Abstract
Background: Previously reported breast invasive carcinoma (BRIC) biomarkers have compromised utility because of their heterogeneity-specific behaviors. The goal of this study was to find BRIC biomarkers that could be used in spite of the heterogeneity barrier. Methods: Previously reported BRIC-linked hub genes were obtained from the literature via a search technique. A protein-protein interaction (PPI) network of the extracted hub genes was constructed, visualized, and analyzed to explore the top six real hub genes. Following this, real hub genes’ expression profiling was carried out using various TCGA data sources and RNA sequencing (RNA-seq) of BT 20 and HMEC cell lines to uncover the tumor-driver roles of the real hub genes. Results: In total, 124 BRIC-linked hub genes were collected from the literature via the search technique. From these collected hub genes, a total of 6 genes, including Centrosomal protein of 55 kDa (CEP55), Kinesin Family Member 2C (KIF2C), kinesin family member 20A (KIF20A), Ribonucleotide Reductase Regulatory Subunit M2 (RRM2), Aurora A Kinase (AURKA), and Protein Regulator of cytokinesis 1 (PRC1) were determined to be the real hub genes. Via expression profiling and validation analyses, we documented the overexpression of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 real hub genes in BRIC patients with different clinical variables. Further correlational analyses showed diverse associations among real hub genes’ expression and other important parameters, including promoter methylation status, genetic alteration, overall survival (OS), relapse-free survival (RFS), tumor purity, CD8+ T, CD4+ T immune cell infiltration, and different mutant genes across BRIC samples. Finally, in this work, we investigated several transcription factors (TFS), microRNAs, and therapeutic medicines related to the real hub genes that have great therapeutic potential. Conclusion: In conclusion, we discovered six real hub genes, which may be employed as novel potential biomarkers for BRIC patients with different clinical parameters.
Keywords: BRIC, biomarker, hub genes
Introduction
Breast invasive carcinoma (BRIC) is the most common women’s malignancy and leads to a large number of cancer-related deaths every year around the globe [1]. According to recent reports, about 3.45 million cancer cases are annually reported in Europe [1]. As a result of ongoing efforts by researchers, the availability of modern technologies has greatly helped to reduce the BRIC-related mortality rate by identifying reliable potential biomarkers for the timely detection, treatment, and monitoring of prognosis. However, due to the heterogeneity-associated nature of the reported BRIC biomarkers [2], the management of disease in BRIC patients of different cancer stages, races, genders, ages, and subclasses has not been addressed completely and remains a major clinical treatment obstacle [2].
With the help of microarray techniques, various disease-associated differentially expressed genes can be recognized simultaneously [3,4]. Besides, this technique also enables researchers to carry out a detailed analysis of specified key genes to explore their potential as molecular biomarkers. Gene Expression Omnibus (GEO) database is a free online available microarray and RNA sequencing-based platform maintained by the National Center for Biotechnology Information (NCBI) [5]. This database is one of the most specialized platforms for researchers to submit, re-evaluate, and re-analyze the already submitted microarray datasets for the identification of disease-specific molecular biomarkers [6].
In this work, we re-analyzed multiple GEO datasets to find a few BRIC-associated biomarkers that could be used to overcome the heterogeneity barrier. The current work may be useful in developing a unique system of biomarkers that can be applied to BRIC patients with various clinical characteristics across the heterogeneity-specific barrier.
Methods
Mining of hub genes
Relevant studies that dealt with the BRIC GEO expression datasets, in order to explore hub genes, until June 2022 were searched via “PubMed”. For search purposes, the two selected keywords were “Hub genes AND Breast cancer” and “Hub genes AND Breast neoplasia” with the “Research article” filter. By doing so, a total of 108 studies appeared at the end of the search process. Those studies were further shortlisted to only 24 studies that collectively used 31 BRIC GEO datasets. Following the search process, all collected hub genes were compiled into a single pool.
Gene enrichment analysis
The GO “(Gene Ontology)” and KEGG “(Kyoto Encyclopedia of Genes and Genomes)” analyses were performed via the DAVID 9th tool [7]. This is an online platform that is publicly available for GO and KEGG analysis of any given gene list. In these analyses, a p-value <0.05 was regarded as significant.
Real hub genes screening
In this study, STRING “Search Tool for the Retrieval of Interacting Genes/Proteins” [8] analysis was conducted for constructing the protein-protein interaction (PPI) network of the hub genes. Later on, Cytoscape [9] plugin applications, including MCODE and Cytohubba, were used to determine the significant module and the top six real hub genes. Based on the 4 different scoring algorithms, “the maximum neighborhood component (MNC), the density of the maximum neighborhood component (DMNC), the maximal clique centrality (MCC), and the Degree of the Cytohubba” [10], the shared top six genes by these 4 algorithms were selected as real hub genes.
GEPIA-based mRNA expression analysis
The TCGA “(Cancer Genome Atlas)” expression data is used to create gene expression plots based on various pathological factors in the GEPIA “(Gene Expression Profiling Interactive Analysis)” database, a new online web-based tool that enables users to perform interactive and customizable analyses between normal-v-normal cancer samples [11]. In this work, we utilized the TCGA BRIC dataset from GEPIA to analyze real hub genes’ expression. The p-value cutoff was selected as 0.05.
mRNA and translation expression validation analysis by other databases
The bc-GenExMiner “(Breast Cancer Gene-Expression Miner)” [12], GENT2 [13], and the UALCAN “(University of ALabama at Birmingham CANcer)” [14] were utilized in this study for the mRNA and translational expression validation of the real hub genes using new independent cohorts of BRIC patients. All these online databases are cancer microarray-based expression analysis platforms, which provide expression analysis results in the form of box plots. Additionally, the UALCAN database was also utilized to measure the expression of real hub genes targeting TFS and miRNAs. The p-value cutoff was selected as 0.05.
Promoter methylation analysis
In this study, the correlations among the real hub genes expression and their promoter methylation levels in BRIC were examined via MEXPRESS through the Pearson correlation method [15]. MEXPRESS database highlight associations among patient clinical information and promoter methylation levels across TCGA datasets. The p-value cutoff was selected as 0.05.
cBioPortal analyses
Multidimensional cancer genomic analysis on TCGA cancer datasets is carried out using the cBioPortal, which is an online open-access platform [16]. In this study, this database was used for analyzing genetic mutations in real hub genes across BIRC samples.
Survival analysis
The Kaplan-Meier plotter [17] tool was used to compute the relapse-free survival (RFS) and overall survival (OS) of real hub genes. To do this, BRIC patient samples were divided into 2 different cohorts in accordance with the median expression of the real hub gene (high vs. low). The p-value cutoff was selected as 0.05.
Hub genes and immune cells infiltration
The TIMER “(Tumor Immune Estimation Resource TIMER)” database [18] was used in this study to find associations between tumor purity, CD8+ T, CD4+ T immune cell infiltration, and real hub gene expression. A variety of algorithms are used in this database to estimate the abundance of immune cells across different cancers.
TFS-miRNA-mRNA network
To construct the TFS-miRNA-mRNA network, The ENCORI “(Encyclopedia of RNA Interactomes)” and transcriptional regulatory relationships unraveled by sentence-based text-mining (TRRUST) were conducted in the present study [19]. These databases are widely utilized for exploring miRNA-ncRNA and mRNA-miRNA interactions from CLIP-seq interactome data.
MuTarget analysis
The MuTarget [20] analysis was conducted in this study with default setting to identify the mutant genes causing expressional changes in the real hub genes across BRIC. The p-value cutoff was selected as 0.05.
Real hub gene associated drugs
The CTD “(Comparative Toxicogenomics Database)” (CTD) database [21] was used in the current work to identify real hub gene-associated different drugs in the current study. Because we believe that the identified real hub genes can be interesting therapeutic targets. This database offers information on drugs that target hub genes from numerous trustworthy sources [21].
