Abstract
Loss-of-function variants in CHAMP1 were recently described as cause of a neurodevelopmental disorder characterized by intellectual disability (ID), autism, and distinctive facial characteristics. By exome sequencing (ES), we identified a truncating variant in CHAMP1, c.1858A > T (p.Lys620*), in a patient who exhibited a similar phenotype of severe ID and dysmorphisms. Whether haploinsufficiency or a dominant negative effect is the underlying pathomechanism in these cases is a question that still needs to be addressed. By array-CGH, we detected a 194 kb deletion in 13q34 encompassing CHAMP1, CDC16 and UPF3, in another patient who presented with borderline neurodevelopmental impairment and with no dysmorphisms. In a further patient suffering from early onset refractory seizures, we detected by ES a missense variant in CHAMP1, c.67 G > A (p.Gly23Ser). Genomic abnormalities were all de novo in our patients. We reviewed the clinical and the genetic data of patients reported in the literature with: loss-of-function variants in CHAMP1 (total 40); chromosome 13q34 deletions ranging from 1.1 to 4 Mb (total 7) and of the unique patient with a missense variant. We could infer that loss-of-function variants in CHAMP1 cause a homogeneous phenotype with severe ID, autism spectrum disorders (ASD) and highly distinctive facial characteristics through a dominant negative effect. CHAMP1 haploinsufficiency results in borderline ID with negligible consequences on the quality of life. Missense variants give rise to a severe epileptic encephalopathy through gain-of-function mechanism, most likely. We tentatively define for the first time distinct categories among the CHAMP1-related disorder on the basis of pathomechanisms.
Subject terms: Disease genetics, Disease genetics
Introduction
Whole genome investigations by either chromosomal microarray analysis (CMA) or exome sequencing (ES) are usually applied as first-tier tests for the genetic diagnosis of unexplained neurodevelopmental disorders. In particular, the recent large-scale application of ES is allowing the discovery of an extraordinary number of new disease-causing genes and new genetic conditions. Nevertheless, an important issue raised by a “genotype first” approach is nosological accuracy, with respect to the proper assessment of the clinical phenotypes associated with different gene variants and their prognostic relevance. That has major consequences in counselling patients and families.
De novo frameshift or stop-gain variants in the CHAMP1 gene (MIM 616327) located on 13q34, encoding a protein that regulates kinetochore-microtubule attachment and chromosome segregation, were first reported by Hempel et al. in 2015 in five unrelated individuals who presented with an overlapping phenotype characterized by ID and dysmorphic features [1]. Following this report, additional patients with de novo loss-of-function mutations in CHAMP1 have been published in the last few years [2–6]. Clinical manifestations in these patients consisted of severe ID, associated with ASD in most, and highly distinctive facial characteristics.
Additionally, one single subject with a de novo missense variant at codon 16 of CHAMP1 was reported, presenting with refractory infantile myoclonic epilepsy [7].
Literature deals with a limited number of 13q33q34 deletions, which encompass not only CHAMP1 but also several additional contiguous genes [8–10]. Although the large extent of the deletion in most cases, and its frequent association with an additional chromosome imbalance make it difficult to infer consistent genotype-phenotype correlations with respect to intragenic variants, on average the associated neurodevelopmental impairment appears to be mild, and the facial dysmorphisms subtle and nonspecific.
Actually, the spectrum of clinical manifestations and pathomechanisms underlying different genomic alterations affecting CHAMP1 are still poorly understood. In reporting three novel individuals we diagnosed with different CHAMP1 gene variants, and through extensive literature analysis, we discuss on potential pathomechanisms of loss-of-function variants, missense variants and single gene deletions affecting CHAMP1, in relation to the corresponding clinical manifestations. Distinct nosological categories of the CHAMP1-relatated disorders are tentatively defined for the first time on the basis of predicted different mechanisms.
