Abstract
Formation of the dorsoventral axis in Drosophila melanogaster is mediated through control of the expression of several genes by the morphogen Dorsal. In the ventral part of the embryo Dorsal activates twist and represses zen amongst others. Recently, several proteins have been shown to assist Dorsal in the repression of zen, one of which is DSP1, a HMG box protein that was isolated as a putative co-repressor of Dorsal. In this report we used a DSP1 null mutant to ascertain in vivo the involvement of DSP1 in Dorsal-mediated repression of zen but not in the activation of twist. We show that Dorsal has the ability to interact with DSP1 in vitro as well as with rat HMG1. Using truncated versions of the proteins we located the domains of interaction as being the HMG boxes for DSP1 and HMG1 and the Rel domain for Dorsal. Finally, studies of the zen DNA binding properties of Dorsal and another related Rel protein (Gambif1 from Anopheles gambiae) revealed that their DNA binding affinities were increased in the presence of DSP1 and HMG1.
INTRODUCTION
Establishment of the dorsoventral axis in Drosophila melanogaster embryos depends upon the maternal morphogen Dorsal (1). Dorsal belongs to the Rel family of transcription factors (2) which includes, among others, the c-rel proto-oncogene (3), NF-κB (reviewed in 4), and the insect Dif (5), Relish (6) and Gambif1 (7). This family of transcription factors has been implicated in many different physiological and cellular processes, such as immunity, inflammation, oncogenesis, apoptosis, and embryonic development, as well as human immunodeficiency virus infection (reviewed in 8–10). The members of this family share a conserved 300 amino acid region, the Rel homology domain (RHD), located at the N-terminus (reviewed in 4,11). The RHD domain mediates DNA binding, homodimer and heterodimer formation and contains the nuclear localisation signal. The C-termini of these proteins are more divergent and are involved in transcriptional activation/repression.
During embryonic development, Dorsal can act as a transcriptional activator or a repressor depending on the promoter content. For example, Dorsal activates the twist and snail promoters but represses the zen (zerknüllt) promoter (12). This repression is mediated through a region called the ventral repression element (VRE) which contains three Dorsal binding sites (dl1–dl3) and four additional elements (AT1–AT4 sites) (13–16). Proteins binding to the AT2 site, namely the Cut and Dri proteins, are necessary for activity of the VRE (17). A fourth protein, Groucho, is unable to bind DNA on its own but is a critical component of Dorsal-mediated repression since Groucho-depleted maternal embryos show nearly complete derepression of zen expression (18). Consequently, ventral repression at the VRE requires the formation of a multiprotein complex whereby Groucho is recruited to the template with the help of additional DNA-binding proteins.
In addition to these proteins, a putative co-repressor of Dorsal, called DSP1 (Dorsal Switch Protein) was identified in a yeast screen for cDNAs encoding proteins that convert Dorsal from an activator to a repressor (19). DSP1 is first deposited by the mother in oocytes during oogenesis, and then expressed uniformly in embryos at the beginning of embryogenesis (20). At later stages of embryonic development it becomes restricted to the ventral nerve cord and brain. In adult flies, DSP1 is detected in heads and ovaries.
DSP1 is a member of the high mobility group (HMG) family of non-histone chromosomal DNA-binding proteins (21,22). DSP1, like the HMG1/2 proteins, has two HMG boxes and a C-terminal acidic tail but also contains an additional N-terminal glutamine-rich region. Alignment of the sequences of the mammalian HMG1/2 and DSP1 proteins shows that the two proteins are strongly similar in the HMG region (19). The HMG1/2 proteins bind to DNA through their HMG box with no sequence specificity but recognise structural DNA features like cruciforms and bent double helices. They also have the property to distort the DNA structure quite dramatically by introducing sharp bends upon binding (23–25). The biological function of the HMG 1/2 proteins remains elusive, but their interaction with several transcription factors and the observation that mouse cell lines depleted of HMG1 are impaired in gene activation by the glucocorticoid receptor both suggest a role in transcription (26–30).
As a way to address the role of DSP1 in Dorsal-mediated repression, we have taken advantage of a DSP1 null mutant to study zen expression in the embryo. Our results suggest that DSP1 is involved in zen repression but not in twist activation. This suggestion is strengthened by the fact that DSP1 and Dorsal interact directly with one another. The two proteins act synergistically on DNA and the end effect of these interactions is an increase in Dorsal affinity for its binding site. Our results are discussed in the context of the transcriptional repression mediated by Dorsal at the zen promoter during early embryogenesis.
MATERIALS AND METHODS
Strains
The homozygous strain dsp1 f was obtained by P element mutagenesis (31). The wild-type Oregon-R strain is described in Lindsey and Zimm (32). The strains were maintained at 22°C on conventional medium.
In situ hybridisation
The in situ hybridisation to whole mount embryos procedure was based on the protocol described by Tautz and Pfeifle (33). The probes used were a zen or twist antisense riboprobe labelled with digoxigenin-11-UTP (Boehringer Mannheim Biochemicals). The plasmids containing zen and twist were kindly provided by B. Limbourg-Bouchon and R. Terracol, respectively.
Construction of vectors expressing full-length and truncated DSP1 proteins
Most of the constructions used were produced by PCR using the original DSP1 cDNA construct corresponding to the a isoform of the protein (20). PCR experiments were performed using 1 U Taq DNA polymerase (Promega), 1 ng of template and 100 pmol of primers for 30 cycles (1 min for 94°C, 1 min for 45°C, 1 min for 70°C). The sequences of the 5′ primers were: (1) 5′-ATG GAA CAC TTT CAT CAA-3′ (DSP1); (2) 5′-ATC AAG GAC CCC AAT GCT-3′ (L5); (3) 5′-AAG CCC AGA GGC CGA ATG-3′ (G81 and M2); (4) 5′-AAT GTC ATC AAT TCG-3′ (F33); (5) 5′-ATC AAG GAC CCC AAT GCT-3′ (J11); (6) 5′-ATG GCC AGC AGT TTT CCG AAT-3′ (B22 and D16). The 3′ primers were: (7) 5′-CTT TTA TTT CCC GCT CGT CTT GTA-3′ (J11 and M2); (8) 5′-CTT TTA TCC CTT GGG CGG CAC ATA-3′ (D16, F33 and G81); (9) 5′-CGA AAC CTC TAA AGC GGG-3′ (DSP1, B22 and L5). A TAA stop codon was added to the sequence when necessary (underlined sequence for primers 7 and 8). The target sequence for primer 9 falls 137 bp downstream from the stop codon within the original vector.
PCR products were blunt ended using the Klenow fragment of DNA polymerase I, purified on agarose gel, electroeluted and ethanol precipitated. The DNA fragments obtained were ligated into pMAL-c2 (New England Biolabs) using the XmnI site in-frame with the upstream MBP (maltose binding protein) gene.
The C8 coding sequence was obtained by digestion of pMAL-c2 B22 (see above) by PstI, giving a 960 bp fragment which was inserted in a PstI-digested pMAL-c2 vector.
pMAL-c2 E5 originated from pMAL-c2 D16. This plasmid was digested with PstI to obtain a 390 bp fragment which was again ligated into pMAL-c2 using the PstI site.
For the N4 truncated protein, pMAL-c2 DSP1 was digested by EcoRI, removing 550 bp from the coding sequence. The digested plasmid was end filled with the Klenow fragment of DNA polymerase I and religated.
All constructions were verified by dideoxy sequencing and all vectors were transformed into an Escherichia coli TB1 strain.
