Abstract
Over the years, much attention has been drawn to antibiotic resistance bacteria, but drug inefficacy caused by a subgroup of special phenotypic variants – persisters – has been largely neglected in both scientific and clinical field. Interestingly, this subgroup of phenotypic variants displayed their power of withstanding sufficient antibiotics exposure in a mechanism different from antibiotic resistance. In this review, we summarized the clinical importance of bacterial persisters, the evolutionary link between resistance, tolerance, and persistence, redundant mechanisms of persister formation as well as methods of studying persister cells. In the light of our recent findings of membrane-less organelle aggresome and its important roles in regulating bacterial dormancy depth, we propose an alternative approach for anti-persister therapy. That is, to force a persister into a deeper dormancy state to become a VBNC (viable but non-culturable) cell that is incapable of regrowth. We hope to provide the latest insights on persister studies and call upon more research interest into this field.
1. Introduction
The introduction of antibiotics into clinical use was inarguably one of the greatest medical breakthroughs of the 20th century. Fascinating as they are, overuse and misuse of these medications caused an antibiotic resistance crisis, threatening our ability to treat common infectious disease. Over the years, much attention has been drawn to resistance caused by gene mutations, but drug inefficacy caused by a subgroup of special phenotypic variants has been largely neglected.
This subgroup of phenotypic variants is called “Persisters”, representing their power of withstanding sufficient antibiotics exposure. When exposing an isogenic cell culture to antibiotics, bulk cells are rapidly killed, but the small subgroup of persisters can survive antibiotic challenge and resuscitate after antibiotic removal (Bigger, 1944). Indeed, the hallmark of antibiotic persistence is illustrated by the biphasic killing curve during antibiotic exposures. In this curve, persisters correspond to the significantly slower killing phase after the bulk of the bacterial population is eliminated during the first phase of rapid killing. Usually, the tolerance of persisters is seen as rooted in a dormant physiological state in which the targets of antibiotic drugs are inactive and thus cannot be poisoned or corrupted by the drug treatment, enabling bacterial survival (Harms et al., 2016; Lewis, 2010; Wood et al., 2013).
Persisters were first described by Joseph Bigger in 1944 when he investigated the action of penicillin on Staphylococcus pyogenes, a human specific bacterial pathogen causing a broad range of symptoms. However, it took a long time for the importance of this transient phenotype to be recognized, and this recognition is still a work in progress. Consequently, and not surprisingly, the study of antibiotic persistence has been in a slow pace.
1.1. Clinical importance of bacterial persisters
Chronic and relapsing infections are often associated with genetically susceptible bacteria that survive even massive and long-lasting antibiotic treatment (Fauvart et al., 2011; Levin & Rozen, 2006). This phenomenon is commonly linked to the formation of specialized “persister” cells. Antibiotics do not kill persisters that survive to live another day, fueling chronic and relapsing infections.
Incomplete eradication of tuberculosis during medical treatment allows pathogens to remain in the patients. This remaining bacterial population can result in refractory tuberculosis (Davies, 2001), where genetic mutation-conferred drug resistance may play a part, but bacterial persistence is believed to be the primary cause (Wayne, 1994). Many persisters are found in a microenvironment known as a granuloma (Davies, 2001; Honer Zu Bentrup & Russell, 2001), which creates difficult conditions such as high oxidative stress, low pH, and limited nutrients that induce persister formation (Davies, 2001; Honer Zu Bentrup & Russell, 2001; Marakalala et al., 2016; Mok et al., 2015; Rubin, 2009). M. tuberculosis cells cease to grow upon encountering this stressful environment, resulting in a non-replicating state that shields them from eradication by antibiotics (Voskuil et al., 2003). The percentage of persister cells can be as high as 1% of the whole M. tuberculosis population (Keren et al., 2011), highlighting the significant role of persisters in drug tolerance.
Salmonella typhimurium, a pathogen that causes systemic and enteric infections, shares similarities with M. tuberculosis in terms of the significance of non-heritable persistence in disease progression. The bacteria reside in professional phagocytes, particularly in macrophage cells, and can be tolerant to adverse conditions such as antibiotic treatment (LaRock et al., 2015; Monack et al., 2004; Rivera-Chavez & Baumler, 2015; Wain et al., 2015). Recent fluorescent single-cell analysis revealed S. typhimurium persisters could form non-replicating cells, which may serve as a reservoir for relapsing infection (Helaine et al., 2014). Some toxins, including sehA, relE-5, relE and vapC, are highly expressed in the cause of S. typhimurium infection of macrophages (Silva-Herzog et al., 2015). The S. typhimurium persisters may be capable of tolerating host environment stress and escaping the antibiotic's killing effects (Helaine & Holden, 2013; Helaine et al., 2014; Silva-Herzog et al., 2015).
Staphylococcus aureus is a pathogenic bacterium that causes infections in hospitals and communities, including wounds, endovascular, and bone infections (Conlon, 2014; Malani, 2013). These chronic infections have a high rate of relapse due to S. aureus's ability to adapt to the microenvironment (Conlon, 2014; Trouillet-Assant et al., 2016). S. aureus can form persister cells that help it survive the stressful environment during human infection (Trouillet-Assant et al., 2016), which may explain the need for long-term antibiotic treatment to eradicate the infection (Conlon, 2014).
Aside from M. tuberculosis, S. typhimurium, and S. aureus, many other pathogenic bacteria can cause chronic infections that are difficult to treat, such as brucellosis (Ahmed et al., 2016; von Bargen et al., 2012), lung infections caused by Pseudomonas aeruginosa in cystic fibrosis patients (Hazan et al., 2014; Mulcahy et al., 2010), melioidosis caused by Burkholderia pseudomallei (Butt et al., 2014; Nierman et al., 2015), and urinary tract infections by uropathogenic Escherichia coli (Niu et al., 2015; Norton & Mulvey, 2012).
1.2. Evolutionary link between resistance, tolerance, and persistence
One reason for the slow pace of persister studies is perhaps that scientific and public attention was more drawn to the crisis of antibiotic resistance. “Antibiotic resistance” is the genetically inherited ability of bacteria to reproduce consecutively in the presence of a certain type of antibiotic. The gold standard for measuring the level of antibiotic resistance is the Minimum Inhibitory Concentration (MIC), which indicates the minimum antibiotic concentration needed to prevent visible growth of bacteria. Higher resistance corresponds to a higher MIC. Resistance can be acquired through horizontal transfer of resistance gene elements between different strains (Du et al., 2018; Jacoby, 2009) or de novo mutations (Blair et al., 2015).
“Antibiotic persistence” enables a subpopulation of phenotypic variants to survive a bactericidal antibiotic treatment. Different from antibiotic resistance, persistence phenotype is not heritable. Persistent cells sub-cultured in fresh medium will demonstrate the same antibiotic susceptibility as the initial population, that is, only a subgroup of the population will exhibit the persistent phenotype. Furthermore, the level of persistence is independent of MIC, as persisters can tolerate antibiotic concentrations that far exceeds the MIC. The hallmark of persistence is the observation of biphasic killing curve during antibiotic exposure, in which the persister subset is represented by the second slow killing phase. The formation of persisters can occur as stochastic phenotypic switch, or triggered by stress, such as starvation or ROS exposure.
Apart from antibiotic resistance and persistence, Balaban et al. classified a third type of cells that can endure antibiotic killing, termed “antibiotic tolerance,” a transient ability to endure high concentrations antibiotic treatment because of long lag phase (Fig. 1) (Balaban et al., 2019; Brauner et al., 2016). The phenotype of tolerance is like persistence. However, tolerance phenotype is for the entire population and is often caused by mutations that disturbs cell growth. The killing process is significantly arrested without a change in MIC due to slow growth rate of tolerant strains. The method of the Minimum Duration of Killing 99% of the bacterial population (MDK99) is introduced to measure antibiotic killing rate of tolerant strains (Fridman et al., 2014).
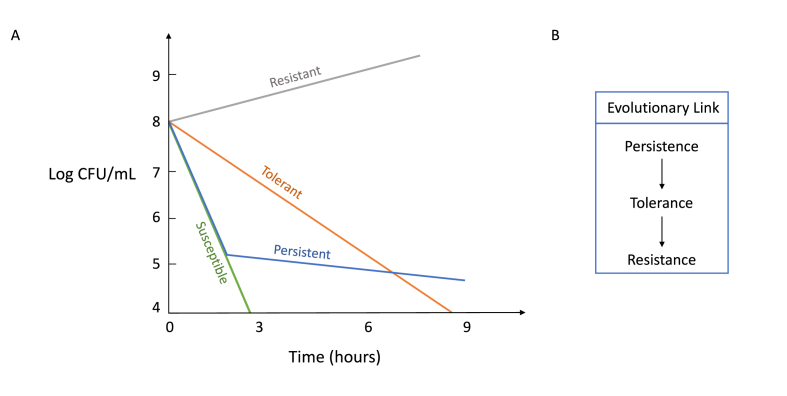
Fig. 1.
