Abstract
Synthetic biology enables design of gene networks to confer specific biological functions, yet it remains a challenge to rationally engineer a biological trait as complex as longevity. A naturally-occurring toggle switch underlies fate decisions toward either nucleolar or mitochondrial decline during aging of yeast cells. We rewired this endogenous toggle to engineer an autonomous genetic clock that generates sustained oscillations between the nucleolar and mitochondrial aging processes in individual cells. These oscillations increased cellular lifespan through delay of the commitment to aging that resulted from either loss of chromatin silencing or depletion of heme. Our results establish a connection between gene network architecture and cellular longevity that could lead to rationally-designed gene circuits that slow aging.
One-Sentence Summary:
Preventing cells from the normal deterioration of aging through synthetic biology can markedly extend lifespan.
The era of genomic sequencing has generated a huge body of knowledge defining molecular components and interactions within gene networks that control cellular functions. However, further advances in understanding how these networks confer biological functions have been hindered by the daunting complexity of related regulatory interactions (1). One strategy in synthetic biology is to build simple orthogonal networks analogous to the core parts of natural systems that can be used to uncover key design principles of biological functions that are embedded in sophisticated network connections (2, 3). For example, synthetic networks have been constructed to enable specific dynamic behaviors or functions, such as toggle switches, genetic oscillators, cellular counters, homeostasis, and multistability (4–12). As technologies for engineering biological systems improve rapidly, synthetic biology also offers a powerful approach to rewire and perturb intricate endogenous networks and interrogate the relationship between network structure and cellular functions (3, 13–19). We engineered an oscillatory gene network that effectively promotes longevity of the cell.
Cellular aging is a fundamental and complex biological process that is an underlying driver for many diseases (20). We studied replicative aging of the yeast Saccharomyces cerevisiae, which has proven to be a genetically tractable model for the aging of mitotic cell types such as stem cells and has led to the identification of well-conserved genetic factors that influence longevity in eukaryotes (21–26). For instance, the lysine deacetylase Sir2 and heme-activated protein (HAP) complex are deeply conserved, well-characterized transcriptional regulators that govern yeast aging and lifespan. Sir2 mediates chromatin silencing at ribosomal DNA (rDNA) to maintain the stability of this fragile genomic locus and the integrity of the nucleolus (27–30). HAP regulates the expression of genes important for heme biogenesis and mitochondrial function (31).
To track rDNA silencing during aging of WT yeast cells, we used a green fluorescent protein (GFP) reporter inserted at the rDNA locus (rDNA-GFP). Its expression and fluorescence reflect the state of rDNA silencing – decreased fluorescence indicates enhanced silencing (32). To track heme abundance, we used a nuclear-anchored infrared fluorescent protein (nuc. iRFP), the fluorescence of which depends on biliverdin, a heme catabolism product, and correlates with the abundance of cellular heme (33, 34). To observe these two reporters, we used microfluidics coupled with time-lapse microscopy of single cells. We saw that isogenic WT cells age toward two discrete terminal states (34): one with decreased rDNA silencing (Fig. 1A, red dots; Fig. S1A), which leads to nucleolar enlargement and fragmentation (34), another with decreased heme abundance (Fig. 1A, blue dots; Fig. S1B) and hence mitochondrial aggregation and dysfunction (34). We further identified a mutual inhibition circuit of Sir2 and HAP that resembles a toggle switch and drives cellular fate decisions and commitment to either of these two detrimental states, contributing to cell deterioration and aging (34) (Fig. 1B).
Fig. 1. Construction of a synthetic gene oscillator to reprogram aging.
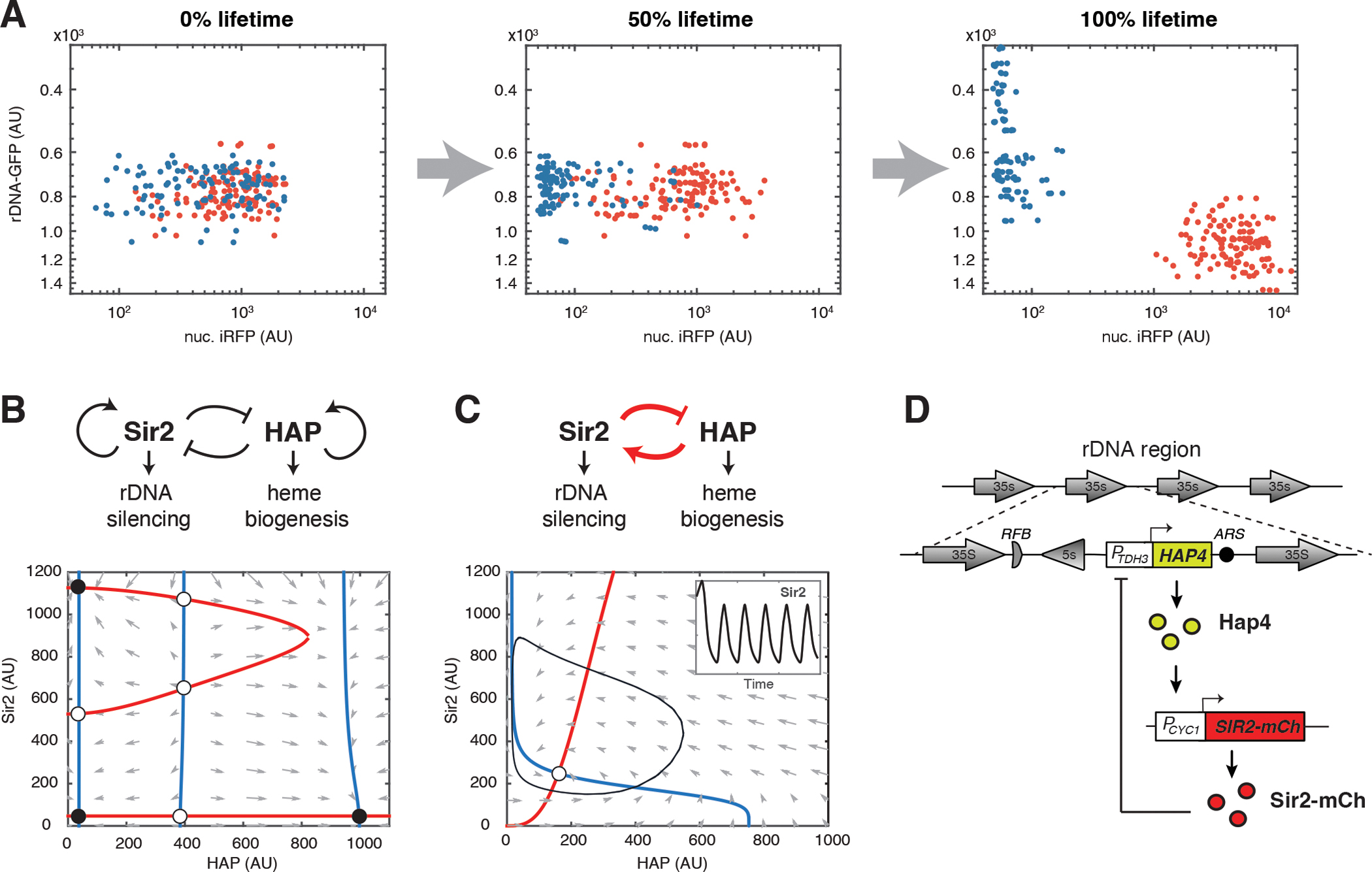