Production of HMG1 proteins and the Gambif1 RHD
Plasmids rHMG1, K82Z, pT7-HMG1bB, pT7 HMG1-B′ and pRNHMG1/M1-V176, directing the synthesis of the entire protein, box A, box B, box B′ and protein A+B, respectively, were kindly provided by M.E. Bianchi (34) and J.O. Thomas (35). Recombinant proteins were produced as described (36).
The Gambif1 RHD was produced from 300 ml of culture of an overexpressing strain (pET-15 Gambif1 in BL21 DE3 pLysS, a generous gift from Dr C. Müller). After growth to an absorbance of 0.4 at 600 nm, protein expression was induced by 0.1 mM IPTG overnight at 23°C. After centrifugation, cells were resuspended in 20 ml of buffer A (20 mM HEPES pH 7.5, 1 mM DTT, 1 mM EDTA, 0.5 mM PMSF, 10% glycerol, 1 µg/ml leupeptin and 1 µg/ml pepstatin) with 150 mM NaCl and lysed by sonication. The crude lysate was centrifuged for 15 min at 8000 g and 1.2 ml of 10% (w/v) polyethyleneimine was added to the supernatant. After stirring for 20 min and centrifugation at 7000 g, ammonium sulphate was added to the supernatant to a final concentration of 55%, followed by stirring for 15 min and centrifugation. The protein-containing pellet was resuspended in 20 ml of buffer A, filtered and loaded on a Uno S column (Bio-Rad). The Gambif1 RHD was eluted from the column with a 20 ml NaCl gradient from 0.1 to 0.5 M NaCl in buffer A at 0.5 ml/min. The protein eluted around 0.25 M NaCl.
Cloning and partial purification of Dorsal
The Dorsal coding sequence was amplified by PCR using the PAR Dorsal construct as template (37; a generous gift from Dr J.M. Reichhart) and with the primers 5′-GCAGCTGGTACC-TTTCCGAACCAGAACAATGGAGCCGCTCC-3′ and 5′-AC-AGCCAAGCTTTTACGTGGATATGGACAGGTTCGATA-TCT-3′.
The PCR product was inserted into the pTrcHisB vector (Invitrogen) using the KpnI and HindIII sites and the plasmid used to transform E.coli strain DH5α. The protein obtained from this recombinant pTrcHisB Dorsal vector contains an N-terminal 6-histidine tag.
Cells (2 l) were grown to an absorbance of 0.6 at 600 nm and protein expression induced by 0.1 mM IPTG for 16 h at 25°C. The purification was performed as proposed by the supplier of the vector. The purity of the protein obtained was estimated at ~20% by Coomassie staining of the bands obtained by SDS–PAGE.
Construction of Dorsal Δ404
The truncated Dorsal Δ404 was obtained by deleting amino acids 404–678 from Dorsal. Briefly, a DNA fragment coding for the region from residue 1 to 403 was recovered by digestion of PAR Dorsal with NdeI and SacI enzymes and inserted into pGEM-3Z (Promega) after mutagenesis of the EcoRI site of the vector to create a stop codon.
Production of 35S-labelled Dorsal and Gambif1 RHD proteins
Synthesis of radiolabelled Dorsal, Dorsal Δ404 and Gambif1 RHD proteins was performed using PAR Dorsal, pGEM Dorsal Δ404 and pET-15 Gambif1 as templates and the TNT coupled reticulocyte lysate system (Promega) in the presence of [35S]methionine. The plasmids were linearised by digestion with XmnI for PAR Dorsal and pGEM Dorsal Δ404 and HindIII for pET-15 Gambif1.
Affinity chromatography
Cells were grown at 37°C in 10 ml of LB medium with ampicillin to an absorbance of 0.5 at 600 nm. Expression of the proteins was induced by 0.3 mM IPTG for 2 h at 37°C. Cells were centrifuged and the bacterial pellet resuspended in 1.5 ml of amylose resin column buffer (20 mM Tris–HCl pH 7.4, 200 mM NaCl, 1 mM EDTA). After sonication for 1 min on ice, the clear lysate was obtained by centrifugation at 8000 g for 20 min at 4°C. An aliquot of 100 µl of the clear lysate was incubated with 50 µl of amylose resin previously equilibrated in column buffer, for 15 min at 4°C with gentle stirring. After centrifugation at 800 g at 4°C, the supernatant was eliminated and the beads washed with 1 ml of column buffer. Another centrifugation and elimination of the supernatant was followed by addition of 250 µl of buffer B (20 mM HEPES pH 7.9, 100 mM NaCl for Dorsal, 50 mM NaCl for Gambif1, 1 mM EDTA, 0.1% NP40, 1 mM DTT, 0.2 mM PMSF) with 1% BSA (w/v). After incubation for 1 h at 4°C, 5 µl of in vitro radiolabelled Dorsal or Gambif1 RHD was added and a further incubation of 1 h was performed. The beads were washed three times with 1 ml of buffer B and, after the last centrifugation, the pellet was resuspended in 50 µl of SDS–PAGE loading buffer. An aliquot of 10 µl of the mixture was used to load an 8% SDS–PAGE gel for Dorsal or a 12% SDS–PAGE gel for the Gambif1 RHD (29:1 acrylamide:bisacrylamide). After migration the gel was dried and revealed by phosphorimager (Molecular Dynamics).
Far-western experiments
After induction (see above), 1 ml of cells were centrifuged and the bacterial pellet resuspended in 200 µl of SDS–PAGE loading buffer. Samples (either lysates or purified proteins) were loaded on an 8% SDS–PAGE (29:1 acrylamide:bisacrylamide) gel. After migration, the proteins were transferred to a nitrocellulose membrane (Hybond C Extra; Amersham) using a semi-dry blotter at 0.6 mA/cm2 for 90 min in 48 mM Tris, 39 mM glycine, 0.04% SDS and 20% methanol. The membrane was incubated in buffer B with 5% skimmed milk for 4 h at 4°C, then overnight in buffer B with 1% skimmed milk and radiolabelled Dorsal. Finally, the membrane was washed with buffer B for 10 min, dried and the bands revealed by phosphorimager.
EMSA
Four 43mer oligonucleotides corresponding to zen strong site dl2 and zen weak site dl3 were purified on a 12% polyacrylamide denaturing gel (19:1 acrylamide:bisacrylamide) and electroeluted. The corresponding top strands were end-labelled using [γ-32P]ATP and T4 kinase and hybridised with their complementary strands. The double-stranded species were purified on a 6% polyacrylamide gel followed by electroelution. Their sequences are as follows (the underlined sequences correspond to the Dorsal binding site):
dl2 5′- ATCCTCTATAACTGGGAGAAACCCAATCAAT-ATTCGTTCACCC-3′
3′-TAGGAGATATTGACCCTCTTTGGGTTAGTTAT-AAGCAAGTGGG-5′
dl3 5′-ATCTGGATGTCCTGGGAAAACCAAGCTCTGA-ATCCATTCTCCC-3′
3′-TAGACCTACAGGACCCTTTTGGTTCGAGACTTAGGTAAGAGGG-5′.
DNA (2 nM) was incubated with increasing quantities of the Gambif1 RHD for 15 min on ice in a total reaction volume of 10 µl containing 50 mM Tris–HCl pH 8, 150 mM NaCl, 1 mM DTT, 1 mM EDTA, 100 µg/ml BSA (binding buffer). To this was added 2 µl of 15% Ficoll with dyes and the mixture was loaded on a 6% polyacrylamide gel containing 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA). Electrophoresis was performed at 4°C for 1 h at 12.5 V/cm using 0.5× TBE buffer. After migration, the gel was dried and exposed on a phosphorimager screen. Bands were quantified and the binding constants were calculated by fitting four sets of data to the following equation:
f = {(1 + KaPT + KaDT) – √[(1 + KaPT + KaDT)2 – (4DTKa2PT)]}/2DTKa
where f is the fraction of bound DNA, Ka the association constant, PT the total concentration of protein and DT the total concentration of DNA (38).