Clinical reports
Subject 1
This is an 18 year-old girl of Italian ancestry, born as the first child to healthy and non consanguineous parents. She has a healthy younger sister. Pregnancy was uneventful until 36 weeks, when the delivery occurred by cesarean section due to fetal bradycardia. Birth weight (BW) was 3050 g (81st centile) and length (BL) 50 cm (90th centile). At birth she presented with posterior plagiocephaly due to unilambdoid synostosis, that was surgically corrected. She experienced motor delay, and could walk unsupported at 22 months. Language development was delayed, she was able to speech her first few words at age 3 years. Speech is currently limited to less than 40 single words. She suffered from three episodes of febrile seizure with onset at the age of 20 months, for which she was treated with valproic acid until the age of 12 years, successfully.
Additional clinical manifestations include severe ID, sleep disturbances characterized by inverted circadian rhythm and frequent nocturnal and early morning awakening, and decreased pain perception. She presented with behavioural abnormalities from early childhood, consisting of stereotyped movements and episodes of aggressiveness, but an overt ASD was never diagnosed. Behavioural and sleep disturbances were partially controlled by treatment with olanzapine, risperidone and melatonin. Major malformations were not detected, brain MRI gave normal results. Menarche occurred at age 11 years, menses were regular. At age 17 years, due to hyperinsulinemia, she started metformin therapy. Myopia was diagnosed in late adolescence. A basic sweet demeanor is reported from early infancy.
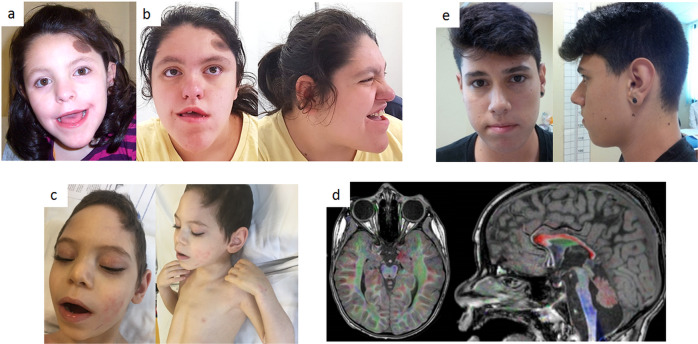
On physical examination, at age 18 years, weight was 75 kg (92nd centile), height 163 cm (40th centile) and occipital-frontal circumference (OFC) 52.8 cm (10th centile). Distinctive facial characteristics were noted, including narrow forehead, round face, short nose with broad nasal bridge, short philtrum, large mouth with tented upper lip, prognathism, morsus inversus, crowded and misplaced teeth, prominent and pointed chin and short neck. She had a single hyperpigmented nevus on the forehead and valgus knees. (Fig. 1 panels “a” and “b”).
Fig. 1. Photographs of reported patients.
a, b subject 1 (stop gain variant) at 8 and 18 ys, respectively. c, d subject 2 (missense variant) clinical phenotype and brain MRI, respectively. e subject 3 (whole gene deletion).
Although her neurodevelopmental impairment was not evaluated with structured assessments due to poor cooperation, ID was unequivocally severe.
By array-CGH (8x60K kit Agilent Technologies, Santa Clara, CA, USA), a 140 Kb interstitial deletion in 10q11.21 (arr[GRCh37/hg19]10q11.21(44282223_44422255) × 1) inherited from the healthy mother was detected and it was considered benign.
Then she was enrolled in the research program for Undiagnosed Diseases supported by Telethon Foundation. ES in the family trio revealed the de novo nonsense variant NM_001164144.3:c.1858A > T (p.Lys620*) in CHAMP1, at a heterozygous state (ClinVar Variant Accession ID: VCV000981896.2).
