Abstract
Background:
Preferred first-line treatment for patients with metastatic EGFR-mutant lung cancer is osimertinib, yet it is not known if patient outcomes may be improved by identifying and intervening upon molecular markers associated with therapeutic resistance.
Patients and Methods:
All patients with metastatic EGFR-mutant lung cancer treated with first-line osimertinib at Memorial Sloan Kettering Cancer Center (n=327) were identified. Available pre-treatment and post-progression tumor samples underwent targeted gene panel sequencing and mutational signature analysis using SigMA algorithm. Progression-free and overall survival were estimated using Kaplan Meier methods.
Results:
Using multivariate analysis, baseline atypical EGFR (mPFS 5.8 mo, p<0.001) and concurrent TP53/RB1 alterations (mPFS 10.5 mo, p=0.015) were associated with shorter progression-free survival on first-line osimertinib. Of 95 patients with post-progression biopsies, acquired resistance mechanisms were identified in 52% (off-target n=24, histological transformation n=14, on-target n=12), with MET amplification (n=9), small cell lung transformation (n=7) and acquired EGFR amplification (n=7) the most frequently identified mechanisms. While there was no difference in post-progression survival based on identified resistance (p=0.07), patients with subsequent second-line therapy tailored to post-progression biopsy results had improved post-progression survival (HR 0.09, p=0.006). Paired post-progression tumors had higher tumor mutational burden (p=0.008) and further dominant APOBEC mutational signatures (p=0.07) compared to pre-treatment samples.
Conclusions:
Patients with EGFR-mutant lung cancer treated with first-line osimertinib have improved survival with treatment adaptation based on identified mechanisms of resistance at time of progression using tissue-based genomic analysis. Further survival gains may be achieved using risk-based treatment adaptation of pre-treatment genomic alterations.
Keywords: EGFR, mechanisms of resistance, osimertinib, tumor evolution, targeted therapies
INTRODUCTION:
Despite remaining the leading cause of cancer deaths globally1, lung cancer mortality in the United States has steadily declined in the last decade.2 While this is partially attributable to population-level declines in cigarette smoking2, the advent of effective targeted therapies for non-small cell lung cancer (NSCLC) harboring oncogenic alterations has also driven significant survival gains.3 Comprising 25% of lung adenocarcinoma cases4, NSCLC with activating epidermal growth factor receptor (EGFR) alterations is the single largest lung cancer genotype treated with molecularly targeted therapies and often considered a paradigm for oncogene-addicted cancers. The FLAURA trial established osimertinib, a highly central nervous system (CNS)-penetrant, third-generation EGFR tyrosine kinase inhibitor (TKI) as the preferred first-line treatment for metastatic EGFR-mutant lung cancer, with improvement in overall survival (OS)5 and progression-free survival (PFS)6 compared to earlier generation EGFR TKIs.
Mediators of resistance to osimertinib differ between first- versus later-line treatment setting, with on-target acquired EGFR alterations more common when osimertinib is used after other EGFR TKIs.7–11 Mechanisms of resistance to first-line osimertinib reported in small, primarily plasma-based series are similar to later-line osimertinib, including emergence of EGFR C797S12, 13, a mutation at the covalent bonding site of third-generation EGFR TKIs14, and off-target mechanisms such as MET amplification9, 15, albeit at differing frequencies. Importantly, although tailoring treatment to an identified mechanism of acquired resistance in small series or individual cases has induced disease responses16–18, the impact of this approach on a larger scale has not been systematically studied. Furthermore, studies focusing on clinicogenomic correlates impacting outcomes on first-line osimertinib are limited, with previous studies including patients treated with later-line osimertinib7–10, or relying upon serial plasma samples rather than tumor tissue.15
In this work, we identified patients with EGFR-mutant NSCLC treated with first-line osimertinib at Memorial Sloan Kettering Cancer Center (MSKCC). We focused on patients with pre- and post-osimertinib progression tumor tissue that underwent next-generation sequencing (NGS) by the FDA-cleared MSK-Integrated Mutation Profiling of Actionable Targets assay (MSK-IMPACT) platform.19 Utilizing the largest reported dataset of patients treated with first-line osimertinib with both clinical and genomic annotation, we sought to characterize pre-treatment clinicogenomic correlates that are associated with time to progression, mechanisms of acquired resistance, and the impact of identifying and treating mechanisms of resistance on post-osimertinib progression survival.
MATERIALS AND METHODS:
Patients
The study was conducted in accordance with the Declaration of Helsinki and good clinical practice guidelines following approval by MSK institutional review board/privacy board (MSK IRB 21–416). Patients with NSCLC and EGFR alterations who received treatment with first-line osimertinib at MSK were retrospectively identified using an automated query20 capturing presence of EGFR alterations from MSK-IMPACT, MSK-ACCESS,21 and Idylla tumor or plasma testing. Inclusion criteria comprised pathological confirmation of metastatic lung cancer, identification of an EGFR sensitizing alteration, and documented first-line osimertinib treatment for metastatic disease. Receipt of concurrent neoplastic agents with first-line osimertinib and/or receipt of prior neoplastic treatments in the adjuvant setting for locally advanced disease were allowed. Patient and tumor characteristics, including baseline EGFR alteration and concurrent genomics, treatment history, imaging, and pathology reports were extracted from the electronic health record.
Genomic analyses
Samples from included patients were queried for somatic tumor genomic testing obtained for routine clinical purposes by the FDA-cleared MSK-IMPACT19, 22 and/or other institutional or commercial gene panels (Supplementary Figure 1). Patients with a post-osimertinib progression tumor sample were retrospectively queried for testing with next-generation RNA sequencing (RNAseq) by Archer FusionPlex Custom Solid Panel, a custom RNAseq panel designed to detect gene fusions in 123 cancer-related genes involved in chromosomal rearrangements.23 Somatic genomic alterations, including mutations, copy number alterations and structural variants, were filtered for “oncogenic”, “likely oncogenic”, and “predicted oncogenic” variants based on OncoKB annotation.24 Mutational signature analyses, fraction genome altered (FGA), whole genome doubling (WGD) and tumor mutational burden (TMB) were only derived from tumor samples with ≥20% purity. TMB was calculated as the total number of nonsynonymous mutations divided by the length of the genomic target region captured with the exome assay, as previously described.22 WGD status was inferred for samples sequenced by MSK-IMPACT panel, as previously reported.25 The FGA was computed as the fraction of log2 copy number variation (gain or loss) >0.2 divided by the size of the genome whose copy number was profiled, as previously described 26
Tailored treatment
Patients were considered to have “tailored treatment” if their second-based on post-progression biopsy results, including treatment for small cell lung cancer transformation or targeted therapy directly against an acquired genomic alteration with or without continued osimertinib. Patients who either did not receive further therapy or their second-line treatment was not based on biopsy results were in the “not tailored” group
Mutational signature analysis
For tumor samples that underwent targeted sequencing by MSK-IMPACT, had at least five somatic synonymous and non-synonymous singles nucleotide variants (SNVs) and tumor purity ≥20%, mutational signatures were computed using Signature Multivariate Analysis (SigMA)27, a tool extensively benchmarked for formalin-fixed paraffin-embedded samples subjected to multi-gene panel sequencing, as previously described.28, 29 SigMA was run with customized settings (i.e., data type: msk, cancer type: lung, add_sig3: true, check msi: true). A dominant signature for each sample was determined based on the proposed category assigned by SigMA, as previously reported.27
Statistical analysis
Comparisons of categorical and continuous variables were performed by Fisher’s exact and Mann-Whitney U tests, respectively. Comparisons of frequencies of genes altered by somatic genetic alterations were performed using Fisher’s exact test and logistic regression. Multiple testing correction using the Benjamini-Hochberg method was applied to control for the false discovery rate whenever appropriate. Kaplan-Meier estimates of real-world progression-free survival (rwPFS), time to treatment discontinuation (TTD), and overall survival (OS) were computed. The log-rank test was used to assess for differential effects in EGFR alteration, concurrent genomics, brain metastases, type of osimertinib treatment received. rwPFS was defined from the start date of osimertinib therapy to the date of progression or death, with dates of progression determined based on the AACR GENIE BPC PRISSMM framework 30 as the time point at which both imaging reports and medical oncology notes concurred with disease progression. TTD was defined as date of osimertinib initiation to last administered dose and OS was defined as the date of osimertinib initiation to date of death or last follow-up. The cutoff date for this analysis was March 1, 2022. Hazard estimates, starting from the time of progression, were used to evaluate the post-progression effect of tailored treatment and the mechanism of resistance on OS. The post-baseline effects were tested using a time-dependent covariate Cox model.
