Abstract
Saliva-secreting and transporting cells are part of the complex cellular milieu of the human salivary gland, where they play important roles in normal glandular physiology and diseased states. However, comprehensive molecular characterization, particularly at single-cell resolution, is still incomplete, in part due to difficulty in procuring normal human tissues. Here, we perform an in-depth analysis of male and female adult human submandibular gland (SMG) samples by bulk RNA sequencing (RNA-seq) and examine the molecular underpinnings of the heterogeneous cell populations by single-cell (sc) RNA-seq. Our results from scRNA-seq highlight the remarkable diversity of clusters of epithelial and nonepithelial cells that reside in the SMG that is also faithfully recapitulated by deconvolution of the bulk-RNA data sets. Our analyses reveal complex transcriptomic heterogeneity within both the ductal and acinar subpopulations and identify atypical SMG cell types, such as mucoacinar cells that are unique to humans and ionocytes that have been recently described in the mouse. We use CellChat to explore ligand–receptor interactome predictions that likely mediate crucial cell–cell communications between the various cell clusters. Finally, we apply a trajectory inference method to investigate specific cellular branching points and topology that offers insights into the dynamic and complex differentiation process of the adult SMG. The data sets and the analyses herein comprise an extensive wealth of high-resolution information and a valuable resource for a deeper mechanistic understanding of human SMG biology and pathophysiology.
Keywords: salivary glands, single-cell RNA-sequencing, transcriptomics, gene expression, transcription factors
Introduction
Salivary glands (SGs) play a vital role in oral health and are affected in diseases such as Sjögren’s syndrome and from radiation therapy side effects. Humans and commonly used genetic models, such as rodents, have 3 pairs of major SGs: parotid gland (PG), sublingual gland (SL), and submandibular gland (SMG), as well as various minor SGs (Maruyama et al. 2019). Indeed, much of our knowledge of SG function is derived from studies that span from reverse genetics to more recent genomic approaches in mice (Chibly et al. 2022). The development and maturation of the SG generates specialized epithelial cells such as the acinar cells that produce salivary proteins and fluids as well as basal and myoepithelial cells that provide support and facilitate the secretion of saliva into the lumens of the branched ducts (Tucker 2007). In addition, the epithelial-rich arborized structure of the SG is surrounded by a fibrous-rich mesenchyme alongside immune cells, nerves, and blood vessels (Holmberg and Hoffman 2014).
A comprehensive molecular characterization of all cell types is crucial to understand SG biology in health and disease. Single-cell and bulk RNA sequencing (RNA-seq) and epigenomic studies in the mouse SGs have begun to offer closeup views of the cellular complexity of this tissue and the underpinning gene regulatory networks (Gluck et al. 2016; Michael et al. 2019; Oyelakin et al. 2019; Hauser et al. 2020; Sekiguchi et al. 2020; Gluck et al. 2021; Horeth et al. 2021). Similar studies in human SGs, however, have been rather limited due to difficulties in procuring tissue samples (Huang et al. 2021; Chen et al. 2022; Costa-da-Silva et al. 2022). A recent study has addressed this issue by performing RNA-seq experiments of human fetal and adult SGs (Saitou et al. 2020). Analysis of such data by Saitou et al. (2020) has revealed regulatory processes that shape SG-specific functions and offered broad insights through transcriptomic and proteomic comparisons. Recent progress notwithstanding, there remains a continuing need for additional SG data sets of high-quality transcriptomics, particularly at single-cell resolution, and subsequent follow-up analysis.
Here we have used normal human SMG to perform bulk transcriptomic analysis and examined the cellular landscape of the SMG via single-cell RNA-seq (scRNA-seq). We show that human SMGs exhibit minimal gender-specific global transcriptomic differences and identify 16 distinct cell populations that represent different types of epithelial, fibroblast, endothelial, and immune cells. Interestingly, we find several subpopulations of ductal and acinar cells that are conserved between human and mouse and novel subtypes that are recently discovered in mouse or unique to human. Finally, we leverage gene expression patterns of signaling ligands and receptors to infer the underlying landscape of the intercellular communication and connectivity network that operates in the human SMG. The comprehensive atlas of the SMG detailed here offers new insights into the molecular identities of cells that inhabit the complex ecosystem of the gland.
Materials and Methods
Patient Samples
Healthy SMGs from 10 male and 5 female adult patients were collected as part of the standard of care for patients undergoing resections for unrelated pathologies at Erie County Medical Center. The University at Buffalo Institutional Review Board (UBIRB) approved all research procedures (STUDY00003370). Data sets can be accessed through accession number GSE199209. Additional details of materials and methods can be found in the Appendix.
Results
Defining the Transcriptome of the Human Submandibular Gland
To better define the global gene expression patterns in the human SMG, we performed RNA-seq–based gene expression profiling of 5 female and 10 male adult glands obtained from patients undergoing head and neck surgery (Appendix Fig. 1). Importantly, histological analysis of these SMGs showed normal glandular architecture with no detectable pathology (Appendix Fig. 2). To substantiate our results, we included 6 adult human SMG RNA-seq data sets (GSE143702) (Saitou et al. 2020), which showed good concordance with our samples based on principal component analysis (PCA) (Appendix Fig. 3A). Interestingly, female and male glands, which we confirmed by sex-specific expression of XIST and ZFY, respectively (Kao et al. 1992; Hoch et al. 2020), showed a high degree of similarity in broad gene expression patterns (Appendix Fig. 3B, C). The lack of any sex-specific pattern was further evident upon comparison between human female and male glands, which revealed only 22 differentially expressed genes (DEGs), representing mostly X and Y sex chromosome–specific genes (Appendix Fig. 3D). This observation is different from mouse SMG, where the sex-based differences in gene expression are more prominent (Maruyama et al. 2019; Mukaibo et al. 2019; Godfrey et al. 2020).