Subject 2
This is a 7 year-old boy, the firstborn of a twin pregnancy from unrelated healthy parents of Italian ancestry. His twin brother was healthy and the couple had another healthy son from a previous pregnancy. Family history was unremarkable for neurodevelopmental disorders. He was born at 32 + 5 weeks of gestation. Birth weight was 2310 g (90th centile), length 42 cm (50th centile) and OFC 29.5 cm (25th centile). Apgar score was 9 at 1 min. In the first months of life he experienced feeding difficulties due to dysphagia, leading to failure to thrive. Physical examination at birth revealed hypotonia and prominence of the metopic suture suggestive of trigonocephaly. The patient was diagnosed with severe developmental delay during the first year of life. Brain MRI at 1 year of age showed periventricular leukoencephalopathy, opto-chiasmatic atrophy, decreased volume and altered signal of basal ganglia, and hypoplasia of the centrum semiovale and corpus callosum, especially in the posterior regions. On physical examination at the age of 7 years, he was able to hold his head but could not sit, and was nonverbal. Lack of visual tracking, strabismus, nystagmus, decreased muscle mass and diffuse spasticity were diagnosed. Dysmorphic features were noted, including prominent metopic ridge, highly arched eyebrows, mild palpebral ptosis, prominent nasal ridge, bulbous nose tip and macrotia (Fig. 1 panel “c”). He also had pectus excavatum, left clubfoot, and two cafè-au-lait spots. Measures were as follows: weight 13 kg (−3 SD), length 101 cm (−4.5 SD), and OFC 45.4 cm (−3 SD). He suffered from early-onset, recurrent seizures (4–5/day) despite treatment with depakin. Brain MRI confirmed the previous findings, consistent with degeneration of the ponto-olivo-cerebellar structures (Fig. 1 panel “d”). EEG showed an irregular background with nonspecific abnormalities.
Array-CGH analysis (8x60K kit Agilent Technologies, Santa Clara, CA, USA) gave normal results.
He was enrolled in the research program for Undiagnosed Diseases supported by Telethon Foundation, for exome sequencing (ES). ES in the family quartet revealed a de novo heterozygous missense variant in CHAMP1, NM_001164144.3:c.67 G > A (p.Gly23Ser) (ClinVar Variant Accession ID: VCV001806448.1).
Subject 3
This is a 26-year-old boy of Italian ancestry, the only child of healthy non-consanguineous parents. Pregnancy, delivery and early infancy were uneventful. Milestones were normal, he walked unsupported at age 12 months. Speech development was slightly delayed with first words at 18 months, but a rich and well-organized language was achieved in the following two years, albeit characterized by stuttering, for which he underwent speech therapy. He attended public schools without support. Although he experienced learning difficulties as a child, he graduated from high school for technicians autonomously. Learning difficulties as a child and azoospermia were the reason of referral at age 19 years. Weight was 66 kg (50–75th centile), height 179 cm (50th centile) and OFC 52.8 cm (10th centile). As referred, during puberty he developed gynecomastia and mild, not clinically relevant varicocele. On repeated physical examination, at 19 and 26 years, no peculiar facial characteristics were noted, with the exception of simplified auricles, apparent hypotelorism and malar hypoplasia, all signs resembling mostly the paternal phenotype (Fig. 1 panel “e”).
By array-CGH (8x60K kit Agilent Technologies, Santa Clara, CA, USA), a de novo heterozygous 13q34 deletion of 194 kb was detected (hg38 chr13:114136484-114330991) (ClinVar Variant Accession ID VCV001878430.1). Included in the deletion interval were the CHAMP1, CDC16 and UPF3 genes.
He currently works in a transport company independently, can drive a car, is good at banking operations and fully autonomous in daily activities.
Methods
Genetics
Genetic analyses were carried out by array-CGH in all patients, by ES in patient 1 and 2 and by cDNA sequencing in patient 1.
Array-CGH analysis on DNA from peripheral blood cells was performed by means of the commercial Agilent 2 × 244 kit (following manufacturer’s instructions, using ADM-2 algorithm for data analysis with Agilent CytoGenomics software) (Santa Clara, CA, USA Agilent Technologies).
ES was enriched using the SureSelect QXT Target Enrichement system (Agilent Technologies, Santa Clara, CA, USA), according to the manufacturer’s instructions. The libraries were then sequenced on the NovaSeq 6000 Sequencing System (Illumina Inc., San Diego, CA, USA) making use of NovaSeq 6000 S4 Reagent Kit (300 cycles) (IlluminaInc., San Diego, CA, USA), generating 150 bp-long paired-end reads. The average exome coverage of the target bases of at least 100X with 90% of the bases covered by at least 40 reads.
Sequencing of cDNA was performed from mRNA extracted from peripheral blood cells, according to standard procedures.
