Abstract
Age-related (AR) hearing loss (HL) is a prevalent sensory deficit in the elderly population. Several studies showed that common variants increase ARHL susceptibility. Here, we demonstrate that rare-variants play a crucial role in ARHL etiology. We analyzed exome and imputed data from white-European UK Biobank volunteers, performing both single-variant and rare-variant aggregate association analyses using self-reported ARHL phenotypes. We identified and replicated associations between ARHL and rare-variants in KLHDC7B, PDCD6, MYO6, SYNJ2, and TECTA. PUS7L and EYA4 also revealed rare-variant associations with ARHL. EYA4, MYO6, and TECTA are all known to underline Mendelian nonsyndromic HL. PDCD6, a new HL gene, plays an important role in apoptosis and has widespread inner ear expression, particularly in the inner hair cells. An unreplicated common variant association was previously observed for KHLDC7B, here we demonstrate that rare-variants in this gene also play a role in ARHL etiology. Additionally, the first replicated association between SYNJ2 and ARHL was detected. Analysis of common variants revealed several previously reported, i.e., ARHGEF28, and new, i.e., PIK3R3, ARHL associations, as well as ones we replicate here for the first time, i.e., BAIAP2L2, CRIP3, KLHDC7B, MAST2, and SLC22A7. It was also observed that the odds ratios for rare-variant ARHL associations, were higher than those for common variants. In conclusion, we demonstrate the vital role rare-variants, including those in Mendelian nonsyndromic HL genes, play in the etiology of ARHL.
Subject terms: Genome-wide association studies, Genetics research
Introduction
Age-related (AR) hearing loss (HL) or presbycusis, is defined as nonsyndromic (NS), bilateral, symmetrical sensorineural HL that advances with age, typically starting in the high frequencies and progressing to the mid-frequencies [1] ARHL is the most prevalent sensory deficit in older adults, i.e., ~50% of individuals >75 years-of-age have ARHL. It has a heritability of 0.35–0.55 [2]. ARHL is a complex trait with several risk factors that include male gender, genetic susceptibility, noise exposure, and ototoxic medication [3] and comorbidities e.g., cognitive decline, depression, diabetes, and hypertension. GWAS have identified many candidate variants/genes for ARHL [4–8], with only ARHGEF2, CHMP4C, CTBP2, EYA4, HLA-DRB1, ILDR1, ISG20, LOXHD1, NID2, SPTBN1, TRIOBP, and ZNF318 being replicated in two or more cohorts [4–6, 8].
GWAS studies of genotype array and imputed data facilitates the analysis of common and low frequency variants, but rare-variants are often missing or are of low imputation quality. Therefore, the analysis of exome data is important to study rare-variants. We examined the role rare-variants play in ARHL etiology, using exome sequence data and self-reported measures of HL obtained from white-European UK Biobank volunteers. Through rare-variant aggregate analysis we detected significant associations with ARHL for EYA4, KLHDC7B, PDCD6, PUS7L, MYO6, TECTA, and SYNJ2.
Materials and methods
Data access and ethical approval
This research was conducted using the UK Biobank Resource (application numbers 32285 and 36827) a longitudinal cohort of ~500,000 individuals who were 40 to 69 years-of-age at recruitment between 2006 and 2010 who are being followed-up [9, 10].The UK Biobank study was conducted under generic approval from the National Health Services’ National Research Ethics Service. The present analyses were approved by the Institutional Review Boards at Yale University (2000026836) and Columbia University (AAAS3494).
Quality control of genetic data
Genotype array data were used to detect inconsistencies in sex, generate principal components (PCs), and to perform ridge regression in the generalized linear mix model (GLMM) analysis. Information on subjects and variant quality control (QC) is provided in Table S1. UK Biobank also imputed SNVs, indels, and structural variants. For secondary-replication, imputed variants with an INFO Score ≥0.3 were analyzed.
Exome sequence data were generated using an IDT xGen Exome Research Panel v1.0 capture-array that targeted 39Mbp of the genome with different oligo lots used for the first (~50 K) and second release (~150 K), with coverage >20× at 95.6% of the targeted bases. Details on sequencing protocols can be found elsewhere [11]. A total of 200,643 samples (release 2) were processed by the UK Biobank: aligning FASTQs to the human genome with BWA. GATK was used to call SNVs and indels, followed by base quality score recalibration [11, 12]. Multiallelic splitting and left normalization were performed using bcftools v.1.12. Variants with an allelic imbalance (SNVs < 0.15 and indels <0.2) and genotypes with a read depth <10× or genotype quality score <20 were removed.
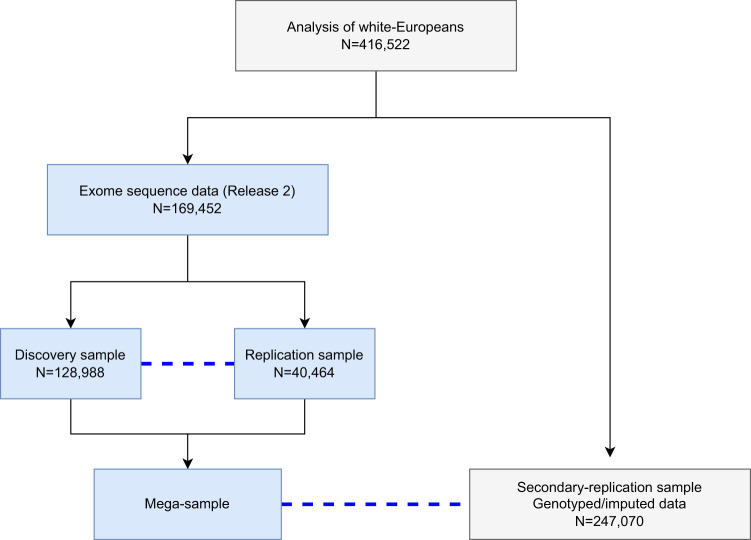
Individuals of self-reported white-European or unspecified ancestry, with both exome (release 2; October 2020) and genotype data, were used for PC analysis (PCA) to detect outliers (Table S1 and Fig. S1). Ten PCs were constructed using unrelated individuals (Θij < 0.0625), related individuals were projected back onto the PC plots, and Mahalanobis distances computed. Samples with a Mahalanobis distance of >0.997 were removed (Table S1). The exome data were divided into discovery and replication samples with those individuals in release 1 assigned to the replication sample and the remaining individuals assigned to the discovery sample. QC as described above was also performed for the secondary-replication sample, which is comprised of white-Europeans who did not have release 2 exome data (Table S1 and Fig. S1) but had available imputed data. There is no overlap of individuals included in the discovery, replication, and secondary-replication samples (Fig. 1).
Fig. 1. An illustration of samples used in the analysis.
We included in the analysis 416,522 samples after excluding individuals who are not white-European, do not meet the inclusion criteria, or provided inconsistent answers. Blue dashed lines indicates the replication sample which was used for the discovery and mega-analysis. Associations identified in the discovery sample are only replicated using the exome replication sample and not the secondary-replication sample. The replication sample consists of individuals included in UK Biobank Release 1. For replication, permutation is used to estimate empirical p-values adjusting for the number of variants/genes and phenotypes brought to replication. There is no overlap of individuals in the discovery, replication, and secondary-replication samples. All samples were used in the single-variant analysis and only those samples highlighted in light blue were used in the rare-variant aggregate analysis. There was no available replication sample for the mega-analysis of rare-variant aggregated association testing.
Variant annotation
ANNOVAR was used to annotate exome variants to the GRCh38 genomic reference. For gene-based annotation, RefSeq was used to annotate variants and to determine gene boundaries. Several bioinformatic tools (e.g., SIFT, Polyphen2, MutationTaster, FATHMM, CADD, and GERP++) were used to perform functional and prediction annotation and gnomAD v2.1.1 was used to annotate AFs.
