Abstract
The cornea is a transparent and avascular tissue located in front of the eye. Its inner surface is lined by a monolayer of corneal endothelial cells (CECs), which maintain the cornea transparency. CECs remain arrested in a non-proliferative state and damage to these cells can compromise their function leading to corneal opacity. The primary culture of donor-derived CECs is a promising cell therapy. It confers the potential to treat multiple patients from a single donor, alleviating the global donor shortage. Nevertheless, this approach has limitations preventing its adoption, particularly culture protocols allow limited expansion of CECs and there is a lack of clear parameters to identify therapy-grade CECs. To address this limitation, a better understanding of the molecular changes arising from the primary culture of CECs is required. Using single-cell RNA sequencing on primary cultured CECs, we identify their variable transcriptomic fingerprint at the single cell level, provide a pseudo-temporal reconstruction of the changes arising from primary culture, and suggest markers to assess the quality of primary CEC cultures. This research depicts a deep transcriptomic understanding of the cellular heterogeneity arising from the primary expansion of CECs and sets the basis for further improvement of culture protocols and therapies.
Subject terms: Cell biology, Molecular biology
Introduction
The cornea is the transparent window transmitting light into the eye. The inner part of this avascular tissue is covered by a monolayer of hexagonal corneal endothelial cells (CECs)1 that maintain corneal transparency and hydration by their pump and barrier function2. Human CECs are arrested in a non-proliferative state and lack regenerative capacity. Consequently, damage to CECs due to surgery, inherited diseases, or acquired conditions results in irreversible corneal oedema, impairing vision3.
Corneal transplantation is the current therapy for treating corneal endothelium dysfunction. Still, only one donor cornea is available for every 70 patients in need, leaving 12.7 million people awaiting treatment worldwide4. A first landmark clinical trial showed that primary cultivated CECs can restore corneal transparency, breaking the one-donor–one-recipient paradigm5,6. Encouraged by the long-term success of this therapy7, clinical trials are ongoing in Japan (UMIN000034334 and UMIN000012534), Mexico (NCT04191629) and Singapore (NCT04319848) to assess the therapeutic potential of cultured CECs.
Nonetheless, the transplantation of cultured CECs has limitations preventing its wider adoption. Primary CEC cultures are only successful when derived from donors younger than 45 years of age, limiting the pool of donor corneas suitable for this technique. Furthermore, cultures become heterogeneous over time, and significant alterations diminishing the cell phenotype and functionality are observed after the second passage8. Notably, there is a lack of clear parameters to identify therapy-grade cells9. Recently, cell morphology10 and a set of markers: CD44, CD105, CD24, and CD133, also referred to as the E-ratio, have been used as exclusion criteria for therapy-grade CECs11. If we were able to identify additional or other cell-specific markers we could selectively assess and enrich for therapy-grade CECs.
To deconstruct the heterogeneity and gain knowledge on the alterations arising from the primary culture of CECs, we used single-cell RNA sequencing (scRNAseq) to profile 42,220 primary human CECs from six corneas of three donors at five time points over three passages in culture. Our analysis revealed that the culture diversified over time into heterogeneous subpopulations including cells less desirable for therapy that were entering a senescent or fibrotic state. We identified markers that can be used in combination to assess for therapy-grade cells and enrich for desired cell populations. Pseudo time analysis further uncovered the different trajectories arising during culture. Together, our study sheds light on the various routes followed by CECs in culture, identifies novel markers to increase culture efficiency, and presents a roadmap to improve culture protocols.
Results
scRNAseq reveals different subpopulations of CECs arising from primary culture
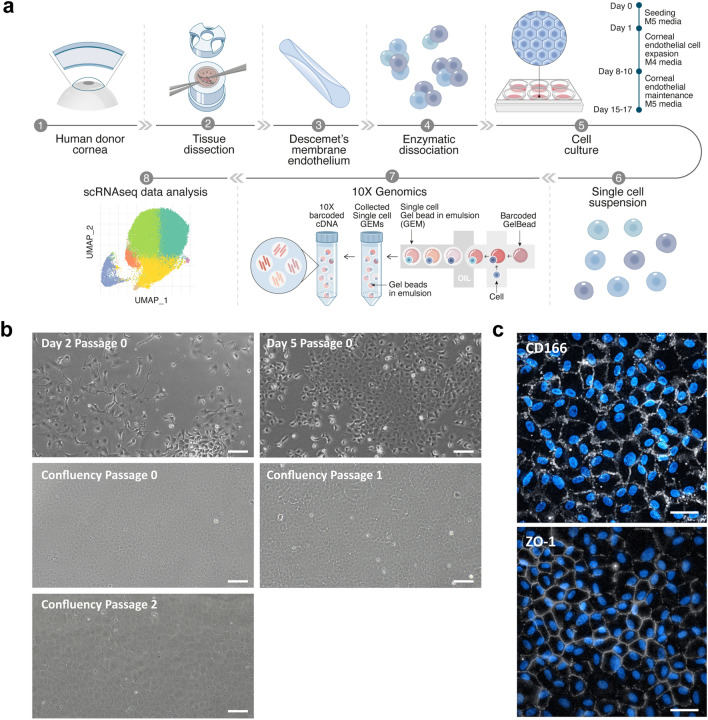
Six paired human corneas, from two male and one female donor (Table 1), were used for isolation, primary culture, and scRNAseq of CECs (Fig. 1A). Cells from five different time points in culture were loaded for sequencing as specified in Table 2. Namely, cells at days 2 and 5 of culture after their isolation in M4 proliferation media at passage 0, and cells at passages 0, 1, and 2, at confluency after 7 days of culture in M5 stabilization media. The cultures showed characteristic CEC morphology over time (Figs. 1B, S1) and expressed the desired CEC proteins such as CD166 and zonula occludens-1 (ZO-1) (Fig. 1c).
Table 1.
Donor cornea information.
| Donor | Sex | Age (years) | Preservation time (days) | COD | ECD OS/OD (cells/mm2) |
|---|---|---|---|---|---|
| Donor 1 | Male | 34 | 12 | Trauma | 3195/2917 |
| Donor 2 | Male | 29 | 11 | GI bleed | 2832/2878 |
| Donor 3 | Female | 27 | 14 | MVA | 2861/3159 |
COD cause of death, GI Gastrointestinal, MVA motor vehicle accident, ECD endothelial cell density, OS oculus sinister, OD oculus dexter.
Figure 1.
Human CECs were successfully isolated and cultured for scRNAseq. (a) Schematic representation of the experimental overview. This illustration was made by Daniela Velasco, scientific illustrator. (b) Phase contrast images of CECs over the different time points selected for scRNAseq confirm the desired hexagonal morphology of the cells. Scale bars represent 100 μm. (c) Immunofluorescence assessment of CEC markers CD166 and ZO-1 confirm the phenotype of the primary cultured cells at passage 2. Scale bars represent 50 μm.
Table 2.
