Abstract
Purpose
Neovascular age-related macular degeneration (nAMD) shows variable treatment response to intravitreal anti-VEGF. This analysis compared the potential of different artificial intelligence (AI)-based machine learning models using OCT and clinical variables to accurately predict at baseline the best-corrected visual acuity (BCVA) at 9 months in response to ranibizumab in patients with nAMD.
Design
Retrospective analysis.
Participants
Baseline and imaging data from patients with subfoveal choroidal neovascularization secondary to age-related macular dengeration.
Methods
Baseline data from 502 study eyes from the HARBOR (NCT00891735) prospective clinical trial (monthly ranibizumab 0.5 and 2.0 mg arms) were pooled; 432 baseline OCT volume scans were included in the analysis. Seven models, based on baseline quantitative OCT features (Least absolute shrinkage and selection operator [Lasso] OCT minimum [min], Lasso OCT 1 standard error [SE]); on quantitative OCT features and clinical variables at baseline (Lasso min, Lasso 1SE, CatBoost, RF [random forest]); or on baseline OCT images only (deep learning [DL] model), were systematically compared with a benchmark linear model of baseline age and BCVA. Quantitative OCT features were derived by a DL segmentation model on the volume images, including retinal layer volumes and thicknesses, and retinal fluid biomarkers, including statistics on fluid volume and distribution.
Main Outcome Measures
Prognostic ability of the models was evaluated using coefficient of determination (R2) and median absolute error (MAE; letters).
Results
In the first cross-validation split, mean R2 (MAE) of the Lasso min, Lasso 1SE, CatBoost, and RF models was 0.46 (7.87), 0.42 (8.43), 0.45 (7.75), and 0.43 (7.60), respectively. These models ranked higher than or similar to the benchmark model (mean R2, 0.41; mean MAE, 8.20 letters) and better than OCT-only models (mean R2: Lasso OCT min, 0.20; Lasso OCT 1SE, 0.16; DL, 0.34). The Lasso min model was selected for detailed analysis; mean R2 (MAE) of the Lasso min and benchmark models for 1000 repeated cross-validation splits were 0.46 (7.7) and 0.42 (8.0), respectively.
Conclusions
Machine learning models based on AI-segmented OCT features and clinical variables at baseline may predict future response to ranibizumab treatment in patients with nAMD. However, further developments will be needed to realize the clinical utility of such AI-based tools.
Financial Disclosure(s)
Proprietary or commercial disclosure may be found after the references.
Keywords: Machine learning, Neovascular age-related macular degenration, Optical coherence tomography, Prediction, Ranibizumab
Intravitreal anti-VEGF therapy can lead to significant improvement in vision and quality of life in patients with neovascular age-related macular degeneration (nAMD).1 However, variable response to anti-VEGF therapy is observed and visual gains may vary across patients with nAMD.
To address the variable clinical response and significant treatment burden experienced with anti-VEGF therapies, several new therapeutic options, such as long-acting delivery technologies, molecules with new mechanisms of action, or gene therapy, are now either available or under clinical development.2, 3, 4 Although exciting, the treatment landscape and clinical management for patients with nAMD is expected to become increasingly complex.
Artificial intelligence (AI) has the potential to benefit both clinical research and clinical practice in the field of retina, predicting functional or anatomical outcomes and supporting the choice for individualized treatment regimens, eventually enabling better patient outcomes. In clinical research, AI can improve clinical trial design and execution, automate or standardize retinal image grading, or assist in the interpretation of complex study results. In clinical practice, AI may be able to facilitate efficient retinal image analysis, quantify pathologic features, predict future outcomes, and empower health care providers to make treatment decisions informed by complex clinical data that cannot be readily analyzed through currently available technologies.5 The first step in realizing future potential of AI-based models is to identify the most suitable models for further development.