When the effect of DSP1 and HMG1 was analysed, proteins were incubated together for 15 min on ice in binding buffer prior to incubation with DNA. The percentage of bound DNA and the error were calculated from a set of four independent experiments.
RESULTS
zen ventral repression is partially suppressed in DSP1 mutant embryos
Recently, we obtained a homozygous strain containing a null allele of DSP1 with a complete deletion of the DSP1 open reading frame (31). This strain is poorly fertile and only 50% of the eggs lead to viable larvae. The embryos that do not give larvae have stopped their development before the cellular blastoderm stage. This strain offers us the opportunity to test in vivo the possibility that DSP1 participates in zen transcriptional repression.
zen expression was monitored by in situ hybridisation to whole mount embryos using a zen antisense RNA as probe. The wild-type strain expressed zen exclusively in the dorsal part of the blastoderm embryo, and the early zen-expressing area covers 40% of the dorsal embryo circumference (Fig. 1A), in agreement with Roth et al. (1). In the DSP1 mutant strain, zen expression expanded moderately towards its ventral side, covering ~60% of the embryo circumference (Fig. 1B). zen expansion was no longer observed in older DSP1 mutant embryos (data not shown). At these stages, zen expression does not depend upon the VRE-dependent Dorsal-mediated repression mode, which implies that DSP1 participates in early zen ventral repression, as does the maternal morphogen Dorsal (14,15). On the other hand, the expression pattern of a Dorsal-dependent activated promoter, the twist promoter, was not affected in DSP1 mutant embryos (Fig. 1C and D).
Figure 1.
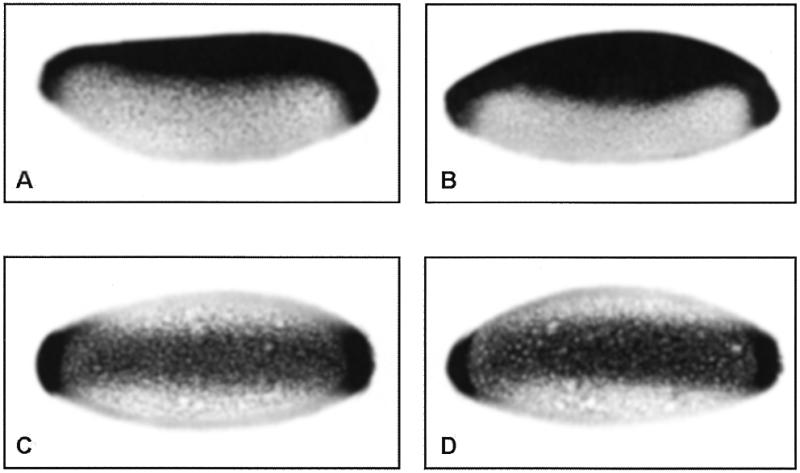
Expression pattern of the zen (A and B) and twist (C and D) genes in blastoderm embryos. Expression is visualised by whole mount in situ hybridisation using a digoxigenin-11-UTP-labelled antisense RNA probe followed by immunological staining. Embryos are oriented with anterior to the left and dorsal up (A and B) or in ventral view (C and D). (A) and (C), wild-type embryos; (B) and (D), DSP1 mutant embryos.
Altogether, these data strongly suggest that DSP1 plays a role, either direct or indirect, in zen transcriptional repression but not in activated twist transcription. The role of DSP1 in this developmental process is probably not predominant since derepression of zen is partial and is observed in only 2% of the DSP1 mutant embryos which have reached the cellular blastoderm stage. Consistent with this interpretation, no obvious defects in formation of the dorso-ventral axis were reported after examination of the cuticle of DSP1 mutant embryos (31).
DSP1 interacts with Dorsal in vitro
To visualise DSP1–Dorsal interactions, we used affinity chromatography whereby DSP1 was immobilised on amylose resin through an N-terminal fusion with MBP and radiolabelled Dorsal was applied to the MBP–DSP1–amylose resin. If Dorsal is able to interact with MBP–DSP1, it will be retained on the resin with its target, thus SDS–PAGE analysis of the material bound to the resin will show a radioactive band of the appropriate size. As shown in Figure 2B, an interaction does take place between radiolabelled Dorsal and full-length DSP1 or truncated DSP1 (B22) (lanes 2 and 3, respectively). Dorsal interacts with DSP1 with considerable selectivity, as seen by the lack of a signal when MBP alone was immobilised on the resin (lane 1).
Figure 2.
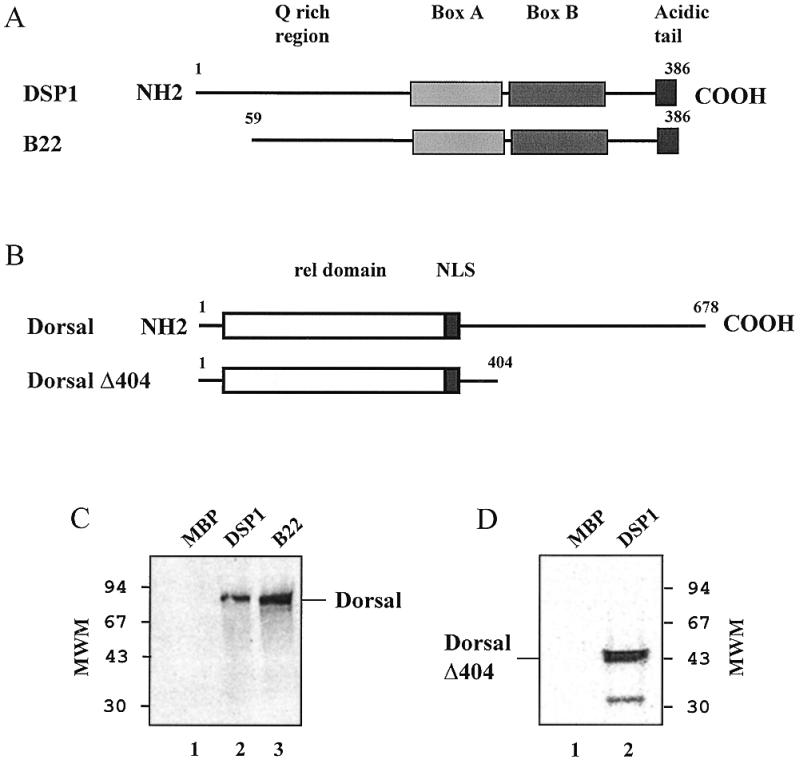
Interaction between DSP1 and full-length Dorsal or Dorsal Δ404 monitored by affinity chromatography. (A) Schematic representation of the proteins DSP1 and B22. The MBP moiety located at the N-terminus is not represented. (B) Schematic representation of the proteins Dorsal and Dorsal Δ404. NLS, nuclear localisation signal. (C) Phosphorimager scan of the gel showing the interaction between Dorsal and MBP–DSP1 (lane 2) or B22 (lane 3); no interaction is seen with unfused MBP (lane 1). (D) Phosphorimager scan of the gel showing the interaction between Dorsal Δ404 and MBP–DSP1 (lane 2, the faster migrating band could be due to abortive synthesis or degradation of Dorsal Δ404); the MBP control is as above (lane 1).