Phenotype-genotype correlations
We made a comparative evaluation, both clinically and genetically, of the following patient series, our patients included: group A: 41 patients with de novo loss-of-function variants in CHAMP1; group B: 2 patients with de novo missense variants; group C: 8 patients with 13q34 deletion encompassing CHAMP1, sized from 0.194 to 4 Mb.
Results
Genetics
Array-CGH and ES results are described in the clinical reports section.
By cDNA sequencing in patient 1, harbouring the truncating variant c.1858A > T (p.Lys620*) in CHAMP1, we found that both the wild-type and the variant alleles were expressed.
Genotype-phenotype correlations
Group A
Evaluating the 41 subjects in this group, we found that, genetically, the loss-of-function mutations (stop-gain and frameshift), all de novo, were interspersed along the entire single coding exon of the gene, with no mutational hot spots.
Clinically, these variants were associated with a homogeneous phenotype characterized by severe neurodevelopmental impairment and morphogenetic anomalies. In detail, clinical features that recurred in all patients were: ID, which was severe in most cases; hypotonia; severe speech impairment; motor delay and distinctive facial dysmorphisms, including broad nasal bridge, short philtrum, tented upper lip and prognathism. Additional component manifestations were ophtalmologic abnormalities (77%), including hyperopia, astigmatism, nystagmus and amblyopia; strabismus (47%); behavioural abnormalities (65%); true ASD (30%); microcephaly (56%); dental anomalies (70%), including prominent upper incisors, retained primary teeth, and crowded, misplaced or spaced teeth; hearing loss (21%), both conductive or sensorineural; seizures (36%); sleep problems (50%); high pain tolerance (55%). A lovely behaviour is described in several patients. These findings are summarized in Table 1.
Table 1.
Data of patients with de novo truncating and frameshift mutations in CHAMP1.
| Hempel 2015 [1] | Tanaka 2016 [3] | Isidor 2016 [2] | Okamoto 2017 [4] | Garrity 2021 [5] | Levy 2022 [6] | Our case | tot | % | |
|---|---|---|---|---|---|---|---|---|---|
| No of subjects | 5 | 5 | 6 | 1 | 12 | 11 | 1 | 41 | |
| Female/male | 2/3 | 5/0 | 3/3 | 0/1 | 7/5 | 8/3 | F | 26/15 | |
| Motor delay | 5/5 | 5/5 | 6/6 | 1/1 | 12/12 | 11/11 | + | 41/41 | 100 |
| Severe Speech impairment | 5/5 | 5/5 | 6/6 | 1/1 | 12/12 | 11/11 | + | 41/41 | 100 |
| ID | 5/5 | 5/5 | 6/6 | 1/1 | 12/12 | 11/11 | + | 41/41 | 100 |
| Facial dysmorphisms: broad nasal bridge, short philtrum, tented upper lip, prognatism | 5/5 | 5/5 | 6/6 | 1/1 | 12/12 | 11/11 | + | 41/41 | 100 |
| Hypotonia | 5/5 | 5/5 | 6/6 | 1/1 | 12/12 | 11/11 | + | 41/41 | 100 |
| Ophtalmologic abnormalities | 2/5 | 4/5 | 5/6 | 0/1 | 11/11 | 8/11 | + | 31/40 | 77.5 |
| Strabismus | 2/4 | 3/5 | 3/6 | 0/1 | 6/12 | 5/11 | - | 19/40 | 47.5 |
| Microcephaly | 3/5 | 4/5 | 3/6 | 1/1 | 7/12 | 5/11 | - | 23/41 | 56 |
| Behavioural problems | 4/5 | 4/5 | 3/6 | NR | 5/12 | 9/11 | + | 26/40 | 65 |
| ASD features | 1/5 | NR | 1/6 | NR | 5/12 | 3/9 | - | 10/33 | 30 |
| Sleep problem | 3/5 | 3/5 | 2/6 | NR | 4/12 | 7/11 | + | 20/40 | 50 |
| Hearing abnormalities | NR | 3/5 | NR | NR | 1/11 | 2/11 | - | 6/28 | 21.4 |
| Seizures | 1/5 | 2/5 | ¼ | 1/1 | 4/12 | 4/11 | + | 14/39 | 36 |
| Dental abnormalities | 4/5 | NR | NR | NR | NR | 7/11 | + | 12/17 | 70.6 |
| High pain tolerance | 4/5 | NR | NR | NR | 5/12 | NR | + | 10/18 | 55.5 |
Group B
The de novo missense variants diagnosed in patients in this group affected early codons of the gene, i.e. the c.46 T > C (p.(Cys16Arg)) variant in the literature patient [7] and the c.67 G > A (p.(Gly23Ser)) variant in our patient.
