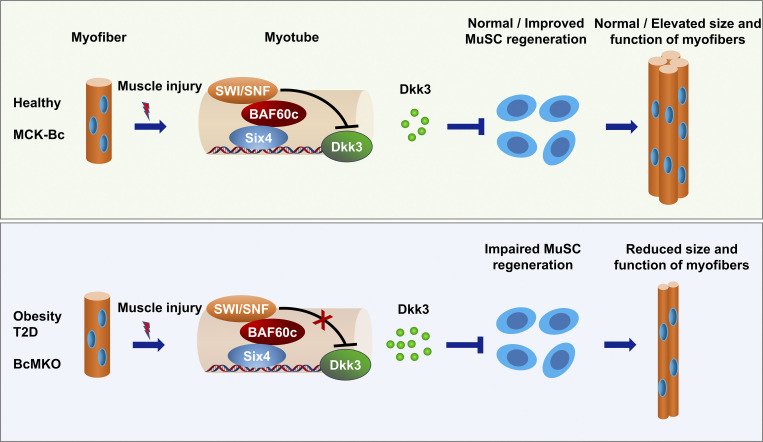
Xu et al. define Baf60c in skeletal myofibers as a critical regulator of muscle regeneration through Dkk3-mediated paracrine signaling on muscle stem cells. They also uncover that Baf60c interacts with transcription factor Six4 to synergistically suppress Dkk3 expression in myocytes.
Abstract
Obesity and type 2 diabetes (T2D) are the leading causes of the progressive decline in muscle regeneration and fitness in adults. The muscle microenvironment is known to play a key role in controlling muscle stem cell regenerative capacity, yet the underlying mechanism remains elusive. Here, we found that Baf60c expression in skeletal muscle is significantly downregulated in obese and T2D mice and humans. Myofiber-specific ablation of Baf60c in mice impairs muscle regeneration and contraction, accompanied by a robust upregulation of Dkk3, a muscle-enriched secreted protein. Dkk3 inhibits muscle stem cell differentiation and attenuates muscle regeneration in vivo. Conversely, Dkk3 blockade by myofiber-specific Baf60c transgene promotes muscle regeneration and contraction. Baf60c interacts with Six4 to synergistically suppress myocyte Dkk3 expression. While muscle expression and circulation levels of Dkk3 are markedly elevated in obese mice and humans, Dkk3 knockdown improves muscle regeneration in obese mice. This work defines Baf60c in myofiber as a critical regulator of muscle regeneration through Dkk3-mediated paracrine signaling.
Graphical Abstract
Introduction
Obesity and its associated chronic metabolic diseases, such as type 2 diabetes (T2D), have become a global pandemic that severely affect human health. Insulin resistance and pancreatic β-cell dysfunction are the major causes of T2D, which possesses a high worldwide prevalence and a series of clinical complications (DeFronzo et al., 2015). In healthy individuals, skeletal muscle is the major site of postprandial glucose uptake (Ferrannini et al., 1988; Thiebaud et al., 1982). As such, impaired glucose sensing and utilization in skeletal muscle commonly feature in the development of obesity-associated metabolic diseases (DeFronzo and Tripathy, 2009; Meng et al., 2017). Skeletal myofibers have remarkable heterogeneity and plasticity in metabolic and contractile functions. Over the past decades, several mechanisms have been elucidated in the pathogenic progression of T2D in which excessive intramyocellular and intermuscular lipid accumulation (Kelley et al., 2002; Muoio and Neufer, 2012; Samuel et al., 2010), reduced mitochondrial oxidative capacity (Mootha et al., 2004; Patti et al., 2003; Petersen et al., 2003), and a switch from oxidative to glycolytic myofibers (Lillioja et al., 1987; Simoneau and Kelley, 1997; Simoneau et al., 1995) have all participated to a different extent in the deterioration of muscle metabolism and function. These studies led to the identification of several key transcriptional factors and cofactors, such as PGC-1α, AMPK, and SIRT1, that are important in oxidative myofiber specification and lipid metabolism (Bassel-Duby and Olson, 2006; Lagouge et al., 2006; Lin et al., 2002).
It is of note that obesity and T2D not only impair muscle insulin sensitivity and glucose metabolism but also muscle maintenance and regeneration after injury, leading to a metabolic syndrome which is termed diabetic myopathy with characteristics of progressive decline in metabolic activity, strength, and muscle mass (D’Souza et al., 2013). Additionally, this muscle complication is believed to further impair systemic energy homeostasis and promote the disease progression of T2D as well as its associated common complications, such as cardiovascular disease and non-alcoholic steatohepatitis. However, the molecular pathways underpinning obesity- and T2D-induced attenuation of muscle regeneration and myopathy remain unsolved (Akhmedov and Berdeaux, 2013; Nguyen et al., 2011).
Muscle satellite cells (MuSCs), also known as muscle stem cells, reside between the basal lamina and plasma membrane of myofibers and keep dormant under normal physiological conditions (Mauro, 1961). MuSCs acquire a robust regenerative capacity in response to injury or other external stimuli, and the regeneration-active form of MuSCs is defined as the primary contributor of muscle maintenance and plasticity. Muscle repair is a precisely orchestrated process involving muscle degeneration, inflammation, regeneration, and remodeling, in which several cell types, such as MuSCs, immune cells, and interstitial cells, are actively engaged (Yin et al., 2013). However, besides serving as the residing site for MuSCs, the role of mature myofiber in controlling muscle stem cell niche and regenerative capacity remains largely unexplored.
Both genetic and epigenetic mechanisms are important for the pathogenesis of T2D and its associated complications. Among the four major ATP-dependent chromatin remodeling complexes in mammals, the switching defective/sucrose nonfermenting (SWI/SNF) chromatin-remodeling complex has recently been shown to play important roles in diverse cellular processes including nutrient sensing, energy metabolism, and cell fate determination under both physiological and disease conditions (Clapier et al., 2017; Dong and Cheung, 2021; Wang et al., 2018). The SWI/SNF complex is composed of up to 11 subunits, among which the Baf60 subunit serves as the linker between the core complex and tissue-specific transcription factors to regulate context-dependent chromatin landscape and transcriptional network (Wu et al., 2009). Baf60 subunit includes three members, namely Baf60a, Baf60b, and Baf60c. Our recent studies have revealed that Baf60a is highly expressed in liver and adipose tissue, functioning as a critical regulator of hepatic fatty acid oxidation, bile acid metabolism, and adipose tissue thermogenesis (Li et al., 2008; Liu et al., 2020; Meng et al., 2015; Tao et al., 2011). Baf60c is preferentially expressed in skeletal muscle, serving as the key component of a glucose-sensing pathway that governs glycolytic muscle fiber formation, postprandial glucose metabolism, and systemic glucose homeostasis (Meng et al., 2017; Meng et al., 2013; Meng et al., 2018; Meng et al., 2014).
Beyond the metabolic and contractile functions, skeletal muscle is emerging as an important secretory organ that produces and releases muscle-secreted factors, namely myokines, to modulate systemic energy metabolism through muscle-tissue crosstalk and regulate muscle tissue homeostasis through autocrine and paracrine signaling (Febbraio and Pedersen, 2020; Pedersen and Febbraio, 2012). The Dickkopf (Dkk) gene family consists of four main members (Dkk1–4) and a unique Dkk3-related gene, Dkkl1 (Niehrs, 2006). They encode secreted proteins that are generally considered antagonists of Wnt/β-catenin signaling. Unlike other members of the Dkk family, Dkk3 is the only member enriched in skeletal muscle (Yin et al., 2018). Recent studies have demonstrated that Dkk3 plays a critical role in controlling smooth muscle cell fate specification and differentiation (Karamariti et al., 2013; Wang et al., 2015). Notably, it has been reported that the plasma level of Dkk3 is increased in the elderly (Zenzmaier et al., 2008) and is involved in the development of age-related muscle atrophy, also known as sarcopenia (Yin et al., 2018).
In this study, we demonstrated that mRNA and protein expression of Baf60c in skeletal muscle is markedly downregulated in obese and diabetic mice and humans, suggesting a potential role of Baf60c in the pathogenesis of impaired muscle regeneration in obesity and T2D. Using myofiber-specific Baf60c knockout and transgenic mice, we uncover the important role of Baf60c in mature muscle cells in regulating muscle stem cell microenvironment and regeneration through Dkk3-mediated paracrine signaling. Baf60c physically interacts with transcription factor Six4 to synergistically inhibit Dkk3 gene expression in skeletal myofibers. In addition, Dkk3 levels in circulation are positively correlated with body mass index (BMI) and are remarkably elevated in obese human subjects, highlighting the translational potential of Dkk3 as a biomarker for the diagnosis and therapeutic evaluation of obesity-associated impairment in muscle regeneration and health.
Results
Skeletal myofiber-specific inactivation of BAF60c impairs muscle regeneration
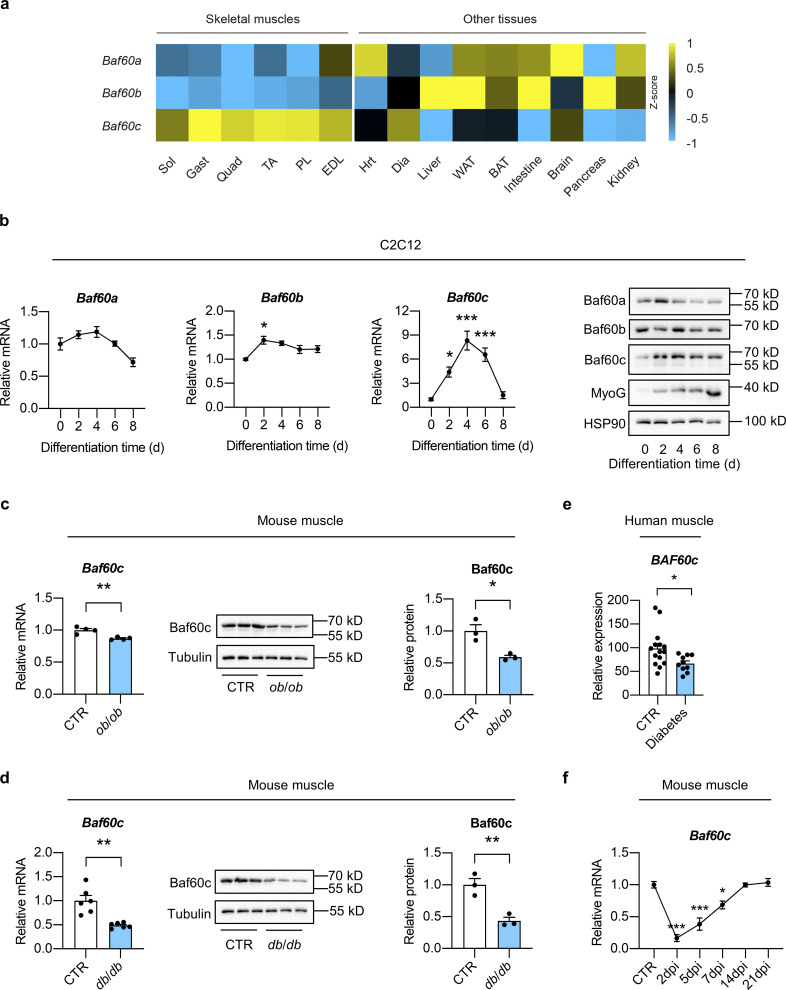
The distinct tissue distribution patterns and regulatory mechanisms of Baf60s confer each protein with specialized functions (Wang et al., 2018). Among the three Baf60 family members, Baf60c is highly expressed in skeletal muscle and is preferentially expressed in glycolytic muscles (Fig. S1 a). During the differentiation of C2C12 myotubes, Baf60c mRNA and protein levels were strongly induced at the beginning followed by a robust decline toward the basal level in fully differentiated myotubes, while no significant change in expression was observed in other Baf60 members in this process (Fig. S1 b). Both mRNA and protein levels of Baf60c were downregulated in skeletal muscle from genetically obese (ob/ob) and diabetic (db/db) mice compared with their respective controls (Fig. S1, c and d). In accordance, analysis of the skeletal muscle gene expression profiles obtained from the previously published microarray datasets (NCBI acccession no. GSE25462; Jin et al., 2011) revealed that BAF60c gene expression was also significantly decreased in the skeletal muscle from subjects with T2D compared with normal controls (Fig. S1 e). In this study, cardiotoxin (CTX) was injected into tibialis anterior (TA) muscles to induce acute muscle injury and regeneration. Notably, we observed a robust decrease of Baf60c expression in the TA muscles of wild-type (WT) mice immediately after CTX injection, followed by a gradual recovery process during the regeneration (Fig. S1 f). Taken together, these data demonstrate that Baf60c expression in skeletal muscle is downregulated in mouse models of obesity and T2D, and human subjects with T2D, suggesting that Baf60c might be involved in the pathogenesis of muscle regeneration impairment in obesity and T2D.
Figure S1.
Identification of Baf60c as a potential regulator of muscle regeneration in obesity and T2D. (a) Heatmap representation of the scaled, normalized FPKM (as Z-score) of Baf60s genes in the indicated mouse tissues. The total RNA sample for each tissue was pooled from three C57BL6/J WT mice. Sol, soleus; Gast, gastrocnemius; Quad, quadriceps; TA, tibialis anterior; PL, plantaris; EDL, extensor digitorum longus; Hrt, heart; Dia, diaphragm; WAT, white adipose tissue; BAT, brown adipose tissue. (b) qPCR analysis of Baf60s gene expression (left) and representative immunoblots (right) of total cell lysates during differentiation of C2C12 myotubes (n = 4 biological replicates at each time point). *P < 0.05, ***P < 0.001; one-way ANOVA with multiple comparisons. (c) qPCR analysis of Baf60c gene expression (left; n = 4 mice per group), immunoblots (middle), and quantification of relative Baf60c protein level (right; n = 3 mice per group) of total Quad muscle protein lysates from ob/ob mice and their respective controls. *P < 0.05, **P < 0.01; two-tailed unpaired Student’s t test. (d) qPCR analysis of Baf60c gene expression (left; n = 6 mice per group), immunoblots (middle), and quantification of relative Baf60c protein level (right; n = 3 mice per group) of total Quad muscle protein lysates from db/db mice and their respective controls. **P < 0.01; two-tailed unpaired Student’s t test. (e) Relative BAF60c mRNA levels in human skeletal muscles. For non-diabetic control (CTR), n = 15 independent biological samples; for diabetes, n = 10 independent biological samples. *P < 0.05; two-tailed unpaired Student’s t test. (f) qPCR analysis of Baf60c gene in TA muscle from WT mice during the process of muscle regeneration (n = 3–4 mice at each time point). *P < 0.05, ***P < 0.001; one-way ANOVA with multiple comparisons. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData FS1.
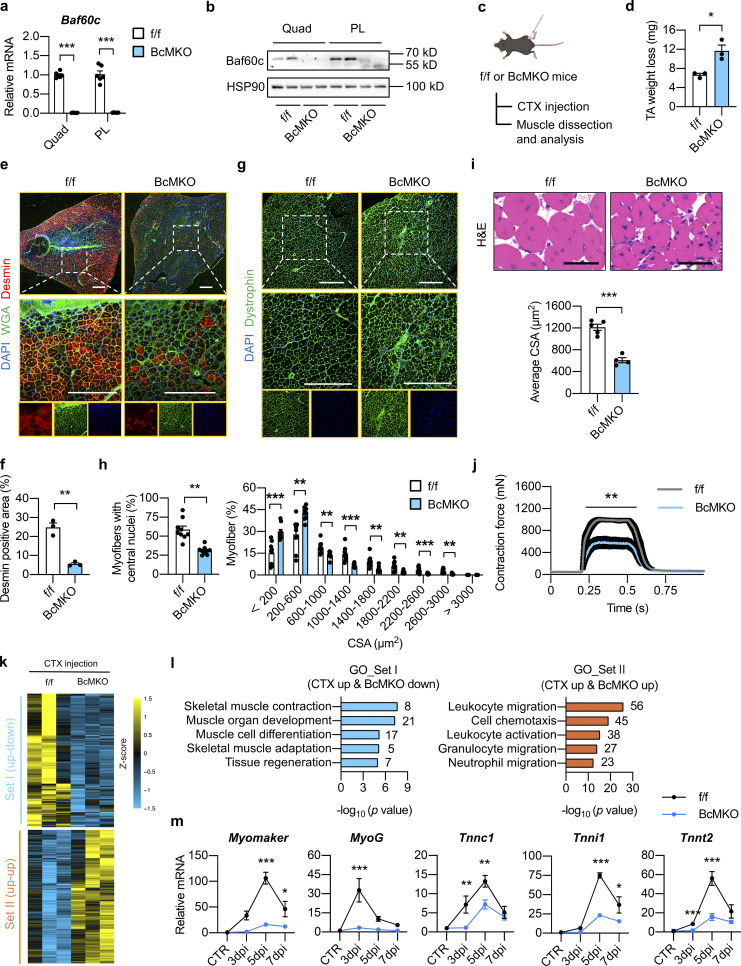
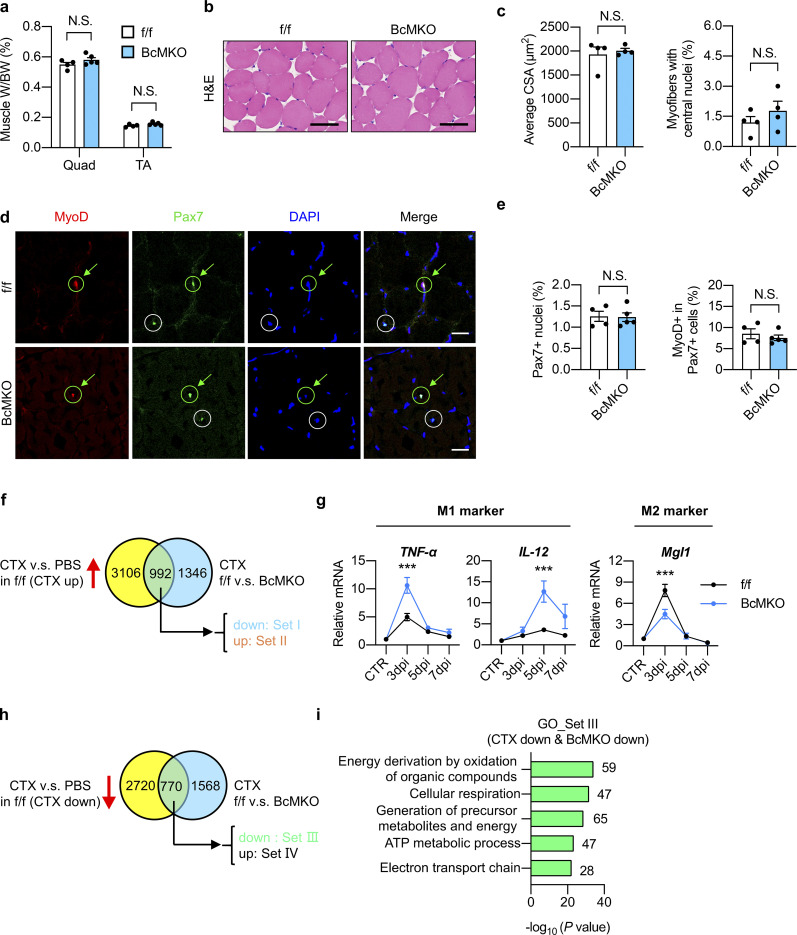
To test this, we generated Baf60c muscle-specific knockout (BcMKO) mice by crossing the Baf60cflox/flox (f/f) mice with Cre recombinase transgenic mice driven by the skeletal muscle–specific myosin light chain promoter (MLC-Cre), as previously described (Bothe et al., 2000; Meng et al., 2017). MLC promoter is mainly expressed in mature skeletal myocytes, and the MLC-Cre mouse line has been widely used to generate mature myocyte-specific KO mouse models (Bothe et al., 2000). Our previous study demonstrated that Baf60c is the preferentially expressed glycolytic myofiber and its protein is barely detectable in oxidative myofiber-enriched skeletal muscles such as soleus muscle (Meng et al., 2013). To more comprehensively examine the deletion efficiency of Baf60c in BcMKO mice, we performed qPCR and Western blotting analyses of Baf60c mRNA and protein in quadriceps (Quad) muscle (a representative muscle contains both glycolytic and oxidative myofibers) and glycolytic myofiber-enriched plantaris (PL) muscle. As shown in Fig. 1, a and b, both mRNA and protein levels of Baf60c were nearly absent in BcMKO mouse muscles. In contrast, Baf60c expression in other metabolic tissues, including the liver, white adipose tissue (WAT), brown adipose tissue (BAT), and heart remained similar between the two groups as described in our previous study (Meng et al., 2017). To examine the effects of Baf60c on non-regenerating muscles, we performed more thorough histological studies on skeletal muscles from control and BcMKO mice under normal physiological conditions. As expected, muscle weight (Fig. S2 a), the morphology of muscle cross sections, as revealed by hematoxylin and eosin (H&E) staining (Fig. S2 b), and myofiber size (Fig. S2 c, left) are indistinguishable between control and BcMKO mice. In addition, the percentage of myofibers with central nuclei, an indicator of regenerating myofiber, is similar between the two groups (Fig. S2 c, right). To further examine the effect of Baf60c deficiency in mature myofibers on the number and activity of satellite cells, we went on to perform immunofluorescence staining of Pax7 (a marker for quiescent and activated satellite cells) and/or MyoD (a marker for activated satellite cells; Dhawan and Rando, 2005) on frozen muscle cross-sections from Baf60cflox/flox and BcMKO mice. As expected, the number of satellite cells in non-regenerating muscles, as revealed by the percentages of Pax7+ nuclei in total nuclei in the muscle cross-sections, were similar between f/f and BcMKO mice (Fig. S2, d and e). Previous studies reported the low activity of satellite cells in the non-regenerating skeletal muscle, as revealed by the immunofluorescence costaining of MyoD and Pax7 (Schwörer et al., 2016; Wang et al., 2022a). Consistently, we observed low and comparable percentages of MyoD and Pax7 double-positive (MyoD+/Pax7+) satellite cells in non-regenerating control and BcMKO muscles (Fig. S2, d and e). Taken together, these data suggest that Baf60c inactivation in skeletal myofibers has no significant prior effects on muscle stem cell number and activation.
