Abstract
Snail family proteins are zinc finger transcriptional regulators first identified in Drosophila which play critical roles in cell fate determination. We identified a novel Snail-related gene from murine skeletal muscle cells designated Smuc. Northern blot analysis showed that Smuc was highly expressed in skeletal muscle and thymus. Smuc contains five putative DNA-binding zinc finger domains in its C-terminal half. In electrophoretic mobility shift assays, recombinant zinc finger domains of Smuc specifically bound to CAGGTG and CACCTG E-box motifs (CANNTG). Because basic helix–loop–helix transcription factors (bHLH) bind to the same E-box sequences, we examined whether Smuc competes with the myogenic bHLH factor MyoD for DNA binding. Smuc inhibited the binding of a MyoD–E12 complex to the CACCTG E-box sequence in a dose-dependent manner and suppressed the transcriptional activity of MyoD–E12. When heterologously targeted to the thymidine kinase promoter as fusion proteins with the GAL4 DNA-binding domain, the non-zinc finger domain of Smuc acted as a transcriptional repressor. Furthermore, overexpression of Smuc in myoblasts repressed transactivation of muscle differentiation marker Troponin T. Thus, Smuc might regulate bHLH transcription factors by zinc finger domains competing for E-box binding, and non-zinc finger repressor domains might also confer transcriptional repression to control differentiation processes.
INTRODUCTION
Snail-related proteins are zinc finger transcription factors with conserved structures among many species including Drosophila and vertebrates, and are thought to be involved in many cell differentiation processes (1,2). They bind to the specific nucleotide sequence CANNTG, called the E-box motif, by highly conserved C2H2 type zinc finger domains (3,4) and suppress the transactivation of downstream target genes. The E-box is also the consensus binding site for the basic helix–loop–helix transcription factors (bHLHs) which participate in cell differentiation processes of various tissues and cell types (5). Thus, the Snail-related proteins function as regulators of a complex gene activation cascade.
Several Snail family members have been reported among several species. In Drosophila, two Snail-related genes, Snail and Escargot have been well characterized (4,6). Snail is indispensable to mesoderm formation during development, because Snail suppresses the differentiation of neuroectoderm by repressing the proneural genes sim and rho (6). Escargot is required for the development of imaginal histoblasts and fusion of tracheal cells (7,8). In the developing nervous system, Escargot inhibits the transcriptional activities of the proneural bHLH factors achaetescute and daughterless via competition for one of the E-box binding sites (9).
In higher vertebrates, murine Sna and Slug and chick cSnR have been identified as Snail family members (10–13). They contain highly conserved C2H2 type zinc finger domains and are expressed in early embryos. Sna is expressed in the mesoderm and somites of early mouse embryos (10,11). Although its role in early embryogenesis has not been elucidated, Sna has been suggested to suppress the transcriptional activities of bHLH proteins E47 and Mash2 in trophoblast development (14). Slug induces cell migration in the epithelial–mesenchymal transition, mesoderm formation and neural crest cell migration (9). Antisense inhibition of Slug impairs fibroblast growth factor-induced scattering and hepatocyte growth factor-mediated tubular morphogenesis of epithelial cell (15). cSnR is expressed in the chick right-hand lateral mesoderm. Antisense disruption of its function randomizes the asymmetrical body organization (15). These three proteins have a highly conserved N-terminal domain. This conservation is also found in another zinc finger protein, Gfi1, which immortalizes IL-2-dependent T cells (16). Therefore, this domain is called the SNAG (Snail/Gfi1) domain and is thought to play roles in nuclear targeting and transcriptional repression (16).
In this study, we searched for a Snail-related gene in developing mouse embryos and adult organs and identified a novel Snail-related gene Smuc (Snail-related transcription factor of muscle cells). Smuc was highly expressed in adult skeletal muscle and thymus in addition to the developing embryos. Smuc bound to particular E-box sequences with high affinity and inhibited E-box-dependent transactivation by the MyoD–E12 complex in C2C12 cells. Therefore, Smuc might function as a regulator of differentiation processes by mechanisms similar to other Snail family members.
MATERIALS AND METHODS
RNA isolation
Total RNA was extracted from skeletal muscle of adult B6 mice (S.L.C., Shizuoka, Japan) by the guanidine thiocyanate/phenol/chloroform method (17) and from mouse embryos with Trizol (Gibco BRL). Poly(A)+ RNA was prepared using Oligotex dT Super (Takara Shuzo, Kusatsu, Japan) according to the manufacturer’s instructions.
cDNA cloning of Smuc
Total RNA of mouse skeletal muscles was reverse transcribed with oligo(dT) primer using the Superscript II cDNA synthesis Kit (Gibco BRL). Synthesized cDNA was used as a template for PCR amplification with a set of sense and antisense primers whose sequences were 5′-GCGTGGATCCATGCCGCGCTCCTTCCTGGTCAA-3′ and 5′-GTA(C/T)TC(C/T)TTNT(C/T)-(A/G)CA(A/G)T(A/C)(C/T)TT(A/G)CA-3′, respectively. The sense primer corresponded to the conserved N-terminal amino acid sequence MPRSFLVK and the antisense primer to CKYC(D/E)KEY, present in one of the conserved zinc finger domains of Sna and Slug. The conditions for the PCR reaction were one cycle of denaturation at 94°C for 3 min, 33 cycles of denaturation at 94°C for 1 min, annealing at 50°C for 1.5 min and synthesis at 72°C for 1.5 min, and one cycle of synthesis at 72°C for 5 min, using a DNA Engine (Perkin-Elmer). The PCR product was separated on a 6% polyacrylamide gel and a DNA fragment of 550 bp in size was isolated with Qiaex II (Qiagen). The fragment was subcloned into pKRX-XcmI (18) and sequenced with a Dye-Terminator Kit (Perkin-Elmer) using an ABI 377 or 310 Genetic Analyzer (Applied Bioscience).
BLAST search analysis of the obtained sequences revealed that two clones in 20 encode a novel sequence containing a putative zinc finger domain. This DNA fragment was excised from the vector, labeled with 32P and used as a probe to obtain the full-length cDNA.
A λgt 11 cDNA library vector (Pharmacia) was constructed using 3 µg of poly(A)+ RNA from skeletal muscle as described (19) and screened by plaque hybridization (20). Briefly, ~5 × 105 phage plaques were transferred onto nylon membranes (NEN). After prehybridization with hybridization buffer (5× Denhardt’s, 600 mM NaCl, 1% SDS, and 50 mM Tris–HCl, pH 7.5) at 68°C for 2 h, filters were hybridized to the 32P-labeled probe at 68°C for 16 h. The filters were washed three times with 0.5× SSC, 0.2% SDS at 68°C for 30 min. Positive phage clones were subcloned into pBluescript SK(+) (Stratagene) and the nucleotide sequences were determined.
