Abstract
A dense and diverse microbial community inhabits the gut and many epithelial surfaces. Referred to as the microbiota, it co-evolved with the host and is beneficial for many host physiological processes. A major function of these symbiotic microorganisms is protection against pathogen colonization and overgrowth of indigenous pathobionts. Dysbiosis of the normal microbial community increases the risk of pathogen infection and overgrowth of harmful pathobionts. The protective mechanisms conferred by the microbiota are complex and include competitive microbial–microbial interactions and induction of host immune responses. Pathogens, in turn, have evolved multiple strategies to subvert colonization resistance conferred by the microbiota. Understanding the mechanisms by which microbial symbionts limit pathogen colonization should guide the development of new therapeutic approaches to prevent or treat disease.
Introduction
The mammalian intestine is colonized by trillions of microorganisms, including bacteria, viruses, fungi, archaea and protozoa, that have co-evolved with the host in a symbiotic relationship. The collection of microbial populations that reside within the host is commonly referred to as the microbiota. Microbial symbionts colonize mammalian hosts immediately after birth (Box 1). In adult individuals, Gram-negative Pseudomonadota (formerly Proteobacteria), Bacteroidota (formerly Bacteroidetes) and Gram-positive Bacillota (formerly Firmicutes) that include Clostridiales and Lactobacillales represent the major phyla among intestinal eubacteria1. The microbial communities differ between the small intestine and the large intestine, and between mucosa-associated and luminal communities at the same intestinal segment2,3, which reflect different nutritional and metabolic requirements of individual bacteria. In the gut, resident bacteria have adapted to the local environments and developed complex interactions with host niches and other symbionts to acquire nutrients for survival4. In addition, microorganisms compete with each other for space and available nutrients, and this intermicrobial competition not only regulates the composition of the community but also limits the ability of indigenous and foreign microorganisms to colonize the gut5.
Box 1. Microbiota maturation and early-life colonization resistance.
The early composition of the microbiota is heavily influenced by maternal transmission, dietary factors and the mode of delivery158,159. Acquisition of an intact microbiota during vaginal delivery seems important because microbial transfer via caesarean section has been associated with increased colonization by pathobionts that can cause opportunistic infections158. Unlike the microbiota of the adult intestine that is dominated by obligate anaerobes, the microbiota of neonates and infants is less diverse and contains large numbers of facultative anaerobes159,160. The difference in microbiota composition between adults and infants reflects a major role for nutrition in shaping microbiota communities, as cessation of breastfeeding is required for maturation into an adult-like microbiota160. This transition coincides with dietary changes from simple sugars and oligosaccharides present in the maternal milk to a more diverse diet rich in complex polysaccharides. Infants are highly susceptible to infections caused by enteric pathogens161. The reason for this increased susceptibility seems multifactorial, including the absence of a mature immune system162. However, the neonatal microbiota is unable to prevent colonization by two common bacterial pathogens independently of the age of the host113. The lack of colonization resistance was caused by the absence of Clostridiales in the neonatal gut, as administration of Clostridiales restored protection against pathogen colonization independently of the host immune system113. However, whether the lack of Clostridiales can account for overall increased susceptibility to enteric infection in neonates remains unclear. The microbiota can also promote the protection of neonates against pathogens indirectly through the stimulation of immune responses. For example, dysbiosis induced by treatment with antibiotics increases the susceptibility to sepsis by impairing IL-17-mediated neutrophil homeostasis163. In humans, perinatal antibiotic treatment augments the incidence of early-onset sepsis, which was associated with a reduction in lactobacilli in the neonatal gut and the vagina of the mother164. Consistent with these findings, bacterial strains from the neonatal microbiota such as those belonging to the Lactobacillus genus and Escherichia coli can protect neonatal mice from late-onset sepsis, but the mechanism remains unknown165. One possibility is that certain bacteria residing in the neonatal gut can elicit immune responses, including production of protective IgG, that can also target pathogens130. Consistent with this notion, the maternal gut microbiota can induce IgG that protects newborn babies from pathogens and enteric E. coli infection166.
Millions of years of co-evolution between the host and microorganisms have led to a mutualistic symbiosis in which the microbiota contributes to host metabolic functions and the host, in turn, provides nutrients and a hospitable space for the microorganisms. In addition to metabolic benefits, the microbiota provides the host with several functions that promote the intestinal epithelial barrier, immune homeostasis and protection against pathogen colonization6. The notion that certain beneficial members of the microbiota could outcompete harmful ones was first proposed by Elie Metchnikoff in the early twentieth century7. Later, studies with antibiotics and germ-free (GF) animals revealed that the symbiotic bacteria have a major role in limiting pathogen colonization in the gut8,9. This phenomenon that is detected within a few hours after pathogen inoculation was later coined ‘colonization resistance’10. In addition to limiting pathogen colonization, the microbiota can also inhibit the expansion of indigenous but potentially dangerous ‘pathobionts’, as well as colonization of innocuous species, such as probiotics11. There are a wide variety of mechanisms now known to participate in colonization resistance6. Many involve direct interactions between microbial cells, whereas others regulate host physiology, and largely host immune responses that limit pathogen colonization and expansion inside and outside the gut. In this Review, we focus on our current understanding of how the microbiota limit colonization by pathogens as well as the strategies used by pathogens to counter colonization resistance.
Direct mechanisms
Direct mechanisms of colonization resistance take place between bacteria, with the host acting as the environment in which competition takes place. Bacteria can directly inhibit the growth of each other by producing inhibitory compounds (‘interference’ competition in ecological terms) or by competing for resources (‘exploitative’ competition)12, in both the gut (Fig. 1) and outside the gastrointestinal tract (Box 2). In the gut, the available nutritional resources are largely provided by the host either from ingested food or from secretions such as mucus. Additional nutrients are also provided by other microorganisms that inhabit the gut.
Fig. 1 |. Direct mechanisms of colonization resistance.
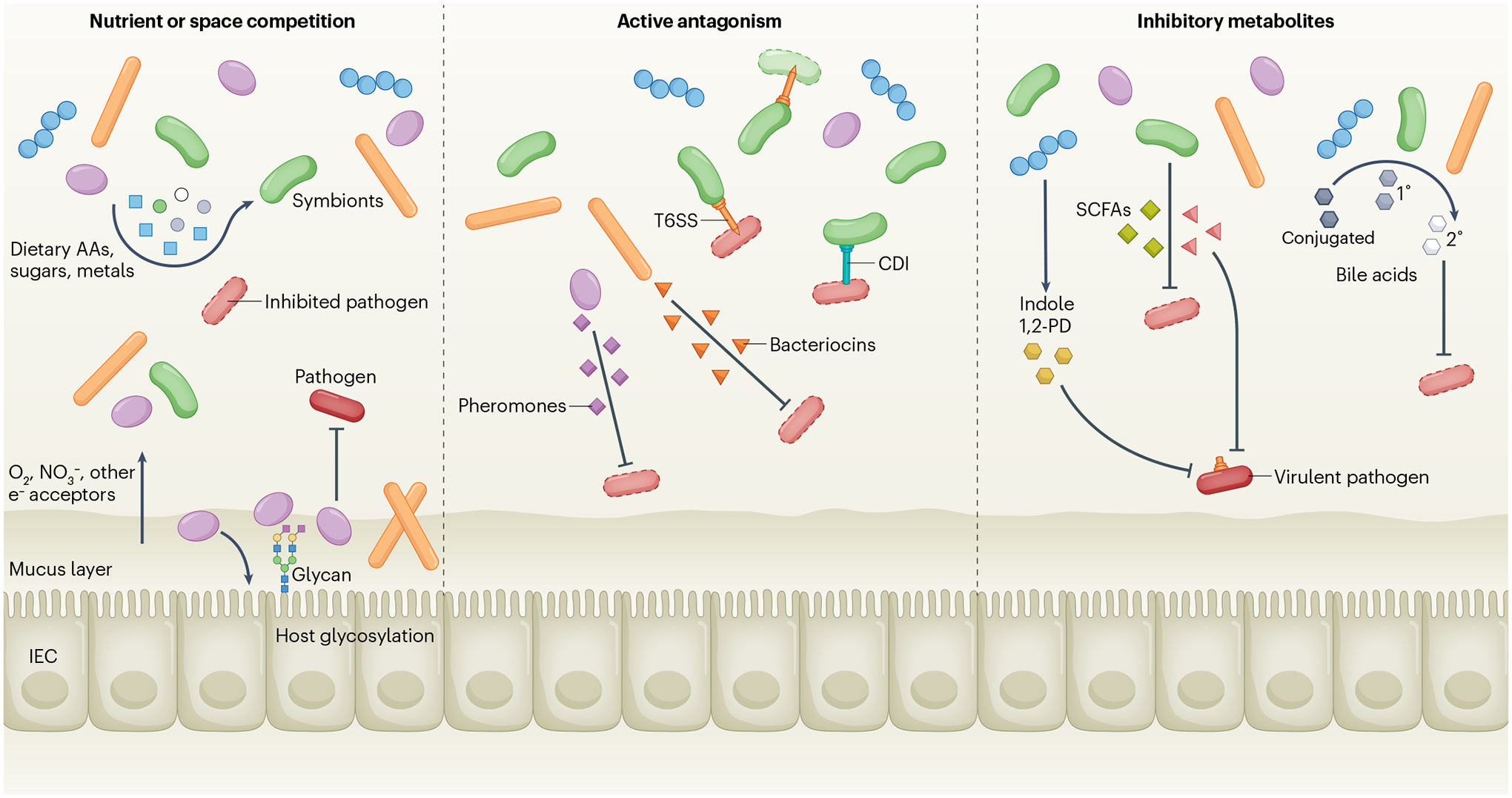
Native symbiotic bacteria consume dietary amino acids (AAs), sugars, metals and respiratory electron acceptors such as O2 and NO3−, thus starving the pathogen of essential nutrients and molecules (left panel). At the epithelial surface, bacteria can modify host glycosylation and/or use it as a nutrient for adhesion, creating a new microscopic niche that blocks pathogen access to the epithelium. Other symbionts adhering to sites on the epithelium or in the mucus can also prevent pathogen access. Symbionts can also directly kill pathogens via contact-dependent inhibition (CDI), the type VI secretion system (T6SS) or secreted molecules, including bacteriocins or pheromone peptides (centre panel). Other inhibitory compounds such as acetic, butyric or propionic acids (short-chain fatty acids (SCFAs)), indole or 1,2-propanediol (1,2-PD) are secreted by the microbiota and inhibit growth or suppress virulence factor expression in pathogens (right panel). Many symbionts can modify bile acids by deconjugation, whereas some (for example, Clostridium scindens) can convert primary to secondary bile acids that inhibit pathogen growth. IEC, intestinal epithelial cell.
Box 2. Colonization resistance outside the gut.
