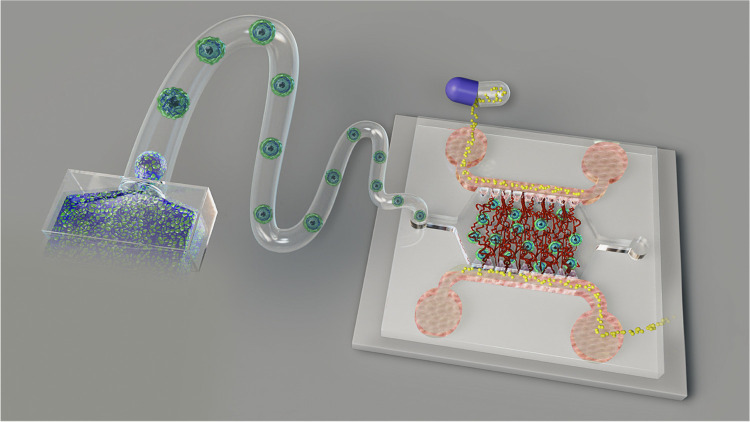
Abstract
High-fidelity in vitro tumor models are important for preclinical drug discovery processes. Currently, the most commonly used model for in vitro drug testing remains the two-dimensional (2D) cell monolayer. However, the natural in vivo tumor microenvironment (TME) consists of extracellular matrix (ECM), supporting stromal cells and vasculature. They not only participate in the progression of tumors but also hinder drug delivery and effectiveness on tumor cells. Here, we report an integrated engineering system to generate vessel-supported tumors for preclinical drug screening. First, gelatin-methacryloyl (GelMA) hydrogel was selected to mimic tumor extracellular matrix (ECM). HCT-116 tumor cells were encapsulated into individual micro-GelMA beads with microfluidic droplet technique to mimic tumor–ECM interactions in vitro. Then, normal human lung fibroblasts were mingled with tumor cells to imitate the tumor–stromal interaction. The tumor cells and fibroblasts reconstituted in the individual GelMA microbead and formed a biomimetic heterotypic tumor model with a core–shell structure. Next, the cell-laden beads were consociated into a functional on-chip vessel network platform to restore the tumor–tumor microenvironment (TME) interaction. Afterward, the anticancer drug paclitaxel was tested on the individual and vessel-supported tumor models. It was demonstrated that the blood vessel-associated TME conferred significant additional drug resistance in the drug screening experiment. The reported system is expected to enable the large-scale fabrication of vessel-supported heterotypic tumor models of various cellular compositions. It is believed to be promising for the large-scale fabrication of biomimetic in vitro tumor models and may be valuable for improving the efficiency of preclinical drug discovery processes.
Keywords: microfluidic droplet, core−shell tumor, vessel-on-a-chip, tumor microenvironment, preclinical drug screening
Introduction
Chemotherapy remains one of the most widely used and effective anticancer treatments.1 For a new molecular entity, it usually takes 10 years from the drug discovery stage to market approval. Most drug candidates that pass the preclinical development fail in phase I clinical trials.2 Particularly, the accurate predictive ability of preclinical physiological models is one of the important factors affecting the efficiency of drug development.3−5 In recent years, multicellular tumor spheroid has been increasingly used as in vitro model for drug screening assays.6−8 Compared with the traditional two-dimensional (2D) cellular monolayer model, the three-dimensional (3D) spheroids can exhibit more similar in vivo physiological characteristics, including hypoxic core, pondus hydrogenii gradient distribution, cellular metabolism, and gene expression, thus providing a more realistic drug response.9 Accordingly, the manufacturing technology of tumor spheroids has also been continuously improved and developed.10−16 Among them, droplet-based techniques stand out for their capability to generate uniform spheroids at a high-throughput manner.17−26 As a relatively automated production method, microfluidic droplet technology breaks the device limitations of traditional well-array plates27 and can mass-produce identical tumor spheroids for drug screening in a time-dependent manner.28 For instance, Kwak et al. reported a microfluidic water-in-oil droplet system for the mass fabrication of uniform tumor spheroids.29 The highest generation rate of the system can reach 1000 droplets/min, and the encapsulated cells in the single droplets can be developed into 3D homogeneous tumor spheroids in 24 h. However, the homotypic tumor spheroids still cannot fully represent the drug response in real tumors. Due to the heterogeneity of tumor tissues, tumor extracellular matrix (ECM), tumor–stromal cells, and tumor–tumor microenvironment (TME) interactions all play important roles in drug resistance.30−34 Notably, as the main component of TME, blood vessels have a special vascular transport function and barrier function, which significantly affect the transport and diffusion of drugs.35−40 Therefore, all of these interactions should be considered when constructing tumor models for preclinical drug discovery.
With the continuous development of microfabrication technology, microfluidic tumor–vessel chips not only help to precisely arrange the spatial distribution of cellular components in vitro but can also apply biomechanical and biochemical stimuli, including blood flow and growth factor gradient, thereby reproducing the interaction between the heterotypic tumor and TME with high fidelity.41−47 For example, Haase et al. reported an on-chip 3D-vascularized tumor model for drug examination in a TME within a large bed of perfusable vasculature.48,49 They demonstrated that the dose-dependent effect of anticancer drugs on tumor activity was significantly affected by the form of vascular infusion administration. Meanwhile, great strides have also been made in the field of biomaterials.50,51 As a representative biomaterial, GelMA has been widely used in the regenerative medicine field for 3D cell culture.52,53 Various organ models have been successfully established in GelMA, which demonstrate its good biocompatibility.5,54,55 Antunes et al. reported an in-air production of tumor spheroid-laden GelMA microgel to expedite drug screening, where GelMA was involved as tumor–ECM.56 In addition, GelMA also has the advantages of stability at room temperature, adjustable properties, processable into arbitrary shapes, and instant curing.57 Based on this, its combination with different processing techniques makes the spatial distribution of cells in tissues highly controllable, which brings broad application prospects for tissue engineering (Figure 1).58,59
Figure 1.
Schematic of the vessel-supported heterotypic tumor model for preclinical drug screening.
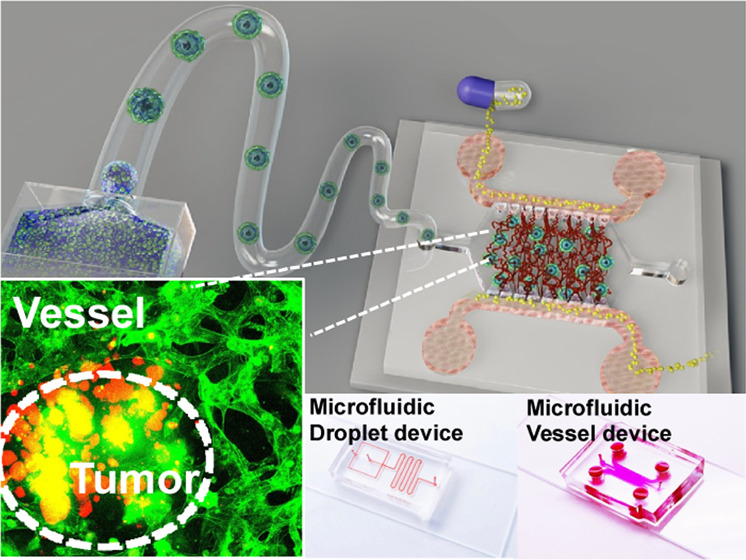
In this study, inspired by the “bottom-up” engineering concept, a vessel-supported heterotypic tumor model was reported for preclinical drug discovery in a large-scale and controllable manner.60 Here, we first mass-produced homotypic GelMA microbeads of tumor cells with a microfluidic droplet generation device to reconstitute tumor–ECM interactions in vitro. Then, by mixing the tumor cells with the fibroblasts, heterotypic GelMA microbeads were established to recreate tumor–stromal interactions. After the cellular interactions and self-reassembly, the heterotypic tumor beads presented a core–shell structure with the core as tumor clusters and the shell as the fibroblast monolayer. Next, the developed cell-laden beads were incorporated into an on-chip vessel network platform to reestablish the tumor–TME interaction. As a proof-of-concept application, the antitumor drug, paclitaxel (PAC), was administrated on the individual tumor beads and vessel-supported tumor beads.61 The drug screening result showed that different tumor components had different responses to drugs. The vigorously proliferating tumor cells were more sensitive to PAC, while the viability of the fibroblasts in the shell layer was not severely affected by PAC. Further, the fibroblast shell had a certain protective effect on the viability of tumor cells in the core area. Afterward, the blood vessel-associated TME further significantly reduced the efficacy of drugs on tumors. By combining these two technologies, microfluidic droplet and on-chip microphysiological vessel platform, we report an integrated engineering system that enables high-fidelity fabrication of vessel-supported biomimetic tumor models for preclinical drug discovery while inheriting the high-throughput properties of droplet technology. It is believed that the reported system can facilitate the drug discovery procedure and contribute to antitumor therapy development.