Northern blot analysis
Five micrograms of poly(A)+ RNA from adult organs was separated on a 1% agarose gel and transferred onto a nylon membrane. Multiple tissue northern blots (MTN; Clontech) were used for mouse embryos. The DNA fragment corresponding to nucleotides 1–540 of the coding sequence of Smuc was 32P-labeled with a Prime-It II random labeling kit (Stratagene) and used as a probe. Comparable amounts of RNA on each lane were confirmed by hybridization of the filters to a glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probe. Hybridization and washing of the filters were performed under the same conditions as for cDNA library screening, as described (21).
Plasmid construction
pCMV-MyoD, pCMV-E12 and pCMV were gifts from Dr E. Hara (Paterson Institute of Cancer Research), pNLS-LacZ from Dr N. Akiyama (Kyoto University), ptk-GALpx3-Luc and pEF-GAL4-DBD from Dr T. Kanno (Kyoto University) (22), and pE7-luc, which contains seven E-box sequences derived from the MCKR promoter upstream of the luciferase gene, from Dr R. Kageyama (Kyoto University).
The entire coding area of Smuc (nucleotides 1–864) was PCR amplified with a set of primers containing 5′ EcoRI sites by Deep Vent DNA polymerase (New England Biolabs) and subcloned into the EcoRI site of pBluescript SK(+) to generate pBluescript-Smuc. The authenticities of the nucleotide sequences of PCR fragments were confirmed as described.
For expression, the EcoRI fragment from pBluescript-Smuc was subcloned into the EcoRI site of pCMV to generate pCMV-Smuc.
To express fusion proteins of the GAL4 DNA-binding domain (GAL4 DBD) with the non-zinc finger region of Smuc, sequences corresponding to amino acids 1–148 (GAL4–ΔZf), 24–148 (GAL4–ΔZfΔN), and 1–21 (GAL4–N) were PCR amplified by Deep Vent DNA polymerase and subcloned in-frame into the XbaI and BamHI sites of pEF-GAL4-DBD to generate pEFGAL4-ΔZf, pEFGAL4-ΔZfΔN, and pEFGAL4-N, respectively.
Electrophoretic mobility shift assay (EMSA)
For EMSA, GST–Smuc-Zf in which the zinc finger domains of Smuc were fused with GST was generated, because the full-length Smuc protein turned out to be unstable. A 405 bp XhoI fragment containing the region encompassing putative zinc finger domains (from amino acid 142 to 287) was subcloned in-frame into pGEX4T2 (Pharmacia). The product plasmid pGEX4T2-Smuc-Zf was transformed into BL21 cells and the protein was generated as per the manufacturer’s recommendations. The purity was >90% by Coomassie Blue staining. MyoD and E12 proteins were generated with the T7 TNT in vitro transcription–translation kit (Promega) using pCMV-MyoD and pCMV-E12 as templates, respectively, according to the manufacturer’s recommendations. To form the MyoD and E12 heterodimer, both plasmids were co-translated in the same reactions. Equal amounts of translation products of E12 and MyoD were confirmed by SDS–PAGE analysis of [35S]methionine-labeled products.
The nucleotide sequences of oligonucleotides used in the gel shift assays were as follows (E-box motifs are underlined).
SMU, 5′-CGCGTGCGGCCTGACAGGTGCTTTGA-3′ (3)
mutant SMU, 5′-CGCGTGCGGCCTGCACACTGCTTTGA-3′
MCK1, 5′-CGCGTCAGGCAGCAGGTGTTGGA-3′ (23)
mutant MCK1, 5′-CGCGTCAGGCAGTGGCATTTGGA-3′
MCK2, 5′-GGATCCCCCAACACCTGCTGCCTGA-3′ (14)
mutant MCK2, 5′-GGATCCCCCAAACCGTCCTGCCTGA-3′
kE2, 5′-GTTCCTGCGAGGCAGGTGGCCCAG-3′ (25)
CGRP, 5′-TCGAAGGACGCAGCTGATCC-3′ (26)
Troponin I, 5′-GTCTGAGGAGACAGCTGCAGCT-3′ (27)
N-box, 5′-ACGCCACGAGCCACAAGGATTGT-3′ (28)
RIPE3, 5′-GCCCCTCTGGCCATCTGCTGAT-3′ (29)
MLCA-1, 5′-GTCCATTTTTGCACCTGCTGCGACTT-3′ (30)
MCKR-1, 5′-GGATCCCCCCAACACCTGCTGCCTGA-3′ (31)
MCKL-1, 5′-AATAACCCAGACATGTGGCTGCC-3′ (24)
EF-1, 5′-GCAAGAACAGATGGTCCCCAGAAATAG-3′ (30)
8701, 5′-CTAGTGGCCAGATGTTTTCAGC-3′ (31)
MLCB-1, 5′-GTTGCTTCGCCAGCTGGTGGGGATT-3′(24)
MLCC-1, 5′-GAGGAATTAGGCACCTGTTGCTTCG-3′ (24)
CE2-1, 5′-GATCCAAAGGGGCAGCTGTGCAAATCA-3′ (32)
The oligonucleotide sequences of m3MCK2–m12MCK2 are identical to MCK2 except that the internal dinucleotides of the E-boxes (CANNTG) are changed as follows: m3, CG; m4, CT; m5, CA; m6, GC; m7, GA; m8, GT; m9, TA; m10, AT; m11, AA; m12, GG.
Each oligonucleotide was annealed with the complementary oligonucleotide in annealing buffer containing 10 mM Tris–HCl (pH 7.5), 1 mM EDTA, 5 mM MgCl2 and 200 mM NaCl to generate 5′-overhangs containing G residues on both strands. Two hundred nanograms of paired oligonucleotides were 32P-labeled with Klenow fragment and purified on Sephadex G 50 Probe-Quant (Pharmacia). Labeled oligonucleotides (~20 000 c.p.m.) and 40 ng of GST–Smuc Zf were incubated in binding buffer (20 mM HEPES–KOH pH 7.9, 50 mM KCl, 1 mM DTT and 20 µM ZnCl2) at room temperature for 15 min and electophoresed in a 6% polyacrylamide gel for 100 min at 150 V. Specific binding of GST–Smuc-Zf to labeled probes was confirmed by adding a 400-fold molar excess of unlabeled competitive oligonucleotide probes. In the competition assays, ~5000 c.p.m. of MCK2 oligonucleotide probe, GST–Smuc Zf and in vitro translated MyoD and E12 proteins were incubated in the same buffer in various combinations. Fixed amounts of in vitro translated MyoD and E12 products and 60 ng GST–Smuc-Zf were incubated with increasing concentrations of the competitive proteins and electophoresed in a 5% polyacrylamide gel.