In addition to the intestine, other body cavities and the skin that are exposed to the external environment are colonized by a diverse population of microbial symbionts. The surface of the epidermis, the outermost multilayer aspect of the skin, consists of the protein-rich and lipid-rich cornified layer dotted with pilosebaceous units and sweat glands. These structures provide the skin not only protection against pathogen invasion but also a unique metabolic niche for microbial survival167. Despite being relatively nutrient-poor and acidic, the skin harbours millions of bacteria, fungi and viruses that can limit pathogen colonization and invasion167,168. Although most skin symbionts are considered commensal or mutualistic, some microorganisms including Staphylococcus aureus and Staphylococcus epidermidis can induce the expression of virulence genes and become pathogenic depending on host or microbial factors that remain poorly understood. Similar to other microbial-rich host sites, skin symbionts have developed strategies to limit competition with other bacteria and to promote their colonization. For example, coagulase-negative Staphylococcus (CoNS) species that are abundant in the skin can reduce the colonization of pathogenic S. aureus through the production of bacteriocins and other antimicrobial peptides (AMPs)169,170. These peptides produced by some strains of CoNS can synergize with host AMPs to inhibit S. aureus growth170–172. Some competitive interactions between skin bacteria are not based on prokaryotic antimicrobials. For example, Corynebacterium accolens produces the LiS1 lipase that acts on skin surface triacylglycerols to release antibacterial free fatty acids that inhibit the growth of the opportunistic pathogen Streptococcus pneumoniae173. CoNS can also antagonize other CoNS species and S. aureus through the accessory gene regulator (agr) quorum-sensing system. The agr system is used by all staphylococci to coordinate cellular behaviour in response to bacterial density through the production of a cyclic peptide known as the autoinducing peptide (AIP)174. Every staphylococcal species contains an agr locus and produces a unique stimulatory AIP molecule, but non-cognate AIPs can inhibit quorum-sensing activation in other species175. Such mechanisms of agr crosstalk can be used to inhibit S. aureus in models of intradermal skin infection and atopic dermatitis176,177. Some members of the skin microbiota can also target pathogens indirectly by inducing AMP production in keratinocytes reducing pathogen invasion178.
Like the skin, the nasal cavity harbours a diverse microbiota that includes opportunistic pathogens such as S. aureus that is present in the nares of up to 40% of healthy individuals179. Nasal CoNS species produce multiple antibiotics against competitors that could regulate pathogen colonization180. Likewise, Staphylococcus lugdunensis, another CoNS that inhabits the nasal cavity, produces the cyclic peptide lugdunin that inhibits S. aureus colonization181. Furthermore, nasal carriage of S. lugdunensis is negatively correlated with that of S. aureus in humans181. S. epidermidis can also interfere with S. aureus virulence and colonization through the production of the serine protease Esp that can degrade proteins that are important for S. aureus biofilm formation and nasal colonization182,183. A negative correlation between abundance of S. epidermidis and several opportunistic pathogens including S. aureus and Moraxella catarrhalis was also observed in the human nasal cavity184, suggesting interbacterial competition. Further analyses revealed that S. epidermidis can stimulate host AMP production that can kill and outcompete S. aureus and M. catarrhalis184. Although the physiological relevance of these intermicrobial interactions in the skin and nasal cavity remains unclear, these studies suggest that CoNS and other bacteria can antagonize pathogens via different mechanisms.
In most women, the cervicovaginal community is dominated by Lactobacillus species185. The replacement of the Lactobacillus species-dominated microbiota by a diverse community with a greater abundance of anaerobes, a clinical condition called bacterial vaginosis, is a risk factor for several common sexually transmitted infections186. The mechanism by which a diverse, non-Lactobacillus species-dominant microbiota increases the risk of infection is poorly understood. The presence of lactic acid-producing Lactobacillus species reduces the pH in the vagina, which can reduce pathogen colonization including bacteria associated with urinary tract infections, for which the optimal pH for growth is neutral187. Furthermore, some lactobacilli can also produce H2O2 and AMPs188,189. Bacterial vaginosis has been associated with increased cytokine production, which could also promote pathogen growth; however, increased inflammatory responses were not observed in some studies190. Further studies are needed to understand the relationship between microbiota dysbiosis and increased risk for infection in bacterial vaginosis.
Spatial competition
The concept of ‘niche’ of a species, and the idea that two species cannot occupy the same niche, has developed from observations made by naturalists, including Darwin, to a fleshed out mathematical concept13. For the purpose of this Review, the niche can be thought of as the combination of many variables that include both physical location and other factors such as oxygen concentration and availability of various nutrients, which define the actual place of a species in an ecosystem. It has long been assumed that two species with identical niches cannot coexist for long. This ‘competitive exclusion’ can be demonstrated when two identical bacterial strains, only differing by a neutral marker, are sequentially given to GF mice. The first strain will take up the entire niche, and the second, identical strain will be unable to colonize14,15. Giving one strain a new functional ability can alter its niche and allow it to coexist with its former competitors. For example, acquisition of a high-affinity transporter for a sugar (gluconate) allowed two otherwise similar Escherichia coli strains to coexist16. Comparing different natural variants of E. coli, which partially overlap in their sugar preferences, suggests that nutrient use may be a large factor in competition between strains17. Freter et al.18 formulated the ‘nutrient niche’ hypothesis, which proposed that the relative abundance of a species in the gut is controlled by levels of just a few nutrients. In general, species compete directly with those species with which they share the most gene functions, usually being their closest evolutionary relatives, for example, Klebsiella, Citrobacter or Clostridioides species can prevent colonization with their often more pathogenic cousins19–21.
The mammalian gut is not a homogeneous environment. There are many different microenvironments that effectively separate it into more niches with different characteristics. Oxygen concentration and pH are both reduced in the large intestine, and many nutrients, such as simple sugars and amino acids are absorbed by the host before reaching the large intestine. The sugars displayed on host cells and mucus can vary in local patches along the intestine22, as can respiratory electron acceptors produced from host inflammation23, which could divide up the physical space into different niches and potentially increase diversity or help maintain species or strains that would otherwise be outcompeted. This has led to a new variation of the nutrient niche idea by Freter that incorporates this spatial heterogeneity of nutrients24. Adhesion of bacteria to host cells, mucus or food particles could be a key factor in their continued survival in the gut. Adhesion can have different effects, either promoting washout or retention15,25,26. Competition for adhesion sites may therefore be another contributor to colonization resistance (Fig. 1). Some bacteria can also construct their own niches by modifying the environment in the host. For instance, some species can induce the host to produce different sugars, which can serve as nutrients or adhesion sites27, or to trigger the induction of specific antibodies, which might enable better colonization15.
Nutritional competition
The gut microbiota is generally stable over time and is presumed to be a climax community that has reached equilibrium, unless disturbed by antibiotic administration or other major events. Therefore, under homeostatic conditions, it is assumed that most of the available niches are filled and that this is a major factor behind colonization resistance. Although bacterial taxa, as defined by 16S rRNA gene sequences, can vary considerably between individuals, the gene functions present are less variable28, at least based on our limited ability to accurately predict the function of bacterial genes. This suggests that the same functional niches are being filled in most individuals. A main consequence of this would be that the gut microorganisms scavenge most available nutrients, keeping them at low levels at the steady state, which limits pathogen expansion (Fig. 1). Indeed, GF or antibiotic-treated mice have increased amounts of sugars, amino acids and other nutrients in the gut lumen and a concomitant reduction in colonization resistance8,29,30. In line with this, a genetic screen revealed that depletion of dietary amino acids by the microbiota is a major factor underlying colonization resistance for the pathogen Citrobacter rodentium29. Notably, administration of a high-protein diet to conventionally raised mice enhanced pathogen colonization by ~3 logs29. To achieve complete colonization resistance against a generalist bacterium such as Salmonella enterica subsp. enterica serovar Typhimurium, a diverse microbial community is probably required to saturate all niches. The altered Schaedler flora, consisting of eight murine bacterial strains that stably colonize and recapitulate some aspects of a normal microbiota, provides little resistance to S. Typhimurium31. A larger collection containing 12 strains from 5 different phyla that dominate mammalian guts provided better protection against S. Typhimurium but not to the extent of a full complex microbiota32. To determine what could be missing from the community, the gene content was examined by metagenomic sequencing. There was a reduction of respiration-related genes in the 12 strains compared with a conventional microbiota. Three additional facultative anaerobes, which can respire oxygen, were able to improve resistance to S. Typhimurium to levels similar to a complete microbiota32. This has also been observed in another study in which native E. coli in mice correlated with protection against S. Typhimurium, and oxygen respiration was required for the protection33. Therefore, oxygen or anaerobic respiratory electron acceptors may be a limiting resource that can mediate resistance to S.Typhimurium. Essential metal cofactors such as iron, zinc and manganese are also limited in the gut and are actively sequestered by the host during inflammation. Competition for iron and zinc contributes to the ability of the probiotic E. coli Nissle strain to colonize the gut and provide resistance to S. Typhimurium34,35.
Prokaryotic antimicrobial peptides
Nutrient competition is certainly a powerful determinant of community composition and colonization resistance in the gut. However, there are also many examples of active growth inhibition or interference that contribute to colonization resistance (Fig. 1). Bacteriocins are a heterogeneous class of peptides produced by bacteria, which have bacteriostatic or bactericidal activity against symbiotic bacteria and pathogens36. In line with the competitive exclusion concept, they often have a narrow target range and, in general, act on species closely related to the producer. They can inhibit growth or kill by various mechanisms; some require a specific receptor, whereas others do not. Bacteriocin-producing strains of E. coli outcompete non-producers in a mouse model37. A bacteriocin also contributes to the ability of the probiotic E. coli Nissle strain to protect from other Enterobacteriaceae38. The Bacteroidota phylum is highly represented in the mammalian gut and its members can produce various types of secreted antibacterial proteins, typically acting within the same genus13,39. In at least one case, a toxin producer was able to outcompete a sensitive strain in vivo40. Enterococcus faecalis and Enterococcus faecium are usually harmless gut residents that can become pathogenic when they acquire antibiotic resistance and become, for instance, vancomycin-resistant enterococcus (VRE), a major problem in the clinic41. An E. faecalis bacteriocin-producing strain was able to colonize mice, presumably by killing native enterococci, and cleared a pathogenic strain from the gut as well42. Gut resident enterococci can also target pathogenic E. faecalis via a different type of secreted small peptides called pheromones43. Blautia producta, a species in the Clostridia class, was identified as part of a minimal group of strains that could resist and eliminate VRE from mice44. The mechanism has been subsequently found to be a secreted bacteriocin, which inhibited growth of VRE as well as various other Gram-positive species45. However, the contribution of prokaryotic antimicrobial peptides (AMPs) to VRE colonization resistance in the intact microbiota of animals or humans remains unclear.
Contact-dependent inhibition
Although bacteriocins and similar molecules can diffuse some distance from the producing cell, some inhibitory mechanisms require direct cell–cell contact (Fig. 1). A cell contact-dependent growth inhibition system, termed contact-dependent inhibition (CDI), was discovered in E. coli and later in other Pseudomonadota46,47. Contact-dependent inhibition requires a specific receptor protein on the target cell and can encode different toxic effector domains with various modes of inhibition48. The genes are usually accompanied by an immunity protein that neutralizes the toxin to protect the producing cell48. A more elaborate contact-dependent system was first discovered in Pseudomonas aeruginosa49.
The type VI secretion system (T6SS) is a large multiprotein complex that can ‘spear’ nearby cells and inject toxic proteins, without the need for a receptor. These systems are found in many Gram-negative bacteria, especially Pseudomonadota and Bacteroidota50. The toxic effectors can have various functions and are usually found with a cognate immunity protein. Clusters of immunity genes have also been found to accumulate on their own, suggesting that protection from T6SS attack is beneficial in vivo51, and other mechanisms have also been developed to protect from T6SS killing52. The T6SS can have a role in competition among species of the Bacteroidales order in the human and mouse gut and prevent invasion of a pathogenic strain53–56. Distinct T4SS and T7SS in Gram-negative and Gram-positive species, respectively, have also been demonstrated to kill cells via translocated toxic proteins57,58. Thus, secreted or cell contact-dependent antagonism probably has an important role in competition in the gut, especially among closely related species in confined physical niches, although their contribution to pathogen colonization resistance remains unclear.