In vitro validation of the hub gene expression
Cell lines
One BRIC cell line (BT 20), as well as one normal mammary gland cell line (HMEC) were purchased from the American Type Culture Collection (ATCC, USA) and cultivated in accordance with the manufacturer’s instructions.
Total RNA extraction
Total RNA extraction from both BRIC and normal cell lines was done by isopycnic centrifugation as described previously [22]. The extracted RNA was then processed for DNA digestion step of incubation with RNase-free DNase I (Roche, Germany) at 37°C for 15 minutes. The quality of the extracted RNA was checked by a 2100 Bioanalyzer (Agilent Technologies, Germany).
RNA-Seq analysis
RNA samples were sent to Macrogen, Korea company for RNA-seq analysis. Following RNA-seq analysis, the gene expression values of the hub genes were normalized using reads per kilo base million reads (RPKM) and fragments per kilo base million reads (FPKM). The obtained FPKM values against real hub genes in BIRC and normal control cell line were compared to identify differences in the expression level.
Statistics details
For GO and KEGG enrichment analysis, we used Fisher’s Exact test for computing statistical difference [23]. Correlational analyses were carried out using the Pearson method. For comparisons, a student t-test was adopted in the current study. All the analyses were carried out in R version 3.6.3 software.
Results
Hub genes collection from the literature
We selected 24 molecular studies that explored hub genes in BRIC GEO datasets. We then performed the extraction of hub genes from these studies and pooled these hub genes after normalizing the duplicated hub genes. Ultimately, a pool of 124 hub genes from 31 GEO BRIC datasets containing 2908 BRIC and 313 normal samples was further explored (Table 1). Original data (without normalization) can be seen in the Supplementary Material.
Table 1.
Detail of BRIC datasets and hub genes obtained from the literature
| BRIC Dataset | samples C/N | Hub genes | Reference |
|---|---|---|---|
| GSE10797 | 28/5 | RPS9, RPL11, RPS14, RPL10A, EPCAM, MELK, KRT8, KRT19, KPNA2, ECT2, TPX2, KIF2C, CDCA8, BUB1B, CCNA2, TOP2A, PCNA, CCNB1, CDC20, BIRC5, PHLPP1, UBC, ACACB, TGFB1, ACTB, CASC5, FAM83D, TFAP2C, KIF23, GINS1, CDCA5, CCNE1, KRT16, MYBL2, AGO2, MCM10, TTK, KIF18B, CDKN2A, MME, IGFBP3, CKAP2L, TGM2, ACTA2, PDGFRβ, SUMO1, FYN, CAV1, COL5A1, SKA1, MMP2, CDK1, NDC80, KRT18, STAMBP, JUN, MCM6, FOS, ATF3, STAT1, COL1A1, FN1, TP53, GAPDH, CCND1, HRAS, CAPG, SPI1, LEF1, PBX3, TCF7L2, VCAM1, PLAGL1, PBX1, EGFR, IGF1, LEP, PTEN, FOXO1, FGF2, PPARG, AURKA, IK3CA, CDH1, CDK1, NOTCH1, MAPK14, SRC, HSPA8, ESR1, PPP2CA, RPL4 | [59-82] |
| GSE15852 | 43/43 | RAC1, KIF20A, RRM2, ASPM, NUSAP1, CEP55 | |
| GSE92697 | 26/0 | PGR, GATA3, ABLIM3, MYC, IL18, CD274, ITGB1, ITGB3, ITGA2B, CXCR4, COL1A2, EGR1, HMOX1, NR3C1, STAT5A, TFF1, FOXA1, HSP90AA1, KIF11, CCNB2, CDKN3, CENPF, PRC1, PTTG1, UBE2C, ZWINT | |
| GSE102484 | 683/0 | ||
| GSE65212 | 130/11 | ||
| GSE43837 | 19/0 | ||
| GSE23988 | 61/0 | ||
| GSE20194 | 230/0 | ||
| GSE42568 | 104/17 | ||
| GSE75333 | 6/3 | ||
| GSE5847 | 95/0 | ||
| GSE22035 | 43/0 | ||
| GSE3744 | 47/0 | ||
| GSE5764 | 10/20 | ||
| GSE21422 | 14/5 | ||
| GSE26910 | 12/12 | ||
| GSE41970 | 270/59 | ||
| GSE8977 | 7/15 | ||
| GSE45827 | 144/11 | ||
| GSE71142 | 10/0 | ||
| GSE86945 | 100/0 | ||
| GSE86946 | 58/0 | ||
| GSE29431 | 66/0 | ||
| GSE65194 | 167/11 | ||
| GSE22093 | 103/0 | ||
| GSE31192 | 22/0 | ||
| GSE9014 | 123/0 | ||
| GSE10780 | 143/62 | ||
| GSE29431 | 54/12 | ||
| GSE61304 | 59/3 | ||
| GSE10810 | 31/27 | ||
| Total = 31 | Total = 2908/313 | Total = 124 |
C = Cancerous, N = Normal.
GO and KEGG analysis
The GO and KEGG enrichment analysis revealed hub the genes that were enriched in different GO and KEGG terms, including “cell division, mitotic nuclear division, and response to drug” biological processes GO terms, and pathways in “cancer, cell cycle, focal adhesion, and proteoglycans in cancer” KEGG terms (Figure 1 and Tables 2, 3).
Figure 1.

A heatmap representing the GO and KEGG terms across identified hub genes related with BRIC. (A) A heatmap of GO terms across identified hub genes, and (B) a heatmap of KEGG terms across identified hub genes.
Table 2.
Details of the GO analysis
| Biological process ID | Biological process | Gene count | P-value | Gene name |
|---|---|---|---|---|
| GO:0051301 | cell division | 24 | 2.8E-16 | UBE2C, CDCA5, CDCA8, BUB1B, KIF11, NR3C1, NDC80, SKA1, ZWINT, AURKA, CCNA2, CDC20, CCNB2, TPX2, CENPF, CCNB1, KIF18B, PTTG1, CCND1, CCNE1, CDK1, BIRC5, KIF2C, FAM83D |
| GO:0045944 | positive regulation of transcription from RNA polymerase II promoter | 36 | 3.5E-16 | FOXA1, TOP2A, SPI1, NOTCH1, LEF1, GATA3, NR3C1, FGF2, FOXO1, EGFR, ABLIM3, EPCAM, MYC, PLAGL1, UBC, MYBL2, HRAS, TCF7L2, EGR1, JUN, TFAP2C, TGFB1, CDKN2A, STAT1, IL18, PBX3, IGF1, FOS, MAPK14, ESR1, PBX1, AGO2, PPARG, PGR, TP53, ATF3 |
| GO:0007067 | mitotic nuclear division | 19 | 1.3E-13 | CDCA5, BUB1B, KIF11, NR3C1, NDC80, SKA1, AURKA, CCNA2, CDC20, ASPM, CCNB2, TPX2, CENPF, PTTG1, CDK1, BIRC5, KIF2C, FAM83D, CEP55 |
| GO:0042493 | response to drug | 18 | 4.2E-11 | JUN, HSP90AA1, TGFB1, STAT1, SRC, PTEN, GATA3, FOS, ACACB, COL1A1, CENPF, CCNB1, CCND1, CDH1, MYC, CDK1, FYN, PPARG |
| 05166 | HTLV-I infection | 19 | 6.7E-11 | EGR1, HRAS, STAT5A, TP53, SPI1, CDC20, PTTG1, TGFB1, VCAM1, FOS, CCND1, ATF3, CDKN2A, JUN, PCNA, BUB1B, PDGFRB, MYC, TP53INP1 |
| GO:0045893 | positive regulation of transcription, DNA-templated | 22 | 8.1E-11 | EGR1, JUN, SPI1, TGFB1, NOTCH1, CDKN2A, STAT1, SRC, LEF1, GATA3, FOS, IGF1, FGF2, ESR1, FOXO1, CCNA2, COL1A1, CCNE1, CDH1, MYC, PPARG, TP53 |
| GO:0008284 | positive regulation of cell proliferation | 21 | 8.8E-11 | ITGB1, RPS9, TGFB1, NOTCH1, LEF1, PTEN, FN1, TTK, IGF1, FGF2, EGFR, PBX1, CDC20, EPCAM, PRC1, MYC, LEP, BIRC5, STAMBP, HRAS, ATF3 |
| GO:0008283 | cell proliferation | 19 | 1.1E-9 | TCF7L2, PCNA, SRC, PTEN, BUB1B, MCM10, IGF1, EGFR, TPX2, CENPF, MELK, KRT16, MYC, CDK1, KIF2C, RAC1, FAM83D, HRAS, TP53 |
| GO:0051726 | regulation of cell cycle | 12 | 2.5E-9 | ITGB1, HSPA8, CCNB2, CENPF, CCNB1, JUN, CCNE1, SRC, LEP, PTEN, MYBL2, FGF2 |
| GO:0000082 | G1/S transition of mitotic cell cycle | 11 | 3.7E-9 | ITGB1, RRM2, PCNA, CCND1, CCNE1, CDKN2A, CDCA5, CDK1, MCM10, MCM6, CDKN3 |
Table 3.
