Abstract
This study proposes a new deep learning-based method that demonstrates high performance in detecting Covid-19 disease from cough, breath, and voice signals. This impressive method, named CovidCoughNet, consists of a deep feature extraction network (InceptionFireNet) and a prediction network (DeepConvNet). The InceptionFireNet architecture, based on Inception and Fire modules, was designed to extract important feature maps. The DeepConvNet architecture, which is made up of convolutional neural network blocks, was developed to predict the feature vectors obtained from the InceptionFireNet architecture. The COUGHVID dataset containing cough data and the Coswara dataset containing cough, breath, and voice signals were used as the data sets. The pitch-shifting technique was used to data augmentation the signal data, which significantly contributed to improving performance. Additionally, Chroma features (CF), Root mean square energy (RMSE), Spectral centroid (SC), Spectral bandwidth (SB), Spectral rolloff (SR), Zero crossing rate (ZCR), and Mel frequency cepstral coefficients (MFCC) feature extraction techniques were used to extract important features from voice signals. Experimental studies have shown that using the pitch-shifting technique improved performance by around 3% compared to raw signals. When the proposed model was used with the COUGHVID dataset (Healthy, Covid-19, and Symptomatic), a high performance of 99.19% accuracy, 0.99 precision, 0.98 recall, 0.98 F1-Score, 97.77% specificity, and 98.44% AUC was achieved. Similarly, when the voice data in the Coswara dataset was used, higher performance was achieved compared to the cough and breath studies, with 99.63% accuracy, 100% precision, 0.99 recall, 0.99 F1-Score, 99.24% specificity, and 99.24% AUC. Moreover, when compared with current studies in the literature, the proposed model was observed to exhibit highly successful performance. The codes and details of the experimental studies can be accessed from the relevant Github page: (https://github.com/GaffariCelik/CovidCoughNet).
Keywords: Deep learning, Covid-19, Pitch shifting, Convolutional neural network, Mel frequency cepstral coefficients, Cough sound, Voice signals, Sound analysis
1. Introduction
Covid-19, caused by Severe acute respiratory syndrome coronavirus 2 (SARS-CoV2) in 2019, was declared a global pandemic by the World Health Organization (WHO) on February 11, 2020. A new type of coronavirus, Covid-19, is similar to SARS-CoV, which caused epidemics and was seen in 2002, and MERS-CoV (Middle East respiratory syndrome coronavirus), which appeared in 2012 [1,2]. WHO has declared that the coronavirus disease (Covid-19) has been seen in more than 661 million confirmed cases worldwide as of 5 January 2023 and has caused the death of more than 6.7 million people [1].
Covid-19 continues to spread rapidly globally in different mutations [3,4]. Regional, regular testing and contact tracing played an important role in reducing the spread of the virus. Especially in the early stages of the pandemic, the Covid-19 outbreak gradually decreased in countries such as China, Singapore, and South Korea with the “watch, follow and treat” method [5,6]. However, the mutation of the Covid-19 virus and the relaxation of the measures taken, caused the epidemic to increase again. With the outbreak of the second wave, the health system in many countries came to a standstill. For this reason, many countries have introduced many strict measures to prevent the spread of the virus, such as promoting social distancing, wearing masks, personal hygiene, and mass vaccination [7].
Symptoms such as fever, dry cough, shortness of breath, muscle pain, fatigue, joint pain, and loss of smell or taste are common in patients with Covid-19 [8,9]. Reverse transcription-polymerase chain reaction (RT-PCR) kits are generally used to detect this disease. However, these test kits cause many problems such as being costly, difficult to obtain, and time-consuming test results. Similarly, it has greatly affected health services in countries where there are not enough test kits due to economic reasons. Delayed test results accelerate the spread of the virus. The development and implementation of rapid, scalable, reliable, economical, and reproducible tools for diagnosing and detecting Covid-19 are extremely important [10,11].
Many Artificial Intelligence (AI) models have been developed for the detection of Covid-19 using RT-PCR test kits, as well as X-rays [[12], [13], [14], [15]] and CT [12,[16], [17], [18]] scans. However, a well-equipped clinic or diagnostic center is required to use RT-PCR, X-ray, and CT-based methods. During this process, the risk of spreading Covid-19, which is a contagious disease transmitted by contact with many people, is accelerated due to the time it takes to obtain results. Techniques that reduce the spread rate of respiratory diseases such as Covid-19, minimize human contact, and provide rapid results need to be designed. AI-based methods have been developed to detect the virus from sound, breath, or cough, minimizing people's exposure to the virus, reducing the spread rate, and obtaining faster results [7,19,20].
Respiratory data such as coughing, sneezing, breathing, and speaking can be processed by AI and machine learning (ML) algorithms to diagnose certain respiratory diseases. Different respiratory diseases such as pertussis, asthma, pulmonary edema, tuberculosis, Parkinson's, and pneumonia have been detected using AI methods based on cough or sound signals [[21], [22], [23], [24], [25], [26]]. In addition, AI techniques have been developed to detect Covid-19 by analyzing cough, breath, and sound data [4,6,7,10,11,19,[27], [28], [29], [30], [31], [32], [33], [34]].
A Deep Neural Network (DNN) was used in the study by Pal and Sankarasubbu [20] to distinguish between Covid-19, asthma, bronchitis, and healthy individuals. Using 328 cough data from 150 patients, the authors achieved an accuracy of 96.83%. In the study carried out by Brown et al. [30], Covid-19 detection was carried out through ML techniques using sample data obtained from cough and breath. The study obtained over 80% AUC (area under the ROC curve). In the study conducted by Bagad et al. [35], they detected Covid-19 using cough samples from 3621 people over the phone. The authors achieved an AUC of 0.72 using the trained Resnet18 architecture. Laguarta et al. [36] proposed a Resnet50-based convolutional neural network (CNN) architecture. Using cough data from 5320 individuals in total, they achieved 0.97, 98.5%, and 94.2% success according to AUC, sensitivity, and specificity metrics, respectively. In another study by Andreu-Perez et al. [37], they proposed two deep learning-based architectures called DeepCough. In their study, they used a total of 8380 cough data, 2339 Covid-19 positive and 6041 Covid-19 negative, and provided a sensitivity of around 96.43% and specificity at the level of 96.20%.
In another study for the diagnosis of Covid-19 from cough and breath, they proposed a Deep Neural Network based on the ResNet architecture. Using a dataset of 517 sample cough and respiratory sound recordings from 355 individuals, they achieved AUC of 0.846 [38]. Pahar et al. [19] used the Coswara and Sarcos datasets for the diagnosis of Covid-19 from cough. The authors solved the imbalance problem between classes in the datasets by using the SMOTE (synthetic minority oversampling technique) technique. They used seven different ML-based techniques for disease detection. They achieved the highest AUC score with long short-term memory (LSTM) and Resnet50 techniques. In another study by Mohammed et al. [39], many applications were performed using CNN, pre-trained CNN, and shallow ML techniques. They scored 0.77, 0.80, 0.71, 0.75, and 0.53 according to the AUC, precision, recall, F1-measure, and kappa metrics, respectively.
Sharma et al. created a dataset called Coswara, which includes breathing sounds such as voice, cough, and breath for the detection of Covid-19. They collected the voice data through a website app that is accessible to everyone around the world. They demonstrated Covid-19 detection with 66.74% accuracy using the random forest algorithm, one of the ML learning methods [32]. In another study by Mouawad et al., they used four different machine learning techniques: K-nearest-neighbor, Random forest (RF), Support vector machines (SVM), and XGBoost. They worked with a dataset containing a total of 1927 sample voice data, 1895 of which were healthy and 32 patients. They proposed a method to eliminate the imbalance problem due to the number of healthy samples in the dataset being higher than the number of patient samples. Among the spectral approaches for feature extraction in the dataset, they achieved the highest success with the XGBoost algorithm (97.0% vs. 99.0% accuracy) using Mel frequency cepstral coefficients (MFCCs) [40]. In another study by Imran et al., they proposed an AI-based model called AI4COVID-19. They obtained mel-spectrogram images from the cough signals. For feature selection, SVM algorithm disease detection was performed using MFCC and principal component analysis (PCA) techniques. In the study, they obtained 95.6% accuracy and 95.6% F1 values [6].
Lee et al. [41] used deep learning-based CNN models, including VGGNet, GoogLeNet, and ResNet architectures, for Covid-19 detection from coughing. They used time derivatives such as speed (V) and acceleration (A) of sound from coughs, spectrograms, mel-scaled spectrograms, and MFCC spectral approaches to extract important information from cough sounds and achieved a 97.2% test accuracy with the GoogLeNet architecture [41]. Bansal et al. [42] used a deep learning-based VGG16 model in their study. They trained the model with MFCC and spectrogram images, and by using the MFCC feature extraction technique, they obtained higher results with a 70.58% accuracy and 81.00% sensitivity rates [42]. Similarly, another study by Dunne et al. [43] used Logistic Regression (LR), SVM, and CNN methods. They converted cough data into image data using MFCC spectrograms and trained the architectures, achieving Covid-19 diagnosis with a 97.5% accuracy rate [43]. Another study by Dentamaro et al. [10] proposed an end-to-end neural network called AUCO ResNet for Covid-19 detection from coughing and breathing data. This neural network has three different attention mechanisms: novel sinusoidal learnable attention, convolutional block attention module, and squeeze and excitation mechanism. The authors achieved AUC scores of 0.8308, 0.9257, and 0.8972 in different tasks using their proposed model.
In another study conducted for detecting Covid-19 from cough sounds, a dataset containing a total of 1457 sound samples, including 755 Covid-19 and 702 healthy ones, was used. First, sound-to-image conversion was performed, which was optimized with the scalogram technique. Then, performance analysis was made using six different deep learning models, namely ResNet18, ResNet50, ResNet101, NasNetmobil, GoogleNet, and MobileNetv2. The ResNet18 model achieved the highest performance with 94.44% sensitivity and 95.37% specificity [4]. In another study by Chowdhury et al., they used four different datasets: NoCoCoDa, Cambridge, Virufy, and Coswara. The authors obtained the highest performance with rates of 0.95, 1.00, and 0.97 for AUC, precision, and recall metrics, respectively, using the Extra-Trees algorithm [7]. Hamdi et al. [11] used the COUGHVID dataset. They proposed an augmentation pipeline consisting of a pitch-shifting technique to enhance the raw signal of cough sounds and spectral data augmentation technique to increase mel-spectrograms. They achieved 91.13% accuracy, 90.93% sensitivity, and 91.13% AUC values using an attention-based hybrid CNN-LSTM method [11].