Inhibitory metabolites
Metabolic byproducts produced by bacteria can exert inhibitory activity on other gut bacteria (Fig. 1). Bile acids (BAs) are synthesized by the host and conjugated to taurine or glycine in the liver, stored in the gallbladder and released into the intestinal tract to aid in the emulsification and absorption of dietary lipids59. Although most conjugated primary BAs are reabsorbed in the small intestine, a small proportion can be deconjugated by bacterial bile salt hydrolases and reach the distal intestine where they undergo several modifications, including 7α/β-dehydroxylation by rare bacteria such as Clostridium scindens, to generate secondary BAs59. Although primary BAs promote spore germination, secondary BAs such as deoxycholic acid and lithocholic acid inhibit the growth of many Gram-positive bacteria, including vegetative forms of Clostridioides difficile60–63. In mice and humans, the presence of secondary BAs is associated with resistance against C. difficile infection and development of colitis63. Likewise, administration of C. scindens to gnotobiotic mice colonized with a simplified microbiota lacking 7α-dehydroxylation activity partially restores secondary BA production, limiting C. difficile infection64. Conversely, selective disruption of the microbiota composition by antibiotics increases susceptibility to C. difficile, correlating with reduced levels of secondary BAs65. The tolerance levels of different clinical C. difficile isolates to lithocholic acid correlate positively with the disease severity in murine models of infection66,67. Notably, C. scindens can also protect against C. difficile infection even in the absence of detectable levels of secondary BAs, suggesting a BA-independent mechanism of protection68. Therefore, additional studies are needed to fully decipher the specific contribution of BAs to C. difficile and other infections.
Short-chain fatty acids (SCFAs) including acetic, propionic and butyric acid are produced by microbial fermentation of plant-derived dietary polysaccharides by obligate anaerobes in the large intestine. SCFAs provide an energy source for intestinal epithelial cells (IECs), but also can contribute to colonization resistance5,6. For example, SCFAs inhibit the growth of pathogenic E. coli, C. rodentium and C. difficile and limit Salmonella species colonization in the mouse gut69–72. SCFAs including propionic acid diffuse freely across bacterial membranes (when protonated) and exert their inhibitory activity by disrupting intracellular pH homeostasis73. However, the overall contribution of SCFAs to protection against intestinal pathogen colonization remains unclear owing to the inability to specifically remove these molecules from an otherwise intact microbial environment.
Indirect mechanisms
In addition to the direct inhibition of pathogen colonization by symbionts or their products, there are also indirect mechanisms of defence that limit the invasion and/or expansion of enteric pathogens in the gut (Fig. 2). These include the mucus layer and oxygen gradients, as well as innate and adaptive immune responses, that are induced or maintained by the gut microbiota to limit pathogen colonization.
Fig. 2 |. Indirect mechanisms of colonization resistance.
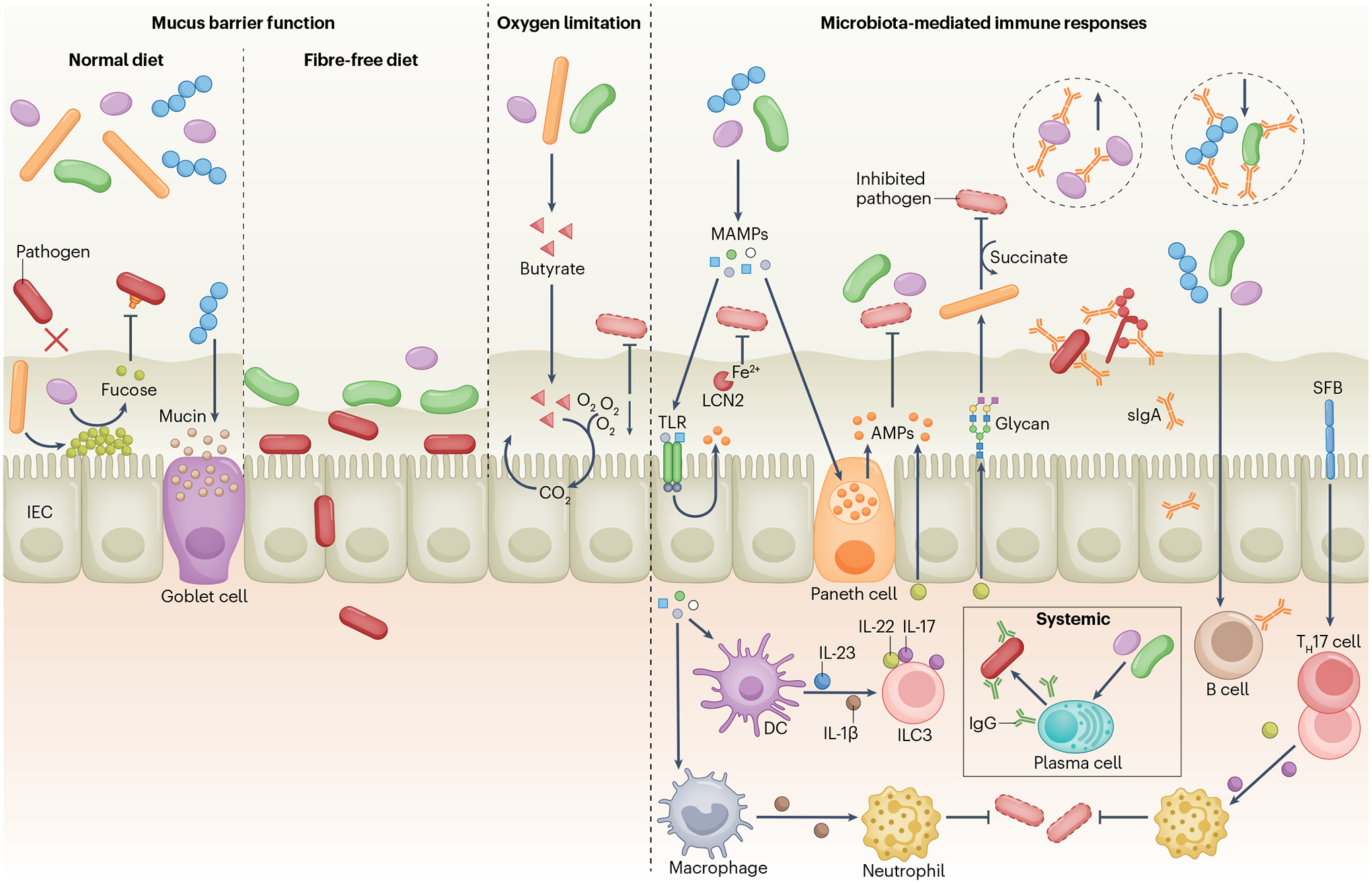
The mucus layer limits pathogen access to the intestinal epithelium (left panel). Microbiota stimulation of host fucosylation protects against pathogens, whereas certain symbionts (such as Bacteroides thetaiotaomicron) release l-fucose from the mucus layer, which reduces virulence gene expression in Citrobacter rodentium, pathogenic Escherichia coli and Enterococcus faecalis. Fibre deprivation leads to an increase in mucus-degrading symbionts that erode the mucus layer, promoting C. rodentium early access to the epithelium. Clostridia produce butyrate that promotes aerobic respiration in intestinal epithelial cells (IECs), reducing oxygen levels in the gut and limiting the expansion of facultative anaerobe pathogens (centre panel). Microorganism-associated molecular patterns (MAMPs) stimulate Toll-like receptors (TLRs) and NOD-like receptors (not shown) in IECs and Paneth cells, leading to the production of antimicrobial peptides (AMPs) that target symbiotic bacteria and pathogens (right panel). Likewise, the microbiota promotes lipocalin 2 (LCN2) production in IECs, limiting pathogen access to iron. Symbiotic bacteria stimulate IL-22 production by type 3 innate lymphoid cells (ILC3s), which regulates AMP production and glycosylation-dependent expansion of symbionts that compete with Clostridioides difficile for succinate. Symbionts also promote pro-IL-1β production by resident macrophages, priming these cells to produce IL-1β in response to Salmonella enterica subsp. enterica serovar Typhimurium and enhancing the recruitment of neutrophils to fight infection. Symbiotic bacteria induce production of polyreactive secretory IgA (sIgA) that can recognize antigens on enteric pathogens, including S. Typhimurium, pathogenic E. coli and virulent Candida albicans. sIgA also limits or promotes expansion of specific symbionts. Microbiota-induced IgG confers protection against pathogens primarily at systemic sites. Segmented filamentous bacteria (SFB) induce IL-17 and IL-22 production by T helper 17 (TH17) cells and ILC3s, promoting neutrophil recruitment and pathogen control. DC, dendritic cell.
The mucosal barrier
The mucus layer forms a physical barrier that limits pathogen interactions with the underlying epithelium (Fig. 2). Pathogens such as C. rodentium, Salmonella species and some pathogenic E. coli strains require attachment to the intestinal epithelium for the initiation of their virulence programmes and effective colonization. Because the mucus layer in GF animals is thinner than in conventionally raised mice74,75, microbial symbionts may limit pathogen colonization by fortifying the mucosal barrier. Mice lacking mucin 2, a major constituent of the intestinal mucus, exhibit increased pathogen burden and disease severity after infection with C. rodentium, S. Typhimurium and Listeria monocytogenes76–78. Similarly, thinning of the mucus layer by administration of a fibre-free diet to gnotobiotic mice harbouring a simple bacterial consortium leads to enhanced C. rodentium colonization and epithelial invasion79. However, in conventionally raised mice fed a comparable fibre-free diet, the increase in pathogen colonization is only marginal and temporal80. These results suggest that a diverse microbiota is important to promote mucus barrier function and colonization resistance in the presence of a disrupted mucus layer.
Under homeostatic conditions, the gut microbiota also releases compounds from the mucus layer and the surface of IECs, which can affect pathogen colonization (Fig. 2). For example, fucosylated proteins shed from IECs can be metabolized by gut bacteria to release l-fucose that reduces pathogen virulence and decreases susceptibility to C. rodentium22. By contrast, disruption of the microbiota by antibiotics induces microbiota-dependent release of carbohydrates from the mucus layer that can fuel pathogen growth. For instance, Bacteroides thetaiotaomicron deploys mucin-degrading enzymes to release and metabolize sialic acid, which can be directly utilized by C. difficile and S. Typhimurium to support their own expansion in the gut81. Thus, the microbiota inhibits or enhances pathogen colonization by promoting the release of specific sugars from the mucus layer or IECs via different mechanisms.
Oxygen limitation
The gut microbiota promotes changes in the gut environment that indirectly lead to colonization resistance against enteric pathogens and pathobionts. A clear example of this effect is the establishment and maintenance of intestinal hypoxia by symbiotic bacteria to limit the expansion of pathogenic facultative anaerobes (Fig. 2). For instance, Clostridia species produce the SCFA butyrate that promotes aerobic respiration in IECs through β-oxidation, reducing the oxygen concentration at the epithelial surface82,83. Accordingly, depletion of Clostridia increases oxygen levels in the gut, which in turn fuels aerobic expansion of S. Typhimurium82,84. Likewise, intestinal inflammation and dysbiosis often coincide with a reduction of butyrate and butyrate producers, which increases luminal oxygen and expansion of pathogenic facultative anaerobes85. Symbiotic facultative anaerobes can in turn limit pathogen colonization by competing for or sequestering residual oxygen in the gut, impairing oxygen-dependent virulence gene expression in Shigella flexneri86 and aerobic respiration in C. rodentium and S. enterica subsp. enterica serovar Enteritidis87,88. In addition, other symbionts such as Mucispirillum schaedleri can also compete for anaerobic respiration substrates such as nitrate in the absence of oxygen, limiting the expansion of E. coli and S. Typhimurium that rely on nitrate respiration to thrive in the inflamed gut89–91.