Details of the KEGG analysis
| ID | Pathway | Genes involved | P-value | Genes |
|---|---|---|---|---|
| 05200 | Pathways in cancer | 30 | 4.9E-15 | HRAS, STAT5A, PPARG, SPI1, FOXO1, CDH1, ITGB1, TCF7L2, PTEN, MMP2, TGFB1, CCNE1, FOS, CDKN2A, CXCR4, RAC1, FGF2, MYC, FN1, EGFR, HSP90AA1, TP53, LEF1, IGF1, BIRC5, STAT1, CCND1, JUN, PDGFRB, ITGA2B |
| 04110 | Cell cycle | 16 | 1.5E-11 | CDK1, TP53, TTK, CDC20, PTTG1, TGFB1, MCM6, CCNB1, CCNE1, CCND1, CDKN2A, CCNB2, PCNA, BUB1B, MYC, CCNA2 |
| 04510 | Focal adhesion | 19 | 2.8E-10 | ACTB, EGFR, HRAS, CAV1, IGF1, ITGB3, PTEN, ITGB1, SRC, COL5A1, CCND1, FYN, JUN, RAC1, COL1A2, PDGFRB, COL1A1, FN1, ITGA2B |
| 05205 | Proteoglycans in cancer | 18 | 1.8E-10 | ACTB, EGFR, HRAS, CAV1, TP53, ESR1, IGF1, ITGB3, ITGB1, MMP2, SRC, TGFB1, CCND1, MAPK14, RAC1, FGF2, MYC, FN1 |
| 05166 | HTLV-I infection | 19 | 5.0E-9 | EGR1, HRAS, STAT5A, TP53, SPI1, CDC20, PTTG1, TGFB1, VCAM1, FOS, CCND1, ATF3, CDKN2A, JUN, PCNA, BUB1B, PDGFRB, MYC, TP53INP1 |
| 05161 | Hepatitis B | 15 | 1.6E-9 | HRAS, STAT5A, TP53, BIRC5, STAT1, PTEN, SRC, TGFB1, CCNE1, FOS, CCND1, JUN, PCNA, MYC, CCNA2 |
| 04151 | PI3K-Akt signaling pathway | 21 | 2.0E-8 | PHLPP1, EGFR, HRAS, HSP90AA1, TP53, IGF1, ITGB3, PTEN, ITGB1, COL5A1, CCNE1, CCND1, PPP2CA, RAC1, COL1A2, PDGFRB, COL1A1, FGF2, MYC, FN1, ITGA2B |
| 04115 | p53 signaling pathway | 11 | 6.0E-8 | CCNB1, CCNE1, CDK1, CCND1, CDKN2A, CCNB2, RRM2, TP53, IGF1, IGFBP3, PTEN |
| 05215 | Prostate cancer | 12 | 8.9E-8 | EGFR, CCNE1, HRAS, CCND1, HSP90AA1, TP53, PDGFRB, FOXO1, LEF1, IGF1, PTEN, TCF7L2 |
| 05219 | Bladder cancer | 9 | 2.4E-7 | EGFR, HRAS, CCND1, CDKN2A, TP53, CDH1, MYC, MMP2, SRC |
Screening of real hub genes
A PPI network of the 124 hub genes was created with the help of STRING. The obtained PPI consisted of 124 nodes and 2110 edges (Figure 2A). Then, the MCODE and Cytohubba analyses via Cytoscape software were performed to identify the most significant module in the PPI and a few more closely BRIC relevant genes (real hub genes) via the degree method. The most significant identified module consisted of 43 hub genes (Figure 2B), and based on the degree method, the screened six real hub genes were CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 (Figure 2C and Table 4).
Figure 2.
(A) A STRING based PPI network of the 124 extracted hub genes associated with BRIC. (B) MCODE based identified most significant module, and (C) degree method based identified real hub genes.
Table 4.
List of real hub genes
| Sr. No | Gene | Degree Score | Node count | Centrality score |
|---|---|---|---|---|
| 1 | CEP55 | 41 | 41 | 1 |
| 2 | KIF2C | 41 | 41 | 1 |
| 3 | KIF20A | 41 | 41 | 1 |
| 4 | RRM2 | 41 | 41 | 1 |
| 5 | AURKA | 41 | 41 | 1 |
| 6 | PRC1 | 41 | 41 | 1 |
Expression analysis and validation
For analyzing and validating real hub gene expression at the mRNA as well as protein levels across BRIC patients of different clinicopathological variables, we utilized four different reliable platforms, including GEPIA, bc-GenExMiner, and UALCAN. Taking together the results of expression analysis and validation, we confirmed the significant up-regulation of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 genes at both mRNA and protein levels in BRIC patients with different cancer stages, races, genders, age groups, and menopause status relative to controls (Figures 3, 4 and 5).
Figure 3.
Transcription expression of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 in control and BRIC samples via GEPIA, bc-GenExMiner, and UALCAN databases. (A) Via GEPIA, (B) via bc-GenExMiner, and (C) via UALCAN.
Figure 4.
Transcription expression of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 across BRIC patients of different clinicopathological features. (A) Expression across different cancer stages, (B) Expression across different races, (C) Expression across different genders, (D) Expression across different age groups and, (E) Expression across different menopause statuses.
Figure 5.
Translation expression of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 in BRIC patients relative to controls. (A) CEP55, (B) KIF2C, (C) KIF20A, (D) RRM2, (E) AURKA, and (F) PRC1.
Promoter methylation level
Promoter methylation participates in the expression regulation and is closely linked with cancer development and progression [24]. We analyzed the promoter methylation level of the real hub genes in BRIC patients relative to controls via the UALCAN platform. Our results revealed that CEP55, RRM2, and PRC1 were significantly hypomethylated, while KIF2C, KIF20A, and AURKA were significantly hypermethylated in BRIC patients relative to controls (Figure 6).
Figure 6.
CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 promoter methylation level in control and BRIC patients. (A) CEP55, (B) KIF2C, (C) KIF20A, (D) RRM2, (E) AURKA, and (F) PRC1.
