Abstract
MITOP (http://www.mips.biochem.mpg.de/proj/medgen/ mitop/ ) is a comprehensive database for genetic and functional information on both nuclear- and mitochondrial-encoded proteins and their genes. The five species files—Saccharomyces cerevisiae, Mus musculus, Caenorhabditis elegans, Neurospora crassa and Homo sapiens—include annotated data derived from a variety of online resources and the literature. A wide spectrum of search facilities is given in the overlapping sections ‘Gene catalogues’, ‘Protein catalogues’, ‘Homologies’, ‘Pathways and metabolism’ and ‘Human disease catalogue’ including extensive references and hyperlinks to other databases. Central features are the results of various homology searches, which should facilitate the investigations into interspecies relationships. Precomputed FASTA searches using all the MITOP yeast protein entries and a list of the best human EST hits with graphical cluster alignments related to the yeast reference sequence are presented. The orthologue tables with cross-listings to all the protein entries for each species in MITOP have been expanded by adding the genomes of Rickettsia prowazeckii and Escherichia coli. To find new mitochondrial proteins the complete yeast genome has been analyzed using the MITOPROT program which identifies mitochondrial targeting sequences. The ‘Human disease catalogue’ contains tables with a total of 110 human diseases related to mitochondrial protein abnormalities, sorted by clinical criteria and age of onset. MITOP should contribute to the systematic genetic characterization of the mitochondrial proteome in relation to human disease.
INTRODUCTION
Mitochondria are essential cell organelles of eukaryotes under the control of both their own and the nuclear genome. They fulfill most of the energy requirements of aerobic cells and are essential for the metabolism of a number of important biological compounds (1). Except for 13 mtDNA encoded proteins in human, all proteins involved in function and structural organization are encoded by the nucleus and imported into mitochondria. Mutations in the mitochondrial genome occur in a wide variety of degenerative diseases, aging and cancer (2). More than 50 different mtDNA base substitution mutations and hundreds of mtDNA deletions and insertions associated with mitochondriopathies have been collected to date (3). Recent discoveries in the field of mitochondrial diseases have demonstrated the enormous ‘nuclear power’ in the aetiology of Mendelian inherited disease (4). The MITOP database aims at the comprehensive identification and characterization of the mitochondrial proteome with a special focus on proteins relevant for human mitochondrial disease (5). MITOP presents data from online resources and the literature annotated by the editors: http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/editors . Information on both nuclear- and mitochondrial-encoded genes and their proteins are combined in one location including cross references to formerly unlinked databases.
GENE AND PROTEIN CATALOGUES
Presently, more than 1150 mitochondria-related genes and corresponding proteins are listed (see Table 1 for breakdown). MITOP includes detailed descriptions of protein structure and function including references. Search facilities for subcellular localization divided into mitochondrial matrix, outer-, inner- or inter-membrane space are offered (http://vms.mips.biochem.mpg. de/htbin/z_filter_subcel/a ). All protein catalogue sections like ‘functional catalogue’ (http://vms.mips.biochem.mpg.de/htbin/z_filter_cat/a&fun ), ‘protein classes’ (http://vms.mips.biochem. mpg.de/htbin/z_filter_cat/a&cla ), ‘protein complexes’ (http://vms. mips.biochem.mpg.de/htbin/z_filter_cat/a&com ), EC number (http://vms.mips.biochem.mpg.de/htbin/z_filter_enzyme/a ) and PROSITE-motifs (http://vms.mips.biochem.mpg.de/htbin/z_filter_prosite/a ) are listed in Table 2. In the ‘gene catalogues’, users may search chromosomes for information about mapped genes (for 219 human genes) and related diseases (http://vms. mips.biochem.mpg.de/htbin/z_filter_chromo/h ), browse lists of genes with related human mitochondriopathies (http://vms. mips.biochem.mpg.de/htbin/z_list_gene/h ) and obtain mitochondrial rRNA and tRNA entries (http://vms.mips.biochem. mpg.de/htbin/z_filter_notorf/h ). MITOP extensively cross references its objects with those in other genetic and molecular databases.