Antimicrobial peptides and proteins
The microbiota can stimulate host epithelial cells to produce various AMPs that can target symbionts and pathogens (Fig. 2). These inhibitory compounds are positively charged and disrupt bacterial membranes by interacting with negative charges on the bacterial surface92. RegIIIβ and RegIIIγ are AMPs contained in the granules of Paneth cells and are released into the intestinal lumen where they promote the spatial segregation of bacterial symbionts from IECs93,94. Impaired production of RegIIIβ and RegIIIγ increases susceptibility to infection with various enteric pathogens in animal models95,96. For example, microbiota depletion by antibiotic administration reduces intestinal expression of RegIIIγ resulting in defective control of VRE97, whereas stimulation with resiquimod (R848), a synthetic ligand for Toll-like receptor 7 (TLR7), restores expression of RegIIIγ in the gut and promotes eradication of VRE in antibiotic-treated mice98. Similarly, NOD1/2 signalling through the receptor-interacting serine–threonine-protein kinase 2 (RIPK2) limits C. rodentium colonization through stimulation of RegIIIβ production at the early phase of infection99. NOD2 stimulation by microbiota-derived peptidoglycan also induces expression of cryptdins, another class of AMPs produced by Paneth cells that promote L. monocytogenes clearance in mice100. In addition to their direct activity against pathogens, AMPs also regulate gut microbiota composition and abundance101, which can also influence colonization resistance.
Lipocalin 2 (LCN2) is another important antimicrobial protein, which sequesters bacterial iron-scavenging siderophores such as enterobactin to prevent pathogen iron acquisition102. LCN2 expression is induced upon inflammation and is dependent on the presence of the gut microbiota103,104. LCN2-deficient mice exhibit elevated levels of enterobactin-expressing gut bacteria and a distinct bacterial community compared with wild type animals, suggesting a role of LCN2 in regulating microbiota composition as well103,104. Like LCN2, calprotectin (a protein released by neutrophils and IECs during inflammation) exerts antimicrobial activity against pathogens by sequestering essential divalent metals such as iron, zinc, calcium and manganese105. However, whether the gut microbiota influences expression and/or release of calprotectin to confer colonization resistance against pathogens remains unclear. Therefore, the gut microbiota has a key role in the production and regulation of host AMPs and proteins that directly inhibit pathogens through different mechanisms. In turn, these molecules can also modify the gut microbiota and thus its colonization resistance effects.
Cytokines
Some cytokines are induced by the microbiota and can exert a protective role in colonization resistance against enteric pathogens (Fig. 2). For example, induction of IL-22 is associated with the presence of segmented filamentous bacteria (SFB) and increased resistance to C. rodentium infection106. Similarly, inoculation of murine norovirus, a persistent and asymptomatic colonizer of the mouse gut, protects young mice against C. rodentium infection in an IL-22-dependent manner as well107,108. IL-22 acts against the pathogen by promoting intestinal barrier function and altering the composition of the gut microbiota109–111, but can also contribute to protection against C. difficile. For example, colonization of human microbiota into GF mice lacking adaptive immunity induces IL-22 production in the gut, which in turn promotes changes in host glycosylation and growth of Phascolarctobacterium species that compete with C. difficile for succinate112. Notably, IL-22 does not confer protection against S. Typhimurium intestinal colonization113 and may even confer this pathogen an advantage over bacterial symbionts in the gut114. Likewise, SFB colonization limits murine rotavirus infection in the gut independently of IL-22, probably by directly altering virus infectivity and/or activity on IECs115. Thus, the role of IL-22 in protection against pathogen colonization is not universal.
The gut microbiota also induces the production of IL-1β in resident macrophages, which primes these cells to respond more rapidly to S. Typhimurium and P. aeruginosa through conversion of pro-IL-1β into IL-1β that enhances pathogen clearance through the recruitment of neutrophils116. Similarly, colonization of GF and antibiotic-treated mice with a defined Bacteroidota consortium protects against Klebsiella pneumoniae via IL-36 signalling117. Butyrate-producing bacteria can also limit pathogen colonization by regulating tight-junction protein expression via IL-10 signalling83,118. Therefore, different cytokines induced by the microbiota can contribute to colonization resistance through multiple mechanisms.
Adaptive immunity
The production of immunoglobulins that contribute to pathogen clearance is also influenced by the gut microbiota (Fig. 2). For example, certain bacterial symbionts induce production of polyreactive, low-affinity IgA antibodies, which can cross-react with antigens expressed by other bacterial species, including enteric pathogens119–121. Therefore, a large proportion of gut symbiotic bacteria are coated with IgA antibodies120,122. Notably, homeostatic secretory IgA derived from human milk or colostrum is protective against necrotoxigenic E. coli infection in GF newborn piglets in a manner dependent on IgA-associated glycans, highlighting the polyreactivity and protective effect of secretory IgA across species123.
Although mucosal antibody responses can also shape microbiota composition in the gut, the degree to which this influences colonization resistance against enteric pathogens remains controversial or largely unexplored. For example, the levels of homeostatic IgA reactive against S. Typhimurium are different between commonly used inbred mouse strains, which correlate with differences in survival after pathogen challenge124. By contrast, mice lacking the polymeric immunoglobulin receptor that cannot transport IgA or IgM into the gut lumen showed increased or reduced susceptibility to S. Typhimurium infection in different studies125,126. Although the reasons for these discrepancies are unclear, they might be explained by the differences in microbiota composition and/or their associated immunoglobulin repertoires in different mice.
Although antibody binding is usually associated with an exclusionary function against pathogens, IgA can also promote the survival of the human symbiont Bacteroides fragilis in monocolonized mice15. Similarly, metagenomic analysis of IgA-bound gut bacteria in healthy individuals and IgA-deficient individuals revealed an unexpected depletion of beneficial symbionts associated with IgA deficiency127. Furthermore, IgA promotes mutualistic interactions between the host and potentially pathogenic fungal symbionts by targeting and suppressing virulent morphotypes, in both mice and humans128. These studies suggest that IgA limits pathogen colonization by promoting the expansion of beneficial symbionts, retaining them in favourable niches or preventing their killing by inhibitory molecules.
Serum IgA and IgG induced by members of the gut microbiota also protect against systemic infection in mice129,130. Notably, depletion of secretory IgT, an ancient immunoglobulin class specialized in mucosal immunity, leads to dysbiosis of the gill microbiota and impaired resistance to mucosal parasite infection in the rainbow trout131. This evidence suggests that steady-state antibody responses against the microbiota emerged early in vertebrate evolution and contribute to host protection against different types of infections.
Pathogen evasion of colonization resistance
Successful pathogens have evolved multiple strategies to overcome colonization resistance. These often involve the use of pathogen virulence programmes to alter the gut environment to better suit them or to overcome the nutritional limitations imposed by the microbiota (Fig. 3). The importance of pathogen virulence in countering colonization resistance is highlighted by the observation that the major virulence programme of C. rodentium is essential for gut colonization in conventionally raised mice, but not in GF mice132. Multiple environmental signals are integrated to decide when and where to express virulence genes after a pathogen enters the gut133. The microbiota can modify some of these signals, and bacterial products such SCFAs134–136, indole137 and 1,2-propanediol138 can also regulate virulence gene expression.
Fig. 3 |. Pathogen evasion of colonization resistance.
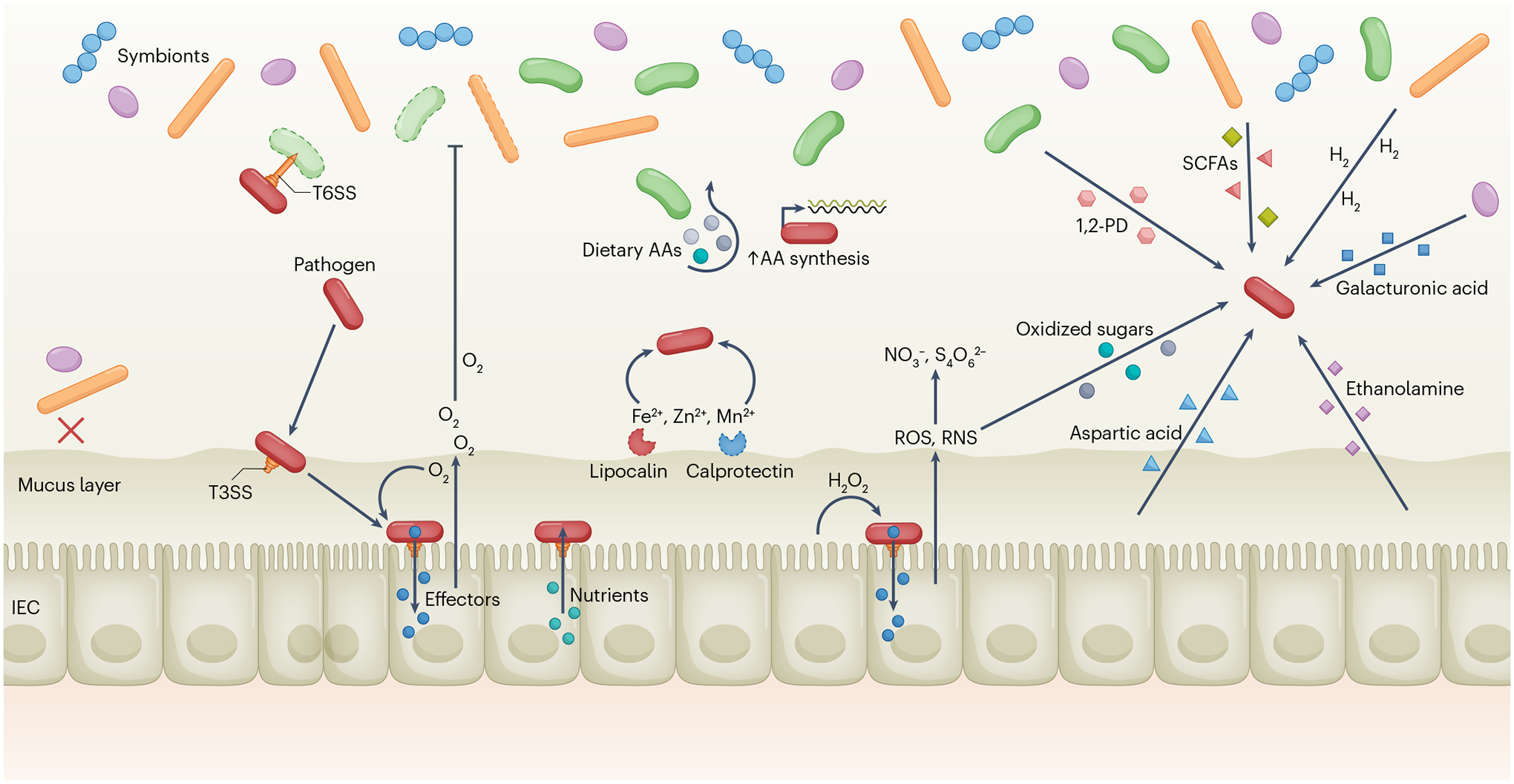
Upon entering the gut, some pathogens can use a type VI secretion system (T6SS) to directly kill competitors and open a niche (upper left). Most symbionts and pathobionts are excluded from the inner mucus layer (lower left). Certain pathogens, such as Citrobacter rodentium, utilize a type III secretion system (T3SS) to adhere to the epithelium and inject effectors into intestinal epithelial cells (IECs) to modify their physiology, including hyperproliferation of epithelial cells, which causes O2 to be released and respired by the pathogen while killing oxygen-sensitive symbiotic bacteria. C. rodentium can also respire hydrogen peroxide (H2O2) produced by IECs during its early, T3SS-mediated access to the epithelium. Enteropathogenic Escherichia coli can also use its T3SS to obtain nutrients directly from the host cell cytoplasm. Dietary amino acids (AAs) are depleted by the microbiota, but pathogens, including C. rodentium, can synthesize AAs de novo to circumvent this nutrient deficiency (centre). Salmonella enterica subsp. enterica serovar Typhimurium has evolved resistant siderophores and higher-affinity transporters to overcome lipocalin 2-mediated and calprotectin-mediated metal (Fe2+, Zn2+, Mn2+) sequestration. Injection of effectors by the T3SS also causes host inflammatory responses, releasing reactive oxygen species (ROS) and reactive nitrogen species (RNS), which produce respiratory electron acceptors such as tetrathionate (S4O62−) and nitrate (NO3−) that can fuel pathogen growth. Oxidized sugars (for example, glucarate and galactarate) are also produced by RNS and used by pathogens. Aspartic acid is released during inflammation and can be utilized for energy production by pathogens. In turn, 1,2-propanediol (1,2-PD), propionic and butyric acids (short-chain fatty acids (SCFAs)), H2 and galacturonic acid are produced by the microbiota and can be metabolized by certain pathogens, as can host-derived ethanolamine, if the appropriate electron acceptors are present.