Genetic changes in real hub genes
Information related to genetic alterations and mutational hotspots in the six real hub genes was obtained from three different TCGA BRIC datasets, Breast Invasive Carcinoma (TCGA, firehose legacy) = 1108 samples, Breast Invasive Carcinoma (TCGA, Nature 2012) = 825 samples, and Breast Invasive Carcinoma (TCGA PanCancer Atlas) = 1984 samples, available via the cBioPortal platform. We have observed a varying degree of genetic variation in the real hub genes, out of which AURKA has shown the highest incidence rate (6%) of genetic variations, followed by PRC1, which has shown the second highest genetic variation rate of 2.5%. While other real hub genes, including RRM2, KIF2C, KIF20A, and CEP55, have shown the genetic variation rates of 1.5%, 1.1%, 0.7%, 0.6% in BRIC samples, respectively. In all the real hub genes, the most frequently observed genetic alteration was deep amplification (Figure 7A). Additionally, we have also observed that mutations in the CEP55 gene, including the most commonly reported Q446Pfs*6 mutation, lie outside the EABR domain (Figure 6B). Similarly, in AURKA, the mutations were also found outside of its most important Pkinase domain (Figure 6B). However, on the other side of the coin, the most important domains, including Kinesen of KI2C and KIF20, and Ribonuc-red-sm and MAP65-ASE1 of AURKA and PRC1, are the major hotspots of the reported mutations (Figure 7B).
Figure 7.
Frequencies of the genetic alterations and mutational hotspots identification CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 across BRIC samples. (A) A view of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene-associated genetic alterations frequencies in BRIC samples, and (B) A view of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene-associated mutational hotspots in BRIC samples.
Survival analysis
Correlations between OS, RFS, and mRNA expression of real hub genes across BRIC patients were explored via KM plotter. We observed that the higher mRNA expressions of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 were significantly (P<0.05) linked with the reduced OS and RFS duration of the BRIC patients, therefore, these genes are supposed to be the good prognostic biomarkers in BRIC patients (Figure 8).
Figure 8.
The prognostic information of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 in BRIC patients via KM plotter. (A) The calculated RFS values of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1, and (B) The calculated OS values of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1. Blue color indicates this low expression while red color indicates the high expression of a gene.
Tumor purity, immune cells, and gene expression
Spearman correlations among tumor purity, CD8+ T, and CD4+ T cell infiltration, and real hub gene expression across BRIC were evaluated via TIMER. Results showed notable positive correlations among CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression and tumor purity, and CD8+ T immune cell infiltration level across BRIC (Figure 9). Moreover, notable (P<0.05) negative correlations among CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression and CD4+ T immune cell infiltration level were also documented across BRIC samples.
Figure 9.
TIMER based Spearman correlational analysis between tumor purity, CD8+ T immune cells infiltration, CD4+ T immune cells infiltration, and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression across BRIC samples. (A) Between CD8+ T immune cells infiltration and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1gene expression, (B) Between CD4+ T immune cells infiltration and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression, and (C) Between tumor purity and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression.
Co-expression network
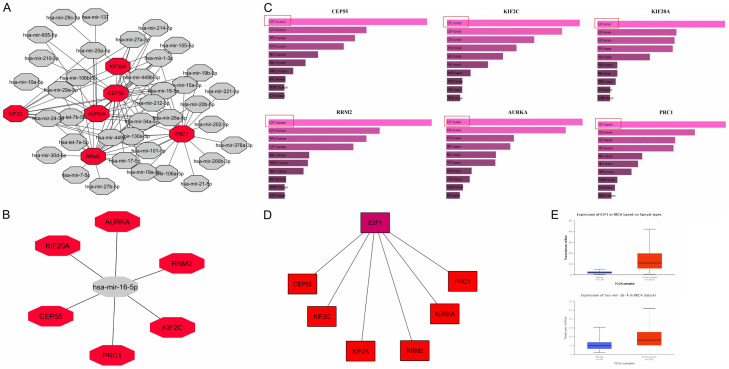
In this study, TRRUST and ENCORI were used to construct the TFS-miRNA-nRNA co-regulatory network. In the obtained network, the total numbers of TFS, miRNAs, and mRNAs were 60, 95, and 6, respectively. In addition, based on the p-value for TFS and degree of centrality for miRNAs, we have identified one TF (E2F1) and one miRNA (hsa-mir-16-5p) that target all the 6 real hub genes. Previous studies reported that the PVT1-miR-16-5p/VEGFA/VEGFR1/AKT TFS-miRNA-hub genes axis, and miR-216-5p-Cx43, and miR-16-1-3p/PGK1 miRNA-hub gene axis are the critical modulators of colorectal and BRIC [25]. However, the identified TFS-miRNAs-mRNA co-regulatory network in the current study has highlighted that the E2F1-has-miR-16-5p/CEP55/KIF2C/KIF20A/RRM2/AURKA/PRC1 axis can also be the potential inducer of the BRIC. To further confirm the participation of identified TF and miRNA in BRIC development via up-regulating real hub genes, we further checked the expression of E2F1 and has-mir-16-5p in BRIC patients via UALCAN. In view of our results, a significant up-regulation of E2F1 and hsa-mir-16-5p was also observed in BRIC samples relative to controls. Finally, we suggested that up-regulated E2F1 and has-miR-16-5p may also exert BRIC-inducing effects by overexpressing their target genes, i.e., CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 (Figure 10).
Figure 10.
Identification of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 targeted potential TFS, miRNAs, and their expression analysis in BRIC. (A) A network of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 targeted miRNAs, (B) A network of has-mir-16-5p miRNA and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1, (C) CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene targeted TFS, (D) A network of E2F1 and CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1, (E) expression analysis of the E2F1 and hsa-mir-16-5p in BRIC samples paired with controls. The red nodes represent the real hub gene, grey nodes represent the miRNAs, while purple node represent the TFS.
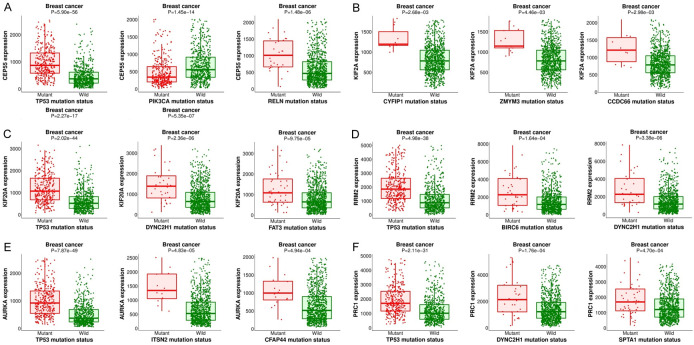
Real hub genes-associated mutant genes
To identify crucial mutant genes associated with real hub genes, the MuTarget analysis was conducted to recognize mutant genes correlated with real hub gene expression. We selected the top 3 mutant genes for each real hub gene. As shown in Figure 11, the top 3 mutant genes that positively correlate with the expression of each real hub gene are TP53, PIK3CA, and RELN with CEP55, CYFIP1, ZMYM3, and CCDC66 with KIF2C, TP53, DYNC2H1, and FAT3 with KIF20A, TP53, BIRC, and DYNC2H1 with RRM2, TP53, ITSN2, and CFAP44 with AURKA, and TP53, DYNC2H1, and SPTA1 with PRC1.
Figure 11.
CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 positively correlated mutant genes in BRIC from MuTarget. (A) Top 3 correlated genes with CEP55, (B) Top 3 correlated genes with KIF2C, (C) Top 3 correlated genes with KIF20A, (D) Top 3 correlated genes with RRM2, (E) Top 3 correlated genes with AURKA, and (F) Top 3 correlated genes with PRC1.
CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene-associated drugs
To identify relationships among CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 and different therapeutic drugs, a gene-drug interaction network was created with the help of CTD and Cytoscape. The expression of identified real hub gene including CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 can potentially be regulated by a variety of drugs. For example, Allyl sulfide and Arsenic trioxide can elevate the expression level of CEP55 while imetidine and bisphenol A can reduce the KIF2C expression level (Figure 12).
Figure 12.
