Abstract
Globally, colorectal cancer (CRC) is the third most prevalent cancer and the second leading cause of cancer-related deaths. Circular RNAs (circRNAs) are single-stranded RNA with covalently closed-loop structures and are highly stable, conserved, and abundantly expressed in various organs and tissues. Recent research found abnormal circRNA expression in CRC patients’ blood/serum, cells, CRC tissues, and exosomes. Furthermore, mounting data demonstrated that circRNAs are crucial to the development of CRC. CircRNAs have been shown to exert biological functions by acting as microRNA sponges, RNA-binding protein sponges, regulators of gene splicing and transcription, and protein/peptide translators. These characteristics make circRNAs potential markers for CRC diagnosis and prognosis, potential therapeutic targets, and circRNA-based therapies. However, further studies are still necessary to improve the understanding of the roles and biological mechanisms of circRNAs in the development of CRC. In this review, up-to-date research on the role of circRNAs in CRC was examined, focusing on their potential application in CRC diagnosis and targeted therapy, which would advance the knowledge of the functions of circRNAs in the development and progression of CRC.
Subject terms: Colorectal cancer, Non-coding RNAs
Facts
CircRNAs are single-stranded RNA that may be used to treat or prevent colorectal cancer.
CircRNAs may service as useful biomarkers for cancer diagnosis and therapy.
CircRNAs are widely expressed in CRC patients’ blood/serum, cells, CRC tissues, and exosomes.
Some circRNAs exert a cancer-promoting effect in CRC, while others are not.
Open Questions
What is the roles and biological mechanisms of circRNAs in the development of CRC?
How do circRNAs contribute to the development and progression of CRC by controlling alternative splicing?
Can circRNAs be utilized as biomarkers to predict chemotherapy resistance in CRC therapies?
Introduction
Globally, colorectal cancer (CRC) is the third most prevalent cancer and the second leading cause of cancer-related deaths [1]. Recent estimates indicate that over 1.9 million new cases and 93,000 CRC-related deaths occurred in 2020, accounting for approximately 10% of all cancer cases and 9.4% of cancer-related deaths [2]. The early detection of CRC can help minimize morbidity and mortality; however, most CRCs are diagnosed at an advanced stage owing to the lack of distinct early symptoms, limiting the opportunity for effective early treatment. Therefore, it is imperative to identify new therapeutic targets and biomarkers for effective early detection, personalized treatment, and monitoring of CRC to improve prognosis.
Both genetic and epigenetic alterations can cause CRC. circRNAs, a novel type of non-coding RNAs, have been identified as tumor-initiating and tumor-progressing factors. Compared with linear RNAs, the closed structure of circRNAs makes them highly stable and conserved [3]. Recently, bioinformatics analysis of RNA-seq has facilitated the identification of several circRNAs in eukaryotes and shown that circRNAs have tissue-specific expression patterns [4]. Ongoing studies have revealed that dysregulation of circRNAs contributes to the development of various cancers, including CRC [5], and lung [6], liver [7], and bladder cancers [8]. Further investigations have identified several dysregulated circRNAs that play important roles in CRC progression [9]. Additionally, circRNAs are abundantly found in exosomes, human peripheral blood, and fluids, making them potential diagnostic biomarkers and therapeutic targets [10]. Therefore, circRNAs may serve as promising biomarkers for CRC.
This review highlights the current research progress on the biogenesis and characteristics of circRNAs and their mechanisms in CRC. Additionally, the therapeutic and diagnostic potentials of circRNAs in CRC were extensively discussed.
Alternative modes of circular RNA splicing
RNA splicing is a fundamental and highly regulated process in the eukaryotic gene. The splicing of pre-messenger RNA (pre-mRNA) is catalyzed via spliceosome, a highly dynamical ribonucleoprotein (RNP) machinery that can remove introns and then join exons together to form mature mRNA [11] (Fig. 1). Alternative splicing involves the transcription of pre-mRNAs to generate different mature mRNAs depending on how they are spliced, thereby increasing protein diversity [12]. The normal pre-mRNA spliceosomal mechanism, which involves the back-splicing of intronic, exonic, or intergenic sequences, is necessary to synthesize circRNAs. By backsplicing pre-mRNAs, which requires the covalent interaction of an upstream 3′ splice site and a downstream 5′ splice site, circRNAs are selectively produced [13]. Through controlling alternative splicing pathways, circRNAs have been demonstrated to play crucial roles in carcinogenesis, according to mounting evidence [14–16]. For example, Wang et al. discovered circURI1, created by the back-splicing of exons 3 and 4 of URI1-14, an unusual prefoldin RPB5 interactor. CircURI1 may control alternative splicing to contribute to developing and spreading gastric cancer [13].
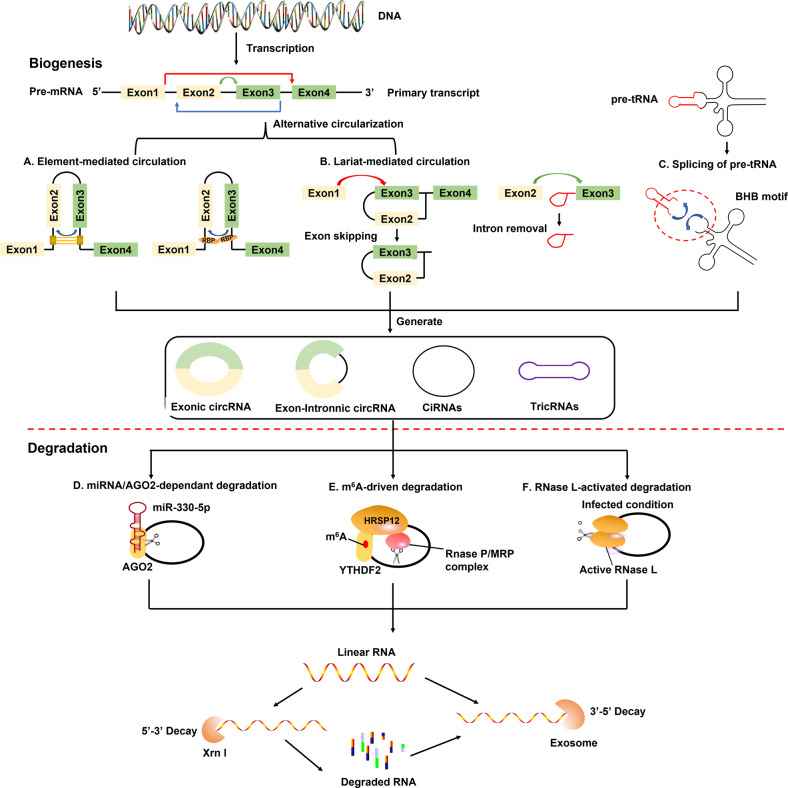
Fig. 1. Biosynthesis and metabolism of circRNA.
A Possibility of looped structures generation either by base-pairing among complementary sequences that flank the circularized exons or via RBPs. To generate EcRNAs and EIciRNAs, the intron sequences can be deleted or kept respectively in the loop structure. B The exon-skipping events generate certain EcRNAs, whereas a lariat is internally spliced to remove intronic sequences. C The intron-containing pre-tRNA is cleaved at the BHB motif into half of the exon and intron part. A mature tRNA is formed by joining the halves of the exons, and a tricRNA is produced by joining the termini of the introns. D miRNA directly binds at the AGO2-dependant cleavage site of targeted mRNA molecules in a complementary way. E RNase mitochondrial RNA processing (RNase MRP) promotes the cleavage of m6A-possessing circRNAs via the activities of YTHDF2 and HRSP12. F When infected by viruses, active RNase L degrades circRNA. AGO2 argonaute 2, BHB motif bulge-helix-bulge motif, ciRNA circular intronic RNA, EcRNAs exonic circRNAs, EIciRNAs exon-intron circRNAs, RBP RNA-binding protein, YTHDF2 YT521-B homology domain family 2, tricRNA tRNA intronic circular RNA.
Numerous RNA-binding splicing factors (RBFOX1/2/3) contain an RNA-recognition motif that binds to this GCAUG element and affects the regulation of various alternate splicing events [14, 15]. Recent studies have shown that RBFOX proteins can either repress or activate alternate splicing, determining the binding location to pre-mRNA exon [16]. Suppression of a splicing regulator RNA binding protein fox-1 homolog 2 (RBFOX2) promoted preferential splicing of the mRNA isoform, such as KIF1B beta [17], TEAD1 [16], and TFRC [18]. Specifically, Zhang et al. found that circRAPGEF5 could interact with RBFOX2 and inhibit its binding to pre-mRNA, thereby causing exon exclusion of TFRC in endometrial cancer [18]. Moreover, although RBFOX2 is known to regulate some of these genes, the role of RBFOX2-mediated splicing events on signaling pathways in cancer remains largely unknown.
circRNAs: biogenesis and characteristics
Based on their biogenesis mechanisms, circRNAs can be classified as EcircRNAs, EIciRNAs, ciRNAs, and mecciRNAs [19–21]. The biogenesis of circRNAs can be facilitated by pre-mRNAs containing a reverse complement of Alu repeat flanking the circularized exons [22]. RNA-seq and bioinformatic analysis has revealed the relationship between flanking introns and reverse complementary Alu repeats in mammalian circRNA biogenesis. CircRNA exons often have long flanking intronic sequences and repetitive Alu elements, both promoting circularisation by base pairing and reducing the distance between potential back-splicing sites [23, 24]. Moreover, loss of flanking Alu repeats inhibited the circularization of circRNA in vitro, including CircERBB2 [25]. ALU repeats present an underestimated risk, while enzymes such as adenosine deaminases acting on RNA (ADARs) and DExH-Box Helicase 9 (DHX9) are critical in destabilizing intron pairing during the biogenesis of circRNAs [26, 27]. A study by Shen et al. characterized ADARs as potent regulators of circular transcriptomes by identifying over a thousand circRNAs in a bidirectional manner [28]. The biogenesis of circRNAs and ADARs has been found to be negatively correlated in recent studies. For example, the knockdown of ADAR1 increased the intracellular circRNA expression in the mammalian brain [29]. Furthermore, ADARs-regulated circRNAs are ubiquitously expressed in numerous cancer types, suggesting high functional relevance to cancer [28]. Additionally, DHX9 deletion increased a subset of circRNA-producing genes and amounts of circular RNA, repeat Alu elements, and transcriptional rewiring of susceptible loci [26]. In addition to the above-mentioned examples, RNA binding proteins (RBPs), including muscleblind (MBL/MBNL1) and quaking (QKI), have been shown to promote the biogenesis of circRNAs [30, 31]. Ashwal-Fluss et al. showed that ectopic expression of MBL/MBNL1 increased the expression of circMbl by binding to flanking introns; however, downregulation of MBL/MBNL1 significantly decreased CircMbl expression [24]. Thus, MBL/MBNL1 was involved in circRNA biogenesis. Another important RBP, QKI, can also positively regulate circRNA biogenesis. For instance, knockdown of QKI inhibited circRNAs expression during circRNA biogenesis, however, overexpression of QKI lead to the circRNA biogenesis in human immortalized mammary epithelial cells [30]. Thus, the biosynthesis of circRNAs is influenced by QKI.
CircRNA mechanisms of action
Recent studies have demonstrated that circRNAs can interact with miRNAs, and RBPs serve as protein baits or antagonists to exert the functions of circRNAs [32]. As regulators of gene expression, circRNAs are involved in several biological processes, including miRNA sponges [33], transcription and translation, RBPs, and translation of peptides and proteins [34] (Fig. 2).
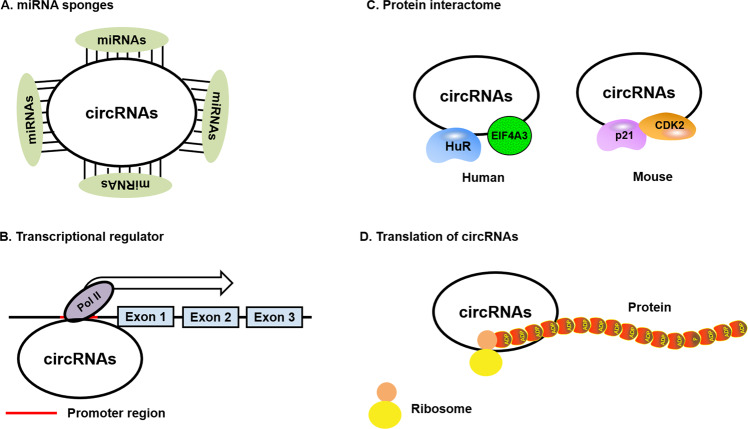
Fig. 2. Mechanisms of circRNA functions.