(A) Divergent aging in isogenic WT cells. Dot plots show the distributions of rDNA-GFP and nuc. iRFP reporter fluorescence in single cells tracked by time-lapse microscopy of single cells over the course of their lifespans. Each dot represents a single cell monitored individually in a microfluidic chamber. The red dots represent aging with rDNA silencing loss, indicated by increased rDNA-GFP fluorescence; the blue dots represent aging with heme depletion, indicated by decreased iRFP fluorescence. Experiments were independently performed at least three times. AU: arbitrary units. (B) The endogenous Sir2-HAP circuit and its simulated dynamic behaviors in WT aging. Top panel shows the diagram of the circuit topology. Bottom panel shows the phase plane diagram illustrating the dynamic changes of Sir2 and HAP activities during aging. The nullclines of Sir2 and HAP are shown in red and blue, respectively. The quivers show the rate and direction of the movement of the system. Fixed points are indicated with open (unstable) and closed (stable) circles. The stable fixed point on the bottom right corresponds to the terminal states of aging cells undergoing rDNA silencing loss and nucleolar decline (red dots in panel A); the stable fixed points on the left correspond to the terminal states of aging cells undergoing heme depletion and mitochondrial decline (blue dots in panel A). (C) The rewired Sir2-HAP circuit and its dynamic behaviors. Top panel shows the circuit topology with the synthetic negative feedback loop in red. Bottom panel shows the phase plane diagram with a limit cycle (black line) arising from the circuit, in which Sir2 and HAP periodically change their levels. Inset shows simulated time traces of oscillatory Sir2 expression. (D) A schematic illustrates the construction of the synthetic circuit. The native promoter of SIR2 was replaced with a HAP-inducible CYC1 promoter (PCYC1). HAP4 under a strong, constitutive TDH3 promoter (PTDH3) was inserted at the rDNA locus, which is subject to transcriptional silencing mediated by Sir2.
Design of a synthetic oscillator for longevity
We considered the possibility of altering the Sir2-HAP circuit to reprogram aging trajectories toward a longer lifespan. Specifically, the introduction of a synthetic negative feedback loop between Sir2 and HAP could lead to sustained oscillations in the abundance of these two factors (Fig. 1C). Such periodic cycling might enable a dynamic balance in Sir2 and HAP during aging, avoiding a prolonged duration or cell fate commitment to either rDNA silencing loss or heme depletion state, and thus slow cell deterioration and extend lifespan.
To guide our network engineering, we devised a simple computational model to generate design specifications. The model consisted of positive transcriptional regulation of SIR2 by HAP and Sir2-mediated transcriptional repression of HAP, forming a delayed negative feedback loop (Fig. S2. A and B; Materials and Methods). With appropriate parameter values, the model generated sustained limit cycle oscillations (Fig. 1C, Fig. S2C). We used Monte Carlo simulations to systematically explore the parameter space and analyze the dependence of sustained oscillatory behaviors on the parameter values (Fig. S3A). Oscillations were favored by strong HAP-activated transcription of SIR2, high capacity of transcription of HAP, and tight transcriptional repression of HAP by Sir2 (Fig. S3). We therefore focused our engineering efforts on fulfilling these specifications.
To enable strong positive transcriptional regulation of SIR2 by HAP, we replaced the native promoter of SIR2 with a CYC1 (Cytochrome C 1) promoter, which is bound and activated by HAP (35–37). To monitor dynamic behaviors of the engineered circuit, SIR2 was C-terminally tagged with the fluorescent reporter protein mCherry, which did not affect cell growth or aging (Fig. S4). To ensure a high capacity for transcription of HAP, we built a construct that contained the HAP4 gene, encoding a major component of the HAP complex, under a strong, constitutive TDH3 (triose-phosphate dehydrogenase 3) promoter. To enable dynamic transcriptional repression of HAP by Sir2, we integrated the HAP4 construct at the non-transcribed spacer (NTS) region within rDNA, which is subject to transcriptional silencing mediated by Sir2 (29, 38) (Fig. 1D). The endogenous copy of HAP4 was deleted in the synthetic strain to minimize leakiness of HAP4 expression. HAP4 was not tagged with a fluorescent reporter because its protein abundance is below the detection limit of fluorescence microscopy. These regulatory parts were selected based on the model-guided design specifications - the CYC1 promoter and transcriptional silencing at rDNA were selected because both of them were previously characterized to have low leakiness (36, 39). The TDH3 promoter was selected to drive HAP4 expression because it is one of the strongest constitutive promoters in yeast (40, 41).