Clinically, both patients presented with a severe epileptic encephalopathy, featuring a refractory infantile myoclonic epilepsy [7], and refractory generalized seizures, respectively. The neurodevelopmental impairment was severe in both cases, brain abnormalities were detected in our patient on brain MRI. Facial dysmorphisms were not fully consistent with those of patients in group A. These findings are summarized in Table 2.
Table 2.
Data of patients with de novo missense variants in CHAMP1.
| Ben-Haim 2020 [7] | Our case | |
|---|---|---|
| Female/male | M | M |
| Gene variant | c.46 T > C p.(Cys16Arg) | c.67 G > A (p.Gly23Ser) |
| Severe Speech impairment/absent speech | + | + |
| Severe ID | + | + |
| Early onset and drug-resistant epilepsy | + | + |
| Hypotonia | + | + |
| Brain abnormalities | + | |
| Facial dysmorphisms | +/− | +/− |
| Microcephaly | +/− (−2SD) | + |
Group C
By comparing clinical manifestations of the 8 patients harbouring an isolated 13q34 deletion comprised between 0.194 Mb (our patient 3, the smallest deletion reported so far) and 3.87 Mb, we found that our patient presented with the mildest neurodevelopmental disorder, in agreement with the very small deletion. Clinical manifestations were limited to learning difficulties as a child, with no ID. Significant facial dysmorphisms were not detected. In addition, he had gynecomastia and azoospermia. The neurodevelopmental impairment was very mild in the other cases reported in the literature. Clinical features included mild ID in 5 cases (62%), borderline ID or normal cognitive abilities in 3 (38%); obesity in 5 (62%); gynecomastia in 2/4 males (50%); microcephaly in 2 (25%); heart defects in 2 (25%), consisting of single atrium and VSD, respectively; behavioural abnormalities in 1 (12%); mild speech issues in 4/6 (67%). These findings are summarized in Table 3.
Table 3.
Data of patients with 13q34 deletions encompassing CHAMP1 sized from 0.194 to 3.87 Mb.
| Yang et al. 2013 [8] | Reinstein Family 1 [9] | Reinstein Family 2 patient 3 [9] | Reinstein Family 2 patient 2 [9] | Reinstein Family 2 patient 5 [9] | Reinstein Family 2 patient 4 [9] | Sagi-Dain patient 1 [10] | Our case | Total (%) | |
|---|---|---|---|---|---|---|---|---|---|
| Genomic | Grch37:113987623-115107157 | Grch38:110647373-114344403 | Grch38:111523866-114344403 | Grch38:111523866-114344403 | Grch38:111523866-114344403 | Grch38:111523866-114344403 | Grch37:111780876-115107733 | Grch38: 114136484-114330991 | |
| Size | 1.1 Mb | 3.87 Mb | 3 Mb | 3 Mb | 3 Mb | 3 Mb | 3.3 Mb | 194 Kb | |
| Gender/age (ys) | M 13 | F 35 | F 24 | F 57 | F 28 | M 32 | M 17 | M 18 | |
| De novo | + | + | Inherited from the affected mother | + mosaic 80% | Inherited from the affected mother | Inherited from the affected mother | NR | + | 4/7 (57) |
| Polydactyly | + | - | - | - | - | - | - | - | 1/8 (12) |
| DD | + | + | NR | NR | + | + | 4/6 (67) | ||
| ID | Borderline (IQ 71) | Mild/borderline (IQ 70) | Mild | Mild | Mild | Mild | + | - | 5/8 (62) |
| Speech problems | Fluent reader spelling problems | Low language ability | Low language ability | NR | NR | Speech apraxia | 4/6 (67%) | ||
| Behavior issues | ADHD | 1/ 8 (12) | |||||||
| Heart defect | + (single atrium) | + VSD (mother also) | 2/8 (25) | ||||||
| Seizures | + | 1/8 (12) | |||||||
| Sleep problems | NR | NR | NR | NR | 0/4 | ||||
| Microcephaly | + | + | 2/8 (25) | ||||||
| Gynecomastia | + | + | 2/4 M (50) | ||||||
| Obesity | + | + | + | + | + | 5/8 (62) |
A summary of the most distinctive features of all patients is provided in Table 4.