RESULTS
Baseline clinicogenomic correlates associated with time to progression on osimertinib Since 2012, tumor samples from 8,221 patients with NSCLC underwent somatic genomic testing at MSK, including 2,925 patients harboring an EGFR driver alteration identified by MSK-IMPACT. Three-hundred twenty-seven patients were treated with first-line osimertinib for metastatic disease (Table 1, Supplementary Figure 1). Most patients were treated with osimertinib alone (n = 252, 77%). Others received concurrent chemotherapy (including antiVEGF therapy, n = 57), a second EGFR TKI (n = 16), or immunotherapy (n = 2, Supplementary Table 1). Twenty-nine patients harbored atypical EGFR alterations, defined as activating EGFR alterations that were neither L858R nor exon 19 deletions (Table 2).
Table 1:
Patient and Tumor Characteristics
| Characteristic | Full Cohort n= 327 n (%) | Paired Biopsies Sub-cohort n=74 n(%) | Post-tx biopsy sub-cohort n=21 n(%) |
|---|---|---|---|
| Age, median (IQR) | 64 (57,71) | 60 (52,69) | 61 (53, 66) |
| Sex | |||
| Female | 220 (67.2) | 50 (67.6) | 15 (71.4) |
| Male | 107 (32.7) | 24 (32.4) | 6 (28.6) |
| Race | |||
| White | 197 (60.2) | 46 (62.2) | 10 (47.6) |
| Asian | 98 (30.0) | 21 (28.4) | 8 (38.1) |
| Black | 17 (5.2) | 3 (4.1) | 3 (14.3) |
| Other | 8 (2.4) | 2 (2.7) | 0 (0) |
| Unknown | 6 (1.8) | 2 (2.7) | 0 (0) |
| Smoking history | |||
| Never smoker | 251 (76.8) | 62 (83.8) | 17 (81.0) |
| <15 py history | 50 (15.3) | 8 (10.8) | 3 (14.3) |
| >15 py history or current | 26 (8.0) | 4 (5.4) | 1 (4.7) |
| Baseline brain metastases | |||
| Present | 135 (41.3) | 25 (33.8) | 7 (33.3) |
| Absent | 183 (56.0) | 49 (66.2) | 14 (66.7) |
| Unknown | 9 (2.8) | 0 (0) | 0 (0) |
| Histology | |||
| Adenocarcinoma | 309 (94.5) | 67 (90.5) | 21 (100.0) |
| Non-adenocarcinoma | 18 (5.5) | 7 (9.5) | 0 (0) |
| EGFR alteration | |||
| Exon 19 deletion | 185 (56.6) | 46 (62.2) | 10 (47.6) |
| L858R | 113 (34.6) | 22 (29.7) | 6 (28.6) |
| Atypical | 29 (8.9) | 6 (8.1) | 5 (23.8) |
| Concurrent pre-tx alterations | |||
| TP53 | 141 (43.1) | 40 (54.1) | 1 (4.8) |
| TP53/RB1 | 50 (15.3) | 15 (20.3) | 2 (9.5) |
| Neither | 92 (28.1) | 19 (25.7) | 2 (9.5) |
| Unknown | 44 (13.5) | 0 (0) | 16 (76.2) |
Patient and tumor characteristics are shown. Patients with unknown brain metastases status did not have pre-treatment brain imaging performed. N, number; tx, treatment; IQR, inter-quartile range; py, pack-years.
Table 2:
Atypical EGFR alterations
| Atypical EGFR | n=29 n (%) |
|---|---|
| G719X | 11 (37.9) |
| L861Q | 11 (37.9) |
| Exon 20 insertion | 1 (3.4) |
| Compound L861Q/G719A | 1 (3.4) |
| H7773R | 1 (3.4) |
| V834L | 1 (3.4) |
| L747P | 1 (3.4) |
| Exon 19 insertion | 1 (3.4) |
| E709-T710>D | 1 (3.4) |
Breakdown of atypical EGFR alterations included in the study.
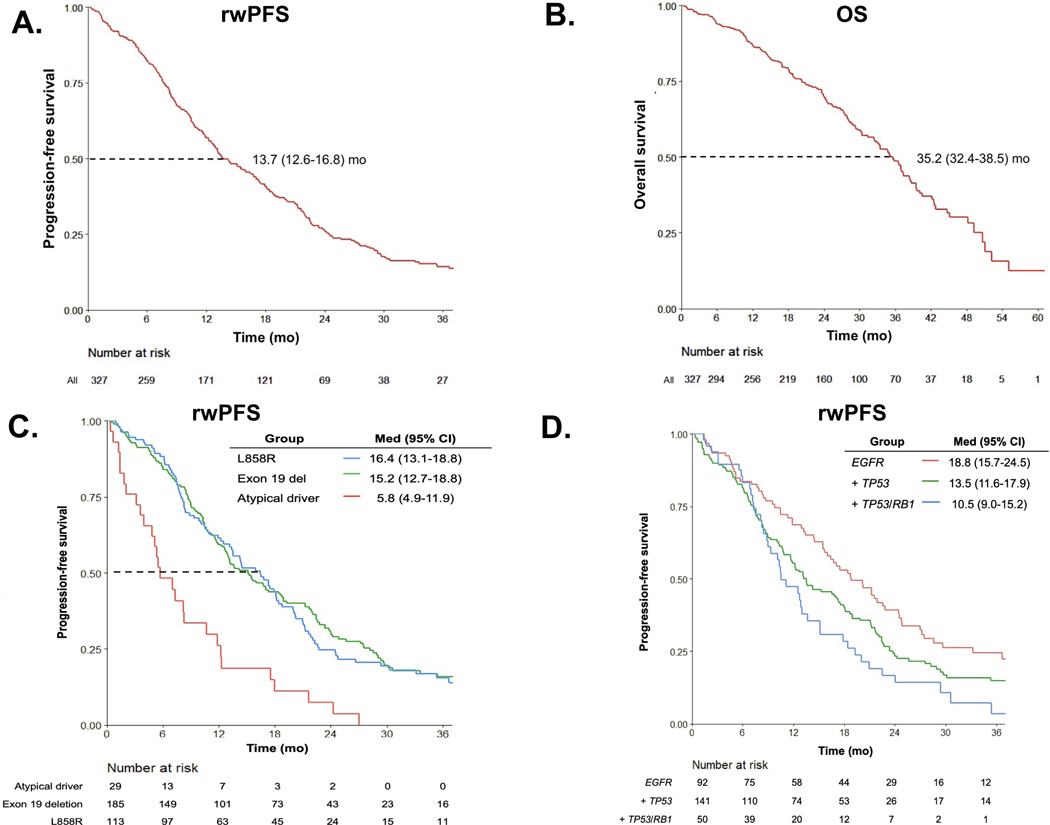
With a median follow-up of 23.9 months (mo) (interquartile range, IQR: 13.6 – 33.9), 250 patients experienced disease progression on osimertinib and 77 patients continued first-line osimertinib at the time of data censoring. Median real-world progression-free survival (rwPFS) was 13.7 mo (95% confidence interval (CI) 12.6–16.8 mo, Fig. 1A). Among all 327 patients, 152 patients had died at data cut-off with a median overall survival of 35.2 mo (95% CI 32.4–38.5 mo, Fig. 1B). Median time to treatment discontinuation (TTD) was 16.4 months (95% CI 13.8–18.4 months, Supplementary Fig. 2A), reflecting the common practice of continuing EGFR TKI beyond initial disease progression.31, 32
Figure 1: Efficacy of osimertinib and baseline factors impact real-world PFS on osimertinib.
