Summary
Equitable SARS-CoV-2 surveillance in low-resource communities lacking centralized sewers is critical as wastewater-based epidemiology (WBE) progresses. However, large-scale studies on SARS-CoV-2 detection in wastewater from low-and middle-income countries is limited because of economic and technical reasons. In this study, wastewater samples were collected twice a month from 186 urban and rural subdistricts in nine provinces of Thailand mostly having decentralized and non-sewered sanitation infrastructure and analyzed for SARS-CoV-2 RNA variants using allele-specific RT-qPCR. Wastewater SARS-CoV-2 RNA concentration was used to estimate the real-time incidence and time-varying effective reproduction number (Re). Results showed an increase in SARS-CoV-2 RNA concentrations in wastewater from urban and rural areas 14–20 days earlier than infected individuals were officially reported. It also showed that community/food markets were “hot spots” for infected people. This approach offers an opportunity for early detection of transmission surges, allowing preparedness and potentially mitigating significant outbreaks at both spatial and temporal scales.
Subject areas: Environmental monitoring, Virology, Applied microbiology, Molecular microbiology
Graphical abstract

Highlights
-
•
Wastewater can provide actionable information in more a clinically relevant time frame
-
•
Study also showed the distribution of Covid-19 variants across different facilities
-
•
We provide an equitable approach to wastewater monitoring with sparse sampling
-
•
Findings advance health equity in low-resource countries with poor sewer system
Environmental monitoring; Virology; Applied microbiology; Molecular microbiology
Introduction
The SARS-CoV-2 pandemic has already resulted in extreme social, economic, and health disruption across the world.1 Vaccination has lessened the risk of severe disease and reduced the need for full-scale lockdowns.2,3,4 As of March 2023, the global death toll from SARS-CoV-2 infections was more than 6.9 million, whereas over 682 million cases had been confirmed worldwide5(Johns Hopkins University Coronavirus Resource Center). With only 25% of the population in low-income countries fully vaccinated, compared to 76% in high-income countries, the vaccine inequity will likely continue to prolong the pandemic and increase the risk of new variant occurrences.2,4,6
Clinical surveillance is impractical for early outbreak identification; expensive and politicized, especially in highly populated low and middle-income countries (LMICs).7,8 Furthermore, the proportion of individuals with asymptomatic SARS-CoV-2 infections are very difficult to trace by clinical surveillance.7,9 Infected people who lack symptoms unknowingly tend to go about their lives, interacting with and potentially infecting many susceptible people. This makes control strategies, such as identification, quarantine, and contact tracing, challenging to implement.10
Wastewater surveillance was critical to many countries’ response to the SARS-CoV-2 pandemic.10,11,12,13,14,15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30 Wastewater-based epidemiology (WBE) is an efficient tool for monitoring the circulation of SARS-CoV-2 in communities, which can play a crucial role in estimating the asymptomatic and symptomatic cases within a population.9,27,31 Numerous studies have shown that detecting SARS-CoV-2 RNA in wastewater and sewage sludge25,27,30,31,32,33,34 can give a snapshot of both asymptomatic and symptomatic circulation ranging from an individual building to an entire community.10,14,16,18,21,22,24,27,28,29,35,36 Overall, the studies highlight the potential of WBE as a complementary tool for monitoring and controlling the spread of infectious diseases.10,14,16,18,21,22,24,27,28,29,35 However, there are still limitations and challenges to be addressed in the implementation of WBE systems, including the need for cost-effectiveness, and testing resilience in low-resource communities with poor sewer infrastructure, especially for LMICs.10,14,16,18,21,22,24,27,28,29,35 Several studies have confirmed the applicability of WBE in Thailand, demonstrating the proof-of-concept of using WBE within LMICs.30,37,38
WBE must be conducted at an optimum frequency to offer the greatest benefits over ‘traditional’ surveillance approaches, especially in LMICs.14,21,22,24,25,28,34 Wastewater sampling frequency must allow for the timely reporting of results to public health authorities, empowering them to direct limited resources to the most effective mitigation efforts.8,32 Consequently, there is great interest in determining the optimal sampling frequency that offers the greatest value to public health authorities and society.
The vast majority of publications on wastewater-based epidemiology report areas with centralized sewer systems,25,27,30,31,32,33,34 reflecting the reality that much of WBE has been done in the global north, which has well-established sewer infrastructure connecting populations, particularly those within cities and large towns.25,27,30,31,32,33,34 However, billions of people, mainly living in LMICs with scarce resources and delicate healthcare systems still use decentralized or non-sewered sanitation systems.11,39,40,41,42 It is becoming increasingly clear that those with the least resources and infrastructure suffer the most under the strain of a global pandemic like COVID-19.42,43,44 Despite the urgent need for solutions, there is a significant lack of research and studies on WBE in these regions.12,15,23,43,45 This raises important questions about whether WBE can be effectively conducted in areas with decentralized and non-sewered sanitation systems and, if so, which sampling frequency offers the optimal balance between cost and public health value.
Here, a large-scale study was conducted in Thailand during the COVID-19 pandemic to examine the use WBE for populations using decentralized and non-sewered sanitation infrastructure and the utility of the data for estimating infection incidence and effective reproduction numbers (Re). Furthermore, wastewater surveillance data was utilized to estimate the lag time between detecting SARS-CoV-2 RNA in wastewater and reporting confirmed cases.
Results
SARS-CoV-2 RNA in wastewater
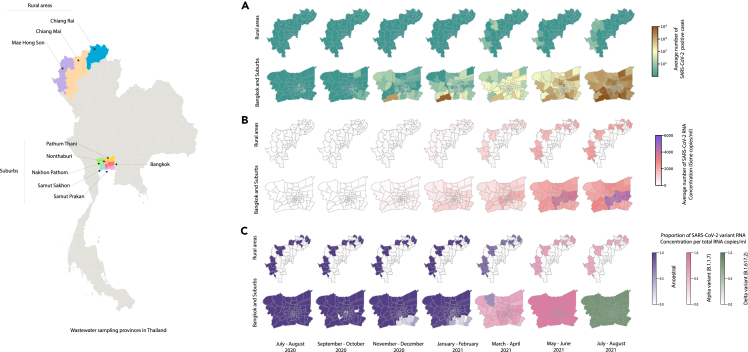
Starting in November 2020, SARS-CoV-2 RNA concentrations in wastewater increased gradually, aligned with the increasing trend in daily newly reported cases in Bangkok (6529.60 copies/mL) and five surrounding provinces (Samut Sakhon 4936.68 copies/mL, Nonthaburi 6050 copies/mL, Samut Prakan 4864 copies/mL, Pathum Thani 1618 copies/mL, and Nakhon Pathom 2281 copies/mL) (Figures 1A, 1B, and 2). Rural areas (1379 copies/mL in November) followed the same direction, even though SARS-CoV-2 RNA concentration remained relatively low compared to Bangkok (p < 0.05) and the five surrounding provinces (p < 0.05) (Figures 1A, 1B, and 2). Thailand reported almost no locally transmitted infections until December 2020, when the second outbreak occurred in Samut Sakhon province and spread to many provinces before partially subsiding in February 2021 (Figures 1A and 2). In April 2021, a third outbreak originated from Bangkok’s Thong Lo–area nightlife venues and rapidly spread in Bangkok and throughout the country.
Figure 1.
Estimation of infection incidence and effective reproductive number for SARS-CoV-2
(A–D) (A) confirmed positive cases, (B) RNA concentration in wastewater, (C) comparison of daily SARS-CoV-2 confirmed cases, infection incidence estimated based on confirmed cases, and infection incidence estimated based on SARS-CoV-2 RNA concentration in wastewater for Bangkok, and (D) comparison of the effective reproductive numbers estimated using the case-based infection incidence (red line) and wastewater-based infection incidence (green line). The ribbons display the standard deviation of 1,000 bootstrap replicates.
Figure 2.
Map for SARS-CoV-2 RNA in wastewater and reported cases
(A–C) (A) confirmed positive SARS-CoV-2 cases, (B) SARS-CoV-2 RNA concentration in wastewater, and (C) percent of variant per total SARS-CoV-2 RNA concentration in wastewater.
Estimating infection incidence and effective reproductive numbers from wastewater measurements
An increase in the estimated incidences based on wastewater SARS-CoV-2 RNA concentrations was observed before infected individuals were officially reported (Figures 1C and S1). Wastewater-based estimated incidence showed a steep increase starting from the last week of November 2020 (Figures 1C and S1). However, case-based incidences remained low until April 2021(Figure 1A). Re derived from confirmed cases came with more uncertainty and fluctuation over time than Re estimated from wastewater SARS-CoV-2 RNA concentration data (Figures 1D and S1).