Lella and Pja [27] claimed that they obtained better results compared to previous studies by performing the Covid-19 prediction process with the deep convolutional neural network (DCNN) method they proposed using a dataset including voice, dry cough, and breath data. The authors extracted the features of the voice data using De-noising Auto Encoder (DAE), Gamma-tone Frequency Cepstral Coefficients (GFCC), and Improved Multi-frequency cepstral coefficients (IMFCC) techniques for training their proposed method. They achieved a success rate of approximately 95.45% with the DCNN method [27]. Sait et al. [28] proposed a new technique called Ai-CovScan for the diagnosis of Covid-19 from chest X-ray images and breath sounds. This method consists of a combination of Inception-v3-based CNN and Multi-layered perceptron (MLP) techniques. The authors showed an accuracy score of 99.6% from chest X-ray images and 80% from breath sounds with the method they suggested. In the study conducted by Erdoğan and Narin, they utilized empirical mode decomposition (EMD) and discrete wavelet transform (DWT) techniques for feature extraction from the sound data. They also provided feature extraction from CNN-based ResNet50 and MobileNet techniques. The features obtained by different methods have been classified using the ReliefF algorithm. The authors showed higher success with the features obtained from the EMD and DWT techniques with 98.4% accuracy and 98.6% F1-score [29].
2. Materials and methods
The general methodology of the system proposed in this study for the detection of Covid-19 from cough, sound, and breath signals is presented in Fig. 1 . This system consists of five stages, namely data collection, preprocessing, feature extraction, deep feature extraction, and estimation. In the data collection phase, sample voice data from Coswara [32] and COUGHVID [34] datasets were used. In the preprocessing stage, firstly, the missing data was cleaned and then the pitch shifting method was used to obtain better-quality sound data. In the feature extraction phase, seven different feature extraction techniques were used from the voice signals. Detailed feature extraction is provided with a proposed model (InceptionFireNet) in the deep feature extraction stage. Features are trained with the proposed CNN network (InceptionFireNet) to obtain deep features and deep features are extracted from the third Fully Connected (FC3) layer of the trained InceptionFireNet architecture. This layer (FC3) is preferred because the information obtained in the previous layers is summarized in the FC layer. Then, the obtained feature maps were estimated with the proposed DeepConvNet architecture. Each stage of the proposed system is given in detail in the next sections.
Fig. 1.
Systematic diagram of the proposed architecture.
2.1. Materials
In this study, two different datasets, Coswara [32] and COUGHVID [34] were used for Covid-19 detection in cough, sound, and breath signals. A summary of the datasets is given in Table 1 .
Table 1.
Number of records in Coswara and COUGHVID data sets.
| Dataset | Healthy | Covid-19 | Symptomatic | No Status | Total |
|---|---|---|---|---|---|
| Coswara | 2065 | 681 | - | - | 2746 |
| COUGHVID | 12479 | 1155 | 2590 | 11326 | 27550 |
The Coswara [32] dataset is a dataset created to contribute to the diagnosis of Covid-19. This dataset consists of cough, breath, and voice signals from Covid-19 and healthy individuals. There are a total of 2746 sample records in the dataset, of which 2065 are healthy and 681 Covid-19. Sample data includes phonation of vowels (/a/ …./i/,/o/), counting numbers fast and slow, breathing sounds fast and slow, and coughing sounds deep and shallow. In addition, participants' meta information (age/gender/country/state or province), health status (healthy/infected/exposed/healed), and Covid-19 test status information are obtained. Data were collected through a publicly accessible web-based data collection platform (https://coswara.iisc.ac.in). The data collected to realize the researchers' projects are made available to everyone on the GitHub platform (https://github.com/iiscleap/Coswara-Data) [32].
The COUGHVID [34] dataset was collected between 1 April 2020 and 1 December 2020 via a web-based application installed on a server at the École Polytechnique Fédérale de Lausanne (EPFL) facilities in Switzerland. In the application, the sound is recorded from the microphone for up to 10 s via the record button. In the application, which is based on the principle of “one record” and “one-click”, the user can optionally send only the cough record. After the sound recording, participants are asked for their metadata (age, gender, current status), geographic location information, and finally, data on Covid-19 status. A summary of the records in the COUGHVID dataset is presented in Table 1. The dataset contains voice data obtained from a total of 27550 coughs, including 12479 healthy, 1155 Covid-19, 2590 symptomatic, and 11326 no status [34].
2.1.1. Preprocessing
2.1.1.1. Data cleaning and preparation
In the preprocessing stage, firstly, data cleaning (without status and missing/incorrect data) operation and rearrangement of datasets were performed. After this process, the distribution of the data in the data sets according to the classes is given in Table 2 . The Coswara dataset (CoswaraCough) produced from cough contains a total of 2702 sample data, including 2024 healthy and 678 Covid-19. In the Coswara dataset (CoswaraBreathing), which was created without deep breathing, there is a total of 2706 sample data, including 2029 healthy and 677 Covid-19. In the Coswara dataset (CoswaraVowel-a), which is created by saying the letter “a", there are a total of 2696 sample data, including 2023 healthy and 673 Covid-19. The COUGHVID2Cls dataset was created by collecting the Covid-19 and Symptomatic records in the COUGHVID dataset under the Covid-19 class. There are 12377 healthy and 3705 Covid-19 (1138 Covid19 and 2567 Symptomatic) records in the post-merger COUGHVID2Cls dataset. In the COUGHVID3Cls dataset, it contains 16082 voice recordings, 12377 healthy, 1138 Covid-19, and 2567 Symptomatic. In this study, the datasets in Table 2, which were obtained after the data cleaning and preparation phase, were used.
Table 2.
Class-based record numbers in datasets after data cleaning and editing.
| Datasets | Number of classes | Healthy | Covid-19 | Symptomatic | Total |
|---|---|---|---|---|---|
| CoswaraCough | 2 | 2024 | 678 | - | 2702 |
| CoswaraBreathing | 2 | 2029 | 677 | - | 2706 |
| CoswaraVowel-a | 2 | 2023 | 673 | - | 2696 |
| COUGHVID2Cls | 2 | 12377 | 3705 | - | 16082 |
| COUGHVID3Cls | 3 | 12377 | 1138 | 2567 | 16082 |
2.1.1.2. Pitch-shifting
Pitch-Shifting is a technique that enables the resampling of a sound or music by raising or lowering its pitch, without changing its speed, to obtain a better sound quality [11,44]. This technique concentrates on preserving the spectral envelope to achieve a natural conversion close to the original signal while preserving as much natural timbre as possible [45]. An example of raw signal (a) and pitch-shifted signal (b) of this signal is given in Fig. 2 .
Fig. 2.
(a) Raw signal, (b) Pitch shifted raw signal.
2.1.2. Feature extraction
In the feature extraction phase, chroma features (CF), root mean square energy (RMSE), spectral centroid (SC), spectral bandwidth (SB), spectral rolloff (SR), zero crossing rate (ZCR) and Mel frequency cepstral coefficients (MFCC) techniques used. The feature extraction techniques provide an efficient computational method for the application for which it is designed, as well as extracting important features from cough, breath, and voice signals. CNN architectures take data of the same size as input. To represent signals of different lengths in a space of the same length, the feature vectors obtained from a signal by feature extraction techniques were averaged. In this way, a 1x1 matrix size feature was obtained from each signal with the CF, RMSE, SC, SB, SR, and ZCR techniques, and a 1x20 matrix size feature was obtained from the MFCC technique, and a total of 1x26 matrix size features were obtained. The resulting averaged feature vector represents a consolidated representation of the input data, incorporating the information captured by each technique. This approach of averaging the feature vectors helps in leveraging the strengths of different techniques and enhancing the overall representation of the data. By combining the features, we can capture a more comprehensive and robust representation that encompasses diverse aspects of the data. A summary of the feature maps obtained for each signal is given in Table 3 . The mathematical formula used to obtain the feature maps of the signals is given below:
| (1) |
Here is the mean, is the signal and is the new representative value of the signal and [] is the merging operation. and , respectively, represent the cepstral coefficient of the features obtained from a signal with the technique and the number of features in different time slots. It demonstrates techniques for obtaining feature vectors from , , , , , and voice signals. Obtaining features from these techniques is given in detail in the next step.
Table 3.
The number of features obtained for each of cough, voice, and speech signals by feature extraction techniques.
| Feature Extraction | Dimension |
|---|---|
| Chroma features | 1x1 |
| Root Mean Square Energy | 1x1 |
| Spectral Centroid | 1x1 |
| Spectral Bandwidth | 1x1 |
| Spectral Rolloff | 1x1 |
| Zero Crossing Rate | 1x1 |
| Mel frequency cepstral coefficients | 1x20 |
| Total number of features | 1x26 |
2.1.2.1. Chroma features
The human hearing system has a system that separates the frequency components of the sound. When analyzing a sound signal, its spectral content, which is the combination of frequency components, is evaluated. Chroma features (CF) represent the tonal properties of this spectral content. Chroma features (CF) are vectors representing the energy distribution of the sound signal in the frequency spectrum of the 12 pitch classes: C, C#, D, D#, E, F, F#, G, G#, A, A# and B. Feature vector, Chroma Energy Normalized (CENS), Constant-Q transform (CQT), short-time Fourier transform (STFT), etc. extracted from the magnitude spectrum. These vectors are then calculated by taking the standard deviation and mean across the frames. Unlike MFCC properties, CF is more sensitive to changes in the timbre of signal and noise. The pitch classes for an example signal are given in Fig. 3 . The pitch is calculated as follows [[46], [47], [48], [49]]:
| (2) |
Where represents the chroma value and represents the ton height.
Fig. 3.
Pitch classes for an example signal.
The calculation of a chroma feature from a given frequency value is given below.
| (3) |
2.1.2.2. Root mean square energy
Root Mean Square Energy (RMSE) represents the overall energy content of the signal. After calculating the energy in a signal squarely, it is found by taking the mean and standard deviation of the squares. This technique is used to determine the overall energy level and strength of the sound signal. The RMSE changes depending on the amplitude and duration of the sound signal. A high RMSE value indicates a higher energy level, while a low RMSE value indicates a lower energy level. After calculating the energy in a signal squarely, it is found by taking the mean and standard deviation of the squares [46]. Finding the energy in a signal is mathematically in Eq. (4), and RMSE in Eq. (5).
| (4) |
| (5) |
Here, denotes the weighted frequency magnitude or value of box . An example signal and its RMSE plot are shown in Fig. 4 .
Fig. 4.
RMS Energy plot for an example signal.
2.1.2.3. Spectral centroid
Spectral centroid (SC) is used in digital signal processing to characterize a spectrum and is strongly associated with the impression of the brightness of the sound. SC shows how the intensity of the frequency components of the sound signal changes with time. It also represents the overall frequency content of the sound and may reflect the perceived “brightness” or “tone color” characteristics of the sound. A higher SC value indicates a brighter or high-pitched sound, while a lower SC value indicates a darker or bass sound. SC is expressed mathematically as in Eq. (6) [46,50,51]. An example signal and its SC graph are given in Fig. 5 .
| (6) |
Here, represents the frequency in box , and represents the spectral magnitude in box .
Fig. 5.
