Abstract
Background and Aims
Polymorphisms near the membrane bound O-acyltransferase domain containing 7 (MBOAT7) genes are associated with worsened nonalcoholic fatty liver (NASH), and nonalcoholic fatty liver disease (NAFLD)/NASH may decrease MBOAT7 expression independent of these polymorphisms. We hypothesized that enhancing MBOAT7 function would improve NASH.
Methods
Genomic and lipidomic databases were mined for MBOAT7 expression and hepatic phosphatidylinositol (PI) abundance in human NAFLD/NASH. Male C57BL6/J mice were fed either choline-deficient high-fat diet or Gubra Amylin NASH diet and subsequently infected with adeno-associated virus expressing MBOAT7 or control virus. NASH histological scoring and lipidomic analyses were performed to assess MBOAT7 activity, hepatic PI, and lysophosphatidylinositol (LPI) abundance.
Results
Human NAFLD/NASH decreases MBOAT7 expression and hepatic abundance of arachidonate-containing PI. Murine NASH models display subtle changes in MBOAT7 expression, but significantly decreased activity. After MBOAT7 overexpression, liver weights, triglycerides, and plasma alanine and aspartate transaminase were modestly improved by MBOAT7 overexpression, but NASH histology was not improved. Despite confirmation of increased activity with MBOAT7 overexpression, content of the main arachidonoylated PI species was not rescued by MBOAT7 although the abundance of many PI species was increased. Free arachidonic acid was elevated but the MBOAT7 substrate arachidonoyl-CoA was decreased in NASH livers compared to low-fat controls, likely due to the decreased expression of long-chain acyl-CoA synthetases.
Conclusion
Results suggest decreased MBOAT7 activity plays a role in NASH, but MBOAT7 overexpression fails to measurably improve NASH pathology potentially due to the insufficient abundance of its arachidonoyl-CoA substrate.
Keywords: NAFLD, NASH, Phosphatidylinositol, Arachidonic Acid, Steatosis
Introduction
Nonalcoholic fatty liver disease (NAFLD)/nonalcoholic fatty liver (NASH) represents a large health care burden due to the risk of progression to cirrhosis and liver failure and lack of approved therapies.1,2 In addition to being considered the hepatic manifestation of the metabolic syndrome, it is becoming more appreciated that genetic factors can play a significant role in NAFLD presence and progression.3,4 One example is a common polymorphism (rs641738) in membrane bound O-acyltransferase domain containing 7 (MBOAT7) associated with increased NAFLD pathology.5, 6, 7 This MBOAT7 rs641738 C>T variant is considered loss-of-function, resulting in decreased MBOAT7 expression.5
MBOAT7 is a membrane-anchored lysophosphatidylinositol (LPI) acyltransferase (LPIAT), which combines an acyl group with LPI to form phosphatidylinositol (PI).8,9 MBOAT7 appears to have selectivity for long-chain polyunsaturated fatty acids such as arachidonic acid,8,10,11 meaning that while total PI levels may not be altered, the abundance of specific unsaturated PI species are significantly regulated by MBOAT7.
In agreement with human genetic studies, cell culture and mouse models suggest that MBOAT7 deficiency leads to hepatocellular lipid accumulation and increased liver injury and fibrosis.12, 13, 14, 15, 16 Several reports suggest this increase in hepatic triglyceride with loss of MBOAT7 stems from enhanced sterol regulatory element binding protein 1 (SREBP1) expression/activity and increased de novo lipogenesis,13,16 while another study suggests a route for PI conversion to diacylglycerol then triglyceride.14
It was reported that obesity, presumably associated with NAFLD, decreases hepatic MBOAT7 expression independent of the rs641738 polymorphism.12 Based on these previous human, rodent, and cell culture studies of MBOAT7 deficiency, we hypothesized that enhancing MBOAT7 expression and activity would improve NASH pathology.
Results
Human NAFLD and NASH are associated with decreased MBOAT7 expression and activity
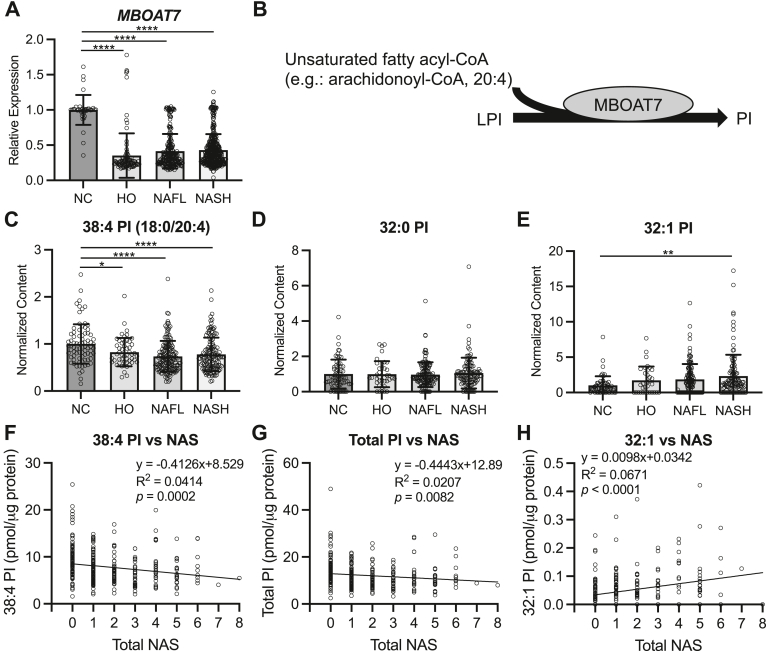
A previous report observed dramatically decreased hepatic MBOAT7 expression in a small set of obese individuals, presumably with NAFLD, compared to lean controls.12 To assess if this decrease in MBOAT7 held true in larger cohorts of patients with confirmed NAFLD or NASH, we mined Gene Expression Omnibus datasets for MBOAT7 expression. Indeed, livers from obese patients without steatosis (healthy obese [HO]), or patients with biopsy confirmed NAFL or NASH displayed significantly reduced MBOAT7 compared to livers from lean normal controls (NC) (Figure 1A). Interestingly, MBOAT7 expression was not significantly different between HO, NAFL, or NASH groups (Figure 1A). MBOAT7 transfers an acyl group, thought to be predominantly arachidonic acid,8,10 to LPI to form PI (Figure 1B). To investigate MBOAT7 activity in human NAFLD we also mined available lipidomic studies in which hepatic PI was measured. The predominant PI species in the liver contains arachidonic acid (20:4), and this 38:4 PI was significantly decreased in HO, NAFL, and NASH livers compared to NC (Figure 1C), suggesting decreased MBOAT7 activity. Other PI species measured, which are not believed to be MBOAT7 products, such as 32:0 PI were unchanged across the spectrum of NASH (Figure 1D). On the other hand, 32:1 PI, which is also not believed to be an MBOAT7 product, was significantly increased in NASH compared to NC livers (Figure 1E). Interestingly, although the MBOAT7 product 38:4 was not significantly reduced between HO and NAFL or NASH livers, both 38:4 PI and total PI levels displayed a significant negative association with total NAFLD activity score (NAS), suggesting that decreased hepatic MBOAT7 activity and certain PI levels associate with worsened NAFLD pathology (Figure 1F and G). On the other hand, 32:1 PI, which is not an MBOAT7 product displayed a significant positive association with total NAS (Figure 1H). Altogether, these results suggest that obesity and NAFLD are associated with reduced hepatic MBOAT7 gene expression and activity.
Figure 1.
Human NAFLD/NASH decreases hepatic MBOAT7 expression and activity. (A) Hepatic MBOAT7 expression combined from GSE89632, GSE126848, GSE135251, GSE167523, and GSE163211 reveals decreased MBOAT7 in humans with biopsy confirmation of healthy obesity (no steatosis; HO, n = 98), “simple steatosis” or NAFL (n = 225), or NASH (n = 391) compared to lean healthy control (HC, n = 38) livers. (B) Schematic showing the catalytic activity of MBOAT7 combining LPI and an unsaturated fatty acyl-CoA to form phosphatidylinositol (PI). (C) The abundant, arachidonoylated PI species (18:0/20:4) is decreased in HO, NAFL, and NASH compared to NC livers (n = 51, 160, 134, and 80, respectively). (D) Saturated 32:0 PI abundance is not altered in NAFL or NASH livers. (E) 32:1 PI is increased in NASH compared to NC livers. (F) The MBOAT7 product, 38:4 PI, measured in Ref. 17, is negatively associated with total NAFLD activity score (NAS). (G) Total PI, measured in Ref. 17, is negatively associated with total NAS. (H) 32:1 PI abundance is positively associated with NAS. Data presented as mean ± SD, analyzed by one-way ANOVA with Tukey’s post hoc correction for multiple comparisons, ∗P < .05, ∗∗P < .01, ∗∗∗∗P < .0001. ANOVA, analysis of variance; SD, standard deviation
Murine NASH also decreases MBOAT7 activity
To investigate whether murine models of NASH were also associated with changes in MBOAT7 expression and activity, we used the common choline-deficient amino acid defined high-fat diet (CDAHFD) and Gubra Amylin NASH (GAN) dietary models of NASH compared to low-fat (LF) control diet. Interestingly, both NASH diets decreased hepatic Mboat7 expression by 10%–20% (Figure A1A and D). However, only in the CDAHFD model was MBOAT7 hepatic protein expression modestly yet significantly reduced (Figure A1B and C), but unchanged in the GAN model (Figure A1E and F). Microsomes isolated from these livers also displayed only minor changes in MBOAT7 protein expression (Figure A1G andH). Despite the small changes in MBOAT7 expression in NASH livers, LPIAT activity with the MBOAT7 substrate, arachidonoyl-CoA, was significantly reduced in microsomes isolated from livers of both NASH diets compared to LF (Figure A1I and J). These NASH livers contained increased abundance of LPI (Figure A2A and D), and decreased abundance of PI (Figure A2B, C, E, and F) compared to LF diet livers, also consistent with decreased MBOAT7/LPIAT activity. Altogether, these results suggest that murine NASH models, despite having only minor reductions in MBOAT7 expression, display decreased MBOAT7 activity like in human NASH.