Single-Cell Characterization of the Adult Human SMG
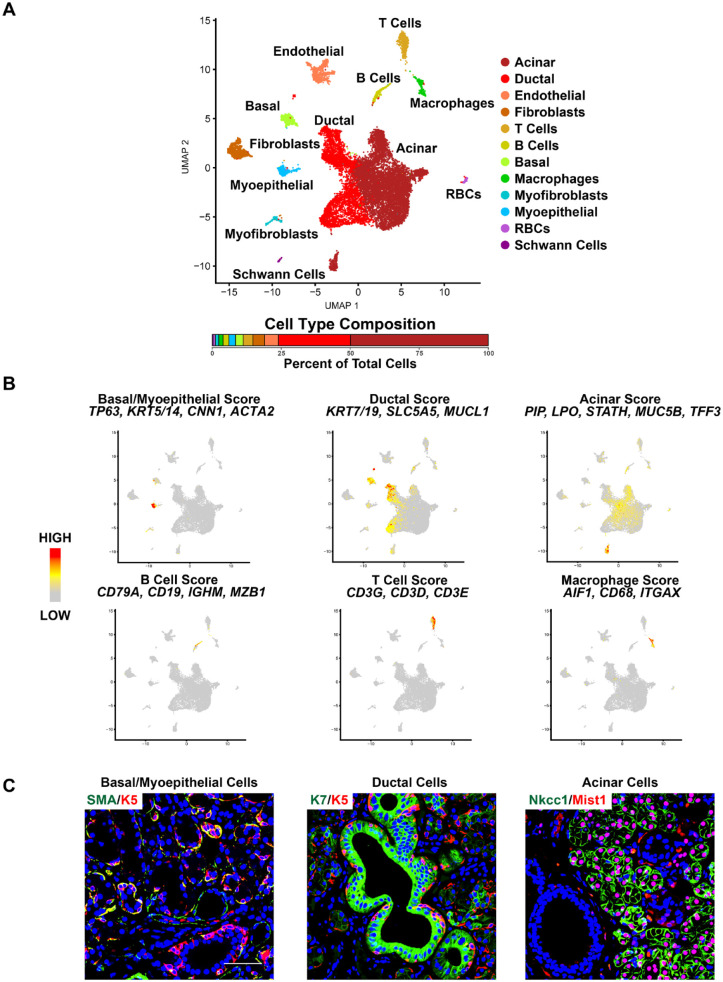
Next, to better elucidate the cellular heterogeneity of the human SMG, we performed scRNA-seq experiments on a female and a male SMG using the 10X Genomics platform. After data processing and quality controls of the scRNA-seq profiles, we obtained transcriptomic profiles for a total of ~15,600 cells. Dimensional reduction and unsupervised clustering with affinity propagation based on the expression of high-variance genes identified 12 transcriptionally distinct cell clusters, as shown via uniform manifold approximation and projection (UMAP) (Fig. 1A). Importantly, all identified clusters contained cells from both female and male samples (Appendix Fig. 4). Comparison of known markers with cluster-specific DEGs revealed the identity of the cell clusters as known constituents of the SG (Fig. 1A, Appendix Fig. 5, and Appendix Table 1) (Song et al. 2018; Oyelakin et al. 2019; Hauser et al. 2020; Min et al. 2020; Sekiguchi et al. 2020; Horeth et al. 2021; Huang et al. 2021). Four clusters comprised the key epithelial cell types of the SMG: basal, myoepithelial cells, ducts, and acinar cells. Basal and myoepithelial cells were defined by the expression of TP63, KRT5/14, CNN1, and ACTA2 (Fig. 1B). While ductal cells expressed markers including KRT7/19, SLC5A5, and MUCL1, acinar cells were enriched for expression of PIP, LPO, STATH, MUC5B, and TFF3 (Fig. 1B). Immunofluorescent images of representative markers of the 4 epithelial cell populations of the human SMG confirmed their identity, as shown in Figure 1C. Additionally, we identified a number of immune cell types, including CD79A, CD19, IGHM, and MZB1 expressing B cells; T cells enriched for expression of CD3G, CD3D, and CD3E; and monocytes marked by the expression of AIF1, CD68, and ITGAX (Fig. 1B). Taken together, these results highlight the degree of cellular heterogeneity and the broad range of cell types of the human SMG.
Figure 1.
Single-cell RNA sequencing reveals the cellular heterogeneity in the human submandibular gland (SMG). (A) Uniform manifold approximation and projection (UMAP) of the human SMG. Cell cluster identities are also shown. RBCs- red blood cells. (B) Feature plots demonstrating the expression of several well-established cell type–specific markers across the various cell populations as indicated. Red indicates maximum gene expression, and gray indicates low or no expression. (C) Representative immunofluorescence images of adult human SMGs stained with K5 and SMA to mark the basal/myoepithelial cells, K7 to mark the ductal cells, and MIST1 and NKCC1 to mark the acinar cells. Blue = nuclear staining. Scale bar: 37.5 µm.