A) Hypothetical antibiotic killing curve is shown graphically. Bactericidal antibiotics (β-lactams, aminoglycosides, fluoroquinolones) are added at time zero, and the log CFU/mL are measured with respect to antibiotic exposure time. A susceptible population (green line) is rapidly killed by the addition of antibiotics. Persistent population (blue line) has the same MIC and antibiotic sensitivity as the susceptible population, yet a subpopulation of which remains viable for extended periods of time. Bacterial resistance and tolerance are attributes of the entire bacterial population. A resistant population (grey line) retains the ability to grow under antibiotic treatment. A tolerant population (orange line) is sensitive to antibiotics, but the kill rate is much higher than that of the susceptible population. B) The proposed evolutionary link between persistence, tolerance, and resistance. Persistence refers to the ability of a subpopulation (typically less than 1%) of bacteria to survive high dose of antibiotics, whereas tolerance refers to the same ability but pertains the entire population. Studies have shown that persistence and tolerance evolve rapidly under intermittent antibiotic exposure, which is of more relevant under clinical settings. Tolerance may also play a crucial role in the evolution of resistance when bacteria are under intermittent exposure to antibiotics.
Most experiments on the evolution of resistance have involved exposing bacterial strains to gradually increasing doses of antibiotics, starting from a sub-MIC level and reaching high MIC levels through the accumulation of resistant mutations over time, as demonstrated by studies conducted by Sun et al. (2009), Toprak et al. (2012), and Baym et al. (2016). However, the pharmacokinetics of many antibiotics in patients often involve intermittent, rather than constant, concentrations. Therefore, understanding the evolution of resistance under intermittent antibiotic exposures is of great interest, as pointed out by Moreillon et al. (1988). When bacteria are exposed to antibiotics intermittently, the most favorable evolutionary outcome is the emergence of a highly tolerant population with extended lag time. This is because cells that remain in the lag phase during each duration of antibiotic exposure are more likely to survive, as demonstrated by studies conducted by Levin-Reisman et al. (2010) and Fridman et al. (2014).
In clinical settings, higher concentrations of antibiotics are often used to achieve levels above the mutation prevention concentration (MPC) and avoid the gradual increase of minimum inhibitory concentration (MIC). In environments with high concentrations of antibiotics, single resistant mutations that arise directly from a susceptible strain are likely to be eliminated. Both in vitro evolution experiments (Levin-Reisman et al., 2017) and in vivo evolution studies conducted on patients undergoing treatment (Liu et al., 2020) have demonstrated that populations of bacteria that become genetically resistant to the antibiotic tend to do so on a foundation of tolerance mutations. Tolerant organisms have a higher likelihood of surviving, and therefore, they have a greater chance of subsequently acquiring resistance mutations. Another reason is the significant variation in the number of genes involved in each process. While only a few genes are responsible for conferring resistance, tolerance involves numerous genes (Levin-Reisman et al., 2017; Lewis & Shan, 2017). As a result, in clinical settings, the accumulation of resistant mutations is only possible after the establishment of antibiotic tolerance. Therefore, the expected evolutionary trajectory under intermittent high concentrations of antibiotics, which is more reflective of pharmacokinetics in patients, begins with the evolution of antibiotic tolerance from antibiotic persistence and subsequently leads to antibiotic resistance, as demonstrated by studies conducted by (Fridman et al., 2014; Levin-Reisman et al., 2017; Liu et al., 2020). Consequently, bacterial persistence is not only clinically important, but also has evolutionary implications.
1.3. Methods in studying persister cells
The slow pace in the study of persisters has been not only due to the late realization of their critical roles in both evolution and clinic, but also due to objective difficulties in studying transient phenotype variants in a heterogeneous population. The recent development of single cell technology, including fluorescence-activated cell sorting (FACS), microfluidics, microscopy, and single-cell RNA-seq, makes in-depth study of antibiotic persistence possible. Yet, the methods of FACS, microfluidics and microscopy strongly depend on successful labeling of persister cells.
2. Persister isolation
Researchers have developed various methods to isolate persistence. One classic way was fusing a degradable green fluorescent protein (GFP) behind a ribosome promoter, rrnBP1-GFP, which is only active in dividing cells (Shah et al., 2006). “Dim” cells in the population correspond to dormant cells, in which persisters are enriched. More markers have been proposed based on the low energy status of persisters. Citrate synthase (GltA), isocitrate dehydrogenase (Icd), and α-ketoglutarate dehydrogenase (SucA) are critical enzymes in the TCA cycle, which links to membrane potential and ATP production. Chromosomal mVenus translational fusions gltA-mVenus, icd-mVenus, sucA-mVenus were constructed (Manuse et al., 2021). When exposed to ciprofloxacin, cells that had “dim” fluorescence intensity had higher survival rate compared to “bright” and “middle” intensity cells, suggesting that low energy production is associated with persister formation. Different from E. coli (Shah et al., 2006), Staphylococcus aureus persisters are produced due to a stochastic entrance into stationary phase (Conlon et al., 2016). Cap5A used to be a stationary phase cell marker, and it now can be a good marker for persister S. aureus. By fusing cap5A promoter region with GFP fluorescent protein, researchers were able to sort these population based on fluorescence intensity (dim, medium, high) which correlates to cap5A expression. Only the cells with the brightest fluorescence with the most expression of cap5A were able to survive antibiotics.
The difficulty in isolating persisters from other cell types to sufficient purity lies in their low abundance, transient nature, and their similarity to the more highly abundant viable but non-culturable cells (VBNCs). The use of FACS based on fluorescence labeling of persisters addressed this technical hurdle and help to quantify persister levels in a population (Allison et al., 2011; Orman and Brynildsen, 2013). This assay provides persister phenotype distributions and can also be used to examine persister heterogeneity. Combining FACS, antibiotic tolerance assay with next generation sequencing, Brynildsen's group developed persister-FACSeq, a method to massively parallelize quantification of persister physiology and its heterogeneity. Persister-FACSeq can be applied to study persistence to any antibiotic in any environment for any bacteria that harbors a fluorescent reporter. Through this method, they found that persistence to ofloxacin is inversely correlated with the capacity of protein synthesis in non-growing cells (Henry & Brynildsen, 2016).
3. Single-cell time-lapse microcopy
Persister cells are phenotypic variants present at low frequency in isogenic populations. Therefore, time-lapse microscopy of single-cell resolution is a gold-standard method to investigate antibiotic persistence. Balaban and colleagues were pioneered in coupling timelapse microcopy with microfluidic device to observe E. coli persister cells. Based on the observation, they modeled two types of persister cells: Type I persisters are stressed induced and type II are stochastically generated in exponentially growing populations (Balaban et al., 2004). Coupling microfluidics with time-lapse microscopy, the study of persisters entered a single-cell era: James Collins' group monitored indole-induced persister formation using microfluidics under the microscope, and proposed that indole signaling ignites a subgroup of cells against antibiotics by activating stress responses, leading to persister formation (Vega et al., 2012); John McKinney's group found that Mycobacterium smegmatis persists by dividing in the presence of the drug isoniazid, negatively correlated with KatG pulsing (Wakamoto et al., 2013); Pu et al. showed that dormant cells have enhanced efflux activity, a double-safe strategy adopted by persister cells to ensure their survival (Pu et al., 2016); Pu et al. also showed that membraneless aggresome formation can induce antibiotic persistence, and eliminating aggresomes is a prerequisite of persister relapsing (Pu et al., 2019).
4. Perspectives of single-cell RNA-seq (scRNA-seq) in persister study
Ever since the method was first published in 2009 (Tang et al., 2009), scRNA-seq has received broad interest in the eukaryote field. One of the biggest advantages of this method is that scRNA-seq can describe transcriptome in individual cells from a heterogeneity population with high resolution and in a genomic scale, which seems quite promising if we can use it in the study of bacterial persisters. However, due to extremely small transciptome size and the absence of effective mRNA polyadenylation tails, the development of scRNA-seq in prokaryotes is exceptionally difficult. Recently, based on the principle of split-pool barcoding, Seelig's group developed microSPLiT, a high throughput scRNA-seq method for bacteria that can resolve heterogeneous transcriptional states (Kuchina et al., 2021). Cai's and Newman's group jointly developed parallel sequential fluorescence in situ hybridization (par-seqFISH) to spy on microbial communities (Dar et al., 2021). These methods paved the way to analyze persister gene expression profiles from a heterogeneity population, which is otherwise not amenable to single-cell analysis in genomic scale.
4.1. Mechanisms underlying persister cell formation
The lack of a comprehensive understanding of the molecular mechanisms and physiological changes that truly underly antibiotic tolerance of persisters has been the current situation in the field. One major obstacle for the development of effective treatments against persisters is that there is no sole target. Furthermore, research progress has been complicated not only by the redundant persister formation pathways, but also by the adoption of different bacterial model systems, methodologies, and definitions in the field (Balaban et al., 2013; Kaldalu et al., 2016).
The origins of persister cells are multifaceted. They can arise spontaneously due to stochastic fluctuations in gene expression within a bacterial population. Furthermore, stress-induced mechanisms such as the SOS and stringent responses, as well as exposure to antibiotics, may also induce persister formation. These stressors can trigger toxin-antitoxin modules, aggresome and biofilm formation, ultimately resulting in reduced ATP levels and protein translation. Nonetheless, persister cells could recover once the stress has been removed or when optimal growth conditions are present. Although other sources have thoroughly explored this topic, we will offer a brief overview and provide some insights into the stress-induced mechanisms that lead to persister formation.