A) Real-world progression-free survival (rwPFS) B) Overall survival (OS) for all patients (n = 327) treated with first-line osimertinib, with median times and 95% CIs noted. C) rwPFS stratified by baseline EGFR alterations (n = 29 atypical, n = 185 exon 19 deletion, and n = 113 L858R). D) rwPFS stratified by baseline concurrent genomic alterations for patients with available data (n = 92 neither TP53/RB1 alterations, n = 141 TP53 co-mutations, n = 50 TP53/RB1 co-mutations). CI, confidence intervals, del, deletion; med, median; mo, months.
We next evaluated pre-treatment biomarkers that may influence time to disease progression on osimertinib. rwPFS was shorter for patients with atypical EGFR alterations (median rwPFS 5.8 mo, 95% CI 4.9–11.9 mo) compared to patients with exon 19 del (15.2 mo, 95% CI 12.7–18.8 mo, p = 0.001) or L858R (16.4 mo, 95% CI 13.1–18.8 mo, p = 0.004, Fig. 1C). No difference in rwPFS between patients with L858R and exon 19 del was observed (p= 0.8). Out of 318 with baseline brain imaging, patients with baseline brain metastases had shorter rwPFS compared to patients without brain metastases (mPFS 11.9 mo, 95% CI 10.5–15.2 mo vs. 16.5 mo, 95% CI 13.1–18.8 mo, p= 0.04, Supplementary Fig. 2B. There was no difference in rwPFS based on whether osimertinib was used alone or in combination, although patients who received combination therapy had numerically longer rwPFS (mPFS 13.7 mo osimertinib plus chemotherapy or ICI vs. 12.5 mo for osimertinib alone, Supplementary Fig. 2C).
To investigate the impact of baseline somatic alterations on osimertinib efficacy, survival analysis was conducted in patients with available pre-treatment NGS (n=283). The presence of baseline TP53/RB1 and TP53 mutations was associated with shorter rwPFS compared to patients without TP53/RB1 (rwPFS 10.5 mo, 95% CI 9.0–15.2 mo), 13.5 mo (95% CI 11.6–17.9 months), 18.8 mo (15.7–24.5 mo, respectively, p= 0.002, Fig. 1D). Concurrent TP53/RB1 alterations were enriched in tumors with EGFR exon19 del (n = 31, 62%) compared to L858R (n = 18, 36%) and atypical (n = 1, 3%). Notably, although concurrent TP3/RB1 alterations increase risk of histological transformation33, only 15/50 (30%) patients with concurrent TP53/RB1 alterations in our series experienced histological transformation, indicating that transformation alone does not fully explain shorter rwPFS for these patients. At univariate analysis, atypical EGFR alterations, concurrent TP53/RB1 alterations and brain metastases were individually associated with rwPFS. Baseline atypical EGFR alterations and concurrent TP53/RB1 alterations remained independently associated with shorter rwPFS in a multivariate model (p<0.001 and 0.015, respectively, Supplemental Table 2).
Mechanisms of acquired resistance to first-line osimertinib
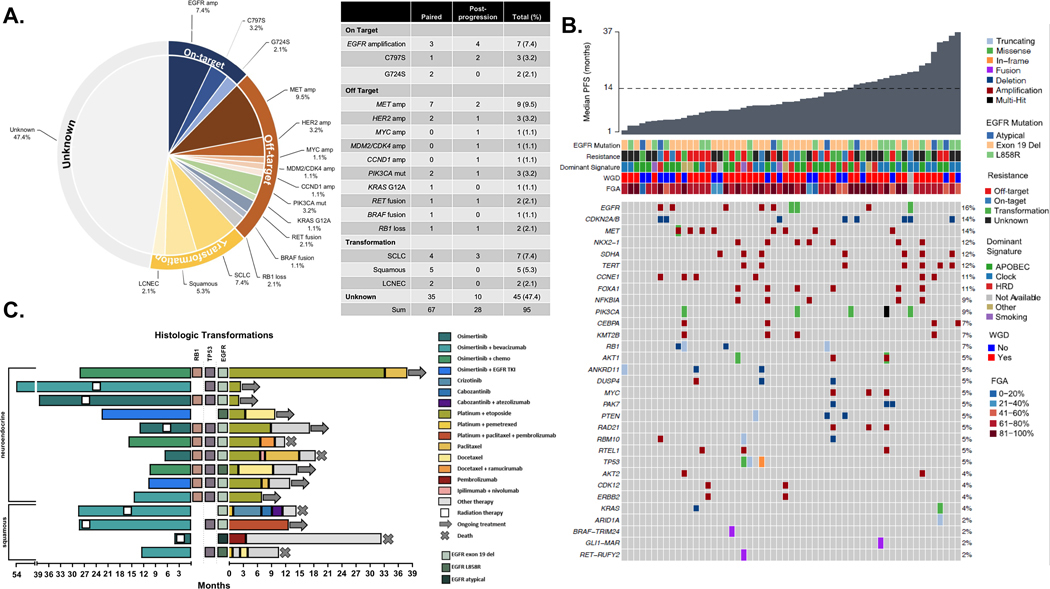
Focusing on patients with a post-progression tumor biopsy sample that underwent NGS (n = 95), genomic mechanisms of acquired resistance to first-line osimertinib were identified in 50/95 (53%) patients (Fig. 2A). Off-target alterations were found in 24 patients (26%) and represented the most frequently identified mechanism of acquired resistance. Gene amplifications (MET n = 9, HER2 n = 3, MYC, MDM2/CDK4, and CCND1 n = 1 each), single gene mutations (PIK3CA n = 3, KRAS G12A n = 1), and acquired fusions (RET fusion n = 2, BRAF fusion n = 1) were detected. In addition, two patients acquired oncogenic loss-of-function (LOF) alterations of RB1. On-target EGFR alterations comprised 13% of acquired alterations (EGFR amplification, n = 7; C797S, n=3; G724S, n=2). Acquired alterations in post-osimertinib samples that occurred in one or more patients in the cohort are shown in Fig. 2B. Oncogenic alterations in pre- and post-progression samples with >5% frequency are shown in Supplementary Fig. 3.