Luciferase assay
C2C12 cells were a gift from Dr S. Yoshida (Department of Pathology, Faculty of Medicine, Kyoto University) and maintained in DMEM (Gibco BRL) with 10% fetal calf serum (BioWhittaker). pE7-luc, which contains seven E-box sequences derived from the MCKR promoter upstream of the luciferase gene, was used as a reporter plasmid. pCMV-MyoD and pCMV-E12 (85 and 90 ng/well, respectively) were co-transfected with various amounts of pCMV-Smuc into subconfluent C2C12 cells in 12-well plates. Total amounts of transfected DNA were normalized with the empty vector. To demonstrate the repressor activity of the non-zinc finger domain of Smuc, 0.6 µg of pEF-GAL4, pEF-GAL4-ΔZf, pEF-GAL4-ΔZfΔN, and pEF-GAL4-N per well were transfected into C2C12 cells in 12-well plates with the reporter plasmid ptk-GALpx3-Luc, which contains three GAL4 DBD sequences upstream of the thymidine kinase (tk) promoter and luciferase gene. After 48 h, cells were lysed and luciferase activity was measured using the Dual-Luciferase assay system and normalized to Renilla luciferase activity as recommended by the manufacturer (Promega).
Cell culture and immunostaining
In the C2C12 differentiation assay, cells were seeded at a density of 3 × 104 cells/well in collagen-coated 6-well plates (Iwaki, Japan) 24 h prior to transfection. Cells in each well were co-transfected with 1.2 µg pNLS-LacZ plus 1.2 µg pCMV-Smuc, PCMV-Id2, or pCMV using lipofectamine for 4 h and then stimulated to differentiate in DMEM containing 2% horse serum. For immunostaining, cells were fixed with 4% paraformaldehyde/phosphate-buffered saline (PBS) (pH 7.4) for 10 min at 4°C. Wells were washed twice with PBS and β-galactosidase-positive cells were detected as described (33). Cells were then washed twice with PBS and incubated in methanol at 4°C each for 5 min. After a blocking/permeabilization step for 30 min with PBS containing 0.2% Triton-X 100, 0.1% bovine serum albumin, and 2% skim milk, cells were stained with an anti-Troponin T monoclonal antibody (JLT20; Sigma). The primary complexes were detected with a peroxidase-labeled anti-mouse immunoglobulin secondary antibody (Amersham) and visualized with diaminobenzidine tetrahydrochloride (Dojin).
Statistical analysis
Data are expressed as means ± SEM. Results were analyzed by one-way ANOVA followed by Fisher’s protected least significant difference test to assess the significance of specific intergroup differences with the software Statview 4.5.
RESULTS
Isolation and characterization of Smuc
In the search for a novel Snail-related gene, we have performed PCR amplifications with a set of primers that were designed according to the conserved amino acid sequences among vertebrate Snail family members. We isolated an expected ~550 bp DNA fragment from the PCR product of adult skeletal muscle and subcloned it into pKRX-Xcm1. Among 20 clones analyzed, two contained a novel cDNA sequence encoding a zinc finger domain. This PCR fragment was used as a probe to screen a mouse skeletal muscle cDNA library. About 500 000 phage plaques were screened and three positive clones were identified. Sequence analyses and sizes of these inserts indicated that they overlapped. The longest cDNA insert was analyzed in detail. The size of the cDNA was 1882 bp and comparable to that of the major transcript detected by northern blot analysis. The nucleotide sequence of the cDNA indicated that this clone contained an open reading frame of 864 bp encoding a 287 amino acid protein with the Kozak consensus sequence for initiation of translation (34; Fig. 1). The deduced amino acid sequence contained five C2H2 zinc finger domains in its C-terminal half, the structure of which is similar to that of Snail family members. The novelty of the sequence was confirmed by BLAST search analysis of the GenBank database. We designated this novel gene Smuc (Snail-related in muscle cells).
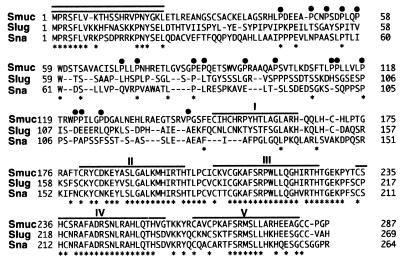
Figure 1.
Amino acid sequence of Smuc compared with those of mouse Sna (6,7) and mouse Slug (8). Identical residues among the three members are indicated by asterisks. The numbers of residues are indicated on both sides. The non-zinc finger domain of Smuc contains a region rich in proline residues, which are indicated by closed circles. Regions of amino acids corresponding to five zinc finger domains of Smuc and Slug and four of Sna are overlined. Conserved amino acid residues in the N-terminal region are indicated by double lines. The DNA sequence corresponding to the amino acids has been deposited in GenBank (accession no. AF133714).
The amino acid sequence indicated that Smuc was highly homologous to mouse Sna and Slug. Amino acid identities between Smuc and Sna and Slug were 43 and 47%, respectively. However, in the zinc finger domains, the identities were 75 and 80% to Sna and Slug, respectively. In addition to the zinc finger motifs, these three proteins show sequence conservation in their N-terminal ends, particularly in the first seven amino acid residues (Fig. 1). Another feature noted in Smuc is abundant proline residues in the N-terminal half (14%), which is also found in other members of the Snail family. These results indicate that Smuc is a new member of the vertebrate Snail family and most related to mouse Slug.
As predicted by PSORT II analysis, Smuc was localized to the nucleus, which was confirmed by co-localization of a GFP–Smuc fusion protein and nuclear staining by Hoechst 33258 (data not shown).
Tissue distribution of Smuc mRNA
To examine the expression patterns of Smuc in mice, northern blot analyses were performed. To avoid possible cross-hybridization to other mRNA species encoding zinc finger proteins, a region encompassing nucleotides 1–540 of the Smuc cDNA was used as a probe. In adult mice, the skeletal muscle and thymus were the dominant place for Smuc expression (Fig. 2A). Much weaker signals were detected in the heart, lung and spleen. The size of the Smuc mRNA was estimated to be 1.9 kb and several larger transcripts were also detected in the thymus, probably due to unprocessed transcripts (Fig. 2A). In developing embryos, Smuc was also expressed in embryonic day 7 (E7) mouse embryos, and higher expression was observed in later developmental stages (E15 and E18) (Fig. 2B).
Figure 2.