Relative abundance of different SARS-CoV-2 viral variants in wastewater
The relative abundance of different SARS-CoV-2 variants in wastewater revealed that the ancestral SARS-CoV-2 was dominant until early March of 2021 in Bangkok and the five surrounding provinces (Figures 2C, 3, and S2) and remained dominant in rural areas until mid-April 2021 (Figures 2 and 3). Notably, the Bangkok province wastewater samples were positive for Alpha (B.1.1.7) variant in the second week of December 2020 and Delta (B.1.617.2) variant in the fourth week of March 2021. This was about two weeks before Thailand reported its first cases of Alpha (3rd January 2021) and Delta (24th April 2021) variants (Figure 3G). The data also showed that Alpha and Delta SARS-CoV-2 variants transmitted rapidly within a short period (Figures 2C and 3). Concordantly, the data also showed that the estimated infection incidences based on Alpha and Delta SARS-CoV-2 variants were greater with more rapid increases than the ancestral virus-based incidences (Figures 3 and S2).
Figure 3.
Relative abundance of different SARS-CoV-2 viral variants in Wastewater
SARS-CoV-2RNA concentration for each variant in wastewater and infection incidence estimated from each variant’s viral loads in wastewater for (A and B) Bangkok, (C and D) Nakhon Pathom, and (E and F) Rural areas. The ribbons show the standard deviation across 1,000 bootstrap replicates.
(G) Relative abundance shows the transition of variants of viral loads in wastewater for each area.
SARS-CoV-2 viral RNA concentrations in wastewater from different facilities
Analysis of SARS-CoV-2 viral RNA concentrations in different facilities revealed that the relative abundances of SARS-CoV-2 viral RNA concentrations in wastewater were relatively high in community/food markets and condominium complexes in Bangkok (Figures 4A–4C). Similarly, higher abundances were found in community/food markets and housing complexes in five surrounding provinces (Figure S3). In rural regions, community malls and community/food markets had similar SARS-CoV-2 viral RNA concentrations in wastewater (Figure 4D). Estimation of infection incidences for each facility showed that community/food markets and condominium complexes tend to carry more infected people (symptomatic or asymptomatic) during the lockdown period in Bangkok (Figures 4B and 4C). A high incidence is reliant on high toilet use, introducing a bias for locations of residence. A similar trend was found in community/food markets and housing complexes in the five surrounding provinces (Figure S3). Community/food markets had higher estimated infection incidences in rural regions than community malls based on viral RNA concentrations in wastewater during the lockdown period (Figures 4E and 4F). Also, entertainment and leisure venues in Bangkok carry a higher number of infected people, and higher relative abundances of SARS-CoV-2 before the third outbreak occurred at an entertainment venue (Figures 4B and 4C). Of interest, a similar trend was observed in the condominium complex in Bangkok for the same period (Figures 4B and 4C). However, the control measures employed by the Thai government seemed to have reduced SARS-CoV-2 circulating in cafeterias, shopping centers, and entertainment and leisure venues (Figures 4B and 4C).
Figure 4.
SARS-CoV-2 RNA concentration in wastewater collected from different facilities
(A–F) (A) Bangkok and (D) Rural areas. Relative abundance in SARS-CoV-2 RNA concentration in wastewater collected from different facilities in (C) Bangkok and (F) Rural areas. Infection incidences based on SARS-CoV-2 RNA concentration in (B) Bangkok and (E) Rural areas. The ribbons show the standard deviation of 1,000 bootstrap replicates. Gray areas show the period during a nationwide ban on large events to ease lockdown (26-Dec-20 to 29-Jan-21, and 17-Apr-21 to 01-Sep-21). The black labels show the superspreading events, and the red labels are the starting of interventions.
The lag time between SARS-CoV-2 RNA loads detected in wastewater and confirmed cases of COVID-19
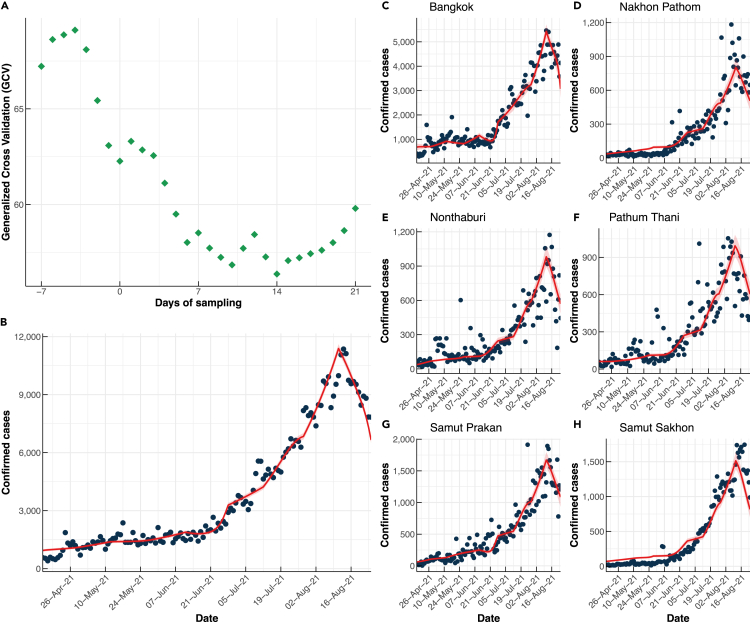
The results of the modeling showed that the confirmed cases of COVID-19 in Bangkok and five surrounding provinces had a lag time of 14 days relative to the changes in RNA concentrations in wastewater (Figures 5, S4, and S5). The lag time increased to 20 days in rural regions, possibly because of a longer delay in reporting infected individuals (Figures S6 and S7). The data suggests that wastewater collected from decentralized and non-sewered sanitation systems in Thailand can offer a minimum of 14 days’ advanced notice of COVID-19 cases ahead of official ‘confirmed cases.’ The advanced warning time increased to approximately 20 days in rural locations.
Figure 5.
Relationship between SARS-CoV-2 RNA concentration in wastewater and reported cases
(A) GCV values of the fitting models, the number of cases obtained from the best model (red line) compared to the reported cases (black dots) with their confidence intervals (ribbons) in (B) Total, (C) Bangkok, (D) Nakhon Pathom, (E) Nonthaburi, (F) Pathum Thani, (G) Samut Prakan, and (H) Samut Sakhon.
Discussion
Wastewater samples were collected from different public spaces which are connected to decentralized wastewater collection systems and non-sewered sanitation systems in Bangkok city, surrounding suburbs and rural areas and SARS-CoV-2 RNA concentrations were monitored for over a year. Re of SARS-CoV-2 transmission was calculated using both confirmed cases and SARS-CoV-2 RNA concentrations detected in wastewater. The data were also used to estimate the lag time between the detection of SARS-CoV-2 RNA in wastewater and the time at which the confirmed cases were reported. The results show that Alpha and Delta SARS-CoV-2 variants existed in Thailand several weeks earlier than the clinical cases are reported. Importantly, the study showed that wastewater surveillance detected SARS-CoV-2 (including variants) 14 days earlier in urban areas and 20 days earlier in rural areas than the confirmed case reports. This is sufficient to provide a substantial improvement over existing public health monitoring and subsequent response times.
Re for SARS-CoV-2 was more stable when generated from twice-a-month wastewater SARS-CoV-2 virus RNA concentration data in Thailand than from official case count data. It has been suggested that three sampling events per week are necessary to estimate the Re from wastewater.21 However, results from this study show that bimonthly sampling can be used to inform a useful measure of Re. Owing to slow confirmed case detection in much of the regions under study, bimonthly wastewater analysis offered a substantial improvement on estimated infection levels (Bibby et al., 2021). However, lack of SARS-CoV-2 case reporting attributed to each variant in Thailand limited the capability to estimate the Re for each variant. The mixing of sewage within a large volume sewer systems will normally disrupt the stool and expose the SARS-CoV-2 RNA to rapid biodegradation, whereas low volume, decentralized systems may extend the half-life of the RNA and minimize the dilution effects and thereby offering greater ability to characterize the health of the community under study.46,47
The analysis of the distribution of different SARS-CoV-2 variants and at different venues provided information on the effectiveness of COVID-19 control measures. Community/food markets were identified as one of the hot spots in the current study. Therefore, WBE can be considered as an indirect proxy for control measure effectiveness. Asymptomatic carriers are largely unquantified contributors to the transmission of SARS-CoV-2 in food or community markets as they are unaware of their illness.9 This risk is captured by the very high concentrations of SARS in the wastewater in these locations. In developing countries, people tend to crowd into community/food markets on a regular basis (often daily). This is regardless of income difference.48 Many of these markets are wet-type markets, where live animals are sold (raw or cooked) and/or slaughtered on site.48 Therefore, community/food markets may play a key element in virus circulation and transmission, including SARS-CoV-249 and Ebola.50 What is clear is that effective infection control with additional community testing in identified hot spots like these markets should become the focus of outbreak prevention for LMICs.23,51
Wastewater sampling from decentralized and non-sewered sanitation systems addresses some of the reported limitations of wastewater-based epidemiology.29 A large sewered catchment with a long hydraulic residence time and the introduction of stormwater into the sewers may produce lower viral concentrations than a smaller non-sewered site, despite disease prevalence being the same in both catchments.29,46 Specifically, the pooled nature of wastewater from large sewer sheds makes it hard to pinpoint subpopulations at risk for targeted interventions.29 However, in many LMICs including Thailand, many, if not most, households, businesses, and rural communities rely on onsite or decentralized sanitation systems.52 As shown in this study, facility-wise wastewater monitoring may be used to pinpoint transmission hotspots and the efficacy of mitigation measures.10,51
Wastewater surveillance data was used to monitor and estimate SARS-CoV-2 transmission dynamics in each venue, community and province at a given time.53,54 Screening wastewater has the potential to identify infection spread to vulnerable communities such as nursing homes, slums, and marginalized villages. Although it is possible to target individual venues connected to centralized sewer systems,20 it is problematic as the sewage will rapidly enter the sewer system making it much more challenging to catch every flush event. The decentralized and non-sewered systems can provide a snapshot of the situation in a given establishment. Connecting the data from such decentralized wastewater testing with human mobility data will help understand how often people travel from wastewater testing areas to other areas and predict where the virus will likely spread.