Figure 1.
Myofiber-specific inactivation of Baf60c impairs muscle regeneration. (a) qPCR analysis of Baf60c gene expression in quadriceps (Quad) or plantaris (PL) muscles from Baf60cflox/flox (f/f) and Baf60cflox/flox MLC-Cre (BcMKO) mice (n = 6 mice per group). ***P < 0.001; two-tailed unpaired Student’s t test. (b) Representative immunoblots of total Quad or PL muscle protein lysates from f/f and BcMKO mice. (c) Schematic outline of cardiotoxin (CTX)-induced muscle injury and regeneration model in f/f and BcMKO mice. CTX was i.m. injected into the TA muscle on one leg, and PBS was injected into the TA muscle on the contralateral leg as control. (d) TA weight loss (calculated by subtraction of tissue weight in CTX-injected TA muscle on one leg from PBS-injected TA muscle on the contralateral leg in the same mouse) in f/f and BcMKO mice at 7 dpi (n = 3 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (e) Representative immunofluorescence images of Desmin (red), WGA (green), and DAPI (blue) of TA muscle cross-sections from f/f and BcMKO mice at 7 dpi. WGA, wheat germ agglutinin. Scale bar: 400 μm. (f) Quantification of the percentage of Desmin positive area in TA muscle cross-sections from f/f and BcMKO mice described in e (n = 3 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (g) Representative immunofluorescence images of Dystrophin (green) and DAPI (blue) of TA muscle cross-sections from f/f and BcMKO mice at 14 dpi. Scale bar: 400 μm. (h) The percentages of myofibers with central nuclei (left) and the percentage distribution of myofiber CSA (right) as described in g (n = 9 mice per group). **P < 0.01, ***P < 0.001, two-tailed unpaired Student’s t test. (i) Representative H&E staining (upper) and average CSA (lower) of TA muscles from f/f and BcMKO mice at 14 dpi (n = 4–5 mice per group; atleast four sections/mouse). Scale bar: 100 μm. ***P < 0.001; two-tailed unpaired Student’s t test. (j) Measurement of tetanic contraction force in TA muscles from f/f and BcMKO mice at 14 dpi (n = 5–7 mice per group). **P < 0.01; area under curve (AUC) for each mouse from f/f and BcMKO group was analyzed with two-tailed unpaired Student’s t test. (k) Heatmap representation of the scaled, normalized FPKM (as Z-score) of significantly changed 992 genes as described in Fig. S2 f (n = 3 mice per group). (l) GO analysis of genes in Set I (CTX up-down) and Set Ⅱ (CTX up-up) described in k. Most significant and nonredundant biological processes with respective gene numbers and −log10 (P value) are shown. (m) qPCR analysis of the dynamic expression patterns of indicated myogenic-related genes in TA muscle from f/f and BcMKO mice at different days post-injury (n = 3–4 mice at each time point from each group). *P < 0.05, **P < 0.01, ***P < 0.001, two-way ANOVA with multiple comparisons. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F1.
Figure S2.
Characterization of non-regenerating and regenerating skeletal muscles from control and BcMKO mice. (a–e) Muscle weight/body weight (BW) ratio (a), representative H&E staining (b), average cross-sectional area (CSA; c, left), and the percentage of myofibers with central nuclei (c, right), representative immunofluorescence images of MyoD (red), Pax7 (green), and DAPI (blue; d), percentage of Pax7+ nuclei to total DAPI+ nuclei (e, left), and percentage of MyoD+ cells in Pax7+ satellite cells (e, right) of TA muscle cross-sections from f/f and BcMKO mice under normal non-injury conditions without CTX injection (n = 4–5 mice per group; at least four sections/mouse). Scale bars represent 100 μm in b and 20 μm in d. Data represent mean ± SEM; N.S., not significant; two-tailed unpaired Student’s t test. (f) Flowchart of RNA-Seq data analysis of TA muscles with PBS or CTX injection at 3 dpi from f/f and BcMKO mice. CTX-induced genes in f/f mice (genes with log2FC (f/f_CTX/f/f_PBS) > −1 and P < 0.05) were divided into two groups: Set I (CTX up & BcMKO down, genes with log2FC (BcMKO_CTX/f/f_CTX) < −0.3 and P < 0.05) and Set II (CTX up & BcMKO up, genes with log2FC (BcMKO_CTX/f/f_CTX) > 0.3 and P < 0.05). Venn plot shows the overlapping genes between the two indicated datasets. (g) qPCR analysis of indicated inflammation-related genes in TA muscles from f/f and BcMKO mice at different days post-injury (n = 3–4 mice at each time point from each group). Data represent mean ± SEM; ***P < 0.001; two-way ANOVA with multiple comparisons. (h) Flowchart of RNA-Seq analysis of TA muscles with or without CTX injection at 3 dpi from f/f and BcMKO mice. CTX-suppressed genes in f/f mice (genes with log2FC (f/f_CTX/f/f_PBS) < −1 and P < 0.05) were divided into two groups: Set III (CTX down & BcMKO down, genes with log2FC (BcMKO_CTX/f/f_CTX) < −0.3 and P < 0.05), and Set Ⅳ (CTX down & BcMKO up, genes with log2FC (BcMKO _CTX/f/f_CTX) > −0.3 and P < 0.05). Venn plot shows the overlapping genes between the two given datasets. (i) GO analysis of genes in Set III as described in h. Most significant and nonredundant biological processes with respective gene numbers and −log10 (P value) are shown. All experimental data were verified in at least two independent experiments.
To test the effect of Baf60c inactivation in myofibers on muscle regeneration, CTX was injected into TA muscle to induce acute muscle injury (Fig. 1 c). Compared with f/f control mice, CTX-injected BcMKO mice exhibited a more severe TA muscle weight loss at 7 d post-injury (dpi; Fig. 1 d), suggesting that muscle regeneration upon injury was impaired in BcMKO mice. We further assessed the expression of Desmin which is a muscle-specific member of the intermediate filament family with remarkable expression in newly formed myofibers during myogenesis and muscle regeneration (Heredia et al., 2013). Immunofluorescence staining of Desmin demonstrated a significant decrease of Desmin positive area in BcMKO muscles compared with controls, indicating that the number and area of newly formed myofibers were significantly reduced in BcMKO muscles (Fig. 1, e and f). In accordance, the percentage of myofibers with central nuclei was much lower in BcMKO muscles at 14 dpi, leading to a significant reduction in myofiber size compared with controls (Fig. 1, g and h). Further H&E staining and cross-sectional area (CSA) quantification of regenerating myofibers revealed that the size of newly formed myofibers is markedly decreased in BcMKO muscles compared with those from f/f control mice (Fig. 1 i). To test the function of muscle with newly synthesized myofibers from regeneration, we measured the contraction force generated by TA muscles in vivo at 14 dpi and observed a robust decrease in muscle contraction force in BcMKO mice compared with f/f control (Fig. 1 j). These data demonstrate that BcMKO attenuates muscle regeneration following injury by impeding the formation of myofibers with normal quantity and size, leading to a significant impairment of muscle contraction in BcMKO mice, suggesting that myofiber Baf60c plays a crucial role in maintaining a functional muscle regeneration against muscle damage.
To evaluate whether the regeneration defect coincided with a transcriptional signature, RNA sequencing analysis (RNA-Seq) was performed to examine the gene expression profiles in TA muscles from control and BcMKO mice following injection with either PBS or CTX for 3 d. Sequencing data were analyzed as follows: in f/f mice, CTX injection significantly upregulated the expression of 4,098 genes (termed as CTX up), in which 992 genes were significantly altered by BcMKO in response to CTX injection (Fig. S2 f). These 992 genes were further divided into two groups: Set I (CTX up and BcMKO down) and Set II (CTX up and BcMKO up; Fig. 1 k). Gene ontology (GO) analysis revealed that genes in Set I were mainly enriched in muscle development and differentiation, while genes in Set II were enriched in inflammation-related pathways (Fig. 1 l). Further qPCR analysis confirmed that the injury-induced elevation of differentiation-related genes, such as Myomaker, MyoG, Tnnc1, Tnni1, and Tnnt2, were robustly suppressed in BcMKO muscles in a time-dependent manner (Fig. 1 m). As for inflammation, macrophage classical activation marker genes (TNF-α, IL-12) were robustly upregulated, while Mgl1, a macrophage alternative activation marker gene, was downregulated in TA muscles from BcMKO compared with those from f/f control mice (Fig. S2 g). Additionally, among the “CTX-suppressed genes” (CTX down; Fig. S2 h), most of which were further downregulated in muscles from BcMKO mice (Set Ⅲ). These genes were mainly enriched in metabolism-related pathways (Fig. S2 i), consistent with our previous findings that Baf60c is highly enriched in glycolytic myofibers and plays a critical role in muscle glucose sensing and utilization (Meng et al., 2017). These results suggest that Baf60c deficiency in mature myocytes severely impairs skeletal muscle regeneration and differentiation process after injury through transcriptional reprogramming.
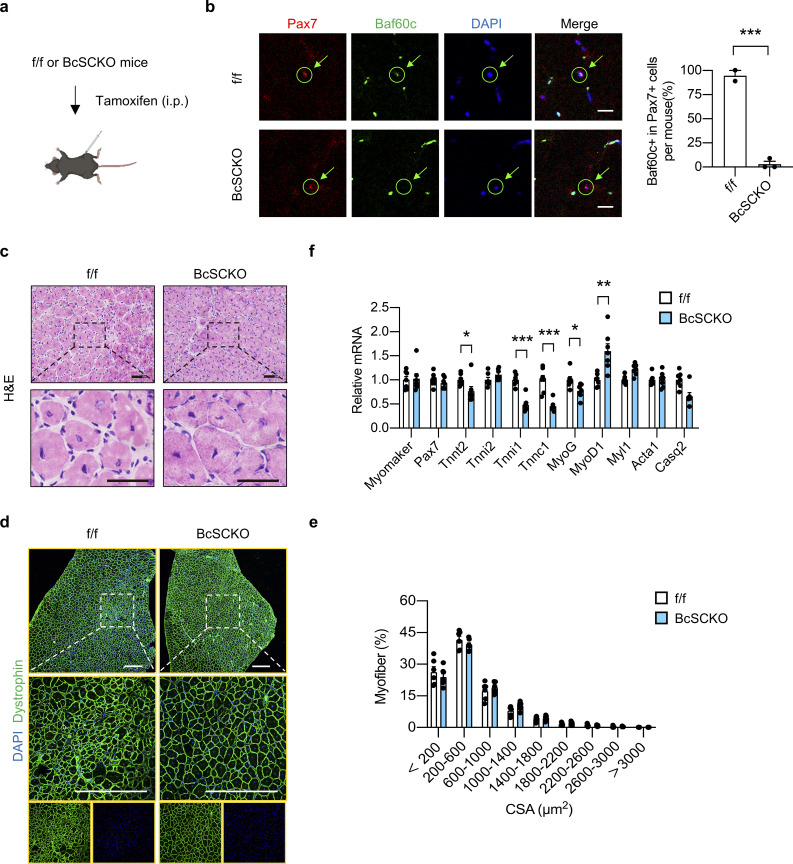
To assess the effect of Baf60c inactivation in MuSCs per se on muscle regeneration in adult mice, muscle stem cell-specific Baf60c KO (BcSCKO) mice were established by crossing the Baf60cf/f mice with the tamoxifen-inducible Cre recombinase expression mice driven by the endogenous Pax7 gene promoter (Pax7-CreERT2; Fig. S3 a). As shown in Fig. S3 b, immunofluorescence staining showed that tamoxifen injection resulted in a successful ablation of Baf60c in MuSCs in BcSCKO mice compared with controls. However, Baf60c inactivation in MuSCs exhibited mild effects on muscle regeneration as revealed by morphological analysis of TA muscles by H&E staining at 7 dpi (Fig. S3 c) and examination of fiber size distribution by immunofluorescence staining of Dystrophin and DAPI on muscle tissue sections at 14 dpi (Fig. S3, d and e). Consistent with previous studies showing that Baf60c might be involved in myoblast differentiation in vitro (Forcales et al., 2012), several differentiation-related genes were downregulated in TA muscle from BcSCKO mice at 14 dpi (Fig. S3 f). These data indicate that Baf60c deficiency in muscle stem cells per se has a modest effect on muscle differentiation and regeneration upon injury in vivo.
Figure S3.
Muscle stem cell-specific Baf60c deficiency exhibits a modest effect on muscle regeneration. (a) Schematic outline of muscle stem cell (MuSC) specific inactivation of Baf60c induced by i.p. injection of tamoxifen in Baf60cflox/flox (f/f) and Baf60cflox/flox Pax7-CreERT2 (BcSCKO) mice. (b) Representative immunofluorescence images (left) of Pax7 (red), Baf60c (green), and DAPI (blue) in TA muscle cross-sections from f/f and BcSCKO mice. Baf60c knockout efficiency in MuSCs (right) was calculated (n = 2–3 mice per group). Scale bar: 20 μm. Data represent mean ± SEM; ***P < 0.001; two-tailed unpaired Student’s t test. (c) Representative H&E staining images of TA muscle cross-sections from f/f and BcSCKO mice at 7 dpi. Scale bar: 50 μm. (d and e) Representative immunofluorescence images (d) of Dystrophin (green) and DAPI (blue) and percentage distribution of muscle fiber CSA (e) in TA muscle cross-sections from f/f and BcSCKO mice at 14 dpi (n = 6–7 mice per group). Scale bar: 200 μm. Data represent mean ± SEM. (f) qPCR analysis of myogenic-related genes in TA muscles from f/f and BcSCKO mice at 14 dpi (n = 7 mice per group). Data represent mean ± SEM; *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. All data are representative of at least two independent experiments.
Baf60c deficiency in skeletal myofiber impairs muscle regeneration through increasing Dkk3 expression
Accumulating evidence suggests that disturbance of the muscle stem cell niche could lead to dysfunction in MuSCs and impairment of muscle repair (Yin et al., 2013). Since Baf60c inactivation in mature myocytes but not in MuSCs impairs muscle regeneration and the muscle has recently emerged as an important endocrine organ through synthesizing and secreting of myokines (Febbraio and Pedersen, 2020; Pedersen and Febbraio, 2012), we speculated that Baf60c in skeletal myocyte could alter the microenvironment of MuSCs by regulating the production and secretion of certain muscle-secreted proteins.
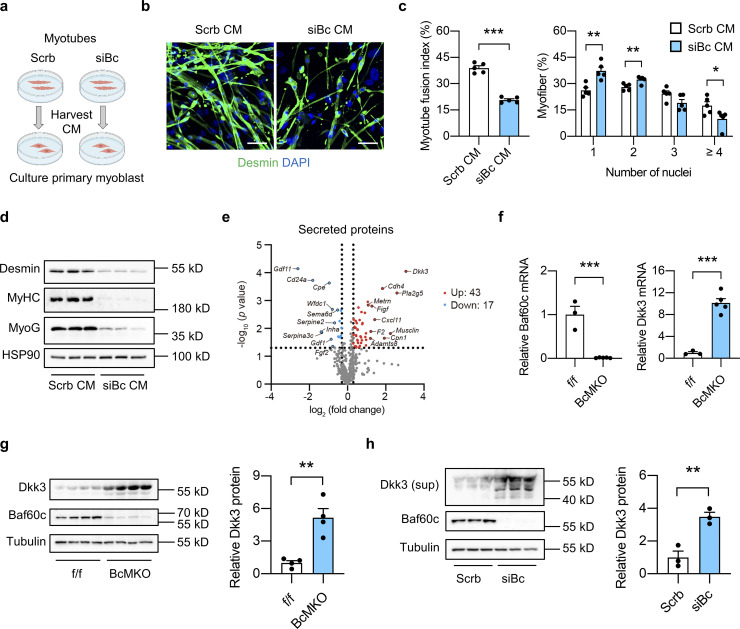
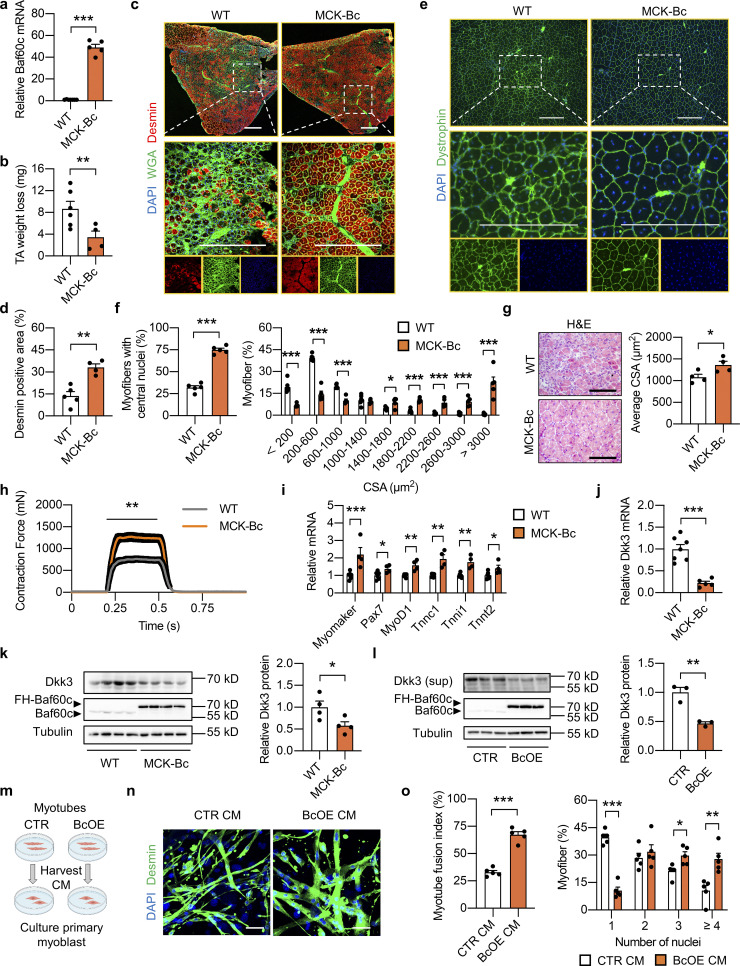
To test this possibility, we generated Baf60c knockdown C2C12 stable cell line by transduction of retroviral vectors expressing scramble (Scrb) shRNA or shRNA targeting Baf60c (siBc) and harvested conditioned medium (CM) from fully differentiated Scrb or siBc myotubes to treat the primary muscle progenitor cells derived from single myofibers of WT mice and monitor their differentiation (Fig. 2 a). During differentiation, myoblasts will fuse together to form multinucleated myotubes. Compared with CM from Scrb control myotubes, treatment with CM from siBc myotubes profoundly attenuated muscle differentiation as revealed by Desmin and DAPI immunofluorescence staining (Fig. 2 b). SiBc CM treatment elicited a much lower percentage of newly generated myofibers with four or more nuclei and a higher percentage of myofibers with one or two nuclei compared with control, suggesting a lower myotube fusion index (Fig. 2 c). In addition, we also performed Western blotting analyses of the regenerative markers, including Desmin, Myogenin (MyoG), and myosin heavy chain (MyHC), in whole-cell protein lysates from fully differentiated primary myotubes treated with Scrb CM or SiBc CM. We observed that protein expression levels of all three muscle differentiation markers were markedly decreased in siBc CM treated cells compared with Scrb CM treated group (Fig. 2 d), further supporting that CM from Baf60c deficient myotubes impaired muscle stem cell differentiation. These results are consistent with our hypothesis that Baf60c in myotubes may produce and secrete certain myokines to regulate muscle stem cell differentiation. To this end, we analyzed our previously generated microarray datasets in Quad muscle from control and BcMKO mice and identified a cluster of secreted-protein encoding genes that are regulated by muscle-specific Baf60c ablation (Fig. 2 e). Among the 43 secreted-protein encoding genes that are upregulated in muscle from BcMKO mice, Dkk3 gene was identified to be top-ranked with the biggest fold change (Fig. 2 e). Unlike other Dkk family members, Dkk3 is the only one preferentially expressed in skeletal muscle and has been recently shown to play an important role in age-related muscle atrophy (Yin et al., 2018). Further qPCR and Western blotting analyses confirmed the elevation of Dkk3 mRNA (Fig. 2 f) and protein (Fig. 2 g) expression in Quad muscles from BcMKO mice compared with controls. As expected, Dkk3 protein levels were also remarkably increased in the CM from Baf60c knockdown C2C12 myotubes compared with CM from Scrb control myotubes (Fig. 2 h). These results suggest that Baf60c ablation in mature myocytes might impair muscle differentiation and regeneration through upregulation of Dkk3 production and secretion, and its mediated paracrine signaling to MuSCs.
Figure 2.
Baf60c inactivation in myocytes may regulate muscle stem cell regeneration by increasing the expression and secretion of Dkk3. (a) Schematic outline of conditioned medium (CM) harvested from C2C12 myotubes stably transduced with retroviral vectors expressing scramble shRNA (Scrb) or shRNA targeting Baf60c (siBc) to treat primary myoblasts derived from freshly isolated single myofibers. (b) Representative immunofluorescence images of Desmin (green) and DAPI (blue) of primary myoblasts treated with CM harvested from Scrb or siBc C2C12 myotubes. Scale bar: 50 μm. (c) Quantification of myotube fusion index and differentiation as described in b. Myotube fusion index was calculated by dividing the number of myocytes with multi-nuclei (two or more nuclei) by the total number of myocytes in each image. Dots represent independent biological replicates (n = 5 per group). Data are representative of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. (d) Immunoblots of total protein lysates from fully differentiated primary myotubes as treated in b. (e) Volcano plot showing the expression differences of genes encoding secreted proteins in the Quad muscle microarray data between f/f and BcMKO mice (n = 3 mice per group). Upregulated (log2FC > −0.3, P < 0.05) or downregulated (log2FC < −0.3, P < 0.05) genes were marked in red and blue, respectively. Top 10 upregulated and top 10 downregulated genes encoding secreted proteins were labeled. (f) qPCR analysis of Baf60c and Dkk3 gene expression in the Quad muscle of f/f and BcMKO mice (n = 3–5 mice per group). ***P < 0.001; two-tailed unpaired Student’s t test. (g) Immunoblots (left) and quantification of relative Dkk3 protein level (right) of total Quad muscle protein lysates from f/f and BcMKO mice (n = 4 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (h) Immunoblots (left) and quantification of relative Dkk3 protein level in the supernatant (right) and Baf60c protein level in the total cell protein lysates from Scrb and siBc C2C12 myotubes (n = 3 biological replicates). Sup, supernatant. **P < 0.01; two-tailed unpaired Student’s t test. Values represent mean ± SEM. All experimental data were verified in at least three independent experiments. Source data are available for this figure: SourceData F2.