Spectral Centroid for an exemplary signal.
2.1.2.4. Spectral bandwidth
Spectral Bandwidth (SB) is a technique used to identify low-bandwidth sounds from high-frequency sound information. It also gives the frequency range within the frame and shows how wide the distribution of a frequency component is. SB can be used to determine the vocal characteristics of the voice or the clarity or softness of speech [[52], [53], [54]]. The graph of a pitch-shifted signal and the Spectral Bandwith is given in Fig. 6 . The mathematical notation of the SB technique is given in Eq. (7):
| (7) |
Fig. 6.
Spectral Bandwidth for an exemplary signal.
2.1.2.5. Spectral rolloff
Spectral Rolloff (SR) is used to represent the density of energy up to a certain percentage in the frequency spectrum of a sound signal. It is defined as the frequency value where the total energy of the spectra is under a certain threshold value (this ratio is usually 85%). For example, 85% SR represents up to 85% of the energy contained in the frequency spectrum. This shows how wide the spectral distribution is and may reflect how energy is distributed in the spectral region [52,55]. The spectral rolloff calculation process of a given signal is shown in Fig. 7 . Calculation of SR is given below [55].
| (8) |
Here, denotes a rectangular window of length .
Fig. 7.
Spectral Rolloff for an exemplary signal.
2.1.2.6. Zero crossing rate
Zero crossing rate (ZCR) is used to calculate how many times a signal crosses the zero axis, ie the rate of change of the signal sign (from positive to negative transition). In other words, it can be defined as the number of signal changes in 1-s intervals [20,46,52]. ZCR is used to detect the nature of voice signals such as speech and cough and to estimate their fundamental frequency [20,52]. In addition, ZCR is a very effective sound activity determination method that distinguishes whether a signal frame is muted or audible. The ZCR plot of a received signal for a certain range is shown in Fig. 8 . The mathematical expression of ZCR is given in Eq. (9) [48].
| (9) |
Where is the length of the frame. The function, which is a sign function, is defined as follows.
| (10) |
Fig. 8.
Zero Crossing Rate in a signal.
2.1.2.7. Mel frequency cepstral coefficients
Mel frequency cepstral coefficients (MFCC) is a sound processing method that is frequently used in the fields of sound processing and analysis and gives successful results. This method plays a very important role in speech and voice recognition systems [46]. MFCC is used to closely represent sound signals to the human hearing system [53]. MFCC techniques have been successfully applied in various fields such as music genre classification [56], speaker recognition [57], music information retrieval [58], language identification systems [59], speaker identification [60], sound similarity measurement [56], speech emotion recognition [61], speech enhancement [62] and vowel detection [63]. In the MFCC method, features are obtained by applying five different techniques, namely framing, windowing, fast Fourier transform (FFT), mel filterbank, and discrete cosine transform (DCT) [52,[64], [65], [66]].
In the Framing stage, the voice signal is divided into overlapping short-time intervals ( frames), and a spectrogram is generated for each time interval. The ideal overlap of frames is between 30% and 50%, and the window length is between 20 and 40 ms. The framing of the signal in short windows removes false detections in voice signals.
It is used to prevent high-frequency noise that may occur at the edges of the frames during the framing process with the windowing process and to obtain stable features. Each frame is multiplied by the same length of the Hanning window, reducing edge effect and signal discontinuity. The windowing process is given in Eq. (11) and the Hanning window is given mathematically in Eq. (12).
| (11) |
| (12) |
Here is the number of samples in each frame, is the window function, and is the input signal.
After converting the signal to the frequency domain with the FFT process, the periodogram is found. The FFT function is given in Eq. (13). The periodogram is found by .
| (13) |
It is used to model the human auditory system with the mel filterbank. For this, by filtering through the mel frequency scale, more sensitivity is gained to low-frequency signals. With a value obtained from each filter, the logarithm of the average values is taken to model the signal in the cepstral area. The spacing between filter banks is found using the Mel scale. Eq. (14) is used to find the Mel filterbanks and Eq. (15) is used to convert to the frequency domain [66].
| (14) |
| (15) |
Discrete cosine transform (DCT) converts discrete signals into the frequency domain for information compression. It is also used to remove noise in the signal. The mathematical function of DCT is given in Eq. (16) [[66], [67], [68]].
| (16) |
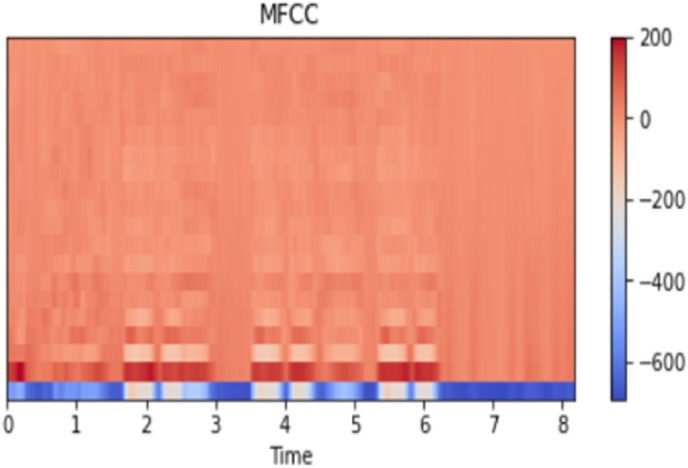
Where N is the number of filter banks. stands for log filterbank amplitudes. The MFCC graph of a pitch-shifted sample signal is given in Fig. 9 .
Fig. 9.
MFC for an exemplary signal.
2.2. Method
Deep learning (DL) architectures consist of interconnected layers and data is processed through layers. The features obtained from the layers are transferred to the next layer, enabling a more detailed examination [69,70]. Convolutional Neural Networks (CNNs) are a sub-branch of deep learning methods with multi-layered deep network architecture inspired by natural visual perception mechanisms [71,72]. CNN architectures generally have three basic categories: convolution layer, pooling layer, and fully connected layers [[73], [74], [75]].
DL architectures perform successfully in many areas such as classification, segmentation, pattern recognition, and object detection, and show remarkable performance with their deep feature extraction feature. In this study, a DL-based architecture is proposed (InceptionFireNet) to extract deep features. The basic structure of the proposed InceptionFireNet architecture is given in Fig. 11. InceptionFireNet architecture has a structure based on Inception and Fire modules.
Fig. 11.
The basic structure of deep feature extraction architecture (InceptionFireNet).
The Inception module (Fig. 10 (a)) is a module used to improve the performance of architectures. This module enables learning of features in different buyer areas and at different scales without increasing the computational cost of architectures. In addition, this structure enables it to capture the fine and coarse features of the input data, making it more resistant to changes in the input data [41,76,77].
Fig. 10.
(a) Inception module and (b) Fire module.
The Fire module (Fig. 10(b)) consists of two phases, squeeze and expansion phases. In the squeeze phase, the input data is reduced in size by a 1x1 convolution operation. In the expansion phase, after the compression phase, it is expanded with 1x1 and 3x3 convolution operations. With the 1x1 convolution process, it is ensured that the number of channels is reduced while preserving important features; With the 3x3 convolution operation, spatial dependencies between features are captured. In addition, these convolution operations increase the representation power of the network [78,79].
The proposed InceptionFireNet architecture for deep feature extraction is given in Fig. 11 . This architecture consists of three consecutive InceptionFireBlocks based on Inception and Fire modules. In InceptionFireNet architecture, a vector value given as an input is applied two times in succession, respectively, convolution1D (CL), Relu (RL), and Batch Normalization (BN). After this process, convolution and size reduction are applied by giving the stride value 2 in the convolution1D operation. The feature maps obtained after these processes are given to the Inception module and combined with the results obtained from this module. Then, the obtained features are subjected to the Fire module, and the obtained feature maps are combined with the feature maps from the Inception module. After these processes are repeated three times in a row, the GlobalAveragePooling (GAP) layer is given. With GAP, it generates a 1x1xC feature map by averaging each of the channels in the feature maps with dimensions H x W x C (Height x Width x Number of channels). In this way, it reduces the computational cost without causing information loss. Feature maps obtained with GAP are subjected to Fully Connected (FC) and RL activation processes three times in succession. Finally, the FC layer and Sigmoid/Softmax activation function are applied to the obtained feature maps.
The convolutional layer applies a convolution operation to the outputs of the input data or previous feature maps. Let represent the input value of the layer's feature map, represent the weights of the input in the layer, and represent the bias value. Accordingly, the convolution operation in the convolutional layer is calculated as follows [80]:
| (17) |
Activation functions such as Relu [81] and LeakyRelu (LR) [82] are used to detect nonlinear features from convolution operations. Sigmoid [83] and Softmax [84] activation functions are generally used for classification in the output layers of the network. The sigmoid function is a function that gives a probabilistic output value between 0 and 1, which is mostly used in binary classification problems. The Softmax activation function gives the probability distribution of classes in multiclass networks. Mathematical functions of Relu in Eq. (18), LeakyRelu in Eq. (19), Sigmoid in Eq. (20), and Softmax activation functions in Eq. (21) are given.
| (18) |
| (19) |
| (20) |
| (21) |
Here, represents the login information in the layer, represents the number of classes, and represents the value up to .
Batch Normalization (BN) provides better learning of the network by preventing the risk of deviation in the network and allows the use of non-linear activation functions without being stuck in a saturated model. BN is calculated by considering the mean and standard deviation in each hidden layer according to Eq. (22), (23), (24), (25)) [80,85].
| (22) |
| (23) |
| (24) |
| (25) |
Where represents the mini-batch information, is the mean, is the standard deviation and ˆ is normalized. and represent the updated offset and scale parameters across the network.
The mathematical expression of the proposed InceptionFireNet architecture to extract deep feature maps is given as follows:
| (26) |
Here represents Convolution, Relu, and Batch Normalization operations and InceptionFireBlock, respectively. denotes number, denotes number and denotes number. denotes the number of repetitions.
The mathematical expression of the InceptionFireBlock structure is as follows:
| (27) |
Where is the Inception function and is the Fire function. represents the process output, ie feature maps, in the relevant layer. gives the value of the attributes obtained by the InceptionFireBlock and [.] represents the join function.
The proposed DeepConvNet architecture for the classification of important features from cough, breath, and voice information using the InceptionFireNet architecture is given in Fig. 12 .
Fig. 12.
The basic structure of the DeepConvNet architecture.
The DeepConvNet architecture takes as input the feature vectors obtained from the FC3 layer of the InceptionFireNet architecture. CL, BN, and LR operations are applied to these feature vectors, respectively. In the next step, size reduction and convolution are applied by giving the stride value as two in the CL process. With size reduction, unnecessary information is eliminated and transaction costs are reduced. Then, after four consecutive CL, BN, and LR operations, the features are combined. The transaction block made at this stage is named ConnectBlock. The feature maps obtained after four consecutive ConnectBlock operations are vectorized with the Flatten layer. Finally, Covid-19 prediction is made with three FC, respectively. After the first two FCs, the R activation function is used. In the last FC, predictions are made using Softmax (for multiple classes) or Sigmoid (for binary classes) activation functions. DeepConvNet mathematical notation is expressed as:
| (28) |
Here denotes convolution operation, denotes convolution, BN, and R operations respectively. {} denotes the number of repetitions.