To date in nAMD, AI has mostly been used to aid in OCT image analysis, with most models using the HARBOR clinical trial and Moorfields Eye Hospital datasets5; AI has also been applied to segmentation and disease classification.6,7 Studies have used AI in a bid to predict response to anti-VEGF treatments. Using OCT images, Feng et al8 showed that the ResNet-50 convolutional neural network was suitable for predicting the effectiveness of anti-VEGF treatment at 21 days after first injection in patients with choroidal neovascularization or cystoid macular edema. Similarly, another study used paired OCT B-scan images plus fluorescein angiography and indocyanine green angiography, and used a network based on a conditional generative adversarial network to predict the presence of anatomical features at 1 month after 3 loading doses of anti-VEGF therapy.9 However, these studies solely focused on anatomical improvements, which may not necessarily correlate with visual functional improvements,10 and were limited by a relatively short time for the predictions. Fu et al11 applied a deep learning (DL) segmentation algorithm to show that models that included both quantitative OCT biomarkers from automatically segmented OCT scans and functional (visual acuity) changes could predict vision outcomes at 3 and 12 months after anti-VEGF therapy, supporting a rationale in using both functional and anatomical features to predict treatment response. It is well known that predicting future best-corrected visual acuity (BCVA) from current morphology is a challenging problem.11 Given the speed of innovation, though, a lot of potential may remain still to be realized, including the development of improved image analysis methods on higher resolution imaging modalities.
In the current study, various machine learning and DL models based on OCT and clinical variables were compared with regard to their ability to accurately predict, at baseline, the functional response to ranibizumab at 9 months in patients with nAMD receiving monthly treatment in a clinical trial setting.
Methods
Source of Dataset
This is a retrospective analysis of OCT data from the HARBOR clinical trial (ClinicalTrials.gov identifier, NCT00891735). The HARBOR trial was a 24-month, phase III, prospective, multicenter, randomized, double-masked, active treatment-controlled clinical trial, with primary results previously published.12,13 In summary, 1097 patients with treatment-naive subfoveal choroidal neovascularization secondary to age-related macular degeneration were randomized to receive ranibizumab intravitreal injections administered monthly or as needed (pro re nata) after 3 monthly loading doses.
For this analysis, data from the monthly ranibizumab 0.5 and 2.0 mg arms were pooled. Study eyes of patients randomized to these arms in the HARBOR trial were included if baseline images were available and in the appropriate format for inclusion in the DL model and if BCVA was recorded for months 8 and 9. For OCT images, the Cirrus HD-OCT III instrument (Carl Zeiss Meditec, Inc) was used, featuring 512 × 128 × 1024 voxels with dimensions of 11.7 × 47.2 × 2.0 μm3 per voxel and covering a total volume of approximately 6 × 6 × 2 mm3. The clinical variables included were baseline age and BCVA.
The trial was conducted in compliance with the principles of the Declaration of Helsinki and the Health Insurance Portability and Accountability Act. This article includes analyses of data from a historical study conducted in patients with nAMD (HARBOR clinical trial, ClinicalTrials.gov identifier, NCT00891735); Institutional Review Board approvals were obtained prior to the HARBOR study initiation.12, 13 Written informed consent was obtained from patients for their data to be used in future medical research and analyses.
Outcome Variables and Fold Definitions
Study eye BCVA of patients randomized to the monthly ranibizumab arms was used as the outcome variable. Because psychophysical tests such as BCVA measurement are found to be highly variable over time, to reduce intrapatient variability, the average of BCVA scores from 2 visits, months 8 and 9, was defined as the outcome of interest. Best-corrected visual acuity at month 9 was selected as the outcome of interest in this analysis, since it is the latest timepoint available across all datasets that are currently available to the authors for potential future validation of the models. Further, as observed in the HARBOR trial,12,13 BCVA gains at these timepoints are similar, with no large variations from month 9 to month 12.