Sustained oscillations during aging
We used microfluidics coupled with time-lapse microscopy to track dynamic changes in Sir2-mCherry fluorescence throughout the lifespan of single cells. Engineered cells (n=113) exhibited oscillations in abundance of Sir2 during aging (Fig. 2A; Fig. S5; Movie S1). WT control cells (n=93) did not show such oscillations (Fig. 2A; Fig. S5). We quantified the amplitude and period of oscillatory pulses in the engineered cells (Fig. S6). The average amplitude of oscillations was 309 ± 108 arbitrary units (Fig. 2B, AU), much larger than fluctuations in WT cells (36 ± 30 AU). The average period was 557 ± 151 minutes (Fig. 2C), longer than the typical cell doubling times (90~120 minutes), indicating that the oscillations were not driven by cell cycle. We also performed spectral analysis of Sir2 time traces (Fig. S7). For the engineered strain, we could clearly see a spectral power peak around frequency 2.33e-5 Hz corresponding to a period of 12 hours. In contrast, the spectrum of WT was flat and white-noise-like, without a clear peak (Fig. S7B).
Fig. 2. Oscillations in the synthetic strain during aging.
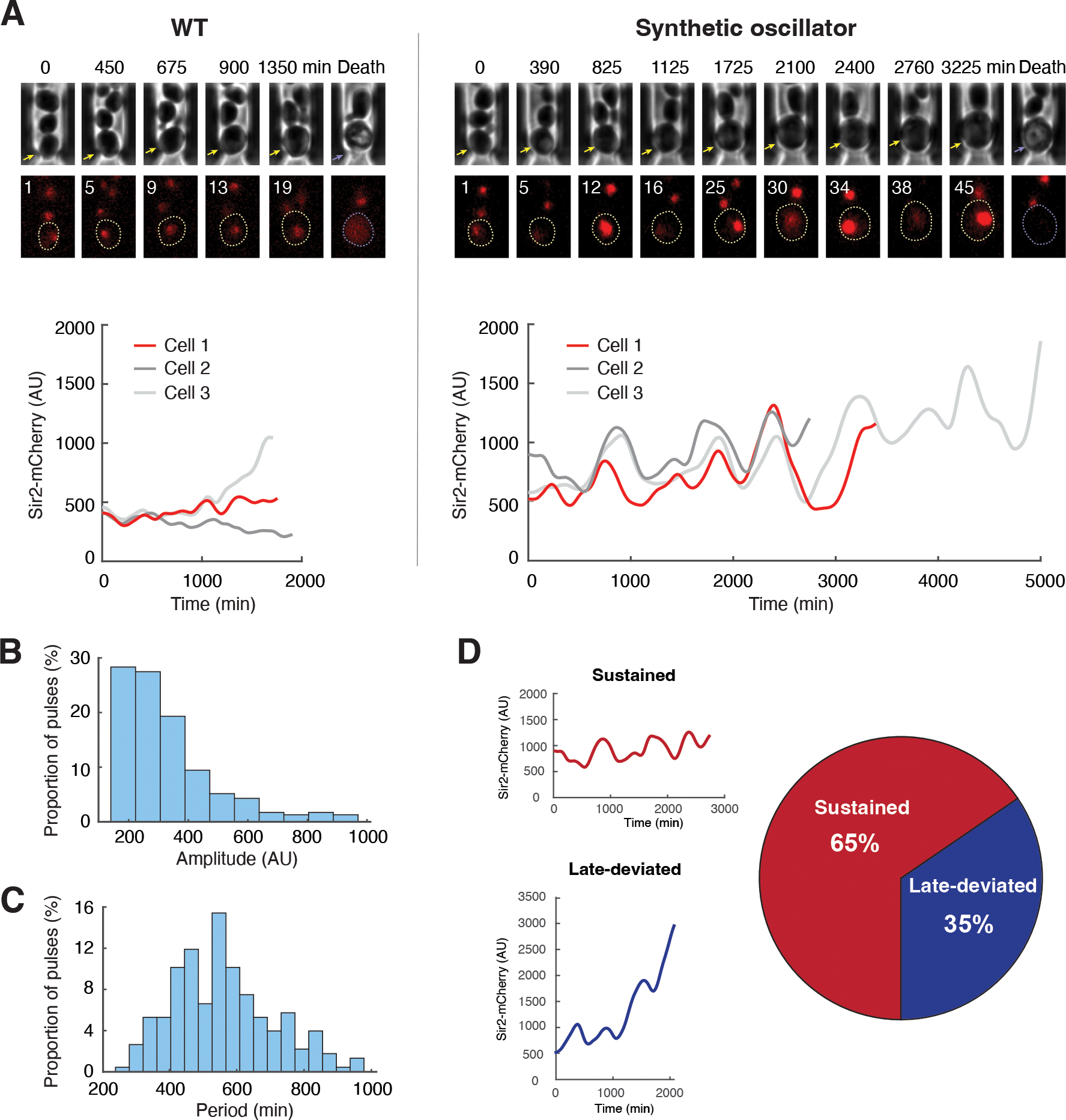
(A) Dynamics of Sir2-mCherry fluorescence in WT (left) and the synthetic strain (right) during aging. Top: representative time-lapse images for phase and Sir2-mCherry from single aging cells in the microfluidic chamber. For phase images, aging and dead mother cells are marked by yellow and purple arrows, respectively. In fluorescence images, replicative age of the mother cell is shown at the top left corner of each image; aging and dead mother cells are circled in yellow and purple, respectively. Bottom: fluorescence time traces throughout the lifespans of representative cells. The time trace in red corresponds to the time-lapse images shown above the plot. Time traces of all the cells measured are included in Fig. S5. (B) Distribution of the amplitudes of Sir2 oscillatory pulses in the engineered cells. (C) Distribution of the periods of Sir2 oscillatory pulses in the engineered cells. Panels B and C show distributions of single pulses. See Materials and Methods and Fig. S6 for the quantification of amplitude and period. (D) Proportions of aging cells from the synthetic strain that show sustained oscillations (Sustained) or a deviation from oscillation late in life (Late-deviated) (n=113). Left: Representative time traces for sustained oscillation (top) and late deviation from oscillation (bottom). See Materials and Methods and Fig. S8 for stability determination for Sir2 oscillations. Experiments were independently performed at least three times. AU, arbitrary units.