Phenotype definition
Using ICD10/ICD9 and self-report codes strict exclusion criteria were applied to avoid inclusion of individuals with congenital, Mendelian, or environmental forms of HL e.g., due to ototoxic drugs usage or viral/bacterial infections. Individuals who had chronic otitis media, salpingitis, mastoiditis, otosclerosis, Meniere’s disease, deafness, labyrinthitis, conductive or ototoxic HL, head, ear or neck trauma, stroke, encephalitis, meningitis, or facial nerve disorders were excluded from the analysis. Individuals with unilateral/bilateral sensorineural or mixed conductive HL were also excluded if they were diagnosed <55 years-of-age or did not have an age-of-diagnosis (Table S2).
After excluding individuals with the above phenotypes, case/control status was determined based on the response to a touchscreen questionnaire during their assessment visit: (i) H-aid self-reported hearing aid use (f.3393: “Do you use a hearing aid most of the time?”); (ii) H-diff self-reported hearing difficulty (f.2247: “Do you have any difficulty with your hearing?”); (iii) H-noise self-reported hearing difficulty with background noise (f.2257: “Do you find it difficult to follow a conversation if there is background noise e.g., TV, radio, children playing)?”; and (iv) H-both individuals with both H-diff and H-noise (Fig. S1). Individuals who provided inconsistent answers for H-aid, H-diff, or H-noise (e.g., reported H-diff on the first visit but not the second visit), or did not provide definite answers, (e.g., answered “Do not know”) were excluded (Table S1 and Fig. S1). For cases, the age used in the analysis was the study subject’s age the first time they answered ‘Yes’ to a specific ARHL question during the assessment. We used a common set of controls that did not have any hearing-related phenotypes. For controls, age at last assessment was used in the analysis.
The number of cases (H-aid, H-diff, H-noise, and H-both) and the common set of controls analyzed in the discovery (exome), replication (exome), and secondary-replication (imputed) samples and their age and sex distributions, as well as the results from score tests (to determine if there are age or sex differences), are shown in Table S3.
Association analysis
Using GLMM as implemented in REGENIE 2.2.4 association analysis was performed for the discovery (n = 128,988), replication (n = 40,464) and mega (n = 169,452) exome data samples, where the mega-sample is the discovery and replication samples combined. Additionally, a secondary-replication sample of imputed data (n = 247,070) was analyzed. REGENIE 2.2.4 implements a multi-stage approach by initially fitting a whole-genome regression model, using ridge regressions, to estimate the polygenic effects parameters to account for population structure and relatedness [13]. A firth correction is used for binary traits, to ensure the results are well calibrated in the presence of rare-variants and unbalanced case-control ratios. The association model included covariates: sex, age, and two PCs. For single-variant analyses, variants with minor allele count ≥4 were analyzed. Rare-variant aggregate association analysis was performed using variants with a MAF < 0.01 (gnomAD non-Finnish European exome AFs) within a gene-region. Two analyses were performed aggregating within a gene-region: (1) predicted loss of function (pLOF) variants and (2) missense and pLOF variants, analyzing genes with ≥2 rare-variants. For every individual a ternary coding was used: 0 - no rare-variants; 1 - one or more heterozygous rare-variants; and 2 - at least one homozygous rare-variant within a gene-region.
Significance thresholds of p < 5.0 × 10−8 for single-variant and p < 2.5 × 10−6 (a Bonferroni correction for testing 20,000 genes) for gene-based rare-variant aggregate tests were used. For variants brought to replication, empirical p-values were obtained using 100,000 permutations. Empirical p-values were estimated by adjusting for the number of variants brought to replication by trait as well as across all ARHL traits. For the rare-variant aggregate tests, the same procedure was used but also adjusted for testing pLoF and missense & pLoF. A finding was considered replicated if an empirical p-value <0.05, adjusted for multiple testing, was obtained.
Conditional and stepwise model selection analysis
Conditional and stepwise model selection analysis was performed using GTCA-COJO to identify independent signals [14]. Linkage disequilibrium (LD) was estimated using a reference panel of unrelated white-European individuals from UK Biobank. Only variants with an r2 < 0.9 were analyzed due to collinearity for variants in strong LD.
In-silico mRNA expression analysis
Human and mouse inner ear expression was assessed in-silico. To study expression during mouse inner ear development an RNA expression dataset of hair cells and surrounding cells of mice cochleae and utricles over developmental stages: embryonic day (E)16, postnatal day (P)0, P4, and P7 and another RNA expression dataset over developmental stages: E12, E13, E16, P0, P6, and P15, that includes the spiral ganglion neurons and vestibular ganglion neurons, both obtained from SHIELD were analyzed [15]. A single-cell RNA sequence data of the cochlear epithelium during mouse developmental stages (E14, E16, P1, and P7) was interrogated and visualized using gEAR [16, 17]. Additionally, human inner ear expression data obtained from the RNA sequencing of inner ear tissue samples were processed, normalized, and visualized using DESeq2 [18, 19].
Results
Rare single-variant association analysis
Analysis of the discovery sample detected four rare-variants that displayed significant (p < 5.0 × 10−8) associations with at least one of the ARHL traits. PDCD6 contained three associated variants within exon 2 that were significantly associated with all four ARHL traits, these variants were replicated with H-diff, H-noise, and H-both (Table 1, Figs. S2 and S3). Rs121912560, a pathogenic MYO6 ADNSHL variant, was significantly associated with H-aid, H-diff, and H-both and replication was observed for H-diff and H-both (Tables 1 and S4).
Table 1.
Discovery single-variant association analysis for age-related hearing loss traits.
| CHR | SNP | EA | EAF | Gene | H-aid | H-diff | H-noise | H-both | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | |||||
| 5 | rs537688122 | G | 6.65 × 10−4 | PDCD6 | 1.99 (7.3) | 0.29 | 2.25 × 10−10 | 1.32 (3.7) | 0.17 | 1.12 × 10−15 | 1.04 (2.8) | 0.16 | 5.50 × 10−11 | 1.27 (3.6) | 0.18 | 1.02 × 10−12 |
| 5 | rs549592074 | C | 5.58 × 10−4 | PDCD6 | 1.99 (7.3) | 0.32 | 1.95 × 10−8 | 1.35 (3.9) | 0.18 | 7.05 × 10−14 | 1.07 (2.9) | 0.18 | 6.69 × 10−10 | 1.28 (3.6) | 0.19 | 5.52 × 10−11 |
| 5 | rs571370281 | G | 7.04 × 10−4 | PDCD6 | 1.92 (6.8) | 0.28 | 6.02 × 10−10 | 1.33 (3.8) | 0.16 | 1.14 × 10−16 | 1.03 (2.8) | 0.16 | 2.26 × 10−11 | 1.29 (3.6) | 0.17 | 9.66 × 10−14 |
| 6 | rs1574430 | C | 6.09 × 10−1 | SLC22A7 | 0.06 (1.1) | 0.01 | 2.77 × 10−8 | |||||||||
| 6 | rs2242416 | G | 6.09 × 10−1 | CRIP3 | 0.06 (1.1) | 0.01 | 2.60 × 10−8 | |||||||||
| 6 | rs121912560 | G | 7.63 × 10−5 | MYO6 | 5.48 (239.8) | 1.12 | 1.79 × 10−10 | 3.54 (34.5) | 0.90 | 3.41 × 10−9 | 3.73 (41.7) | 0.90 | 3.76 × 10−10 | |||
Genome-wide-significant variants (p < 5 × 10−8) with hearing aid (H-aid), hearing difficulty (H-diff), hearing difficulty with background noise (H-noise), and the combined hearing trait (H-both) in the analysis of the discovery sample of white-European individuals from the UK Biobank. The p-values for replicated associations (empirical p-values <0.05 adjusting for variants and traits brought to replication) are shown in bold (See Table S4 for replication analytical and empirical values).
CHR chromosome, EA effect allele, EAF effect allele frequency, OR odds ratio, SE standard error, P p-value.