Experimental Section
Microfluidic Device Fabrication
All of the microfluidic devices were designed with AutoCAD and fabricated with standard lithography technology (Figure S7). For the microfluidic droplet generation, the chip is based on a flow-focusing design (Figure 2A).62,63 The channel width and height were designed as 400 and 300 μm, respectively. The microfluidic vessel platform design was designed based on the previous literature.64,65 Generally, the device consists of a central gel-loading channel and flow channels on the left and right sides. The gel channel and flow channel were separated by a row of trapezoidal micropillars.66 The distance between micropillars was 200 μm to provide sufficient surface tension to maintain infused gel solution within the central channel. The device was designed to be 350 μm in height and fabricated with soft lithography. An SU8-2150 (Kayaku Advanced Materials, Inc.) photoresist layer was spin-coated on a 3 in. silicon wafer, followed by UV exposure, post baking and development procedures. All of the fabrication processes were finished in the Center for Photonics and Nanoelectronics (CPN) at Lehigh University. Poly(dimethylsiloxane) (PDMS) (SYLGARD184, Dow Corning) presolution was poured on a solid SU8 mold and cured at 80 °C for 1 h. After being peeled off, the solid PDMS devices were plasma-bonded to glass substrates. All of the devices were autoclaved for sterilization prior to usage.
Figure 2.
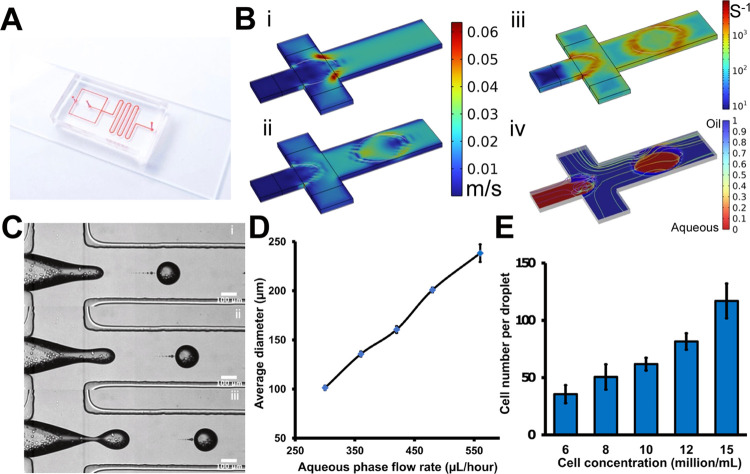
Cell-laden microfluidic droplet generation. (A) Image of the microfluidic droplet generation device. (B) Computational simulation results of microfluidic droplet generation at the flow-focusing zone: the velocity (i, ii), shear rate (iii), and volume fraction (iv). (C) Cell-laden microdroplet generation process. The time interval is 26 μs. (D) Microfluidic droplet size variation according to the aqueous flow rate. (E) Average cell number per individual microdroplet with respect to the original cell concentration. The scale bar is 100 μm.
Computational Simulation
To get the optimized flow condition, finite element method (FEM) was combined with the laminar two-phase level set simulation in COMSOL Multiphysics for the study.67 The motion of the continuous and dispersed phase interfaces is governed as described in eq 1
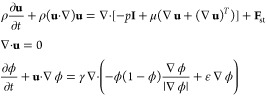 |
1 |
For simplicity, we assumed the continuous phase as HFE7500 engineered fluid and the dispersed phase as water. In the above equation, u is the velocity (m/s), ρ denotes the density of the fluid, t is time (s), p is pressure (Pa), μ is the dynamic viscosity (Pa·s), and Fst is the surface tension force (N/m3). Then, the effective droplet diameter is calculated as described in eq 2
| 2 |
Cell Culture
Human umbilical vein endothelial cells (HUVECs) and normal human lung fibroblasts (NHLFs) were purchased from LONZA. HUVECs were cultured in an endothelial cell growth medium supplemented with EGM-2 SingleQuot kit supply and growth factors (LONZA). NHLFs were cultured in fibroblast growth basal medium supplemented with FGM-2 BulletKit (LONZA). HCT-116 cell line was purchased from ATCC. Dulbecco’s modified Eagle’s medium (DMEM, Life Technology) was supplemented with 10% fetal bovine serum (FBS, Invitrogen) and 1% antibiotic and antimycotic (ThermoFisher). All cells were cultured at 37 °C in a humidified 5% CO2 environment.
Fabrication of Cell-Laden Hydrogel Beads with Microfluidic Droplet Technique
For microfluidic droplet generation, HFE7500 engineered fluid (3 M, Maplewood) was mixed with 2% of 008-FluoroSurfactant (RAN Biotech) as the continuous phase.68 The cells were optimized to the desired concentration and resuspended in 5% w/v of GelMA solution supplemented with 0.5% w/v of lithium phenyl-2,4,6-trimethylbenzoylphosphinate (LAP) (CELLINK, Sweden) as the dispersed phase. Both the continuous and dispersed phases were loaded into the microfluidic droplet generation device with two syringe pumps. The generated cell-laden droplets were transferred out of the microfluidic droplet device and collected in a 35 mm Petri dish containing the culture medium. An ultraviolet light (UV) source (405 nm wavelength) was activated for 15 s for photo-cross-linking. The dish was placed on a shaker for half an hour to realize oil–water separation and release the beads into culture medium for the following culture.69
On-Chip Vessel Network Construction and Vascularization of the Cell-Laden Beads
For vessel seeding, the HUVECs (8 million/mL) were resuspended in bovine fibrinogen (CAS-9001-32-5, Sigma) solution. After being mixed with thrombin (CAS-9002-044, Sigma-Aldrich), the cell-laden fibrin presolution was injected into the on-chip vessel network platform and cured for 20 min. Hydrostatic pressure was provided to promote vessel network formation (Figure 5A). The left flow channel was applied with a high liquid column (300 μL), and the right flow channel was applied with a low liquid column (100 μL), so that there was a hydrostatic pressure gradient across the central 3D fibrin hydrogel.70 The preformed cell-laden beads were mixed in the HUVEC suspension for vascularization. After 3 days of culture, the on-chip vessel system could be constructed, within which the predeveloped cell-laden beads could be integrated. Red blood cells (RBCs) from healthy donors were collected and sent from the University of Maryland School of Medicine. The sample collection process has followed the approved institutional review board protocols with donors providing informed consent.
Figure 5.
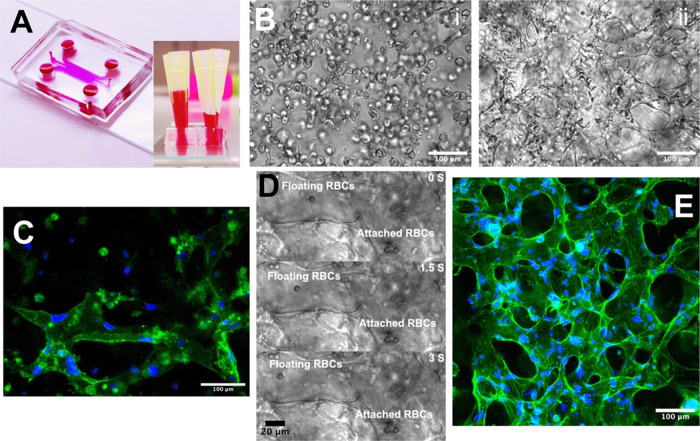
Vessel network-on-a-chip. (A) The microfluidic vessel network-on-a-chip platform. (B) The suspended HUVECs began the self-assembly on day 0 (i), and the perfusable vessel network formed on day 3 (ii). (C) Immunostaining of the vessel tube. CD31 was conjugated with FITC, and cell nucleus was stained with DAPI (blue). (D) RBCs were flowed through the vessel network. (E) Confocal scanning of the 3D vessel network. The F-actin of the vessel network was stained by FITC, and cell nucleus was stained with DAPI (blue).