5. Toxin-antitoxin (TA) system
TA systems are small genetic modules comprising a stable toxic protein and a labile antitoxin that neutralizes its cognate toxin. There are six types of TA system, and the above mentioned hipBA locus belongs to type II TA system. In steady-state conditions, the antitoxin forms a complex with the toxin in which the toxic activity is neutralized. This complex also autorepresses the expression of its own system. Under stress conditions, environmental or physiological stimuli induces an imbalanced toxin:antitoxin ratio, which releases toxin activity and halt cell growth. However, except for some TAs with indisputable roles in persistence, such as HipBA and TisB/istR, many TA systems did not show any persistence phenotype in a substantial number of studies (Bernier et al., 2013; Goormaghtigh et al., 2018a; Harms et al., 2017; Norton & Mulvey, 2012). Therefore, the hypothesis of TA systems on bacterial persistence remains ambiguous. Nevertheless, gain of function mutations of TA system granting a hyper-persistent phenotype might confer a selective advantage under specific conditions (Levin-Reisman et al., 2017; Moyed & Bertrand, 1983), suggesting that wild-type TA systems may constitute a feasible reservoir for the ontogenesis of new functions, especially in persistence.
Frequent isolation of hipBA mutants from patients with chronic and relapsing infections is the most prominent evidence linking TA systems to persisters. The hipBA locus evolves rapidly, and hipA mutations frequently result in a significant increase in persistence (Moyed and Bertrand, 1983; Moyed and Broderick, 1986; Black et al., 1991; 1994). The majority of prokaryotic genomes contain toxin/antitoxin (TA) gene pairs, including hipBA. HipA is the stable toxin, while HipB is the labile antitoxin. HipA acts as a serine/threonine kinase, phosphorylating glutamyl-tRNA synthetase GltX, which prevents glutamate charging on its cognate tRNA (Germain et al., 2013; Kaspy et al., 2013). This results in the incorporation of uncharged tRNA at the ribosomal A site, which activates (p)ppGpp synthesis by releasing RelA enzyme from the ribosome (Potrykus and Cashel, 2008). The hipA7 mutant is a gain-of-function phenotype resulting from two-point mutations in the hipA open reading frame. Although HipA is normally inactive as a dimer, the hipA7 gain-of-function mutation occurs in the interface between the HipA dimers, allowing ATP to access the active site, activate the kinase, and lead to significantly more persisters in these mutants (Schumacher et al., 2015).
Clinical studies have shown that sequencing analysis of P. aeruginosa isolates obtained from patients with cystic fibrosis, one of the most challenging chronic infections, revealed hip gain-of-function mutants. These mutants accounted for approximately 25%–40% of all isolates. Screening of E. coli isolates from patients with frequent relapsed urinary tract infection also associates high persistence with hip mutants, with about half carrying mutations in hipA, and 5% of these being hipA7 (Schumacher et al., 2015). Introduction of hipA7 UTI isolates into bladder cells demonstrated significantly higher persistence compared to an isogenic wild-type strain. Sequencing clinical isolates of Mycobacterium tuberculosis from patients with long-term antibiotic medication also showed the common occurrence of hip isolates (Torrey et al., 2016). These studies strongly suggested that persisters are the primary cause of recalcitrant chronic infections.
6. Stringent response
The stringent response is a ubiquitous stress signaling pathway that enables bacteria to respond to amino acid starvation. Upregulation of cellular levels of the alarmone (p)ppGpp controlled gene expression profile switch is the hallmark of the response (Boutte & Crosson, 2013). Together with the transcription factor DksA, (p) ppGpp binds to the RNA polymerase, thereby downregulating genes necessary for rapid growth and upregulating genes important for survival (Boutte & Crosson, 2013). A considerable number of studies showed correlation between stringent response and persistence. The first line of evidence is the increased level of second messenger (p)ppGpp positively correlated with persistence (Amato et al., 2013; Fung et al., 2010; Korch et al., 2003; Pu et al., 2016), and (p)ppGpp is a key factor for the induction of various persister mechanisms (Hauryliuk et al., 2015; Korch et al., 2003; Verstraeten et al., 2015). The second line of evidence is that by deleting amino acid biosynthesis genes, hence inducing amino acid starvation, showed a strong increase in persistence (Bernier et al., 2013; Girgis et al., 2012). In addition, several downstream stringent response genes play a role in persistence, such as cspD (Yamanaka & Inouye, 1997) and phoU (Rice et al., 2009). However, to establish this link is unequivocally challenging, since in E. coli, disruption of (p)ppGpp synthetase gene relA and spoT are generally pleiotropic. In addition, knocking out all (p)ppGpp synthetases has no obvious effect on the growth and persister formation in S. aureus (Conlon et al., 2016).
7. SOS response
The bacterial SOS response coordinates the expression of the SOS regulon genes in response to DNA damage (Kreuzer, 2013). In E. coli, LexA and RecA are the major regulators, in which LexA act as a repressor by binding to the promoter region of multiple SOS genes (Brent & Ptashne, 1980; Giese et al., 2008; Luo et al., 2001), and RecA is a de-repressor for DNA damage (Little, 1991). The link between SOS response and persistence has been reviewed by mutagenetic studies of recA and lexA (Luidalepp et al., 2011; Debbia et al., 2001; Dö;rr et al., 2009). One of the mechanisms underlying this linkage is that the expression of tisB, a gene part of the SOS regulon, leads to the collapse of the membrane potential and a drop in ATP levels by punching holes on the inner membrane, thereby inducing persistence, and preventing further DNA damage (Dö;rr et al., 2010). In addition, many genes from the SOS response system may play a role in persistence, as these genes were upregulated in ciprofloxacin induced persisters (Pu et al., 2016) and the corresponding mutants display lower persister ratio. Particularly, the SOS response in persistence induction is significant in exponential phase while modest in stationary phase (Völzing & Brynildsen, 2015).
As mentioned above, mechanisms underlying antibiotic persistence is quite redundant and complex. Other mechanisms, such as general stress response, i.e., heat shock (Wallace et al., 2015) and starvation (Pu et al., 2019), may also induce persistence. However, none of these can represent a general mechanism of persister formation. Kim Lewis proposed that relative dormancy, with low levels of metabolic activity and ATP, is a possible general mechanism (Conlon et al., 2016). This hypothesis would be intuitive because the killing processes caused by bactericidal antibiotics generally depend on ATP as energy source and low ATP level could induce cellular proteostasis disruption (Pu et al., 2019). Moreover, other researchers in the field leaned more frequently towards the concept of “Persistence As Stuff Happens (PASH)” (Johnson, Levin, Levin, Li, & Karger, 2013; Levin et al., 2014; Vazquez-Laslop et al., 2006). That is, various types of heterogeneous persisters in a population are formed via different molecular mechanisms by chance (Levin et al., 2014). The existence of PASH seems undisputable since all attempts in creating a non-persister mutant have failed (Johnson et al., 2013). It's worth noting that PASH is not mutually exclusive with ATP depletion as the actual cause of persistence, and this is indeed the mechanism by which TisB produces persisters (Dorr et al., 2010), and bacterial persisters are a stochastically formed subpopulation of low-energy cells (Manuse et al., 2021).
7.1. A manifesto of aggresomes in antibiotic persistence
7.1.1. Dormancy depth: the physiological origin of lag time
Lag time is a temporal period of delayed growth when bacteria are adjusting to a new environment before starting exponential growth. The antibiotic persistence could be explained by the extension of the lag time of individual cells (Balaban et al., 2004). That is, when most cells in a population reach to exponential growth, a fraction of cells are still in lag phase and therefore tolerant to antibiotic treatment (Jõ;ers et al., 2010; Levin-Reisman et al., 2010). To quantitatively establish correlation between lag time and bacterial antibiotic persistence, we observed the regrowth process of survivors at a single cell resolution under the microscope. After antibiotic removal, some survivors resumed growth very soon, meeting the classic definition of persisters, which we referred to as persister-fast-recovery (persister-FR). Some survivors used a longer time to escape from dormancy, which we defined as persister-slow-recovery (persister-SR). The remainder of the surviving cells remained dormant even after 3 days (viable but non-culturable cells, VBNC cells) (Ayrapetyan et al., 2015; Pu et al., 2019). These results revealed that drug-tolerant cells showed a broad distribution of lag times. Based on these observations, we postulated that the degree to which drug-tolerant cells are dormant can be measured by a parameter we term ‘‘dormancy depth” (Fig. 2). As dormancy shields cells from antibiotic killing effects, dormancy depth regulates whether survival cells will resuscitate as well as the lag time duration. Although lag time is a direct and measurable quantity, with ‘‘dormancy depth’’ we can provide a unified framework for understanding the formation of both persisters and VBNC cells and revealing the physiological origin of lag time. This dormancy depth model has important value in demonstrating that instead of a simple binary active-dormant state, as has been a common assumption, dormancy itself is a heterogeneous physiology state which can be characterized quantitatively as having differing depths. This dormancy depth analysis does not touch directly upon the underlying molecular mechanisms of cell resuscitation as such, since the underlying reasons are clearly varied and manifold.