Oscillations in the synthetic strain were heterogeneous among individual cells. 65% of the engineered cells exhibited sustained oscillations throughout their entire lifespans, whereas 35% deviated from oscillations late in lifespan and showed increased accumulation of Sir2 before cell death (Fig. 2D; Fig. S8). This deviation might arise from an age-induced decrease in Sir2-mediated silencing activity (32, 42, 43) in some cells, which could lead to increased HAP expression from the rDNA locus and in turn a continuous increase in Sir2 expression driven by HAP.
During the process of circuit engineering, we also constructed and characterized versions of the synthetic circuit with broken or weakened feedback interactions. These include: (1) a circuit without HAP-activated expression of Sir2; (2) a circuit without Sir2-mediated repression of HAP; and (3) a circuit with a weaker transcriptional capacity of HAP. None of these circuits enabled sustained oscillations in a major fraction of cells (Fig. S9), demonstrating the importance of connectivity and strength of feedback interactions in generating oscillations.
The synthetic oscillator extends lifespan
The synthetic oscillator strain indeed showed an 82% increase in lifespan compared to that of WT control cells (Fig. 3A). This is the most pronounced lifespan extension in yeast that we have observed with genetic perturbations. Among the engineered cells, those aging with sustained oscillations had greater lifespan extension (105% increase in lifespan, doubling that of WT) than those that deviated from oscillations late in life (45% increase relative to that of WT) (Fig. 3A, red vs blue dashed curves). Thus, maintaining Sir2 oscillation appears to be important for maximally extending lifespan.
Fig. 3. Lifespan extension by the synthetic oscillator.
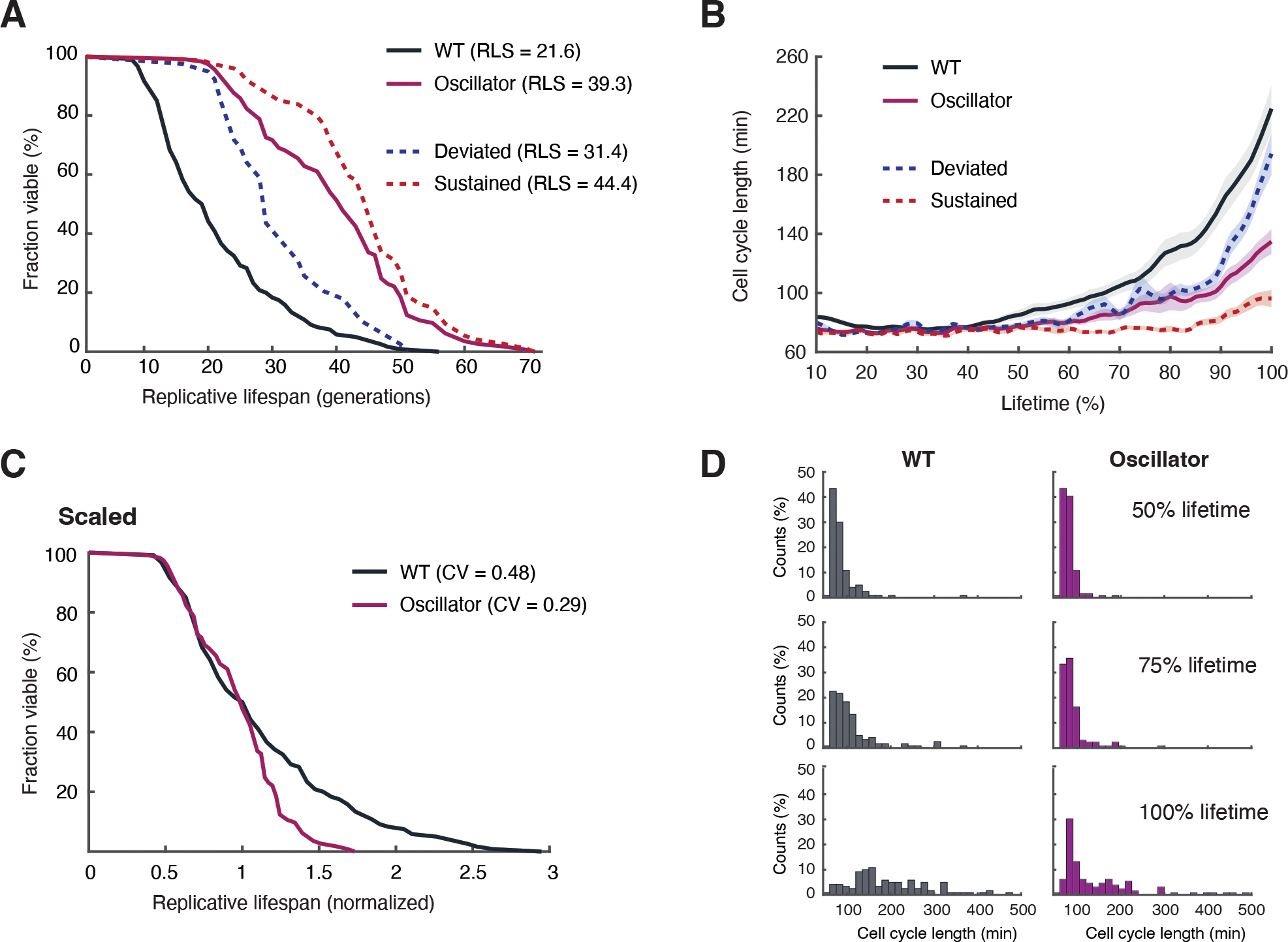
(A) Replicative lifespans for WT (black; n=131) and the synthetic oscillator strain (purple; n=120). Among the cells in the synthetic oscillator strain, the lifespans for those that deviated from oscillations (n=39) and those with sustained oscillations (n=74) were shown as blue and red dashed curves, respectively. p<0.0001with Gehan-Breslow-Wilcoxon test. (B) Changes of cell cycle length during aging for WT (black), the synthetic oscillator strain (purple), the oscillator cells that deviated from oscillations (blue dashed curve), and the oscillator cells with sustained oscillations (red dashed cells). Shaded areas represent standard errors of the mean (SEM). (C) The lifespan curves for WT and the synthetic oscillator strain, scaled by the median. The Coefficient of Variation (CV) of lifespans among cells was calculated for WT and the synthetic oscillator strain. (D) The histograms show distributions of cell cycle lengths at different stages of aging for WT and the synthetic oscillator strain. Experiments were independently performed at least three times.
