Abstract
Bacterial opportunistic pathogens make diverse secondary metabolites both in the natural environment and when causing infections, yet how these molecules mediate microbial interactions and their consequences for antibiotic treatment are still poorly understood. Here, we explore the role of three redox-active secondary metabolites, pyocyanin, phenazine-1-carboxylic acid, and toxoflavin, as interspecies modulators of antibiotic resilience. We find that these molecules dramatically change susceptibility levels of diverse bacteria to clinical antibiotics. Pyocyanin and phenazine-1-carboxylic acid are made by Pseudomonas aeruginosa, while toxoflavin is made by Burkholderia gladioli, organisms that infect cystic fibrosis and other immunocompromised patients. All molecules alter the susceptibility profile of pathogenic species within the “Burkholderia cepacia complex” to different antibiotics, either antagonizing or potentiating their effects, depending on the drug’s class. Defense responses regulated by the redox-sensitive transcription factor SoxR potentiate the antagonistic effects these metabolites have against fluoroquinolones, and the presence of genes encoding SoxR and the efflux systems it regulates can be used to predict how these metabolites will affect antibiotic susceptibility of different bacteria. Finally, we demonstrate that inclusion of secondary metabolites in standard protocols used to assess antibiotic resistance can dramatically alter the results, motivating the development of new tests for more accurate clinical assessment.
Keywords: antibiotic resistance, antibiotic tolerance, Burkholderia cepacia complex, Burkholderia gladioli, efflux systems, interspecies interactions, polymicrobial infections, Pseudomonas aeruginosa, pyocyanin, toxoflavin
1 |. INTRODUCTION
The use of antibiotics revolutionized medicine in the 20th century, but the evasion of antibiotic treatment by pathogens is a pressing health concern in the 21st century (Fair & Tor, 2014; Hutchings et al., 2019; MacLean & San Millan, 2019). Without new approaches, it is estimated that by 2050 our ability to treat infections with antibiotics will dramatically decrease, resulting in up to 10 million deaths per year (O’Neill, 2016). Bacteria can withstand antibiotic treatment in many ways, by acquiring resistance mutations and/or by developing physiological tolerance, which together enable antibiotic resilience (Blair et al., 2015; Levin-Reisman et al., 2017; Windels, Michiels, Fauvart, et al., 2019; Windels, Michiels, Van den Bergh, et al., 2019). Following recent terminology guidelines (Balaban et al., 2019; Brauner et al., 2016; Kester & Fortune, 2014), we define (i) antibiotic resistance as “the ability to grow in the presence of an antibiotic at a given concentration”; (ii) antibiotic tolerance as “the ability to survive transient antibiotic exposure”; and (iii) antibiotic resilience as “the ability of a bacterial population to be refractory to antibiotic treatment” (Perry et al., 2021). When physicians face situations where treatments do not work in the clinic, that is, bacteria are resilient to treatment, this is typically caused due to increased tolerance and/or resistance levels (Windels, Michiels, Van den Bergh, et al., 2019).
One important and underappreciated factor that promotes both resistance and tolerance is the production of secondary metabolites, a topic we have recently reviewed with examples detailing the multiple mechanisms involved (Perry et al., 2021). These are a broad category of molecules generally produced under slow-growth conditions (i.e., “stationary phase” in the laboratory batch cultures) that are secreted extracellularly or kept inside the cell (Davies, 2013; Price-Whelan et al., 2006). When secreted, secondary metabolites have the potential to affect their surroundings, including other microbes (Davies, 2013; Tyc et al., 2017). Even though it is well-known that microbes can influence each other through secondary metabolite production (Tyc et al., 2017), only recently has this concept been considered in the context of antibiotic susceptibility (Meirelles et al., 2021; Orazi et al., 2019; Orazi & O’Toole, 2017; Perry et al., 2021; Radlinski et al., 2017; Schiessl et al., 2019; Zhu et al., 2019). Secondary metabolites typically affect antibiotic susceptibility by (i) inducing efflux systems or (ii) modulating redox homeostasis or oxidative stress responses (Perry et al., 2021). It stands to reason that they might significantly affect our ability to treat polymicrobial infections.
Certain secondary metabolites are redox-active molecules that promote multifaceted benefits for their producers, from acquiring nutrients to controlling redox homeostasis and promoting anaerobic survival (Glasser et al., 2014; Jo et al., 2017; Price-Whelan et al., 2006). Because they readily react with oxygen (leading to reactive oxygen species generation) and with Fe-S clusters inside the cells (directly oxidizing proteins), redox-active secondary metabolites also display high toxicity levels (Gu & Imlay, 2011; Imlay, 2013; Laursen & Nielsen, 2004; Singh et al., 2013). For this reason, they are often called “natural antibiotics”. Their toxicity puts selective pressure on producers and other microbes that are commonly found together with producing species to develop defense mechanisms against their toxic effects (Davies & Davies, 2010; Martinez, 2009; Waglechner et al., 2019). We have recently proposed that, through the induction of such defense mechanisms, redox-active metabolites can promote collateral resilience to certain clinical drugs (Meirelles et al., 2021). This is particularly relevant when the redox-active metabolite and drug are directly recognized by and/or induce the same cellular machinery that helps the cell manage their toxicity (Meirelles et al., 2021). It is still unclear, however, how broad this phenomenon is when considering distinct redox-active secondary metabolites, the molecular mechanisms involved, and the drugs affected.
Here, we explore the role of distinct redox-active secondary metabolites as modulators of antibiotic resilience. We focus on pyocyanin and toxoflavin, structurally similar compounds made by diverse opportunistic pathogens, including Pseudomonas aeruginosa and Burkholderia species. Our mechanistic investigation of representatives of strains that are commonly co-isolated from cystic fibrosis infections reveals that the production of these metabolites and conserved machinery that senses them can affect the antibiotic susceptibility levels of neighboring species in a significant and predictable fashion. Our findings motivate the development of more nuanced antibiotic resistance diagnostic protocols to take the biologically-altered chemical milieu of infections into account, which is necessary to optimize more effective application of available drugs.
2 |. RESULTS
2.1 |. Pyocyanin produced by P. aeruginosa induces complex defense responses in Burkholderia
To further explore the idea that redox-active secondary metabolite production has the potential to alter antibiotic susceptibility levels in polymicrobial infections, we started by exploring interactions between two opportunistic pathogens relevant to the cystic fibrosis (CF) lung environment: P. aeruginosa and Burkholderia multivorans. P. aeruginosa is a global opportunist pathogen that causes serious infections in patients with CF, chronic wounds, and compromised immune systems (Driscoll et al., 2007). B. multivorans is part of the Burkholderia cepacia complex (Bcc). Species in this group can cause severe chronic infections and are associated with dire prognoses in patients with CF (Coutinho et al., 2011; Lipuma, 2010). Even though P. aeruginosa and Bcc species already display high levels of antibiotic resistance (Driscoll et al., 2007; Rhodes & Schweizer, 2016), recent evidence indicated that the production of pyocyanin (PYO) by P. aeruginosa can modulate susceptibility levels to fluoroquinolone antibiotics in Bcc (Meirelles et al., 2021) (Figure 1a), but the mechanisms involved in this process were unknown. PYO is part of a diverse group of molecules classified as phenazines, which are important virulence factors during P. aeruginosa infections and have been detected in patients (Cruickshank & Lowbury, 1953; Lau et al., 2004; Wilson et al., 1988).
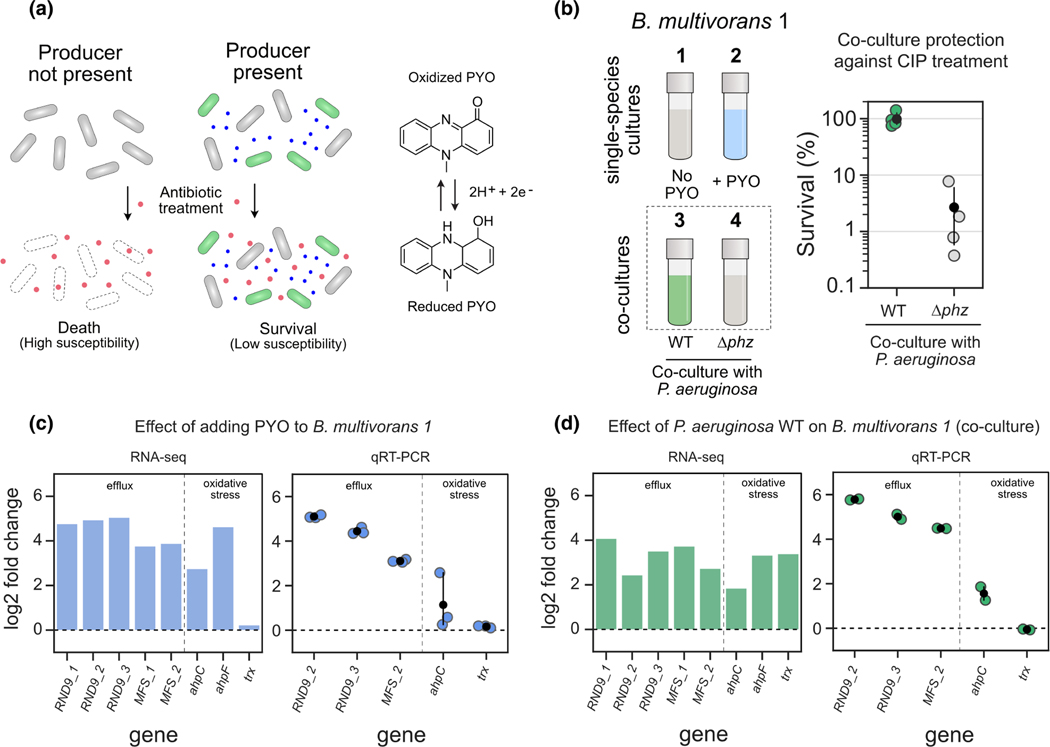
FIGURE 1.
PYO produced by Pseudomonas aeruginosa induces complex defense responses in Burkholderia multivorans. (a) Left: Model of secondary metabolite-mediated induction of survival against antibiotics in microbial populations. Blue dots represent secondary metabolites made by producer species (green cells); red dots represent the antibiotic. Right: One example of such metabolites is PYO, a redox-active molecule produced by P. aeruginosa. (b) Left: Conditions used during RNA-seq and qRT-PCR experiments (1 to 4). In all cases, responses were measured using B. multivorans 1 WT (see Materials and methods). The strain was either grown as a single-species culture and exposed or not to PYO (conditions 1 and 2), or co-cultured with P. aeruginosa that can (WT) or cannot (Δphz) make the phenazine (conditions 3 and 4). Right: B. multivorans 1 tolerance against ciprofloxacin (CIP, 10 μg/ml) when in co-culture with WT or Δphz P. aeruginosa (n = 4). (c and d) representative genes highlighting the responses induced by exogenously added PYO (c) or by co-culturing with PYO-producing P. aeruginosa (d). RNA-seq and qRT-PCR results are shown as bar (left) and strip (right) plots respectively. Genes are displayed in two categories (efflux and oxidative stress) and named accordingly to their draft annotation or to their respective homolog in the P. aeruginosa genome. For qRT-PCR data, n = 3 in panel c, and n = 2 in panel d (see “experiment 1” in the material and methods). For full dataset for each comparison that includes transcriptional changes across B. multivorans genome and their respective loci tags, see Table S2. For additional qRT-PCR results, see Figure S1.
To screen PYO-mediated molecular responses induced in B. multivorans, we first performed RNA-seq experiments either by (i) exposing the cells to exogenously added PYO or (ii) by co-culturing B. multivorans with P. aeruginosa (Figure 1b-left). Control treatments involved sequencing RNA of B. multivorans alone or in co-culture with a P. aeruginosa strain that cannot make phenazines (Δphz), including PYO. We used the B. multivorans strain AU42096 (referred to here as B. multivorans 1, see Table S1 for details). Notably, when in the presence PYO-producing P. aeruginosa, B. multivorans 1 is highly tolerant of fluoroquinolones such as ciprofloxacin (Figure 1b-right). We found that PYO, either when added directly or when produced by P. aeruginosa, induces a set of responses in B. multivorans 1 (Table S2) that can be grouped into two broad classes: (i) induction of specific efflux systems, including a resistance-nodulation-division (RND) efflux system and a potential major facility superfamily (MFS) transporter, and (ii) oxidative stress responses, including alkyl hydroperoxide reductases (Figure 1c). These results were confirmed by qRT-PCR (Figure 1c,d). Not surprisingly, exposing B. multivorans to P. aeruginosa lead to more complex transcriptional responses than exposure to PYO alone, likely due to the wide variety of molecules secreted by P. aeruginosa (Bartell et al., 2017). Importantly, in our experiments with exogenously added PYO, we used a concentration of 100 μM because it is comparable to what our P. aeruginosa wild-type (WT) strain makes in these growth conditions. This is a bulk concentration that has been detected in infections before (Cruickshank & Lowbury, 1953; Wilson et al., 1988), but we have previously observed susceptibility modulator effects even at concentrations as low as 10 μM (Meirelles et al., 2021). Moreover, we still lack the ability to measure local concentrations of such metabolites within infection microenvironments, and actual PYO concentrations inside cellular aggregates might be even higher than 100 μM due to its affinity for extracellular DNA (Saunders et al., 2020).
Despite the complexity of the B. multivorans transcriptomic responses, the induction of one operon called our attention: an efflux system commonly known as RND-9 (Podnecky et al., 2015). All the genes of this operon (Bmul_3930, Bmul_3931, and Bmul_3932) were induced in the presence of PYO, either when added exogenously or via production by co-cultured P. aeruginosa WT (Figures 1c,d, S1, Table S2). Given the importance of RND efflux systems in antibiotic tolerance and resistance (Li et al., 2015), especially when these systems are induced by PYO (Meirelles et al., 2021), we were motivated to investigate whether the RND-9 efflux system in B. multivorans and other Burkholderia species is required to confer antibiotic tolerance in the presence of PYO.