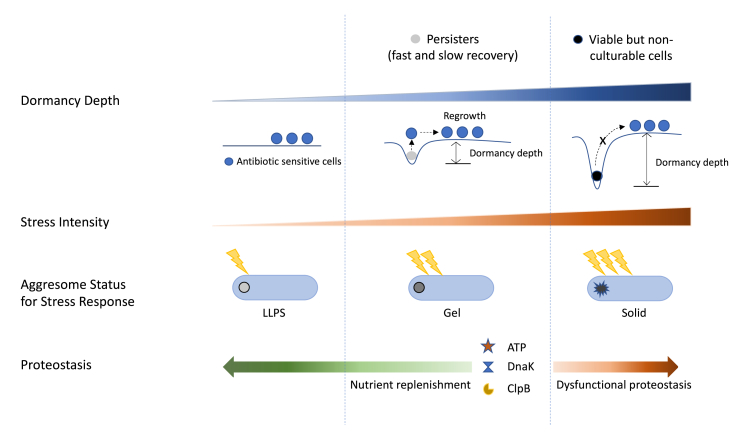
Fig. 2.
The persister phase is indicated in the middle vertical section. Viable but non-culturable (VBNC) phase is indicated in the right vertical section. Dormancy depth and stress intensity increases from left to right. Persister cells have shallow dormancy depth, therefore they can easily recover and regrow. VBNC cells cannot escape from deep dormancy state to achieve regrowth. Aggresome is an indicator for dormancy depth. The aggresome status (LLPS, gel, solid) in response to different stresses are illustrated under each vertical section. In nutrient rich environment, ATP is replenished, and functional protease complexes are recruited to disaggregate aggresomes, therefore facilitate bacterial resuscitation. On the contrary, dysfunctional protease machinery generates more VBNCs.
7.1.2. Aggresome: an indicator of dormancy depth
In bright-field microscopy observations, we noticed that a novel phenotypic feature—dark foci— is often associated with dormant cells but is not present in actively growing cells. By using special fluorescent labeling method and mass spectrometry, the dark foci were proved to be protein aggresomes, a collection of proteome-wide protein aggregates formed when intracellular proteostasis is disrupted. A time-lapse mass spectrometry analysis revealed that the partition of proteins in different biological processes into aggresomes may abide some level of sequential order. By using a time-lapse microscopy, Jin et al. followed the process of how proteins HslU, Kbl, and AcnB were incorporated into aggresomes. They found that the incorporation process happened at different characteristic time scales. Above evidence supports the existence of cell dormancy depth. By using TC-FlAsH labeling method, of which the signal of fluorescence is proportional to the degree of protein aggregation, we further established the connection between dormancy depth and the degree of protein aggregation in aggresomes. Induction of aggresomes by heat shock, streptomycin or hydrogen peroxide can promote cell entry into dormant state. Particularly, aggresomes are highly heterogeneous in terms of protein/RNA composition and physical state when induced by different stresses (Jin et al., 2021; Pu et al., 2019), implying that by tuning the material composition and properties of aggresomes, bacterial cells set the strength of stress response and determine dormancy depth. In contrast, preventing aggresome formation by impairing protein synthesis would stop a cell from sliding into a dormant state. Taken together, these results suggested that the extent of aggresome formation is a good indicator of bacterial cell dormancy depth.
7.1.3. The mechanisms underlying aggresome formation and clearance
We revealed that aggresomes are dynamic, reversible structures that is formed under stressed conditions and disassemble when stress is removed (Pu et al., 2019). Jin et al. exploited experiment and multiscale modeling in tandem to determine the molecular biophysics of aggresomes in a spatiotemporal manner. Through model-interpreted time-resolved microscopy, they revealed that the aggresomes are membraneless liquid droplets that will phase separate following clustering of diffusing proteins under thermal equilibrium in the bacterial cytoplasm (Jin et al., 2021). The findings implied that bacterial cells potentially exploit liquid-liquid phase separation (LLPS) processes in their stress-adaptive system to increase their fitness in changing environment (Fig. 2). LLPS is reported to be highly sensitive to changes of certain parameters, such as pH, temperature, and salt and molecular interactions (Alberti et al., 2019; Kroschwald et al., 2018; Wallace et al., 2015). Therefore, LLPS could perhaps serves as a highly reversible and sensitive way to compartmentalize cell cytoplasm in the absence of membranes. In our work, we demonstrated that the formation of aggresome is driven by depletion of intracellular ATP, which not only energizes cellular processes but also acts as a hydrotrope to increase specific protein solubility (Patel et al., 2017; Pu et al., 2019).
By tracking the resuscitation process of persister cells, we observed that the aggresomes disappear before persisters re-enter the growth cycle but persevere in VBNC cells (Pu et al., 2019). This phenomenon implies that aggresome dissolution and proteostasis restoration are critical steps for persisters to escape dormancy and resume growth. However, at physiological concentrations, the hydrotropic activity of ATP alone is not able to facilitate aggresome disintegration. Through mutagenesis, we revealed that during nutrient replenishment, the DnaK-ClpB bichaperone system played a critical role in the clearance of aggresomes in an ATP-dependent manner (Fig. 2). Through time-lapse microscopy, we demonstrated that the ability to recruit functional DnaK-ClpB machineries determines the lag time for bacterial regrowth. In contrast, dysfunction of the bichaperone system generates more VBNCs but fewer persisters, indicating that dormant cells with deficient disaggregation apparatus, regardless of shallow or deep dormancy depth, encounter difficulties in escaping the dormancy trap. Based on these results, we postulated the relationship between dormancy depth and lag time: lag time = dormancy depth/escaping speed = quantity of aggresomes present/rate of aggresome disaggregation.
7.2. Therapeutic perspectives against persisters
Due to the transient metabolic inactive phenotype, persisters are largely responsible for the ineffective elimination by conventional antibiotics, fueling chronic and relapsing infections. Persisters could also serve as “steppingstone” to antibiotic resistance in clinical settings where bacterial pathogens encounter intermittent high-dose antibiotic exposures (Fig. 1) (Levin-Reisman et al., 2017). Hence, in the battle against bacterial infection, development of effective anti-persister treatment regime is of great importance, yet this urgency is often overlooked in clinical settings (Table 1).
Table 1.
Various anti-persister approaches.
| Approaches | Mechanisms |
|---|---|
| Addition of carbon source with antibiotics (Allison et al., 2011; Orman & Brynildsen, 2013; Taber et al., 1987) | Resuscitates persisters and allows cells to increase intake of conventional antibiotics |
| Addition of efflux pump inhibitors with antibiotics (phenylalanine arginyl β-naphthylamide (PAbN) or 1-(1-Naphthylmethyl) piperazine (NMP) (Pu et al., 2016) | Blocks efflux of conventional antibiotics |
| Acyldepsipeptide (ADEP 4 (Carney et al., 2014; Conlon et al., 2013; Lee et al., 2010; Li et al., 2010) | Activates ClpP to be a non-specific protease and kills persister cells by uncontrollable protein degradation |
| Defensins, cathelicidins (Defraine et al., 2018; Mohammad et al., 2015) HT61 (Hu et al., 2010; Hubbard et al., 2017a) |
Disrupts bacterial cell membrane in a growth-independent manner |
| KCN (Respiration inhibitor) (Orman & Brynildsen, 2015) | Impairs stationary phase respiratory activity and prevents self-digestion of endogenous proteins and RNA, which yields bacteria that are more capable of translation, replication and concomitantly cell death when exposed to antibiotics |
| M64 (MvfR inhibitor) (Allegretta et al., 2017; Starkey et al., 2014; Vieira et al., 2022) | Targets quorum sensing regulator and inhibits persister formation |
| Relacin (Wexselblatt et al., 2012) | Affects entry into stationary phase in several Gram-positive bacteria, leading reduction in cell survival |
| Cis-2-decenoic acid (Marques et al., 2014) | Increases metabolic activity and reverts persisters into an antibiotic-susceptible state |
7.2.1. Sensitizing persister cells before antibiotic killing
In response to this urgency, several strategies and drugs against persisters have been developed in the scientific field (Table 1). One approach is by adding carbon sources: to resuscitate persisters and allow cells to be susceptible to conventional antibiotics. This approach is to potentiate the killing effects of antibiotics, thus considered as a quick route. For example, the transportation of aminoglycosides through the cell membrane is dependent upon proton motive force (PMF) (Taber et al., 1987), which is considerably low in persister cells. Allison et al. demonstrated that carbon sources such as mannitol, glucose, fructose, or pyruvate can activate PMF and significantly increase the uptake of gentamicin by E. coli persisters. As a result, the survival of persisters decreased by 99.9% (Allison et al., 2011). The effectiveness of this adjuvant-antibiotic pair was further verified in a chronic urinary tract infection mouse model. Interestingly, among these carbon sources, only fructose is effective against S. aureus in combination with gentamycin. In Orman and Brynildsen’ study, 60 mM of glycerol, pyruvate, mannitol, glucose, or fructose in combination with kanamycin (25 μg/mL) decreased the survival rate of E. coli persister cells by 99.9% (Orman & Brynildsen, 2013).