It is imperative to minimize the transmission in areas with limited access to facilities like COVID-19 vaccines and health care. During this study, it became apparent that in rural sites, where people live far away from each other with limited mobility, there was a longer lag time between wastewater SARS-CoV-2 RNA detection and clinical case reports. Previous studies have shown different lag times between the case reports and wastewater SARS-CoV-2 RNA detection.21,37,55,56,57 Under the situation where clinical testing is accessible on-demand with a rapid time to results, the lead time of WBE was predicted as 4 days.55 Therefore, the current study highlights the potential to use WBE in more rural areas with decentralized systems and non-sewered sanitation to rapidly predict and respond to the disease transmission.
LMICs have experienced disproportionately worse health outcomes during the pandemic as compared to high-income countries.58 The impact has been exacerbated by less access to health care and a higher prevalence of underlying conditions associated with severe COVID-19. Inequitable access to reliable and convenient COVID-19 testing, new drugs and vaccines, the high cost of over-the-counter rapid antigen tests, very densely populated cities, and marginalization have combined to intensify the impact of COVID-19.58 SARS-CoV-2 monitoring based on WBE is shown to be significantly cheaper than random clinical testing.36 However, implementation of WBE in LMICs has to overcome many barriers including the lack of economic resources, dysfunctional sewer systems, and sophisticated laboratories which can be clearly seen by the limited number of studies reported from LMICs in Global SARS-CoV-2 Wastewater Monitoring Dashboards like COVIDPoops19.40,41 A recent study has calculated the cost of wastewater surveillance of SARS-CoV-2 in Blantyre, Malawi and Kathmandu, Nepal ranged from $25 to $74 (Blantyre) and $120 to $175 (Kathmandu) per sample8 which is expensive considering the GDP of these countries. Outcomes of this study show that some of the privileges afforded by WBE can be realized in LMICs despite limited resources and decentralized and non-sewered sanitation infrastructure systems by identifying the hot spots and optimizing the sampling frequency.
Limitations of the study
This paper reports a year-long analysis of SARS-CoV-2 RNA in wastewater in urban and rural areas of Thailand. The sampling covered a range of provinces and venues; most importantly, many were utilizing decentralized or non-sewered sanitation systems which are typical in LMICs. Despite limited sampling frequency (bimonthly), the ability to detect SARS-CoV-2 variants in wastewater well in advance of clinical reporting was confirmed. The data can be used to model transmission and estimate Re in these communities, which are in line with other studies based on daily measurements and powerful genomic sequencing.14,22,24,28 The finding of SARS-CoV-2 variants in wastewater samples 14 days earlier than clinical reports provides strong evidence for the surveillance of large populations using WBE with the benefit of economies of scale.8,35 Bangkok is the fifth largest city in East Asia and the ninth largest in terms of its population, approaching 10 million.59 Owing to its status as a prominent destination for the sex tourism industry, Thailand is considered a high-risk area for emerging infectious diseases.60 Infections can arise and propagate rapidly in this context, potentially spreading throughout the region.60 In addition, the rural sampling areas were located along one of the most porous border communities in the world (Myanmar and Laos), and marginalized communities (hill tribes) were also included in this study. Because SARS-CoV-2 detection centers and self-detection kits are becoming widespread, individuals with symptoms are more likely to isolate, potentially shifting the proportion of transmissions from symptomatic individuals to those who are asymptomatic. Also, real-world transmission dynamics are not entirely dependent on the individual-level dynamics of infectiousness over time.7 What is clear is that WBE will be a cost-effective tool to estimate the efficacy of any interventions to reduce disease transmission.8,31,35,61
Conclusions
In LMICs with densely populated communities and limited access to therapeutics and vaccines that can shorten or eliminate, the successful control of SARS-CoV-2 cannot solely rely on identifying and isolating symptomatic or asymptomatic cases.58 WBE can help local public health officials bypass access and affordability issues associated with diagnostic testing and provide action-oriented insights about where to focus public health resources, such as booster vaccination, mask-wearing, and social distancing and also to evaluate the progress of these infection control programs. Wastewater can also help reduce the risk of transmission from people with asymptomatic infections. On top of this, it is our assertion that WBE discipline itself would benefit from more studies based on decentralized and non-sewered sanitation systems in LMICs.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and virus strains | ||
| Murine hepatitis virus; Strain: MHV-A59 | ATCC | ATC.VR-764 |
| Chemicals, peptides, and recombinant proteins | ||
| RNase-Free Water | Qiagen | 129112 |
| TaqPath™ qPCR Master Mix, CG | Thermo Fisher Scientific | A15297 |
| Critical commercial assays | ||
| RNeasy PowerSoil Total RNA Kit | Qiagen | 12866-25 |
| iTaq Universal Probes One-Step Kit | Bio-Rad | 1725141 |
| Deposited data | ||
| Confirmed COVID-19 cases in Thailand | Department of Disease Control, Ministry of Public Health, Thailand | https://data.go.th/dataset/covid-19-daily |
| Oligonucleotides | ||
| 2019-nCoV_N1-F 5’-GACCCCAAAA TCAGCGAAAT-3’ 3 |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N1-R 5’-TCTGGTTACTGC CAGTTGAATCTG-3’ 3 |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N1-P 5’-FAM-ACCCCGCATTA CGTTTGGTGGACC-ZEN/Iowa Black-3’ 3 |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N2-F 5’-TTACAAACATTG GCCGCAAA-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N2-R 5’-GCGCGACATTCC GAAGAA-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N2-P 5’-FAM-ACAATTTGC CCCCAGCGCTTCAG- ZEN/Iowa Black-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N3-F 5’-GGGAGCCTTGA ATACACCAAAA-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N3-R 5’-TGTAGCACGAT TGCAGCATTG-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| 2019-nCoV_N3-P 5’-FAM-AYCACATTGG CACCCGCAATCCTG- ZEN/Iowa Black-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| E_Sarbeco_F 5’-ACAGGTACGTTAATAGT TAATAGCGT-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| E_Sarbeco_R 5’-ATATTGCAGCAGTA CGCACACA-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| E_Sarbeco_P1 5’-FAM-ACACTAGCCATCC TTACTGCGCTTCG-ZEN/Iowa Black-3’ |
U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| RP-F- 5’-AGA TTT GGA CCT GCG AGC G -3’ | U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| RP-R- 5’-GAG CGG CTG TCT CCA CAA GT -3’ | U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| RP-P- 5’- FAM – TTC TGA CCT GAA GGC TCT GCG CG – BHQ-1 -3’ | U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| RP-P- 5’- FAM-TTC TGA CCT /ZEN/ GAA GGC TCT GCG CG-3 - IABkFQ -3’ | U.S. Centers for Disease Control and Prevention (CDC) N1, N2, E and human RP gene primers sets | https://www.cdc.gov/coronavirus/2019-ncov/lab/multiplex.html |
| Ancestral ACAATTTGGCAGAGACATCGC | Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Ancestral AGAACATGGTGTAATGTCAA GAATC |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Ancestral /56- FAM/ACTGATGCTGT CCGTGATCCACAG/3BHQ_1/ |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Alpha (B.1.1.7) ACAATTTGGC AGAGACATCGA |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Alpha (B.1.1.7) AGAACATGGTGTA ATGTCAAGAATC |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Alpha (B.1.1.7) /56- FAM/ACTGATGCTG TCCGTGATCCA CAG/3BHQ_1/ |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Delta (B.1.617.2) 5′ GGTTGGTGG TAATTATAATTCCCG |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Delta (B.1.617.2) 5′ CCTTCAACAC CATTACAACGTT |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Delta (B.1.617.2) 5′ FAM-TCTCTCAAAAG GTTTGAGATTAGACTTCC-BHQ |
Lee, W.L. et al. and Garson, J.A. et al.25,62 | N/A |
| Recombinant DNA | ||
| Synthetic full-length SARS-CoV- 2 RNA | USA-WA1/2020 | ATCC-VR-1986D |
| Software and algorithms | ||
| Infection incidences estimate code | Huisman et al.20 | https://github.com/JSHuisman/wastewaterRe |
| mgcv | R software package | https://www.r-project.org |
| dplyr 1.0.7 | R software package | https://www.r-project.org |
| tidyverse 1.3.1 | R software package | https://www.r-project.org |
| splines 4.1.0 | R software package | https://www.r-project.org |
| zoo 1.8-9 | R software package | https://www.r-project.org |
| astsa 1.14 | R software package | https://www.r-project.org |
| lubridate 1.7.10 | R software package | https://www.r-project.org |
| patchwork 1.1.1 | R software package | https://www.r-project.org |
| ggplot2 3.3.5 | R software package | https://www.r-project.org |
| dslabs 0.7.4 | R software package | https://www.r-project.org |
| scales 1.2.1 | R software package | https://www.r-project.org |
| ggalt 0.4.0 | R software package | https://www.r-project.org |
| ggpubr 0.4.0 | R software package | https://www.r-project.org |
| Other | ||
| Centricon® Plus-70 centrifugal ultrafilters | Merck Millipore | UFC710008 |
Resource availability
Lead contact
Further information and requests for resources, raw data, and code should be directed to and will be fulfilled by the lead contact, Dhammika Leshan Wannigama (Dhammika.L@chula.ac.th).