2.2 |. Redox-regulated efflux mediates Burkholderia susceptibility to pyocyanin and its collateral effects on antibiotic resilience
The PYO-mediated induction of RND-9 in B. multivorans seen in our RNA-seq and qRT-PCR experiments led us to investigate this efflux system more carefully. When searching the genomic region where RND-9 is present, we noticed a flanking gene annotated as a “MerR family transcriptional regulator” (Bmul_3929, Figure 2a). BLASTing the Bmul_3929 protein sequence from our B. multivorans 1 strain against proteins encoded in the P. aeruginosa PA14 and P. aeruginosa PAO1 genomes (Winsor et al., 2016) resulted in their respective SoxR as first hits (gene locus tags PA2273 and PA14_35170, respectively, Table S3). SoxR is primarily studied in P. aeruginosa and E. coli; in both cases, the transcription factor contains a Fe-S cluster that can be directly oxidized by redox-active molecules, such as PYO (Figure 2a) (Dietrich et al., 2006; Gu & Imlay, 2011). This results in an induction of stress responses, including efflux systems (Dietrich et al., 2006; Meirelles et al., 2021; Meirelles & Newman, 2018). However, the role of SoxR in other organisms is much less understood.
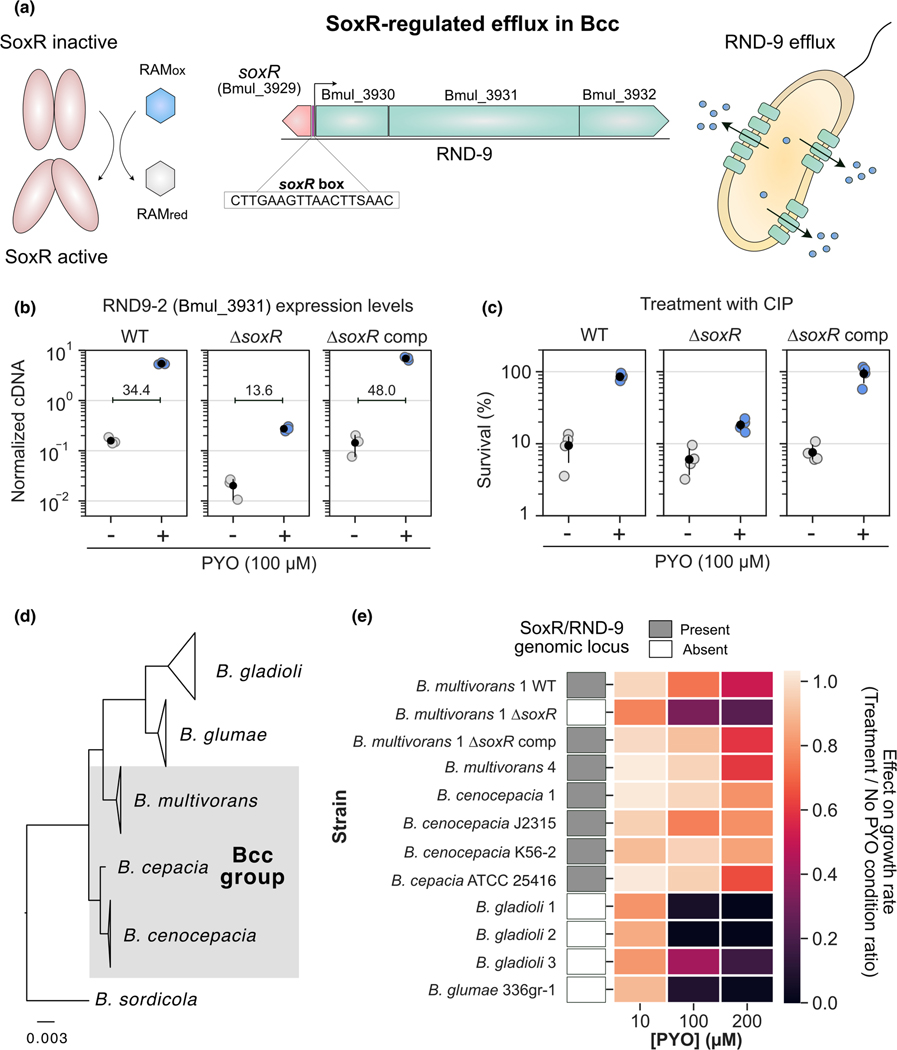
FIGURE 2.
Redox-regulated efflux mediates Burkholderia susceptibility to PYO and its collateral effects on antibiotic resilience. (a) SoxR-mediated regulation of RND-9 in bcc. The example is based on the Burkholderia multivorans ATCC 17616 genome locus structure and orientation. SoxR is oxidized by redox active metabolites (RAMs), triggering its activation (left) (Dietrich et al., 2008; Imlay, 2013; Perry et al., 2021). In bcc, SoxR is commonly found adjacent to the RND-9 efflux system; the SoxR box (Dietrich et al., 2008) is found upstream to RND-9 genes, and the sequence displayed is the consensus found in this genomic region for the bcc strains studied (center). Following other examples of RND efflux systems, proteins derived from Bmul_3930 and Bmul_3931 are likely associated with the inner membrane, while the one from Bmul_3932 is likely an outer membrane protein (Du et al., 2018). SoxR-mediated induction of the system allows efflux of toxic molecules by the bacterial cell (right). (b) Expression levels of the second gene in the RND-9 operon (Bmul_3931) measured by qRT-PCR in different B. multivorans strains in the presence or absence of PYO (n = 3). ΔsoxR comp means complementation of ΔsoxR. Data are shown as normalized cDNA (see Materials and methods). Mean fold differences between control (− PYO) and treatment (+ PYO) are also displayed for all three strains. For additional qRT-PCR results, see Figure S2. (c) Effect of PYO on tolerance to ciprofloxacin (CIP, 10 μg/ml) in the same three B. multivorans strains (n = 4). (d) Phylogenetic relationship between the Burkholderia species used in this study (gray shading highlights species within the bcc group). For full tree detailing different strains, see Figure S4. For broader phylogenetic placement of these species within the Burkholderia genus, see Depoorter et al. (2016). (e) PYO effect on growth rates of distinct strains of the different Burkholderia species studied. Data for three different concentrations are shown (10, 100, and 200 μM). The results are shown as a ratio of the growth rates for each strain under different PYO concentrations by their growth rates in the “no PYO” condition (i.e., values close to 1 mean no inhibition, while values close to 0 mean severe growth inhibition by PYO). Presence or absence of the genomic locus containing SoxR and RND-9 next to each other in these strains is indicated by the gray or white boxes respectively. Growth rates were estimated based on growth curves under the different conditions (Figure S5). In panels b and c, the black dots mark the means and error bars represent 95% confidence intervals.
First, we hypothesized that, by sensing the activity of redox-active metabolites such as PYO, SoxR might regulate the expression of RND-9, which is then used to export them. We confirmed that in our model Bcc strain, B. multivorans 1, the RND-9 promoter region has a SoxR box (Figure 2a). The SoxR box has been shown to be a strong predictor for the operon’s regulation by SoxR (Dietrich et al., 2008), suggesting that SoxR regulates RND-9 expression in B. multivorans. To test whether SoxR is necessary for PYO-mediated increase in RND-9 expression in B. multivorans 1, we made a ΔsoxR deletion mutant. PYO did not increase RND-9 expression in the ΔsoxR strain to the extent it did in the WT (~21 to 34-fold increase in the WT, while ~7 to 13-fold increase in the ΔsoxR mutant, depending on the gene analyzed; see Figures 2b and S2), indicating that SoxR is involved in the pump’s regulation. However, the remaining PYO-mediated induction of RND-9 in the ΔsoxR mutant suggests the existence of an additional uncharacterized regulatory system that is responsive to this metabolite. This is not surprising as PYO, depending on the conditions, has been shown to induce at least two different pumps regulated by two distinct redox sensors in P. aeruginosa (Meirelles et al., 2021). Notably, overall RND-9 expression levels in the ΔsoxR in the presence of PYO were significantly lower and comparable to background levels (i.e., no PYO in the WT) (Figures 2b and S2). Complementation of the soxR gene restored patterns of PYO-mediated activation of RND-9, similar to that observed for the WT (Figures 2b and S2).
We hypothesized that the PYO-mediated increase in resilience against fluoroquinolones in B. multivorans was mediated by SoxR-induction of RND-9. Supporting our hypothesis, the ΔsoxR strain was much less tolerant to ciprofloxacin in the presence of PYO than the WT (Figure 2c). Even though there was an apparent slight increase in the survival percentage for ΔsoxR when PYO is present, we attribute this to a normalization artifact: because PYO affected the growth of ΔsoxR, the strain reached a lower cell density when PYO was present, causing the normalization (i.e., % survival) to be calculated by a smaller CFU number. Raw CFU values showed that PYO only mildly increased the number of surviving cells upon treatment with ciprofloxacin in the ΔsoxR strain, while the effect was significantly higher in the WT strain (Figure S3). Complementation of soxR in the mutant restored WT levels of tolerance (Figures 2c and S3). Notably, due to (i) the expression patterns observed in the ΔsoxR mutant in the presence of PYO (i.e., PYO still increased expression of RND-9 in ΔsoxR, but the overall expression levels were much lower—Figure 2b), and (ii) the fact that PYO induces a broad range of responses in B. multivorans, including oxidative stress (Figure 1c, Table S2), we cannot rule out that PYO might still benefit the ΔsoxR mutant under less stringent conditions, such as when exposed to different concentrations of the metabolite and/or ciprofloxacin, or when treated with different antibiotics.
Because PYO significantly increased the expression of RND-9 in B. multivorans, we decided to investigate the importance of this system for PYO resistance in other Burkholderia species. Specifically, we hypothesized that the presence of the SoxR-regulated RND-9 efflux system might be used to predict how well different Burkholderia species can manage PYO toxicity. Export is crucial for handling PYO’s toxic effects (Dietrich et al., 2006; Meirelles et al., 2021; Meirelles & Newman, 2018). In this scenario, Burkholderia species containing SoxR-mediated RND-9 would sense the presence of PYO secreted by P. aeruginosa and quickly induce the efflux system, increasing their fitness in the presence of the molecule. We searched the Burkholderia Genome Database (www.burkholderia.com) (Winsor et al., 2008) for species commonly found in CF patients as well as in the environment (e.g., plant-associated strains). These included species within the Bcc (such as B. multivorans, B. cepacia, B. cenocepacia) and non-Bcc species (such as B. glumae and B. gladioli). We obtained several strains within these species, many of which are clinical isolates from lung respiratory infections (see Table S1), compared their phylogenetic relationship (Figures 2d and S4), and assessed whether the SoxR-regulated RND-9 efflux system was present (Figure 2e). The system’s presence was determined by (i) directly inspecting the genome (available in databases or through whole-genome sequencing), by (ii) PCR-amplifying a fragment containing part of the SoxR/RND-9 genome locus, or by (iii) by inference based on the genomic content of closely-related strains within the same species in genome databases, such as the Burkholderia Genome Database (Winsor et al., 2008) and Integrated Microbial Genomes and Microbiomes IMG/M (Chen et al., 2021; Mukherjee et al., 2021) (see Materials and Methods for details). In parallel, we measured the growth rates of these strains in the presence of different concentrations of PYO (Figures 2e and S5). We saw a strong correlation between the presence of SoxR/RND-9 locus and the strain’s ability to handle PYO toxicity. The system was present in all Bcc species tested, and growth of these strains was only mildly affected by PYO, even at concentrations as high as 200 μM (Figures 2e and S5). However, growth of strains lacking the regulator/efflux system (B. glumae, different B. gladioli strains, and the B. multivorans ΔsoxR) was dramatically inhibited by PYO (Figures 2e and S5). This indicates that SoxR-regulated efflux plays a conserved role in how well Burkholderia species can tolerate PYO, potentially affecting their ability to inhibit habitats where P. aeruginosa is present. Moreover, this system also modulates P. aeruginosa’s impact on Burkholderia susceptibility to antibiotics because the P. aeruginosa-mediated increase in antibiotic tolerance on B. multivorans is SoxR dependent (Figure S6).
Overall, these results highlight the importance of SoxR and its redox-regulated efflux systems during interspecies interactions mediated by redox-active secondary metabolites, demonstrating their antagonistic effect on fluoroquinolone susceptibility.
2.3 |. Redox-active secondary metabolites as interspecies modulators of antibiotic susceptibility: The toxoflavin example
Our results with PYO motivated us to ask whether other redox-active secondary metabolites might similarly be capable of modulating antibiotic susceptibility in polymicrobial infections (Perry et al., 2021). Accordingly, we searched for other potential candidate molecules known to be made by pathogens that have been found to be infecting CF patients. We found that phenazine-1-carboxylic acid (PCA), another phenazine made by P. aeruginosa, also dramatically increased B. multivorans tolerance against ciprofloxacin in a SoxR-dependent manner (Figure S7). But to test the generality of the phenomenon, we looked for non-phenazine molecules made by species other than Pseudomonas. One example we found was toxoflavin (TOX) (Figure 3a). TOX is made by different Burkholderia species, including B. glumae and B. gladioli, and has been proposed to increase the fitness and virulence of its producers when infecting plants (Chen et al., 2012; Jeong et al., 2003; Lee et al., 2016). B. glumae is a plant pathogen, while B. gladioli can cause disease in plants and humans (Ham et al., 2011; Jones et al., 2021; Lipuma, 2010). In fact, B. gladioli is among the most prevalent Burkholderia species in CF patients (Lipuma, 2010). Though TOX detection in CF sputum has not been attempted to our knowledge, clinical B. gladioli strains commonly produce TOX in vitro (Jones et al., 2021). TOX is redox-active and thought to induce efflux and oxidative stress response in bacteria (Figure 3a,b) (Latuasan & Berends, 1961; Stern, 1935). Therefore, TOX is a potentially clinically relevant molecule and a good candidate for testing the hypothesis that redox-active secondary metabolites have broad potential to modulate antibiotic resilience.