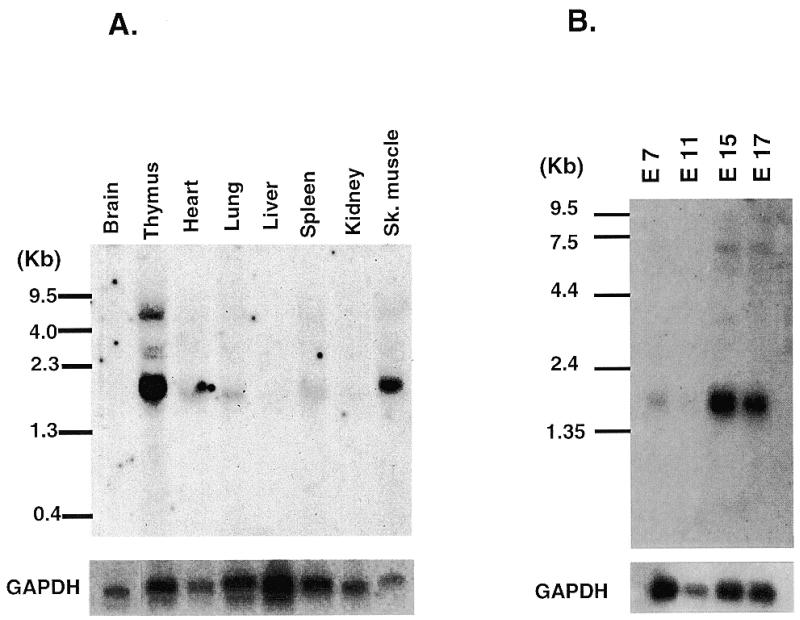
Tissue distribution of Smuc mRNA. (A) Northern blot analysis of adult organs. Approximately 5 µg of poly(A)+ RNA derived from the indicated adult organs were hybridized with 32P-labeled Smuc probe as described in Materials and Methods. The size of Smuc was estimated to be 1.9 kb. Smuc was dominantly expressed in thymus and skeletal muscle with weak expression in lung, heart and spleen. (B) Expression of Smuc in developing mouse embryos. Each lane contains 2 µg of embryo poly(A)+ RNA. Smuc expression was detected at embryonic day 7 (E7) and up-regulated at later stages (E15 and E17). The presence of adequate amounts of RNA was confirmed by hybridization with GAPDH.
DNA binding property of Smuc
Drosophila Snail, Escargot and mouse Sna proteins have been shown to bind E-box sequences of DNA by their zinc finger domains (3,4,9,14). We examined whether Smuc also has an ability to bind E-box motifs by the highly conserved zinc finger domains among the family. EMSA assays were performed with a GST fusion protein, GST–Smuc-Zf, which contains five zinc finger domains of Smuc (amino acids 142–287). Oligonucleotide probes with the CAGGTG E-box sequence (SMU and MCK1) were first examined because CAGGTG is the core sequence for high affinity binding of Drosophila Snail and Escargot. As expected, GST–Smuc-Zf bound specifically to these oligonucleotide probes (Fig. 3A).
Figure 3.
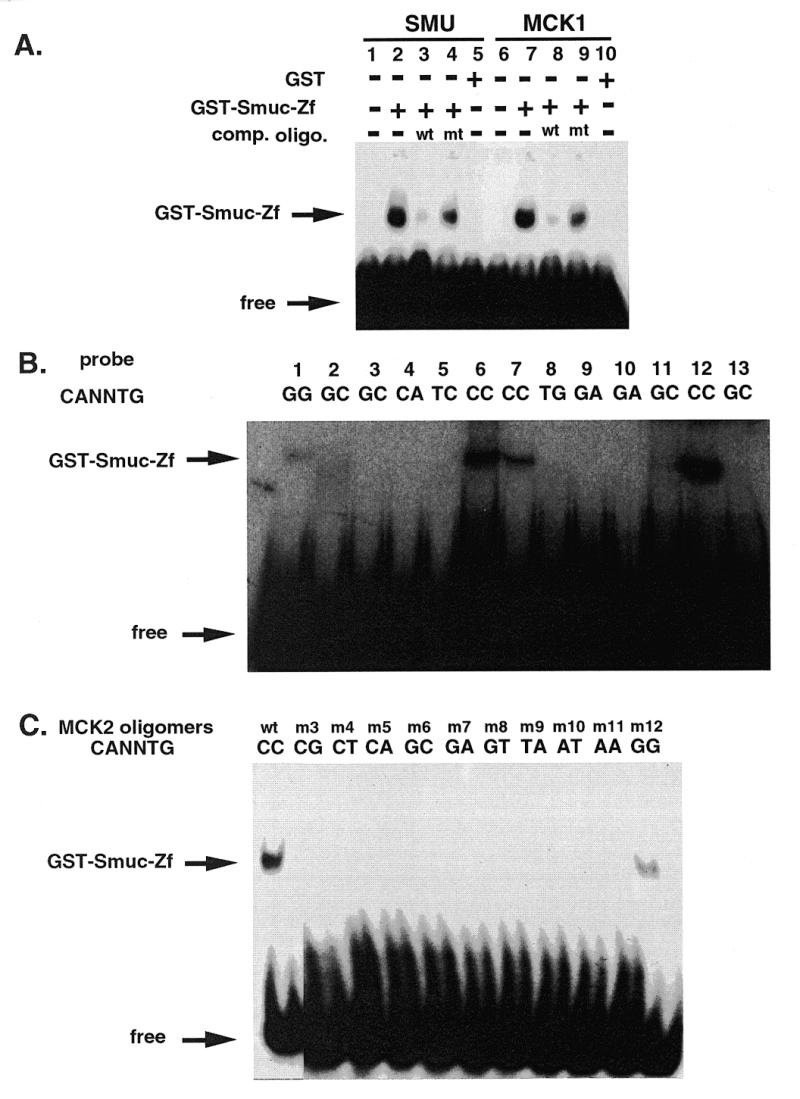
DNA binding properties of Smuc. (A) Binding of Smuc protein to CAGGTG E-box sites. EMSA was performed with GST–Smuc-Zf and double-stranded oligonucleotide probes containing the CAGGTG sequence. GST–Smuc-Zf was shown to bind to oligonucleotide probe SMU or MCK1, specifically to the CAGGTG sites in these two oligonucleotide probes. Competitive wild-type or mutant (mutant SMU or MCK1) oligonucleotide probes were added at 400 molar excess (lanes 3, 4, 8, and 9). (B) Preferential binding of Smuc zinc finger domains to CACCTG and CAGGTG E-box sequences. Various E-box and one N-box sequences were used as probes for binding of GST–Smuc-Zf. Oligonucleotide probes used in each lane were as follows: lane 1, kE2; lane 2, CGRP; lane 3, Troponin I; lane 4, N-box; lane 5, RIPE 3; lane 6, MLCA; lane 7, MCKR-1; lane 8, MCKL; lane 9, EF-1; lane 10, 8701; lane 11, MLCB; lane 12, MLCC; lane 13, CE2. The sequences of these oligonucleotides are described in Materials and Methods. GST–Smuc-Zf was shown to bind only to the E-box sequences containing CACCTG or CAGGTG (lane 1, 6, 7, and 12). One of four independent experimental results is shown. (C) Binding of GST–Smuc-Zf specifically to CACCTG and CAGGTG core E-box sequences. With other backbone sequences fixed, only the internal dinucleotides (CANNTG) of the MCK2 oligonucleotide probe were mutated and used in the EMSAs. GST–Smuc-Zf bound with high affinity only to the probes whose E-box sequences were CACCTG or CAGGTG.