A 10× Genomics sample loading and library information.
| Library | Donor and time point (cell count ratio) |
|---|---|
| G1 | Donor 3 day 5 proliferation + donor 2 confluency passage 0 (1:3) |
| G2 | Donor 3 confluency passage 0 + donor 2 day 5 proliferation (3:1) |
| G3 | Donor 3 confluency passage 1 + donor 1 day 2 proliferation (3:1) |
| G4 | Donor 3 confluency passage 2 + donor 2 day 2 proliferation (3:1) |
| G5 | Donor 1 confluency passage 0 |
| G6 | Donor 1 confluency passage 1 |
| G7 | Donor 1 confluency passage 2 |
| G8 | Donor 2 confluency passage 1 |
| G9 | Donor 2 confluency passage 2 |
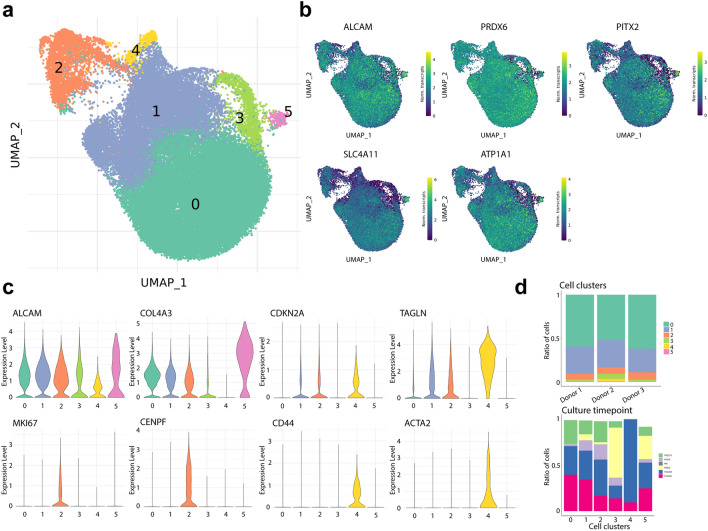
After filtering for cells with a minimum of 1,000 transcripts, the transcriptome profiles of 42,220 cells were embedded in a uniform manifold approximation and projection (UMAP). Cells from all donors were distributed homogeneously across the UMAP (Fig. S2). Unbiased low-resolution clustering revealed six major cell clusters (Fig. 2A), all of which expressed CEC markers ALCAM (CD166), PRDX6, SLC4A11, PITX2, and ATP1A19,12,13, confirming their endothelial identity (Fig. 2B). The absence of keratocyte markers CD34, KERA, and ALDH1A114,15, and epithelial markers KRT12, KRT14, and PAX616,17 confirmed the absence of contaminating cell types from corneal stroma and epithelium (Fig. S3). Differential gene expression profiling was used for cluster identification.
Figure 2.
scRNAseq analysis reveals distinct clusters of primary cultured CECs. (a) UMAP of the 42,220 sequenced cells reveals six cell clusters. (b) Gene expression UMAP of typical CEC markers namely ALCAM (CD166), PRDX6, PITX2, SLC4A11, and ATP1A1 confirms the endothelial identity of the sequenced cells. (c) Violin plots show gene expression for markers of endothelium (ALCAM, COL4A3), senescence (CDKN2A, TAGLN), proliferation (MKI67, CENPF), and fibrosis (CD44, ACTA2). (d) Bar charts show the distribution of cells of each donor per cluster (top) and the time point composition of each cluster (bottom).
Clusters 0 and 5 presented a statistically significant differential gene expression of typical functional CEC markers namely, SLC4A11 and phenotypical CEC markers, namely COL4A312, CDH218, and ALCAM, compared to the other clusters. Based on this, we identified clusters 0 and 5 as high-quality and therapy-grade CECs (Figs. 2C and S4). Cluster 1 was identified as CECs transitioning towards a senescent phenotype due to the high differential expression of senescence markers such as MT2A19, CDKN2A (p16)20, and TAGLN21 (Figs. 2C and S4). Cluster 2 was composed of highly proliferative CECs expressing MKI6722, CENPF23, and PTTG124 (Fig. 2C). Cluster 4 was composed of fibrotic CECs with increased differential expression of ACTA2 [α-smooth muscle actin (SMA)]25, CD4426, and COL6A127 (Figs. 2C and S4). Finally, our analysis revealed that cluster 3 was mainly composed of cells in passage 0 (at both day 2 and day 5) (Fig. 2D) that differentially expressed ribosomal-associated genes (Fig. S5). This finding suggested that cells at an early culture stage clustered together due to the necessary adaptation to in vitro culture conditions and the use of proliferation media, which led us to further explore the cells that had been cultured to confluency in passages 0, 1 and 2.
In line with previous findings at both gene and protein level11,28, our dataset demonstrates that senescence marker CDKN2A, and fibrosis marker ACTA2 increase over the sequenced culture time points (Fig. S6) reaching the highest expression at passage 2. These findings suggest that in vitro expanded CECs are susceptible to senescence and fibrotic transition in agreement with previous research6,11,28. Notably, the expression of CEC marker ALCAM, proliferation markers MKI67 and CENPF, and CD44 appear to remain constant across the sequenced time points (Fig. S6). While CEC marker COL4A3 was not detected on day 2 of culture, it remained constant across other time points (Fig. S6), further supporting the hypothesis that CECs at an early culture stage such as day 2 (passage 0) require adaptation to in vitro culture conditions.
scRNAseq reveals seven distinct subpopulations of primary cultured CECs at therapeutically relevant time points
To identify the meaningful differences at therapeutically relevant time points and remove the clustering bias introduced by the adaptation to primary culture conditions after cell isolation, we separately analyzed the CECs at confluency in passages 0, 1, and 2. These time points are the most therapeutically relevant, as CECs are most suitable for therapy after 7 days in M5 stabilization media up to passage 28,29.
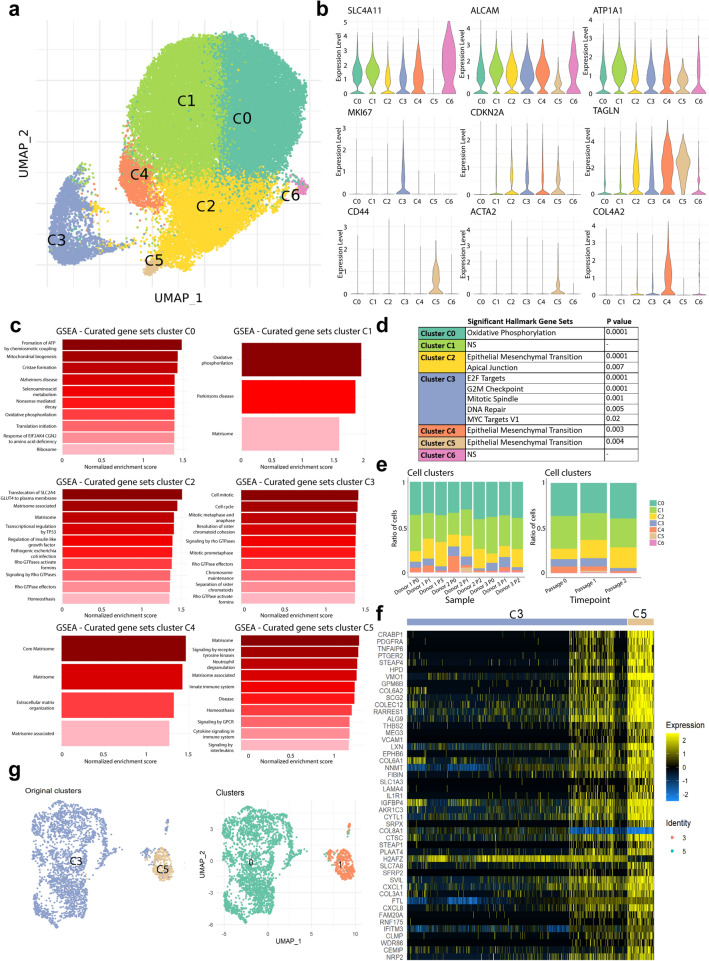
After removing sub-confluent cells from day 2 and day 5 (in passage 0) of culture, the transcriptome profiles of 37,158 CECs at confluency in passages 0, 1, and 2 were embedded in a UMAP. Unbiased low-resolution clustering revealed seven cell clusters (Fig. 3A) with distinct transcriptomic signatures. Cells from all donors were distributed homogeneously across the UMAP (Fig. S8). Differential gene expression analysis was used for identification of each cell cluster (C0–C6).
Figure 3.
scRNAseq analysis reveals seven distinct CEC clusters at therapeutically relevant time points. (a) UMAP of the 37,158 cells at confluency time points reveals seven cell clusters. (b) Violin plots show gene expression for markers of endothelium (SLC4A11, ALCAM, ATP1A1), proliferation (MKI67), senescence (CDKN2A, TAGLN), fibrosis (CD44, ACTA2), and extracellular matrix production (COL4A2). (c) Gene set enrichment analysis (GSEA) reveal differentially expressed gene sets across cell clusters (p < 0.05). (d) Significant differentially expressed hallmark gene sets across clusters (p < 0.05) show an endothelial to mesenchymal transition in lower quality CEC clusters C2, C4 and C5; and proliferation hallmarks in cluster C3. (e) Bar charts show the composition of cell clusters across the different sequenced samples (left), and across the different time points (right). (f) Heatmap of the top 50 differentially expressed genes across clusters C3 and C5 (p < 0.01) shows a subpopulation of CECs within cluster C3 that highly resembles the cells comprising cluster C5. Heatmap was performed in R (version 4.2.0). (g) UMAP of the reclustering analysis of cell clusters C3 and C5 with original clusters (left) and newly detected clusters (right).
