as the extreme gradient boosting (XGBoost) model, random-forest (RF) tree model, generalized linear model (GLM), and the support vector machine (SVM) model, were then use to construct the model, and the reverse cumulative distribution of the residuals and pROC package were used to plot the 4 method receiver operating characteristic (ROC) curve area under the curve (AUC). The values of the above results were combined to determine the best machine learning feature model, and rms packages were used to plot the column line plots for the best prediction models (23). Finally, the ROC curves were used in the GSE68015 data set to verify the diagnostic power of the model (the area under the ROC curve is greater than 0.5 and the diagnostic test has some diagnostic value. Also, the closer the area under the ROC curve is to 1, the better the authenticity of the diagnostic test.).
Immune cell infiltration analysis
We used the single-sample gene set enrichment analysis (GSEA) calculated by the GSVA package and the inverse convolution algorithm provided by the TIMER (http://timer.cistrome.org/) database for the analysis (24). The immune infiltration of ACP and the correlation between the model genes and immune cells were calculated separately. The “ggplot2” package was used to draw small maps of the distribution of immune cells and their differences.
GSEA
The c2.all.v7.0.symbols.gmt from MSigDB Collections was selected as the reference gene set, and a GSEA was performed using the clusterProfiler package to determine the significant functional and pathway differences between the high and low expression groups to explore the effect of ACP and its diagnostic genes on disease development mechanisms, and Limma package was used to visualize the results (19).
Drug sensitivity analysis of diagnostic markers
The CellMiner database (https://discover.nci.nih.gov/cellminer/home.do) was used to find the relevant drugs for the model genes (25). Using Limma package, the Pearson correlation coefficients between each gene expression and different drugs were calculated separately, and the results were visualized using ggplot2.
Statistical analyses
The data were analyzed using the same methodology as that described previously, one-way ANOVA was performed to compare the differences among multiple groups (≥2 groups). The Student’s t-test was used to compare the differences between the two groups. Statistical significance was set at P<0.05 (*, P<0.05; **, P<0.01; ***, P<0.001) (23).
Results
Study protocol
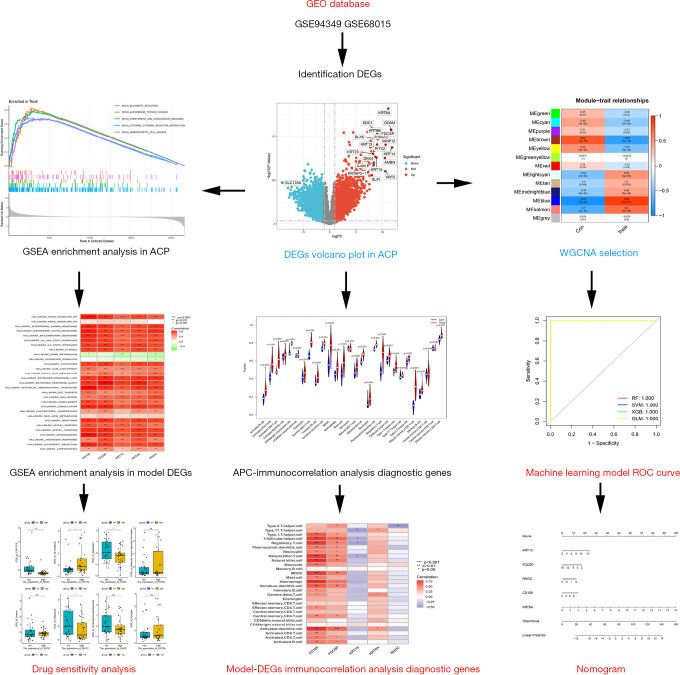
Figure 1 shows the general research process. First, the DEGs was screened using the GSE94349 data set, and the DEGs causing the disease and the possible mechanisms were explored by GO and KEGG analyses. A WGCNA was then conducted to identify the genes most closely correlated with the clinical traits of ACP, and four machine learning methods were used to screen model genes using lasso regression analysis and validate the diagnostic model using GSE68015. Finally, correlations between the individual model genes and immune cells were explored and the CellMiner database was used to predict the drug sensitivity of the final model markers and to screen for possible drugs to treat APC.
Figure 1.
Flow chart of the systematic analysis of the diagnostic genes and pharmacological treatment of ACP. GEO, Gene Expression Omnibus; DEGs, differentially expressed genes; GSEA, gene set enrichment analyse; ACP, adamantinomatous craniopharyngioma; WGANA, weighted correlation network; APC, adamantinomatous craniopharyngioma; ROC, receiver operating characteristic; SVM, support vector machine; GLM, generalized linear model; RF, random forest; XGB, extreme gradient boosting; ns, no statistical difference.
Identification of ACP-EDG
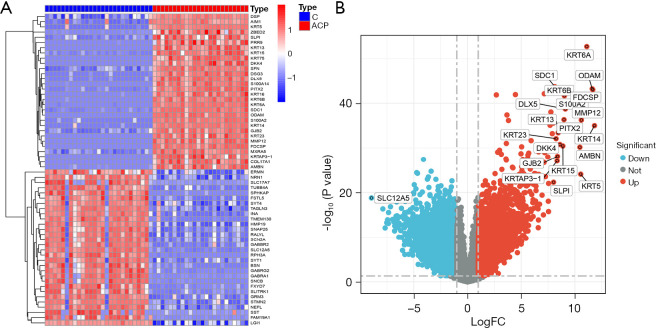
The 9 ACP samples in the GSE94349 data set were compared to the 17 control normal brain tissue samples, and 4,899 genes (of which 2,563 were upregulated and 2,336 were downregulated) were found to be significantly differentially expressed in ACP. Among which, KRT14 (type I cytoskeletal 14), FDCSP (follicular dendritic cell secreted peptide), and ODAM (odontogenic ameloblast-associated protein) were the top 3 most upregulated genes based on the ploidy changes, and SLC12A5 (solute carrier family 12 member 5), INA (class-IV neuronal intermediate filament that is able to self-assemble), and STMN2 (stathmin-2) were the top 3 most downregulated genes. The results are shown in a heat map (Figure 2A) and volcano map (Figure 2B).
Figure 2.
DEGs in ACP. (A) Heat maps of the DEGs in ACP; (B) the DEGs volcano plot in ACP. DEGs, differentially expressed genes; ACP, adamantinomatous craniopharyngioma; FC, fold change; C, control group.
