Abstract
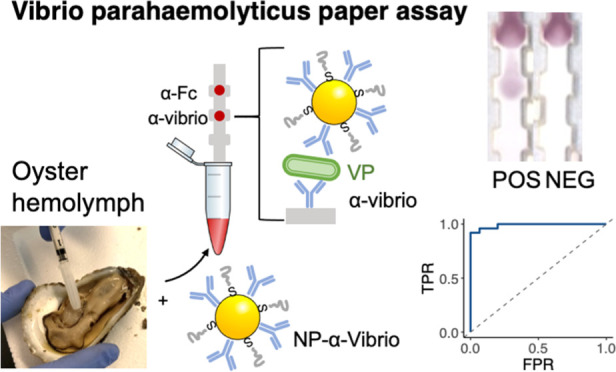
Seafood contamination with Vibrio bacteria is a problem for aquaculture, especially with oysters, which are often consumed raw. Current methods for diagnosing bacterial pathogens in seafood involve lab-based assays such as polymerase chain reaction or culturing, which are time consuming and must occur in a centralized location. Detection of Vibrio in a point-of-care assay would be a significant tool for food safety control measures. We report here a paper immunoassay that can detect the presence of Vibrio parahaemolyticus (Vp) in buffer and oyster hemolymph. The test uses gold nanoparticles conjugated to polyclonal anti-Vibrio antibodies in a paper-based sandwich immunoassay. A sample is added to the strip and wicked through by capillary action. If Vp is present, it results in a visible color at the test area that can be read out by eyes or a standard mobile phone camera. The assay has a limit of detection of 6.05 × 105 cfu/mL and a cost estimate of $5 per test. Receiver operating characteristic curves with validated environmental samples showed a test sensitivity of 0.96 and a specificity of 1.00. Because the assay is inexpensive and can be used on Vp directly without the requirement for culturing, or sophisticated equipment, it has the potential to be used in fieldable settings.
Introduction
Technological and food safety advances are instrumental to secure the food demand to feed the world’s expected population of 9.1 billion by 2050.1 Fish and shellfish can be a good source to cope with these food requirements efficiently.2−4 To meet this ever-growing demand, aquaculture production has been increasing and in 2020, provided nearly an equivalent amount of food as did fisheries.5 However, disease outbreaks in aquaculture have also increased, hindering shellfish production and challenging the control measures required to ensure safe food consumption.6 The burden of this foodborne illness is poorly addressed since only a small fraction of these cases are reported and diagnosed. Surveillance tools and surveys provide data that can be analyzed with novel models to obtain estimates of episodes and aid in case management, treatment, and resource allocation.7 Because of the complexity of infections by food sources and transmission by nonfood consumption, testing is crucial for health agencies to track potential outbreaks for disease control.8 The challenge resides in identifying the specific pathogen from the wide span of agent sources that can cause foodborne illnesses as well as the number of concurrent cases that must be reported.
Here, we focus our attention on the detection of vibrios in shellfish, one of the 31 major foodborne pathogens in the United States.8,9 Vibrios are Gram-negative bacteria commonly found in coastal waters during the warmer months. Vibrio parahaemolyticus (Vp) is the pathogenic species most frequently reported from this genus in the United States.10 This pathogen is spread through consumption of raw seafood11 and causes acute gastroenteritis or vibriosis. Symptoms usually appear within 24 h after ingestion and include watery or bloody diarrhea, abdominal cramping, nausea, vomiting, and fever. These symptoms are largely nonspecific and common to many other diseases. Only a small fraction of sufferers with vibriosis seek medical care and, more importantly, many cases are not identified because vibrios do not grow on routine enteric media. The Centers for Disease Control (CDC) estimates 34,664 annual episodes caused by Vp, from which only 287 cases were laboratory-confirmed. In 2014 in the United States, 48% of confirmed vibriosis cases were due to Vp, from which 15% were hospitalized and 1% died.10
The predominant control measures for a positive Vp report are for oyster farms to be shutdown, which typically last from weeks to months, until the bacteria are ostensibly cleared from the environment. Consequently, Vp-caused illnesses have significant economic repercussions, leading to an estimated cost of $319M in the United States. Vp is also an acute problem for shrimp aquaculture as it causes early mortality syndrome, resulting in 100% shrimp mortality as soon as 24 h post-exposure.12 Major outbreaks occurred from 2011 to 2013 in Southeast Asia, producing annual losses estimated to be more than $1 billion USD, and these outbreaks continued to spread into 2016.13−15
Current Vp detection methods involve taking sample to the laboratory and either performing the polymerase chain reaction (PCR) on DNA extracts from the sample or growing the bacterium in selective agar plates. Both procedures require bringing the sample back to a centralized lab, and the process of growing cultures can take several days to up to a week, which is too long to effectively respond against a potential outbreak as it hinders a rapid response. There are other innovative sensors for bacteria such as electrochemical sensors, but these require specialized instrumentation and expertise to analyze the data. Thus, providing a point of consumption (POC) assay that can be read out without special training or instruments is critical to contribute to the prevention of vibriosis. Nanotechnology has enabled novel sensing methods for various diseases and biomarkers and thus has the potential to facilitate new modes of detection of these pathogens.16−19 Novel methods recent in the literature include colorimetric aggregation assays20 and fluorescent nanoclusters in enzyme-linked immuno sorbent assay (ELISA).21
Here, we describe an inexpensive and ready-to-use immunochromatographic assay to detect Vp in raw oysters (Figure 1). These assays possess the advantages of being low-cost and easy to use and can be administered at POC.22−26 Paper test strips have proven to be a unique format for assays due to their low cost, ability to be manufactured at scale level, and robustness. This test exploits the unique optical properties of gold NPs (Au NPs), which provide the color that can be simply read out visually without the need for additional instrumentation.27−31 Au NPs were conjugated to a commercial anti-Vibrio polyclonal antibody (NP-Ab).32 The same antibody immobilized on the nitrocellulose allowed the formation of the sandwich immunoassay. The retention of NP-Ab conjugates at the test line led to a signal readable by eyes. We demonstrated that tests can be run in the oyster circulatory fluid (hemolymph), a realistic sample matrix, without the need for sample preparation or bacterial culturing. We measured the limit of detection (LOD) of the test against a reference pathogenic Vp strain. While others have reported immunochromatographic Vp detection,33 we went further to test the cross reactivity of the assay against bacteria isolated from oyster hemolymph. Additionally, the performance of the test was further validated with environmental Vp strains isolated from local coastal waters and oysters, demonstrating function beyond commercial lab-grown Vibrio strains.34 The rapid test has an estimated cost of $5, can be used for foodborne illness control, does not show cross reactivity with the non-Vibrio bacteria tested, and does not require external instrumentation for readout. We hypothesize that these results can bring testing capabilities closer to POC formats without the need for culturing.
Figure 1.