Dkk3 attenuates muscle regeneration in vivo and in cultured myocytes
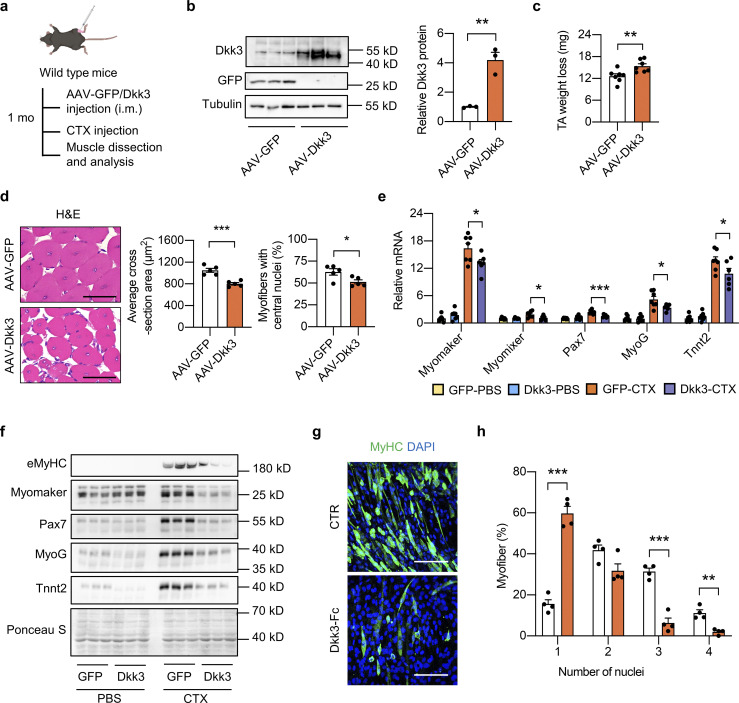
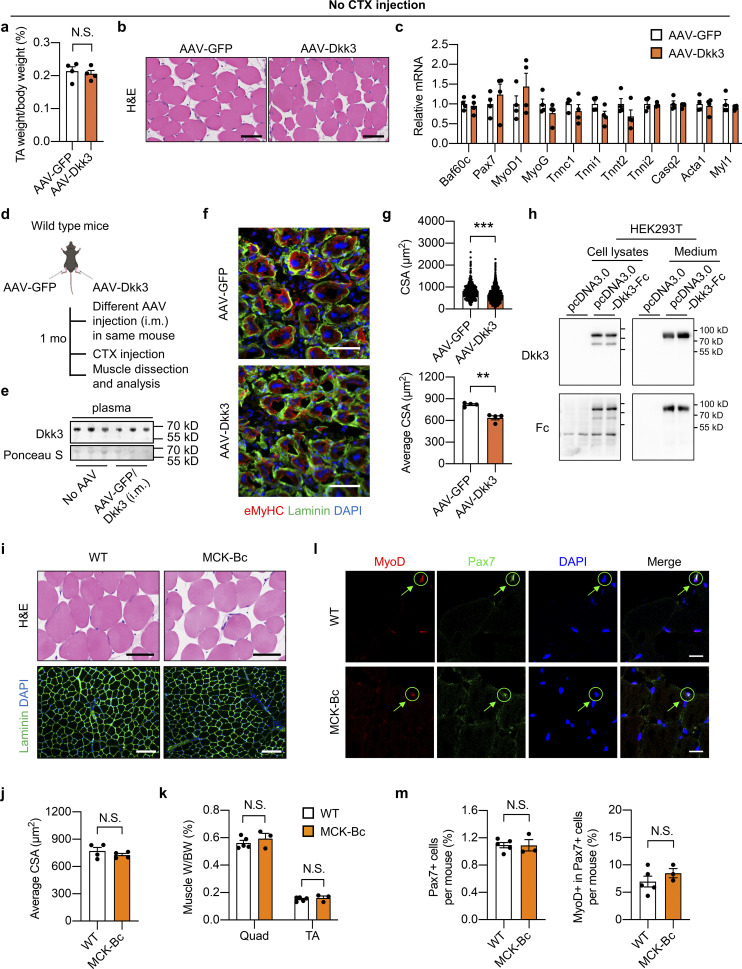
To further delineate the mechanism by which Dkk3 impedes muscle regeneration, the adeno-associated virus (AAV)-Dkk3 virus was intramuscularly (i.m.) injected into TA muscles of WT adult mice (Fig. 3 a). Western blotting analysis validated the successful overexpression (OE) of Dkk3 in AAV-Dkk3 injected TA muscles (Fig. 3 b). Muscle weight loss in the AAV-Dkk3 injected group was significantly higher than that in the AAV-GFP injected group at 5 dpi (Fig. 3 c). In addition, myofiber size and percentage of myofibers with central nuclei, as revealed by H&E staining and quantification, were much smaller in AAV-Dkk3 injected group at 14 dpi (Fig. 3 d), suggesting that local muscular elevation of Dkk3 levels impaired muscle regeneration upon injury. Consistently, the induction of muscle differentiation–related genes by CTX was remarkably attenuated by AAV-mediated Dkk3 OE (Fig. 3 e). In addition, we also performed Western blotting analyses of several regeneration markers including embryonic myosin heavy chain (eMyHC), Myomaker, Pax7, MyoG, and Tnnt2. Consistent with gene expression changes shown in Fig. 3 e, while the protein levels of these regeneration markers in the quiescent state (PBS injected group) remained largely unaltered, their induction by CTX treatment was robustly attenuated by Dkk3 OE (Fig. 3 f). However, no overt difference in muscle weight, histology, and gene expression was observed between AAV-GFP and AAV-Dkk3 injected groups under basal condition without CTX injection (Fig. S4, a–c).
Figure 3.
Dkk3 impairs muscle regeneration in mice and in cultured myocytes. (a) Schematic representation of mouse model using used AAV system to achieve GFP or Dkk3 overexpression in TA muscles in WT mice and the following experimental setup. (b) Immunoblots (left) and quantification of relative Dkk3 protein level (right) of total protein lysates of AAV-GFP or AAV-Dkk3 injected TA muscles in WT mice (n = 3 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (c) Muscle weight loss in AAV-GFP or AAV-Dkk3 injected TA muscles in WT mice at 5 dpi (n = 7 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (d) Representative H&E staining (left), average CSA (middle), and percentages of myofibers with central nuclei (right) of AAV-GFP or AAV-Dkk3 injected TA muscles at 14 dpi (n = 5 mice per group; at least four sections/mouse). Scale bar: 50 μm. *P < 0.05, ***P < 0.001; two-tailed unpaired Student’s t test. (e) qPCR analysis of muscle differentiation-related genes in TA muscles from AAV-GFP or AAV-Dkk3 injected WT mice at 5 dpi (n = 6–7 mice per group). *P < 0.05, ***P < 0.001; one-way ANOVA with multiple comparisons. (f) Immunoblots of total protein lysates from AAV-GFP or AAV-Dkk3 injected TA muscles in WT mice at 5 dpi. (g and h) Representative immunofluorescence images on MyHC (green) and DAPI (blue) of Dkk3-Fc protein treated C2C12 myotubes (g), and quantification of myotube differentiation (h). C2C12 myoblasts were seeded into 12-well plates at around 90–100% confluency. The culture medium was then switched to a differentiation medium (DMEM plus 2% FBS) in the presence of vehicle (CTR) or purified Dkk3-Fc protein. The differentiation medium was replaced every 2 d for a total of 6 d, followed by immunofluorescence staining. Dots represent independent biological replicates. Data are representative of three independent experiments. MyHC, myosin heavy chain. Scale bar: 100 μm. **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F3.
Figure S4.
Effects of Dkk3 overexpression on muscle regeneration and characterization of Baf60c transgene on non-regenerating skeletal muscles. (a–c) TA weight/body weight ratio (a), representative H&E staining of TA muscle cross-sections (b), and qPCR analysis of muscle differentiation-related genes (c) in AAV-GFP or AAV-Dkk3 i.m. injected mice under the normal physiological condition without CTX injection (n = 4 mice per group). Scale bar in b: 100 μm. N.S., not significant; two-tailed unpaired Student’s t test. (d) Schematic representation of mouse model using AAV to achieve GFP overexpression in TA muscle in one leg and Dkk3 overexpression in TA muscle in the contralateral leg in one mouse, and the following experimental setup. (e) Representative immunoblots of plasma samples from WT mice without (no AAV) or with AAV injection as described in d. Mouse plasma samples were filtered with the Amicon centrifugal concentrator with a 100 kD molecular weight cutoff (MWCO) prior to Western blotting analysis of Dkk3 (7.5 μl original plasma/lane). (f) Representative immunofluorescence images of eMyHC (red), laminin (green), and DAPI (blue) of AAV-GFP or AAV-Dkk3 injected TA muscle cross-sections. eMyHC, embryonic myosin heavy chain. Scale bar: 50 μm. (g) CSA distribution (upper) and average CSA (lower) of AAV-GFP or AAV-Dkk3 injected side TA muscles. Dots in the upper panel represent individual myofibers; dots in the lower panel represent four mice per group; at least three sections/mouse. **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. (h) Immunoblots of total protein lysates and conditioned medium from HEK293T cells transiently transfected with pcDNA3.0 or pcDNA3.0-Dkk3-Fc plasmids. (i) Representative H&E staining (upper) and immunofluorescence images (lower) of laminin (green) and DAPI (blue) of the non-regenerating TA muscle cross-sections from WT and MCK-Bc mice under the normal physiological condition without CTX injection. Scale bar: 100 μm for H&E staining, 200 μm for laminin staining. (j) Average CSA of TA muscles calculated using laminin/DAPI immunofluorescence staining images described in (i, lower panel; n = 4 mice per group; at least four sections/mouse). N.S., not significant; two-tailed unpaired Student’s t test. (k–m) Muscle weight/BW ratio (k), representative immunofluorescence images of MyoD (red), Pax7 (green), and DAPI (blue; l), percentage of Pax7+ nuclei to total DAPI+ nuclei (m, left) and percentage of MyoD+ cells in Pax7+ satellite cells (m, right) of TA muscle cross-sections from WT and MCK-Bc mice under the normal un-injury condition without CTX injection (n = 3–5 mice per group; atleast four sections/mouse). Scale bar: 20 μm. N.S., not significant; two-tailed unpaired Student’s t test. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData FS4.
To further examine the effect of local intramuscular Dkk3 concentration on muscle regeneration, we subjected the same mice to i.m. injection of AAV-GFP into the TA muscle in one leg and AAV-Dkk3 into the TA muscle in the contralateral leg, followed by CTX injection to induce muscle injury and evaluation of regenerative capacity (Fig. S4 d). As shown in Fig. S4 e, plasma Dkk3 protein levels in AAV-GFP+AAV-Dkk3 injected mice remained unaltered compared to those in their littermate control mice without AAV i.m. injection, suggesting that local i.m. injection of AAV-Dkk3 has a mild effect on systemic Dkk3 levels in the circulation. Remarkably, AAV-Dkk3 injection robustly impaired muscle regeneration as revealed by eMyHC/laminin/DAPI immunofluorescence staining of frozen muscle cross-sections (Fig. S4 f). The regenerative myofiber size was significantly decreased in AAV-Dkk3-treated TA muscles compared with AAV-GFP treated control group (Fig. S4 g). Taken together, these results support the local inhibitory effect of Dkk3 on muscle stem cell regeneration upon injury in mice, while having a mild effect on muscle fitness in normal physiological conditions.
To examine whether Dkk3 has a direct effect on C2C12 cells, we generated a pcDNA3.0 vector expressing Dkk3 protein of which the C terminus was fused with the crystallizable fragment (Fc) domain of immunoglobulin G (IgG; Dkk3-Fc). Western blotting analysis of CM from transiently transfected HEK293T cells confirmed that Dkk3-Fc was able to be secreted and detected in the cultured medium (Fig. S4 h). Then, we transiently transfected Expi293F cells for the expression and purification of Dkk3-Fc protein from the cultured medium. Purified Dkk3-Fc protein was added to the differentiation medium to observe its effects on C2C12 myotube differentiation. The differentiation medium with or without Dkk3-Fc protein was replaced every 2 d for a total of 6 d. Dkk3-Fc treatment led to the formation of shorter MyHC-positive myotubes with fewer nuclei (Fig. 3, g and h). Taken together, these results demonstrate that Dkk3 cell autonomously suppresses muscle differentiation and regenerative capacity in vitro and in vivo.
Dkk3 knockdown rescues the regeneration defect in muscle from BcMKO mice
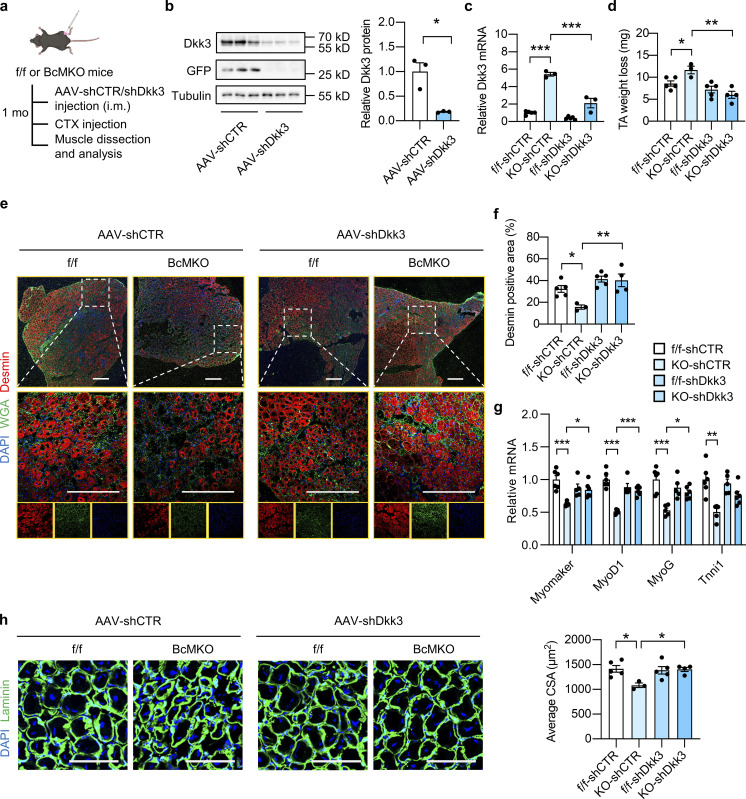
To further confirm the role of Dkk3 in mediating the effect of BcMKO on muscle regeneration capacity after injury, an AAV-mediated shRNA expression system was employed to achieve Dkk3 knockdown in vivo. AAV viral vectors expressing control shRNA (AAV-shCTR) or Dkk3 shRNA (AAV-shDkk3) were i.m. injected into TA muscles of f/f and BcMKO adult mice, followed by CTX injection. TA muscles were harvested at around 1 mo following AAV injection (Fig. 4 a). The higher mRNA and protein levels of Dkk3 in BcMKO muscle were markedly reduced by AAV-shDkk3 treatment, whereas the basal gene expression level of Dkk3 in muscle from f/f mice was relatively low and remained largely unaltered by AAV-shDkk3 treatment compared with shCTR-treated group (f/f-shCTR; Fig. 4, b and c). Dkk3 knockdown markedly reduced muscle mass loss in BcMKO mice at 7 dpi, while no further benefits were observed in f/f mice (Fig. 4 d). In the AAV-shCTR injected groups, Desmin positive area in TA muscle was significantly smaller from BcMKO mice than that from f/f mice (Fig. 4, e and f), and this finding was consistent with the result shown in Fig. 1, e and f. In the AAV-shDkk3 groups, the regenerating area as revealed by Desmin staining of TA muscle from BcMKO mice was largely recovered to a level comparable with that from f/f mice (Fig. 4 f). In accordance, Dkk3 knockdown rescued the lower expression of muscle differentiation-related genes, such as Myomaker, MyoD1, MyoG, and Tnni1 in BcMKO mice at 7 dpi (Fig. 4 g). Furthermore, the decrease in regenerative myofiber size in KO-shCTR muscles compared with f/f-shCTR muscles was largely reversed by Dkk3 knockdown (KO-shDkk3 vs. KO-shCTR; Fig. 4 h), as revealed by laminin/DAPI immunofluorescence staining of muscle cross-sections at 3 dpi. These results suggest that the upregulation of Dkk3 expression may contribute to muscle regeneration defects in BcMKO mice.
Figure 4.
Dkk3 knockdown rescues the regeneration defect in muscles from BcMKO mice. (a) Schematic representation of mouse model using AAV system to achieve local muscle Dkk3 knockdown in f/f and BcMKO mice and the following experimental setup. (b) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total protein lysates from AAV-shCTR or AAV-shDkk3 injected TA muscles in BcMKO mice (n = 3 mice per group). AAV-shCTR, AAV-control shRNA; AAV-shDkk3, AAV-Dkk3 shRNA. *P < 0.05; two-tailed unpaired Student’s t test. (c) qPCR analysis of Dkk3 gene expression in the AAV-shCTR or AAV-shDkk3 injected TA muscles of f/f and BcMKO mice (n = 3–5 mice per group). ***P < 0.001; one-way ANOVA with multiple comparisons. (d) TA weight loss in indicated groups at 7 dpi (n = 3–5 mice per group). *P < 0.05, **P < 0.01; one-way ANOVA with multiple comparisons. (e) Representative immunofluorescence images on Desmin (red), WGA (green), and DAPI (blue) of TA muscle cross-sections from AAV-shCTR or AAV-shDkk3 injected f/f and BcMKO mice at 7 dpi. Scale bar: 400 μm. (f) Quantification of the percentage of Desmin positive area percentage of TA muscle cross-sections from AAV-shCTR or AAV-shDkk3 injected f/f and BcMKO mice as described in e (n = 3–5 mice per group). *P < 0.05, **P < 0.01; one-way ANOVA with multiple comparisons. (g) qPCR analysis of muscle differentiation-related genes in indicated groups at 3 dpi (n = 5–6 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with multiple comparisons. (h) Representative immunofluorescence images (left) of laminin (green) and DAPI (blue) and average CSA (right) of TA muscle cross-sections from indicated groups (n = 3–5 mice per group; at least three sections/mouse). Scale bar: 100 μm. One-way ANOVA with multiple comparisons. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F4.
Inactivation of Dkk3 by myofiber-specific Baf60c transgenic expression improves muscle regeneration
We next investigated whether Dkk3 inactivation by muscle transgenic expression of Baf60c could rescue muscle regeneration. We generated muscle-specific Baf60c transgenic mice driven by muscle creatine kinase (MCK) promoter (MCK-Bc) as previously described (Meng et al., 2013). Muscle gene expression of Baf60c was successfully increased in MCK-Bc mice compared with littermate controls (Fig. 5 a). Similar to the studies performed in BcMKO mice, we first examined the effect of Baf60c transgenic expression on non-regenerating muscle in MCK-Bc mice under normal physiological conditions. Histological analysis of muscle cross-sections by H&E and laminin/DAPI immunofluorescence staining revealed that the tissue morphology and muscle fiber size are indistinguishable between WT and MCK-Bc mice (Fig. S4, i and j). Muscle weight was also similar between the two groups (Fig. S4 k). To further examine the effect of Baf60c gain-of-function on the number and activity of satellite cells, we went on to perform immunofluorescence staining of Pax7 and/or MyoD on frozen muscle cross-sections from control and MCK-Bc mice. Similar to the results obtained in control and BcMKO mice, the number of satellite cells in non-regenerating muscles, as revealed by the percentages of Pax7+ nuclei in total nuclei in the muscle cross-sections, were similar between control and MCK-Bc mice (Fig. S4, l and m). In addition, we also observed low and comparable percentages of MyoD and Pax7 double-positive (MyoD+/Pax7+) satellite cells in non-regenerating muscles from control and MCK-Bc mice (Fig. S4, l and m). These data suggest that Baf60c transgenic expression in skeletal myofibers has a mild prior effect on muscle stem cell number and activation.
Figure 5.