Table 4.
CHAMP1-related disorders: clinical-genetic categories.
| Dominant negative (frameshift, stop gain variants) | Gain-of-function (missense variants) | Haploinsufficiency (whole gene deletions; other gene variants?) | |
|---|---|---|---|
| ID | Severe/moderate | Severe | Absent/very mild |
| Hypotonia | Yes | Yes | Unusual |
| Severe speech impairment | Yes | Yes | No |
| Early onset and drug-resistant epilepsy | No | Yes | No |
| Facial dysmorphisms broad nasal bridge, short philtrum, tented upper lip, prognatism | Yes | ? | No |
| Brain abnormalities | Unusual | Possible | No |
| Microcephaly | Yes | Yes | No |
Discussion
We describe three individuals with different CHAMP1 genomic alterations, consisting in a truncating variant, a whole-gene deletion and a missense variant, respectively, who presented with markedly different clinical phenotypes.
Nonsense or frameshift de novo mutations in “chromosome alignment maintaining phosphoprotein 1” gene (CHAMP1, MIM 616327), first described in 2015 [1], define an emerging neurodevelopmental disorder, characterized by severe ID with high risk of ASD, severe speech impairment, microcephaly, epilepsy, high pain tolerance, ophthalmologic issues and distinctive facial dysmorphisms [2–6]. The most specific facial characteristics include a short nose with depressed and large nasal bridge; a peculiar mouth conformation that appears large, with short philtrum and tented upper lip; prognathism; pointed chin and short neck. Since all variants are predicted to be loss-of-function mutations, haploinsufficiency was first considered the underlying pathomechanism in these cases. Our patient 1 harbouring the c.1858A > T (p.Lys620*) truncating variant exhibited a similar phenotype of severe ID and morphologic anomalies. Subject 3 in the present report was diagnosed with the smallest 13q34 deletion reported so far. The deletion encompasses CHAMP1 and only two additional contiguous gene. Thus, CHAMP1 unequivocally undergoes haploisufficiency in our patient. However he presented with a very mild neurodevelopmental impairment, limited to learning difficulties as a child. In addition, no consistent facial dysmorphisms were noted. A very limited number of small and isolated 13q34 deletions encompassing CHAMP1 are described in the literature. Despite the rearrangements were larger than in our patient, evaluating the seven reported patients with chromosome deletions ranging from 1.1 to 3.87 Mb, we found that they all presented with borderline ID or very mild neurodevelopmental issues. Of note, nonspecific and mild facial dysmorphisms were described as well, consisting of high forehead, prominent nose with long nasal bridge and small chin [8–10]. It is worth noting that in three out seven cases the 13q34 deletion was inherited from an affected parent, leading us to consider that the associated neurodevelopmental disorder did not limited the social skills and the personal autonomy of the carrier individuals.
Thus, we can assume with high confidence that haploinsufficiency is no longer to be considered the pathomechanism underlying truncating gene variants. On the other hand, the nonsense and frameshift mutations identified so far are located in the only coding exon of the gene, and they might reasonably escape nonsense-mediated mRNA decay, as already suggested [2]. By cDNA sequencing in our subject 1, we could demonstrated that the mutated mRNA is stable. Therefore, the resulting truncated protein is expected to be expressed and someway functional. Of relevance, Isidor and collaborators [2] demonstrated that the loss-of-function mutations identified in CHAMP1 gave rise to truncated proteins lacking the C-terminal region where the zinc finger domains, which are essential for protein-protein interactions, are localized. Confirming this assumption, they observed that CHAMP1 truncated proteins were unable to interact with the POGZ (POGO transposable element with ZNF domain) and the HP1 (Heterochromatin protein 1) proteins, which represent two of the direct partners of CHAMP1 [2]. We can infer that the truncated proteins resulting from heterozygous truncating mutations in CHAMP1 act through a dominant-negative effect, since they interfere with the wild-type protein, most likely.