Hepatic MBOAT7 overexpression in mice modestly improves plasma and gene expression markers for NASH liver injury
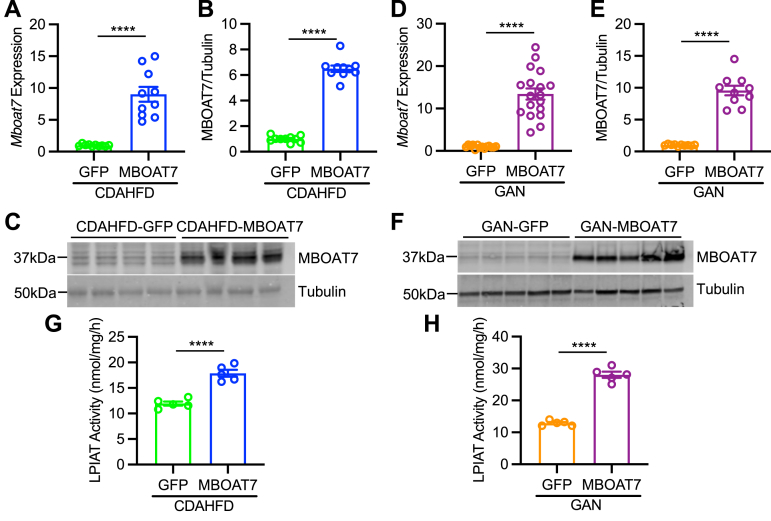
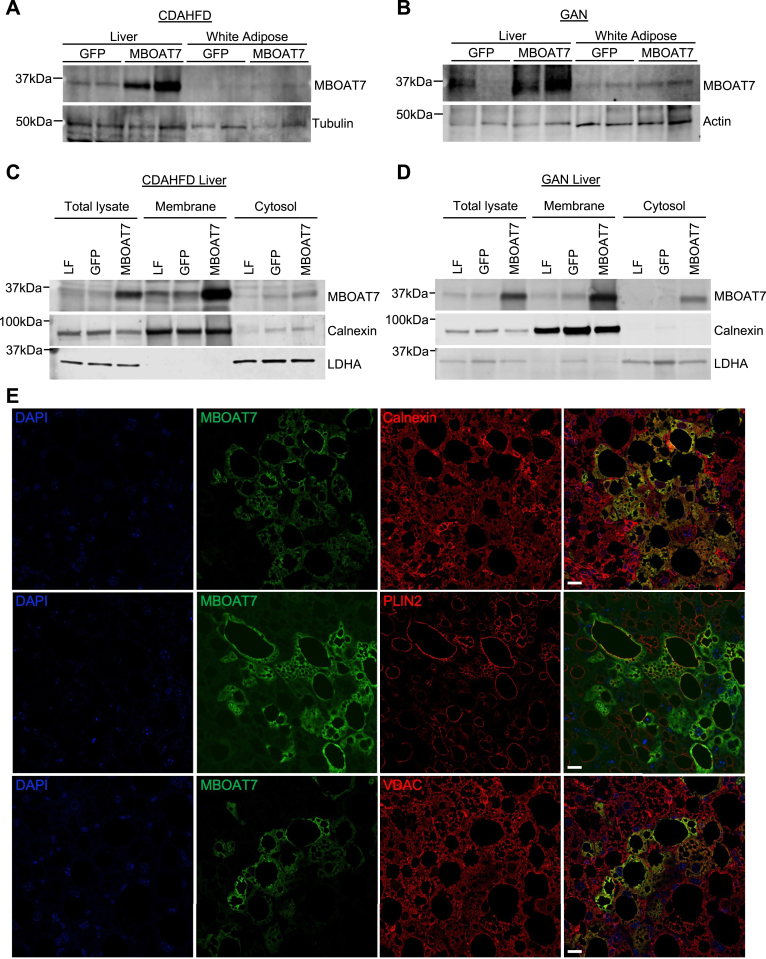
If reduced MBOAT7 expression or activity contributes to NAFLD pathology, we hypothesized that increasing MBOAT7 expression may improve NASH. To test this, we used the CDAHFD and GAN diet models of murine NASH, and infected mice with adeno-associated virus (AAV) to stably express MBOAT7 or green fluorescent protein (GFP) control in the liver. Adeno-associated virus serotype 8 (AAV8)-thyroid binding globulin (TBG)-MBOAT7 significantly enhanced hepatic Mboat7 RNA (Figure 2A and D) and MBOAT7 protein expression (Figure 2B, C, E, and F) compared to mice treated with AAV8-TBG-GFP control virus. This MBOAT7 overexpression led to a 2- to 3-fold increase in LPIAT activity in microsomes isolated from these livers, indicating enhanced MBOAT7 activity (Figure 2G and H). To assess liver-specific MBOAT7 overexpression, we also measured MBOAT7 expression in gonadal white adipose tissue and observed no increase in MBOAT7 in adipose of AAV8-TBG-MBOAT7 treated mice (Figure 3A and B). We next tested whether the overexpressed MBOAT7 localized properly to hepatic endomembranes (endoplasmic reticulum, mitochondrial-associated membranes, and lipid droplets).5,9 In both NASH models, the majority of overexpressed MBOAT7 was localized to the membrane fraction containing the endoplasmic reticulum marker calnexin (Figure 3C and D). Only small amounts of the overexpressed MBOAT7 appeared in the cytosolic fraction marked by lactate dehydrogenase (Figure 3C and D). We also assessed colocalization of MBOAT7 to endomembrane markers by immunostaining liver sections from AAV8-TBG-MBOAT7 treated mice. Hepatocytes with increased MBOAT7 staining displayed substantial colocalization with the endoplasmic reticulum marker calnexin, the lipid droplet membrane marker perilipin 2 (PLIN2), and to some extent the mitochondrial outer membrane marker voltage-dependent anion channe (VDAC) (Figure 3E). Thus, these results suggest the overexpressed MBOAT7 properly localized the endomembranes allowing it to increase LPIAT activity.
Figure 2.
MBOAT7 overexpression in murine NASH. Mice consuming NASH diets were infected with AAV to express either control GFP or MBOAT7. (A and D) Hepatic Mboat7 gene expression is overexpressed by AAV8-MBOAT7 in CDAHFD and GAN models, respectively. (B, C, E and F) Hepatic MBOAT7 expression in CDAHFD and GAN models, respectively (n = 10). (G and H) LPIAT activity, measured with 17:1 LPI and 20:4 acyl-CoA suggests increased MBOAT7 activity with MBOAT7 overexpression in the CDAHFD and GAN models, respectively. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18, except for G-H which n = 5 each. Data analyzed by two-tailed unpaired t-test, ∗∗∗∗P < .0001. SEM, standard error of the mean.
Figure 3.
MBOAT7 overexpression is liver-specific and properly localized to endomembranes. (A and B) Representative western blots from liver or gonadal white adipose tissue from CDAHFD and GAN studies, respectively, suggest liver-specific MBOAT7 overexpression. (C and D) Representative western blots from liver tissue homogenized into total lysate or purified into membrane and cytosolic fractions by differential centrifugation. MBOAT7 is predominantly localized to the membrane fraction, marked by the endoplasmic reticulum marker calnexin. A small amount of the overexpressed MBOAT7 appears in the cytosolic fraction, marked by lactate dehydrogenase A (LDHA) expression. For A–D, 3–4 independent western blots were performed. (E) Representative liver sections from MBOAT7 overexpressing mice immunostained for MBOAT7, the endoplasmic reticulum marker calnexin, the lipid droplet membrane marker PLIN2, and the outer mitochondrial membrane marker VDAC, suggesting colocalization with these endomembrane markers. Scale bar = 10 μm.
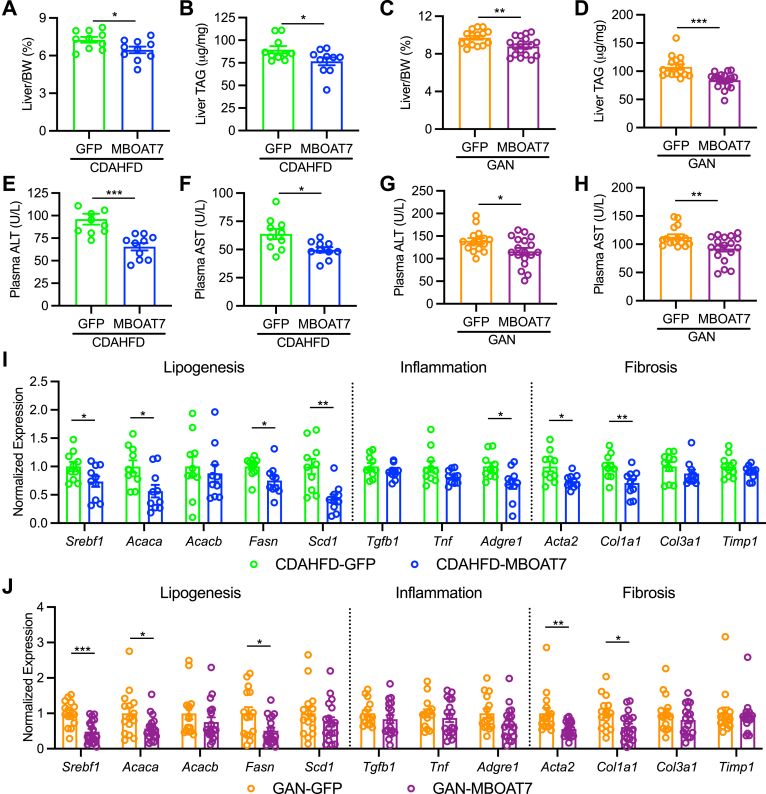
MBOAT7 overexpression in either NASH diet modestly but significantly improved liver weights and hepatic triglyceride (TAG) accumulation (Figure 4A–D). Likewise, the plasma markers for liver injury, alanine transaminase (ALT) and aspartate transaminase (AST), were significantly improved by MBOAT7 overexpression (Figure 4E–H). Previous studies have suggested MBOAT7 deficiency leads to hepatic steatosis from the activation of SREBP and lipogenesis.13,16 Indeed, MBOAT7 overexpression decreased the expression of Srebf1 and several lipogenic target genes (Figure 4I and J), suggesting decreased lipogenesis could explain the small but significant reductions in hepatic TAG with MBOAT7 overexpression. Despite significant improvements in serum markers of hepatic injury, gene expression for inflammatory cytokines and monocyte/macrophage markers were not improved by MBOAT7 overexpression other than Adgre1 in the CDAHFD model (Figure 4I and J). Lastly, some hepatic gene expression markers of stellate cell activation and fibrogenesis were significantly reduced in livers with MBOAT7 overexpression (Acta2 and Col1a1), while others were not improved in MBOAT7 livers (Col3a1 and Timp1, Figure 4I and J). Altogether, these results suggest that hepatic MBOAT7 overexpression may have small but significant beneficial effects on NASH pathology.
