Abstract
To accelerate the translation of cancer nanomedicine, we used an integrated genomic approach to improve our understanding of the cellular processes governing nanoparticle trafficking. We developed a massively parallel screen leveraging barcoded, pooled cancer cell lines annotated with multi-omic data to investigate cell association patterns across a nanoparticle library spanning a range of formulations with clinical potential. We identified both materials properties and cell-intrinsic features mediating nanoparticle-cell association. Using machine learning algorithms, we constructed genomic nanoparticle trafficking networks and identified nanoparticle-specific biomarkers. We validated one such biomarker, gene expression of SLC46A3, which inversely predicts lipid-based nanoparticle uptake in vitro and in vivo. Our work establishes the power of integrated screens for nanoparticle delivery and enables the identification and utilization of biomarkers to rationally design nanoformulations.
One-Sentence Summary:
Pooled human cell line screening identifies genomic nanoparticle trafficking networks and predictive biomarkers.
Nanoparticle (NP)-based therapeutics have enormous potential for personalized cancer therapy as they can encapsulate a range of therapeutic cargos including small molecules, biologics and, more recently, nucleic acids. Therapy-loaded NPs can be designed to prevent undesired degradation of the cargo, increase circulation time, and direct drugs specifically to target tumors.(1–3) There have been notable successes in clinical translation of nanomedicines, including liposomal formulations of doxorubicin (Doxil) and irinotecan (Onivyde).(4) These formulations extend the half-life of the active agent and have the potential to lower toxicity, but do not efficiently accumulate in tumors.(5, 6)
Delivery challenges attributed to circulation, immune detection and clearance, as well as extravasation and diffusion through tissue all influence NP accumulation at target disease sites. Efforts to improve NP accumulation in tumors via active targeting motifs have been met with limited success, both in the laboratory and the clinic.(1, 7) Fewer efforts have focused on gaining a fundamental understanding of the biological features mediating successful NP-cell interaction and uptake. While progress has been made in understanding how specific physical and chemical NP properties affect trafficking and uptake, comprehensive evaluation of multiple NP parameters in combination has thus far been elusive. Additionally, the biologic diversity of cancer targets makes it prohibitively challenging to gain a holistic understanding of which NP properties dictate successful trafficking and drug delivery.(8, 9) Once NP parameters are considered in combination, the number of unique formulations to test increases exponentially, particularly as comparisons across several systems need to be drawn. A further barrier is the need to adapt the nanoparticle formulation of each encapsulated therapy for a given drug or target, as each formulation has its own unique biological fate.(9) As therapies continue to increase in molecular complexity, new nanocarrier formulations capable of delivering such entities will need to be developed and examined for their unique trafficking properties.
We and others have designed panels of NPs to elucidate the structure-function relationships to cellular targeting and uptake.(10–13) However, there is a need to equally consider the influence of biological heterogeneity on interactions at the NP-cell interface, for example by probing cells across cancer cell lineages with a range of genetic drivers and cell states. In the era of precision medicine, with the desire to deliver molecularly targeted and gene-based therapies to specific subcellular compartments within cancer cells, it is imperative to holistically probe the structure-function relationship of NPs as they relate to cellular interactions.
Inspired by recent advancements in cancer genomics,(14) we postulated that applying similar techniques to the study of cancer nanomedicine would uncover both the cell- and NP-specific features mediating efficient targeting and delivery. The combination of pooled screening with multi-omic annotation has accelerated target discovery and uncovered previously unrecognized mechanisms of action in small molecule screens. Specifically, in the Profiling Relative Inhibition in Mixtures (PRISM) method, DNA-barcoded mixtures of cells have recently been used for multiplexed viability screening. In cell line pools grouped by doubling time, 500 barcoded cell lines have been screened against tens of thousands of compounds to identify genotype-specific cancer vulnerabilities.(15, 16)
To comprehensively capture pan-cancer complexities and enable the statistical power to link NP association with cell intrinsic characteristics, we developed a competitive phenotypic screen to assess associations of a curated NP library with hundreds of cancer cell lines simultaneously. NP-cell association was correlated with genomic features to identify candidate biomarkers. Coupling our biomarker findings with k-means clustering, we constructed genomic interaction networks associated with NP engagement, enabling the identification of genes associated with the binding, recognition, and subcellular trafficking of distinct NP formulations. Moreover, through the use of univariate analyses and random forest algorithms, we identified that the gene SLC46A3 holds value as a predictive, NP-specific biomarker. We further validated SLC46A3 as a negative regulator of liposomal NP uptake in vitro and in vivo. The strategy outlined herein identifies cellular features underlying nanoparticle engagement in cancer nanomedicine.
Results
nanoPRISM: screening nanoparticle association with pooled cell lines
To screen hundreds of cancer cell lines simultaneously for NP-cancer cell line association patterns, we cultured pooled PRISM cells and incubated them with fluorescent NPs. We then implemented a fluorescence-activated cell sorting (FACS) adaptive gating strategy to sort cell populations into four bins (quartiles, A-D) based on fluorescence signal as a proxy for the extent of NP-cell association (Figure 1A). Experimental parameters were optimized to ensure sufficient cell number and barcode representation post-cell sorting (Figure S1), and NPs were incubated for 4 and 24 hours.
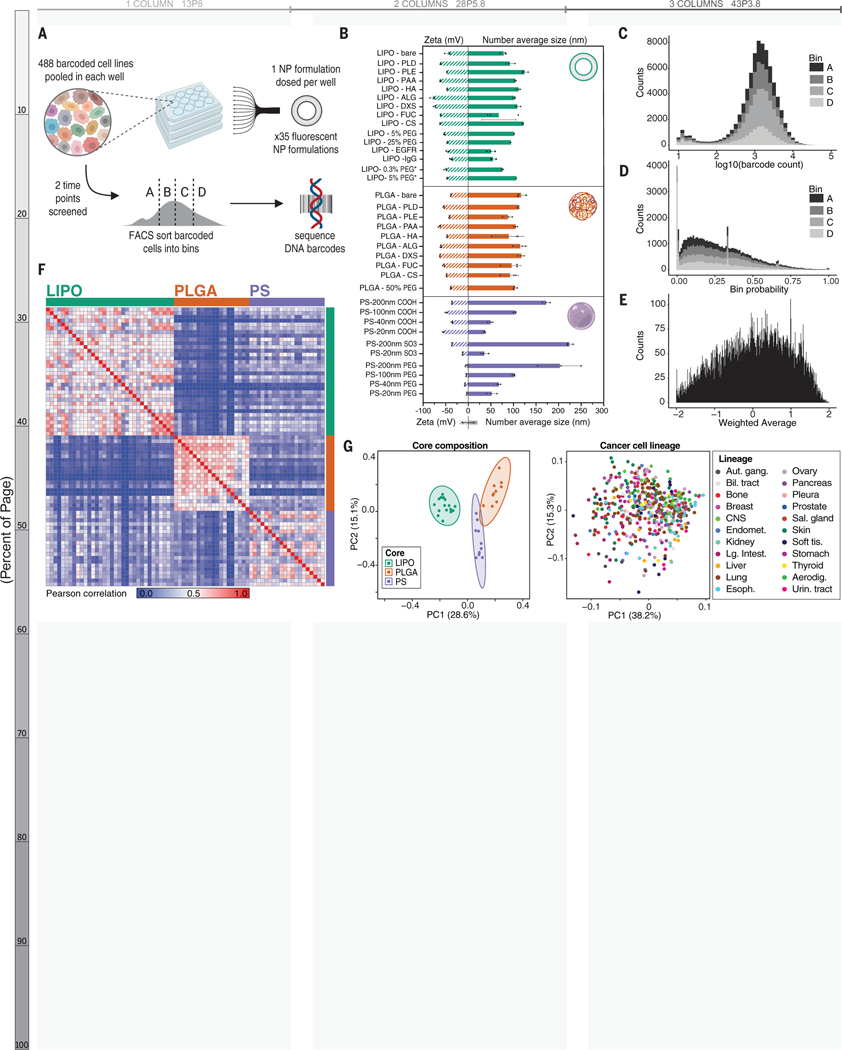
Figure 1. Assessing NP-cell interactions across hundreds of cancer cell lines simultaneously.
(A) Schematic of the nanoPRISM assay: Fluorescently-labeled NPs are incubated with pooled cancer cells before fluorescence-activated cell sorting (FACS) by NP-association and sequencing of DNA barcodes for downstream analyses. (B) Characterization of the diameter and zeta potential of the NP library via dynamic light scattering. Data are represented as the mean and standard deviation of three technical repeats. Formulations marked with an asterisk represent drug-free analogs of clinical liposomal formulations as described in the text. (C) Raw data from the screen were obtained in the form of barcode counts, with similar numerical distribution of barcodes in each bin, represented as a stacked histogram. (D) Accounting for baseline differences in barcode representation yields the probability (P) that each cell line will be found in a particular bin. (E) Probabilities are collapsed into a single weighted average (WA) for each NP-cell line pair. (F) A similarity matrix collapsing WA values for 488 cell lines reveals clusters of NP formulations with the same core formulation. (G) Principal component analysis (PCA) of NP-cell line WA values at 24 h confirms distinct clustering of NP formulations based on core composition (left) but cell lines do not form clusters (right).