scRNA-seq Reveals Distinct Ductal and Acinar Cell Populations
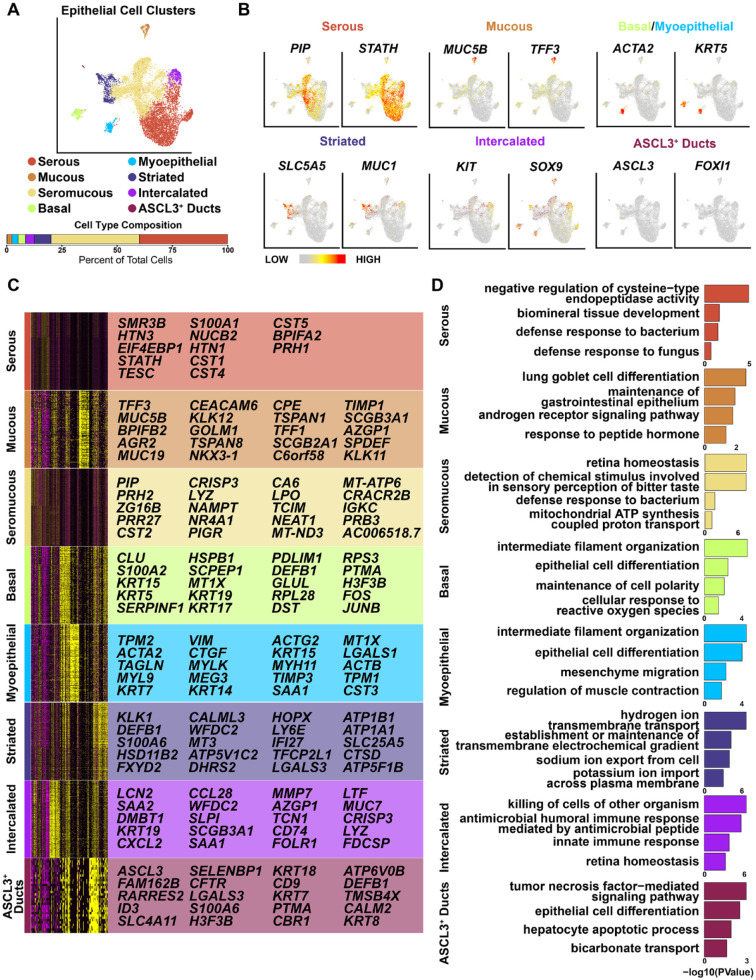
We next probed deeper into the cellular heterogeneity of the epithelial cells, which revealed 8 distinct clusters upon hierarchical clustering (Fig. 2A, Appendix Fig. 6, and Appendix Table 1). While the basal and myoepithelial cell populations persisted as single clusters after reclustering, the ductal and acinar cells segregated into discrete subclusters, highlighting their inherent heterogeneity (Fig. 2A). Top 20 DEGs per cluster and representative feature plots of select genes from each cluster are shown (Fig. 2B, C, and Appendix Table 1). Our analysis identified 3 acinar cell clusters consisting of serous acinar cells, which were defined by enriched expression levels of PIP and STATH; mucous acinar cells with high expression levels of MUC5B andTFF3; and seromucous acinar cells that expressed genes common to both mucous and serous acinar cells (Fig. 2B, C) (Hauser et al. 2020; Saitou et al. 2020; Huang et al. 2021; Costa-da-Silva et al. 2022). We also identified 3 ductal cell clusters, of which 2 were prominent: striated ducts, defined by enriched expression of MUC1 and SLC5A5, and intercalated ducts with enriched expression levels of KIT and SOX9 (Liu et al. 2002; Chenevert et al. 2012; Ohtomo et al. 2013; Zinn et al. 2015; Jhiang and Sipos 2021). Interestingly, we also identified a relatively small population of ASCL3-marked ductal cells, similar to what has been reported in the mouse SMG (Fig. 2A–C) (Hauser et al. 2020; Mauduit et al. 2022). Gene Ontology (GO) analysis of the top enriched genes per cluster is shown in Figure 2D.
Figure 2.
Analysis of human submandibular gland (SMG) epithelium. (A) Uniform manifold approximation and projection visualization of the epithelial cell populations based on hierarchical clustering analysis performed in Figure 1A. Epithelial cell clusters are shown. (B) Feature plots illustrating the expression patterns of select marker genes representing the various clusters. (C) Heatmap depicts the top 20 genes enriched in each epithelial cell cluster as identified in panel A. (D) Gene Ontology (GO) analysis of genes enriched in each epithelial cell cluster as identified in panel A/B.
To explore potential lineage relationships and branching trajectories of these cell populations, we used Potential of Heat-diffusion for Affinity-based Trajectory Embedding (PHATE) (Moon et al. 2019). This confirmed the UMAP-based findings and highlighted transcriptional similarity between the various cell types within the acinar and ductal clusters (Fig. 2 and Appendix Fig. 7). Furthermore, cell trajectory analyses using Velocyto (La Manno et al. 2018) revealed that while the acinar cells show relatively little connectivity between the basal and ASCL3+ duct clusters, the differentiation trajectories of the myoepithelial, intercalated ducts and striated ducts were closely intertwined with those of the serous and seromucous acinar cells, respectively (Appendix Fig. 7). Taken together, our results highlight the level of cellular heterogeneity of the human SMG, most prominently displayed by the acinar cells (Hauser et al. 2020; Costa-da-Silva et al. 2022).
Deconvolution of Bulk RNA-seq Samples Reveals the Existence and Proportions of Different Cell Populations
Having identified a total of 16 cell populations within the SMG, we next investigated whether these cell types were present in the bulk RNA-seq data sets generated from the 21 human SMG samples. Toward this end, we used BayesPrism-based deconvolution that allowed us to use our scRNA-seq data to characterize cell-type compositions from the bulk RNA-seq data (Chu et al. 2022). Hierarchical clustering of the samples with the 16 scRNA-seq annotated clusters revealed a clear separation between the epithelial and immune cell types, except for the ASCL3+ cell population, which might be due to the limited number of cells (Appendix Fig. 8A). Pearson correlation analysis of gene expression profiles of the bulk and scRNA-seq data sets was positively and highly correlated and the 21 bulk samples shared similar estimated cell-type proportions (Appendix Fig. 8B, C), reaffirming the cellular diversity of the SMG.