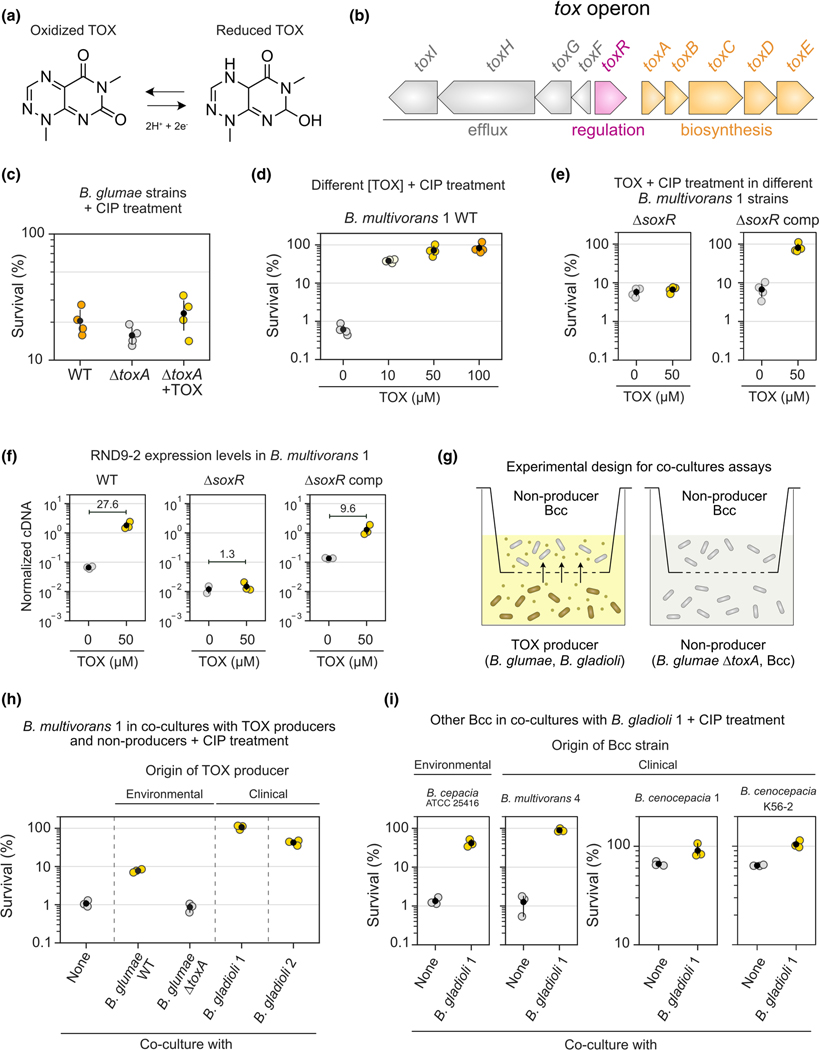
FIGURE 3.
TOX increases tolerance against ciprofloxacin in bcc. (a) TOX molecule and its redox states. (b) Genomic structure of the tox operon present in the TOX producer B. glumae (Chen et al., 2012; Kim et al., 2004). Similar genomic context is found in other TOX producers (Jones et al., 2021). (c) Effect of TOX on tolerance to ciprofloxacin (CIP, 1 μg/ml) in different B. glumae strains (n = 4). TOX was either produced endogenously by the WT strain or added exogenously (50 μM) to the ΔtoxA mutant. (d) Effect of different TOX concentrations on tolerance to CIP (10 μg/ml) in the Burkholderia multivorans 1 WT strain (n = 4). (e) Effect of TOX on tolerance to CIP (10 μg/ml) in the B. multivorans 1 ΔsoxR or ΔsoxR comp strains (n = 4). This experiment was performed separately from that shown in panel D. (f) Expression levels of the second gene in the RND-9 operon (Bmul_3931) measured by qRT-PCR in different B. multivorans strains in the presence or absence of TOX (n = 3 for all except “ΔsoxR + 0 μM TOX”, where n = 2). Data are shown as normalized cDNA (see Materials and methods). Mean fold differences between control (0 μM TOX) and treatment (50 μM TOX) are also displayed for all three strains. Also, see Figure S9 for additional qRT-PCR data. (g) Experimental design used during the co-culture antibiotic assays (see Materials and methods for details). (h) Effect of TOX produced by different producer species (B. glumae WT and several B. gladioli strains) on the tolerance of B. multivorans 1 to CIP (10 μg/ml). Conditions where TOX was not produced were: “none” (i.e., only B. multivorans 1 is present) or co-culture with the B. glumae ΔtoxA strain that cannot make TOX. B. glumae was originally isolated from environmental sample (Chen et al., 2012), while B. gladioli strains are derived from CF patients (see Table S1). Only the top part of the co-culture plate (i.e., containing B. multivorans 1) was plated for CFUs for survival assessment (n = 3). (i) Effect of TOX produced by B. gladioli 1 on the tolerance of multiple Bcc species (isolated from environmental and clinical samples) to CIP (10 μg/ml, n = 3). Note that the scales on the two plots assessing tolerance in B. cenocepacia (two on the right) are different from the ones on the left, since the background tolerance levels in these strains are much higher than the ones in B. cepacia or B. multivorans 4 (left). In panels c–f and h–i, the black dots mark the means and error bars represent 95% confidence intervals.
We started by determining whether TOX can increase Ciprofloxacin tolerance in the producing species B. glumae, just like PYO does in P. aeruginosa (Meirelles et al., 2021; Schiessl et al., 2019). TOX induces the efflux system ToxFGHI in B. glumae (Kim et al., 2004), and we confirmed that the WT strain makes TOX under the studied conditions (Figure S8). However, when comparing WT ciprofloxacin survival levels to a ΔtoxA strain that cannot make TOX (Lelis et al., 2019), we did not see a significant increase in tolerance (Figure 3c); similarly, adding TOX exogenously to ΔtoxA did not significantly increase tolerance (Figure 3c). Although we observed a slight tolerance increase trend when TOX was present, the magnitude of the effect is much smaller than what PYO provides to P. aeruginosa (Meirelles et al., 2021), revealing that ToxFGHI and the overall responses induced by TOX in B. glumae do not confer ciprofloxacin tolerance to this strain under these conditions.
Even though TOX did not provide significant protection against ciprofloxacin in B. glumae, we reasoned that it might act as an interspecies modulator of antibiotic resilience in another organism. For example, B. gladioli strains (potentially TOX producers) (Graves et al., 1997; Jones et al., 2021) can infect CF patients together with other Bcc pathogens (Kennedy et al., 2007). We thus decided to test if Bcc species might benefit from the presence of TOX when treated with ciprofloxacin. Tolerance assays using our Bcc model organism, B. multivorans 1, showed that TOX increased its survival against ciprofloxacin in a concentration-dependent manner (Figure 3d). Levels of 50 μM or above made the strain completely tolerant to the antibiotic treatment (Figure 3d). Importantly, concentrations of TOX within the 10–100 μM range are physiologically relevant since they fall within the amounts produced by B. gladioli and B. glumae under the studied conditions (Figure S8). We next hypothesized that this tolerance phenotype is mediated by the SoxR-regulated RND-9 efflux system, like it is for PYO. Consistent with this prediction, the TOX-mediated increase in tolerance against ciprofloxacin disappeared in the ΔsoxR strain but was restored in the SoxR-complemented strain (Figure 3e). Moreover, using qRT-PCR to quantify the expression of RND-9 in B. multivorans 1, we observed that TOX increased transcription of RND-9 in B. multivorans 1 and its induction disappeared in the ΔsoxR mutant but was restored in the SoxR-complemented strain (Figures 3f and S9). This indicates that SoxR in B. multivorans 1 works as a broader sensor for the activity of a wide range of redox-active molecules, with direct consequences for antibiotic efficacy.
We next evaluated the effect of TOX in co-culture assays (Figure 3g). First, we tested if B. multivorans 1 becomes more tolerant to ciprofloxacin when grown with environmental or clinical TOX producers (Figure 3h). Co-culture with B. glumae WT (i.e., TOX present) increased B. multivorans 1 tolerance to ciprofloxacin. The phenotype disappeared when the strain was co-cultured with the B. glumae ΔtoxA (i.e., no TOX). Notably, co-culture with two different clinical strains of B. gladioli isolated from CF patients dramatically increased tolerance against ciprofloxacin by B. multivorans 1 (Figure 3h). These two clinical strains produce TOX (Figure S8), and the molecule was present in the co-cultures, evident by its yellow pigmentation. To evaluate the generality of these results, we compared the Ciprofloxacin tolerance of multiple Bcc strains from different species (B. cepacia, B. multivorans, and B. cenocepacia) to that seen when they were co-cultured with a TOX-producing clinical strain of B. gladioli. These Bcc strains were derived either from environmental samples or from patients. In all cases, co-culture with B. gladioli increased ciprofloxacin tolerance levels in the Bcc species (Figure 3i). All co-cultures were yellow, indicating TOX was present. The effect was dramatic for B. cepacia and B. multivorans, increasing tolerance levels more than an order of magnitude; although B. cenocepacia strains showed higher background tolerance levels when grown alone in the absence of TOX, co-culturing these strains with TOX-producing B. gladioli still made them more tolerant of ciprofloxacin (Figure 3i).
Overall, our results show that phenazines made by P. aeruginosa are not the only redox-active secondary metabolites that can modulate antibiotic resilience. TOX, a redox-active molecule produced by different Burkholderia species, can do the same, suggesting generality for the phenomenon.
2.4 |. Assessing the effects of redox-active secondary metabolites on antimicrobial susceptibility testing
These mechanistically oriented laboratory results raised an important practical question: how might redox-active secondary metabolites impact standard clinical antibiotic susceptibility testing (AST)? Recent work indicates that PYO and other phenazines may affect such tests (Gerstel et al., 2020; Zhu et al., 2019). However, current AST methods are blind to potential modulating effects of redox-active secondary metabolites. This happens because the synthesis of these metabolites is usually controlled by quorum sensing, being made only at cells densities that are much higher than what is used for typical AST inocula (Davies, 2013; Jorgensen & Ferraro, 2009). However, redox-active secondary metabolites have been detected in infections (Cruickshank & Lowbury, 1953; Wilson et al., 1988) and presumably could dramatically change the performance of certain drugs during treatment (Perry et al., 2021). To take this into account, we modified the protocol of a traditional AST assay that determines the minimum inhibitory concentration (MIC) for specific drugs. Using PYO and TOX as examples, we tested how these metabolites affect susceptibility to several antibiotics from different classes.
Our experimental design is shown in Figures 4a and S10a. In brief, MIC assays following the EUCAST guidelines (see Materials and Methods) were performed in the absence or presence of exogenously added PYO or TOX. For these assays, we used our model organism, B. multivorans 1. We tested several different (sub)classes of drugs, including fluoroquinolones (ciprofloxacin and levofloxacin), tetracyclines (tetracycline and doxycycline), amphenicols (chloramphenicol), sulfonamides (sulfamethoxazole in combination with trimethoprim), aminoglycosides (tobramycin), carbapenems (meropenem), cephalosporins (ceftazidime), and polymyxins (colistin). PYO and TOX altered the MIC of B. multivorans for several types of drugs. Both PYO and TOX had overall antagonistic effects on fluoroquinolones, tetracyclines, and chloramphenicol, increasing their MICs (Figure 4b,c). In some cases, although not enough to change the MIC, PYO, and TOX had drug-antagonistic effects that could be measured by an increase in optical density at the pre-MIC concentration (see antibiotics marked with “+” in Figure 4b–d). Examples include exposure to PYO together with tetracycline or sulfamethoxazole/trimethoprim (Figure 4d). Similar antagonistic effects were observed for cells exposed to TOX and levofloxacin (Figure 4d). On the other hand, PYO and TOX acted synergistically with other drugs, potentiating their toxicity, resulting in lower MICs or lower optical densities at the pre-MIC concentrations when the metabolites were present. This was the case for tobramycin and meropenem in the presence of PYO (Figure 4b,d), and for sulfamethoxazole/trimethoprim, tobramycin, meropenem, and ceftazidime in the presence of TOX (Figure 4c,d). Cases where the synergistic effect was only visible at the pre-MIC concentrations are marked as “-” in Figure 4b–d. We also tested the effect of PYO and TOX on colistin susceptibility, with both enhancing its toxicity. In the absence of these metabolites, B. multivorans 1 was completely resistant to colistin at all the concentrations tested (MIC > 4.096 mg/ml, Table S4), and therefore colistin is not included in Figure 4b,c. However, addition of PYO caused a decrease in optical density (Figure S11c), and TOX caused a dramatic drop in the MIC (Table S4), indicating that these metabolites can act synergistically with polymyxins (Meirelles et al., 2021; Schiessl et al., 2019; Zhu et al., 2019). Overall, these results show that the effects of redox-active secondary metabolites on antibiotic susceptibility can be dramatic and are distinct for different classes of drugs.
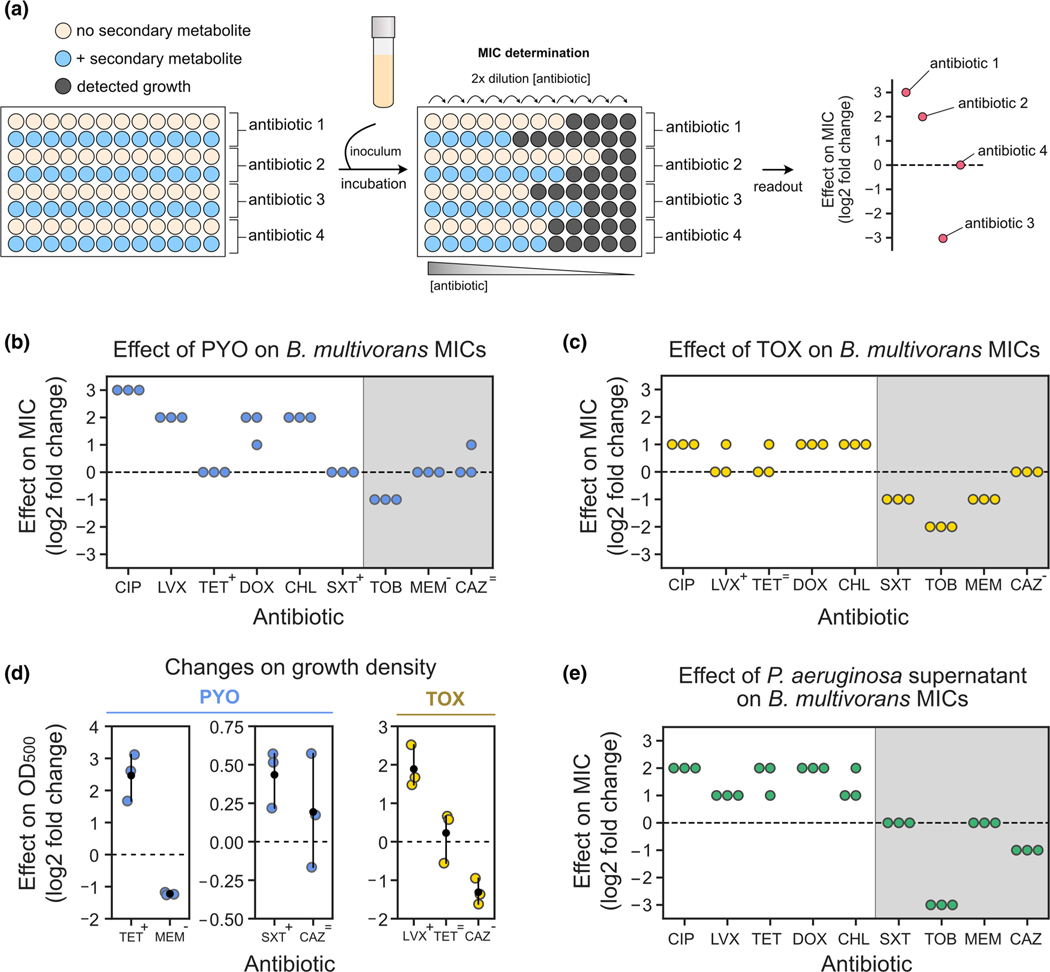
FIGURE 4.