The synthetic oscillator strain exhibited a fast cell cycle rate and the elongation of cell cycles during aging was delayed and decreased, compared to that in WT cells (Fig. 3B). Engineered cells with sustained oscillations retained a fast cell cycle rate (70 to 90 mins/cell cycle) throughout their entire lifespans, whereas those that deviated from oscillations had much slower cell cycles late in life (Fig. 3B, red vs blue dashed curves). Thus, maintaining Sir2 oscillation appears to slow age-induced cell deterioration.
WT cells show a large cell-to-cell variation in lifespan (coefficient of variation, CV=0.48), in part due to the stochasticity and divergence of the Sir2 and HAP deterioration pathways (34). The synthetic negative feedback loop in our engineered strain could function to avoid or delay such pathway divergence. In agreement with this, the synthetic oscillator strain showed a more uniform lifespan among cells (CV=0.29) and less increase in cell cycle length during aging, compared to WT (Fig. 3, C and D).
In the synthetic oscillator strain, the abundance of Sir2, averaged over the lifetime, was elevated by about 2-fold relative to that of WT (Fig. S10). To test whether the lifespan extension is simply due to the increased Sir2 abundance, we examined the strain with 2-fold constitutive overexpression of Sir2. We observed a ~23% increase in the lifespan compared to WT (Fig. S11A). 2-fold overexpression of Sir2 in combination with Hap4 overexpression resulted in a more dramatic lifespan extension (~42% increase compared to WT), which is still substantially less than the lifespan extension from the oscillator strain (82% increase compared to WT) (Fig. S11). The oscillator strain also has a faster cell cycle rate than the overexpression mutants (Fig. S11C). These results confirm that the oscillatory dynamics of Sir2, in addition to its increased expression, contribute to the lifespan extension and fast cell cycle rate in the synthetic strain. In line with this, the oscillator strain is also much more long-lived than strains with engineered Sir2-HAP circuits that cannot generate oscillations due to broken or weakened feedback interactions (Fig. S12).
To further assess the performance of the synthetic oscillator strain, we compared it with longest-lived single and double mutants identified from genetic screens (44–46). These include the deletion mutants fob1Δ (‘forkblocking less’, encoding a protein required for replication fork blocking), sgf73Δ (SAGA associated Factor 73, encoding a component of the SAGA/SLIK complex deubiquitination module), fob1Δ hxk2Δ (the double mutant of genes encoding ‘forkblocking less’ and hexokinase 2), and fob1Δ sch9Δ (the double mutant of genes encoding ‘forkblocking less’ and an ortholog of the mammalian S6 kinase). Under the genetic background and experimental conditions used (Materials and Methods) (32, 34, 47), the synthetic oscillator strain had a longer and more uniform lifespan than most mutants (Fig. S13, A and B). Moreover, some longevity mutants displayed impaired cell cycle progression even in young cells, suggesting moderate physiological defects associated with the genetic perturbations. In contrast, the oscillator strain has faster cell cycles than WT and mutants throughout the entire aging process, indicating a healthier cellular lifespan (Fig. S13C).
The synthetic oscillator avoids fate commitment to deterioration states
To test whether sustained oscillations in the engineered Sir2-HAP circuit could prevent aging cells from committing to either the rDNA silencing loss or heme depletion state, we simultaneously monitored rDNA silencing and heme abundance in the synthetic strain with the rDNA-GFP and iRFP reporters.
In accord with previous results (34), in WT cells, about half of the cells showed continuously increased GFP fluorescence at the later stages of aging, indicating sustained loss of rDNA silencing, and ended life in a state with low rDNA silencing and high abundance of heme. The other cells showed decreased iRFP fluorescence, indicating that heme was depleted, and ended life in a state with high rDNA silencing and low abundance of heme (Fig. 4A). In contrast, most synthetic oscillator cells exhibited short, intermittent pulses of rDNA-GFP and iRFP signals throughout the lifespan without a prolonged commitment to either a state of rDNA silencing loss or of heme depletion (Fig. 4B). We further quantified the continuous times in the states of rDNA silencing loss and heme depletion during aging of each individual cells (Fig. S14). Almost all of WT aging cells experienced a prolonged duration in silencing loss or heme depletion, whereas the oscillator cells showed shorter durations in either state (Fig. 4C; Fig. S15). Thus, the engineered negative feedback loop in the Sir2-HAP circuit enabled a time-based balance between rDNA silencing and heme biogenesis that promoted longevity. In further support of this balance, synthetic Sir2-HAP circuits with broken or weakened feedback interactions failed to maintain such a balance, resulting in prolonged commitments to detrimental states (Fig. S16) and thereby shorter lifespans (Fig. S12).