Figure 2: Mechanisms of acquired resistance to first-line osimertinib.
A) Identified mechanisms of resistance for 95 patients with post-progression tumor biopsy that underwent NGS with MSK-IMPACT B) Oncoprint of oncogenic, acquired alterations in post-progression samples with frequency C) Swimmers plot detailing clinical course of patients with histological transformation. Amp, amplification; SCLC, small cell lung cancer; LCNEC, large cell neuroendocrine cancer; FGA, fraction of genome altered; NGS, next-generation sequencing; WGD, whole genome doubling; HRD, homologous recombination deficiency.
Lineage plasticity is of particular interest in EGFR-mutant lung cancer. Fourteen patients (15%) initially had adenocarcinoma histology and underwent histological transformation (small cell lung cancer [SCLC] n = 6, squamous cell carcinoma [SQCC] n = 4, large cell neuroendocrine [LCNEC] n = 2, combined LCNEC/SCLC n = 1, combined LCNEC/SQCC n = 1) at the time of disease progression. Eight of these patients had baseline TP53/RB1 alterations (SCLC n = 7 and LCNEC/SCLC n = 1), three TP53 alterations only (SQCC n = 2, LCNEC n = 1) and three had neither (SQCC n = 2, LCNEC/SQCC n = 1). Clinical courses are shown in Fig. 2C. Median time to treatment discontinuation of osimertinib in this subgroup was 15.2 mo. Median survival after transformation was 24.4 mo. Six patients were treated with local therapy; three of these cases continued osimertinib after local therapy. Ten patients received platinum-etoposide after transformation, with time on treatment ranging from 2 to 33 months.
We assessed whether identified resistance mechanisms differed when patients received first-line osimertinib monotherapy vs combination therapy. The proportion of patients with transformation initially treated with osimertinib combination therapy was higher than those treated with osimertinib monotherapy (27% vs 7%, p=0.02 Supplemental Fig. 4A-B). Ten patients receiving combination treatment and 8 patients receiving osimertinib monotherapy had baseline concurrent EGFR/TP53/RB1 alterations. Conversely, gene amplifications and fusions were identified as mechanisms of resistance almost exclusively in patients treated with osimertinib monotherapy (p<0.001). We further considered whether patients with baseline atypical EGFR alterations may have unique mechanisms of resistance to first-line osimertinib. In total, 15 patients with atypical EGFR alterations had post-progression biopsies (n=8 paired, n=7 post-progression only). Off target [n=6 total, MET amp (3), acquired RB1 loss, PIK3CA E545K, MDM2/CDK4 amp], on target (n=2, EGFR amp) and transformation (n=1 squamous) were identified mechanisms of resistance. Although sample size was too small for formal comparison, the distribution of identified resistance mechanisms resembled that of patients with classical EGFR alterations.
Site of biopsy may impact identification of resistance mechanisms, particularly if non-progressive disease areas are biopsied for safety or accessibility. Of the 95 patients with post-progression biopsies, 89 were performed at a progressive disease site (Supplemental Table 3). Of the six patients with biopsies at non-progressive disease sites, five did not have an identified resistance mechanism. We further considered that certain biopsy sites may limit identification of resistance mechanism due to tissue quality or quantity or that resistance mechanisms may differ by site of metastases, but no patterns emerged between category of resistance and biopsy site (Supplemental Fig. 4C).
Role of identifying and treating resistance mechanisms
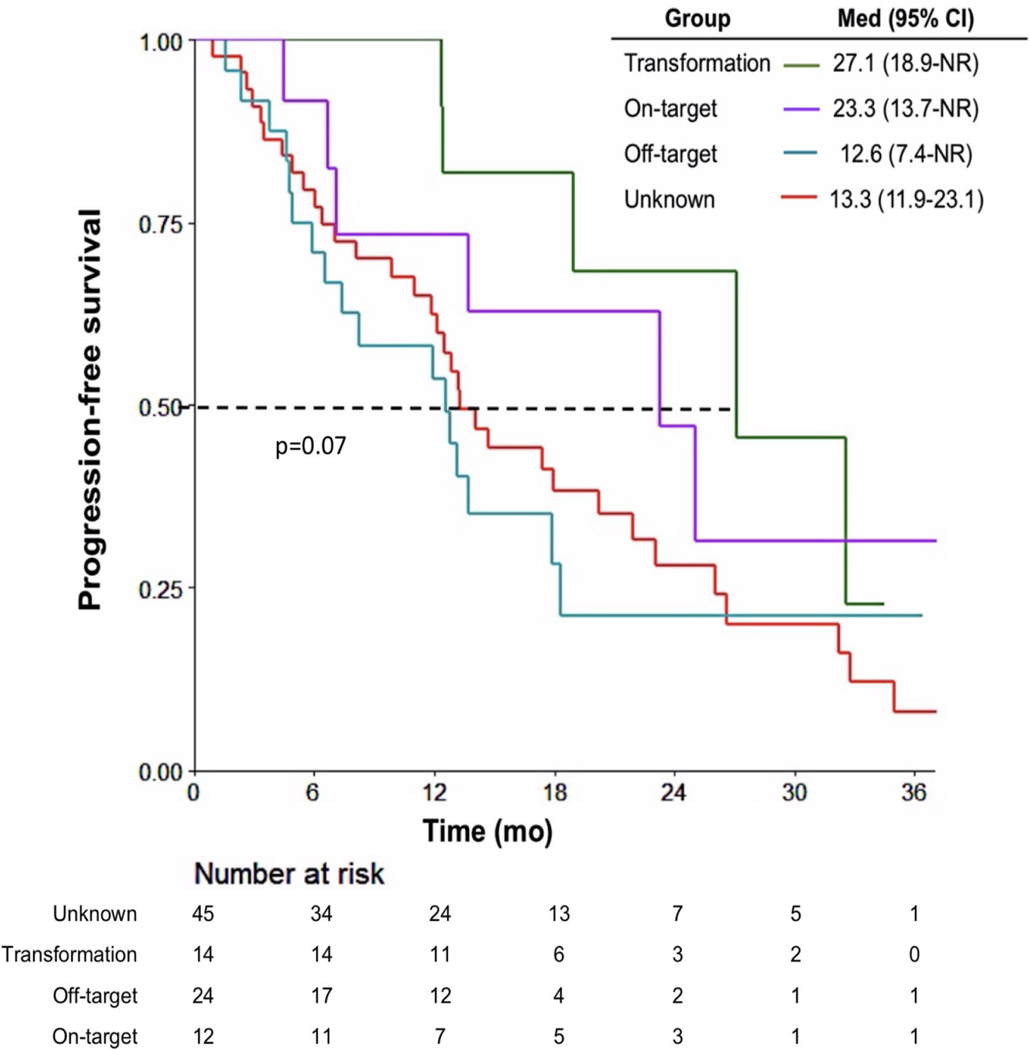
There was no statistical difference in post-osimertinib progression survival between patients with on-target, off-target, histological transformation, or unknown resistance mechanisms (transformation 27.1 mo, on-target 23.3 mo, unknown 13.3 mo, off-target 12.6 mo; p = 0.07, Fig. 3). In contrast, when using a time-dependent Cox model, there was a difference in post-osimertinib survival between patients who received no subsequent therapy (n=10), subsequent tailored second-line therapy based on biopsy results (n=26) and subsequent second-line therapy not tailored to biopsy results (n=59, p<0.001, Supplemental Fig. 5A). The difference in survival was most notable between the tailored therapy cohort and those who received nontailored therapy (24-mo HR 0.09). For the above analyses, small cell-directed therapies were included in tailored second-line therapies; the survival benefit with tailored therapy was still present when histologic transformation cases were considered separately. (24-month HR 0.02, Supplemental Fig. 5B, Supplemental Table 4).