Increased differential expression of typical CEC markers, such as COL4A612, SLC4A11, ATP1A1, COL4A3, and CDH2 suggested that clusters C0 and C1 were composed of therapy-grade CECs (Figs. 3B and S8). Further analysis by gene set enrichment analysis (GSEA) revealed that highly metabolically active cells comprised these clusters (Fig. 3C), with a distinct hallmark for oxidative phosphorylation in cluster C0 (Fig. 3D). These markers are also found in native functioning human CECs, suggesting these cells were therapy-grade CECs. Cluster C3 was composed of proliferating CECs with differential high expression of MKI67 (KI-67), CENPF, and PTTG1 (Figs. 3B and S8). GSEA further confirmed enriched gene sets and significant hallmarks related to cell proliferation (Fig. 3C,D).
Clusters C2 and C4 were composed of CECs with increased differential expression of senescence related genes. Namely, CDKN2A (p16), TAGLN, and MT2A (Figs. 3B and S8) in cluster C2 and CDKN1A (p21)30, CDKN2A (p16), and TAGLN in cluster C4 (Figs. 3B and Fig. 8), suggesting these cells were transitioning towards an undesirable senescent and fibrotic phenotype. Cluster C4 had a high extracellular matrix production suggested by the differential high expression of COL4A1, COL4A2, COL5A1, and FBLN5 in cells that maintained the expression of endothelial markers such as SLC4A11, and COL4A3 (Figs. 3B and S8). In contrast, cells in cluster C2 presented a low expression of CEC markers such as ALCAM, SLC4A11, CDH2, and ATP1A1 (Figs. 3B and S8). GSEA revealed that both clusters C2 and C4 had a significant hallmark for epithelial to mesenchymal transition (Fig. 3D), confirming these cells were transitioning towards senescence and fibrosis. GSEA revealed that cluster C2 was enriched for genes related to alterations in the matrisome production, upregulation of p53 pathway, and upregulation on Rho GTPase pathway, which are known to regulate cellular senescence31,32 (Fig. 3C), and cluster C4 expression was enriched for genes related to matrisome, extracellular matrix organization and matrisome associated genes suggesting a remodeling of the extracellular matrix (Fig. 3C). Cluster C5 was composed of fibrotic cells differentially expressing the fibrosis-associated markers COL6A1, COL6A3, CD44, and ACTA2 (Fig. 3B). The cells in cluster C5 had diminished expression of ALCAM (CD166) and lacked SLC4A11 expression, two CEC markers. GSEA further suggested enriched expression of genes related to the matrisome and matrisome associated processes and increased signaling by G-coupled protein receptors and receptor tyrosine kinase (Fig. 3C), with a significant hallmark for epithelial to mesenchymal transition for cells in cluster C5 (Fig. 3D). Finally, Cluster C6 was composed of CECs with differential high expression of typical endothelial markers such as SLC4A11, COL4A3, and CDH2 (Fig. 3B) suggesting they were therapy-grade CECs. These cells differentially expressed higher GOLGA8A and GOLGA8B, suggesting a possible increase in secretory pathways. GSEA did not reveal a significant upregulation of gene sets nor significant hallmarks compared to other clusters.
Longer culture times decrease proliferation and increase transitioning to senescence
Our scRNAseq data analysis revealed that at longer culture time points, namely confluency in passage 2, there was an increase in the number of cells transitioning to a senescent/fibrotic phenotype (cluster C2 and C4) and a decrease in the number of proliferative cells (cluster C3) compared to earlier time points (Fig. 3E). Interestingly, the number of cells in cluster C4 decreased over culture time (Fig. 3E) while cells in cluster C2 increased over time. This could be because the cells in C4 are early senescent cells and transition to later senescent cells in cluster C2. Regarding fibrotic CECs in cluster C5, these cells were detected as early as passage 1, but were also present in passage 2 at lower prevalence compared to passage 1. Finally, the prevalence ratio of therapy grade CECs (cluster C0 and C1) was maintained over time points (Fig. 3E) showing the presence of therapy-grade CECs over all culture passages.
Consistent with prior research at both the gene and protein levels, our dataset demonstrates that the expression of CEC functional markers SLC4A11, and ATP1A1 and phenotype marker COL4A2 decreases over extended culture durations (Fig. S9)6,11,28. Furthermore, the senescence marker CDKN2A, and the fibrosis marker ACTA2 increase over the sequenced culture time points (Fig. S9), with the highest expression observed at passage 2, in agreement with previous research6,11,20,28. Interestingly, expression of TAGLN, senescence marker, is elevated at passage 1 compared to the other passages. Additionally, the expression levels of CD44, a lower-quality marker of CECs remain relatively constant across passages, suggesting its involvement in the early cell transition to a fibrotic state.
scRNAseq subclustering analysis revealed two distinct transcriptomic profiles of proliferating cells
The CEC marker ALCAM, and the fibrotic markers CD44 and ACTA2 were heterogeneously expressed across different cells comprising proliferative cluster C3 (Fig. S10), suggesting it contained a subcluster of highly proliferative fibrotic CECs. Correlation and differential expression analysis revealed similarities between a subcluster of C3 with the fibrotic cells present in cluster C5 (Fig. 3F). This was confirmed by a reclustering analysis of only clusters C3 and C5, which showed that a small subpopulation of cells originating from cluster C3 clustered together with the cells originating from cluster C5 (Fig. 3G). This finding confirms the presence of a subpopulation of highly proliferative fibrotic CECs within cluster C3.
Primary cultured CECs resemble native human CECs
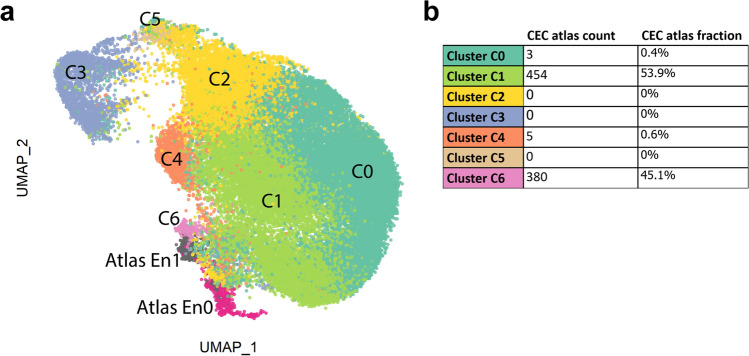
To assess how comparable primary cultured CECs are to native human CECs, we integrated the transcriptome of cells cultured to confluency in passages 0, 1, and 2 with a previously published cornea scRNAseq atlas33. To do so, the cluster information from both the CECs in the cornea cell atlas and this study were overlaid in 2D space (Fig. 4A). The clustering analysis revealed that the CEC clusters originating from the native human corneal endothelium (Atlas En0 and Atlas En1) clustered adjacent to primary CEC clusters C6, C1 and C0, suggesting these cell clusters share comparable transcriptomic profiles (Fig. 4A).
Figure 4.
Therapy-grade primary cultured CECs resemble human native CECs. (a) Integrated data UMAP of primary cultured CECs at confluency and native human CECs from the previously published cornea cell atlas33. The integration analysis shows that native CECs (Atlas 0 and Atlas 1) cluster adjacent to high quality primary cultured CEC clusters (clusters C0, C1, and C6) suggesting clusters C0, C1 and C6 can be considered therapy-grade primary cultured CECs. (b) The cell type of the atlas CECs was predicted using the CEC confluency dataset using the cell label transfer functionality from Seurat. 99.4% of the atlas cells were attributed to clusters C0, C1 and C6, further suggesting the therapeutic standard of these clusters.