The ConnectBlock module mathematical methodology is as follows:
| (29) |
Where represents the attributes of the layer.
For the training of the proposed InceptionFireNet and DeepConvNet architectures, Categorical CrossEntropy (CE) for multi-class applications and Binary CrossEntropy (BCE) for two-class problems are used as cost functions. The CE cost function is as follows.
| (30) |
Here is the number of classes, is the actual class, is the predicted class. The Binary CrossEntropy cost function is defined as follows.
| (31) |
3. Performance metrics
Six evaluation metrics, namely accuracy (Acc), precision (Pre), recall (Rec), F1-Score (F1), specificity (Spe), and AUC, were used for performance measurement in experimental studies. The mathematical formulas of these metrics are as follows:
| (32) |
| (33) |
| (34) |
| (35) |
| (36) |
Here, (True Positives) refers to correctly defined patient samples, (True Negatives) refers to correctly defined healthy cases, (False Positives) refers to misidentified patients, (False Negatives) refers to incorrectly defined healthy cases [86,87]. AUC (Area under the curve) represents the area under the ROC curve, which is a probability curve [87]. ROC (The receiver operating characteristic), (true positive rate). It is expressed as a fraction of (false positive rate).
| (37) |
| (38) |
4. Experimental study results and discussion
In this study, different experimental studies were carried out for the detection of Covid-19 from cough, breath, and sound signals. Python programming language and Keras deep learning library were used in the studies. The experimental study process of this study is given in Fig. 1. This process consists of five stages: data preparation, preprocessing, feature extraction, deep feature extraction, and estimation. Each stage is given in detail in Section-2. Coswara and COUGHVID datasets specified in Table 2 were used as datasets. In the experimental studies, 80% of these datasets were used for training and 20% for testing. The class distribution of the training and test datasets used in the applications is given in Table 4 in detail. The architectures proposed in the experimental studies were trained in 32 batches of 100 epochs using the Adam [88] optimization algorithm. In the applications, Binary CrossEntropy (BCE) is used as a cost function for training two-class datasets, and the Sigmoid algorithm is used as an activation algorithm for classification in the last layer. Categorical CrossEntropy (CE) was used as a cost function for training three-class COUGHVID3Cls and the Softmax algorithm was used as the activation algorithm for estimation in the last layer.
Table 4.
Number of records of training and test datasets used in applications by classes.
| Application (s) | Data set(s) | Number of classes | Train/Test | Healthy | Covid-19 | Symptomatic | Total |
|---|---|---|---|---|---|---|---|
| Application1 |
COUGHVID3Cls | 3 | Train Set | 9887 | 925 | 2053 | 12865 |
| Test Set | 2490 | 213 | 514 | 3217 | |||
| COUGHVID2Cls |
2 |
Train Set | 9887 | 2998 | - | 12865 | |
| Test Set |
2490 |
727 |
- |
3217 |
|||
| Application2 |
CoswaraCough | 2 | Train Set | 1615 | 546 | - | 2161 |
| Test Set | 409 | 132 | - | 541 | |||
| CoswaraBreathing | 2 | Train Set | 1634 | 530 | 2164 | ||
| Test Set | 395 | 147 | 542 | ||||
| CoswaraVowel-a |
2 |
Train Set | 1615 | 541 | - | 2156 | |
| Test Set |
408 |
132 |
- |
540 |
|||
| Application3 | COUGHVID2Cls + CoswaraCough | 2 | Train Set | 11502 | 3524 | - | 15026 |
| Test Set | 2899 | 859 | - | 3758 |
In the first experimental study, the effect of the pitch-shifting technique on the success of the proposed CovidCoughNet model was examined. For this, the Pitch shifting technique of the Librosa library is used. In the pitch shifting technique, the datasets were reconstructed by giving different values to the n_steps parameter (−4,-8,-10,-12,-20, and −30). In the tables, the pitch-shifting technique is not applied to the original signals (Raw). However, for all signals (original signal and pitch-shifted signal), the feature extraction, deep feature extraction, and estimation processes indicated in Fig. 1 were applied respectively. The results of the first application using the COUGHVID datasets are presented in Table 5 and Table 6 . The COUGHVID3Cls (Healthy, Covid-19, Symptomatic) dataset is used in Table 5 and the COUGHVID2Cls dataset is used in Table 6.
Table 5.
The test results were obtained with the proposed CovidCoughNet architecture on the COUGHVID (Healthy, Covid-19, and Symptomatic) dataset according to the different n_step values of the pitch shift technique.
| Dataset | pitch_shift (n_steps) | Acc (%) | Pre | Rec | F1 | Spe (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| COUGHVID3Cls | Raw | 97.92 | 0.97 | 0.95 | 0.96 | 96.82 | 96.56 |
| −4 | 99.19 | 0.99 | 0.98 | 0.98 | 97.77 | 98.44 | |
| −8 | 98.91 | 0.99 | 0.98 | 0.98 | 97.37 | 98.22 | |
| −10 | 98.94 | 0.99 | 0.98 | 0.98 | 98.18 | 98.23 | |
| −12 | 97.73 | 0.95 | 0.96 | 0.96 | 96.23 | 97.01 | |
| −20 | 92.04 | 0.88 | 0.81 | 0.84 | 82.41 | 86.31 | |
| −30 | 93.78 | 0.91 | 0.86 | 0.89 | 87.80 | 89.92 |
Table 6.
Test results were obtained with the proposed CovidCoughNet architecture on the COUGHVID2Cls dataset according to the different n_step values of the pitch shift technique.
| Dataset | pitch_shift (n_steps) | Acc (%) | Pre | Rec | F1 | Spe (%) | AUC (%) |
|---|---|---|---|---|---|---|---|
| COUGHVID2Cls | Raw | 93.97 | 0.93 | 0.89 | 0.91 | 89.43 | 89.43 |
| −4 | 96.86 | 0.96 | 0.95 | 0.95 | 95.05 | 95.05 | |
| −8 | 95.24 | 0.94 | 0.92 | 0.93 | 92.45 | 92.45 | |
| −10 | 95.62 | 0.95 | 0.92 | 0.93 | 91.86 | 91.86 | |
| −12 | 93.97 | 0.94 | 0.89 | 0.91 | 88.75 | 88.75 | |
| −20 | 89.21 | 0.89 | 0.79 | 0.83 | 79.20 | 79.20 | |
| −30 | 83.21 | 0.83 | 0.65 | 0.68 | 65.20 | 65.20 |
When the findings in Table 5 are examined, when the n_steps parameter takes the value of −4, the highest performance was obtained in the Acc, Pre, Rec, F1, Spe, and AUC metrics at the rates of 99.19%, 0.99, 0.98, 0.98, 97.77% and 98.44%, respectively. We can say that using the Pitch shifting (n_steps = −4) technique in the preprocessing stage increases the success compared to Raw signals (not using the Pitch shifting technique). It is possible to say that when the n_steps value is given a value smaller than −4, the success gradually decreases, that is, it causes information loss in the signals.
In another study conducted to examine the effect of pitch shifting technique on success, the COUGHVID2Cls dataset was used and the findings are presented in Table 6.
When examining the results in Tables 6 and it can be stated that the highest performance is achieved when the n_steps parameter for the pitch shifting technique is set to −4, with 96.86% Acc, 0.96 Pre, 0.95 Rec, 0.95 F1, 95.05% Spe, and 95.05% AUC. Examining the studies using COUGHVID2Cls and COUGHVID3Cls datasets to examine the effect of pitch shifting technique on success, it was seen that the pitch shifting technique significantly increased the success. Considering the effect of the pitch shifting technique on the success, the pitch shifting technique was used in the preprocessing stage in the following applications, and the applications were carried out by giving the n_steps parameter a value of −4.
A summary of the test findings obtained with the CovidCoughNet architecture using COUGHVID datasets containing cough data for Covid-19 detection is presented in Table 7 . In addition, the Confusion matrix results showing the class performance of the proposed architecture are given in Fig. 13 , and the ROC curve graph is given in Fig. 14 . When we examine the results given in Table 7, we can say that a higher performance was achieved with 99.19% Acc, 0.99 Pre, 0.98 Rec, 0.98 F1, 97.77 Spe, and 98.44 AUC using the COUGHVID3Cls (Healthy, Covid-19, and Symptomatic) dataset. The Covid-19 records in the COUGHVID2Cls dataset are a combination of the Covid-19 and Symptomatic records in the COUGHVID3Cls dataset. We can say that the lower performance from the COUGHVID2Cls dataset is due to the differences in the sound signals of the Covid-19 and Symptomatic recordings in the Covid-19 class. Similarly, it is possible to say that higher success was achieved with the COUGHVID3Cls dataset and that the proposed architecture better distinguishes the differences in voice characteristics in Healthy, Covid-19, and Symptomatic patients.
Table 7.
Findings obtained as a result of using the COUGHVID dataset for Covid-19 detection from cough signals with the proposed architecture.
| Dataset | Acc (%) | Pre | Rec | F1 | Spe (%) | AUC (%) |
|---|---|---|---|---|---|---|
| COUGHVID2Cls | 96.86 | 0.96 | 0.95 | 0.95 | 95.05 | 95.05 |
| COUGHVID3Cls | 99.19 | 0.99 | 0.98 | 0.98 | 97.77 | 98.44 |
Fig. 13.
Confusion Matrix results for COUGHVID datasets with the proposed architecture. (a) for COUGHVID2Cls (Healthy and Covid-19), (b) for COUGHVID3Cls (Healthy, Covid-19, and Symptomatic).
Fig. 14.
ROC-Curve graph for COUGHVID datasets with the proposed architecture. (a) for COUGHVID2Cls (Healthy and Covid-19), (b) for COUGHVID3Cls (Healthy, Covid-19, and Symptomatic).
Fig. 13(a) shows the results of 3217 subjects in the COUGHVID2Cls (Healthy and Covid-19) test dataset. It can be seen that 2449 out of 2490 healthy subjects were correctly predicted with a rate of 98.35%. Additionally, 667 out of 727 Covid-19 (Covid-19 and Symptomatic) patients were correctly diagnosed with a rate of 91.75%. Similarly, when the results obtained with the COUGHVID3Cls dataset, which includes Healthy, Covid-19, and Symptomatic data, using the proposed architecture are examined (see Fig. 13(b)), it can be seen that only 6 out of 2490 healthy subjects were wrongly predicted, and only 9 out of 213 Covid-19 patients were wrongly predicted. Additionally, only 11 out of 514 Symptomatic patients were wrongly predicted, and 503 out of 514 patients were correctly predicted with a success rate of 97.86%. The findings obtained for the detection of Covid-19 from cough data show that the proposed model performs extremely successfully. In addition, we can say that the findings in Fig. 13 predict records in the multiclass COUGHVID3Cls dataset with higher performance. It is possible to say that the proposed architecture is due to the characteristic differences in the sound signals of Covid-19 and Symptomatic patient samples and that the proposed architecture can distinguish these differences well.