Immunofluorescence Staining and Imaging
The vessel sample was fixed with 4% of paraformaldehyde in PBS overnight at 4 °C and incubated with a 1:50 diluted Alexa Fluor 488 antihuman CD31 antibody (BioLegend) and 50 μg/mL of DAPI overnight at 4 °C. The fibroblast-laden beads were fixed and incubated with a human α-smooth muscle actin (α-SMA) Alexa Fluor 594-conjugated antibody (R&D System).71 The HCT-116-laden beads were fixed and incubated with an antihuman CD326 (EpCAM) antibody (BioLegend).72 The vessel network sample was fixed and incubated with an Alexa Fluor 488 antimouse CD31 (PECAM) antibody (BioLegend).73 All of the fluorescent images and confocal scanning were acquired with a Nikon C2+ laser scanning confocal microscope in the Health Research Hub Center, Lehigh University.
Drug Treatment and Cell Viability Assay
The viability of the cell-laden beads was verified at days 0, 1, 3, and 5 to demonstrate the GelMA biocompatibility. The live/dead staining solution was prepared in complete medium with a concentration of 5 and 10 μM of calcein AM and propidium iodide (PI, Biotium), respectively, and incubated with the sample for 30 min at 37 °C prior to imaging. The viability was quantified from the fluorescence images and processed with the NIH ImageJ software. The therapeutic drug paclitaxel (PAC, LC Labs) was dissolved in DMSO at a 1 mM stock solution and then diluted into a concentration of 100 and 1000 nM with complete medium.74 For the individual bead testing, the PAC solution was deposited to the GelMA beads on day 7. For the vessel-supported condition, PAC was introduced into the microfluidic device from one side channel and flowed through a central vessel network for 3 h on day 3. After 72 h, the viability was examined by the calcein AM (Corning) and propidium iodide (PI, Biotium).
Statistical Analyses
The calcein AM and PI staining fluorescent images of the samples were analyzed to quantify the viability after drug treatment. All of the samples were examined within the same view of the field. Each experimental group was repeated three times. In each experimental group, 20 samples were counted. The error bars of all of the figures represented the standard deviation. The quantified results were analyzed using an independent, two-tailed Student t-test, in which the P-value <0.05 was considered statistically significant.
Results and Discussion
Microfluidic Droplet Generation On-Chip
The microfluidic droplet generation device has two inlets for the oil and aqueous phase loadings, respectively. Briefly, the aqueous phase and the oil phase with different flow rates joined at the flow junction area (Figure 2B). Since these two-phase solutions were immiscible with each other, under the action of viscous shear stress (Figure 2, Biii), the aqueous phase dissociates into a dispersed phase (Figure 2, Biv).25 Specifically, the surfactant-added fluorinated oils are selected to provide sufficient interfacial tension to avoid the coalescence of contacting droplets.24 The size of the droplets depends on the relative flow rates of the oil and aqueous phases (Figure 2C). By fixing the oil flow rate and adjusting the aqueous phase flow rate, droplets of different sizes could be produced (Figure 2D). Here, the diameter of the droplets was set around 250 μm. Meanwhile, the average cell load per droplet can be controlled by regulating the cell concentration in the original suspension (Figure 2E). To generate uniform cell-laden droplets, the flow rates of aqueous and oil phases were set as 480 and 3360 μL/h, respectively. The concentration of the original cell suspension was set at 10 million/mL, and the number of cells per drop was about 65. In the end, the generated cell-laden microdroplets were collected in a 35 mm Petri dish and packed into stable arrays for potential gelation operation (Figure 2E).
Fabrication and Characterization of Homotypic and Heterotypic Cell-Laden Beads
GelMA was selected as a tumor-localized matrix scaffold for encapsulating cells into droplets due to its ease of engineering and good biocompatibility.75,76 Initially, the cells were resuspended in the GelMA presolution and encapsulated into individual microdroplets. After being collected in the Petri dish, the cell-laden droplets were cross-linked by the 25 mW/cm2 UV source into the solid GelMA beads (Figure 3A). Then, a sufficient culture medium was added to the dish. Due to the interfacial tension, the solid beads were encapsulated in the oil layer, floating on top of the medium. While being shaken slightly, the beads were detached from the oil layer and gradually settled into the underlying medium. In this oil–water separation process, the superficial oil shell on the surface of the beads was removed, leaving only the beads immersed in the medium for long-term cell culture (Figure 3B). For the homotypic cell-laden bead fabrication, the fibroblasts and HCT-116 tumor cells were loaded into the GelMA beads separately. In a week-long in vitro culture, the cell viability was well maintained (Figure 3F), which is the basis of the proposed biofabrication system. The cells of two different types exhibited different growth behaviors in the GelMA matrix (Figure 3C–E). For the fibroblast-laden beads, the cells migrated to the bead surface and formed a confluent layer. This fibroblast monolayer appeared to have contractile properties as the contour of the bead was slightly compressed. Besides, the cellular layer also exhibited adhesion properties, as the two adjacent beads could be easily bonded together (Figure 3Ciii). αSMA was stained on the selected sample to identify that the fibroblasts were localized on the bead surface (Figure 4A). It is worth mentioning that the morphology of the fibroblasts was related to GelMA composition (Figure S3). In soft (3% w/v) GelMA, the fibroblasts self-assembled into spheroids after overnight culture. In stiff (8% w/v) GelMA, the fibroblasts grew slowly and self-assembled into a network after 1 week of culture.
Figure 3.
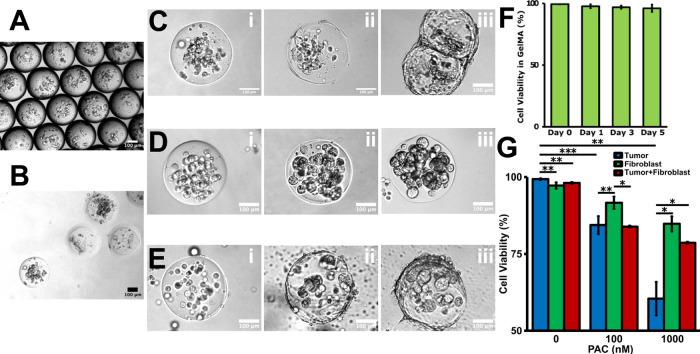
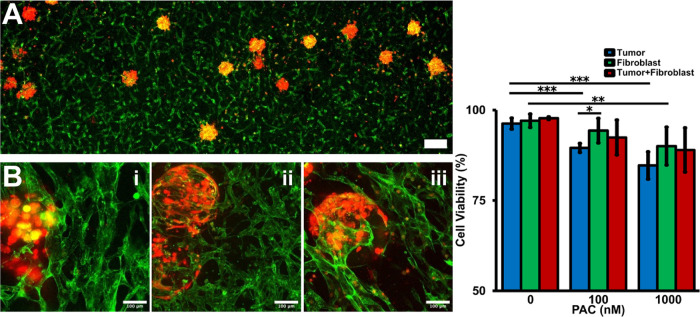
In vitro culture of the homotypic and heterotypic cell-laden microbeads. (A) Cell-laden droplet recollection from the microfluidic droplet generation device. (B) The cell-laden microbeads fall to the medium layer during oil/water separation process. (C) Fibroblasts gradually migrated to the bead surface from D0 (i) to D3 (ii) and form a confluent monolayer on D7 (iii). (D) HCT-116 tumor cells just proliferated in situ from D0 (i) to D3 (ii), and solid clusters formed inside the GelMA beads on D7 (iii). (E) Coculture of the fibroblast cells and tumor cells was encapsulated into the GelMA beads on D0 (i). The fibroblasts gradually migrated to the bead surface and the tumor cells proliferated in situ on D3 (ii). A core–shell tumor model formed on D7 (iii). (F) Viability characterization of the cell-laden GelMA beads during the in vitro culture. (G) Drug screening result on the homotypic and heterotypic beads. P-values were calculated using a two-sample t-test with respect to control. *p < 0.05; **p < 0.01; ***p < 0.001. The scale bar is 100 μm.