7.2.2. Adding adjuvants with antibiotics to kill persisters
An attractive approach for new therapies to address persister cells is to develop antibiotic adjuvants that work effectively when combined with antibiotics but have no effect on their own. Maiden et al. discovered that when the FDA-approved compound triclosan, which targets bacteria primarily by inhibiting fatty acid synthesis, was highly effective in killing persister cells of P. aeruginosa when used in combination with tobramycin. Within 8 h, the combination resulted in a 100-fold reduction in persister cells, and complete eradication was achieved within 24 h. Triclosan also improved tobramycin's ability to kill multiple Burkholderia cenocepacia and Staphylococcus aureus clinical isolates grown as biofilms, which have a high proportion of persister cells. Moreover, triclosan demonstrated synergy with other aminoglycosides, including streptomycin or gentamicin (Maiden et al., 2018). Another adjuvant is by adding efflux pump inhibitors. Pu et al. demonstrated that persisters which possess active efflux and passive dormancy could survive antibiotic exposure. Therefore, efflux pump inhibitors, such as phenylalanine arginyl β-naphthylamide (PAbN) or 1-(1-Naphthylmethyl) piperazine (NMP), in combination with antibiotics have an anti-persister efficacy, exhibiting therapeutic promise (Pu et al., 2016).
7.2.3. Direct killing of persister cells
The direct elimination of persisters by targeting growth-independent elements is another anti-persister therapy. The most successful example is the utilization of bacterial serine protease ClpP activators, ADEPs (Carney et al., 2014; Conlon et al., 2013; Lee et al., 2010; Li et al., 2010). ClpP in combination with its ATPase chaperone can degrade proteins in an ATP-dependent manner (Brö;tz-Oesterhelt et al., 2005). However, after binding to ADEP4, the protease machinery becomes ATP-independent, resulting in uncontrolled protein degradation (Conlon et al., 2013). ADEP4 (5 μg/mL) is effective against methicillin-resistant Staphylococcus aureus (MRSA) persisters from stationary phase. In combination with minimal rifampicin (0.4 μg/mL), ADEP4 can completely eradicate persister cells (Conlon et al., 2013). Furthermore, antimicrobial peptides (AMPs) secreted by host immune system, such as defensins and cathelicidins, can efficiently disrupt bacterial cell membrane in a growth-independent manner, thus having activity against persisters (Defraine et al., 2018; Mohammad et al., 2015). Based on the findings, systemic approaches to identify membrane-active small molecules against persisters were carried out by several groups. Coates’ group identified HT61 as a promising candidate with persister killing capacity from 952,601 compounds (Hu et al., 2010), which is currently in clinical trials to determine the efficacy as a therapeutic agent (Hubbard, Coates, & Harvey, 2017a). The action mode of HT61 is to directly interact with bacterial lipid bilayers and induce the alternation of lipid bilayer structure, which consequently leads to membrane disruptions and ATP leakage.
7.2.4. Reducing persister formation
The identification of various approaches that disrupt the processes involved in persister formation has been facilitated by an increased understanding of the mechanisms that underlie persistence. This has resulted in a reduction in the number of persisters. For instance, MfvR, a quorum sensing regulator in P. aeruginosa, has been inhibited, leading to a decrease in persister numbers (Allegretta et al., 2017; Starkey et al., 2014). Inhibitors that prevent the accumulation of (p)ppGpp have also been developed for several Gram-positive species (Wexselblatt et al., 2012), and the phage-encoded expression of LexA3 and SoxR has been used to inhibit the SOS- and oxidative-stress responses in E. coli (Lu & Collins, 2009). Additionally, treatment with KCN has been shown to reduce the formation of E. coli type I persisters when fresh medium is added by inhibiting stationary phase respiration (Orman & Brynildsen, 2015). Although the elimination of existing persister cells can be effective in treating infections, inhibiting, or reducing their formation before antibiotic treatment can potentially prevent chronic infections.
7.2.5. Proposal of an alternative anti-persister therapy
Using extensive single-cell observation data, we have introduced a model of dormancy depth that portrays it as a continuum, rather than a singular homogeneous state. This model suggests that fast-recovered persisters exist in shallow depths of dormancy, while slow-recovered persisters reside in medium depths, and VBNC cells are found in very deep depths where the obstacle to re-initiate growth is insurmountable. In light of the discovery of the aggresome and its role in regulating dormancy depth, we propose an alternative approach to anti-persister therapy. Our proposal involves inducing a persister to enter a deeper state of dormancy, thereby transforming into a VBNC cell incapable of growth. One way to achieve this is by increasing the level of protein aggregation to an irreversible point, or by disrupting disaggregation machineries, such as by employing DnaK ATPase activity inhibitors to prevent persisters from escaping dormancy. Both strategies present new avenues for potential drug targets in the development of future antibiotics.
Declaration of competing interest
The authors declare no competing interests.
Acknowledgements
The work by the authors is supported by National Natural Science Foundation of China (31970089), Key Technologies Research and Development Program (2021YFC2701600), Natural Science Foundation of Hubei Province (2022CFB693), Hubei Province National Science Fund for Distinguished Young Scholars (2022CFA077) and National Key Research and Development Program of China (2022YFA0912200).
References
- Ahmed W., Zheng K., Liu Z.F. Establishment of chronic infection: Brucella's stealth strategy. Frontiers in Cellular and Infection Microbiology. 2016;6:30. doi: 10.3389/fcimb.2016.00030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberti S., Gladfelter A., Mittag T. Considerations and challenges in studying liquid-liquid phase separation and biomolecular condensates. Cell. 2019;176:419–434. doi: 10.1016/j.cell.2018.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allegretta G., Maurer M., Eberhard J., Maura D., Hartmann R., Rahme L., Empting M. In-depth profiling of MvfR-regulated small molecules in Pseudomonas aeruginosa after quorum sensing inhibitor treatment. Frontiers in Microbiology. 2017;8(1):924. doi: 10.3389/fmicb.2017.00924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allison K.R., Brynildsen M.P., Collins J.J. Metabolite-enabled eradication of bacterial persisters by aminoglycosides. Nature. 2011;473:216–220. doi: 10.1038/nature10069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amato S.M., Orman M.A., Brynildsen M.P. Metabolic control of persister formation in Escherichia coli. Molecular Cell. 2013;50:475–487. doi: 10.1016/j.molcel.2013.04.002. [DOI] [PubMed] [Google Scholar]
- Ayrapetyan M., Williams T.C., Baxter R., Oliver J.D. Viable but nonculturable and persister cells coexist stochastically and are induced by human serum. Infection and Immunity. 2015;83(11):4194–4203. doi: 10.1128/IAI.00404-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N.Q., Gerdes K., Lewis K., Mckinney J.D. A problem of persistence: Still more questions than answers? Nature Reviews Microbiology. 2013;11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- Balaban N.Q., Helaine S., Lewis K., Ackermann M., Aldridge B., Andersson D.I., Brynildsen M.P., Bumann D., Camilli A., Collins J.J., Dehio C., Fortune S., Ghigo J.-M., Hardt W.-D., Harms A., Heinemann M., Hung D.T., Jenal U., Levin B.R.…Zinkernagel A. Definitions and guidelines for research on antibiotic persistence. Nature Reviews Microbiology. 2019;1:441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaban N.Q., Merrin J., Chait R., Kowalik L., Leibler S. Bacterial persistenceas a phenotypic switch. Science. 2004;305:1622–1625. doi: 10.1126/science.1099390. [DOI] [PubMed] [Google Scholar]
- von Bargen K., Gorvel J.P., Salcedo S.P. Internal affairs: Investigating the Brucella intracellular lifestyle. FEMS Microbiology Reviews. 2012;36:533–562. doi: 10.1111/j.1574-6976.2012.00334.x. [DOI] [PubMed] [Google Scholar]
- Baym M., Lieberman T.D., Kelsic E.D., Chait R., Gross R., Yelin I., Kishony R. Spatiotemporal microbial evolution on antibiotic landscapes. Science. 2016;353:1147–1151. doi: 10.1126/science.aag0822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernier S.P., Lebeaux D., DeFrancesco A.S., Valomon A., Soubigou G., Coppee J.-Y., Ghigo J.-M., Beloin C. Starvation, together with the SOS Response, mediates high biofilm-specific tolerance to the fluoroquinolone ofloxacin. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigger J. Treatment of staphylococcal infections with penicillin by intermittent sterilisation. The Lancet. 1944;244:497–500. [Google Scholar]