Figure 3. Mechanisms of resistance and post-progression survival.
Post-progression survival for patients stratified by mechanism of identified resistance.
Resistance beyond somatic NGS testing
Resistance mechanisms were not identified for nearly half of patients with somatic tumor NGS testing, prompting exploration of whether additional methods for identifying drivers of resistance are warranted. Given that structural rearrangements mediate resistance in patients with EGFR-mutant NSCLC8, 9, RNAseq may identify fusion events not captured by NGS.23 In the post-progression biopsy cohort, 40/95 (42%) patients underwent Archer FusionPlex, including 25/45 (56%) patients without an identified resistance mechanism (Fig. 4A). Three of the 25 patients with unknown resistance mechanisms had acquired fusions of unknown significance on Archer not identified by MSK-IMPACT (TGF-GPR128, MARS-GLI1 and GOLGA3-STAT6 fusions). Among patients with identified mechanisms of resistance on MSK-IMPACT (n = 15), Archer testing confirmed the one fusion, a TRIM24-BRAF fusion, also detected on MSK-IMPACT. Resistance mechanisms can be heterogeneous with multiple present in the same patient and may change over time. In Fig. 4B, we illustrate a case with initial ERBB2 amplification preosimertinib, with acquired MET amplification post-osimertinib. A subsequent sample after progression on osimertinib with capmatinib was positive for a CD74-ROS1 fusion identified on Archer. This case demonstrates clonal evolution in response to the selective pressures of targeted therapies that may be missed with somatic NGS DNA testing alone.
Figure 4. Methods for investigating unknown resistance.
A) Results of RNAseq testing among 95 patients with a post-progression tumor biopsy sample. B) Single patient with serial biopsies analyzed using Archer, ROS1 IHC staining and MSK-IMPACT. Figure illustrating clonality of each variant, total copy number of ERBB2/MET, and the presence of ROS1 fusion. Clonality is presented by RAF. T1=tumor biopsy 1; T2=tumor biopsy 2; T3=tumor biopsy 3; PD, progressive disease; RT, radiation. C) Sankey plot demonstrating evolution of mutational signatures derived from MSK-IMPACT before and after osimertinib treatment. D) Patterns of mutational signatures and TMB in patients with known and unknown mechanisms of resistance. IHC, immunohistochemistry; RAF, relative allele frequency; RNAseq, RNA sequencing; TMB, tumor mutational burden.
Alternatively, resistance to first-line osimertinib may be mediated by tumor evolution beyond acquisition of somatic single-gene alterations, including large-scale genomic events. Work from our group has validated identification of mutational signatures from targeted NGS data34. The most frequent dominant mutational signatures identified in paired post-progression tumor samples were APOBEC (28%) and clock/aging mutational signatures (36%) (Fig. 4C). There was a non-statistically significant enrichment in APOBEC dominant signature in the post-treatment compared to pre-treatment samples (28% vs 10%, p=0.07), suggesting that larger scale mutational processes may contribute to the accumulation of somatic alterations driving osimertinib resistance. In addition, among 39 paired samples with tumor purity of >20%, TMB was significantly higher in post-progression tumors compared to paired pre-treatment tumors (median 5.3 vs 4.4 mut/Mb, p=0.008, Supplemental Fig. 6A). Interestingly, four post-progression tumor samples had ≥ 10 mut/Mb (Fig. 4D), including three cases with histological transformation, suggesting an association between genomic instability and lineage plasticity. Neither fraction of genome altered (FGA, p=0.12, Supplemental Fig. 6B) nor whole genome doubling (WGD, Supplemental Fig. 6C) was significantly different between pre- and post-progression tumors, although the proportion of WGD in post-progression samples was higher (WGD in 71% vs. 61%, p=0.3, Supplemental Fig. 6C).
DISCUSSION:
The approval of first-line osimertinib was a significant paradigm shift in EGFR-mutant lung cancers. Rather than treating patients sequentially with multiple EGFR TKIs, the FLAURA trial demonstrated that upfront treatment with osimertinib prolongs OS, presumably due to better on-target inhibition and avoiding development of heterogenous resistant subclones selected for by treatment with sequential EGFR TKIs11, 35. Despite this success, further understanding of the evolutionary pressures driving resistance to first-line osimertinib is needed, as the approach of first-line osimertinib monotherapy for all patients is likely suboptimal. Further, there are ongoing studies assessing the efficacy of osimertinib combinations as first-line treatment. Little is known whether resistance mechanisms may differ with combination treatment. Using post-progression tumor samples that underwent NGS with MSK-IMPACT, as well as the largest retrospective dataset to date of patients treated with first-line osimertinib, this work highlights the importance of identification of biomarkers mediating response and resistance and the limitations of current methods of identifying resistance.
The study utilized a real-world cohort of patients with EGFR-mutant lung cancers receiving first-line osimertinib with a median rwPFS shorter than what was reported in prospective trials of first-line osimertinib6. Reasons for this include no official RECIST evaluation of disease progression, inclusion of patients with atypical EGFR alterations (not included in FLAURA), as well as capturing a broader, real-world population likely less fit and more diverse than patients on clinical trials36. For example, brain metastases were twice as high in this real-world cohort than the FLAURA study (41% vs. 21%, respectively). Our cohort reinforced that baseline genomic alterations, namely concurrent TP53/RB1 alterations and atypical EGFR mutations, are associated with shorter rwPFS. While limited gene panel sequencing or immunohistochemistry may rapidly identify EGFR alterations, comprehensive sequencing with NGS may risk stratify patients with poor prognostic genomic features at baseline. Patients with these pre-treatment alterations may benefit from escalation of care such as adding chemotherapy to osimertinib and/or may prompt clinicians to pursue a change in treatment earlier at first signs of disease progression.
While the landscape of acquired resistance to osimertinib has been reported in small series10, 11, this is the largest dataset of patients receiving first-line osimertinib who underwent a tumor tissue biopsy at progression and demonstrates that resistance to osimertinib is characterized primarily by accumulation of off-target events, histological transformation, and unknown mechanisms of resistance. Interestingly, the landscape of mechanisms of resistance may differ for osimertinib monotherapy compared to combination treatment. Incidence of histologic transformation was higher with combination therapy while gene amplifications and gene fusions were more commonly seen with osimertinib monotherapy. With several osimertinib combination studies ongoing including FLAURA2,37 (NCT04035486), it will be paramount to understand how combination therapy alters the selective pressures that drives resistance. The majority (93%) of tumor biopsies obtained in this series were from progressive disease sites, reducing the likelihood of missing subclonal events perpetuating resistance. Further, mechanisms of resistance identified did not seem to be associated with site of metastatic disease biopsied.