For this screen, we designed a modular NP library to capture the effects of NP core composition, surface chemistry, and size on cell interactions. This panel of 35 NPs encompassed both clinical and experimental formulations. Specifically, anionic liposomes were formulated and electrostatically coated with cationic poly-L-arginine (PLR) followed by a series of polyanions.(17–21) The polyanions were selected for their synthetic (polyacrylic acid, PAA), semisynthetic (poly-L-aspartate, PLD; poly-L-glutamate, PLE), or natural (hyaluronate, HA; dextran sulfate, DXS; fucoidan, FUC; alginate, ALG; chondroitin sulfate, CS) origin as well as the inclusion of both carboxylate and sulfate ions.(22–24) These same electrostatic coatings were used to modify polymeric NP cores (polylactide-co-glycolide, PLGA) to test the effects of core composition on NP-cell interactions. We optimized formulations to obtain a diameter of approximately 100 nm for the liposome and PLGA formulations as the similar sizes would enable cross-core comparisons. We also included commercially manufactured fluorescent carboxylate- and sulfate-modified polystyrene (PS) nanoparticles in a range of diameters from 20–200 nm, enabling study of particle size and surface chemistry. Because of the clinical importance of polyethylene glycol (PEG)-containing formulations,(25) PEGylated versions of liposome, PLGA, and PS particles were prepared, including the drug-free versions of two commercial formulations, liposomal doxorubicin (Doxil) and liposomal irinotecan (Onyvide). The latter two formulations are denoted as LIPO-5% PEG* and LIPO-0.3% PEG*, respectively. All of the nanoparticles examined exhibited negative or neutral net charge, as the focus of this work is on systemic nanoparticle delivery systems. Positively charged nanoparticles have been shown to undergo nonspecific charge interactions with cells and proteins, leading to toxicity and premature clearance in vivo.(26) Dynamic light scattering (DLS) was used to characterize the diameter, zeta potential, and polydispersity index (Figure 1B, Tables S1–S3) of this NP library.
To ensure that our methods led to robust and meaningful data we selected an anti-epidermal growth factor receptor (EGFR) antibody as an active targeting control. We hypothesized that the design of our screen would allow us to identify features relevant to EGFR expression with a high level of confidence. A nonlethal EGFR antibody or IgG isotype control was covalently incorporated onto a liposome via a PEG tether.(27) We elected to focus on EGFR due to the wide range of native EGFR expression of the 488 cell lines included in our screen as well as prior evaluation of EGFR-targeting compounds via the PRISM assay (Figure S2).(15)
After incubating the cells with the NP library, we utilized fluorescence-activated cell sorting to bin cells into quartiles according to fluorescence intensity (Figure S3). Cells were then lysed, and the DNA barcodes were amplified, sequenced, and deconvoluted according to previously detailed protocols.(15, 28) After quality control analysis of technical (n=2) and biological (n=3) replicates, all 488 cell lines met quality control measures and were carried forward for downstream analyses (Figure S4). This dynamic gating strategy was used to enable comparison of cell line representation per bin (quartile) independent of fluorophore identity or amount incorporated into each tested formulation.
A probabilistic model was developed and applied to the data to infer the relative distribution of each cell line into the pre-determined bins (A-D) for each NP formulation. The probability of a cell from a given cell line falling into a given bin is used to represent those distributions, i.e., PA+ PB+ PC+ PD = 1 (Figure 1C–D). The technical details and the model’s implementation are presented in the Supplementary Materials (29). Given the concordance of the inferred probabilities among the biologic replicates (Figure S5), we collapsed the replicates through their arithmetic average. Probabilities were then summarized using a weighting factor alpha (α) to calculate a weighted average (WA) for each NP-cell line pair: WA = -αPA-PB+PC+αPD in which a higher WA implies higher NP-cell association and vice versa (Figure 1E). We trialed a range of weighting factors (α = 2, 10, 20, and 100) and found that downstream results were unchanged with the higher α values (Figure S6), and therefore, α = 2 was used for subsequent analyses.
Cancer cells distinguish nanoparticles based on core composition
Pearson-based unsupervised hierarchical clustering of pairwise WAs identified NP core material as a strong determinant of cell association, with the three core materials tested (liposomal, PLGA, and PS) forming distinct clusters (Figure 1F and S7A). This result was unexpected as we hypothesized surface chemistry to be a larger predictor of NP-cell interactions. Principal component analysis (PCA) similarly identified core specific trends at both the 4 and 24 hour time points (Figures 1G and S7B–C. Further analysis within each core material did reveal surface chemistry dependent trends, though they were more subtle than core-based clustering (Figure S8).
In contrast, no clusters were apparent when PCA was performed based on cell line, indicating that cancer cells of the same lineage did not have similar NP-association trends (Figure 1H, Figure S7B–C). Heterogeneity in NP-cell association in proliferating cells has been attributed to various aspects of cell growth and metabolism.(30–33) To ensure that differential cell proliferation did not confound our results, we performed a parallel growth experiment with the same pooled cells and found no correlation between estimated doubling time and WA (Figure S9).
Cell-intrinsic features mediate nanoparticle trafficking
We applied data from the Cancer Cell Line Encyclopedia (CCLE)(34, 35) to identify genomic features that act as predictive biomarkers for NP-cell association. To do this, we used both univariate analyses and a random forest algorithm to correlate the baseline molecular features of each cell line (cell lineage; gene copy number; messenger RNA, microRNA, protein or metabolite abundance; function-damaging, hotspot, or missense mutations) with NP association (Figure S10A–B).
EGFR-targeting compounds identified relevant biomarkers with high confidence
Using univariate analysis for all CCLE features, we identified EGFR gene expression and protein abundance as the two most significantly correlated hits (q = 4× 10−100 and q= 4×10−76, respectively) with anti-EGFR antibody, but much less significantly (q = 6 × 10−9 and q = 4 × 10−10, respectively) associated with the isotype control (Figure 2A, top panels). We also confirmed that fluorophore identity does not impact biomarker identification, demonstrating that both AlexaFluor 488 and Cy5 conjugated anti-EGFR antibodies perform similarly (Figure S10C–E).
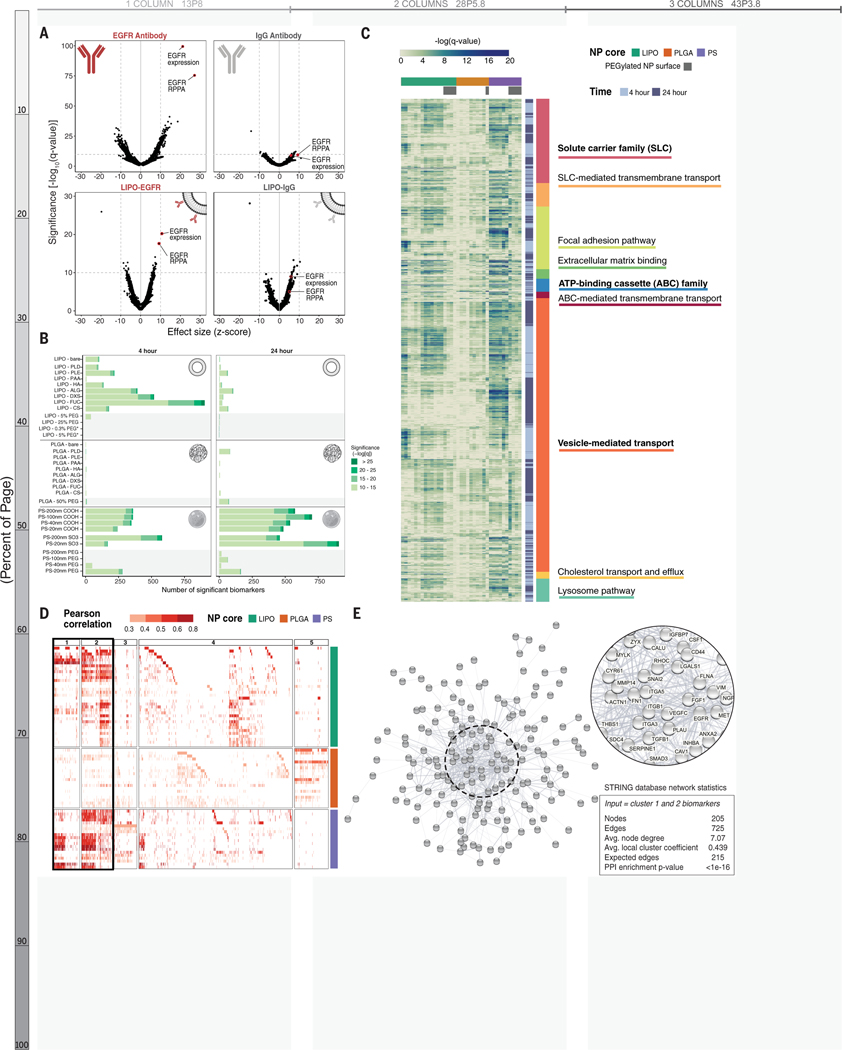
Figure 2. Correlative genomic analysis identifies expected validation biomarkers as well as hundreds of formulation- and time-dependent biomarkers.