Fig. 4. The synthetic oscillator maintains a balance between rDNA silencing and heme biogenesis.
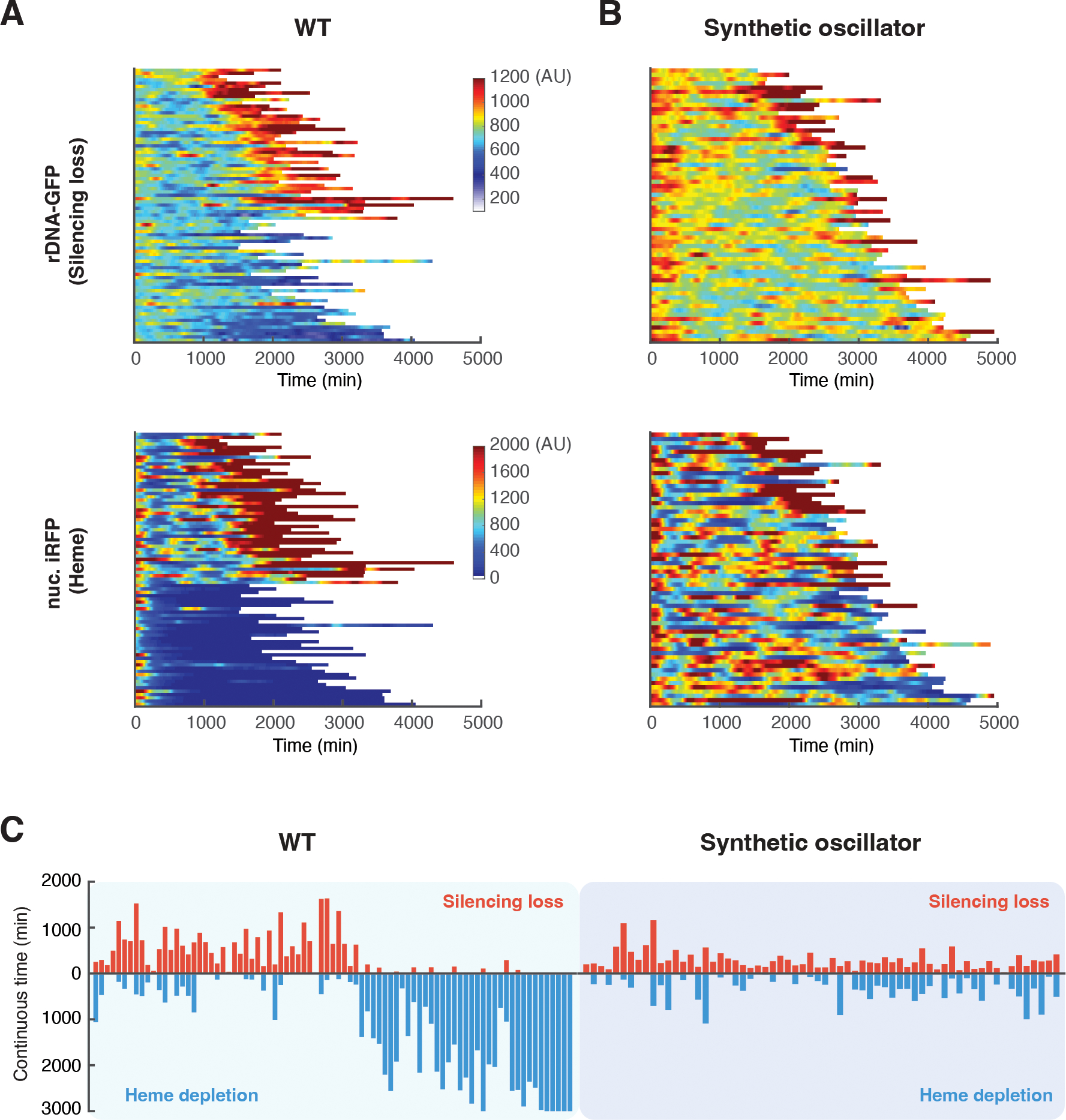
(A) Single-cell color map trajectories of rDNA-GFP (top) and nuclear-anchored iRFP (bottom) in WT aging cells (n=83). Each row represents the time trace of a single cell throughout its lifespan. Color represents the fluorescence intensity as indicated in the color bar. Color maps for rDNA-GFP and iRFP are from the same cells with the same top to bottom order. Cells are classified into two groups. The top half in the color maps – WT cells that showed continuous high GFP and iRFP signals, indicating rDNA silencing loss and high heme abundance, at the later stage of lifespan. These cells also produced elongated daughters at the later stage of lifespan and were previously designated as “mode 1” aging (34). The bottom half in the color maps – WT cells that showed constant or gradually decreased GFP fluorescence and sharply decreased iRFP fluorescence, indicating high rDNA silencing and heme depletion, at the late stage of aging. These cells produced small round daughters throughout the lifespan, previously designated as “mode 2” aging (34). (B) Single-cell color map trajectories of rDNA-GFP (top) and nuc. iRFP (bottom) in aging cells of the synthetic oscillator strain (n=64). Color maps for rDNA-GFP and iRFP are from the same cells. Color maps used the same color bars as those in panel A. (C) Bar graphs showing continuous times of the rDNA silencing loss or heme depletion state for WT (left) and the synthetic oscillator strain (right). Each bidirectional bar represents a single cell, in which the red upward portion indicates its continuous time of the rDNA silencing loss state and the blue downward portion indicates the continuous time of the heme depletion state. The graphs were quantified using the data from panels A and B (Fig. S14; Materials and Methods). Experiments were independently performed at least three times.