Table 1. Overview of mitochondria-related genes and proteins in the five MITOP species files.
| Species | Nuclear-coded | Mitochondrial-coded |
|---|---|---|
| Homo sapiens | 303 | 37 |
| Mus musculus | 138 | 37 |
| Saccharomyces cerevisiae | 380 | 58 |
| Caenorhabditis elegans | 63 | 34 |
| Neurospora crassa | 59 | 45 |
Table 2. Current entries in the protein and disease catalogues.
| MITOP catalogues | Number of protein entries |
|---|---|
| Functional categories | 2400 |
| Protein classes | 149 |
| Protein complexes | 458 |
| EC number catalogue | 676 |
| PROSITE motifs catalogue | 703 |
| Disease catalogue | 110 |
Please note that proteins can occur more than once in the same catalogue.
HOMOLOGIES
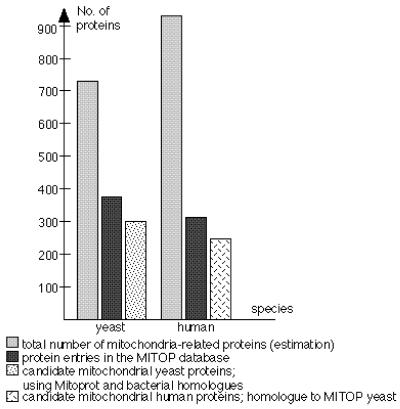
Physiological mechanisms in mitochondria are highly conserved in evolution among eukaryotes. Therefore, cross-species homology searches are particularly powerful to identify new mitochondria-related human proteins (6). The dataset for Saccharomyces cerevisiae, the most completely-described eukaryotic genome (7), currently consists of 438 entries (http://vms.mips.biochem.mpg.de/htbin/z_list_gene/y ), representing 11.8% of the yeast genes with known function. So far, >36% of the MITOP yeast proteins share significant homologies with human mitochondrial proteins. Current estimates (Fig. 1) of the total number of mitochondrial proteins range between 700 and 1000 for yeast and humans, respectively (4). To identify new human mitochondrial genes, we ran FASTA (8) searches using all MITOP yeast protein entries as query sequences and dbEST as the target database. We detected human EST matches with significant scores [expect() <0.001] for 243 MITOP yeast sequences. A link to a list of best EST hits (page cut-off 30 ESTs) and graphical alignments of EST clusters to the related yeast reference sequence are included. EST match scores, amino acid alignments of overlapping regions and hyperlinks to EMBL and Unigene are presented. For example, for the mitochondrial yeast protein YLR168c (http://vms.mips. biochem.mpg.de/htbin/z_get_entry/y&YLR168c ), MITOP presents 12 human ESTs with an identity >30% on protein level for overlapping regions (http://www.mips.biochem. mpg.de/cgi-bin/proj/medgen/est_main?YLR168c ). The link to Unigene leads to mapping information on chromosome 20. For each protein entry in MITOP, a list of homologous proteins is provided using the FASTA database at MIPS (http://vms. mips.biochem.mpg.de/htbin/z_pir_fasta/S57749 ). Homology to bacterial proteins has been postulated for mitochondrial proteins according to the hypothesis of endosymbiotic mitochondrial evolution. Therefore, we searched the yeast genome for homologies to the Escherichia coli and Rickettsia prowazeckii genomes (9). The orthologue tables with cross listings to all MITOP entries for each species generated using the FASTA program were expanded by adding these two species (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/ orthologs?y ). A program (MITOPROT) is available to identify mitochondrial targeting sequences (10) under MITOP-Tools (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/mitofilter ). The complete set of yeast open reading frames (ORFs) has been analyzed using this program (http://www.mips.biochem. mpg.de/cgi-bin/proj/medgen/mitoprot_yeast ). We identified >300 highly significant ORFs (scores > 0.9) potentially coding for mitochondrial proteins.
Figure 1.
Estimated number of mitochondria-related proteins in human and yeast, current and candidate protein entries in MITOP.
PATHWAYS AND METABOLISM
The graphical depiction of selected mitochondrial processes includes the respiratory chain (http://www.mips.biochem. mpg.de/cgi-bin/proj/medgen/rescha?h ), mitochondrial membrane transporters (http://www.mips.biochem.mpg.de/cgi-bin/proj/medgen/meta?h/transport ) and an overview of mitochondrial metabolic pathways (http://www.mips.biochem.mpg.de/cgi-bin/ proj/medgen/meta?h/overview ). Cross-links to individual mito-chondria-related protein entries are provided.