Figure 4.
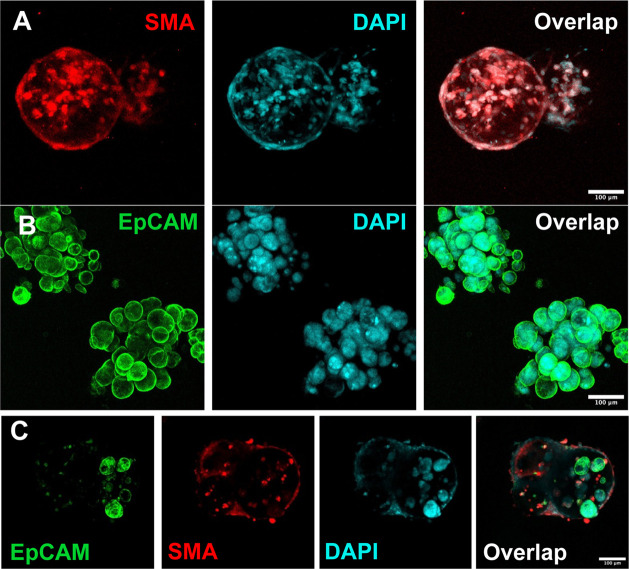
Immunostaining of the homotypic and heterotypic cell-laden beads. (A) The fibroblast-laden beads were characterized by SMA (red), and the nuclei were characterized by DAPI (cyan). (B) The tumor-laden beads were characterized by EPCAM (green), and the nuclei were characterized by DAPI (cyan). (C) The heterotypic beads were characterized by SMA (red) and EPCAM (green), and the nuclei were characterized by DAPI (cyan).
For the tumor-laden beads, the cells did not show any migration behavior but just proliferated in situ into the aggregates. Likewise, EPCAMs were stained on the selected samples to outline the cellular aggregate morphology (Figure 4B). As for the heterotypic cell-laden beads, the fibroblasts were mixed with tumor cells mixed in a 1:3 ratio to better facilitate tumor–vessel interactions.38 Likewise, most of the fibroblasts egressed to the bead surface, with a small amount of them remaining in the core, while the tumor cells proliferated in situ. The immunostaining confirmed that a core–shell compartmentalization was formed by the cellular self-assembly, with the fibroblast sheet covering the surface and the tumor clusters in the center (Figure 4C). This separated structure of the two cell types also facilitated the calculation of the viability of the two cells in subsequent drug experiments. If cultured longer, the surface fibroblast sheets could bring adjacent tumor beads together to form macroscopic tumor tissues (Figure S2F). Hence, the tumor–ECM and tumor–stromal interactions were remodeled in vitro on the individual microbeads. Beads of other cell compositions and sizes can also be precisely mass-produced as needed based on the capability of the microfluidic droplet technique.77
On-Chip Vessel System Construction and Engineering Tumor–TME Interaction
After fabrication and development, the cell-laden beads were integrated into the on-chip vessel network platform for reorganizing the tumor–TME interaction.78 First, the vessel network was formed on the on-chip vessel platform. The high-concentration HUVECs were seeded in the fibrinogen hydrogel. Then, cell culture medium columns of different heights were set at the left and right flow channels of the device to generate interstitial flow (IF). The IF conducted hydraulic pressure across the central gel zone and activated the HUVECs to form the capillary vessel network79,80 (Figure 5A). The medium columns were refreshed every 24 h so that there was always hydraulic pressure across the gel zone. On day 0, vacuoles were observed in every single HUVEC, which is often considered a hallmark of vascular tubulogenesis initiation81 (Figure 5Bi). On day 4, the self-organized vessel network was fully developed on-chip (Figure 5Bii). Biomarker immunostaining shows a strong CD31 signal, indicating tight cell-to-cell junctions (Figure 5C). To better characterize the 3D vessel morphology, filamentous actin (F-actin) of the selected sample was stained and confocal scanned (Figure 5D). As one of the basic properties of blood vessels, the vascular perfusability was demonstrated by flowing suspended RBCs (Supplementary Videos 1 and 2) from the side endothelialized channel through the artificial anastomosis zone and into the central network area (Figure S2). Figure 5E shows a floating RBC flowing through the vessel tube, while some RBCs got attached to the vessel surface. Therefore, a functional vessel network system was established on-chip.
To engineer the tumor–TME interaction, the predeveloped homotypic and heterotypic cell-laden beads were reseeded into the on-chip vessel network platform separately. The beads were cultured 1 week prior to being incorporated into the vessel system. The beads were settled to the bottom of a centrifuge tube by centrifugation and then resuspended into the fibrinogen solution. After being mixed with the thrombin at an appropriate concentration, the solution was injected into the vessel device. On day 3, the functional vessel network formed around the microbeads as well (Figure 6B). The blood vessel with the fibroblasts was slightly smaller in diameter, and the blood vessel with pure tumors was slightly thicker (Figure S6). This may imply that cancer-associated fibroblasts (CAFs) play an important role that cannot be ignored in real tumor vascularization cases.82,83
Figure 6.
Vessel-supported tumor models for preclinical drug screening. (A) Large-scale integration of the cell-laden beads into the on-chip vessel network platform. (B) The homotypic beads, tumor-laden beads (i), and fibroblast-laden beads (ii) were integrated into the vessel network separately. The heterotypic tumor bead was integrated into the vessel network (iii). (C) The drug screening result on the vessel-supported tumor models. P-values were calculated using a two-sample t-test with respect to control. *p < 0.05; **p < 0.01; ***p < 0.001.
In Vitro Antitumor Treatment Testing
To validate the drug resistance brought by the tumor–stromal and tumor–TME interactions, the anticancer drug treatment was administrated on the individual tumor bead model and vessel-supported tumor model, respectively. Paclitaxel is widely employed as an anticancer drug, and the mechanism is to stop the mitotic cells.84 By directly administering the beads under static conditions, a concentration dependence of drug efficacy was observed on different cellular components (Figure 3G). The tumor beads were more sensitive to drug than the fibroblasts (Figure S4). The higher the drug concentration, the lower the viability of tumor beads. Although the highest concentration of PAC was the most lethal for tumor cells in 3D tumor beads, the fibroblast beads still maintained about 80% viability. Literatures report that PAC may affect some functions of the stromal cells without significant cell loss.85,86 For the heterotypic tumor beads, the viability of the inner tumor clusters was largely preserved compared with the homotypic tumor beads. A possible hypothesis is that the fibroblasts in the shell absorbed part of the drug, reducing the real concentration of the drug that actually reached the tumor cells in the core area.87 As for the drug testing on the vessel-supported models, PAC was loaded into the side endothelialized channel and flowed through the artificial anastomosis and went into the capillary vessel networks (Figure S5). Then, the drug diffused across the capillary vessel wall and permeated into the surrounding matrix. The live/dead staining showed that the tumor cell viability was further preserved even at the most lethal drug concentration groups (Figure 6C). In conclusion, the TME composed of different cellular components could bring a non-negligible mitigating effect on the actual killing effect of drugs, and the real drug efficiency decreased.88 It is believed that the involvement of heterogeneous TME can import both pharmacodynamic (cell sensitivity-related) resistance and pharmacokinetics (drug delivery-related).89 For example, the blood vessels might have transported some of the drugs away before they diffused laterally into the surrounding tissue.90 As a result, the drug concentration might not be the same for all of the cells in the vessel network-based TME. In practical drug particle designs, the drug delivery efficiency to overcome the barrier of solid tumor tissues needs extra attention. It may be necessary to define a quantitative model to estimate resistance from different tumor component sources.
Conclusions
In summary, we report an integrated engineering system to fabricate a vessel-supported heterotypic tumor model for preclinical drug discovery by coupling microfluidic droplet and microfluidic vessel-on-a-chip techniques. After loading into the GelMA microbeads, the tumor cells and the fibroblasts reassembled into a core–shell spatial structure to mimic the tumor–stromal interaction in the ECM. Inspired by the concept of bottom-up engineering, the micro-cell-laden beads were applied as building blocks and consolidated into an on-chip vessel network platform to mimic the tumor–TME interaction. Further, the antitumor drug paclitaxel was tested on the individual tumor model and vessel-supported heterotypic tumor model. The results showed that the vasculature-associated TME had a significantly attenuating effect on the effectiveness of chemotherapy. The reported system could be beneficial for large-scale preclinical antitumor drug discovery.