To further understand the similarity of native human CEC to primary cultured cells, we performed a cluster prediction analysis of the atlas CECs using the clustering analysis of cells at confluency. The prediction analysis revealed that 99.4% of the atlas CECs were associated to clusters C1 (53.9%), C6 (45.1%), and C0 (0.4%) (Fig. 4B), suggesting these clusters are similar to native human CECs and could be used for therapeutic purposes. Furthermore, our prediction analysis found that no native CECs were associated with the senescent and fibrotic clusters C2 and C5, or the proliferative cluster C3 (Fig. 4B).
Pseudo time reconstruction and evaluation reveal the dynamics of CEC profiles arising from primary expansion
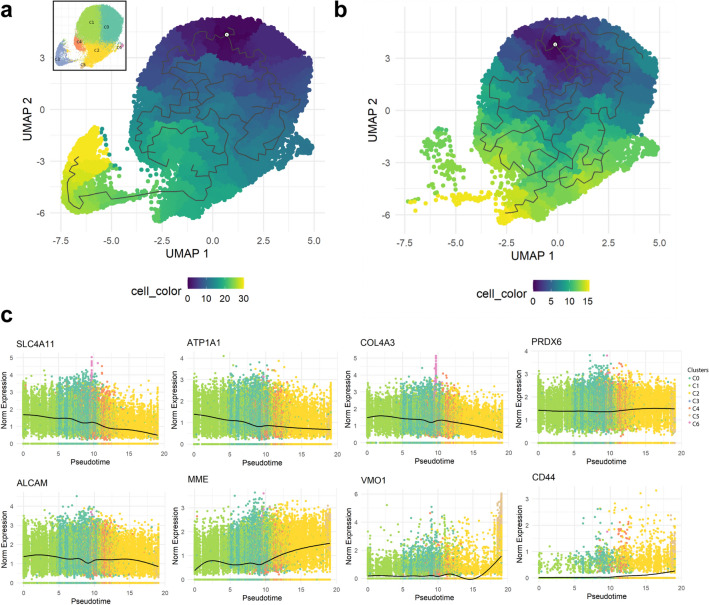
To assess how the cells transition between clusters over time, we performed a pseudo time reconstruction of the cells at confluency in passages 0, 1, and 2. Our first analysis revealed that the CECs originating from clusters C0, C1, and C6, transitioned into the senescent cells in cluster C2, and then became the fibrotic cells in cluster C5 (Fig. 5A). Interestingly, the last cluster the pseudo time trajectory identified were the proliferative cells in cluster C3. We hypothesize this is due to the presence of a side population of fibrotic proliferative cells within cluster C3 (Fig. 3F,G), which might interfere with the pseudo time trajectory analysis. To reduce such bias and identify gene trends over the pseudo time trajectory, we performed a pseudo time analysis of the confluent cells excluding cluster C3 (Fig. 5B). In line with the first analysis, the pseudo time trajectory revealed that the CECs from clusters C0, C1, and C6 transitioned into the transitioning senescent cells in cluster C2, and then the fibrotic cells in cluster C5 (Fig. 5B) showing that primary cultured CECs transition towards senescence and fibrosis over culture time.
Figure 5.
Pseudo temporal trajectory reconstruction reveals the dynamics of primary cultured CECs. (a) Monocle 3 pseudo temporal trajectory reconstruction on UMAP reduction of the scRNAseq confluency time points reveals the CEC cluster dynamics over primary culture. UMAP reduction is colored by pseudo time bins with dark blue being the earliest and yellow corresponding to late. (b) Monocle 3 pseudo temporal trajectory reconstruction on UMAP reduction of the scRNAseq confluency time points excluding proliferative cluster C3 reduces bias and reveals the temporal dynamics of the CEC clusters at confluence level. UMAP reduction is colored by pseudo time bins with dark blue being the earliest and yellow corresponding to late. (c) Pseudo time reconstruction reveals differential gene expression trends over CEC clusters. Our analysis revealed a reduction over time of CEC markers SLC4A11, ATP1A1, COL4A3, and ALCAM, while PRDX6 expression was constant. Conversely, CD44, MME, and VMO1 expression significantly increased over time.
The pseudo time reconstruction revealed that the expression of functional markers such as SLC4A11 and ATP1A1 was reduced over time, showing that senescent (cluster C2 and C4) and fibrotic (cluster C5) cells had a highly reduced expression of crucial functional markers (Fig. 5C). Moreover, pseudo time reconstruction also revealed reduced expression of COL4A3 and ALCAM over time (Fig. 5C). Interestingly, the expression of PRDX6, a known marker of CECs, remained constant and did not decrease over time in the senescent (C2 and C4) and fibrotic (C5) clusters (Fig. 5C). Besides crucial CEC markers, our analysis also revealed a significant increase of CD44 expression over time in clusters C2 and C5 CECs. Furthermore, the expression of MME (CD10) and VMO1 in the pseudo time analysis was increased in the senescent and fibrotic clusters C2 and C5 (Fig. 5C). These genes were also differentially expressed in lower quality clusters C2 and C5, respectively, and can be candidates to assess quality of primary cultured CECs.
scRNAseq transcriptomic profiles for quality assessment of primary cultured CEC correlate with protein level expression
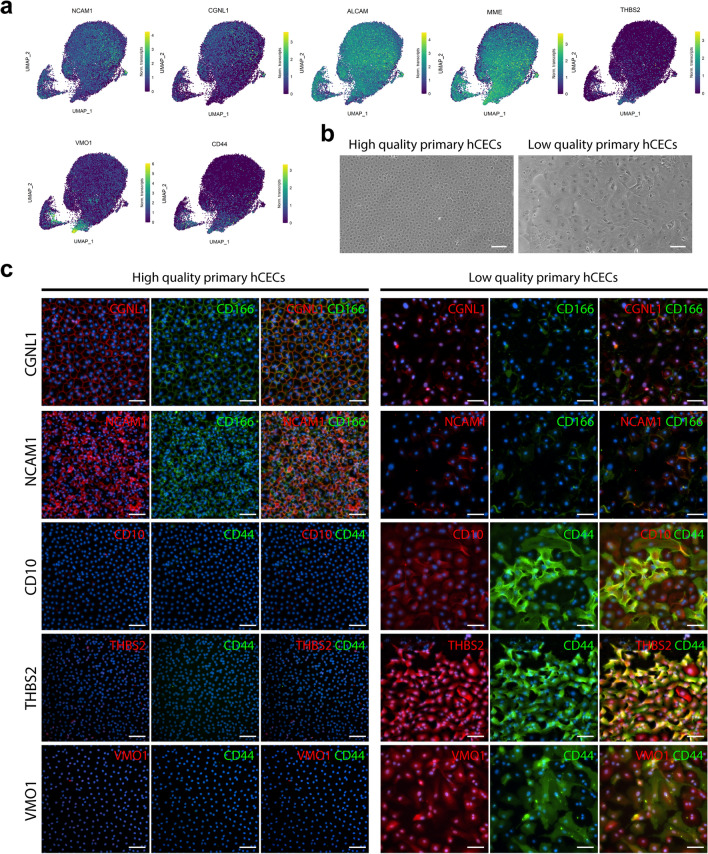
The differential gene expression across CEC clusters and the pseudo time reconstruction both showed that the quality of the primary cultured CEC could be evaluated with a specific set of markers to differentiate therapy-grade CEC (clusters C0, C1, and C6), from lower quality CECs transitioning towards a senescent or fibrotic phenotype (clusters C2 and C5). The expression of ALCAM (CD166), CGNL1 (cingulin-like protein 1), and NCAM1 (CD56) were higher in the clusters comprising therapy-grade CECs (clusters C0 and C1), and lower in the cells in clusters C2 and C5 (Fig. 6A). Additionally, the expression of CD44, MME (CD10), VMO1, and THBS2 were higher in the fibrotic cells in cluster C5 and lower in the cells in clusters C0, C1, and C6 (Fig. 6A), representing markers that could be used to exclude low quality CECs.