A CircRNAs can act as sponges or decoys for miRNAs. MiRNA binding to circRNAs may release target mRNAs from miRNA-dependent degradation, resulting in more effective translation. B circRNAs containing RBP motifs (such as HuR, EIF4A3, P21, and CDK2) may act either as sponges or decoys for the aforementioned proteins while regulating their functions. C circRNAs may interact with the RNA polymerase II (Pol II) complex containing the U1 snRNP in the promoter region of targeted genes, and significantly enhance its function. D circRNAs contain ribosome entry sites that may be translated to produce unique peptides under specific conditions.
miRNA sponges
Little non-coding RNAs called miRNAs play an important role in physiological and pathological processes. They typically have a base length of 21–25 nucleotides. CircRNAs may behave as miRNA sponges, limiting miRNA action in the transcriptional and post-transcriptional control of gene expression (for example, mRNA stability) [35]. Cancer cells’ proliferation, migration, and angiogenesis have all been linked to this sponge process. For instance, by functioning as a sponge for miR-328-5p and reversing its repression of E2F1 [36], circSHKBP1 facilitated the advancement of CRC. By activating the miR-142-3p/miR-506-3p-TGF-1 axis, CRC-derived exosomal circPACRGL promoted cell proliferation, migration, invasion, and neutrophil differentiation [8]. This method has also been described in other fields. For instance, circHIPK3 promoted the development of retinal vascular dysfunction in diabetes mellitus by sponging miR-30a-3p [37] and modulated the autophagy in STK11 mutant lung cancer by sponging miR-124-3p [38]. In addition, circHIPK3 also suppressed CRC growth and metastasis by sponging miR-7 [35]. However, some circRNAs have been shown to function in multiple roles by sponging different miRNAs. CircSLC8a1 exacerbated myocardial injury by sponging miR-214-5p [36] and inhibited the progression of non-small cell lung cancer (NSCLC) by sponging miR-106b-5p [39].
Transcription and translation
CircRNAs can regulate gene transcription in both a direct and indirect manner. Moreover, certain circRNAs have been reported to modulate gene transcription by interacting with the RNA polymerase II complex and translating associated proteins. For instance, circEIF3J and circPAIP2 promoted PAIP2 and EIF3J transcription by interacting with U1 small nuclear ribonucleoprotein (snRNP) and RNA polymerase II [40]. However, circRNAs are mainly cis-regulators of transcription in various physiological and pathological processes. For instance, circRHOT1 inhibited the progression of hepatocellular Carcinoma (HCC) via recruiting TIP60 to the NR2F6 promoter and subsequently initiating the transcription of NR2F6 [41]. circMEMO1 can regulate TCF21 promoter methylation and its gene expression to promote the progression of HCC [42]. In another study, circAmotl1 promoted skin wound repair by increasing STAT3 expression and nuclear translocation [43]. It may be possible to provide clinical insight into skin wound healing by the ectopic application of circ-Amotl1.
Recently, Pamudurti et al. demonstrated that translating ribosomes is associated with a set of circRNAs through ribosome footprinting from fly heads [44]. Mass spectrometry also detected a protein encoded by circRNA generated from the muscleblind locus [44]. Additionally, exosomal circLPAR1 directly bound to eIF3h and specifically suppressed the Interaction between METTL3 and eIF3h, which caused BRD4 translation to decrease [45]. CircVAMP3 interacted with CAPRIN1 and G3BP1 to trigger phase separation of CAPRIN1 and promoted stress granule formation [46]. CircVAMP3 can reduce the protein level of Myc proto‐oncogene protein by inhibiting c‐Myc translation. Hence, circVAMP3 suppressed tumor growth in HCC by inhibiting the translation of c‐Myc.
RBPs
Besides their miRNA sponge and transcription and translation function, circRNAs with RBP binding sites may function as protein sponges or decoys in regulating gene expression [47]. CircRNAs can directly interact with one or different proteins and act as “scaffolding” protein complexes to form an RNA-protein complex. For example, circRHOBTB3 suppressed lung metastasis by binding to the HuR protein and promoting β-Trcp1-mediated ubiquitination of HuR, improving the stability of polypyrimidine tract-binding protein 1 (PTBP1) [48]. In cancer cells, Chen et al. found circAGO2 promoted cancer progression by interacting with HuR protein and inhibiting the functions of AGO2-miRNA complexes [49]. In addition, circRNA can also affect their biological function by sequestering proteins [50]. For example, circPABPN1 reduced ATG16L1 production by inhibiting HuR binding to Atg16l1 mRNA [51]. However, scaffolding enhanced direct protein interactions in contrast to their sequestering function. For example, circFOXO3 functioned as a scaffolding molecule that enhanced the Interaction between CDK2 and p21 [52]. CircACC1 can directly bind to the β and γ subunits of AMPK, enhancing its stability and enzymatic activity [53].
Peptides and proteins
Based on bioinformatics platforms and computational analysis, some circRNAs have open reading frames (ORFs) and can encode proteins [44]. Some other circRNAs also encode proteins or peptides with tumor-suppressive or oncogenic properties. Presently, hundreds of peptides encoded by circRNAs have been detected by liquid chromatography coupled with mass spectrometry (LC-MS), indicating that circRNAs can translate proteins. CircPLCE1 encoded a novel circPLCE1-411 protein that inhibited tumor proliferation and metastasis in CRC cells [54]. Exosomal CircATG4B encoded a novel protein and induced oxaliplatin resistance in CRC by promoting autophagy [55]. In addition, circular RNAs can be classified as IRES-dependent or IRES-independent translational machinery. For example, circSHPRH can encode an SHPRH-146aa protein in an IRES-dependent manner [56]. It was found that SHPRH-146aa is a tumor suppressor protein that prevents SHPRH full-length protein from degradation [56], suggesting that aberrant translation of circSHPRH affects tumor malignancy. Lastly, some circRNAs are capable of encoding peptides without requiring IRES. Recent studies have discovered that consensus N6-methyladenine (m6A) modification motifs are enriched in circRNAs, and one m6A site can initiate translation [57]. For example, YTHDF3, an m6A reader protein, modulates circ-ZNF609 to translate two proteins using two alternative START codons [58]. Overall, the protein-coding capacity of circRNA is of great significance to human disease diagnosis and treatment.
Dysregulation of circRNA in CRC
Recently, the roles of circRNAs in tumorigenesis and other diseases have received considerable research attention. Accumulating evidence indicates that abnormalities in circRNAs are associated with colorectal malignancies (Table 1; Fig. 3). For instance, circHERC4 was highly elevated in CRC tissues and positively associated with lymph node metastasis and advanced tumor [59]. In contrast, circPLCE1 was downregulated in CRC tissues and was linked to poor survival and advanced clinical stages [54]. Interestingly, the expression levels of circRNAs vary in different cancer types, indicating their distinct biological roles. For example, circHIPK3 was significantly upregulated in gastric cancer, HCC, breast cancer, CRC, and lung cancer tissues and cell lines [35, 60–62], but downregulated in bladder cancer [63]. Although several studies have reported abnormal expression of circRNAs in various cancers [6]; the possible causes of circRNA dysregulation remain largely unknown and require further investigation.
Table 1.
Summary of dysregulated circRNAs in colorectal cancer.
| Mechanism | CircRNAs | Annotated number | Expression | Samples | Targets/ Regulators | Function | Ref |
|---|---|---|---|---|---|---|---|
| miRNA sponge | CircPACRGL | has_circ_0069313 | Up | CRC cells | miR-142-3p/506-3p | Promotes proliferation, migration, and invasion. | [8] |
| CircCCDC66 | Up | Tissues | miR-33b/93 | Promotes proliferation, and metastasis. | [137] | ||
| hsa_circ_0001313 | miR-370 | Promotes proliferation, migration, and invasion | [138] | ||||
| Circ_0000392 | hsa_circ_0000392 | Up | Tissues | miR-193a-5p | Promotes proliferation, and invasion. | [74] | |
| ciRS-122 | hsa_circ_0005963 | Up | CRC cells | miR-122 | Promotes chemoresistance. | [98] | |
| Circ_101277 | hsa_circ_101277 | Up | Tissues | miR-370 | Promotes proliferation and cisplatin resistance. | [108] | |
| CircATAD1 | hsa_circRNA_100641 | Up | Tissues | miR-618 | Promotes proliferation. | [139] | |
| CircMET | hsa_circ_0082002 | Up | Tissues; CRC cells | miR-410-3p | Promotes proliferation and growth. | [140] | |
| Circ_0000467 | hsa_circ_0000467 | Up | Tissues; CRC cells | miR-330-5p | Promotes proliferation, migration, and invasion. | [141] | |
| CircN4BP2l2 | hsa_circ_0000471 | Up | Tissues; CRC cells | miR-340-5p | Promotes migration, invasion, proliferation | [103] | |
| Circ_0068464 | hsa_circ_0068464 | Up | Tissues; CRC cells | miR-383 | Promotes tumor growth and lung metastasis | [86] | |
| Circ_0000231 | hsa_circ_0000231 | Up | Tissues | miR-375 | Promotes proliferation, and tumorigenesis. | [142] | |
| CircRAD23b | hsa_circ_0087862 | Up | Tissues; CRC cells | miR-1205 | Promotes proliferation, and metastasis | [143] | |
| Circ_0011385 | hsa_circ_0011385 | Up | Tissues | miR-330-3p | Promotes proliferation, migration, and invasion. | [144] | |
| Circ_0006732 | hsa_circ_0006732 | Up | Tissues; CRC cells | miR-127-5p | Promotes proliferation, migration, invasion, and EMT | [102] | |
| Circ_0007919 | hsa_circ_0007919 | Up | Tissues; CRC cells | miR-942-5p | Promotes growth and migration | [145] | |
| Circ_0014130 | hsa_circ_0014130 | Up | Tissues; cells | miR-197-3p | Promotes proliferation. | [146] | |
| Circ_0101802 | hsa_circ_0101802 | Up | Tissues; cells | miR-665 | Promotes proliferation, migration, and tube formation. | [147] | |
| CircALG1 | hsa_circ_0037777 | Up | Tissues; blood | miR-342-5p | Promotes metastasis, migration, and invasion | [81] | |
| CircSMAD4a | hsa_circ_0004846 | Up | Tissues | miR-545-3p | Promotes proliferation. | [148] | |
| Circ_0001955 | hsa_circ_0001955 | Up | Tissues | miR-583 | Promotes tumor growth. | [149] | |
| CircFAT1 | hsa_circ_0001461 | Up | Tissues | miR-619-5p | Promotes migration, invasion, and angiogenesis. | [150] | |
| CircLHFPL2 | hsa_circ_7431 | Up | Tissues | miR-556-5p/1322 | Promotes CRC progression. | [109] | |
| CircTMEM59 | hsa_circ_0012634 | Down | Tissues; cells | miR-668-3p | Inhibits growth and metastasis. | [151] | |
| Circ-PLXNB1 | hsa_circ_0065378 | Down | Tissues | miR-4701-5p | Inhibits proliferation, cell invasion, migration, and EMT. | [104] | |
| Protein decoy | CircACC1 | hsa_circ_001391 | Up | Tissues | AMPK | Promotes proliferation. | [53] |
| CircREEP3 | hsa_circ_400564 | Up | Tissues | FKBP10 | Promotes tumorigenesis and metastasis. | [152] | |
| CircLPAR1 | hsa_circ_0087960 | Down | Tissues | BRD4 | Inhibits tumor growth. | [45] | |
| CircPTEN1 | hsa_circ_0002232 | Down | Tissues | TGF-β/Smad signaling | Inhibits metastasis, and invasion. | [85] | |
| Circ-LECRC | has_circ_0004140 | Down | Tissues | YAP signaling | Inhibits proliferation, migration, and invasion and promotes apoptosis. | [153] | |
| CircRHOBTB3 | hsa_circ_00074444 | Down | Tissues; cells | HuR | Suppresses tumor metastasis | [48] | |
| Circ_0014717 | hsa_circ_0014717 | Down | Tissues; cells | p16 | Inhibits cell proliferation and colony formation | [154] | |
| CircRIP2 | has_circ_0005777 | Up | Tissues; cells | CBFB | Aggravates CRC deterioration | [155] | |
| Coding peptides | CircPLCE1 | hsa_circ_0019223 | Down | Tissues; cells | NF-κB, HSP90α/RPS3 complex | Inhibits CRC proliferation and metastasis. | [54] |
| Circ_0000725; circ_0008826 | hsa_circ_0000725; hsa_circ_0008826 | Up | Tissues | 306-a.a. and 475-a.a. peptide | Promotes proliferation, and invasion. | [156] | |
| Circ_0006401 | hsa_circ_0006401 | Up | Tissues | circ_0006401 peptide | Promotes CRC proliferation and migration | [157] | |
| CircPPP1R12a | hsa_circ_0000423 | Up | Tissues | circPPP1R12A-73aa | Promotes cell proliferation, migration, and invasion. | [100] |
AMPK AMP-activated protein kinase, BRD4 bromodomain containing 4, CBFB core-binding factor subunit beta, CRC colorectal cancer.