In the analysis of the mega-sample additional genome-wide statistically significant associations were observed between H-both and SYNJ2 (rs146694394) which was replicated in the secondary-replication sample (Tables 2 and S4; Figs. S4 and S5). FILIP1 (rs765264064) was also observed to be associated with ARHL trait H-aid in the mega-sample but was not present in the secondary-replication sample (Table 2). This variant is in LD with rs121912560 (MYO6) and conditional analysis ruled out that FILIP1 is associated with ARHL.
Table 2.
Mega-sample single variant association analysis with age-related hearing loss traits.
| CHR | SNP | EA | EAF | Gene | H-aid | H-diff | H-noise | H-both | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | |||||
| 1 | rs11589562 | C | 0.424 | MAST2 | −0.05 (0.95) | 0.01 | 2.25 × 10−8 | |||||||||
| 1 | rs2275426 | A | 0.431 | MAST2 | −0.05 (0.95) | 0.01 | 3.39 × 10−8 | |||||||||
| 1 | rs1707336 | G | 0.435 | MAST2 | −0.05 (0.95) | 0.01 | 3.63 × 10−8 | |||||||||
| 1 | rs1707304 | A | 0.436 | PIK3R3 | −0.05 (0.95) | 0.01 | 2.34 × 10−8 | −0.05 (0.95) | 0.01 | 3.30 × 10−8 | ||||||
| 5 | rs537688122a | G | 7 × 10−4 | PDCD6 | 1.79 (6.0) | 0.25 | 7.06 × 10−11 | 1.35 (3.9) | 0.14 | 1.04 ×10−21 | 1.1 (3.0) | 0.14 | 4.96×10−16 | 1.32 (3.8) | 0.15 | 1.11 × 10−18 |
| 5 | rs549592074a | C | 6 × 10−4 | PDCD6 | 1.70 (5.5) | 0.28 | 2.48 × 10−8 | 1.37 (3.9) | 0.16 | 5.19 × 10−19 | 1.08 (3.0) | 0.15 | 2.19×10−13 | 1.32 (3.8) | 0.16 | 6.63 × 10−16 |
| 5 | rs571370281 | G | 7 × 10−4 | PDCD6 | 1.71 (5.5) | 0.24 | 1.34 × 10−10 | 1.31 (3.7) | 0.14 | 1.00 × 10−21 | 1.04 (2.8) | 0.14 | 1.83×10−15 | 1.28 (3.6) | 0.15 | 8.00 × 10−19 |
| 5 | rs7714670 | C | 0.467 | ARHGEF28 | 0.11 (1.1) | 0.02 | 9.99 × 10−9 | 0.05 (1.05) | 0.01 | 1.63 × 10−9 | 0.05 (1.05) | 0.01 | 1.06 × 10−8 | |||
| 5 | rs11949860 | A | 0.462 | ARHGEF28 | 0.11 (1.1) | 0.02 | 3.87 × 10−8 | 0.05 (1.05) | 0.01 | 9.92 × 10−9 | ||||||
| 5 | rs35525194 | G | 0.471 | ARHGEF28 | 0.11 (1.1) | 0.02 | 7.03 × 10−9 | 0.05 (1.05) | 0.01 | 2.19 × 10−9 | 0.05 (1.05) | 0.01 | 1.21 × 10−8 | |||
| 5 | rs6453022 | A | 0.501 | ARHGEF28 | 0.11 (1.1) | 0.02 | 7.30 × 10−9 | 0.05 (1.05) | 0.01 | 2.75 × 10−10 | 0.06 (1.06) | 0.01 | 4.13 × 10−10 | |||
| 5 | rs7716253 | C | 0.524 | ARHGEF28 | 0.11 (1.1) | 0.02 | 8.82 × 10−9 | 0.05 (1.05) | 0.01 | 6.29 × 10−9 | 0.05 (1.05) | 0.01 | 2.19 × 10−8 | |||
| 5 | rs2973549 | A | 0.478 | ARHGEF28 | 0.11 (1.1) | 0.02 | 1.23 × 10−8 | 0.05 (1.05) | 0.01 | 2.22 × 10−8 | ||||||
| 5 | rs2973548 | T | 0.478 | ARHGEF28 | 0.11 (1.1) | 0.02 | 2.61 × 10−8 | 0.05 (1.05) | 0.01 | 4.90 × 10−8 | ||||||
| 6 | rs146694394 | T | 0.005 | SYNJ2 | 0.33 (1.4) | 0.06 | 1.72 × 10−8 | |||||||||
| 6 | rs1574430 | C | 0.608 | SLC22A7 | 0.05 (1.05) | 0.01 | 2.10 × 10−10 | 0.05 (1.05) | 0.01 | 4.22×10−10 | 0.06 (1.06) | 0.01 | 8.06 × 10−12 | |||
| 6 | rs2242416 | G | 0.606 | CRIP3 | 0.05 (1.05) | 0.01 | 2.25 × 10−10 | 0.05 (1.05) | 0.01 | 3.89×10−10 | 0.06 (1.06) | 0.01 | 8.13 × 10−12 | |||
| 6 | rs2254303 | A | 0.606 | CRIP3 | 0.05 (1.05) | 0.01 | 1.49 × 10−9 | 0.05 (1.05) | 0.01 | 1.90×10−9 | 0.06 (1.06) | 0.01 | 3.88 × 10−11 | |||
| 6 | rs765264064a | C | 6 × 10−5 | FILIP1 | 3.01(20.3) | 0.48 | 2.81 × 10−9 | |||||||||
| 6 | rs121912560a | G | 0.005 | MYO6 | 5.28(196.3) | 0.98 | 5.15 × 10−14 | 3.73 (41.9) | 0.87 | 2.26 × 10−12 | 3.26 (26.1) | 0.86 | 1.09×10−9 | 3.86 (47.7) | 0.88 | 8.72 × 10−13 |
| 7 | rs2286276 | T | 0.284 | TBL2 | 0.05 (1.05) | 0.01 | 4.66 × 10−8 | |||||||||
| 7 | rs61010704 | G | 0.283 | MLXIPL | 0.05 (1.05) | 0.01 | 3.16 × 10−8 | 0.05 (1.05) | 0.01 | 2.72 × 10−8 | ||||||
| 22 | rs371997714 | G | 0.293 | BAIAP2L2 | 0.05 (1.05) | 0.01 | 1.40 × 10−8 | |||||||||
| 22 | rs36062310 | A | 0.043 | KLHDC7B | 0.12 (1.1) | 0.02 | 1.32 × 10−9 | 0.12 (1.3) | 0.02 | 6.66 × 10−9 | ||||||
Genome-wide significant variants (p < 5 × 10−8) with hearing aid (H-aid), hearing difficulty (H-diff), hearing difficulty with background noise (H-noise) and the combined hearing trait (H-both) in the analysis of the mega-sample of white-European individuals from the UK Biobank. The p-values for replicated associations (empirical p-values <0.05 adjusting for variants and traits brought to replication) are shown in bold (See Table S4 for replication analytical and empirical values).
CHR chromosome, EA effect allele, EAF effect allele frequency, OR odds ratio, SE standard error, P p-value.
aVariant not found present in the replication sample.