Figure 6.
scRNAseq analysis suggests specific markers for CEC quality assessment. (a) Gene expression UMAP of differentially expressed markers in clusters of therapy-grade CECs (clusters C0 and C1) and clusters of senescent/fibrotic CECs (cluster C5). NCAM1, CGNL1, and ALCAM were differentially expressed in clusters CO and C1 (p < 0.01). MME and CD44 were differentially expressed in clusters C2 and C5. THBS2 and VMO1 were differentially expressed in cluster C5 (p < 0.01). (b) Phase contrast images of a high quality therapy-grade CEC culture showing the typical hexagonal cell morphology and a low quality culture of primary human CECs showing the characteristic morphological alterations of an endothelial to mesenchymal transition. Scale bars represent 100 μm. (c) Immunofluorescence analysis shows expression of CD166 (green), NCAM1 (red), and CGNL (red) in high quality CEC cultures (N = 2) and absence of expression in lower quality CEC cultures (N = 2). Immunofluorescence analysis shows expression of CD44 (green), MME (CD10) (red), THBS2 (red), and VMO1 (red) in lower quality CEC cultures (N = 2) and the absence of expression in high quality CEC cultures (N = 2). Cell nuclei were stained with Hoechst 33342 (blue). Scale bars represent 100 μm.
To confirm the findings at the transcriptomic level were also present at the protein level, we assessed protein expression on primary CEC cultures with a characteristic hexagonal morphology associated with therapy-grade CECs and on primary CEC cultures composed of cells with spindle shape morphology, a characteristic of cells undergoing an endothelial to mesenchymal transition, referred as low quality primary CECs (Fig. 6B). Immunofluorescence analysis confirmed that CGLN1, NCAM1 (CD56) and ALCAM (CD166) were exclusively expressed by good quality CECs and not expressed in the cultures containing CECs with an altered morphology (Fig. 6C). Furthermore, immunofluorescence analysis also confirmed that CD44, MME (CD10), THBS2, and VMO1 were exclusively expressed by low quality CECs and not expressed in high quality CEC cultures (Fig. 6C).
Discussion
In this study, we present a single-cell roadmap of human CECs in culture, revealing the diverse trajectories of individual cells. Our scRNAseq census of 42,200 primary cultured CECs revealed the presence of 7 clusters in therapeutically relevant time points, including therapy-grade CECs expressing SLC4A11, ALCAM, and COL4A3 (clusters C0, C1 and C6); highly proliferative CECs expressing MKI67 and CENPF (cluster C3); lower quality CECs entering senescence and EMT expressing CDKN1A, CDKN2A, and TAGLN (clusters C2 and C4); as well as fibrotic CECs expressing CD44, and ACTA2 (cluster C5). We assessed to which extent CECs in culture resemble native human CECs and analyzed how these CEC populations diverge over culture time, giving insights into the alterations arising during primary culture. Moreover, our transcriptomic profiling provides an array of combinatorial markers to differentiate therapy-grade CEC from cells undergoing senescence and EMT, thereby paving the way for improving culture protocols and guiding the selection of cells for therapy. The transcriptomic data we obtained will help better our understanding of the mechanisms involved in the alterations occurring during primary culture of CECs leading to a loss of function and phenotype.
Our analysis showed that proliferating sub-confluent CECs sequenced at day 2 and day 5 clustered together (cluster 3), with high differential expression of ribosomal-related genes. We hypothesize that this is most likely due to their necessary adaptation to in vitro culture conditions and the use of proliferation media, which biased the first clustering. These findings led us to separately explore the 37,158 cells at confluency in passages 0, 1, and 2. Our analysis revealed three clusters of therapy-grade CEC (clusters C0, C1, and C6) based on the high differential expression of functional CEC markers SLC4A11 and ATP1A1 and the CEC markers ALCAM, PRDX6, and COL4A3. These cells were the majority in all passages, comprising 70% of all the sequenced cells. GSEA revealed that cells in C0 and C1 were metabolically active with increased expression of genes related to oxidative phosphorylation, in line with the crucial activity and function of CECs28.
The integration analysis of the dataset from this study with native CECs from a previously published cornea cell atlas32 revealed that native CECs clustered close to clusters C0, C1 and C6, suggesting the similarity of these cells. Cluster prediction analysis on native CECs revealed that these cells would group within clusters C1 (53.9%), C6 (45.1%), and C0 (0.4%). The low 0.4% prediction for cluster C0 is interesting because the primary cultured cells still expressed high levels of endothelial markers, namely SLC4A11, ATP1A1, ALCAM, and PRDX6. We hypothesize the low prediction might be due to a slight increase in ribosomal protein expression or a difference in the number of genes/cell which can be a technical sampling variation compared to clusters C1 and C6, which skews the cell clustering. Overall, our data shows that CECs in clusters C0, C1, and C6 are good quality CECs and are a suitable source for therapy. And while our dataset shows that cells in clusters C1 and C6 more closely resemble native human CECs that cells in cluster C0, it does not mean that cells in cluster C0 are unsuitable for therapy, but that they are distinguishable from native corneal endothelium.
Our scRNAseq analysis also revealed the presence of two clusters composed of senescent cells that were transitioning towards a mesenchymal phenotype (clusters C2 and C4). Our pseudo time reconstruction showed these cells were originating from the therapy-grade clusters C0, C1, and C6. This finding shows the transition to senescence of CECs during primary culture. Senescent cells in cluster C2 had reduced expression of key functional markers such as SLC4A11, ATP1A1, and ALCAM. This finding is in line with a recently published report that demonstrated a decrease of SLC4A11 in lower-quality primary CECs28. The total number of senescent cells (clusters C2 and C4) increased over culture time, suggesting the CECs transitioned towards senescence over extended culture times. Cells comprising cluster C4 decreased over culture time, indicating that they either transition into senescent cells in cluster C2 or represent an end-point cluster, where cells tend to die over time. GSEA revealed that cells in cluster C2 had differential gene expression, specifically an increase in genes involved in the p53 and Rho GTPase pathways, suggesting these pathways might play a key role on the senescence and endothelial to mesenchymal transition of primary cultured CECs. P53 is a known senescence regulator31,34, and its inhibition could delay the cellular senescence in primary CECs. A study in 2013 revealed that the inhibition of p53 was associated with improved morphology and higher expression of CEC markers, namely collagen type 8, Na/K ATPase, and N-cadherin, in primary cultured CECs35. Furthermore, our findings suggest that the inhibition of the Rho GTPase pathway can play a key role in delaying cellular senescence, further confirming that the use of Rho-associated protein kinase (ROCK) inhibitors such as Y-27632 might be a pivotal factor in the protocols for primary expansion of therapy-grade CECs. Indeed, previous studies have used Y-27632 for the primary expansion of CECs6,36–38. Based on these findings, we therefore recommend the use of ROCK inhibitors during the primary expansion of CECs.
Our scRNAseq analysis revealed that a cluster of CECs (cluster C5) expressing characteristic fibrotic markers ACTA2 and CD44 originated from the senescent cells in clusters C2, suggesting a transition from senescence to an EMT phenotype. While fibrotic markers were found in later time points, we did observe that cells comprising cluster C5 appeared as early as passage 1, but were then reduced by passage 2. This finding could be due to the sequencing sampling limitation of 10,000 cells per sample, making it highly possible that this small fibrotic cell population was not sequenced from a culture of hundreds of thousands of cells. Our second hypothesis is that after passaging, the fibrotic cells could not successfully adhere, causing a reduction of this cell population and enriching for good quality cells.
Our findings showed that extended culture times decreased the proliferation potential of CECs, shown by a reduction in the number of cells in the proliferation cluster C3 across passages. Moreover, we detected the presence of a subpopulation of undesired proliferative fibrotic cells that could potentially overgrow the culture of CECs over extended culture periods. In our view, these results show that with the current protocols, culturing CECs further than passage 2 is incompatible with their therapeutic use, a recommendation in line with previous studies that suggested passage 2 as the threshold time point to assure the therapeutic suitability of primary cultured CECs8,29,39.