Gene-drug interaction network of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1. (A) CEP55, (B) KIF2C, (C) KIF20A, (D) RRM2, (E) AURKA, and (F) PRC1. Red arrows: drugs that increase the real hub genes expression, Green arrows: drug that decrease the real hub genes expression while the numbers of arrows represent the supported numbers of studies by literature.
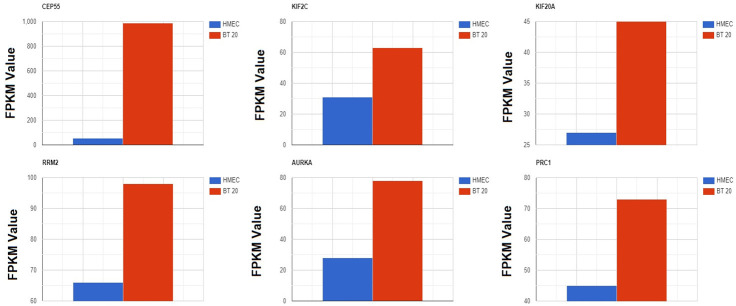
Experimental in vitro validation of the hub gene expression and methylation status
In this work, by performing RNA-seq analysis of one BRIC cell line (BT 20), as well as one normal mammary gland cell line (HMEC), the expression levels of the hub gene were validated. The expression levels of the CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 genes were validated using FPKM values. The FPKM is a quantitative value with widespread use in RNA-seq analysis. As shown in Figure 13, it was noticed that hub genes express in both normal and BRIC cell lines, and FPKM values of the hub genes were notably higher in BRIC cell line (BT 20) as compared to a normal cell line (HMEC) (Figure 13).
Figure 13.
Validating CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 gene expression using BT 20 and HMEC cell lines via RNA-seq analysis.
Discussion
This study was launched to discover the BRIC biomarkers that could be employed over the heterogeneity barrier. To do so, initially we extracted 124 hub genes from the literature. Later, the creation of a PPI network of the hub genes and a significant module identification from this network have helped us to obtain six real hub genes, including CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1.
The CEP55 gene encodes a 55 kDa protein consisting of 464 amino acids, which is initially described as a midbody-related protein. CEP55 is a key regulator of physical cytokinesis [26]. The overexpression of CEP55 was earlier linked to the pathogenesis and poor prognosis of lung [27], oral [28], cervical [29], breast cancers [28], and osteosarcoma [30]. In addition, CEP55 knockdown was also found to inhibit tumor cell proliferation [31]. In sum, the CEP55 up-regulation can result in disordered cytokinesis and lead to the enhancement in multinucleated cells, which is a key phenomenon of tumorigenesis [32].
The KIF2C belongs to the kinesin-13 family, which plays key roles in cell cycle regulation and progression [33]. Thus, KIF2C is supposed to be involved in tumorigenesis. However, the KIF2C role in tumorigenesis has not been explored deeply so far. Previously, Nakamura et al. have shown that KIF2C overexpression is linked to nodal metastasis and poor prognosis in gastric cancer [34]. Wei et al. have revealed the significant up-regulation of KIF2C in hepatocellular carcinoma promoting cell proliferation, cell migration, cell invasion, and metastasis [35]. Moreover, a study by Abdel-Fatah et al. has also reported the overexpression of KIF2C across BRIC patients and associated this overexpression with unfavorable clinicopathological variables [36].
KIF20A is another important member of the kinesin-13 family and is involved in chromosome segregation and mitosis. Previous studies have implicated KIF20A overexpression in different human cancers including pancreatic cancer [37], bladder cancer [38], gastric cancer [39], head and neck cancer [40], lung cancer [41], melanoma [42], and breast cancer [43]. However, to date, only a few studies have explored the KIF20A role in breast cancer. Moreover, a decreased level of KIF20A has also been documented in pancreatic ductal adenocarcinoma [44].
Ribonucleotide reductase (RR) is a key enzyme, mainly involved in DNA replication and repair processes [45]. RRM2 is an important subunit of RR, and it has recently gained much attention in cancer research because of its significant dysregulation in different human cancers, including BRIC [46]. It was earlier reported that cancer patients with overexpressed RRM2 suffer from poor prognoses and tumor recurrence in different cancers such as BRIC, lung cancer, and cancers of the colorectum and crevices [47]. Furthermore, it is also observed that RRM2 overexpression enhances BRIC cell proliferation and inhibits apoptosis [48]. However, the mechanisms behind the involvement of RRM2 in BRIC development and progression are not completely understood.
AURKA is a member of the serine/threonine kinase family, which is very important for activating cell division processes through mitosis regulation [49]. Apart from these functions, when AURKA is differentially expressed, it could act as an oncogene and participate in cancer development and progression [49]. The aberrant expression of AURKA across human cancers was previously reported by various studies. For example, the overexpression of AURKA was revealed in colon, breast, and lung cancer patients [50].
The PRC1 gene, also known as MAP65, is the substrate of cyclin-dependent kinases (CDKs). PRC1 up-regulation has already been seen in different human cancers, including the cancers of the breast [51], bladder [52], and kidney [53]. Additionally, a study by Kanehira et al. has reported that knockdown of PRC1 using siRNA can inhibit the proliferation of breast cancer cells [52]. Recently, a study by Chen et al. explored that PRC1 promotes metastasis and tumorigenesis of hepatocellular carcinoma by dysregulating the Wnt signaling pathway [54]. In our study, we found that CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1 were significantly overexpressed in BRIC patients with diverse clinical parameters compared to non-cancer samples. Taken together, the expression profiling of CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1, it was spectated that overexpression of these real hub genes may serve as potential biomarkers of BRIC regardless of different clinical parameters.
Moreover, the identified real hub genes were found to be altered in a minor proportion of the BRIC patients. Additionally, it was also explored in the current study that genetic mutations can alter amino acids at different positions in the resultant proteins from the real hub genes. Furthermore, this study documented significant negative correlations among real hub gene promoter methylation levels and expressions. This study also revealed that real hub gene overexpression was associated with worse prognosis of BRIC patients.
We also documented correlations among tumor purity, CD8+ T, CD4+ T immune cell infiltration levels, and the expression of real hub genes across BRIC. Results showed that real hub genes have positive correlations with the tumor purity in BRIC, which further confirmed that a higher proportion of tumor cells in BRIC is linked with the real hub gene overexpression. The CD8+ T and CD4+ T immune cells are the core constituents of immunotherapy [55]. Earlier, Trojan et al. in their trial study successfully used CD8+ T immune and CD4+ T immune cells infiltration levels for the immunotherapy of LSCC patients [56]. In the current study, our results showed positive correlations among real hub genes (CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1) expressions at mRNA level and CD8+ T immune cells infiltration in BRIC. In addition to this, in the current study, we also explored the significant negative correlations among real hub genes (CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1) expressions at the mRNA level and CD4+ T immune cell infiltration in BRIC. Collectively, the observed correlations shed light on the new possible aspects of the real hub genes in BRIC tumorigenesis by regulating CD8+ T and CD4+ T immune cells. To the best of our knowledge, we are the first to explore such correlations among real hub genes (CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1) and CD8+ T and CD4+ T immune cells across BRIC.
By constructing a TFS-miRNA-real hub genes co-regulatory network, it was observed that one TF (E2F1) and one miRNA (miR-16-5p) target all six real hub genes for regulating their expressions. It is noted by previous studies that PVT1-miR-16-5p/VEGFA/VEGFR1/AKT TFS-miRNA-hub genes axis, and miR-216-5p-Cx43, miR-16-1-3p/PGK1 miRNA-hub genes axis are the critical modulators of colorectal cancer and BRIC [25,57,58]. In view of our results, we suggested that E2F1-has-miR-16-5p/CEP55/KIF2C/KIF20A/RRM2/AURKA/PRC1 TFS-miRNAsmRNA co-regulatory networks can also be used as novel therapeutic targets for treating BRIC.