Dkk3 blockade by myocyte-specific Baf60c overexpression promotes muscle regeneration. (a) qPCR analysis of Baf60c gene expression in Quad muscle from WT and MCK-Baf60c (MCK-Bc) mice (n = 5–7 mice per group). ***P < 0.001; two-tailed unpaired Student’s t test. (b) TA weight loss in WT and MCK-Bc mice at 7 dpi (n = 4–6 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (c) Representative immunofluorescence images on Desmin (red), WGA (green), and DAPI (blue) of TA muscle cross-sections from WT and MCK-Bc mice at 7 dpi. Scale bar: 400 μm. (d) Quantification of the percentage of Desmin positive area as described in c (n = 4–5 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (e) Representative immunofluorescence images on Dystrophin (green) and DAPI (blue) of TA muscle cross-sections from WT and MCK-Bc mice at 14 dpi. Scale bar: 200 μm. (f) The percentages of myofibers with central nuclei (left), and the percentage distribution of myofiber CSA (right) as described in e (n = 4–5 mice per group). *P < 0.05, ***P < 0.001; two-tailed unpaired Student’s t test. (g) Representative H&E staining (left) and average CSA (right) of TA muscles from WT or MCK-Bc mice at 14 dpi (n = 4 mice per group). Scale bar: 100 μm. *P < 0.05; two-tailed unpaired Student’s t test. (h) Measurement of muscle tetanic contraction of TA muscles from WT and MCK-Bc mice at 14 dpi (n = 5–6 per group). **P < 0.01; AUC for each mouse from WT and MCK-Bc group was analyzed with two-tailed unpaired Student’s t test. (i) qPCR analysis of muscle differentiation-related genes in WT and MCK-Bc mice at 3 dpi (n = 4–6 mice per group). *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. (j) qPCR analysis of Dkk3 gene expression in WT and MCK-Bc mice Quad muscle (n = 5–7 mice per group). ***P < 0.001; two-tailed unpaired Student’s t test. (k) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total Quad muscle protein lysates from WT and MCK-Bc mice (n = 4 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (l) Immunoblots of Dkk3 protein in the supernatant and Baf60c protein in the whole cell protein lysates of C2C12 myotubes stably transduced with control retroviral vector (CTR) or retroviral vector expressing Baf60c (BcOE; left) and quantification of relative Dkk3 protein levels in the supernatant (right; n = 3 biological replicates). **P < 0.01; two-tailed unpaired Student’s t test. (m) Schematic outline for CM harvested from C2C12 myotubes as described in l to treat primary myoblasts derived from freshly isolated single myofibers. (n) Representative immunofluorescence images on Desmin (green) and DAPI (blue) of primary myoblasts treated with CM harvested from CTR or BcOE C2C12 myotubes. Scale bar: 50 μm. (o) Quantification of myotube fusion index and differentiation as described in n. Dots represent independent biological replicates (n = 5 per group). Data are representative of two independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001; two-tailed unpaired Student’s t test. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F5.
We then subjected the control and MCK-Bc mice to CTX injection and muscle regeneration evaluation. Consistent with our expectations, we observed less muscle weight loss (Fig. 5 b) and higher expression of Desmin (Fig. 5, c and d) at 7 dpi in MCK-Bc mice compared with WT control mice. In addition, more myofibers with central nuclei were observed in MCK-Bc mice at 14 dpi, leading to a dramatically larger myofiber size, as revealed by Dystrophin/DAPI immunofluorescence staining (Fig. 5, e and f) and H&E staining (Fig. 5 g) of muscle cross sections. Remarkably, muscle contraction force was also greatly enhanced in TA muscle from MCK-Bc mice compared with controls (Fig. 5 h). The expression of muscle regeneration- and differentiation-related genes, such as Myomaker, Pax7, MyoD1, Tnnc1, Tnni1, and Tnnt2, were also significantly upregulated in MCK-Bc mice at 3 dpi (Fig. 5 i). These results suggest that skeletal muscle-specific transgenic expression of Baf60c accelerates muscle regeneration in vivo.
We next went on to examine whether Baf60c OE in myotubes promotes muscle regeneration by regulating the production and secretion of Dkk3. As expected, muscle-specific Baf60c transgene significantly reduced Dkk3 expression, both at mRNA and protein levels (Fig. 5, j and k). We then generated C2C12 myoblast stable cell lines without (CTR) or with Baf60c OE (BcOE) by transduction of retroviruses expressing vector control or Baf60c, respectively, and harvested CM from fully differentiated myotubes. As shown in Fig. 5 l, the Dkk3 protein level in the supernatant of BcOE C2C12 myotubes was significantly downregulated. CMs from CTR and BcOE myotubes were employed to treat the primary muscle progenitor cells derived from WT mice to monitor their effects on muscle differentiation (Fig. 5 m). As expected, compared with CM from CTR myotubes, treatment with CM from BcOE myotubes led to much thicker and longer characteristics of differentiated primary myotubes as revealed by Desmin and DAPI immunofluorescence staining (Fig. 5 n), accompanied by higher myotube fusion index, and a significantly higher percentage of myofibers with multinuclei (Fig. 5 o).
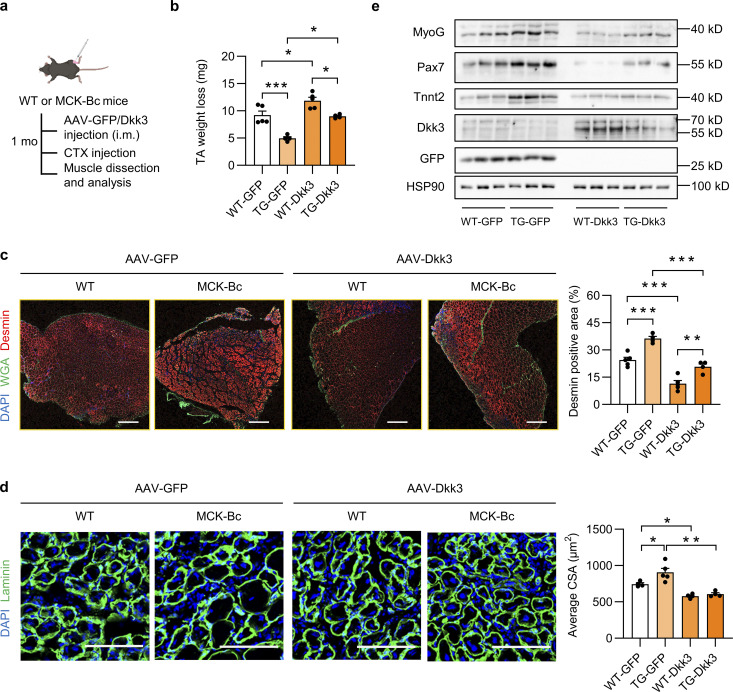
The above cell culture experiments suggested that Baf60c OE in myotubes may regulate muscle stem cell differentiation and regenerative capacity by suppressing the expression and secretion of Dkk3. To further confirm this in vivo, we performed the Dkk3 rescue study in MCK-Bc mice. As shown in Fig. 6 a, WT and MCK-Bc mice were subjected to i.m. injection of AAV-GFP or AAV-Dkk3 into the TA muscles, followed by CTX injection and assays for the evaluation of muscle regenerative activity. As expected, the lower TA muscle weight loss in the TG-GFP group compared with the WT-GFP group was significantly abolished by AAV-mediated Dkk3 overexpression in the TG-Dkk3 group (Fig. 6 b). Consistently, Desmin/WGA staining of the newly generated myofiber revealed that the improvement of muscle regenerative activity in muscles from TG-GFP mice was largely attenuated by Dkk3 overexpression (Fig. 6 c). Moreover, the increase in regenerative myofiber size in TG-GFP muscles compared with WT-GFP muscles was robustly decreased by Dkk3 overexpression (TG-Dkk3 vs. TG-GFP; Fig. 6 d). In addition, Western blotting analyses further confirmed the successful upregulation of Dkk3, and Baf60c-induced upregulation of muscle regeneration markers, including MyoG, Pax7, and Tnnt2, in CTX-treated muscles from TG-GFP mice was markedly abolished by Dkk3 overexpression (TG-Dkk3 vs. TG-GFP; Fig. 6 e). Collectively, these new data further support that Baf60c in myofibers controls muscle stem cell regenerative capacity through Dkk3-mediated paracrine signaling.
Figure 6.
Local intramuscular Dkk3 overexpression abolishes the beneficial effect of MCK-Bc on muscle regeneration. (a) Schematic representation of mouse model using AAV system to achieve local intramuscular GFP or Dkk3 overexpression in WT and MCK-Bc mice and the following experimental setup. (b) TA weight loss in indicated groups at 5 dpi (n = 4–5 mice per group). *P < 0.05, ***P < 0.001; one-way ANOVA with multiple comparisons. (c) Representative immunofluorescence images (left) on Desmin (red), WGA (green), and DAPI (blue), and quantification (right) of the percentage of Desmin positive area percentage of TA muscle cross-sections from indicated groups at 5 dpi. Scale bar: 400 μm. ***P < 0.001; one-way ANOVA with multiple comparisons. (d) Representative immunofluorescence images (left) of laminin (green) and DAPI (blue) and average CSA (right) of TA muscle cross sections from indicated groups (n = 4–5 mice per group; at least three sections/mouse were calculated). *P < 0.05, **P < 0.01; one-way ANOVA with multiple comparisons. Scale bar: 100 μm. (e) Immunoblots of total protein lysates of TA muscles from indicated groups at 5 dpi. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F6.
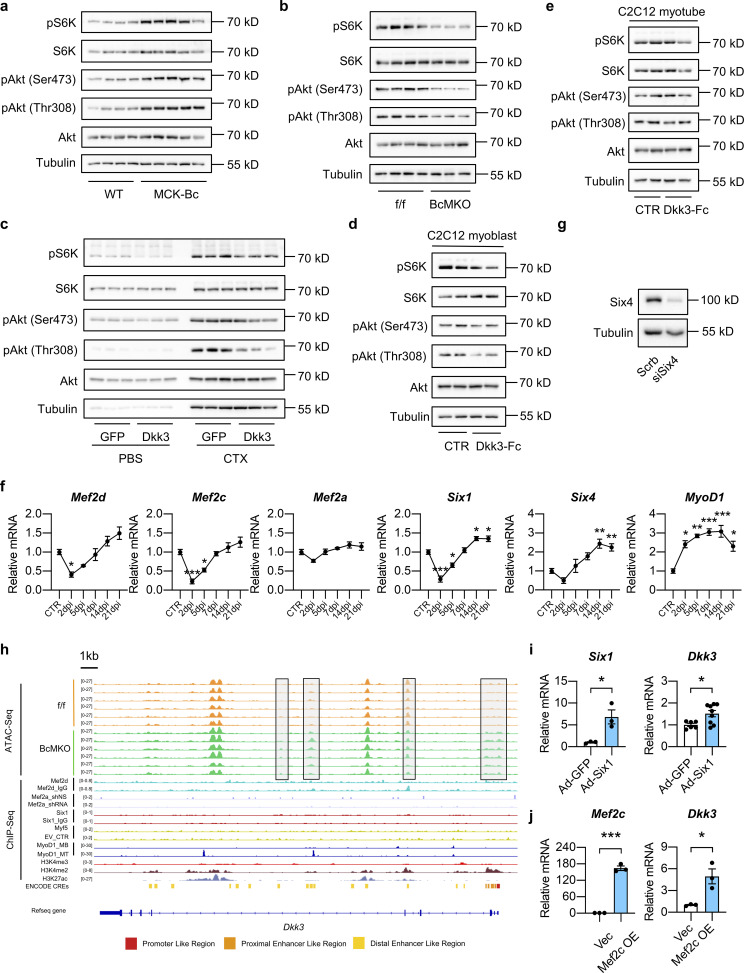
Akt/mTOR signaling pathway plays an important role in muscle regeneration and maintenance (Bodine et al., 2001). Intriguingly, the activities of Akt/mTOR signaling pathway as indicated by the phosphorylation levels of S6K, Akt (Ser473), and Akt (Thr308) were significantly elevated in MCK-Bc skeletal muscle compared with WT controls (Fig. S5 a). Conversely, muscle-specific inactivation of Baf60c resulted in a robust attenuation of Akt/mTOR signaling pathway (Fig. S5 b). These results suggest that the Akt/mTOR pathway might be involved in the regulation of muscle differentiation and regeneration by Baf60c-Dkk3 signaling. Consistent with this hypothesis, we observed that AAV-mediated Dkk3 OE in TA muscle dramatically diminished the activation of Akt/mTOR signaling pathway in response to CTX injection as seen in the AAV-GFP injected groups (Fig. S5 c). Similarly, treatment with Dkk3-Fc protein (5 μg/ml) markedly decreased the phosphorylation levels of S6K, Akt (Ser473), and Akt (Thr308) in C2C12 myoblasts during the differentiation process (Fig. S5 d), while having modest effects on fully differentiated myotubes (Fig. S5 e). This finding is consistent with the observation that AAV-Dkk3 i.m. injection had a mild effect on muscle weight, morphology, and transcriptional profile of genes associated with muscle differentiation under non-injury basal conditions (Fig. S4, a–c). Taken together, these data suggest that Baf60c-mediated production and secretion of Dkk3 might control muscle regeneration by modulating the Akt/mTOR signaling pathway.
Figure S5.
Effects of Baf60c-Dkk3 axis on the Akt/mTOR signaling pathway and validation of transcription factors mediating the regulation of Dkk3 expression by Baf60c in skeletal myocytes. (a) Immunoblots total protein lysates of CTX-injected TA muscles from WT and MCK-Bc mice at 5 dpi. (b) Immunoblots of total protein lysates of CTX-injected TA muscles from f/f and BcMKO mice at 5 dpi. (c) Immunoblots of total protein lysates of AAV-GFP or AAV-Dkk3 injected TA muscles following PBS or CTX injection at 5 dpi. (d) Immunoblots of total protein lysates of undifferentiated C2C12 myoblasts at 90–100% confluency treated with or without Dkk3-Fc protein for 30 min (n = 2 biological replicates). (e) Immunoblots of total protein lysate of fully differentiated C2C12 myotubes treated with or without Dkk3-Fc protein for 30 min (n = 2 biological replicates). (f) qPCR analysis of gene expression of indicated transcription factors (in Fig. 7 c) in TA muscles from WT mice during the process of muscle regeneration (n = 3–4 mice at each time point). *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with multiple comparison. (g) Immunoblots of whole cell protein lysates from C2C12 myotubes stably transduced with retroviral vectors expressing Scramble (Scrb) shRNA or Six4 targeting shRNA (siSix4). (h) Genome browser track view of ATAC-Seq peaks of quadriceps muscles from f/f and BcMKO mice in Dkk3 gene locus. Chip-Seq peaks of Mef2d, Mef2a, Six1, Myf5, MyoD1, H3K4me3, H3K4me2, and H3K27ac, as well as Encyclopedia of DNA Elements annotated candidate regulatory elements (ENCODE CREs) are also displayed. EV_CTR, empty vector control; MB, myoblast; MT, myotube. (i) qPCR analysis of Six1 and Dkk3 gene expression in C2C12 myotubes infected with adenoviruses expressing GFP (Ad-GFP) or Six1 (Ad-Six1; n = 3 biological replicates). *P < 0.05; two-tailed unpaired Student’s t test. (j) qPCR analysis of Mef2c and Dkk3 gene expression in myotubes differentiated from C2C12 myoblasts stably transduced with retroviruses expressing vector control (Vec) or Mef2c (n = 3 biological replicates). *P < 0.05, ***P < 0.001; two-tailed unpaired Student’s t test. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData FS5.
Baf60c physically interacts with Six4 to suppress Dkk3 expression
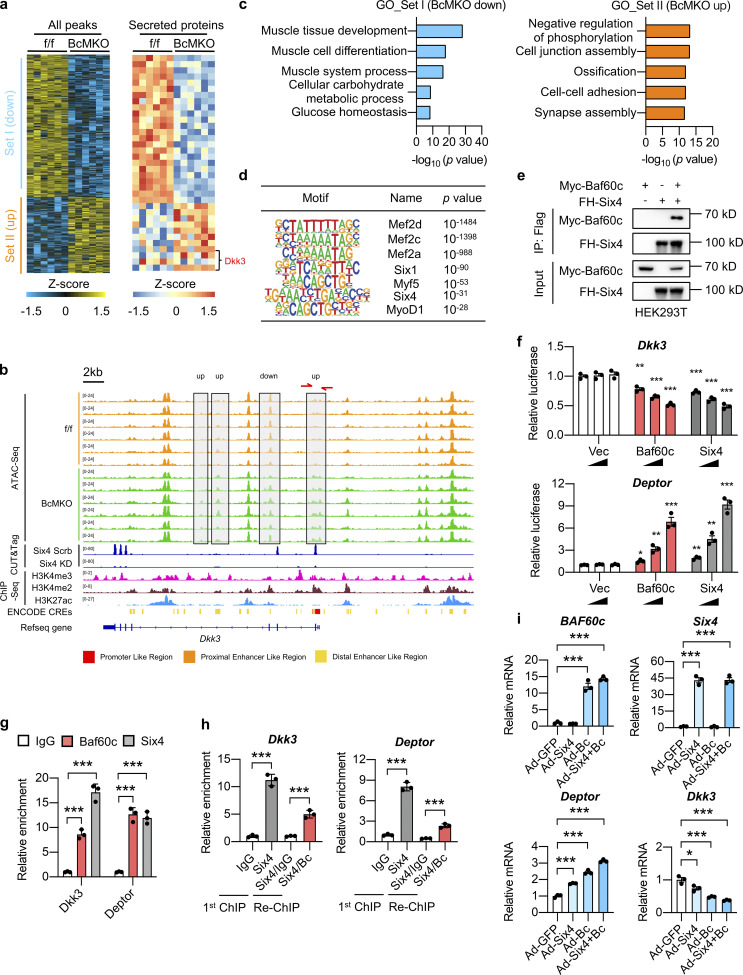
As transcription cofactors, Baf60 subunits have to interact with tissue-specific transcription factors to recruit the SWI/SNF complexes to selective genomic loci to alter the local chromatin accessibility, thereby orchestrating the transcriptional profiles of target genes (Wang et al., 2018). To examine the effects of Baf60c on the genome-wide chromatin landscape in skeletal muscle, we performed the assay for transposase-accessible chromatin using sequencing (ATAC-Seq) on nuclei isolated from Quad muscles of f/f and BcMKO mice. We obtained 11,542 ATAC-Seq peaks that are significantly altered (differential peaks) in BcMKO muscles compared with controls, among which 7,520 peaks were significantly downregulated (Set I) and 4,022 peaks were upregulated (Set II) by Baf60c inactivation in skeletal muscle (Fig. 7 a, left). To investigate whether changes in chromatin openness result in changes in the expression of genes encoding the secreted proteins, we analyzed the chromatin accessibility near the genes encoding the secreted proteins which were regulated by BcMKO as shown in Fig. 2 e. Among the peaks near the top 10 upregulated and top 10 downregulated genes encoding secreted proteins, we found 35 differential peaks, directly annotated to 16 genes encoding secreted proteins (Fig. 7 a, right). Intriguingly, a set of differential peaks were found near the Dkk3 gene, of which three upregulated peaks and one downregulated peak were statistically significant (Fig. 7 a, right; Fig. 7 b). GO analysis of genes annotated to peaks in Set I (downregulated in BcMKO muscles) revealed that these genes are mainly involved in muscle tissue development and differentiation, muscle glucose metabolism, while genes annotated to peaks in Set II (upregulated in BcMKO muscles) were mainly enriched in cell junction assembly and cell–cell adhesion (Fig. 7 c).
Figure 7.
Baf60c physically interacts with Six4 to regulate Dkk3 expression. (a) Heatmap representation of the scaled, normalized read-counts (as Z-score) of all differential peaks (left) or differential peaks near the top 10 upregulated and top 10 downregulated genes encoding secreted-proteins (labeled in Fig. 2 e; right) regulated by BcMKO in Quad muscles as revealed by ATAC-Seq (|log2FC| > 0.5, P < 0.05, n = 6 mice per group). Peaks in Set I and Set II were down-upregulated or upregulated in BcMKO muscles compared to f/f control mice. (b) Representative browser tracks of ATAC-Seq displaying Dkk3 gene locus regulated by BcMKO in Quad muscles as described in a. CUT&Tag-Seq tracks of Six4 in C2C12 myotubes stably transduced with retroviral vectors expressing Scramble shRNA (Six4 Scrb) or Six4 targeting shRNA to knock down Six4 expression (Six4 KD), and ChIP-Seq tracks of H3K4me3, H3K4me2, and H3K27ac, as well as ENCODE annotated CREs are also displayed. (c) GO analysis of genes annotated from differential peaks of Set I and Set II as described in a. Most significant and nonredundant biological processes with −log10 (P value) are shown. (d) Known motif analysis of differential ATAC-Seq peaks in Quad muscles between f/f and BcMKO mice (|log2FC| > 1, P < 0.05). Consensus muscle-related motif (Motif), transcription factor names (Name), and −log10 (P value) are shown. (e) Physical interaction between Baf60c and Six4 in transiently transfected HEK293T cells. IP, immunoprecipitation; Myc-Baf60c, Myc-tagged Baf60c; FH-Six4, Flag HA-tagged Six4. (f) Dual luciferase reporter assay with PGL3 basic constructs containing Dkk3 or Deptor promoters (n = 3 biological replicates). *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA with multiple comparisons. (g) Chromatin immunoprecipitation (ChIP) assay in C2C12 myotubes using antibodies against Baf60c, Six4, or control IgG (n = 3 technical replicates). ***P < 0.001; one-way ANOVA with multiple comparisons. (h) ChIP-ReChIP assay in C2C12 myotubes. The first ChIP assay was performed using antibodies against Six4 or control IgG in chromatin lysates from C2C12 myotubes. The second ChIP (ReChIP) assay was performed using antibodies against Baf60c or control IgG in ChIP samples using the antibody against Six4 from the first ChIP assay (n = 3 technical replicates). ***P < 0.001; one-way ANOVA with multiple comparisons. (i) qPCR analysis of BAF60c, Six4, Deptor, and Dkk3 gene expression in C2C12 myotubes infected with adenoviruses expressing GFP (Ad-GFP), Six4 (Ad-Six4), and BAF60c (Ad-Bc; n = 3 biological replicates). *P < 0.05, ***P < 0.001; one-way ANOVA with multiple comparisons. Data are shown as mean ± SD (g and h) or mean ± SEM (f and i) and are representative of at least two independent experiments. Source data are available for this figure: SourceData F7.