Rare-variant aggregate association analysis
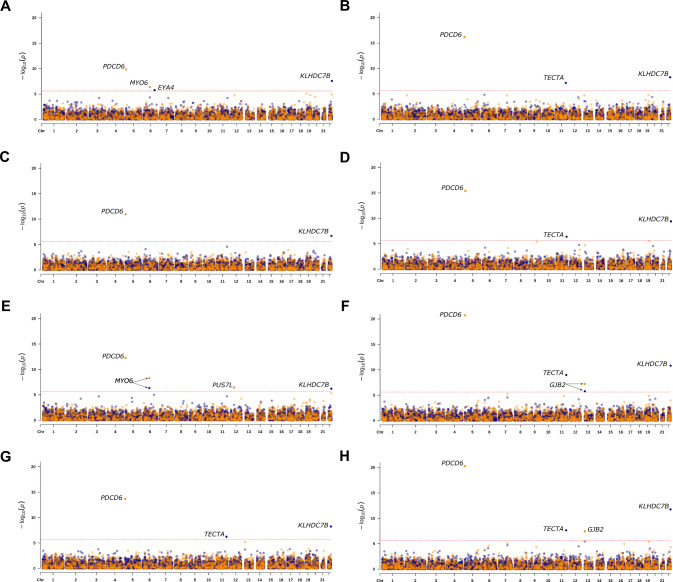
Rare-variant aggregate analysis of 18,012 genes revealed five exome-wide significant (p < 2.5 × 10−6) associations between ARHL and PDCD6, MYO6, KLHDC7B, TECTA, and EYA4 (Table 3, Fig. 2A–D and S6) in the discovery sample. Table S5 provides information on the rare-variants included in the aggregate analysis including: bioinformatic annotation e.g., CADD score, GERP++, PhyloP scores, NSHL ClinVar annotation, and MAFs. Aggregate association analysis of missense and pLoF variants showed significant associations between PDCD6 and all ARHL phenotypes and these associations were also replicated (Tables 3 and S6). When the two rare PDCD6 missense variants (rs549592074 and rs571370281) that were statistically significant in the single-variant analysis were removed from the aggregate analysis, PDCD6 remained exome-wide significant for H-diff and H-both and was also replicated (Tables 3 and S6). Rare missense and pLoF in MYO6 were associated with ARHL trait H-aid in the discovery sample and were also replicated (Table 3). KLHDC7B was significantly associated with all four ARHL traits in the discovery when only rare pLoF variants were included in aggregate association analysis and these findings were replicated, with exception of H-aid (Tables 3 and S6). For the analysis of TECTA, a Mendelian NSHL gene, rare-variant aggerate analysis of pLoF variants were found to be associated with H-diff and H-both and these findings were also replicated. EYA4, a Mendelian ADNSHL gene, rare pLoF variants were associated with H-aid, however, this finding could not be replicated due to no pLoF variants being present in the replication sample (Tables 3, S5, and S6).
Table 3.
Discovery and mega rare-variant aggregate association analysis with age-related hearing loss traits.
| Sample | Type of variation | Gene | H-aid | H-diff | H-noise | H-both | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | Beta (OR) | SE | P | |||
| Discovery | pLoF | KLHDC7B | 1.29 (3.6) | 0.21 | 2.65 × 10−8 | 0.69 (1.9) | 0.12 | 5.59 × 10−9 | 0.56 (1.8) | 0.11 | 2.01 × 10−7 | 0.77 (2.2) | 0.12 | 3.99 × 10−10 |
| TECTA | 0.84 (2.3) | 0.16 | 7.08 × 10−8 | 0.84 (2.3) | 0.16 | 4.18 × 10−7 | ||||||||
| EYA4 | 3.30 (27.1) | 0.61 | 1.74 × 10−6 | |||||||||||
| pLoF + missense | PDCD6 | 1.06 (2.9) | 0.15 | 1.57 × 10−10 | 0.67 (2.0) | 0.08 | 6.22 × 10−17 | 0.50 (1.7) | 0.07 | 1.08 × 10−11 | 0.69 (2.0) | 0.08 | 4.07 × 10−16 | |
| PDCD6a | 0.76 (2.1) | 0.19 | 1.33 × 10−4 | 0.45 (1.6) | 0.09 | 1.07 × 10−6 | 0.34 (1.4) | 0.08 | 4.27 × 10−5 | 0.50 (1.7) | 0.10 | 2.26 × 10−7 | ||
| MYO6 | 0.44 (1.6) | 0.08 | 4.54 × 10−7 | |||||||||||
| MYO6b | 0.40 (1.5) | 0.08 | 7.30 × 10−6 | |||||||||||
| Mega | pLoF | KLHDC7B | 1.10 (3.0) | 0.20 | 6.23 × 10−7 | 0.71 (2.0) | 0.10 | 1.55 × 10−11 | 0.56 (1.8) | 0.10 | 5.72 × 10−9 | 0.77 (2.2) | 0.11 | 1.73 × 10−12 |
| TECTA | 0.85 (2.3) | 0.14 | 1.09 × 10−9 | 0.66 (1.9) | 0.13 | 5.86 × 10−7 | 0.83 (2.3) | 0.15 | 2.26 × 10−8 | |||||
| MYO6 | 1.43 (4.2) | 0.25 | 4.67 × 10−7 | |||||||||||
| GJB2 | 0.21 (1.2) | 0.04 | 1.77 × 10−6 | |||||||||||
| GJB2c | 0.11 (1.1) | 0.14 | 4.3 × 10−1 | |||||||||||
| pLoF + missense | PDCD6 | 1.03 (2.8) | 0.13 | 5.41×10−13 | 0.66 (1.9) | 0.07 | 1.93 × 10−21 | 0.49 (1.6) | 0.06 | 1.99 × 10−14 | 0.68 (2.0) | 0.07 | 5.38 × 10−21 | |
| PDCD6a | 0.78 (2.2) | 0.16 | 4.94 × 10−6 | 0.43 (1.1) | 0.08 | 6.53 × 10−8 | 0.32 (1.4) | 0.07 | 1.89 × 10−5 | 0.48 (1.6) | 0.08 | 8.69 × 10−9 | ||
| MYO6 | 0.43 (1.5) | 0.07 | 5.20 × 10−9 | |||||||||||
| MYO6b | 0.38 (1.5) | 0.07 | 2.45 × 10−7 | |||||||||||
| PUS7L | −0.34 (0.7) | 0.07 | 3.85 × 10−7 | |||||||||||
| GJB2 | 0.15 (1.2) | 0.03 | 6.42 × 10−8 | 0.17 (1.2) | 0.03 | 3.50 × 10−8 | ||||||||
| GJB2c | 0.11 (1.1) | 0.03 | 1.60 × 10−3 | 0.13 (1.4) | 0.04 | 4.40 × 10−4 | ||||||||
Genes associated to an exome-wide significance level (p < 2.5 × 10−6) with hearing aid (H-aid), hearing difficulty (H-diff), hearing difficulty with background noise (H-noise), and the combined trait (H-both). Using rare-variant aggregate association tests pLoF or missense + pLoF variants with a MAF < 0.01 in gnomAD v2.1.1 were analyzed in the discovery and mega-samples of white-European individuals from the UK Biobank. The p-values for replicated associations [empirical p-values <0.05 adjusting for genes (pLoF and missense & pLof) and traits brought to replication], are shown in bold (See Table S6 for replication analytical and empirical values).
pLoF putative loss of function, CHR chromosome, EA effect allele, EAF effect allele frequency, OR odds ratio, SE standard error, P p-value.
aResults when variants: rs549592074 and rs571370281 in PDCD6 were removed.
bResults when rs121912560 in MYO6 was removed.
cResults when rs80338939 in GJB2 was removed.
Fig. 2. Manhattan plots for the discovery and mega-sample rare-variant aggregate association analysis.
Results for the discovery sample are shown for the analysis of H-aid (A), H-diff (B), H-noise (C), and H-both (D). Results for the mega-sample are shown for the analysis of H-aid (E), H-diff (F), H-noise (G), and H-both (H). For each Manhattan plot, the blue dots display the result for the analysis of predicted loss of function (pLoF) variants and the orange dots for missense and pLoF variants. The threshold for exome-wide significance (p < 2.5 × 10−6) is indicated by a red dotted line. Genes that reached exome-wide significance are annotated.
Analysis of the mega-sample revealed additional associations (Table 3, Figs. 2E–H and S7). Rare-variant aggregate analysis of pLoF variants provided exome-wide significant results for TECTA with an additional ARHL trait, H-noise. For Mendelian NSHL gene, GJB2, a statistically significant association was observed with H-diff and H-both. Single-variant association analysis for c.35delG showed suggestive association with H-diff (p-value = 1.11 × 10−6) and H-both (p-value = 5.23 × 10−6). Given the role of this variant in autosomal recessive NSHL, we excluded it from the aggregate analysis and the associations with H-diff and H-both were no longer significant (Table 3). Analyzing both missense and pLoF variants resulted in the observation of an exome-wide significant association between PUS7L and H-aid (Table 3) with a negative beta indicating that PUS7L may be protective.