Another performance metric used to measure the success of the CovidCoughNet architecture on the COUGHVID dataset is the ROC-Curve graph, which is shown in Fig. 14. In Fig. 14 (a), a 95.05% AUC value was achieved for binary classification (Healthy and Covid-19). For the COUGHVID3Cls dataset (Fig. 14(b)), it can be seen that a higher performance was obtained with AUC values of 98.64% in healthy samples, 97.85% in Covid-19 patients, and 98.82% in symptomatic patients.
Looking at the test results of the COUGHVID dataset with the proposed architecture, it can be seen that higher performance is achieved for the three-class (Healthy, Covid-19, and Symptomatic) data compared to the two-class data. This indicates that the proposed architecture is quite successful in diagnosing different diseases. This shows that the proposed architecture performs very well in diagnosing different diseases.
The proposed CovidCoughNet architecture was also applied to the Coswara dataset for Covid-19 detection. The Coswara dataset includes cough, breath signals, and signals in different phonation of voice (/a/ …./i/,/o/). The results obtained using datasets containing cough, breath, and the “a" sound phonation are presented in Table 8 , where an average accuracy of over 99% was achieved for Covid-19 detection. It can be seen that an accuracy of 99.26% was obtained with the CoswaraCough dataset, while an accuracy of 98.52% was achieved with the CoswaraBreathing dataset. However, the dataset containing the “a" sound phonation (CoswaraVowel-a) achieved higher accuracy with 99.63% accuracy, 1.00 precision, 0.99 recall, 0.99 F1 score, 99.24% specificity, and 99.24% AUC rate. In the experimental application using the Coswara dataset for Covid-19 detection, an average performance of more than 99% was achieved with the proposed architecture. We can say that achieving higher success with the CoswaraVowel-a dataset is due to saying the letter “a" without hesitation. Obtaining a performance higher than 99% in the COUGHVID and Coswara datasets clearly shows that the proposed model has a strong structure.
Table 8.
Results obtained with CovidCoughNet architecture using different types of Coswara datasets for Covid-19 detection from cough, breath, and voice signals.
| Dataset | Acc (%) | Pre | Rec | F1 | Spe (%) | AUC (%) |
|---|---|---|---|---|---|---|
| CoswaraCough | 99.26 | 1.00 | 0.98 | 0.99 | 98.48 | 98.48 |
| CoswaraBreathing | 98.52 | 0.98 | 0.98 | 0.98 | 97.92 | 97.92 |
| CoswaraVowel-a | 99.63 | 1.00 | 0.99 | 0.99 | 99.24 | 99.24 |
The results obtained using the CovidCoughNet architecture with different types of the Coswara dataset for Covid-19 detection are given in Confusion Matrix in Fig. 15 and the ROC-Curve graph in Fig. 16 . When the results obtained from the cough data in Fig. 15 (a) are analyzed, it can be seen that all healthy subjects were correctly predicted with 100% accuracy, and Covid-19 patients were classified with 96.97% accuracy. In Fig. 15 (b), when the test results obtained from the breath data are analyzed, it can be said that only 3 out of 395 healthy subjects were incorrectly predicted, and out of 147 Covid-19 patients, only 5 were incorrectly predicted, while 142 were correctly predicted. When the results obtained for the “a" sound for Covid-19 detection are examined, it is possible to say that healthy individuals were correctly predicted with 100% accuracy, and Covid-19 patients were correctly predicted with a high success rate of 98.48%. When the Confusion Matrix results were examined in general, it was seen that a higher success was achieved with the dataset (CoswaraVowel-a) obtained from the “a" sound. It is seen that cough and breathing results are slightly lower. The reason for this is thought to be caused by the noise created by the air in cough and breath sounds.
Fig. 15.
Confusion Matrix results were obtained using CovidCoughNet architecture and test data from the Coswara dataset: (a) for the CoswaraCough dataset, (b) for the Coswarabreathing dataset, (c) for the CoswaraVowel-a dataset.
Fig. 16.
The ROC-Curve graph was obtained using test data from the Coswara dataset with the proposed architecture: (a) CoswaraCough, (b) CoswaraBreathing, (c) CoswaraVowel-a.
When we examine the ROC-Curve results obtained with the Coswara dataset in Fig. 16, we can see that when the CoswaraCough dataset (Fig. 16(a)) is used, a performance of 98.48% AUC is achieved in both healthy and Covid-19. Similarly, with the CoswaraBreathing dataset (Fig. 16(b)), a performance of 97.92% AUC is obtained. When the CoswaraVowel-a (Fig. 16(c)) test data is used, we can say that a higher success rate of 99.24% AUC is achieved.
In another application, the COUGHVID2Cls and CoswaraCough datasets, which contain cough data, were combined to detect Covid-19 from cough. The findings obtained are presented in Table 9 . In addition, the Confusion Matrix results and the ROC-Curve graph are given in Fig. 17 . When we examine the results in Table 9, we can say that the CovidCoughNet architecture achieved results of 97.13%, 0.97, 0.95, 0.96, 94.86%, and 94.86% in Acc, Pre, Rec, F1, Spe, and AUC metrics, respectively. It can be stated that the results obtained by combining both datasets (as seen in Table 7) achieve higher success compared to using only the COUGHVID2Cls dataset. The higher success is thought to be due to the CoswaraCough dataset having less noisy cough signals. Additionally, when examining the results obtained by combining the two datasets (as seen in Table 8), it is observed that it exhibits lower performance compared to using only the CoswaraCough dataset. This is believed to be due to the cough signals in the COUGHVID2Cls dataset.
Table 9.
Covid-19 detection from cough with proposed architecture using data from the combination of COUGHVID2Cls and CoswaraCough datasets.
| Datasets | Acc (%) | Pre | Rec | F1 | Spe (%) | AUC (%) |
|---|---|---|---|---|---|---|
| COUGHVID2Cls + CoswaraCough | 97.13 | 0.97 | 0.95 | 0.96 | 94.86 | 94.86 |
Fig. 17.
Confusion Matrix (a) results and ROC-Curve plot (b) from COUGHVID2Cls and CoswaraCough datasets containing cough data with CovidCoughNet architecture.
According to the results of the Confusion Matrix given in Fig. 17 (a), it can be seen that the proposed architecture correctly predicted 99.03% of 2871 individuals out of 2899 healthy individuals. Additionally, 779 out of 859 Covid-19 patients were correctly predicted. It is seen that Covid-19 patients are estimated at a slightly lower rate (90.69%). This is thought to be due to Covid-19 and Symptomatic recordings with different sound characteristics in the Covid-19 class in the COUGHVID2Cls dataset. When the ROC-Curve graph in Fig. 17 (b) is examined, we can say that both healthy and Covid-19 subjects were correctly predicted with 94.86% AUC.
In addition, a summary of the studies using the same dataset as ours and using a different dataset for the detection of Covid-19 from cough, breath, or sound signals is given in Table 10 . When studies using data sets different from ours are examined, we can say that Laguarta et al. [89] achieved a higher success rate of 98.5%, 98.5%, 94.2%, and 97% in their study using cough signals, according to Acc, Rec, Spe and AUC metrics, respectively. According to the Pre metric, the highest success was obtained by Imran et al. [6] with a success rate of 91.43%. According to the F1 metric, it is possible to say that the highest success was achieved by Lella and Pja [27] with a value of 96.96%.
Table 10.
Summary of studies for detection of Covid-19 from cough, breath, and sound signals. UC stands for the University of Cambridge and Sen stands for Sensitivity.
| Study |
Dataset |
Method |
Modality |
Acc (%) |
Pre |
Rec/Sen |
F1 |
Spe (%) |
AUC (%) |
|---|---|---|---|---|---|---|---|---|---|
| Studies using datasets different from ours | |||||||||
| Lella ve Pja, 2022 [27] | UC | DCCN | Breath + Cough + Voice | 95.45 | - | - | 96.96 | - | - |
| Imran et al., 2020 [6] | Privately collected | DTL-BC | Cough | 92.85 | 91.43 | 94.57 | 92.97 | 91.14 | - |
| Brown ve arkadaşları, 2020 [30] | UC: University of Cambridge | SVM | Cough | 0.80 | 0.72 | 82.00 | |||
| Mohammed et al., 2021 [39] | Collected from Github | Scratch CNN | Cough | 0.77 | 0.80 | 0.71 | 0.75 | 0.77 | |
| Chaudhari et al., 2020 [90] | Data is collected from Ruijin Hospital | AI-based method | Cough | - | - | - | - | - | 77.1 |
| Hassan et al., 2020 [91] | Data collected from COVID affected 14 patients | Recurrent Neural Network (RNN) | Cough | 97 | - | - | - | - | - |
| Hassan et al., 2020 [91] | Data collected from COVID affected 14 patients | RNN | Breath | 98.2 | - | - | - | - | - |
| Laguarta et al., 2020 [89] |
MIT open voice data set |
CNN based method |
Cough |
98.5 |
- |
98.5 |
- |
94.2 |
97.0 |
|
Studies using the same datasets as ours | |||||||||
| Harvill et al., 2021 [92] | COUGHVID | LSTM | Cough | - | - | - | - | - | 85.35 |
| Hamdi et al., 2022 [11] | COUGHVID | CNN-LSTM | Cough | 91.13 | 90.47 | 90.93 | 90.71 | 91.31 | 91.13 |
| Orlandic et al., 2021 [34] | COUGHVID | eXtreme Gradient Boosting (XGB) | Cough | 88.1 | 95.5 | 80.8 | - | 95.5 | 96.5 |
| Our Model | COUGHVID | CovidCoughNet (CNN based) | Cough | 99.19 | 0.99 | 0.98 | 0.98 | 97.77 | 98.44 |
| Pahar et al., 2021 [19] | Coswara | ResNet50 | Cough | 95.33 | - | 93 | - | 98.00 | 97.6 |
| Tena et al., 2021 [93] | UC + UL + Coswara + Pertussis + Virufy | Random Forest | Cough | 98.79 | 90.97 | 93.81 | 92.10 | 81.54 | 96.04 |
| Xue ve Salim, 2021 [94] | UC + Coswara | Transformer-CP + Transformer-CP | Cough | 84.43 | 84.57 | 73.24 | 78.50 | 90.03 | |
| Chowdhury et al., 2022 [7], | NoCoCoDa + Cambridge + Virufy+ Coswara |
Extra-Trees Algorithm | Cough | - | 1.00 | 0.97 | - | - | 95 |
| Sharma et al., 2020 [32] | Coswara | Random Forest | Breath + Cough + Voice | 66.74 | - | - | - | - | - |
| Our Model | Coswara | CovidCoughNet (CNN based) | Cough | 99.26 | 1.00 | 0.98 | 0.99 | 98.48 | 98.48 |
| Our Model | Coswara | CovidCoughNet (CNN based) | Breathing | 98.52 | 0.98 | 0.98 | 0.98 | 97.92 | 97.92 |
| Our Model | Coswara | CovidCoughNet (CNN based) | Voice (Vowel-a) | 99.63 | 1.00 | 0.99 | 0.99 | 99.24 | 99.24 |
When examining studies that use the same COUGHVID dataset as us, it can be said that our study showed the highest performance with 99.19% ACC, 99% Pre, 98% Rec, 98% F1, 97.77% Spe, and 98.44% AUC. Similarly, when examining studies that use the same Coswara dataset as us, it can be said that the proposed architecture showed the highest success according to the Acc (99.63%), Rec (99%), F1 (99%), Spe (99.24%), and AUC (99.24%) metrics. Additionally, in terms of the Pre metric, the study conducted by Chowdhury et al. [7] showed the highest success with 100% accuracy compared to our study.