Acknowledgments
This work was supported by the National Institute of Health grants R01HL131750 and R21EB033102, the National Science Foundation grants CBET 2039310 and OAC 2215789, the Pennsylvania Department of Health Commonwealth Universal Research Enhancement Program (CURE), and Pennsylvania Infrastructure Technology Alliance (PITA).
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsami.2c23305.
(Figure S1) Cell viability test in GelMA; (Figure S2) cell-laden bead generation by microfluidic droplet technique; (Figure S3) fibroblast morphologies in the different GelMA constructs; (Figure S4) live/dead staining of the cell-laden beads after the drug testing; (Figure S5) confocal staining of the functional on-chip vessel network; (Figure S6) confocal staining of the vessel-supported tumor-laden beads; and (Figure S7) snapshot of the microfluidic device designs in AutoCAD (PDF)
The suspended RBCs were perfused through the on-chip vessel network (Video 1) (AVI)
Bright-field scanning of the fibroblast-laden beads (Video 2) (AVI)
Author Contributions
Y.L. conceived and supervised the study. Y.W., Y.Z., Y.Z., and Y. L. designed and performed the experiments. Y.W. collected and analyzed the data. Y.Z. and Y.Z. assisted with data analysis. K.I. assisted with simulation. Y.W. wrote the manuscript. Y.W., Y.Z., Y.Z., K.I., and Y.L. revised the manuscript. All authors discussed the results and approved the submission.
The authors declare no competing financial interest.
Supplementary Material
References
- Wong C. H.; Siah K. W.; Lo A. W. Estimation of Clinical Trial Success Rates and Related Parameters. Biostatistics 2019, 20, 273–286. 10.1093/biostatistics/kxx069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Gao W.; Hu H.; Zhou S. Why 90% of Clinical Drug Development Fails and How to Improve It?. Acta Pharm. Sin. B. 2022, 12, 3049–3062. 10.1016/j.apsb.2022.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.; Yu M.; Tie C.; Deng Y.; Wang J.; Yi Y.; Zhang F.; Huang C.; Zheng H.; Mei L.; Wu M. Tumor Microenvironment-Specific Chemical Internalization for Enhanced Gene Therapy of Metastatic Breast Cancer. Research 2021, 2021, 9760398 10.34133/2021/9760398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y.; Ma X.; Zhou W.; Liu C.; Zhang H. Reregulated Mitochondrial Dysfunction Reverses Cisplatin Resistance Microenvironment in Colorectal Cancer. Smart Med. 2022, 1, e20220013 10.1002/smmd.20220013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.; Zhang X.; Fu X.; Zu Y.; Sun W.; Zhao Y. Liver Spheroids on Chips as Emerging Platforms for Drug Screening. Eng. Regener. 2021, 2, 246–256. 10.1016/j.engreg.2021.10.003. [DOI] [Google Scholar]
- Moffat J. G.; Vincent F.; Lee J. A.; Eder J.; Prunotto M. Opportunities and Challenges in Phenotypic Drug Discovery: An Industry Perspective. Nat. Rev. Drug Discov. 2017, 16, 531–543. 10.1038/nrd.2017.111. [DOI] [PubMed] [Google Scholar]
- Chen B.; Wu Y.; Ao Z.; Cai H.; Nunez A.; Liu Y.; Foley J.; Nephew K.; Lu X.; Guo F. High-Throughput Acoustofluidic Fabrication of Tumor Spheroids. Lab Chip 2019, 19, 1755–1763. 10.1039/C9LC00135B. [DOI] [PubMed] [Google Scholar]
- Uhl C. G.; Liu Y. Microfluidic Device for Expedited Tumor Growth towards Drug Evaluation. Lab Chip 2019, 19, 1458–1470. 10.1039/C8LC01250D. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa E. C.; Moreira A. F.; de Melo-Diogo D.; Gaspar V. M.; Carvalho M. P.; Correia I. J. 3D Tumor Spheroids: An Overview on the Tools and Techniques Used for Their Analysis. Biotechnol. Adv. 2016, 34, 1427–1441. 10.1016/j.biotechadv.2016.11.002. [DOI] [PubMed] [Google Scholar]
- Popova A. A.; Tronser T.; Demir K.; Haitz P.; Kuodyte K.; Starkuviene V.; Wajda P.; Levkin P. A. Facile One Step Formation and Screening of Tumor Spheroids Using Droplet-Microarray Platform. Small 2019, 15, e1901299 10.1002/smll.201901299. [DOI] [PubMed] [Google Scholar]
- Kingsley D. M.; Roberge C. L.; Rudkouskaya A.; Faulkner D. E.; Barroso M.; Intes X.; Corr D. T. Laser-Based 3D Bioprinting for Spatial and Size Control of Tumor Spheroids and Embryoid Bodies. Acta Biomater. 2019, 95, 357–370. 10.1016/j.actbio.2019.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patra B.; Peng C.-C.; Liao W.-H.; Lee C.-H.; Tung Y.-C. Drug Testing and Flow Cytometry Analysis on a Large Number of Uniform Sized Tumor Spheroids Using a Microfluidic Device. Sci. Rep. 2016, 6, 21061 10.1038/srep21061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi W.; Kwon J.; Huang Y.; Tan J.; Uhl C. G.; He R.; Zhou C.; Liu Y. Facile Tumor Spheroids Formation in Large Quantity with Controllable Size and High Uniformity. Sci. Rep. 2018, 8, 6837 10.1038/s41598-018-25203-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Ao Z.; Chen B.; Muhsen M.; Bondesson M.; Lu X.; Guo F. Acoustic Assembly of Cell Spheroids in Disposable Capillaries. Nanotechnology 2018, 29, 504006 10.1088/1361-6528/aae4f1. [DOI] [PubMed] [Google Scholar]
- LeSavage B. L.; Suhar R. A.; Broguiere N.; Lutolf M. P.; Heilshorn S. C. Next-Generation Cancer Organoids. Nat. Mater. 2022, 21, 143–159. 10.1038/s41563-021-01057-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z.; Cai H.; Wu Z.; Hu L.; Nunez A.; Zhou Z.; Liu H.; Bondesson M.; Lu X.; Lu X.; Dao M.; Guo F. Microfluidics Guided by Deep Learning for Cancer Immunotherapy Screening. Proc. Natl. Acad. Sci. U.S.A. 2022, 119, e2214569119 10.1073/pnas.2214569119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. M.; Choi J. W.; Ahrberg C. D.; Choi H. W.; Ha J. H.; Mun S. G.; Mo S. J.; Chung B. G. Generation of Tumor Spheroids Using a Droplet-Based Microfluidic Device for Photothermal Therapy. Microsyst. Nanoeng. 2020, 6, 52 10.1038/s41378-020-0167-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z.; Gong Z.; Ao Z.; Xu J.; Cai H.; Muhsen M.; Heaps S.; Bondesson M.; Guo S.; Guo F. Rapid Microfluidic Formation of Uniform Patient-Derived Breast Tumor Spheroids. ACS Appl. Bio Mater. 2020, 3, 6273–6283. 10.1021/acsabm.0c00768. [DOI] [PubMed] [Google Scholar]
- Sun Q.; Tan S. H.; Chen Q.; Ran R.; Hui Y.; Chen D.; Zhao C.-X. Microfluidic Formation of Coculture Tumor Spheroids with Stromal Cells as a Novel 3D Tumor Model for Drug Testing. ACS Biomater. Sci. Eng. 2018, 4, 4425–4433. 10.1021/acsbiomaterials.8b00904. [DOI] [PubMed] [Google Scholar]