A similar experiment was performed with a truncated version of Dorsal (Dorsal Δ404) which was obtained by deletion of 275 residues in the C-terminus of Dorsal. Dorsal Δ404 contains the Rel domain, the nuclear localisation signal plus 63 residues of the Dorsal C-terminal part. This truncated protein was retained on the MBP–DSP1 resin (Fig. 2C), suggesting that the N-terminal part of Dorsal participates in the interaction.
Each HMG box of DSP1 participates in the interaction
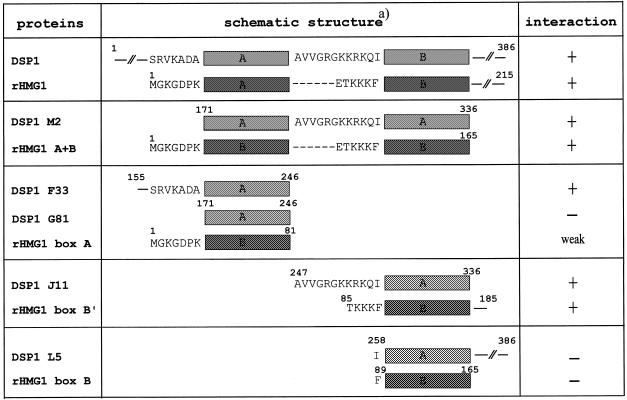
In order to investigate the domain(s) of interaction in DSP1 protein, we used several truncated forms of MBP–DSP1 (Fig. 3A) and analysed their binding to Dorsal by far-western blotting (Fig. 3C). As a control for the amounts of protein, a Coomassie stained SDS–PAGE gel of the lysates is shown in Figure 3B. Several points can be drawn from these data. The N-terminal part of DSP1, containing several runs of glutamines, seems to be dispensable for the interaction. Indeed, deleting an increasing number of N-terminal residues did not alter Dorsal binding (proteins B22, C8, M2, D16, E5 and F33 in Fig. 3C, lanes 1, 2, 9 and 3–5, respectively). The C-terminal part of DSP1 which contains the acidic tail does not appear to be crucial for recognition. No obvious difference can be seen between the truncated proteins C8 (lane 2), E5 (lane 4) and M2 (lane 9). The inability of the N4 truncated protein, where only one-third of box A is present, to interact with Dorsal shows the importance of the box in this interaction (lane 10). However, the HMG box is not sufficient for full recognition. The G81 protein for box A and the L5 protein for box B do not interact with Dorsal (lanes 6 and 8, respectively). Only when flanking sequences are present at the N-terminus of each box can the interaction be restored. Indeed, F33 (lane 5) and J11 (lane 7) are able to bind Dorsal (the weaker signal obtained with J11 correlates with a low amount of protein loaded on the gel, Fig. 3B, lane 7).
Figure 3.
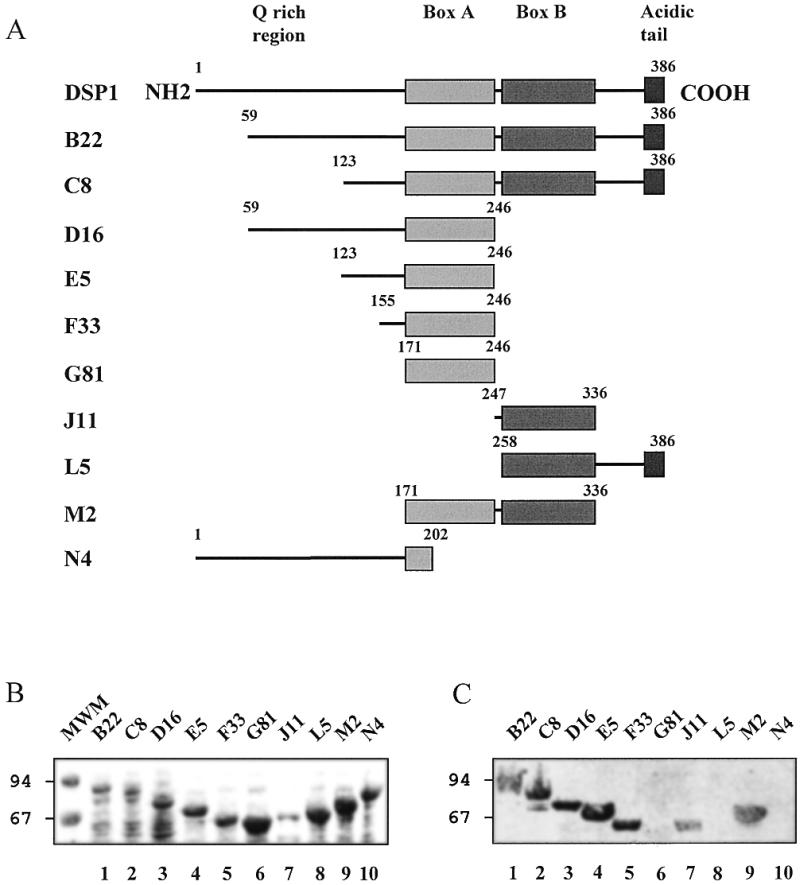
Interaction between Dorsal and truncated MBP–DSP1 proteins monitored by far-western blotting. (A) Schematic representation of the truncated proteins (the MBP moiety located at the N-terminus is omitted). (B) Coomassie stained SDS–PAGE of the lysates. (C) Phosphorimager scan of the membrane after transfer of the proteins and incubation with radiolabelled Dorsal.
The HMG boxes of HMG1 interact with Dorsal in vitro
The results presented above point to the HMG boxes of DSP1 as the main providers of interacting residues. Sequence comparison between DSP1 and HMG1 in this region shows numerous similarities as well as some differences. The two HMG A boxes share a high degree of identity (69.3% identity) while the B boxes are more divergent (46.2% identity), and DSP1 contains six additional residues between the two boxes (19). In order to analyse, at least qualitatively, the effect of these differences on the interaction, we performed far-western assays using several truncated forms of HMG1 as targets (Fig. 4). When compared to the results presented above, a remarkable similitude can be seen between HMG1 and DSP1. HMG1 is able to bind Dorsal (Fig. 4C, lane 1; the faster migrating band corresponds to degradation products present in the HMG1 sample, Fig. 4B, lane 1). Again, the C-terminal part of the protein (from the end of box B to the C-terminus) does not seem to participate in the interaction (A+B protein interacts with Dorsal; lane 2). Box A (residues 1–81) interacts very weakly with Dorsal (lane 3) and box B (residues 89–165) does not show any binding (lane 4). Only when flanking sequences are present can the interaction take place (box B′, residues 85–185; lane 5). Again, the critical residues are located at the N-terminus. A+B protein, the equivalent of truncated DSP1 M2 protein which lacks the C-terminal residues, is still able to interact with Dorsal (Fig. 4C, lane 2). Table 1 gives a summary of the results obtained on DSP1 and HMG1 and clearly shows the importance of the N-terminal flanking sequences of the boxes in the interaction.
Figure 4.