To elucidate the specificity of Smuc–DNA interaction, the same experiment was performed with different E-box oligonucleotides containing various core sequences. Among the oligonucleotide probes which represent the upstream E-box sequences of various genes, GST–Smuc-Zf bound only to the oligonucleotides with CACCTG or CAGGTG core sequences but not to other oligonucleotides (Fig. 3B). To further confirm the specificity of the target sequences, the MCK2 probe was mutated only in the internal dinucleotides of the E-box sequence. GST–Smuc-Zf bound with high affinity only to the probes that contained CACCTG and CAGGTG sequences (Fig. 3C). The preferential binding of Smuc zinc finger domains to CACCTG and CAGGTG motifs is similar to that of Drosophila Snail and Escargot, suggesting conservation of targets of transcriptional regulation by Snail-related genes.
Smuc and MyoD compete for a common DNA binding site
It is well known that myogenic bHLH factors such as MyoD bind to E-box sequences and strongly induce transcriptions of target genes (5). The E-box sequences CACCTG and CAGGTG, with which Smuc specifically interacts, are motifs often found in muscle-specific gene promoters, including muscle creatinine kinase (MCK). These findings raise the possibility that Smuc interferes with MyoD binding to E-box motifs. To confirm this possibility, the ability of Smuc to inhibit MyoD binding to the E-box was examined. In vitro translated MyoD–E12 heterodimer was incubated with a 32P-labeled MCK2 oligonucleotide probe containing the CACCTG sequence in the presence of various amounts of GST–Smuc-Zf (Fig. 4A). GST–Smuc-Zf inhibited MyoD–E12 complex binding to the CACCTG sequence dose-dependently. In another experiment, GST–Smuc-Zf was displaced from the E-box by the addition of increasing amounts of MyoD–E12 complex (Fig. 4B). These results suggest that Smuc and MyoD–E12 compete for the same E-box site. Similar competition results were obtained with Myogenin–E12 (data not shown).
Figure 4.
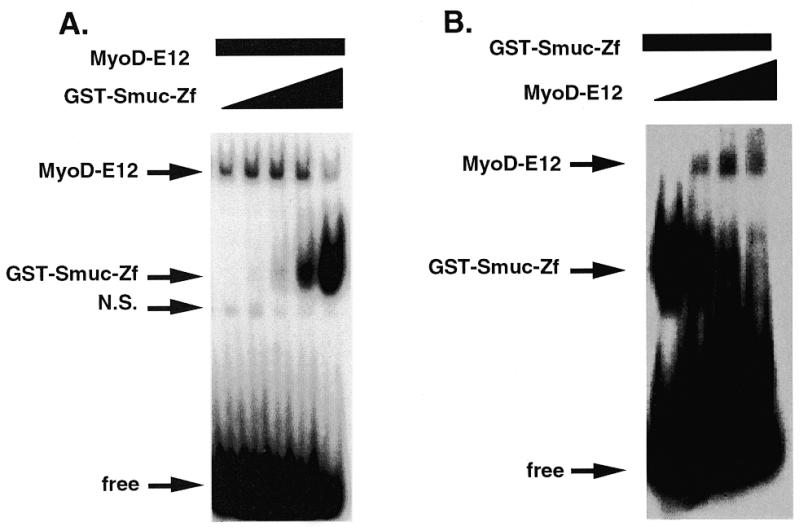
Competition between Smuc and MyoD–E12 for the E-box DNA binding site. (A) Competition between GST–Smuc-Zf and bHLH proteins for a CACCTG site in the MCK2 oligonucleotide probe. The in vitro translated MyoD–E12 heterodimer (3 µl) was incubated with increasing amounts of GST–Smuc-Zf (0, 15, 50, 150 and 450 ng). Binding of MyoD–E12 heterodimer was decreased when increasing amounts of GST–Smuc-Zf were added. (B) Sixty nanograms of GST–Smuc-Zf was incubated with 32P-labeled MCK2 oligonucleotide with MyoD–E12 complex. GST–Smuc-Zf was displaced from the binding site by increasing amounts of MyoD–E12 complex (0, 2, 4 and 8 µl).
Non-zinc finger region of Smuc as a repressor domain
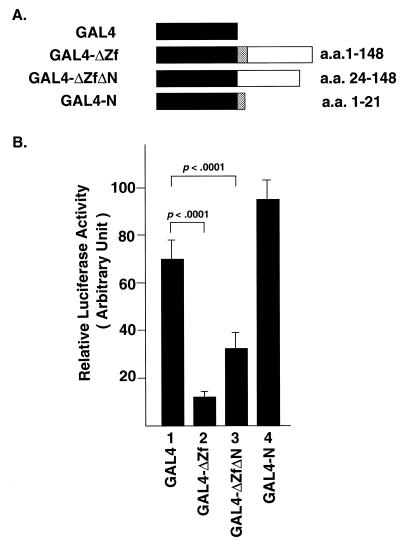
Many transcription factors have been shown to contain distinct functional regions, serving as DNA-binding, activation or repressor domains. The zinc finger domain of Smuc was shown to act as a DNA-binding domain. We further examined if the non-zinc finger domain of Smuc might serve as a repressor or activation domain in C2C12 cells, by targeting this domain to the heterologous tk promoter as fusion proteins with the GAL4 DBD (Fig. 5A). When the whole non-zinc finger domain of Smuc (amino acids 1–148) was expressed with the GAL4 DBD and targeted to the GAL4 binding sites upstream of the tk promoter, luciferase activity was repressed to 17% of the control. About 20 N-terminal amino acids, termed the SNAG domain, are conserved among vertebrate Snail family and Gfi1 proteins. The SNAG domain of Gfi1 has been shown to be sufficient for transcriptional repression (16). A GAL4 fusion protein which lacked the conserved N-terminal amino acids among Smuc, Slug and Sna (amino acids 24–148) led to 50% suppression, suggesting some importance of this region. However, when only the conserved N-terminal amino acids were expressed as a GAL4 fusion protein, they did not confer transcriptional repression (Fig. 5B). Therefore, the non-zinc finger domain of Smuc as a whole possesses repressor activity and the N-terminal region is important but not sufficient to mediate this repression.
Figure 5.
Non-zinc finger domain of Smuc as repressor domain. The non-zinc finger region of Smuc was expressed as a fusion protein with the GAL4 DBD and targeted heterologously to the tk promoter. An expression vector expressing only the GAL4 DBD was used as control (column 1). The reporter plasmid contained three GAL4 binding sites upstream of the luciferase gene. (A) Schematic illustrations of the GAL4 fusion constructs. Hatched and white boxes indicate the conserved N-terminal amino acids and the rest of the non-zinc finger domain, respectively. (B) The whole non-zinc finger domain (amino acids 1–148) repressed the transcriptional activity to 17% of the control level (column 2). Further deletion of the conserved N-terminal amino acids partially abolished the repression (column 3). However, the highly conserved 21 amino acids were insufficient to confer repression (column 4). Experiments were repeated four times in triplicate wells. One of the representative luciferase activities was statistically analyzed and is shown.