Materials availability
This study did not generate new unique reagents.
Experimental model and study participant details
Sample collection
Wastewater samples (250ml) were collected twice a month (second and fourth week of each month) for 12 months at 63 sampling points in Bangkok province (covering 50 subdistricts), 134 sampling points in five adjacent provinces (covering 117 subdistricts), and 31 sampling points in 3 rural provinces (covering 19 subdistricts) between July 2020 and August 2021. Sampling locations were selected to represent residents and public spaces based on access to the transportation system, population density (Table S1), and popularity (Arifwidodo and Chandrasiri, 2020) (supplemental information). Wastewater from condominium complexes (n=10), cafeteria and shopping centers (n=11), Community/Food markets (n=13), office complexes (n=10), wastewater treatment plants (n=9), and entertainment/leisure venues (n=10) were sampled in the Bangkok province. In Nakhon Pathom, Nonthaburi, Pathum Thani, Samut Prakan, and Samut Sakhon provinces, wastewater was collected from community/food markets (n=40), housing complexes (n=30), cafeterias and shopping centers (30), work sites (construction camps) (n=20), and office complexes (n=14). For rural communities, samples were collected from community/food markets (n=20) and community malls (n=11). All those locations have closed non-sewered storage systems, except wastewater treatment plants, and raw or untreated sewage was sampled by grab sampling.
Method details
Sample collection and RNA extraction
In total, 1512 samples were collected from Bangkok, 2976 from five adjacent provinces, and 744 from rural provinces. Samples were transported to the laboratory on dry ice. Larger particles, including debris and bacteria, were removed from the samples by pelleting using centrifugation of the sample in 50- or 250-ml conical centrifuge tubes at 4654×g for 30 mins without brake. About 100 to 200 ml supernatant volume was filtered through Centricon® Plus-70 centrifugal ultrafilters with a cut-off value of 100 kDa (Merck Millipore) by centrifugation at 1500×g for 15 minutes, with a resulting concentrate ranging between 0.39-1.80 g. RNA was extracted, and SARS-CoV-2 gene markers (N1, N2, N3, and E) were quantified by Real-time qPCR immediately or within one week after RNA extraction (storage at −80°C) following the same procedure described in a previous study.30 The Ancestral, Alpha variant (B.1.1.7), and Delta variant (B.1.617.2) were quantified9 using allele-specific RT-qPCR as described previously to evaluate the prevalence of different SARS-CoV-2 variants in the wastewater.26,66
To quantify SARS-CoV-2 RNA concentration, 2.5 ml of well-mixed Centricon® concentrates were added directly to a commercial kit optimized for isolation of total RNA from environmental samples according to the manufacturers protocol (RNeasy PowerSoil Total RNA Kit, Qiagen).33 Two replicate RNA extractions and analyses were performed for each sample. Isolated RNA pellets were dissolved in 50 μl of ribonuclease-free water, and total RNA was measured by spectrophotometry (NanoDrop, Thermo Fisher Scientific) as previously described.33 RNA samples were stored at -80°C until virus quantification.
SARS-CoV-2 quantification by real-time qPCR
SARS-CoV-2 RNA was quantified by one-step qRT–PCR using the U.S. Centers for Disease Control and Prevention (CDC) primer N1, N2, and N3 sets that each target a different region of the nucleocapsid (N)gene33,67,68 and the set targeting the envelope protein (E) gene from Medema et al. to include targets against two separate SARS-CoV-2 genes (Table S1).68 The specificity of these primer/probe sets against other respiratory viruses, including human coronaviruses, had been confirmed by several other studies.33,68 For control and in accordance with the CDC protocol, analysis was also conducted for the human RP gene,1 and SARS-CoV-2 results were reported only if RP gene detection was positive. Samples were analyzed using the Bio-Rad iTaq Universal Probes One-Step Kit in 20-μl reactions run at 50°C for 10 min and 95°C for 1 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s per the manufacturer’s recommendations. SARS-CoV-2 RNA concentrations were determined using a standard curve as previously described and presented as virus RNA copies.33,67 For the standard curve, complementary DNA synthesized from full-length SARS-CoV-2 RNA (WA1-USA strain) was used as a template to generate SARS-CoV-2 N gene transcripts as previously described.33,67 To validate N1, N2, N3 and E primers sets, standard curves using the ten-fold series dilution of the N and E gene transcripts were analyzed as previously described.33,67 The primer sets generated a standard curve with N1 primer values of R2 0.99, efficiency: 97.1%, N2 primer R2: 0.99, efficiency: 98.4%, N3 primer R2: 0.99, efficiency: 96.1%, and E primer R2: 0.98, efficiency: 94.3%, Ancestral R2: 0.99, efficiency: 96.7%, Alpha (B.1.1.7) R2: 0.99, efficiency 99.4%, Delta R2: 0.99, efficiency 98.6%. The SARS-CoV-2 concentration results were adjusted to the total RNA extracted by multiplying sample concentrations by the ratio of the maximum RNA concentration to the sample RNA concentration. This accounts for week-to-week variations in wastewater and RNA extraction efficiency. SARS-CoV-2 variant concentrations were measured using the primers given in key resources table.
Virus concentration control
In a subset of 100 samples, the concentration of F-specific RNA phages was also measured by the Double Agar Layer plaque assay method according to ISO 10705, before and after the centrifugation and ultrafiltration step, to determine the virus recovery of these steps as previously described.68 The recovery of F-specific RNA phages by the purification and concentration steps was 80.09 ± 18.68% (n = 100).
Estimation of infection incidence
Infection incidence was estimated using the SARS-CoV-2 viral loads found in wastewater. As these viral loads in wastewater at a specific time reflect a cumulative number of infections that occurred before the measurement, a deconvolution method was used to estimate the rate of infections at a particular time. This approach assumes that earlier infections will impact the measured accumulated viral loads in wastewater with a time-delay weight function, ω(τ), where τ is the time since infection. By following the method described in ref.17, the relationship between the measured accumulated viral loads in the wastewater on day t (At) and the number of infections on day t (It) can be written as
| (Equation 1) |
The weight of the deconvolution, ω(τ), can be estimated from the shedding load profile, which describes the average amount of virus shed by an infected individual τ days after infection. The detailed estimation of ω(τ) has been done in ref.17 It was assumed that assumed that all SARS-CoV-2 variants have the same ω(τ), and daily viral loads were estimated using linear interpolation.
In addition, the infection incidence based on the number of confirmed COVID-19 cases was also estimated; in this case, Equation 1 could still be used, with the viral loads replaced with the daily number of confirmed cases. In this case, the weight of the deconvolution, ω(τ), can be estimated from the delay from infection to symptom onset and the delay from symptom onset to case confirmation.26 In the calculations, it was assumed that the mean incubation period (the time from infection to symptom onset) is 5.3 days with a standard deviation of 3.2 days,17,69 and the mean delay from symptom onset to case confirmation is 2.0 days with a standard deviation of 1.0 days. The number of confirmed COVID-19 cases in Thailand attributed to each province was obtained from the Department of Disease Control, Ministry of Public Health, Thailand (https://data.go.th/dataset/covid-19-daily).