Assessing the effects of secondary metabolites on antimicrobial susceptibility testing. (a) Experimental design used during MIC tests that account for the effect of secondary metabolites on resistance levels. (b) Effects of PYO (100 μM) on Burkholderia multivorans 1 MICs (for each antibiotic, n = 3). (c) Effects of TOX (50 μM) on B. multivorans 1 MICs (for each antibiotic, n = 3). In b and c, symbols above the antibiotic names represent the effects of PYO and TOX on growth density displayed in D (“+” represents increase in density, “-” represents decrease in density, and “=” represents no consistent change in density). (d) Effects of PYO (two plots on the left) and TOX (right) on the growth density at the pre-MIC antibiotic concentrations during MIC assays. Antibiotics shown are the ones previously highlighted by the symbols in panels b and c. Note that scales are different for each plot. For normalized absorbance values, see Figure S11a,b. (e) Effects of Pseudomonas aeruginosa WT supernatant (i.e., PYO present) on B. multivorans 1 MICs (for each antibiotic, n = 3). For experimental design, see Figure S10b. Gray shading in b, c, and e represent antibiotics for which metabolite-mediated increase in resilience was not observed under the studied conditions. In panels d, the black dots mark the means and error bars represent 95% confidence intervals. CIP, ciprofloxacin; LVX, levofloxacin; TET, tetracycline, DOX, doxycycline, CHL, chloramphenicol, SXT, sulfamethoxazole/trimethoprim; TOB, tobramycin; MEM, meropenem; CAZ, ceftazidime.
Recognizing that these effects might be generalizable for a wide range of unknown molecules for which purified compounds are unavailable, we sought to determine whether modified MIC protocols could be agnostic to any specific metabolite secreted by a pathogen. Accordingly, we used spent media from grown cultures (i.e., containing secondary metabolites) mixed with fresh media (Figure S10b and Materials and Methods for experimental design) to modify the traditional MIC assay. As proof of principle, we tested the susceptibility of our model organism, B. multivorans 1, to all the drugs mentioned above, using spent medium from P. aeruginosa WT (Figure 4e). We predicted that the phenazine-producing P. aeruginosa cultures would change the MIC like exogenously added PYO did in the previous experiments. As expected, P. aeruginosa spent medium increased the MIC of B. multivorans against fluoroquinolones, tetracyclines, and chloramphenicol (Figure 4e). We did not see changes in MIC against sulfamethoxazole/trimethoprim (Figure 4e), likely because our experimental design led to a fourfold dilution of the active metabolite(s) from its initial concentration in the spent medium. For instance, the PYO concentration was ~80–100 μM in the original culture from which the spent medium was taken, but only ~20–25 μM in the final assay (see Materials and Methods). Because the impact of exogenously adding 100 μM PYO was already small (Figure 4d), it is not surprising that we observed no effect using the diluted spent media. Still, in agreement with our experiments using purified PYO, P. aeruginosa spent medium potentiated the efficacy of tobramycin, reducing the MIC (Figure 4e). On the other hand, unlike our findings with purified PYO, P. aeruginosa spent medium decreased the MIC against ceftazidime (Figure 4e). To evaluate if the results observed for P. aeruginosa spent medium were caused by phenazines, we also tested spent medium from the P. aeruginosa Δphz mutant (Figure S11d). We saw no significant increase in the B. multivorans MICs when Δphz spent medium was used (Figure S11d), suggesting that the antagonistic effects against these drugs were due to phenazines. Finally, we also tested the impact of spent medium from B. multivorans on its own MICs (Figure S11e). We saw no effect on MICs for fluoroquinolones or sulfamethoxazole/trimethoprim, but we did see changes for the other drugs: B. multivorans spent medium increased MICs against tetracyclines, chloramphenicol, and meropenem but decreased MICs against tobramycin and ceftazidime (Figure S11e). Altogether, our results suggest that metabolites secreted by opportunistic pathogens can substantially alter MIC levels in ways that are overlooked by current protocols.
3 |. DISCUSSION
Redox-active secondary metabolites, produced by organisms throughout the tree of life (Jacob et al., 2011), are of particular interest due to their multifaceted and nuanced effects, which depend on the environmental and physiological conditions experienced by the cells when exposed to them (Glasser et al., 2017; Meirelles & Newman, 2018). Within the clinical context, their dynamic functions range from the support of biofilm development to serving as virulence factors that are toxic to host cells (Dietrich et al., 2013; Lau et al., 2004; Liu & Nizet, 2009; Ramos et al., 2010; Saunders et al., 2020). In this study, we found that these molecules can also modulate antibiotic resilience, altering susceptibility levels to drugs commonly used to treat infections. Together with recent evidence demonstrating that these molecules can increase mutation rates to antibiotic resistance against certain drugs (Meirelles et al., 2021), our results point toward the production of redox-active secondary metabolites by opportunistic pathogens as an underappreciated route for the evolution of antimicrobial resistance.
To better predict contexts where redox-active secondary metabolites might affect antimicrobial susceptibility, it is necessary to understand the molecular mechanisms involved in the process. Our results support the role of SoxR as a broad redox sensor in nature (Dietrich et al., 2008). This transcription factor has been primarily studied in enteric bacteria (E. coli and Salmonella sp.) and in P. aeruginosa, where it senses redox-active molecules through oxidation of its Fe-S cluster (Dietrich et al., 2006; Gerstel et al., 2020; Gu & Imlay, 2011; Lee et al., 2015; Sheplock et al., 2013; Singh et al., 2013). Biogenesis of such clusters is essential for the cells to appropriately respond to the presence of these molecules (Gerstel et al., 2020). However, SoxR is widely distributed throughout the bacterial domain, with homologs enriched in the Actinobacteria and Proteobacteria (Glasser et al., 2017). Many of these organisms include well-established or emerging opportunistic pathogens, such as species within the genus Mycobacterium, Nocardia, Burkholderia (including the Bcc species studied here), Ralstonia, Acinetobacter, and Stenotrophomonas, among others (Dietrich et al., 2008; Glasser et al., 2017). However, we know little about the redox-active molecules SoxR might sense and the responses it might control in these organisms. Our work demonstrates that, if focused on antibiotic susceptibility, special attention should be given to SoxR-regulated efflux systems and to the drugs these systems might be able to transport. SoxR homologs in other pathogens might control the transport of yet-to-be-discovered redox-active secondary metabolites that affect antibiotic susceptibility, whether the metabolite is produced endogenously or by a neighboring species.
While we have shown that two phenazines and toxoflavin can dramatically modulate antibiotic susceptibility in vitro, it is important to recognize that these effects have yet to be confirmed in vivo. Testing the relevance of this phenomenon in the host context is an essential next step, and we hope our study will stimulate future work on this topic. Moreover, there is still much to learn about interspecies interactions during infections, mostly due to our still limited understanding of the spatial and temporal colonization dynamics in polymicrobial infections. This is true even for CF, a well-studied infection context. Although we know certain species can be found infecting patients at the same time, our understanding of the frequency of these interactions is much less clear. For example, P. aeruginosa can be found with Staphylococcus or Bcc (Chmiel et al., 2014; Folescu et al., 2015; Lipuma, 2010; Schwab et al., 2014), but the prevalence of such co-infections among patients is not well-documented. Similarly, B. gladioli is among the most common Burkholderia species isolated from CF sputum (Lipuma, 2010), but we do not know which other microbes typically co-reside with this species. Moreover, how these organisms co-exist within infections is poorly constrained due to very limited data on their distribution within patients, though methods for accessing this are improving (DePas et al., 2016; Earle et al., 2015; Shi et al., 2020; Wilbert et al., 2020). While our study suggests that redox-active metabolites such as PYO and TOX may be broad modulators of antibiotic susceptibility, to accurately predict how/which neighboring species might be affected by such molecules, future work must characterize community dynamics and spatial proximity between co-infecting organisms within individual patients.
Our results also motivate more nuanced design of antimicrobial susceptibility tests to make them better mimic the context of infections. While there is room for improvement across a range of parameters, inclusion of secondary metabolites from spent supernatants of infecting strains in AST protocols could be a simple and constructive first step, even without knowledge of the identity of the metabolite(s) that impact the results. Because the vast majority of secondary metabolites are still uncharacterized (Skinnider et al., 2020), it is important to use phenotypic screens to identify the existence of an unknown whose identity can later be determined through follow-up research. Our case study using PYO and TOX as models for evaluating the impact of redox-active secondary metabolites on antibiotic resilience serves as a proof of principle that awareness and understanding of these interactions has the potential to inform drug selection for more effective treatment. For example, our results suggest that when in the presence of P. aeruginosa, treating Bcc with fluoroquinolones, tetracyclines, chloramphenicol, or sulfamethoxazole/trimethoprim would likely be less effective, increasing the chances of the evolution of resistance (Meirelles et al., 2021). For these situations, drugs like meropenem or ceftazidime would possibly be more appropriate. TOX-producing B. gladioli might also significantly impact Bcc susceptibility to antibiotics, mainly decreasing the efficacy of fluoroquinolones. Finally, B. multivorans appears to produce unknown metabolites that reduce its susceptibility to certain drugs, such as tetracyclines, chloramphenicol, and meropenem, which could be investigated in more detail in the future.
More generally, we end by observing that approaches that have been employed to identify polymicrobial and/or microbe-host interactions mediated by secondary metabolites in the context of the gut microbiome (Agus et al., 2021; Vernocchi et al., 2016) can and should be leveraged more strongly in the context of infectious disease. While some studies have begun to light the way (Bauermeister et al., 2021; Garg et al., 2017), there remains tremendous potential for discovery of secondary metabolite-mediated effects that shape clinical outcomes. To identify endogenously produced molecules that impact antimicrobial susceptibility, we must ask: Who is there? What are they making? What are the responses induced in the community by the presence of specific molecules? And, finally, which drugs are affected by these responses? High-throughput methods combined with various “systems approaches” used for studying how drugs interact with the gut microbiome (Brochado et al., 2018; Maier et al., 2018; Maier et al., 2021; Zimmermann et al., 2021) could be adapted to study how metabolites produced by pathogens in different infection contexts might affect their interactions with each other and their susceptibility to antibiotics. We anticipate that our findings with PYO and TOX are likely just the tip of the iceberg, and we hope growing awareness of the potential ubiquity of these type of interactions will enhance our understanding of and ability to control polymicrobial infections.
4 |. MATERIALS AND METHODS
4.1 |. Media and incubation conditions
Most of the media and conditions used followed previous descriptions (Meirelles et al., 2021). The defined medium mostly used in this work was the glucose minimal medium (GMM), comprising 20 mM glucose (or 10 mM, if specified), 50 mM KH2PO4/K2HPO4 (pH 7.2), 42.8 mM NaCl, 9.35 mM NH4Cl, 1 mM MgSO4, 1× MEM Amino Acids (AA) solution (MilliporeSigma, Cat. No. M5550), and a trace elements solution (Widdel & Pfennig, 1981). As described previously (Meirelles et al., 2021), the medium was prepared by autoclaving all the components together, except for the glucose and the 1000× trace elements stock solution, which were sterilized through filtration and added after the autoclave step. Autoclave step proceeded for 20 min at 121°C. As previously noted (Meirelles et al., 2021), autoclaving MgSO4 together with other media components is essential for consistent production of PYO by our WT P. aeruginosa UCBPP-PA14 strain in this medium. Two other media were used: Luria–Bertani (LB) Miller broth (BD Biosciences) and BBL cation-adjusted Mueller–Hinton II (MH) broth (BD Biosciences). Their preparation followed the manufacturer’s instructions. Importantly, for the MH broth medium, the autoclave step was 10 min. 1.5% Bacto agar (BD Biosciences) was used for solid media unless mentioned otherwise (see strains construction section below). Pyocyanin (PYO) was synthesized and purified following published protocols (Cheluvappa, 2014; Costa et al., 2017), and 10 mM stock solutions were prepared using 20 mM HCl and stored at −20°C. Toxoflavin (TOX) (MedChemExpress) was dissolved in dimethyl sulfoxide (DMSO) to make 10 mM stock solutions that were prepared on the same day of the experiment. Phenazine-1-carboxylic acid (PCA) (Princeton Bio) was dissolved in 20 mM NaOH and stored at −20°C. Finally, experiments using PYO, TOX, or PCA always had negative controls with equivalent volumes of the solvents used (20 mM HCl, DMSO, or 20 mM NaOH respectively). Unless otherwise specified, incubations were performed at 37°C, with shaking for liquid cultures at 250 rpm.