HUMAN MITOCHONDRIAL DISEASE CATALOGUE
In addition to the individual disease descriptions included in the protein files, a table of human diseases related to mitochondrial protein abnormalities was created (http://vms.mips.biochem. mpg.de/htbin/z_table_disease ). A total of 110 human diseases are associated with mutations in 85 MITOP-human proteins and 11 mtDNA tRNAs are presently listed. The table is sorted by clinical criteria and age of onset (neonatal, infantile, juvenile and adult), with short descriptions, causally-related mitochondria or nuclear-encoded gene products, links to the relevant MITOP protein entries and hyperlinks to OMIM. In addition, a hyperlink to a table with yeast homologues of human-disease associated genes is given in the yeast section of MITOP (http://www.mips. biochem.mpg.de/proj/yeast/reviews/human_diseases.html ). Recently discovered examples for mitochondriopathies include Friedreich’s ataxia and X-linked sideroblastic anemia and ataxia (XLSA/A) with mutated proteins, frataxin and ABC7, involved in mitochondrial iron homeostasis (11,12). Spastic paraplegia has been shown to be caused by defects in a mitochondrial metalloprotease, SPG7 (13). The protein mutated in Wilson’s disease, a copper transporting P-type ATPase (ATP7B) and the deafness-dystonia-peptide (DDP1) which is mutated in patients with Mohr–Tranebjaerg syndrome could be localized to the mitochondrial inner membrane (14,15).
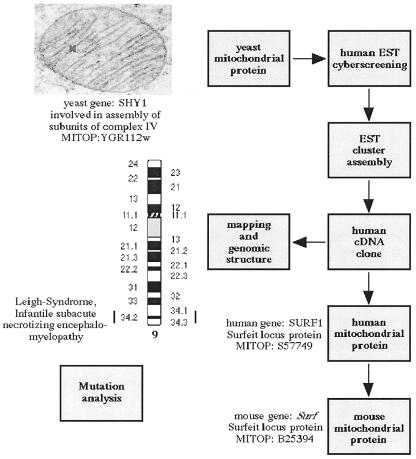
Identification of human proteins homologous to yeast mitochondrial proteins is a method to find new candidates involved in human mitochondriopathies. An example is presented in Figure 2 using the mitochondrial yeast protein SHY1 (http://vms.mips.biochem.mpg.de/htbin/z_get_entry/y&YGR112w ). By screening dbEST the human SURF1 cDNA was identified as a homologue of the yeast SHY1 protein, a mitochondrial protein necessary for maintenance of cytochrome-c-oxidase activity and respiration (16). SURF1 has been mapped to chromosome 9q34.3 thereby becoming a candidate gene for Leigh syndrome, a subacute necrotizing encephalomyelopathy commonly associated with a deficiency of complex IV of the respiratory chain. Sequence analysis of SURF1 revealed loss-of-function mutations in a majority of these families (17,18). By clicking ‘respiration’ in the MITOP sections of ‘functional categories’ (http://vms.mips.biochem.mpg.de/htbin/z_search_cat/a&u&002.013.000.000.000.000 ) the user receives information for the yeast, human and homologous murine protein named Surf1 (http://vms.mips.biochem.mpg.de/htbin/z_get_entry/m&B25394 ). The discovery of SURF1 as the disease gene underlying a respiratory chain deficiency has a major impact for further disease studies in this field, since the genetic basis of most of these cases is unknown and efforts to find mutations in the structural subunits themselves have been only partially successful for complexes I and II (19–21).
Figure 2.
In silico approach for identification of nuclear gene defects in human mitochondriopathies. The human SURF1 protein is highly homologous to the yeast SHY1 protein which is necessary for the maintenance of cytochrome-c-oxidase activity and respiration.
A description of MITOP’s features is provided at the main page in ‘About the database’ (http://www.mips.biochem.mpg. de/cgi-bin/proj/medgen/description ). We welcome inquiries regarding specific subjects or concerns.
Acknowledgments
ACKNOWLEDGEMENTS
We wish to thank the MIPS yeast crew for providing the yeast protein catalogues. The MITOPROT program was kindly provided by Pierre Vincens and Manuel G. Claros. The MITOP project is supported by a grant from the Federal Ministry of Education, Science, Research and Technology (BMBF) and by the Ludwig-Maximilians-University, Munich.