(A) Cells with high EGFR antibody association are strongly correlated with EGFR gene expression and protein abundance (via reverse phase protein array; RPPA) (top left). These correlations are diminished in the isotype control-treated sample. The same EGFR-related hits, in addition to NP specific markers, are observed for cells treated with antibody-conjugated liposomes (bottom row). (B) Univariate analysis identifies genomic features correlated with NP association. All biomarkers meeting a significance threshold of -log10(q-value) >10 are shown as stacked bar graphs separated by NP formulation and time point. PEGylated NP formulations are highlighted with a gray background. (C) A heatmap showing the significance of biomarkers associated with established transport, uptake, and adhesion gene sets. Gene set headings are bolded and subsections are listed below respective headings. (D) A heatmap showing all gene- and protein- expression features with positive correlation identified by random forest algorithm in columns, and NP formulations in rows. Features are colored based on their Pearson correlation and clustered using k-means clustering, with clusters 1+2 highlighted as features present across multiple NP formulations. (E) Visual representation of the STRING network generated by inputting the 205 features from clusters 1+2, with network statistics. Each node represents a feature, and the edges represent predicted functional associations. The most interconnected nodes are labeled in the zoomed inset.
In EGFR-conjugated liposomes, the same hits were also identified more significantly (q=6×10−21 and q=2×10−18, respectively) than the IgG control (q = 3 × 10−9 and q = 3 × 10−6, respectively) (Figure 2A, bottom panels).
The statistical significance of EGFR biomarkers was lower for the antibody-conjugated liposome than the free antibody, which may be due to changes in protein concentration across samples or steric blockage introduced by covalently linking an antibody to a NP surface that may interfere with binding to its target.(36) Thus, we demonstrated the ability to quantitatively compare expected biomarker targets of both free antibodies and antibody-conjugated NPs using our platform.
Biomarker number and identity are influenced by nanoparticle properties
We applied univariate analysis to correlate association and CCLE features for each NP formulation, both quantitatively and qualitatively using curated gene sets. First, we thresholded q-values at less than 1×10−10 to compare the absolute number of candidate biomarkers at varying degrees of significance (Figure 2B). Selection of this cutoff was guided by the IgG-conjugated antibody analysis, which returned few hits above this threshold. For liposomal NPs, we observed that the number of significant biomarkers was higher at 4 h than 24 h. We believe this may be indicative of active uptake processes, established to take place within the first few hours of NP-cell interactions, whereas at 24 hours, we may be capturing features associated with less specific interactions.(37, 38) We next investigated biomarkers associated with established uptake, transport, and adhesion gene sets (Figure 2C). (39–41) To examine the distribution of biomarker significance across curated gene sets and NP formulations, each gene was visualized using the -log(q-value) for gene expression. As expected, we identified highly significant biomarkers from gene sets important in drug import and export such as solute carrier (SLC) transporter family and ATP-binding cassette (ABC) family. Our screen provides data on both the significance and the relationship to NP delivery. For example, we found that ABCA1, which plays a role in cholesterol transport, has a positive relationship with liposomal NPs, while several members of the multidrug resistance subfamily (ABCB1/P-GP, ABCC1/MRP, ABCC4/MRP4) have a negative relationship with PLGA NPs (Figure S11).(42) We also identified biomarkers important for cell engagement (focal adhesion, extracellular matrix) as well as intracellular trafficking (vesicular transport, lysosome, and cholesterol transport). This highlights the ability of our screen to identify expected biomarkers and enable comparison between drug delivery modalities.
We also observed that liposome surface modification influences the number and significance of biomarkers. Specifically, liposomes electrostatically coated with polysaccharides (HA, ALG, DXS, FUC, CS) had the highest amount of associated biomarkers, which we hypothesize is due to the high degree of interactions between sugars and cell surface proteins as well as the potential for naturally occurring polysaccharides to interact with a wide range of cell surface elements.(23, 43, 44) In line with this hypothesis, the addition of PEG, a well-established antifouling polymer, reduces the number and significance of associated biomarkers almost to zero. In light of the highly specific hits generated from EGFR-conjugated liposomes (formulated using 25% PEG liposomes), this abrupt decrease in significant biomarkers further indicates the ability of our platform to identify specific NP binding and recognition elements. In contrast to the liposomal formulations, PLGA formulations, regardless of surface modification, resulted in few biomarkers at either time point. Lastly, a high number of significant biomarkers was associated with both carboxylated and sulfated PS NPs included in our screen, though there was no time dependence, in contrast to the liposomal formulation. While this result was initially surprising, as the PS formulations are made of synthetic polystyrene polymers, meaningful biological interactions with anionic polystyrenes, both in polymer and particle form, have been reported. Specifically, it was described that nanoparticles bearing anionic polystyrene motifs have the appropriate mix of hydrophobicity and anionic charge character to interact favorably with trafficking proteins, including the caveolins.(45)
NP biomarkers are connected and create trafficking networks
We then used an unbiased approach to identify predictive biomarkers using a random-forest algorithm, annotated by feature set: gene expression, gene copy number, and protein abundance. Data from the 4 h time point were chosen for this analysis based on the EGFR-related hits for liposomes, which were more significant at 4 h than at 24 h. As we were interested in applying this approach to identify cellular features positively correlated with uptake (for example, increased expression of trafficking proteins), hits negatively correlated with NP association were removed from this analysis. Next, we used K-means clustering to visualize biomarkers based on their relative importance and presence across formulations (Figure 2D). Clusters 1 and 2 contained 205 hits shared across NP formulations and were especially enriched for liposomal and PS NPs. These genes and proteins were input into the STRING database(46–48) to generate a protein-protein interaction (PPI) network that was found to be highly interconnected (PPI enrichment p-value <1×10−16) (Figure 2E). Notably, the network is enriched in proteins found in the plasma membrane, extracellular region, and extracellular matrix (false discovery rate [FDR] = 8×10−12, 3×10−9, and 3×10−8, respectively) based on enrichment analysis with gene ontology (GO) localization datasets (Figure S12).(49–51) The identification of overlapping biomarkers that are localized to the cell surface and have established protein-protein interactions led us to hypothesize that these proteins are important in early NP trafficking. Enrichment analyses using GO molecular functions datasets showed enrichment in numerous binding processes (Data S1, Figure S12), giving further credence to this theory.
SLC46A3 is a negative regulator of liposomal NP uptake
Evaluating univariate results across NP formulations, we identified one biomarker with a strong, inverse relationship with liposomal NP association: expression of solute carrier family 46 member 3 (SLC46A3). A member of the solute carrier (SLC) transporter family, SLC46A3, is a relatively unstudied transporter that has been localized to the lysosome.(52, 53) SLC46A3 was recently identified as a modulator of cytosolic copper homeostasis in hepatocytes, connecting hepatic copper concentrations with lipid catabolism and mitochondrial function.(54) This reported relationship between SLC46A3 and lipid catabolism may help to explain why SLC46A3 found to have a strong relationship with liposomal NP uptake and not uptake of polymeric NPs. In the context of cancer, SLC46A3 was recently shown to transport non-cleavable antibody-drug conjugate (ADC) catabolites from the lysosome to the cytosol, thereby being necessary for therapeutic efficacy.(55) Further, downregulation of SLC46A3 was identified as a resistance mechanism for antibody-drug conjugate delivery in cancer cells, including in patient samples of multiple myeloma.(55–58) While the biologic function of SLC46A3 in cancer is not yet clear, given the potential therapeutic implications and the unusual inverse relationship between SLC46A3 expression and NP delivery, we sought to validate the predictive power of SLC46A3 as a biomarker for liposomal NP association.
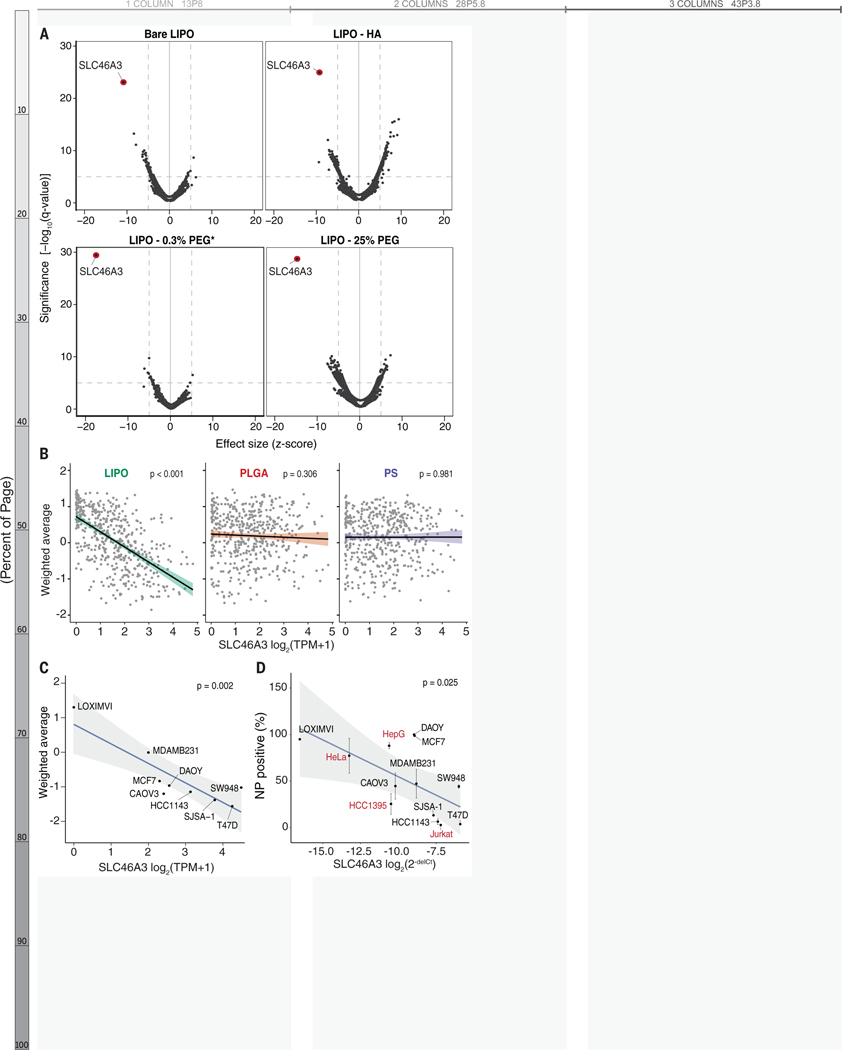
SLC46A3 expression was the most significant hit on univariate analysis and also the top ranked random forest feature for each liposomal NP tested at 24 h, regardless of surface modification (q-values < 10−20, Figures 3A and S13). This inverse relationship between SLC46A3 expression and NP association was found to be specific to liposomal NPs, and not observed with PLGA or PS NPs, and was maintained regardless of cancer cell lineage (Figures 3B and S13).