GO KEGG enrichment analyses of ACP-EDG
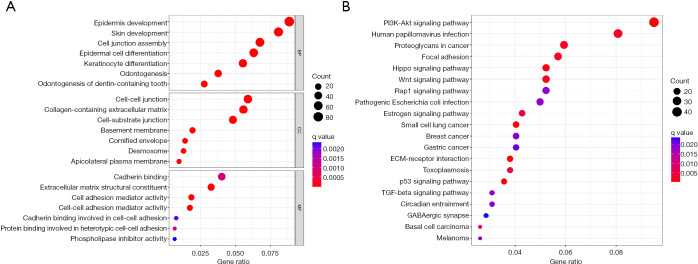
GO and KEGG enrichment analyses were then performed on the 1,045 common DEGs using R software (Figure 3). The GO analysis revealed that the biological process mainly included epidermis development, epidermal cell differentiation, cell junction assembly and keratinocyte differentiation, the cellular component mainly included desmosome, collagen-containing extracellular matrix, cell-cell junction, cornified envelope, basement membrane, and cell-substrate junction, and the molecular function mainly included cell-cell adhesion mediator activity, extracellular matrix structural constituent, cell adhesion mediator activity, cadherin binding and protein binding involved in heterotypic cell-cell adhesion, cadherin binding involved in cell-cell adhesion, and phospholipase inhibitor activity.
Figure 3.
Enrichment analysis of clinically relevant DEGs. (A) GO analysis of DEGs; (B) KEGG pathway analysis of DEGs. GO, gene ontology; BP, biological process; CC, cellular component; MF, molecular function; PI3K, phosphatidylinositol 3-kinase; ECM, extracellular matrix; TGF, transforming growth factor; KEGG, Kyoto Encyclopedia of Genes and Genomes; DEGs, differentially expressed genes.
In addition, these 1,045 genes were present in a variety of signaling pathways, such as the PI3K-Akt signaling pathway, p53 signaling pathway, Hippo signaling pathway, Rap1 signaling pathway, TGF-beta signaling pathway, estrogen signaling pathway, Wnt signaling pathway and proteoglycans in cancer. These signaling pathways are all associated with tumor development.
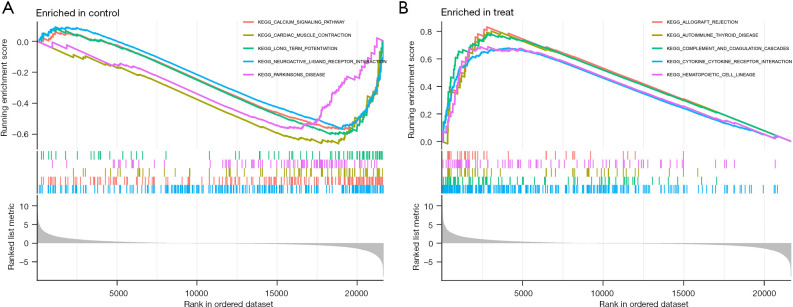
We also conducted a pathway enrichment analysis of the ACP and normal brain tissues (Figure 4). We found that the following signaling pathways were active in the normal tissues: cardiac muscle contraction, calcium signaling pathway, long-term potentiation, Parkinson’s disease, and neuroactive ligand receptor interaction. While the following signaling pathways were active in the ACP tissues: allograft rejection, autoimmune thyroid disease, complement and coagulation cascades, cytokine-cytokine receptor interaction, and hematopoietic cell lineage. These results may direct future research on the pathogenesis of APC.
Figure 4.
Pathway correlation exploration in ACP and normal brain tissues. (A) GSEA of Normal brain tissue in the pathway. (B) GSEA of ACP in the pathway. GSEA, gene set enrichment analysis; ACP, adamantinomatous craniopharyngioma; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Potential core proteins of the ACP
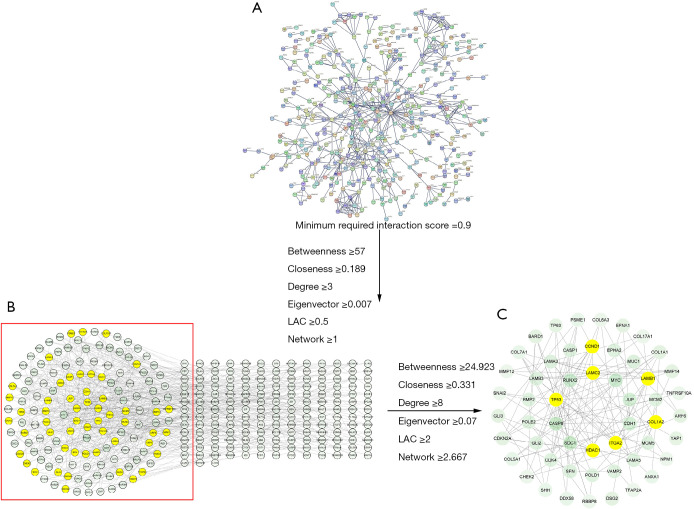
The interaction information of the proteins with a minimum required interaction score of >0.9 was obtained by importing the 1,045 common genes from the WGCNA analysis of the ACP samples into the STRING database (Figure 5A), The protein-protein interactions included 315 nodes and 607 edges (Figure 5B). A total of 262 nodes and 1,204 edges were included in the network diagram based on the following selection criteria: betweenness ≥57; closeness ≥0.189; degree ≥3; eigenvector ≥0.007; LAC (local average connectivity) ≥0.5; and network ≥1. The core network, which comprised 53 nodes and 132 edges, was screened using the following selection criteria: betweenness ≥24.923; closeness ≥0.331; degree ≥8; eigenvector ≥0.069; LAC ≥2; and network ≥2.667. The core genes of the network were laminin subunit beta-1 (LAMB1), histone deacetylase 1 (HDAC1), G1/S-specific cyclin-D1 (CCND1), integrin alpha-2 (ITGA2), and collagen alpha-2(I) chain (COL1A2), cellular tumor antigen p53 (TP53), laminin subunit gamma-2 (LAMC2) (Figure 5C).
Figure 5.
Protein-protein interaction networks of the clinically relevant DEGs. (A) The protein-protein interaction network comprised 1,049 clinically relevant DEGs based on minimum required interaction score of >0.9; (B) initially censored protein-protein interaction network (315 nodes, and 607 edges); (C) core DEG protein-protein interaction network (53 nodes, and 132 edges). LAC, local average connectivity; DEGs, differentially expressed genes.