- Black D.S., Irwin B., Moyed H.S. Autoregulation of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. Journal of Bacteriology. 1994;176:4081. doi: 10.1128/jb.176.13.4081-4091.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black D.S., Kelly A.J., Mardis M.J., Moyed H.S. Structure and organization of hip, an operon that affects lethality due to inhibition of peptidoglycan or DNA synthesis. Journal of Bacteriology. 1991;173:5732. doi: 10.1128/jb.173.18.5732-5739.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair J.M.A., Webber M.A., Baylay A.J., Ogbolu D.O., Piddock L.J.V. Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology. 2015;13:42–51. doi: 10.1038/nrmicro3380. [DOI] [PubMed] [Google Scholar]
- Boutte C.C., Crosson S. Bacterial lifestyle shapes stringent response activation. Trends in Microbiology. 2013;21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauner A., Fridman O., Gefen O., Balaban N.Q. Distinguishing between resistance, tolerance and persistence to antibiotic treatment. Nature Reviews Microbiology. 2016;14:320–330. doi: 10.1038/nrmicro.2016.34. [DOI] [PubMed] [Google Scholar]
- Brent R., Ptashne M. The lexA gene product represses its own promoter. Proceedings of the National Academy of Sciences. 1980;77:1932–1938. doi: 10.1073/pnas.77.4.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt A., Higman V.A., Williams C., Crump M.P., Hemsley C.M., Harmer N., Titball R.W. The HicA toxin from Burkholderia pseudomallei has a role in persister cell formation. Biochemical Journal. 2014;459:333–344. doi: 10.1042/BJ20140073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carney D.W., Schmitz K.R., Truong J.V., Sauer R.T., Sello J.K. Restriction of the conformational dynamics of the cyclic acyldepsipeptide antibiotics improves their antibacterial activity. Journal of the American Chemical Society. 2014;136:1922–1929. doi: 10.1021/ja410385c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B.P. Staphylococcus aureus chronic and relapsing infections: Evidence of a role for persister cells: An investigation of persister cells, their formation and their role in S. aureus disease. BioEssays. 2014;36:991–996. doi: 10.1002/bies.201400080. [DOI] [PubMed] [Google Scholar]
- Conlon B.P., Nakayasu E.S., Fleck L.E., Lafleur M.D., Isabella V.M., Coleman K., Leonard S.N., Smith R.D., Adkins J.N., Lewis K. Activated ClpP kills persisters and eradicates a chronic biofilm infection. Nature. 2013;503:365–370. doi: 10.1038/nature12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conlon B.P., Rowe S.E., Gandt A.B., Nuxoll A.S., Donegan N.P., Zalis E.A., Clair G., Adkins J.N., Cheung A.L., Lewis K. Persister formation in Staphylococcus aureus is associated with ATP depletion. Nature Microbiology. 2016;1 doi: 10.1038/nmicrobiol.2016.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar D., Dar N., Cai L., Newman D.K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science. 2021;373(6556) doi: 10.1126/science.abi4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies P.D. Drug-resistant tuberculosis. Journal of the Royal Society of Medicine. 2001;94:261–263. doi: 10.1177/014107680109400601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debbia E.A., Roveta S., Schito A.M., Gualco L., Marchese A. Antibiotic persistence: The role of spontaneous DNA repair response. Microbial Drug Resistance. 2001;7:335–342. doi: 10.1089/10766290152773347. [DOI] [PubMed] [Google Scholar]
- Defraine V., Fauvart M., Michiels J. Fighting bacterial persistence: Current and emerging anti-persister strategies and therapeutics. Drug Resistance Updates. 2018;38:12–26. doi: 10.1016/j.drup.2018.03.002. [DOI] [PubMed] [Google Scholar]
- Dörr T., Lewis K., Vulić M. SOS response induces persistence to fluoroquinolones in Escherichia coli. PLoS Genetics. 2009;5 doi: 10.1371/journal.pgen.1000760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr T., Vulić M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biology. 2010;8 doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du D., Wang-kan X., Neuberger A., van Veen H.W., Pos K.M., Piddock L.J.V., Luisi B.F. Multidrug efflux pumps: Structure, function and regulation. Nature Reviews Microbiology. 2018;16:523–539. doi: 10.1038/s41579-018-0048-6. [DOI] [PubMed] [Google Scholar]
- Fauvart M., De Groote V.N., Michiels J. Role of persister cells in chronic infections: Clinical relevance and perspectives on anti-persister therapies. Journal of Medical Microbiology. 2011;60:699–709. doi: 10.1099/jmm.0.030932-0. [DOI] [PubMed] [Google Scholar]
- Fridman O., Goldberg A., Ronin I., Shoresh N., Balaban N.Q. Optimization of lag time underlies antibiotic tolerance in evolved bacterial populations. Nature. 2014;513:418–421. doi: 10.1038/nature13469. [DOI] [PubMed] [Google Scholar]
- Fung D.K., Chan E.W., Chin M.L., Chan R.C. Delineation of a bacterial starvation stress response network which can mediate antibiotic tolerance development. Antimicrobial Agents and Chemotherapy. 2010;54:1082–1093. doi: 10.1128/AAC.01218-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain E., Castro-Roa D., Zenkin N., Gerdes K. Molecular mechanism of bacterial persistence by HipA. Molecular Cell. 2013;52:248–254. doi: 10.1016/j.molcel.2013.08.045. [DOI] [PubMed] [Google Scholar]
- Giese K.C., Michalowski C.B., Little J.W. RecA-dependent cleavage of LexA dimers. Journal of Molecular Biology. 2008;377:148–161. doi: 10.1016/j.jmb.2007.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girgis H.S., Harris K., Tavazoie S. Large mutational target size for rapid emergence of bacterial persistence. Proceedings of the National Academy of Sciences. 2012;109:12740–12745. doi: 10.1073/pnas.1205124109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goormaghtigh F., Fraikin N., Putrins M., Hallaert T., Hauryliuk V., Garcia-Pino A., Sjodin A., Kasvandik S., Udekwu K., Tenson T., Kaldalu N., Van Melderen L. Reassessing the role of type II toxin-antitoxin systems in formation of Escherichia coli type II persister cells. mBio. 2018;9 doi: 10.1128/mBio.00640-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Fino C., Sørensen M.A., Semsey S., Gerdes K. Prophages and growth dynamics confound experimental results with antibiotic-tolerant persister cells. mBio. 2017;8 doi: 10.1128/mBio.01964-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms A., Maisonneuve E., Gerdes K. Mechanisms of bacterial persistence during stress and antibiotic exposure. Science. 2016;354 doi: 10.1126/science.aaf4268. [DOI] [PubMed] [Google Scholar]
- Hauryliuk V., Atkinson G.C., Murakami K.S., Tenson T., Gerdes K. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nature Reviews Microbiology. 2015;13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazan R., Maura D., Que Y.A., Rahme L.G. Assessing Pseudomonas aeruginosa persister/antibiotic tolerant cells. Methods in Molecular Biology. 2014;1149:699–707. doi: 10.1007/978-1-4939-0473-0_54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S., Cheverton A.M., Watson K.G., Faure L.M., Matthews S.A., Holden D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helaine S., Holden D.W. Heterogeneity of intracellular replication of bacterial pathogens. Current Opinion in Microbiology. 2013;16:184–191. doi: 10.1016/j.mib.2012.12.004. [DOI] [PubMed] [Google Scholar]
- Henry T.C., Brynildsen M.P. Development of persister-FACSeq: A method to massively parallelize quantification of persister physiology and its heterogeneity. Scientific Reports. 2016;6 doi: 10.1038/srep25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honer Zu Bentrup K., Russell D.G. Mycobacterial persistence: Adaptation to a changing environment. Trends in Microbiology. 2001;9:597–605. doi: 10.1016/s0966-842x(01)02238-7. [DOI] [PubMed] [Google Scholar]
- Hubbard A.T., Coates A.R., Harvey R.D. Comparing the action of HT61 and chlorhexidine on natural and model Staphylococcus aureus membranes. Journal of Antibiotics. 2017;70:1020–1025. doi: 10.1038/ja.2017.90. [DOI] [PubMed] [Google Scholar]