EMT epithelial-mesenchymal transition, FKBP10 FKBP prolyl isomerase 10, hnRNP A1 heterogeneous nuclear ribonucleoprotein A1, HSP90a heat shock protein 90 alpha. HuR human antigen R, NF-κB nuclear factor kappa B, RPS3 ribosomal protein S3, TGF-β transforming growth factor-beta.
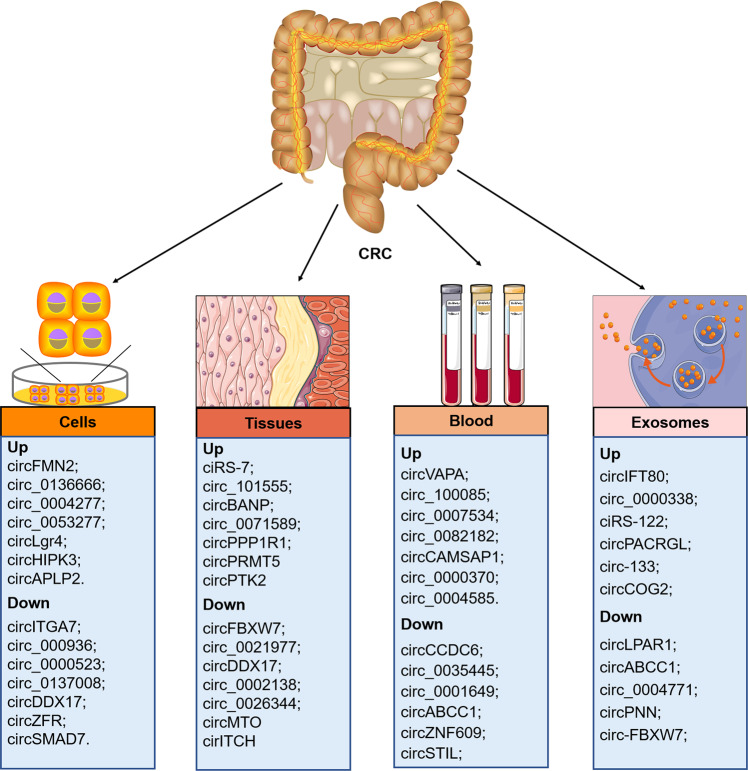
Fig. 3. Aberrant expressions of circRNAs in CRC patients.
circRNAs are aberrantly expressed in blood/serum, cells, CRC tissues, and exosomes from CRC patients. circRNA dysregulations are highlighted in the figure; ‘Up’ indicates upregulated, ‘Down’ indicates downregulated.
The dysregulated circRNA expression in CRC could be due to abnormal expression of host genes, such as chromosomal amplification, depletion, or translocation. CircPRKCI is a proto-oncogenic circRNA located in the 3q26.2 amplicon in several cancers, including lung cancer, glioma, and esophageal cancer [64]. Therefore, circPRKCI upregulation in cancers may be due to PRKCI amplification. Fusion-circRNAs (F-circRNAs) are products of cancer-associated chromosomal translocations in host genes [65]. For example, two novel circRNAs (F-circSR1 and F-circSR2) generated from oncogenic SLC34A2-ROS1 fusion gene, promoted cell migration in non-small cell lung cancer [66]. Additionally, F-circEA generated from oncogenic EML4-ALK fusion facilitated cell migration and invasion in lung cancer [67]. The fusion genes produce functional proteins that contribute to oncogenesis.
Several studies have demonstrated the elimination of circRNAs by RNase L following the release of extracellular vesicles, viral infection, or poly I: C stimulation. Degradation of circRNAs by RNase L activation can increase protein kinase R (PKR) phosphorylation. Apart from degradation, circRNAs can also be eliminated from cells via exocytosis. Additionally, circRNAs in exosomes may be involved in the communication mechanism, indicating the need for further studies on the degradation and extracellular transportation of circRNAs. Moreover, circRNAs are abundant in the cytoplasm and can be transported to exosomes from the cytoplasm.
Recently, some studies reported that RNA modification at the m6A site was associated with circRNA degradation, translation, and expression in cancer cells [68]. The most prevalent internal alteration associated with eukaryotic ncRNAs is m6A, which influences RNA stability, splicing, export, translation, and degradation, all of which affect biological activities. MALAT1 is highly methylated at m6A, and its two residues can block local RNA structure formation and facilitate the recognition and binding of heterogeneous nuclear ribonucleoprotein C (HNRNPC) through an “m6A switch” mechanism [68]. m6A alteration accelerated circNSUN2 transport to the cytoplasm and increased the stability of HMGA2 mRNA to induce CRC metastasis by creating a circNSUN2/IGF2BP2/HMGA2 complex in the cytoplasm [69]. Recently, m6A-modified circRNAs have been identified using cell-type-specific methylation patterns. m6A recruits YTHDF3 and eIF4G2 to regulate protein synthesis from circRNAs. ALKBH5 (m6A eraser) and METTL3 (m6A writer) can affect circRNA biosynthesis by altering the m6A levels. For instance, the m6A levels of circRNA-SORE were elevated in sorafenib-resistant cells and downregulated when m6A modification was suppressed [70]. Recent findings showed that circCUX1 expression was stabilized by METTL3-mediated m6A modification [71]. Thus, the loss of m6A sites or removal of m6A from circRNAs can decrease their methylation levels, resulting in the dysregulation of circRNAs. Furthermore, the deregulation of critical components during the degradation of circRNAs might result in abnormal circRNA expression.
Super enhancers (SEs) comprises of large putative enhancer clusters that are enriched to bind key master transcription factors. These enhancer clusters play key roles in driving tumorigenesis and act as causal mechanisms for regeneration by regulating circRNA expression. SEs are frequently dysregulated in cancer and are central to the maintenance of cancer cell identity. SEs are important for controlling tumor metastasis, proliferation, and chemoresistance. SEs are abnormally activated in various tumors, regulate key target genes in cancer, and promote tumorigenesis and development. For example, EphA2-SE at core active regions contains an E1-enhancing component that induces cell proliferation and metastasis via the involvement of TCF7L2 and FOSL2 to upregulate EphA2 expression [72]. RNA-seq combined with in vitro functional experiments revealed that EphA2-SE deletion mediated the suppression of cell growth and metastasis in HCT-116, HeLa, and MCF-7 cells, whereas EphA2 overexpression in EphA2-SE−/− clones reversed EphA2-SE knockdown-induced effects on cell proliferation and metastasis [72]. Recent studies have shown that circRNAs are potential SEs that modulate gene expression and are involved in the pathogenesis of several diseases. Thus, SEs are likely to control tumor metastasis and chemoresistance by controlling circRNA expression.
CircRNAs participate in the progression of CRC
Tumor invasion and metastasis are multistep, complex dynamic processes involving growth, invasion, metastasis, and intravasation, and are responsible for most CRC-associated mortalities. Transcriptome analysis identified 80 differentially expressed circRNAs, including 33 upregulated and 47 downregulated circRNAs, between CRC and para-cancerous tissues. Circ3823 and circRNA_0000392 were significantly upregulated in CRC tissues and cell lines, indicating that higher circ3823 and circRNA_0000392 expression levels could predict poor prognosis in CRC patients [73, 74]. Several studies have shown that circRNAs can regulate CRC metastasis primarily by influencing key factors that regulate several pathways closely associated with CRC metastasis. The mechanism of circRNAs in CRC metastasis is diverse and includes acting as miRNA sponges, interacting with RBPs, regulating gene splicing or transcription, translating proteins, and regulating epigenetics.
CircRNAs are stable in tissues and cells owing to their closed-loop structure with no 5′ or 3′ ends, thereby preventing ribonuclease degradation. Additionally, circular sequences include several miRNA response elements that facilitate the binding of circRNAs and miRNAs. CircRNAs might therefore function as natural miRNA sponges to modulate target gene expression. For example, circ3823 acted as a sponge for miR-30c-5p and regulated its target TCF7 expression, which increased the expression of MYC and CCND1 and promoted CRC progression [73]. Additionally, circRNA_0000392 promoted the proliferation and invasion of CRC cells through the miR-193a-5p/PIK3R3/Akt axis [74], indicating the potential of circRNA_0000392 as a prospective therapeutic target for CRC therapy as well as a prediction marker. However, most circRNAs are less abundant than miRNAs and may fail to meet the stoichiometric requirement for a sponge effect.
In addition to acting as miRNA sponges, certain circRNAs with RBP binding sites may act as protein sponges or decoys. Studies have shown that circRNAs can bind to RBPs, such as Quaking (QKI), HuR (ELAVL1), eukaryotic translation initiation factor 4A3 (EIF4A3), and AlkB homolog H5 (ALKBH5), to play important roles in tumor progression. circRNAs can interact with regulatory RBPs to influence the destiny of their target mRNAs. For example, circRHOBTB3 acted as a HuR sponge and facilitated HuR-mediated PTBP1mRNA stability [48], indicating that circRHOBTB3 exerted a suppressive effect on CRC. An increase in AMPK activation in CRC tissues is related to elevated expression of circACC1, and circACC1 has been demonstrated to stabilize and enhance AMPK holoenzyme activity by forming a complex with regulatory β and γ subunits [53]. RNA pull-down and RNA immunoprecipitation (RIP) assays showed that hsa_circ_0068631 can bind to EIF4A3 and recruit EIF4A3 to increase c-Myc mRNA stability in breast cancer [75]. Additionally, cIARS (hsa_circ_0008367) physically interacted with ALKBH5 and markedly promoted sorafenib-induced ferroptosis in HCC via inhibition of ALKBH5-mediated autophagy [76]. Recent research suggests that distinct RBPs may play varied or opposing functions in the back-splicing process. For instance, circRNA synthesis can be enhanced by the RBPs highlighted above; in contrast, circRNA synthesis can be inhibited by the RNA-editing enzyme ADAR1. ADAR1 significantly and specifically inhibited the biogenesis of circHIPK3, which altered the precursor of circHIPK3 secondary structure [77].
CircRNAs as potential biomarkers for CRC diagnosis and prognosis
CRC screening and early detection are crucial in enhancing treatment effectiveness and reducing CRC-related mortalities. CRC can be undetected over a long period, and only a few cases are diagnosed after presenting classic symptoms, such as weight loss, change in bowel habits, and perirectal bleeding. However, although stool occult blood tests, electronic colonoscopy, and digital rectal examination have improved the detection of CRC, effective biomarkers for CRC are necessary for early detection. Identifying either blood/serum or urine-based epigenetic biomarkers could be a promising diagnostic tool, as it would be noninvasive and inexpensive (Fig. 4).