WGCNA analysis
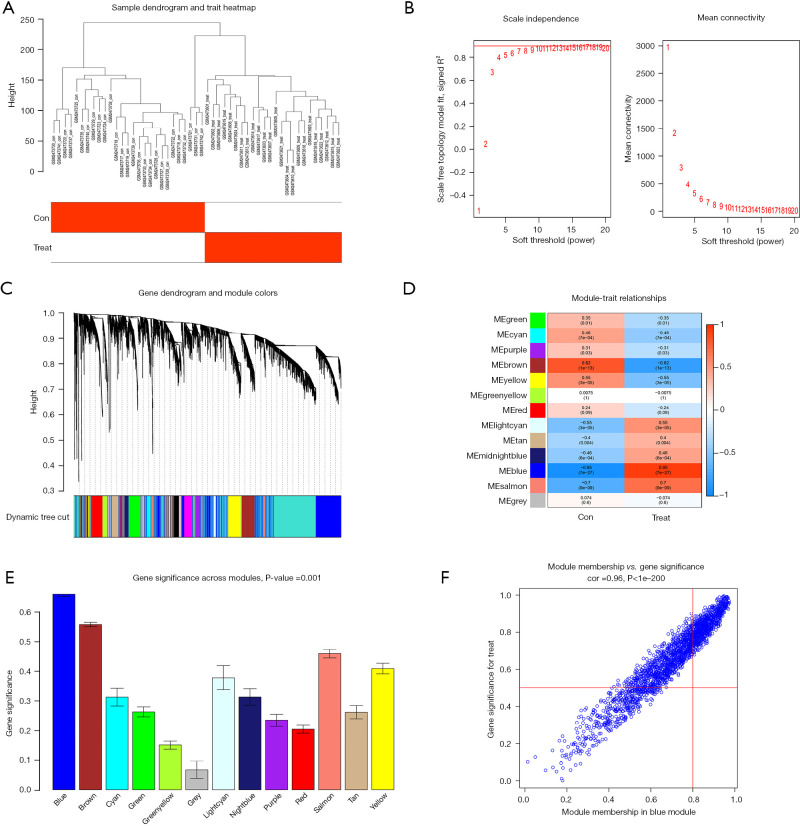
We first performed a sample clustering analysis of the GSE94349 data set and then set the correlation soft threshold to select the top 25% of genes with the largest fluctuations for the WGCNA analysis. We removed the outlier samples and included the remaining samples in the analysis (Figure 6A). As Figure 6B shows, at a power of 8, the scale independence reached 0.9, and the average connectivity was high. The co-expression modules were constructed using a power of 8, and the preliminary module delineation results were obtained with different modules identified by different colors (Figure 6C). The expression correlation with the ACP region in the blue module was 0.95 (P=7.0E-27) (Figure 6D), and there were 1,046 genes co-expressed in this module of the hub genes. The blue module was identified as the most important module. The horizontal coordinate represents the module gene classification type, and the vertical coordinate represents the module gene importance score. The blue module had a score of >0.6 (Figure 6E). A correlation plot of the modules and diseases is shown in Figure 6F, and the correlation between the blue module and diseases was 0.96 (P<1e-200).
Figure 6.
WGCNA selection of the ACP disease-related modules. (A) Outliers were detected in the sample cluster. (B) The cut-off point was set as 0.9, and the soft threshold power was set as β=8. (C) Tree diagram of all the DEGs based on the cluster of difference measurement. The colored bands show the results from the automated monolithic analysis. (D) Correlation of blue modules with disease. (E) Importance analysis of module signature genes and phenotype correlations. We selected the MEblue module for subsequent analysis (the vertical values are the correlation coefficients for the feature modules). (F) Relevance of the MEblue to the disease. The vertical coordinate indicates the importance of the module gene in the disease, and the horizontal coordinate indicates the relevance of the module gene to the disease; the higher the score, the more important and more relevant the module in the disease. ME, module eigengene; WGCNA, weighted gene co-expression network analysis; ACP, adamantinomatous craniopharyngioma; DEGs, differentially expressed genes.
Identification and validation of the diagnostic markers for ACP
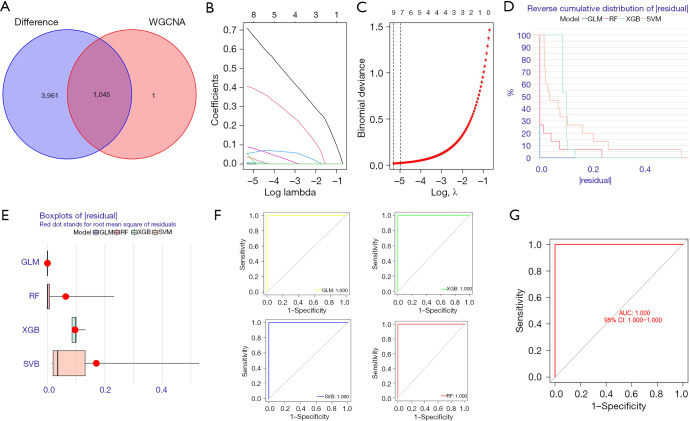
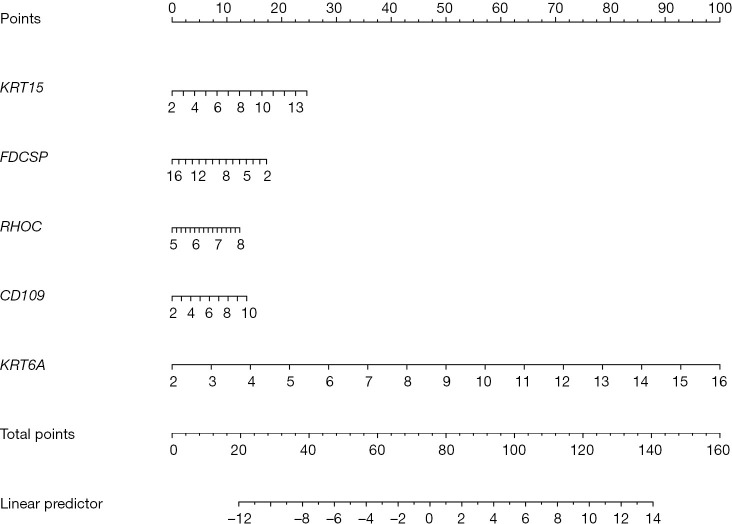
A total of 1045 trait genes were identified by cross-tabulating the appealed clinical trait gene modules with the DEGs (Figure 7A). A LASSO regression cross-validation analysis was conducted to identify the most relevant genes (Figure 7B,7C), and a total of 9 trait genes were identified. Next, 4 machine-learning algorithms were used to construct a diagnostic model of the traits (Figure 7D). The box plots of the residuals of the 4 algorithms (Figure 7E), in which the magnitude of the residuals of the samples were ranked as GLM < RF < XGB < SVM (the red dots indicate the root mean square of the residuals), were validated against the ROC curves of the 4 models (Figure 7F,7G), with AUC values of 1 for each of the 4 models. Based on the above combined results, the GLM model was chosen, and the final model was validated using the GSE68015 gene set with an AUC value of 1. Finally, the genes in the model were plotted in a Nomo plot (Figure 8), which showed that the genes in the model were KRT15, FDCSP, RHOC, CD109, and KRT6A.