- Hu Y., Shamaei-Tousi A., Liu Y., Coates A. A new approach for the discovery of antibiotics by targeting non-multiplying bacteria: A novel topical antibiotic for staphylococcal infections. PLoS One. 2010;5 doi: 10.1371/journal.pone.0011818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby G.A. AmpC β-lactamases. Clinical Microbiology Reviews. 2009;22:161–182. doi: 10.1128/CMR.00036-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Lee J.E., Schaefer C., Luo X., Wollman A.J.M., Payne-Dwyer A.L., Tian T., Zhang X., Chen X., Li Y., McLeish T.C.B., Leake M.C., Bai F. Membraneless organelles formed by liquid-liquid phase separation increase bacterial fitness. Science Advances. 2021;7(43) doi: 10.1126/sciadv.abh2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jõers A., Kaldalu N., Tenson T., Jo A. The frequency of persisters in Escherichia coli reflects the kinetics of awakening from dormancy. Journal of Bacteriology. 2010;192:3379–3384. doi: 10.1128/JB.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P.J.T., Levin B.R., Levin B.R., Li L., Karger B. Pharmacodynamics, population dynamics, and the evolution of persistence in Staphylococcus aureus. PLoS Genetics. 2013;9 doi: 10.1371/journal.pgen.1003123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldalu N., Hauryliuk V., Tenson T. Persisters – as elusive as ever. Applied Microbiology and Biotechnology. 2016;100:6545–6553. doi: 10.1007/s00253-016-7648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaspy I., Rotem E., Weiss N., Ronin I., Balaban N.Q., Glaser G. HipA-mediated antibiotic persistence via phosphorylation of the glutamyl-tRNA-synthetase. Nature Communications. 2013;4:3001. doi: 10.1038/ncomms4001. [DOI] [PubMed] [Google Scholar]
- Keren I., Minami S., Rubin E., Lewis K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio. 2011;2 doi: 10.1128/mBio.00100-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch S.B., Henderson T.A., Hill T.M. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Molecular Microbiology. 2003;50:1199–1213. doi: 10.1046/j.1365-2958.2003.03779.x. [DOI] [PubMed] [Google Scholar]
- Kreuzer K.N. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harbor Perspectives in Biology. 2013;5 doi: 10.1101/cshperspect.a012674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Munder M.C., Maharana S., Franzmann T.M., Richter D., Ruer M., Hyman A.A., Alberti S. Different material states of Pub1 condensates define distinct modes of stress adaptation and recovery. Cell Reports. 2018;23:3323–3339. doi: 10.1016/j.celrep.2018.05.041. [DOI] [PubMed] [Google Scholar]
- Kuchina A., Brettner L.M., Paleologu L., Roco C.M., Rosenberg A.B., Carignano A., Kibler R., Hirano M., DePaolo R.W., Seelig G. Microbial single-cell RNA sequencing by split-pool barcoding. Science. 2021;371(6531) doi: 10.1126/science.aba5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaRock D.L., Chaudhary A., Miller S.I. Salmonellae interactions with host processes. Nature Reviews Microbiology. 2015;13:191–205. doi: 10.1038/nrmicro3420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee B.G., Park E.Y., Lee K.-E., Jeon H., Sung K.H., Paulsen H., Rübsamen-Schaeff H., Brötz- Oesterhelt H., Song H.K. Structures of ClpP in complex with acyldepsipeptide antibiotics reveal its activation mechanism. Nature Structural & Molecular Biology. 2010;17:471–478. doi: 10.1038/nsmb.1787. [DOI] [PubMed] [Google Scholar]
- Levin-Reisman I., Gefen O., Fridman O., Ronin I., Shwa D., Sheftel H., Balaban N.Q. Automated imaging with ScanLag reveals previously undetectable bacterial growth phenotypes. Nature Methods. 2010;7:737–739. doi: 10.1038/nmeth.1485. [DOI] [PubMed] [Google Scholar]
- Levin-Reisman I., Ronin I., Gefen O., Braniss I., Shoresh N., Balaban N.Q. Antibiotic tolerance facilitates the evolution of resistance. Science. 2017;355:826–830. doi: 10.1126/science.aaj2191. [DOI] [PubMed] [Google Scholar]
- Levin B.R., Concepción-Acevedo J., Udekwu K.I. Persistence: A copacetic and parsimonious hypothesis for the existence of non-inherited resistance to antibiotics. Current Opinion in Microbiology. 2014;21:18–21. doi: 10.1016/j.mib.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin B.R., Rozen D.E. Non-inherited antibiotic resistance. Nature Reviews Microbiology. 2006;4:556–562. doi: 10.1038/nrmicro1445. [DOI] [PubMed] [Google Scholar]
- Lewis K. Persister cells. Annual Review of Microbiology. 2010;64:357–372. doi: 10.1146/annurev.micro.112408.134306. [DOI] [PubMed] [Google Scholar]
- Lewis K., Shan Y. Why tolerance invites resistance. Science. 2017;355(6327):796. doi: 10.1126/science.aam7926. [DOI] [PubMed] [Google Scholar]
- Li D.H.S., Chung Y.S., Gloyd M., Joseph E., Ghirlando R., Wright G.D., Cheng Y.-Q., Maurizi M.R., Guarné A., Ortega J. Acyldepsipeptide antibiotics induce the formation of a structured axial channel in ClpP: A model for the ClpX/ClpA-bound state of ClpP. Chemistry & Biology. 2010;17:959–969. doi: 10.1016/j.chembiol.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Little J.W. Mechanism of specific LexA cleavage: Autodigestion and the role of RecA coprotease. Biochimie. 1991;73:411–421. doi: 10.1016/0300-9084(91)90108-d. [DOI] [PubMed] [Google Scholar]
- Liu J., Gefen O., Ronin I., Bar-Meir M., Balaban N.Q. Effect of tolerance on the evolution of antibiotic resistance under drug combinations. Science. 2020;367(6474):200–204. doi: 10.1126/science.aay3041. [DOI] [PubMed] [Google Scholar]
- Lu T.K., Collins J.J. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proceedings of the National Academy of Sciences. 2009;106(12):4629–4634. doi: 10.1073/pnas.0800442106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luidalepp H., Jõers A., Kaldalu N., Tenson T. Age of inoculum strongly influences persister frequency and can mask effects of mutations implicated in altered persistence. Journal of Bacteriology. 2011;193:3598–3605. doi: 10.1128/JB.00085-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Pfuetzner R.A., Mosimann S., Paetzel M., Frey E.A., Cherney M., Kim B., Little J.W., Strynadka N.C.J. Crystal structure of LexA: A conformational switch for regulation of self-cleavage. Cell. 2001;106:585–594. doi: 10.1016/s0092-8674(01)00479-2. [DOI] [PubMed] [Google Scholar]
- Maiden M.M., Hunt A.M.A., Zachos M.P., Gibson J.A., Hurwitz M.E., Mulks M.H., Waters C.M. Triclosan is an aminoglycoside adjuvant for eradication of Pseudomonas aeruginosa biofilms. Antimicrobial Agents and Chemotherapy. 2018;62(6) doi: 10.1128/AAC.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malani P.N. Preventing postoperative Staphylococcus aureus infections: The search continues. JAMA. 2013;309:1408–1409. doi: 10.1001/jama.2013.3382. [DOI] [PubMed] [Google Scholar]
- Manuse S., Shan Y., Canas-Duarte S.J., Bakshi S., Sun W.S., Mori H., Paulsson J., Lewis K. Bacterial persisters are a stochastically formed subpopulation of low-energy cells. PLoS Biology. 2021;19 doi: 10.1371/journal.pbio.3001194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marakalala M.J., Raju R.M., Sharma K., Zhang Y.J., Eugenin E.A., Prideaux B., Daudelin I.B., Chen P.Y., Booty M.G., Kim J.H., Eum S.Y., Via L.E., Behar S.M., Barry C.E., 3rd, Mann M., Dartois V., Rubin E.J. Inflammatory signaling in human tuberculosis granulomas is spatially organized. Nature Medicine. 2016;22:531–538. doi: 10.1038/nm.4073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques C.N.H., Morozov A., Planzos P., Zelaya H.M. The fatty acid signaling molecule cis-2-decenoic acid increases metabolic activity and reverts persister cells to an antimicrobial-susceptible state. Applied and Environmental Microbiology. 2014;80(22):6976–6991. doi: 10.1128/AEM.01576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammad H., Thangamani S., Seleem M.N. Antimicrobial peptides and peptidomimetics – potent therapeutic allies for staphylococcal infections. Current Pharmaceutical Design. 2015;21:2073–2088. doi: 10.2174/1381612821666150310102702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mok W.W., Orman M.A., Brynildsen M.P. Impacts of global transcriptional regulators on persister metabolism. Antimicrobial Agents and Chemotherapy. 2015;59:2713–2719. doi: 10.1128/AAC.04908-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monack D.M., Mueller A., Falkow S. Persistent bacterial infections: The interface of the pathogen and the host immune system. Nature Reviews Microbiology. 2004;2:747–765. doi: 10.1038/nrmicro955. [DOI] [PubMed] [Google Scholar]
- Moreillon P., Tomasz A., Tomasz A. Penicillin resistance and defective lysis in clinical isolates of pneumococci: Evidence for two kinds of antibiotic pressure operating in the clinical environment. The Journal of Infectious Diseases. 1988;157:1150–1157. doi: 10.1093/infdis/157.6.1150. [DOI] [PubMed] [Google Scholar]