4.2 |. Strain construction
We performed genetics in the B. multivorans AU42096 strain, our model Bcc organism. We refer to this strain as B. multivorans 1 (Meirelles et al., 2021) (Table S1). An unmarked deletion of the Bmul_3929 gene (soxR homolog) was made through homologous recombination using the pEX18Tc plasmid (Hoang et al., 1998; Huang & Wilks, 2017). Briefly, ~800 bp fragments upstream and downstream of the gene were amplified and cloned into the pEX18Tc suicide vector using Gibson assembly (Gibson et al., 2009). Amplifications were done using B. multivorans 1 genomic DNA (gDNA, extracted with DNeasy Blood & Tissue Kit, Qiagen), cleaned up using the Monarch PCR Purification kit (New England Biolabs), and used in a Gibson assembly reaction (New England Biolabs) together with a PCR-amplified fragment containing the entire pEX18Tc sequence. The assembled construct was transformed into E. coli S17 (Simon et al., 1983), with selection in LB with 10 μg/ml tetracycline and incubation at 30°C. The assembled plasmid was identified by colony PCA, verified by Sanger sequencing (Laragen), and inserted into B. multivorans 1 genome by biparental conjugation using a modified version of previously published protocols (Choi & Schweizer, 2006; Dubarry et al., 2010). Briefly, E. coli S17 (containing the pEX18Tc-based deletion plasmid) and the B. multivorans 1 WT strain were grown overnight in LB (with 10 μg/ml tetracycline for the E. coli strain) and then diluted in the same medium to OD600 of 0.05 and 0.025, respectively. Cultures were grown until OD600 of ~0.5; if one of the cultures reached the final OD earlier, this culture was moved to a non-shaking incubator (also 37°C) until the other was ready for the next steps. When ready, 400 μl of each culture were mixed into a Falcon tube and left stand (non-shaking) for 1 h. Next, 100–200 μl of the mixed cultures were plated on top of polycarbonate membranes (MilliporeSigma, Cat. No. WHA10417006) on LB plates, followed by an incubation step of ~15 h at 37°C. Next, cells were scrapped from the filters, resuspended in 0.9% NaCl, plated on to VBMM agar plates (3 g/L trisodium citrate, 2 g/L citric acid, 10 g/L K2HPO4, 3.5 g/L NaNH4PO4·4H2O, 1 mM MgSO4, 100 μM CaCl2, pH 7) containing 100 μg/ml of tetracycline (Choi & Schweizer, 2006), and incubated for 24–48 h at 30°C. This resulted in colonies of B. multivorans 1 that were merodiploids containing the construct integrated in their genomes, which were then plated on to LB containing 10% sucrose. Importantly, LB plates lacked NaCl and contained only 1.05% agar. This was relevant because we noticed that even though pEX18Tc contains a sacB cassette, the sacB counter selection was not effective in our B. multivorans 1 strain, as has been observed for other Burkholderia strains (Barrett et al., 2008). However, these conditions still allowed the screening of colonies that lost the integrated plasmid. We noticed that such colonies presented a spreading “flat” morphology under these conditions, while the merodiploids were round “thick” colonies (Figure S12). The flat colonies were rare (roughly 1 in ~200 colonies) and, as expected, ~50% of them were WT genotype and ~ 50% were unmarked deletions. These were screened by PCR and verified by both Sanger sequencing and whole-genome sequencing.
Complementation of the ΔsoxR mutant was also done through homologous recombination with the re-insertion of the gene in its native site following the same protocols described above. The only difference was that, in this case, a unique fragment from ~800 bp fragments upstream to ~800 downstream region of soxR (i.e., containing the soxR gene) was amplified from B. multivorans 1 WT gDNA and cloned into the pEX18Tc. This was then inserted in the ΔsoxR mutant through homologous recombination as described above, with PCR and whole-genome sequencing verifications. Information on all primers and strains used is available in Table S1.
4.3 |. Whole-genome sequencing and draft genome assembly and annotation
Genomic DNA (gDNA) was extracted from the following strains using the DNeasy Blood and Tissue kit (Qiagen): B. multivorans 1 WT; B. multivorans 1 ΔsoxR; B. multivorans 1 ΔsoxR comp (complementation of soxR in the ΔsoxR background). Library preparation was performed by the Microbial Genome Sequencing Center (MiGS) (Pittsburgh, Pennsylvania, USA) and included (i) 2 × 150 bp paired-end Illumina sequencing for all these strains (minimum of 400-Mb sequencing output per sample, with approximately 50–60× coverage), and (ii) an additional Nanopore sequencing for the B. multivorans 1 WT strain (minimum of 300 Mb Long Reads) for draft genome assembly and annotation of this strain.
Draft genome assembly and annotation of the B. multivorans 1 WT strain was performed by the MiGS analysis pipeline, and included: (i) quality control and adapter trimming performed using bcl2fastq (for Illumina reads) and porechop (for Nanopore reads, available at: https://github.com/rrwick/Porechop); (ii) assembly (using both Illumina and Nanopore reads) performed using Unicycler (Wick et al., 2017); (iii) assembly statistics assessed using QUAST (Gurevich et al., 2013), which can be found in the “BM1_assembly_metrics.tsv” file in the supplementary material; and (iv) draft assembly annotation performed using Prokka (Seemann, 2014). The assembly and annotation files can also be found in the supplementary material as “BM1_assembly.fasta” and “BM1_annotation.gff”, together with a summary of the assembled contigs “BM1_contigs_ summary.tsv”.
To check for the ΔsoxR and ΔsoxR comp strains, quality control was performed using Trimmomatic (version 0.39) (Bolger et al., 2014) with the following settings: LEADING:27 TRAILING:27 SLIDINGWINDOW:4:20 MINLEN:35. Mutations were then identified using breseq (version 0.35.7) (Deatherage & Barrick, 2014) using the draft annotation we described above. This was important because we observed dozens of potential non-related mutations that appeared in the genome during the process of making these strains. This was not surprising since B. multivorans 1 is a clinical strain that has not been extensively used in the laboratory, with the exception of our previous work that did not include genetic manipulation (Meirelles et al., 2021). Despite these additional potential mutations, we were able to confirm the correct soxR deletion and complementation, which resulted in the phenotypes described in our results. Whole-genome sequence data for the strains studied were submitted to the NCBI Sequence Read Archive under the accession number PRJNA787476.
4.4 |. Assessment of the presence of SoxR/RND-9 locus in the studied strains
The SoxR/RND-9 locus presence was determined by three complementary methods. First, we directly inspected the genomes of the Burkholderia species studied that are available at the Burkholderia Genome Database (BGD) (Winsor et al., 2008) and Integrated Microbial Genomes and Microbiomes IMG/M (Chen et al., 2021; Mukherjee et al., 2021). These included B. cenocepacia J2315, B. cenocepacia K56–2, B. cepacia ATCC 25416, and B. glumae 336gr-1. We also inspected the draft genomes for the strains we performed whole-genome sequencing, which included all the genotypes of our B. multivorans 1 strain. Second, we performed PCR amplification attempts for a fragment containing part of the SoxR/RND-9 genome locus in all strains. Primers were designed using consensus sequences of the region known from the available genomes (see primers in Table S1). As positive controls for this screen, we used primers for 16S rRNA amplification. For all PCRs, overnight cultures for all strains were grown in LB and 1 μl of the cultures were used during amplification attempts. Finally, we also checked annotated genome of closely related strains available at the BGD or IMG/M. We could not find strains of B. glumae or B. gladioli that had the SoxR/RND-9 genome locus. It is important to mention that this does not mean that such species do not have SoxR homologs. However, it is unlikely that those are regulating RND efflux systems that can protect against PYO. In addition, we cannot exclude the presence of other SoxR-like MerR family transcriptional regulators in the genomes of the studied strains, but it is unlikely such regulators respond to PYO activity.
4.5 |. Phylogenetically analyses
Phylogenetic analysis included 15 Burkholderia strains for which 16S sequences were either generated by Sanger sequencing (Laragen; primers available in Table S1) or retrieved from the BGD database or GenBank. For generated sequences, contigs were prepared using MacVector (version 18.2.0). Sequences were aligned with MAFFT (version 7.490, alignment available in the supplementary material) (Katoh et al., 2002; Katoh & Standley, 2013), and phylogeny was reconstructed through Bayesian inference using MrBayes (version 3.2.7) (Ronquist et al., 2012). Briefly, GTR was used as initial model for two independent analyses, each consisting of three heated chains and one cold chain. Markov Chain Monte Carlo sampling ran for one million generations (additional parameters: diagnfreq = 1000; samplefreq = 100; relburnin = yes; and burninfrac = 0.25), which was enough to reach convergency (i.e., average standard deviation of split frequencies < 0.01). The final tree was edited in FigTree (version 1.4.4, available at: https://github.com/rambaut/figtree/releases) and polished in Affinity Designer (Serif, version 1.10.4).
4.6 |. RNA-seq experiment and data analysis
Four different conditions were prepared for the RNA-seq experiment: (i) B. multivorans (no PYO added); (ii) B. multivorans + 100 μM PYO; (iii) B. multivorans + P. aeruginosa WT (co-culture, PYO and other phenazines produced); and (iv) B. multivorans + P. aeruginosa Δphz (co-culture, no PYO or phenazines produced). For these experiments, we used our B. multivorans 1 WT strain. From freshly streaked LB plates (<2 days old), overnight cultures of B. multivorans, P. aeruginosa WT and P. aeruginosa Δphz were grown in GMM (20 mM glucose) + AA. Cells were washed (12,500 rpm for 2 min) and resuspended in the same medium. OD500 values were measured and adjusted for the start of the experiment. Due to growth differences, the conditions involving co-cultures started with four times more Burkholderia cells than Pseudomonas cell. ODs used were as follows: conditions (i) and (ii) had initial OD500 = 0.04 (only Burkholderia was present); conditions (iii) and (iv) had final OD500 = 0.05, composed from 0.04 of Burkholderia cells and 0.01 of Pseudomonas cells. Cultures were prepared in 7 ml media using GMM (20 mM glucose) + AA, with either 100 μM PYO (used in condition 2) or the respective amount of HCl added to the cultures. The four tubes were incubated for ~9 h, and then collected for RNA extraction. For this, 0.7 ml of cultures were spun down (14,000 rpm for 2 min), supernatants were removed, and the pellets were immediately frozen using liquid nitrogen and stored at −80°C. At this same time, PYO concentration produced by the co-culture containing the WT P. aeruginosa was measured from the supernatant using absorbance at OD691 (Reszka et al., 2004), and ~ 60 μM was detected at the time of sampling.
Next, RNA extraction was performed using the RNeasy kit (Qiagen) following the manufacturer’s instructions. Samples were thawed on ice and resuspended in TE buffer (30 mM Tris.Cl, 1 mM EDTA, pH 8.0) containing 15 mg/ml of lysozyme and proteinase K solution (20 mg/ml, Qiagen), followed by an incubation with vortex and lysis steps described in the kit. Purified RNA was then sent for sequencing at the MiGS Center as done for whole-genome sequencing. The facility pipeline included: (i) DNase treatment (RNAse free) (Invitrogen); (ii) library preparation using Qiagen FastSelect and Library Prep kits (Qiagen) and Ribo-Zero Plus kit (Illumina); (iii) sequencing using a NextSeq500, with 1 × 75 bp reads for the four conditions, with a minimum of 16 M reads for conditions 1 and 2 (single Burkholderia cultures), or 24 M reads for conditions 3 and 4 (co-cultures); (iv) demultiplexing and adaptors trimming using bcl2fastq (version 2.20.0.422). Next, low-quality bases were removed using Trimmomatic (version 0.39) with the following settings: LEADING: 27 TRAILING: 27 SLIDINGWINDOW: 4:20 MINLEN: 35 (Bolger et al., 2014). Genome mapping and calculation of number of reads per gene was performed through Rockhopper (version 2.03) (Tjaden, 2015) using the Burkholderia multivorans ATCC 17616 genome available in the software as reference. Mapping was done against all three chromosomes (NC_010084; NC_010086; NC_010087), as well as against the pBMUL01 and pTGL1 plasmids (NC_010070 and NC_010802, respectively). Settings within the software were: 0.15 for allowed mismatches; 0.33 for minimum seed length; 500 for max bases between paired reads; 0.5 for minimum expression of UTRs and ncRNAs; and the “strand specific” option was unmarked. Even though the library preparation pipeline included an rRNA depletion step, we retrieved a large amount of rRNA sequences, which were manually deleted from the read-count table exported by Rockhopper. Data exploration and analysis were performed using the online Degust tool (Powell, 2015). With Degust, counts per million (CPM) for each gene were used for sequencing depth normalization and log2 fold changes are presented for two different comparison. In comparison 1, condition 1 (B. multivorans—no PYO added) was used as a baseline control during fold-changes calculation detected for condition 2 (B. multivorans + 100 μM PYO). In comparison 2, condition 4 (B. multivorans + P. aeruginosa Δphz) was used as baseline control during fold-changes calculation detected for condition 3 (B. multivorans + P. aeruginosa WT). See Table S2 for the full results. Because our samples consisted of one replicate, no statistical tests were performed. Instead, we used this RNA-seq data as a screen where specific genes of interest (particularly related to efflux) were next explored by qRT-PCR experiments. RNA-seq sequence data were submitted to the NCBI Sequence Read Archive under the accession number PRJNA787476.
4.7 |. Quantitative reverse transcription PCR (qRT-PCR)
4.7.1 |. Experiment 1: Validation of RNA-seq
The set up for this experiment was exactly the same as the RNA-seq described above, with the exception that three independent replicates were prepared. For each strain used in this experiment (B. multivorans 1, P. aeruginosa WT, P. aeruginosa Δphz), three independent overnight cultures were grown in GMM (20 mM glucose) + AA. Each of overnights were inoculated from three different spots from the freshly streaked LB plates of each strain. Next, each one of these overnight cultures was used in the inoculum preparation of the four different conditions, as described in the RNA-seq section. Again, the conditions were: (i) B. multivorans (no PYO); (ii) B. multivorans + 100 μM PYO; (iii) B. multivorans + P. aeruginosa WT (co-culture, PYO and other phenazines produced); and (iv) B. multivorans + P. aeruginosa Δphz (co-culture, no PYO or phenazines produced). A total of 12 tubes were prepared (three for each conditions). These were grown for ~9 h, after which the pellets were collected (from 0.7 ml of culture), immediately frozen using liquid nitrogen, and stored at −80°C for later RNA extraction.
4.7.2 |. Experiment 2: Measuring the SoxR effect on PYO-mediated induction of RND-9
Four different conditions were prepared for this qRT-PCR experiment: (i) B. multivorans ΔsoxR; (ii) B. multivorans ΔsoxR + 100 μM PYO; (iii) B. multivorans ΔsoxR comp; (iii) B. multivorans ΔsoxR comp +100 μM PYO. Three independent overnight cultures of each strain were grown in GMM (20 mM glucose) + AA, cells were then washed and resuspended at an OD500 of 0.04 in fresh GMM (20 mM glucose, 7 ml culture). Three replicates were prepared for each condition, with a total of 12 samples. Depending on the condition, either 100 μM PYO or the respective amount of HCl was added to the cultures. Cultures were then incubated for ~10 h, after which the pellets were collected (from 0.7–1 ml of culture), immediately frozen using liquid nitrogen, and stored at −80°C for later RNA extraction.