Discussion
Most studies of aging focus on measuring lifespan as a static endpoint assay and on identifying genes, whose deletion or overexpression affects lifespan. These investigations have led to identification of many conserved genes that influence aging (24, 48–50). Building on the knowledge of aging factors and pathways from genetic studies, we used engineering principles to rationally optimize aging dynamics toward extended longevity. Specifically, based on the understanding of Sir2 and HAP pathways in the aging of WT cells (34, 45), we rewired their interactions into a negative feedback loop and created a gene oscillator that functions to maintain cellular homeostasis. This synthetic system is advantageous in its robustness and effectiveness on lifespan extension over longevity mutants from genetic screens and simple overexpression of Sir2 or HAP or both (Fig. S17). The latter led to variations in gene expression that inevitably drive cell fate commitment and deterioration in a fraction of cells (Fig. S18), leading to short-lived cell subpopulations (34). We established a causal connection between gene network architecture and longevity and further validated the mechanistic understanding of aging in the natural system.
Using engineering principles to modulate biological functions is one of the major goals of synthetic biology (2, 3). Many studies have succeeded in generating specific spatiotemporal dynamics and functions with synthetic gene circuits, yet it remains a challenge to rationally engineer a biological trait as complex as longevity. Our work represents a proof-of-concept example, demonstrating the successful application of synthetic biology to reprogram the cellular aging process, and may set the stage for designing synthetic gene circuits to effectively promote longevity in more complex organisms.
Supplementary Material
Acknowledgments:
Funding:
National Institutes of Health R01AG056440 (NH, JH, LP, LST)
National Institutes of Health R01GM144595 (NH, JH, LP, LST)
National Institutes of Health R01AG068112 (NH)
National Institutes of Health R01GM111458 (NH)
Footnotes
Competing interests: Authors declare that they have no competing interests.
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at http://www.sciencemag.org/. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Data and materials availability:
All data are available in the main text or the supplementary materials. The code from this work is available at https://github.com/zhoutopo/science_aging_model.git and Zenodo (51).
References and Notes
- 1.Milo R et al. , Network motifs: simple building blocks of complex networks. Science 298, 824–827 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Elowitz M, Lim WA, Build life to understand it. Nature 468, 889–890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashor CJ, Collins JJ, Understanding biological regulation through synthetic biology. Annu Rev Biophys 47, 399–423 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Gardner TS, Cantor CR, Collins JJ, Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Elowitz MB, Leibler S, A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000). [DOI] [PubMed] [Google Scholar]
- 6.Stricker J et al. , A fast, robust and tunable synthetic gene oscillator. Nature 456, 516–519 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Friedland AE et al. , Synthetic gene networks that count. Science 324, 1199–1202 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Danino T, Mondragon-Palomino O, Tsimring L, Hasty J, A synchronized quorum of genetic clocks. Nature 463, 326–330 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Becskei A, Serrano L, Engineering stability in gene networks by autoregulation. Nature 405, 590–593 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Zhu R, Del Rio-Salgado JM, Garcia-Ojalvo J, Elowitz MB, Synthetic multistability in mammalian cells. Science 375, eabg9765 (2022). [DOI] [PubMed] [Google Scholar]
- 11.Wu F, Su RQ, Lai YC, Wang X, Engineering of a synthetic quadrastable gene network to approach Waddington landscape and cell fate determination. eLife 6, (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tigges M, Marquez-Lago TT, Stelling J, Fussenegger M, A tunable synthetic mammalian oscillator. Nature 457, 309–312 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Bashor CJ, Horwitz AA, Peisajovich SG, Lim WA, Rewiring cells: synthetic biology as a tool to interrogate the organizational principles of living systems. Annu Rev Biophys 39, 515–537 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu F, Bethke JH, Wang M, You L, Quantitative and synthetic biology approaches to combat bacterial pathogens. Curr Opin Biomed Eng 4, 116–126 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bintu L et al. , Dynamics of epigenetic regulation at the single-cell level. Science 351, 720–724 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keung AJ, Bashor CJ, Kiriakov S, Collins JJ, Khalil AS, Using targeted chromatin regulators to engineer combinatorial and spatial transcriptional regulation. Cell 158, 110–120 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Toda S, Blauch LR, Tang SKY, Morsut L, Lim WA, Programming self-organizing multicellular structures with synthetic cell-cell signaling. Science 361, 156–162 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang S et al. , Coupling spatial segregation with synthetic circuits to control bacterial survival. Mol Syst Biol 12, 859 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ng AH et al. , Modular and tunable biological feedback control using a de novo protein switch. Nature 572, 265–269 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Belikov AV, Age-related diseases as vicious cycles. Ageing Res Rev 49, 11–26 (2019). [DOI] [PubMed] [Google Scholar]
- 21.Fontana L, Partridge L, Longo VD, Extending healthy life span--from yeast to humans. Science 328, 321–326 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Longo VD, Kennedy BK, Sirtuins in aging and age-related disease. Cell 126, 257–268 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Wasko BM, Kaeberlein M, Yeast replicative aging: a paradigm for defining conserved longevity interventions. FEMS Yeast Res 14, 148–159 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaeberlein M, Kennedy BK, Large-scale identification in yeast of conserved ageing genes. Mech Ageing Dev 126, 17–21 (2005). [DOI] [PubMed] [Google Scholar]