Niche construction via virulence factors
Pathogens depend on virulence factors for successful infection of a host (Fig. 3). Prime examples are the T3SS used by Salmonella species to inject effector proteins into host cells and to modify their physiology, or the toxins secreted by C. difficile or Vibrio cholerae. Most of the virulence programmes used by enteric pathogens are dedicated to modifying the niche to better enable the growth of the pathogen, also known as ‘niche construction’. C. rodentium, S. Typhimurium and other Enterobacteriaceae are facultative anaerobes that are not only able to grow aerobically but also anaerobically by fermenting or respiring various substrates. Most of the symbiotic bacteria in the large intestine, by contrast, are fermenters and highly oxygen-sensitive. Thus, facultative anaerobe pathogens gain a growth advantage if oxygen or other respiratory substrates became available. For instance, on infecting a mouse, C. rodentium attaches to the caecal or colonic epithelium, where oxygen is highest. Using its T3SS, the pathogen injects effectors into host epithelial cells, which results in the characteristic hyperplasia of crypt cells. This changes the overall metabolism of the epithelium, resulting in decreased oxygen consumption. The excess oxygen can then be respired by C. rodentium87. In fact, even before hyperplasia develops, C. rodentium can respire hydrogen peroxide produced by epithelial NADPH oxidase (NOX1) during early T3SS-mediated attachment139. To circumvent the amino acid limitation imposed by the microbiota, this pathogen induces amino acid biosynthesis genes to make its own amino acids, which is essential for gut colonization29. C. rodentium can also use alternative carbon sources for its initial expansion in the gut, including dietary-derived metabolites that are made available by commensal bacteria activity140. In addition, this pathogen uses its T3SS to localize to or near the epithelium, avoiding spatial and nutritional competition from the microbiota as well132. Furthermore, enteropathogenic E. coli and presumably C. rodentium might use their T3SS to extract nutrients from epithelial cells when attached to IECs to subvert nutritional limitations imposed by the microbiota141.
S. Typhimurium uses its Spi1 T3SS to trigger inflammation in the ileum and caecum of mice after overcoming initial colonization resistance142. Spi1 effectors cause the host to produce reactive oxygen and nitrogen species, which create the byproducts tetrathionate and nitrate that S. Typhimurium can respire143,144, as well as oxidized sugars that it can metabolize145. This pathogen also uses oxygen that becomes more abundant when inflammation represses certain native anaerobes and disrupts epithelial cell oxygen consumption84. Certain carbon sources in the gut are able to be metabolized by S. Typhimurium in the presence of appropriate respiratory substrates, for example, host-derived ethanolamine and the bacterial metabolite 1,2-propanediol146,147. Hydrogen (H2) produced by certain symbionts can be used by the pathogen as a respiratory electron donor148. Notably, Salmonella species can even make use of some of the bacterially produced compounds that normally inhibit its growth, such as the SCFAs propionate and butyrate, to promote its survival149,150. Moreover, S. Typhimurium has evolved ways to outcompete the host and other bacteria for scarce and essential metals such as iron, zinc and manganese, including high-affinity transporters and siderophores that are resistant to host factors151–153. Thus, enteric pathogens can deploy an arsenal of specialized strategies to overcome the colonization resistance imposed by the microbiota.
Direct antagonism of native bacteria
Just as the native microbiota uses direct inhibition against outsiders, pathogens can also use direct methods to compete with the native microbiota and carve out a niche to invade (Fig. 3). The T6SS is used by pathogens as well as by symbionts to compete with one another by injecting various toxic effectors into nearby cells. V. cholerae can use its T6SS to kill competing E. coli in a neonatal mouse model, allowing it to better infect and cause disease154. S. Typhimurium also uses its T6SS to colonize adult mice, in which it can kill a competing Enterobacteriaceae strain: Klebsiella oxytoca155. Shigella sonnei, which causes dysentery in humans, also has a T6SS that can kill the related species S. flexneri or E. coli, which enhances its colonization in the mouse gut, although it is still rapidly cleared by the microbiota156. The T6SS of C. rodentium has been recently shown to have a role during early invasion, through killing of, and avoiding killing by, symbiotic E. coli that also possess a T6SS157.
Conclusions and future perspectives
Symbiotic microorganisms colonize the gut and every epithelial surface in humans and animals and provide many benefits to the host including protection against pathogen colonization. In fact, the native gut microbiota can be harnessed to prevent or treat infections (Box 3). The different mechanisms by which the microbiota mediate colonization resistance are influenced by a wide range of factors including the baseline composition of the microbiota, diet, sex, age and circadian rhythm, as well as pathogen dosage and developmental stage of the pathogen. The protection against pathogen colonization is accomplished largely through multiple mechanisms that indigenous symbionts have evolved to compete with related microorganisms for niches in the gut and other epithelial surfaces. Thus, the direct mechanisms underlying colonization resistance do not target pathogens specifically, in that they are used for constant microbial–microbial competition and microbial warfare in the gut. Although most of the mechanisms of direct colonization resistance are not pathogen-specific, they often exhibit narrow specificity for their targets. This is the case for bacteriocins or contact-dependent killing such as the T6SS. In addition, the microbiota can produce metabolites such as secondary BAs that kill Gram-positive bacteria. Much of the evidence supporting a role for particular microorganisms or microbial molecules in colonization resistance has relied on the use of simplified consortia of microorganisms in gnotobiotic mice. Although the use of such consortia has provided important mechanistic insights in that biological context, the contribution of particular microorganisms or microbial molecules to colonization resistance in the presence of a complete microbiota is unclear. Thus, studies to assess the role of such microorganisms or molecules in conventionally raised animals or humans are needed. Future studies should also provide new insight into the complexity of microbial interactions in their natural niches, particularly those that are occupied by incoming pathogens or opportunistic pathogens that reside in the gut. In addition, the precise location in the intestine where the inhibition of pathogens by the resident microorganisms takes place remains unknown and warrants further investigation. The presence of a dense and diverse population of gut microorganisms that consume most available nutrients, keeping them at low levels at steady state, also limits pathogen colonization and expansion. The limitation of nutrients and physical space is likely to have a broad and important role in colonization resistance against various pathogens. It is also to be expected that consumption of different diets may affect protection against enteric pathogens by altering the amounts of nutrients, although the effect of diet on pathogen colonization resistance remains poorly understood and warrants further investigation as well. Indirect mechanisms of colonization resistance involve mechanisms by which the microbiota limits pathogen colonization primarily through the induction of immune responses in the host. Such mechanisms are not pathogen-specific either, in that epithelial-derived AMPs or polyreactive antibodies can also target bacterial symbionts. Further understanding of the mechanisms of colonization resistance can guide future approaches for preventing and treating infections.
Box 3. Harnessing the microbiota to fight infections.
The use of faecal microbiota transplantation and other microbiota-based therapeutics to treat infectious and inflammatory disorders has been recently reviewed191. The concept that the intestinal microbiota can be harnessed to treat infectious disease was first tested in the 1950s and later demonstrated to be effective in the treatment of recurrent colitis caused by Clostridioides difficile192,193. Infection with C. difficile is associated with the use of broad-spectrum antibiotics that remove symbiotic bacteria that mediate colonization resistance against the bacterium in the gut. The engraftment of the donor microbiota can be predicted largely from the abundance and phylogeny of bacteria in the donor and recipient faeces194. Although faecal microbiota transplantation is highly effective in treating recurrent C. difficile infection, the variable nature and potential pathogen transmission of the faecal microbiota make the implementation of the approach in the clinic challenging. Thus, current efforts are devoted to identifying and characterizing defined consortia of bacterial isolates that mediate protection against C. difficile and other pathogens195. The assembly of effective consortia for the treatment of disease is also challenging because of our lack of knowledge about the gut microbial ecosystem. Increased understanding of the metabolic requirements and microbial–microbial interactions should lead to the development of consortia of microorganisms that can function cooperatively to target pathogens more effectively. However, administration of isolated live microorganisms in the clinic is problematic because probiotics typically have limited effects on the overall composition of the microbiota, as these exogenous bacterial species are often unable to persistently colonize hosts11. Concurrent supplementation with prebiotics may be useful in that regard because it can enhance the colonization of exogenous microorganisms, including enhanced colonization resistance against C. difficile196. Furthermore, understanding of the mechanisms that are used by symbionts to outcompete pathogens may lead to the genetic engineering of microbial species with increased capacity to limit pathogen colonization. For instance, symbiotic bacteria can be engineered to gain a competitive edge over related resident bacteria enhancing their colonization in the gut197. Such approaches could improve our armamentarium to treat intestinal infections such as those caused by C. difficile. Diet and prebiotics can alter the composition of the microbiota and could potentially be effective in the treatment of infectious disease as well. However, such approaches can only be effective if sufficient protective microorganisms are present in the microbiota of the patient, which is not always the case in clinical situations.
Acknowledgements
The authors thank B. Lo for critical review of the manuscript. The authors apologize to their colleagues whose work was not cited because of space limitations. G.C.-F. is supported by a K99/R00 award from the US National Institutes of Health. Work in the authors’ laboratory is supported by US National Institutes of Health grants (G.N.).
Glossary
- Bacteriocins
Antimicrobial peptides produced by bacteria to inhibit the growth of similar or closely related bacteria
- Colonization resistance
Mechanisms by which the intestinal microbiota limits the colonization of pathogens and pathobionts
- Cytokine
Small immunoregulatory protein released by cells that mediates cell-to-cell communication in immune responses
- Germ-free (GF) animals
Animals that are born and raised in isolators without exposure to microorganisms
- Pathobionts
Microorganisms that under normal circumstances live as non-harming symbionts, but under certain conditions, usually involving environmental or genetic alterations, induce disease
- Secretion system
Molecular structure assembled by bacteria that enables them to inject effector proteins directly into the host cytoplasm or other microbial cells
- Prebiotics
Compounds in food that promote the growth or activity of beneficial symbionts
- Probiotics
Live microorganisms that upon administration could confer a health benefit on the host
- Secretory IgA
(sIgA). A dimer of IgA molecules bound by a joining peptide (secretory component). sIgA is the predominant immunoglobulin in mucosal surfaces, including the gut
- Symbionts
Organisms of different species living together in a long-term relationship or symbiosis. Many microbial symbionts are mutualistic in that they and the host benefit from each other, whereas commensals receive benefits from the host, but the host is not helped or harmed
Footnotes
Competing interests
The authors declare no competing interests.