Importantly, we demonstrate that categories of resistance are not prognostic; patients without an identified mechanism of resistance do not have shorter post-progression survival compared to patients with an identified mechanism of resistance. Nevertheless, intervening upon an acquired mechanism improves post-progression survival, including recognition of and treatment for small cell transformation. Although small cell transformation is associated with a poor prognosis38, several patients in this study did remarkably well with a median post-transformation survival of 24 months, in part due to local therapy interventions and systemic therapy directed at transformation. Patients presumably benefit from early identification of histologic transformation when the disease is still histologically heterogeneous and local therapy can address subclones that have transformed and osimertinib can continue to control the remaining adenocarcinoma. This is a strong rationale to encourage tumor tissue biopsies to identify cases with histologic transformation earlier.
The improvement in post-progression survival gained from intervening upon identified mechanisms of resistance in the second-line setting underscores the importance of expanding methods for identifying and treating mediators of resistance. Post-progression tumor biopsies have distinct advantages over plasma testing in EGFR-mutant lung cancer for several reasons. Not only is tissue necessary to diagnose histological transformation, but tissue may capture mechanisms not detected in plasma alone and can be used for additional exploratory analyses such as RNAseq, immunohistochemistry, and/or epigenetic analyses. In our cohort, four patients had increased TMB post-osimertinib (TMB ≥ 10 mutations/megabase), potentially selecting for patients that may benefit from immunotherapy39, in contrast to the EGFR-mutant lung cancer population at large where immunotherapy has not demonstrated efficacy40.
In lung cancer, there are no therapeutic interventions that target mutational signatures, but homologous repair deficiency (HRD) signals sensitivity to PARP inhibitors in ovarian cancer41–43, and microsatellite instability portends sensitivity to immunotherapy44, 45, establishing a paradigm where mutational signatures dictate therapeutic strategies. Further exploration is needed to see if mutational signatures will result in additional treatment options in lung cancers, but changes in mutational signatures reveal larger-scale drivers of evolution under the selective pressures of targeted therapies. Post-progression samples were enriched for APOBEC or clock/aging mutational signatures, which are both associated with acquisition of subclonal events during tumor evolution and.35 While clock/aging-like mutational signatures naturally occur in either normal and tumor cells over time and are affected by the rate of cell division46APOBEC mutagenesis is particularly linked to large-scale genomic events and chromosomal instability47 that fuel intratumoral heterogeneity and linked to worse clinical outcomes.35 Concurrent inhibition of APOBEC enzymatic activity is therefore a possible avenue for addressing drug resistance to osimertinib treatment and worthy of prospective evaluation, although at present, no methods of direct inhibition of APOBEC enzymes are clinically available. Synthetic lethality-based therapeutic approaches targeting APOBEC mutagenesis are an area of active research in preclinical tumor models, including lung cancer48, while other preclinical work demonstrates activation of APOBEC mutagenesis may sensitize cell lines to ATR49 and PARP inhibition50. In our view, indeed, EGFR-mutant lung cancers could represent an ideal subset where the inhibition of APOBEC enzymes activity should be explored to seek to prevent the acquisition of subclonal mutations and intratumor heterogeneity and enhance the antitumor efficacy of available TKIs.
This work has several important limitations. First, rwPFS, although consistent with how clinicians define disease progression in real-world settings, is more subjective than RECIST-based PFS. Concurrent treatment with osimertinib and other cancer therapies was also allowed in this study, which may limit the generalizability of some results; however, it also provides valuable information on how mechanisms of acquired resistance differ when more intensive therapy (i.e, addition of chemotherapy) is used upfront. The sample size of patients with post-progression biopsies, although significantly larger than previously reported studies, may be underpowered to detect differences in post-progression survival among the different resistance subtypes. Finally, although patient and tumor characteristics of patients in the tumor biopsy cohort were similar to the larger clinical cohort treated with first-line osimertinib, there may be intrinsic differences in patients who underwent post-progression tumor biopsies compared to those who did not.
In conclusion, with only 50% of resistance to osimertinib driven by single gene alterations, the mechanisms by which tumors escape EGFR-directed therapies are multifaceted and not fully characterizable by somatic NGS testing alone. Exploration should begin at the genome-wide level and narrow to the epigenome, gene, RNA transcript, and protein level as each analysis may reveal relevant mediators of resistance to validate and therapeutically
Supplementary Material
Supplemental Table 1: Treatment Details
Supplemental Table 2: Clinicogenomic factors associated with PFS to first-line osimertinib treatment
Supplemental Table 3: Post-progression biopsy details
Supplemental Table 4: Post-progression osimertinib treatment details
Supplemental Figure 1: Patient selection.
Supplemental Figure 2: Impact of clinical factors on osimertinib treatment.
Supplemental Figure 3: Alterations in pre-osimertinib and post-progression biopsy samples.
Supplemental Figure 4: Impact of osimertinib treatment and biopsy site on identified resistance mechanisms.
Supplemental Figure 5: Impact of tailored therapy on survival.
Supplemental Figure 6: Large-scale genomic analysis in pre-osimertinib and post-progression biopsy samples with tumor purity >20%.
FUNDING:
HAY supported in part by the Haussler fund, EGFR Resisters/LUNGevity Lung Cancer Research Award, STARR foundation, and R01CA264078. MSK authors supported by National Cancer Institute Cancer Center Core Grant No. P30-CA008748.
FINANCIAL DISCLOSURES:
NJC: Institutional research fundings from Merck, AbbVie, Monte Rosa Therapeutics; royalties from Wolters Kluwer (Pocket Oncology). AM: reports royalties from ililli. AJS: reports consulting/advising role to J&J, KSQ therapeutics, Merck, Enara Bio, Perceptive Advisors, and Heat Biologics. Research funding: GSK (Inst), PACT pharma (Inst), Iovance Biotherapeutics (Inst), Achilles therapeutics (Inst), Merck (Inst), BMS (Inst), Harpoon Therapeutics (Inst). DG: reports grants from Varian, AstraZeneca, Merck, and Bristol Myers Squibb, personal fees from Varia, AstraZeneca, Merck, US Oncology, Bristol Myers Squibb, Relfexion, WebMD, Vindico, and Medscape; and has served on the advisory board for AstraZeneca. ML: reports consulting or advisory roles for Takeda Oncology, Janssen Pharmaceuticals. MB: reports personal fees from Roche, grants from Illumina, grants from Grail, outside the submitted work; in addition, M.B. has a patent Systems and Methods for Detecting Cancer Via cfDNA Screening pending. MA: reports personal fees for ad-hoc board participation from AstraZeneca, Bristol Myers Squibb, Jansen Global services, speaker fees from Biocartis, Invivoscribe, RMEI Medical Education, Physician Education Resources, Peerview Institute for Medical Education, I3 Health Education on Point outside the submitted work. CMR: reports personal fees from AbbVie, Amgen, Ascentage, AstraZeneca, Bicycle, Celgene, Daiichi Sankyo, Genentech/Roche, Ipsen, Jansen, Jazz, Lilly/Loxo, Pfizer, PharmaMar, Syros, Vavotek, Bridge Medicines, and Harpoon Therapeutics, outside the submitted work GJR: reports grants from National Institutes of Health/National Cancer Institute, has been an uncompensated consultant to Daiichi, Pfizer, and Mirati, he has institutional research support from Mirati, Takeda, Merck, Roche, Pfizer, and Novartis. GJR has pending patents US20170273982A1 and WO2017164887A8. MGK: receives personal fees from Novartis, Genentech, Sanofi-Genzyme, AstraZeneca, Pfizer, Janssen, and Daiichi-Sankyo; received honoraria for participation in educational programs from WebMD, OncLive, Physicians Education Resources, Prime Oncology, Intellisphere, Creative Educational Concepts, Peerview, i3 Health, Paradigm Medical Communications, AXIS, Carvive Systems, AstraZeneca, and Research to Practice; received travel support from AstraZeneca, Pfizer, Regeneron, Daiichi-Sankyo, and Genentech; received editorial support from Hoffman La-Roche. Memorial Sloan Kettering has received research funding from The National Cancer Institute (USA), The Lung Cancer Research Foundation and Genentech Roche for research conducted by MGK. JSF-F: reports receiving personal/consultancy fees from Goldman Sachs, REPARE Therapeutics, Paige.AI and Eli Lilly, membership of the scientific advisory boards of VolitionRx, REPARE Therapeutics and Paige.AI, membership of the Board of Directors of Grupo Oncoclinicas, and ad hoc membership of the scientific advisory boards of Roche Tissue Diagnostics, Ventana Medical Systems, Novartis, Genentech and InVicro, outside the scope of this study and owns Paige.AI and REPARE Therapeutics stocks. HAY: consultant or advisory role for AstraZeneca, Janssen, Blueprint Med, Black Diamond, C4 Therapeutics, and Daiichi. Research funding: AstraZeneca (Inst), Pfizer (Inst), Daiichi (Inst), Cullinan (Inst), Lilly (Inst), and Novartis (Inst), ERASCA (Inst).