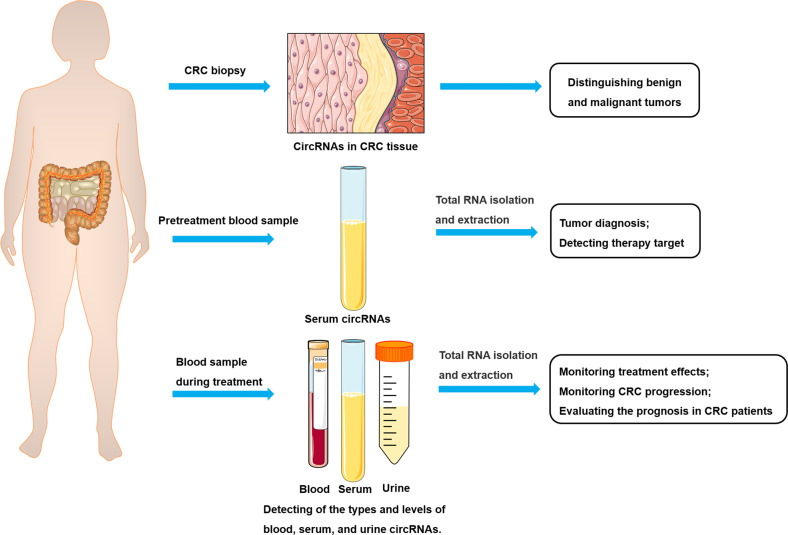
Fig. 4. circRNAs as potential biomarkers for the diagnosis and treatment of CRC.
circRNAs can act as an indicator of the differentiation of benign and malignant tumors. Serum circRNAs levels can act as potential biomarkers in CRC diagnosis, providing information for the selection of appropriate therapeutic strategies by clinicians. The circRNA levels of blood, serum, and urine samples could also serve as important clinical markers for CRC monitoring, treatment, and prediction of patient outcomes.
The sensitivity and specificity of circRNAs provide a valuable biomarker for CRC diagnosis for several reasons. First, due to their lack of 5′ or 3′ prime ends, circRNAs are highly resistant to exonuclease degradation and are extremely stable; thus, they are highly specific for tissues and diseases [78]. Secondly, circRNAs are found in cancer cells, solid tumors, peripheral blood, exosomes, and body fluids such as serum, plasma, and saliva [79]. For example, circ1662 and circPACRGL were significantly higher in patients with CRC, implying their specificity to cancer [8, 80]. Due to their resistance to degradation and presence in body fluids, circRNAs are the perfect candidate for noninvasive liquid biopsy, and therefore they have a high diagnostic potential. Recently, circ-KLDHC10 was successfully detected in serum samples, which can be used to distinguish patients with and without CRC [78]. A high level of CircALG1 expression was observed in CRC patients’ peripheral blood and tumor tissues and correlated with CRC metastasis [81], suggesting it may be an important biomarker for cancer. The importance of circRNAs as biomarkers for CRC diagnosis and prognosis is emphasized in this research because they regulate cancer signaling pathways. Numerous studies have demonstrated the critical roles that circRNAs play in cancer signaling pathways, including the PI3K/Akt, JAK/STAT, GEF-H1/RhoA, Wnt/-Catenin, and TGF-/Smad pathways, by upregulating oncogene expression, downregulating tumor suppressor genes, and/or downregulating downstream proteins [82–86]. Thus, circRNAs may be a valuable biomarker for CRC diagnosis.
CircRNAs possess potential applications as clinical biomarkers for liquid biopsies considering their resistance to RNase R digestion, presence as covalently closed continuous loops, and wide existence in eukaryotes. Particularly, circRNAs are promising biomarkers for the clinical diagnosis and prognosis of cancer because they can easily be detected using qualitative real-time PCR (qRT-PCR), and are highly stable and abundant in bodily fluids, such as serum/blood, saliva, and urine (Fig. 4). Current findings have shown that circRNAs exhibit aberrant expression, increased disease specificity, and clinical implications, making them potential candidates for CRC diagnosis. For example, circ3823 upregulation in CRC tissues was correlated with increased cell proliferation, metastasis, and angiogenesis and was an independent predictor of poor prognosis in patients with CRC [73]. Human circRNA microarray analysis indicated an increase in the expression of 30 circRNAs between CRC and normal tissues, which may be used as prognostic biomarkers for overall survival [87]. Additionally, five circRNAs (circ_0003906, circCDC66, circITGA7, circ_0000567, and circ_0001649) have been identified in CRC tissues and clinically validated using qRT-PCR and RNA-seq [88], and area under the curve (AUC) values were 0.818, 0.884, 0.879, 0.865, and 0.857, respectively, indicating their potential as diagnostic biomarkers. Notably, circ_0001178 had an AUC value of 0.945 [88], indicating a highly effective value for accurate diagnosis. Additionally, circPTK2 overexpression in CRC tissue and blood serum was positively linked to metastasis, clinical stage, and chemoresistance [87]. Moreover, circ5615 upregulation in CRC tissues was highly correlated with T stage and poor prognosis in CRC patients [89]. However, circPTEN1 is downregulated in the peritumoral and tumor tissues of patients with CRC [85]. Decreased expression of circRNA has been reported to facilitate metastasis and promote cell invasion in PDX models and is an independent predictor of poor survival in patients [85]. These findings suggest that circRNAs possess promising applications as diagnostic biomarkers for CRC.
Furthermore, certain circRNAs possess potential applications as prognostic biomarkers for CRC. circHIPK3 significantly promoted CRC cell proliferation, migration, and invasion, induced apoptosis in vitro, and facilitated CRC growth and metastasis in vivo [62]. Additionally, circHIPK3 was highly upregulated in CRC tissues and cell lines, and positively correlated with metastasis and advanced clinical stages, indicating the potential of circHIPK3 as a predictive biomarker of CRC. Similarly, Geng et al. observed that circ_0009361 downregulation promoted proliferation, migration, invasion, and epithelial-mesenchymal transition (EMT) in CRC cells [90]. In contrast, circ_0009361 overexpression significantly inhibited CRC growth and metastasis, indicating that circ_0009361 could act as a prognostic biomarker for CRC. Additionally, circSPARC expression was upregulated in CRC cells and positively correlated with advanced tumor node metastasis (TNM) stage, lymph node metastases, and poor survival in patients [83]. Furthermore, correlation analysis indicated that circSPARC expression was associated with tumor size, invasion, lymphatic metastasis, distant metastasis, and clinical stage [83]. Kaplan-Meier analysis showed that high circSPARC levels were associated with a decrease in overall survival [83], indicating its potential as a predictive biomarker for CRC.
These findings confirm that certain circRNAs possess promising potential as diagnostic biomarkers for CRC. However, although these circRNAs are differentially expressed in tissues, they cannot be detected in the blood/plasma or serum. Therefore, detecting circRNAs circulating in liquid biopsies, such as blood/plasma and/or serum, could facilitate the development of valid test procedures to distinguish between CRC patients and healthy individuals.
However, several limitations are associated with the clinical application of circRNAs for cancer as biomarkers. Circular RNAs are difficult to detect since their sequences are nearly identical to linear RNAs. Circular RNAs must be distinguished from linear RNA species using appropriate methods, and these methods need to be sensitive enough to detect the closed-loop structure of circRNAs efficiently. For example, circular RNAs can be detected with qRT-PCR, but when primers are designed using a linear genome as a template, circRNAs cannot be distinguished from linear RNAs. Microarray technology is an effective and relatively sensitive technique for quantifying circRNA expression, but it can only detect known circRNAs and cannot detect unknown circRNAs [91]. Apart from the qRT-PCR and Microarray technology, high-throughput sequencing techniques have become increasingly popular, such as second-generation high-throughput sequencing (NGS) and third-generation high-throughput sequencing (HTS). Thousands of circRNAs in human cells have recently been identified by applying high-throughput RNA sequencing technology and bioinformatics methods [92].
Although several circRNAs can be potential tumor biomarkers, studies investigating circRNAs as CRC biomarkers are limited. Due to their lack of sensitivity or specificity, most circRNA biomarkers are unlikely to be suitable for clinical application. Importantly, numerous clinical studies should be conducted on circRNAs as biomarkers will require standardized techniques and bioinformatics methods for their detection. For example, sophisticated large-scale prospective studies involving collecting serial samples and establishing time points and intervals are necessary before circRNAs can be used as biomarkers. Additionally, circRNA standardization is a crucial factor in liquid biopsies. Repeated experiments are necessary to determine the optimal time and cutoff value for circRNAs to be consistent with patient demands. Regardless of the challenges, these findings suggest that circRNAs possess promising applications as diagnostic biomarkers for CRC.
CircRNAs as potential therapeutic targets
More than 70 upregulated circRNAs are actively involved in CRC tumorigenesis and progression, and silencing them exerts opposite effects in vitro and in vivo [93]. Thus, these oncogenic circRNAs may serve as potential therapeutic targets, and target oncogenic circRNAs’ unique back-splice junctions for degradation by siRNAs may have anti-tumor properties. Numerous animal studies have revealed that siRNAs or short hairpin RNAs (shRNAs) specifically targeting oncogenic circRNAs have been shown to effectively inhibit the growth, proliferation, and metastasis of CRC [81, 94, 95]. For example, treatment with an shRNA targeting circMETTL3 inhibited tumor growth and metastasis in nude mice xenograft models [96], suggesting that the oncogenic circMETTL3 may serve as a potential therapeutic target. A PDX model for tumor metastasis was used by Chen et al. to confirm that the knockdown of circNSUN2 significantly reduced tumor metastasis in either liver or lung metastasis models [69].
Similarly, targeting circLONP2 by antisense oligonucleotide (ASO) significantly reduced the penetrance of CRC metastasis to foreign organs in vivo, including a reduction in both nodule size and number [97]. Interestingly, Wang et al. confirmed that an exosome-delivered siRNA targeting hsa_circ_0005963 sensitized CRC-resistant mice [98], implying a novel approach for reversing oxaliplatin resistance in CRC. Furthermore, certain drugs and compounds may exert anticancer activity through circRNA-associated pathways. For instance, lidocaine treatment inhibited the proliferation, and metastasis and induced cell apoptosis via regulating the circITFG2/miR-1204/SOCS2 axis, providing a novel treatment in improving CRC therapy [99]. Peptide 17 is a YAP-specific inhibitor that significantly inhibited the proliferation and metastasis-promoting effect of circPPP1R12A-73aa on colon cancer cells [100].
It is well known that numerous downregulated circRNAs negatively regulate CRC growth and metastasis. Due to their high stability and long half-life, tumor suppressor circRNAs may have significant anti-tumor effects when expressed in CRC cells or tissues. Zheng et al. observed that circLPAR1 was downregulated in CRC tissues and circLPAR1 overexpression treatment reduced tumor weight and size, implying that it portends poor prognosis [45]. Moreover, exogenous circRNAs may be delivered by specific vectors containing gene expression cassettes designed to express circRNAs or by transfection of purified in vitro-generated circRNAs. Recent studies have confirmed the synthesis and cloning of circRNA sequences into special vectors (such as lentiviruses vectors [LV] and recombinant adeno-associated viral [AAV] vectors) to produce LV or AAV to transfect CRC cell lines or animal model and constitutively overexpress the desired circRNAs [101]; the exogenous circRNA then acted as a tumor suppressor by sponging multiple miRNAs [102–104]. Engineered circRNAs could serve as sponges for specific oncogenic miRNAs in CRC cells or tissues, representing an efficient and innovative treatment approach for the disease in the future.
Some circRNAs such as circRS-122, circ_001680, circ_0002813, circ_101277, circ_0000236, and circ-ZEB1 are associated with chemotherapy resistance (e.g., fluorouracil (5-Fu), oxaliplatin, cisplatin, and irinotecan) in CRC [55, 98, 105–108]. For this reason, detecting the expression of these circRNAs may be useful for predicting the sensitivity of patients with CRC to chemoradiotherapy in the clinic. In addition, some circRNAs, such as circIFNGR2 and circLHFPL2, are related to drug resistance (e.g., cetuximab and MEK inhibitor) in CRC [95, 109]. Circular RNAs may be useful for predicting drug resistance in patients with CRC. Moreover, therapy targeting these circRNAs may also improve chemoradiotherapy and drug resistance in patients with CRC. For the treatment of CRC, antisense oligonucleotides (ASOs) were also developed that target circularization and secretion elements of circRNAs, including circRHOBTB3 [22]. Immunotherapy for CRC patients might be more effective with interventions targeting circular RNA CDR1-AS by PD-1/PD-L1 blocking therapies [110]. Finally, a combination of sh-circQSOX1 and anti-T-lymphocyte-associated antigen-4 (CTLA-4) could be more effective for overcoming the resistance to immune therapies mediated by Treg cells in CRC [111].