Selecting and assessing the quality of the primary cultured CECs are a crucial aspect to ensure a safe and efficacious therapy. Based on our differential expression analysis and pseudo time reconstruction, we show that therapy-grade CECs should be identified by the expression of CD166 and NCAM1 membrane proteins together with CGNL1, a membrane-associated protein to cellular tight junctions; lack of expression of altered extracellular matrix, namely VMO1 and THBS2; and lack of expression of membrane proteins CD44 and CD10. While the membrane protein markers suggested by our analysis would allow sorting for therapy-grade CECs, we believe is equally important to characterize CEC culture quality based on expression of other fundamental proteins such as aberrant extracellular matrix production. Future studies are required to understand how the expression of these markers correlate to therapeutic success. Similar to our suggestion to analyze markers for therapy-grade CECs (CD166+, NCAM1+, CGNL1+, CD44−, CD10−, VMO1−, and THBS2−), Kinoshita and colleagues proposed the combinatorial marker expression referred as the E-ratio (CD166+, CD44−, CD133−, CD24−, and CD105−) to assess for therapy-grade CECs. We and they both detected increased CD166 expression in therapy-grade CECs and CD44 exclusively expressed in lower quality senescent and transitioning CECs. By contrast, our analysis revealed that CD24 and ENG (CD105) were heterogeneously and minimally expressed across clusters in some CECs, and we did not detect expression of PROM1 (CD133) in any cluster. These differences might be due to the lack of correlation between transcript and protein detection. Future studies analyzing such differences can shed light on the suitability of markers to assess or enrich for therapy-grade CECs.
While primary cultured CECs have been traditionally assessed as bulk entities without accounting for their heterogeneity, our study analyses them at the single-cell level over five culture time points in three different passages. Our study provides significant information to help understand the changes arising from the culture of human CECs, portraying their cellular heterogeneity as well as characterizing their variability over extended culture times. These results provide a pivotal dataset that can help identify and characterize the undesired cell populations arising from primary culture in the attempt to improve current protocols. Our results also show the importance of supplementing media for CEC expansion with ROCK inhibitors to reduce cellular senescence. Furthermore, based on the results reported in this study, we propose a combination of markers to assess the quality of primary cultured CECs. Overall, this transcriptomic cell analysis offers a baseline for future studies with the aim of improving CEC-based therapies.
Materials and methods
Research-grade donor human corneas and ethical statement
This study was performed in compliance with the tenets of the Declaration of Helsinki. All research-grade human donor corneas used for primary culture were obtained from the Lions Eye Institute for Transplant & Research (Tampa, USA), with informed consent from the next of kin. The research involving human-derived corneas was performed in accordance to Maastricht University and Dutch national regulations. All experiment protocols involving human donor corneas were approved by the Maastricht University ethics review committee for non-WMO research (UM-REC). All corneas had an endothelial cell density of at least 2800 cells/mm2, deemed unsuitable for transplantation, and were preserved in Optisol-GS at 4 °C for up to 14 days prior to their use (Table 1). No corneas were procured from prisoners.
Isolation and culture of primary human corneal endothelial cells
Six paired corneas, from two male and one female donor aged 27 to 34 years, were used for isolation and primary culture of endothelial cells. Donors had no history of ocular disease, chronic systemic disease, or pathological infection such as HIV, Hepatitis B and C, HTLV-I/II, syphilis, or SARS-CoV-2.
Prior to isolation, the endothelial–Descemet’s corneal layer was manually stripped as follows: the corneas were vacuum fixed in a punch base (e.janach) endothelial cell side up and trephined with a 10 mm Ø corneal punch at a fixed depth of 100 μm (e.janach). To delimit the endothelial trephined line, corneas were stained with a trypan blue solution (0.4%) for 30 s, and washed with balanced salt solution sterile irrigating solution (BSS; Alcon). The corneal endothelium was then gently lifted using a DMEK cleavage hook (e.janach) and fully stripped using angled McPherson tying forceps.
Human corneal endothelial cells were isolated and cultured as previously reported5. Briefly, the stripped endothelium–Descemet’s layer was incubated with 2 mg/ml collagenase A (Roche) solution in human endothelial serum free media (SFM) (Thermo Fisher Scientific) for 2–5 h at 37 °C followed by a 5 min incubation in TrypLE express (Thermo Fisher Scientific) to generate small clumps of corneal endothelial cells. Cells from each cornea were seeded equally across 2 wells of a 24-well plate coated with fibronectin collagen (FNC) coating mix (Athena Enzyme Systems) in M5 stabilization media [human endothelial SFM (Thermo Fisher Scientific) supplemented with 5% fetal bovine serum (FBS), 100 U/mL penicillin–streptomycin, and 0.25 μg/mL amphotericin B] supplemented with 10 μM Y-27632 (STEMCELL Technologies). Subsequently, corneal endothelial cells were cultured in M4 proliferation medium [1:1 Ham’s F12 (Thermo Fisher Scientific) and M199 (Thermo Fisher Scientific) supplemented with 5% FBS, 20 μg/mL ascorbic acid (Sigma), 1 × ITS (Thermo Fisher Scientific), 10 ng/mL human recombinant bFGF (Sigma), and 10 μM Y-27632 (STEMCELL Technologies)]; media was refreshed every other day. Upon reaching 90% confluency, after approximately 8–10 days of culture, cells were cultured in M5 stabilization media for 7 days. After this, corneal endothelial cells were treated with TripLE express and passaged into wells pre-coated with FNC-coating mix at a seeding density of 10,500 cells/cm2 in M5 stabilization medium. All cell culture was performed in incubators with humidified atmosphere of 37 °C and 5% CO2.
Preparation of single cell suspension and methanol fixation
Cells from five different culture time points were methanol fixed for sequencing. Namely, cells at days 2 and 5 of culture after isolation in M4 proliferation media at passage 0, and cells at confluency after 7 days of culture in M5 stabilization media, at passages 0, 1, and 2.
To generate a single cell suspension, primary cultured corneal endothelial cells were treated with TripLE express for approximately 30 min at 37 °C. Then cells were centrifuged for 5 min at 800 × g and resuspended in 1 mL ice-cold Dulbecco’s phosphate-buffered saline (DPBS). Next, the cells were centrifuged for 5 min at 800 × g and resuspended in ice-cold DPBS at a ratio of 200 μL DPBS/1 × 106 cells, followed by the dropwise addition of ice-cold methanol at a ratio of 800 μL DPBS/1 × 106 cells. The fixed cell suspensions were stored at − 80 °C until sequencing.
Single-cell RNA sequencing
scRNAseq of primary cultured CECs was performed at Single Cell Discoveries (Utrecht, the Netherlands) following standard 10 × Genomics 3′ V3.1 chemistry protocol. Cells were rehydrated and loaded on the 10 × Chromium controller as follows. Approximately 10,000 cells were loaded per each sample specified in Table 2. The resulting sequencing libraries were prepared following a standard 10 × Genomics protocol and sequenced with an Illumina NovaSeq 6000 platform; read length: 150 bp, paired-end.