References
- 1.Peterson J et al. The NIH human microbiome project. Genome Res 19, 2317–2323 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gu S et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS ONE 8, e74957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nava GM, Friedrichsen HJ & Stappenbeck TS Spatial organization of intestinal microbiota in the mouse ascending colon. ISME J 5, 627–638 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clemente JC, Ursell LK, Parfrey LW & Knight R The impact of the gut microbiota on human health: an integrative view. Cell 148, 1258–1270 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada N, Chen GY, Inohara N & Núñez G Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol 14, 685–690 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pickard JM, Zeng MY, Caruso R & Núñez G Gut microbiota: role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev 279, 70–89 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavaillon J-M & Legout S Centenary of the death of Elie Metchnikoff: a visionary and an outstanding team leader. Microbes Infect 18, 577–594 (2016). [DOI] [PubMed] [Google Scholar]
- 8.Bohnhoff M, Drake BL & Miller CP Effect of streptomycin on susceptibility of intestinal tract to experimental Salmonella infection. Proc. Soc. Exp. Biol. Med 86, 132–137 (1954). [DOI] [PubMed] [Google Scholar]
- 9.Freter R The fatal enteric cholera infection in the guinea pig, achieved by inhibition of normal enteric flora. J. Infect. Dis 97, 57–65 (1955). [DOI] [PubMed] [Google Scholar]
- 10.van der Waaij D, Berghuis-de Vries JM & Lekkerkerk-Van Der Wees JEC Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. J. Hyg 69, 405–411 (1971). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kristensen NB et al. Alterations in fecal microbiota composition by probiotic supplementation in healthy adults: a systematic review of randomized controlled trials. Genome Med 8, 52 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stecher B Establishing causality in Salmonella-microbiota-host interaction interaction: the use of gnotobiotic mouse models and synthetic microbial communities. Int. J. Med. Microbiol 311, 151484 (2021). [DOI] [PubMed] [Google Scholar]
- 13.Hutchinson GE An Introduction to Population Ecology (Yale Univ. Press, 1978). [Google Scholar]
- 14.Lee SM et al. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature 501, 426–429 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Donaldson GP et al. Gut microbiota utilize immunoglobulin A for mucosal colonization. Science 360, 795–800 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that microbiota-specific IgA antibodies can enhance colonization of their target bacteria in the gut.
- 16.Sweeney NJ et al. The Escherichia coli K-12 gntP gene allows E. coli F-18 to occupy a distinct nutritional niche in the streptomycin-treated mouse large intestine. Infect. Immun 64, 3497–3503 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maltby R, Leatham-Jensen MP, Gibson T, Cohen PS & Conway T Nutritional basis for colonization resistance by human commensal Escherichia coli strains HS and Nissle 1917 against E. coli O157:H7 in the mouse intestine. PLoS ONE 8, e53957 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Freter R, Brickner H, Botney M, Cleven D & Aranki A Mechanisms that control bacterial populations in continuous-flow culture models of mouse large intestinal flora. Infect. Immun 39, 676–685 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mullineaux-Sanders C et al. Citrobacter amalonaticus inhibits the growth of Citrobacter rodentium in the gut lumen. mBio 12, e0241021 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leslie JL et al. Protection from lethal Clostridioides difficile infection via intraspecies competition for cogerminant. mBio 12, e00522–21 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Osbelt L et al. Klebsiella oxytoca causes colonization resistance against multidrug-resistant K. pneumoniae in the gut via cooperative carbohydrate competition. Cell Host Microbe 29, 1663–1679.e7 (2021). [DOI] [PubMed] [Google Scholar]
- 22.Pickard JM et al. Rapid fucosylation of intestinal epithelium sustains host-commensal symbiosis in sickness. Nature 514, 638–641 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liou MJ et al. Host cells subdivide nutrient niches into discrete biogeographical microhabitats for gut microbes. Cell Host Microbe 30, 836–847.e6 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]; This recent work shows how the microscopic location of a nutrient can determine its availability to a bacterial species in the intestine.
- 24.Leatham-Jensen MP et al. The streptomycin-treated mouse intestine selects Escherichia coli envZ missense mutants that interact with dense and diverse intestinal microbiota. Infect. Immun 80, 1716–1727 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moor K et al. High-avidity IgA protects the intestine by enchaining growing bacteria. Nature 544, 498–502 (2017). [DOI] [PubMed] [Google Scholar]
- 26.McLoughlin K, Schluter J, Rakoff-Nahoum S, Smith AL & Foster KR Host selection of microbiota via differential adhesion. Cell Host Microbe 19, 550–559 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Bry L, Falk PG, Midtvedt T & Gordon JI A model of host–microbial interactions in an open mammalian ecosystem. Science 273, 1380–1383 (1996). [DOI] [PubMed] [Google Scholar]
- 28.Turnbaugh PJ et al. A core gut microbiome in obese and lean twins. Nature 457, 480–484 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Caballero-Flores G, Pickard JM, Fukuda S, Inohara N & Núñez G An enteric pathogen subverts colonization resistance by evading competition for amino acids in the gut. Cell Host Microbe 28, 526–533.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study performed a genome-wide screen to identify pathogen genes required for gut colonization in conventionally raised and GF mice.
- 30.Theriot CM et al. Antibiotic-induced shifts in the mouse gut microbiome and metabolome increase susceptibility to Clostridium difficile infection. Nat. Commun 5, 3114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stecher B et al. Like will to like: abundances of closely related species can predict susceptibility to intestinal colonization by pathogenic and commensal bacteria. PLoS Pathog 6, e1000711 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brugiroux S et al. Genome-guided design of a defined mouse microbiota that confers colonization resistance against Salmonella enterica serovar Typhimurium. Nat. Microbiol 2, 16215 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Velazquez EM et al. Endogenous Enterobacteriaceae underlie variation in susceptibility to Salmonella infection. Nat. Microbiol 4, 1057–1064 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deriu E et al. Probiotic bacteria reduce Salmonella Typhimurium intestinal colonization by competing for iron. Cell Host Microbe 14, 26–37 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Behnsen J et al. Siderophore-mediated zinc acquisition enhances enterobacterial colonization of the inflamed gut. Nat. Commun 12, 7016 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Heilbronner S, Krismer B, Brötz-Oesterhelt H & Peschel A The microbiome-shaping roles of bacteriocins. Nat. Rev. Microbiol 19, 726–739 (2021). [DOI] [PubMed] [Google Scholar]
- 37.Gillor O, Giladi I & Riley MA Persistence of colicinogenic Escherichia coli in the mouse gastrointestinal tract. BMC Microbiol 9, 165 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sassone-Corsi M et al. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature 540, 280–283 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coyne MJ et al. A family of anti-Bacteroidales peptide toxins wide-spread in the human gut microbiota. Nat. Commun 10, 3460 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Roelofs KG, Coyne MJ, Gentyala RR, Chatzidaki-Livanis M & Comstock LE Bacteroidales secreted antimicrobial proteins target surface molecules necessary for gut colonization and mediate competition in vivo. mBio 7, e01055–16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Reyes K, Bardossy AC & Zervos M Vancomycin-resistant Enterococci: epidemiology, infection prevention, and control. Infect. Dis. Clin. North Am 30, 953–965 (2016). [DOI] [PubMed] [Google Scholar]
- 42.Kommineni S et al. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526, 719–722 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gilmore MS et al. Pheromone killing of multidrug-resistant Enterococcus faecalis V583 by native commensal strains. Proc. Natl Acad. Sci. USA 112, 7273–7278 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Caballero S et al. Cooperating commensals restore colonization resistance to vancomycin-resistant Enterococcus faecium. Cell Host Microbe 21, 592–602.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim SG et al. Microbiota-derived lantibiotic restores resistance against vancomycin-resistant Enterococcus. Nature 572, 665–669 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identifies a precise molecular mechanism behind the colonization resistance conferred by a defined group of bacterial species.