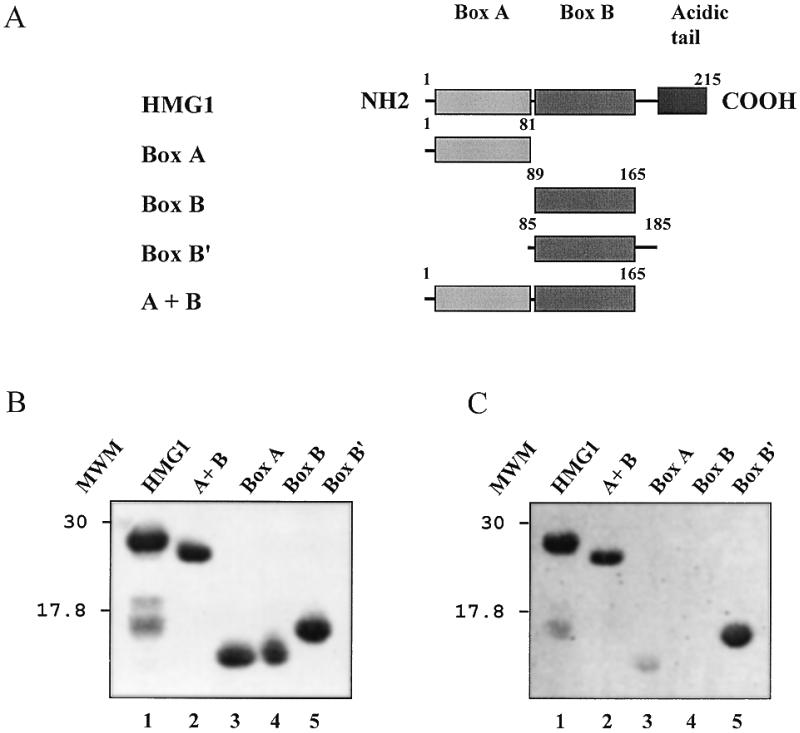
Interaction between Dorsal and full-length or truncated HMG1 proteins monitored by far-western blotting. (A) Schematic representation of the proteins. (B) Coomassie stained SDS–PAGE of the proteins. (C) Phosphorimager scan of the membrane after transfer of the proteins and incubation with radiolabelled Dorsal.
Table 1. Summary of the interactions between Dorsal and truncated versions of DSP1 and HMG1.
aOnly the flanking sequences of the boxes are shown. Dashes correspond to residues missing in HMG1.
DSP1 stimulates Dorsal binding on DNA
The results presented above show that DSP1 is involved in Dorsal-mediated repression of the zen gene, that the two proteins are able to interact with each other and that this interaction is a property shared with HMG1. One of the key questions concerns the effects of such a cooperation on Dorsal DNA binding properties. We have analysed the effect of DSP1 on Dorsal DNA binding by EMSA. Figure 5A shows that DSP1 causes a significant increase in the amount of DNA bound by Dorsal, reaching 1.5 times for 50 nM and 2.4 times for 150 nM DSP1. The presence of DSP1 slows down migration of the Dorsal–DNA complex, suggesting that a ternary complex is formed.
Figure 5.
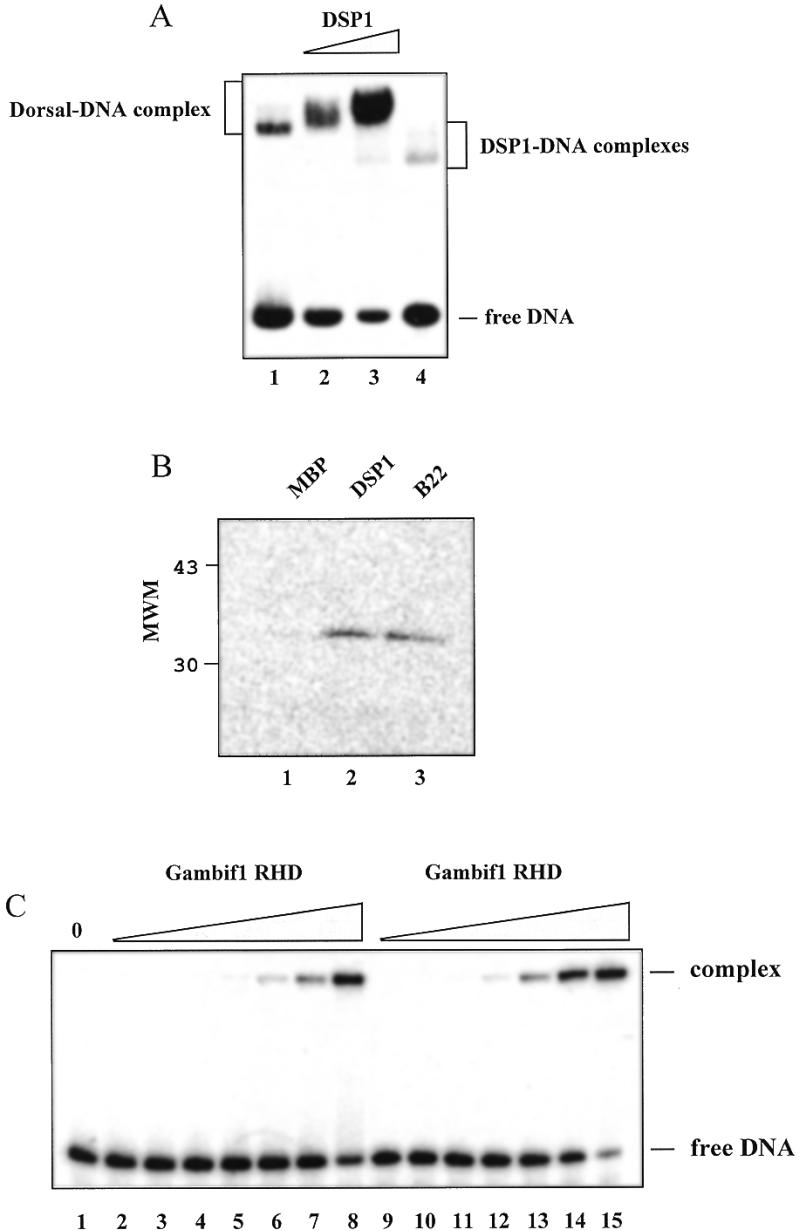
Effect of DSP1 on Dorsal DNA binding and characterisation of the Gambif1 RHD. (A) Phosphorimager scan of an EMSA experiment whereby partially purified Dorsal was incubated with 50 (lane 2) or 150 nM (lane 3) DSP1 prior to incubation with 2 nM Dorsal strong site dl2. In that instance DSP1 was used as an unfused protein. In lane 1 no DSP1 was added to Dorsal and lane 4 reflects the binding on DNA of 150 nM DSP1 alone. (B) Affinity chromatography experiment showing that MBP–DSP1 (lane 2), MBP–truncated DSP1 (B22, lane 3), but not MBP alone (lane 1), interact with the Gambif1 RHD. (C) EMSA on the Gambif1 RHD and 2 nM Dorsal weak site dl3 (lanes 1–8) or Dorsal strong site dl2 (lanes 9–15). The Gambif1 RHD dimer concentrations are identical for both DNA sites and are as follows: lane 1, no protein; lanes 2 and 9, 0.05 nM; lanes 3 and 10, 0.15 nM; lanes 4 and 11, 0.5 nM; lanes 5 and 12, 1.5 nM; lanes 6 and 13, 5 nM; lanes 7 and 14, 15 nM; lanes 8 and 15, 50 nM.