Infection incidence was estimated using the published computer code by Huisman et al. (https://github.com/JSHuisman/wastewaterRe)1 with the parameterization shown below.
getCountParams <- function(obs_type){
switch(obs_type,
incubation = getGammaParams(5.3, 3.2),
zero = list(shape = 0, scale = 0),
#confirmed = getGammaParams(5.5, 3.8),
confirmed_zh = getGammaParams(5.0, 4.0), #2.83, 2.96),
confirmed_cali = getGammaParams(4.51, 3.16),
confirmed_bkk = getGammaParams(2.0, 1.0),
death = getGammaParams(15.0, 6.9),
han = getGammaParams(4.7, 1.7),
wolfel = getGammaParams(8.6, 0.9),
benefield = list(shape = 0.929639, scale = 7.241397))
}
The effective reproduction number (Re) is a measure of the average number of secondary cases caused by one infected individual. A Re value greater than 1 means that the number of infections is increasing and the disease is spreading, while a value less than 1 indicates that the number of infections is decreasing. In this study, a statistical method developed by Cori et al.62 was used to estimate Re using infection incidence estimated from both the number of confirmed cases and viral loads in wastewater. The serial interval distribution, i.e., the time between infections of successive cases in a chain of transmission, was assumed to be a discretized Gamma distribution with a mean of 3.96 days and a standard deviation of 4.75 days.70
Estimation of the time lag between SARS-CoV-2 RNA loads detected in wastewater and confirmed cases of COVID-19
To determine the lag time between SARS-CoV-2 RNA loads and confirmed cases of COVID-19, a regression analysis was conducted utilising quasi-Poisson models commonly employed in environmental epidemiology for handling over-dispersed data.71,72,73 LOESS regression was applied to smooth out the time series. Based on the LOESS regression, a natural cubic spline (f) with four degrees of freedom between the number of cases and viral RNA counts was selected, except for the Delta variant viral counts, for which a linear spline was employed. The following form of the model was used:
where is the expected number of cases, t is the day of sampling, k is the number of days before and after sampling (-7 to 21 days), VAp,t is the Alpha variant concentration in province p at day t (gc/ml), VD p,t is the Delta variant concentration in province p at day t (gc/ml), and VW p,t is the Ancestral viral concentration in province p at day t (gc/ml). Models were fitted using the number of confirmed cases within the Bangkok metropolitan region from seven days prior to wastewater sampling (t = −7) to 28 days after sampling (t = 28) and identified the best-fit models using the GCV (Generalized Cross Validation) criterion in the ‘mgcv’ R software package65 as detailed below.
leadmax=21
lagmin=7
k=leadmax+lagmin+1
leads=c(1:leadmax)
lead_names = paste('lead', formatC(leads, width = nchar(max(leads)), flag = "0"),sep='.')
lead_fun=setNames(paste("dplyr::lead(., ", leads, ")"), lead_names)
data_cases_lead=data_frame(data_cases)%>%
group_by(Province)%>%
mutate_at(vars(case),funs_(lead_fun))
lags=c(lagmin:1)
lag_names = paste('lag', formatC(lags, width = nchar(max(lags)), flag = "0"),sep='.')
lag_fun=setNames(paste("dplyr::lag(., ", lags, ")"), lag_names)
We used the GCV (Generalized Cross Validation) criterion in the ‘mgcv’5 R software package to find the best-fit model, as detailed below.3
models_gamProvinceV2toV4=list()
for (y in c(lag_names,'case', lead_names)) {
form <- formula(paste0(y, "∼", 'fac.Province+ns(V2,4)+sc.V3+ns(V4,4)'))
#(V2,2)+ns(V3,2)+ns(V4,2)
models_gamProvinceV2toV4[[y]] <- gam(form,family = quasipoisson,
data=data_test_Province) }
Acknowledgments
We thank all the volunteers who kindly supported with sample collection. Also, thanks to Dr. Ong-orn Prasarnphanich at the United States Centers for Disease Control and Prevention (CDC) Thailand for technical support and the previous Chargé d'Affaires of the United States of America to Thailand (US Embassy & Consulate in Thailand), Mr. Michael Heath, for facilitating collaboration with the CDC and Armed Forces Research Institute of Medical Sciences (AFRIMS). We also thank the LGBTQIA+ community in Thailand for generous support with sample collection, TEDxChiangMai team and Martin venzky-Stalling for facilitating platform for collaboration, and marginalized, vulnerable indigenous communities in northern Thailand for support with sample collection. Special thanks to Nuttawut Kietchaiyakorn for helping with the illustrations.
Ethics approval was not required for this type of environmental wastewater surveillance study.
Funding Sources: D.L.W. was supported by Balvi Filantropic Fund and Chulalongkorn University (Second Century Fund- C2F Postdoctoral Fellowship), University of Western Australia (Overseas Research Experience Fellowship) and Yamagata Prefectural Central Hospital, Yamagata, Japan (Clinical Residency Fellowship). C.M. was supported by the Centre of Excellence in Mathematics, Ministry of Higher Education, Science, Research and Innovation, Thailand, Center of Excellence on Medical Biotechnology (CEMB), and Thailand Center of Excellence in Physics (ThEP). A.K. is a Rothwell Family Fellow. The funder(s) had no role in study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Author contributions
D.L.W., conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
M.A., conception, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
P.H., conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
C.H., conception, funding acquisition, investigation, data curation, formal analysis, supervision, writing the original draft of the manuscript.
C.M., data curation, formal analysis, supervision, writing the original draft of the manuscript.
S.C., data curation, formal analysis, supervision, writing the original draft of the manuscript.
S.A., data curation, formal analysis, supervision, writing the original draft of the manuscript.
P.P., supervision, critical review and editing of the manuscript.
S.A., supervision, critical review and editing of the manuscript.
A.H.R.S.M., supervision, critical review and editing of the manuscript.
E.D.L., supervision, critical review and editing of the manuscript.
A.T.H., supervision, critical review and editing of the manuscript.
P.V., data curation, formal analysis.
T.S., data curation, formal analysis.
S.L-I., supervision, critical review and editing of the manuscript.
R.J.S., supervision, critical review and editing of the manuscript.
P.O., supervision, critical review and editing of the manuscript.
N.K.D.R., supervision, critical review and editing of the manuscript.
P.K., supervision, critical review and editing of the manuscript.
D.S., supervision, critical review and editing of the manuscript.
T.F., supervision, critical review and editing of the manuscript.
K.S., supervision, critical review and editing of the manuscript.
A.L., supervision, critical review and editing of the manuscript.
T.K., supervision, critical review and editing of the manuscript.
N.H., supervision, critical review and editing of the manuscript.
P.G.H., supervision, critical review and editing of the manuscript.
A.K., supervision, critical review and editing of the manuscript.
A.S., supervision, critical review and editing of the manuscript.
T.C., critical review and editing of the manuscript.
S.T., supervision, critical review and editing of the manuscript.
A.DM., supervision, critical review and editing of the manuscript.
H.I., supervision, critical review and editing of the manuscript.
Declaration of interests
No author declares any potential conflict of interest or competing financial or non-financial interest in relation to the manuscript.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research. One or more of the authors of this paper self-identifies as an underrepresented ethnic minority in their field of research or within their geographical location. One or more of the authors of this paper self-identifies as a gender minority in their field of research. One or more of the authors of this paper self-identifies as a member of the LGBTQIA+ community. One or more of the authors of this paper self-identifies as living with a disability. While citing references scientifically relevant for this work, we also actively worked to promote gender balance in our reference list.
Published: June 9, 2023
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2023.107019.
Contributor Information
Dhammika Leshan Wannigama, Email: dhammika.l@chula.ac.th.
Charin Modchang, Email: charin.mod@mahidol.edu.
Tanittha Chatsuwan, Email: tanittha.c@chula.ac.th.
Alexander D. McLellan, Email: alex.mclellan@otago.ac.nz.
Supplemental information
Data and code availability
Data generated and analyzed during this study are included in this published article and its supplemental information file. As this study is ongoing, additional wastewater details will be available upon reasonable request from the corresponding author Dhammika Leshan Wannigama. The cumulative number of confirmed COVID-19 cases in Thailand attributed to each province was obtained from the Department of Disease Control, Ministry of Public Health, Thailand (https://data.go.th/dataset/covid-19-daily).