Conclusion
This detailed, systematic study has led us to the identification of six real hub genes (CEP55, KIF2C, KIF20A, RRM2, AURKA, and PRC1), that may be utilized as a novel diagnostic and prognostic biomarkers, and therapeutic targets for the precise treatment of BRIC.
Acknowledgements
The authors extend their appreciation to the Researchers Supporting Project number (RSPD2023R725) King Saud University, Riyadh, Saudi Arabia.
Disclosure of conflict of interest
None.
Supporting Information
References
- 1.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D, Bray F. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer. 2013;49:1374–1403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 2.Rivenbark AG, O’Connor SM, Coleman WB. Molecular and cellular heterogeneity in breast cancer: challenges for personalized medicine. Am J Pathol. 2013;183:1113–1124. doi: 10.1016/j.ajpath.2013.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen YJ, Guo YN, Shi K, Huang HM, Huang SP, Xu WQ, Li ZY, Wei KL, Gan TQ, Chen G. Down-regulation of microRNA-144-3p and its clinical value in non-small cell lung cancer: a comprehensive analysis based on microarray, miRNA-sequencing, and quantitative real-time PCR data. Respir Res. 2019;20:48. doi: 10.1186/s12931-019-0994-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, Chiao PJ, Huang P, Xie D, Li YH, Ju HQ, Xu RH. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7:4836–4849. doi: 10.7150/thno.20942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barrett T, Edgar R. Mining microarray data at NCBI’s gene expression omnibus (GEO)*. Methods Mol Biol. 2006;338:175–190. doi: 10.1385/1-59745-097-9:175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu Y, Gu HY, Zhu J, Niu YM, Zhang C, Guo GL. Identification of hub genes and key pathways associated with bipolar disorder based on weighted gene co-expression network analysis. Front Physiol. 2019;10:1081. doi: 10.3389/fphys.2019.01081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sherman BT, Hao M, Qiu J, Jiao X, Baseler MW, Lane HC, Imamichi T, Chang W. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update) Nucleic Acid Res. 2022;50:W216–W221. doi: 10.1093/nar/gkac194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607–D613. doi: 10.1093/nar/gky1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pan X, Chen S, Chen X, Ren Q, Yue L, Niu S, Li Z, Zhu R, Chen X, Jia Z, Zhen R, Ban J. UTP14A, DKC1, DDX10, PinX1, and ESF1 modulate cardiac angiogenesis leading to obesity-induced cardiac injury. J Diabetes Res. 2022;2022:2923291. doi: 10.1155/2022/2923291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45:W98–W102. doi: 10.1093/nar/gkx247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jézéquel P, Campone M, Gouraud W, Guérin-Charbonnel C, Leux C, Ricolleau G, Campion L. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat. 2012;131:765–775. doi: 10.1007/s10549-011-1457-7. [DOI] [PubMed] [Google Scholar]
- 13.Park SJ, Yoon BH, Kim SK, Kim SY. GENT2: an updated gene expression database for normal and tumor tissues. BMC Med Genomics. 2019;12(Suppl 5):101. doi: 10.1186/s12920-019-0514-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chandrashekar DS, Bashel B, Balasubramanya SAH, Creighton CJ, Ponce-Rodriguez I, Chakravarthi B, Varambally S. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19:649–658. doi: 10.1016/j.neo.2017.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch A, De Meyer T, Jeschke J, Van Criekinge W. MEXPRESS: visualizing expression, DNA methylation and clinical TCGA data. BMC Genomics. 2015;16:636. doi: 10.1186/s12864-015-1847-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, Cerami E, Sander C, Schultz N. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6:pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Szász AM, Lánczky A, Nagy Á, Förster S, Hark K, Green JE, Boussioutas A, Busuttil R, Szabó A, Győrffy B. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget. 2016;7:49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48:W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, Yang S, Kim CY, Lee M, Kim E, Lee S, Kang B, Jeong D, Kim Y, Jeon HN, Jung H, Nam S, Chung M, Kim JH, Lee I. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46:D380–D386. doi: 10.1093/nar/gkx1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nagy Á, Győrffy B. muTarget: a platform linking gene expression changes and mutation status in solid tumors. Int J Cancer. 2021;148:502–511. doi: 10.1002/ijc.33283. [DOI] [PubMed] [Google Scholar]
- 21.Mattingly CJ, Colby GT, Forrest JN, Boyer JL. The comparative toxicogenomics database (CTD) Environ Health Perspect. 2003;111:793–795. doi: 10.1289/ehp.6028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tong D, Schneeberger C, Leodolter S, Zeillinger R. Quantitative determination of gene expression by competitive reverse transcription-polymerase chain reaction in degraded RNA samples. Anal Biochem. 1997;251:173–177. doi: 10.1006/abio.1997.2280. [DOI] [PubMed] [Google Scholar]
- 23.Kim HY. Statistical notes for clinical researchers: Chi-squared test and fisher’s exact test. Restor Dent Endod. 2017;42:152–155. doi: 10.5395/rde.2017.42.2.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurkjian C, Kummar S, Murgo AJ. DNA methylation: its role in cancer development and therapy. Curr Probl Cancer. 2008;32:187–235. doi: 10.1016/j.currproblcancer.2008.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu H, Wei M, Jiang X, Tan J, Xu W, Fan X, Zhang R, Ding C, Zhao F, Shao X, Zhang Z, Shi R, Zhang W, Wu G. lncRNA PVT1 promotes tumorigenesis of colorectal cancer by stabilizing miR-16-5p and interacting with the VEGFA/VEGFR1/AKT axis. Mol Ther Nucleic Acids. 2020;20:438–450. doi: 10.1016/j.omtn.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fabbro M, Zhou BB, Takahashi M, Sarcevic B, Lal P, Graham ME, Gabrielli BG, Robinson PJ, Nigg EA, Ono Y, Khanna KK. Cdk1/Erk2-and Plk1-dependent phosphorylation of a centrosome protein, Cep55, is required for its recruitment to midbody and cytokinesis. Dev Cell. 2005;9:477–488. doi: 10.1016/j.devcel.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 27.Liu L, Mei Q, Zhao J, Dai Y, Fu Q. Suppression of CEP55 reduces cell viability and induces apoptosis in human lung cancer. Oncol Rep. 2016;36:1939–1945. doi: 10.3892/or.2016.5059. [DOI] [PubMed] [Google Scholar]
- 28.Kalimutho M, Sinha D, Jeffery J, Nones K, Srihari S, Fernando WC, Duijf PH, Vennin C, Raninga P, Nanayakkara D, Mittal D, Saunus JM, Lakhani SR, López JA, Spring KJ, Timpson P, Gabrielli B, Waddell N, Khanna KK. CEP 55 is a determinant of cell fate during perturbed mitosis in breast cancer. EMBO Mol Med. 2018;10:e8566. doi: 10.15252/emmm.201708566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qi J, Liu G, Wang F. High levels of centrosomal protein 55 expression is associated with poor clinical prognosis in patients with cervical cancer. Oncol Lett. 2018;15:9347–9352. doi: 10.3892/ol.2018.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu L, Xia C, Sheng F, Sun Q, Xiong J, Wang S. CEP55 promotes the proliferation and invasion of tumour cells via the AKT signalling pathway in osteosarcoma. Carcinogenesis. 2018;39:623–631. doi: 10.1093/carcin/bgy017. [DOI] [PubMed] [Google Scholar]