The Gambif1 RHD as a model for the Dorsal RHD
Due to the low purity (~20%) of the full-length Dorsal, we used another protein from the Rel family, Gambif1 from Anopheles gambiae (A.gambiae immune factor 1), to verify the formation of a ternary complex. Gambif1 recognises NF-κB like sites such as those found in the A.gambiae defensin and D.melanogaster diptericin and cecropin genes (7). Its RHD bears 69% identity with the Dorsal RHD and it has been suggested that these two proteins are orthologues (7). The rest of the protein being more divergent, we decided to limit ourselves to the Gambif1 RHD, namely the region from residue 47 to 330. Beforehand, we verified if the Gambif1 RHD was a satisfactory model for Dorsal in its interaction with DSP1 and its binding to DNA. Figure 5B shows that the Gambif1 RHD can interact with MBP–DSP1 and MBP–truncated DSP1 (B22) but not with MBP. Using EMSA, we analysed binding of the Gambif1 RHD to two Dorsal zen sites (dl2 and dl3). The Gambif1 RHD is able to recognise both sites (Fig. 5C) with an apparent dissociation constant of 28 ± 4 nM for the stronger dl2 site and 80 ± 3 nM for the weaker dl3 site (the protein is in the form of a dimer on DNA; 39). Therefore, the Gambif1 RHD is able, like Dorsal, to differentiate between the two kinds of sites. Taken together these results suggest that the Gambif1 RHD behaves like the Dorsal RHD and constitutes a good model for Dorsal.
DSP1 and HMG1 increase Gambif1 RHD binding to a Dorsal zen site
EMSA experiments were performed with the Gambif1 RHD and Dorsal dl2 binding site in the presence of increasing amounts of DSP1 (Fig. 6A) or HMG1 (Fig. 6B). As obtained for full-length Dorsal, Gambif1 DNA binding capability is increased by the presence of DSP1. A slower migrating band appears, again suggesting the formation of a ternary complex. HMG1 causes a stimulation of Dorsal binding that seems to be even stronger than that by DSP1, and again slower migrating bands appear.
Figure 6.
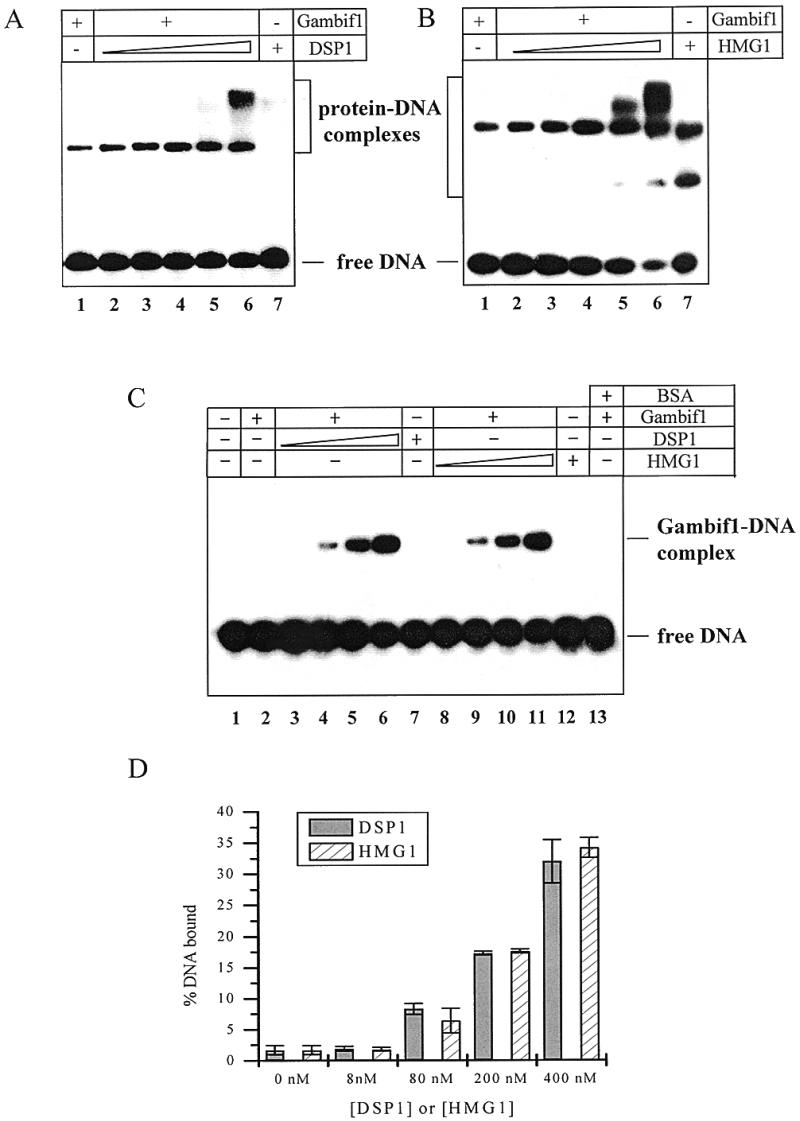
Effect of DSP1 and HMG1 on Gambif1 RHD DNA binding with and without poly(dI-dC)·(dI-dC). (A) EMSA with 5 nM Gambif1 RHD and increasing quantities of DSP1 in the absence of poly(dI-dC)·(dI-dC). The DNA is Dorsal strong site dl2 (2 nM). DSP1 concentrations are as follows: lane 1, no DSP1; lane 2, 10 nM; lane 3, 25 nM; lane 4, 50 nM; lane 5, 100 nM; lane 6, 200 nM; lane 7, 200 nM DSP1 alone. (B) EMSA with 5 nM Gambif1 RHD and increasing quantities of HMG1 in the absence of poly(dI-dC)·(dI-dC). Concentrations of DNA and proteins are identical to those used for DSP1. (C) EMSA with 8 nM Gambif1 RHD, 2 nM Dorsal strong site dl2 and increasing quantities of DSP1 (lanes 2–6) or HMG1 (lanes 8–11) in the presence of 1.5 µg/ml poly(dI-dC)·(dI-dC). In lane 1, no protein was added to the DNA. Lane 2 corresponds to binding of the Gambif1 RHD alone. DSP1 and HMG1 concentrations are as follows: lanes 3 and 8, 8 nM; lanes 4 and 9, 80 nM; lanes 5 and 10, 200 nM; lanes 6 and 11, 400 nM. Lanes 7 and 12 correspond, respectively, to binding of DSP1 and HMG1 alone. In lane 13, 400 nM BSA was added to the Gambif1 RHD prior to binding of DNA. (D) Graphic representation.
DSP1 and HMG1 both bind DNA on their own, making the interpretation of these gels difficult. This led us to wonder whether these supershifts were really due to the formation of a ternary complex or whether they originated from independent binding of HMG1 and DSP1 to the Gambif1–DNA specific complex. One way to answer this question is to decrease DSP1- and HMG1-independent binding by use of a competitor. When 1.5 µg/ml of poly(dI-dC)·poly(dI-dC) was added to the reaction mixture, no supershift corresponding to a ternary complex was observed. It seems therefore that the retarded species observed in Figure 6A and B originate from independent binding of HMG1 and DSP1. One can note that an unrelated protein such as BSA is unable to produce any effect (Fig. 6C, lane 13).
The main result of this experiment is that the amount of Gambif1 RHD–DNA complex increases on addition of DSP1 or HMG1. The Gambif1 Rel domain DNA binding affinity has increased 40-fold for a ratio of 20 between this Rel domain and the HMG box proteins. Remarkably, DSP1 and HMG1 are indistinguishable in the effect they produce on Gambif1 RHD binding, as they were when protein–protein interactions were analysed.
DISCUSSION
The aim of the work presented here was to investigate the involvement of DSP1 in Dorsal-mediated zen repression. We first studied zen repression in vivo in a DSP1 null mutant of Drosophila and then analysed the interactions between Dorsal, DSP1 and DNA in vitro.