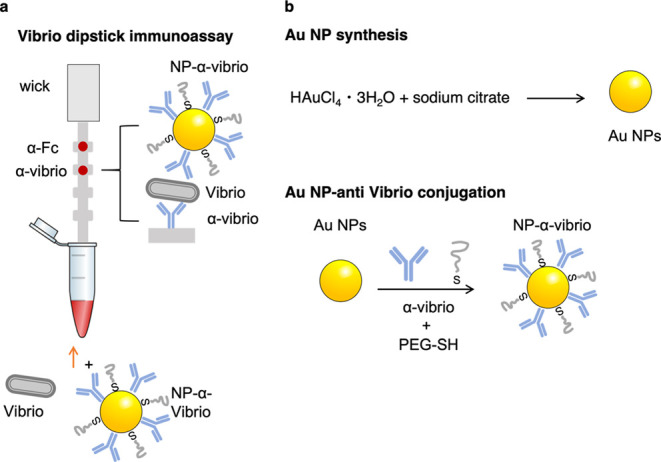
Schematic of the vibrio paper sensor and its synthesis. (a) Presence of Vibrio results in the formation of a sandwich immunoassay at the test line, where anti-Vibrio antibodies are immobilized. NP-anti-Vibrio antibody conjugates bind to the Vibrio. (b) synthesis of the gold nanoparticles (NPs) and antibody conjugation.
Experimental Section
Reagents
Gold chloride (HAuCl4·3H2O) (99.9%) (CAS: 16961-25-4), sucrose (CAS: 57-50-1), bovine serum albumin (BSA) (CAS: 9048-46-8), Tween-20 (CAS: 9005-64-5), trisodium citrate (Na3C6H5O7) (CAS: 6132-04-3), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (C8H18N2S) (≥99.5%) (CAS 7365-45-9), serum from human male AB plasma sterile-filtered, anti-rabbit IgG (Fc), and gel blotting paper used as an absorbent pad (GB003 Gel Blot paper) were all purchased from Millipore Sigma, Burlington MA. The BacTrace rabbit polyclonal anti-Vibrio species antibody was purchased from SeraCare, Milford MA. UniStart nitrocellulose CN140 with plastic backing was purchased from Sartorius, Bohemia, New York. The backing paper (MIBA-010 Backing Card, 0.020″ thickness) was purchased through DCN Diagnostics, Carlsbad, CA. 5 kDa mPEG was purchased from nanocs, New York, NY. Phosphate-buffered saline (1× PBS, pH 7.4) and tryptone peptone were purchased from Gibco. Sodium chloride (≥99%) (CAS: 7647-14-5) was purchased from Honeywell, Charlotte, NC. Technical agar solidifying agent, technical yeast extract, and thiosulfate-citrate-bile salts-sucrose (TCBS) agar were purchased from Difco. Vibrio parahaemolyticus ATCC 17802 was obtained from the American Type Culture Collection (ATCC), Manassas, VA. Escherichia coli with plasmid Bsrs078-LuxR (Addgene #85142) grown in DIAL strain (pir) was obtained from Addgene, Watertown, MA.
Gold NP Synthesis
Gold NPs (Au NPs) were synthesized using citrate reduction of gold(III) chloride.26 In short, 1 mL of a 6.8 mM sodium citrate solution was added to a 50 mL solution of HAuCl4 at 0.25 mM while boiling. The solution was left to boil for 15 min while stirring to allow the Au NPs to form and then cooled down to room temperature with continued agitation.
NP Characterization
The optical properties of the NPs were characterized by UV–vis spectroscopy (Agilent Cary 5000 UV–vis NIR). The morphology of the NPs was verified by transmission electron microscopy (TEM) (FEI Tecnai G2 at 120 kV), and ImageJ35 was used to determine their size. The nNP analyzer SZ-100 from Horiba was used to measure the hydrodynamic diameter (DH) and the zeta potential (ζ) of the bare and Ab-conjugated NPs. Agarose gel electrophoresis was also used to confirm the antibody conjugation by running a mixture of 8 μL of concentrated NPs with 4 μL of 50% glycerol in a 1% agarose gel.
Antibody Conjugation
Bioconjugation of the antibody to AuNPs was performed by electrostatic binding. Aliquots of 1 mL of the Au NPs synthesis solution were first centrifuged at 12,000 rcf for 12 min, and the pellet was resuspended in a mixture of 100 μL of 40 mM HEPES at pH 7.7 and 300 μL of MilliQ water. Then, 5 μL of a 1 mg/mL Ab solution reconstituted in phosphate buffer 10 mM at pH 7.4 was added to the solution and agitated overnight at room temperature to avoid the binding of the Ab to the NP surface. A polyethylene glycol (PEG) backfill was used to reduce nonspecific binding on the NP, which was achieved by the addition of 10 μL of 0.1 mM mPEG to the NP–Ab mixture. The solution was left under agitation at room temperature for 15 min. The antibody and PEG excess was separated from the NPs by centrifuging at 8000 rcf, and the 25 μL pellet was resuspended and used in the dipstick immunoassays.
Vibrio Culture
V. parahaemolyticus ATCC 17802 was grown on TCBS agar plates. A single colony was inoculated in 5 mL of saline lactose broth (SLB) and grown at 30 °C overnight. To better control bacterial growth, an inoculum from this solution was taken with an inoculation loop and added to 5 mL of fresh sterile SLB broth and left for 5 h at 30 °C to reach the desired concentration. To calculate the concentration of the sample, a 1:100,000 dilution was made in sterile 1× PBS and 50 μL of the diluted sample was spread in SLB agar plates grown overnight at 30 °C. The counting on the plates was done in triplicate. Samples were kept at 4 °C to prevent the growth until they were run in the rapid tests.
Hemolymph Extraction
Hemolymph was obtained from oysters, which were either purchased locally at a market or obtained from local coastal areas. Oysters were shucked, and hemolymph was extracted from the oyster with a syringe. For use in later trials, the hemolymph was sterilized by filtering it through a sterile 0.22 μm pore-size filter and kept at −20 °C until use.
Negative Control Isolation from Environmental Samples (V. splendidus and Marinomonas)
Hemolymph extracted from market oysters was directly plated on TCBS and SLB agar and incubated for 24 h at 37 °C. Isolated colonies were streaked for the isolation of pure culture on TCBS and SLB agar. Pure colonies were inoculated into 5 mL of SLB broth and incubated overnight at 37 °C. Nucleic acid extractions were performed on SLB cultures using the Qiagen DNeasy UltraClean Microbial Kit.
Vibrio parahaemolyticusIsolation from Environmental Samples
Hemolymph extracted from local coastal oysters was enriched for Vibrio isolation following a procedure modified from Hartnell et al.36 Briefly, 300 μL of hemolymph was added to 2.7 mL of alkaline peptone water (APW) and incubated for 6 h at 37 °C. Following incubation, 300 μL of the initial culture was inoculated into 2.7 mL of APW and incubated for 18 h at 37 °C. Enrichments were streaked for isolation of pure culture on TCBS agar. Individual colonies characteristic of Vp growth (dark blue-green) were picked into SLB broth and incubated at 37 °C overnight. Nucleic acid extractions of SLB cultures were performed using PrepMan Ultra Sample Preparation Reagent (ThermoFisher).
Isolates were archived in 75% SLB broth/25% glycerol and stored at −80 °C. Archives were regrown by streaking on TCBS plates and followed the same protocol with the ATCC Vp to let them grow to the concentrations used to run them in the immunochromatography tests.