Data were analysed and plotted using the tidyverse 1.3.1,63 dplyr 1.0.7, splines 4.1.0, zoo 1.8-9, astsa 1.14, lubridate 1.7.10, patchwork 1.1.1, ggplot2 3.3.5, dslabs 0.7.4, scales 1.2.1, ggalt 0.4.0 and ggpubr 0.4.0 packages of R program version 4.1.0.64
The computer code for estimating the infection incidence is available at the published computer code by Huisman et al. (https://github.com/JSHuisman/wastewaterRe)21 with the parameterization and also original code is available in this paper’s supplemental information. To evaluate the data n days before sampling (t = − 7) to 21 days after sampling (t = 21), we used the dplyr package.62 GCV (Generalized Cross Validation) criterion in the ‘mgcv’5 R software package was used to find the best-fit model.65
References
- 1.Pinilla J., Barber P., Vallejo-Torres L., Rodríguez-Mireles S., López-Valcárcel B.G., Serra-Majem L. The economic impact of the SARS-COV-2 (COVID-19) pandemic in Spain. Int. J. Environ. Res. Public Health. 2021;18:4708. doi: 10.3390/ijerph18094708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu F., Zhao Z., Ma C., Nie X., Wu A., Li X. Return to normal pre-COVID-19 life is delayed by inequitable vaccine allocation and SARS-CoV-2 variants. Epidemiol. Infect. 2022;150:e46. doi: 10.1017/s0950268822000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews N., Stowe J., Kirsebom F., Toffa S., Sachdeva R., Gower C., Ramsay M., Lopez Bernal J. Effectiveness of COVID-19 booster vaccines against COVID-19-related symptoms, hospitalization and death in England. Nat. Med. 2022;28:831–837. doi: 10.1038/s41591-022-01699-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mills E.J., Reis G. Evaluating COVID-19 vaccines in the real world. Lancet. 2022;399:1205–1206. doi: 10.1016/S0140-6736(22)00194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.University J.H. Johns Hopkins University Coronavirus Resource Center; 2023. COVID 19 Dashboard. [Google Scholar]
- 6.Gavi Covax data brief. 2023. gavi.org
- 7.Johansson M.A., Quandelacy T.M., Kada S., Prasad P.V., Steele M., Brooks J.T., Slayton R.B., Biggerstaff M., Butler J.C. SARS-CoV-2 transmission from people without COVID-19 symptoms. JAMA Netw. Open. 2021;4:e2035057. doi: 10.1001/jamanetworkopen.2020.35057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ngwira L.G., Sharma B., Shrestha K.B., Dahal S., Tuladhar R., Manthalu G., Chilima B., Ganizani A., Rigby J., Kanjerwa O., et al. Cost of wastewater-based environmental surveillance for SARS-CoV-2: evidence from pilot sites in Blantyre, Malawi and Kathmandu, Nepal. PLOS Glob. Public Health. 2022;2:e0001377. doi: 10.1371/journal.pgph.0001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmitz B.W., Innes G.K., Prasek S.M., Betancourt W.Q., Stark E.R., Foster A.R., Abraham A.G., Gerba C.P., Pepper I.L. Enumerating asymptomatic COVID-19 cases and estimating SARS-CoV-2 fecal shedding rates via wastewater-based epidemiology. Sci. Total Environ. 2021;801:149794. doi: 10.1016/j.scitotenv.2021.149794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Islam M.A., Hossen F., Rahman M.A., Sultana K.F., Hasan M.N., Haque M.A., Sosa-Hernández J.E., Oyervides-Muñoz M.A., Parra-Saldívar R., Ahmed T., et al. An opinion on Wastewater-Based Epidemiological Monitoring (WBEM) with Clinical Diagnostic Test (CDT) for detecting high-prevalence areas of community COVID-19 infections. Curr. Opin. Environ. Sci. Health. 2023;31:100396. doi: 10.1016/j.coesh.2022.100396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Adhikari S., Halden R.U. Opportunities and limits of wastewater-based epidemiology for tracking global health and attainment of UN sustainable development goals. Environ. Int. 2022;163:107217. doi: 10.1016/j.envint.2022.107217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ahmed F., Islam M.A., Kumar M., Hossain M., Bhattacharya P., Islam M.T., Hossen F., Hossain M.S., Islam M.S., Uddin M.M., et al. First detection of SARS-CoV-2 genetic material in the vicinity of COVID-19 isolation Centre in Bangladesh: variation along the sewer network. Sci. Total Environ. 2021;776:145724. doi: 10.1016/j.scitotenv.2021.145724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur. J. Clin. Microbiol. Infect. Dis. 2021;40:2665–2667. doi: 10.1007/s10096-021-04304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amman F., Markt R., Endler L., Hupfauf S., Agerer B., Schedl A., Richter L., Zechmeister M., Bicher M., Heiler G., et al. Viral variant-resolved wastewater surveillance of SARS-CoV-2 at national scale. Nat. Biotechnol. 2022;40:1814–1822. doi: 10.1038/s41587-022-01387-y. [DOI] [PubMed] [Google Scholar]
- 15.Chaudhuri A., Pangaria A., Sodhi C., Kumar V N., Harshe S., Parikh N., Shridhar V. Building health system resilience and pandemic preparedness using wastewater-based epidemiology from SARS-CoV-2 monitoring in Bengaluru, India. Front. Public Health. 2023;11:1064793. doi: 10.3389/fpubh.2023.1064793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Daigle J., Racher K., Hazenberg J., Yeoman A., Hannah H., Duong D., Mohammed U., Spreitzer D., Gregorchuk B.S.J., Head B.M., et al. A sensitive and rapid wastewater test for SARS-COV-2 and its use for the early detection of a cluster of cases in a remote community. Appl. Environ. Microbiol. 2022;88:e0174021. doi: 10.1128/aem.01740-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fernandez-Cassi X., Scheidegger A., Bänziger C., Cariti F., Tuñas Corzon A., Ganesanandamoorthy P., Lemaitre J.C., Ort C., Julian T.R., Kohn T. Wastewater monitoring outperforms case numbers as a tool to track COVID-19 incidence dynamics when test positivity rates are high. Water Res. 2021;200:117252. doi: 10.1016/j.watres.2021.117252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitzgerald S.F., Rossi G., Low A.S., McAteer S.P., O’Keefe B., Findlay D., Cameron G.J., Pollard P., Singleton P.T.R., Ponton G., et al. Site specific relationships between COVID-19 cases and SARS-CoV-2 viral load in wastewater treatment plant influent. Environ. Sci. Technol. 2021;55:15276–15286. doi: 10.1021/acs.est.1c05029. [DOI] [PubMed] [Google Scholar]
- 19.Girón-Guzmán I., Díaz-Reolid A., Truchado P., Carcereny A., Garcia-Pedemonte D., Hernaez B., Bosch A., María Pintó R., Guix S., Allende A., et al. Wastewater based epidemiology beyond SARS-CoV-2: Spanish wastewater reveals the current spread of Monkeypox virus. medRxiv. 2022 doi: 10.1101/2022.09.19.22280084. Preprint at. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hewitt J., Trowsdale S., Armstrong B.A., Chapman J.R., Carter K.M., Croucher D.M., Trent C.R., Sim R.E., Gilpin B.J. Sensitivity of wastewater-based epidemiology for detection of SARS-CoV-2 RNA in a low prevalence setting. Water Res. 2022;211:118032. doi: 10.1016/j.watres.2021.118032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huisman J.S., Scire J., Caduff L., Fernandez-Cassi X., Ganesanandamoorthy P., Kull A., Scheidegger A., Stachler E., Boehm A.B., Hughes B., et al. Wastewater-based estimation of the effective reproductive number of SARS-CoV-2. Environ. Health Perspect. 2022;130:057011. doi: 10.1289/EHP10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jahn K., Dreifuss D., Topolsky I., Kull A., Ganesanandamoorthy P., Fernandez-Cassi X., Bänziger C., Devaux A.J., Stachler E., Caduff L., et al. Early detection and surveillance of SARS-CoV-2 genomic variants in wastewater using COJAC. Nat. Microbiol. 2022;7:1151–1160. doi: 10.1038/s41564-022-01185-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jakariya M., Ahmed F., Islam M.A., Al Marzan A., Hasan M.N., Hossain M., Ahmed T., Hossain A., Reza H.M., Hossen F., et al. Wastewater-based epidemiological surveillance to monitor the prevalence of SARS-CoV-2 in developing countries with onsite sanitation facilities. Environ. Pollut. 2022;311:119679. doi: 10.1016/j.envpol.2022.119679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karthikeyan S., Levy J.I., De Hoff P., Humphrey G., Birmingham A., Jepsen K., Farmer S., Tubb H.M., Valles T., Tribelhorn C.E., et al. Wastewater sequencing reveals early cryptic SARS-CoV-2 variant transmission. Nature. 2022;609:101–108. doi: 10.1038/s41586-022-05049-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.La Rosa G., Iaconelli M., Veneri C., Mancini P., Bonanno Ferraro G., Brandtner D., Lucentini L., Bonadonna L., Rossi M., Grigioni M., SARI network. Suffredini E. The rapid spread of SARS-COV-2 Omicron variant in Italy reflected early through wastewater surveillance. Sci. Total Environ. 2022;837:155767. doi: 10.1016/j.scitotenv.2022.155767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee W.L., Imakaev M., Armas F., McElroy K.A., Gu X., Duvallet C., Chandra F., Chen H., Leifels M., Mendola S., et al. Quantitative SARS-CoV-2 Alpha variant B.1.1.7 tracking in wastewater by allele-specific RT-qPCR. Environ. Sci. Technol. Lett. 2021;8:675–682. doi: 10.1021/acs.estlett.1c00375. [DOI] [Google Scholar]