Figure 3. Native expression of the lysosomal transporter SLC46A3 predicts NP-cell interaction for liposome formulations.
(A) Univariate analysis identified SLC46A3 expression as strongly inversely correlated with liposome association, regardless of liposomal surface modification. (B) Using linear regression to evaluate the biomarker relationship across core formulations revealed SLC46A3 expression was inversely correlated with NP association in liposome-cell line pairs (p < 0.001) but not PLGA- and PS-cell line pairs (p > 0.05); n=488 for each plot. (C) Cell lines in the nanoPRISM pool exhibited a range of native SLC46A3 expression and a log linear correlation with uptake of bare liposomes. (D) This same correlation was also exhibited when assessing liposome-cell associations via flow cytometry in a non-pooled fashion (p = 0.025). Cell lines in red were not part of the pooled PRISM screen. Data represented in D are shown as the mean and standard deviation of four biological replicates. Error bars are not shown when smaller than data points.
We selected nine cancer cell lines from the nanoPRISM pool and four additional cell lines, spanning multiple lineages, with a range of native SLC46A3 expression levels for screening in a non-pooled fashion (Figures 3C–D, S3, S14–S15). Analogous to the pooled screen, individual cell lines were profiled using flow cytometry and NP-associated fluorescence was quantified after 24 h incubation; here SLC46A3 expression was concurrently quantified using quantitative polymerase chain reaction (qPCR) (Figures 3D and S9). In line with observations from pooled screening, the inverse relationship between liposome association and native SLC46A3 expression was maintained, suggesting that SLC46A3 may play a key role in regulating the degree of liposomal NP uptake.
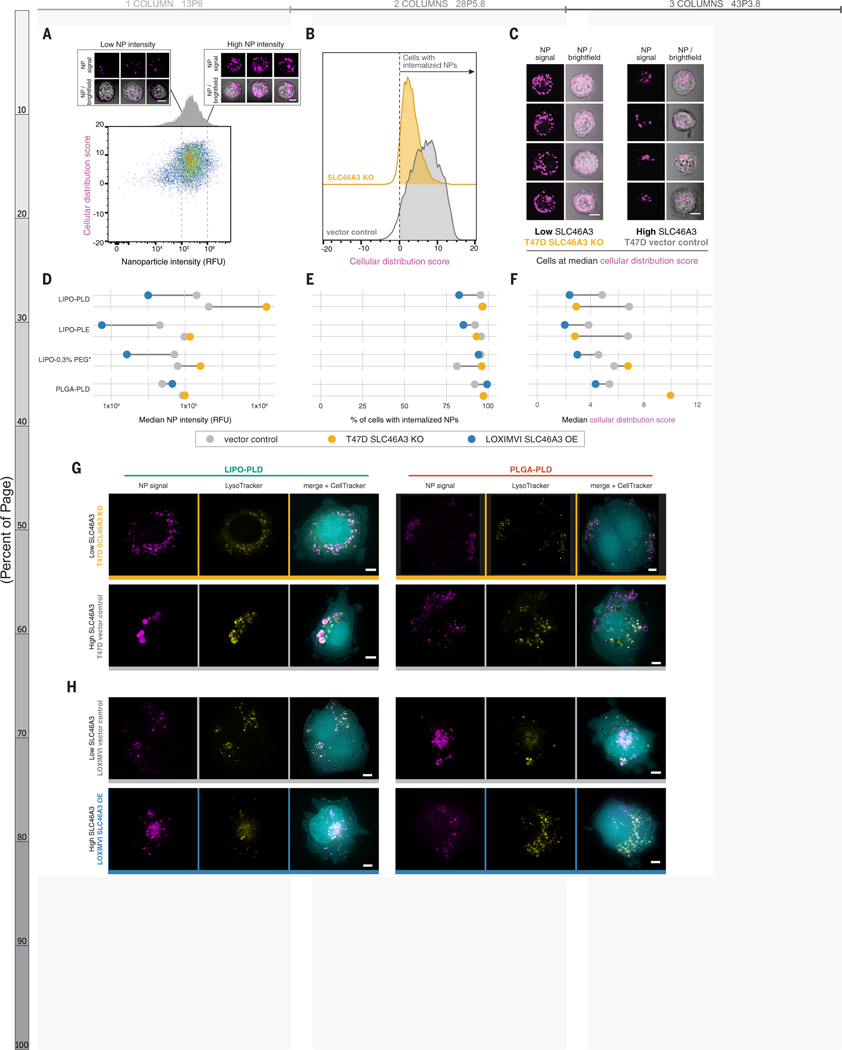
To probe whether SLC46A3 governs cellular association with NPs, we selected the breast cancer cell line T47D, which exhibited high native SLC46A3 (Figure 4A). We knocked down SLC46A3 with siRNA and evaluated the effect on liposomal NP association. We observed that T47D cells with reduced SLC46A3 had higher NP-cell association with both tested formulations, suggesting that modulating SLC46A3 expression alone can regulate NP-cell association (Figure 4B).
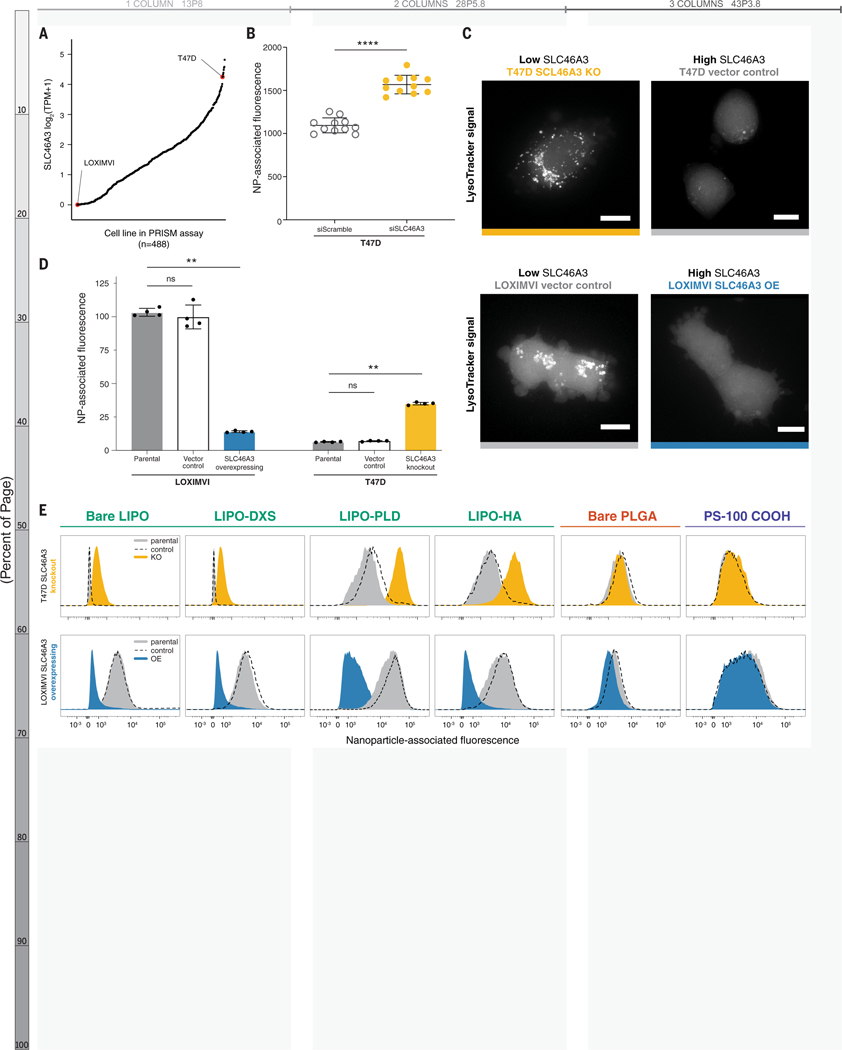
Figure 4. Modulating SLC46A3 expression in cancer cell lines is sufficient to negatively regulate interaction with liposome NP formulations.
(A) T47D and LOXIMVI cells have high and low SLC46A3 expression, respectively, among the cells in the nanoPRISM cell line pool. (B) T47D cells treated with siRNA to knock down SLC46A3 have higher uptake of Lipo-PLD compared to T47D cells treated with a scrambled siRNA control (**** p < 0.0001, Mann-Whitney test). (C) Representative micrographs of Lysotracker signal in engineered cell lines showed endolysosomal compartments. Scale bars = 10 μm. (D) Using lentivirus to overexpress SLC46A3 in LOXIMVI cells and CRISPR/Cas9 to knock out SLC46A3 in T47D cells, we showed that modulation results in significantly changed liposome association, as determined by flow cytometry (** p < 0.001, Kruskal-Wallis test). NP-associated fluorescence is defined as median fluorescence intensity normalized to untreated cells. Data are represented as the mean and standard deviation of four biological replicates. (E) Shifts in NP association were consistently observed across all tested liposomes, independent of surface modification. No shifts were observed with PLGA or PS formulations.