Figure 7.
LASSO and machine-learning build models. (A) A WGCNA was conducted to analyze the disease characteristic module and differential gene crossover. (B) The screening was carried out using LASSO regression analysis. (C) Cross validation: for each λ value, around the mean value of the target parameter shown in red, we obtained a confidence interval of the target parameter. The 2 dashed lines indicate 2 special λ values, respectively. (D) Reverse cumulative distribution of the residuals of the SVM, RF tree, GLM, and XGB. (E) Reverse cumulative distribution of the residuals of the SVM, RF tree, GLM, and XGB. The Y-axis value represents the percentile of the outliers. (F) ROC analysis of the SVM, RF tree, GLM and XGB. (G) ROC analysis of the GLM model in the GSE68015 validation set. WGCNA, weighted gene co-expression network analysis; GLM, generalized linear models; RF, random-forest; XGBoost, extreme gradient boosting; SVM, support vector machine; AUC, area under the curve; ROC, receiver operating characteristic; CI, confidence interval; LASSO, least absolute shrinkage and selection operator.
Figure 8.
Nomograms were constructed to predict the progression of ACP patients. The values for each variable (KRT15, FDCSP, RHOC, CD109, and KRT6A) were summed to obtain a total score. By drawing a vertical line from the total score axis to the probability scale, the probability can be calculated. ACP, adamantinomatous craniopharyngioma.
Correlation and tissue expression analysis of ACP diagnostic markers
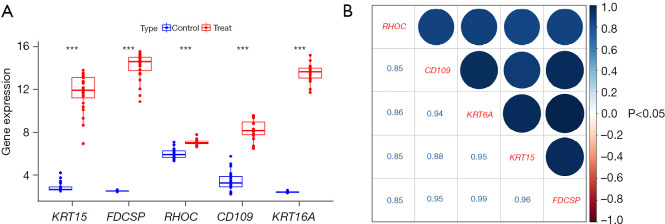
We visualized the KRT15, FDCSP, RHOC, CD109, and KRT6A genes using the Limma package, and all 5 genes were highly expressed in ACP (Figure 9A), the correlation between each gene was positive, and the correlation coefficients were as high as 0.85. For example, KRT15 was positively correlated with FDCSP and had a correlation coefficient of 0.96, CD109 was positively correlated with KRT15 and had a correlation coefficient of 0.88, and KRT15 was positively correlated with RHOC and had a correlation coefficient of 0.85 (Figure 9B).
Figure 9.
Expression and correlation of model genes. (A) Expression levels of the model genes in the ACP and normal brain tissues; (B) correlation analysis of the model genes, such as KRT15, FDCSP, RHOC, CD109, and KRT6A. ACP, adamantinomatous craniopharyngioma; ***, P<0.001.
Analysis of immune cell infiltration and its correlation with the central genetic diagnostic markers
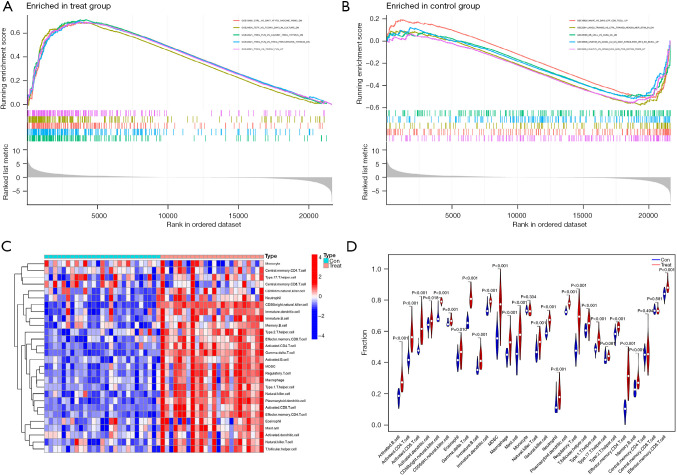
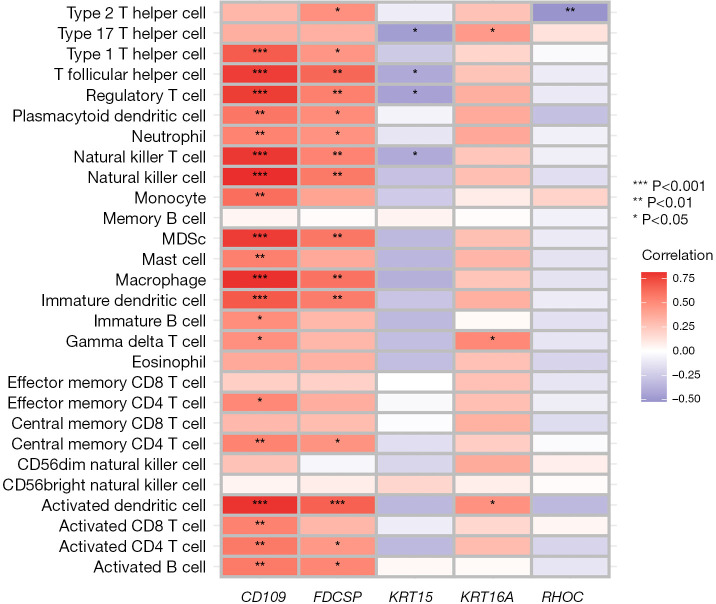
We explored the specific mechanism of actions of the diagnostic genes in ACP. We performed an enrichment analysis of the GSE94349 data set in the ACP tissue, mainly enriched in the GSE42021 data set TREG PLN VS CD24INT TREG THYMUS DN, GSE13485 CTRL VS DAY7 YF17D VACCINE PBMC DN, GSE42021 TREG VS TCONV PLN UP, GSE24634 TEFF VS TCONV DAY3 IN CULTURE DN, GSE42021 TREG PLN VS TREG PRECURSORS THYMUS DN. The immune gene set of normal brain tissue mainly enriched in the GSE22611 data set NOD2 TRANSD VS CTRL TRANSD HEK293 MDP STIM 2H DN, GSE19825 NAIVE VS DAY3 EFF CD8 TCELL UP, GSE45365 NK CELL VS CD8A DC DN, GSE9960 HEALTHY VS GRAM NEG AND POS SEPSIS PBMC UP, GSE46606 UNSTIM VS CD40L IL2 IL5 3DAY STIMULATED IRF4 KO BCELL UP (Figure 10A,10B). We analyzed the GSE94349 data set, and found that the immune cells were correlated with activated CD4 T cells, activated B cells, activated CD8 T cells, CD56 bright natural-killer cells, activated dendritic cells, CD56dim natural-killer cells, gamma delta T cells, eosinophils, immature B cells, myeloid-derived suppressor cells (MDSCs), immature dendritic cells, macrophages, natural-killer T cells, mast cells, natural-killer cells, plasmacytoid dendritic cells, neutrophils, regulatory T cells, type 1 T helper cells, T follicular helper cells, type 1 T helper cells, T helper cells, memory B cells, effector memory CD4 T cells, and effector memory CD8 T cells in the ACP tissue (Figure 10C,10D). We also performed an immune cell correlation analysis of our diagnostic genes KRT15, FDCSP, RHOC, CD109, and KRT6A (Figure 11), and found that both CD109 and FDCSP were positively correlated with natural-killer T cells, natural-killer cells, neutrophils, plasmacytoid dendritic cells, T follicular helper cells, regulatory T cells, type 1 T helper cells, MDSCs, immature dendritic cells, activated dendritic cells, macrophages, activated CD4 T cells, central memory CD4 T cells, activated B cell, and other immune cells were. Further, KRT6A expression was positively correlated with activated dendritic cells, gamma delta T cells, and type 17 T helper cells. While KRT15 was negatively correlated with regulatory T cells, natural-killer T cells, T follicular helper cells, and RHOC was negatively correlated with type 2 T helper cells.