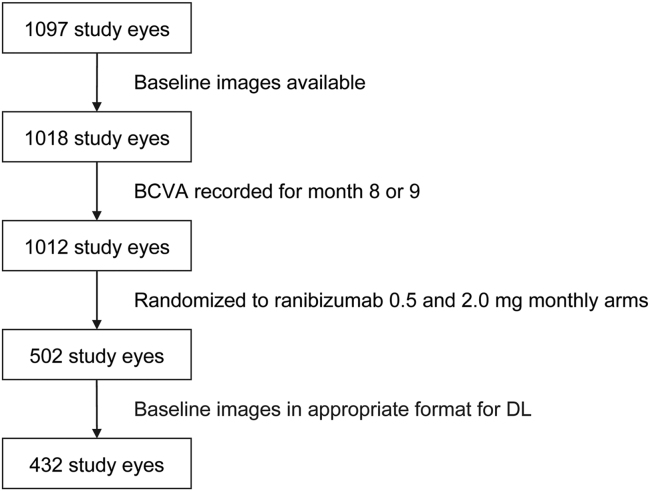
Of the total patient population from the HARBOR trial, 502 study eyes were available for model training. However, only a subset of 432 study eyes were deemed appropriate for DL predictions due to the input image processing pipeline relying on a device-provided retinal pigment epithelium segmentation that was either unavailable or of a poor quality that would result in significant cropping errors14 (Fig 1).
Figure 1.
Diagram showing patient flow. BCVA = best-corrected visual acuity; DL = deep learning.
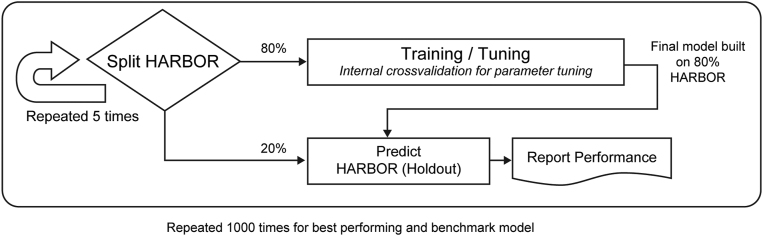
The data were split 1000 times into 5-fold cross-validation (CV), making sure the treatment arms were stratified across folds. To limit computational effort, the first split was used to compare all models. The best performing and benchmark models were further evaluated on the remaining splits (Fig 2). A visual assessment further confirmed that the first data split was balanced across folds for baseline BCVA and age. Therefore, the first CV split was evaluated on the subset of 432 study eyes, but further assessment not involving the DL model was carried out on the full subset of 502 study eyes.
Figure 2.
Diagram showing model training and validation scheme.
OCT Segmentation Model
A pretrained OCT segmentation model was used as previously described15 using the U-Net16 architecture (Appendix S1; available at www.ophthalmologyscience.org). It was trained with annotations by certified graders from the Liverpool Ophthalmology Reading Center and can segment 4 retinal pathology biomarkers (intraretinal fluid [IRF], subretinal fluid [SRF], pigment epithelium detachment, and subretinal hyper-reflective material), along with 6 retinal layers (internal limiting membrane, interface between outer plexiform layer and Henle’s fiber layer, external limiting membrane, inner and outer boundary of retinal pigment epithelium, and Bruch’s membrane) in a fully automated fashion.
In total, 105 quantitative features were automatically extracted from these OCT segmentations in 3 topographic locations (0.5 mm, 1.5 mm, and 3.0 mm radius circle). Specifically, 36 volumetric retinal pathology-related features (4 retinal pathology biomarkers × 3 biomarker measurements × 3 topographic locations), 54 retinal layer-related thickness features (6 pairs of layers × 3 biomarker measurements × 3 topographic locations), and 15 retinal layer-related volumetric features (5 layer combinations × 3 topographic locations) were extracted. (Table S1; available at www.ophthalmologyscience.org). Biomarker measurements include width, height, and volume of the retinal pathology features, as well as minimum, maximum, and average thickness of the space between 2 retinal layers.