PCR and Sequencing
Bacterial isolates were identified using PCR amplification and sequencing of the 16S rRNA gene and the gyrase B subunit gene (gyrB) using primers 8F (5′-CCTACGGGAGGCAGCAG), 534R (5′-ATTACCGCGGCTGCTGG) and Up1E (5′-GAAGT CATCA TGACC GTTCT GCAYG CNGGN GGNAA RTTYR A), and UP2AR (5′-AGCA G GGTAC GGATG TGCGA GCCRT CNACR TCNGC RTCNG YCAT), respectively (Muyzer et al.;37 Green et al.38). DNA sequencing was performed by Eton Biosciences, Boston, MA. Identification of all isolates was done by 16S rRNA and gyrB sequence matches using BLASTn (NCBI) against the nucleotide collection (nr/nt) database.
Paper Immunoassays
CN140 nitrocellulose strips were laser cut (LaserPro Spirit LS) at power 85% and a speed of 100%. Strips were attached to the absorbent pad with the help of a backing paper. Antibodies were spotted manually in 0.3 μL of aliquots to obtain 1.2 μg of Ab spotted. The wider areas of the strips provided a visual indication of where the spot would potentially appear. For the positive control, an antirabbit Fc-specific antibody was used in the upper spot. A blocking step was added to prevent false positives. Strips with the spotted antibody were left to dry at least 30 min before adding them to a 1.5 mL microcentrifuge tube with 50 μL of BSA solution at 1 mg/mL and left to dry at room temperature overnight. Tests were run in sequential steps to get the best results by immersing the strip in different solutions and letting the solution migrate through the strip by capillary action. First, a solution containing 12 μL of a sucrose/tween mixture (4 μL 50% w/v sucrose in water and 8 μL of 1% v/v Tween 20 in 1× PBS) and 30 μL of Vp spiked in sterile hemolymph or PBS 1× was run. Then, the strips were washed with 50 μL of PBS-Tween 20 0.1% (PBST) to avoid nonspecific binding of the bacteria with nitrocellulose. Once the strip was washed, a solution containing 12 μL of sucrose/tween, 3 μL of conjugated NPs, and 30 μL of human serum (HS) was run to prevent false positives.39 A final step with 50 μL of PBST was run to wash the nitrocellulose from nonspecific binding. The strips were left to dry at room temperature and scanned for quantitative analysis. ImageJ was used to measure the gray intensity of the test area.
Image Analysis: LOD
For the LOD analysis, five independent
replicates were run at different concentrations of bacteria. Gray
values of the test area were obtained by subtracting the test intensity
of the background and normalized with the following equation: 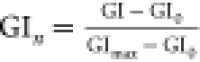 , where GI is the gray intensity at a given
concentration, GI0 is the gray intensity of the blank,
and GImax is the gray value at saturation. These values
at different concentrations (GIn) were
plotted and fitted in a Langmuir equation using a Matlab script (R2018a).
The LOD of the test was calculated as the concentration at which the
test showed a signal 3× the value of the standard deviation of
the blank.
, where GI is the gray intensity at a given
concentration, GI0 is the gray intensity of the blank,
and GImax is the gray value at saturation. These values
at different concentrations (GIn) were
plotted and fitted in a Langmuir equation using a Matlab script (R2018a).
The LOD of the test was calculated as the concentration at which the
test showed a signal 3× the value of the standard deviation of
the blank.
Image Analysis: ROC Curve
The test was validated with the PCR-identified Vp isolates obtained from hemolymph and run in sterile PBS in duplicates. The negative tests were also run in PBS to address the sensitivity and specificity of the test. Gray intensity values at the test line and background outside of the test area were measured using ImageJ and subtracted the background for each test. The receiver operating characteristic (ROC) curve and the area under the curve (AUC) were obtained with the R libraries plotROC and ggplot2 (RStudio version 1.0.153). The script varies the theoretical gray intensity threshold and classifies the tests as positive if they are found above this threshold. With the PCR-validated samples, true positive rate (TPR) (sensitivity) and false positive rate (FPR) (1-specificity) are calculated and plotted to obtain the ROC curve. The optimal cut-off value was obtained as the one that leads to the highest value for the sum of specificity and selectivity.
Results and Discussion
NP–Ab Conjugates
Spherical Au NPs were synthesized using citrate reduction of gold chloride (Figure 1), resulting in NPs with a mean diameter of 18.8 ± 2.5 nm as measured by TEM (Figure 2a). We chose spherical gold NPs for the particle due to their ease of synthesis, surface functionalization, and stability in biological fluids.34 The hydrodynamic diameter (DH) determined by dynamic light scattering (DLS) was 27.5 ± 1.8 nm (Figure 2d), and zeta potential showed that the NPs had a negative charge of −50.7 ± 8.7 mV (Figure 2d). Optical absorption spectroscopy determined that the NPs had a surface plasmon resonance (SPR) peak wavelength of 524 nm (Figure 2b).
Figure 2.
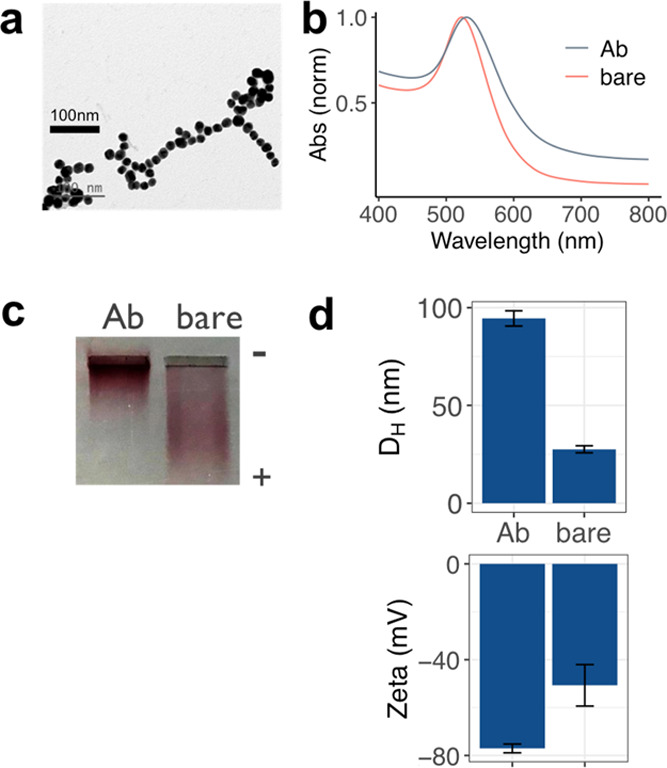
NP synthesis and Ab conjugation. (a) TEM image of Au NPs, (b) optical absorption of bare NPs (red) and NPs conjugated to anti-Vibrio Abs (black), (c) agarose gel electrophoresis of bare (right) and NP–Ab conjugates (left), and (d) DH (upper) and zeta potential (lower) of bare (right) and NP–Ab conjugates (right) measured by DLS. Error bars are averages of three independent measurements.
NPs were conjugated to a commercial polyclonal anti-Vibrio species antibody by electrostatic attachment. After Ab adsorption, the NP surface was passivated with a thiolated PEG (MW = 5 kDa) to avoid nonspecific interactions that can lead to false positives in complex biological media.40,41 The conjugation was confirmed with a higher retention of the conjugates when run in a 1% agarose gel as a result of their increased size (Figure 2c). This size increase was also confirmed by DLS, leading to a bioconjugate DH of 94.5 ± 3.4 nm (Figure 2d). Also, a decrease of the bioconjugate charge was observed compared to the bare NPs, with a zeta potential of −77.1 ± 1.8 mV (Figure 2d). The bioconjugate SPR peak exhibited a redshift and a slight broadening, also suggesting a successful NP conjugation (Figure 2b).