Footnotes
All other authors have declared no conflicts of interest.
CRediT Statement:
Choudhury, Noura: conceptualization, data curation, formal analysis, investigation, original draft writing, review and editing
Marra, Antonio: conceptualization, data curation, formal analysis, methodology, investigation, original draft writing, review and editing
Sui, Jane: conceptualization, data curation, formal analysis, investigation, original draft writing, review and editing
Flynn, Jessica: formal analysis, methodology, writing: review and editing
Yang, Soo-Ryum: formal analysis, writing: review and editing
Falcon, Christina: data curation, project administration, writing: review and editing
Selenica, Pier: formal analysis, writing: review and editing
Schoenfeld, Adam: investigation, writing: review and editing
Rekhtman, Natasha: resources, writing: review and editing
Gomez, Daniel: resources, writing: review and editing
Berger, Mike: resources, writing: review and editing
Ladanyi, Marc: resources, writing: review and editing
Arcila, Maria: resources, writing: review and editing
Rudin, Charles: resources, writing: review and editing
Riely, Gregory: resources, writing: review and editing
Kris, Mark: resources, writing: review and editing
Heller, Glenn: formal analysis, supervision, methodology, writing: review and editing
Reis-Filho, Jorge: supervision, methodology, writing: review and editing
Yu, Helena: conceptualization, funding acquisition, investigation, methodology, supervision, writing: original draft, writing: review and editing
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES:
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA: A Cancer Journal for Clinicians 2021;71:209–249. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fuchs HE, et al. Cancer statistics, 2022. CA: A Cancer Journal for Clinicians 2022;72:7–33. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Forjaz G, Mooradian MJ, et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. The New England journal of medicine 2020;383:640–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jordan EJ, Kim HR, Arcila ME, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer discovery 2017:CD-16–1337. [DOI] [PMC free article] [PubMed]
- 5.Ramalingam SS, Vansteenkiste J, Planchard D, et al. Overall Survival with Osimertinib in Untreated, EGFR-Mutated Advanced NSCLC. New England Journal of Medicine 2019;382:41–50. [DOI] [PubMed] [Google Scholar]
- 6.Soria JC, Ohe Y, Vansteenkiste J, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. The New England journal of medicine 2018;378:113–125. [DOI] [PubMed] [Google Scholar]
- 7.Le X, Puri S, Negrao MV, et al. Landscape of EGFR-Dependent and -Independent Resistance Mechanisms to Osimertinib and Continuation Therapy Beyond Progression in EGFR-Mutant NSCLC. Clin Cancer Res 2018;24:6195–6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piotrowska Z, Isozaki H, Lennerz JK, et al. Landscape of Acquired Resistance to Osimertinib in EGFR-Mutant NSCLC and Clinical Validation of Combined EGFR and RET Inhibition with Osimertinib and BLU-667 for Acquired RET Fusion. Cancer discovery 2018;8:1529–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schoenfeld AJ, Chan JM, Kubota D, et al. Tumor Analyses Reveal Squamous Transformation and Off-Target Alterations As Early Resistance Mechanisms to First-line Osimertinib in EGFR-Mutant Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2020;26:2654–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leonetti A, Sharma S, Minari R, et al. Resistance mechanisms to osimertinib in EGFRmutated non-small cell lung cancer. British journal of cancer 2019;121:725–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Passaro A, Jänne PA, Mok T, et al. Overcoming therapy resistance in EGFR-mutant lung cancer. Nature Cancer 2021;2:377–391. [DOI] [PubMed] [Google Scholar]
- 12.Yu HA, Tian SK, Drilon AE, et al. Acquired Resistance of EGFR-Mutant Lung Cancer to a T790M-Specific EGFR Inhibitor: Emergence of a Third Mutation (C797S) in the EGFR Tyrosine Kinase Domain. JAMA oncology 2015;1:982–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Thress KS, Paweletz CP, Felip E, et al. Acquired EGFR C797S mutation mediates resistance to AZD9291 in non-small cell lung cancer harboring EGFR T790M. Nature medicine 2015;21:560–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou W, Ercan D, Chen L, et al. Novel mutant-selective EGFR kinase inhibitors against EGFR T790M. Nature 2009;462:1070–1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ramalingam SS, Cheng Y, Zhou C, et al. Mechanisms of acquired resistance to first-line osimertinib: Preliminary data from the phase III FLAURA study. Annals of Oncology 2018;29:viii740. [Google Scholar]
- 16.Rotow J, Patel J, Hanley M, et al. FP14.07 Combination Osimertinib plus Selpercatinib for EGFR-mutant Non-Small Cell Lung Cancer (NSCLC) with Acquired RET fusions. Journal of Thoracic Oncology 2021;16:S230. [Google Scholar]
- 17.Deng L, Kiedrowski LA, Ravera E, et al. Response to Dual Crizotinib and Osimertinib Treatment in a Lung Cancer Patient with MET Amplification Detected by Liquid Biopsy Who Acquired Secondary Resistance to EGFR Tyrosine Kinase Inhibition. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2018;13:e169–e172. [DOI] [PubMed] [Google Scholar]
- 18.York ER, Varella-Garcia M, Bang TJ, et al. Tolerable and Effective Combination of Full-Dose Crizotinib and Osimertinib Targeting MET Amplification Sequentially Emerging after T790M Positivity in EGFR-Mutant Non-Small Cell Lung Cancer. J Thorac Oncol 2017;12:e85–e88. [DOI] [PubMed] [Google Scholar]
- 19.Cheng DT, Mitchell TN, Zehir A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD 2015;17:251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eubank MH, Hyman DM, Kanakamedala AD, et al. Automated eligibility screening and monitoring for genotype-driven precision oncology trials. Journal of the American Medical Informatics Association 2016;23:777–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rose Brannon A, Jayakumaran G, Diosdado M, et al. Enhanced specificity of clinical high-sensitivity tumor mutation profiling in cell-free DNA via paired normal sequencing using MSKACCESS. Nature communications 2021;12:3770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zehir A, Benayed R, Shah RH, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nature medicine 2017;23:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Benayed R, Offin M, Mullaney K, et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25:4712–4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chakravarty D, Gao J, Phillips SM, et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol 2017;2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bielski CM, Zehir A, Penson AV, et al. Genome doubling shapes the evolution and prognosis of advanced cancers. Nat Genet 2018;50:1189–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou J, Sanchez-Vega F, Caso R, et al. Analysis of Tumor Genomic Pathway Alterations Using Broad-Panel Next-Generation Sequencing in Surgically Resected Lung Adenocarcinoma. Clin Cancer Res 2019;25:7475–7484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gulhan DC, Lee JJ, Melloni GEM, et al. Detecting the mutational signature of homologous recombination deficiency in clinical samples. Nature genetics 2019;51:912–919. [DOI] [PubMed] [Google Scholar]