Figure 4.
MBOAT7 overexpression improves liver triglycerides and circulating ALT and AST markers of NASH liver injury. (A and C) Liver weights normalized to body weight (BW) are modestly but significantly decreased by MBOAT7 overexpression. (B and D) Liver triglyceride (TAG) concentrations are modestly yet significantly decreased by MBOAT7 overexpression. (E–H) Plasma levels of the liver injury markers alanine transaminase (ALT) and aspartate transaminase (AST) were significantly improved by MBOAT7 overexpression in both the CDAHFD and GAN models of NASH. (I and J) Hepatic gene expression for Srebf1 and lipogenic target genes was improved by MBOAT7 overexpression. Gene expression for inflammatory cytokines or macrophage/monocyte markers was unaffected by MBOAT7. Acta2 and Col1a1 expression are significantly improved in livers with MBOAT7 overexpression, while Col3a1 and Timp1 expression are not improved by MBOAT7 overexpression. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed unpaired t-test, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001. SEM, standard error of the mean
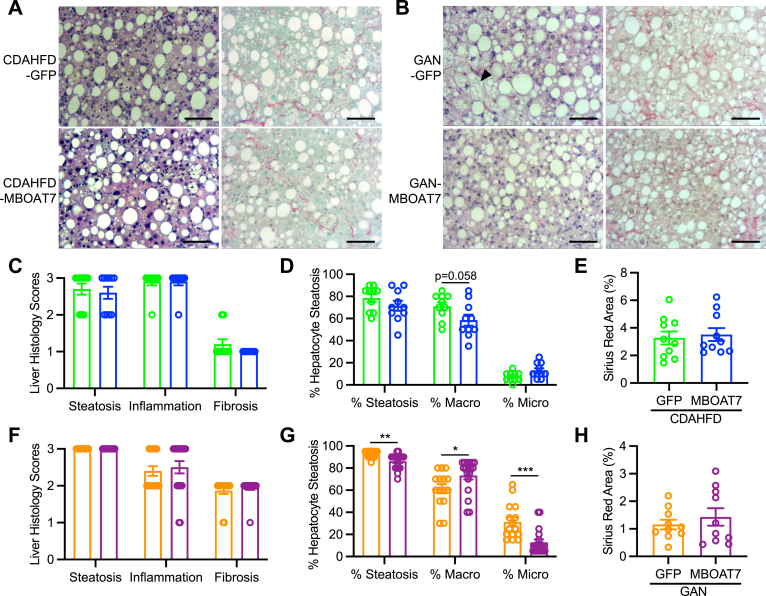
Hepatic MBOAT7 overexpression does not measurably improve liver histology
Histologic analyses confirmed the presence of extensive steatosis, inflammation, and fibrosis in both models of NASH (Figure 5A and B). Histologic scoring revealed that MBOAT7 overexpression did not improve any of these NAFLD histology indices in either NASH diet models (Figure 5C and F). Additionally, the percentage of hepatic steatosis was estimated, and characterized as either macrovesicular or microvesicular lipid droplets. The CDAHFD NASH model produced almost entirely macrovesicular steatosis (Figure 5A and D); however, the GAN diet resulted in nearly 40% microvesicular steatosis which was significantly reduced by MBOAT7 overexpression (Figure 5B and G). Digital quantification of the Picrosirius red staining validated the histology scores in that fibrosis was not improved by MBOAT7 overexpression (Figure 5E and H). In summary, hepatic MBOAT7 overexpression did not markedly improve NASH pathology.
Figure 5.
MBOAT7 overexpression does not improve categorical scoring of NASH histology. (A and B) Representative hematoxylin and eosin and Picrosirius red stains of livers from mice fed CDAHFD or GAN diet, respectively with GFP or MBOAT7 overexpression (10× magnification, scale bar = 100 μm). (C and F) Broad categorical liver histology scoring for steatosis, inflammation, and fibrosis in CDAHFD and GAN models, respectively suggests no improvement in histology with MBOAT7 overexpression. (D and G) Histological assessment of the percentage of hepatocytes with any steatosis, or macrosteatosis vs microsteatosis indicates that GAN diet induces a notable amount of microsteatosis (arrowhead in B) which is significantly reduced by MBOAT7 overexpression. (E and H) Digitally quantified Sirius Red-stained area suggests no improvement in fibrosis with MBOAT7 overexpression. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed unpaired t-test, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001.
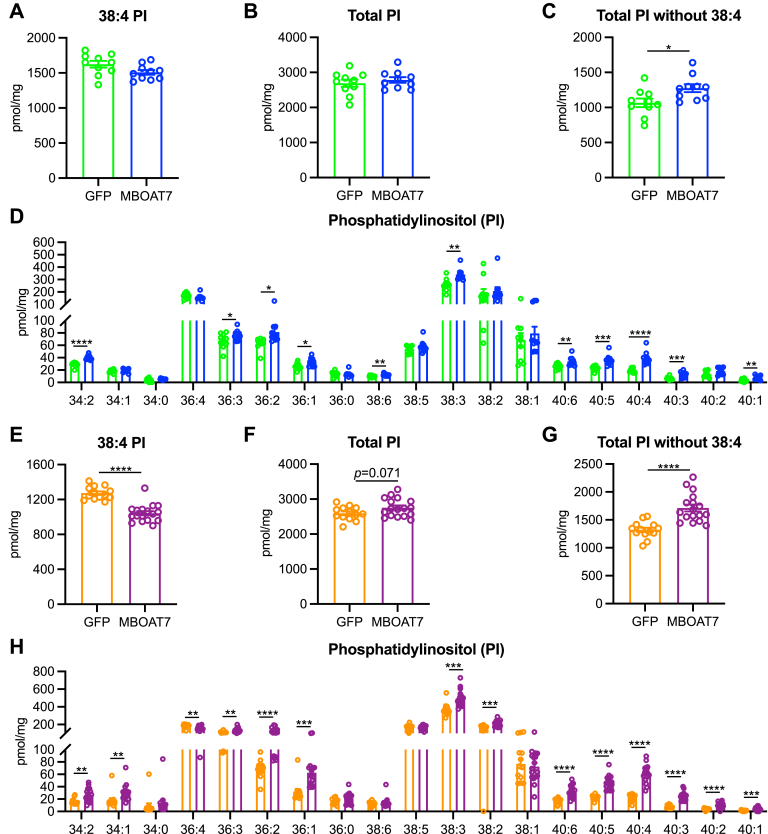
MBOAT7 overexpression alters various hepatic phosphatidylinositol levels
To further characterize the hepatic lipid changes with MBOAT7 overexpression, we measured liver LPI and PI concentrations by shotgun lipidomics. Increased MBOAT7 activity should reduce the abundance of LPI, yet to our surprise, total LPI levels were not affected by MBOAT7 (Figure A3A and C), which was driven by the abundant 18:0 LPI not being reduced by MBOAT7 overexpression (Figure A3B and D). However, a few less abundant LPI species were significantly reduced by MBOAT7 overexpression (Figure A3B and D). Arachidonate-containing 38:4 PI accounts for roughly half of the total PI in the liver. Since MBOAT7 is suggested to be a relatively specific acyltransferase for arachidonic acid,8,10,11 we expected this species to be increased by MBOAT7 overexpression. Surprisingly, 38:4 PI was not increased by MBOAT7 in either NASH model and was even significantly decreased in livers with MBOAT7 overexpression in the GAN diet (Figure 6A and E), which also lead to no significant increase in total hepatic PI levels with MBOAT7 overexpression (Figure 6B and F). However, many lesser-abundant PI species were significantly elevated by MBOAT7 overexpression in both NASH models, and if the predominant 38:4 PI specie is excluded, total PI levels were significantly increased by MBOAT7 overexpression (Figure 6C, D, G, and H). These data suggest that while MBOAT7 did not rescue the major arachidonate-containing PI species, other PI levels were significantly elevated by MBOAT7 overexpression in both NASH models.
Figure 6.
MBOAT7 overexpression increases many phosphatidylinositol species. (A and E) The most abundant PI specie (38:4) is not increased by MBOAT7 overexpression, and significantly decreased with MBOAT7 overexpression in the GAN model of NASH. (B and F) Total PI concentrations are not altered by MBOAT7 overexpression in the CDAHFD and GAN models of NASH, respectively. (C and G) If the abundant 38:4 PI specie is removed, total PI levels are increased by MBOAT7 in the CDAHFD and GAN models of NASH, respectively. (D and H) Many PI species are significantly increased by MBOAT7 overexpression in both CDAHFD and GAN models, respectively. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed unpaired t-test, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. SEM, standard error of the mean.