Figure 10.
Analysis of the immune infiltration patterns in ACP. (A) Immunogenomic analysis of the ACP samples. (B) Immunogenomic analysis of the normal brain tissue samples. (C) Heat map of the immune cell infiltration analysis of the ACP and normal brain tissues. (D) Violin plot showing differences in infiltrating immune cells between the 2 groups. MDSC, myeloid-derived suppressor cell; ACP, adamantinomatous craniopharyngioma.
Figure 11.
Correlation analysis of the immune cells in the model genes. Heat map of the KRT15, FDCSP, RHOC, CD109, and KRT6A correlations with the immune cells. MDSC, bone marrow-derived suppressor cells.
Model gene pathway correlation analysis
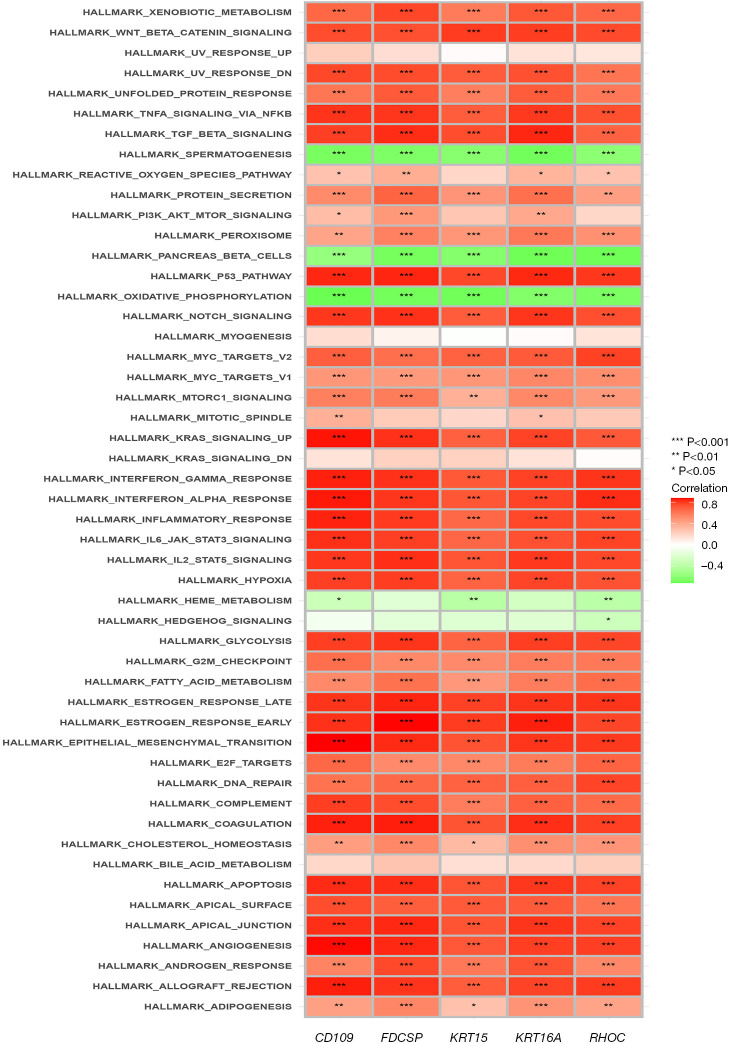
The results of the single-sample GSEA of pairs of genes, such as KRT15, FDCSP, RHOC, CD109, and KRT6A (Figure 12), suggested that these genes expression were negatively correlated with oxidative phosphorylation. Conversely, positive correlations were found between these genes and notch signaling, Wnt-beta catenin signaling, xenobiotic metabolism, TGF-beta signaling, p53 pathway, pi3k AKT mTOR signaling, DNA repair, and other signaling pathways.
Figure 12.
Pathway enrichment correlation analysis of the model genes. Pathway enrichment correlation analysis heat map of the KRT15, FDCSP, RHOC, CD109, and KRT6A genes.
Drug sensitivity analysis
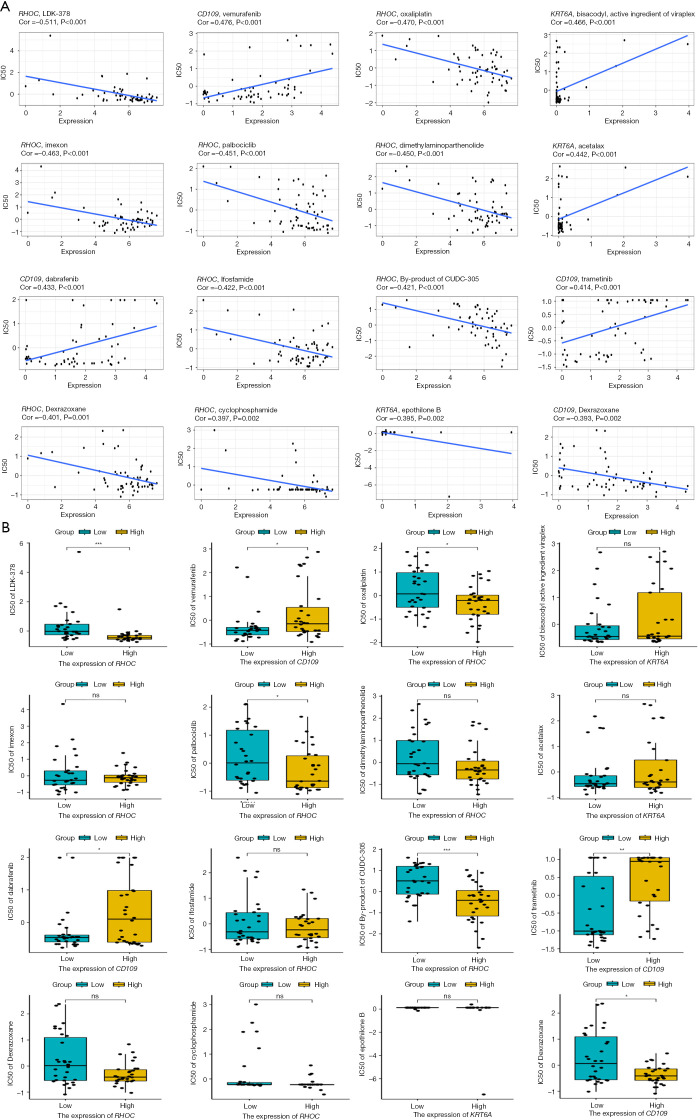
Our drug sensitivity analysis revealed Greater sensitivity to these drugs to LDK-378, Oxaliplatin, Palbociclib, CUDC-305 when RHOC was highly expressed and to Dexrazoxane when CD109 was highly expressed (Figure 13). The results are marked as “***” for P values <0.001, “**” for P values <0.01, “*” for P values <0.05, and “ns” for P values >0.05.