- Moyed H.S., Bertrand K.P. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. Journal of Bacteriology. 1983;155:768–775. doi: 10.1128/jb.155.2.768-775.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyed H.S., Broderick S.H. Molecular cloning and expression of hipA, a gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. Journal of Bacteriology. 1986;166(2):399–403. doi: 10.1128/jb.166.2.399-403.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulcahy L.R., Burns J.L., Lory S., Lewis K. Emergence of Pseudomonas aeruginosa strains producing high levels of persister cells in patients with cystic fibrosis. Journal of Bacteriology. 2010;192:6191–6199. doi: 10.1128/JB.01651-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nierman W.C., Yu Y., Losada L. The in vitro antibiotic tolerant persister population in Burkholderia pseudomallei is altered by environmental factors. Frontiers in Microbiology. 2015;6:1338. doi: 10.3389/fmicb.2015.01338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H., Cui P., Shi W., Zhang S., Feng J., Wang Y., Sullivan D., Zhang W., Zhu B., Zhang Y. Identification of anti-persister activity against uropathogenic Escherichia coli from a clinical drug library. Antibiotics. 2015;4:179–187. doi: 10.3390/antibiotics4020179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton J.P., Mulvey M.A. Toxin-antitoxin systems are important for niche-specific colonization and stress resistance of uropathogenic Escherichia coli. PLoS Pathogens. 2012;8 doi: 10.1371/journal.ppat.1002954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M.A., Brynildsen M.P. Dormancy is not necessary or sufficient for bacterial persistence. Antimicrobial Agents and Chemotherapy. 2013;57:3230–3239. doi: 10.1128/AAC.00243-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orman M.A., Brynildsen M.P. Inhibition of stationary phase respiration impairs persister formation in E. coli. Nature Communications. 2015;6:7983. doi: 10.1038/ncomms8983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Malinovska L., Saha S., Wang J., Alberti S., Krishnan Y., Hyman A.A. ATP as a biological hydrotrope. Science. 2017;356(6339):753–756. doi: 10.1126/science.aaf6846. [DOI] [PubMed] [Google Scholar]
- Potrykus K., Cashel M. ppGpp: Still magical? Annual Review of Microbiology. 2008;62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- Pu Y., Li Y., Jin X., Tian T., Ma Q., Zhao Z., Lin S., Chen Z., Yao G., Leake M., Lo C., Bai F. ATP-dependent dynamic protein aggregation regulates bacterial dormancy depth critical for antibiotic tolerance. Molecular Cell. 2019;73(1):143–156. doi: 10.1016/j.molcel.2018.10.022. [DOI] [PubMed] [Google Scholar]
- Pu Y., Zhao Z., Li Y., Zou J., Ma Q., Zhao Y., Ke Y., Zhu Y., Chen H., Baker M., Ge H., Sun Y., Xie X., Bai F. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Molecular Cell. 2016;62:284–294. doi: 10.1016/j.molcel.2016.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice C.D., Pollard J.E., Lewis Z.T., McCleary W.R. Employment of a promoter- swapping technique shows that PhoU modulates the activity of the PstSCAB2ABC transporter in Escherichia coli. Applied and Environmental Microbiology. 2009;75:573–582. doi: 10.1128/AEM.01046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera-Chavez F., Baumler A.J. The pyromaniac inside you: Salmonella metabolism in the host gut. Annual Review of Microbiology. 2015;69:31–48. doi: 10.1146/annurev-micro-091014-104108. [DOI] [PubMed] [Google Scholar]
- Rubin E.J. The granuloma in tuberculosis-friend or foe? New England Journal of Medicine. 2009;360:2471–2473. doi: 10.1056/NEJMcibr0902539. [DOI] [PubMed] [Google Scholar]
- Schumacher M.A., Balani P., Min J., Chinnam N.B., Hansen S., Vulic M., Lewis K., Brennan R.G. HipBA-promoter structures reveal the basis of heritable multidrug tolerance. Nature. 2015;524:59–64. doi: 10.1038/nature14662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah D., Zhang Z., Khodursky A., Kaldalu N., Kurg K., Lewis K. Persisters: A distinct physiological state of E. coli. BMC Microbiology. 2006;6:53. doi: 10.1186/1471-2180-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Herzog E., McDonald E.M., Crooks A.L., Detweiler C.S. Physiologic stresses reveal a Salmonella persister state and TA family toxins modulate tolerance to these stresses. PLoS One. 2015;10 doi: 10.1371/journal.pone.0141343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starkey M., Lepine F., Maura D., Bandyopadhaya A., Lesic B., He J., Kitao T., Righi V., Milot S., Tzika A., Rahme L. Identification of anti-virulence compounds that disrupt quorum-sensing regulated acute and persistent pathogenicity. PLoS Pathogens. 2014;10 doi: 10.1371/journal.ppat.1004321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun S., Berg O.G., Roth J.R., Andersson D.I. Contribution of gene amplification to evolution of increased antibiotic resistance in Salmonella typhimurium. Genetics. 2009;182:1183–1195. doi: 10.1534/genetics.109.103028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H.W., Mueller J.P., Miller P.F., Arrow A.S. Bacterial uptake of aminoglycoside antibiotics. Microbiological Reviews. 1987;51:439–457. doi: 10.1128/mr.51.4.439-457.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., Wang X., Bodeau, Tuch B.B., Siddiqui A., Lao K. mRNA-Seq whole-transcriptome analysis of a single cell. Nature Methods. 2009;6:377–382. doi: 10.1038/nmeth.1315. [DOI] [PubMed] [Google Scholar]
- Toprak E., Veres A., Michel J.-B., Chait R., Hartl D.L., Kishony R. Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nature Genetics. 2012;44:101–105. doi: 10.1038/ng.1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torrey H.L., Keren I., Via L.E., Lee J.S., Lewis K. High persister mutants in Mycobacterium tuberculosis. PLoS One. 2016;11 doi: 10.1371/journal.pone.0155127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillet-Assant S., Lelievre L., Martins-Simoes P., Gonzaga L., Tasse J., Valour F., Rasigade J.P., Vandenesch F., Muniz Guedes R.L., Ribeiro de Vasconcelos A.T., Caillon J., Lustig S., Ferry T., Jacqueline C., Loss de Morais G., Laurent F. Adaptive processes of Staphylococcus aureus isolates during the progression from acute to chronic bone and joint infections in patients. Cellular Microbiology. 2016;18:1405–1414. doi: 10.1111/cmi.12582. [DOI] [PubMed] [Google Scholar]
- Vazquez-Laslop N., Lee H., Neyfakh A.A. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. Journal of Bacteriology. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vega N.M., Allison K.R., Khalil A.S., Collins J.J. Signaling-mediated bacterial persister formation. Nature Chemical Biology. 2012;8:431–433. doi: 10.1038/nchembio.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verstraeten N., Knapen W.J., Kint C.I., Liebens V., Van Den Bergh B., Dewachter L., Michiels J.E., Fu Q., David C.C., Fierro A.C., Marchal K., Beirlant J., Versees W., Hofkens J., Jansen M., Fauvart M., Michiels J. Obg and membrane depolarization are part of a microbial bet-hedging strategy that leads to antibiotic tolerance. Molecular Cell. 2015;59:9–21. doi: 10.1016/j.molcel.2015.05.011. [DOI] [PubMed] [Google Scholar]
- Vieira T.F., Magalhaes R.P., Simoes M., Sousa S.F. Drug repurposing targeting Pseudomonas aeruginosa MvfR using docking, virtual screening, molecular dynamics, and free-energy calculations. Antibiotics. 2022;11(2):185. doi: 10.3390/antibiotics11020185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Völzing K.G., Brynildsen & M.P. Stationary-phase persisters to ofloxacin sustain DNA damage and require repair systems only during recovery. mBio. 2015;6 doi: 10.1128/mBio.00731-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil M.I., Schnappinger D., Visconti K.C., Harrell M.I., Dolganov G.M., Sherman D.R., Schoolnik G.K. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. Journal of Experimental Medicine. 2003;198:705–713. doi: 10.1084/jem.20030205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wain J., Hendriksen R.S., Mikoleit M.L., Keddy K.H., Ochiai R.L. Typhoid fever. The Lancet. 2015;385:1136–1145. doi: 10.1016/S0140-6736(13)62708-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakamoto Y., Dhar N., Chait R., Schneider K., Signorino-Gelo F., Leibler S., Mckinney J.D. Dynamic persistence of antibiotic-stressed mycobacteria. Science. 2013;339:91–95. doi: 10.1126/science.1229858. [DOI] [PubMed] [Google Scholar]
- Wallace E.W., Kear-Scott J.L., Pilipenko E.V., Schwartz M.H., Laskowski P.R., Rojek A.E., Katanski C.D., Riback J.A., Dion M.F., Franks A.M., Airoldi E.M., Pan T., Budnik B.A., Drummond D.A. Reversible, specific, active aggregates of endogenous proteins assemble upon heat stress. Cell. 2015;162:1286–1298. doi: 10.1016/j.cell.2015.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne L.G. Dormancy of Mycobacterium tuberculosis and latency of disease. European Journal of Clinical Microbiology & Infectious Diseases. 1994;13:908–914. doi: 10.1007/BF02111491. [DOI] [PubMed] [Google Scholar]
- Wexselblatt E., Oppenheimer-Shaanan Y., Kaspy I., London N., Schueler-Furman O., Yavin E., Glaser G., Katzhendler J., Ben-Yehuda S. Relacin, a novel antibacterial agent targeting the stringent response. PLoS Pathogens. 2012;8(9) doi: 10.1371/journal.ppat.1002925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood T.K., Knabel S.J., Kwan B.W. Bacterial persister cell formation and dormancy. Applied and Environmental Microbiology. 2013;79:7116–7121. doi: 10.1128/AEM.02636-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka K., Inouye M. Growth-phase-dependent expression of cspD, encoding a member of the CspA family in Escherichia coli. Journal of Bacteriology. 1997;176:5126–5130. doi: 10.1128/jb.179.16.5126-5130.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]