Our shotgun lipidomic analyses also measured hepatic lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylserine (PS), cholesteryl esters, ceramides, and sphingomyelins (Figures A4–A7). As MBOAT7 has been previously described to have specific LPIAT activity, unsurprisingly there were extremely few significant changes between GFP and MBOAT7 overexpression for these other lipids. Specifically, only 40:8 PC, 34:1 PE, 38:4 PS, and 18:3 cholesteryl ester were significantly altered in MBOAT7 livers in the CDAHFD model (Figures A4B–D and A5A), while only 34:3 PC, 16:1 and 16:0 cholesteryl esters, and 16:1 sphingomyelin were significantly altered by MBOAT7 in the GAN diet model (Figures A6B and A7A and C). Altogether, these results suggest that as expected, MBOAT7 overexpression predominantly affected the abundance of LPI and PI.
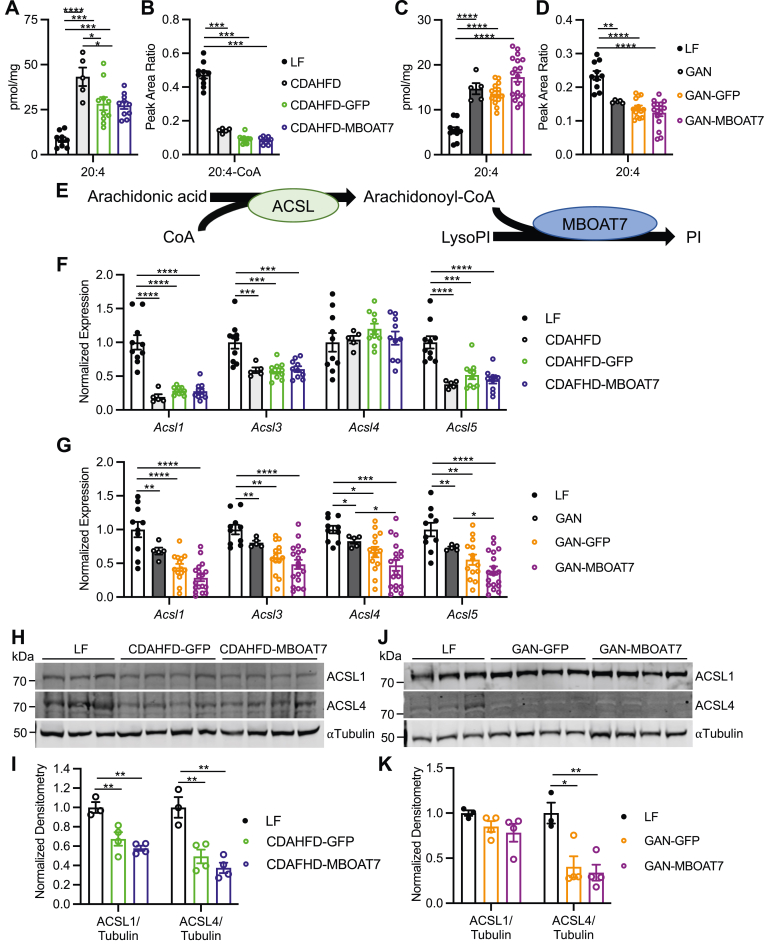
Murine NASH decreases arachidonic acid availability via decreased long-chain acyl-CoA synthetase expression
While MBOAT7 overexpression increased activity, this was not reflected in the hepatic LPI and PI levels. We hypothesized that arachidonic acid levels may be limited in NASH, helping to explain why MBOAT7 overexpression did not increase the main arachidonate-containing PI species. We first measured hepatic free fatty acid levels which uncovered an increase in 20:4 arachidonic acid in both NASH diets with or without AAV infection compared to LF diet (Figure 7A and C), and several other long-chain free fatty acids (Figure A8A and C). MBOAT7 overexpression did not affect this increase in arachidonic acid (Figure 7A and C). However, for arachidonate to become a substrate of MBOAT7, it needs to be activated by condensation with CoA, converting it into arachidonoyl-CoA. Therefore, we also measured long-chain acyl-CoAs in these livers, and 20:4 arachidonoyl-CoA, as well as most other acyl-CoAs, was significantly decreased by NASH diets with and without AAV infection compared to LF (Figures 7B and D and A8B and D). Thus, all NASH livers displayed relatively limited amounts of arachidonoyl-CoA, potentially explaining why MBOAT7 failed to enhance the abundance of the arachidonoylated 38:4 and 36:4 PI species.
Figure 7.
NASH decreases hepatic arachidonoyl-CoA concentrations due to decreased ACSL4. (A and C) Hepatic abundance shows increased levels of arachidonic acid (20:4) in the CDAHFD and GAN livers, respectively, which are unaltered by MBOAT7 overexpression. (B and D) Abundance of arachidonoyl-CoA (20:4-CoA) is decreased in the CDAHFD and GAN models of NASH, respectively, and not altered by MBOAT7 overexpression. (E) Schematic pathway of arachidonic acid incorporation into PI. (F and G) Hepatic gene expression of Acsl1, Acsl3, Acsl4, and Acsl5 are almost all significantly decreased in the CDAHFD and GAN models of NASH, respectively, and unaltered by MBOAT7 overexpression. (H–K) ACSL1 is decreased in the CDAHFD model of NASH and ACSL4 expression is decreased in both the CDAHFD and GAN NASH models compared to LF (n = 3–4 each). Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5, CDAHFD-GFP n = 10, CDAHFD-MBOAT7 n = 10; GAN study: LF n = 10, GAN n = 5, GAN-GFP n = 15, GAN-MBOAT7 n = 18. Data analyzed by one-way ANOVA with Tukey’s post hoc correction for multiple comparisons, ∗P < .05, ∗∗P < .01, ∗∗∗P < .001, ∗∗∗∗P < .0001. ANOVA, analysis of variance.
The enzymes responsible for ligating long-chain fatty acids with CoA belong to the long-chain acyl-CoA synthetase (ACSL) family (Figure 7E). Increased arachidonic acid levels and decreased arachidonoyl-CoA suggests decreased ACSL activity in NASH livers. There are several ACSL isoenzymes, with ACSL4 described as particularly important for arachidonic acid ligase activity.18 We first measured the hepatic gene expression of Acsl1, Acsl3, Acsl4, and Acsl5 in these livers, which all displayed significantly reduced expression in livers from both NASH diets compared to LF, except for Acsl4 in the CDAHFD model which was unchanged (Figure 7F and G). Protein expression for ACSL1 and ACSL4 was significantly decreased in the CDAHFD model (Figure 7H and I), while the GAN model of NASH decreased ACSL4 but not ACSL1 expression compared to LF (Figure 7J and K). In summary, these data from two dietary NASH models suggests that arachidonoyl-CoA is decreased in NASH potentially due to decreased ACSL expression and activity.
Discussion
A common polymorphism near MBOAT7 (rs641738), associates with increased hepatic steatosis and fibrosis,5, 6, 7 and is now widely considered a genetic risk allele for NASH.4,19 This C>T variant decreases MBOAT7 expression;5 however, another study suggested obesity decreases hepatic MBOAT7 independent of the rs641738 polymorphism.12 In this present study, we mined publicly available genomic and lipidomic databases to confirm that livers from humans with obesity, NAFLD, or NASH display decreased MBOAT7 expression and activity compared to lean controls. A wealth of recent data from cell and rodent models suggests that MBOAT7-deficiency induces hepatic steatosis and NASH injury and fibrosis.12, 13, 14, 15, 16 The exact mechanism for increased steatosis and injury with MBOAT7 deficiency remains unclear; however, several studies suggest increased SREBP1 activity and de novo lipogenesis.13,16 If decreased MBOAT7 activity is a driving factor for NASH liver injury, we postulated that increasing MBOAT7 expression and activity would improve NASH. We addressed this question by overexpressing MBOAT7 in the liver with AAV in two diet-induced models of NASH in mice. Unfortunately, while liver triglycerides, plasma ALT and AST, and some hepatic gene expression markers for fibrosis were improved, histologic measures of NASH were not improved by MBOAT7 in either model.
Since MBOAT7 is a LPIAT with a preference for arachidonic acid,8,10 we performed lipidomic analyses to measure LPI and PI species in these livers to assess the functional implications of MBOAT7 overexpression. Both NASH models resulted in decreased MBOAT7 activity and increased LPI and decreased PI, despite only minor decreases in MBOAT7 expression in these models compared to LF control livers. Therefore, these mouse models of NASH do not entirely recapitulate human NASH which involves more significant reductions in hepatic MBOAT7 expression. Two of the most abundant PI species contain arachidonic acid (36:4 and 38:4 PI), and would be expected to be increased by MBOAT7, yet to our surprise, the levels of 36:4 and 38:4 PI were not enhanced by MBOAT7 overexpression. However, many other PI species, including non-arachidonate containing species, were significantly elevated by MBOAT7 overexpression. In agreement with these results, lipidomic analyses performed in MBOAT7 knockout models have identified non-arachidonate containing PI species that are reduced with MBOAT7 deficiency.12, 13, 14,16 Additionally, a previous study observed significant LPIAT activity for MBOAT7 with 20:5 eicosapentaenoyl-CoA,10 and our lipidomic results of increased 40:5 and 40:6 PI with MBOAT7 overexpression support this observation. Thus, one conclusion that can be made from the present study is that in vivo, MBOAT7 may not be entirely specific to arachidonic acid.