- 25.He C, Zhou C, Kennedy BK, The yeast replicative aging model. Biochim Biophys Acta, (2018). [DOI] [PubMed] [Google Scholar]
- 26.Smith ED et al. , Quantitative evidence for conserved longevity pathways between divergent eukaryotic species. Genome Res 18, 564–570 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gartenberg MR, Smith JS, The nuts and bolts of transcriptionally silent chromatin in Saccharomyces cerevisiae. Genetics 203, 1563–1599 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M, McVey M, Guarente L, The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev 13, 2570–2580 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Saka K, Ide S, Ganley AR, Kobayashi T, Cellular senescence in yeast is regulated by rDNA noncoding transcription. Curr Biol 23, 1794–1798 (2013). [DOI] [PubMed] [Google Scholar]
- 30.Sinclair DA, Guarente L, Extrachromosomal rDNA circles--a cause of aging in yeast. Cell 91, 1033–1042 (1997). [DOI] [PubMed] [Google Scholar]
- 31.Buschlen S et al. , The S. cerevisiae HAP complex, a key regulator of mitochondrial function, coordinates nuclear and mitochondrial gene expression. Comp Funct Genomics 4, 37–46 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li Y et al. , Multigenerational silencing dynamics control cell aging. Proc Natl Acad Sci U S A 114, 11253–11258 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filonov GS et al. , Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol 29, 757–761 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li Y et al. , A programmable fate decision landscape underlies single-cell aging in yeast. Science 369, 325–329 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olesen J, Hahn S, Guarente L, Yeast HAP2 and HAP3 activators both bind to the CYC1 upstream activation site, UAS2, in an interdependent manner. Cell 51, 953–961 (1987). [DOI] [PubMed] [Google Scholar]
- 36.Guarente L, Mason T, Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell 32, 1279–1286 (1983). [DOI] [PubMed] [Google Scholar]
- 37.Hahn S, Guarente L, Yeast HAP2 and HAP3: transcriptional activators in a heteromeric complex. Science 240, 317–321 (1988). [DOI] [PubMed] [Google Scholar]
- 38.Li C, Mueller JE, Bryk M, Sir2 represses endogenous polymerase II transcription units in the ribosomal DNA nontranscribed spacer. Mol Biol Cell 17, 3848–3859 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallo CM, Smith DL Jr., Smith JS, Nicotinamide clearance by Pnc1 directly regulates Sir2-mediated silencing and longevity. Mol Cell Biol 24, 1301–1312 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ho B, Baryshnikova A, Brown GW, Unification of protein abundance datasets yields a quantitative Saccharomyces cerevisiae proteome. Cell Syst 6, 192–205 e193 (2018). [DOI] [PubMed] [Google Scholar]
- 41.Xiong L et al. , Condition-specific promoter activities in Saccharomyces cerevisiae. Microb Cell Fact 17, 58 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dang W et al. , Histone H4 lysine 16 acetylation regulates cellular lifespan. Nature 459, 802–807 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smeal T, Claus J, Kennedy B, Cole F, Guarente L, Loss of transcriptional silencing causes sterility in old mother cells of S. cerevisiae. Cell 84, 633–642 (1996). [DOI] [PubMed] [Google Scholar]
- 44.McCormick MA et al. , The SAGA histone deubiquitinase module controls yeast replicative lifespan via Sir2 interaction. Cell Rep 8, 477–486 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kaeberlein M, Kirkland KT, Fields S, Kennedy BK, Sir2-independent life span extension by calorie restriction in yeast. PLoS Biol 2, E296 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaeberlein M et al. , Regulation of yeast replicative life span by TOR and Sch9 in response to nutrients. Science 310, 1193–1196 (2005). [DOI] [PubMed] [Google Scholar]
- 47.Jin M et al. , Divergent aging of isogenic yeast cells revealed through single-cell phenotypic dynamics. Cell Syst 8, 242–253 e243 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guarente L, Kenyon C, Genetic pathways that regulate ageing in model organisms. Nature 408, 255–262 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Kuningas M et al. , Genes encoding longevity: from model organisms to humans. Aging Cell 7, 270–280 (2008). [DOI] [PubMed] [Google Scholar]
- 50.McCormick MA et al. , A comprehensive analysis of replicative lifespan in 4,698 single-gene deletion strains uncovers conserved mechanisms of aging. Cell Metab 22, 895–906 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.DOI: 10.5281/zenodo.7407339. [DOI]
- 52.Buck SW, Sandmeier JJ, Smith JS, RNA polymerase I propagates unidirectional spreading of rDNA silent chromatin. Cell 111, 1003–1014 (2002). [DOI] [PubMed] [Google Scholar]
- 53.Pusnik Z, Mraz M, Zimic N, Moskon M, Computational analysis of viable parameter regions in models of synthetic biological systems. J Biol Eng 13, 75 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dhooge A, Govaerts W, Kuznetsov YA, Meijer HGE, Sautois B, New features of the software MatCont for bifurcation analysis of dynamical systems. Mathematical and Computer Modelling of Dynamical Systems 14, 147–175 (2008). [Google Scholar]
- 55.Paxman J et al. , Age-dependent aggregation of ribosomal RNA-binding proteins links deterioration in chromatin stability with challenges to proteostasis. eLife 11, (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O’Laughlin R et al. , Advances in quantitative biology methods for studying replicative aging in Saccharomyces cerevisiae. Transl Med Aging 4, 151–160 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lavielle M, Using penalized contrasts for the change-point problem. Signal Processing 85, 1501–1510 (2005). [Google Scholar]
- 58.Killick R, Fearnhead P, Eckley IA, Optimal detection of changepoints with a linear computational cost. Journal of the American Statistical Association 107, 1590–1598 (2012). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available in the main text or the supplementary materials. The code from this work is available at https://github.com/zhoutopo/science_aging_model.git and Zenodo (51).