REFERENCES
- 1.Kiberstis P.A. (1999) Science, 283, 1475. [Google Scholar]
- 2.Wallace D.C. (1999) Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 3.Kogelnik A.M., Lott,M.T., Brown,M.D., Navathe,S.B. and Wallace,D.C. (1998) Nucleic Acids Res., 26, 112–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DiMauro S. and Schon,E.A. (1998) Nature Genet., 19, 214–215. [DOI] [PubMed] [Google Scholar]
- 5.Scharfe C., Zaccaria,P., Hoertnagel,K., Jaksch,M., Klopstock,T., Lill,R., Prokisch,H., Gerbitz,K.D., Mewes,H.W. and Meitinger,T. (1999) Nucleic Acids Res., 27, 153–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hieter P. (1998) Clin. Genet., 54, 113–116. [DOI] [PubMed] [Google Scholar]
- 7.Goffeau A. et al. (1997) Nature, Suppl., 387. [Google Scholar]
- 8.Pearson W.R. and Lipman,D.J. (1988) Proc. Natl Acad. Sci. USA, 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Andersson S.G., Zomorodipour,A., Andersson,J.O., Sicheritz-Ponten,T., Alsmark,U.C., Podowski,R.M., Naslund,A.K., Eriksson,A.S., Winkler,H.H. and Kurland,C.G. (1998) Nature, 396, 133–140. [DOI] [PubMed] [Google Scholar]
- 10.Claros M.G. and Vincens,P. (1996) Eur. J. Biochem., 241, 779–786. [DOI] [PubMed] [Google Scholar]
- 11.Koutnikova H., Campuzano,V., Foury,F., Dolle,P., Cazzalini,O. and Koenig,M. (1997) Nature Genet., 16, 345–351. [DOI] [PubMed] [Google Scholar]
- 12.Allikmets R., Raskind,W.H., Hutchinson,A., Schueck,N.D., Dean,M. and Koeller,D.M. (1999) Hum. Mol. Genet., 8, 743–749. [DOI] [PubMed] [Google Scholar]
- 13.Casari G., DeFusco,M., Ciarmatori,S., Zeviani,M., Mora,M., Fernandez,P., DeMichele,G., Filla,A., Cocozza,S., Marconi,R., Durr,A., Fontaine,B. and Ballabio,A. (1998) Cell, 93, 973–983. [DOI] [PubMed] [Google Scholar]
- 14.Lutsenko S. and Copper,M.J. (1998) Proc. Natl Acad. Sci. USA, 95, 6004–6009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koehler C.M., Leuenberger,D., Merchant,S., Renold,A., Junne,T. and Schatz,G. (1999) Proc. Natl Acad. Sci. USA, 96, 2141–2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mashkevich G., Repetto,B., Glerum,D.M., Jin,C. and Tzagoloff,A. (1997) J. Biol. Chem., 272, 14356–14364. [DOI] [PubMed] [Google Scholar]
- 17.Tiranti V., Hoertnagel,K., Carrozzo,R., Galimberti,C., Munaro,M., Granatiero,M., Zelante,L., Gasparini,P., Marzell,R., Rocchi,M., Bayona-Baafaluy,M.P., Enriquez,A., Uziel,G., Bertini,E., Dionisi-Vici,C., Meitinger,T. and Zeviani,M. (1998) Am. J. Hum. Genet., 63, 1609–1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhu Z., Yao,J., Johns,T., Fu,K., De Bie,I., Macmillan,C., Cuthbert,A.P., Newbold,R.F., Wang,J., Chevrette,M., Brown,G.K., Brown,R.M. and Shoubridge,E.A. (1998) Nature Genet., 20, 337–343. [DOI] [PubMed] [Google Scholar]
- 19.Bougeron T., Rustin,P., Chretien,D., Birch-Machin,M., Bourgeois,M., Viegas-Pequignot,E., Munnich,A. and Rotig,A. (1995) Nature Genet., 11, 144–149. [DOI] [PubMed] [Google Scholar]
- 20.Jaksch M., Hofmann,S., Kleinle,S., Liechti-Gallati,S., Pongratz,D.E., Müller-Höcker,J., Jedele,K.D., Meitinger,T. and Gerbitz,K.D. (1998) J. Med. Genet., 35, 895–900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smeitink J. and van den Heuvel,L. (1999) Am. J. Hum. Genet., 64, 1505–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]