CircRNA therapies
The use of RNA-based therapeutics may provide a potential treatment for various human diseases, including infectious diseases, cancers, and lipid-related diseases. For instance, mRNA vaccines can induce specific immune responses to protect against infectious diseases and cancers in animal models and humans [112]. Various RNA-based therapies, such as antisense oligonucleotides (ASOs), siRNAs, ASO anti-microRNAs (anti-miRs), miRNA mimics, miRNA sponges, circRNA therapies and CRISPR-Cas9-based gene editing, are performed and found to improve the quality of life and prolongs the lifespan of patients with various disease [113, 114]. The FDA and/or European Medicines Agency (EMA) have approved 11 RNA-based therapeutics targeting multiple patient tissues and organs [113]. Furthermore, their study and ours indicated that siRNA is a useful tool for silencing genes [115, 116]; four siRNA drug candidates (Patisiran, Givosiran, Lumasiran, and Inclisiran) have been approved by FDA and/or EMA [114]. Yu et al. designed synthesized chrysotile nanotubes (SCNTs) to encapsulate siRNA (SCNTs/si-circPRMT5) against the oncogenic circPRMT5 expression and then inhibited bladder cancer growth and metastasis [117]. Hence, SCNTs/si-circPRMT5 may have therapeutic value in treating bladder cancer.
Like other RNA therapeutics, circRNA has the potential effects of modulating gene expression or carrying out modular functions. CircRNAs served as miRNA sponges, further broadening the possibilities for inhibiting the oncogenic RNA function. For example, hsa_circ_001783 promoted breast cancer progression via sponging miR-200c-3p [118]. Synthetic circRNAs have attracted more attention due to their strong and stable translation in eukaryotic cells [119, 120]. Recently, Li et al. developed a novel circRNA vaccine platform to stimulate robust innate and adaptive immune responses for the anti-tumor effect in multiple mouse tumor models [121]. Qu et al. presented the circRNA vaccine against SARS-CoV-2 encoding the spike protein to protect against SARSCoV-2 infection [122]. Moreover, they also demonstrated the use of synthetic circRNAs to produce neutralizing antibodies against SARS-CoV-2 and hACE2 decoys to neutralize pseudovirus particles [122]. However, synthesized circRNAs still face many challenges for their development as therapeutic agents, such as avoiding sustained overexpression due to their exceptional properties, the production of highly purified artificial circRNAs, and their specific delivery. Therefore, further research should be required to address and overcome these challenges.
In another application, engineered circRNA purified by high-performance liquid chromatography demonstrated outstanding protein production quality in both quantity and stability production in eukaryotic cells [123]. Thus, circRNA is a viable alternative to linear mRNA. In addition to engineered circRNAs, CircaRNA-based aptamers are produced by twister-optimized RNA for durable overexpression (Tornado) that circularizes RNA to produce aptamers capable of binding proteins [124]. Despite significant progress in the research and application of circRNAs, most candidates are currently in the discovery or preclinical stages. So far, no circRNA therapeutic candidate has entered clinical trials. Numerous medical and research applications exist for CircRNA, including cancer therapy, protein replacement therapy, and prophylactic vaccines. In addition, it is important to note that circRNAs can also be targeted for modulation using other methods, such as CRISPR-Cas9 or siRNA, and native circRNAs can be used as biomarkers or sponging agents for various diseases, especially cancer. Nevertheless, RNA therapeutics, like mRNA-based therapies, can be translatable to circRNA-based therapies and provide valuable insights into the development of circRNAs as therapeutic agents.
Challenges and future perspectives
Although the biological role of circRNA in CRC has received increased research attention recently, the role of circRNAs in CRC treatment has not yet been extensively explored, indicating the need for further studies. For instance, the biological and molecular mechanisms of only a few circRNAs in CRC have been elucidated. Further research is needed to determine the exact mechanisms of circRNA circularization, degradation, extracellular transport, and subcellular localization in CRC. Owing to the potential for alternative splicing of pre-mRNA [125], the internal structure of circRNAs remains unclear. Recently, a novel algorithm called CircSplice has been developed and is capable of identifying alternate splicing in circRNAs and comparing distinct circRNA splicing events [126]. The patterns of cancer-specific circRNA alternative splicing (circ-AS) could be characterized using CircSplice, providing a promising resource for elucidating the regulation and functional implications of circRNAs in cancers. Several studies on circRNAs mainly focus on their role as miRNA sponges, RBP sponges, and regulators of mRNA expression. However, circRNA function may also be regulated by mechanisms other than those mentioned above. Additionally, studies are yet to clarify whether circRNAs can be simultaneously regulated by different molecular mechanisms.
Furthermore, several studies have shown that TME is closely related to colorectal tumor initiation and progression [127]. In several cancers, including CRC, the characteristics of the TME strongly influence tumor invasion, metastasis, proliferation, and drug resistance [128]. Therefore, research on the role of circRNAs in TME in patients with colorectal cancer may provide potential novel biomarkers and therapeutic targets for CRC treatment. Currently, the clinical applications of circRNA biomarkers are limited because of their lack of sensitivity and specificity, indicating the need for further studies using standardized techniques and bioinformatics approaches.
Although some circRNAs are located in the nucleus (such as intronic and exon-intron circRNAs) [40], most circRNAs accumulate in the cytoplasm [23]. Numerous studies have demonstrated that circRNAs can be exported from the nucleus to the cytoplasm to perform regulatory functions [129, 130]. However, it is unclear how circRNAs are exported from the nucleus since they lack characteristics used by RNA export pathways. Surprisingly, Huang et al. discovered a novel, evolutionarily conserved pathway for circRNA export [131]. Deleting the Drosophila DExH/D-box helicase Hel25E accumulates long (more than 800 nucleotides) circular RNAs in the nucleus [131]. In recent studies, DDX39B and DDX39A, components of TREX, Exportin 4 (XPO4), are shown to regulate the export of exonic circular RNAs, while NXF1/NXT1 determines the export of cytoplasmic circular RNAs [132]. The NXF1-NXT1 pathway modulates toxic DPR production through the nuclear export of circular introns [129]. Researchers have identified DDX39A and DDX39B as regulators of ecircRNA nuclear export [131]. Recent studies have shown that DDX39B could unwind R-loops, and DDX39 participated in ecircRNA export by resolving ciR-loops [133]. In addition, Chen et al. reported that the conserved XPO4 is linked to the nuclear export of circRNAs in metazoans [134]. Knockout of XPO4 led to the ecircRNA accumulation in the nucleus [134]. Moreover, Chen et al. identified an N6-methyladenosine (m6A) modified circRNA, which can affect cytoplasmic export and CRC development. For example, m6A modification of circNSUN2 increased its export to the cytoplasm, forming a cirNSUN2/IGF2BP2/ HMGA2 RNA-protein ternary complex in the cytoplasm and promoting CRC by enhancing the stability of HMGA2 mRNA [69]. Thus, export of circRNA from the nucleus to the cytoplasm is required for its proper function.
Finally, further studies are required to develop effective circRNA delivery systems to tumor cells to regulate cancer progression without immune rejection and with sustained long-term effects. The applications of circRNAs may be remarkably improved by exosomes. Several eukaryotic cells and cancerous cells release extracellular vesicles, known as exosomes, that mediate intercellular communication via the transport of signaling molecules, including circRNAs, to cancer cells [135]. Exosomes have been confirmed to contain circRNAs, including circPACRGL, circSHKBP1, circUHRF1, and ciRS-122 [136]. Interestingly, serum exosomal circSATB2 levels can be used to identify patients with lung cancer with high sensitivity and specificity [78]; however, the biological functions of exosomal circRNAs in CRC requires further study.
Conclusions
This review extensively discusses the biogenesis, characteristics, and mechanisms of circRNA in CRC. Particularly, the potential clinical applications of circRNA as biomarkers for CRC diagnosis and prognosis and as therapeutic targets for CRC treatment were highlighted. However, the understanding of the activities of circRNAs and how they initiate CRC is lacking, indicating the need for further research.
Author contributions
YY Zhang and WC Ye conceived the structure of the manuscript; YY Zhang and WC Ye drafted the initial manuscript; YY Zhang, JY Luo, WK Yang and WC Ye revised and edited the manuscript. All authors contributed to the preparation of this manuscript. All the authors approved the final manuscript.
Funding
This work was supported by the grants from National Natural Science Foundation of China (No. 82000407 and 81802551), Shenzhen Science and Technology Program (No. JCYJ20220531091200001) and Natural Science Foundation of Guangdong Province (2023A1515010425).
Data availability
There are no experimental datasets, given that this is a review article that is prepared based on a literature review.
Competing interests
The authors declare no competing interests.
Consent for publication
Not applicable.