Bioinformatic analysis of scRNA-seq data
The BCL files resulting from sequencing were transformed to FASTQ files with 10 × Genomics Cell Ranger mkfastq following its mapping with Cell Ranger count. During sequencing, Read 1 was assigned 28 bp, and were used for identification of the Illumina library barcode, cell barcode and unique molecular identifier (UMI). R2 was used to map the human reference genome GRCh38. Filtering of empty barcodes was done in Cell Ranger. The data from all samples were loaded in R (version 4.2.0)40 and processed using the Seurat package (version 4.1.1)41. More specifically, for each library a UMI cutoff was used to filter out low quality cells because of the differences between the libraries (i.e. g1—500, g2—3000, g3—500, g4—1313, g5—4000, g6—4000, g7—4000, g8—4000, g9—4000) (Table 2). Additionally, cells with less than 10% mitochondrial gene content were retained for analysis. The data of all 10 × libraries were merged and processed together. The merged dataset was normalized for sequencing depth per cell and log-transformed using a scaling factor of 10,000. The multiplexed samples were demultiplexed based on their snp profile using Souporcell42. Briefly, the bam file and barcodes of each library were used as input together with the reference genome GRCh38. Besides the default parameters, the number of clusters was set to the number of multiplexed samples per library. The demultiplexing information for each cell was added to the metadata object in Seurat. The patient and library effect was corrected using Harmony43, as implemented in Seurat and used for dimensionality reduction and clustering of all cells. Cells were clustered using graph-based clustering and the original Louvain algorithm was utilized for modularity optimization. The differentially expressed genes per cluster were calculated using the Wilcoxon rank sum test and used to identify cell types. Putative doublets were computationally identified using scDblFinder (version 1.2.0)44 but did not compose a separate cluster and therefore were not removed from the dataset (Fig. S11). Pseudo time analysis was performed using the Monocle-3 package (version 1.0.0)45. Gene set enrichment analysis was performed on lists of differentially regulated genes without prefiltering step. Gene lists were preranked using the signed -log10 (P adj) and subjected to enrichment analysis using fgsea package (version 1.22.0)46 and gage (version 2.46.1)47 with curated and hallmark gene sets from MSigDB Collections (version 7.5.1)48,49. To prune selectively the resulting pathways and GO terms, enrichment was considered when up- or downregulated gene sets were detected using both methods. The CEC dataset was integrated with the previously published cornea atlas33. The library effect was corrected for using harmony, followed by dimensionality reduction. The cluster information from the separate analysis was used to overlay in 2D space. The cell type of the atlas cells was predicted using the CEC confluency dataset using the cell label transfer functionality from Seurat.
Immunofluorescence
Primary cultured CECs at time point confluency passage 2 deriving from all three donors were used for immunofluorescence analysis. CECs were fixed in 4% PFA for 15 min at ambient temperature and the cells were permeabilized with 0.1% (v/v) Triton X-100 in phosphate buffered saline (PBS) for 10 min. After permeabilization, non-specific antibody interactions were blocked with blocking buffer (2% (w/v) BSA solution in PBS) for 1 h at ambient temperature. CECs were incubated overnight at 4 °C with primary antibodies mouse monoclonal anti-CD166 [3A6] (1:200 dilution, BD Biosciences), rabbit polyclonal anti-ZO1 (1:100 dilution, Thermo Fisher Scientific), mouse monoclonal anti-CD44 [Hermes-3] (1:400 dilution, Abcam), rabbit polyclonal anti-VMO1 (1:100 dilution, Prestige Antibodies), rabbit polyclonal anti-THBS2 (1:100 dilution, Abcam), rabbit monoclonal anti-CD10 [EPR22867-118] (1:100 dilution, Abcam), rabbit monoclonal anti-NCAM1 [CAL53] (1:100 dilution, Abcam), and rabbit polyclonal anti-CGNL1 (1:200 dilution, Atlas antibodies) diluted in blocking buffer. After primary antibody incubation, tissues were washed three times in PBS and then incubated with secondary antibodies goat anti-mouse A488 (1:400 dilution; Thermo Fisher Scientific), and donkey anti-rabbit A568 (1:400 dilution; Thermo Fisher Scientific) diluted in blocking buffer for 50 min at ambient temperature in the dark. Cell nuclei were stained with 1 μg/mL Hoechst 33342 (Thermo Fisher Scientific) for 10 min. The CEC samples were then washed three times in PBS and examined on an Eclipse Ti-E inverted microscope (Nikon) equipped with an X-Light V2-TP spinning disk (Crest Optics).
Statistical and quantitative analysis of scRNAseq data
All the statistical analysis and graphs for scRNAseq were performed in R (version 4.2.0) with the packages described in the methods detail section. Briefly, differentially expressed genes were detected using a Wilcoxon Rank-Sum test, statistical significance was defined as p < 0.01. GSEA revealed differentially enriched gene sets from MSigDB Collections, statistical significance was defined as p < 0.05.
Supplementary Information
Acknowledgements
The authors thank Single Cell Discoveries (Utrecht, the Netherlands) for the single cell sequencing services provided. The authors thank the Lions Eye Institute for Transplant & Research (Tampa, FL, USA) for providing research-grade human corneas.
Author contributions
Conceptualization: P.C., N.G., V.L.S.L., M.M.D.; Investigation: P.C., N.G.; Resources: P.C., N.G., V.L.S.L., M.M.D.; Data Curation: N.G.; Writing—Original Draft: P.C.; Writing—Review & Editing: N.G., V.L.S.L., M.M.D.; Supervision: V.L.S.L., M.M.D.; Funding acquisition: P.C., V.L.S.L., M.M.D. All authors ensured that questions on the accuracy or integrity of all parts of the study were appropriately researched and resolved.
Funding
This research was partly funded by the Bayer Ophthalmology Research Awards 2021 and Chemelot InSciTe under the EyeSciTe consortium.
Data availability
The sequencing data regarding this study has been uploaded to the Gene Expression Omnibus (GEO) with Accession Number GSE227942 and is accessible via this link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE227942.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Pere Català and Nathalie Groen.
These authors jointly supervised this work: Vanessa L. S. LaPointe and Mor M. Dickman.
Contributor Information
Vanessa L. S. LaPointe, Email: mor.dickman@mumc.nl
Mor M. Dickman, Email: v.lapointe@maastrichtuniversity.nl
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-36567-6.
References
- 1.DelMonte DW, Kim T. Anatomy and physiology of the cornea. J. Cataract Refract. Surg. 2011;37:588–598. doi: 10.1016/j.jcrs.2010.12.037. [DOI] [PubMed] [Google Scholar]
- 2.Bonanno JA. Molecular mechanisms underlying the corneal endothelial pump. Exp. Eye Res. 2012;95:2–7. doi: 10.1016/j.exer.2011.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Price MO, Mehta JS, Jurkunas UV, Price FW. Corneal endothelial dysfunction: Evolving understanding and treatment options. Prog. Retin. Eye Res. 2021;82:100904. doi: 10.1016/j.preteyeres.2020.100904. [DOI] [PubMed] [Google Scholar]
- 4.Gain P, et al. Global survey of corneal transplantation and eye banking. JAMA Ophthalmol. 2016;134:167–173. doi: 10.1001/jamaophthalmol.2015.4776. [DOI] [PubMed] [Google Scholar]
- 5.Peh GSL, et al. Propagation of human corneal endothelial cells: A novel dual media approach. Cell Transpl. 2015;24:287–304. doi: 10.3727/096368913X675719. [DOI] [PubMed] [Google Scholar]
- 6.Kinoshita S, et al. Injection of cultured cells with a ROCK inhibitor for bullous keratopathy. N. Engl. J. Med. 2018;378:995–1003. doi: 10.1056/NEJMoa1712770. [DOI] [PubMed] [Google Scholar]
- 7.Numa K, et al. Five-year follow-up of first eleven cases undergoing injection of cultured corneal endothelial cells for corneal endothelial failure. Ophthalmology. 2021;128:504–514. doi: 10.1016/j.ophtha.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 8.Frausto RF, et al. Phenotypic and functional characterization of corneal endothelial cells during in vitro expansion. Sci. Rep. 2020;10:7402. doi: 10.1038/s41598-020-64311-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Català P, et al. Approaches for corneal endothelium regenerative medicine. Prog. Retin. Eye Res. 2022;87:100987. doi: 10.1016/j.preteyeres.2021.100987. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto A, et al. A physical biomarker of the quality of cultured corneal endothelial cells and of the long-term prognosis of corneal restoration in patients. Nat. Biomed. Eng. 2019;3:953–960. doi: 10.1038/s41551-019-0429-9. [DOI] [PubMed] [Google Scholar]