Transcriptional regulation by Smuc
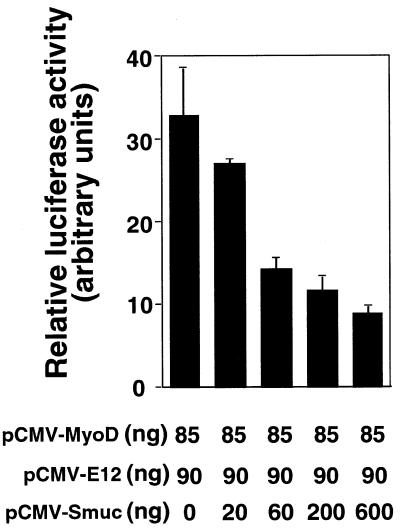
Smuc was shown to bind to E-box DNA sequences, thereby competing with MyoD–E12. Because MyoD is a powerful transcriptional activator by binding to E-box motifs, this competition might lead to repression of the transcriptional activity of MyoD. This possibility was examined in C2C12 cells by the luciferase assay using pE7-luc as a reporter plasmid. Transfection of pCMV-MyoD and pCMV-E12 induced expression of luciferase, and co-transfection of the Smuc expression vector repressed this transactivation by MyoD–E12 in a dose-dependent manner (Fig. 6). In this assay expression of only Smuc did not augment the basal luciferase activity (data not shown). This result suggested a repressive effect of Smuc on E-box-dependent transactivation of downstream genes by myogenic bHLHs.
Figure 6.
Function of Smuc as a bHLH regulator. Inhibition of the transcriptional activity of MyoD–E12 by Smuc was determined by luciferase reporter assays. C2C12 myoblast cells were transfected with expression vectors for MyoD and E12 (pCMV-MyoD, 85 ng/well; pCMV-E12, 90 ng/well). The reporter plasmid contains the luciferase gene under the control of 7× E-box sequences. Co-transfection of a Smuc expression vector suppressed the luciferase activity dose-dependently. Experiments were performed independently four times in triplicate wells. One of four independent results is shown.
Smuc suppresses the expression of Troponin T in myoblasts
In myogenesis, bHLH transcription factors function as cell fate determinants and differentiation inducers by E-box-dependent activation of muscle-specific downstream genes. Therefore, repression of the transcriptional activity of MyoD by Smuc might interfere with the differentiation of muscle cells. To evaluate this, we overexpressed Smuc in C2C12 myoblasts and its inhibitory effect on differentiation to muscles was examined. pNLS-LacZ was used to detect the cells that incorporated exogenous DNA. After 4 days culture in differentiation medium, differentiation of C2C12 cells into myocytes was determined by counting cells positive for Troponin T expression among the LacZ-positive cells. Only 7% of Smuc-expressing C2C12 cells differentiated to muscles, while 27% of mock-transfected cells differentiated (Fig. 7A and B). An inactivator of bHLH transcription factors, Id2, tested as a positive control, inhibited the differentiation of myoblasts more potently (<1%) (Fig. 7B). These results suggest that Smuc represses the differentiation of C2C12 myoblast cells in vitro and that Smuc could regulate myogenic bHLHs in muscle cell differentiation.
Figure 7.
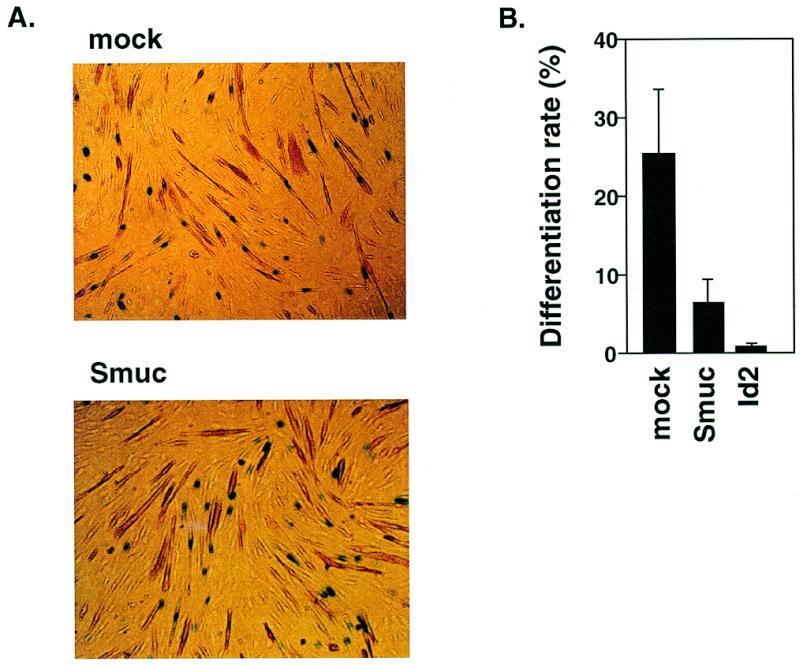
Inhibition of myoblast differentiation by Smuc. (A) Smuc suppressed the expression of a muscle differentiation marker, Troponin T, in C2C12 myoblast cells in differentiation. C2C12 cells were transiently transfected with pCMV, p-CMV-Id2 or pCMV-Smuc, along with pNLS-LacZ, and then differentiated to muscles. Overexpression of Id2, a representative bHLH inhibitor, was considered as a positive control for negative regulation of the differentiation process. Nuclear staining of LacZ was used as a marker of incorporation of exogenous DNA. Percentages of Troponin T-positive cells (brown) among cells with LacZ nuclear staining (blue) [LacZ(+)] were measured in each well within several different microscopic fields. In total, 500 LacZ(+) cells were evaluated in each well. The percentages were defined as differentiation rates (percent Troponin T(+) + LacZ(+)/LacZ(+) = differentiation rate). (B) Differentiation rate of C2C12 cells was decreased with transfection of pCMV-Smuc. Bars indicate the percentages of differentiated Troponin T-positive cells among LacZ-positive cells. Experiments were performed three times in duplicate wells. The averages of these three independent results are indicated with SEM error bars.