Footnotes
Edited by Giovanni Blandino
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Global, regional, and national burden of colorectal cancer and its risk factors, 1990-2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet Gastroenterol Hepatol 2022. [DOI] [PMC free article] [PubMed]
- 3.Pritchard CC, Grady WM. Colorectal cancer molecular biology moves into clinical practice. Gut. 2011;60:116–29. doi: 10.1136/gut.2009.206250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo JU, Agarwal V, Guo H, Bartel DP. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 2014;15:409. doi: 10.1186/s13059-014-0409-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–91. doi: 10.1038/s41576-019-0158-7. [DOI] [PubMed] [Google Scholar]
- 6.Kristensen LS, Jakobsen T, Hager H, Kjems J. The emerging roles of circRNAs in cancer and oncology. Nat Rev Clin Oncol. 2022;19:188–206. doi: 10.1038/s41571-021-00585-y. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Wang C, Sun H, Wang J, Liang Y, Wang Y, et al. The bioinformatics toolbox for circRNA discovery and analysis. Brief Bioinform. 2021;22:1706–28. doi: 10.1093/bib/bbaa001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shang A, Gu C, Wang W, Wang X, Sun J, Zeng B, et al. Exosomal circPACRGL promotes progression of colorectal cancer via the miR-142-3p/miR-506-3p- TGF-β1 axis. Mol Cancer. 2020;19:117. doi: 10.1186/s12943-020-01235-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang C, Tan S, Li J, Liu W-R, Peng Y, Li W. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett. 2020;492:106–15. doi: 10.1016/j.canlet.2020.08.013. [DOI] [PubMed] [Google Scholar]
- 10.Liu H, Lan T, Li H, Xu L, Chen X, Liao H, et al. Circular RNA circDLC1 inhibits MMP1-mediated liver cancer progression via interaction with HuR. Theranostics. 2021;11:1396–411. doi: 10.7150/thno.53227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6:319–36. doi: 10.1016/j.trecan.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 12.Yang X, Ye T, Liu H, Lv P, Duan C, Wu X, et al. Expression profiles, biological functions and clinical significance of circRNAs in bladder cancer. Mol Cancer. 2021;20:4. doi: 10.1186/s12943-020-01300-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang X, Li J, Bian X, Wu C, Hua J, Chang S, et al. CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc Natl Acad Sci USA. 2021;118:e2012881118. doi: 10.1073/pnas.2012881118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jangi M, Boutz PL, Paul P, Sharp PA. Rbfox2 controls autoregulation in RNA-binding protein networks. Genes Dev. 2014;28:637–51. doi: 10.1101/gad.235770.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim KK, Nam J, Mukouyama Y-S, Kawamoto S. Rbfox3-regulated alternative splicing of Numb promotes neuronal differentiation during development. J Cell Biol. 2013;200:443–58. doi: 10.1083/jcb.201206146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi S, Lee HS, Cho N, Kim I, Cheon S, Park C, et al. RBFOX2-regulated TEAD1 alternative splicing plays a pivotal role in Hippo-YAP signaling. Nucleic Acids Res. 2022;50:8658–73. doi: 10.1093/nar/gkac509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gordon MA, Babbs B, Cochrane DR, Bitler BG, Richer JK. The long non-coding RNA MALAT1 promotes ovarian cancer progression by regulating RBFOX2-mediated alternative splicing. Mol Carcinog. 2019;58:196–205. doi: 10.1002/mc.22919. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Chen S, Wei S, Cheng S, Shi R, Zhao R, et al. CircRAPGEF5 interacts with RBFOX2 to confer ferroptosis resistance by modulating alternative splicing of TFRC in endometrial cancer. Redox Biol. 2022;57:102493. doi: 10.1016/j.redox.2022.102493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Huang C, Shan G. Circular RNAs in physiology and non-immunological diseases. Trends Biochem Sci. 2022;47:250–64. doi: 10.1016/j.tibs.2021.11.004. [DOI] [PubMed] [Google Scholar]
- 20.Ren B, Guan M-X, Zhou T, Cai X, Shan G. Emerging functions of mitochondria-encoded noncoding RNAs. Trends Genet. 2023;39:125–39. doi: 10.1016/j.tig.2022.08.004. [DOI] [PubMed] [Google Scholar]
- 21.Liu X, Wang X, Li J, Hu S, Deng Y, Yin H, et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci China Life Sci. 2020;63:1429–49. doi: 10.1007/s11427-020-1631-9. [DOI] [PubMed] [Google Scholar]
- 22.Chen C, Yu H, Han F, Lai X, Ye K, Lei S, et al. Tumor-suppressive circRHOBTB3 is excreted out of cells via exosome to sustain colorectal cancer cell fitness. Mol Cancer. 2022;21:46. doi: 10.1186/s12943-022-01511-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–57. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ashwal-Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, et al. circRNA biogenesis competes with pre-mRNA splicing. Mol Cell. 2014;56:55–66. doi: 10.1016/j.molcel.2014.08.019. [DOI] [PubMed] [Google Scholar]
- 25.Huang X, He M, Huang S, Lin R, Zhan M, Yang D, et al. Circular RNA circERBB2 promotes gallbladder cancer progression by regulating PA2G4-dependent rDNA transcription. Mol Cancer. 2019;18:166. doi: 10.1186/s12943-019-1098-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aktaş T, Avşar Ilık İ, Maticzka D, Bhardwaj V, Pessoa Rodrigues C, Mittler G, et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature. 2017;544:115–9. doi: 10.1038/nature21715. [DOI] [PubMed] [Google Scholar]
- 27.Ivanov A, Memczak S, Wyler E, Torti F, Porath HT, Orejuela MR, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10:170–7. doi: 10.1016/j.celrep.2014.12.019. [DOI] [PubMed] [Google Scholar]
- 28.Shen H, An O, Ren X, Song Y, Tang SJ, Ke X-Y, et al. ADARs act as potent regulators of circular transcriptome in cancer. Nat Commun. 2022;13:1508. doi: 10.1038/s41467-022-29138-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell. 2015;58:870–85. doi: 10.1016/j.molcel.2015.03.027. [DOI] [PubMed] [Google Scholar]
- 30.Conn SJ, Pillman KA, Toubia J, Conn VM, Salmanidis M, Phillips CA, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160:1125–34. doi: 10.1016/j.cell.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 31.Gupta SK, Garg A, Bär C, Chatterjee S, Foinquinos A, Milting H, et al. Quaking inhibits doxorubicin-mediated cardiotoxicity through regulation of cardiac circular RNA expression. Circ Res. 2018;122:246–54. doi: 10.1161/CIRCRESAHA.117.311335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333–8. doi: 10.1038/nature11928. [DOI] [PubMed] [Google Scholar]
- 33.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wong CH, Lou UK, Fung FK-C, Tong JHM, Zhang C-H, To K-F, et al. CircRTN4 promotes pancreatic cancer progression through a novel CircRNA-miRNA-lncRNA pathway and stabilizing epithelial-mesenchymal transition protein. Mol Cancer. 2022;21:10. doi: 10.1186/s12943-021-01481-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zeng K, Chen X, Xu M, Liu X, Hu X, Xu T, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9:417. doi: 10.1038/s41419-018-0454-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Lan Z, Wang T, Zhang L, Jiang Z, Zou X. CircSLC8A1 exacerbates hypoxia-induced myocardial injury via interacting with MiR-214-5p to upregulate TEAD1 expression. Int Heart J. 2022;63:591–601. doi: 10.1536/ihj.21-547. [DOI] [PubMed] [Google Scholar]
- 37.Shan K, Liu C, Liu B-H, Chen X, Dong R, Liu X, et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation. 2017;136:1629–42. doi: 10.1161/CIRCULATIONAHA.117.029004. [DOI] [PubMed] [Google Scholar]
- 38.Chen X, Mao R, Su W, Yang X, Geng Q, Guo C, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p-STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16:659–71. doi: 10.1080/15548627.2019.1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yong W, Deng S, Tan Y, Li S. Circular RNA circSLC8A1 inhibits the proliferation and invasion of non-small cell lung cancer cells through targeting the miR-106b-5p /FOXJ3 axis. Cell Cycle. 2021;20:2597–606. doi: 10.1080/15384101.2021.1995968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–64. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 41.Wang L, Long H, Zheng Q, Bo X, Xiao X, Li B. Circular RNA circRHOT1 promotes hepatocellular carcinoma progression by initiation of NR2F6 expression. Mol Cancer. 2019;18:119. doi: 10.1186/s12943-019-1046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dong Z-R, Ke A-W, Li T, Cai J-B, Yang Y-F, Zhou W, et al. CircMEMO1 modulates the promoter methylation and expression of TCF21 to regulate hepatocellular carcinoma progression and sorafenib treatment sensitivity. Mol Cancer. 2021;20:75. doi: 10.1186/s12943-021-01361-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang Z-G, Awan FM, Du WW, Zeng Y, Lyu J, Wu D, et al. The circular RNA interacts with STAT3, increasing its nuclear translocation and wound repair by modulating Dnmt3a and miR-17 function. Mol Ther. 2017;25:2062–74. doi: 10.1016/j.ymthe.2017.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pamudurti NR, Bartok O, Jens M, Ashwal-Fluss R, Stottmeister C, Ruhe L, et al. Translation of CircRNAs. Mol Cell. 2017;66:9–21.e7. doi: 10.1016/j.molcel.2017.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zheng R, Zhang K, Tan S, Gao F, Zhang Y, Xu W, et al. Exosomal circLPAR1 functions in colorectal cancer diagnosis and tumorigenesis through suppressing BRD4 via METTL3-eIF3h interaction. Mol Cancer. 2022;21:49. doi: 10.1186/s12943-021-01471-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen S, Cao X, Zhang J, Wu W, Zhang B, Zhao F. circVAMP3 drives CAPRIN1 phase separation and inhibits hepatocellular carcinoma by suppressing c-Myc translation. Adv Sci (Weinh) 2022;9:e2103817. doi: 10.1002/advs.202103817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang Z, Lei X. Prediction of RBP binding sites on circRNAs using an LSTM-based deep sequence learning architecture. Brief Bioinform. 2021;22:bbab342. doi: 10.1093/bib/bbab342. [DOI] [PubMed] [Google Scholar]
- 48.Chen J, Wu Y, Luo X, Jin D, Zhou W, Ju Z, et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics. 2021;11:7507–26. doi: 10.7150/thno.59546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen Y, Yang F, Fang E, Xiao W, Mei H, Li H, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26:1346–64. doi: 10.1038/s41418-018-0220-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Du WW, Zhang C, Yang W, Yong T, Awan FM, Yang BB. Identifying and characterizing circRNA-Protein interaction. Theranostics. 2017;7:4183–91. doi: 10.7150/thno.21299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li X-X, Xiao L, Chung HK, Ma X-X, Liu X, Song J-L, et al. Interaction between HuR and circPABPN1 modulates autophagy in the intestinal epithelium by altering ATG16L1 translation. Mol Cell Biol. 2020;40:e00492–19. doi: 10.1128/MCB.00492-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–58. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li Q, Wang Y, Wu S, Zhou Z, Ding X, Shi R, et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 2019;30:157–73.e7. doi: 10.1016/j.cmet.2019.05.009. [DOI] [PubMed] [Google Scholar]
- 54.Liang Z-X, Liu H-S, Xiong L, Yang X, Wang F-W, Zeng Z-W, et al. A novel NF-κB regulator encoded by circPLCE1 inhibits colorectal carcinoma progression by promoting RPS3 ubiquitin-dependent degradation. Mol Cancer. 2021;20:103. doi: 10.1186/s12943-021-01404-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pan Z, Zheng J, Zhang J, Lin J, Lai J, Lyu Z, et al. A novel protein encoded by exosomal circatg4b induces oxaliplatin resistance in colorectal cancer by promoting autophagy. Adv Sci (Weinh) 2022;9:e2204513. doi: 10.1002/advs.202204513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang M, Huang N, Yang X, Luo J, Yan S, Xiao F, et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene. 2018;37:1805–14. doi: 10.1038/s41388-017-0019-9. [DOI] [PubMed] [Google Scholar]
- 57.Yang Y, Fan X, Mao M, Song X, Wu P, Zhang Y, et al. Extensive translation of circular RNAs driven by N-methyladenosine. Cell Res. 2017;27:626–41. doi: 10.1038/cr.2017.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Di Timoteo G, Dattilo D, Centrón-Broco A, Colantoni A, Guarnacci M, Rossi F, et al. Modulation of circRNA metabolism by m6A modification. Cell Rep. 2020;31:107641. doi: 10.1016/j.celrep.2020.107641. [DOI] [PubMed] [Google Scholar]
