Abstract
The Homeodomain Resource is an annotated collection of non-redundant protein sequences, three-dimensional structures and genomic information for the homeodomain protein family. Release 2.0 contains 765 full-length homeodomain-containing sequences, 29 experimentally derived structures and 116 homeobox loci implicated in human genetic disorders. Entries are fully hyperlinked to facilitate easy retrieval of the original records from source databases. A simple search engine with a graphical user interface is provided to query the component databases and assemble customized data sets. A new feature for this release is the addition of more automated methods for database searching, maintenance and implementation of efficient data management. The Homeodomain Resource is freely available through the WWW at http://genome.nhgri.nih.gov/homeodomain
INTRODUCTION
The homeodomain is a common DNA-binding structural motif found in many eukaryotic regulatory proteins (1,2). The homeodomain proteins control transcription of a wide range of developmentally important genes. A number of human genetic and genomic disorders have been linked to mutations in homeodomain-containing genes. X-ray crystallographic and NMR spectroscopic studies (3–11) on several members of this family revealed that the homeodomain contains three helical regions folded into a compact globular structure, with an N-terminal extension. Helices I and II lie parallel to each other and across from the third helix which is also known as the recognition helix. The third helix, in conjunction with the N-terminal arm, confers the DNA-binding specificity of individual homeodomain proteins. The homeodomain has been evolutionary conserved in sequence, structure and mode of DNA binding (12).
The Homeodomain Resource represents a comprehensive collection of information about the homeodomain family. The database contains all available full-length and homeodomain-only sequence data and structures as of October 1999. The genetic information on this family includes human diseases in which homeodomain containing proteins are implicated, cytogenetic map locations and specific mutation data underlying the disease condition. Since its last release (13), 139 new entries have been added to the database which is updated monthly. Each entry in this database is rigorously selected to assure non-redundancy, annotated and cross-referenced. We suggest that the Homeodomain Resource provides a model framework around which a variety of related information about a large family of proteins can be organized.
DATABASE DESCRIPTION
The current version of the database contains 765 full-length homeodomain protein sequences isolated from 77 different species (Table 1). The complete full-length sequence data as well as the homeodomain portion of the sequence is available in FASTA format. The database can be searched on the basis of SWISS-PROT ID, GenBank accession number, gene names, protein description, sequence and organism name. A new feature introduced in the search criteria is the inclusion of an alternative gene names field, since one of the major problems encountered during data acquisition is related to the discrepancy in naming conventions. The addition of alternative gene names/ symbols to each entry will help to alleviate this problem.
Table 1. Homeodomain Resource statistics.
| Number of entries | |
|---|---|
| Total sequences available | 3796 |
| Non-redundant full-length sequences | 765 |
| Genes/gene symbols | 300 |
| Distinct organisms | 77 |
| Three-dimensional structures | 29 |
| Homeobox loci implicated in genetic disorders | 116 |
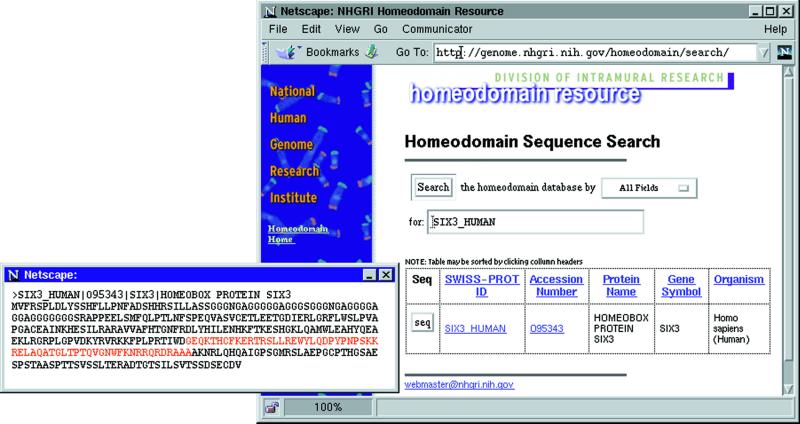
A number of features for the Web-based user interface have been improved in this release. The new search engine now supports Boolean queries and allows users to search on individual fields or on all fields at once. The search results are returned in a new tabular format with hyperlinks to the original records in GenBank and SWISS-PROT, respectively (Fig. 1). Individual sequences can also be retrieved in FASTA format from a pop-up window.