Paper Immunoassay for Vp Detection
We constructed a paper immunoassay test to detect Vp directly from oyster hemolymph. Hemolymph is a complex biological fluid with challenging physico-chemical properties that can compromise the colloidal stability of gold NPs due to its high salt content. Therefore, the immunoassay was tested first with a standard solution to detect a reference Vp. Due to the high salinity of this biological fluid, 1× PBS was chosen to optimize the test with the reference Vp stock from ATCC. Vp was cultured in selective agar plates TCBS and was grown to high concentrations by inoculating a single colony in SLB broth, and the concentration was determined by cell counting on SLB plates.
The dipstick assay consisted of a nitrocellulose strip attached to an absorbent pad that acted as a wick (Figure 1).42,43 The anti-Vibrio species antibody was immobilized on the test line. A control antibody that binds to the anti-Vibrio Ab conjugated to the Au NPs was immobilized on the control line (anti-Fc IgG). The bottom of the test strips was immersed in a pretreatment solution of 1 mg/mL BSA, which migrated through the test strip by capillary action. This pretreatment allowed minimization of false positives by preventing nonspecific interactions of the NPs with immobilized capture antibodies on the nitrocellulose.35 The test strips were dried overnight. Tests were run in sequential steps, which prevented false positives and also increased the spot intensity in the test line (Supporting Information, Figure S1). Briefly, the bacterium spiked in PBS at a concentration of 1.8 × 107 cfu/mL was run first with a sucrose/Tween solution. An intermediate washing step was added to eliminate any nonspecific binding between the bacteria and the immobilized antibodies. The conjugated NPs were run with HS and the sucrose/tween solution. The purpose of HS was to provide a protein corona (PC) that prevents false positives.39 Finally, a last washing step was added to eliminate nonspecific interactions of the Au NPs with nitrocellulose.
An intense spot at the test area was observed when Vp was present in the sample (Figure 3a). This indicated that the NP–Ab conjugates were retained at the test line due to the specific interaction of both the capture and detection antibodies with the bacteria. The successful generation of this sandwich immunoassay showed that the adsorbed antibody was not displaced from the NP surface by the PEG backfill or the PC formation. When PBS without Vp present was run, no spot appeared at the test line, suggesting that the Vp binding was not due to specific interactions of the probes with the immobilized antibodies (Figure 3a). A second colored spot appeared at the control line due to the binding of the control antibody to the anti-Vibrio Ab conjugated to the NPs and therefore validating the test. This was indicative of a proper flow of the NPs as well as the correct Ab conjugation of the Au NPs while migrating through the test strip. Strips were run five times (error bars, Figure 3b) which were indicative of interday reproducibility.
Figure 3.
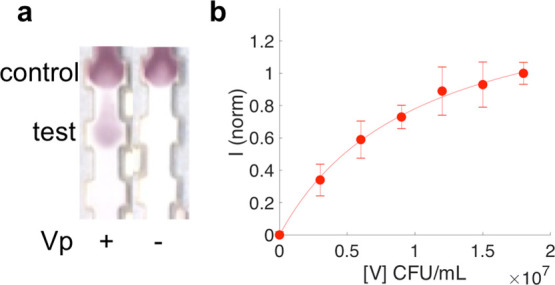
Paper sandwich immunoassay test with Vp spiked into 1× PBS. (a) Vp present (+) or absent (−). (b) Test line intensity as a function of Vp concentration in 1× PBS (circles). Error bars indicate averages of five independent strips. Fit to a modified Langmuir curve (line) to obtain an LOD.
To determine the LOD of the test in 1× PBS, we ran the strips at different Vp concentrations ranging from 0 to 1.8 × 107 cfu/mL. The signal increased with increasing concentration and then started to reach saturation. Using image analysis, we obtained test line signal intensities as a function of Vp concentration (Figure 3b). The titration curve was fit to a modified Langmuir equation to obtain the LOD of the test. The fit resulted in an LOD of 4.0 × 105 cfu/mL (R2 = 0.997).
Performance of the Test in Hemolymph
Because operation of the test in real settings would require deployment in oysters, we ran the assay in oyster hemolymph. Hemolymph was obtained from oysters and filtered to remove any bacteria present in the real samples (Figure 4a). Single colonies from TCBS plates were grown in SLB broth and determined the concentration of the new stocks in SLB plates. Bacteria were spiked into the filtered hemolymph at concentrations ranging from 0 to 3.1 × 107 cfu/mL to determine the test LOD in hemolymph. Accumulation of the NP–Ab conjugates at the tests area only occurred when Vp was present in the sample and increased the gray intensity with higher bacteria concentrations (Figure 4b). This denoted that this complex biological fluid did not trigger nonspecific binding between the NPs and the immobilized antibody. The titration curve fitted into a modified Langmuir equation led to an LOD of 6.0 × 105 cfu/mL (R2 = 0.996) (Figure 4c). The pathogenic concentration of Vp is reported at 108 cfu per serving in hemolymph for an ID50.44 Given the fact that the volume of hemolymph in an oyster is on the order of ∼1 mL, the optimized test can identify the presence of Vp directly in hemolymph below this threshold. Furthermore, this challenging matrix of the oyster hemolymph did not impact the LOD of the test, leading to detection limits comparable to the standard solution used to develop it. More importantly, these results sustain that the designed test does not require any sample preparation, which eases its use in the field by nonexperts. The variability for the test in hemolymph has a standard deviation on the order of 23–35%, and while test precision is much less compared to commercial assays for other antigens, we believe some of this can be attributed to the challenging sample matrix of the hemolymph, which has a high salinity and thus prone to inducing NP aggregation.40,45
Figure 4.
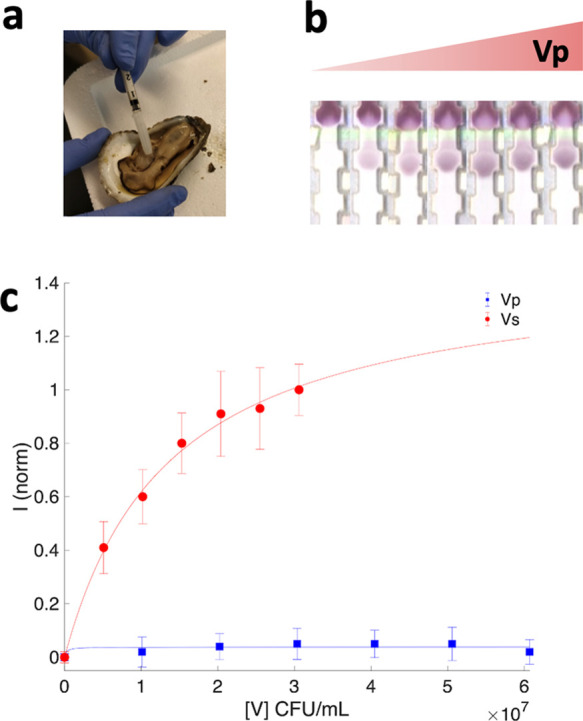
Test run in hemolymph. (a) Extraction of hemolymph from oyster (b) test strips with increasing Vp concentration spiked into hemolymph and (c) intensity of test area as a function Vp concentration (red) andVibrio splendidus (blue). Error bars indicate averages of five and seven independent measurements, respectively. Fits to a modified Langmuir curve (lines) to obtain LODs.