- 59.He J, Chu Z, Lai W, Lan Q, Zeng Y, Lu D, et al. Circular RNA circHERC4 as a novel oncogenic driver to promote tumor metastasis via the miR-556-5p/CTBP2/E-cadherin axis in colorectal cancer. J Hematol Oncol. 2021;14:194. doi: 10.1186/s13045-021-01210-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang D, Hu Z, Zhang Y, Zhang X, Xu J, Fu H, et al. CircHIPK3 promotes the tumorigenesis and development of gastric cancer through miR-637/AKT1 pathway. Front Oncol. 2021;11:637761. doi: 10.3389/fonc.2021.637761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z-G, Zhao H-J, Lin L, Liu J-B, Bai J-Z, Wang G-S. Circular RNA CirCHIPK3 promotes cell proliferation and invasion of breast cancer by sponging miR-193a/HMGB1/PI3K/AKT axis. Thorac Cancer. 2020;11:2660–71. doi: 10.1111/1759-7714.13603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen G, Shi Y, Liu M, Sun J. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9:175. doi: 10.1038/s41419-017-0204-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang C, et al. CircHIPK3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18:1646–59. doi: 10.15252/embr.201643581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qiu M, Xia W, Chen R, Wang S, Xu Y, Ma Z, et al. The circular RNA circPRKCI promotes tumor growth in lung adenocarcinoma. Cancer Res. 2018;78:2839–51. doi: 10.1158/0008-5472.CAN-17-2808. [DOI] [PubMed] [Google Scholar]
- 65.Guarnerio J, Bezzi M, Jeong JC, Paffenholz SV, Berry K, Naldini MM, et al. Oncogenic role of fusion-circRNAs derived from cancer-associated chromosomal translocations. Cell. 2016;165:289–302. doi: 10.1016/j.cell.2016.03.020. [DOI] [PubMed] [Google Scholar]
- 66.Wu K, Liao X, Gong Y, He J, Zhou J-K, Tan S, et al. Circular RNA F-circSR derived from SLC34A2-ROS1 fusion gene promotes cell migration in non-small cell lung cancer. Mol Cancer. 2019;18:98. doi: 10.1186/s12943-019-1028-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan S, Gou Q, Pu W, Guo C, Yang Y, Wu K, et al. Circular RNA F-circEA produced from EML4-ALK fusion gene as a novel liquid biopsy biomarker for non-small cell lung cancer. Cell Res. 2018;28:693–5. doi: 10.1038/s41422-018-0033-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang L, Hou C, Chen C, Guo Y, Yuan W, Yin D, et al. The role of N-methyladenosine (mA) modification in the regulation of circRNAs. Mol Cancer. 2020;19:105. doi: 10.1186/s12943-020-01224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen R-X, Chen X, Xia L-P, Zhang J-X, Pan Z-Z, Ma X-D, et al. N-methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat Commun. 2019;10:4695. doi: 10.1038/s41467-019-12651-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Xu J, Wan Z, Tang M, Lin Z, Jiang S, Ji L, et al. N-methyladenosine-modified CircRNA-SORE sustains sorafenib resistance in hepatocellular carcinoma by regulating β-catenin signaling. Mol Cancer. 2020;19:163. doi: 10.1186/s12943-020-01281-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wu P, Fang X, Liu Y, Tang Y, Wang W, Li X, et al. N6-methyladenosine modification of circCUX1 confers radioresistance of hypopharyngeal squamous cell carcinoma through caspase1 pathway. Cell Death Dis. 2021;12:298. doi: 10.1038/s41419-021-03558-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cui S, Wu Q, Liu M, Su M, Liu S, Shao L, et al. EphA2 super-enhancer promotes tumor progression by recruiting FOSL2 and TCF7L2 to activate the target gene EphA2. Cell Death Dis. 2021;12:264. doi: 10.1038/s41419-021-03538-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Guo Y, Guo Y, Chen C, Fan D, Wu X, Zhao L, et al. Circ3823 contributes to growth, metastasis and angiogenesis of colorectal cancer: involvement of miR-30c-5p/TCF7 axis. Mol Cancer. 2021;20:93. doi: 10.1186/s12943-021-01372-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39:283. doi: 10.1186/s13046-020-01799-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang X, Chen M, Fang L. hsa_circ_0068631 promotes breast cancer progression through c-Myc by binding to EIF4A3. Mol Ther Nucleic Acids. 2021;26:122–34. doi: 10.1016/j.omtn.2021.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu Z, Wang Q, Wang X, Xu Z, Wei X, Li J. Circular RNA regulates ferroptosis in HCC cells through interacting with RNA binding protein ALKBH5. Cell Death Discov. 2020;6:72. doi: 10.1038/s41420-020-00306-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Omata Y, Okawa M, Haraguchi M, Tsuruta A, Matsunaga N, Koyanagi S, et al. RNA editing enzyme ADAR1 controls miR-381-3p-mediated expression of multidrug resistance protein MRP4 via regulation of circRNA in human renal cells. J Biol Chem. 2022;298:102184. doi: 10.1016/j.jbc.2022.102184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25:981–4. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J, Zhang X, Yan M, Li H. Emerging Role of Circular RNAs in Cancer. Front Oncol. 2020;10:663. doi: 10.3389/fonc.2020.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen C, Yuan W, Zhou Q, Shao B, Guo Y, Wang W, et al. N6-methyladenosine-induced circ1662 promotes metastasis of colorectal cancer by accelerating YAP1 nuclear localization. Theranostics. 2021;11:4298–315. doi: 10.7150/thno.51342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lin C, Ma M, Zhang Y, Li L, Long F, Xie C, et al. The N-methyladenosine modification of circALG1 promotes the metastasis of colorectal cancer mediated by the miR-342-5p/PGF signalling pathway. Mol Cancer. 2022;21:80. doi: 10.1186/s12943-022-01560-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jiang T, Wang H, Liu L, Song H, Zhang Y, Wang J, et al. CircIL4R activates the PI3K/AKT signaling pathway via the miR-761/TRIM29/PHLPP1 axis and promotes proliferation and metastasis in colorectal cancer. Mol Cancer. 2021;20:167. doi: 10.1186/s12943-021-01474-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wang J, Zhang Y, Song H, Yin H, Jiang T, Xu Y, et al. The circular RNA circSPARC enhances the migration and proliferation of colorectal cancer by regulating the JAK/STAT pathway. Mol Cancer. 2021;20:81. doi: 10.1186/s12943-021-01375-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Yang H, Zhang H, Yang Y, Wang X, Deng T, Liu R, et al. Hypoxia induced exosomal circRNA promotes metastasis of colorectal cancer via targeting GEF-H1/RhoA axis. Theranostics. 2020;10:8211–26. doi: 10.7150/thno.44419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zheng L, Liang H, Zhang Q, Shen Z, Sun Y, Zhao X, et al. circPTEN1, a circular RNA generated from PTEN, suppresses cancer progression through inhibition of TGF-β/Smad signaling. Mol Cancer. 2022;21:41. doi: 10.1186/s12943-022-01495-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zou Y, Liu L, Meng J, Dai M. Circular RNA circ_0068464 combined with microRNA-383 regulates Wnt/β-catenin pathway to promote the progression of colorectal cancer. Bioengineered. 2022;13:5113–25. doi: 10.1080/21655979.2022.2036905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang H, Li X, Meng Q, Sun H, Wu S, Hu W, et al. CircPTK2 (hsa_circ_0005273) as a novel therapeutic target for metastatic colorectal cancer. Mol Cancer. 2020;19:13. doi: 10.1186/s12943-020-1139-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal Cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18:8. doi: 10.1186/s12943-018-0932-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ma Z, Han C, Xia W, Wang S, Li X, Fang P, et al. circ5615 functions as a ceRNA to promote colorectal cancer progression by upregulating TNKS. Cell Death Dis. 2020;11:356. doi: 10.1038/s41419-020-2514-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Geng Y, Zheng X, Hu W, Wang Q, Xu Y, He W, et al. Hsa_circ_0009361 acts as the sponge of miR-582 to suppress colorectal cancer progression by regulating APC2 expression. Clin Sci (Lond) 2019;133:1197–213. doi: 10.1042/CS20190286. [DOI] [PubMed] [Google Scholar]
- 91.Li S, Teng S, Xu J, Su G, Zhang Y, Zhao J, et al. Microarray is an efficient tool for circRNA profiling. Brief Bioinform. 2019;20:1420–33. doi: 10.1093/bib/bby006. [DOI] [PubMed] [Google Scholar]
- 92.Xia S, Feng J, Lei L, Hu J, Xia L, Wang J, et al. Comprehensive characterization of tissue-specific circular RNAs in the human and mouse genomes. Brief Bioinform. 2017;18:984–92. doi: 10.1093/bib/bbw081. [DOI] [PubMed] [Google Scholar]
- 93.Long F, Lin Z, Li L, Ma M, Lu Z, Jing L, et al. Comprehensive landscape and future perspectives of circular RNAs in colorectal cancer. Mol Cancer. 2021;20:26. doi: 10.1186/s12943-021-01318-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Fu Z, Zhang P, Zhang R, Zhang B, Xiang S, Zhang Y, et al. Novel hypoxia-induced HIF1α-circTDRD3-positive feedback loop promotes the growth and metastasis of colorectal cancer. Oncogene. 2023;42:238–52. doi: 10.1038/s41388-022-02548-8. [DOI] [PubMed] [Google Scholar]
- 95.Zhang Q, Zheng Y, Liu J, Tang X, Wang Y, Li X, et al. CircIFNGR2 enhances proliferation and migration of CRC and induces cetuximab resistance by indirectly targeting KRAS via sponging to MiR-30b. Cell Death Dis. 2023;14:24. doi: 10.1038/s41419-022-05536-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zhang F, Su T, Xiao M. RUNX3-regulated circRNA METTL3 inhibits colorectal cancer proliferation and metastasis via miR-107/PER3 axis. Cell Death Dis. 2022;13:550. doi: 10.1038/s41419-022-04750-8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 97.Han K, Wang F-W, Cao C-H, Ling H, Chen J-W, Chen R-X, et al. CircLONP2 enhances colorectal carcinoma invasion and metastasis through modulating the maturation and exosomal dissemination of microRNA-17. Mol Cancer. 2020;19:60. doi: 10.1186/s12943-020-01184-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X, Zhang H, Yang H, Bai M, Ning T, Deng T, et al. Exosome-delivered circRNA promotes glycolysis to induce chemoresistance through the miR-122-PKM2 axis in colorectal cancer. Mol Oncol. 2020;14:539–55. doi: 10.1002/1878-0261.12629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang H, Zhang X, Li Y, Li Y, Pang T. Lidocaine hampers colorectal cancer process via circITFG2/miR-1204/SOCS2 axis. Anticancer Drugs. 2022;33:235–44. doi: 10.1097/CAD.0000000000001091. [DOI] [PubMed] [Google Scholar]
- 100.Zheng X, Chen L, Zhou Y, Wang Q, Zheng Z, Xu B, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019;18:47. doi: 10.1186/s12943-019-1010-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Meganck RM, Borchardt EK, Castellanos Rivera RM, Scalabrino ML, Wilusz JE, Marzluff WF, et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors In Vivo. Mol Ther Nucleic Acids. 2018;13:89–98. doi: 10.1016/j.omtn.2018.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yang T, Sun J, Wang W, Li D, Yang X, Jia A, et al. Hsa_circ_0006732 regulates colorectal cancer cell proliferation, invasion and EMT by miR-127-5p/RAB3D axis. Mol Cell Biochem. 2022;477:2751–60. doi: 10.1007/s11010-022-04458-5. [DOI] [PubMed] [Google Scholar]
- 103.Yang K-D, Wang Y, Zhang F, Luo B-H, Feng D-Y, Zeng Z-J. CircN4BP2L2 promotes colorectal cancer growth and metastasis through regulation of the miR-340-5p/CXCR4 axis. Lab Invest. 2022;102:38–47. doi: 10.1038/s41374-021-00632-3. [DOI] [PubMed] [Google Scholar]
- 104.Yan D, Liu W, Liu Y, Zhu X. Circular RNA circ_0065378 upregulates tumor suppressor candidate 1 by competitively binding with miR-4701-5p to alleviate colorectal cancer progression. J Gastroenterol Hepatol. 2022;37:1107–18. doi: 10.1111/jgh.15862. [DOI] [PubMed] [Google Scholar]
- 105.Jian X, He H, Zhu J, Zhang Q, Zheng Z, Liang X, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19:20. doi: 10.1186/s12943-020-1134-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chen H, Zhang J, Yang L, Li Y, Wang Z, Ye C. circ-ZEB1 regulates epithelial-mesenchymal transition and chemotherapy resistance of colorectal cancer through acting on miR-200c-5p. Transl Oncol. 2023;28:101604. doi: 10.1016/j.tranon.2022.101604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cheng P-Q, Liu Y-J, Zhang S-A, Lu L, Zhou W-J, Hu D, et al. RNA-Seq profiling of circular RNAs in human colorectal cancer 5-fluorouracil resistance and potential biomarkers. World J Gastrointest Oncol. 2022;14:678–89. doi: 10.4251/wjgo.v14.i3.678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lv Q, Xia Q, Li A, Wang Z. circRNA_101277 influences cisplatin resistance of colorectal cancer cells by modulating the miR-370/IL-6 Axis. Genet Res (Camb) 2022;2022:4237327. doi: 10.1155/2022/4237327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chong X, Chen J, Zheng N, Zhou Z, Hai Y, Chen S, et al. PIK3CA mutations-mediated downregulation of circLHFPL2 inhibits colorectal cancer progression via upregulating PTEN. Mol Cancer. 2022;21:118. doi: 10.1186/s12943-022-01531-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Tanaka E, Miyakawa Y, Kishikawa T, Seimiya T, Iwata T, Funato K, et al. Expression of circular RNA CDR1‑AS in colon cancer cells increases cell surface PD‑L1 protein levels. Oncol Rep. 2019;42:1459–66. doi: 10.3892/or.2019.7244. [DOI] [PubMed] [Google Scholar]