In agreement with other studies,8,12, 13, 14, 15, 16 we observed extremely few alterations in other hepatic phospholipids, cholesteryl esters, ceramides, or sphingomyelin species from modulating MBOAT7 expression, suggesting it is specifically an LPI acyltransferase. Since MBOAT7 overexpression increased LPIAT activity but did not lead to the expected increase in hepatic 38:4 PI, we hypothesized that NASH livers had limited availability of arachidonic acid to incorporate into phospholipids. Indeed, although free arachidonic acid was significantly elevated, the activated form, arachidonoyl-CoA, which is a required intermediate for incorporation into phospholipids, was significantly reduced by both NASH diets compared to the LF diet. To our knowledge hepatic arachidonoyl-CoA levels have not previously been measured in human or rodent models of NAFLD/NASH. Several rodent studies also identified increased free arachidonic acid in NAFLD/NASH vs control livers;20,21 however, human data suggest decreased free arachidonate in NASH.20,22, 23, 24 Nevertheless, reduced arachidonate content in hepatic phospholipids and/or triglycerides has been observed in several studies of human NASH, in agreement with our current findings.17,20,24,25 Increased omega oxidation products of arachidonate have been demonstrated in plasma lipidomic studies of NASH patients, suggesting increased disposal may reduce arachidonate availability for incorporation into phospholipid species.26 ACSL4 is believed to play a specific role in arachidonoyl-CoA synthesis.18 Hepatic ACSL4 was significantly decreased in both of our NASH models and also decreased by high-fat diet feeding in mice.27 Interestingly, free arachidonic acid causes ACSL4 to be ubiquitinated and degraded, leading to decreased ACSL4 protein abundance.27,28 However, this decrease in hepatic ACSL4 in NAFLD/NASH may be restricted to rodent models, as increased ACSL4 has been identified in human NAFLD.29,30 Although we and others27 observed decreased ACSL4 in rodent models of NAFLD/NASH, a recent report described that ACSL4 deletion or pharmacologic inhibition in mice protected from NASH primarily by enhancing fat oxidation.30 Thus, more studies are required to gain a better understanding of the importance of ACSL4 and arachidonoyl-CoA in both humans and rodent models of NASH.
Aside from the rs641738 variant associated with NASH, several homozygous nonsense mutations in MBOAT7 have been identified which cause intellectual disability, epilepsy, and autism.31, 32, 33, 34, 35, 36 Whole-body MBOAT7 knockout mice may have similar phenotypes and die within the first month of life with atrophy of the cerebral cortex and hippocampus due to defective cortical lamination.8 While it remains to be tested, it is exciting to speculate on the potential utility of neuronal-targeting viral gene therapies to overcome these MBOAT7 mutations.
Conclusion
In summary, in this study we observed lipid changes suggesting a loss of MBOAT7 activity in two diet-induced models of murine NASH and confirmed decreased MBOAT7 activity. However, hepatic MBOAT7 overexpression was unable to substantially improve NASH pathology although liver triglyceride content and markers of liver injury were improved. Broad categorical histologic scoring may not be sensitive to these small changes, and indeed, when steatosis was stratified as microsteatosis vs macrosteatosis, there were identifiable changes with MBOAT7 overexpression. This lack of measurable histologic improvement may be due to the inability of MBOAT7 to enhance the major arachidonoylated PI (38:4 and 36:4) species, likely due to the low arachidonoyl-CoA levels from decreased ACSL4 expression in NASH livers. These findings suggest that MBOAT7 may not be an actionable target in NASH but set the stage for further studies related to the activity of ACSL4 and arachidonoyl-CoA in NASH.
Materials and methods
Human gene expression and metabolomic database analysis
We mined publicly available Gene Expression Omnibus datasets for studies of human liver microarray and RNA sequencing expression profiles in NAFLD and/or NASH and extracted expression data for MBOAT7. Data for MBOAT7 expression were combined from GSE89632,37 GSE126848,38 GSE135251,39 GSE167523,40 and GSE163211,41 and normalized to healthy control liver expression levels. In total, MBOAT7 expression was measured in livers from individuals with biopsy-determined status as NC (n = 38), HO (n = 98), “simple steatosis” or NAFL (n = 225), or NASH with varying degrees of fibrosis, including NASH cirrhosis (n = 391). We also mined publicly available lipidomic analyses for PI measured in human NAFLD/NASH compared to control livers. These included data deposited in Metabolomics Workbench ST000915,25 as well as supplemental data from an open access publication.17 Total PI levels as well as the predominant arachidonoylated PI specie (38:4) and several non-arachidonoylated species (32:0 and 32:1) were assessed in a total of 80 NC livers, 51 HO livers, 160 NAFL livers, and 134 NASH livers. Since the two studies were performed using different mass spectrometry methods and normalized in a different manner (protein concentration vs starting tissue weight), data for each study was normalized to NC prior to combining.
Animal studies
Animal care and experimentation was approved by the Institutional Animal Care and Use Committee of Saint Louis University and complied with criteria outlined in the Guide for the Care and Use of Laboratory Animals. Five-week-old male C57BL/6J mice were purchased from Jackson Laboratories (000664, The Jackson Laboratories, Bar Harbor, ME). Mice were housed in specific-pathogen-free, climate-controlled rooms with a 6:00–18:00 light on/off cycle. Mice were group housed, up to 5 per cage, with ad libitum access to water and food. At 6-weeks of age, mice were randomly chosen to consume either LF diet (D12450K, Research Diets, New Brunswick, NJ; 10% kcal fat, 20% kcal protein, and 70% kcal carbohydrate) or CDAHFD (Research Diets A06071309; 46% kcal fat, 18% kcal protein, and 36% kcal carbohydrate). After consuming diets for 6 weeks, a subset of CDAHFD-fed mice were infected with 7 × 1011 genome copies of AAV8 via intravenous injection of the tail vein. AAV8 was purchased from Vector Biolabs (Malvern, PA), and mice were randomly chosen to be infected with AAV8 expressing either a control GFP (catalog VB1743) or untagged murine Mboat7 (RefSeq BC023417, catalog AAV-264332), both under the control of the hepatocyte-specific thyroxine binding globulin promoter. Mice continued to consume special diets for another 4 weeks at which point they were fasted for 2–3 hours and euthanized by CO2 asphyxiation. Blood was collected from the abdominal aorta into an ethylenediaminetetraacetic acid (EDTA) coated tube and spun at 2000g for 10 minutes to collect plasma. The liver was excised, weighed, divided, and either snap frozen in liquid nitrogen, fixed in 10% neutral buffered formalin, or placed in 1 mL of RNAlater (Ambion, Austin, TX). A second set of 6-week old mice was fed either LF diet (D12450K) or GAN (Research Diets D09100310; 40% kcal fat from mostly palm oil, 20% kcal protein, and 40% kcal carbohydrate containing fructose, as well as 2% wt cholesterol). After 19-weeks on diet, a subset of GAN-fed mice were randomized to infection with AAV8-GFP or AAV8-MBOAT7 viral infection as described above. These mice consumed diets for 28 weeks total, then euthanized by CO2 asphyxiation after a 2–3 hour fast and plasma/tissue processed as described above. All diets used contained only trace amounts of 20:4 arachidonic acid (LF: 0.06 g of 20:4 in 45 g fat total, CDAHFD: 0.5 g of 20:4 in 202.5 g fat total, and GAN: 0.06 g of 20:4 in 180 g of fat total).
Gene expression analyses
Total RNA was isolated from ∼5 mg frozen liver tissue by homogenization in 1 mL RNA-STAT (TelTest, Friendswood, TX) with isopropanol and ethanol precipitation and analyzed as performed previously.42 Target gene cycle threshold (Ct) values were normalized to reference gene (Rplp0) Ct values by the 2−ΔΔCt method. Oligonucleotide primer sequences are listed in Table A1.
Protein expression analyzes
Protein extracts were prepared by homogenizing ∼50 mg liver tissue in lysis buffer (15 mM NaCl, 25 mM Tris base, 1 mM EDTA, 0.2% NP-40, and 10% glycerol), supplemented with protease and phosphatase inhibitors. To assess MBOAT7 localization, liver samples were fractionated by dounce homogenization and centrifuged 1,000g × 5 minutes at 4°C to pellet whole cells and nuclei, and then the supernatant centrifuged at 21,100g × 20 minutes at 4°C to separate membrane and cytosolic fractions. Protein concentration was measured by a MicroBCA kit (ThermoFisher Scientific, Waltham, MA). A total of 50 μg of protein was electrophoresed on Criterion 4%–15% precast polyacrylamide gels (Bio-Rad, Hercules, CA) and transferred onto polyvinyldifluoride membranes. Protein lysates were not boiled prior to electrophoresis for MBOAT7 immunoblots. Membranes were blocked for at least 1 hour in 5% bovine serum albumin (BSA)(MilliporeSigma, Burlington, MA) in tris-buffered saline with Tween-20 (TBST). Membranes were incubated with primary antibodies at 1:1000 dilution overnight at 4 °C. Membranes were then washed 5 × 5 minutes in TBST, and then incubated with secondary antibodies at 1:10,000 dilution in 5%-BSA-TBST for 1 hour. After washing 5 × 5 minutes in TBST, membranes were developed on an Odyssey imaging system and analyzed with Image Studio software (Li-Cor Biosciences, Lincoln, NE). Information for all antibodies used in western blotting and immunofluorescence imaging are listed in Table A2.
Plasma ALT and AST and hepatic triglyceride measurement
Plasma ALT and AST were measured with commercial assays as previously described.43 Liver TAG concentrations were measured as previously described43 by homogenizing ∼100 mg frozen liver in saline to provide 0.1 mg/μL. This liver homogenate was combined 1:1 with 1% sodium deoxycholate, vortexed, and incubated at 37°C for 5 minutes to solubilize lipids. TAG was then measured by colorimetric assay (TR22421, ThermoFisher Scientific, Waltham, MA).