Our subject 2 harbouring a de novo missense variant at level of an early codon of the gene (p.Gly23Ser) presented with a severe and drug-resistant epileptic encephalophay, and with a severe neurodevelopmental delay as well, that was significantly more severe with respect to that of our patient with the loss-of-function variant. Literature deals with only one patient that was diagnosed with a de novo missense variant in CHAMP1 [7]. Also in this case an early codon of the gene was affected (p.Cys16Arg) and the associated clinical phenotype was a refractory infantile myoclonic epilepsy. Although the very limited number of observations prevents to draw definite conclusions, we can infer that pathogenic missense variants in CHAMP1 define another distinct category of the CHAMP1-related disorders, reflecting a gain-of-function effect, most likely.
Of importance, for the precise genetic diagnosis, we must specify that, considering the gnomAD database (https://gnomad.broadinstitute.org/) gene constraint metrics, CHAMP1 shows a high likelihood of being a loss-of-function intolerant gene, while the number of observed missense variants is similar to what is expected, suggesting that evaluation of the pathogenicity of missense variants should require specific investigations and caution.
In conclusion, we made the first attempt to define distinct nosological categories among the CHAMP1-related disorders on the basis of alternative pathomechanisms. Truncating and frameshift gene variants cause moderate to severe ID, high risk for ASD usually associated with a sweet behaviour, and distinctive facial characteristics, acting through a dominant negative effect. Pathogenic missense variants give rise to a severe neurodevelopmental impairment with refractory seizures, reflecting a gain of function of the mutated protein. CHAMP1 haploinsufficiency, as a consequence of whole gene deletions, is associated with a very mild neurodevelopmental impairment with negligible consequences on the quality of life, and it can be easily tolerated.
Further observations, along with functional studies, are needed to reinforce these conclusions.
Acknowledgements
We thank Giovanni Neri, for participating to the discussion. We are grateful to the families for their support.
Author contributions
SA is involved in manuscript writing and literature revision. GM collaborated in manuscript writing and literature revision and performed bioinformatics analyses. DO performed array-CGH. SF performed cDNA analysis. FG, MS, FR and VC contributed patient cases and contributed to manuscript revision. EP contributed in literature revision and in created tables and patient figures. TUDP performed ES analysis. VN performed interpretation and validation of ES results. MZ was responsible for the study design and supervised all the activities.
Funding
This study was supported by Fondazione Telethon (Italy) (GSP15001- Telethon Undiagnosed Program). This project has also received funding from the European Union’s Horizon 2020 research and innovation program under grant agreement No. 779257 (Solve‐RD): “SOLVE RD”.
Data availability
All data relevant to the study are included in the article. The three CHAMP1 variants described in this study were submitted to the ClinVar Database (https://www.ncbi.nlm.nih.gov/clinvar/) and received the following codes: SCV002817425 (Accession: VCV001878430.1, Variation ID: 1878430), SCV002766597 (Accession: VCV001806448.1, Variation ID: 1806448), and SCV002765127 (Accession: VCV000981896.2, Variation ID: 981896).
Competing interests
The authors declare no competing interests.
Ethical approval
All procedures performed in this study were in accordance with the ethical standards of the institutional research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. They were specifically approved by the Department of Life Sciences and Public Health of Catholic University of Roma, Italy, with protocol nr. DIPUVSP-PD-10-2231.
Consent for publication
Patient/parental consent obtained.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Simona Amenta, Giuseppe Marangi.
A list of authors and their affiliations appears at the end of the paper.
Contributor Information
Marcella Zollino, Email: marcella.zollino@unicatt.it.