When examining the results in Table 10 in a general manner, it can be said that our proposed model showed the highest success in all metrics when using the sound data (Vowel-a) of the Coswara dataset.
In the literature, Covid-19 detection is frequently performed by deep learning techniques using X-ray and CT images [12,[95], [96], [97], [98], [99], [100], [101], [102], [103]]. However, the images must be taken from a well-equipped clinic or a hospital through a specialist. The risk of spreading Covid-19, which is a contagious disease, increases during this process and threatens the health status of healthcare personnel. Considering these disadvantages, it is extremely important to detect Covid-19 from cough, breath, and sound signals, which is a faster and more reliable method.
A summary of the studies conducted using the proposed model, which utilizes X-ray/CT images and sound signals for Covid-19 detection, is provided in Table 11 . In Table 11, Celik [12] presents the results of their study using X-ray images, employing the same hyperparameters and the same dataset as other studies [[95], [96], [97], [98], [99]], showing the training durations and accuracy metrics obtained. Additionally, some studies conducted using X-ray/CT images [[100], [101], [102], [103]] do not provide any information about their training durations. Upon examining the results in the table, it is evident that studies conducted using X-ray images had significantly longer training durations. However, in this study utilizing cough, breath, and voice signals, the proposed architecture was trained in a very short time, approximately 9 and 20 min. Therefore, it can be concluded that utilizing sound signal data is the correct approach for the detection of Covid-19. It is possible to state that using different feature extraction methods in this study allowed the representation of sound signals in a smaller-dimensional space, providing a significant advantage in terms of time. Furthermore, when evaluating the performances of the architectures based on accuracy metrics, it is observed that the proposed method exhibited higher performance with a 99.63% accuracy rate compared to other studies that used image-based approaches. This study, which utilized cough, breath, and voice signals for Covid-19 detection, offers a significant advantage in terms of time, performance, speed, cost, and reducing transmission.
Table 11.
Successful results of some studies for the detection of Covid-19 from X-ray, CT images, and cough and sound signals.
| Model | Hardware (GPU) | Input (Sound/Image) | Input_size | Number of records | Epoch | Train Time (hour) | Acc (%) |
|---|---|---|---|---|---|---|---|
| Wang et al. [95] | Yes | X-ray | 224x224 | 21165 | 200 | 6.00 | 82.63 |
| Mahmud et al. [96] | Yes | X-ray | 128x128 | 21165 | 200 | 15.33 | 92.44 |
| Khan et al. [97] | Yes | X-ray | 150x150 | 21165 | 200 | 6.33 | 92.11 |
| Monshi et al. [98] | Yes | X-ray | 256x256 | 21165 | 200 | 8.33 | 95.39 |
| Ozturk et al. [99] | Yes | X-ray | 256x256 | 21165 | 200 | 7.54 | 94.33 |
| Celik [12] | Yes | X-ray | 128x128 | 21165 | 200 | 7.24 | 96.81 |
| Barnawi et al. [100] | - | X-ray | 224x224 | 2905 | 50 | - | 94.92 |
| Chaddad et al. [101] | - | X-ray + CT | 512x512 | 1271 | 10 | - | 99.09 |
| CHHIKARA et al. [102] | Yes | X-ray | 224x224 | 6432 | 50 | - | 97.70 |
| Tan et al. [103] | Yes | CT | 512x512 | 5007 | 100 | - | 91.98 |
| Our Model | Yes | Cough | 1x26 | 16082 | 100 | 0.20 | 99.19 |
| Our Model | Yes | Voice (Vowel-a) | 1x26 | 2696 | 100 | 0.09 | 99.63 |
5. Conclusion
In this study, we propose a deep learning-based model for Covid-19 detection using cough, breath, and voice signals. The proposed model consists of preprocessing, feature extraction, deep feature extraction, and prediction stages. In the preprocessing stage, unnecessary data is removed and a higher-quality sound is obtained using the pitch-shifting technique. In the feature extraction stage, CF, RMSE, SC, SB, ZR, ZCR, and MFCC techniques are used to extract important features of cough, breath, and voice signals. In the deep feature extraction stage, a model named InceptionFireNet is proposed based on Inception and Fire models. Inception and Fire modules are used to learn features at different scales. In the prediction phase, the disease prediction was carried out with the DeepConvNet architecture, which we proposed, using the feature vectors obtained from the FC3 layer of the pre-trained InceptionFireNet architecture. Coswara and COUGHVID datasets are used as the dataset. Six different metrics, namely accuracy, precision, recall, F1-Score, specificity, and AUC, are used to evaluate the performance in experimental studies. Additionally, the Confusion Matrix and ROC-Curve are used to analyze the class performance of the proposed method.
When examining the experimental studies, it was found that using the Pitch Shift technique resulted in a 3% increase in the performance of the proposed architecture (CovidCoughNet). In an experimental study using only cough data from the COUGHVID dataset (Healthy, Covid-19, and Symptomatic), higher success was achieved with the three-class COUGHVID3Cls dataset compared to the two-class COUGHVID2Cls dataset. Using the three-class COUGHVID3Cls dataset, success rates of 99.19%, 0.99, 0.98, 0.98, 97.77%, and 98.44% were achieved in accuracy, precision, recall, F1-Score, specificity, and AUC metrics, respectively. Additionally, in an experimental study using the Coswara dataset which includes cough, breath, and voice signals, higher success was achieved compared to cough and breath data alone with an accuracy of 99.63%, precision of 1.00, recall of 0.99, F1-Score of 0.99, specificity of 99.24%, and AUC of 99.24%. Also, It is possible to say that higher success has been achieved with our proposed model when examining the literature studies conducted for Covid-19 detection using cough, breath, or voice signals similarly.
In future studies on detecting Covid-19, we plan to work with different and larger datasets and test our work with experts in healthcare institutions. We also aim to move our application to a web and mobile-based platform for faster access and diagnosis.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.World Health Organization WHO coronavirus (COVID-19) dashboard. https://covid19.who.int/
- 2.Miyata R., et al. Oxidative stress in patients with clinically mild encephalitis/encephalopathy with a reversible splenial lesion (MERS) Brain Dev. Feb. 2012;34(2):124–127. doi: 10.1016/j.braindev.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 3.Khalifa N.E.M., Smarandache F., Manogaran G., Loey M. A study of the neutrosophic set significance on deep transfer learning models: an experimental case on a limited COVID-19 chest X-ray dataset. Cognit. Comput. 2021 doi: 10.1007/s12559-020-09802-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loey M., Mirjalili S. COVID-19 cough sound symptoms classification from scalogram image representation using deep learning models. Comput. Biol. Med. 2021;139(November) doi: 10.1016/j.compbiomed.2021.105020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marcel S., et al. COVID-19 epidemic in Switzerland: on the importance of testing, contact tracing and isolation. Swiss Med. Wkly. 2020;150(11–12):4–6. doi: 10.4414/smw.2020.20225. [DOI] [PubMed] [Google Scholar]
- 6.Imran A., et al. AI4COVID-19: AI enabled preliminary diagnosis for COVID-19 from cough samples via an app. Inform. Med. Unlocked. 2020;20 doi: 10.1016/j.imu.2020.100378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chowdhury N.K., Kabir M.A., Rahman M.M., Islam S.M.S. Machine learning for detecting COVID-19 from cough sounds: an ensemble-based MCDM method. Comput. Biol. Med. 2022;145(March) doi: 10.1016/j.compbiomed.2022.105405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang D., et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus–infected pneumonia in wuhan, China. JAMA. Mar. 2020;323(11):1061. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carfì A., Bernabei R., Landi F. Persistent symptoms in patients after acute COVID-19. JAMA. Aug. 2020;324(6):603. doi: 10.1001/jama.2020.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dentamaro V., Giglio P., Impedovo D., Moretti L., Pirlo G. AUCO ResNet: an end-to-end network for Covid-19 pre-screening from cough and breath. Pattern Recogn. 2022;127 doi: 10.1016/j.patcog.2022.108656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hamdi S., Oussalah M., Moussaoui A., Saidi M. Attention-based hybrid CNN-LSTM and spectral data augmentation for COVID-19 diagnosis from cough sound. J. Intell. Inf. Syst. 2022;59(2):367–389. doi: 10.1007/s10844-022-00707-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Celik G. Detection of Covid-19 and other pneumonia cases from CT and X-ray chest images using deep learning based on feature reuse residual block and depthwise dilated convolutions neural network. Appl. Soft Comput. Jan. 2023;133 doi: 10.1016/j.asoc.2022.109906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Calderon-Ramirez S., Yang S., Elizondo D., Moemeni A. Dealing with distribution mismatch in semi-supervised deep learning for COVID-19 detection using chest X-ray images: a novel approach using feature densities. Appl. Soft Comput. 2022;123 doi: 10.1016/j.asoc.2022.108983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gupta A., Anjum, Gupta S., Katarya R. InstaCovNet-19: a deep learning classification model for the detection of COVID-19 patients using Chest X-ray. Appl. Soft Comput. 2021;99 doi: 10.1016/j.asoc.2020.106859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feki I., Ammar S., Kessentini Y., Muhammad K. Federated learning for COVID-19 screening from Chest X-ray images. Appl. Soft Comput. 2021;106 doi: 10.1016/j.asoc.2021.107330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bandyopadhyay R., Basu A., Cuevas E., Sarkar R. Harris Hawks optimisation with Simulated Annealing as a deep feature selection method for screening of COVID-19 CT-scans. Appl. Soft Comput. 2021;111 doi: 10.1016/j.asoc.2021.107698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou T., Lu H., Yang Z., Qiu S., Huo B., Dong Y. The ensemble deep learning model for novel COVID-19 on CT images. Appl. Soft Comput. 2021;98 doi: 10.1016/j.asoc.2020.106885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ye Q., et al. Robust weakly supervised learning for COVID-19 recognition using multi-center CT images. Appl. Soft Comput. 2022;116 doi: 10.1016/j.asoc.2021.108291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pahar M., Klopper M., Warren R., Niesler T. COVID-19 cough classification using machine learning and global smartphone recordings. Comput. Biol. Med. 2021;135(May) doi: 10.1016/j.compbiomed.2021.104572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pal A., Sankarasubbu M. Proceedings of the 36th Annual ACM Symposium on Applied Computing. Mar. 2021. Pay attention to the cough: early diagnosis of COVID-19 using interpretable symptoms embeddings with cough sound signal processing; pp. 620–628. [DOI] [Google Scholar]