To identify the transcription factors that interact with Baf60c for regulation of Dkk3 transcription, we performed transcription factor binding motif analysis on the differential peaks of ATAC-Seq between f/f and BcMKO muscles and obtained seven muscle-enriched transcription factors including the Mef2 family proteins such as Mef2a, Mef2c, and Mef2d; six family members such as Six1 and Six4, as well as Myf5 and MyoD1 (Fig. 7 d). All these transcription factors except for Mef2a and Myf5 exhibited profound dynamic expression patterns during the process of CTX-induced muscle injury and regeneration (Fig. S5 f). Integrative analysis of our ATAC-Seq data with previously published chromatin immunoprecipitation sequencing (ChIP-Seq) datasets on genome-wide binding profiles of H3K4me3 in C2C12 myotubes, H3K4me2, and H3K27ac in skeletal muscle as well as ENCODE annotated cis-regulatory elements (CREs; Moore et al., 2020) revealed that the Baf60c-dependent differential peaks near Dkk3 gene are colocalized with the active promoter and enhancer regions (Fig. 7 b). ChIP-Seq datasets for all the aforementioned transcription factors, except for Six4 and Mef2c, obtained in skeletal muscles or C2C12 myotubes are available to download from GEO or ENCODE databases. We then performed Cleavage Under Targets and Tagmentation (CUT&Tag) assay on Six4 in nuclei isolated from C2C12 myotubes stably transduced with retroviral vectors expressing Scrb shRNA or shRNA targeting Six4 (siSix4) to identify the global binding sites of Six4 in myocytes. As shown in Fig. S5 g, protein levels of Six4 were markedly decreased in Six4 knockdown (Six4 KD) myotubes stably transduced with siSix4 compared with controls. We next performed further integrative analyses of the ATAC-Seq data with our Six4 CUT&Tag-Seq data in C2C12 myotubes and previously published ChIP-Seq data of the above-mentioned other transcription factors in skeletal muscles or C2C12 myotubes. We observed that one of the BcMKO upregulated peaks (with higher chromatin accessibility) near the promoter region flanking the transcription start site (TSS) of the Dkk3 gene was colocalized with Six4 binding sites (Fig. 7 b) rather than other transcription factors including Mef2a, Mef2d, Six1, Myf5, and MyoD1 (Fig. S5 h). Co-immunoprecipitation (Co-IP) experiment further validated the physical interaction between Baf60c and Six4 (Fig. 7 e). These results suggest that Baf60c may interact with Six4 to regulate Dkk3 gene expression in myocytes.
To test this, luciferase reporter assays were performed to evaluate the effect of Baf60c and Six4 on the transcription activity of the Dkk3 promoter. The proximal Dkk3 promoter (−3,000 to +300 relative to the TSS) was cloned into the pGL3 basic luciferase reporter vector (pGL3-Dkk3). HEK293T cells were transiently transfected with pGL3-Dkk3 and renilla luciferase internal control vectors, as well as increasing concentrations of control, Baf60c, or Six4 expressing vectors, followed by dual luciferase reporter assay. We showed that both Baf60c and Six4 overexpression dose-dependently inhibited Dkk3 reporter gene expression (Fig. 7 f, upper). Consistent with our previous report (Meng et al., 2013), Baf60c and Six4 synergistically upregulated the reporter gene expression of Deptor, encoding a protein involved in the regulation of Akt/mTOR pathway (Fig. 7 f, lower). To further confirm that Baf60c and Six4 can be recruited to the same promoter regions of the Dkk3 gene to regulate its transcription, we performed ChIP-qPCR assays using antibodies targeting endogenous Baf60c and Six4 in C2C12 myocytes and revealed that both Baf60c and Six4 are recruited to the same region on the proximal Dkk3 promoter (Fig. 7 g), corresponding to the aforementioned open region near the TSS in muscle from BcMKO mice compared with f/f controls as revealed by ATAC-Seq (Fig. 7 d). Next, we went on to perform ChIP-ReChIP assay to provide more evidence supporting the physical interaction and direct recruitment of Baf60c and Six4 to the same promoter regions of Dkk3 to regulate its gene transcription. ReChIP-qPCR assay with antibodies against Baf60c or IgG (as control) in the ChIP samples using Six4 antibody revealed a robust co-enrichment of Baf60c and Six4 on the Dkk3 promoter region flanking the TSS of the Dkk3 gene (Fig. 7 h). In addition, to examine whether Baf60c and Six4 could synergistically regulate Dkk3 gene expression in muscle cells, we infected C2C12 myotubes with adenoviruses expressing GFP (as control), Baf60c, and/or Six4. Consistent with our previous report (Meng et al., 2013), Baf60c and Six4 synergistically upregulated the mRNA expression of the Deptor gene (Fig. 7 i). Interestingly, both Six4 and Baf60c individually repressed Dkk3 mRNA expression, and the inhibitory effects became more robust with simultaneous OE of Six4 and Baf60c (Fig. 7 i). Notably, opposite to the effect of Six4 on Dkk3 expression, viral-mediated OE of Six1 or Mef2c significantly increased Dkk3 expression in C2C12 myotubes (Fig. S5, i and j), further excluding the possibility of their involvement in the repression of Dkk3 expression by Baf60c. These results provide evidence suggesting that Baf60c physically interacts with Six4 to synergistically suppress Dkk3 gene transcription likely through alteration of the local chromatin landscape in a cell-autonomous manner.
Elevation of Dkk3 expression in skeletal muscle contributes to the declined muscle regeneration capacity in obese and diabetic mice
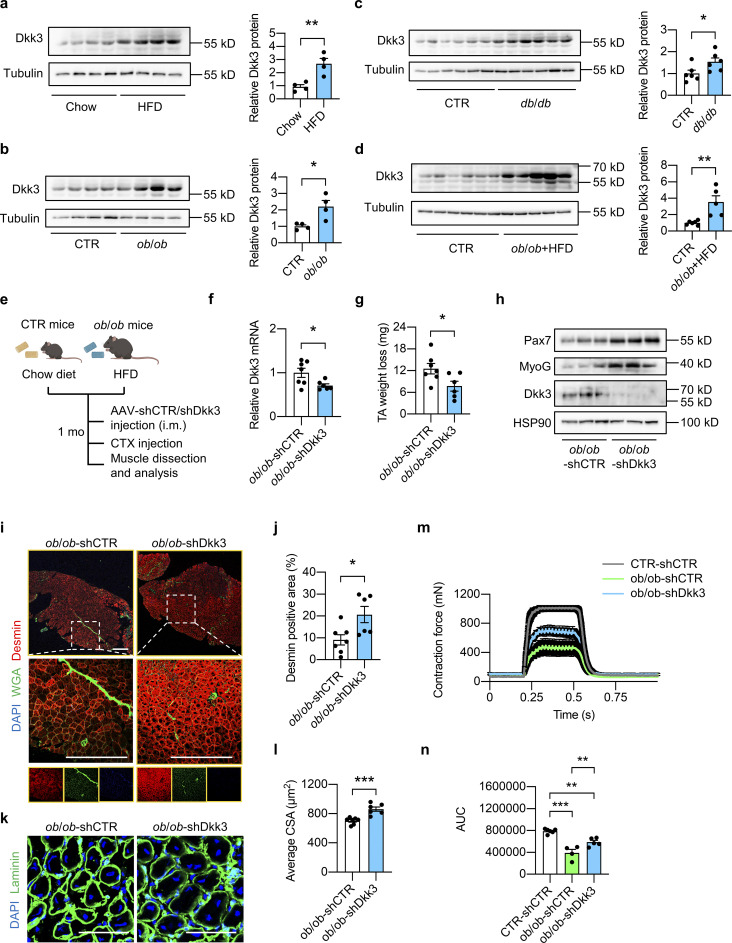
Obesity and T2D are complex metabolic states that are generally accompanied by impaired muscle regeneration capacity and severe muscle loss. However, the underlying molecular mechanism is largely unknown. We found that the Dkk3 protein levels were markedly elevated in skeletal muscle from diet-induced obesity, ob/ob, and db/db mouse models compared with their respective controls (Fig. 8, a–c). It was further upregulated to a greater extent in skeletal muscle from ob/ob mice fed with a high-fat diet (HFD) compared with their littermate controls fed with a chow diet (Fig. 8 d). Given that Dkk3 OE impaired muscle regeneration capacity in vivo and in vitro (Fig. 3), these data suggest that the elevation of Dkk3 levels might be responsible for the decline in muscle regenerative capacity in obesity and T2D. As such, we tested whether Dkk3 knockdown could rescue the muscle regeneration defects in obesity through i.m. injection of AAV-shCTR or AAV-shDkk3 into TA muscles of HFD-fed ob/ob mice (their littermate controls were fed with chow diet and injected with AAV-shCTR as control; Fig. 8 e). Both mRNA and protein levels of Dkk3 in TA muscles were significantly reduced by AAV-shDkk3 treatment compared with AAV-shCTR treatment in ob/ob mice (Fig. 8, f and h). Remarkably, TA muscle weight loss was significantly lower in AAV-shDkk3 injected group compared with the AAV-shCTR group (Fig. 8 g), accomplished by pronounced upregulation of the myogenic regeneration-related marker proteins, Pax7 and MyoG, in ob/ob-shDkk3 muscles compared with ob/ob-shCTR group (Fig. 8 h). In accordance, the newly generated myofiber area, as revealed by Desmin positive staining, was markedly increased by Dkk3 knockdown (approximately twofold increase, Fig. 8, i and j). Furthermore, laminin/DAPI immunofluorescence staining of muscle cross sections at 7 dpi also showed that the newly generated myofiber size was significantly increased by Dkk3 knockdown (Fig. 8, k and l). Moreover, muscle contraction force in ob/ob mice was reduced to almost half of that in control mice and Dkk3 knockdown in ob/ob mice restored contraction ability to ∼75% of that in control mice (Fig. 8, m and n). These results uncover Dkk3 as a key driver in mediating the deterioration of muscle regeneration capacity in obesity and T2D in mice.
Figure 8.
Elevation of Dkk3 expression in skeletal muscle contributes to the declined muscle regeneration capacity in obese and diabetic mice. (a) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total Quad muscle protein lysates from HFD-fed WT mice and their respective chow diet-fed control (n = 4 mice per group). HFD, high-fat diet. **P < 0.01; two-tailed unpaired Student’s t test. (b) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total Quad muscle protein lysates from ob/ob mice and their respective controls (n = 4 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (c) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total Quad muscle protein lysates from db/db mice and their respective controls (n = 6 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (d) Immunoblots (left) and quantification of relative Dkk3 protein levels (right) of total Quad muscle protein lysates from HFD-fed ob/ob mice and their respective controls (n = 5–6 mice per group). **P < 0.01; two-tailed unpaired Student’s t test. (e) Schematic depicting mouse models using AAV i.m. injection to achieve Dkk3 knockdown in TA muscles in chow diet-fed lean control (CTR) mice and HFD-fed ob/ob mice. (f) qPCR analysis of Dkk3 gene expression in AAV-shCTR or AAV-shDkk3 injected TA muscles in ob/ob mice (n = 6–7 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (g) TA weight loss in AAV-shCTR or AAV-shDkk3 injected ob/ob mice at 7 dpi (n = 6–7 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (h) Representative immunoblots of total TA muscle protein lysates from AAV-shCTR or AAV-shDkk3 i.m. injected ob/ob mice at 7 dpi. (i) Representative immunofluorescence images of Desmin (red), WGA (green), and DAPI (blue) in TA muscle cross-sections from AAV-shCTR or AAV-shDkk3 i.m. injected ob/ob mice at 7 dpi. Scale bar: 400 μm. (j) Quantification of the percentages of Desmin positive area in TA muscle cross-sections from AAV-shCTR or AAV-shDkk3 injected ob/ob mice as described in i (n = 6–7 mice per group). *P < 0.05; two-tailed unpaired Student’s t test. (k) Representative immunofluorescence images of laminin (green) and DAPI (blue) of TA muscle cross sections from AAV-shCTR or AAV-shDkk3 injected ob/ob mice at 7 dpi. Scale bar: 100 μm. (l) Average CSA of TA muscles described in k (n = 6–7 mice per group; at least three sections/mouse). ***P < 0.001; two-tailed unpaired Student’s t test. (m) Measurement of muscle tetanic contraction of TA muscles from AAV-shCTR-injected CTR and AAV-shCTR/shDkk3-injected ob/ob mice at 14 dpi (n = 4–6 per group). (n) Comparison of muscle tetanic contraction of TA muscles by calculating the AUC of each mouse from shCTR-injected CTR and shCTR/shDkk3-injected ob/ob group described in m. **P < 0.01, ***P < 0.001; one-way ANOVA with multiple comparisons. All values represent mean ± SEM. All experimental data were verified in at least two independent experiments. Source data are available for this figure: SourceData F8.
Skeletal muscle expression and circulation levels of DKK3 are elevated in obese human subjects
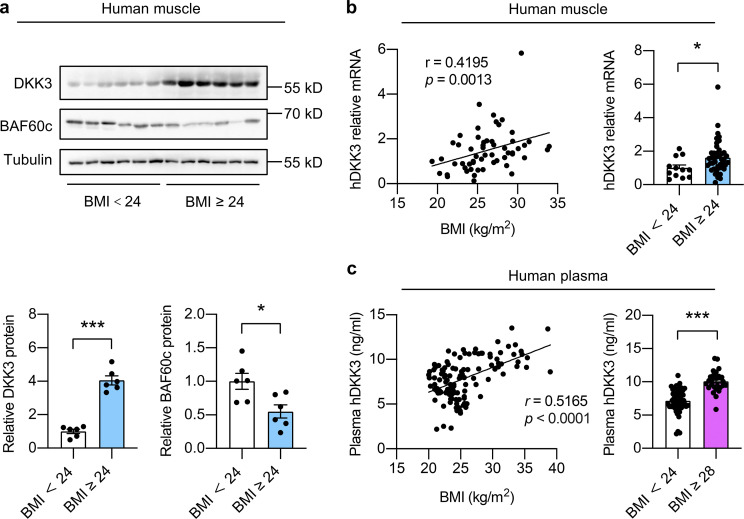
We next explored the significance of the Baf60c-Dkk3 axis in impaired muscle regeneration and maintenance in human subjects with obesity. Similar to the observations in mouse models of obesity, the protein levels of DKK3 and BAF60c were robustly increased and decreased, respectively, in skeletal muscle from overweight (BMI ≥ 24) human subjects compared with normal controls (BMI < 24; Fig. 9 a). In addition, DKK3 mRNA levels in human skeletal muscle were positively correlated with BMI (r = 0.4195, P = 0.0013; Fig. 9 b, left) and were significantly elevated in overweight human subjects (Fig. 9 b, right; Table S1 a). Further measurement of DKK3 protein levels in the circulation by enzyme-linked immunosorbent assay (ELISA) revealed a strong positive correlation between plasma concentrations of DKK3 and BMI in humans (r = 0.5165, P < 0.0001, Fig. 9 c, left). Consistently, the plasma levels of DKK3 were robustly elevated in obese (BMI ≥ 28) human subjects compared with controls (Fig. 9 c, right; Table S1 b). These data indicate that the dysregulation of the BAF60c-DKK3 axis, leading to elevated production and secretion of DKK3 in skeletal muscle, may play an important causal role in the pathogenesis of impaired muscle stem-cell function and reparative capacity in obese humans. The profound elevation of DKK3 levels in the circulation of obese human subjects highlights its translational potential as a potential biomarker for the diagnosis and therapeutic evaluation of obesity and T2D-associated human skeletal muscle diseases.
Figure 9.
Skeletal muscle expression and circulation levels of DKK3 are elevated in obese human subjects. (a) Immunoblots (upper) and quantification of relative DKK3 and BAF60c protein levels (lower) of human skeletal muscle protein lysates. For BMI < 24, n = 6 biological independent samples; for BMI ≥ 24, n = 6 biological independent samples. BMI, body mass index. *P < 0.05, ***P < 0.001; two-tailed unpaired Student’s t test. (b) Correlation between relative DKK3 mRNA levels in human skeletal muscles and BMI as analyzed by Spearman correlation analysis (left). Relative DKK3 mRNA levels in muscles from human subjects (BMI ≥ 24 vs. BMI < 24; right). For BMI < 24, n = 44 biological independent samples; for BMI ≥ 24, n = 12 biological independent samples. *P < 0.05; two-tailed unpaired Student’s t test. (c) Correlation between plasma DKK3 levels and BMI in human subjects as analyzed by Pearson correlation analysis (left). Plasma DKK3 levels in obese (BMI ≥ 28) and control (BMI < 24) human subjects (right). For BMI < 24, n = 51 biological independent samples; for 24 < BMI < 28, n = 38 biological independent samples; for BMI ≥ 28, n = 28 biological independent samples. ***P < 0.001, two-tailed unpaired Student’s t test. Source data are available for this figure: SourceData F9.
Discussion
Skeletal muscle functions not only as a motor for locomotion but also as a master regulator of whole-body metabolism (Meng et al., 2017). As an important endocrine organ, muscle regulates systemic metabolic homeostasis mainly via myokine-mediated interorgan crosstalk (Severinsen and Pedersen, 2020). The concept of “myokine” was first proposed in 2003 (Pedersen et al., 2003), defined as the bioactive molecules produced by muscles and secreted to act on other body parts. In this study, we identified Dkk3 as a downstream target of Baf60c in skeletal myofibers mediating its effects on muscle reparative capacity and contractile function through paracrine signaling to muscle stem cells.
Baf60c is highly expressed in skeletal muscle, heart, and brain, and has been previously demonstrated to play important roles in heart and retina development (Lamba et al., 2008; Lickert et al., 2004; Takeuchi and Bruneau, 2009). Our previous studies have also revealed that Baf60c functions as the core component of the chromatin remodeling pathway in glycolytic myofiber specification and glucose homeostasis (Meng et al., 2017; Meng et al., 2013; Meng et al., 2018; Meng et al., 2014). In this study, we identified Baf60c in skeletal myofibers as a positive regulator of muscle regeneration through Dkk3-mediated paracrine signaling to muscle stem cells. Notably, a previous study has demonstrated that forced expression of Baf60c promotes the epigenetic reprogramming of human embryonic stem cells (hESCs) into skeletal muscle cells in vitro (Albini et al., 2013). In addition, it has been reported that Baf60c plays an important role in myoblast differentiation by facilitating MyoD binding to target genes and enabling the signal-dependent recruitment of the SWI/SNF core complex to muscle genes (Forcales et al., 2012). Consistent with these observations, we also noticed significant impacts of Baf60c overexpression or knockdown on C2C12 myoblast differentiation. These results support the important roles of Baf60c in regulating muscle specification and differentiation in cell culture models. However, the role of Baf60c in MuSCs in adult mice remains unclear. Here, using Pax7-Cre ERT2 mice, we successfully inactivated Baf60c in MuSCs in adult mice and revealed that Baf60c deficiency in MuSCs per se exhibited modest effects on muscle regeneration and function following CTX injection in vivo (Fig. S3). Taken together, these findings suggest that Baf60c may exert distinct regulatory functions on muscle progenitor cell fate commitment and differentiation depending on the local environment and physiological conditions.
In this study, Dkk3 was identified as a downstream muscle-secreted factor that mediates the effects of myofiber Baf60c on muscle stem cell reparative function. We showed that AAV-mediated overexpression of Dkk3 impaired muscle regeneration following CTX injection. Moreover, purified protein Dkk3-Fc inhibited C2C12 myoblast differentiation in a cell-autonomous manner. Through i.m. injection of AAV-GFP into the TA muscle in one leg and AAV-Dkk3 into the TA muscle in the contralateral leg in the same mouse, we provided further evidence suggesting the local inhibitory effect of Dkk3 on muscle stem cell regeneration in mice (Fig. S4). It is of note that, in this study, we also showed that plasma Dkk3 levels are robustly elevated in obese human subjects (Fig. 9), indicating an endocrine function of Dkk3 in vivo. DKK3 was shown to play a critical role in controlling smooth muscle cell fate specification and differentiation (Karamariti et al., 2013; Wang et al., 2015). Recent studies also suggested a role for DKK3 in renal and cardiovascular diseases (Piek et al., 2021). Consistent with our observations, it has been reported that the plasma level of DKK3 is increased in the elderly and is involved in the development of aging-associated muscle sarcopenia (Yin et al., 2018; Zenzmaier et al., 2008). Further, significantly increased DKK3 levels in plasma and cerebrospinal fluid were observed for Alzheimer’s disease (AD) patients compared with healthy subjects, indicating that elevated DKK3 levels might serve as a potential non-invasive AD biomarker in plasma (Zenzmaier et al., 2009). A recent study has also examined the biomarker potential of the circulating DKK3 in evaluating muscle fitness in the elderly with respiratory diseases and revealed that plasma DKK3 level could serve as a useful tool to assess accelerated sarcopenia phenotype in the elderly with respiratory diseases (Qaisar et al., 2020). In addition, elevated plasma concentrations of DKK3 were observed in patients with chronic graft-versus-host disease (GVHD) after allogeneic hematopoietic cell transplantation (Inamoto et al., 2020). It has also been reported that higher plasma DKK3 levels were associated with cardiovascular risk factors and might serve as a predictor for new-onset chronic kidney disease (Piek et al., 2021). Together, these data suggest that DKK3 in circulation may exert endocrine functions on target tissues/cells and play a causal role in the pathogenesis and progression of the aforementioned human diseases. However, unlike other members in the DKK family, DKK3 is unable to bind Wnt coreceptor Lrp and Kremen proteins, and its receptor remains to be identified. Further studies are clearly required to elucidate the cause-and-effect relationships and the underlying mechanisms mediating the endocrine roles of DKK3 under both physiological and disease conditions.
The Akt/mammalian target of rapamycin (mTOR) pathway is a well-known intracellular signaling pathway that plays important roles in the regulation of multiple cellular functions including the growth and maintenance of functional muscle mass (Bodine et al., 2001; Egerman and Glass, 2014; Kim and Guan, 2019; Laplante and Sabatini, 2012). Here, we observed that the activity of Akt/mTOR signaling pathway was significantly elevated in skeletal muscles from MCK-Bc mice and was markedly attenuated in skeletal muscles from BcMKO mice compared with their respective controls. Intriguingly, AAV-mediated overexpression of Dkk3 in TA muscles dramatically diminished the activation of the Akt/mTOR signaling pathway in response to CTX injection as seen in the AAV-GFP injected groups. Moreover, treatment with purified Dkk3-Fc protein markedly decreased the phosphorylation levels of S6K, Akt (Ser473), and Akt (Thr308) in C2C12 myoblasts during the differentiation process. These results suggest that the Akt/mTOR pathway might be involved in the regulation of muscle differentiation and regeneration by the Baf60c-Dkk3 axis. Future studies on the identification of the Dkk3 receptor and its antagonist in muscle cells may help the development of novel treatments for the impairment of muscle regeneration and metabolic fitness in obesity and T2D.