In a DSP1 null context, the expression domain of zen is extended into the ventral part of the embryo, suggesting that DSP1 is indeed involved in the repression of zen during embryonic development. However, DSP1 does not appear to be essential: the expansion of the expression domain is weak (60% of the embryo in DSP1 mutants versus 40% for the wild-type) and is limited to a subset of the embryos (2% of embryos which have reached the blastoderm stage). This limited effect could have several origins. (i) Some other protein(s) could substitute for DSP1. Indeed, proteins with a similar HMG domain and an overlapping expression pattern during embryogenesis have been described in Drosophila (HMG D, DssRP1, Pangolin and Dichaete; 40–45). (ii) There could be redundant repression regions outside the VRE that function independently of DSP1, but not of Dorsal and Groucho. Such a redundancy is common to many developmentally important genes in Drosophila, such as Ubx (46), twist (47,48) and tailless (49). (iii) Partial derepression of zen could correspond to a limited role for DSP1, which could be only an accessory protein in this system.
DSP1 and Dorsal interact with each other in vitro as shown by affinity chromatography and far-western experiments. These results strengthen the idea that DSP1 is involved in Dorsal-mediated repression of zen, via direct interactions with Dorsal. To further characterise the Dorsal–DSP1 interactions, we mapped the interacting domains in each partner. The domain organisation of Dorsal and the role of these domains have been the subject of several studies in the literature. The N-terminal part of Dorsal, the Rel domain, mediates DNA binding (50). The C-terminal part is required for transcriptional activation and repression (51,52) but even deleting the entire C-terminal domain does not abolish repression activity totally (51). In keeping with this result, we show that the N-terminal part of Dorsal (residues 1–404) or an even shorter related protein (the Gambif1 RHD, residues 47–330) are both able to interact with DSP1.
As to the DSP1 partner, analysis of the binding to Dorsal of several truncated proteins has revealed two principal facts: the HMG boxes of DSP1, or at least their fold, are necessary for the interaction but are not sufficient; N-terminal flanking residues are essential. It would be tempting to involve these N-terminal residues in the interaction with Dorsal, but one cannot exclude a more structural effect such as a stabilisation of the extremities of the boxes. Provided that these N-terminal residues are present, the A box and the B box of DSP1 are both able to interact with Dorsal, despite their differences in sequence. This suggests that other Drosophila HMG box proteins could perhaps substitute for DSP1 in the DSP1 null mutant. This is further suggested by the ability of rat HMG1 to interact with Dorsal in a very similar way to DSP1.
Concerning Dorsal binding to DNA, DSP1 causes a significant increase in the amount of protein–DNA complex and slows down its gel migration, which could suggest the appearance of a ternary complex. A similar result was obtained with Gambif1 from A.gambiae, whose Rel domain sequence is very close to Dorsal. Rat HMG1, like DSP1, stimulates Gambif1 RHD binding and again causes the appearance of slow migrating bands. The appearance of these supershifts could not be taken as proof of the existence of a ternary complex because DSP1 and HMG1 are capable of binding DNA in a Dorsal-independent manner. The addition of competitor to the binding buffer, by eliminating DSP1- and HMG1-independent DNA binding, totally abrogates the supershifts. This absence of a ternary complex in gel shift assays is not uncommon for these proteins, as observed in the interactions between HMG1 and Hox proteins (26) or HMG1 and p53 (28). Two hypotheses have been proposed: (i) HMG1 may not participate in the ternary complex, but could facilitate binding of the target by altering the DNA structure; (ii) a ternary complex does exist but it dissociates prior to entry into the gel. Nevertheless, DSP1 and HMG1 cause a significant increase in Dorsal DNA binding and their effect is indistinguishable.
By stimulating Dorsal DNA binding, DSP1 would increase the occupancy of Dorsal sites, particularly in the lateral parts of the embryo where the concentration of nuclear Dorsal is lower. This might explain the extension in zen expression that we observed for the DSP1 null mutant in these regions. In this model, the DSP1 role in zen repression is limited to an effect on Dorsal. Alternatively, DSP1 could bring about a repression state of the chromatin at the VRE by recruiting other proteins necessary for repression. In keeping with this idea, Lehming et al. (53) have shown that DSP1 can interact with the mammalian protein SP100B which, in turn, can interact with HP1, a protein required for the form of repression called position effect variegation.
HMG1 has been shown to interact with several other proteins implicated in regulation of transcription: steroid hormone receptors (29,54), p53 (28), Hox proteins (26), octamer transcription factors (27), the adenovirus major late promoter transcription factor MTLF (55,56) and TATA box-binding protein TBP (57,58). Strikingly, in all systems which have been looked at, the protein–protein interactions take place within the HMG box itself, i.e. the DNA-binding domain, and DSP1 is no exception. Results on the effect of HMG1 box A on Hox (26) or TBP (58) functions seem to argue that a single box is sufficient for both interactions. As these proteins contain two HMG boxes, one could wonder about the function of the second HMG box. By use of the Drosophila DSP1 null mutant, specifically designed transgenic strains are under study with the hope of answering this intriguing question.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to A. Soulas and M. Martineau for excellent technical assistance. This work was supported in part by la Ligue Contre le Cancer, la Fondation pour la Recherche Médicale, l’Association pour la Recherche Contre le Cancer and EU contract ERB4061PL97028.
REFERENCES
- 1.Roth S., Stein,D. and Nüsslein-Volhard,C. (1989) Cell, 59, 1189–1202. [DOI] [PubMed] [Google Scholar]
- 2.Steward R. (1987) Science, 238, 692–694. [DOI] [PubMed] [Google Scholar]
- 3.Gilmore T.D., Koedood,M., Piffat,K.A. and White,D.W. (1996) Oncogene, 13, 1367–1378. [PubMed] [Google Scholar]
- 4.Siebenlist U., Franzoso,G. and Brown,K. (1994) Annu. Rev. Cell Biol., 10, 405–455. [DOI] [PubMed] [Google Scholar]
- 5.Ip Y.T., Reach,M., Engstrom,Y., Kadalayil,L., Cai,H., Gonzalez-Crespo,S., Tatei,K. and Levine,M. (1993) Cell, 75, 753–763. [DOI] [PubMed] [Google Scholar]
- 6.Dushay M.S., Asling,B. and Hultmark,D. (1996) Proc. Natl Acad. Sci. USA, 93, 10343–10347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barillas-Mury C., Charlesworth,A., Gross,I., Richman,A., Hoffman,J.A. and Kafatos,F.C. (1996) EMBO J., 15, 4691–4701. [PMC free article] [PubMed] [Google Scholar]
- 8.Baeuerle P.A. and Henkel,T. (1994) Annu. Rev. Immunol., 12, 141–179. [DOI] [PubMed] [Google Scholar]
- 9.Baldwin A.S. (1996) Annu. Rev. Immunol., 14, 649–683. [DOI] [PubMed] [Google Scholar]
- 10.Belvin M.P. and Anderson,K.V. (1996) Annu. Rev. Cell Dev. Biol., 12, 393–416. [DOI] [PubMed] [Google Scholar]
- 11.Liou H.C. and Baltimore,D. (1993) Curr. Opin. Cell Biol., 5, 477–487. [DOI] [PubMed] [Google Scholar]
- 12.Courey A.J. and Huang,J. (1995) Biochim. Biophys. Acta, 1261, 1–18. [DOI] [PubMed] [Google Scholar]
- 13.Pan D.J. and Courey,A.J. (1992) EMBO J., 11, 1837–1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang J., Rushlow,C.A., Zhou,Q., Small,S. and Levine,M. (1992) EMBO J., 11, 3147–3154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang J., Cai,H., Zhou,Q. and Levine,M. (1993) EMBO J., 12, 3201–3209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kirov N., Zhelnin,L., Shah,J. and Rushlow,C. (1993) EMBO J., 12, 3193–3199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Valentine S.A., Chen,G., Shandala,T., Fernandez,J., Mische,S., Saint,R. and Courey,A.J. (1998) Mol. Cell. Biol., 18, 6584–6594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubnicoff T., Valentine,S.A., Chen,G., Shi,T., Lengyel,J.A., Paroush,Z. and Courey,A.J. (1997) Genes Dev., 11, 2952–2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lehming N., Thanos,D., Brickman,J.M., Ma,J., Maniatis,T. and Ptashne,M. (1994) Nature, 371, 175–179. [DOI] [PubMed] [Google Scholar]
- 20.Canaple,L., Decoville,M., Leng,M. and Locker,D. (1997) Gene, 184, 285–290. [DOI] [PubMed] [Google Scholar]
- 21.Bianchi M.E., Beltrame,M. and Falciola,L. (1992) In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, Germany, Vol. 6, pp. 112–128.