DISCUSSION
In this study, we cloned a novel Snail family protein, Smuc, from adult mouse skeletal muscles and analyzed its function. This protein belongs to the Snail family, a subfamily of the zinc finger transcription factors, the members of which play important roles in various differentiation processes during development of many species, including Drosophila and higher vertebrates (1,2,6). Smuc could bind DNA at E-box sequences with high specificity in EMSA analyses. Smuc competed with MyoD–E12 heterodimer in binding to the same E-box sequence. Moreover, the non-zinc finger domain of Smuc acted as a repressor when heterologously targeted to the tk promoter. Consistent with this, Smuc suppressed transactivation by MyoD–E12 heterodimers in luciferase assays in a dose-dependent manner. Moreover, overexpression of Smuc in C2C12 myoblasts inhibited their terminal differentiation into myocytes. These results suggest that Smuc is a regulator of E-box-dependent differentiation processes.
The deduced amino acid sequence of Smuc indicates five C2H2 type zinc finger domains in its C-terminal half (Fig. 1). This region is highly homologous to members of the Snail family. Because mouse Sna contains only four zinc finger domains, and Slug contains five, Smuc is more closely related to Slug. Furthermore, more than half of the about 20 N-terminal amino acid residues are well conserved among mouse Sna, Slug and Smuc (Fig. 1). This conservation is also found in Snail-related proteins of other species such as chick and zebrafish (2), suggesting some important role of the region (16).
Smuc is localized in the nucleus and binds DNA at the E-box sequences present in the promoter region of MCK (Fig. 3A), similarly to other Snail family members. Detailed analyses by EMSA with oligonucleotides containing different E-box sequences indicate specificity of Smuc. Smuc bound to only two sequences, CACCTG and CAGGTG, but not to those with other E-box motifs (Fig. 3B and C). Because Drosophila Snail and Escargot bind to the E-box sequence CAGGTG with high affinity (3,4,9), and mouse Sna to the CACCTG E-box site (14), it is conceivable that these two E-box motifs are the consensus core target sequences for Snail family members. On the other hand, it is well known that E-box sequences are also the consensus binding sites of bHLHs such as MyoD and E2A (5,35). This raised the possibility that Smuc competes for binding to E-box sequences with bHLHs. In fact, Smuc competed for the binding site with MyoD in EMSA analyses (Fig. 4). Moreover, the non-zinc finger domain of Smuc possesses a repressor activity (Fig. 5). In luciferase assays, E-box-mediated transactivation induced by the MyoD–E12 complex was suppressed by Smuc in a dose-dependent manner (Fig. 6). Smuc inhibited terminal differentiation of C2C12 myoblasts, a process which is activated by myogenic bHLHs (Fig. 7) (35). Taken together, these results indicate that Smuc can inhibit the function of bHLHs by the zinc finger domains competing for their common E-box binding sites. Smuc also mediates inhibition through a repressor domain located in the non-zinc finger region.
Generally, bHLHs bind to a wide range of E-box core sequences and induce transactivation, although there is some preference for the internal dinucleotides. MyoD, for example, binds not only to CACCTG and CAGGTG but also to other E-box motifs (5). Since Smuc binds only to CACCTG and CAGGTG E-box sequences, competition between bHLHs and Smuc for DNA binding is limited to a subset of E-box sequences. Therefore, Smuc is not a general inhibitor of bHLHs and regulates a subset of target genes of bHLHs, the promoter of which has an E-box sequence CACCTG or CAGGTG. Examples of bHLH inhibitors specific to a subset of E-box motifs have been reported. ZEB is a non-Snail homeodomain/zinc finger transcriptional regulator with zinc finger domains in both its C- and N-terminal regions (36,37). δ-EF1 is another zinc finger protein with a homeodomain between the zinc finger motifs (38). Both of them have been shown to bind specifically to the core sequences CAGGTG and CACCTG, and suppress the activities of bHLHs. Therefore, in terms of specificity for target E-box motifs, Smuc is similar to ZEB and δ-EF1.
In addition to these inhibitory factors which have specificity for their targets, there exist different inhibitory molecules of bHLHs in terms of their structures and functions. Id, an inhibitor of DNA binding/differentiation, is an example of such molecules (39). It possesses HLH domains but lacks the basic region that is important for DNA binding. Heterodimerization of Id with bHLH such as E12 results in the formation of an inactive heterodimer and causes the sequestration of bHLHs. Thus, Id negatively regulates activities of bHLHs at the protein level without any specificity for downstream genes. Orchestration of bHLHs and these regulatory molecules would be important for the proper execution of developmental programs and for the functional regulation of organs and tissues (40).
In contrast to Gfi1, the N-terminal conserved amino acids of Smuc are important but not sufficient for repressor activity (Fig. 5B). However, the whole non-zinc finger domain of Smuc contains repressor activity. This raises the possibility that transcriptional repression by Smuc might be mediated by a dual mechanism: competition with bHLH transcription factors for the E-box motif by the zinc finger domains and the repressor activity conferred by the non-zinc finger region. It should be determined if the non-zinc finger repressor domain of Smuc might interact with the recently identified co-repressors, CtBPs, which are important in transcriptional repression by Drosophila Snail (41,42).
Although we have demonstrated an in vitro function of Smuc, its physiological function in vivo is still unclear. In the adult, Smuc mRNA is highly expressed in skeletal muscle and thymus, as demonstrated by northern blot analysis. The heart, lung and spleen also express the gene at a lower level. Collectively, skeletal muscle and the cardiovascular and immune systems are sites of Smuc expression. Development and the functions of all of these tissues are well known to be regulated by bHLHs. In adult skeletal muscle, MyoD and Myogenin are reported to regulate the contents of contractile proteins in fast and slow skeletal muscles (43). Moreover, lymphocyte development is dependent on bHLHs such as E2A and HEB (44,45). Smuc, therefore, may participate in the functional regulation of bHLHs in these organs. During embryogenesis, on the other hand, Smuc is detected in E7 mouse embryos by northern blot analysis and shows increased expression in embryos at later stages. This embryonic expression implies that Smuc is involved in pattern formation and cell differentiation processes. Further studies such as detailed in situ expression analysis in both embryos and adult tissues and gene targeting are required to elucidate the biological functions of Smuc.
Acknowledgments
ACKNOWLEDGEMENTS
We thank H. Horiuchi, H. Arai and K. Ishii for critically reading the manuscript, Dr T. Kanno for pEF-BOS-GAL4 and ptk-3XGAL-Luc, Dr S. Yoshida for C2C12 cells, and R. Kageyama for the pE7-Luc construct. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (nos. 08407026, 08670788, 09044293, 09281104 and 09281103) and a Research Grant for Cardiovascular Diseases (A8-1) from the Ministry of Health and Welfare of Japan.