There is a growing body of evidence suggesting that immune cells within the muscle stem cell niche may play an important role in muscle regeneration (Oprescu et al., 2020). Macrophages are considered to be the main effector of immune cells involved in this process (Chazaud, 2020). They participate in cell debris removal and release of cytokines and growth factors which promote satellite cell activation and accelerate the process of muscle regeneration (Bentzinger et al., 2013; Latroche et al., 2017; Perdiguero et al., 2011; Saclier et al., 2013; Tidball, 2017). However, chronic macrophage infiltration inversely correlates with muscle health thereby leading to chronic muscle damage (Webster et al., 2020). In fact, our RNA-Seq analysis of TA muscles at 3 dpi has also revealed elevated immune responses in muscles from BcMKO mice compared with those from controls (Fig. 1 l). Further studies are necessary to elucidate the role of immune cells, macrophages in particular, in the regulation of muscle regeneration by Baf60c in skeletal myofibers. Notably, in our microarray analysis of Baf60c-regulated secreted proteins (Fig. 2 e), besides Dkk3, the most robustly upregulated secreted protein by Baf60c, 17 secreted protein–encoding genes were also identified to be downregulated by BcMKO in skeletal muscles. Among these downregulated genes, Gdf11, Cd24a, and Cpe are top ranked with higher statistical significance and bigger fold changes. Gdf11 (growth differentiation factor 11) was identified by McPherron et al. in 1999, who cloned the human and mouse GDF11 and characterized its function in pattering the axial skeleton (McPherron et al., 1999). Two years prior, the same group also discovered and characterized the GDF8, also known as Myostatin (Mstn; McPherron et al., 1997). Both Gdf11 and Mstn are members of the transforming growth factor β (TGFβ) superfamily. Mstn and GDF11 share the most homology among TGFβ family members, showing ∼90% sequence identity in the mature ligand form McPherron (2010). Mstn is preferentially expressed in cardiac and skeletal muscle and was identified as an important negative regulator of muscle growth in an autocrine/paracrine fashion. Intriguingly, Mstn was recently shown to be expressed in BAT, and its mediated BAT-muscle crosstalk plays a critical role in the regulation of muscle mass and exercise capacity (Kong et al., 2018). However, Gdf11 is expressed in multiple tissues, including skeletal muscle, kidney, spleen, and pancreas (McPherron, 2010). Notably, Gdf11 has gained more attention in the last decade due to numerous reports demonstrating its functions in multiple systems, such as skeletal muscle, cardiac myocyte, and bone homeostasis, with inconsistent or even controversial results (Harper et al., 2016; Loffredo et al., 2013; Walker et al., 2016). In particular, Gdf11 has been shown to have positive, negative, or neutral effects on muscle growth and mass (Egerman et al., 2015; Egerman and Glass, 2019; Honda et al., 2022; Rodgers and Eldridge, 2015; Sinha et al., 2014), indicating that further careful studies with greater scientific rigor and mechanistic approaches are required to elucidate the exact roles of Gdf11 in muscle regeneration and function. Cd24a (also known as Cd24), a small glycosylphosphatidylinositol (GPI)-anchored cell surface glycoprotein, was initially identified as the first natural ligand of Siglec-G/10 (Chen et al., 2009). The Cd24-Siglec-G interaction selectively suppresses the inflammatory response to damage-associated molecular patterns (DAMPs) in tissue injuries (Chen et al., 2011; Chen et al., 2009). Cd24 was also defined as a “don’t eat me” signal in cancer pathogenesis and suggested as a new target for cancer immunotherapy (Barkal et al., 2019). In this case, tumor-expressed Cd24 interacts with macrophage-expressed Siglec-10 to promote tumor immune escape (Barkal et al., 2019). In addition, a recent study identified the Cd24-Siglec axis as an innate immune checkpoint against metaflammation and metabolic disorders (Wang et al., 2022c).Remarkably, in a randomized, double-blind, placebo-controlled, phase III study, CD24FC was observed to exert statistically significant and clinically meaningful therapeutic effects in patients with COVID-19 (Welker et al., 2022). Intriguingly, Cd24 was reported to be regulated by Myogenin in skeletal muscle (Meadows et al., 2008) and identified as a marker for human regenerating muscle (Figarella-Branger et al., 1993). Carboxypeptidase E (Cpe) was discovered in 1982 and has been well characterized for the biosynthesis of a variety of peptide hormones and neuropeptides in the endocrine tissue and nervous system. In the past two decades, using Cpefat/fat mice with reduced Cpe enzyme activity (Naggert et al., 1995) and Cpe KO mice (Cawley et al., 2004), additional enzymatic and nonenzymatic roles of Cpe have been identified in protein internalization, vesicle transport, and signal transduction (Cawley et al., 2012; Ji et al., 2017). Accumulating evidence suggests that Cpe serves critical roles in the regulation of metabolic and glucose homeostasis, obesity, diabetes, bone remodeling, reproduction, and memory (Cawley et al., 2012; Ji et al., 2017). Intriguingly, Cpe KO mice also displayed reduced muscle strength and coordination, indicating a potentially important role of Cpe in regulating muscle growth, regeneration, and fitness (Cawley et al., 2004). Taken together, the known functions of these Baf60c downregulated factors in muscle biology and immune regulation indicate their potentially important roles in muscle regeneration through either directly acting on myocytes and muscle stem cells (Gdf11, Cd24a, and Cpe) or indirectly modulating the immune microenvironment during muscle damage and repair process (CD24a). Their roles in orchestrating muscle microenvironment and muscle reparative capacity warrant future studies.
In summary, this work uncovers an unexpected role of Baf60c in mature skeletal myocytes in modulating muscle stem cell function and regeneration through Dkk3-mediated paracrine signaling. In obesity and T2D, the elevation of Dkk3 in the muscle microenvironment downstream of Baf60c downregulation in skeletal myocytes impairs muscle stem cell differentiation and regeneration in a cell-autonomous manner. Moreover, the strong positive correlation of circulating levels of Dkk3 with BMI in human subjects shed light on the attractive translational potential of using circulating Dkk3 levels as a biomarker for the diagnosis and therapeutic evaluation of metabolic disease–associated muscle regeneration defect and myopathy. These findings provide the molecular basis for developing novel strategies for the clinical prevention and treatment of obesity and T2D-associated decline in skeletal muscle regeneration capacity and muscle mass.
Materials and methods
Animal models
The generation of skeletal muscle-specific Baf60c transgenic mice (MCK-Bc) and Baf60cflox/flox (f/f) mice and skeletal muscle-specific Baf60c KO (BcMKO) mice have been previously described (Meng et al., 2017; Meng et al., 2013). BcMKO were generated by crossing the Baf60cflox/flox mice with the transgenic mice expressing the Cre recombinase driven by the skeletal muscle-specific MLC-Cre (Bothe et al., 2000; Meng et al., 2017). Muscle stem cell–specific Baf60c KO (BcSCKO) mice were generated by crossing the Baf60cflox/flox mice with Pax7-CreERT2 (#017763; Jackson) mice. Cre expression in Pax7-CreERT2 mice was induced through intraperitoneal (i.p.) administration of tamoxifen at the dose of 0.1g kg−1 body weight for 7 consecutive days. Leptin-deficient ob/ob (#000632; Jackson) mice on C57BL6/J background were kindly provided by Dr. Jingya Li (Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China). WT mice on C57BL/6J background were purchased from GemPharmatech Co. Ltd. All mouse strains used in this study were backcrossed with mice on C57BL/6J background for at least six generations. All mouse strains were born at the expected Mendelian ratios and showed normal fertility. Littermates were used as controls for animal studies.
Mice were housed in 12/12 h light/dark cycles and fed with either a normal rodent chow diet or HFD containing 60% of calories from fat (D12492; Research Diets). At the end of the experiments, mice were sacrificed. Tissues were dissected and immediately transferred to liquid nitrogen and stored at −80°C for future RNA and protein analyses. For the histological analysis, skeletal muscles were either embedded in the Tissue-Tek optimal cutting temperature (OCT) compound, frozen with liquid nitrogen-cooled isopentane, and sectioned on a cryostat microtome for future immunofluorescence staining, or fixed in formalin for paraffin embedding and H&E staining.
Human skeletal muscle and plasma analyses
The human muscle samples analyzed were gracilis and semitendinosus muscles collected from patients with ligamentous injury of the knee joint or patella injury at the First People’s Hospital of Lianyungang City during 2018–2021. The human plasma samples were collected from obesity clinic patients at the Second Affiliated Hospital of Zhejiang University during 2020–2021 and health examination at the Second Affiliated Hospital Soochow University during 2021. Patients with secondary or syndromic obesity and lean subjects with previously diagnosed diabetes or impaired glucose regulation were excluded. All the tissue samples were immediately frozen in liquid nitrogen after dissection.
Human blood samples were collected from subjects with different BMI. Donors with tumors, organic lesions, or surgery within several months to half a year were excluded. Plasma samples were harvested after centrifugation and were then stored at −80°C before use. Plasma Dkk3 concentrations were measured using a commercial enzyme-linked immunosorbent assay (ELISA) kit purchased from Shenjiying Co. Ltd.
Plasmid construction
shRNAs targeting mouse Dkk3, Baf60c, and Six4 genes were synthesized and cloned into pX601 plasmid (#61591; Addgene), pSuper-Retro-Puro plasmid (OligoEngine), and pSuper-Retro-Neo plasmid (OligoEngine), respectively. The shRNA targeting sites are listed in Table S3.
Muscle injury and regeneration model
To induce acute muscle injury and regeneration, the mouse was anesthetized with isoflurane followed by intramuscular administration of 35 μl of 20 μM CTX into the TA muscle of one leg, or an equal volume of PBS into the TA muscle of the contralateral leg as control. Muscles were harvested on different dpi, and TA weight loss was calculated by subtracting the weight of CTX-injected TA muscle from the weight of PBS-injected TA muscle of the contralateral leg. For frozen sections, TA muscles were immediately embedded in Tissue-Tek O.C.T. compound in liquid nitrogen-cooled isopentane after dissection and cut into 9-μm transverse sections using a cryostat microtome. For the evaluation of local muscle overexpression of Dkk3 on muscle regeneration, the same mouse was subjected to i.m. injection of AAV-GFP into the TA muscle in one leg and AAV-Dkk3 into the TA muscle in the contralateral leg, followed by CTX injection to induce muscle injury and evaluation of regenerative capacity.
In vivo muscle contraction force measurement
In vivo TA muscle force analysis was performed with 1300 A 3-in-1 whole animal system (Aurora Scientific) as previously described (Cosgrove et al., 2014; Llewellyn et al., 2010; Sacco et al., 2010). Mice were anesthetized by 3-bromo-2-fluoro-phenyl methanol (31.2 g/kg body weight) through intraperitoneal injection. TA muscles should be completely separated from tendon to tendon. A bread silk suture was used to tie the patella ligament of the distal TA muscles. Then the distal TA tendon suture loop was attached to the lever arm hook of the instrument and the contractile force was measured. During the whole process, TA muscles were constantly superfused with prewarmed 37°C PBS solution. The results were analyzed by DMA software (Aurora Scientific). For each treatment, five independent experiments were performed.
C2C12 cell culture, differentiation, and adenoviral transduction
C2C12 myoblasts were obtained from ATCC and cultured in a growth medium (DMEM containing 5% FBS). For the establishment of C2C12 stable cell lines, myoblasts were transduced with control retroviruses (vector or scramble shRNA), retroviruses expressing Baf60c or shRNAs targeting Baf60c (siBaf60c) or Six4 (siSix4), and subjected to puromycin or neomycin selection and differentiation. Myotube differentiation was initiated by switching C2C12 myoblast reaching ∼80–90% confluency to a differentiation medium (DMEM containing 2% FBS). The differentiation medium was replaced every 2 d until terminal differentiation. For adenovirus transduction, cells were infected with adenoviral vectors expressing GFP, Six4, and Baf60c after differentiation for 3 d, and were then subjected to full differentiation. Cells were then harvested for qPCR analysis of gene expression.
Single myofiber isolation and culture
Single myofibers were isolated from the skeletal muscles of C57BL6/J WT adult mice as described (Pasut et al., 2013). In brief, intact muscles were dissected from tendon to tendon and digested with a solution containing 0.2% collagenase type I (Sigma-Aldrich, C0130) in DMEM at 37°C for 2 h. After digestion, individual fibers were separated from each other by pipetting under a dissecting microscope and transferred to horse serum (HS)-coated dishes. Fibers were gently washed in DMEM before being replaced with culture medium (DMEM supplemented with 20% FBS, 10% HS, 1% chicken embryo extract [C3999; US Biological], and 50 μg/ml streptomycin and 50 U/ml penicillin). During the culture, muscle progenitor cells underwent activation and proliferation and reached 80–90% confluency in 7–9 d.
Conditioned medium collection and treatment
For the collection of conditioned medium (CM), C2C12 myoblasts stably transduced with retroviruses expressing vector (Vec), Baf60c, scrambled shRNA (Scrb), or Baf60c-targeting shRNA (siBaf60c) were seeded in culture dishes and allowed to grow until reaching ∼80–90% confluency. Myotube differentiation was initiated by switching the growth medium to the C2C12 differentiation medium (DMEM containing 2% FBS). The differentiation medium was replaced every 2 d for 6 d. Then, the C2C12 differentiation medium was switched to serum-free DMEM plus 0.1% BSA. 2 d later, the CM was harvested, filtered with a 0.2-μm filter unit, aliquoted, and stored at −80°C for future treatment. For the CM treatment, primary muscle stem cells or C2C12 myoblasts were allowed to grow until reaching 80–90% confluency, the culture media were then switched to “CM treatment media” containing the regulator differentiation media (DMEM containing 2% HS for primary muscle stem cells; DMEM containing 2% FBS for C2C12) and CM in a 1:1 (vol./vol.) ratio. The media were replaced every 2 d for a total of 6 d, followed by assays for the evaluation of muscle cell differentiation.
Immunoblotting analysis
Protein lysates from skeletal muscles and whole cell lysates from cultured cells were quantified using BCA protein assay kit (Beyotime), separated by SDS-PAGE gels, and transferred to a PVDF membrane (Millipore), followed by immunoblotting with the following primary antibodies: eMyHC (1:500; F1.652.b), MyHC (1:500; MF 20-c), Tnnt2 (1:1,000; CT3), Pax7 (1:1,000; Pax7-c) from DSHB; HSP90 (1:1,000; 4877s), Desmin (1:1,000; 5332s), GFP (1:5,000; 2956s), Akt (1:1,000; 4691S), phospho-Akt (Ser473; 1:1,000; 4058L), phospho-Akt (Thr308; 1:1,000; 9275L), p70 S6 kinase (1:1,000; 2708S), and phospho-p70 S6 kinase (Thr389; 1:1,000; 9234S) were purchased from CST; MyoG (1:1,000; sc-52903) and Six4 (1:5,000; sc-390779 X) were purchased from Santa Cruz; Dkk3 (1:1,000, 10365-1-AP) was purchased from Proteintech; Myomaker (1:1,000; A18158) was purchased from ABclonal; α-tubulin (1:1,000; T6199) and Baf60a (1:1,000; HPA004101) were from Sigma-Aldrich; Baf60b (1:1,000; ab220164) and Fc (1:1,000; A10648) from Abcam and Invitrogen, respectively. Rabbit polyclonal antibody against Baf60c (1:500) was generated with the recombinant GST fusion mouse Baf60c protein and affinity purified. Secondary HRP-conjugated antibodies were applied as follows: goat anti-mouse (1:5,000; A4416; Sigma-Aldrich) and goat anti-rabbit (1:5,000; A6154; Sigma-Aldrich).
Luciferase reporter assay
The proximal Dkk3 promoter (−3,000 to +300 relative to the TSS) was synthesized and cloned into the pGL3 basic luciferase reporter vector (pGL3-Dkk3). HEK293T cells cultured in 24-well plates were transiently transfected with pGL3-Dkk3 vector (100 ng/well) and renilla luciferase internal control vector (10 ng/well), as well as increasing concentrations of pcDNA3 control vector, pcDNA3-FH (Flag/HA)-Baf60c, or pcDNA3-FH-Six4 expressing vectors (from 20 ng, 50–100 ng/well, and the amount of DNA was supplemented to 100 ng/well with pcDNA3 control vector) using PEI (Cat# 23966-2; Polysciences) followed by dual luciferase reporter assay. All the reporter assays were repeated at least three times in triplicates.
Co-IP
HEK293T cells were transiently transfected with Myc-tagged Baf60c and Flag HA-tagged Six4 for 48 h. Total lysates or immunoprecipitated proteins were analyzed by immunoblotting using antibodies to Flag (1:1,000, M2) from Sigma-Aldrich.
ChIP and ChIP-ReChIP assays
ChIP was performed according to the protocol developed by Upstate Biotechnology as described (Meng et al., 2013). Briefly, chromatin lysates were prepared from C2C12 myotubes following crosslinking with 1% formaldehyde. The samples were precleared with protein A/G agarose beads (Yeasen) and immunoprecipitated using antibodies against BAF60c (generated in our lab), Six4 (sc-390779 X; Santa Cruz), or control IgG in the presence of BSA and salmon sperm DNA. The next day, Protein A/G agarose beads were added to each immunoprecipitation reaction for 1 h, followed by extensive washing and ChIP sample elution from the beads. Following reverse crosslinking, DNA in the ChIP sample was purified using a PCR Purification Kit (Qiagen) and subsequently analyzed by qPCR using primers located on the proximal Dkk3 promoter (Table S2) or Deptor promoter as described (Meng et al., 2013). For the Re-ChIP assay, ChIP samples using Six4 antibody from the first ChIP assays were subjected to a second round of immunoprecipitation using antibodies against BAF60c or control IgG in the presence of BSA and salmon sperm DNA, followed by beads washing, elution, reverse crosslinking, DNA purification, and qPCR analysis as described above.
Immunofluorescence staining
Frozen sections of the muscle were then fixed in 4% paraformaldehyde/PBS for 10 min on ice and permeabilized by 0.3% Triton X-100/PBS for 20 min. Then, they were blocked with 10% BSA/PBS for 1 h at room temperature and incubated overnight at 4°C with anti-Desmin (1:200, 5332s; CST), anti-Dystrophin (1:100, ab15277; Abcam), anti-Pax7 (1:200, Pax7-c; DSHB), anti-MyoD (1:500, sc-32758; Santa Cruz), anti-eMyHC (1:200, F1.652.b; DSHB), and anti-Baf60c (1:100, homemade as described above), followed by incubation with conjugated secondary antibodies with DAPI for nuclear staining on mounted slides. The images were collected on LSM880 Meta laser-scanning confocal microscope (Carl Zeiss). Once set, the detector gain value was kept constant throughout the image acquisition process. Images were analyzed with Zeiss LSM Image Examiner Software.
Myotube fusion index calculation
Myotube fusion index (the percentage of multinucleated myotube) was calculated by dividing the number of myocytes with multinuclei (two or more nuclei) by the total number of myocytes in each immunofluorescence image, as previously described (Park and Chen, 2005).
RNA and RT-qPCR
Total RNA from muscle and cultured cells was extracted using the TRIzol method. For RT-qPCR analysis, an equal amount of RNA was reversed transcribed using HiScript II Q RT SuperMix for qPCR (R222-01; Vazyme), followed by qPCR reactions using SYBR Green (Roche). The relative abundance of mRNA was normalized to ribosomal protein, large, P0 (Rplp0) and was calculated using the 2−ΔΔCt method. The qPCR primers used are listed in Table S2.
RNA-Seq and analysis
For RNA-Seq analysis, total muscle RNA samples were sent for library preparation and sequencing by the BGI group. In brief, mRNAs were enriched from total RNA and fragmentized and used for reverse transcription and second-strand cDNA synthesis. The cDNAs were then tailed with adenine and ligated with adaptors for PCR amplification and sequencing. PCR-amplified cDNA libraries were subjected to paired-end sequencing on BGISEQ-500 system. Data were processed following the standard BGI mRNA analysis pipeline. The expression level of mRNA was computed as FPKM for statistical analysis performed with Deseq2 (v.1.20.0) package (Love et al., 2014). GO and pathway grouping and enrichment studies were performed by clusterProfiler (V3.12.0), and pathway visualization was conducted by pathview (V1.26.0; Luo and Brouwer, 2013; Yu et al., 2012).
ATAC-Seq and analysis
Quadriceps were dissected from six mice for each genotype and snap-frozen individually in liquid nitrogen. Nuclei from the quadriceps were isolated as previously described (Corces et al., 2017). 5 × 104 nuclei were used in each transposase reaction (Ilumina), followed by barcoding and amplification of sequencing libraries. HiSeq 4000 instrument with 150-bp paired-end reads was used for DNA sequencing in quadriceps. The reads were trimmed to 38-bp paired-end reads by fastx_trimmer (http://hannonlab.cshl.edu/fastx_toolkit) for further processing.
The downstream analysis pipeline was adapted from previous studies (Kong et al., 2022; Orchard et al., 2019). Briefly, broad peaks were called by Macs2 (2.1.1.20160309) package (Zhang et al., 2008) using the parameter of (--nomodel --shift −100 --extsize 200 -B --broad) and differential accessed peaks were called using Deseq2 (v.1.20.0) package (Love et al., 2014). Motif enrichment analysis and peak-associated-gene annotation were performed by HOMER (V4.10; Heinz et al., 2010) using peaks filtered by corresponding criteria mentioned previously. Browser tracks were visualized by IGV browser (V2.4.14; Robinson et al., 2011) after normalizing the reads from each individual sample to its own library size. Ataqv (V1.0.0, https://github.com/ParkerLab/ataqv) package developed by Parker’s lab from the University of Michigan was used to perform the ATAC-Seq data quality control analysis.