Cell–Cell Communication Network Structure in Human SMGs
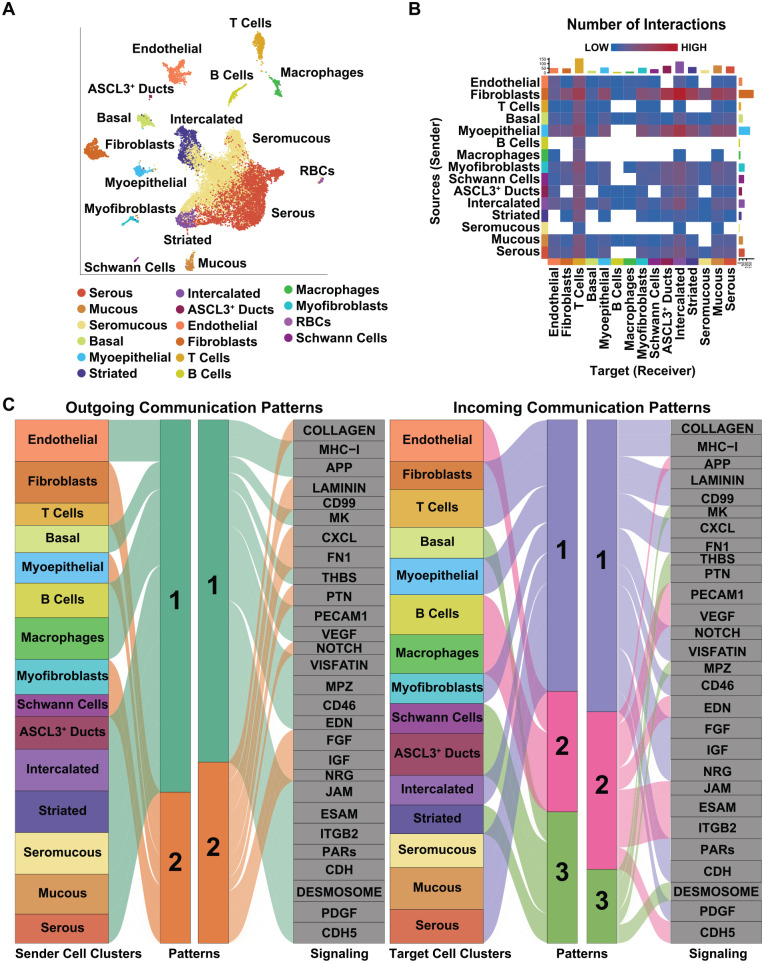
Communication between cells plays important roles during development, tissue homeostasis, and regeneration. To probe the potential incoming and outgoing cellular signaling communications, as well as to identify potential ligand–receptor pairs between 2 interacting cell groups between the 16 different cell types of the gland, we used CellChat (Jin et al. 2021) (Fig. 3A). Among the 16 cell types, CellChat predicted 90 significant ligand–receptor pairs, which were further classified into 28 signaling pathways. Closer examination revealed fibroblasts; myoepithelial, intercalated ducts; and serous acinar cells as the cellular sources (senders) with the highest number of putative signaling interactions (Fig. 3B). Conversely, T cells, intercalated ducts, and mucous and ASCL3+ ducts were ranked as the highest putative targets (receivers) (Fig. 3B). Similarly, we examined the source and target interaction strengths of the various populations, which revealed the fibroblasts and myoepithelial cells as the highest sources and T cells, ASCL3+, and intercalated ducts to be the highest target cell type (Appendix Fig. 9). Having uncovered the different cell–cell communications between the various cell types, we next explored how multiple cell groups and signaling pathways act coordinately. Interestingly, we identified 2 patterns of outgoing signals and 3 patterns of incoming signals (Fig. 3C). The outgoing communication patterns revealed that most of the epithelial, immune, and endothelial cells, with the exception of fibroblasts, myofibroblasts, myoepithelial cells, and T cells, were characterized by signaling pattern 1, including MHC-I, PCAM1, VEGF, PDGF, and PARs signaling (Fig. 3C). The remaining outgoing signaling from fibroblasts, T cells, and Schwann cells was characterized by pattern 2, representing such pathways as collagen, laminin, CXCL, and FGF (Fig. 3C). Conversely, incoming communication patterns of target cells revealed that incoming T-cell, fibroblast, and epithelial signaling is dominated by patterns 1 and 3, which include signaling pathways such as CXCL, MPZ, desmosome, and IGF (Fig. 3C). Interestingly, incoming B-cell, macrophage, and endothelial cell signaling is characterized by pattern 2, driven by CDH5, ITGB2, JAM, and PECAM1 (Fig. 3C). Taken together, these results highlight the degree of cellular communication between the different cell types of the SMG.
Figure 3.
Network and cell–cell communication signaling patterns of submandibular gland (SMG) cell populations. (A) Uniform manifold approximation and projection visualization of the 16 human cell population cluster identities. (B) Heatmap visualization of the number of possible interactions between any 2 cell populations. Red (high) and blue (low) represent stronger and lower interaction strengths, respectively. (C) Alluvial plot showing outgoing signaling patterns of sender (secreting) cells, which demonstrates the correspondence between the inferred latent patterns and cell clusters, as well as signaling pathways. The thickness of the flow indicates the contribution of the cell cluster or cell signaling pathway to each latent pattern. Height of each pattern is proportional to the number of associated cell clusters or signaling pathways. The right panel shows incoming signaling patterns of target cells demonstrating how target cells coordinate with each other and certain signaling pathways, to respond to incoming signaling.