Liver histology and scoring
Formalin-fixed liver was embedded in paraffin blocks and sectioned onto glass slides. Slides were stained by hematoxylin and eosin and Picrosirius red and were evaluated by a histopathologist blinded to diet and treatment groups. Slides were scored for steatosis, inflammation, and fibrosis using typical NAS criteria.44 In addition, total percentage of hepatocytes with steatosis, and stratification of macrosteatosis or microsteatosis was recorded. Percentage of Picrosirius red-stained fibrosis was also quantified digitally by averaging the red-stained area from at least 10 independent pictures of each slide using Image J software as performed previously.42
Liver immunofluorescence imaging
To assess localization of the overexpressed MBOAT7 protein, liver sections from MBOAT7 overexpression mice were immunostained for MBOAT7, as well as the endoplasmic reticulum marker calnexin, the lipid droplet membrane marker PLIN2, or the VDAC marker of the outer mitochondrial membrane using procedures similar to previously described.45 Antibody details are provided in Table A2. Primary antibodies for MBOAT7, calnexin, and VDAC were used at 1:100, while PLIN2 was used at 1:25 dilution. Fluorescent-conjugated secondary antibodies were all used at 1:400 dilution. Slides were imaged on an Andor Dragonfly spinning disk confocal microscope (OXFORD Instruments, Abingdon, United Kingdom) using a 63× objective with lasers for excitation of 405, 488, 594 nm.
Liver lipidomic analyses
Lipids were extracted by pulverizing ∼100 mg frozen liver and subjecting ∼15 mg tissue to a modified Bligh-Dyer extraction as performed previously46 after spiking in a cocktail of internal standards including di-20:0 PC, di-14:0 PE, di-14:0 PS, N17:0 sphingomyelin, 17:0 cholesteryl ester, 17:0 fatty acid, N17:0 ceramide, 17:0 lysophosphatidylcholine, 17:0–18:1-d5 phosphatidylinositol (PI; 850,111, Avanti Polar Lipids, Alabaster, AL), and 17:0-d5 LPI (LPI; 850,108, Avanti Polar Lipids, Alabaster, AL). Lipid extracts were diluted in methanol/chloroform (4/1, v/v) and lipid species quantified using shotgun lipidomics by electrospray ionization-mass spectrometry on a triple-quadrupole Quantum Ultra (ThermoFisher Scientific, Waltham, MA) as performed previously.47 Individual molecular species were quantified by comparing the ion intensities of the individual molecular species to that of the lipid class internal standard with additional corrections for type I and type II 13C isotope effects. 38:4 and 36:4 PI were confirmed as 18:0/20:4 and 16:0/20:4 by product ion scanning for individual fatty acid constituents for oleate, palmitate, and arachidonate at m/z of 281.3, 255.2, and 303.4, respectively, at collision energy of +35 eV.
Measurement of free fatty acids was performed as previously described.46 Briefly, 50 μL lipid extract was dried and resuspended in 100 μL 2.5% diisopropylethylamine and 5% pentafluorobenzyl bromide in acetonitrile. Samples were incubated at 45°C for 1 hour, allowed to cool at room temperature, and twice dried and resuspended in 1 mL ethyl acetate. After drying one final time, samples were suspended in 200 μL ethyl acetate and free fatty acids detected by gas chromatography-mass spectrometry and selected ion monitoring as previously performed.46
Acyl-CoAs were measured by homogenizing ∼100 mg frozen liver in water (0.25 g/mL), and protein precipitation performed to extract acyl-CoAs from 50 μL of homogenate in the presence of a d4-palmitoyl-CoA internal standard. Analysis of acyl-CoA was performed with a high-performance liquid chromatography system (Shimadzu 20A, Kyoto, Japan) coupled to a 6500QTRAP+ mass spectrometer (AB Sciex LLC, Framingham, MA) operated in positive multiple reaction monitoring mode. Data processing were conducted with Analyst 1.6.3 software. Quality control (QC) samples were prepared by pooling aliquots of study samples and this QC sample was injected between every ten study samples to monitor instrument performance. Only acyl-CoA species with coefficient of variance <15% of QC injections are reported. Relative quantification of acyl-CoA is reported as the peak area ratios of the analytes to the corresponding internal standard.
MBOAT7/LPIAT activity assay
LPIAT activity was measured using similar procedures as described previously.11 Briefly, ∼50 mg of liver tissue was homogenized in a glass dounce homogenizer in 1 mL of microsome isolation buffer (250 mM sucrose, 50 mM Tris-HCl, 1 mM EDTA, 20% w/v glycerol, pH∼7.4, plus 1X complete protease inhibitor). Lysates were centrifuged at 1000g for 5 minutes at 4°C to pellet unbroken cells and nuclei. Supernatant was transferred to new tubes and centrifuged at 100,000g for 1 hour at 4°C to pellet microsomes. This pellet was resuspended in 400 μL of assay buffer (150 mM NaCl, 10 mM Tris-HCl, 1 mM EDTA, pH ∼ 7.4), and protein concentration measured by a MicroBCA kit (ThermoFisher Scientific, Waltham, MA). For the activity assay, 20 μg of microsomes were placed in a total volume of 200 μL assay buffer containing 10 μM 17:1 LPI (850,103, Avanti Polar Lipids, Alabaster, AL), 30 μM arachidonoyl-CoA (20:4-CoA, 870,721, Avanti Polar Lipids, Alabaster, AL), and 12.5 μM fatty-acid free BSA, and incubated for 30 minutes at 37°C. Reaction was then terminated by modified Bligh-Dyer extraction as described previously with the addition of 100 ng 17:0–18:1-d5 PI internal standard (850,111, Avanti Polar Lipids, Alabaster, AL). PI was measured by mass spectrometry as described above and peak area of the product 17:1/20:4 PI normalized to 17:0–18:1-d5 PI standard, and mass converted to nmol/mg/h.
Statistical analyzes
Unless stated, data are presented as individual data points with mean ± standard error of the mean (S.E.M.). All datasets were analyzed for statistical significance by two-tailed unpaired t test for comparison of two groups, or one-way analysis of variance with post hoc analysis by Tukey’s correction for multiple comparisons using GraphPad Prism, Version 9.3.1. Adjusted P values < 0.05 were considered significant.
Acknowledgments:
We thank Drs. Hiroyuki Arai and Nozomu Kono for the gracious gift of the MBOAT7 antibody.
Authors' Contributions:
Martin C. Sharpe: data curation, formal analysis, investigation, methodology, writing–review and editing; Kelly D. Pyles: data curation, formal analysis, investigation, methodology, writing–review and editing; Taylor Hallcox: data curation, formal analysis, investigation, methodology, writing–review and editing; Dakota R. Kamm: data curation, formal analysis, investigation, methodology, writing–review and editing; Michaela Piechowski: data curation, formal analysis, investigation, methodology, writing–review and editing; Bryan Fisk: formal analysis, software, writing–review and editing; Carolyn J. Albert: data curation, formal analysis, investigation, methodology, supervision, validation, writing–review and editing; Danielle H. Carpenter: formal analysis, writing–review and editing; Barbara Ulmasov: conceptualization, data curation, formal analysis, investigation, methodology, project administration, supervision, visualization, writing–review and editing; David A. Ford: data curation, formal analysis, methodology, resources, project administration, supervision, writing–review and editing; Brent A. Neuschwander-Tetri: conceptualization, project administration, supervision, visualization, writing–review and editing; Kyle S. McCommis: conceptualization, data curation, formal analysis, funding acquisition, investigation, methodology, project administration, supervision, visualization, writing–original draft, writing–review and editing.
Footnotes
Conflicts of Interest: These authors disclose the following: Dr Neuschwander-Tetri declares COI, all nonrelevant to this project. Dr Tetri is an advisor/consultant for Akero, Alimentiv, Allergan, Allysta, Alnylam, Amgen, Arrowhead, Axcella, Boehringer Ingelheim, BMS, Coherus, Cymabay, Durect, Enanta, Fortress, Genfit, Gilead, Glympse, Hepeon, HighTide, HistoIndex, Innovo, Intercept, Ionis, LG Chem, Lipocine, Madrigal, Medimmune, Merck, Mirum, NGM, NovoNordisk, Novus Therapeutics, pH-Pharma, Sagimet, Target RWE, Theratechnologies, 89Bio; holds stock options for HepGene; and institutional research grants from Allergan, BMS, Celgene, Cirius Therapeutics, Enanta, Genfit, Gilead, HighTide, Intercept, Madrigal, NGM. The remaining authors disclose no conflicts.
Funding: Study funded by: Friends of the SLU Liver Center seed grant, NIHS10 OD025246, UL1 TR002345, and R00 HL136658.
Ethical Statement: The corresponding author, on behalf of all authors, jointly and severally, certifies that their institution has approved the protocol for any investigation involving humans or animals and that all experimentation was conducted in conformity with ethical and humane principles of research.
Data Transparency Statement: The data associated with this paper are included within the text, figures, and supplementary materials, and are also available upon request to the corresponding author.
Reporting Guidelines: ARRIVE/Care and Use of Laboratory Animals.
Preprint: An earlier version of this manuscript is posted to bioRxiv: https://doi.org/10.1101/2022.03.24.485677.
Material associated with this article can be found in the online version at https://doi.org/10.1016/j.gastha.2023.02.004.
Supplementary materials
Hepatic MBOAT7 activity is reduced in murine NASH models. (A) Hepatic Mboat7 expression in mice fed either low fat (LF) control or choline-deficient high-fat diet (CDAHFD). (B and C) MBOAT7 expression in livers from LF- or CDAHFD-fed mice. (D) Hepatic Mboat7 expression in mice fed either LF or Gubra Amylin NASH diet (GAN). (E and F) MBOAT7 expression in livers from LF- or GAN-fed mice. (G and H) Hepatic microsomal MBOAT7 expression in the CDAHFD and GAN models of NASH compared to LF, respectively. (I and J) Hepatic microsomal lysophosphatidylinositol (LPI) acyltransferase (LPIAT) activity, measured with 17:1 LPI and 20:4 acyl-CoA suggests decreased MBOAT7 activity in the CDAHFD and GAN models of NASH, respectively. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5; GAN study: LF n = 10, GAN n = 5. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Murine NASH livers display altered LPI and PI abundance. (A and D) Hepatic LPI levels are increased in the CDAHFD and GAN models of NASH, respectively. (B, C, E and F) Hepatic PI species are decreased in the CDAHFD and GAN models of NASH, respectively. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5; GAN study: LF n = 10, GAN n = 5. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
MBOAT7 overexpression causes minor decreases in LPI species. (A and C) Total LPI concentrations are not altered by MBOAT7 overexpression in the CDAHFD and GAN models of NASH, respectively. (B and D) Individual LPI species measured in CDAHFD and GAN models, respectively, suggest MBOAT7 overexpression decreases only a few minor LPI species. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in the CDAHFD model of NASH. (A–D) Hepatic LPC, PC, PE, and PS molecular species concentrations, respectively, in livers from CDAHFD-fed mice, with few significant changes between GFP control and MBOAT7 overexpression. Data presented as mean ± SEM, n = 10 each. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic cholesteryl ester (CE), ceramide, and sphingomyelin (SM) concentrations in the CDAHFD model of NASH. (A) Individual CE species not altered other than 18:3 CE increased by MBOAT7 overexpression. (B) Individual ceramide species are not altered by MBOAT7 overexpression compared to GFP control. (C) Individual SM species are not altered by MBOAT7 overexpression compared to control. Data presented as mean ± SEM, n = 10 each. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05.