Machine Learning and DL Models Versus Benchmark Model
The machine learning models analyzed herein are least absolute shrinkage and selection operator (Lasso) OCT minimum (min), Lasso OCT 1 standard error (SE), Lasso min, Lasso 1SE, CatBoost, and random forest (RF). Least absolute shrinkage and selection operator17 was used for fitting linear regression models with various configurations of OCT features. The degree of regularization was tuned such that the error on the training set was either at min or within 1 SE of the min using the parameter lambda. Least absolute shrinkage and selection operator min corresponds to the value of lambda that gives min mean CV error, whereas Lasso 1SE is the largest value of lambda such that the error is within 1 SE of the CV errors for Lasso min, yielding a more regularized model.
CatBoost, a gradient boosting model, was used for fitting regression models using all baseline OCT and clinical features (age and BCVA). Two parameters (number of iterations and L2 leaf regression) were tuned on the training set. Learning rate and tree depth were optimized in a similar way, and the best parameter combination found (as measured by coefficient of determination [R2]) was used to build the model with the full training data. Similarly, RF was tuned for the number of variables randomly sampled as candidates at each split of a leaf node of a tree.
The DL model was first developed using 50 275 OCT B-scans from 1071 patients to predict BCVA at the concurrent visit using the ResNet-50 v2 convolutional neural network as previously described,14 with patients being split in the manner of nested 5-fold CV. After training for a single epoch to predict BCVA at the concurrent visit, model weights from each nested fold were then transferred to predict BCVA at month 9, using baseline images from study and fellow eyes in all arms by training for up to 1000 epochs. Using the weights from each nested fold from the epoch with the lowest validation loss, the models were further fine-tuned using only study eyes from patients in the monthly arms for up to an additional 5000 epochs. After identifying the epoch with the lowest validation loss from each nested fold, the models were then retrained to include the nested fold and stopped early at the same number of steps as determined by the nested training.
Least absolute shrinkage and selection operator OCT min, Lasso OCT 1SE, and the DL models were based on baseline OCT quantitative features or the OCT images only, whereas Lasso min, Lasso 1SE, CatBoost, and RF were based on baseline OCT quantitative features and baseline clinical variables (age and BCVA).
In addition, a linear regression model including the baseline clinical variables of age and BCVA, which have previously been associated with prognostic utility,18 was fitted and used as a benchmark to compare other models systematically.