Comparisons between Human and Mouse SMGs at Single-Cell Resolution
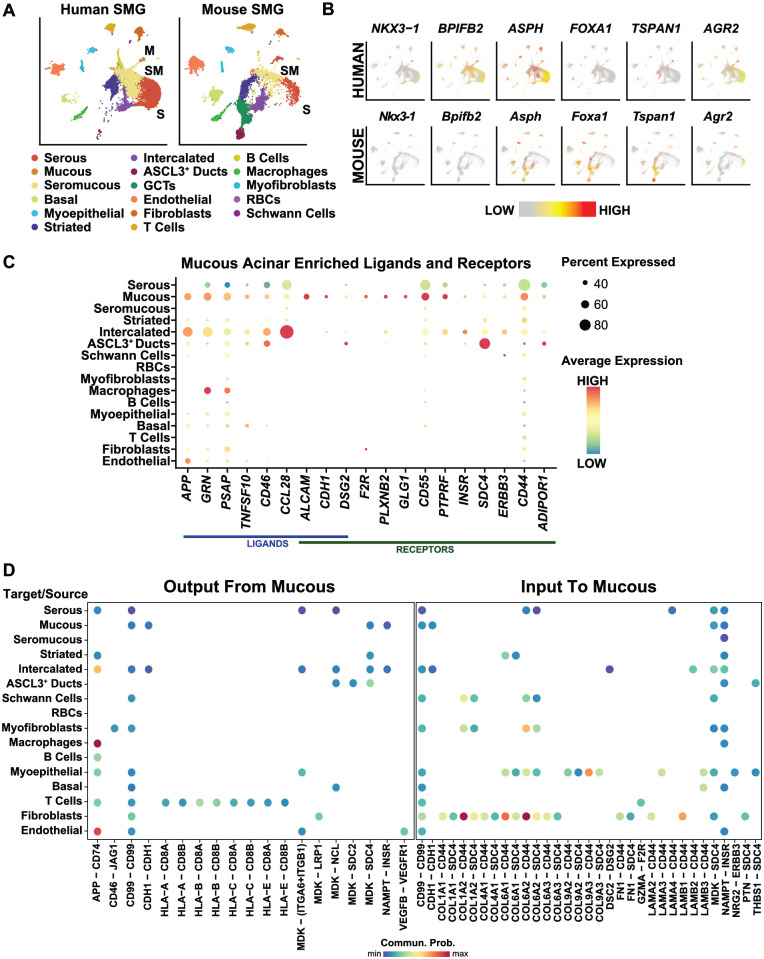
To better appreciate the degree of similarity between the glands, we next performed a comparative analysis of the human and mouse SMG scRNA-seq data sets (Fig. 4A) (Horeth et al. 2021). While all the major cell types identified in the human SMG were quite similar to those of the mouse, some prominent differences were notable, particularly among the epithelial cells. For instance, we were unable to identify the human counterparts of mucous acinar cells in the mouse (Hauser et al. 2020; Horeth et al. 2021). Indeed, closer examination of the 71 genes enriched in the human mucous acinar cells showed that many of the mouse counterparts were broadly expressed in different epithelial cell populations, or, as in the case of NKX3.1 and BPIFB2, these gene were not expressed in the mouse SMG (Fig. 4B). Conversely, as expected, the granular convoluted tubules (GCTs), which represent a population of ductal cells, were exclusively represented in the mouse SMG (Fig. 4A) (Maruyama et al. 2019).
Figure 4.
Comparative analysis and associated signaling of a unique population of human mucous acinar cells. (A) Uniform manifold approximation and projection visualization of human (left panel) and mouse (right panel) submandibular gland (SMG) cell populations (Horeth et al. 2021). Cell cluster identities are shown. (B) Feature plot visualization of genes enriched in human mucous acinar cells with corresponding expression in mouse SMG. (C) Dot plot showing ligands and receptors significantly enriched in human mucous cells. (D) Dot plot showing outgoing communication probability of ligand–receptor pairs contributing to the signaling from mucous cells to other cell types (left panel). Right panel is a dot plot showing incoming communication probability of ligand–receptor pairs contributing to the signaling from other cell types to mucous cells. M, mucous; S, serous; SM, seromucous.
To gain additional insight into the human mucous acinar cell population, we performed cell–cell communication analysis and identified mucous acinar cell–enriched ligands and receptors in the SMG. As shown in the dot plot, several ligands and receptors were found to be enriched in mucous acinar cells, including ALCAM, CDH1, APP, and GRN, while enriched receptors included CD55, CD44, SDC4, and ERBB3 (Fig. 4C). Next, to investigate the predicted outgoing signals from mucous acinar cells, we performed cell–cell interaction analysis, which identified 19 ligand–receptor pairs implicating several signaling pathways, including APP, HLA, MDK, and VEGF (Fig. 4D). As expected, our results inferred outgoing ligand–receptor pairs from mucous cells to other epithelial cell types, including other acinar cells and ductal cells, not surprising given the close proximity of the acinar cells to the intercalated ducts (Fig. 4D). To our surprise, however, we observed the highest number of predicted outgoing ligand–receptor interactions occurred between the mucous cells and T cells (Fig. 4D). Predicted incoming ligand–receptor interactions in mucous cells revealed 33 ligand–receptor pairs. Interestingly, most of the predicted signaling interactions were found between the fibroblasts and myoepithelial cells with the most abundant ligands expressed on these cells being collagens and laminins (Fig. 4D).
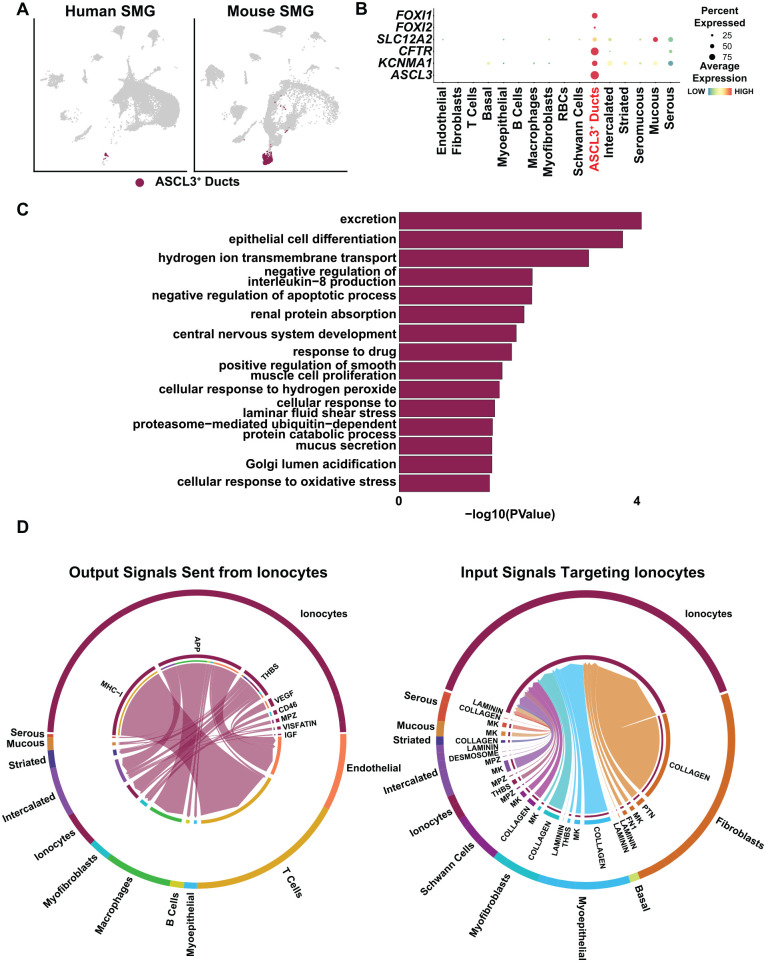
Given the recent identification of ionocytes, a specialized cell type representing the Ascl3+ ducts of the mouse SMG, we wondered if these were also present in the human SMG (Fig. 5A) (Mauduit et al. 2022). We therefore interrogated our human scRNA-seq data sets for cell types enriched for expression of ionocyte markers FOXI1, FOXI2, CFTR, SLC12A2, and KCNMA1 (Hsu et al. 2014; Montoro et al. 2018; Martin et al. 2019) and identified ASCL3+ cells as potential ionocytes (Fig. 5B). GO analysis of the top enriched genes in the ionocyte cluster revealed biological processes associated with excretion, transmembrane transport, and mucus secretion, in good agreement with the ion-regulatory roles of these cells in different organs (Fig. 5C) (Dymowska et al. 2012; Plasschaert et al. 2018). To decipher the intercellular communication networks, we performed cell–cell interaction analysis to examine outgoing signals from ionocytes. Our findings predicted 35 ligand–receptor pairs from 8 signaling pathways, including MHC-1, APP, VEGF, and THBS (Fig. 5D). Similar to the mucous cells, the highest number of predicted outgoing ligand–receptor interactions occurred between ionocytes and T cells, as observed through MHC-1 signaling (Fig. 5D, left panel). This was followed by ionocyte interactions with endothelial, intercalated, myoepithelial, macrophage, and