- 46.Aoki SK et al. Contact-dependent inhibition of growth in Escherichia coli. Science 309, 1245–1248 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Aoki SK et al. A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hayes CS, Koskiniemi S, Ruhe ZC, Poole SJ & Low DA Mechanisms and biological roles of contact-dependent growth inhibition systems. Cold Spring Harb. Perspect. Med 4, a010025 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hood RD et al. A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Coyne MJ, Roelofs KG & Comstock LE Type VI secretion systems of human gut Bacteroidales segregate into three genetic architectures, two of which are contained on mobile genetic elements. BMC Genomics 17, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ross BD et al. Human gut bacteria contain acquired interbacterial defence systems. Nature 575, 224–228 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Flaugnatti N et al. Human commensal gut Proteobacteria withstand type VI secretion attacks through immunity protein-independent mechanisms. Nat. Commun 12, 5751 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Russell AB et al. A type VI secretion-related pathway in Bacteroidetes mediates interbacterial antagonism. Cell Host Microbe 16, 227–236 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chatzidaki-Livanis M, Geva-Zatorsky N & Comstock LE Bacteroides fragilis type VI secretion systems use novel effector and immunity proteins to antagonize human gut Bacteroidales species. Proc. Natl Acad. Sci. USA 113, 3627–3632 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wexler AG et al. Human symbionts inject and neutralize antibacterial toxins to persist in the gut. Proc. Natl Acad. Sci. USA 113, 3639–3644 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hecht AL et al. Strain competition restricts colonization of an enteric pathogen and prevents colitis. EMBO Rep 17, 1281–1291 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Souza DP et al. Bacterial killing via a type IV secretion system. Nat. Commun 6, 6453 (2015). [DOI] [PubMed] [Google Scholar]
- 58.Whitney JC et al. A broadly distributed toxin family mediates contact-dependent antagonism between Gram-positive bacteria. eLife 6, e26938 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Guzior DV & Quinn RA Review: microbial transformations of human bile acids. Microbiome 9, 140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sannasiddappa TH, Lund PA & Clarke SR In vitro antibacterial activity of unconjugated and conjugated bile salts on Staphylococcus aureus. Front. Microbiol 8, 1581 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Watanabe M, Fukiya S & Yokota A Comprehensive evaluation of the bactericidal activities of free bile acids in the large intestine of humans and rodents. J. Lipid Res 58, 1143–1152 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sun X et al. Microbiota-derived metabolic factors reduce Campylobacteriosis in mice. Gastroenterology 154, 1751–1763.e2 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Buffie CG et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517, 205–208 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Studer N et al. Functional intestinal bile acid 7α-dehydroxylation by Clostridium scindens associated with protection from Clostridium difficile infection in a gnotobiotic mouse model. Front. Cell. Infect. Microbiol 6, 191 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Theriot CM, Bowman AA & Young VB Antibiotic-induced alterations of the gut microbiota alter secondary bile acid production and allow for Clostridium difficile spore germination and outgrowth in the large intestine. mSphere 1, e00045–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lewis BB, Carter RA & Pamer EG Bile acid sensitivity and in vivo virulence of clinical Clostridium difficile isolates. Anaerobe 41, 32–36 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lewis BB et al. Pathogenicity locus, core genome, and accessory gene contributions to Clostridium difficile virulence. mBio 8, e00885–17 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aguirre AM et al. Bile acid-independent protection against Clostridioides difficile infection. PLoS Pathog 17, e1010015 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Bohnhoff M, Miller CP & Martin WR Resistance of the mouse’s intestinal tract to experimental Salmonella infection. I. Factors which interfere with the initiation of infection by oral inoculation. J. Exp. Med 120, 805–816 (1964). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shin R, Suzuki M & Morishita Y Influence of intestinal anaerobes and organic acids on the growth of enterohaemorrhagic Escherichia coli O157:H7. J. Med. Microbiol 51, 201–206 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Rolfe RD Role of volatile fatty acids in colonization resistance to Clostridium difficile. Infect. Immun 45, 185–191 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Osbelt L et al. Variations in microbiota composition of laboratory mice influence Citrobacter rodentium infection via variable short-chain fatty acid production. PLoS Pathog 16, e1008448 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sorbara MT et al. Inhibiting antibiotic-resistant Enterobacteriaceae by microbiota-mediated intracellular acidification. J. Exp. Med 216, 84–98 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Petersson J et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am. J. Physiol. Gastrointest. Liver Physiol 300, 327–333 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Johansson MEV et al. Normalization of host intestinal mucus layers requires long-term microbial colonization. Cell Host Microbe 18, 582–592 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Bergstrom KSB et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog 6, e1000902 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zarepour M et al. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun 81, 3672–3683 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang T, Sasabe J, Hullahalli K, Sit B & Waldor MK Increased Listeria monocytogenes dissemination and altered population dynamics in Muc2-deficient mice. Infect. Immun 89, e00667–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Desai MS et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 167, 1339–1353.e21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Neumann M et al. Deprivation of dietary fiber in specific-pathogen-free mice promotes susceptibility to the intestinal mucosal pathogen Citrobacter rodentium. Gut Microbes 13, 1966263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ng KM et al. Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 502, 96–99 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Byndloss MX et al. Microbiota-activated PPAR-γ signaling inhibits dysbiotic Enterobacteriaceae expansion. Science 357, 570–575 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kelly CJ et al. Crosstalk between microbiota-derived short-chain fatty acids and intestinal epithelial HIF augments tissue barrier function. Cell Host Microbe 17, 662–671 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Rivera-Chávez F et al. Depletion of butyrate-producing Clostridia from the gut microbiota drives an aerobic luminal expansion of Salmonella. Cell Host Microbe 19, 443–454 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Litvak Y, Byndloss MX, Tsolis RM & Bäumler AJ Dysbiotic Proteobacteria expansion: a microbial signature of epithelial dysfunction. Curr. Opin. Microbiol 39, 1–6 (2017). [DOI] [PubMed] [Google Scholar]
- 86.Marteyn B et al. Modulation of Shigella virulence in response to available oxygen in vivo. Nature 465, 355–358 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lopez CA et al. Virulence factors enhance Citrobacter rodentium expansion through aerobic respiration. Science 353, 1249–1253 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Litvak Y et al. Commensal Enterobacteriaceae protect against Salmonella colonization through oxygen competition. Cell Host Microbe 25, 128–139.e5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Winter SE et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339, 708–711 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Spees AM et al. Streptomycin-induced inflammation enhances Escherichia coli gut colonization through nitrate respiration. mBio 4, e00430–13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Herp S et al. Mucispirillum schaedleri antagonizes Salmonella virulence to protect mice against colitis. Cell Host Microbe 25, 681–694.e8 (2019). [DOI] [PubMed] [Google Scholar]
- 92.Gong T, Fu J, Shi L, Chen X & Zong X Antimicrobial peptides in gut health: a review. Front. Nutr 8, 751010 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vaishnava S et al. The antibacterial lectin RegIIIγ promotes the spatial segregation of microbiota and host in the intestine. Science 334, 255–258 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Cash HL, Whitham CV, Behrendt CL & Hooper LV Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science 313, 1126–1130 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Brandl K, Plitas G, Schnabl B, DeMatteo RP & Pamer EG MyD88-mediated signals induce the bactericidal lectin RegIIIγ and protect mice against intestinal Listeria monocytogenes infection. J. Exp. Med 204, 1891–1900 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.van Ampting MTJ et al. Intestinally secreted C-type lectin Reg3b attenuates salmonellosis but not listeriosis in mice. Infect. Immun 80, 1115–1120 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Brandl K et al. Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Abt MC et al. TLR-7 activation enhances IL-22-mediated colonization resistance against vancomycin-resistant enterococcus. Sci. Transl Med 8, 327ra25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Waldschmitt N et al. The regenerating family member 3 β instigates IL-17A-mediated neutrophil recruitment downstream of NOD1/2 signalling for controlling colonization resistance independently of microbiota community structure. Gut 68, 1190–1199 (2019). [DOI] [PubMed] [Google Scholar]
- 100.Kobayashi KS et al. Nod2-dependent regulation of innate and adaptive immunity in the intestinal tract. Science 307, 731–734 (2005). [DOI] [PubMed] [Google Scholar]
- 101.Bosch TCG & Zasloff M Antimicrobial peptides — or how our ancestors learned to control the microbiome. mBio 12, e0184721 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Mayneris-Perxachs J, Moreno-Navarrete JM & Fernández-Real JM The role of iron in host–microbiota crosstalk and its effects on systemic glucose metabolism. Nat. Rev. Endocrinol 18, 683–698 (2022). [DOI] [PubMed] [Google Scholar]
- 103.Singh V et al. Microbiota-inducible innate immune siderophore binding protein lipocalin 2 is critical for intestinal homeostasis. Cell. Mol. Gastroenterol. Hepatol 2, 482–498.e6 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Klüber P et al. Depletion of lipocalin 2 (LCN2) in mice leads to dysbiosis and persistent colonization with segmented filamentous bacteria. Int. J. Mol. Sci 22, 13156 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Zygiel EM & Nolan EM Transition metal sequestration by the host-defense protein calprotectin. Annu. Rev. Biochem 87, 621–643 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Ivanov II et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell 139, 485–498 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kernbauer E, Ding Y & Cadwell K An enteric virus can replace the beneficial function of commensal bacteria. Nature 516, 94–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Neil JA et al. IFN-I and IL-22 mediate protective effects of intestinal viral infection. Nat. Microbiol 4, 1737–1749 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zheng Y et al. Interleukin-22 mediates early host defense against attaching and effacing bacterial pathogens. Nat. Med 14, 282–289 (2008). [DOI] [PubMed] [Google Scholar]
- 110.Guo X et al. Innate lymphoid cells control early colonization resistance against intestinal pathogens through ID2-dependent regulation of the microbiota. Immunity 42, 731–743 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen J, Waddell A, Lin YD & Cantorna MT Dysbiosis caused by vitamin D receptor deficiency confers colonization resistance to Citrobacter rodentium through modulation of innate lymphoid cells. Mucosal Immunol 8, 618–626 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Nagao-Kitamoto H et al. Interleukin-22-mediated host glycosylation prevents Clostridioides difficile infection by modulating the metabolic activity of the gut microbiota. Nat. Med 26, 608–617 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work describes that IL-22 modulates glycosylation of host N-linked glycans in response to microbiota colonization, whereas host-derived glycans promote growth of protective symbionts that compete with C. difficile for a nutritional niche in the gut.
- 113.Kim YG et al. Neonatal acquisition of Clostridia species protects against colonization by bacterial pathogens. Science 356, 315–319 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that the lack of colonization resistance in the neonatal gut is caused by the absence of Clostridiales independently of the host immune system.
- 114.Behnsen J et al. The cytokine IL-22 promotes pathogen colonization by suppressing related commensal bacteria. Immunity 40, 262–273 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Shi Z et al. Segmented filamentous bacteria prevent and cure rotavirus infection. Cell 179, 644–658.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Franchi L et al. NLRC4-driven production of IL-1β discriminates between pathogenic and commensal bacteria and promotes host intestinal defense. Nat. Immunol 13, 449–456 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Sequeira RP, McDonald JAK, Marchesi JR & Clarke TB Commensal Bacteroidetes protect against Klebsiella pneumoniae colonization and transmission through IL-36 signalling. Nat. Microbiol 5, 304–313 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study unravels distinct microbiota-mediated immune responses that limit K. pneumoniae colonization in different niches, showing that protection in the gut depends on the development of Bacteroidota and IL-36, whereas in the upper airways, Pseudomonadota prime immunity through IL-17A.
- 118.Zheng L et al. Microbial-derived butyrate promotes epithelial barrier function through IL-10 receptor-dependent repression of claudin-2. J. Immunol 199, 2976–2984 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Macpherson AJ et al. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science 288, 2222–2226 (2000). [DOI] [PubMed] [Google Scholar]
- 120.Bunker JJ et al. Innate and adaptive humoral responses coat distinct commensal bacteria with immunoglobulin A. Immunity 43, 541–553 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Bunker JJ et al. Natural polyreactive IgA antibodies coat the intestinal microbiota. Science 358, eaan6619 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van der Waaij LA, Limburg PC, Mesander G & van der Waaij D In vivo IgA coating of anaerobic bacteria in human faeces. Gut 38, 348–354 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Raskova Kafkova L et al. Secretory IgA N-glycans contribute to the protection against E. coli O55 infection of germ-free piglets. Mucosal Immunol 14, 511–522 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Fransen F et al. BALB/c and C57BL/6 mice differ in polyreactive IgA abundance, which impacts the generation of antigen-specific IgA and microbiota diversity. Immunity 43, 527–540 (2015). [DOI] [PubMed] [Google Scholar]
- 125.Wijburg OLC et al. Innate secretory antibodies protect against natural Salmonella typhimurium infection. J. Exp. Med 203, 21–26 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Betz KJ et al. Enhanced survival following oral and systemic Salmonella enterica serovar Typhimurium infection in polymeric immunoglobulin receptor knockout mice. PLoS ONE 13, e0198434 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Fadlallah J et al. Microbial ecology perturbation in human IgA deficiency. Sci. Transl Med 10, eaan1217 (2018). [DOI] [PubMed] [Google Scholar]
- 128.Ost KS et al. Adaptive immunity induces mutualism between commensal eukaryotes. Nature 596, 114–118 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]; This study reveals that intestinal IgA preferentially targets and suppresses pathogenic (hyphal) morphotypes of Candida albicans, promoting commensalism between this opportunistic pathogen and the host during homeostatic conditions.
- 129.Wilmore JR et al. Commensal microbes induce serum IgA responses that protect against polymicrobial sepsis. Cell Host Microbe 23, 302–311.e3 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Zeng MY et al. Gut microbiota-induced immunoglobulin G controls systemic infection by symbiotic bacteria and pathogens. Immunity 44, 647–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu Z et al. Specialization of mucosal immunoglobulins in pathogen control and microbiota homeostasis occurred early in vertebrate evolution. Sci. Immunol 5, eaay3254 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This is a good example of how secretory immunoglobulins protect against mucosal pathogens and preserve microbiota homeostasis in organisms other than mammals, suggesting a functionally convergent evolution between phylogenetically distant secretory immunoglobulins.