Common single-variant analysis
Genome-wide significant associations were observed for two common variants in the discovery sample which also replicated, between H-both with rs1574430 in SLC22A7 and with rs2242416 in CRIP3 (Tables 1 and S4A). These variants are in LD (r2 = 0.99), and it was not possible to disentangle which is the gene involved in ARHL. Analysis of the mega-sample revealed more variants that were statistically significant (Table 2). SLC22A7 (rs1574430) and CRIP3 (rs2242416) were also associated with H-diffand H-noise. An additional CRIP3 variant, rs2254303, was found to be associated with H-noise, H-diff, and H-both. These findings were all replicated in the secondary-replication sample (Table S4B). A total of seven variants in ARHGEF28 were associated with ARHL and replicated (Tables 2 and S4B). There were statistically significant associations between three variants in MAST2 and H-diff and these findings were replicated. For MAST2 the observed betas are negative, suggesting that MAST2 is protective (Tables 2 and S4B). We identified statistically significant associations between PIK3R3 (rs1707304) H-diff and H-both which were replicated. The beta for PIK3R3 is also negative. The variants in PIK3R3 and MAST2 are in high LD (r2 = 0.99). We found a statistically significant association between rs371997714 in BAIAP2L2 and H-diff that was replicated (Tables 2 and S4B). In addition to our replicated rare-variant aggregate association findings with KLHDC7B, we found a significant common variant association between rs36062310 (MAF = 0.04) with H-diff and H-both, that we replicated. We also found significant associations with two variants which are in LD (r2 = 0.98) rs2286276 in TBL2, with H-both and rs61010704 which lies in MLXIPL with H-diff and H-both. Only the association between H-diff and rs61010704 in MLXIPL was replicated when correcting for variants brought to replication (Tables 2 and S4B).
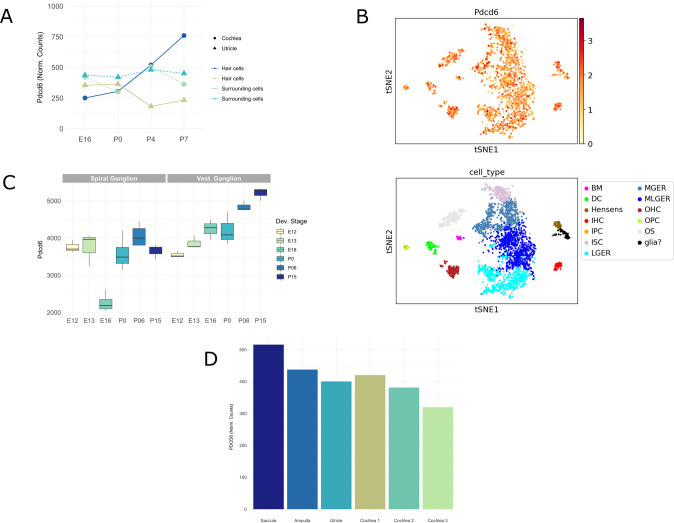
In-silico mRNA expression of PDCD6/Pdcd6 in mice and humans
We further investigated mRNA inner ear expression for PDCD6/Pdcd6 since it is a new HL candidate gene. In mice, expression of the Pdcd6 occurs in the hair cells and surrounding cells of the cochlea and utricle over several developmental stages (E16, P0, P4, and P7) (Figs. 3A, 4B, S8, S9, and S10). For the hair cells of the utricle, the highest expression level occurs at P0. While for cochlea hair cells Pdcd6 expression is upregulated continuously until reaching its maximum levels at P7 (Fig. 3A).
Fig. 3. Expression of PDCD6/Pdcd6 in the human and mouse inner ear.
Pdcd6 expression levels (normalized counts) from RNA sequencing of hair cells (GFP+) and surrounding cells (GFP-) from the cochleae and utricles of mice at four developmental stages: E16, P0, P4, and P7. Pdcd6 is expressed in cochlea and utricle cells over all four developmental stages (A). Pdcd6 expression levels (top panel: red = high expression, yellow = low expression) and location in the cochlear floor epithelium at P7 developmental stage (bottom panel: colors indicate the cell type). The scale bar represents gene expression based on log transformed and normalized expression data. Abbreviations for bottom panel: BM basilar membrane cells, DC dieter cells, Hensen Hensen cells, IHC inner hair cells, IPC inner pillar cells, ISC inner sulcus cells, LGER lateral great epithelial ridge cells, MGER medial greater epithelial ridge cells, MLGER medial lateral greater epithelial ridge cells, OHC outer hair cells, OPC outer pillar cells, OS outer sulcus cells, Glia glial cells (B). RNA expression of Pdcd6 obtained from microarray data based on perfect match and mismatch probe differences (PM/MM) in spiral ganglion neurons and vestibular ganglion neurons from mice at six developmental stages: E12, E13, E16, P0, P06, and P15 (C). PDCD6 expression in the adult human inner ear. PDCD6 expression data (normalized counts) were obtained from RNA sequencing of adult human inner ear tissues (D).
RNA expression of Pdcd6 in ganglion cells (Fig. 3C) displays downregulation in the spiral ganglion neurons of mice at stage E16. In contrast, Pdcd6 expression is consistently upregulated during development in the vestibular ganglion neurons reaching its maxima at P15 (Fig. 3C). Single-cell RNA experiments of the developing mouse cochlear epithelium (Figs. 3B and S8) shows that Pdcd6 is widely expressed in the cochlear epithelium during all developmental stages. In general, both the expression intensity of Pdcd6 and the percentage of cells that express Pdcd6 increase as inner ear development progresses (Fig. S9). A prominent upregulation of Pdcd6 occurs for both the inner and outer hair cells from E16 to P7 (Figs. S9 and S10). In humans, PDCD6 expression was found in all tissues sampled from the cochlear duct and the vestibular labyrinth including the ampulla of the semicircular canals, the utricle, and saccule, as shown in Fig. 3D.
Discussion
UK Biobank data has been widely used to understand the genetic etiology of a variety of complex traits. Here we studied self-reported ARHL, concentrating on rare-variant associations. Although pure-tone audiometry is the gold standard to assess hearing, it is challenging to evaluate large cohorts that are necessary for well-powered GWAS. Recent studies have shown that using self-reports for hearing are a valid measure [5].
We identified a new ARHL association, with PDCD6, programmed cell death 6 (also known as ALG2, apoptosis-linked gene 2), which has not been previously reported to be associated with HL. It is likely that this finding was not previously observed since rare-variants are driving the association. The odds ratios (ORs) observed for rare-variants in PDCD6 ranged from 1.4 to 7.3 (Tables 1–3). We show that PDCD6 is widely expressed in both the mouse and human inner ear. PDCD6 is a cytoplasmic Ca2+ binding protein with an important role in apoptotic cell death [20]. The dysregulation of programmed cell death (PCD), typically mediated by apoptosis, has been implicated in the pathogenesis of monogenic forms of HL [21]. Several PCD genes have been shown to underlie HL, e.g., TJP2, DFNA5, and MSRB3 [21]. Gain-of-function DFNA5 variants have been shown to induce PCD in vitro [22]. Individuals with pathogenic or likely pathogenic (PLP) variants in DFNA5 display progressive ADNSHL starting in the high frequencies, with age-of-onset from 0 to 50 years-of-age [23]. PCD has been implicated in noise-induced HL, and ototoxicity to aminoglycoside and Cisplatins [21]. Studies in CBA/CaJ mice have shown that multiple cell-death pathways, potentially linked to oxidative stress, are activated in auditory hair cells in aging mice [24]. Due to hair cells not having regenerative capacity, activation of the apoptosis-inducing events leads to progressive and permanent cell death and HL. Basal hair cells, responsible for high-frequency hearing, are more susceptible to apoptotic cell death, which is consistent with the observation of HL starting in the high frequencies for ARHL and DFNA5 [21].