B and T cells, via APP signaling (Fig. 5D, left panel). Additionally, we identified THBS signaling between ionocytes and the majority of epithelial cells, including ionocytes themselves (Fig. 5D, left panel). Conversely, predicted incoming ligand–receptor interactions in ionocytes revealed 17 ligand–receptor pairs representing 8 signaling pathways. Prominent extracellular matrix–related pathways included collagen, laminin, fibronectin 1 (FN1), and THBS. The major sources of these signaling pathways were from different fibroblasts while the epithelial cells comprised a modest number (Fig. 5D, right panel). Overall, these findings shed light on the degree of cell–cell communications between the various cell types and the robust signaling networks and pathways at play.
Figure 5.
Ionocyte-associated signaling in human submandibular gland (SMG). (A) Uniform manifold approximation and projection visualization of the ASCL3+ cell population in human (left panel) and mouse (right panel) (Horeth et al. 2021). (B) Dot plot showing expression of genes known to be highly expressed in ionocytes and that are enriched in the ASCL3+ cell cluster. (C) Gene Ontology analysis showing biological processes associated with the top enriched genes in the human ionocyte cell cluster. (D) Chord diagrams of inferred outgoing signaling pathways sent from ionocytes to other cell types (left panel). Right panel shows inferred incoming signaling pathways from other cell types targeting ionocytes. Signaling sources are zoomed out and shown on the outer ring with the colored segments representing cell identity information. Segment size is proportional to the total outgoing or incoming interaction strength associated with each pathway in the corresponding cell population.
Discussion
The complex cellular ecosystems of SGs are being unraveled at a remarkable pace and depth driven by technological advances. However, such data sets have been somewhat lacking for the human SMG, and most studies have been skewed toward minor and parotid SGs that can be more easily accessed. Filling this knowledge gap, however, is important given that mouse models for SG studies continue to predominantly focus on the SMG. Here, we generate a bulk and scRNA-seq–based atlas of the human SMG to examine the considerable level of cellular heterogeneity and reaffirm a multifaceted differentiation program that is more complex than previously appreciated.
While acinar cells could be broadly separated based on various mucous (MUC5B, TFF3) and serous-specific markers (PIP and STATH), distinct seromucous cells were found to be of a hybrid molecular nature expressing markers for both cell types. Of particular interest were mucous acinar cells, a cell population marked by NKX3-1, a transcription factor that so far has not been associated with SMG biology. Notably, this mucous acinar cell cluster seems to be missing from the mouse SMG, raising the tantalizing prospect for a possible species-specific role. Additional studies to investigate such neo-cell types and their function will likely reveal interesting insights. In a similar vein, ductal cells of the human SMG also show molecularly distinct clusters, suggesting the existence hitherto of unappreciated cell types that could be important for SMG homeostasis and regeneration. These observations, overall, dovetail well with the findings from the scRNA-seq data for the human parotid and minor glands (Huang et al. 2021; Chen et al. 2022; Costa-da-Silva et al. 2022). Finally, it is worth noting that our scRNA-seq analysis provides direct support for the evolutionarily conserved ASCL3+ ionocytes, a recently described cell population in the mouse SMG (Mauduit et al. 2022).
Another significant aspect of our bioinformatics-based analysis has been on the cell-to-cell communication via various signaling pathways. Among the interesting revelations include mucous cell signaling that operate via MHCI (HLA) to B and T cells, macrophages and other epithelial cells, and conversely mucous cells receiving reciprocal signaling from fibroblasts in the SMG. The discovery of specific cell-to-cell communication signaling is likely to be of added value to rare and less well-described cell types, such as the aforementioned ionocytes. Indeed, in addition to confirming that VEGF signaling plays a prominent role in ionocytes, similar to what has been described for mouse, we found that interestingly, the highest number of predicted outgoing ligand–receptor interactions was between ionocytes and T cells, and conversely, the incoming ligand–receptor interactions were primarily from fibroblasts. These examples typify the signaling events that are at play in the human SMG and can be a starting point to probe the mechanistic basis of cell behavior in vivo.