4.7.3 |. Experiment 3: Measuring the SoxR effect on TOX-mediated induction of RND-9
Six different conditions were prepared for this qRT-P CR experiment. Each of the three tested strains (B. multivorans WT, ΔsoxR and ΔsoxR comp) were grown with and without 50 μM TOX. Three independent overnight cultures of each strain were grown in GMM (20 mM glucose) + AA, cells were then washed and resuspended at an OD500 of 0.05 in fresh GMM + AA (20 mM glucose, 5 ml culture). Again, three replicates were prepared for each condition, with a total of 18 samples. Either 50 μM TOX or the respective amount of DMSO was added to the cultures. These cultures were incubated for ~20 h, cells were pelleted (from 0.7 ml of culture), immediately frozen using liquid nitrogen, and stored at −80°C for later RNA extraction.
For the next steps, we followed previously published protocols (Babin et al., 2016; Meirelles et al., 2021; Meirelles & Newman, 2018). RNA extraction was performed as described in these studies and in the RNA-seq experiment using the RNeasy kit (Qiagen). Contaminant gDNA was removed using TURBO DNA-free kit (Invitrogen, Waltham, Massachusetts, USA), and cDNA was synthesized using the iScript cDNA Synthesis kit (Bio-Rad, Hercules, California, USA) (a total of 0.8 μg of total RNA was used), following the manufacturer’s instructions. Then, qRT-PCR reactions were performed using iTaq Universal SYBR Green Supermix (Bio-Rad) (total of 20 μl per reaction) using a 7500 Fast Real-Time PCR System machine (Applied Biosystems, Waltham, Massachusetts, USA). For additional details on the protocol, see Meirelles and Newman (2018). Finally, within each run, standard curves for each primer pair were prepared using known concentration of B. multivorans WT gDNA to calculate amounts of cDNA for each of the target genes. The gene Bmul_2161 (annotated as uvrC, coding for excinuclease ABC subunit C) was used as housekeeping gene during normalizations. As an additional control, we also ran reactions with the housekeeping gene Bmul_1456 (annotated as rumA gene, coding for the 23S rRNA 5-methyluridine methyltransferase) (Schnetterle et al., 2021). Primer pairs sequences are available in Table S1.
Data showing total uvrC-normalized cDNA levels and/or the log2 fold change in expression are shown in Figures 1c,d, 2b, 3f, S1, S2, and S9. Normalizations for cDNA measurement followed what we have previously described for oprI-normalized cDNA in P. aeruginosa (Meirelles et al., 2021) but using the uvrC gene instead. In brief, the cDNA estimated for a certain gene in a certain sample was divided by the respective cDNA estimated for uvrC in the same sample (Meirelles et al., 2021). In some occasions, we also present the data as fold changes. These were calculated for each replicate relative to the mean cDNA value of the replicates within the negative control. The “no treatment” samples (i.e., “no PYO” or “no TOX”) were used as negative controls for conditions where B. multivorans was in single-species cultures (in experiments 1, 2 and 3); meanwhile, the “B. multivorans + P. aeruginosa Δphz” samples was used as negative control for the “B. multivorans + P. aeruginosa WT” condition (in experiment 1). Importantly, we noticed that the one replicate for three following conditions had significantly lower amounts of cDNA by the end of our qRT-PCR protocol: “B. multivorans + P. aeruginosa WT” and “B. multivorans + P. aeruginosa Δphz” within experiment 1, and “ΔsoxR no TOX” in experiment 3. These were removed from the analysis as noted in the legends of figures where these data are displayed.
4.8 |. Antibiotics used for tolerance and resistance assays
The following antibiotics were used in this study: ciprofloxacin (Fluka), levofloxacin (Sigma-Aldrich), tetracycline (tetracycline hydrochloride, Sigma-Aldrich), doxycycline (doxycycline hyclate, Sigma-Aldrich), tobramycin (TCI), colistin (colistin sulfate salt, Sigma-Aldrich), ceftazidime (TCI, containing ca. 10% Na2CO3), chloramphenicol (Sigma-Aldrich), meropenem (meropenem trihydrate, Sigma-Aldrich), trimethoprim (Sigma-Aldrich), and sulfamethoxazole (Sigma-Aldrich). Stock solutions of the antibiotics used were prepared in different solvents. Ciprofloxacin was dissolved in 20 mM HCl; levofloxacin, tetracycline, doxycycline, tobramycin, colistin, and ceftazidime were dissolved in deionized water; chloramphenicol, meropenem, and trimethoprim-sulfamethoxazole (mixed at 1:1 ratio) were dissolved in DMSO. Stock concentrations used were of 1 mg/ml for ciprofloxacin and levofloxacin, and 10 mg/ml for all other antibiotics.
4.9 |. Antibiotic tolerance assays with single species
A growth curve assay was used to measure resistance levels to PYO, following a previously described protocol (Meirelles et al., 2021). Briefly, cells of the respective strain used were grown from a fresh plate into overnight cultures in GMM + AA with 20 mM glucose. Cells were then pelleted, washed, and resuspended in four independent cultures per treatment at an OD500 of 0.05 in GMM + AA, with 10 mM glucose. Treatments involved addition or not of the redox-active secondary metabolite (PYO, PCA, or TOX, at the respective concentration displayed in the figures). These four independent replicates were prepared in 7 ml (for PYO and PCA) or 5 ml cultures (for TOX), always using 18 × 150 mm glass tubes, and incubated for around 20 h. This was enough for cells to reach stationary phase. Next, each individual culture was then split into a “no antibiotic” negative control, or an “+ antibiotic” treatment as done before (Meirelles et al., 2021), using 2 ml of culture per treatment in plastic Falcon tubes (VWR, Cat. No. 352059). After antibiotic treatment was added, cultures were then incubated for four hours under shaking conditions. Finally, cultures were serially diluted using Minimum Phosphate Buffer (50 mM KH2PO4/K2HPO4, 42.8 mM NaCl, pH 7.2) and plated for CFUs using LB agar plates. These plates were then incubated at room temperature, with CFUs counted after 36–48 h. To check for slow-growing/late-arising colonies, plates were also checked after 5–7 days. This protocol was used for all the strains mentioned in the figures containing antibiotic assays with single species cultures. These included: B. multivorans 1 (WT, ΔsoxR or ΔsoxR comp strains), and B. glumae (WT and ΔtoxA strains). The antibiotic used was ciprofloxacin (at 10 μg/ml), as indicated in the figure legends.
4.10 |. Growth curves for measuring resistance to PYO and data analysis
A growth curves assay was used to measure resistance levels to PYO, following conditions previously described (Meirelles et al., 2021). Briefly, the several Burkholderia strains listed in Figures 2e and S4 (also see Table S1 for additional details) were used in this experiment. Each strain was grown overnight from a fresh plate in GMM with 20 mM glucose + AA. Cells were then pelleted, washed, and resuspended at an OD500 = 0.05 using the same medium (this was the initial OD used in the experiment). The experiment was set up in 96-well plates, with different concentration of PYO being used (0, 10, 100, or 200 μM). Three wells were prepared for each treatment, and each well was considered an independent replicate. The total volume within each well contained 150 μl of culture and 70 μl of mineral on top (used for evaporation prevention). Incubation proceeded for 24 h at 37°C under shaking conditions. A Spark 10 M plate reader was used (Tecan), with absorbance readings at an OD500 every 15 min.
Data analysis was performed using a custom Python software containing tools for data processing/analysis/visualization designed for the datasets exported from our plate readers (available at: https://github.com/jciemniecki/dknlab_tools , version 1.1.0). Growth curves for all the strains and treatments can be seen in Figure S5. Next, growth rates were determined using linear fits of the linear range of these growth curves (time range used: 3–8 h for all strains except the slow growers B. cenocepacia J2315 and B. gladioli 3; for which a range of 3–15 h was used). This was done using a linear regression function (also available at the same GitHub repository). Means of the growth rates were calculated for each strain/treatment based on the data from the three replicates. Finally, for each strain, these growth rate means obtained for treatments containing the different concentrations of PYO (10, 100, and 200 μM) were normalized (i.e., divided by) the growth rate means obtained for the “No PYO” control to calculate the “growth rate ratio” displayed in Figure 2e.
4.11 |. Antibiotic tolerance assays with co-cultures
Antibiotic tolerance assays using co-cultures were performed using a membrane-separated 12-well tissue plate cultures containing 0.1 μm pore PET membranes (VWR, Cat. No. 10769–226), following previously protocols (Meirelles et al., 2021). These were used (i) to study how P. aeruginosa can have affect antibiotic tolerance levels of different strains B. multivorans 1; (ii) to study how multiple TOX producers (B. glumae and B. gladioli strains) can affect antibiotic tolerance levels of several Bcc species. Overnight cultures were grown for each strain in GMM (20 mM glucose) + AA, cells were pelleted, washed, and resuspended in the same medium at different OD500: 0.05 for redox-active metabolites producers; and 0.025 for the Bcc species (in which antibiotic susceptibility was evaluated). Producers (P. aeruginosa, B. glumae, and B. gladioli strains) were cultured in the bottom part of the well using a volume of 600 μl, while Bcc were cultured in the upper part of the well using a volume of 100 μl. The experimental design can be seen in Figure 3g. When grown alone (i.e., no producer), the Bcc strain was added to both bottom and upper part of the well. Cells were grown under shaking conditions (175 rpm) for ~20 h at 37°C, with the plates placed within airtight plastic container. Several wet paper towels were used to maintain humidity. Next, ciprofloxacin was then added (10 μg/ml), and cultures were incubated for additional four hours. Within each plate, three replicates (wells) were used for “no antibiotic” (negative control) and three replicates for “+ ciprofloxacin” treatment. After incubation, the upper part of the co-cultures (containing the Bcc cells) was plated for CFUs on LB agar plates using serial dilution as described above.
In addition to the co-cultures grown on the membrane-separated 12-well tissue plate, we also performed a co-culture assay using 18 × 150 mm glass tubes (Figure 1b, right). Here, B. multivorans 1 WT was grown together with P. aeruginosa WT or Δphz in 7 ml cultures with a total starting OD500 of 0.05 (0.025 for each species). Four independent replicates were prepared, cultures were grown for 20 h, treated with ciprofloxacin (10 μg/ml), and plated for CFUs as described before. Differently from the membrane-separated plates used before, the two species were mixed in this assay. However, P. aeruginosa and B. multivorans colonies display different morphologies and growth rates, allowing for visual distinction in the “no antibiotic” control treatment. Moreover, P. aeruginosa is orders of magnitude more susceptible to the drug than B. multivorans under the concentration used (10 μg/ml), allowing for easy counting of the B. multivorans colonies in the “+ ciprofloxacin” treatments (Figure 1b).
4.12 |. Determination of minimum inhibitory concentrations (MICs)
4.12.1 |. MIC assays using pure redox-active molecules
MICs were determined following the EUCAST standard clinical methods suggested for broth microdilution assays (EUCAST, 2003), with modifications to account for the effects of redox-active molecules (e.g., PYO or TOX). Briefly, B. multivorans 1 cultures was grown from a fresh LB plate into MH broth, and were then diluted 1:100 into three independent replicates (5 ml) and grown for 14–1 8 h until stationary phase. These three independent replicates were individually resuspended and used as inocula in twofold dilution series assays using 96-well microtiter plates, the 10 antibiotics mentioned before, and the redox-active molecules. The final concentrations used for PYO and TOX were 100 μM and 50 μM, respectively; controls involved adding the equivalent amount of the solvents used to solubilize these molecules (20 mM HCl for PYO, or DMSO for TOX). One antibiotic was used per plate, and the plate design can be seen in Figure S10a, with each well having a final volume of 100 μl. Appropriate “no antibiotic” and “no cells” controls were always prepared (Figure S10a). After inoculation, the microtiter plates were sealed with a plastic film (to avoid evaporation), wrapped in aluminum foil, and incubated without shaking at 37°C for 18 h. Microtiter plates were always incubated in a single layer. After incubation, the wells were assessed for growth (turbidity) using a BioTek Synergy 4 plate reader (BioTek) or a Spark 10 M plate reader (Tecan). Growth assessment was also done by naked eye as suggested by the reading guide for broth microdilutions by EUCAST (version 3.0, January 1st, 2021), and used for the OD500 thresholding (see the data analysis section below).
4.12.2 |. MIC assays using spent media from pathogens’ cultures
These assays followed the same design described above, with the exception that instead of using pure redox-active molecules, a filtered-sterilized spent media from stationary cultures was mixed to fresh media and used during twofold dilution series assays. In summary, cultures of the strains in which spent media were analyzed (P. aeruginosa WT, P. aeruginosa Δphz, and B. multivorans WT) were grown in MH broth, diluted back 1:100 into 50 ml cultures (initial OD500 = ~0.05), and grown for around 12 h for metabolite production. Cells were then spun down (5000 rpm for 10 min, twice), and the supernatant containing the spent media was collected and filter-sterilized through a large bottle top filter (Millipore, Cat. No. SCGPS02RE). The filtered spent media samples were plated on LB agar for contamination verification. Then, spent media samples were mixed with fresh MH broth (1/4 of the final volume was spent media) during twofold dilution series assays using the antibiotics indicated in Figures 4e and S11d,e. Importantly, spent medium from P. aeruginosa WT contained ~80–100 μM PYO measured by absorbance at 691 nm, meaning that the final PYO concentration on microtiter plates were around 20–25 μM. Incubation and growth assessment protocol followed what was described above.