Telethon Undiagnosed Diseases Program (TUDP) Study Group:
Annalaura Torella, Gerarda Cappuccio, Francesco Musacchia, Margherita Mutarelli, Diego Carrella, Giuseppina Vitiello, Giancarlo Parenti, Vincenzo Leuzzi, Angelo Selicorni, Silvia Maitz, Nicola Brunetti-Pierri, Sandro Banfi, Martino Montomoli, Donatella Milani, Corrado Romano, Albina Tummolo, Daniele De Brasi, Antonietta Coppola, and Claudia Santoro
References
- 1.Hempel M, Cremer K, Ockeloen CW, Lichtenbelt KD, Herkert JC, Denecke J, et al. De novo mutations in CHAMP1 cause intellectual disability with severe speech impairment. Am J Hum Genet. 2015;97:493–500. doi: 10.1016/j.ajhg.2015.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Isidor B, Küry S, Rosenfeld JA, Besnard T, Schmitt S, Joss S, et al. De novo truncating mutations in the kinetochore-microtubules attachment gene CHAMP1 cause syndromic intellectual disability. Hum Mutat. 2016;37:354–8. doi: 10.1002/humu.22952. [DOI] [PubMed] [Google Scholar]
- 3.Tanaka AJ, Cho MT, Retterer K, Jones JR, Nowak C, Douglas J, et al. De novo pathogenic variants in CHAMP1 are associated with global developmental delay, intellectual disability, and dysmorphic facial features. Cold Spring Harb Mol Case Stud. 2016;2:a000661. doi: 10.1101/mcs.a000661.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Okamoto N, Tsuchiya Y, Kuki I, Yamamoto T, Saitsu H, Kitagawa D, et al. Disturbed chromosome segregation and multipolar spindle formation in a patient with CHAMP1 mutation. Mol Genet Genom Med. 2017;5:585–91. doi: 10.1002/mgg3.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Garrity M, Kavus H, Rojas-Vasquez M, Valenzuela I, Larson A, Reed S, et al. Neurodevelopmental phenotypes in individuals with pathogenic variants in CHAMP1. Cold Spring Harb Mol Case Stud. 2021;7:a006092. doi: 10.1101/mcs.a006092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Levy T, Lerman B, Halpern D, Frank Y, Layton C, Zweifach J, et al. CHAMP1 disorder is associated with a complex neurobehavioral phenotype including autism, ADHD, repetitive behaviors and sensory symptoms. Hum Mol Genet. 2022:ddac018. 10.1093/hmg/ddac018. [DOI] [PMC free article] [PubMed]
- 7.Ben-Haim R, Heyman E, Benyamini L, Shapira D, Lev D, Tzadok M, et al. CHAMP1 mutations cause refractory infantile myoclonic epilepsy. J Pediatr Neurol. 2020;18:027–32. doi: 10.1055/s-0039-1683449.. [DOI] [Google Scholar]
- 8.Yang YF, Ai Q, Huang C, Chen JL, Wang J, Xie L, et al. A 1.1 Mb deletion in distal 13q deletion syndrome region with congenital heart defect and postaxial polydactyly: additional support for a CHD locus at distal 13q34 region. Gene. 2013;528:51–4. doi: 10.1016/j.gene.2013.03.145.. [DOI] [PubMed] [Google Scholar]
- 9.Reinstein E, Liberman M, Feingold-Zadok M, Tenne T, Graham JM., Jr Terminal microdeletions of 13q34 chromosome region in patients with intellectual disability: Delineation of an emerging new microdeletion syndrome. Mol Genet Metab. 2016;118:60–3. doi: 10.1016/j.ymgme.2016.03.007. [DOI] [PubMed] [Google Scholar]
- 10.Sagi-Dain L, Goldberg Y, Peleg A, Sukenik-Halevy R, Sofrin-Drucker E, Appelman Z, et al. The rare 13q33-q34 microdeletions: eight new patients and review of the literature. Hum Genet. 2019;138:1145–53. doi: 10.1007/s00439-019-02048-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data relevant to the study are included in the article. The three CHAMP1 variants described in this study were submitted to the ClinVar Database (https://www.ncbi.nlm.nih.gov/clinvar/) and received the following codes: SCV002817425 (Accession: VCV001878430.1, Variation ID: 1878430), SCV002766597 (Accession: VCV001806448.1, Variation ID: 1806448), and SCV002765127 (Accession: VCV000981896.2, Variation ID: 981896).