To further functionally evaluate the relationship of SLC46A3 expression and NP-cell association, we selected two cancer cell lines from the pooled screen (Figure 4A): the T47D cell line and the melanoma cell line LOXIMVI. We developed a toolkit using these two cell lines by knocking out SLC46A3 in T47D cells and inducing SLC46A3 overexpression in LOXIMVIs (Figures S16A–D).
As SLC46A3 is a protein associated with lysosomal membranes(55, 56, 59), we used LysoTracker dye to evaluate the effect of SLC46A3 modulation on endolysosomal compartments in both T47D and LOXIMVI engineered cell lines (Figure 4C). We observed an SLC46A3-dependent change: cells with lower SLC46A3 expression (T47D-SLC46A3 knockout, LOXIMVI-vector control) exhibited more brightly dyed endolysosomal compartments compared to their high SLC46A3 expression counterparts (T47D-vector control, LOXIMVI-SLC46A3 OE).
Overexpression of SLC46A3 in LOXIMVI cells significantly abrogated interaction with bare liposomes (p = 0.006) using flow cytometry profiling (Figure 4D). The T47D-SLC46A3 knockout cell line demonstrated significantly increased association with bare liposomes compared to parental or vector control lines (p = 0.0017, Figure 4D). We further confirmed that these trends are generalizable across a range of surface functionalized liposomes (Figure 4E, S16E). Moreover, no significant changes in NP association were observed for PLGA and PS NPs (Figures 4E, S16F–G). We also confirmed that the presence of serum proteins in cell culture media does not abrogate this trend (Figure S16H). Taken together, these data indicate modulation of SLC46A3 alone in cancer cells is sufficient to negatively regulate association and uptake of liposomal NPs.
Because flow cytometry does not provide spatial information with respect to NP-cell interactions, we used imaging cytometry to characterize NP localization in a high throughput manner (Figure 5A–F). We selected four representative formulations: three liposomal NPs to probe the relationship of SLC46A3 expression with liposome trafficking and one PLGA NP formulation with the same outer layer (PLD).
Figure 5. High-throughput imaging cytometry confirmed NP internalization and revealed SLC46A3-dependent changes to intracellular trafficking.
(A) Imaging cytometry was used to investigate the intensity (x-axis) and distribution (y-axis) of NPs in a high-throughput manner. Bivariate density plot of n=10,000 cells (T47D-vector control) after 24 h incubation with LIPO-PLD NPs, with representative cell images at low and high NP signal. (B) Cellular distribution patterns of NPs were scored such that scores greater than 0 indicate cells with internalized NPs. Representative data from LIPO-PLD NPs in engineered T47D cells are shown. (C) Representative cell images at the median cellular distribution score for engineered T47D cells treated with LIPO-PLD NPs. (D) Quantification of median intensity of tested NP formulations in engineered T47D and LOXIMVI cell lines demonstrated SLC46A3-dependent changes. (E) NPs remained predominantly internalized independent of SLC46A3 expression. (F) Shifts in the median cellular distribution scores were observed in response to SLC46A3 modulation. Live cell micrographs of (G) T47D-vector control and T47D-SLC46A3 knockout cells and (H) LOXIMVI-vector control and LOXIMVI-SLC46A3 OE cells incubated with LIPO-PLD and PLGA-PLD NPs for 24h. NP signal is pseudo-colored magenta, LysoTracker signal yellow, and CellTracker cyan. For A and C, scale bar = 7 μm. For G-H, scale bar = 5 μm.
Consistent with trends observed by flow cytometry, we observed an inverse relationship between NP intensity and SLC46A3 expression for liposomal, but not PLGA, NPs (Fig 5A, D, S11). Using brightfield images, we applied a mask to investigate cellular localization of NPs. All tested formulations were internalized, and this did not change with SLC46A3 modulation (Figure 5B, E).
We investigated localization of NPs by scoring NP signal based on distribution within each cell (Figure 5C, F, Figure S17). We observed stark differences in median cellular distribution scores of liposomal NPs in relation to SLC46A3 expression in T47D cells. This was not observed for PLGA NPs, mimicking the previously observed core-specific relationship between NP-cell association and SLC46A3 expression. Changes in this score, though less pronounced, were also observed for liposomal NPs in LOXIMVI cells.
To confirm our findings with higher spatial resolution, we used deconvolution microscopy of live cells and incorporated a lysosomal stain to observe changes in intracellular trafficking (Figure 5G–H). NPs appeared uniformly distributed within T47D-SLC46A3 KO cells, co-localizing with endolysosomal vesicles. In contrast, LIPO-PLD NPs were localized to large endolysosomal clusters in T47D-vector control cells. This trend was also observed for LIPO-PLE and LIPO-0.3% PEG* NPs and at the earlier time point of 4 h (Figure S18). Changes in localization were not observed for the tested PLGA PLD NPs. This again indicates a NP core-dependent relationship with SLC46A3.
In the engineered LOXIMVI cell lines, we also observed co-localization of liposomal NPs with endolysosomal signal (Figure 5H). However, predictable changes in NP localization were not detected, in line with smaller changes in median cellular distribution scores.
Impact of SLC46A3 expression on endolysosomal maturation is minimal
To further probe the relationship between intracellular liposomal NP trafficking and SLC46A3 expression, we used imaging cytometry to spatially interrogate markers of endolysosomal transport. We elected to study markers of early (EEA1, RAB5A), late (RAB7), and recycling endosomes (RAB11) as well as lysosomes (LAMP1) in engineered LOXIMVI cells (Figures S19–S20, Table S4). While no apparent differences in endolysosomal marker signal strength, size, and shape were observed when comparing LOXIMVI-SLC46A3 OE and LOXIMVI-vector control cells both in the absence and in the presence of liposomal NPs, modest changes in EEA1, RAB7, and LAMP1 texture were noted (Figure S19A–B).
We then assigned values to the colocalization between each endolysosomal marker and NP signals and observed increasing colocalization from EEA1 to RAB5 to RAB7, consistent with liposome trafficking from early to late endosomes (Figure S19C–F). Colocalization between RAB7 and liposomal NPs was higher in LOXIMVI-SLC46A3 OE cells compared to vector control and the opposite relationship was observed for LAMP1 colocalization.
Liposome retention and accumulation remains SLC46A3-dependent in vivo
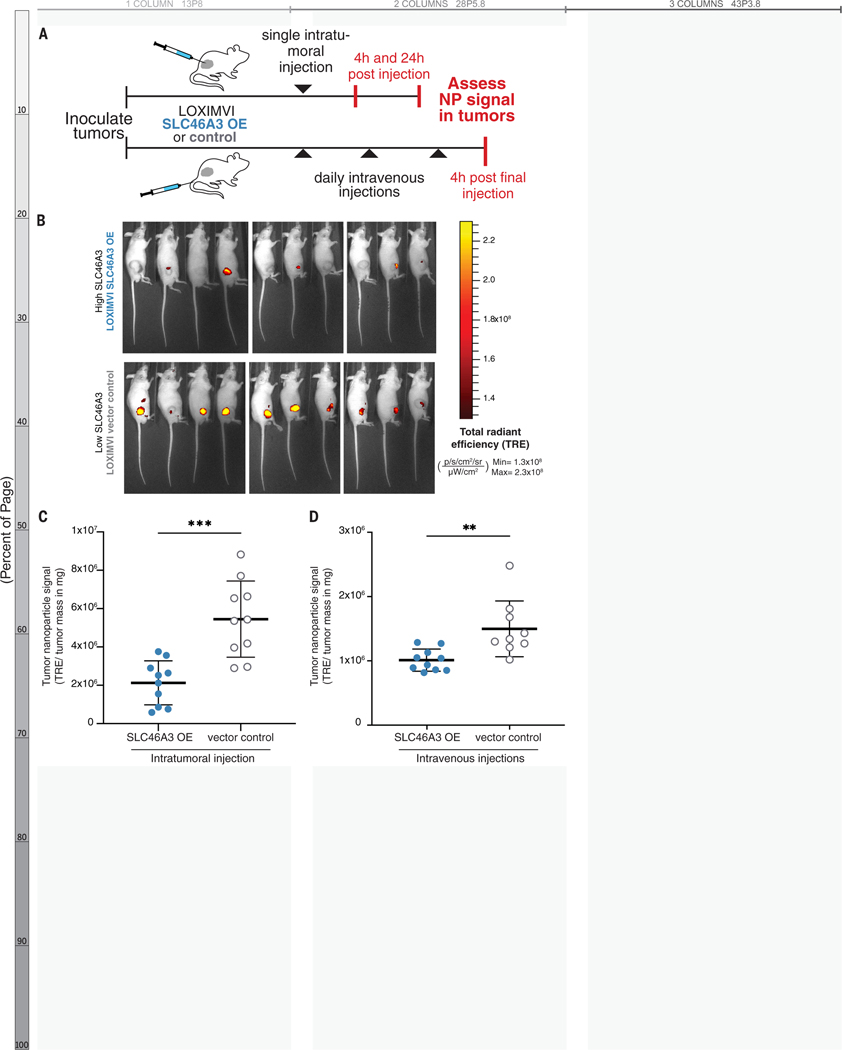
To evaluate the potential clinical utility of SLC46A3 as a negative regulator of liposomal NP delivery, we tested in vivo delivery of an FDA-approved nanoparticle analog, the drug-free version of liposomal irinotecan (LIPO-0.3% PEG*), in mice bearing subcutaneous LOXIMVI flank tumors. Fluorescently-labeled NPs were administered via a one-time intratumoral (IT) injection or repeat intravenous (IV) administration to evaluate tumor retention and accumulation, respectively (Figure 6A, Figure S21).