- 27.Lodder W., de Roda Husman A.M. SARS-CoV-2 in wastewater: potential health risk, but also data source. Lancet. Gastroenterol. Hepatol. 2020;5:533–534. doi: 10.1016/S2468-1253(20)30087-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth D.S., Trujillo M., Gregory D.A., Cheung K., Gao A., Graham M., Guan Y., Guldenpfennig C., Hoxie I., Kannoly S., et al. Tracking cryptic SARS-CoV-2 lineages detected in NYC wastewater. Nat. Commun. 2022;13:635. doi: 10.1038/s41467-022-28246-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wade M.J., Lo Jacomo A., Armenise E., Brown M.R., Bunce J.T., Cameron G.J., Fang Z., Farkas K., Gilpin D.F., Graham D.W., et al. Understanding and managing uncertainty and variability for wastewater monitoring beyond the pandemic: lessons learned from the United Kingdom national COVID-19 surveillance programmes. J. Hazard Mater. 2022;424:127456. doi: 10.1016/j.jhazmat.2021.127456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wannigama D.L., Amarasiri M., Hurst C., Phattharapornjaroen P., Abe S., Hongsing P., Rad S.M.A.H., Pearson L., Saethang T., Luk-In S., et al. Tracking COVID-19 with wastewater to understand asymptomatic transmission. Int. J. Infect. Dis. 2021;108:296–299. doi: 10.1016/j.ijid.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahmed W., Tscharke B., Bertsch P.M., Bibby K., Bivins A., Choi P., Clarke L., Dwyer J., Edson J., Nguyen T.M.H., et al. SARS-CoV-2 RNA monitoring in wastewater as a potential early warning system for COVID-19 transmission in the community: a temporal case study. Sci. Total Environ. 2021;761:144216. doi: 10.1016/j.scitotenv.2020.144216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McMahan C.S., Self S., Rennert L., Kalbaugh C., Kriebel D., Graves D., Colby C., Deaver J.A., Popat S.C., Karanfil T., Freedman D.L. COVID-19 wastewater epidemiology: a model to estimate infected populations. Lancet Planet. Health. 2021;5:e874–e881. doi: 10.1016/s2542-5196(21)00230-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peccia J., Zulli A., Brackney D.E., Grubaugh N.D., Kaplan E.H., Casanovas-Massana A., Ko A.I., Malik A.A., Wang D., Wang M., et al. Measurement of SARS-CoV-2 RNA in wastewater tracks community infection dynamics. Nat. Biotechnol. 2020;38:1164–1167. doi: 10.1038/s41587-020-0684-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Radu E., Masseron A., Amman F., Schedl A., Agerer B., Endler L., Penz T., Bock C., Bergthaler A., Vierheilig J., et al. Emergence of SARS-CoV-2 Alpha lineage and its correlation with quantitative wastewater-based epidemiology data. Water Res. 2022;215:118257. doi: 10.1016/j.watres.2022.118257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weidhaas J., Aanderud Z.T., Roper D.K., VanDerslice J., Gaddis E.B., Ostermiller J., Hoffman K., Jamal R., Heck P., Zhang Y., et al. Correlation of SARS-CoV-2 RNA in wastewater with COVID-19 disease burden in sewersheds. Sci. Total Environ. 2021;775:145790. doi: 10.1016/j.scitotenv.2021.145790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wright J., Driver E.M., Bowes D.A., Johnston B., Halden R.U. Comparison of high-frequency in-pipe SARS-CoV-2 wastewater-based surveillance to concurrent COVID-19 random clinical testing on a public U.S. university campus. Sci. Total Environ. 2022;820:152877. doi: 10.1016/j.scitotenv.2021.152877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sangsanont J., Rattanakul S., Kongprajug A., Chyerochana N., Sresung M., Sriporatana N., Wanlapakorn N., Poovorawan Y., Mongkolsuk S., Sirikanchana K. SARS-CoV-2 RNA surveillance in large to small centralized wastewater treatment plants preceding the third COVID-19 resurgence in Bangkok, Thailand. Sci. Total Environ. 2022;809:151169. doi: 10.1016/j.scitotenv.2021.151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wannigama D.L., Amarasiri M., Hongsing P., Hurst C., Modchang C., Chadsuthi S., Anupong S., Phattharapornjaroen P., Sm A.H.R., Fernandez S., et al. Multiple traces of monkeypox detected in non-sewered wastewater with sparse sampling from a densely populated metropolitan area in Asia. Sci. Total Environ. 2023;858:159816. doi: 10.1016/j.scitotenv.2022.159816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Deshpande A., Miller-Petrie M.K., Lindstedt P.A., Baumann M.M., Johnson K.B., Blacker B.F., Abbastabar H., Abd-Allah F., Abdelalim A., Abdollahpour I., et al. Mapping geographical inequalities in access to drinking water and sanitation facilities in low-income and middle-income countries, 2000–17. Lancet Global Health. 2020;8:e1162–e1185. doi: 10.1016/S2214-109X(20)30278-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Street R., Malema S., Mahlangeni N., Mathee A. Wastewater surveillance for covid-19: an African perspective. Sci. Total Environ. 2020;743:140719. doi: 10.1016/j.scitotenv.2020.140719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Naughton C.C., Roman F.A., Jr., Alvarado A.G.F., Tariqi A.Q., Deeming M.A., Kadonsky K.F., Bibby K., Bivins A., Medema G., Ahmed W., et al. Show us the data: global COVID-19 wastewater monitoring efforts, equity, and gaps. FEMS Microbes. 2023;4:xtad003. doi: 10.1093/femsmc/xtad003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lefrançois T., Malvy D., Atlani-Duault L., Benamouzig D., Druais P.-L., Yazdanpanah Y., Delfraissy J.-F., Lina B. After 2 years of the COVID-19 pandemic, translating One Health into action is urgent. Lancet. 2023;401:789–794. doi: 10.1016/S0140-6736(22)01840-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murakami M., Kitajima M., Endo N., Ahmed W., Gawlik B.M. The growing need to establish a global wastewater surveillance consortium for future pandemic preparedness. J. Travel Med. 2023:taad035. doi: 10.1093/jtm/taad035. [DOI] [PubMed] [Google Scholar]
- 44.Xiao K., Zhang L. Wastewater pathogen surveillance based on One Health approach. Lancet. Microbe. 2023;4:e297. doi: 10.1016/S2666-5247(23)00039-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Islam M.A., Rahman M.A., Jakariya M., Bahadur N.M., Hossen F., Mukharjee S.K., Hossain M.S., Tasneem A., Haque M.A., Sera F., et al. A 30-day follow-up study on the prevalence of SARS-COV-2 genetic markers in wastewater from the residence of COVID-19 patient and comparison with clinical positivity. Sci. Total Environ. 2023;858:159350. doi: 10.1016/j.scitotenv.2022.159350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Li X., Zhang S., Shi J., Luby S.P., Jiang G. Uncertainties in estimating SARS-CoV-2 prevalence by wastewater-based epidemiology. Chem. Eng. J. 2021;415:129039. doi: 10.1016/j.cej.2021.129039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monteiro S., Rente D., Cunha M.V., Gomes M.C., Marques T.A., Lourenço A.B., Cardoso E., Álvaro P., Silva M., Coelho N., et al. A wastewater-based epidemiology tool for COVID-19 surveillance in Portugal. Sci. Total Environ. 2022;804:150264. doi: 10.1016/j.scitotenv.2021.150264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin B., Dietrich M.L., Senior R.A., Wilcove D.S. A better classification of wet markets is key to safeguarding human health and biodiversity. Lancet Planet. Health. 2021;5:e386–e394. doi: 10.1016/s2542-5196(21)00112-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Worobey M., Levy J.I., Malpica Serrano L., Crits-Christoph A., Pekar J.E., Goldstein S.A., Rasmussen A.L., Kraemer M.U.G., Newman C., Koopmans M.P.G., et al. The huanan seafood wholesale market in Wuhan was the early epicenter of the COVID-19 pandemic. Science. 