Figure 13.
The drug sensitivity of model genes was detected based on the CellMiner database. (A) Correlation diagram of RHOC, CD109, KRT6A, and other genes in terms of drug sensitivity. (B) Differences in the expression of RHOC, CD109, KRT6A, and drug sensitivity. IC50, half maximal inhibitory concentration; *, P<0.05; **, P<0.01; ***, P<0.001; ns, no statistical difference.
Discussion
CPs are rare benign epithelial tumors located in the hypothalamus/pituitary gland, account for 2–5% of all primary intracranial tumors, and have an overall incidence of 1.6–2.14 million/year (11). Approximately 30–50% of CPs occur in children (4). Children with CP develop a range of neuroendocrine disorders, including hypothalamic obesity, pituitary hormone deficiency, visual impairment, and cognitive deficits due to tumor hypothalamus, pituitary visual pathway, and limbic system invasion (4). Because of this site specificity, surgical resection is also highly likely to result in adverse postoperative outcomes, including permanent diabetes, obesity, and cranial nerve damage (26). Additionally, because the corresponding anatomy is narrower in children than in adults, pediatric patients are more prone to developmental abnormalities and neurological and endocrine disorders, particularly in cases involving CPs in the third ventricular wall, which can affect the cognitive and neuroendocrine processes of patients (27) and lead to developmental disorders in children. Thus, conventional surgery and radiotherapy for ACP may result in complex complications and a poor patient prognosis. This study sought to explore the biological mechanisms of ACP and to identify novel and meaningful molecular markers that could provide new ideas for diagnostic methods and safe therapeutic approaches for ACP.
In this study, ACP sequencing data, including 2 data sets (i.e., GSE94349 and GSE68015), were comprehensively analyzed, and the genes most relevant to ACP were found by a WGCNA analysis. Clinically relevant modular genes and differentially expressed gene intersections, the GO enrichment analysis revealed that these DEGs were associated with epidermal development, epidermal cell differentiation, and the cell junction. The DEGs were also associated with CCs, such as cell junctions and, stromal junctions, and MFs, such as cell-cell adhesion mediator activity, extracellular matrix structural components, and cell adhesion mediator activity. It has been suggested that epithelial cell adhesion molecule expression in CP may be a predictive marker for recurrence (28) and that CP tissue originates from the remnants of the Rathke’s cleft, which is the neuroepithelium, similar to the results of our GO enrichment analysis.
The KEGG enrichment analysis results suggested that the DEGs were associated with the PI3K-Akt signaling pathway, p53 signaling pathway, Hippo signaling pathway, TGF-beta signaling pathway, estrogen signaling pathway, Wnt signaling pathway, and other signaling pathways. In the genome-wide mapping WGS analysis of CP, a novel mutation was identified in exon 3 of CTNNB1. This novel mutation promotes ACP progenitor cell proliferation by increasing the stability of β-catenin to activate the Wnt signaling pathway (29). Conversely, the overexpression of CXCL12/CXCR4 significantly promoted PI3K/AKT signaling pathway activation, thereby promoting the proliferation, migration, and invasion of primary ACP cells (30).
The core proteins of our protein-protein interaction network were LAMB1, HDAC1, CCND1, ITGA2, COL1A2, TP53, and LAMC2. A previous study examined the immunohistochemistry of 15 CP patients to determine the cytoarchitectonic features expressed by members of the p53 family that may be required for CP histogenesis (31). Radmanesh et al. found that LAMB1 is thought to mediate cell attachment, migration, and organization into tissues by interacting with other extracellular matrix components during embryonic development (32). LAMB1 acts as an anchor point for the radial glial cell end-foot and as a physical barrier for migrating neurons. Radial glial cells play a central role in the development of the cerebral cortex (32). CCND1 is a regulatory component of the cell cycle protein D1-CDK4 (DC) complex, which phosphorylates and inhibits members of the retinoblastoma (RB) protein family, including RB1, and regulates the cell cycle during the G1/S transition (33,34). These core proteins may affect the development of ACP; thus, further experimental studies need to be conducted.
Next, a WGCNA was conducted to identify the genes most associated with clinical traits in ACP, and a LASSO cross-validation analysis was performed to screen the signature marker genes. Finally, diagnostic models with signatures were constructed using algorithms, such as RF trees, SVM, XGB, and GLM. The following GLMs were selected as marker genes for ACP: KRT15, FDCSP, RHOC, CD109, and KRT6A. We explored the correlation among these genes and immune cells and possible targeting drugs. Our drug sensitivity analysis showed that RHOC was more sensitive to LDK-378, oxaliplatin, Palbociclib and CUDC-305 when highly expressed.
A previous study showed that both AQB (HOTAIR-EZH2 inhibitor AC1Q3QWB) and palbociclib inhibited Wnt/β-linked protein signaling, and that the combination of both inhibitors had a stronger inhibitory effect on glioma metastasis (35). Another study showed that the use of low doses of poly-L-glutamate-paboxinib couples in pediatric gliomas improved the ability of paboxinib alone to killing diffuse pontine glioma cells (36). LDK-378, also known as Ceritinib, is a small molecule inhibitor of mesenchymal lymphoma kinase, insulin-like growth factor 1 receptor, and focal adhesion kinase, and is highly expressed in glioblastomas and many brain metastases, and has anti-tumor activity in central nervous system malignancies (37).