Isolation of Negative Controls from Hemolymph
Hemolymph contains a wide variety of bacteria46 that may result in false positives in the present test. Different bacterial species were isolated from hemolymph to explore the cross reactivity of the paper-based assay following a single colony purification method. Sequencing for the conserved 16S rRNA gene was performed to identify them, and concentration was determined in SLB plates prior to running the tests. Two purified samples were identified as Vibrio splendidus (Vs) and Marinomonas. Vs is a nonpathogenic Vibrio species that is common in coastal waters but grows optimally at colder temperatures compared to Vp. Thus, it is critical to understand the specificity of the test against this close-related bacterium to better address its performance with environmental samples.
Study of Cross Reactivity with Negative Controls
We ran these purified negative controls from hemolymph as well as E. coli. Similar to the previous samples, each bacteria was grown in SLB broth and determined their concentration in SLB plates. The obtained pellets were spiked in hemolymph and run as previously described.
Vs resulted in a faint signal at the test line when run at a concentration of 6.07 × 107 cfu/mL (Figure 5a), showing that the antibodies were able to pair with this other vibrio species. Vertical line scans across the strips showed that the gray intensity in the test line for Vs was 6× less intense compared to Vp but greater than the other negative controls (Figure 5c). This indicated that the Ab binding to this other nonpathogenic bacterium is lower compared to the reference Vp. To better address this binding, a titration curve was run with Vs spiked in filtered hemolymph (Figure 5b). The test area intensity remained low across the Vs concentrations, and due to the low signal, an LOD could not be reliably quantified for these nonpathogenic bacteria (Figure 4b). This confirmed that the antibody did not have as high an affinity for this nonpathogenic species.
Figure 5.
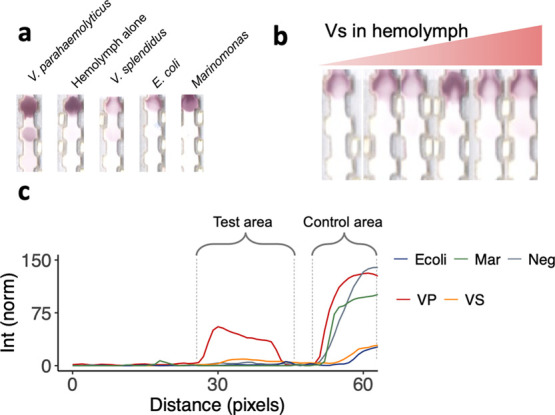
Negative controls. (a) Different negative controls compared to Vp: hemolymph alone, Vs, E. coli, and Marinomonas. (b) Test strips run with Vs in hemolymph. (c) Line scans of strips in b for E. coli (blue), Marinomonas (green), hemolymph alone (gray), Vp (red), and Vs (orange).
We also examined the response of the assay against different negative controls spiked in hemolymph, i.e., the same sample matrix. No signal was observed at the test line, while a red spot was obtained for all of them at the control line. This confirmed that the absence of signal at the test line was due to the inability of the NPs to bind to the negative controls (Figure 5a).
Validation of the Test with Environmental Samples
We then investigated the performance of the developed test with environmental strains. Oysters were collected from coastal sites, and Vp was isolated from hemolymph following a protocol selective for this Vibrio species, even when found at low concentrations.27 This protocol allowed us to obtain 24 environmental PCR-confirmed (16S RNA sequence) Vp isolates that were kept at −80 °C until use.
The environmental isolates and the reference Vp were grown for 5 h at 30 °C, spiked in sterile 1× PBS, and run through the tests. Six samples were chosen arbitrarily to determine the concentration range of the isolates, which was found to be between 9.4 × 107 and 1.7 × 108 cfu/mL. The concentration of the reference Vp was 1.8 × 107 cfu/mL. Gray value intensities of the test lines were at least 5 and 10 times greater than the standard deviation and average obtained for the negative control, respectively (Figure 6a). Test line intensities varied among the environmental isolates probably due to the distinct binding capability of the antibodies against the isolated strains. All environmental isolates gave rise to gray intensities at least 1.7 times lower than the one obtained for the reference Vp from ATCC grown under the same conditions.
Figure 6.
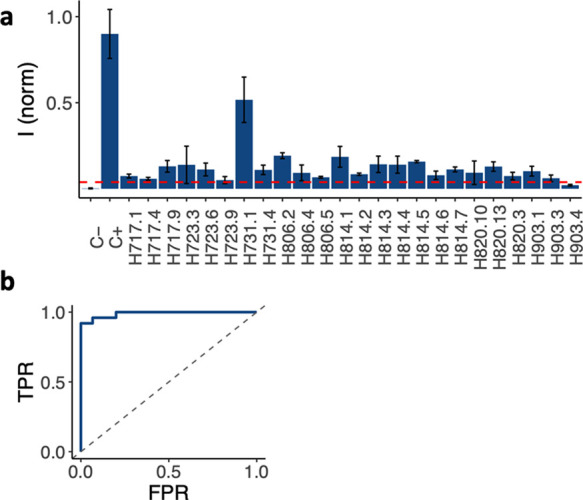
Environmental samples from oysters collected from a Massachusetts harbor. (a) Test line intensities for different oysters that contained Vp. Optimal test intensity cutoff value (red dotted line) and (b) ROC curve analysis for environmental samples.
The LOD of the test in environmental samples was obtained for the isolate H731.1, which gave rise to higher intensities. The isolate was run at concentrations between 0 and 9.4 × 107 cfu/mL and resulted in an LOD of 4.7 × 106 cfu/mL (R2 = 0.993) (Supporting Information, Figure S2).47 This confirms that the binding of the Ab pair against environmental Vp is weaker than the ATCC reference, but it is still below the dose with 50% probability to cause food borne infection (ID50) reported in the literature.44 While the reference Vp is a pathogenic reference strain, this may not be the case for the isolated environmental samples. Thus, further experiments need to be conducted to address the virulence of the environmental vibrios isolated in this work, such as PCR to detect the presence of Vp toxin genes tdh and trh, encoding thermostable direct hemolysin and TDH-related hemolysin, respectively.
The ROC curve was constructed to determine the performance of the test with environmental samples.48,49 To that end, negative controls run in 1× PBS were added to the data set used to determine the sensitivity and specificity of the paper-based test. Gray intensities of the test areas were obtained by subtracting the background for each test run for this data set, yielding intensities ranging from −0.06 to 64.9. ROC curves were obtained by plotting the TPR vs the FPR as a function of a varied threshold in this range The resulting AUC determines the performance of the test where 1.0 represents a perfect test and 0.5 represents a random indicator. The AUC for the tests was 0.989 (Figure 6b). The optimal cutoff was determined as the one that led to the highest sum of sensitivity and specificity of the test. Thus, the optimal performance of the test was obtained with a cutoff of 2.44, leading to a sensitivity of 0.96 and a specificity of 1.00. The optimal cutoff was also used to discriminate negative from positive tests from this paper-based assay. From the environmental data set, only one Vp sample was found to be negative (H903.4); in other words, one false negative was obtained (Supporting Information, Table S1).