- 21.Adhi Pramono R.X., Anas Imtiaz S., Rodriguez-Villegas E. Automatic identification of cough events from acoustic signals. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. EMBS. 2019:217–220. doi: 10.1109/EMBC.2019.8856420. [DOI] [PubMed] [Google Scholar]
- 22.Botha G.H.R., et al. Detection of tuberculosis by automatic cough sound analysis. Physiol. Meas. Apr. 2018;39(4) doi: 10.1088/1361-6579/aab6d0. [DOI] [PubMed] [Google Scholar]
- 23.Xiong Y., Lu Y. Deep feature extraction from the vocal vectors using sparse autoencoders for Parkinson's classification. IEEE Access. 2020;8:27821–27830. doi: 10.1109/ACCESS.2020.2968177. [DOI] [Google Scholar]
- 24.Monge-Alvarez J., Hoyos-Barcelo C., San-Jose-Revuelta L.M., Casaseca-de-la-Higuera P. A machine hearing system for robust cough detection based on a high-level representation of band-specific audio features. IEEE Trans. Biomed. Eng. Aug. 2019;66(8):2319–2330. doi: 10.1109/TBME.2018.2888998. [DOI] [PubMed] [Google Scholar]
- 25.Kumar A., et al. Towards cough sound analysis using the Internet of things and deep learning for pulmonary disease prediction. Trans. Emerg. Telecommun. Technol. Oct. 2022;33(10) doi: 10.1002/ett.4184. [DOI] [Google Scholar]
- 26.Sakar C.O., et al. A comparative analysis of speech signal processing algorithms for Parkinson's disease classification and the use of the tunable Q-factor wavelet transform. Appl. Soft Comput. Jan. 2019;74:255–263. doi: 10.1016/j.asoc.2018.10.022. [DOI] [Google Scholar]
- 27.Lella K.K., Pja A. Automatic diagnosis of COVID-19 disease using deep convolutional neural network with multi-feature channel from respiratory sound data: cough, voice, and breath. Alex. Eng. J. 2022;61(2):1319–1334. doi: 10.1016/j.aej.2021.06.024. [DOI] [Google Scholar]
- 28.Sait U., et al. A deep-learning based multimodal system for Covid-19 diagnosis using breathing sounds and chest X-ray images. Appl. Soft Comput. 2021;109 doi: 10.1016/j.asoc.2021.107522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Erdoğan Y.E., Narin A. COVID-19 detection with traditional and deep features on cough acoustic signals. Comput. Biol. Med. 2021;136(August) doi: 10.1016/j.compbiomed.2021.104765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown C., et al. Exploring automatic diagnosis of COVID-19 from crowdsourced respiratory sound data. Proc. ACM SIGKDD Int. Conf. Knowl. Discov. Data Min. 2020:3474–3484. doi: 10.1145/3394486.3412865. [DOI] [Google Scholar]
- 31.Chaudhari G., et al. Virufy: global applicability of crowdsourced and clinical datasets for AI detection of COVID-19 from cough audio samples. 2020. http://arxiv.org/abs/2011.13320 [Online]. Available:
- 32.Sharma N., et al. Coswara - a database of breathing, cough, and voice sounds for COVID-19 diagnosis. Proc. Annu. Conf. Int. Speech Commun. Assoc. INTERSPEECH. 2020;2020-Octob:4811–4815. doi: 10.21437/Interspeech.2020-2768. [DOI] [Google Scholar]
- 33.Fakhry A., Jiang X., Xiao J., Chaudhari G., Han A., Khanzada A. Virufy: a multi-branch deep learning network for automated detection of COVID-19. Mar. 2021. http://arxiv.org/abs/2103.01806 [Online]. Available:
- 34.Orlandic L., Teijeiro T., Atienza D. The COUGHVID crowdsourcing dataset, a corpus for the study of large-scale cough analysis algorithms. Sci. Data. 2021;8(1):2–11. doi: 10.1038/s41597-021-00937-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bagad P., et al. 2020. Cough against COVID: evidence of COVID-19 signature in cough sounds.http://arxiv.org/abs/2009.08790 no. Ml. [Online]. Available: [Google Scholar]
- 36.Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andreu-Perez J., et al. A generic deep learning based cough analysis system from clinically validated samples for point-of-need covid-19 test and severity levels. IEEE Trans. Serv. Comput. 2022;15(3):1220–1232. doi: 10.1109/TSC.2021.3061402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Coppock H., Gaskell A., Tzirakis P., Baird A., Jones L., Schuller B. End-to-end convolutional neural network enables COVID-19 detection from breath and cough audio: a pilot study. BMJ Innov. 2021;7(2):356–362. doi: 10.1136/bmjinnov-2021-000668. [DOI] [PubMed] [Google Scholar]
- 39.Mohammed E.A., Keyhani M., Sanati-Nezhad A., Hejazi S.H., Far B.H. An ensemble learning approach to digital corona virus preliminary screening from cough sounds. Sci. Rep. 2021;11(1):1–11. doi: 10.1038/s41598-021-95042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mouawad P., Dubnov T., Dubnov S. Robust detection of COVID-19 in cough sounds: using recurrence dynamics and variable markov model. SN Comput. Sci. 2021;2(1):1–13. doi: 10.1007/s42979-020-00422-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lee G.T., Nam H., Kim S.H., Choi S.M., Kim Y., Park Y.H. Deep learning based cough detection camera using enhanced features. Expert Syst. Appl. 2022;206(June) doi: 10.1016/j.eswa.2022.117811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bansal V., Pahwa G., Kannan N. 2020 IEEE Int. Conf. Comput. Power Commun. Technol. GUCON 2020. 2020. Cough classification for COVID-19 based on audio mfcc features using convolutional neural networks; pp. 604–608. [DOI] [Google Scholar]
- 43.Dunne R., Morris T., Harper S. 2020. High Accuracy Classification of COVID-19 Coughs Using Mel-Frequency Cepstral Coefficients and a Convolutional Neural Network with a Use Case for Smart Home Devices. [Google Scholar]
- 44.Juillerat N., Hirsbrunner B. Low latency audio pitch shifting in the frequency domain. ICALIP 2010 - 2010 Int. Conf. Audio, Lang. Image Process. Proc. 2010:16–24. doi: 10.1109/ICALIP.2010.5685027. [DOI] [Google Scholar]
- 45.Santacruz J.L., Tardón L.J., Barbancho I., Barbancho A.M. Spectral envelope transformation in singing voice for advanced pitch shifting. Appl. Sci. 2016;6(11) doi: 10.3390/app6110368. [DOI] [Google Scholar]
- 46.Bahuleyan H. Apr. 2018. Music Genre Classification Using Machine Learning Techniques; pp. 1–4.http://arxiv.org/abs/1804.01149 [Online]. Available: [Google Scholar]
- 47.Kronvall T., Juhlin M., Swärd J., Adalbjörnsson S.I., Jakobsson A. Sparse modeling of chroma features. Signal Process. 2017;130:105–117. doi: 10.1016/j.sigpro.2016.06.020. [DOI] [Google Scholar]
- 48.Chittaragi N.B., Koolagudi S.G. Dialect identification using chroma-spectral shape features with ensemble technique. Comput. Speech Lang. 2021;70 doi: 10.1016/j.csl.2021.101230. [DOI] [Google Scholar]
- 49.Kattel M., Nepal A., Shah A.K., Shrestha D.C. Encycl. GIS; 2019. Chroma Feature Extraction; pp. 1–9. no. January. [Google Scholar]
- 50.Nam U. Special area exam Part II. https://ccrma.stanford.edu/~unjung/AIR/areaExam.pdf
- 51.Xu X., Cai J., Yu N., Yang Y., Li X. Effect of loudness and spectral centroid on the music masking of low frequency noise from road traffic. Appl. Acoust. 2020;166 doi: 10.1016/j.apacoust.2020.107343. [DOI] [Google Scholar]
- 52.Sharma G., Umapathy K., Krishnan S. Trends in audio signal feature extraction methods. Appl. Acoust. 2020;158 doi: 10.1016/j.apacoust.2019.107020. [DOI] [Google Scholar]
- 53.Karatana A., Yıldız O. 2017. Music Genre Classification with Machine Learning Techniques; pp. 2–5. [Google Scholar]
- 54.Xie Z., Liu Y., Li J., Zhang R., He M., Miao D. Influence of the interferometric spectral bandwidth on the precision of large-scale dual-comb ranging. Measurement. 2023;215(January) doi: 10.1016/j.measurement.2023.112842. [DOI] [Google Scholar]
- 55.Rajesh S., Nalini N.J. Musical instrument emotion recognition using deep recurrent neural network. Procedia Comput. Sci. 2020;167(Iccids 2019):16–25. doi: 10.1016/j.procs.2020.03.178. [DOI] [Google Scholar]
- 56.Müller M. Vol. 2. Heidelb. Springer; 2007. (Information Retrieval for Music and Motion). [Google Scholar]
- 57.Sahidullah M., Saha G. Design, analysis and experimental evaluation of block based transformation in MFCC computation for speaker recognition. Speech Commun. May 2012;54(4):543–565. doi: 10.1016/j.specom.2011.11.004. [DOI] [Google Scholar]
- 58.Hu Ning, Dannenberg R.B., Tzanetakis G. IEEE Workshop on Applications of Signal Processing to Audio and Acoustics. IEEE Cat. No.03TH8684; 2003. Polyphonic audio matching and alignment for music retrieval; pp. 185–188. [DOI] [Google Scholar]
- 59.Mukherjee H., Obaidullah S.M., Santosh K.C., Phadikar S., Roy K. A lazy learning-based language identification from speech using MFCC-2 features. Int. J. Mach. Learn. Cybern. Jan. 2020;11(1):1–14. doi: 10.1007/s13042-019-00928-3. [DOI] [Google Scholar]