We identified and replicated rare-variant aggregate associations for KLHDC7B (Kelch-like domain containing 7B). We also identified and replicated the association with a higher frequency variant rs36062310 which was previously reported to be associated with ARHL [5, 8, 25, 26], although this finding had not been replicated in previous studies. The rare-variant aggregate analysis for KLHDC7B displayed ORs between 1.75 and 3.63, with the highest effect size for H-aid. Klhdc7b has been found to be exclusively expressed in inner ear hair cells of the mouse ear and homozygous Klhdc7b knock-out mice from the International Mouse Phenotyping Consortium showed abnormal auditory brainstem response.
We also identified an association between H-both and rare-variant rs146694394 in SYNJ2 (OR = 1.4) in the mega-sample which was replicated in the secondary-replication sample. An association between SYNJ2 was previously reported but not replicated [5, 8]. SYNJ2 encodes synaptojanin2, an important enzyme in clathrin-mediated endocytosis, that is known to cause high-frequency progressive HL in mice [27, 28]. Although the mechanisms underlying the HL caused by Synj2 in mice are not fully understood, outer hair cell degeneration seems a plausible origin for the HL [27]. In humans, SYNJ2 is expressed in the outer spiral bundles and in the spiral ganglion neuronal cells, possibly increasing the risk of HL by altered synaptic activity between the spiral neurons and the outer hair cells [29].
We furthermore identified a rare-variant aggregation association with H-aid and PUS7L that encodes pseudouridylate synthase 7 homolog-like protein and is a paralog of PUS7. In humans, pseudouridine synthases are encoded by 13 genes and are thought to increase RNA stability by pseudouridylation, that is the isomerization of uridine into 5-ribosyluracil (pseudouridine, Ψ) [30, 31]. PUS have not been very well studied in humans, however some Mendelian diseases have been recently linked to PUS1, PUS3, and PUS7 [32–35]. An OR = 0.71 for PUS7L suggests rare-variants in this gene are protective for ARHL. The association for this gene was identified in the mega-analysis but a replication sample was not available therefore additional study is necessary.
Additionally, rare-variant aggregate analysis demonstrated the important contribution of Mendelian HL genes, i.e. MYO6, TECTA, and EYA4 to the genetics of ARHL. It is of note, that these Mendelian HL genes found to be associated with ARHL here either have shown to mimic ARHL and/or show a variable onset/severity of HL. For MYO6 we identified and replicated both single-variant and rare-variant aggregate associations for ARHL. MYO6 underlies ADNSHL (DFNA22) and autosomal recessive NSHL (DFNB37) [36, 37]. For MYO6 inherited with an autosomal recessive mode of inheritance the NSHL is prelingual, profound, and affects all frequencies [37]. On the other hand, ADNSHL due to MYO6 is postlingual progressive and can range from moderate to profound [36, 38]. The ultra-rare missense variant we found in MYO6, rs121912560: p.(His246Arg) which is classified as PLP in CLinVar was identified in a large family with AD progressive sensorineural HL, with reduced penetrance and variable age-of-onset [39]. PLP MYO6 variants have been observed to cause late-onset HL. The effect size for rs121912560 (MYO6) ranged from OR = 26 (H-noise) to OR = 200 (H-aid) (Table 2). After removing rs121912560 from the rare-variant aggregate analysis, the effect size decreased considerably (H-aid mega-sample OR = 1.46) but remained significant (Table 3). The MYO6 rare-variant aggregate analyses included three PLP variants for NSHL amongst other variants (see Table S5), suggesting that additional MYO6 rare-variants may play a role in ARHL etiology.
TECTA, that encodes alpha-tectorin, underlies ADNSHL (DFNA12) and autosomal recessive NSHL (DFNB21). Aggregate analysis of rare pLoF variants in TECTA displayed ORs ranging from 1.93 to 2.35. ADNSHL due to TECTA is progressive, affects mid-high frequencies, and previous reports have described that HL due to AD TECTA variants may mimic ARHL [40].
EYA4 (Eyes Absent Homolog 4) participates in the development of different organs including the inner ear. In humans, EYA4 variants cause ADNSHL (DFNA10) with early to late age-of-onset and severe phenotype that ultimately progresses to profound HL [41]. In our rare-variant aggregate analyses, EYA4 was only associated with H-aid (OR = 27.1), possibly indicating that this gene could cause a more severe form of ARHL.
GJB2, which was associated with ARHL in the rare-variant aggregate mega-analysis, is a known gene for autosomal recessive NSHL (DFNB1A), accounting for up to 50% of the cases. GJB2 variants have also been described for families with ADNSHL (DFNA3A) [42]. For a range of GJB2 pLoF/missense variants, phenotype variability has been described. Even in the presence of c.35delG, the most common cause of autosomal recessive NSHL in European populations, some individuals can display mild HL in the presence of a second non-truncating variant [43]. Additionally, pathogenic missense variants, p.(Met34Thr) and p.(Val37Ile) are known to have reduced penetrance, variable expressivity, and cause a milder HL with onset <18 years-of-age that can progress [44]. The association between ARHL and GJB2 is most likely due to milder forms of HL that went undiagnosed and therefore lacked ICD10/ICD9 or self-report codes (Table S2). It is unlikely that GJB2 contributes to ARHL. A previously reported association between GJB2 and ARHL may also be due to individuals with early-onset HL being included in the analysis [26].
Through analysis of our mega-sample, we also identified new common variant associations for TBL2, MLXIPL, and PIK3R3 genes, which have not been previously associated with ARHL. For TBL2 and MLXIPL that are in LD only the variant in MLXIPL was replicated adjusting by variants and not by traits. TBL2 and MLXIPL are deleted/duplicated in individuals with Williams-Beuren syndrome a developmental disorder caused by deletion of multiple genes at 7q11.23 with some patients presenting with sensorineural HL that resembles noise-induced HL [45, 46]. Although there are no studies investigating the role of TBL2 or MLXIPL in human hearing, MLXIPL has been proposed as a hair-cell-specific transcription factor in the utricle hair cell regeneration process of the chicken inner ear [47].
The PIK3R3 association was replicated in the secondary-replication sample. PIK3R3 has been shown to be differentially expressed in the murine cochlea, with the highest levels of expression in the inner hair cells [48]. Additionally, PI3K signaling pathway has been shown to promote hair cell survival in mouse cochlea cell cultures after treatment with ototoxic medications (e.g., aminoglycoside) and it is hypothesized that lack of signaling could be a cause of HL [49]. PIK3R3 is in strong LD with MAST2 and they are unlikely to be separate signals. Based on the importance of PIK3R3 in hair cell survival this gene is more likely to contribute to hearing etiology than MAST2. The UTR variant in PIK3R3 could possibly increase gene expression and have a protective effect (OR = 0.95) via the promotion of hair cell survival.
In this study we evaluated the contribution of rare-variants to the etiology of ARHL. We demonstrated that not only do Mendelian HL genes (EYA4, MYO6, and TECTA) play a role in ARHL, but also rare-variants in PDCD6, KLHDC7B, and SYNJ2 which are not known to cause Mendelian forms of HL. The role of PDCD6 in HL was first reported here, and this finding strengthens the role of programmed cell death in ARHL. It was also observed that the effect sizes for rare-variants were much higher than those observed for common variants including those genes i.e., PDCD6, SYNJ2, and KLHDC7B which do not cause Mendelian HL. Due to rare-variants having larger effect sizes, they should play a crucial role in risk prediction by increasing accuracy. Although most findings were replicated in independent samples of white-Europeans, additional studies are necessary to elucidate whether these variants/genes play a role in the genetic etiology of ARHL in other populations.