Conclusions
In conclusion, we developed a point of consumption paper immunoassay test that can detect the presence of Vibrio parahaemolyticus without the need of doing sample preparation or culturing, supporting our hypothesis. The test was operable directly in oyster hemolymph, the fluid encountered in oyster consumption. The LOD of the test in this biological fluid was 6.05 × 105 cfu/mL, which is below the ID50 reported in the literature for values at which infection can occur.44 Because the use case we envisioned was for food safety, we used this benchmark as opposed to clinical LODs as the clinical scenario is different from food consumption. We validated the immunochromatography test with environmental Vp strains isolated from hemolymph, demonstrating its use beyond commercially available Vp strains. These environmental strains were also used to determine the specificity and sensitivity of the test against strains found in local coastal areas, which was found to be 1.00 and 0.96, respectively.
Typical of visually read out immunoassay tests, its sensitivity is not comparable to alternative techniques such as PCR, ELISA, or electrochemistry. While these other approaches can detect Vp with lower LODs and determine its potential virulence, they all require bringing the sample back to a central lab for culturing and analysis on complex instrumentation and thus cannot provide immediate answers at point of consumption.50−52 Furthermore, the assay presented here is much lower in cost, estimated at $5 per strip, based on estimates for the antibodies, NP reagents, and paper materials.47 At this cost level, the assay has potential for cost-effective on-site monitoring. Alternative enhancement methods such as surface-enhanced Raman spectroscopy, nanozymes, and silver staining can be applied to this format to lower the LOD by increasing the test line intensity.53−56 The assay does exhibit slight cross reactivity to otherVibriospecies, but this could be improved by using monoclonal antibodies rather than polyclonals. The stability of the strip was tested out to a few weeks and determined to be stable when stored under dry conditions, but longer stability studies will be part of future efforts. Therefore, this approach could be deployed as a low-cost initial screening tool to flag samples that should be investigated further via PCR to determine species, strain, and virulence.
Acknowledgments
We thank Chris Schillaci for advice. We thank the Center for Materials Science and Engineering (CMSE) facilities at MIT for use of TEM. This work was supported by funding from Canon Virginia, Inc.
Glossary
Abbreviations
- Ab
antibody
- APW
alkaline peptone water
- AUC
area under the curve
- DH
hydrodynamic diameter
- DLS
dynamic light scattering
- FPR
false positive rate
- HS
human serum
- LOD
limit of detection
- NP
nanoparticle
- PC
protein corona
- PEG
polyethylene glycol
- POC
point of consumption
- ROC
receiver operating characteristic
- SLB
saline lactose broth
- TCBS
thiosulfate-citrate-bile salts-sucrose
- TEM
transmission electron microscopy
- TPR
true positive rate
- Vp
Vibrio parahaemolyticus
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c00853.
Additional sensitivity and specificity measurements and details on workflow (PDF)
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported by Canon Virginia, Inc. CRQ was supported by a Rafael del Pino Fellowship and a UMass Boston College of Science and Mathematics Dean’s Doctoral Research Fellowship from Oracle. BML was funded by a fellowship from Ronald E. McNair grant (P217A170241), Beacon Student Success Fellowship, and an Oracle/Sanofi student fellowship.
The authors declare no competing financial interest.
Supplementary Material
References
- How To Feed the World in 2050. In Expert meeting on How to Feed the World in 2050, FAO Headquarters, Rome, 24-26 June 2009; Office of Assistant Director-General (Economic and Social Department), Food and Agriculture Organization, United Nations, 2009; p 514.
- Tlusty M. F. Trophic level links seafood sustainability to human health. Front. Ecol. Environ. 2013, 11, 121–122. 10.1890/13.wb.006. [DOI] [Google Scholar]
- Stevens J. R.; Newton R. W.; Tlusty M.; Little D. C. The rise of aquaculture by-products: Increasing food production, value, and sustainability through strategic utilisation. Mar. Pol. 2018, 90, 115–124. 10.1016/j.marpol.2017.12.027. [DOI] [Google Scholar]
- Thilsted S. H.; Thorne-Lyman A.; Webb P.; Bogard J. R.; Subasinghe R.; Phillips M. J.; Allison E. H. Sustaining healthy diets: The role of capture fisheries and aquaculture for improving nutrition in the post-2015 era. Food Pol. 2016, 61, 126–131. 10.1016/j.foodpol.2016.02.005. [DOI] [Google Scholar]
- FISH to 2030: Prospects for Fisheries and Aquaculture, Report Number 83177-GLB; World Bank: Washington DC, 2013.
- Stentiford G. D.; Neil D. M.; Peeler E. J.; Shields J. D.; Small H. J.; Flegel T. W.; Vlak J. M.; Jones B.; Morado F.; Moss S.; Lotz J.; Bartholomay L.; Behringer D. C.; Hauton C.; Lightner D. V. Disease will limit future food supply from the global crustacean fishery and aquaculture sectors. J. Invertebr. Pathol. 2012, 110, 141–157. 10.1016/j.jip.2012.03.013. [DOI] [PubMed] [Google Scholar]
- Busa L. S. A.; Mohammadi S.; Maeki M.; Ishida A.; Tani H.; Tokeshi M. Advances in microfluidic paper-based analytical devices for food and water analysis. Micromachines 2016, 7, 86. 10.3390/mi7050086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scallan E.; Hoekstra R. M.; Angulo F. J.; Tauxe R. V.; Widdowson M.-A.; Roy S. L.; Jones J. L.; Griffin P. M. Foodborne illness acquired in the United States--major pathogens. Emerg. Infect. Dis. 2011, 17, 7–15. 10.3201/eid1701.p11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bintsis T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. 10.3934/microbiol.2017.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Enteric Disease Surveillance: COVIS Annual Summary, 2014; Centers for Disease Control and Prevention, National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Division of Foodborne, Waterborne, and Environmental Diseases, Centers for Disease Control and Prevention, 2016.