Supplemental Figure 7. Hepatic cholesteryl ester, ceramide, and sphingomyelin concentrations in the GAN model of NASH. (A) Individual cholesteryl ester species in GAN livers with GFP control or MBOAT7 overexpression. (B) Individual ceramide species in GAN livers with GFP control or MBOAT7 overexpression. (C) Individual sphingomyelin species in GAN livers with GFP or MBOAT7 overexpression. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.Hepatic lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in the GAN model of NASH. (A–D) Hepatic LPC, PC, PE, and PS molecular species concentrations in GAN livers, with only 34:3 PC displaying a significant difference between MBOAT7 overexpression and GFP control. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05.
Hepatic cholesteryl ester, ceramide, and sphingomyelin concentrations in the GAN model of NASH. (A) Individual cholesteryl ester species in GAN livers with GFP control or MBOAT7 overexpression. (B) Individual ceramide species in GAN livers with GFP control or MBOAT7 overexpression. (C) Individual sphingomyelin species in GAN livers with GFP or MBOAT7 overexpression. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic free fatty acids and long-chain acyl-CoAs are altered in NASH. (A and C) Hepatic free fatty acid species are predominantly increased in both the CDAHFD and GAN NASH livers compared to LF livers. Free fatty acids are not altered by MBOAT7 overexpression. (B and D) Long chain acyl-CoAs are mostly decreased in NASH livers compared to LF control livers and are not altered by MBOAT7 overexpression. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5, CDAHFD-GFP n = 10, CDAHFD-MBOAT7 n = 10; GAN study: LF n = 10, GAN n = 5, GAN-GFP n = 15, GAN-MBOAT7 n = 18. Data analyzed by one-way ANOVA with Tukey’s post-hoc correction for multiple comparisons, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
References
- 1.Ferguson D., Finck B.N. Emerging therapeutic approaches for the treatment of NAFLD and type 2 diabetes mellitus. Nat Rev Endocrinol. 2021;17:484–495. doi: 10.1038/s41574-021-00507-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cariou B., Byrne C.D., Loomba R., et al. Nonalcoholic fatty liver disease as a metabolic disease in humans: a literature review. Diabetes Obes Metab. 2021;23:1069–1083. doi: 10.1111/dom.14322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meroni M., Longo M., Tria G., et al. Genetics is of the essence to face NAFLD. Biomedicines. 2021;9:1359. doi: 10.3390/biomedicines9101359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luukkonen P.K., Qadri S., Ahlholm N., et al. Distinct contributions of metabolic dysfunction and genetic risk factors in the pathogenesis of non-alcoholic fatty liver disease. J Hepatol. 2022;76:526–535. doi: 10.1016/j.jhep.2021.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mancina R.M., Dongiovanni P., Petta S., et al. The MBOAT7-TMC4 variant rs641738 increases risk of nonalcoholic fatty liver disease in individuals of European descent. Gastroenterology. 2016;150:1219–1230.e6. doi: 10.1053/j.gastro.2016.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Luukkonen P.K., Zhou Y., Hyotylainen T., et al. The MBOAT7 variant rs641738 alters hepatic phosphatidylinositols and increases severity of non-alcoholic fatty liver disease in humans. J Hepatol. 2016;65:1263–1265. doi: 10.1016/j.jhep.2016.07.045. [DOI] [PubMed] [Google Scholar]
- 7.Teo K., Abeysekera K.W.M., Adams L., et al. rs641738C>T near MBOAT7 is associated with liver fat, ALT and fibrosis in NAFLD: a meta-analysis. J Hepatol. 2021;74:20–30. doi: 10.1016/j.jhep.2020.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee H.C., Inoue T., Sasaki J., et al. LPIAT1 regulates arachidonic acid content in phosphatidylinositol and is required for cortical lamination in mice. Mol Biol Cell. 2012;23:4689–4700. doi: 10.1091/mbc.E12-09-0673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Caddeo A., Jamialahmadi O., Solinas G., et al. MBOAT7 is anchored to endomembranes by six transmembrane domains. J Struct Biol. 2019;206:349–360. doi: 10.1016/j.jsb.2019.04.006. [DOI] [PubMed] [Google Scholar]
- 10.Caddeo A., Hedfalk K., Romeo S., et al. LPIAT1/MBOAT7 contains a catalytic dyad transferring polyunsaturated fatty acids to lysophosphatidylinositol. Biochim Biophys Acta Mol Cell Biol Lipids. 2021;1866:158891. doi: 10.1016/j.bbalip.2021.158891. [DOI] [PubMed] [Google Scholar]
- 11.Gijon M.A., Riekhof W.R., Zarini S., et al. Lysophospholipid acyltransferases and arachidonate recycling in human neutrophils. J Biol Chem. 2008;283:30235–30245. doi: 10.1074/jbc.M806194200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helsley R.N., Varadharajan V., Brown A.L., et al. Obesity-linked suppression of membrane-bound O-acyltransferase 7 (MBOAT7) drives non-alcoholic fatty liver disease. Elife. 2019;8 doi: 10.7554/eLife.49882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Meroni M., Dongiovanni P., Longo M., et al. Mboat7 down-regulation by hyper-insulinemia induces fat accumulation in hepatocytes. EBioMedicine. 2020;52 doi: 10.1016/j.ebiom.2020.102658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tanaka Y., Shimanaka Y., Caddeo A., et al. LPIAT1/MBOAT7 depletion increases triglyceride synthesis fueled by high phosphatidylinositol turnover. Gut. 2021;70:180–193. doi: 10.1136/gutjnl-2020-320646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thangapandi V.R., Knittelfelder O., Brosch M., et al. Loss of hepatic Mboat7 leads to liver fibrosis. Gut. 2021;70:940–950. doi: 10.1136/gutjnl-2020-320853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia M., Chandrasekaran P., Rong S., et al. Hepatic deletion of Mboat7 (LPIAT1) causes activation of SREBP-1c and fatty liver. J Lipid Res. 2021;62:100031. doi: 10.1194/jlr.RA120000856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vvedenskaya O., Rose T.D., Knittelfelder O., et al. Nonalcoholic fatty liver disease stratification by liver lipidomics. J Lipid Res. 2021;62:100104. doi: 10.1016/j.jlr.2021.100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soupene E., Kuypers F.A. Mammalian long-chain acyl-CoA synthetases. Exp Biol Med (Maywood) 2008;233:507–521. doi: 10.3181/0710-MR-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mann J.P., Pietzner M., Wittemans L.B., et al. Insights into genetic variants associated with NASH-fibrosis from metabolite profiling. Hum Mol Genet. 2020;29:3451–3463. doi: 10.1093/hmg/ddaa162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chiappini F., Coilly A., Kadar H., et al. Metabolism dysregulation induces a specific lipid signature of nonalcoholic steatohepatitis in patients. Sci Rep. 2017;7:46658. doi: 10.1038/srep46658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Garcia-Jaramillo M., Spooner M.H., Lohr C.V., et al. Lipidomic and transcriptomic analysis of western diet-induced nonalcoholic steatohepatitis (NASH) in female Ldlr -/- mice. PLoS One. 2019;14 doi: 10.1371/journal.pone.0214387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Araya J., Rodrigo R., Videla L.A., et al. Increase in long-chain polyunsaturated fatty acid n - 6/n - 3 ratio in relation to hepatic steatosis in patients with non-alcoholic fatty liver disease. Clin Sci (Lond) 2004;106:635–643. doi: 10.1042/CS20030326. [DOI] [PubMed] [Google Scholar]
- 23.Allard J.P., Aghdassi E., Mohammed S., et al. Nutritional assessment and hepatic fatty acid composition in non-alcoholic fatty liver disease (NAFLD): a cross-sectional study. J Hepatol. 2008;48:300–307. doi: 10.1016/j.jhep.2007.09.009. [DOI] [PubMed] [Google Scholar]
- 24.Puri P., Baillie R.A., Wiest M.M., et al. A lipidomic analysis of nonalcoholic fatty liver disease. Hepatology. 2007;46:1081–1090. doi: 10.1002/hep.21763. [DOI] [PubMed] [Google Scholar]
- 25.Gorden D.L., Myers D.S., Ivanova P.T., et al. Biomarkers of NAFLD progression: a lipidomics approach to an epidemic. J Lipid Res. 2015;56:722–736. doi: 10.1194/jlr.P056002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Loomba R., Quehenberger O., Armando A., et al. Polyunsaturated fatty acid metabolites as novel lipidomic biomarkers for noninvasive diagnosis of nonalcoholic steatohepatitis. J Lipid Res. 2015;56:185–192. doi: 10.1194/jlr.P055640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sen P., Kan C.F.K., Singh A.B., et al. Identification of p115 as a novel ACSL4 interacting protein and its role in regulating ACSL4 degradation. J Proteomics. 2020;229:103926. doi: 10.1016/j.jprot.2020.103926. [DOI] [PubMed] [Google Scholar]
- 28.Kan C.F., Singh A.B., Stafforini D.M., et al. Arachidonic acid downregulates acyl-CoA synthetase 4 expression by promoting its ubiquitination and proteasomal degradation. J Lipid Res. 2014;55:1657–1667. doi: 10.1194/jlr.M045971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Westerbacka J., Kolak M., Kiviluoto T., et al. Genes involved in fatty acid partitioning and binding, lipolysis, monocyte/macrophage recruitment, and inflammation are overexpressed in the human fatty liver of insulin-resistant subjects. Diabetes. 2007;56:2759–2765. doi: 10.2337/db07-0156. [DOI] [PubMed] [Google Scholar]