- Khan A. H.; Zhou S. P.; Moe M.; Ortega Quesada B. A.; Bajgiran K. R.; Lassiter H. R.; Dorman J. A.; Martin E. C.; Pojman J. A.; Melvin A. T. Generation of 3D Spheroids Using a Thiol-Acrylate Hydrogel Scaffold to Study Endocrine Response in ER+ Breast Cancer. ACS Biomater. Sci. Eng. 2022, 8, 3977–3985. 10.1021/acsbiomaterials.2c00491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K.; Sui C.; Wu Y.; Ao Z.; Guo S.-S.; Guo F. A Digital Acoustofluidic Device for On-Demand and Oil-Free Droplet Generation. Nanotechnology 2019, 30, 084001 10.1088/1361-6528/aaf3fd. [DOI] [PubMed] [Google Scholar]
- Seeto W. J.; Tian Y.; Pradhan S.; Minond D.; Lipke E. A. Droplet Microfluidics-Based Fabrication of Monodisperse Poly(Ethylene Glycol)-Fibrinogen Breast Cancer Microspheres for Automated Drug Screening Applications. ACS Biomater. Sci. Eng. 2022, 8, 3831–3841. 10.1021/acsbiomaterials.2c00285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojek K. O.; Ćwiklińska M.; Kuczak J.; Guzowski J. Microfluidic Formulation of Topological Hydrogels for Microtissue Engineering. Chem. Rev. 2022, 122, 16839–16909. 10.1021/acs.chemrev.1c00798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q.; Utech S.; Chen D.; Prodanovic R.; Lin J.-M.; Weitz D. A. Controlled Assembly of Heterotypic Cells in a Core-Shell Scaffold: Organ in a Droplet. Lab Chip 2016, 16, 1346–1349. 10.1039/C6LC00231E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohamed M. G. A.; Kheiri S.; Islam S.; Kumar H.; Yang A.; Kim K. An Integrated Microfluidic Flow-Focusing Platform for on-Chip Fabrication and Filtration of Cell-Laden Microgels. Lab Chip 2019, 19, 1621–1632. 10.1039/C9LC00073A. [DOI] [PubMed] [Google Scholar]
- Sart S.; Ronteix G.; Jain S.; Amselem G.; Baroud C. N. Cell Culture in Microfluidic Droplets. Chem. Rev. 2022, 122, 7061–7096. 10.1021/acs.chemrev.1c00666. [DOI] [PubMed] [Google Scholar]
- Chao C.; Ngo Le P.; Engelward B. P. SpheroidChip: Patterned Agarose Microwell Compartments Harboring HepG2 Spheroids Are Compatible with Genotoxicity Testing. ACS Biomater. Sci. Eng. 2020, 6, 2427–2439. 10.1021/acsbiomaterials.9b01951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepers A.; Li C.; Chhabra A.; Seney B. T.; Bhatia S. Engineering a Perfusable 3D Human Liver Platform from IPS Cells. Lab Chip 2016, 16, 2644–2653. 10.1039/C6LC00598E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak B.; Lee Y.; Lee J.; Lee S.; Lim J. Mass Fabrication of Uniform Sized 3D Tumor Spheroid Using High-Throughput Microfluidic System. J. Control. Release 2018, 275, 201–207. 10.1016/j.jconrel.2018.02.029. [DOI] [PubMed] [Google Scholar]
- Chi C.-W.; Lao Y.-H.; Ahmed A. H. R.; Benoy E. C.; Li C.; Dereli-Korkut Z.; Fu B. M.; Leong K. W.; Wang S. High-Throughput Tumor-on-a-Chip Platform to Study Tumor-Stroma Interactions and Drug Pharmacokinetics. Adv. Healthcare Mater. 2020, 9, e2000880 10.1002/adhm.202000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro M. V.; Rocha M.; Gaspar V. M.; Mano J. F. Programmable Living Units for Emulating Pancreatic Tumor-Stroma Interplay. Adv. Healthcare Mater. 2022, 11, e2102574 10.1002/adhm.202102574. [DOI] [PubMed] [Google Scholar]
- Wu Z.; Chen B.; Wu Y.; Xia Y.; Chen H.; Gong Z.; Hu H.; Ding Z.; Guo S. Scaffold-Free Generation of Heterotypic Cell Spheroids Using Acoustofluidics. Lab Chip 2021, 21, 3498–3508. 10.1039/d1lc00496d. [DOI] [PubMed] [Google Scholar]
- Asghar W.; El Assal R.; Shafiee H.; Pitteri S.; Paulmurugan R.; Demirci U. Engineering Cancer Microenvironments for in Vitro 3-D Tumor Models. Mater. Today 2015, 18, 539–553. 10.1016/j.mattod.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmondson R.; Broglie J. J.; Adcock A. F.; Yang L. Three-Dimensional Cell Culture Systems and Their Applications in Drug Discovery and Cell-Based Biosensors. Assay Drug Dev. Technol. 2014, 12, 207–218. 10.1089/adt.2014.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang M.; Soon R. H.; Lim C. T.; Khoo B. L.; Han J. Microfluidic Modelling of the Tumor Microenvironment for Anti-Cancer Drug Development. Lab Chip 2019, 19, 369–386. 10.1039/C8LC00970H. [DOI] [PubMed] [Google Scholar]
- Shi W.; Reid L.; Huang Y.; Uhl C. G.; He R.; Zhou C.; Liu Y. Bi-Layer Blood Vessel Mimicking Microfluidic Platform for Antitumor Drug Screening Based on Co-Culturing 3D Tumor Spheroids and Endothelial Layers. Biomicrofluidics 2019, 13, 044108 10.1063/1.5108681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y.; Zhou Y.; Qin X.; Liu Y. From Cell Spheroids to Vascularized Cancer Organoids: Microfluidic Tumor-on-a-Chip Models for Preclinical Drug Evaluations. Biomicrofluidics 2021, 15, 061503 10.1063/5.0062697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nashimoto Y.; Okada R.; Hanada S.; Arima Y.; Nishiyama K.; Miura T.; Yokokawa R. Vascularized Cancer on a Chip: The Effect of Perfusion on Growth and Drug Delivery of Tumor Spheroid. Biomaterials 2020, 229, 119547 10.1016/j.biomaterials.2019.119547. [DOI] [PubMed] [Google Scholar]
- Li C.; Li S.; Du K.; Li P.; Qiu B.; Ding W. On-Chip Replication of Extremely Early-Stage Tumor Behavior. ACS Appl. Mater. Interfaces 2021, 13, 19768–19777. 10.1021/acsami.1c03740. [DOI] [PubMed] [Google Scholar]
- Thomas A.; Daniel Ou-Yang H.; Lowe-Krentz L.; Muzykantov V. R.; Liu Y. Biomimetic Channel Modeling Local Vascular Dynamics of Pro-Inflammatory Endothelial Changes. Biomicrofluidics 2016, 10, 014101 10.1063/1.4936672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Z.; Cao Y.; Galan E. A.; Hao L.; Zhao H.; Tang J.; Sang G.; Wang H.; Xu B.; Ma S. Vascularized Tumor Spheroid-on-a-Chip Model Verifies Synergistic Vasoprotective and Chemotherapeutic Effects. ACS Biomater. Sci. Eng. 2022, 8, 1215–1225. 10.1021/acsbiomaterials.1c01099. [DOI] [PubMed] [Google Scholar]
- Kim Y.; Ko J.; Shin N.; Park S.; Lee S.-R.; Kim S.; Song J.; Lee S.; Kang K.-S.; Lee J.; Jeon N. L. All-in-One Microfluidic Design to Integrate Vascularized Tumor Spheroid into High-Throughput Platform. Biotechnol. Bioeng. 2022, 119, 3678–3693. 10.1002/bit.28221. [DOI] [PubMed] [Google Scholar]
- Wu Y.; Zhou Y.; Paul R.; Qin X.; Islam K.; Liu Y. Adaptable Microfluidic Vessel-on-a-Chip Platform for Investigating Tumor Metastatic Transport in Bloodstream. Anal. Chem. 2022, 94, 12159–12166. 10.1021/acs.analchem.2c02556. [DOI] [PubMed] [Google Scholar]
- Wan Z.; Floryan M. A.; Coughlin M. F.; Zhang S.; Zhong A. X.; Shelton S. E.; Wang X.; Xu C.; Barbie D. A.; Kamm R. D. New Strategy for Promoting Vascularization in Tumor Spheroids in a Microfluidic Assay. Adv. Healthcare Mater. 2022, e2201784 10.1002/adhm.202201784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J.; Kim S.; Hong J.; Jeon J. S. Enabling Perfusion through Multicellular Tumor Spheroids Promoting Lumenization in a Vascularized Cancer Model. Lab Chip 2022, 22, 4335–4348. 10.1039/D2LC00597B. [DOI] [PubMed] [Google Scholar]