- 22.Read C.M., Cary,P.D., Crane-Robinson,C., Driscoll,P.C., Carrillo,M.O.M. and Norman,D.G. (1995) In Eckstein,F. and Lilley,D.M.J. (eds), Nucleic Acids and Molecular Biology. Springer-Verlag, Berlin, Germany, Vol. 9, pp. 222–250.
- 23.Pöhler G.R.J., Norman,D.G., Bramham,J., Bianchi,M.E. and Lilley,D.M.J. (1998) EMBO J., 17, 817–826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ohndorf U.M., Rould,M.A., He,Q., Pabo,C.O. and Lippard,S.J. (1999) Nature, 399, 708–712. [DOI] [PubMed] [Google Scholar]
- 25.Bianchi M.E. and Beltrame,M. (1998) Am. J. Hum. Genet., 63, 1573–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zappavigna V., Falciola,L., Citterich,M.H., Mavilio,F. and Bianchi,M.E. (1996) EMBO J., 15, 4981–4991. [PMC free article] [PubMed] [Google Scholar]
- 27.Zwilling S., König,H. and Wirth,T. (1995) EMBO J., 14, 1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayaraman L., Moorthy,N.C., Murthy,K.G., Manley,J.L., Bustin,M. and Prives,C. (1998) Genes Dev., 12, 462–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Boonyaratanakornkit V., Melvin,V., Prendergast,P., Altman,M., Ronfani,L., Bianchi,M.E., Taraseviciene,L., Nordeen,S.K., Allegretto,E.A. and Edwards,D.P. (1998) Mol. Cell. Biol., 18, 4471–4487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Calogero S., Grassi,F., Aguzzi,A., Voigtlander,T., Ferrier,P., Ferrari,S. and Bianchi,M.E. (1999) Nature Genet., 22, 276–280. [DOI] [PubMed] [Google Scholar]
- 31.Mosrin-Huaman C., Canaple,L., Locker,D. and Decoville,M. (1998) Dev. Genet., 23, 324–334. [DOI] [PubMed] [Google Scholar]
- 32.Lindsley D.L. and Zimm,G.G. (1992) The Genome of Drosophila melanogaster. Academic Press, San Diego, CA.
- 33.Tautz D. and Pfeifle,C. (1989) Chromosoma, 98, 81–85. [DOI] [PubMed] [Google Scholar]
- 34.Bianchi M.E., Falciola,L., Ferrari,L. and Lilley,D.M.J. (1992) EMBO J., 11, 1055–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Teo S.O., Grasser,K.D. and Thomas,J.O. (1995) Eur. J. Biochem., 230, 943–950. [DOI] [PubMed] [Google Scholar]
- 36.Locker D., Decoville,M., Maurizot,J.C., Bianchi,M.E. and Leng,M. (1995) J. Mol. Biol., 246, 243–247. [DOI] [PubMed] [Google Scholar]
- 37.Rushlow C.A., Han,K., Manley,J.L. and Levine,M. (1989) Cell, 59, 1165–1177. [DOI] [PubMed] [Google Scholar]
- 38.Giraud-Panis M.J.E. and Lilley,D.M.J. (1996) J. Biol. Chem., 271, 33148–33155. [DOI] [PubMed] [Google Scholar]
- 39.Cramer P., Varrot,A., Barillas-Mury,C., Kafatos,F.C. and Müller,C.W. (1999) Structure, 7, 841–852. [DOI] [PubMed] [Google Scholar]
- 40.Stroumbakis N.D. and Tolias,P.P. (1994) Biochim. Biophys. Acta, 1218, 245–249. [DOI] [PubMed] [Google Scholar]
- 41.Hsu T., King,D.L., LaBonne,C. and Kafatos,F.C. (1993) Biochemistry, 90, 6488–6492. [Google Scholar]
- 42.Brunner E., Peter,O., Schweizer,L. and Basler,K. (1997) Nature, 385, 829–833. [DOI] [PubMed] [Google Scholar]
- 43.van de Wetering M., Cavallo,R., Dooijes,D., van Beest,M., van Es,J., Loureiro,J., Ypma,A., Hursh,D., Jones,T., Bejsovec,A., Peifer,M., Mortin,M. and Clevers,H. (1997) Cell, 88, 789–799. [DOI] [PubMed] [Google Scholar]
- 44.Russel S.R.H., Sanchez-Soriano,N., Wright,C.R. and Ashburner,M. (1996) Development, 122, 3669–3676. [DOI] [PubMed] [Google Scholar]
- 45.Nambu P.A. and Nambu,J.R. (1996) Development, 122, 3467–3475. [DOI] [PubMed] [Google Scholar]
- 46.Simon J., Peifer,M., Bender,W. and O’Connor,M. (1990) EMBO J., 9, 3945–3956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang J., Kosman,D., Ip,Y.T. and Levine,M. (1991) Genes Dev., 5, 1881–1891. [DOI] [PubMed] [Google Scholar]
- 48.Pan D.J., Valentine,S.A. and Courey,A.J. (1994) Mech. Dev., 46, 41–53. [DOI] [PubMed] [Google Scholar]
- 49.Liaw G.J. and Lengyel,J.A. (1993) Mech. Dev., 40, 47–61. [DOI] [PubMed] [Google Scholar]
- 50.Ip Y.T., Kraut,R., Levine,M. and Rushlow,C.A. (1991) Cell, 64, 438–446. [Google Scholar]
- 51.Isoda K., Roth,S. and Nüsslein-Volhard,C. (1992) Genes Dev., 6, 619–630. [DOI] [PubMed] [Google Scholar]
- 52.Richardson P.M. and Gilmore,T.D. (1991) J. Virol., 65, 3122–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lehming N., LeSaux,A., Schüller,J. and Ptashne,M. (1998) Proc. Natl Acad. Sci. USA, 95, 7322–7326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Oñate S.A., Prendergast,P., Wagner,J.P., Nissen,M., Reeves,R., Pettijohn,D.E. and Edwards,D.P. (1994) Mol. Cell. Biol., 14, 3376–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremethick D.J. and Molloy,P.L. (1989) FEBS Lett., 242, 346–350. [DOI] [PubMed] [Google Scholar]
- 56.Watt F. and Molloy,P.L. (1989) Nucleic Acids Res., 16, 1471–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ge H. and Roeder,R.G. (1994) J. Biol. Chem., 269, 17136–17140. [PubMed] [Google Scholar]
- 58.Sutrias-Grau M., Bianchi,M.E. and Bernués,J. (1999) J. Biol. Chem., 274, 1628–1634. [DOI] [PubMed] [Google Scholar]