Our scRNA-seq–based results described here have some limitations that need to be mentioned. First, challenges of sample preparation such as the long enzymatic digestion times for the SMG might affect cellular transcriptomes and prevent efficient capture of some cell populations. Although our procedure was skewed toward more efficient recovery of primarily the epithelial cells, the reasonably strong representation of various immune and fibroblast cell populations in our human SMG scRNA-seq data sets suggests otherwise. This notion is further supported by our recovery of the neural Schwann cells, which typically have not been reported in published SG scRNA-seq data sets. However, we cannot rule out missed minor cell populations, such as rare tuft cells that have been recently discovered (Tavares Dos Santos et al. 2022). Finally, it should be noted that our work offers a limited snapshot into the cellular milieu of only the adult and comparatively older human SMG. We suspect similar studies of younger individuals might reveal interesting novel cellular features and patterns that are not affected by aging. These shortcomings notwithstanding, the stage is set for future studies of mechanisms that shape the diverse cellular landscape of the human SMG and dictate its versatile functions.
Author Contributions
E. Horeth, J. Bard, M. Che, T. Wrynn, E.A.C. Song, B. Marzullo, contributed to data analysis, critically revised the manuscript; M.S. Burke, S. Popat, T. Loree, J. Zemer, J.L. Tapia, J. Frustino, J.M. Kramer, contributed to data acquisition, critically revised the manuscript; S. Sinha, contributed to conception and design, data analysis, drafted and critically revised the manuscript; R.A. Romano, contributed to conception and design, data acquisition and analysis, drafted and critically revised the manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Supplemental Material
Supplemental material, sj-docx-1-jdr-10.1177_00220345221147908 for High-Resolution Transcriptomic Landscape of the Human Submandibular Gland by E. Horeth, J. Bard, M. Che, T. Wrynn, E.A.C. Song, B. Marzullo, M.S. Burke, S. Popat, T. Loree, J. Zemer, J.L. Tapia, J. Frustino, J.M. Kramer, S. Sinha and R.A. Romano in Journal of Dental Research
Footnotes
A supplemental appendix to this article is available online.
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by National Institutes of Health/National Institute of Dental and Craniofacial Research (NIH/NIDCR) training grant DE023526 to the State University of New York at Buffalo, School of Dental Medicine, Department of Oral Biology to support E. Horeth, M. Che, and E.A.C. Song.
ORCID iDs: E. Horeth  https://orcid.org/0000-0002-1376-2042
https://orcid.org/0000-0002-1376-2042
J. Bard  https://orcid.org/0000-0001-7433-0086
https://orcid.org/0000-0001-7433-0086
R.-A. Romano  https://orcid.org/0000-0001-9652-9555
https://orcid.org/0000-0001-9652-9555
References
- Chen M, Lin W, Gan J, Lu W, Wang M, Wang X, Yi J, Zhao Z. 2022. Transcriptomic mapping of human parotid gland at single-cell resolution. J Dent Res. 101(8):972–982. [DOI] [PubMed] [Google Scholar]
- Chenevert J, Duvvuri U, Chiosea S, Dacic S, Cieply K, Kim J, Shiwarski D, Seethala RR. 2012. DOG1: a novel marker of salivary acinar and intercalated duct differentiation. Mod Pathol. 25(7):919–929. [DOI] [PubMed] [Google Scholar]
- Chibly AM, Aure MH, Patel VN, Hoffman MP. 2022. Salivary gland function, development and regeneration. Physiol Rev. 102(3):1495–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu T, Wang Z, Pe’er D, Danko CG. 2022. Cell type and gene expression deconvolution with BayesPrism enables Bayesian integrative analysis across bulk and single-cell RNA sequencing in oncology. Nat Cancer. 3(4):505–517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa-da-Silva AC, Aure MH, Dodge J, Martin D, Dhamala S, Cho M, Rose JJ, Bassim CW, Ambatipudi K, Hakim FT, et al. 2022. Salivary ZG16B expression loss follows exocrine gland dysfunction related to oral chronic graft-versus-host disease. iScience. 25(1):103592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dymowska AK, Hwang PP, Goss GG. 2012. Structure and function of ionocytes in the freshwater fish gill. Respir Physiol Neurobiol. 184(3):282–292. [DOI] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Che M, Horeth E, Song EAC, Bard J, Lamb N, Sinha S, Romano RA. 2021. A global vista of the epigenomic state of the mouse submandibular gland. J Dent Res. 100(13):1492–1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gluck C, Min S, Oyelakin A, Smalley K, Sinha S, Romano RA. 2016. RNA-seq based transcriptomic map reveals new insights into mouse salivary gland development and maturation. BMC Genomics. 17(1):923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey AK, Naqvi S, Chmatal L, Chick JM, Mitchell RN, Gygi SP, Skaletsky H, Page DC. 2020. Quantitative analysis of Y-chromosome gene expression across 36 human tissues. Genome Res. 30(6):860–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser BR, Aure MH, Kelly MC; Genomics, Computational Biology Core, Hoffman MP, Chibly AM. 2020. Generation of a single-cell RNAseq atlas of murine salivary gland development. iScience. 23(12):101838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch D, Novakovic B, Cvitic S, Saffery R, Desoye G, Majali-Martinez A. 2020. Sex matters: XIST and DDX3Y gene expression as a tool to determine fetal sex in human first trimester placenta. Placenta. 97:68–70. [DOI] [PubMed] [Google Scholar]
- Holmberg KV, Hoffman MP. 2014. Anatomy, biogenesis and regeneration of salivary glands. Monogr Oral Sci. 24:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horeth E, Oyelakin A, Song EC, Che M, Bard J, Min S, Kiripolsky J, Kramer JM, Sinha S, Romano RA. 2021. Transcriptomic and single-cell analysis reveals regulatory networks and cellular heterogeneity in mouse primary Sjogren’s syndrome salivary glands. Front Immunol. 12:729040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu HH, Lin LY, Tseng YC, Horng JL, Hwang PP. 2014. A new model for fish ion regulation: identification of ionocytes in freshwater- and seawater-acclimated medaka (Oryzias latipes). Cell Tissue Res. 357(1):225–243. [DOI] [PubMed] [Google Scholar]