- 11.Ueno M, et al. Superiority of mature differentiated cultured human corneal endothelial cell injection therapy for corneal endothelial failure. Am. J. Ophthalmol. 2022;237:267–277. doi: 10.1016/j.ajo.2021.11.012. [DOI] [PubMed] [Google Scholar]
- 12.Van den Bogerd B, et al. Corneal endothelial cells over the past decade: Are we missing the mark(er)? Transl. Vis. Sci. Technol. 2019;8:6–13. doi: 10.1167/tvst.8.6.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Català P, et al. Transport and preservation comparison of preloaded and prestripped-only DMEK grafts. Cornea. 2020;39:1407–1414. doi: 10.1097/ICO.0000000000002391. [DOI] [PubMed] [Google Scholar]
- 14.Foster JW, Gouveia RM, Connon CJ. Low-glucose enhances keratocyte-characteristic phenotype from corneal stromal cells in serum-free conditions. Sci. Rep. 2015;5:10839. doi: 10.1038/srep10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fernández-Pérez J, Ahearne M. Influence of biochemical cues in human corneal stromal cell phenotype. Curr. Eye Res. 2019;44:135–146. doi: 10.1080/02713683.2018.1536216. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi R, et al. Co-ordinated ocular development from human iPS cells and recovery of corneal function. Nature. 2016;531:376–380. doi: 10.1038/nature17000. [DOI] [PubMed] [Google Scholar]
- 17.Ouyang H, et al. WNT7A and PAX6 define corneal epithelium homeostasis and pathogenesis. Nature. 2014;511:358–361. doi: 10.1038/nature13465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He Z, et al. 3D map of the human corneal endothelial cell. Sci. Rep. 2016;6:29047. doi: 10.1038/srep29047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Malavolta M, et al. Changes in Zn homeostasis during long term culture of primary endothelial cells and effects of Zn on endothelial cell senescence. Exp. Gerontol. 2017;99:35–45. doi: 10.1016/j.exger.2017.09.006. [DOI] [PubMed] [Google Scholar]
- 20.Enomoto K, Mimura T, Harris DL, Joyce NC. Age differences in cyclin-dependent kinase inhibitor expression and Rb hyperphosphorylation in human corneal endothelial cells. Investig. Ophthalmol. Vis. Sci. 2006;47:4330–4340. doi: 10.1167/iovs.05-1581. [DOI] [PubMed] [Google Scholar]
- 21.Benadjaoud MA, et al. Deciphering the dynamic molecular program of radiation-induced endothelial senescence. Int. J. Radiat. Oncol. Biol. Phys. 2022;112:975–985. doi: 10.1016/j.ijrobp.2021.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Sun X, Kaufman PD. Ki-67: More than a proliferation marker. Chromosoma. 2018;127:175–186. doi: 10.1007/s00412-018-0659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varis A, Salmela AL, Kallio MJ. Cenp-F (mitosin) is more than a mitotic marker. Chromosoma. 2006;115:288–295. doi: 10.1007/s00412-005-0046-0. [DOI] [PubMed] [Google Scholar]
- 24.Ersvær E, et al. Prognostic value of mitotic checkpoint protein BUB3, cyclin B1, and pituitary tumor-transforming 1 expression in prostate cancer. Mod. Pathol. 2020;33:905–915. doi: 10.1038/s41379-019-0418-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rao BK, Malathi N, Narashiman S, Rajan ST. Evaluation of myofibroblasts by expression of alpha smooth muscle actin: A marker in fibrosis, dysplasia and carcinoma. J. Clin. Diagn. Res. 2014;8:14–17. doi: 10.7860/JCDR/2014/7820.4231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hamuro J, et al. Metabolic plasticity in cell state homeostasis and differentiation of cultured human corneal endothelial cells. Investig. Ophthalmol. Vis. Sci. 2016;57:4452–4463. doi: 10.1167/iovs.16-19807. [DOI] [PubMed] [Google Scholar]
- 27.Oouchi Y, Watanabe M, Ida Y, Ohguro H, Hikage F. Rosiglitasone and ROCK inhibitors modulate fibrogenetic changes in TGF-β2 treated human conjunctival fibroblasts (HconF) in different manners. Int. J. Mol. Sci. 2021;22:7335. doi: 10.3390/ijms22147335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deguchi H, et al. Intracellular pH affects mitochondrial homeostasis in cultured human corneal endothelial cells prepared for cell injection therapy. Sci. Rep. 2022;12:1–14. doi: 10.1038/s41598-022-10176-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peh GSL, et al. Functional evaluation of two corneal endothelial cell-based therapies: tissue-engineered construct and cell injection. Sci. Rep. 2019;9:6087. doi: 10.1038/s41598-019-42493-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthaei M, Meng H, Meeker AK, Eberhart CG, Jun AS. Endothelial Cdkn1a (p21) overexpression and accelerated senescence in a mouse model of Fuchs endothelial corneal dystrophy. Investig. Ophthalmol. Vis. Sci. 2012;53:6718–6727. doi: 10.1167/iovs.12-9669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rufini A, Tucci P, Celardo I, Melino G. Senescence and aging: The critical roles of p53. Oncogene. 2013;32:5129–5143. doi: 10.1038/onc.2012.640. [DOI] [PubMed] [Google Scholar]
- 32.Orgaz JL, Herraizy C, Sanz-Moreno V. Rho GTPases modulate malignant transformation of tumor cells. Small GTPases. 2014;5:e983867. doi: 10.4161/sgtp.29019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Català P, et al. Single cell transcriptomics reveals the heterogeneity of the human cornea to identify novel markers of the limbus and stroma. Sci. Rep. 2021;11:21727. doi: 10.1038/s41598-021-01015-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules. 2020;10:420. doi: 10.3390/biom10030420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sha X, Liu Z, Song L, Wang Z, Liang X. Human amniotic epithelial cell niche enhances the functional properties of human corneal endothelial cells via inhibiting P53-survivin-mitochondria axis. Exp. Eye Res. 2013;116:36–46. doi: 10.1016/j.exer.2013.08.008. [DOI] [PubMed] [Google Scholar]
- 36.Peh GSL, et al. The effects of rho-associated kinase inhibitor Y-27632 on primary human corneal endothelial cells propagated using a dual media approach. Sci. Rep. 2015;5:1–10. doi: 10.1038/srep09167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Parekh M, et al. Human corneal endothelial cells from older donors can be cultured and passaged on cell-derived extracellular matrix. Acta Ophthalmol. 2020;1:1–11. doi: 10.1111/aos.14614. [DOI] [PubMed] [Google Scholar]
- 38.Okumura N, et al. Enhancement on primate corneal endothelial cell survival in vitro by a rock inhibitor. Investig. Ophthalmol. Vis. Sci. 2009;50:3680–3687. doi: 10.1167/iovs.08-2634. [DOI] [PubMed] [Google Scholar]
- 39.Frausto RF, Le DJ, Aldave AJ. Transcriptomic analysis of cultured corneal endothelial cells as a validation for their use in cell replacement therapy. Cell Transplant. 2016;25:1159–1176. doi: 10.3727/096368915X688948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.R Core Team. R: A Language and Environment for Statistical Computing. (2013).
- 41.Stuart T, et al. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heaton H, et al. Souporcell: Robust clustering of single-cell RNA-seq data by genotype without reference genotypes. Nat. Methods. 2020;17:615–620. doi: 10.1038/s41592-020-0820-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korsunsky I, et al. Fast, sensitive and accurate integration of single-cell data with Harmony. Nat. Methods. 2019;16:1289–1296. doi: 10.1038/s41592-019-0619-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Germain, P.-L. scDblFinder: scDblFinder. R package version 1.2.0. (2020).
- 45.Cao J, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. 2019;566:496–502. doi: 10.1038/s41586-019-0969-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Korotkevich G, et al. Fast gene set enrichment analysis. BioRxiv. 2021 doi: 10.1101/060012. [DOI] [Google Scholar]
- 47.Luo W, Friedman MS, Shedden K, Hankenson KD, Woolf PJ. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009;10:161. doi: 10.1186/1471-2105-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liberzon A, Birger C, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The sequencing data regarding this study has been uploaded to the Gene Expression Omnibus (GEO) with Accession Number GSE227942 and is accessible via this link: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE227942.