Figure 6. Retention and accumulation of PEGylated liposomes (LIPO-0.3% PEG*) in LOXIMVI tumors is dependent on SLC46A3 expression.
(A) Fluorescently labeled LIPO-0.3% PEG* NPs were administered to mice bearing LOXIMVI flank tumors via a one-time intratumoral injection or repeat intravenous injections. (B) Whole animal fluorescence images of mice (4 males, 6 females per group) 24 h after being intratumorally injected with LIPO-0.3% PEG* NPs. (C) Quantification of LIPO-0.3% PEG* NP retention 24 h after intratumoral administration to LOXIMVI flank tumors. (D) Quantification of LIPO-0.3% PEG* NP accumulation after repeat IV injections. In panels C-D, nanoparticle signal is expressed on the y-axis as total radiant efficiency divided by tumor mass, units provided in figure. The mean and standard deviation of n = 10 are shown with the exception of the LOXIMVI-vector control, repeat IV injection group, where n = 9 (** < 0.01, *** < 0.001, Mann-Whitney test).
NP signal was quantified at both 4 and 24 h after IT administration. In line with our hypothesis, as well as with in vitro NP-associated fluorescence data (Figure S21A), we observed an inverse relationship between SLC46A3 expression and LIPO-0.3% PEG* NP retention that became more pronounced over time (p = 0.0115, 4 h; p = 0.0002, 24 h) (Figure 6B–C, Figure S21B–E). Moreover, these findings also align with our initial nanoPRISM findings, in which SLC46A3 expression was a more significant biomarker at 24 h (q-value=3.49×10−30, Data S2, Figure S13A) than at 4 h (q-value=1.47×10−4, Data S2, Figure S13A).
To determine if SLC46A3 expression predictably governs accumulation of nontargeted NPs, which bear no specific functional ligands on their surface, after systemic administration, we quantified NP signal after IV injections. We observed a significant relationship between SLC46A3 and NP accumulation (p = 0.0019) (Figure 6D, Figure S21F). This demonstrates that baseline tumor expression of SLC46A3 may influence NP delivery in a physiologic setting.
Together, these data highlight the real-world relevance of the nanoPRISM screening assay in general as well as the utility of SLC46A3 in particular as a potential biomarker.
Solid lipid nanoparticle uptake and transfection are dependent on SLC46A3 expression
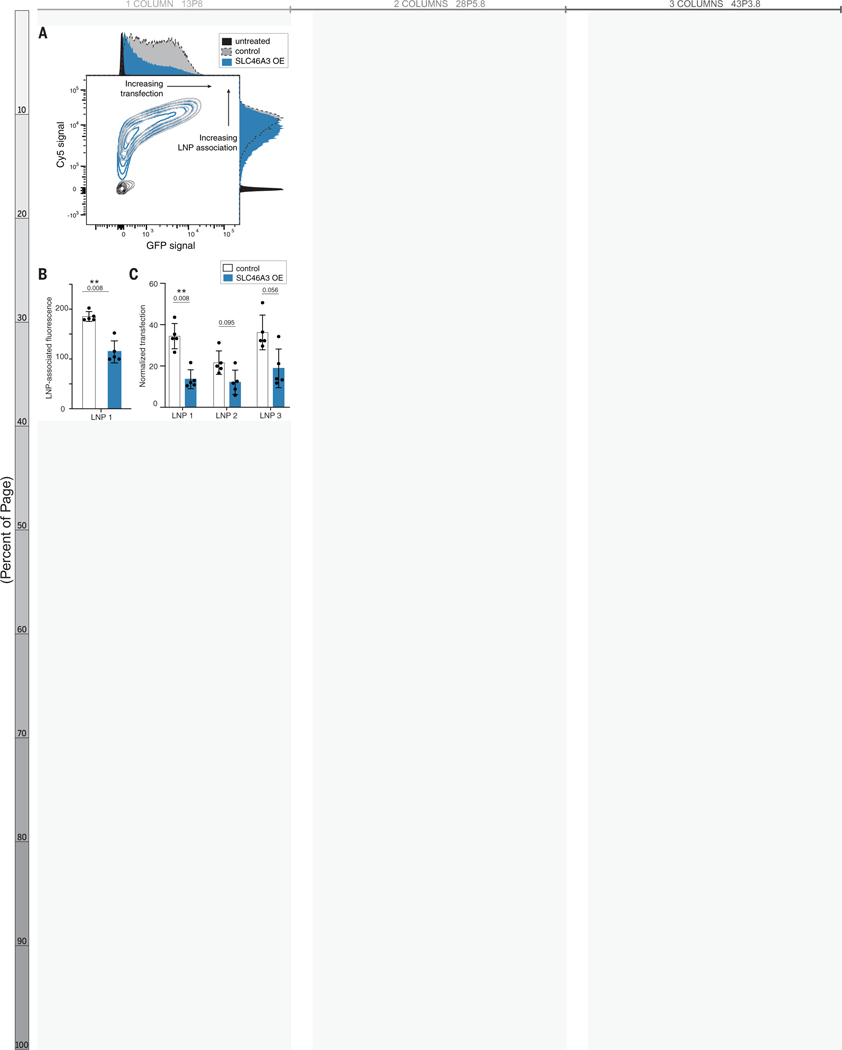
Given the recent translational success and promising potential of nucleic acid-carrying solid lipid nanoparticles (LNPs),(60, 61) we sought to determine if the relationship of SLC46A3 expression extends to LNP association as well as transfection efficiency. We generated fluorescently (Cy5) labeled LNPs containing messenger RNA (mRNA) encoding green fluorescent protein (GFP) (LNP 1) and incubated these particles with engineered LOXIMVI cell lines (Tables S3, S5).
LNP association, as quantified by Cy5 signal, was significantly lower for LOXIMVI-SLC46A3 OE cells than LOXIMVI-vector control cells, showing the same relationship (lower SLC46A3 expression correlating with higher association) for LNPs as for liposomal NPs (p = 0.008, Figure 7A–B). A similarly inverse relationship with SLC46A3 expression was seen for transfection, as quantified by GFP signal of formulation LNP 1 (Figure 7C). Taken together, these findings suggest that SLC46A3 regulates cytosolic delivery of mRNA cargo by way of LNP uptake. Expanding on this, we generated two additional LNPs, analogous to commercial formulations (Table S5).(62–65) While we observed lower transfection in LOXIMVI-SLC46A3 OE cells than LOXIMVI-vector control cells, these differences were not statistically significant (p > 0.05). Nevertheless, the inverse relationship between SLC46A3 expression and cell association in multiple LNP formulations supports the relevance of SLC46A3 as a predictive biomarker for lipid-based nanoparticle formulations.
Figure 7. Solid lipid nanoparticle-cell association and transfection are SLC46A3-dependent, as determined via flow cytometry.
(A) Contour plot of Cy5 signal and GFP signal indicating decreased LNP-cell association and transfection efficacy in LOXIMVI cells overexpressing SLC46A3. (B) Quantification of LNP signal revealed a significant change in LNP-cell association across control and SLC46A3-overexpressing LOXIMVI cells (** p = 0.008, Mann-Whitney). LNP-associated fluorescence is defined as median fluorescence intensity normalized to untreated cells. (C) LOXIMVI-SLC46A3 OE cells exhibited lower transfection efficiency than LOXIMVI-vector control cells following dosing of three different LNP formulations (Mann-Whitney). Normalized transfection is defined as median GFP intensity normalized to untreated cells.
Discussion
This work represents high-throughput interrogation of NP-cancer cell interactions through the lens of multi-omics. Harnessing the power of pooled screening and high-throughput sequencing, we developed and validated a platform to identify predictive biomarkers for NP interactions with cancer cells. We used this platform to screen a 35 member NP library against a panel of 488 cancer cell lines. This enabled the comprehensive study and identification of key parameters mediating NP-cell interactions, highlighting the importance of considering both nanomaterials and cellular features in concert.
While pooled screening is a powerful tool, we also note several important limitations. First, we primarily focused on lipid-based and polymeric NP formulations with translational drug delivery potential. We recognize that there are several additional categories of nanomaterials with wide-ranging properties, such as inorganic systems, that can be useful for both therapeutic and diagnostic applications(66, 67) and believe additional biomarkers mediating the trafficking of inorganic NPs may be identified using similar screening approaches. Second, the results of in vitro screens are often met with limited success when translated in vivo, as NP-mediated delivery is dependent on many factors beyond the nano-cell interface.(8) However, the level of molecular characterization and statistical/computational power afforded by annotated biological datasets, such as the Cancer Cell Line Encyclopedia, is currently unrivaled. Therefore, existing in vivo screens cannot yet provide this breadth or statistical power. Keeping translational barriers in mind is key to successful validation of candidate biomarkers, and for this reason we used multiple isogenic models and tested a range of lipid-based nanoparticles across in vitro and in vivo conditions. Third, an additional limitation of this screen is related to the availability of genomic datasets for each cell line tested, as dataset completeness contributes to the power of detection for both univariate and multivariate analyses. At the time of analysis, ten feature sets were available for the majority of cell lines in our pool. However, as datasets expand over time, it will be possible to re-analyze our data in the future. Especially for emerging fields such as proteomics and metabolomics, the opportunity to intersect nanoparticle delivery metrics with additional datasets could add a new dimension to our existing findings.