Oxaliplatin is a third-generation platinum anti-cancer agent, and a platinum analogue of diamino cyclohexane. A previous study found that Oxaliplatin reduced STAT3 (signal transducer and activator of transcription 3) activity, decreased MGMT (O-6-methylguanine-DNA methyltransferase) levels, and increased glioma temozolomide (38). CUDC-305 is a novel synthetic HSP90 (heat shock protein 90) inhibitor with unique cancer therapeutic pharmacological properties that crosses the blood-brain barrier to reach therapeutic levels in brain tissue. CUDC-305 demonstrated dose-dependent anti-tumor activity in a subcutaneous xenograft model of U87MG glioblastoma and significantly prolonged animal survival in an in-situ model of U87MG (39). A previous study found that CD109 physically interacts with glycoprotein 130 to promote the activation of the interlukin-6/STAT3 pathway in GSC (glioma stem cell) and sensitizes GSC to chemotherapy, which suggests that targeting the CD109/STAT3 axis may overcome treatment resistance in glioblastoma (40).
We also explored the downstream signaling pathways of the identified 5 genes, which had positively associated downstream pathways, such as Wnt-beta catenin signaling, TGF-beta signaling, pi3k AKT mTOR signaling, p53 pathway, notch signaling, and DNA repair. The Wnt-beta catenin signaling pathway, p53 pathway, and other pathways regulate the occurrence and development of ACP (29,31). These model molecules and related drugs have not been reported in CP; however, they play an important role in glioma and may become targets for subsequent treatments of CP in the future.
The immune microenvironment in ACP is currently poorly understood, which limits the further application of immune-targeted therapy in clinical practice. Thus, we comprehensively analyzed the immune infiltration in ACP, and found that many immune cells, such as activated CD4 T cells and CD8 T cells, are elevated in ACP tissues, while our marker genes. The expression of our marker genes, CDCD109 and FDCSP, correlated positively with immune cells such as Natural killer cell, Natural killer T cell, Activated B cell and Activated CD4 T cell. A comparative study found that M2 macrophages, CD8+ T cells and PD-L1 were more distributed in papillary CPs than in enzymatic CPs (ACPs), while M2 macrophages, CD8+ T cells and PD-L1 were more highly expressed in an adult ACP group than in a pediatric ACP group, and the high expression of M2 macrophages in primary CPs suggested a shorter time to tumor recurrence (41). These results suggest that immune cells may play an important role in ACP, which we will explore further in subsequent studies.
This study had some shortcomings. We undertook a comprehensive computer-based analysis of ACP, but due to the low incidence of clinical CP, the sample size of the study was small. It was difficult for us to obtain a large number of tissue samples for validation in a short period. There was also a lack of stable ACP cell lines, so experimental studies could not be conducted with the cells. We will expand the sample size in the future to further validate our results.
Conclusions
In this study, we found that the PI3K-Akt signaling pathway, p53 signaling pathway, TGF-beta signaling pathway, and Wnt signaling pathway are important signaling pathways in the development of ACP. KRT15, FDCSP, RHOC, CD109, and KRT6A may regulate the formation of ACP through these pathways. Moreover, we explored the targeting drugs of these model genes through the CellMiner database, and found that RHOC high expression showed significant drug sensitivity to LDK-378, Oxaliplatin, Palbociclib, and CUDC-305, while Sensitive to Dexrazoxane when CD109 is highly expressed. We will examine these in the future in our next in-depth study.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: None.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the TRIPOD reporting checklist. Available at https://tp.amegroups.com/article/view/10.21037/tp-23-152/rc
Peer Review File: Available at https://tp.amegroups.com/article/view/10.21037/tp-23-152/prf
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tp.amegroups.com/article/view/10.21037/tp-23-152/coif). The authors have no conflicts of interest to declare.
(English Language Editor: L. Huleatt)
References
- 1.Louis DN, Perry A, Burger P, et al. International Society Of Neuropathology--Haarlem consensus guidelines for nervous system tumor classification and grading. Brain Pathol 2014;24:429-35. 10.1111/bpa.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fernandez-Miranda JC, Gardner PA, Snyderman CH, et al. Craniopharyngioma: a pathologic, clinical, and surgical review. Head Neck 2012;34:1036-44. 10.1002/hed.21771 [DOI] [PubMed] [Google Scholar]
- 3.Muller HL, Merchant TE, Warmuth-Metz M, et al. Craniopharyngioma. Nat Rev Dis Primers 2019;5:75. 10.1038/s41572-019-0125-9 [DOI] [PubMed] [Google Scholar]
- 4.Nielsen EH, Feldt-Rasmussen U, Poulsgaard L, et al. Incidence of craniopharyngioma in Denmark (n = 189) and estimated world incidence of craniopharyngioma in children and adults. J Neurooncol 2011;104:755-63. 10.1007/s11060-011-0540-6 [DOI] [PubMed] [Google Scholar]
- 5.Zacharia BE, Bruce SS, Goldstein H, et al. Incidence, treatment and survival of patients with craniopharyngioma in the surveillance, epidemiology and end results program. Neuro Oncol 2012;14:1070-8. 10.1093/neuonc/nos142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Daubenbuchel AM, Muller HL. Neuroendocrine Disorders in Pediatric Craniopharyngioma Patients. J Clin Med 2015;4:389-413. 10.3390/jcm4030389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Norris GA, Garcia J, Hankinson TC, et al. Diagnostic accuracy of neuroimaging in pediatric optic chiasm/sellar/suprasellar tumors. Pediatr Blood Cancer 2019;66:e27680. 10.1002/pbc.27680 [DOI] [PubMed] [Google Scholar]
- 8.Mortini P, Gagliardi F, Bailo M, et al. Magnetic resonance imaging as predictor of functional outcome in craniopharyngiomas. Endocrine 2016;51:148-62. 10.1007/s12020-015-0683-x [DOI] [PubMed] [Google Scholar]
- 9.Zucchini S, Di Iorgi N, Pozzobon G, et al. Management of Childhood-onset Craniopharyngioma in Italy: A Multicenter, 7-Year Follow-up Study of 145 Patients. J Clin Endocrinol Metab 2022;107:e1020-31. 10.1210/clinem/dgab784 [DOI] [PubMed] [Google Scholar]
- 10.Muller HL. The Diagnosis and Treatment of Craniopharyngioma. Neuroendocrinology 2020;110:753-66. 10.1159/000504512 [DOI] [PubMed] [Google Scholar]
- 11.Lesueur P, Calugaru V, Nauraye C, et al. Proton therapy for treatment of intracranial benign tumors in adults: A systematic review. Cancer Treat Rev 2019;72:56-64. 10.1016/j.ctrv.2018.11.004 [DOI] [PubMed] [Google Scholar]