- Zhou Y.; Wu Y.; Paul R.; Qin X.; Liu Y. Hierarchical Vessel Network-Supported Tumor Model-on-a-Chip Constructed by Induced Spontaneous Anastomosis. ACS Appl. Mater. Interfaces 2023, 15, 6431–6441. 10.1021/acsami.2c19453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y.; Richardson K.; Yang R.; Bousraou Z.; Lee Y. K.; Fasciano S.; Wang S. Notch Signaling and Fluid Shear Stress in Regulating Osteogenic Differentiation. Front. Bioeng. Biotechnol. 2022, 10, 1007430 10.3389/fbioe.2022.1007430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase K.; Offeddu G. S.; Gillrie M. R.; Kamm R. D. Endothelial Regulation of Drug Transport in a 3D Vascularized Tumor Model. Adv. Funct. Mater. 2020, 30, 2002444 10.1002/adfm.202002444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf M. N. S.; Vivas A.; Kasi D. G.; van den Hil F. E.; van den Berg A.; van der Meer A. D.; Mummery C. L.; Orlova V. V. Multiplexed Fluidic Circuit Board for Controlled Perfusion of 3D Blood Vessels-on-a-Chip. Lab Chip 2022, 23, 168–181. 10.1039/D2LC00686C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gungor-Ozkerim P. S.; Inci I.; Zhang Y. S.; Khademhosseini A.; Dokmeci M. R. Bioinks for 3D Bioprinting: An Overview. Biomater. Sci. 2018, 6, 915–946. 10.1039/C7BM00765E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iovene A.; Zhao Y.; Wang S.; Amoako K. Bioactive Polymeric Materials for the Advancement of Regenerative Medicine. J. Funct. Biomater. 2021, 12, 14. 10.3390/jfb12010014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue K.; Trujillo-de Santiago G.; Alvarez M. M.; Tamayol A.; Annabi N.; Khademhosseini A. Synthesis, Properties, and Biomedical Applications of Gelatin Methacryloyl (GelMA) Hydrogels. Biomaterials 2015, 73, 254–271. 10.1016/j.biomaterials.2015.08.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. B.; Srigunapalan S.; Wheeler A. R.; Simmons C. A. A 3D Microfluidic Platform Incorporating Methacrylated Gelatin Hydrogels to Study Physiological Cardiovascular Cell-Cell Interactions. Lab Chip 2013, 13, 2591–2598. 10.1039/c3lc00051f. [DOI] [PubMed] [Google Scholar]
- Chen K.; Jiang E.; Wei X.; Xia Y.; Wu Z.; Gong Z.; Shang Z.; Guo S. The Acoustic Droplet Printing of Functional Tumor Microenvironments. Lab Chip 2021, 21, 1604–1612. 10.1039/d1lc00003a. [DOI] [PubMed] [Google Scholar]
- Ao Z.; Cai H.; Wu Z.; Ott J.; Wang H.; Mackie K.; Guo F. Controllable Fusion of Human Brain Organoids Using Acoustofluidics. Lab Chip 2021, 21, 688–699. 10.1039/D0LC01141J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antunes J.; Gaspar V. M.; Ferreira L.; Monteiro M.; Henrique R.; Jerónimo C.; Mano J. F. In-Air Production of 3D Co-Culture Tumor Spheroid Hydrogels for Expedited Drug Screening. Acta Biomater. 2019, 94, 392–409. 10.1016/j.actbio.2019.06.012. [DOI] [PubMed] [Google Scholar]
- Chen Y.-C.; Lin R.-Z.; Qi H.; Yang Y.; Bae H.; Melero-Martin J. M.; Khademhosseini A. Functional Human Vascular Network Generated in Photocrosslinkable Gelatin Methacrylate Hydrogels. Adv. Funct. Mater. 2012, 22, 2027–2039. 10.1002/adfm.201101662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pepelanova I.; Kruppa K.; Scheper T.; Lavrentieva A. Gelatin-Methacryloyl (GelMA) Hydrogels with Defined Degree of Functionalization as a Versatile Toolkit for 3D Cell Culture and Extrusion Bioprinting. Bioengineering 2018, 5, 55. 10.3390/bioengineering5030055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velasco D.; Tumarkin E.; Kumacheva E. Microfluidic Encapsulation of Cells in Polymer Microgels. Small 2012, 8, 1633–1642. 10.1002/smll.201102464. [DOI] [PubMed] [Google Scholar]
- Agarwal P.; Wang H.; Sun M.; Xu J.; Zhao S.; Liu Z.; Gooch K. J.; Zhao Y.; Lu X.; He X. Microfluidics Enabled Bottom-up Engineering of 3D Vascularized Tumor for Drug Discovery. ACS Nano 2017, 11, 6691–6702. 10.1021/acsnano.7b00824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitaeva K. V.; Rutland C. S.; Rizvanov A. A.; Solovyeva V. V. Cell Culture Based in Vitro Test Systems for Anticancer Drug Screening. Front. Bioeng. Biotechnol. 2020, 8, 322. 10.3389/fbioe.2020.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Q.-Q.; Zhao S.-K.; Cai B.; He R.-X.; Rao L.; Wu Y.; Guo S.-S.; Liu Q.-Y.; Liu W.; Zhao X.-Z. Biocompatible Fabrication of Cell-Laden Calcium Alginate Microbeads Using Microfluidic Double Flow-Focusing Device. Sens. Actuators, A 2018, 279, 313–320. 10.1016/j.sna.2018.06.006. [DOI] [Google Scholar]
- Cai B.; Ji T.-T.; Wang N.; Li X.-B.; He R.-X.; Liu W.; Wang G.; Zhao X.-Z.; Wang L.; Wang Z. A Microfluidic Platform Utilizing Anchored Water-in-Oil-in-Water Double Emulsions to Create a Niche for Analyzing Single Non-Adherent Cells. Lab Chip 2019, 19, 422–431. 10.1039/C8LC01130C. [DOI] [PubMed] [Google Scholar]
- Hajal C.; Offeddu G. S.; Shin Y.; Zhang S.; Morozova O.; Hickman D.; Knutson C. G.; Kamm R. D. Engineered Human Blood-Brain Barrier Microfluidic Model for Vascular Permeability Analyses. Nat. Protoc. 2022, 17, 95–128. 10.1038/s41596-021-00635-w. [DOI] [PubMed] [Google Scholar]
- Wan Z.; Zhong A. X.; Zhang S.; Pavlou G.; Coughlin M. F.; Shelton S. E.; Nguyen H. T.; Lorch J. H.; Barbie D. A.; Kamm R. D. A Robust Method for Perfusable Microvascular Network Formation in Vitro. Small Methods 2022, 6, e2200143 10.1002/smtd.202200143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelman M. A.; Kim D. Y.; Kakarla S.; Grath A.; Silvia N.; Dai G. Interstitial Flow Enhances the Formation, Connectivity, and Function of 3D Brain Microvascular Networks Generated within a Microfluidic Device. Lab Chip 2021, 22, 170–192. 10.1039/D1LC00605C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikfar M.; Paul R.; Islam K.; Razizadeh M.; Jagota A.; Liu Y. Respiratory Droplet Resuspension near Surfaces: Modeling and Analysis. J. Appl. Phys. 2021, 130, 024702 10.1063/5.0050447. [DOI] [Google Scholar]
- Choi C.-H.; Wang H.; Lee H.; Kim J. H.; Zhang L.; Mao A.; Mooney D. J.; Weitz D. A. One-Step Generation of Cell-Laden Microgels Using Double Emulsion Drops with a Sacrificial Ultra-Thin Oil Shell. Lab Chip 2016, 16, 1549–1555. 10.1039/C6LC00261G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang T.; Zhang H.; Zhou W.; Jiang K.; Liu C.; Wang R.; Zhou Y.; Zhang Z.; Mei Q.; Dong W.-F.; Sun M.; Li H. One-Step Generation and Purification of Cell-Encapsulated Hydrogel Microsphere with an Easily Assembled Microfluidic Device. Front. Bioeng. Biotechnol. 2021, 9, 816089 10.3389/fbioe.2021.816089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walji N.; Kheiri S.; Young E. W. K. Angiogenic Sprouting Dynamics Mediated by Endothelial-Fibroblast Interactions in Microfluidic Systems. Adv. Biol. 2021, 5, e2101080 10.1002/adbi.202101080. [DOI] [PubMed] [Google Scholar]