One strength of our screening approach is the use of robust analytical tools, such as univariate analyses and random forest algorithms, enabling us to identify biomarkers correlated with NP association. The robust and quantitative manner in which we detected EGFR hits for antibodies as well as antibody-targeted NPs shows the utility of this platform for the development and optimization of targeted drug delivery platforms, including antibody-targeted NPs, and its potential to apply to other targeted therapeutics, including ADCs. This method of analysis will provide therapeutic insights in the design of antibody-drug conjugates, specifically in evaluating the effects of conjugation site or linker chemistry.
By clustering NP-specific biomarkers across formulations, we constructed interaction networks, identifying and connecting genes associated with NP binding, recognition, and subcellular trafficking. This provides the scientific community with a blueprint for the fundamental study of cellular processes mediating NP engagement, with applications for both basic and translational research.
We identified expression of SLC46A3, a lysosomal transporter, to be a negative regulator and potential biomarker for lipid-based nanoparticle uptake and downstream functional efficacy. While SLC46A3 has recently been implicated in hepatic copper homeostasis as well as sensitivity to ADCs in cancer cells,(54–56) its role in NP delivery was previously unexplored. We first validated SLC46A3 as a negative regulator of lipid-based NP uptake in a panel of non-pooled cell lines, as well as engineered isogenic cell lines with modulated SLC46A3 expression. Because all current FDA-approved NPs for anticancer applications are liposomal formulations, there is notable potential for this biomarker to be quickly implemented in clinical studies with existing, approved formulations. To this end, we recapitulated our findings in an in vivo model using an analog of an FDA-approved liposomal NP formulation.
Moreover, we demonstrated that SLC46A3 has potential as a predictive biomarker beyond liposomal nanoparticles by investigating solid lipid nanoparticles. Both LNP-cell association and mRNA transfection were inversely correlated with SLC46A3 expression. These preliminary findings suggest that SLC46A3 expression may serve as a predictive biomarker for functional delivery of nucleic acid cargo via lipid nanoparticles. Our findings support the continued exploration of SLC46A3 as a potential biomarker for therapeutic nanoparticle delivery.
In summary, we present a platform to study NP-cancer cell interactions simultaneously through the use of pooled screening, genomics, and machine learning algorithms. Application of this integrated platform should advance the rational design of nanocarriers.
Supplementary Material
Acknowledgments:
We thank the Koch Institute’s Robert A. Swanson (1969) Biotechnology Center for technical support, specifically the Flow Cytometry, High Throughput Sciences, Genomics Core, Microscopy, and Preclinical Modeling, Imaging & Testing cores, the Hope Babette Tang (1983) Histology Facility, and the Peterson (1957) Nanotechnology Materials Core Facility.
We would like to thank Todd Golub and Alex Burgin for formative feedback and helpful discussion.
We also gratefully acknowledge Thomas Diefenbach and the Ragon Institute Microscopy Core for assistance with imaging cytometry.
The LOXIMVI and T47D cell lines were gifts from the F. Gertler Lab (MIT), and the Jurkat cell line was a gift from the D. Sabatini Lab (previously of the Whitehead Institute for Biomedical Research).
We would like to thank Chris Straehla for help in figure design.
Figures 1A and 6A were created in part using Biorender.com.
Funding:
This work was supported in part by SPARC funding at The Broad Institute.
This work was also supported by grants from the Koch Institute’s Marble Center for Cancer Nanomedicine and Frontier Research Program.
This work was supported in part by the Koch Institute Support (core) Grant P30-CA14051 from the National Cancer Institute.
NB was supported by a Department of Defense Congressionally Directed Medical Research Programs Peer Reviewed Cancer Research Program Horizon Award (W81XWH-19-1-0257) and the NIH-NCI (K99CA255844).
JPS was supported as a National Institutes of Health grant T32 trainee (CA136432-08) and by the Helen Gurley Brown Presidential Initiative of Dana-Farber Cancer Institute.
Fellowship support for CHA was from the NIH (NRSA F31 CA228241-01).
RRC is a fellow of the Parker B. Francis Foundation.
NN was supported by a grant from the Gates Foundation.
Fellowship support for AGB was from the NIH (F30 DK130564) and a Termeer Fellowship of Medical Engineering and Science.
NGL was supported by Cancer Research UK and the Brain Tumour Charity (C42454/A28596) and a fellowship from the Ludwig Center at the Koch Institute for Integrative Cancer Research.
HL was supported by the Charles W. (1955) and Jennifer C. Johnson Cancer Research Fund and NIH (K08DK123414).
Footnotes
Competing interests: NB, JPS, ANK, and PTH have submitted a patent application for the SLC46A3 biomarker discovery. ANK is a founder and member of the board of 76Bio. PTH is a member of the science advisory board at Moderna, board member at Alector, Advanced Chemotherapy Technologies, and Burroughs-Wellcome Fund, and board member and co-founder of LayerBio.
Data and materials availability: All data are available in the main text or the supplementary materials.
References and Notes
- 1.Shi JJ, Kantoff PW, Wooster R, Farokhzad OC, Cancer nanomedicine: progress, challenges and opportunities. Nat. Rev. Cancer 17, 20–37 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mitchell MJ, Billingsley MM, Haley RM, Wechsler ME, Peppas NA, Langer R, Engineering precision nanoparticles for drug delivery. Nat. Rev. Drug Discov, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boehnke N, Hammond PT, Power in Numbers: Harnessing Combinatorial and Integrated Screens to Advance Nanomedicine. JACS Au 2, 12–21 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tran S, DeGiovanni PJ, Piel B, Rai P, Cancer nanomedicine: a review of recent success in drug delivery. Clin. Transl. Med 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wilhelm S, Tavares AJ, Dai Q, Ohta S, Audet J, Dvorak HF, Chan WCW, Analysis of nanoparticle delivery to tumours. Nat. Rev. Mater 1, (2016). [Google Scholar]
- 6.Cheng YH, He CL, Riviere JE, Monteiro-Riviere NA, Lin ZM, Meta-Analysis of Nanoparticle Delivery to Tumors Using a Physiologically Based Pharmacokinetic Modeling and Simulation Approach. ACS Nano 14, 3075–3095 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Youn YS, Bae YH, Perspectives on the past, present, and future of cancer nanomedicine. Adv. Drug Deliver. Rev 130, 3–11 (2018). [DOI] [PubMed] [Google Scholar]
- 8.Poon W, Kingston BR, Ouyang B, Ngo W, Chan WCW, A framework for designing delivery systems. Nat. Nanotechnol 15, 819–829 (2020). [DOI] [PubMed] [Google Scholar]
- 9.Poon W, Zhang YN, Ouyang B, Kingston BR, Wu JLY, Wilhelm S, Chan WCW, Elimination Pathways of Nanoparticles. ACS Nano 13, 5785–5798 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Correa S et al. , Tuning Nanoparticle Interactions with Ovarian Cancer through Layer-by-Layer Modification of Surface Chemistry. ACS Nano 14, 2224–2237 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boehnke N, Correa S, Hao L, Wang W, Straehla JP, Bhatia SN, Hammond PT, Theranostic Layer-by-Layer Nanoparticles for Simultaneous Tumor Detection and Gene Silencing. Angew. Chem. Int. Ed. Engl 59, 2776–2783 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Boehnke N, Dolph KJ, Juarez VM, Lanoha JM, Hammond PT, Electrostatic Conjugation of Nanoparticle Surfaces with Functional Peptide Motifs. Bioconjug. Chem 31, 2211–2219 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlman JE et al. , Barcoded nanoparticles for high throughput in vivo discovery of targeted therapeutics. P. Natl. Acad. Sci. USA 114, 2060–2065 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nogrady B, How cancer genomics is transforming diagnosis and treatment. Nature 579, S10–S11 (2020). [DOI] [PubMed] [Google Scholar]
- 15.Yu CN et al. , High-throughput identification of genotype-specific cancer vulnerabilities in mixtures of barcoded tumor cell lines. Nat. Biotechnol 34, 419–423 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corsello SM et al. , Discovering the anticancer potential of non-oncology drugs by systematic viability profiling. Nature Cancer, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Correa S, Boehnke N, Deiss-Yehiely E, Hammond PT, Solution Conditions Tune and Optimize Loading of Therapeutic Polyelectrolytes into Layer-by-Layer Functionalized Liposomes. ACS Nano 13, 5623–5634 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Correa S et al. , Highly Scalable, Closed-Loop Synthesis of Drug-Loaded, Layer-by-Layer Nanoparticles. Adv. Funct. Mater 26, 991–1003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deng ZJ, Morton SW, Ben-Akiva E, Dreaden EC, Shopsowitz KE, Hammond PT, Layer-by-Layer Nanoparticles for Systemic Codelivery of an Anticancer Drug and siRNA for Potential Triple-Negative Breast Cancer Treatment. ACS Nano 7, 9571–9584 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Morton SW, Poon ZY, Hammond PT, The architecture and biological performance of drug-loaded LbL nanoparticles. Biomaterials 34, 5328–5335 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Decher G, Fuzzy nanoassemblies: Toward layered polymeric multicomposites. Science 277, 1232–1237 (1997). [Google Scholar]
- 22.Dreaden EC et al. , Tumor-Targeted Synergistic Blockade of MAPK and PI3K from a Layer-by-Layer Nanoparticle. Clin. Cancer Res 21, 4410–4419 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreaden EC, Morton SW, Shopsowitz KE, Choi JH, Deng ZJ, Cho NJ, Hammond PT, Bimodal Tumor-Targeting from Microenvironment Responsive Hyaluronan Layer-by-Layer (LbL) Nanoparticles. ACS Nano 8, 8374–8382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oommen OP, Duehrkop C, Nilsson B, Hilborn J, Varghese OP, Multifunctional Hyaluronic Acid and Chondroitin Sulfate Nanoparticles: Impact of Glycosaminoglycan Presentation on Receptor Mediated Cellular Uptake and Immune Activation. ACS Appl. Mater. Inter 8, 20614–20624 (2016). [DOI] [PubMed] [Google Scholar]
- 25.Suk JS, Xu QG, Kim N, Hanes J, Ensign LM, PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Adv. Drug Deliver. Rev 99, 28–51 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Frohlich E, The role of surface charge in cellular uptake and cytotoxicity of medical nanoparticles. Int. J. Nanomed 7, 5577–5591 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berg EA, Fishman JB, Labeling Antibodies Using N-Hydroxysuccinimide (NHS)-Fluorescein. Cold Spring Harbor Protocols 3, 229–231 (2019). [DOI] [PubMed] [Google Scholar]
- 28.Jin X et al. , A metastasis map of human cancer cell lines. Nature 588, 331–336 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Detailed materials and methods are available online in the provided Supplementary Materials.