4.12.3 |. Data analysis
After turbidity measurements by OD500 absorbance and inspection by naked eye, we adopted OD500 = 0.11 as a threshold value for the indication of growth in the microtiter plates. This means that this was the lowest absorbance where growth could be detected by naked eye; any reading value lower than this was determined “no growth”. This threshold was applied to the raw absorbance values, and no normalizations by the “no cells” controls wells were done at this stage. MIC values for each treatment tested are available in Table S4. Next, calculation of the molecules’ “Effect on MIC (log2 fold change)” in Figure 4b,c were performed as follows: for each antibiotic, MIC values detected for treatments containing PYO or TOX were divided by the MIC detected in respective negative controls (where HCl or DMSO were added, respectively). The plate design was always the same, and individual ratios were calculated within each replicate (for each antibiotic, all replicates were prepared in parallel on the same day and plate—see Figure S10a for design). These ratios were then log2-transformed and are shown in Figure 4b,c. Finally, for MICs detected in experiments containing spent media (from the P. aeruginosa strains or from B. multivorans), the plates were prepared on different days from the original experiments with the pure molecules. However, because MIC values were consistent (i.e., HCl or DMSO did not seem to affect the MICs measured at the amounts used), the same negative controls used for ratios’ calculation in the experiments with pure molecules were also used for the ratios’ calculation in experiments with spent media. Specifically, the “No PYO” controls (i.e., where HCl was added) were used for this purpose. Again, these ratios were log2-transformed and are shown in Figures 4e and S11d,e.
Finally, we realized that sometimes, even though the MIC detected itself did not change, treatments could alter the growth detected at the pre-MIC concentrations. For this reason, we have also included plots with the OD500 absorbance measurements and the log2 fold change for OD500 absorbances between different treatment. For this, the absorbances measurements were normalized by their respective “no cells” control wells within the same plate. Then, as done for the MICs values, the fold changes were determined by calculating the ratio for each treatment (PYO or TOX) by their respective negative control within each replicate (e.g., absorbance for condition “+PYO replicate 1 in TET” divided by “+HCl replicate 1 in TET”), and these values were log2 transformed. This was only done for experiments using pure molecules, since these were performed in parallel (i.e., same day) with their respective solvent control. Both OD500 normalized absorbance measurements and their log2 fold change are shown (Figures S11a,b and 4d respectively).
4.13 |. Measurements of PYO and TOX concentrations
PYO concentrations in cultures containing P. aeruginosa WT were estimated by absorbance of the culture supernatant at 691 nm (Reszka et al., 2004) using standard curves with the pure molecule. A similar approach was used for estimation of TOX concentrations within cultures containing B. glumae and B. gladioli. UV–Vis spectra (200–800 nm) were recorded using a spectrophotometer (Beckman Coulter DU 800) for solutions containing pure TOX (25, 50, and 100 μM), and supernatants of overnight cultures (grown GMM, 20 mM glucose + AA) of the following strains: B. glumae WT, B. gladioli 1 and B. gladioli 2. Moreover, supernatants of B. glumae ΔtoxA and B. multivorans 1 strains grown in the same medium were used as negative controls since these strains cannot make TOX. Concentration estimations were done for each sample based on characteristic absorbance of the molecule at 393 nm (Chen et al., 2012; Kim et al., 2004), calibrated by the absorbance values obtained for the standards using the pure molecule. Because only oxidized TOX absorbs at 393 nm, after being taken out of the shaking incubator, cultures were immediately spun down for supernatant collection. Absorbance spectra within the 300–500 nm range for these samples are shown in Figure S8.
4.14 |. Data wrangling, analysis, and visualization
Data wrangling and analysis involved a combination of processing in (i) Python (version 3.8), using the Pandas (version 1.3.1) (McKinney, 2010; The Pandas Development Team, 2020) and NumPy (version 1.20.3) (Harris et al., 2020) libraries; or in (ii) Microsoft Excel (version 16.39), as described throughout the Materials and Methods section. All the visualization presented in the manuscript was performed using Matplotlib (version 3.4.2) (Hunter, 2007) and Seaborn (version 0.11.1) (Waskom, 2021). 95% confidence intervals presented in the figures were estimated with Seaborn while plotting the respective data using 10,000 bootstraps. Plots legends and their display organization within each figure were adjusted using Affinity Designer (Serif, version 1.10.4). The same software was used for drawing all the illustrations shown in the manuscript.
Supplementary Material
ACKNOWLEDGMENTS
We thank Newman lab members for feedback and advice throughout the development of this project. In particular, we thank Megan Bergkessel for assistance and discussions about the RNA-seq, John Ciemniecki for developing and sharing the “dknlab_tools” package used during plate reader data analysis, John LiPuma (University of Michigan, CFF Burkholderia cepacia Research Laboratory and Repository) for providing the clinical Burkholderia strains, and the Microbial Genome Sequencing Center (MiGS) at Pittsburgh for sequencing of the samples. Finally, we thank Jong Ham and Inderjit Barphagha (Louisiana State University) for providing the B. glumae 336gr-1 strains (WT and ΔtoxA), and Joanna Goldberg (Emory University) for providing the pEX18Tc plasmid. This work was supported by grants to D.K.N from the NIH (1R01AI127850-01A1, 1R01HL152190-01), and the Doren Family Foundation.
Funding information
Doren Family Foundation; National Heart, Lung, and Blood Institute, Grant/Award Number: 1R01HL152190-01; National Institute of Allergy and Infectious Diseases, Grant/Award Number: 1R01AI127850-01A1
Footnotes
ETHICS STATEMENT
Usage of Burkholderia gladioli and Burkholderia glumae was permitted by the USDA-APHIS.
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of the article at the publisher’s website.
CONFLICT OF INTEREST
The authors declare no competing interests.
DATA AVAILABILITY STATEMENT
Data that supports the findings of this study has been deposited at the NCBI Sequence Read Archive under the accession number PRJNA787476.
REFERENCES
- Agus A, Clément K. & Sokol H. (2021) Gut microbiota-derived metabolites as central regulators in metabolic disorders. Gut, 70, 1174–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babin BM, Bergkessel M, Sweredoski MJ, Moradian A, Hess S, Newman DK et al. (2016) SutA is a bacterial transcription factor expressed during slow growth in Pseudomonas aeruginosa. Proceedings of the National Academy of Sciences of the United States of America, 113, E597–E605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban NQ, Helaine S, Lewis K, Ackermann M, Aldridge B, Andersson DI et al. (2019) Definitions and guidelines for research on antibiotic persistence. Nature Reviews Microbiology, 17, 441–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett AR, Kang Y, Inamasu KS, Son MS, Vukovich JM & Hoang TT (2008) Genetic tools for allelic replacement in Burkholderia species. Applied and Environmental Microbiology, 74, 4498–4508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartell JA, Blazier AS, Yen P, Thøgersen JC, Jelsbak L, Goldberg JB et al. (2017) Reconstruction of the metabolic network of Pseudomonas aeruginosa to interrogate virulence factor synthesis. Nature Communications, 8, 14631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauermeister A, Mannochio-Russo H, Costa-Lotufo LV, Jarmusch AK & Dorrestein PC (2021) Mass spectrometry-based metabolomics in microbiome investigations. Nature Reviews Microbiology, 20, 143–160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair JMA, Webber MA, Baylay AJ, Ogbolu DO & Piddock LJV (2015) Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology, 13, 42–51. [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M. & Usadel B. (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A, Fridman O, Gefen O. & Balaban NQ (2016) Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature Reviews Microbiology, 14, 320–330. [DOI] [PubMed] [Google Scholar]
- Brochado AR, Telzerow A, Bobonis J, Banzhaf M, Mateus A, Selkrig J. et al. (2018) Species-specific activity of antibacterial drug combinations. Nature, 559, 259–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheluvappa R. (2014) Standardized chemical synthesis of Pseudomonas aeruginosa pyocyanin. MethodsX, 1, 67–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Barphagha IK, Karki HS & Ham JH (2012) Dissection of quorum-sensing genes in Burkholderia glumae reveals non-canonical regulation and the new regulatory gene tofM for toxoflavin production. PLoS One, 7, e52150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen IMA, Chu K, Palaniappan K, Ratner A, Huang J, Huntemann M. et al. (2021) The IMG/M data management and analysis system v.6.0: new tools and advanced capabilities. Nucleic Acids Research, 49, D751–D763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chmiel JF, Aksamit TR, Chotirmall SH, Dasenbrook EC, Elborn JS, LiPuma JJ et al. (2014) Antibiotic management of lung infections in cystic fibrosis. I. the microbiome, methicillin-resistant Staphylococcus aureus, gram-negative bacteria, and multiple infections. Annals of the American Thoracic Society, 11, 1120–1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi K-H & Schweizer HP (2006) Mini-Tn7 insertion in bacteria with single attTn7 sites: example Pseudomonas aeruginosa. Nature Protocols, 1, 153–161. [DOI] [PubMed] [Google Scholar]
- Costa KC, Glasser NR, Conway SJ & Newman DK (2017) Pyocyanin degradation by a tautomerizing demethylase inhibits Pseudomonas aeruginosa biofilms. Science, 355, 170–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coutinho CP, Dos Santos SC, Madeira A, Mira NP, Moreira AS & Sá-Correia I. (2011) Long-term colonization of the cystic fibrosis lung by Burkholderia cepacia complex bacteria: epidemiology, clonal variation, and genome-wide expression alterations. Frontiers in Cellular and Infection Microbiology, 1, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cruickshank CN & Lowbury EJ (1953) The effect of pyocyanin on human skin cells and leucocytes. British Journal of Experimental Pathology, 34, 583–587. [PMC free article] [PubMed] [Google Scholar]
- Davies J. (2013) Specialized microbial metabolites: functions and origins. The Journal of Antibiotics, 66, 361–364. [DOI] [PubMed] [Google Scholar]
- Davies J. & Davies D. (2010) Origins and evolution of antibiotic resistance. Microbiology and Molecular Biology Reviews, 74, 417–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deatherage DE & Barrick JE (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods in Molecular Biology, 1151, 165–188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePas WH, Starwalt-Lee R, Van Sambeek L, Ravindra Kumar S, Gradinaru V. & Newman DK (2016) Exposing the three-dimensional biogeography and metabolic states of pathogens in cystic fibrosis sputum via hydrogel embedding, clearing, and rRNA labeling. mBio, 7, e00796–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depoorter E, Bull MJ, Peeters C, Coenye T, Vandamme P. & Mahenthiralingam E. (2016) Burkholderia: an update on taxonomy and biotechnological potential as antibiotic producers. Applied Microbiology and Biotechnology, 100, 5215–5229. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Okegbe C, Price-Whelan A, Sakhtah H, Hunter RC & Newman DK (2013) Bacterial community morphogenesis is intimately linked to the intracellular redox state. Journal of Bacteriology, 195, 1371–1380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich LE, Price-Whelan A, Petersen A, Whiteley M. & Newman DK (2006) The phenazine pyocyanin is a terminal signalling factor in the quorum sensing network of Pseudomonas aeruginosa. Molecular Microbiology, 61, 1308–1321. [DOI] [PubMed] [Google Scholar]
- Dietrich LEP, Teal TK, Price-Whelan A. & Newman DK (2008) Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science, 321, 1203–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll JA, Brody SL & Kollef MH (2007) The epidemiology, pathogenesis and treatment of Pseudomonas aeruginosa infections. Drugs, 67, 351–368. [DOI] [PubMed] [Google Scholar]
- Du D, Wang-Kan X, Neuberger A, van Veen HW, Pos KM, Piddock LJV et al. (2018) Multidrug efflux pumps: structure, function and regulation. Nature Reviews. Microbiology, 16, 523–539. [DOI] [PubMed] [Google Scholar]
- Dubarry N, Du W, Lane D. & Pasta F. (2010) Improved electrotransformation and decreased antibiotic resistance of the cystic fibrosis pathogen Burkholderia cenocepacia strain J2315. Applied and Environmental Microbiology, 76, 1095–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earle KA, Billings G, Sigal M, Lichtman JS, Hansson GC, Elias JE et al. (2015) Quantitative imaging of gut microbiota spatial organization. Cell Host & Microbe, 18, 478–488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- European Committee for Antimicrobial Susceptibility Testing (EUCAST) of the European Society of Clinical Microbiology and Infectious Diseases (ESCMID). (2003) Determination of minimum inhibitory concentrations (MICs) of antibacterial agents by broth dilution. Clinical Microbiology and Infection, 9, ix–xv. [Google Scholar]
- Fair RJ & Tor Y. (2014) Antibiotics and bacterial resistance in the 21st century. Perspectives in Medicinal Chemistry, 6, 25–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folescu TW, da Costa CH, Cohen RWF, da Conceição Neto OC, Albano RM & Marques EA (2015) Burkholderia cepacia complex: clinical course in cystic fibrosis patients. BMC Pulmonary Medicine, 15, 158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg N, Wang M, Hyde E, da Silva RR, Melnik AV, Protsyuk I. et al. (2017) Three-dimensional microbiome and metabolome cartography of a diseased human lung. Cell Host & Microbe, 22, 705–716.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel A, Zamarreño Beas J, Duverger Y, Bouveret E, Barras F. & Py B. (2020) Oxidative stress antagonizes fluoroquinolone drug sensitivity via the SoxR-SUF Fe-S cluster homeostatic axis. PLoS Genetics, 16, e1009198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson DG, Young L, Chuang R-Y, Venter JC, Hutchison CA & Smith HO (2009) Enzymatic assembly of DNA molecules up to several hundred kilobases. Nature Methods, 6, 343–345. [DOI] [PubMed] [Google Scholar]
- Glasser NR, Kern SE & Newman DK (2014) Phenazine redox cycling enhances anaerobic survival in Pseudomonas aeruginosa by facilitating generation of ATP and a proton-motive force. Molecular Microbiology, 92, 399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glasser NR, Saunders SH & Newman DK (2017) The colorful world of extracellular electron shuttles. Annual Review of Microbiology, 71, 731–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves M, Robin T, Chipman AM, Wong J, Khashe S. & Janda JM (1997) Four additional cases of Burkholderia gladioli infection with microbiological correlates and review. Clinical Infectious Diseases, 25, 838–842. [DOI] [PubMed] [Google Scholar]