- Margolis E. A.; Cleveland D. S.; Kong Y. P.; Beamish J. A.; Wang W. Y.; Baker B. M.; Putnam A. J. Stromal Cell Identity Modulates Vascular Morphogenesis in a Microvasculature-on-a-Chip Platform. Lab Chip 2021, 21, 1150–1163. 10.1039/D0LC01092H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thege F. I.; Lannin T. B.; Saha T. N.; Tsai S.; Kochman M. L.; Hollingsworth M. A.; Rhim A. D.; Kirby B. J. Microfluidic Immunocapture of Circulating Pancreatic Cells Using Parallel EpCAM and MUC1 Capture: Characterization, Optimization and Downstream Analysis. Lab Chip 2014, 14, 1775–1784. 10.1039/C4LC00041B. [DOI] [PubMed] [Google Scholar]
- Lee S.; Kim S.; Koo D.-J.; Yu J.; Cho H.; Lee H.; Song J. M.; Kim S.-Y.; Min D.-H.; Jeon N. L. 3D Microfluidic Platform and Tumor Vascular Mapping for Evaluating Anti-Angiogenic RNAi-Based Nanomedicine. ACS Nano 2021, 15, 338–350. 10.1021/acsnano.0c05110. [DOI] [PubMed] [Google Scholar]
- Lin L.; Lin X.; Lin L.; Feng Q.; Kitamori T.; Lin J.-M.; Sun J. Integrated Microfluidic Platform with Multiple Functions to Probe Tumor-Endothelial Cell Interaction. Anal. Chem. 2017, 89, 10037–10044. 10.1021/acs.analchem.7b02593. [DOI] [PubMed] [Google Scholar]
- Nichol J. W.; Koshy S. T.; Bae H.; Hwang C. M.; Yamanlar S.; Khademhosseini A. Cell-Laden Microengineered Gelatin Methacrylate Hydrogels. Biomaterials 2010, 31, 5536–5544. 10.1016/j.biomaterials.2010.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X.; Ashfaq R.; Cheng F.; Maharjan S.; Li J.; Ying G.; Hassan S.; Xiao H.; Yue K.; Zhang Y. S. A Tumor-on-a-chip System with Bioprinted Blood and Lymphatic Vessel Pair. Adv. Funct. Mater. 2019, 29, 1807173 10.1002/adfm.201807173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Estefan J. H.; Gonidec M.; Vu T. T.; Daro N.; Chastanet G. In Situ Fine-tuning of Microfluidic Chips by Swelling and Its Application to Droplet Microfluidics. Adv. Mater. Technol. 2019, 4, 1900232 10.1002/admt.201900232. [DOI] [Google Scholar]
- Hajal C.; Ibrahim L.; Serrano J. C.; Offeddu G. S.; Kamm R. D. The Effects of Luminal and Trans-Endothelial Fluid Flows on the Extravasation and Tissue Invasion of Tumor Cells in a 3D in Vitro Microvascular Platform. Biomaterials 2021, 265, 120470 10.1016/j.biomaterials.2020.120470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Wan Z.; Pavlou G.; Zhong A. X.; Xu L.; Kamm R. D. Interstitial Flow Promotes the Formation of Functional Microvascular Networks in Vitro through Upregulation of Matrix Metalloproteinase-2. Adv. Funct. Mater. 2022, 32, 2206767 10.1002/adfm.202206767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sano H.; Watanabe M.; Yamashita T.; Tanishita K.; Sudo R. Control of Vessel Diameters Mediated by Flow-Induced Outward Vascular Remodeling in Vitro. Biofabrication 2020, 12, 045008 10.1088/1758-5090/ab9316. [DOI] [PubMed] [Google Scholar]
- Calderon G. A.; Thai P.; Hsu C. W.; Grigoryan B.; Gibson S. M.; Dickinson M. E.; Miller J. S. Tubulogenesis of Co-Cultured Human IPS-Derived Endothelial Cells and Human Mesenchymal Stem Cells in Fibrin and Gelatin Methacrylate Gels. Biomater. Sci. 2017, 5, 1652–1660. 10.1039/C7BM00223H. [DOI] [PubMed] [Google Scholar]
- Sahai E.; Astsaturov I.; Cukierman E.; DeNardo D. G.; Egeblad M.; Evans R. M.; Fearon D.; Greten F. R.; Hingorani S. R.; Hunter T.; Hynes R. O.; Jain R. K.; Janowitz T.; Jorgensen C.; Kimmelman A. C.; Kolonin M. G.; Maki R. G.; Powers R. S.; Puré E.; Ramirez D. C.; Scherz-Shouval R.; Sherman M. H.; Stewart S.; Tlsty T. D.; Tuveson D. A.; Watt F. M.; Weaver V.; Weeraratna A. T.; Werb Z. A Framework for Advancing Our Understanding of Cancer-Associated Fibroblasts. Nat. Rev. Cancer 2020, 20, 174–186. 10.1038/s41568-019-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunz-Schughart L. A.; Schroeder J. A.; Wondrak M.; van Rey F.; Lehle K.; Hofstaedter F.; Wheatley D. N. Potential of Fibroblasts to Regulate the Formation of Three-Dimensional Vessel-like Structures from Endothelial Cells in Vitro. Am. J. Physiol. Cell Physiol. 2006, 290, C1385–98. 10.1152/ajpcell.00248.2005. [DOI] [PubMed] [Google Scholar]
- Pradhan S.; Smith A. M.; Garson C. J.; Hassani I.; Seeto W. J.; Pant K.; Arnold R. D.; Prabhakarpandian B.; Lipke E. A. A Microvascularized Tumor-Mimetic Platform for Assessing Anti-Cancer Drug Efficacy. Sci. Rep. 2018, 8, 3171 10.1038/s41598-018-21075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai B. F. L.; Lu R. X. Z.; Hu Y.; Davenport Huyer L.; Dou W.; Wang E. Y.; Radulovich N.; Tsao M. S.; Sun Y.; Radisic M. Recapitulating Pancreatic Tumor Microenvironment through Synergistic Use of Patient Organoids and Organ-on-a-chip Vasculature. Adv. Funct. Mater. 2020, 30, 2000545 10.1002/adfm.202000545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axel D. I.; Kunert W.; Göggelmann C.; Oberhoff M.; Herdeg C.; Küttner A.; Wild D. H.; Brehm B. R.; Riessen R.; Köveker G.; Karsch K. R. Paclitaxel Inhibits Arterial Smooth Muscle Cell Proliferation and Migration in Vitro and in Vivo Using Local Drug Delivery. Circulation 1997, 96, 636–645. 10.1161/01.CIR.96.2.636. [DOI] [PubMed] [Google Scholar]
- Castells M.; Thibault B.; Delord J.-P.; Couderc B. Implication of Tumor Microenvironment in Chemoresistance: Tumor-Associated Stromal Cells Protect Tumor Cells from Cell Death. Int. J. Mol. Sci. 2012, 13, 9545–9571. 10.3390/ijms13089545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junttila M. R.; de Sauvage F. J. Influence of Tumour Micro-Environment Heterogeneity on Therapeutic Response. Nature 2013, 501, 346–354. 10.1038/nature12626. [DOI] [PubMed] [Google Scholar]
- Abdul-Aziz M. H.; Lipman J.; Mouton J. W.; Hope W. W.; Roberts J. A. Applying Pharmacokinetic/Pharmacodynamic Principles in Critically Ill Patients: Optimizing Efficacy and Reducing Resistance Development. Semin. Respir. Crit. Care Med. 2015, 36, 136–153. 10.1055/s-0034-1398490. [DOI] [PubMed] [Google Scholar]
- Uhl C. G.; Muzykantov V. R.; Liu Y. Biomimetic Microfluidic Platform for the Quantification of Transient Endothelial Monolayer Permeability and Therapeutic Transport under Mimicked Cancerous Conditions. Biomicrofluidics 2018, 12, 014101 10.1063/1.5000377. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.