- Huang N, Perez P, Kato T, Mikami Y, Okuda K, Gilmore RC, Conde CD, Gasmi B, Stein S, Beach M, et al. 2021. SARS-CoV-2 infection of the oral cavity and saliva. Nat Med. 27(5):892–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jhiang SM, Sipos JA. 2021. Na+/I– symporter expression, function, and regulation in non-thyroidal tissues and impact on thyroid cancer therapy. Endocr Relat Cancer. 28(10):T167–T177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, Nie Q. 2021. Inference and analysis of cell-cell communication using Cellchat. Nat Commun. 12(1):1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao SM, Tang GC, Hsieh TT, Young KC, Wang HC, Pao CC. 1992. Analysis of peripheral blood of pregnant women for the presence of fetal Y chromosome-specific ZFY gene deoxyribonucleic acid sequences. Am J Obstet Gynecol. 166(3):1013–1019. [DOI] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. 2018. RNA velocity of single cells. Nature. 560(7719):494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu B, Lague JR, Nunes DP, Toselli P, Oppenheim FG, Soares RV, Troxler RF, Offner GD. 2002. Expression of membrane-associated mucins MUC1 and MUC4 in major human salivary glands. J Histochem Cytochem. 50(6):811–820. [DOI] [PubMed] [Google Scholar]
- Martin KE, Ehrman JM, Wilson JM, Wright PA, Currie S. 2019. Skin ionocyte remodeling in the amphibious mangrove rivulus fish (Kryptolebias marmoratus). J Exp Zool A Ecol Integr Physiol. 331(2):128–138. [DOI] [PubMed] [Google Scholar]
- Maruyama CL, Monroe MM, Hunt JP, Buchmann L, Baker OJ. 2019. Comparing human and mouse salivary glands: a practice guide for salivary researchers. Oral Dis. 25(2):403–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauduit O, Aure MH, Delcroix V, Basova L, Srivastava A, Umazume T, Mays JW, Bellusci S, Tucker AS, Hajihosseini MK, et al. 2022. A mesenchymal to epithelial switch in Fgf10 expression specifies an evolutionary-conserved population of ionocytes in salivary glands. Cell Rep. 39(2):110663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael DG, Pranzatelli TJF, Warner BM, Yin H, Chiorini JA. 2019. Integrated epigenetic mapping of human and mouse salivary gene regulation. J Dent Res. 98(2):209–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min S, Oyelakin A, Gluck C, Bard JE, Song EC, Smalley K, Che M, Flores E, Sinha S, Romano RA. 2020. P63 and its target follistatin maintain salivary gland stem/progenitor cell function through TGF-β/activin signaling. iScience. 23(9):101524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montoro DT, Haber AL, Biton M, Vinarsky V, Lin B, Birket SE, Yuan F, Chen S, Leung HM, Villoria J, et al. 2018. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 560(7718):319–324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon KR, van Dijk D, Wang Z, Gigante S, Burkhardt DB, Chen WS, Yim K, Elzen AVD, Hirn MJ, Coifman RR, et al. 2019. Visualizing structure and transitions in high-dimensional biological data. Nat Biotechnol. 37(12):1482–1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukaibo T, Gao X, Yang NY, Oei MS, Nakamoto T, Melvin JE. 2019. Sexual dimorphisms in the transcriptomes of murine salivary glands. FEBS Open Bio. 9(5):947–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohtomo R, Mori T, Shibata S, Tsuta K, Maeshima AM, Akazawa C, Watabe Y, Honda K, Yamada T, Yoshimoto S, et al. 2013. SOX10 is a novel marker of acinus and intercalated duct differentiation in salivary gland tumors: a clue to the histogenesis for tumor diagnosis. Mod Pathol. 26(8):1041–1050. [DOI] [PubMed] [Google Scholar]
- Oyelakin A, Song EAC, Min S, Bard JE, Kann JV, Horeth E, Smalley K, Kramer JM, Sinha S, Romano RA. 2019. Transcriptomic and single-cell analysis of the murine parotid gland. J Dent Res. 98(13):1539–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plasschaert LW, Zilionis R, Choo-Wing R, Savova V, Knehr J, Roma G, Klein AM, Jaffe AB. 2018. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 560(7718):377–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitou M, Gaylord EA, Xu E, May AJ, Neznanova L, Nathan S, Grawe A, Chang J, Ryan W, Ruhl S, et al. 2020. Functional specialization of human salivary glands and origins of proteins intrinsic to human saliva. Cell Rep. 33(7):108402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi R, Martin D, Genomics and Computational Biology Core, Yamada KM. 2020. Single-cell RNA-seq identifies cell diversity in embryonic salivary glands. J Dent Res. 99(1):69–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song EC, Min S, Oyelakin A, Smalley K, Bard JE, Liao L, Xu J, Romano RA. 2018. Genetic and scRNA-seq analysis reveals distinct cell populations that contribute to salivary gland development and maintenance. Sci Rep. 8(1):14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares Dos Santos H, Nam K, Maslow FM, Small T, Galloway TLI, Dooley LM, Tassone PT, Zitsch RP, III, Weisman GA, Baker OJ. 2022. Tuft cells are present in submandibular glands across species. J Histochem Cytochem. 70(9):659–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker AS. 2007. Salivary gland development. Semin Cell Dev Biol. 18(2):237–244. [DOI] [PubMed] [Google Scholar]
- Zinn VZ, Khatri A, Mednieks MI, Hand AR. 2015. Localization of cystic fibrosis transmembrane conductance regulator signaling complexes in human salivary gland striated duct cells. Eur J Oral Sci. 123(3):140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-jdr-10.1177_00220345221147908 for High-Resolution Transcriptomic Landscape of the Human Submandibular Gland by E. Horeth, J. Bard, M. Che, T. Wrynn, E.A.C. Song, B. Marzullo, M.S. Burke, S. Popat, T. Loree, J. Zemer, J.L. Tapia, J. Frustino, J.M. Kramer, S. Sinha and R.A. Romano in Journal of Dental Research