- Gu M. & Imlay JA (2011) The SoxRS response of Escherichia coli is directly activated by redox-cycling drugs rather than by superoxide. Molecular Microbiology, 79, 1136–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurevich A, Saveliev V, Vyahhi N. & Tesler G. (2013) QUAST: quality assessment tool for genome assemblies. Bioinformatics, 29, 1072–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ham JH, Melanson RA & Rush MC (2011) Burkholderia glumae: next major pathogen of rice? Molecular Plant Pathology, 12, 329–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris CR, Millman KJ, van der Walt SJ, Gommers R, Virtanen P, Cournapeau D. et al. (2020) Array programming with NumPy. Nature, 585, 357–362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoang TT, Karkhoff-Schweizer RR, Kutchma AJ & Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene, 212, 77–86. [DOI] [PubMed] [Google Scholar]
- Huang W. & Wilks A. (2017) A rapid seamless method for gene knock-out in Pseudomonas aeruginosa. BMC Microbiology, 17, 199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter JD (2007) Matplotlib: A 2D graphics environment. Computing in Science & Engineering, 9, 90–95. [Google Scholar]
- Hutchings MI, Truman AW & Wilkinson B. (2019) Antibiotics: past, present and future. Current Opinion in Microbiology, 51, 72–80. [DOI] [PubMed] [Google Scholar]
- Imlay JA (2013) The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature Reviews Microbiology, 11, 443–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Jamier V. & Ba LA (2011) Redox active secondary metabolites. Current Opinion in Chemical Biology, 15, 149–155. [DOI] [PubMed] [Google Scholar]
- Jeong Y, Kim J, Kim S, Kang Y, Nagamatsu T. & Hwang I. (2003) Toxoflavin produced by Burkholderia glumae causing rice grain rot is responsible for inducing bacterial wilt in many field crops. Plant Disease, 87, 890–895. [DOI] [PubMed] [Google Scholar]
- Jo J, Cortez KL, Cornell WC, Price-Whelan A. & Dietrich LE (2017) An orphan cbb3-type cytochrome oxidase subunit supports Pseudomonas aeruginosa biofilm growth and virulence. eLife, 6, e30205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones C, Webster G, Mullins AJ, Jenner M, Bull MJ, Dashti Y. et al. (2021) Kill and cure: genomic phylogeny and bioactivity of Burkholderia gladioli bacteria capable of pathogenic and beneficial lifestyles. Microbial Genomics, 7, 000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen JH & Ferraro MJ (2009) Antimicrobial susceptibility testing: a review of general principles and contemporary practices. Clinical Infectious Diseases, 49, 1749–1755. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K. & Miyata T. (2002) MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30, 3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K. & Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennedy MP, Coakley RD, Donaldson SH, Aris RM, Hohneker K, Wedd JP et al. (2007) Burkholderia gladioli: five year experience in a cystic fibrosis and lung transplantation center. Journal of Cystic Fibrosis, 6, 267–273. [DOI] [PubMed] [Google Scholar]
- Kester JC & Fortune SM (2014) Persisters and beyond: mechanisms of phenotypic drug resistance and drug tolerance in bacteria. Critical Reviews in Biochemistry and Molecular Biology, 49, 91–101. [DOI] [PubMed] [Google Scholar]
- Kim J, Kim J-G, Kang Y, Jang JY, Jog GJ, Lim JY et al. (2004) Quorum sensing and the LysR-type transcriptional activator ToxR regulate toxoflavin biosynthesis and transport in Burkholderia glumae. Molecular Microbiology, 54, 921–934. [DOI] [PubMed] [Google Scholar]
- Latuasan HE & Berends W. (1961) On the origin of the toxicity of toxoflavin. Biochimica et Biophysica Acta, 52, 502–508. [DOI] [PubMed] [Google Scholar]
- Lau GW, Hassett DJ, Ran H. & Kong F. (2004) The role of pyocyanin in Pseudomonas aeruginosa infection. Trends in Molecular Medicine, 10, 599–606. [DOI] [PubMed] [Google Scholar]
- Laursen JB & Nielsen J. (2004) Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chemical Reviews, 104, 1663–1686. [DOI] [PubMed] [Google Scholar]
- Lee J, Park J, Kim S, Park I. & Seo YS (2016) Differential regulation of toxoflavin production and its role in the enhanced virulence of Burkholderia gladioli. Molecular Plant Pathology, 17, 65–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K-L, Singh AK, Heo L, Seok C. & Roe J-H (2015) Factors affecting redox potential and differential sensitivity of SoxR to redox-active compounds. Molecular Microbiology, 97, 808–821. [DOI] [PubMed] [Google Scholar]
- Lelis T, Peng J, Barphagha I, Chen R. & Ham JH (2019) The virulence function and regulation of the metalloprotease gene prtA in the plant-pathogenic bacterium Burkholderia glumae. Molecular Plant-Microbe Interactions, 32, 841–852. [DOI] [PubMed] [Google Scholar]
- Levin-Reisman I, Ronin I, Gefen O, Braniss I, Shoresh N. & Balaban NQ (2017) Antibiotic tolerance facilitates the evolution of resistance. Science, 355, 826–830. [DOI] [PubMed] [Google Scholar]
- Li XZ, Plésiat P. & Nikaido H. (2015) The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clinical Microbiology Reviews, 28, 337–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipuma JJ (2010) The changing microbial epidemiology in cystic fibrosis. Clinical Microbiology Reviews, 23, 299–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GY & Nizet V. (2009) Color me bad: microbial pigments as virulence factors. Trends in Microbiology, 17, 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacLean RC & San Millan A. (2019) The evolution of antibiotic resistance. Science, 365, 1082–1083. [DOI] [PubMed] [Google Scholar]
- Maier L, Goemans CV, Wirbel J, Kuhn M, Eberl C, Pruteanu M. et al. (2021) Unravelling the collateral damage of antibiotics on gut bacteria. Nature, 599, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier L, Pruteanu M, Kuhn M, Zeller G, Telzerow A, Anderson EE et al. (2018) Extensive impact of non-antibiotic drugs on human gut bacteria. Nature, 555, 623–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL (2009) The role of natural environments in the evolution of resistance traits in pathogenic bacteria. Proceedings of the Biological Sciences, 276, 2521–2530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney W. (2010) Data structures for statistical computing in python. In: Proceedings of the 9th Python in Science Conference. SciPy, pp. 56–61. Available from: https://conference.scipy.org/proceedings/scipy2010/mckinney.html [Accessed 14th May 2022]. [Google Scholar]
- Meirelles LA & Newman DK (2018) Both toxic and beneficial effects of pyocyanin contribute to the lifecycle of Pseudomonas aeruginosa. Molecular Microbiology, 110, 995–1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meirelles LA, Perry EK, Bergkessel M. & Newman DK (2021) Bacterial defenses against a natural antibiotic promote collateral resilience to clinical antibiotics. PLoS Biology, 19, e3001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Stamatis D, Bertsch J, Ovchinnikova G, Sundaramurthi JC, Lee J. et al. (2021) Genomes OnLine database (GOLD) v.8: overview and updates. Nucleic Acids Research, 49, D723–D733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill J. (2016) Tackling drug-resistant infections globally: final report and recommendations. Available from: https://apo.org.au/node/63983 [Accessed 14th May 2022].
- Orazi G. & O’Toole GA (2017) Pseudomonas aeruginosa alters Staphylococcus aureus sensitivity to vancomycin in a biofilm model of cystic fibrosis infection. mBio, 8, e00873–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orazi G, Ruoff KL & O’Toole GA (2019) Pseudomonas aeruginosa increases the sensitivity of biofilm-grown Staphylococcus aureus to membrane-targeting antiseptics and antibiotics. mBio, 10, e01501–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry EK, Meirelles LA & Newman DK (2021) From the soil to the clinic: the impact of microbial secondary metabolites on antibiotic tolerance and resistance. Nature Reviews Microbiology, 20, 129–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podnecky NL, Rhodes KA & Schweizer HP (2015) Efflux pump-mediated drug resistance in Burkholderia. Frontiers in Microbiology, 6, 305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell DR (2015) Degust: interactive RNA-seq analysis. Zenodo. Available from: https://zenodo.org/record/3501067#.Yn2T0y8RqqA [Accessed 14th May 2022]. [Google Scholar]
- Price-Whelan A, Dietrich LEP & Newman DK (2006) Rethinking “secondary” metabolism: physiological roles for phenazine antibiotics. Nature Chemical Biology, 2, 71–78. [DOI] [PubMed] [Google Scholar]
- Radlinski L, Rowe SE, Kartchner LB, Maile R, Cairns BA, Vitko NP et al. (2017) Pseudomonas aeruginosa exoproducts determine antibiotic efficacy against Staphylococcus aureus. PLoS Biology, 15, e2003981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos I, Dietrich LE, Price-Whelan A. & Newman DK (2010) Phenazines affect biofilm formation by Pseudomonas aeruginosa in similar ways at various scales. Research in Microbiology, 161, 187–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reszka KJ, O’Malley Y, McCormick ML, Denning GM & Britigan BE (2004) Oxidation of pyocyanin, a cytotoxic product from Pseudomonas aeruginosa, by microperoxidase 11 and hydrogen peroxide. Free Radical Biology & Medicine, 36, 1448–1459. [DOI] [PubMed] [Google Scholar]
- Rhodes KA & Schweizer HP (2016) Antibiotic resistance in Burkholderia species. Drug Resistance Updates, 28, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S. et al. (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders SH, Tse ECM, Yates MD, Otero FJ, Trammell SA, Stemp EDA et al. (2020) Extracellular DNA promotes efficient extracellular electron transfer by pyocyanin in Pseudomonas aeruginosa biofilms. Cell, 182, 919–932.e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiessl KT, Hu F, Jo J, Nazia SZ, Wang B, Price-Whelan A. et al. (2019) Phenazine production promotes antibiotic tolerance and metabolic heterogeneity in Pseudomonas aeruginosa biofilms. Nature Communications, 10, 762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnetterle M, Gorgé O, Nolent F, Boughammoura A, Sarilar V, Vigier C. et al. (2021) Genomic and RT-qPCR analysis of trimethoprim-sulfamethoxazole and meropenem resistance in Burkholderia pseudomallei clinical isolates. PLoS Neglected Tropical Diseases, 15, e0008913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab U, Abdullah LH, Perlmutt OS, Albert D, Davis CW, Arnold RR et al. (2014) Localization of Burkholderia cepacia complex bacteria in cystic fibrosis lungs and interactions with Pseudomonas aeruginosa in hypoxic mucus. Infection and Immunity, 82, 4729–4745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. (2014) Prokka: rapid prokaryotic genome annotation. Bioinformatics, 30, 2068–2069. [DOI] [PubMed] [Google Scholar]
- Sheplock R, Recinos DA, Mackow N, Dietrich LEP & Chander M. (2013) Species-specific residues calibrate SoxR sensitivity to redox-active molecules. Molecular Microbiology, 87, 368–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H, Shi Q, Grodner B, Lenz JS, Zipfel WR, Brito IL et al. (2020) Highly multiplexed spatial mapping of microbial communities. Nature, 588, 676–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon R, Priefer U. & Pühler A. (1983) A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nature Biotechnology, 1, 784–791. [Google Scholar]
- Singh AK, Shin J-H, Lee K-L, Imlay JA & Roe J-H (2013) Comparative study of SoxR activation by redox-active compounds. Molecular Microbiology, 90, 983–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider MA, Johnston CW, Gunabalasingam M, Merwin NJ, Kieliszek AM, MacLellan RJ et al. (2020) Comprehensive prediction of secondary metabolite structure and biological activity from microbial genome sequences. Nature Communications, 11, 6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern KG (1935) Oxidation-reduction potentials of toxoflavin. The Biochemical Journal, 29, 500–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Pandas Development Team. (2020) pandas-dev/pandas: Pandas 1.0.3. Zenodo. [Google Scholar]
- Tjaden B. (2015) De novo assembly of bacterial transcriptomes from RNA-seq data. Genome Biology, 16, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc O, Song C, Dickschat JS, Vos M. & Garbeva P. (2017) The ecological role of volatile and soluble secondary metabolites produced by soil bacteria. Trends in Microbiology, 25, 280–292. [DOI] [PubMed] [Google Scholar]
- Vernocchi P, Del Chierico F. & Putignani L. (2016) Gut microbiota profiling: metabolomics based approach to unravel compounds affecting human health. Frontiers in Microbiology, 7, 1144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waglechner N, McArthur AG & Wright GD (2019) Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nature Microbiology, 4, 1862–1871. [DOI] [PubMed] [Google Scholar]
- Waskom M. (2021) Seaborn: statistical data visualization. Journal of Open Source Software, 6, 3021. [Google Scholar]
- Wick RR, Judd LM, Gorrie CL & Holt KE (2017) Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Computational Biology, 13, e1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widdel F. & Pfennig N. (1981) Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. Archives of Microbiology, 129, 395–400. [DOI] [PubMed] [Google Scholar]
- Wilbert SA, Mark Welch JL & Borisy GG (2020) Spatial ecology of the human tongue dorsum microbiome. Cell Reports, 30, 4003–4015.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson R, Sykes DA, Watson D, Rutman A, Taylor GW & Cole PJ (1988) Measurement of Pseudomonas aeruginosa phenazine pigments in sputum and assessment of their contribution to sputum sol toxicity for respiratory epithelium. Infection and Immunity, 56, 2515–2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, Michiels JE, Fauvart M, Wenseleers T, Van den Bergh B. & Michiels J. (2019) Bacterial persistence promotes the evolution of antibiotic resistance by increasing survival and mutation rates. The ISME Journal, 13, 1239–1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Windels EM, Michiels JE, Van den Bergh B, Fauvart M. & Michiels J. (2019) Antibiotics: combatting tolerance to stop resistance. mBio, 10, e02095–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Griffiths EJ, Lo R, Dhillon BK, Shay JA & Brinkman FSL (2016) Enhanced annotations and features for comparing thousands of pseudomonas genomes in the pseudomonas genome database. Nucleic Acids Research, 44, D646–D653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winsor GL, Khaira B, Van Rossum T, Lo R, Whiteside MD & Brinkman FSL (2008) The Burkholderia genome database: facilitating flexible queries and comparative analyses. Bioinformatics, 24, 2803–2804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu K, Chen S, Sysoeva TA & You L. (2019) Universal antibiotic tolerance arising from antibiotic-triggered accumulation of pyocyanin in Pseudomonas aeruginosa. PLoS Biology, 17, e3000573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann M, Patil KR, Typas A. & Maier L. (2021) Towards a mechanistic understanding of reciprocal drug-microbiome interactions. Molecular Systems Biology, 17, e10116. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data that supports the findings of this study has been deposited at the NCBI Sequence Read Archive under the accession number PRJNA787476.