- 30.Kim JA, Aberg C, Salvati A, Dawson KA, Role of cell cycle on the cellular uptake and dilution of nanoparticles in a cell population. Nat. Nanotechnol 7, 62–68 (2012). [DOI] [PubMed] [Google Scholar]
- 31.Aberg C, Kim JA, Salvati A, Dawson KA, Reply to ‘The interface of nanoparticles with proliferating mammalian cells’. Nat. Nanotechnol 12, 600–603 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Panet E et al. , The interface of nanoparticles with proliferating mammalian cells. Nat. Nanotechnol 12, 598–600 (2017). [DOI] [PubMed] [Google Scholar]
- 33.Rees P, Wills JW, Brown MR, Barnes CM, Summers HD, The origin of heterogeneous nanoparticle uptake by cells. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barretina J et al. , The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity (vol 483, pg 603, 2012). Nature 492, 290–290 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ghandi M et al. , Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tsuchikama K, An ZQ, Antibody-drug conjugates: recent advances in conjugation and linker chemistries. Protein Cell 9, 33–46 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rejman J, Oberle V, Zuhorn IS, Hoekstra D, Size-dependent internalization of particles via the pathways of clathrin-and caveolae-mediated endocytosis. Biochem. J 377, 159–169 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Behzadi S et al. , Cellular uptake of nanoparticles: journey inside the cell. Chem. Soc. Rev 46, 4218–4244 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Subramanian A et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. P. Natl. Acad. Sci. USA 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP, Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dean M, Hamon Y, Chimini G, The human ATP-binding cassette (ABC) transporter superfamily. J. Lipid Res 42, 1007–1017 (2001). [PubMed] [Google Scholar]
- 43.Shamay Y et al. , P-selectin is a nanotherapeutic delivery target in the tumor microenvironment. Sci. Transl. Med 10, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saravanakumar G, Jo DG, Park JH, Polysaccharide-Based Nanoparticles: A Versatile Platform for Drug Delivery and Biomedical Imaging. Curr. Med. Chem 19, 3212–3229 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Voigt J, Christensen J, Shastri VP, Differential uptake of nanoparticles by endothelial cells through polyelectrolytes with affinity for caveolae. P. Natl. Acad. Sci. USA 111, 2942–2947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Szklarczyk D et al. , STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.von Mering C, Huynen M, Jaeggi D, Schmidt S, Bork P, Snel B, STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 31, 258–261 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Snel B, Lehmann G, Bork P, Huynen MA, STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res. 28, 3442–3444 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin D, Brun C, Remy E, Mouren P, Thieffry D, Jacq B, GOToolBox: functional analysis of gene datasets based on Gene Ontology. Genome Biol. 5, (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ashburner M et al. , Gene Ontology: tool for the unification of biology. Nat. Genet 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carbon S et al. , The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res. 49, D325–D334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin L, Yee SW, Kim RB, Giacomini KM, SLC transporters as therapeutic targets: emerging opportunities. Nat. Rev. Drug Discov 14, 543–560 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chapel A et al. , An Extended Proteome Map of the Lysosomal Membrane Reveals Novel Potential Transporters. Mol. Cell Proteomics 12, 1572–1588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim JH et al. , Lysosomal SLC46A3 modulates hepatic cytosolic copper homeostasis. Nat. Commun 12, 290 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hamblett KJ et al. , SLC46A3 Is Required to Transport Catabolites of Noncleavable Antibody Maytansine Conjugates from the Lysosome to the Cytoplasm. Cancer Res. 75, 5329–5340 (2015). [DOI] [PubMed] [Google Scholar]
- 56.Kinneer K et al. , SLC46A3 as a Potential Predictive Biomarker for Antibody-Drug Conjugates Bearing Noncleavable Linked Maytansinoid and Pyrrolobenzodiazepine Warheads. Clin. Cancer Res 24, 6570–6582 (2018). [DOI] [PubMed] [Google Scholar]
- 57.Zhao Q et al. , Increased expression of SLC46A3 to oppose the progression of hepatocellular carcinoma and its effect on sorafenib therapy. Biomed. Pharmacother 114, (2019). [DOI] [PubMed] [Google Scholar]
- 58.Li GM et al. , Mechanisms of Acquired Resistance to Trastuzumab Emtansine in Breast Cancer Cells. Mol. Cancer Ther 17, 1441–1453 (2018). [DOI] [PubMed] [Google Scholar]
- 59.Tsui CK et al. , CRISPR-Cas9 screens identify regulators of antibody-drug conjugate toxicity. Nat Chem Biol 15, 949–958 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hou XC, Zaks T, Langer R, Dong YZ, Lipid nanoparticles for mRNA delivery. Nat. Rev. Mater 6, 1078–1094 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Samaridou E, Heyes J, Lutwyche P, Lipid nanoparticles for nucleic acid delivery: Current perspectives. Adv. Drug Deliver. Rev 154, 37–63 (2020). [DOI] [PubMed] [Google Scholar]
- 62.Jayaraman M et al. , Maximizing the Potency of siRNA Lipid Nanoparticles for Hepatic Gene Silencing In Vivo. Angew. Chem. Int. Edit 51, 8529–8533 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Whitehead KA et al. , Degradable lipid nanoparticles with predictable in vivo siRNA delivery activity. Nat. Commun 5, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kauffman KJ et al. , Optimization of Lipid Nanoparticle Formulations for mRNA Delivery in Vivo with Fractional Factorial and Definitive Screening Designs. Nano Lett. 15, 7300–7306 (2015). [DOI] [PubMed] [Google Scholar]
- 65.Hassett KJ et al. , Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucl. Acids 15, 1–11 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Liu QQ, Kim YJ, Im GB, Zhu JT, Wu YZ, Liu YJ, Bhang SH, Inorganic Nanoparticles Applied as Functional Therapeutics. Adv. Funct. Mater 31, (2021). [Google Scholar]
- 67.Paul W, Sharma CP, Inorganic nanoparticles for targeted drug delivery. Woodhouse Publishing Series in Biomaterials, 333–373 (2020). [Google Scholar]
- 68.Adelmann CH et al. , MFSD12 mediates the import of cysteine into melanosomes and lysosomes. Nature 588, 699–704 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Stern S, Neun B, NCL Method GTA-12. NCI Hub, doi: 10.17917/YPTH-N396. [DOI] [Google Scholar]
- 70.Boehnke N, Straehla JP, Kocak M, Ronan M, Roth J, Data and processing scripts for PRISM barcode sequencing data used in “Massively parallel pooled screening reveals genomic determinants of nanoparticle-cell interactions”. Zenodo, DOI: 10.5281/zenodo.6642633. [DOI] [Google Scholar]
- 71.Stephens M, False discovery rates: a new deal. Biostatistics 18, 275–294 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.R Core Team, R: A language and envrionemnt for statistical computing. R Foundation for Statistical Compouting. https://www.R-project.org/. [Google Scholar]
- 73.Wickham H, ggplot2: Elegant Graphics for Data Analysis. Use R, 1–212 (2009). [Google Scholar]
- 74.Kocak M, Boghossian A, Linear association function used in the manuscript “Massively parallel pooled screening reveals genomic determinants of nanoparticle-cell interactions”. Zenodo, DOI: 10.5281/zenodo.6558445. [DOI] [Google Scholar]
- 75.Tang Y, Horikoshi M, Li WX, ggfortify: Unified Interface to Visualize Statistical Results of Popular R Packages. R. J 8, 474–485 (2016). [Google Scholar]
- 76.Horikoshi M, Tang Y, ggfortify: Data Visualization Tools for Statistical Analysis Results. https://CRAN.R-project.org/package=ggfortify (2016).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.