CUT&Tag and analysis
The library preparation for CUT&Tag was performed as previously reported (Kaya-Okur et al., 2019). In brief, ∼1 × 105 single nuclei from fully differentiated C2C12 myotubes stably transduced with retroviral vectors expressing Scrb shRNA or shRNA targeting Six4 (siSix4) were washed twice in 1 ml PBS. Nuclei were incubated with 10 μl of activated concanavalin A coated magnetic beads (N515-C1; Vazyme) in 500 μl Wash Buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM spermidine, and protease inhibitor cocktail; Roche) for 10 min under room temperature. Cell-bound beads were collected and resuspended with 50 μl Dig-Wash Buffer (20 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM spermidine, protease inhibitor cocktail, and 0.05% Digitonin) containing 2 mM EDTA, 0.1% BSA and a 1:50 dilution of the primary antibody (mouse monoclonal anti-Six4, Santa Cruz, sc-390779 X; or normal mouse IgG), and incubated at 4°C overnight. A secondary antibody (donkey anti-mouse IgG, SAB3701101; Sigma-Aldrich) diluted at 1:100 in 100 μl of Dig-Wash buffer was then administered into the beads, and incubated for 60 min at room temperature following primary antibody removal with a magnet stand (CM101; Vazyme). The preparation of pG-Tn5 adapter complex was performed according to the manufacturer’s instruction with Hyperactive pG-Tn5 Transposase for CUT&Tag (S602; Vazyme). Standard tagmentation and amplification were performed as reported previously (Kaya-Okur et al., 2019). Amplified DNA libraries were purified with VAHTS DNA Clean Beads (N411; Vazyme) and shipped for NGS sequencing by Annoroad Gene Technology. Reads were filtered and mapped to the mm10 genome, and the pipeline before generating the peak matrix was adapted from the workflow established by Ye Zheng on the website: https://yezhengstat.github.io/CUTTag_tutorial/. Peaks were then called to generate a peak matrix with a pipeline similar to the ATAC-Seq analysis mentioned above, as described in our recent studies (Kong et al., 2022; Wang et al., 2022b).
AAV production and transduction
AAV production and purification were performed by ChuangRui Bio. Briefly, AAV293 cells were maintained at 37°C with a humidified incubator containing 5% CO2 in Dulbecco’s modified Eagle medium (11995065; Gibco) and 10% (vol./vol.) FBS (SE100-011; VISTECH). On the day before transfection, AAV293 cells were seeded on 20 × 15 cm culture dishes and grown to appropriate 90% confluency before transfection. For each 15 cm dish, 7 μg of AAV shuttle vector carrying target genes, 20 μg of Delta F6 helper vector, 7 μg of RC2/9 vector, and 250 μl of PEI (Cat# 23966-2; Polysciences) were added to 1 ml of DMEM for 15 min incubation at room temperature before adding to the cells. The culture medium was replaced with DMEM containing 0.5% FBS at 24 h after transfection. Culture medium and cells were collected at 72 h after transfection, and centrifuged at 1,000 rpm for 10 min to pellet the cells. Cells were then resuspended in 20 ml PBS and centrifuged at 1,000 rpm for 10 min. The cell pellet was resuspended in 5 ml cell lysis buffer (150 mM NaCl, 20 mM Tris, pH 8.0).
AAV was purified via a discontinuous iodixanol gradient (D1556-250ML; Sigma-Aldrich). In brief, cells in lysis buffer were frozen and thawed for three times, followed by adding stock solutions containing 1 M MgCl2 and 25 KU/ml Benzonase (E8263-25k; Sigma-Aldrich) to achieve the final concentrations of 1 mM and 250 U/ml, respectively. Cell lysates were incubated at 37°C water bath for 15 min, and centrifuged at 4,000 rpm at 4°C for 30 min. Iodixanol solutions were prepared in four different concentrations, then added into the ultracentrifuge tube (361625; Beckman) in the order of 60, 40, 25, and 17%. Cell lysates were transferred onto the top layer gently, and the remaining volume of the ultracentrifuge tube was filled up with cell lysis buffer. The virus was isolated from the lysate by centrifugation at 53,000 rpm (Beckman OptimaX100 Ultracentrifuge with Type 70Ti rotor) at 14°C for 2 h and 40 min, and the viral titer was determined by qPCR assay with the standard curve generated by serial dilutions of the AAV shuttle vector. C57BL6/J WT mice were transduced with AAV-GFP or AAV-Dkk3 (∼1 × 1013 viral particles (vp)/ml in 20 μl per TA muscle) through intramuscular injection or intraperitoneal injection. BcMKO and their littermate control mice were transduced with AAV-GFP or AAV-Dkk3 shRNA (∼2 × 1013 viral particles (vp)/ml in 60 μl per TA muscle) through intramuscular injection.
Preparation of Fc-tagged Dkk3 protein
Expi293F cells were transfected with pcDNA3.0-Dkk3-Fc using standard methods. 24 h after transfection, 1 M sodium butyrate (1:1,000) was added to suppress cell proliferation, followed by incubation for another 4–5 d. Media were collected and centrifuged at 1,500 rpm (5 min) and 12,000 g (5 min) to pellet residual cells and cell debris respectively, followed by concentration with an Ultrafiltration concentration tube (Millipore) to 100 ml.
Evaluation of Dkk3-Fc on C2C12 cell differentiation
C2C12 myoblasts were seeded into six-well plates. When cells reached 90% confluency, the growth medium was switched to a differentiation medium with or without Dkk3-Fc protein (5 μg/ml). The purified protein was supplemented when the differentiation medium was replaced every 2 d. Myotube differentiation was evaluated by myosin heavy chain, sarcomere (MyHC) immunofluorescence staining using a monoclonal antibody against MyHC from DSHB (MF 20-c). The size and the number of nuclei in each myofiber were quantified using ImageJ software and compared between groups.
Statistics
All statistical analyses were performed using GraphPad Prism8 software. Statistical differences were evaluated using the two-tailed unpaired Student’s t test for two-group comparisons. One-way analysis of variance (ANOVA) and appropriate post-hoc analyses were applied when more than two groups were compared. Two-way ANOVA was applied for multiple comparisons involving two independent variables. For muscle contraction force comparison, the area under the curve was calculated for each mouse and evaluated by the two-tailed unpaired Student’s t test for statistical differences between the two groups. For human sample studies, Spearman correlation analysis was used to investigate the correlation between muscle Dkk3 gene expression and BMI, while Pearson correlation analysis was used to investigate the correlation between plasma DKK3 concentration and BMI. A P value of < 0.05 (*P < 0.05, **P < 0.01, and ***P < 0.001) was considered statistically significant.
Study approval
The studies on human muscles were approved by the First People’s Hospital of Lianyungang City (approval number: LW-20180321001), while the studies on human plasma were approved by the Second Affiliated Hospital Zhejiang University School of Medicine (approval number: A2020001353) and the Second Affiliated Hospital of Soochow University (approval number: JD-LK-2020-038-01), respectively. The donors’ characteristics have been described in Table S1. Written informed consent was provided by each individual before inclusion in the study. All participants received medical history inquiries and blood biochemical analyses after overnight starvation.
All animal studies were performed according to procedures approved by the University Committee on Use and Care of Animals at Zhejiang University and conducted in accordance with the policies of institutional guidelines on the care and use of laboratory animals.
Online supplemental material
Fig. S1 shows Baf60s tissue distribution and Baf60c expression pattern under both physiological and pathological conditions. Fig. S2 provides an additional characterization of non-regenerating muscles and the transcriptomic analysis of regenerating muscles from control and BcMKO mice. Fig. S3 demonstrates that muscle stem cell-specific inactivation of Baf60c exhibits a modest effect on muscle regeneration. Fig. S4 includes the effect of Dkk3 overexpression on skeletal muscle morphology and differentiation under physiological conditions, the effect of local AAV-GFP/AAV-Dkk3 i.m. injection into TA muscles in the contralateral legs of the same mouse on muscle regeneration, additional information for validation and production of Dkk3-Fc fusion protein. The effect of Baf60c transgenic expression (MCK-Bc) on muscle stem cell number and activation in non-regenerating muscles was also included. Fig. S5 shows the effects of the Baf60c-Dkk3 axis on the activity of Akt/mTOR signaling pathway in myocytes, and provides additional information for the validation of potential transcription factors mediating the regulation of Dkk3 expression by Baf60c in skeletal muscle. Table S1 includes the basic information on human subjects for skeletal muscle and plasma samples. Table S2 lists qPCR primers. Table S3 lists shRNA targeting sites.
Supplementary Material
shows information of human subjects.
shows sequences of qPCR primers used in this study.
shows sequences of shRNA targeting sites used in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S4.
is the source file for Fig. S5.
Acknowledgments
The authors thank Dr. Zhe Yu Zhang for the critical reading of the manuscript. We thank Dr. Ping Hu (Chinese Academy of Sciences) and Dr. Yiting Zhou and Tizhong Shan (Zhejiang University, Hangzhou, China) for providing important reagents for this study. We thank the Meng lab members for the helpful discussion and technical support for this study. We also thank Qiong Huang from Core Facilities of Zhejiang University School of Medicine for technical support.
This work was supported by grants from the National Key Research and Development Programme of China (2018YFA0800403, 2021YFC2701903), the Training Program of the Major Research Plan of the National Natural Science Foundation of China (91857110), the National Natural Science Fund for Excellent Young Scholars of China (81722012), the National Natural Science Foundation of China (81670740), Zhejiang Provincial Natural Science Foundation of China (LZ21H070001, LHDMD22H02001), the Innovative Institute of Basic Medical Sciences of Zhejiang University, the Fundamental Research Funds for the Central Universities, and the Construction Fund of Key Medical Disciplines of Hangzhou (No. OO20200055) to Z.X. Meng. This study was supported by grants from the National Natural Science Foundation of China (81870564 and 81670744) and the Science Technology Department of Zhejiang Province of China (2017C33037) to P. Shan. This study was also supported by grants from the National Natural Science Foundation of China (82070838) and the “National Tutor System” Training Program for Youth Talents of Suzhou Health Care System (Qngg2021007) to Y. Feng. The authors gratefully acknowledge the support of K.C. Wong Education Foundation.
Author contributions: Z.-X. Meng initiated and supervised the project. Z.-X. Meng conceived and designed the research. J. Xu, W. Chen, Y. Zhou, C. Lv, Y. Gou, L. Jin, Q. Wu, L. Mi, Z. Yang, T. Yu, and X. Pan performed the experiments. Z. Zhang carried out most of the bioinformatics analysis with help from X. Qiu and T. Liu. X. Li, S. Ma, Y. Feng, and P. Shan contributed to human studies. P. Shan contributed to the discussion and data interpretation. Z.-X. Meng, J. Xu, and Y. Gou wrote the manuscript with help from other authors.
Data availability
RNA-Seq datasets are available for the data presented in Fig. 1, k and l; and Fig. S2, f–i; as well as microarray datasets presented in Fig. 2 e, as well as ATAC-Seq and Six4 CUT&Tag-Seq datasets presented in Fig. 7, a–d. These processed data have been deposited to Mendeley data (https://doi.org/10.17632/28ks32hwxh.1). The bigwig files for H3K4me2 (GSE123879), H3K27ac (GSE123879), Mef2d (GSE43223), Six1 (GSE175999), Myf5 (GSE24852), and Mef2a (GSE61207) ChIP-Seq data were downloaded from GEO database. H3K4me3 (ENCSR000AJR) and MyoD1 (ENCSR000AJS) Chip-Seq data was downloaded from ENCODE database. Microarray datasets for BAF60c in healthy and diabetic human muscles (presented in Fig. S1 e) have been previously described (Jin et al., 2011), with GEO accession number GSE25462. All other data and image files are available from the corresponding author upon reasonable request. All packages and codes used in this study are open-source and publicly available.
References
- Akhmedov, D., and Berdeaux R.. 2013. The effects of obesity on skeletal muscle regeneration. Front. Physiol. 4:371. 10.3389/fphys.2013.00371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albini, S., Coutinho P., Malecova B., Giordani L., Savchenko A., Forcales S.V., and Puri P.L.. 2013. Epigenetic reprogramming of human embryonic stem cells into skeletal muscle cells and generation of contractile myospheres. Cell Rep. 3:661–670. 10.1016/j.celrep.2013.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkal, A.A., Brewer R.E., Markovic M., Kowarsky M., Barkal S.A., Zaro B.W., Krishnan V., Hatakeyama J., Dorigo O., Barkal L.J., and Weissman I.L.. 2019. CD24 signalling through macrophage Siglec-10 is a target for cancer immunotherapy. Nature. 572:392–396. 10.1038/s41586-019-1456-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassel-Duby, R., and Olson E.N.. 2006. Signaling pathways in skeletal muscle remodeling. Annu. Rev. Biochem. 75:19–37. 10.1146/annurev.biochem.75.103004.142622 [DOI] [PubMed] [Google Scholar]
- Bentzinger, C.F., Wang Y.X., Dumont N.A., and Rudnicki M.A.. 2013. Cellular dynamics in the muscle satellite cell niche. EMBO Rep. 14:1062–1072. 10.1038/embor.2013.182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodine, S.C., Stitt T.N., Gonzalez M., Kline W.O., Stover G.L., Bauerlein R., Zlotchenko E., Scrimgeour A., Lawrence J.C., Glass D.J., and Yancopoulos G.D.. 2001. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat. Cell Biol. 3:1014–1019. 10.1038/ncb1101-1014 [DOI] [PubMed] [Google Scholar]
- Bothe, G.W., Haspel J.A., Smith C.L., Wiener H.H., and Burden S.J.. 2000. Selective expression of Cre recombinase in skeletal muscle fibers. Genesis. 26:165–166. [DOI] [PubMed] [Google Scholar]
- Cawley, N.X., Wetsel W.C., Murthy S.R.K., Park J.J., Pacak K., and Loh Y.P.. 2012. New roles of carboxypeptidase E in endocrine and neural function and cancer. Endocr. Rev. 33:216–253. 10.1210/er.2011-1039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cawley, N.X., Zhou J., Hill J.M., Abebe D., Romboz S., Yanik T., Rodriguiz R.M., Wetsel W.C., and Loh Y.P.. 2004. The carboxypeptidase E knockout mouse exhibits endocrinological and behavioral deficits. Endocrinology. 145:5807–5819. 10.1210/en.2004-0847 [DOI] [PubMed] [Google Scholar]
- Chazaud, B. 2020. Inflammation and skeletal muscle regeneration: Leave it to the macrophages!. Trends Immunol. 41:481–492. 10.1016/j.it.2020.04.006 [DOI] [PubMed] [Google Scholar]
- Chen, G.-Y., Chen X., King S., Cavassani K.A., Cheng J., Zheng X., Cao H., Yu H., Qu J., Fang D., et al. 2011. Amelioration of sepsis by inhibiting sialidase-mediated disruption of the CD24-SiglecG interaction. Nat. Biotechnol. 29:428–435. 10.1038/nbt.1846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, G.-Y., Tang J., Zheng P., and Liu Y.. 2009. CD24 and Siglec-10 selectively repress tissue damage-induced immune responses. Science. 323:1722–1725. 10.1126/science.1168988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clapier, C.R., Iwasa J., Cairns B.R., and Peterson C.L.. 2017. Mechanisms of action and regulation of ATP-dependent chromatin-remodelling complexes. Nat. Rev. Mol. Cell Biol. 18:407–422. 10.1038/nrm.2017.26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corces, M.R., Trevino A.E., Hamilton E.G., Greenside P.G., Sinnott-Armstrong N.A., Vesuna S., Satpathy A.T., Rubin A.J., Montine K.S., Wu B., et al. 2017. An improved ATAC-seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods. 14:959–962. 10.1038/nmeth.4396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosgrove, B.D., Gilbert P.M., Porpiglia E., Mourkioti F., Lee S.P., Corbel S.Y., Llewellyn M.E., Delp S.L., and Blau H.M.. 2014. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat. Med. 20:255–264. 10.1038/nm.3464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Souza, D.M., Al-Sajee D., and Hawke T.J.. 2013. Diabetic myopathy: Impact of diabetes mellitus on skeletal muscle progenitor cells. Front. Physiol. 4:379. 10.3389/fphys.2013.00379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeFronzo, R.A., Ferrannini E., Groop L., Henry R.R., Herman W.H., Holst J.J., Hu F.B., Kahn C.R., Raz I., Shulman G.I., et al. 2015. Type 2 diabetes mellitus. Nat. Rev. Dis. Primers. 1:15019. 10.1038/nrdp.2015.19 [DOI] [PubMed] [Google Scholar]
- DeFronzo, R.A., and Tripathy D.. 2009. Skeletal muscle insulin resistance is the primary defect in type 2 diabetes. Diabetes Care. 32:S157–S163. 10.2337/dc09-S302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan, J., and Rando T.A.. 2005. Stem cells in postnatal myogenesis: Molecular mechanisms of satellite cell quiescence, activation and replenishment. Trends Cell Biol. 15:666–673. 10.1016/j.tcb.2005.10.007 [DOI] [PubMed] [Google Scholar]
- Dong, A., and Cheung T.H.. 2021. Deciphering the chromatin organization and dynamics for muscle stem cell function. Curr. Opin. Cell Biol. 73:124–132. 10.1016/j.ceb.2021.08.001 [DOI] [PubMed] [Google Scholar]
- Egerman, M.A., Cadena S.M., Gilbert J.A., Meyer A., Nelson H.N., Swalley S.E., Mallozzi C., Jacobi C., Jennings L.L., Clay I., et al. 2015. GDF11 increases with age and inhibits skeletal muscle regeneration. Cell Metab. 22:164–174. 10.1016/j.cmet.2015.05.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerman, M.A., and Glass D.J.. 2014. Signaling pathways controlling skeletal muscle mass. Crit. Rev. Biochem. Mol. Biol. 49:59–68. 10.3109/10409238.2013.857291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egerman, M.A., and Glass D.J.. 2019. The role of GDF11 in aging and skeletal muscle, cardiac and bone homeostasis. Crit. Rev. Biochem. Mol. Biol. 54:174–183. 10.1080/10409238.2019.1610722 [DOI] [PubMed] [Google Scholar]
- Febbraio, M.A., and Pedersen B.K.. 2020. Who would have thought: Myokines two decades on. Nat. Rev. Endocrinol. 16:619–620. 10.1038/s41574-020-00408-7 [DOI] [PubMed] [Google Scholar]
- Ferrannini, E., Simonson D.C., Katz L.D., Reichard G. Jr, Bevilacqua S., Barrett E.J., Olsson M., and DeFronzo R.A.. 1988. The disposal of an oral glucose load in patients with non-insulin-dependent diabetes. Metabolism. 37:79–85. 10.1016/0026-0495(88)90033-9 [DOI] [PubMed] [Google Scholar]
- Figarella-Branger, D., Moreau H., Pellissier J.F., Bianco N., and Rougon G.. 1993. CD24, a signal-transducing molecule expressed on human B lymphocytes, is a marker for human regenerating muscle. Acta Neuropathol. 86:275–284. 10.1007/BF00304142 [DOI] [PubMed] [Google Scholar]
- Forcales, S.V., Albini S., Giordani L., Malecova B., Cignolo L., Chernov A., Coutinho P., Saccone V., Consalvi S., Williams R., et al. 2012. Signal-dependent incorporation of MyoD-BAF60c into Brg1-based SWI/SNF chromatin-remodelling complex. EMBO J. 31:301–316. 10.1038/emboj.2011.391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, S.C., Brack A., MacDonnell S., Franti M., Olwin B.B., Bailey B.A., Rudnicki M.A., and Houser S.R.. 2016. Is growth differentiation factor 11 a realistic therapeutic for aging-dependent muscle defects? Circ. Res. 118:1143–1150. 10.1161/CIRCRESAHA.116.307962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz, S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H., and Glass C.K.. 2010. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell. 38:576–589. 10.1016/j.molcel.2010.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heredia, J.E., Mukundan L., Chen F.M., Mueller A.A., Deo R.C., Locksley R.M., Rando T.A., and Chawla A.. 2013. Type 2 innate signals stimulate fibro/adipogenic progenitors to facilitate muscle regeneration. Cell. 153:376–388. 10.1016/j.cell.2013.02.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honda, M., Makino T., Zhao X., Matsuto M., Sakurai H., Takahashi Y., Shimizu M., Sato R., and Yamauchi Y.. 2022. Pathophysiological levels of GDF11 activate Smad2/Smad3 signaling and induce muscle atrophy in human iPSC-derived myocytes. Am. J. Physiol. Cell Physiol. 323:C1402–C1409. 10.1152/ajpcell.00341.2022 [DOI] [PubMed] [Google Scholar]
- Inamoto, Y., Martin P.J., Lee S.J., Momin A.A., Tabellini L., Onstad L.E., Pidala J., Flowers M.E.D., Lawler R.L., Katayama H., et al. 2020. Dickkopf-related protein 3 is a novel biomarker for chronic GVHD after allogeneic hematopoietic cell transplantation. Blood Adv. 4:2409–2417. 10.1182/bloodadvances.2020001485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji, L., Wu H.-T., Qin X.-Y., and Lan R.. 2017. Dissecting carboxypeptidase E: Properties, functions and pathophysiological roles in disease. Endocr. Connect. 6:R18–R38. 10.1530/EC-17-0020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, W., Goldfine A.B., Boes T., Henry R.R., Ciaraldi T.P., Kim E.-Y., Emecan M., Fitzpatrick C., Sen A., Shah A., et al. 2011. Increased SRF transcriptional activity in human and mouse skeletal muscle is a signature of insulin resistance. J. Clin. Invest. 121:918–929. 10.1172/JCI41940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karamariti, E., Margariti A., Winkler B., Wang X., Hong X., Baban D., Ragoussis J., Huang Y., Han J.-D.J., Wong M.M., et al. 2013. Smooth muscle cells differentiated from reprogrammed embryonic lung fibroblasts through DKK3 signaling are potent for tissue engineering of vascular grafts. Circ. Res. 112:1433–1443. 10.1161/CIRCRESAHA.111.300415 [DOI] [PubMed] [Google Scholar]