Figure 1.
Searching the database for the SWISS-PROT identifier SIX3_HUMAN. The search result is returned in a tabular format with hyperlinks to the corresponding records in the SWISS-PROT and GenBank databases. Clicking the seq button will retrieve the searched sequence in FASTA format, presenting the sequence in a pop-up window as illustrated.
The genetic information available for the homeodomain protein family has increased ~15% since the last release. The genetic data are compiled from both the literature and from the Online Mendelian Inheritance in Man database at NCBI (http:// www.ncbi.nlm.nih.gov/Omim/ ). A search engine is provided in this release to query these genomic data. Search results are presented in a tabular format with hyperlinks to the original records and can be sorted by disease name, map location, gene symbol, protein name or OMIM ID. With the rapid increase in genomic information on these proteins, this value-added format allows users easy access to related information.
IMPROVEMENTS IN DATA STORAGE AND RETRIEVAL
The homeodomain data was previously stored as a series of flat-files and HTML files; these data have now been imported into a Sybase database. The new organization of the data allows for faster, more powerful searching and it is now possible to perform data integrity tests (e.g., double-checking for redundancy and detecting unreported domains). A Perl program was written to automatically search SWISS-PROT through the Swiss Institute of Bioinformatics ExPASy SRS server (http://www.expasy.ch/srs5/ ). The search results are first compared to the existing data and a list of new entries is generated. Next, the new entries are examined manually and either added to the database or eliminated as false positives. The search program is run automatically each month.
REFERENCES
- 1.Gehring W., Qian,Y., Billeter,M., Furukubo-Tokunaga,K., Schier,A., Resendez-Perez,D., Affolter,M., Otting,G. and Wuthrich,K. (1994) Cell, 78, 211–223. [DOI] [PubMed] [Google Scholar]
- 2.Laughon A. (1991) Biochemistry, 30, 11357–11367. [DOI] [PubMed] [Google Scholar]
- 3.Dekker N., Cox,M., Boelens,R., Verrijzer,C., van der Vliet,P. and Kaptein,R. (1993) Nature, 362, 852–855. [DOI] [PubMed] [Google Scholar]
- 4.Endo T., Ohta,K., Saito,T., Haraguchi,K., Nakazato,M., Kogai,T. and Onaya,T. (1994) Biochem. Biophys. Res. Commun., 204, 31358–31363. [DOI] [PubMed] [Google Scholar]
- 5.Gruschus J., Tsao,D., Wang,L., Nirenberg,M. and Ferretti,J. (1997) Biochemistry, 36, 5372–5380. [DOI] [PubMed] [Google Scholar]
- 6.Kissinger C., Liu,B., Martin-Blanco,E., Kornberg,T. and Pabo,C. (1990) Cell, 63, 579–590. [DOI] [PubMed] [Google Scholar]
- 7.Ceska T., Lamers,M., Monaci,P., Nicosia,A., Cortese,R. and Suck,D. (1993) EMBO J., 12, 1805–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu B., Kissinger,C. and Pabo,C. (1990) Biochem. Biophys. Res. Commun., 171, 257–259. [DOI] [PubMed] [Google Scholar]
- 9.Qian Y., Billeter,M., Otting,G., Muller,M., Gehring,W. and Wuthrich,K. (1989) Cell, 59, 573–580. [DOI] [PubMed] [Google Scholar]
- 10.Qian Y., Furukubo-Tokunaga,K., Resendez-Perez,D., Muller,M., Gehring,W. and Wuthrich,K. (1994) J. Mol. Biol., 238, 333–345. [DOI] [PubMed] [Google Scholar]
- 11.Wolberger C., Vershon,A., Liu,B., Johnson,A. and Pabo,C. (1991) Cell, 67, 517–528. [DOI] [PubMed] [Google Scholar]
- 12.Buerglin T. (1994) In Duboule,D. (ed.) Guidebook to the Homeobox Genes. Oxford University Press, Oxford, UK, pp. 25–71.
- 13.Banerjee-Basu S. and Baxevanis,A. (1999) Nucleic Acids Res., 27, 336–337. [DOI] [PMC free article] [PubMed] [Google Scholar]