DDBJ/EMBL/GenBank accession no. AF133714
REFERENCES
- 1.Boulay J.L., Dennefeld,C. and Alberga,A. (1987) Nature, 330, 395–398. [DOI] [PubMed] [Google Scholar]
- 2.Sefton M., Sanchez,S. and Nieto,M.A. (1998) Development, 125, 3111–3121. [DOI] [PubMed] [Google Scholar]
- 3.Mauhin V., Lutz,Y., Dennefeld,C and Alberga,A. (1993) Nucleic Acids Res., 21, 3951–3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fuse N., Hirose,S. and Hayashi,S. (1996) Development, 122, 1059–1067. [DOI] [PubMed] [Google Scholar]
- 5.Braun T. and Arnold,H.H. (1991) Nucleic Acids Res., 19, 5645–5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kosman D., Ip,Y.T., Levine,M. and Arora,K. (1991) Science, 254, 118–122. [DOI] [PubMed] [Google Scholar]
- 7.Hayashi S., Hirose,S., Metcalfe,T. and Shirras,A.D. (1993) Development, 118, 105–115. [DOI] [PubMed] [Google Scholar]
- 8.Samakovlis C., Manning,G., Steneberg,P., Hacohen,N., Cantera,R. and Krasnow,M.A. (1996) Development, 122, 3531–3536. [DOI] [PubMed] [Google Scholar]
- 9.Fuse N., Hirose,S. and Hayashi,S. (1994) Genes Dev., 8, 2270–2281. [DOI] [PubMed] [Google Scholar]
- 10.Smith D.E., Franco del Amo,F. and Gridley,T. (1992) Development, 116, 1033–1039. [DOI] [PubMed] [Google Scholar]
- 11.Nieto M.A., Bennett,M.F., Sargent,M.G. and Wilkinson,D.G. (1992) Development, 116, 227–237. [DOI] [PubMed] [Google Scholar]
- 12.Nieto M.A., Sargent,M.G., Wilkinson,D.G. and Cooke,J. (1994) Science, 264, 835–839. [DOI] [PubMed] [Google Scholar]
- 13.Isaac A., Sargent,M.G. and Cooke,J. (1997) Science, 275, 1301–1304. [DOI] [PubMed] [Google Scholar]
- 14.Nakayama H., Scott,I.C. and Cross,J.C. (1998) Dev. Biol., 199, 150–163. [DOI] [PubMed] [Google Scholar]
- 15.Savagner P., Yamada,K.M. and Thiery,J.P. (1997) J. Cell Biol., 137, 1403–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grimes H.L., Chan,T.O., Zweidler-McKay,P.A., Tong,B. and Tsichlis,P.N. (1996) Mol. Cell. Biol., 16, 6263–6272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chomczynski P. and Sacchi,N. (1987) Anal. Biochem., 162, 156–159. [DOI] [PubMed] [Google Scholar]
- 18.Schutte B., Ranade,K., Pruessner,J. and Dracopoli,N. (1996) Biotechniques, 22, 40–44. [DOI] [PubMed] [Google Scholar]
- 19.Kakizuka A., Yu,R., Evans,R. and Umesono,K. (1992) In Essential Developmental Biology, A Practical Approach. IRL Press, Oxford, UK, pp. 223–231.
- 20.Benton W.D. and Davis,R.W. (1977) Science, 196, 180–182. [DOI] [PubMed] [Google Scholar]
- 21.Alwine J.C., Kemp,D.J. and Stark,G.R. (1977) Proc. Natl Acad. Sci. USA, 74, 5350–5354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanno T., Kanno,Y., Chen,L.F., Ogawa,E., Kim,W.Y. and Ito,Y. (1998) Mol. Cell. Biol., 18, 2444–2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kunne A.G., Meierhans,D. and Allemann,R.K. (1996) FEBS Lett., 391, 79–83. [DOI] [PubMed] [Google Scholar]
- 24.Apone S. and Hauschka,S.D. (1995) J. Biol. Chem., 270, 21420–21427. [DOI] [PubMed] [Google Scholar]
- 25.Akimenko M.A., Ekker,M., Doyen,N., Biben,C. and Rougeon,F. (1989) Nucleic Acids Res., 17, 4745–4755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tverberg L.A. and Russo,A.F. (1993) J. Biol. Chem., 268, 15965–15973. [PubMed] [Google Scholar]
- 27.Lemercier C., To,R.Q., Carrasco,R.A. and Konieczny,S.F. (1998) EMBO J., 17, 1412–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sasai Y., Kageyama,R., Tagawa,Y., Shigemoto,R. and Nakanishi,S. (1992) Genes Dev., 6, 2620–2634. [DOI] [PubMed] [Google Scholar]
- 29.Peyton M., Stellrecht,C.M., Naya,F.J., Huang,H.P., Samora,P.J. and Tsai,M.J. (1996) Mol. Cell. Biol., 16, 626–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nielsen A.L., Pallisgaard,N., Pedersen,F.S. and Jorgensen,P. (1992) Mol. Cell. Biol., 12, 3449–3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Elnitski L., Miller,W. and Hardison,R. (1997) J. Biol. Chem., 272, 369–378. [DOI] [PubMed] [Google Scholar]
- 32.Klein E.S., Simmons,D.M., Swanson,L.W. and Rosenfeld,M.G. (1993) Genes Dev., 7, 55–71. [DOI] [PubMed] [Google Scholar]
- 33.Yoshida,H., Hayashi,S., Shultz,L.D., Yamamura,K., Nishikawa,S. and Kunisada,T. (1996) Dev. Dyn., 207, 222–232. [DOI] [PubMed] [Google Scholar]
- 34.Kozak M. (1984) Nucleic Acids Res., 12, 857–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson E. (1992) Symp. Soc. Exp. Biol., 46, 331–341. [PubMed] [Google Scholar]
- 36.Postigo A.A. and Dean,D.C. (1997) EMBO J., 16, 3935–3943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Postigo A.A., Sheppard,A.M., Mucenski,M.L. and Dean,D.C. (1997) EMBO J., 16, 3924–3934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sekido R., Murai,K., Funahashi,J., Kamachi,Y., Fujisawa-Sehara,A., Nabeshima,Y. and Kondoh,H. (1994) Mol. Cell. Biol., 14, 5692–5700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sun X.H., Copeland,N.G., Jenkins,N.A. and Baltimore,D. (1991) Mol. Cell. Biol., 11, 5603–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krempler A. and Brenig,B. (1999) Mol. Gen. Genet., 261, 209–215. [DOI] [PubMed] [Google Scholar]
- 41.Nibu Y., Zhang,H. and Levine,M. (1998) Science, 280, 101–104. [DOI] [PubMed] [Google Scholar]
- 42.Turner J. and Crossley,M. (1998) EMBO J., 17, 5129–5140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hughes S.M., Taylor,J.M., Tapscott,S.J., Gurley,C.M., Carter,W.J. and Peterson,C.A. (1993) Development, 118, 1137–1147. [DOI] [PubMed] [Google Scholar]
- 44.Sawada S. and Littman,D.R. (1993) Mol. Cell. Biol., 13, 5620–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J.S., Olson,E.N. and Kingston,R.E. (1992) Mol. Cell. Biol., 12, 1031–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]