- 12.Otterlei OM, Indelicato DJ, Toussaint L, et al. Variation in relative biological effectiveness for cognitive structures in proton therapy of pediatric brain tumors. Acta Oncol 2021;60:267-74. 10.1080/0284186X.2020.1840626 [DOI] [PubMed] [Google Scholar]
- 13.Zheng J, Fang Y, Cai BW, et al. Intracystic bleomycin for cystic craniopharyngiomas in children. Cochrane Database Syst Rev 2014;(9):CD008890. [DOI] [PubMed] [Google Scholar]
- 14.Deopujari C, Behari S, Shroff K, et al. Intraventricular Craniopharyngiomas-Overcoming Their Relative Inaccessibility: Institutional Experience With a Review of Literature. Front Neurol 2021;12:755784. 10.3389/fneur.2021.755784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen C, Wang Y, Zhong K, et al. Frequent B7-H3 overexpression in craniopharyngioma. Biochem Biophys Res Commun 2019;514:379-85. 10.1016/j.bbrc.2019.04.142 [DOI] [PubMed] [Google Scholar]
- 16.Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43:e47. 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012;16:284-7. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kanehisa M, Furumichi M, Tanabe M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45:D353-D61. 10.1093/nar/gkw1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinformatics 2013;14:7. 10.1186/1471-2105-14-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res 2003;31:258-61. 10.1093/nar/gkg034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chin CH, Chen SH, Wu HH, et al. cytoHubba: identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8 Suppl 4:S11. 10.1186/1752-0509-8-S4-S11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J, Chen ZJ, Liang J, et al. Identifying and validating key genes mediating intracranial aneurysm rupture using weighted correlation network analysis and exploration of personalized treatment. Ann Transl Med 2022;10:1057. 10.21037/atm-22-4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu J, Qin C, Cai Y, et al. Machine learning screening for Parkinson's disease-related cuproptosis-related typing development and validation and exploration of personalized drugs for cuproptosis genes. Ann Transl Med 2023;11:11. 10.21037/atm-22-5756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakraborty H, Hossain A. R package to estimate intracluster correlation coefficient with confidence interval for binary data. Comput Methods Programs Biomed 2018;155:85-92. 10.1016/j.cmpb.2017.10.023 [DOI] [PubMed] [Google Scholar]
- 25.Shankavaram UT, Varma S, Kane D, et al. CellMiner: a relational database and query tool for the NCI-60 cancer cell lines. BMC Genomics 2009;10:277. 10.1186/1471-2164-10-277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jensterle M, Jazbinsek S, Bosnjak R, et al. Advances in the management of craniopharyngioma in children and adults. Radiol Oncol 2019;53:388-96. 10.2478/raon-2019-0036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Graffeo CS, Perry A, Link MJ, et al. Pediatric Craniopharyngiomas: A Primer for the Skull Base Surgeon. J Neurol Surg B Skull Base 2018;79:65-80. 10.1055/s-0037-1621738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tena-Suck ML, Ortiz-Plata A, Galan F, et al. Expression of epithelial cell adhesion molecule and pituitary tumor transforming gene in adamantinomatous craniopharyngioma and its correlation with recurrence of the tumor. Ann Diagn Pathol 2009;13:82-8. 10.1016/j.anndiagpath.2008.12.004 [DOI] [PubMed] [Google Scholar]
- 29.He J, Zeng Z, Wang Y, et al. Characterization of novel CTNNB1 mutation in Craniopharyngioma by whole-genome sequencing. Mol Cancer 2021;20:168. 10.1186/s12943-021-01468-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yin X, Liu Z, Zhu P, et al. CXCL12/CXCR4 promotes proliferation, migration, and invasion of adamantinomatous craniopharyngiomas via PI3K/AKT signal pathway. J Cell Biochem 2019;120:9724-36. 10.1002/jcb.28253 [DOI] [PubMed] [Google Scholar]
- 31.Momota H, Ichimiya S, Ikeda T, et al. Immunohistochemical analysis of the p53 family members in human craniopharyngiomas. Brain Tumor Pathol 2003;20:73-7. 10.1007/BF02483450 [DOI] [PubMed] [Google Scholar]
- 32.Radmanesh F, Caglayan AO, Silhavy JL, et al. Mutations in LAMB1 cause cobblestone brain malformation without muscular or ocular abnormalities. Am J Hum Genet 2013;92:468-74. 10.1016/j.ajhg.2013.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Santra MK, Wajapeyee N, Green MR. F-box protein FBXO31 mediates cyclin D1 degradation to induce G1 arrest after DNA damage. Nature 2009;459:722-5. 10.1038/nature08011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Febres-Aldana CA, Chang JC, Ptashkin R, et al. Rb Tumor Suppressor in Small Cell Lung Cancer: Combined Genomic and IHC Analysis with a Description of a Distinct Rb-Proficient Subset. Clin Cancer Res 2022;28:4702-13. 10.1158/1078-0432.CCR-22-1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi J, Lv S, Wu M, et al. HOTAIR-EZH2 inhibitor AC1Q3QWB upregulates CWF19L1 and enhances cell cycle inhibition of CDK4/6 inhibitor palbociclib in glioma. Clin Transl Med 2020;10:182-98. 10.1002/ctm2.21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melnyk T, Masia E, Zagorodko O, et al. Rational design of Poly-L-glutamic acid-palbociclib conjugates for pediatric glioma treatment. J Control Release 2023;355:385-94. 10.1016/j.jconrel.2023.01.079 [DOI] [PubMed] [Google Scholar]
- 37.Mehta S, Fiorelli R, Bao X, et al. A Phase 0 Trial of Ceritinib in Patients with Brain Metastases and Recurrent Glioblastoma. Clin Cancer Res 2022;28:289-97. 10.1158/1078-0432.CCR-21-1096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roberts NB, Alqazzaz A, Hwang JR, et al. Oxaliplatin disrupts pathological features of glioma cells and associated macrophages independent of apoptosis induction. J Neurooncol 2018;140:497-507. 10.1007/s11060-018-2979-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bao R, Lai CJ, Qu H, et al. CUDC-305, a novel synthetic HSP90 inhibitor with unique pharmacologic properties for cancer therapy. Clin Cancer Res 2009;15:4046-57. 10.1158/1078-0432.CCR-09-0152 [DOI] [PubMed] [Google Scholar]
- 40.Filppu P, Tanjore Ramanathan J, Granberg KJ, et al. CD109-GP130 interaction drives glioblastoma stem cell plasticity and chemoresistance through STAT3 activity. JCI Insight 2021;6:e141486. 10.1172/jci.insight.141486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lin D, Wang Y, Zhou Z, et al. Immune Microenvironment of Primary and Recurrent Craniopharyngiomas: A Study of the Differences and Clinical Significance. World Neurosurg 2019;127:e212-20. 10.1016/j.wneu.2019.02.212 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as