- Morris J. G. Jr.; Acheson D. Cholera and Other Types of Vibriosis: A Story of Human Pandemics and Oysters on the Half Shell. Clin. Infect. Dis. 2003, 37, 272–280. 10.1086/375600. [DOI] [PubMed] [Google Scholar]
- Choi M.; Stevens A. M.; Smith S. A.; Taylor D. P.; Kuhn D. D. Strain and dose infectivity of Vibrio parahaemolyticus: the causative agent of early mortality syndrome in shrimp. Aquacult. Res. 2017, 48, 3719–3727. 10.1111/are.13197. [DOI] [Google Scholar]
- Zorriehzahra M. J.; Banaederakhshan R. Early Mortality Syndrome (EMS) as new Emerging Threat in Shrimp Industry. Adv. Anim. Vet. Sci. 2015, 3, 64–72. 10.14737/journal.aavs/2015/3.2s.64.72. [DOI] [Google Scholar]
- Prachumwat A.; Taengchaiyaphum S.; Mungkongwongsiri N.; Aldama-Cano D. J.; Flegel T. W.; Sritunyalucksana K. Update on early mortality syndrome/acute hepatopancreatic necrosis disease by April 2018. J. World Aquacult. Soc. 2019, 50, 5–17. 10.1111/jwas.12559. [DOI] [Google Scholar]
- Thadtapong N.; Salinas M. B. S.; Charoensawan V.; Saksmerprome V.; Chaturongakul S. Genome Characterization and Comparison of Early Mortality Syndrome Causing Vibrio parahaemolyticus pirABvp– Mutant From Thailand With V. parahaemolyticus pirABvp+ AHPND Isolates. Front. Mar. Sci. 2020, 7, 290. 10.3389/fmars.2020.00290. [DOI] [Google Scholar]
- Russo L.; Leva Bueno J.; Bergua J. F.; Costantini M.; Giannetto M.; Puntes V.; de la Escosura-Muñiz A.; Merkoçi A. Low-Cost Strategy for the Development of a Rapid Electrochemical Assay for Bacteria Detection Based on AuAg Nanoshells. ACS Omega 2018, 3, 18849–18856. 10.1021/acsomega.8b02458. [DOI] [Google Scholar]
- Rong-Hwa S.; Shiao-Shek T.; Der-Jiang C.; Yao-Wen H. Gold nanoparticle-based lateral flow assay for detection of staphylococcal enterotoxin B. Food Chem. 2010, 118, 462–466. 10.1016/j.foodchem.2009.04.106. [DOI] [Google Scholar]
- Prompamorn P.; Sithigorngul P.; Rukpratanporn S.; Longyant S.; Sridulyakul P.; Chaivisuthangkura P. The development of loop-mediated isothermal amplification combined with lateral flow dipstick for detection of Vibrio parahaemolyticus. Lett. Appl. Microbiol. 2011, 52, 344–351. 10.1111/j.1472-765x.2011.03007.x. [DOI] [PubMed] [Google Scholar]
- Liu Y.; Zhang Z.; Wang Y.; Zhao Y.; Lu Y.; Xu X.; Yan J.; Pan Y. A highly sensitive and flexible magnetic nanoprobe labeled immunochromatographic assay platform for pathogen Vibrio parahaemolyticus. Int. J. Food Microbiol. 2015, 211, 109–116. 10.1016/j.ijfoodmicro.2015.07.005. [DOI] [PubMed] [Google Scholar]
- Fu K.; Zheng Y.; Li J.; Liu Y.; Pang B.; Song X.; Xu K.; Wang J.; Zhao C. Colorimetric Immunoassay for Rapid Detection of Vibrio parahemolyticus Based on Mn2+ Mediates the Assembly of Gold Nanoparticles. J. Agric. Food Chem. 2018, 66, 9516–9521. 10.1021/acs.jafc.8b02494. [DOI] [PubMed] [Google Scholar]
- Yang G.; Wei C.; Tang Y.; Zhang J.; Zhang X.; Chi H.; Tao L.; Kong C. A novel fluorescence immunoassay based on inner filter effect and gold nanoclusters for Vibrio parahaemolyticus determination. Results Chem. 2021, 3, 100208. 10.1016/j.rechem.2021.100208. [DOI] [Google Scholar]
- Martinez A. W.; Phillips S. T.; Whitesides G. M.; Carrilho E. Diagnostics for the developing world: microfluidic paper-based analytical devices. Anal. Chem. 2010, 82, 3–10. 10.1021/ac9013989. [DOI] [PubMed] [Google Scholar]
- Posthuma-Trumpie G. A.; Korf J.; van Amerongen A. Lateral flow (immuno)assay: its strengths, weaknesses, opportunities and threats. A literature survey. Anal. Bioanal. Chem. 2009, 393, 569–582. 10.1007/s00216-008-2287-2. [DOI] [PubMed] [Google Scholar]
- Cordeiro M.; Ferreira Carlos F.; Pedrosa P.; Lopez A.; Baptista P. Gold Nanoparticles for Diagnostics: Advances towards Points of Care. Diagnostics 2016, 6, 43. 10.3390/diagnostics6040043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X.; Shi D.; Zhu S.; Wang B.; Zhang X.; Wang G. Portable Aptasensor of Aflatoxin B1 in Bread Based on a Personal Glucose Meter and DNA Walking Machine. ACS Sens. 2018, 3, 1368–1375. 10.1021/acssensors.8b00304. [DOI] [PubMed] [Google Scholar]
- Han T.; Wang G. Peroxidase-like activity of acetylcholine-based colorimetric detection of acetylcholinesterase activity and an organophosphorus inhibitor. J. Mater. Chem. B 2019, 7, 2613–2618. 10.1039/C8TB02616E. [DOI] [PubMed] [Google Scholar]
- Zhao W.; Brook M. A.; Li Y. Design of Gold Nanoparticle-Based Colorimetric Biosensing Assays. ChemBioChem 2008, 9, 2363–2371. 10.1002/cbic.200800282. [DOI] [PubMed] [Google Scholar]
- Murphy C. J.; Gole A. M.; Stone J. W.; Sisco P. N.; Alkilany A. M.; Goldsmith E. C.; Baxter S. C. Gold Nanoparticles in Biology: Beyond Toxicity to Cellular Imaging. Acc. Chem. Res. 2008, 41, 1721–1730. 10.1021/ar800035u. [DOI] [PubMed] [Google Scholar]
- Doane T. L.; Burda C. The unique role of nanoparticles in nanomedicine: imaging, drug delivery and therapy. Chem. Soc. Rev. 2012, 41, 2885–2911. 10.1039/C2CS15260F. [DOI] [PubMed] [Google Scholar]
- Ge X.; Asiri A. M.; Du D.; Wen W.; Wang S.; Lin Y. Nanomaterial-enhanced paper-based biosensors. Trends Anal. Chem. 2014, 58, 31–39. 10.1016/j.trac.2014.03.008. [DOI] [Google Scholar]
- Hristov D. R.; Rodríguez-Quijada C.; Gomez-Marquez J.; Hamad-Schifferli K. Designing Paper-Based Immunoassays for Biomedical Applications. Sensors 2019, 19, 554. 10.3390/s19030554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada-González D.; Merkoçi A. Nanoparticle-based lateral flow biosensors. Biosens. Bioelectron. 2015, 73, 47–63. 10.1016/j.bios.2015.05.050. [DOI] [PubMed] [Google Scholar]
- Sakata J.; Kawatsu K.; Iwasaki T.; Kumeda Y. Development of a rapid and simple immunochromatographic assay to identify Vibrio parahaemolyticus. J. Microbiol. Methods 2015, 116, 23–29. 10.1016/j.mimet.2015.06.009. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Quijada C.; Lyons C.; Santamaria C.; Quinn S.; Tlusty M.; Shiaris M.; Hamad-Schifferli K. Optimization of paper-based nanoparticle immunoassays for direct detection of the bacterial pathogen V. parahaemolyticus in oyster hemolymph. Anal. Methods 2020, 12, 3056–3063. 10.1039/d0ay00725k. [DOI] [PubMed] [Google Scholar]
- Abramoff M. D.; Magalhães P. J.; Ram S. J. Image Processing with ImageJ. Biophot. Int. 2004, 11, 36–42. [Google Scholar]
- Hartnell R. E.; Stockley L.; Keay W.; Rosec J. P.; Hervio-Heath D.; Van den Berg H.; Leoni F.; Ottaviani D.; Henigman U.; Denayer S.; Serbruyns B.; Georgsson F.; Krumova-Valcheva G.; Gyurova E.; Blanco C.; Copin S.; Strauch E.; Wieczorek K.; Lopatek M.; Britova A.; Hardouin G.; Lombard B.; in’t Veld P.; Leclercq A.; Baker-Austin C. A pan-European ring trial to validate an International Standard for detection of Vibrio cholerae, Vibrio parahaemolyticus and Vibrio vulnificus in seafoods. Int. J. Food Microbiol. 2019, 288, 58–65. 10.1016/j.ijfoodmicro.2018.02.008. [DOI] [PubMed] [Google Scholar]
- Muyzer G.; de Waal E. C.; Uitterlinden A. G. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. 10.1128/aem.59.3.695-700.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green T. J.; Siboni N.; King W. L.; Labbate M.; Seymour J. R.; Raftos D. Simulated Marine Heat Wave Alters Abundance and Structure of Vibrio Populations Associated with the Pacific Oyster Resulting in a Mass Mortality Event. Microb. Ecol. 2019, 77, 736–747. 10.1007/s00248-018-1242-9. [DOI] [PubMed] [Google Scholar]
- de Puig H.; Bosch I.; Carre-Camps M.; Hamad-Schifferli K. Effect of the protein corona on antibody-antigen binding in nanoparticle sandwich immunoassays. Bioconjugate Chem. 2017, 28, 230–238. 10.1021/acs.bioconjchem.6b00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hristov D.; Pimentel A. J.; Ujialele G.; Hamad-Schifferli K. The immunoprobe aggregation state is central to dipstick immunoassay performance. ACS Appl. Mater. Interfaces 2020, 12, 34620–34629. 10.1021/acsami.0c08628. [DOI] [PubMed] [Google Scholar]
- Rodríguez-Quijada C.; Gomez-Marquez J.; Hamad-Schifferli K. Repurposing old antibodies for new diseases by exploiting cross reactivity and multicolored nanoparticles. ACS Nano 2020, 14, 6626–6635. 10.1021/acsnano.9b09049. [DOI] [PubMed] [Google Scholar]
- Bahadır E. B.; Sezgintürk M. K. Lateral flow assays: Principles, designs and labels. Trends Anal. Chem. 2016, 82, 286–306. 10.1016/j.trac.2016.06.006. [DOI] [Google Scholar]
- Pratt G. W.; Fan A.; Melakeberhan B.; Klapperich C. M. A competitive lateral flow assay for the detection of tenofovir. Anal. Chim. Acta 2018, 1017, 34–40. 10.1016/j.aca.2018.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quantitative Risk Assessment on the Public Health Impact of Pathogenic Vibrio parahaemolyticus In Raw Oysters; Food and Drug Administration, 2005.