- 111.Liu Z, Zheng N, Li J, Li C, Zheng D, Jiang X, et al. N6-methyladenosine-modified circular RNA QSOX1 promotes colorectal cancer resistance to anti-CTLA-4 therapy through induction of intratumoral regulatory T cells. Drug Resist Updat. 2022;65:100886. doi: 10.1016/j.drup.2022.100886. [DOI] [PubMed] [Google Scholar]
- 112.Pardi N, Hogan MJ, Porter FW, Weissman D. mRNA vaccines - a new era in vaccinology. Nat Rev Drug Discov. 2018;17:261–79. doi: 10.1038/nrd.2017.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Winkle M, El-Daly SM, Fabbri M, Calin GA. Noncoding RNA therapeutics - challenges and potential solutions. Nat Rev Drug Discov. 2021;20:629–51. doi: 10.1038/s41573-021-00219-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Zhang C, Zhang B. RNA therapeutics: updates and future potential. Sci China Life Sci. 2023;66:12–30. doi: 10.1007/s11427-022-2171-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huang S-F, Zhao G, Peng X-F, Ye W-C. The pathogenic role of long non-coding RNA H19 in atherosclerosis via the miR-146a-5p/ANGPTL4 pathway. Front Cardiovasc Med. 2021;8:770163. doi: 10.3389/fcvm.2021.770163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang J, Zhao X, Wang Y, Ren F, Sun D, Yan Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11:32. doi: 10.1038/s41419-020-2230-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Yu C, Zhang Y, Wang N, Wei W, Cao K, Zhang Q, et al. Treatment of bladder cancer by geoinspired synthetic chrysotile nanocarrier-delivered circPRMT5 siRNA. Biomater Res. 2022;26:6. doi: 10.1186/s40824-022-00251-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Liu Z, Zhou Y, Liang G, Ling Y, Tan W, Tan L, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10:55. doi: 10.1038/s41419-018-1287-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Zhang Y, Xue W, Li X, Zhang J, Chen S, Zhang J-L, et al. The biogenesis of nascent circular RNAs. Cell Rep. 2016;15:611–24. doi: 10.1016/j.celrep.2016.03.058. [DOI] [PubMed] [Google Scholar]
- 120.Obi P, Chen YG. The design and synthesis of circular RNAs. Methods. 2021;196:85–103. doi: 10.1016/j.ymeth.2021.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Li H, Peng K, Yang K, Ma W, Qi S, Yu X, et al. Circular RNA cancer vaccines drive immunity in hard-to-treat malignancies. Theranostics. 2022;12:6422–36. doi: 10.7150/thno.77350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Qu L, Yi Z, Shen Y, Lin L, Chen F, Xu Y, et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell. 2022;185:1728–44.e16. doi: 10.1016/j.cell.2022.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wesselhoeft RA, Kowalski PS, Anderson DG. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat Commun. 2018;9:2629. doi: 10.1038/s41467-018-05096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Litke JL, Jaffrey SR. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat Biotechnol. 2019;37:667–75. doi: 10.1038/s41587-019-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions. Development. 2016;143:1838–47. doi: 10.1242/dev.128074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Feng J, Chen K, Dong X, Xu X, Jin Y, Zhang X, et al. Genome-wide identification of cancer-specific alternative splicing in circRNA. Mol Cancer. 2019;18:35. doi: 10.1186/s12943-019-0996-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Viralippurath Ashraf J, Sasidharan Nair V, Saleh R, Elkord E. Role of circular RNAs in colorectal tumor microenvironment. Biomed Pharmacother. 2021;137:111351. doi: 10.1016/j.biopha.2021.111351. [DOI] [PubMed] [Google Scholar]
- 128.Liu K, Cui J-J, Zhan Y, Ouyang Q-Y, Lu Q-S, Yang D-H, et al. Reprogramming the tumor microenvironment by genome editing for precision cancer therapy. Mol Cancer. 2022;21:98. doi: 10.1186/s12943-022-01561-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Wang S, Latallo MJ, Zhang Z, Huang B, Bobrovnikov DG, Dong D, et al. Nuclear export and translation of circular repeat-containing intronic RNA in C9ORF72-ALS/FTD. Nat Commun. 2021;12:4908. doi: 10.1038/s41467-021-25082-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Lee ES, Wolf EJ, Ihn SSJ, Smith HW, Emili A, Palazzo AF. TPR is required for the efficient nuclear export of mRNAs and lncRNAs from short and intron-poor genes. Nucleic Acids Res. 2020;48:11645–63. doi: 10.1093/nar/gkaa919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Huang C, Liang D, Tatomer DC, Wilusz JE. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 2018;32:639–44. doi: 10.1101/gad.314856.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Talhouarne GJS, Gall JG. Lariat intronic RNAs in the cytoplasm of vertebrate cells. Proc Natl Acad Sci USA. 2018;115:E7970–E7977. doi: 10.1073/pnas.1808816115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pérez-Calero C, Bayona-Feliu A, Xue X, Barroso SI, Muñoz S, González-Basallote VM, et al. UAP56/DDX39B is a major cotranscriptional RNA-DNA helicase that unwinds harmful R loops genome-wide. Genes Dev. 2020;34:898–912. doi: 10.1101/gad.336024.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Chen L, Wang Y, Lin J, Song Z, Wang Q, Zhao W, et al. Exportin 4 depletion leads to nuclear accumulation of a subset of circular RNAs. Nat Commun. 2022;13:5769. doi: 10.1038/s41467-022-33356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wang S, Dong Y, Gong A, Kong H, Gao J, Hao X, et al. Exosomal circRNAs as novel cancer biomarkers: challenges and opportunities. Int J Biol Sci. 2021;17:562–73. doi: 10.7150/ijbs.48782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Zhang P-F, Gao C, Huang X-Y, Lu J-C, Guo X-J, Shi G-M, et al. Cancer cell-derived exosomal circUHRF1 induces natural killer cell exhaustion and may cause resistance to anti-PD1 therapy in hepatocellular carcinoma. Mol Cancer. 2020;19:110. doi: 10.1186/s12943-020-01222-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Hsiao K-Y, Lin Y-C, Gupta SK, Chang N, Yen L, Sun HS, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–50. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mo Y, Lu Q, Zhang Q, Chen J, Deng Y, Zhang K, et al. Circular RNA CCDC66 improves murine double minute 4 (MDM4) expression through targeting miR-370 in colorectal cancer. Comput Math Methods Med. 2022;2022:7723995. doi: 10.1155/2022/7723995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Cao L, Dong G, Li H. CircRNA circ-ATAD1 suppresses miR-618 maturation to participate in colorectal cancer. BMC Gastroenterol. 2022;22:215. doi: 10.1186/s12876-022-02183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Liu Y, Chen L, Liu T, Su X, Peng L, Chen J, et al. Genome-wide circular RNA (circRNA) and mRNA profiling identify a circMET-miR-410-3p regulatory motif for cell growth in colorectal cancer. Genomics. 2022;114:351–60. doi: 10.1016/j.ygeno.2021.11.038. [DOI] [PubMed] [Google Scholar]
- 141.Huang Y, Chen Z, Zhou X, Huang H. Circ_0000467 exerts an oncogenic role in colorectal cancer via miR-330-5p-dependent regulation of TYRO3. Biochem Genet. 2022;60:1488–510. doi: 10.1007/s10528-021-10171-7. [DOI] [PubMed] [Google Scholar]
- 142.Zhang W, Wang B, Lin Y, Yang Y, Zhang Z, Wang Q, et al. hsa_circ_0000231 promotes colorectal cancer cell growth through upregulation of CCND2 by IGF2BP3/miR-375 dual pathway. Cancer Cell Int. 2022;22:27. doi: 10.1186/s12935-022-02455-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Han B, Wang X, Yin X. Knockdown of circRAD23B exerts antitumor response in colorectal cancer via the regulation of miR-1205/TRIM44 axis. Dig Dis Sci. 2022;67:504–15. doi: 10.1007/s10620-021-06859-w. [DOI] [PubMed] [Google Scholar]
- 144.Wang J, Ke S, Gong Y, Cai Y, Xia L, Shi Z, et al. Circ_0011385 knockdown inhibits cell proliferation, migration and invasion, whereas promotes cell apoptosis by regulating miR-330-3p/MYO6 axis in colorectal cancer. Biomed J. 2022;46:110–21. doi: 10.1016/j.bj.2022.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Zhao X, Cui D, Yan F, Yang L, Huang B. Circ_0007919 exerts an anti-tumor role in colorectal cancer through targeting miR-942-5p/TET1 axis. Pathol Res Pr. 2022;229:153704. doi: 10.1016/j.prp.2021.153704. [DOI] [PubMed] [Google Scholar]
- 146.Wang W, Zhou L, Li Z, Lin G. Circ_0014130 is involved in the drug sensitivity of colorectal cancer through miR-197-3p/PFKFB3 axis. J Gastroenterol Hepatol. 2022;37:908–18. doi: 10.1111/jgh.15829. [DOI] [PubMed] [Google Scholar]
- 147.Li J, Liu X, Dong S, Liao H, Huang W, Yuan X. Circ_0101802 facilitates colorectal cancer progression depending on the regulation of miR-665/DVL3 signaling. Biochem Genet. 2022;60:2250–67. doi: 10.1007/s10528-022-10207-6. [DOI] [PubMed] [Google Scholar]
- 148.Gao Y, Liu C, Xu X, Wang Y, Jiang Y. Circular RNA sterile alpha motif domain containing 4A contributes to cell 5-fluorouracil resistance in colorectal cancer by regulating the miR-545-3p/6-phosphofructo-2-kinase/fructose-2,6-bisphosphataseisotype 3 axis. Anticancer Drugs. 2022;33:553–63. doi: 10.1097/CAD.0000000000001285. [DOI] [PubMed] [Google Scholar]
- 149.Gao X, Yin J, Yao Y. hsa_circ_0001955 promotes colorectal cancer progression by regulating miR-583/FGF21 axis. J Oncol. 2022;2022:4288474. doi: 10.1155/2022/4288474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ma W, Niu Z, Han D, Wang B, Wang X. Circ-FAT1 Up-regulates FOSL2 expression by sponging miR-619-5p to facilitate colorectal cancer progression. Biochem Genet. 2022;60:1362–79. doi: 10.1007/s10528-021-10148-6. [DOI] [PubMed] [Google Scholar]
- 151.Feng Y, Wang X, Huang C, Zhang D, Liu T, Zhang C, et al. Upregulated circTMEM59 inhibits cell growth and metastasis by miR-668-3p/ID4 axis in colorectal cancer. Oxid Med Cell Longev. 2022;2022:7242124. doi: 10.1155/2022/7242124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Chen Z, He L, Zhao L, Zhang G, Wang Z, Zhu P, et al. circREEP3 drives colorectal cancer progression via activation of FKBP10 transcription and restriction of antitumor immunity. Adv Sci (Weinh) 2022;9:e2105160. doi: 10.1002/advs.202105160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.An Y, Xu B, Yan G, Wang N, Yang Z, Sun M. YAP derived circ-LECRC functions as a “brake signal” to suppress hyperactivation of oncogenic YAP signalling in colorectal cancer. Cancer Lett. 2022;532:215589. doi: 10.1016/j.canlet.2022.215589. [DOI] [PubMed] [Google Scholar]
- 154.Wang F, Wang J, Cao X, Xu L, Chen L. Hsa_circ_0014717 is downregulated in colorectal cancer and inhibits tumor growth by promoting p16 expression. Biomed Pharmacother. 2018;98:775–82. doi: 10.1016/j.biopha.2018.01.015. [DOI] [PubMed] [Google Scholar]
- 155.Hou M, Zhang LJ, Liu J, Hu HX, Zhao YL. CircRIP2 aggravates the deterioration of colorectal carcinoma by negatively regulating CBFB. Eur Rev Med Pharm Sci. 2022;26:3514–21. doi: 10.26355/eurrev_202205_28846. [DOI] [PubMed] [Google Scholar]
- 156.Chen Z, Qi Z, He D, Liu J, Xu E, Li B, et al. Strategy for scanning peptide-coding circular RNAs in colorectal cancer based on bioinformatics analysis and experimental assays. Front Cell Dev Biol. 2021;9:815895. doi: 10.3389/fcell.2021.815895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Zhang C, Zhou X, Geng X, Zhang Y, Wang J, Wang Y, et al. Circular RNA hsa_circ_0006401 promotes proliferation and metastasis in colorectal carcinoma. Cell Death Dis. 2021;12:443. doi: 10.1038/s41419-021-03714-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
There are no experimental datasets, given that this is a review article that is prepared based on a literature review.