Web resources
Genome aggregation database (gnomAD): https://gnomad.broadinstitute.org/
Gene expression analysis resource (gEAR) https://umgear.org/
Hereditary hearing loss homepage: https://hereditaryhearingloss.org/
International mouse phenotyping consortium (IMPC): https://www.mousephenotype.org/
Online inheritance in man (OMIM): http://www.omim.org/
Shared Harvard inner-ear laboratory database (SHIELD): https://shield.hms.harvard.edu/
UK Biobank: https://www.ukbiobank.ac.uk/
Supplementary information
Table S1 .UK Biobank - Quality control of UK Biobank Study Participants
Table S2 Inclusion/Exclusion criteria for cases and controls
Table S4. Analytical and empirical permutation p-values for single variant analysis of the variants brought to replication from the discovery (A) and the mega sample (B)
Table S5. Rare-variants included in the aggregate analysis
Table S6. Analytical and empirical permutation p-values for rare-variant aggegrate analysis of the genes brought to replication from the discovery sample
Acknowledgements
The research conducted using the UK Biobank Resource was performed under application numbers 32285 ATD and 36827 SML.
Author contributions
DMCS: analyzed UK Biobank data and drafted the manuscript; GL analyzed UK Biobank data; TB, RW, and YH: extracted and prepared data for analysis; AA: analyzed expression data and aided in drafting the manuscript; MKK and JLE: analyzed expression data, performed data extraction and preparation; IS and GW: performed data interpretation and reviewed the manuscript; ATD: designed experiments; revised the manuscript; and obtained funding; SML designed experiments, drafted and finalized manuscript and obtained funding. All the authors agreed to the final version of the manuscript.
Funding
This work was supported by a grant from the National Institute of Deafness and other Communication Disorders (NIDCD) DC017712 to SML and ATD.
Data availability
Individual-level sequence and phenotype data, from which we derived the traits studied here, are available to approved researchers in the UK Biobank repository. Instructions for access to UK Biobank data are available at https://www.ukbiobank.ac.uk/enable-your-research. Summary statistics for the rare-variants tested in this study are also available in the GWAS Catalog (accession ID is GCP000519).
Code availability
The code used to run the analyses and create the figures presented here is available at https://github.com/statgenetics/ARHL-UKB-Rare-Variants.
Competing interests
The authors declare no competing interests.
Ethical approval
The UK Biobank study was conducted under generic approval from the National Health Services’ National Research Ethics Service. The present analyses were approved by the Human Investigations Committee at Columbia University (institutional review board protocol number IRB-AAAS3494) and Yale University (2000026836).
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41431-023-01302-2.
References
- 1.Salvi R, Ding D, Jiang H, Di Chen G, Greco A, Manohar S, et al. Hidden age-related hearing loss and hearing disorders: current knowledge and future directions. Hear Balanc Commun. 2018;16:74–82. doi: 10.1080/21695717.2018.1442282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duan H, Zhang D, Liang Y, Xu C, Wu Y, Tian X, et al. Heritability of age-related hearing loss in middle-aged and elderly chinese: a population-based twin study. Ear Hear. 2019;40:253–9. doi: 10.1097/AUD.0000000000000610. [DOI] [PubMed] [Google Scholar]
- 3.Bowl MR, Dawson SJ. Age-related hearing loss. Cold Spring Harb Perspect Med. 2019;9:a033217. doi: 10.1101/cshperspect.a033217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffmann TJ, Keats BJ, Yoshikawa N, Schaefer C, Risch N, Lustig LR. A large genome-wide association study of age-related hearing impairment using electronic health records. PLoS Genet. 2016;12:1–20. doi: 10.1371/journal.pgen.1006371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wells HRR, Freidin MB, Zainul Abidin FN, Payton A, Dawes P, Munro KJ, et al. GWAS identifies 44 independent associated genomic loci for self-reported adult hearing difficulty in UK Biobank. Am J Hum Genet. 2019;105:788–802. doi: 10.1016/j.ajhg.2019.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nagtegaal AP, Broer L, Zilhao NR, Jakobsdottir J, Bishop CE, Brumat M, et al. Genome-wide association meta-analysis identifies five novel loci for age-related hearing impairment. Sci Rep. 2019;9:1–10. doi: 10.1038/s41598-019-51630-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalra G, Milon B, Casella AM, Herb BR, Humphries E, Song Y, et al. Biological insights from multi-omic analysis of 31 genomic risk loci for adult hearing difficulty. PLoS Genet. 2020;16:e1009025. doi: 10.1371/journal.pgen.1009025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ivarsdottir EV, Holm H, Benonisdottir S, Olafsdottir T, Sveinbjornsson G, Thorleifsson G, et al. The genetic architecture of age-related hearing impairment revealed by genome-wide association analysis. Commun Biol. 2021;4:706. doi: 10.1038/s42003-021-02224-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, et al. UK Biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12:1–10. doi: 10.1371/journal.pmed.1001779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bycroft C, Freeman C, Petkova D, Band G, Elliott LT, Sharp K, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–9. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szustakowski JD, Balasubramanian S, Kvikstad E, Khalid S, Bronson PG, Sasson A, et al. Advancing human genetics research and drug discovery through exome sequencing of the UK Biobank. Nat Genet. 2021;53:942–8. doi: 10.1038/s41588-021-00885-0. [DOI] [PubMed] [Google Scholar]
- 12.Regier AA, Farjoun Y, Larson DE, Krasheninina O, Kang HM, Howrigan DP, et al. Functional equivalence of genome sequencing analysis pipelines enables harmonized variant calling across human genetics projects. Nat Commun. 2018;9:1–8. doi: 10.1038/s41467-018-06159-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mbatchou J, Barnard L, Backman J, Marcketta A, Kosmicki JA, Ziyatdinov A, et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat Genet. 2021;53:1097–103. doi: 10.1038/s41588-021-00870-7. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, Ferreira T, Morris AP, Medland SE, Madden PAF, Heath AC, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen J, Scheffer DI, Kwan KY, Corey DP. SHIELD: an integrative gene expression database for inner ear research. Database. 2015;2015:bav071. doi: 10.1093/database/bav071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Orvis J, Gottfried B, Kancherla J, Adkins RS, Song Y, Dror AA, et al. gEAR: Gene Expression Analysis Resource portal for community-driven, multi-omic data exploration. Nat Methods. 2021;18:843–4. doi: 10.1038/s41592-021-01200-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kolla L, Kelly MC, Mann ZF, Anaya-Rocha A, Ellis K, Lemons A, et al. Characterization of the development of the mouse cochlear epithelium at the single cell level. Nat Commun. 2020;11:2389. doi: 10.1038/s41467-020-16113-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schrauwen I, Hasin-Brumshtein Y, Corneveaux JJ, Ohmen J, White C, Allen AN, et al. A comprehensive catalogue of the coding and non-coding transcripts of the human inner ear. Hear Res. 2014;333:266–74. doi: 10.1016/j.heares.2015.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon JH, Choi YJ, Kim SG, Nam SW, Lee JY, Park WS. Programmed cell death 6 (PDCD6) as a prognostic marker for gastric cancers. Tumor Biol. 2012;33:485–94. doi: 10.1007/s13277-011-0280-4. [DOI] [PubMed] [Google Scholar]