2022;377:951–959. doi: 10.1126/science.abp8715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Onyekuru N.A., Ume C.O., Ezea C.P., Chukwuma Ume N.N. Effects of ebola virus disease outbreak on bush meat enterprise and environmental health risk behavior among households in south-east Nigeria. J. Prim. Prev. 2020;41:603–618. doi: 10.1007/s10935-020-00619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gonçalves J., Torres-Franco A., Rodriguéz E., Diaz I., Koritnik T., Silva P.G.D., Mesquita J.R., Trkov M., Paragi M., Muñoz R., García-Encina P.A. Centralized and decentralized wastewater-based epidemiology to infer COVID-19 transmission - a brief review. One Health. 2022;15:100405. doi: 10.1016/j.onehlt.2022.100405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ferronato N., Torretta V. Waste mismanagement in developing countries: a review of global issues. Int. J. Environ. Res. Public Health. 2019;16:1060. doi: 10.3390/ijerph16061060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Scott L.C., Aubee A., Babahaji L., Vigil K., Tims S., Aw T.G. Targeted wastewater surveillance of SARS-CoV-2 on a university campus for COVID-19 outbreak detection and mitigation. Environ. Res. 2021;200:111374. doi: 10.1016/j.envres.2021.111374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gibas C., Lambirth K., Mittal N., Juel M.A.I., Barua V.B., Roppolo Brazell L., Hinton K., Lontai J., Stark N., Young I., et al. Implementing building-level SARS-CoV-2 wastewater surveillance on a university campus. Sci. Total Environ. 2021;782:146749. doi: 10.1016/j.scitotenv.2021.146749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bibby K., Bivins A., Wu Z., North D. Making waves: plausible lead time for wastewater based epidemiology as an early warning system for COVID-19. Water Res. 2021;202:117438. doi: 10.1016/j.watres.2021.117438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barua V.B., Juel M.A.I., Blackwood A.D., Clerkin T., Ciesielski M., Sorinolu A.J., Holcomb D.A., Young I., Kimble G., Sypolt S., et al. Tracking the temporal variation of COVID-19 surges through wastewater-based epidemiology during the peak of the pandemic: a six-month long study in Charlotte, North Carolina. Sci. Total Environ. 2022;814:152503. doi: 10.1016/j.scitotenv.2021.152503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reynolds L.J., Gonzalez G., Sala-Comorera L., Martin N.A., Byrne A., Fennema S., Holohan N., Kuntamukkula S.R., Sarwar N., Nolan T.M., et al. SARS-CoV-2 variant trends in Ireland: wastewater-based epidemiology and clinical surveillance. Sci. Total Environ. 2022;838:155828. doi: 10.1016/j.scitotenv.2022.155828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Egger D., Miguel E., Warren S.S., Shenoy A., Collins E., Karlan D., Parkerson D., Mobarak A.M., Fink G., Udry C., et al. Falling living standards during the COVID-19 crisis: quantitative evidence from nine developing countries. Sci. Adv. 2021;7:eabe0997. doi: 10.1126/sciadv.abe0997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Worldbank Thailand economic monitor inequality, opportunity and human capital. 2019. worldbank.org
- 60.Kueakulpattana N., Wannigama D.L., Luk-in S., Hongsing P., Hurst C., Badavath V.N., Jenjaroenpun P., Wongsurawat T., Teeratakulpisan N., Kerr S.J., et al. Multidrug-resistant Neisseria gonorrhoeae infection in heterosexual men with reduced susceptibility to ceftriaxone, first report in Thailand. Sci. Rep. 2021;11:21659. doi: 10.1038/s41598-021-00675-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Betancourt W.Q., Schmitz B.W., Innes G.K., Prasek S.M., Pogreba Brown K.M., Stark E.R., Foster A.R., Sprissler R.S., Harris D.T., Sherchan S.P., et al. COVID-19 containment on a college campus via wastewater-based epidemiology, targeted clinical testing and an intervention. Sci. Total Environ. 2021;779:146408. doi: 10.1016/j.scitotenv.2021.146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cori A., Ferguson N.M., Fraser C., Cauchemez S. A new framework and software to estimate time-varying reproduction numbers during epidemics. Am. J. Epidemiol. 2013;178:1505–1512. doi: 10.1093/aje/kwt133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wickham H., Averick M., Bryan J., Chang W., McGowan L., François R., Grolemund G., Hayes A., Henry L., Hester J., et al. Welcome to the tidyverse. J. Open Source Softw. 2019;4:1686. doi: 10.21105/joss.01686. [DOI] [Google Scholar]
- 64.Team R.C. R Foundation for Statistical Computing; 2021. R: A Language and Environment for Statistical Computing. [Google Scholar]
- 65.Magnusson A., Skaug H., Nielsen A., Berg C.W., Kristensen K., Mächler M., van Benthem K., Bolker B., Brooks M. 2017. glmmTMB: Generalized Linear Mixed Models Using Template Model Builder. [Google Scholar]
- 66.Garson J.A., Badru S., Badhan A., Dustan S., Tedder R.S. Development of highly specific singleplex and multiplex real-time reverse transcription PCR assays for the identification of SARS-CoV-2 Omicron BA.1, BA.2 and Delta variants. medRxiv. 2022 doi: 10.1101/2022.04.07.22273168. Preprint at. [DOI] [Google Scholar]
- 67.Vogels C.B.F., Brito A.F., Wyllie A.L., Fauver J.R., Ott I.M., Kalinich C.C., Petrone M.E., Casanovas-Massana A., Catherine Muenker M., Moore A.J., et al. Analytical sensitivity and efficiency comparisons of SARS-CoV-2 RT–qPCR primer–probe sets. Nat. Microbiol. 2020;5:1299–1305. doi: 10.1038/s41564-020-0761-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Medema G., Heijnen L., Elsinga G., Italiaander R., Brouwer A. Presence of SARS-coronavirus-2 RNA in sewage and correlation with reported COVID-19 prevalence in the early stage of the epidemic in The Netherlands. Environ. Sci. Technol. Lett. 2020;7:511–516. doi: 10.1021/acs.estlett.0c00357. [DOI] [PubMed] [Google Scholar]
- 69.Linton N.M., Kobayashi T., Yang Y., Hayashi K., Akhmetzhanov A.R., Jung S.M., Yuan B., Kinoshita R., Nishiura H. Incubation Period and other epidemiological characteristics of 2019 novel Coronavirus infections with right truncation: a statistical analysis of publicly available case data. J. Clin. Med. 2020;9:538. doi: 10.3390/jcm9020538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wilasang C., Sararat C., Jitsuk N.C., Yolai N., Thammawijaya P., Auewarakul P., Modchang C. Reduction in effective reproduction number of COVID-19 is higher in countries employing active case detection with prompt isolation. J. Travel Med. 2020;27:taaa095. doi: 10.1093/jtm/taaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bhaskaran K., Gasparrini A., Hajat S., Smeeth L., Armstrong B. Time series regression studies in environmental epidemiology. Int. J. Epidemiol. 2013;42:1187–1195. doi: 10.1093/ije/dyt092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodríguez Rasero F.J., Moya Ruano L.A., Rasero Del Real P., Cuberos Gómez L., Lorusso N. Associations between SARS-CoV-2 RNA concentrations in wastewater and COVID-19 rates in days after sampling in small urban areas of Seville: a time series study. Sci. Total Environ. 2022;806:150573. doi: 10.1016/j.scitotenv.2021.150573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zeileis A., Kleiber C., Jackman S. Regression models for count data in R. J. Stat. Softw. 2008;27:1–25. doi: 10.18637/jss.v027.i08. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data generated and analyzed during this study are included in this published article and its supplemental information file. As this study is ongoing, additional wastewater details will be available upon reasonable request from the corresponding author Dhammika Leshan Wannigama. The cumulative number of confirmed COVID-19 cases in Thailand attributed to each province was obtained from the Department of Disease Control, Ministry of Public Health, Thailand (https://data.go.th/dataset/covid-19-daily).
Data were analysed and plotted using the tidyverse 1.3.1,63 dplyr 1.0.7, splines 4.1.0, zoo 1.8-9, astsa 1.14, lubridate 1.7.10, patchwork 1.1.1, ggplot2 3.3.5, dslabs 0.7.4, scales 1.2.1, ggalt 0.4.0 and ggpubr 0.4.0 packages of R program version 4.1.0.64
The computer code for estimating the infection incidence is available at the published computer code by Huisman et al. (https://github.com/JSHuisman/wastewaterRe)21 with the parameterization and also original code is available in this paper’s supplemental information. To evaluate the data n days before sampling (t = − 7) to 21 days after sampling (t = 21), we used the dplyr package.62 GCV (Generalized Cross Validation) criterion in the ‘mgcv’5 R software package was used to find the best-fit model.65