- 30.Duan J., Wang Z., Duan R., et al. Therapeutic targeting of hepatic ACSL4 ameliorates NASH in mice. Hepatology. 2022;75:140–153. doi: 10.1002/hep.32148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johansen A., Rosti R.O., Musaev D., et al. Mutations in MBOAT7, encoding lysophosphatidylinositol acyltransferase I, lead to intellectual disability accompanied by epilepsy and autistic features. Am J Hum Genet. 2016;99:912–916. doi: 10.1016/j.ajhg.2016.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yalnizoglu D., Ozgul R.K., Oguz K.K., et al. Expanding the phenotype of phospholipid remodelling disease due to MBOAT7 gene defect. J Inherit Metab Dis. 2019;42:381–388. doi: 10.1002/jimd.12016. [DOI] [PubMed] [Google Scholar]
- 33.Jacher J.E., Roy N., Ghaziuddin M., et al. Expanding the phenotypic spectrum of MBOAT7-related intellectual disability. Am J Med Genet B Neuropsychiatr Genet. 2019;180:483–487. doi: 10.1002/ajmg.b.32749. [DOI] [PubMed] [Google Scholar]
- 34.Farne M., Tedesco G.M., Bedetti C., et al. A patient with novel MBOAT7 variant: the cerebellar atrophy is progressive and displays a peculiar neurometabolic profile. Am J Med Genet A. 2020;182:2377–2383. doi: 10.1002/ajmg.a.61773. [DOI] [PubMed] [Google Scholar]
- 35.Sun L., Khan A., Zhang H., et al. Phenotypic characterization of intellectual disability caused by MBOAT7 mutation in two consanguineous Pakistani families. Front Pediatr. 2020;8:585053. doi: 10.3389/fped.2020.585053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heidari E., Caddeo A., Zarabadi K., et al. Identification of novel loss of function variants in MBOAT7 resulting in intellectual disability. Genomics. 2020;112:4072–4077. doi: 10.1016/j.ygeno.2020.07.008. [DOI] [PubMed] [Google Scholar]
- 37.Arendt B.M., Comelli E.M., Ma D.W., et al. Altered hepatic gene expression in nonalcoholic fatty liver disease is associated with lower hepatic n-3 and n-6 polyunsaturated fatty acids. Hepatology. 2015;61:1565–1578. doi: 10.1002/hep.27695. [DOI] [PubMed] [Google Scholar]
- 38.Suppli M.P., Rigbolt K.T.G., Veidal S.S., et al. Hepatic transcriptome signatures in patients with varying degrees of nonalcoholic fatty liver disease compared with healthy normal-weight individuals. Am J Physiol Gastrointest Liver Physiol. 2019;316:G462–G472. doi: 10.1152/ajpgi.00358.2018. [DOI] [PubMed] [Google Scholar]
- 39.Govaere O., Cockell S., Tiniakos D., et al. Transcriptomic profiling across the nonalcoholic fatty liver disease spectrum reveals gene signatures for steatohepatitis and fibrosis. Sci Transl Med. 2020;12 doi: 10.1126/scitranslmed.aba4448. [DOI] [PubMed] [Google Scholar]
- 40.Kozumi K., Kodama T., Murai H., et al. Transcriptomics identify thrombospondin-2 as a biomarker for NASH and advanced liver fibrosis. Hepatology. 2021;74:2452–2466. doi: 10.1002/hep.31995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subudhi S., Drescher H.K., Dichtel L.E., et al. Distinct hepatic gene-expression patterns of NAFLD in patients with obesity. Hepatol Commun. 2022;6:77–89. doi: 10.1002/hep4.1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ulmasov B., Noritake H., Carmichael P., et al. An inhibitor of arginine-glycine-aspartate-binding integrins reverses fibrosis in a mouse model of nonalcoholic steatohepatitis. Hepatol Commun. 2019;3:246–261. doi: 10.1002/hep4.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kamm D.R., Pyles K.D., Sharpe M.C., et al. Novel insulin sensitizer MSDC-0602K improves insulinemia and fatty liver disease in mice, alone and in combination with liraglutide. J Biol Chem. 2021;296:100807. doi: 10.1016/j.jbc.2021.100807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liang W., Menke A.L., Driessen A., et al. Establishment of a general NAFLD scoring system for rodent models and comparison to human liver pathology. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zaqout S., Becker L.L., Kaindl A.M. Immunofluorescence staining of paraffin sections step by step. Front Neuroanat. 2020;14:582218. doi: 10.3389/fnana.2020.582218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pike D.P., Vogel M.J., McHowat J., et al. 2-Chlorofatty acids are biomarkers of sepsis mortality and mediators of barrier dysfunction in rats. J Lipid Res. 2020;61:1115–1127. doi: 10.1194/jlr.RA120000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rong X., Wang B., Palladino E.N., et al. ER phospholipid composition modulates lipogenesis during feeding and in obesity. J Clin Invest. 2017;127:3640–3651. doi: 10.1172/JCI93616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Hepatic MBOAT7 activity is reduced in murine NASH models. (A) Hepatic Mboat7 expression in mice fed either low fat (LF) control or choline-deficient high-fat diet (CDAHFD). (B and C) MBOAT7 expression in livers from LF- or CDAHFD-fed mice. (D) Hepatic Mboat7 expression in mice fed either LF or Gubra Amylin NASH diet (GAN). (E and F) MBOAT7 expression in livers from LF- or GAN-fed mice. (G and H) Hepatic microsomal MBOAT7 expression in the CDAHFD and GAN models of NASH compared to LF, respectively. (I and J) Hepatic microsomal lysophosphatidylinositol (LPI) acyltransferase (LPIAT) activity, measured with 17:1 LPI and 20:4 acyl-CoA suggests decreased MBOAT7 activity in the CDAHFD and GAN models of NASH, respectively. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5; GAN study: LF n = 10, GAN n = 5. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
Murine NASH livers display altered LPI and PI abundance. (A and D) Hepatic LPI levels are increased in the CDAHFD and GAN models of NASH, respectively. (B, C, E and F) Hepatic PI species are decreased in the CDAHFD and GAN models of NASH, respectively. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5; GAN study: LF n = 10, GAN n = 5. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.
MBOAT7 overexpression causes minor decreases in LPI species. (A and C) Total LPI concentrations are not altered by MBOAT7 overexpression in the CDAHFD and GAN models of NASH, respectively. (B and D) Individual LPI species measured in CDAHFD and GAN models, respectively, suggest MBOAT7 overexpression decreases only a few minor LPI species. Data presented as mean ± SEM, CDAHFD study: n = 10 each; GAN study: GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in the CDAHFD model of NASH. (A–D) Hepatic LPC, PC, PE, and PS molecular species concentrations, respectively, in livers from CDAHFD-fed mice, with few significant changes between GFP control and MBOAT7 overexpression. Data presented as mean ± SEM, n = 10 each. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic cholesteryl ester (CE), ceramide, and sphingomyelin (SM) concentrations in the CDAHFD model of NASH. (A) Individual CE species not altered other than 18:3 CE increased by MBOAT7 overexpression. (B) Individual ceramide species are not altered by MBOAT7 overexpression compared to GFP control. (C) Individual SM species are not altered by MBOAT7 overexpression compared to control. Data presented as mean ± SEM, n = 10 each. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05.
Supplemental Figure 7. Hepatic cholesteryl ester, ceramide, and sphingomyelin concentrations in the GAN model of NASH. (A) Individual cholesteryl ester species in GAN livers with GFP control or MBOAT7 overexpression. (B) Individual ceramide species in GAN livers with GFP control or MBOAT7 overexpression. (C) Individual sphingomyelin species in GAN livers with GFP or MBOAT7 overexpression. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.Hepatic lysophosphatidylcholine (LPC), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylserine (PS) in the GAN model of NASH. (A–D) Hepatic LPC, PC, PE, and PS molecular species concentrations in GAN livers, with only 34:3 PC displaying a significant difference between MBOAT7 overexpression and GFP control. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05.
Hepatic cholesteryl ester, ceramide, and sphingomyelin concentrations in the GAN model of NASH. (A) Individual cholesteryl ester species in GAN livers with GFP control or MBOAT7 overexpression. (B) Individual ceramide species in GAN livers with GFP control or MBOAT7 overexpression. (C) Individual sphingomyelin species in GAN livers with GFP or MBOAT7 overexpression. Data presented as mean ± SEM, GFP n = 15, MBOAT7 n = 18. Data analyzed by two-tailed, unpaired t-test, ∗p < 0.05, ∗∗p < 0.01.
Hepatic free fatty acids and long-chain acyl-CoAs are altered in NASH. (A and C) Hepatic free fatty acid species are predominantly increased in both the CDAHFD and GAN NASH livers compared to LF livers. Free fatty acids are not altered by MBOAT7 overexpression. (B and D) Long chain acyl-CoAs are mostly decreased in NASH livers compared to LF control livers and are not altered by MBOAT7 overexpression. Data presented as mean ± SEM, CDAHFD study: LF n = 10, CDAHFD n = 5, CDAHFD-GFP n = 10, CDAHFD-MBOAT7 n = 10; GAN study: LF n = 10, GAN n = 5, GAN-GFP n = 15, GAN-MBOAT7 n = 18. Data analyzed by one-way ANOVA with Tukey’s post-hoc correction for multiple comparisons, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001.