- Mata Calidonio J.; Gomez-Marquez J.; Hamad-Schifferli K. Nanomaterial and interface advances in immunoassay biosensors. J. Phys. Chem. C 2022, 126, 17804–17815. 10.1021/acs.jpcc.2c05008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokmer A.; Mathias Wegner K. Hemolymph microbiome of Pacific oysters in response to temperature, temperature stress and infection. ISME J. 2015, 9, 670–682. 10.1038/ismej.2014.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam J. O.; de Puig H.; Yen C.-w.; Bosch I.; Gómez-Márquez J.; Clavet C.; Hamad-Schifferli K.; Gehrke L. A Comparison of Nanoparticle-Antibody Conjugation Strategies in Sandwich Immunoassays. J. Immunoassay Immunochem. 2017, 38, 355–377. 10.1080/15321819.2016.1269338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fawcett T. An introduction to ROC analysis. Pattern Recogn. Lett. 2006, 27, 861–874. 10.1016/j.patrec.2005.10.010. [DOI] [Google Scholar]
- Bosch I.; de Puig H.; Hiley M.; Carré-Camps M.; Perdomo-Celis F.; Narvaez C.; Salgado D. M.; Senthoor D.; O’Grady M.; Phillips E.; Durbin A.; Fandos D.; Miyazaki H.; Yen C.-W.; Gélvez-Ramírez M.; Warke R.; Ribeiro L. S.; Teixeira M. M.; Almeida R.; Muñoz-Medina J. E.; Ludert J.; Nogueira M. L.; Colombo T. E.; Terzian A. C. B.; Bozza P.; Calheiros A. S.; Vieira Y. R.; Barbosa-Lima G.; Vizzoni A.; Cerbino-Neto J.; Bozza F. A.; Souza T. M. L.; Trugilho M. R. O.; de Filippis A. M. B.; de Sequeira P. C.; Marques E. T. A.; Magalhaes T.; Díaz F. J.; Restrepo B. N.; Marín K.; Mattar S.; Olson D.; Asturias E. J.; Lucera M.; Singla M.; Medigeshi G. R.; de Bosch N.; et al. Rapid Antigen Tests for Dengue Virus Serotypes and Zika Virus in Patient Serum. Sci. Transl. Med. 2017, 9, eaan1589 10.1126/scitranslmed.aan1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki W.; Ishibashi M.; Kawahara R.; Inoue K. Development of a loop-mediated Isothermal amplification assay for sensitive and rapid detection of Vibrio parahaemolyticus. BMC Microbiol. 2008, 8, 163. 10.1186/1471-2180-8-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar B. K.; Raghunath P.; Devegowda D.; Deekshit V. K.; Venugopal M. N.; Karunasagar I.; Karunasagar I. Development of monoclonal antibody based sandwich ELISA for the rapid detection of pathogenic Vibrio parahaemolyticus in seafood. Int. J. Food Microbiol. 2011, 145, 244–249. 10.1016/j.ijfoodmicro.2010.12.030. [DOI] [PubMed] [Google Scholar]
- Kampeera J.; Pasakon P.; Karuwan C.; Arunrut N.; Sappat A.; Sirithammajak S.; Dechokiattawan N.; Sumranwanich T.; Chaivisuthangkura P.; Ounjai P.; Chankhamhaengdecha S.; Wisitsoraat A.; Tuantranont A.; Kiatpathomchai W. Point-of-care rapid detection of Vibrio parahaemolyticus in seafood using loop-mediated isothermal amplification and graphene-based screen-printed electrochemical sensor. Biosens. Bioelectron. 2019, 132, 271–278. 10.1016/j.bios.2019.02.060. [DOI] [PubMed] [Google Scholar]
- Guo Z.; Jia Y.; Song X.; Lu J.; Lu X.; Liu B.; Han J.; Huang Y.; Zhang J.; Chen T. Giant Gold Nanowire Vesicle-Based Colorimetric and SERS Dual-Mode Immunosensor for Ultrasensitive Detection of Vibrio parahemolyticus. Anal. Chem. 2018, 90, 6124–6130. 10.1021/acs.analchem.8b00292. [DOI] [PubMed] [Google Scholar]
- Duan D.; Fan K.; Zhang D.; Tan S.; Liang M.; Liu Y.; Zhang J.; Zhang P.; Liu W.; Qiu X.; Kobinger G. P.; Fu Gao G.; Yan X. Nanozyme-strip for rapid local diagnosis of Ebola. Biosens. Bioelectron. 2015, 74, 134–141. 10.1016/j.bios.2015.05.025. [DOI] [PubMed] [Google Scholar]
- Sánchez-Purrà M.; Carré Camps M.; de Puig H.; Bosch I.; Gehrke L.; Hamad-Schifferli K. Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers. ACS Infect. Dis. 2017, 3, 767–776. 10.1021/acsinfecdis.7b00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez-Purrà M.; Roig-Solvas B.; Versiani A.; Rodriguez-Quijada C.; de Puig H.; Bosch I.; Gehrke L.; Hamad-Schifferli K. Design of SERS nanotags for multiplexed lateral flow immunoassays. Mol. Syst. Des. Eng. 2017, 2, 401–409. 10.1039/c7me00052a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