- Kaya-Okur, H.S., Wu S.J., Codomo C.A., Pledger E.S., Bryson T.D., Henikoff J.G., Ahmad K., and Henikoff S.. 2019. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 10:1930. 10.1038/s41467-019-09982-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley, D.E., Goodpaster B.H., and Storlien L.. 2002. Muscle triglyceride and insulin resistance. Annu. Rev. Nutr. 22:325–346. 10.1146/annurev.nutr.22.010402.102912 [DOI] [PubMed] [Google Scholar]
- Kim, J., and Guan K.L.. 2019. mTOR as a central hub of nutrient signalling and cell growth. Nat. Cell Biol. 21:63–71. 10.1038/s41556-018-0205-1 [DOI] [PubMed] [Google Scholar]
- Kong, Q., Zou J., Zhang Z., Pan R., Zhang Z.Y., Han S., Xu Y., Gao Y., and Meng Z.X.. 2022. BAF60a deficiency in macrophage promotes diet-induced obesity and metabolic inflammation. Diabetes. 71:2136–2152. 10.2337/db22-0114 [DOI] [PubMed] [Google Scholar]
- Kong, X., Yao T., Zhou P., Kazak L., Tenen D., Lyubetskaya A., Dawes B.A., Tsai L., Kahn B.B., Spiegelman B.M., et al. 2018. Brown adipose tissue controls skeletal muscle function via the secretion of Myostatin. Cell Metab. 28:631–643.e3. 10.1016/j.cmet.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagouge, M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., et al. 2006. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1α. Cell. 127:1109–1122. 10.1016/j.cell.2006.11.013 [DOI] [PubMed] [Google Scholar]
- Lamba, D.A., Hayes S., Karl M.O., and Reh T.. 2008. Baf60c is a component of the neural progenitor-specific BAF complex in developing retina. Dev. Dyn. 237:3016–3023. 10.1002/dvdy.21697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laplante, M., and Sabatini D.M.. 2012. mTOR signaling in growth control and disease. Cell. 149:274–293. 10.1016/j.cell.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latroche, C., Weiss-Gayet M., Muller L., Gitiaux C., Leblanc P., Liot S., Ben-Larbi S., Abou-Khalil R., Verger N., Bardot P., et al. 2017. Coupling between myogenesis and angiogenesis during skeletal muscle regeneration is stimulated by restorative macrophages. Stem Cell Rep. 9:2018–2033. 10.1016/j.stemcr.2017.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, S., Liu C., Li N., Hao T., Han T., Hill D.E., Vidal M., and Lin J.D.. 2008. Genome-wide coactivation analysis of PGC-1α identifies BAF60a as a regulator of hepatic lipid metabolism. Cell Metab. 8:105–117. 10.1016/j.cmet.2008.06.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert, H., Takeuchi J.K., Von Both I., Walls J.R., McAuliffe F., Adamson S.L., Henkelman R.M., Wrana J.L., Rossant J., and Bruneau B.G.. 2004. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 432:107–112. 10.1038/nature03071 [DOI] [PubMed] [Google Scholar]
- Lillioja, S., Young A.A., Culter C.L., Ivy J.L., Abbott W.G., Zawadzki J.K., Yki-Järvinen H., Christin L., Secomb T.W., and Bogardus C.. 1987. Skeletal muscle capillary density and fiber type are possible determinants of in vivo insulin resistance in man. J. Clin. Invest. 80:415–424. 10.1172/JCI113088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin, J., Wu H., Tarr P.T., Zhang C.-Y., Wu Z., Boss O., Michael L.F., Puigserver P., Isotani E., Olson E.N., et al. 2002. Transcriptional co-activator PGC-1 α drives the formation of slow-twitch muscle fibres. Nature. 418:797–801. 10.1038/nature00904 [DOI] [PubMed] [Google Scholar]
- Liu, T., Mi L., Xiong J., Orchard P., Yu Q., Yu L., Zhao X.-Y., Meng Z.-X., Parker S.C.J., Lin J.D., and Li S.. 2020. BAF60a deficiency uncouples chromatin accessibility and cold sensitivity from white fat browning. Nat. Commun. 11:2379. 10.1038/s41467-020-16148-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llewellyn, M.E., Thompson K.R., Deisseroth K., and Delp S.L.. 2010. Orderly recruitment of motor units under optical control in vivo. Nat. Med. 16:1161–1165. 10.1038/nm.2228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo, F.S., Steinhauser M.L., Jay S.M., Gannon J., Pancoast J.R., Yalamanchi P., Sinha M., Dall’Osso C., Khong D., Shadrach J.L., et al. 2013. Growth differentiation factor 11 is a circulating factor that reverses age-related cardiac hypertrophy. Cell. 153:828–839. 10.1016/j.cell.2013.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love, M.I., Huber W., and Anders S.. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, W., and Brouwer C.. 2013. Pathview: An R/bioconductor package for pathway-based data integration and visualization. Bioinformatics. 29:1830–1831. 10.1093/bioinformatics/btt285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauro, A. 1961. Satellite cell of skeletal muscle fibers. J. Biophys. Biochem. Cytol. 9:493–495. 10.1083/jcb.9.2.493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron, A.C. 2010. Metabolic functions of Myostatin and GDF11. Immunology‚ Endocr. Metab. Agents Med. Chem. 10:217–231. 10.2174/187152210793663810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPherron, A.C., Lawler A.M., and Lee S.-J.. 1997. Regulation of skeletal muscle mass in mice by a new TGF-beta superfamily member. Nature. 387:83–90. 10.1038/387083a0 [DOI] [PubMed] [Google Scholar]
- McPherron, A.C., Lawler A.M., and Lee S.-J.. 1999. Regulation of anterior/posterior patterning of the axial skeleton by growth/differentiation factor 11. Nat. Genet. 22:260–264. 10.1038/10320 [DOI] [PubMed] [Google Scholar]
- Meadows, E., Cho J.-H., Flynn J.M., and Klein W.H.. 2008. Myogenin regulates a distinct genetic program in adult muscle stem cells. Dev. Biol. 322:406–414. 10.1016/j.ydbio.2008.07.024 [DOI] [PubMed] [Google Scholar]
- Meng, Z.-X., Gong J., Chen Z., Sun J., Xiao Y., Wang L., Li Y., Liu J., Xu X.S., and Lin J.D.. 2017. Glucose sensing by skeletal myocytes couples nutrient signaling to systemic homeostasis. Mol. Cell. 66:332–344.e334. 10.1016/j.molcel.2017.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.-X., Li S., Wang L., Ko H.J., Lee Y., Jung D.Y., Okutsu M., Yan Z., Kim J.K., and Lin J.D.. 2013. Baf60c drives glycolytic metabolism in the muscle and improves systemic glucose homeostasis through Deptor-mediated Akt activation. Nat. Med. 19:640–645. 10.1038/nm.3144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.-X., Tao W., Sun J., Wang Q., Mi L., and Lin J.D.. 2018. Uncoupling exercise bioenergetics from systemic metabolic homeostasis by conditional inactivation of Baf60 in skeletal muscle. Diabetes. 67:85–97. 10.2337/db17-0367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.-X., Wang L., Chang L., Sun J., Bao J., Li Y., Chen Y.E., and Lin J.D.. 2015. A diet-sensitive BAF60a-mediated pathway links hepatic bile acid metabolism to cholesterol absorption and atherosclerosis. Cell Rep. 13:1658–1669. 10.1016/j.celrep.2015.10.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng, Z.-X., Wang L., Xiao Y., and Lin J.D.. 2014. The Baf60c/Deptor pathway links skeletal muscle inflammation to glucose homeostasis in obesity. Diabetes. 63:1533–1545. 10.2337/db13-1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore, J.E., Purcaro M.J., Pratt H.E., Epstein C.B., Shoresh N., Adrian J., Kawli T., Davis C.A., Dobin A., Kaul R., et al. 2020. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 583:699–710. 10.1038/s41586-020-2493-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha, V.K., Handschin C., Arlow D., Xie X., St Pierre J., Sihag S., Yang W., Altshuler D., Puigserver P., Patterson N., et al. 2004. Erralpha and Gabpa/b specify PGC-1alpha-dependent oxidative phosphorylation gene expression that is altered in diabetic muscle. Proc. Natl. Acad. Sci. USA. 101:6570–6575. 10.1073/pnas.0401401101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muoio, D.M., and Neufer P.D.. 2012. Lipid-induced mitochondrial stress and insulin action in muscle. Cell Metab. 15:595–605. 10.1016/j.cmet.2012.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naggert, J.K., Fricker L.D., Varlamov O., Nishina P.M., Rouille Y., Steiner D.F., Carroll R.J., Paigen B.J., and Leiter E.H.. 1995. Hyperproinsulinaemia in obese fat/fat mice associated with a carboxypeptidase E mutation which reduces enzyme activity. Nat. Genet. 10:135–142. 10.1038/ng0695-135 [DOI] [PubMed] [Google Scholar]
- Nguyen, M.-H., Cheng M., and Koh T.J.. 2011. Impaired muscle regeneration in ob/ob and db/db mice. ScientificWorldJournal. 11:1525–1535. 10.1100/tsw.2011.137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niehrs, C. 2006. Function and biological roles of the Dickkopf family of Wnt modulators. Oncogene. 25:7469–7481. 10.1038/sj.onc.1210054 [DOI] [PubMed] [Google Scholar]
- Oprescu, S.N., Yue F., Qiu J., Brito L.F., and Kuang S.. 2020. Temporal dynamics and heterogeneity of cell populations during skeletal muscle regeneration. iScience. 23:100993. 10.1016/j.isci.2020.100993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orchard, P., White J.S., Thomas P.E., Mychalowych A., Kiseleva A., Hensley J., Allen B., Parker S.C.J., and Keegan C.E.. 2019. Genome-wide chromatin accessibility and transcriptome profiling show minimal epigenome changes and coordinated transcriptional dysregulation of hedgehog signaling in Danforth’s short tail mice. Hum. Mol. Genet. 28:736–750. 10.1093/hmg/ddy378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, I.-H., and Chen J.. 2005. Mammalian target of rapamycin (mTOR) signaling is required for a late-stage fusion process during skeletal myotube maturation. J. Biol. Chem. 280:32009–32017. 10.1074/jbc.M506120200 [DOI] [PubMed] [Google Scholar]
- Pasut, A., Jones A.E., and Rudnicki M.A.. 2013. Isolation and culture of individual myofibers and their satellite cells from adult skeletal muscle. J. Vis. Exp.:e50074. 10.3791/50074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti, M.E., Butte A.J., Crunkhorn S., Cusi K., Berria R., Kashyap S., Miyazaki Y., Kohane I., Costello M., Saccone R., et al. 2003. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc. Natl. Acad. Sci. USA. 100:8466–8471. 10.1073/pnas.1032913100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedersen, B.K., Steensberg A., Fischer C., Keller C., Keller P., Plomgaard P., Febbraio M., and Saltin B.. 2003. Searching for the exercise factor: Is IL-6 a candidate? J. Muscle Res. Cell Motil. 24:113–119. 10.1023/A:1026070911202 [DOI] [PubMed] [Google Scholar]
- Pedersen, B.K., and Febbraio M.A.. 2012. Muscles, exercise and obesity: Skeletal muscle as a secretory organ. Nat. Rev. Endocrinol. 8:457–465. 10.1038/nrendo.2012.49 [DOI] [PubMed] [Google Scholar]
- Perdiguero, E., Sousa-Victor P., Ruiz-Bonilla V., Jardí M., Caelles C., Serrano A.L., and Muñoz-Cánoves P.. 2011. p38/MKP-1-regulated AKT coordinates macrophage transitions and resolution of inflammation during tissue repair. J. Cell Biol. 195:307–322. 10.1083/jcb.201104053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen, K.F., Befroy D., Dufour S., Dziura J., Ariyan C., Rothman D.L., DiPietro L., Cline G.W., and Shulman G.I.. 2003. Mitochondrial dysfunction in the elderly: Possible role in insulin resistance. Science. 300:1140–1142. 10.1126/science.1082889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piek, A., Smit L., Suthahar N., Bakker S.J.L., de Boer R.A., and Silljé H.H.W.. 2021. The emerging plasma biomarker Dickkopf-3 (DKK3) and its association with renal and cardiovascular disease in the general population. Sci. Rep. 11:8642. 10.1038/s41598-021-88107-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qaisar, R., Karim A., Muhammad T., and Shah I.. 2020. Circulating biomarkers of accelerated sarcopenia in respiratory diseases. Biology (Basel). 9:322. 10.3390/biology9100322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., and Mesirov J.P.. 2011. Integrative genomics viewer. Nat. Biotechnol. 29:24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers, B.D., and Eldridge J.A.. 2015. Reduced circulating GDF11 is unlikely responsible for age-dependent changes in mouse heart, muscle, and brain. Endocrinology. 156:3885–3888. 10.1210/en.2015-1628 [DOI] [PubMed] [Google Scholar]
- Sacco, A., Mourkioti F., Tran R., Choi J., Llewellyn M., Kraft P., Shkreli M., Delp S., Pomerantz J.H., Artandi S.E., and Blau H.M.. 2010. Short telomeres and stem cell exhaustion model Duchenne muscular dystrophy in mdx/mTR mice. Cell. 143:1059–1071. 10.1016/j.cell.2010.11.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saclier, M., Yacoub-Youssef H., Mackey A.L., Arnold L., Ardjoune H., Magnan M., Sailhan F., Chelly J., Pavlath G.K., Mounier R., et al. 2013. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells. 31:384–396. 10.1002/stem.1288 [DOI] [PubMed] [Google Scholar]
- Samuel, V.T., Petersen K.F., and Shulman G.I.. 2010. Lipid-induced insulin resistance: Unravelling the mechanism. Lancet. 375:2267–2277. 10.1016/S0140-6736(10)60408-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwörer, S., Becker F., Feller C., Baig A.H., Köber U., Henze H., Kraus J.M., Xin B., Lechel A., Lipka D.B., et al. 2016. Epigenetic stress responses induce muscle stem-cell ageing by Hoxa9 developmental signals. Nature. 540:428–432. 10.1038/nature20603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severinsen, M.C.K., and Pedersen B.K.. 2020. Muscle–organ crosstalk: The emerging roles of myokines. Endocr. Rev. 41:594–609. 10.1210/endrev/bnaa016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simoneau, J.-A., and Kelley D.E.. 1997. Altered glycolytic and oxidative capacities of skeletal muscle contribute to insulin resistance in NIDDM. J. Appl. Physiol. 83:166–171. 10.1152/jappl.1997.83.1.166 [DOI] [PubMed] [Google Scholar]
- Simoneau, J.A., Colberg S.R., Thaete F.L., and Kelley D.E.. 1995. Skeletal muscle glycolytic and oxidative enzyme capacities are determinants of insulin sensitivity and muscle composition in obese women. FASEB J. 9:273–278. 10.1096/fasebj.9.2.7781930 [DOI] [PubMed] [Google Scholar]
- Sinha, M., Jang Y.C., Oh J., Khong D., Wu E.Y., Manohar R., Miller C., Regalado S.G., Loffredo F.S., Pancoast J.R., et al. 2014. Restoring systemic GDF11 levels reverses age-related dysfunction in mouse skeletal muscle. Science. 344:649–652. 10.1126/science.1251152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, J.K., and Bruneau B.G.. 2009. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature. 459:708–711. 10.1038/nature08039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao, W., Chen S., Shi G., Guo J., Xu Y., and Liu C.. 2011. SWItch/sucrose nonfermentable (SWI/SNF) complex subunit BAF60a integrates hepatic circadian clock and energy metabolism. Hepatology. 54:1410–1420. 10.1002/hep.24514 [DOI] [PubMed] [Google Scholar]
- Thiebaud, D., Jacot E., DeFronzo R.A., Maeder E., Jequier E., and Felber J.-P.. 1982. The effect of graded doses of insulin on total glucose uptake, glucose oxidation, and glucose storage in man. Diabetes. 31:957–963. 10.2337/diacare.31.11.957 [DOI] [PubMed] [Google Scholar]
- Tidball, J.G. 2017. Regulation of muscle growth and regeneration by the immune system. Nat. Rev. Immunol. 17:165–178. 10.1038/nri.2016.150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, R.G., Poggioli T., Katsimpardi L., Buchanan S.M., Oh J., Wattrus S., Heidecker B., Fong Y.W., Rubin L.L., Ganz P., et al. 2016. Biochemistry and biology of GDF11 and Myostatin: Similarities, differences, and questions for future investigation. Circ. Res. 118:1125–1141. 10.1161/CIRCRESAHA.116.308391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R., Chen F., Chen Q., Wan X., Shi M., Chen A.K., Ma Z., Li G., Wang M., Ying Y., et al. 2022a. MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 13:205. 10.1038/s41467-021-27865-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.-R., Pan R., Zhang W., Fu J., Lin J.D., and Meng Z.-X.. 2018. The SWI/SNF chromatin-remodeling factors BAF60a, b, and c in nutrient signaling and metabolic control. Protein Cell. 9:207–215. 10.1007/s13238-017-0442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, R.R., Qiu X., Pan R., Fu H., Zhang Z., Wang Q., Chen H., Wu Q.Q., Pan X., Zhou Y., et al. 2022b. Dietary intervention preserves β cell function in mice through CTCF-mediated transcriptional reprogramming. J. Exp. Med. 219:219. 10.1084/jem.20211779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Karamariti E., Simpson R., Wang W., and Xu Q.. 2015. Dickkopf homolog 3 induces stem cell differentiation into smooth muscle lineage via ATF6 signalling. J. Biol. Chem. 290:19844–19852. 10.1074/jbc.M115.641415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., Liu M., Zhang J., Brown N.K., Zhang P., Zhang Y., Liu H., Du X., Wu W., Devenport M., et al. 2022c. CD24-Siglec axis is an innate immune checkpoint against metaflammation and metabolic disorder. Cell Metab. 34:1088–1103.e6. 10.1016/j.cmet.2022.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster, J.M., Kempen L.J.A.P., Hardy R.S., and Langen R.C.J.. 2020. Inflammation and skeletal muscle wasting during cachexia. Front. Physiol. 11:597675. 10.3389/fphys.2020.597675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welker, J., Pulido J.D., Catanzaro A.T., Malvestutto C.D., Li Z., Cohen J.B., Whitman E.D., Byrne D., Giddings O.K., Lake J.E., et al. 2022. Efficacy and safety of CD24Fc in hospitalised patients with COVID-19: A randomised, double-blind, placebo-controlled, phase 3 study. Lancet Infect. Dis. 22:611–621. 10.1016/S1473-3099(22)00058-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, J.I., Lessard J., and Crabtree G.R.. 2009. Understanding the words of chromatin regulation. Cell. 136:200–206. 10.1016/j.cell.2009.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, H., Price F., and Rudnicki M.A.. 2013. Satellite cells and the muscle stem cell niche. Physiol. Rev. 93:23–67. 10.1152/physrev.00043.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin, J., Yang L., Xie Y., Liu Y., Li S., Yang W., Xu B., Ji H., Ding L., Wang K., et al. 2018. Dkk3 dependent transcriptional regulation controls age related skeletal muscle atrophy. Nat. Commun. 9:1752. 10.1038/s41467-018-04038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, G., Wang L.-G., Han Y., and He Q.-Y.. 2012. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS. 16:284–287. 10.1089/omi.2011.0118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenzmaier, C., Marksteiner J., Kiefer A., Berger P., and Humpel C.. 2009. Dkk-3 is elevated in CSF and plasma of Alzheimer’s disease patients. J. Neurochem. 110:653–661. 10.1111/j.1471-4159.2009.06158.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zenzmaier, C., Sklepos L., and Berger P.. 2008. Increase of Dkk-3 blood plasma levels in the elderly. Exp. Gerontol. 43:867–870. 10.1016/j.exger.2008.05.012 [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Liu T., Meyer C.A., Eeckhoute J., Johnson D.S., Bernstein B.E., Nusbaum C., Myers R.M., Brown M., Li W., and Liu X.S.. 2008. Model-based analysis of ChIP-seq (MACS). Genome Biol. 9:R137. 10.1186/gb-2008-9-9-r137 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows information of human subjects.
shows sequences of qPCR primers used in this study.
shows sequences of shRNA targeting sites used in this study.
is the source file for Fig. 1.
is the source file for Fig. 2.
is the source file for Fig. 3.
is the source file for Fig. 4.
is the source file for Fig. 5.
is the source file for Fig. 6.
is the source file for Fig. 7.
is the source file for Fig. 8.
is the source file for Fig. 9.
is the source file for Fig. S1.
is the source file for Fig. S4.
is the source file for Fig. S5.
Data Availability Statement
RNA-Seq datasets are available for the data presented in Fig. 1, k and l; and Fig. S2, f–i; as well as microarray datasets presented in Fig. 2 e, as well as ATAC-Seq and Six4 CUT&Tag-Seq datasets presented in Fig. 7, a–d. These processed data have been deposited to Mendeley data (https://doi.org/10.17632/28ks32hwxh.1). The bigwig files for H3K4me2 (GSE123879), H3K27ac (GSE123879), Mef2d (GSE43223), Six1 (GSE175999), Myf5 (GSE24852), and Mef2a (GSE61207) ChIP-Seq data were downloaded from GEO database. H3K4me3 (ENCSR000AJR) and MyoD1 (ENCSR000AJS) Chip-Seq data was downloaded from ENCODE database. Microarray datasets for BAF60c in healthy and diabetic human muscles (presented in Fig. S1 e) have been previously described (Jin et al., 2011), with GEO accession number GSE25462. All other data and image files are available from the corresponding author upon reasonable request. All packages and codes used in this study are open-source and publicly available.