- 132.Kamada N et al. Regulated virulence controls the ability of a pathogen to compete with the gut microbiota. Science 336, 1325–1329 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bajaj V, Lucas RL, Hwang C & Lee CA Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol 22, 703–714 (1996). [DOI] [PubMed] [Google Scholar]
- 134.Hung CC et al. The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol. Microbiol 87, 1045–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Nakanishi N et al. Regulation of virulence by butyrate sensing in enterohaemorrhagic Escherichia coli. Microbiology 155, 521–530 (2009). [DOI] [PubMed] [Google Scholar]
- 136.Durant JA, Corrier DE & Ricke SC Short-chain volatile fatty acids modulate the expression of the hilA and invF genes of Salmonella typhimurium. J. Food Prot 63, 573–578 (2000). [DOI] [PubMed] [Google Scholar]
- 137.Kohli N et al. The microbiota metabolite indole inhibits Salmonella virulence: involvement of the PhoPQ two-component system. PLoS ONE 13, e0190613 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Connolly JPR et al. Host-associated niche metabolism controls enteric infection through fine-tuning the regulation of type 3 secretion. Nat. Commun 9, 4187 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Miller BM et al. Anaerobic respiration of NOX1-derived hydrogen peroxide licenses bacterial growth at the colonic surface. Cell Host Microbe 28, 789–797.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]; This work demonstrates that host NOX1-derived H2O2 fuels anaerobic growth of C. rodentium during early infection, whereas the pathogen relies on aerobic respiration for its expansion in the inflamed gut.
- 140.Jimenez AG, Ellermann M, Abbott W & Sperandio V Diet-derived galacturonic acid regulates virulence and intestinal colonization in enterohaemorrhagic Escherichia coli and Citrobacter rodentium. Nat. Microbiol 5, 368–378 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Pal RR et al. Pathogenic E. coli extracts nutrients from infected host cells utilizing injectisome components. Cell 177, 683–696.e18 (2019). [DOI] [PubMed] [Google Scholar]; This work reports that host-attached enteropathogenic E. coli extract nutrients from infected cells in vitro using core components of its T3SS, an intriguing possible explanation behind the characteristic virulence programme of attaching or effacing pathogens in the gut.
- 142.Fattinger SA, Sellin ME & Hardt WD Salmonella effector driven invasion of the gut epithelium: breaking in and setting the house on fire. Curr. Opin. Microbiol 64, 9–18 (2021). [DOI] [PubMed] [Google Scholar]
- 143.Winter SE et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467, 426–429 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lopez CA et al. Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3, e00143–12 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Faber F et al. Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 534, 697–699 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Thiennimitr P et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl Acad. Sci. USA 108, 17480–17485 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Faber F et al. Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13, e1006129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Maier L et al. Microbiota-derived hydrogen fuels Salmonella Typhimurium invasion of the gut ecosystem. Cell Host Microbe 14, 641–651 (2013). [DOI] [PubMed] [Google Scholar]
- 149.Shelton CD et al. Salmonella enterica serovar Typhimurium uses anaerobic respiration to overcome propionate-mediated colonization resistance. Cell Rep 38, 110180 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bronner DN et al. Genetic ablation of butyrate utilization attenuates gastrointestinal Salmonella disease. Cell Host Microbe 23, 266–273.e4 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper shows that an inhibitory compound produced by the gut microbiota, butyrate, is also exploited by Salmonella species for better colonization and infection.
- 151.Raffatellu M et al. Lipocalin-2 resistance confers an advantage to Salmonella enterica serotype Typhimurium for growth and survival in the inflamed intestine. Cell Host Microbe 5, 476–486 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Liu JZ et al. Zinc sequestration by the neutrophil protein calprotectin enhances Salmonella growth in the inflamed gut. Cell Host Microbe 11, 227–239 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Diaz-Ochoa VE et al. Salmonella mitigates oxidative stress and thrives in the inflamed gut by evading calprotectin-mediated manganese sequestration. Cell Host Microbe 19, 814–825 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Zhao W, Caro F, Robins W & Mekalanos JJ Antagonism toward the intestinal microbiota and its effect on Vibrio cholerae virulence. Science 359, 210–213 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Sana TG et al. Salmonella Typhimurium utilizes a T6SS-mediated antibacterial weapon to establish in the host gut. Proc. Natl Acad. Sci. USA 113, 5044–5051 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Anderson MC, Vonaesch P, Saffarian A, Marteyn BS & Sansonetti PJ Shigella sonnei encodes a functional T6SS used for interbacterial competition and niche occupancy. Cell Host Microbe 21, 769–776.e3 (2017). [DOI] [PubMed] [Google Scholar]
- 157.Serapio-Palacios A et al. Type VI secretion systems of pathogenic and commensal bacteria mediate niche occupancy in the gut. Cell Rep 39, 110731 (2022). [DOI] [PubMed] [Google Scholar]
- 158.Shao Y et al. Stunted microbiota and opportunistic pathogen colonization in caesarean-section birth. Nature 574, 117–121 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Dominguez-Bello MG et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl Acad. Sci. USA 107, 11971–11975 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bäckhed F et al. Dynamics and stabilization of the human gut microbiome during the first year of life. Cell Host Microbe 17, 690–703 (2015). [DOI] [PubMed] [Google Scholar]; This study characterizes the gut microbiota during the first year of life and assesses the impact of mode of delivery and feeding on its establishment in a large patient cohort.
- 161.Lanata CF et al. Global causes of diarrheal disease mortality in children. PLoS ONE 8, e72788 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Prabhudas M et al. Challenges in infant immunity: implications for responses to infection and vaccines. Nat. Immunol 12, 189–194 (2011). [DOI] [PubMed] [Google Scholar]
- 163.Deshmukh HS et al. The microbiota regulates neutrophil homeostasis and host resistance to Escherichia coli K1 sepsis in neonatal mice. Nat. Med 20, 524–530 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou P et al. Perinatal antibiotic exposure affects the transmission between maternal and neonatal microbiota and is associated with early-onset sepsis. mSphere 5, e00984–19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Singer JR et al. Preventing dysbiosis of the neonatal mouse intestinal microbiome protects against late-onset sepsis. Nat. Med 25, 1772–1782 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Zheng W et al. Microbiota-targeted maternal antibodies protect neonates from enteric infection. Nature 577, 543–548 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Chen YE, Fischbach MA & Belkaid Y Skin microbiota–host interactions. Nature 553, 427–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Grice EA et al. Topographical and temporal diversity of the human skin microbiome. Science 324, 1190–1192 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.O’Sullivan JN, Rea MC, O’Connor PM, Hill C & Ross RP Human skin microbiota is a rich source of bacteriocin-producing staphylococci that kill human pathogens. FEMS Microbiol. Ecol 95, fiy241 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Cogen AL et al. Selective antimicrobial action is provided by phenol-soluble modulins derived from Staphylococcus epidermidis, a normal resident of the skin. J. Invest. Dermatol 130, 192–200 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Bitschar K et al. Lugdunin amplifies innate immune responses in the skin in synergy with host- and microbiota-derived factors. Nat. Commun 10, 2730 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Nakatsuji T et al. Antimicrobials from human skin commensal bacteria protect against Staphylococcus aureus and are deficient in atopic dermatitis. Sci. Transl Med 9, eaah4680 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Bomar L, Brugger SD, Yost BH, Davies SS & Lemon KP Corynebacterium accolens releases antipneumococcal free fatty acids from human nostril and skin surface triacylglycerols. mBio 7, e01725–15 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Novick RP & Geisinger E Quorum sensing in staphylococci. Annu. Rev. Genet 42, 541–564 (2008). [DOI] [PubMed] [Google Scholar]
- 175.Ji G, Beavis R & Novick RP Bacterial interference caused by autoinducing peptide variants. Science 276, 2027–2030 (1997). [DOI] [PubMed] [Google Scholar]
- 176.Paharik AE et al. Coagulase-negative staphylococcal strain prevents Staphylococcus aureus colonization and skin infection by blocking quorum sensing. Cell Host Microbe 22, 746–756.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Williams MR et al. Quorum sensing between bacterial species on the skin protects against epidermal injury in atopic dermatitis. Sci. Transl Med 11, eaat8329 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Naik S et al. Compartmentalized control of skin immunity by resident commensals. Science 337, 1115–1119 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Bassis CM, Tang AL, Young VB & Pynnonen MA The nasal cavity microbiota of healthy adults. Microbiome 2, 27 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Janek D, Zipperer A, Kulik A, Krismer B & Peschel A High frequency and diversity of antimicrobial activities produced by nasal Staphylococcus strains against bacterial competitors. PLoS Pathog 12, e1005812 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Zipperer A et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 535, 511–516 (2016). [DOI] [PubMed] [Google Scholar]
- 182.Iwase T et al. Staphylococcus epidermidis Esp inhibits Staphylococcus aureus biofilm formation and nasal colonization. Nature 465, 346–349 (2010). [DOI] [PubMed] [Google Scholar]
- 183.Sugimoto S et al. Staphylococcus epidermidis Esp degrades specific proteins associated with Staphylococcus aureus biofilm formation and host–pathogen interaction. J. Bacteriol 195, 1645–1655 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Liu Q et al. Staphylococcus epidermidis contributes to healthy maturation of the nasal microbiome by stimulating antimicrobial peptide production. Cell Host Microbe 27, 68–78.e5 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Ravel J et al. Vaginal microbiome of reproductive-age women. Proc. Natl Acad. Sci. USA 108, 4680–4687 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Fredricks DN, Fiedler TL & Marrazzo JM Molecular identification of bacteria associated with bacterial vaginosis. N. Engl. J. Med 353, 1899–1911 (2005). [DOI] [PubMed] [Google Scholar]
- 187.Turovskiy Y, Sutyak Noll K & Chikindas ML The aetiology of bacterial vaginosis. J. Appl. Microbiol 110, 1105–1128 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Klebanoff SJ, Hillier SL, Eschenbach DA & Waltersdorph M Control of the microbial flora of the vagina by H2O2-generating lactobacilli. J. Infect. Dis 164, 94–100 (1991). [DOI] [PubMed] [Google Scholar]
- 189.Voravuthikunchai SP, Bilasoi S & Supamala O Antagonistic activity against pathogenic bacteria by human vaginal lactobacilli. Anaerobe 12, 221–226 (2006). [DOI] [PubMed] [Google Scholar]
- 190.Murphy K & Mitchell CM The interplay of host immunity, environment and the risk of bacterial vaginosis and associated reproductive health outcomes. J. Infect. Dis 214, S29–S35 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Sorbara MT & Pamer EG Microbiome-based therapeutics. Nat. Rev. Microbiol 20, 365–380 (2022). [DOI] [PubMed] [Google Scholar]
- 192.Eiseman B, Silen W, Bascom GS & Kauvar AJ Fecal enema as an adjunct in the treatment of pseudomembranous enterocolitis. Surgery 44, 854–859 (1958). [PubMed] [Google Scholar]
- 193.van Nood E et al. Duodenal infusion of donor feces for recurrent Clostridium difficile. N. Engl. J. Med 368, 407–415 (2013). [DOI] [PubMed] [Google Scholar]
- 194.Smillie CS et al. Strain tracking reveals the determinants of bacterial engraftment in the human gut following fecal microbiota transplantation. Cell Host Microbe 23, 229–240.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Dsouza M et al. Colonization of the live biotherapeutic product VE303 and modulation of the microbiota and metabolites in healthy volunteers. Cell Host Microbe 30, 583–598.e8 (2022). [DOI] [PubMed] [Google Scholar]
- 196.Mills JP, Rao K & Young VB Probiotics for prevention of Clostridium difficile infection. Curr. Opin. Gastroenterol 34, 3–10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Shepherd ES, Deloache WC, Pruss KM, Whitaker WR & Sonnenburg JL An exclusive metabolic niche enables strain engraftment in the gut microbiota. Nature 557, 434–438 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]