- 28.Schwartz CJ, da Silva EM, Marra A, et al. Morphologic and Genomic Characteristics of Breast Cancers Occurring in Individuals with Lynch Syndrome. Clin Cancer Res 2022;28:404–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pareja F, Ptashkin RN, Brown DN, et al. Cancer-Causative Mutations Occurring in Early Embryogenesis. Cancer Discov 2022;12:949–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrag D. GENIE: Real-World Application. ASCO Annual Meeting. Chicago, IL: 2018. Available at https://meetinglibrary.asco.org/record/166165/video. [Google Scholar]
- 31.Yap TA, Macklin-Doherty A, Popat S. Continuing EGFR inhibition beyond progression in advanced non-small cell lung cancer. Eur J Cancer 2017;70:12–21. [DOI] [PubMed] [Google Scholar]
- 32.Goto Y, Tanai C, Yoh K, et al. Continuing EGFR-TKI beyond radiological progression in patients with advanced or recurrent, EGFR mutation-positive non-small-cell lung cancer: an observational study. ESMO Open 2017;2:e000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Offin M, Chan JM, Tenet M, et al. Concurrent RB1 and TP53 Alterations Define a Subset of EGFR-Mutant Lung Cancers at risk for Histologic Transformation and Inferior Clinical Outcomes. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2019;14:1784–1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Selenica P, Marra A, Choudhury NJ, et al. APOBEC mutagenesis, kataegis, chromothripsis in EGFR-mutant osimertinib-resistant lung adenocarcinomas. Ann Oncol 2022. [DOI] [PMC free article] [PubMed]
- 35.Jamal-Hanjani M, Wilson GA, McGranahan N, et al. Tracking the Evolution of Non–SmallCell Lung Cancer. New England Journal of Medicine 2017;376:2109–2121. [DOI] [PubMed] [Google Scholar]
- 36.Burstein HJ, Krilov L, Aragon-Ching JB, et al. Clinical Cancer Advances 2017: Annual Report on Progress Against Cancer From the American Society of Clinical Oncology. Journal of Clinical Oncology 2017;35:1341–1367. [DOI] [PubMed] [Google Scholar]
- 37.Planchard D, Feng PH, Karaseva N, et al. Osimertinib plus platinum-pemetrexed in newly diagnosed epidermal growth factor receptor mutation-positive advanced/metastatic non-small-cell lung cancer: safety run-in results from the FLAURA2 study. ESMO Open 2021;6:100271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marcoux N, Gettinger SN, O’Kane G, et al. EGFR-Mutant Adenocarcinomas That Transform to Small-Cell Lung Cancer and Other Neuroendocrine Carcinomas: Clinical Outcomes. J Clin Oncol 2019;37:278–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 2020;21:1353–1365. [DOI] [PubMed] [Google Scholar]
- 40.Lisberg A, Cummings A, Goldman JW, et al. A Phase II Study of Pembrolizumab in EGFR-Mutant, PD-L1+, Tyrosine Kinase Inhibitor Naïve Patients With Advanced NSCLC. Journal of thoracic oncology : official publication of the International Association for the Study of Lung Cancer 2018;13:1138–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.González-Martín A, Pothuri B, Vergote I, et al. Niraparib in Patients with Newly Diagnosed Advanced Ovarian Cancer. New England Journal of Medicine 2019;381:2391–2402. [DOI] [PubMed] [Google Scholar]
- 42.Longo DL. Personalized Medicine for Primary Treatment of Serous Ovarian Cancer. New England Journal of Medicine 2019;381:2471–2474. [DOI] [PubMed] [Google Scholar]
- 43.Mirza MR, Monk BJ, Herrstedt J, et al. Niraparib Maintenance Therapy in PlatinumSensitive, Recurrent Ovarian Cancer. New England Journal of Medicine 2016;375:2154–2164. [DOI] [PubMed] [Google Scholar]
- 44.André T, Shiu K-K, Kim TW, et al. Pembrolizumab in Microsatellite-Instability–High Advanced Colorectal Cancer. New England Journal of Medicine 2020;383:2207–2218. [DOI] [PubMed] [Google Scholar]
- 45.Marcus L, Lemery SJ, Keegan P, et al. FDA Approval Summary: Pembrolizumab for the Treatment of Microsatellite Instability-High Solid Tumors. Clinical cancer research : an official journal of the American Association for Cancer Research 2019;25:3753–3758. [DOI] [PubMed] [Google Scholar]
- 46.Alexandrov LB, Jones PH, Wedge DC, et al. Clock-like mutational processes in human somatic cells. Nature genetics 2015;47:1402–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Swanton C, McGranahan N, Starrett GJ, et al. APOBEC Enzymes: Mutagenic Fuel for Cancer Evolution and Heterogeneity. Cancer discovery 2015;5:704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Petljak M, Green AM, Maciejowski J, et al. Addressing the benefits of inhibiting APOBEC3-dependent mutagenesis in cancer. Nature genetics 2022;54:1599–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Buisson R, Lawrence MS, Benes CH, et al. APOBEC3A and APOBEC3B Activities Render Cancer Cells Susceptible to ATR Inhibition. Cancer Research 2017;77:4567–4578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Green AM, Budagyan K, Hayer KE, et al. Cytosine Deaminase APOBEC3A Sensitizes Leukemia Cells to Inhibition of the DNA Replication Checkpoint. Cancer Research 2017;77:4579–4588. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: Treatment Details
Supplemental Table 2: Clinicogenomic factors associated with PFS to first-line osimertinib treatment
Supplemental Table 3: Post-progression biopsy details
Supplemental Table 4: Post-progression osimertinib treatment details
Supplemental Figure 1: Patient selection.
Supplemental Figure 2: Impact of clinical factors on osimertinib treatment.
Supplemental Figure 3: Alterations in pre-osimertinib and post-progression biopsy samples.
Supplemental Figure 4: Impact of osimertinib treatment and biopsy site on identified resistance mechanisms.
Supplemental Figure 5: Impact of tailored therapy on survival.
Supplemental Figure 6: Large-scale genomic analysis in pre-osimertinib and post-progression biopsy samples with tumor purity >20%.