- 31.Wang Y, Jin T, Dai X, Xu J. Lentivirus-mediated knockdown of CEP55 suppresses cell proliferation of breast cancer cells. Biosci Trends. 2016;10:67–73. doi: 10.5582/bst.2016.01010. [DOI] [PubMed] [Google Scholar]
- 32.Xu ZY, Ma XS, Qi ST, Wang ZB, Guo L, Schatten H, Sun QY, Sun YP. Cep55 regulates spindle organization and cell cycle progression in meiotic oocyte. Sci Rep. 2015;5:16978. doi: 10.1038/srep16978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ritter A, Kreis NN, Louwen F, Wordeman L, Yuan J. Molecular insight into the regulation and function of MCAK. Crit Rev Biochem Mol Biol. 2015;51:228–245. doi: 10.1080/10409238.2016.1178705. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura Y, Tanaka F, Haraguchi N, Mimori K, Matsumoto T, Inoue H, Yanaga K, Mori M. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. Br J Cancer. 2007;97:543–549. doi: 10.1038/sj.bjc.6603905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wei S, Dai M, Zhang C, Teng K, Wang F, Li H, Sun W, Feng Z, Kang T, Guan X, Xu R, Cai M, Xie D. KIF2C: a novel link between Wnt/β-catenin and mTORC1 signaling in the pathogenesis of hepatocellular carcinoma. Protein Cell. 2021;12:788–809. doi: 10.1007/s13238-020-00766-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Abdel-Fatah TMA, Green AR, Lemetre C, Moseley P, Chan S, Ellis IO, Balls G. P4-09-11: kinesin family member 2C (KIF2C) is a new surrogate prognostic marker in breast cancer (BC) Cancer Res. 2011;71:P4-09-11. [Google Scholar]
- 37.Taniuchi K, Furihata M, Saibara T. KIF20A-mediated RNA granule transport system promotes the invasiveness of pancreatic cancer cells. Neoplasia. 2014;16:1082–1093. doi: 10.1016/j.neo.2014.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lu Y, Liu P, Wen W, Grubbs CJ, Townsend RR, Malone JP, Lubet RA, You M. Cross-species comparison of orthologous gene expression in human bladder cancer and carcinogen-induced rodent models. Am J Transl Res. 2010;3:8–27. [PMC free article] [PubMed] [Google Scholar]
- 39.Claerhout S, Lim JY, Choi W, Park YY, Kim K, Kim SB, Lee JS, Mills GB, Cho JY. Gene expression signature analysis identifies vorinostat as a candidate therapy for gastric cancer. PLoS One. 2011;6:e24662. doi: 10.1371/journal.pone.0024662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tomita Y, Yuno A, Tsukamoto H, Senju S, Kuroda Y, Hirayama M, Irie A, Kawahara K, Yatsuda J, Hamada A. Identification of promiscuous KIF20A long peptides bearing both CD4+ and CD8+ T-cell epitopes: KIF20A-specific CD4+ T-cell immunity in patients with malignant tumor. Clin Cancer Res. 2013;19:4508–4520. doi: 10.1158/1078-0432.CCR-13-0197. [DOI] [PubMed] [Google Scholar]
- 41.Yew P, Alachkar H, Yamaguchi R, Kiyotani K, Fang H, Yap K, Liu H, Wickrema A, Artz A, Van Besien K, Imoto S, Miyano S, Bishop MR, Stock W, Nakamura Y. Quantitative characterization of T-cell repertoire in allogeneic hematopoietic stem cell transplant recipients. Bone Marrow Transplant. 2015;50:1227–1234. doi: 10.1038/bmt.2015.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamashita J, Fukushima S, Jinnin M, Honda N, Makino K, Sakai K, Masuguchi S, Inoue Y, Ihn H. Kinesin family member 20A is a novel melanoma-associated antigen. Acta Derm Venereol. 2012;92:593–597. doi: 10.2340/00015555-1416. [DOI] [PubMed] [Google Scholar]
- 43.Wonsey DR, Follettie MT. Loss of the forkhead transcription factor FoxM1 causes centrosome amplification and mitotic catastrophe. Cancer Res. 2005;65:5181–5189. doi: 10.1158/0008-5472.CAN-04-4059. [DOI] [PubMed] [Google Scholar]
- 44.Imai K, Hirata S, Irie A, Senju S, Ikuta Y, Yokomine K, Harao M, Inoue M, Tomita Y, Tsunoda T, Nakagawa H, Nakamura Y, Baba H, Nishimura Y. Identification of HLA-A2-restricted CTL epitopes of a novel tumour-associated antigen, KIF20A, overexpressed in pancreatic cancer. Br J Cancer. 2011;104:300–307. doi: 10.1038/sj.bjc.6606052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Torrents E. Ribonucleotide reductases: essential enzymes for bacterial life. Front Cell Infect Microbiol. 2014;4:52. doi: 10.3389/fcimb.2014.00052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang H, Liu X, Warden CD, Huang Y, Loera S, Xue L, Zhang S, Chu P, Zheng S, Yen Y. Prognostic and therapeutic significance of ribonucleotide reductase small subunit M2 in estrogen-negative breast cancers. BMC Cancer. 2014;14:664. doi: 10.1186/1471-2407-14-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang CC, Lin CC, Wang CH, Huang CC, Ke TW, Wei PL, Yeh KT, Hsu KC, Hsu NY, Cheng YW. miR-211 regulates the expression of RRM2 in tumoral metastasis and recurrence in colorectal cancer patients with a k-ras gene mutation. Oncology Lett. 2018;15:8107–8117. doi: 10.3892/ol.2018.8295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang WH, Li N, Yuan ZQ, Qian XL, Wang ZH. DSCAM-AS1 promotes tumor growth of breast cancer by reducing miR-204-5p and up-regulating RRM2. Mol Carcinog. 2019;58:461–473. doi: 10.1002/mc.22941. [DOI] [PubMed] [Google Scholar]
- 49.Du R, Huang C, Liu K, Li X, Dong Z. Targeting AURKA in cancer: molecular mechanisms and opportunities for cancer therapy. Mol Cancer. 2021;20:15. doi: 10.1186/s12943-020-01305-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Twu NF, Yuan CC, Yen MS, Lai CR, Chao KC, Wang PH, Wu HH, Chen YJ. Expression of aurora kinase A and B in normal and malignant cervical tissue: high aurora A kinase expression in squamous cervical cancer. Eur J Obstet Gynecol Reprod Biol. 2009;142:57–63. doi: 10.1016/j.ejogrb.2008.09.012. [DOI] [PubMed] [Google Scholar]
- 51.Subramanian R, Wilson-Kubalek EM, Arthur CP, Bick MJ, Campbell EA, Darst SA, Milligan RA, Kapoor TM. Insights into antiparallel microtubule crosslinking by PRC1, a conserved nonmotor microtubule binding protein. Cell. 2010;142:433–443. doi: 10.1016/j.cell.2010.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kanehira M, Katagiri T, Shimo A, Takata R, Shuin T, Miki T, Fujioka T, Nakamura Y. Oncogenic role of MPHOSPH1, a cancer-testis antigen specific to human bladder cancer. Cancer Res. 2007;67:3276–3285. doi: 10.1158/0008-5472.CAN-06-3748. [DOI] [PubMed] [Google Scholar]
- 53.Wang SM, Ooi LLP, Hui KM. Upregulation of Rac GTPase-activating protein 1 is significantly associated with the early recurrence of human hepatocellular carcinoma. Clin Cancer Res. 2011;17:6040–6051. doi: 10.1158/1078-0432.CCR-11-0557. [DOI] [PubMed] [Google Scholar]