- 60.Tirumala S.S., Shahamiri S.R., Garhwal A.S., Wang R. Speaker identification features extraction methods: a systematic review. Expert Syst. Appl. Dec. 2017;90:250–271. doi: 10.1016/j.eswa.2017.08.015. [DOI] [Google Scholar]
- 61.Peerzade G.N., Deshmukh R.R., Waghmare S.D. A review speech emotion recognition. Int. J. Comput. Sci. Eng. Mar. 2018;6(3):400–402. doi: 10.26438/ijcse/v6i3.400402. [DOI] [Google Scholar]
- 62.Krueger A., Haeb-Umbach R. Model-based feature enhancement for reverberant speech recognition. IEEE Trans. Audio Speech Lang. Process. Sep. 2010;18(7):1692–1707. doi: 10.1109/TASL.2010.2049684. [DOI] [Google Scholar]
- 63.Korkmaz Y., Boyacı A., Tuncer T. Turkish vowel classification based on acoustical and decompositional features optimized by Genetic Algorithm. Appl. Acoust. Nov. 2019;154:28–35. doi: 10.1016/j.apacoust.2019.04.027. [DOI] [Google Scholar]
- 64.Solana-Lavalle G., Rosas-Romero R. Analysis of voice as an assisting tool for detection of Parkinson's disease and its subsequent clinical interpretation. Biomed. Signal Process Control. 2021;66 doi: 10.1016/j.bspc.2021.102415. [DOI] [Google Scholar]
- 65.Prabha A., Yadav J., Rani A., Singh V. Design of intelligent diabetes mellitus detection system using hybrid feature selection based XGBoost classifier. Comput. Biol. Med. 2021;136(July) doi: 10.1016/j.compbiomed.2021.104664. [DOI] [PubMed] [Google Scholar]
- 66.Hawi S., Alhozami J., AlQahtani R., AlSafran D., Alqarni M., El Sahmarany L. Automatic Parkinson's disease detection based on the combination of long-term acoustic features and Mel frequency cepstral coefficients (MFCC) Biomed. Signal Process Control. 2022;78(May) doi: 10.1016/j.bspc.2022.104013. [DOI] [Google Scholar]
- 67.Abbasi Baharanchi S., Vali M., Modaresi M. Noise reduction of lung sounds based on singular spectrum analysis combined with discrete cosine transform. Appl. Acoust. 2022;199 doi: 10.1016/j.apacoust.2022.109005. [DOI] [Google Scholar]
- 68.Muroi Y., Suda S. Binomial tree method for option pricing: discrete cosine transform approach. Math. Comput. Simulat. 2022;198:312–331. doi: 10.1016/j.matcom.2022.02.032. [DOI] [Google Scholar]
- 69.LeCun Y., Bengio Y., Hinton G. Deep learning,” Nature. May 2015;521(7553):436–444. doi: 10.1038/nature14539. [DOI] [PubMed] [Google Scholar]
- 70.Behrad F., Saniee Abadeh M. An overview of deep learning methods for multimodal medical data mining. Expert Syst. Appl. Aug. 2022;200 doi: 10.1016/j.eswa.2022.117006. [DOI] [Google Scholar]
- 71.Sharma A., Lysenko A., Boroevich K.A., Vans E., Tsunoda T. DeepFeature: feature selection in nonimage data using convolutional neural network. Briefings Bioinf. Nov. 2021;22(6) doi: 10.1093/bib/bbab297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Li Z., Liu F., Yang W., Peng S., Zhou J. A survey of convolutional neural networks: analysis, applications, and prospects. IEEE Transact. Neural Networks Learn. Syst. Dec. 2022;33(12):6999–7019. doi: 10.1109/TNNLS.2021.3084827. [DOI] [PubMed] [Google Scholar]
- 73.Subramanian N., Elharrouss O., Al-Maadeed S., Chowdhury M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. Apr. 2022;143 doi: 10.1016/j.compbiomed.2022.105233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.No Title,” doi: 10.1186/s12942-019-0193-9.
- 75.Sarıgül M., Ozyildirim B.M., Avci M. Differential convolutional neural network. Neural Network. Aug. 2019;116:279–287. doi: 10.1016/j.neunet.2019.04.025. [DOI] [PubMed] [Google Scholar]
- 76.Gu J., et al. Recent advances in convolutional neural networks. Pattern Recogn. 2018;77:354–377. doi: 10.1016/j.patcog.2017.10.013. [DOI] [Google Scholar]
- 77.Szegedy C., et al. Going deeper with convolutions. IEEE Comput. Soc. Conf. Comput. Vis. Pattern Recogn. 2015;07–12(June):1–9. doi: 10.1109/CVPR.2015.7298594. [DOI] [Google Scholar]
- 78.Ucar F., Korkmaz D. COVIDiagnosis-Net: deep Bayes-SqueezeNet based diagnosis of the coronavirus disease 2019 (COVID-19) from X-ray images. Med. Hypotheses. 2020;140(April) doi: 10.1016/j.mehy.2020.109761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Iandola F.N., Han S., Moskewicz M.W., Ashraf K., Dally W.J., Keutzer K. 2016. SqueezeNet: AlexNet-level accuracy with 50x fewer parameters and <0.5MB model size.http://arxiv.org/abs/1602.07360 1–13. [Online]. Available: [Google Scholar]
- 80.Budak Ü., Cömert Z., Çıbuk M., Şengür A. DCCMED-Net: densely connected and concatenated multi Encoder-Decoder CNNs for retinal vessel extraction from fundus images. Med. Hypotheses. Jan. 2020;134 doi: 10.1016/j.mehy.2019.109426. [DOI] [PubMed] [Google Scholar]
- 81.Nair V., Hinton G.E. Proceedings of the International Conference on Machine Learning. ICML); 2010. Rectified linear units improve restricted Boltzmann machines; pp. 807–814. [Google Scholar]
- 82.Xu B., Wang N., Chen T., Li M. Empirical evaluation of rectified activations in convolutional network. 2015. http://arxiv.org/abs/1505.00853 [Online]. Available:
- 83.Dombi J., Jónás T. Generalizing the sigmoid function using continuous-valued logic. Fuzzy Set Syst. 2022;449:79–99. doi: 10.1016/j.fss.2022.02.010. [DOI] [Google Scholar]
- 84.Gao B., Pavel L. Apr. 2017. On the Properties of the Softmax Function with Application in Game Theory and Reinforcement Learning; pp. 1–10.http://arxiv.org/abs/1704.00805 [Online]. Available: [Google Scholar]
- 85.Ioffe S., Szegedy C. Batch normalization: accelerating deep network training by reducing internal covariate shift. Feb. 2015. http://arxiv.org/abs/1502.03167 [Online]. Available:
- 86.Ihsanto E., Ramli K., Sudiana D., Gunawan T.S. An efficient algorithm for cardiac arrhythmia classification using ensemble of depthwise separable convolutional neural networks. Appl. Sci. Jan. 2020;10(2):483. doi: 10.3390/app10020483. [DOI] [Google Scholar]
- 87.Subramanian N., Elharrouss O., Al-Maadeed S., Chowdhury M. A review of deep learning-based detection methods for COVID-19. Comput. Biol. Med. Apr. 2022;143 doi: 10.1016/j.compbiomed.2022.105233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kingma D.P., Ba J.L. Adam: a method for stochastic optimization. 3rd Int. Conf. Learn. Represent. ICLR 2015 - Conf. Track Proc. 2015:1–15. [Google Scholar]
- 89.Laguarta J., Hueto F., Subirana B. COVID-19 artificial intelligence diagnosis using only cough recordings. IEEE Open J. Eng. Med. Biol. 2020;1:275–281. doi: 10.1109/OJEMB.2020.3026928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chaudhari G., et al. Virufy: global applicability of crowdsourced and clinical datasets for AI detection of COVID-19 from cough. Nov. 2020. http://arxiv.org/abs/2011.13320 [Online]. Available:
- 91.Hassan A., Shahin I., Alsabek M.B. 2020 International Conference on Communications, Computing, Cybersecurity, and Informatics (CCCI) Nov. 2020. COVID-19 detection system using recurrent neural networks; pp. 1–5. [DOI] [Google Scholar]
- 92.Harvill J., Wani Y.R., Hasegawa-Johnson M., Ahuja N., Beiser D., Chestek D. Interspeech 2021. Aug. 2021. Classification of COVID-19 from cough using autoregressive predictive coding pretraining and spectral data augmentation; pp. 926–930. [DOI] [Google Scholar]
- 93.Tena A., Clarià F., Solsona F. Automated detection of COVID-19 cough. Biomed. Signal Process Control. 2022;71 doi: 10.1016/j.bspc.2021.103175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Xue H., Salim F.D. Exploring self-supervised representation ensembles for COVID-19 cough classification. Association for Computing Machinery. 2021;1(1) [Google Scholar]
- 95.Wang C.-Y., Mark Liao H.-Y., Wu Y.-H., Chen P.-Y., Hsieh J.-W., Yeh I.-H. 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW) Jun. 2020. CSPNet: a new backbone that can enhance learning capability of CNN; pp. 1571–1580. [DOI] [Google Scholar]
- 96.Mahmud T., Rahman M.A., Fattah S.A. CovXNet: a multi-dilation convolutional neural network for automatic COVID-19 and other pneumonia detection from chest X-ray images with transferable multi-receptive feature optimization. Comput. Biol. Med. Jul. 2020;122 doi: 10.1016/j.compbiomed.2020.103869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Khan A.I., Shah J.L., Bhat M.M. CoroNet: a deep neural network for detection and diagnosis of COVID-19 from chest x-ray images. Comput. Methods Progr. Biomed. Nov. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Monshi M.M.A., Poon J., Chung V., Monshi F.M. CovidXrayNet: optimizing data augmentation and CNN hyperparameters for improved COVID-19 detection from CXR. Comput. Biol. Med. Jun. 2021;133 doi: 10.1016/j.compbiomed.2021.104375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Rajendra Acharya U. Automated detection of COVID-19 cases using deep neural networks with X-ray images. Comput. Biol. Med. Jun. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barnawi A., Chhikara P., Tekchandani R., Kumar N., Boulares M. A CNN-based scheme for COVID-19 detection with emergency services provisions using an optimal path planning. Multimed. Syst. Jul. 2021 doi: 10.1007/s00530-021-00833-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Chaddad A., Hassan L., Desrosiers C. Deep CNN models for predicting COVID-19 in CT and x-ray images. J. Med. Imaging. Apr. 2021;8(S1) doi: 10.1117/1.JMI.8.S1.014502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Chhikara P., Gupta P., Singh P., Bhatia T. A deep transfer learning based model for automatic detection of COVID-19 from chest X-ray. Turk. J. Electr. Eng. Comput. Sci. Oct. 2021;29(SI-1):2663–2679. doi: 10.3906/elk-2104-184. [DOI] [Google Scholar]
- 103.Tan W., Liu J. 2021 IEEE/CVF International Conference on Computer Vision Workshops (ICCVW) Oct. 2021. A 3D CNN network with BERT for automatic COVID-19 diagnosis from CT-scan images; pp. 439–445. [DOI] [Google Scholar]