- 21.Op de Beeck K, Schacht J, Van Camp G. Apoptosis in acquired and genetic hearing impairment: The programmed death of the hair cell. Hear Res. 2011;281:18–27. doi: 10.1016/j.heares.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Op de Beeck K, Van Camp G, Thys S, Cools N, Callebaut I, Vrijens K, et al. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur J Hum Genet. 2011;19:965–73. doi: 10.1038/ejhg.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang H, Guan J, Guan L, Yang J, Wu K, Lin Q, et al. Further evidence for “gain-of-function” mechanism of DFNA5 related hearing loss. Sci Rep. 2018;8:8424. doi: 10.1038/s41598-018-26554-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sha S-H, Chen F-Q, Schacht J. Activation of cell death pathways in the inner ear of the aging CBA/J mouse. Hear Res. 2009;254:92–9. doi: 10.1016/j.heares.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trpchevska N, Freidin MB, Broer L, Oosterloo BC, Yao S, Zhou Y, et al. Genome-wide association meta-analysis identifies 48 risk variants and highlights the role of the stria vascularis in hearing loss. Am J Hum Genet. 2022;109:1077–91. doi: 10.1016/j.ajhg.2022.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Praveen K, Dobbyn L, Gurski L, Ayer AH, Staples J, Mishra S, et al. Population-scale analysis of common and rare genetic variation associated with hearing loss in adults. Commun Biol. 2022;5:540. doi: 10.1038/s42003-022-03408-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martelletti E, Ingham NJ, Houston O, Pass JC, Chen J, Marcotti W, et al. Synaptojanin2 mutation causes progressive high-frequency hearing loss in mice. Front Cell Neurosci. 2020;14:1–17. doi: 10.3389/fncel.2020.561857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manji SSM, Williams LH, Miller KA, Ooms LM, Bahlo M, Mitchell CA, et al. A Mutation in Synaptojanin 2 causes progressive hearing loss in the ENU-mutagenised mouse strain mozart. PLoS ONE. 2011;6:e17607. doi: 10.1371/journal.pone.0017607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu W, Johansson A, Rask-Andersen H, Rask-Andersen M. A combined genome wide association and molecular study of age-related hearing loss in man. SSRN Electron J. 2021;19:1–23. doi: 10.1186/s12916-021-02169-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spenkuch F, Motorin Y, Helm M. Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol. 2014;11:1540–54. doi: 10.4161/15476286.2014.992278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–51. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 32.Shaheen R, Han L, Faqeih E, Ewida N, Alobeid E, Phizicky EM, et al. A homozygous truncating mutation in PUS3 expands the role of tRNA modification in normal cognition. Hum Genet. 2016;135:707–13. doi: 10.1007/s00439-016-1665-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fernandez-Vizarra E, Berardinelli A, Valente L, Tiranti V, Zeviani M. Nonsense mutation in pseudouridylate synthase 1 (PUS1) in two brothers affected by myopathy, lactic acidosis and sideroblastic anaemia (MLASA) J Med Genet. 2007;44:173–80. doi: 10.1136/jmg.2006.045252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shaheen R, Tasak M, Maddirevula S, Abdel-Salam GMH, Sayed ISM, Alazami AM, et al. PUS7 mutations impair pseudouridylation in humans and cause intellectual disability and microcephaly. Hum Genet. 2019;138:231–9. doi: 10.1007/s00439-019-01980-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.de Brouwer APM, Abou Jamra R, Körtel N, Soyris C, Polla DL, Safra M, et al. Variants in PUS7 cause intellectual disability with speech delay, microcephaly, short stature, and aggressive behavior. Am J Hum Genet. 2018;103:1045–52. doi: 10.1016/j.ajhg.2018.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Melchionda S, Ahituv N, Bisceglia L, Sobe T, Glaser F, Rabionet R, et al. MYO6, the human homologue of the gene responsible for deafness in Snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet. 2001;69:635–40. doi: 10.1086/323156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ahmed ZM, Morell RJ, Riazuddin S, Gropman A, Shaukat S, Ahmad MM, et al. Mutations of MYO6 are associated with recessive deafness, DFNB37. Am J Hum Genet. 2003;72:1315–22. doi: 10.1086/375122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanggaard KM, Kjaer KW, Eiberg H, Nürnberg G, Nürnberg P, Hoffman K, et al. A novel nonsense mutation in MYO6 is associated with progressive nonsyndromic hearing loss in a Danish DFNA22 family. Am J Med Genet Part A. 2008;146:1017–25. doi: 10.1002/ajmg.a.32174. [DOI] [PubMed] [Google Scholar]
- 39.Mohiddin SA, Ahmed ZM, Griffith AJ, Tripodi D, Friedman TB, Fananapazir L, et al. Novel association of hypertrophic cardiomyopathy, sensorineural deafness, and a mutation in unconventional myosin VI (MYO6) J Med Genet. 2004;41:309–14. doi: 10.1136/jmg.2003.011973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yasukawa R, Moteki H, Nishio S, Ishikawa K, Abe S, Honkura Y, et al. The prevalence and clinical characteristics of TECTA-associated autosomal dominant hearing loss. Genes. 2019;10:744. doi: 10.3390/genes10100744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Neill ME, Marietta J, Nishimura D, Wayne S, Van Camp G, Van, et al. A gene for autosomal dominant late-onset progressive non-syndromic hearing loss, DFNA10, maps to chromosome 6. Hum Mol Genet. 1996;5:853–6. doi: 10.1093/hmg/5.6.853. [DOI] [PubMed] [Google Scholar]
- 42.Denoyelle F, Lina-Granade G, Plauchu H, Bruzzone R, Chaïb H, Lévi-Acobas F, et al. Connexin 26 gene linked to a dominant deafness. Nature. 1998;393:319–20. doi: 10.1038/30639. [DOI] [PubMed] [Google Scholar]
- 43.Snoeckx RL, Huygen PLM, Feldmann D, Marlin S, Denoyelle F, Waligora J, et al. GJB2 mutations and degree of hearing loss: a multicenter study. Am J Hum Genet. 2005;77:945–57. doi: 10.1086/497996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shen J, Oza AM, del Castillo I, Duzkale H, Matsunaga T, Pandya A, et al. Consensus interpretation of the p.Met34Thr and p.Val37Ile variants in GJB2 by the ClinGen Hearing Loss Expert Panel. Genet Med. 2019;21:2442–52. doi: 10.1038/s41436-019-0535-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cherniske EM, Carpenter TO, Klaiman C, Young E, Bregman J, Insogna K, et al. Multisystem study of 20 older adults with Williams syndrome. Am J Med Genet A. 2004;131:255–64. doi: 10.1002/ajmg.a.30400. [DOI] [PubMed] [Google Scholar]
- 46.Gothelf D, Farber N, Raveh E, Apter A, Attias J. Hyperacusis in Williams syndrome: characteristics and associated neuroaudiologic abnormalities. Neurology. 2006;66:390–5. doi: 10.1212/01.wnl.0000196643.35395.5f. [DOI] [PubMed] [Google Scholar]
- 47.Ku YC, Renaud NA, Veile RA, Helms C, Voelker CCJ, Warchol ME, et al. The transcriptome of utricle hair cell regeneration in the avian inner ear. J Neurosci. 2014;34:3523–35. doi: 10.1523/JNEUROSCI.2606-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Xue N, Song L, Song Q, Santos-Sacchi J, Wu H, Navaratnam D. Genes related to SNPs identified by Genome-wide association studies of age-related hearing loss show restriction to specific cell types in the adult mouse cochlea. Hear Res. 2021;410:108347. doi: 10.1016/j.heares.2021.108347. [DOI] [PubMed] [Google Scholar]
- 49.Jadali A, Kwan KY. Activation of PI3K signaling prevents aminoglycoside-induced hair cell death in the murine cochlea. Biol Open. 2016;5:698–708. doi: 10.1242/bio.016758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1 .UK Biobank - Quality control of UK Biobank Study Participants
Table S2 Inclusion/Exclusion criteria for cases and controls
Table S4. Analytical and empirical permutation p-values for single variant analysis of the variants brought to replication from the discovery (A) and the mega sample (B)
Table S5. Rare-variants included in the aggregate analysis
Table S6. Analytical and empirical permutation p-values for rare-variant aggegrate analysis of the genes brought to replication from the discovery sample
Data Availability Statement
Individual-level sequence and phenotype data, from which we derived the traits studied here, are available to approved researchers in the UK Biobank repository. Instructions for access to UK Biobank data are available at https://www.ukbiobank.ac.uk/enable-your-research. Summary statistics for the rare-variants tested in this study are also available in the GWAS Catalog (accession ID is GCP000519).
The code used to run the analyses and create the figures presented here is available at https://github.com/statgenetics/ARHL-UKB-Rare-Variants.