- 54.Chen J, Rajasekaran M, Xia H, Zhang X, Kong SN, Sekar K, Seshachalam VP, Deivasigamani A, Goh BK, Ooi LL, Hong W, Hui KM. The microtubule-associated protein PRC1 promotes early recurrence of hepatocellular carcinoma in association with the Wnt/β-catenin signalling pathway. Gut. 2016;65:1522–1534. doi: 10.1136/gutjnl-2015-310625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xia A, Zhang Y, Xu J, Yin T, Lu XJ. T cell dysfunction in cancer immunity and immunotherapy. Front Immunol. 2019;10:1719. doi: 10.3389/fimmu.2019.01719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Trojan A, Urosevic M, Dummer R, Giger R, Weder W, Stahel RA. Immune activation status of CD8+ T cells infiltrating non-small cell lung cancer. Lung Cancer. 2004;44:143–147. doi: 10.1016/j.lungcan.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 57.Xia C, Jiang H, Ye F, Zhuang Z. The multifunction of miR-218-5p-Cx43 axis in breast cancer. OncoTargets Ther. 2019;12:8319–8328. doi: 10.2147/OTT.S218524. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 58.Ye T, Liang Y, Zhang D, Zhang X. MicroRNA-16-1-3p represses breast tumor growth and metastasis by inhibiting PGK1-mediated warburg effect. Front Cell Dev Biol. 2020;8:615154. doi: 10.3389/fcell.2020.615154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fang E, Zhang X. Identification of breast cancer hub genes and analysis of prognostic values using integrated bioinformatics analysis. Cancer Biomark. 2017;21:373–381. doi: 10.3233/CBM-170550. [DOI] [PubMed] [Google Scholar]
- 60.Qiu J, Du Z, Wang Y, Zhou Y, Zhang Y, Xie Y, Lv Q. Weighted gene co-expression network analysis reveals modules and hub genes associated with the development of breast cancer. Medicine (Baltimore) 2019;98:e14345. doi: 10.1097/MD.0000000000014345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cai Y, Mei J, Xiao Z, Xu B, Jiang X, Zhang Y, Zhu Y. Identification of five hub genes as monitoring biomarkers for breast cancer metastasis in silico. Hereditas. 2019;156:20. doi: 10.1186/s41065-019-0096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liu F, Wu Y, Mi Y, Gu L, Sang M, Geng C. Identification of core genes and potential molecular mechanisms in breast cancer using bioinformatics analysis. Pathol Res Pract. 2019;215:152436. doi: 10.1016/j.prp.2019.152436. [DOI] [PubMed] [Google Scholar]
- 63.Lu X, Gao C, Liu C, Zhuang J, Su P, Li H, Wang X, Sun C. Identification of the key pathways and genes involved in HER2-positive breast cancer with brain metastasis. Pathol Res Pract. 2019;215:152475. doi: 10.1016/j.prp.2019.152475. [DOI] [PubMed] [Google Scholar]
- 64.Fu Y, Zhou QZ, Zhang XL, Wang ZZ, Wang P. Identification of hub genes using co-expression network analysis in breast cancer as a tool to predict different stages. Med Sci Monit. 2019;25:8873–8890. doi: 10.12659/MSM.919046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shao N, Yuan K, Zhang Y, Yun Cheang T, Li J, Lin Y. Identification of key candidate genes, pathways and related prognostic values in ER-negative/HER2-negative breast cancer by bioinformatics analysis. J BUON. 2018;23:891–901. [PubMed] [Google Scholar]
- 66.Zhou Q, Ren J, Hou J, Wang G, Ju L, Xiao Y, Gong Y. Co-expression network analysis identified candidate biomarkers in association with progression and prognosis of breast cancer. J Cancer Res Clin Oncol. 2019;145:2383–2396. doi: 10.1007/s00432-019-02974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vastrad B, Vastrad C, Tengli A, Iliger S. Identification of differentially expressed genes regulated by molecular signature in breast cancer-associated fibroblasts by bioinformatics analysis. Arch Gynecol Obstet. 2018;297:161–183. doi: 10.1007/s00404-017-4562-y. [DOI] [PubMed] [Google Scholar]
- 68.Chai F, Liang Y, Zhang F, Wang M, Zhong L, Jiang J. Systematically identify key genes in inflammatory and non-inflammatory breast cancer. Gene. 2016;575:600–614. doi: 10.1016/j.gene.2015.09.025. [DOI] [PubMed] [Google Scholar]
- 69.Wang Y, Zhang Y, Huang Q, Li C. Integrated bioinformatics analysis reveals key candidate genes and pathways in breast cancer. Mol Med Rep. 2018;17:8091–8100. doi: 10.3892/mmr.2018.8895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang Y, Xu H, Zhu B, Qiu Z, Lin Z. Systematic identification of the key candidate genes in breast cancer stroma. Cell Mol Biol Lett. 2018;23:44. doi: 10.1186/s11658-018-0110-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Peng C, Ma W, Xia W, Zheng W. Integrated analysis of differentially expressed genes and pathways in triple-negative breast cancer. Mol Med Rep. 2017;15:1087–1094. doi: 10.3892/mmr.2017.6101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zheng T, Wang A, Hu D, Wang Y. Molecular mechanisms of breast cancer metastasis by gene expression profile analysis. Mol Med Rep. 2017;16:4671–4677. doi: 10.3892/mmr.2017.7157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Liu Z, Liang G, Tan L, Su AN, Jiang W, Gong C. High-efficient screening method for identification of key genes in breast cancer through microarray and bioinformatics. Anticancer Res. 2017;37:4329–4335. doi: 10.21873/anticanres.11826. [DOI] [PubMed] [Google Scholar]
- 74.Wang YW, Zhang W, Ma R. Bioinformatic identification of chemoresistance-associated microRNAs in breast cancer based on microarray data. Oncology Rep. 2018;39:1003–1010. doi: 10.3892/or.2018.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen J, Liu C, Cen J, Liang T, Xue J, Zeng H, Zhang Z, Xu G, Yu C, Lu Z, Wang Z, Jiang J, Zhan X, Zeng J. KEGG-expressed genes and pathways in triple negative breast cancer: protocol for a systematic review and data mining. Medicine (Baltimore) 2020;99:e19986. doi: 10.1097/MD.0000000000019986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lin Y, Fu F, Lv J, Wang M, Li Y, Zhang J, Wang C. Identification of potential key genes for HER-2 positive breast cancer based on bioinformatics analysis. Medicine (Baltimore) 2020;99:e18445. doi: 10.1097/MD.0000000000018445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wu JR, Zhao Y, Zhou XP, Qin X. Estrogen receptor 1 and progesterone receptor are distinct biomarkers and prognostic factors in estrogen receptor-positive breast cancer: evidence from a bioinformatic analysis. Biomed Pharmacother. 2020;121:109647. doi: 10.1016/j.biopha.2019.109647. [DOI] [PubMed] [Google Scholar]
- 78.Zhou Q, Sun E, Ling L, Liu X, Zhang M, Yin H, Lu C. Bioinformatic analysis of computational identified differentially expressed genes in tumor stoma of pregnancy-associated breast cancer. Mol Med Rep. 2017;16:3345–3350. doi: 10.3892/mmr.2017.6947. [DOI] [PubMed] [Google Scholar]
- 79.He L, Wang D, Wei N, Guo Z. Integrated bioinformatics approach reveals crosstalk between tumor stroma and peripheral blood mononuclear cells in breast cancer. Asian Pac J Cancer Prev. 2016;17:1003–1008. doi: 10.7314/apjcp.2016.17.3.1003. [DOI] [PubMed] [Google Scholar]
- 80.Zhang BH, Liu J, Zhou QX, Zuo D, Wang Y. Analysis of differentially expressed genes in ductal carcinoma with DNA microarray. Eur Rev Med Pharmacol Sci. 2013;17:758–766. [PubMed] [Google Scholar]
- 81.Deng JL, Xu YH, Wang G. Identification of potential crucial genes and key pathways in breast cancer using bioinformatic analysis. Front Genet. 2019;10:695. doi: 10.3389/fgene.2019.00695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Jin H, Huang X, Shao K, Li G, Wang J, Yang H, Hou Y. Integrated bioinformatics analysis to identify 15 hub genes in breast cancer. Oncology Lett. 2019;18:1023–1034. doi: 10.3892/ol.2019.10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.