Statistical Analysis
The prognostic ability of the models was evaluated using R2 and median absolute error (MAE; in letters).
Results
Both the full subset of 502 study eyes and the subset of 432 study eyes used for the first CV split were comparable in terms of baseline characteristics and treatment outcomes (Table S2; available at www.ophthalmologyscience.org).
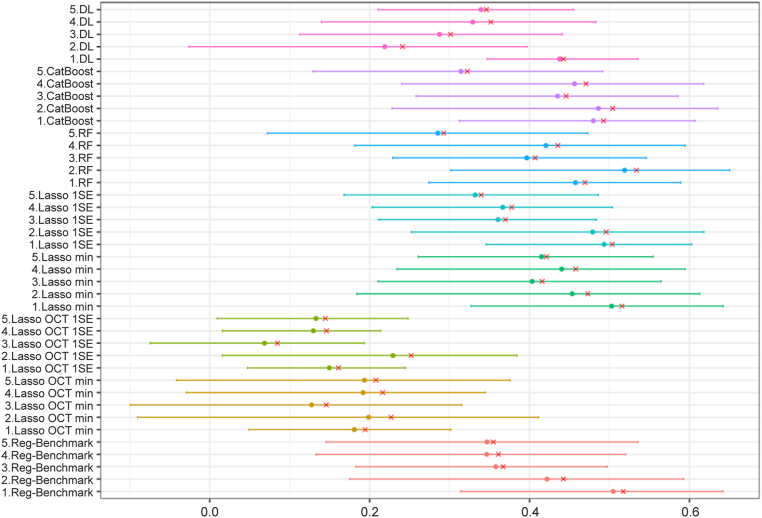
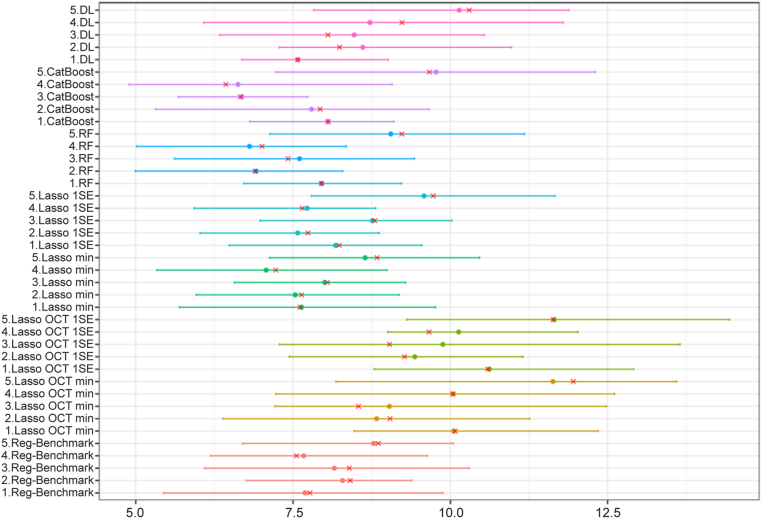
Table 1 includes the mean R2 for all models on the first CV split. Compared with the benchmark model (mean R2 = 0.41), models without clinical variables (Lasso OCT min, Lasso OCT 1SE, and DL) had lower performance, with mean R2 of 0.20, 0.16, and 0.34, respectively. The DL model demonstrated improved performance compared with Lasso OCT features only. Models based on OCT segmentation and clinical variables (Lasso min, Lasso 1SE, CatBoost, and RF) had mean R2 of 0.46, 0.42, 0.45, and 0.43, respectively. Variability across folds can be relatively large. Bootstrapped R2 by CV folds are presented in Figure 3. Median absolute error, which uses the same scale as the predictions, provides a more interpretable metric. Model selection was determined by R2 results; thus, Lasso min was chosen as the optimal model to further explore. Bootstrapped MAE values by CV folds are presented in Figure 4. Of note, using MAE as a metric (Table 2), the DL model seems to perform better compared with the benchmark model, with the RF model being ranked highest.
Table 1.
Coefficient of Determination of Models on First Cross-Validation Split
| Fold | Benchmark | Lasso OCT Min | Lasso OCT 1SE | Lasso Min | Lasso 1SE | RF | CatBoost | DL |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.52 | 0.19 | 0.16 | 0.52 | 0.50 | 0.47 | 0.49 | 0.44 |
| 2 | 0.44 | 0.23 | 0.25 | 0.47 | 0.50 | 0.53 | 0.50 | 0.24 |
| 3 | 0.37 | 0.15 | 0.08 | 0.42 | 0.37 | 0.41 | 0.45 | 0.30 |
| 4 | 0.36 | 0.22 | 0.15 | 0.46 | 0.38 | 0.44 | 0.47 | 0.35 |
| 5 | 0.35 | 0.21 | 0.14 | 0.42 | 0.34 | 0.29 | 0.32 | 0.35 |
| Mean | 0.41 | 0.20 | 0.16 | 0.46 | 0.42 | 0.43 | 0.45 | 0.34 |
DL = deep learning; Lasso = least absolute shrinkage and selection operator; Min = minimun; RF = random forest; SE = standard error.
Figure 3.
Graph showing bootstrapped coefficient of determination by cross-validation fold. Shown is 1000 bootstrap of the predictions of each fold. Red cross is the point estimate of the single data split. DL = deep learning; Lasso = least absolute shrinkage and selection operator; min = minimum; RF = random forest; SE = standard error.
Figure 4.
Graph showing bootstrapped median absolute error by cross-validation fold. Shown is 1000 bootstrap of the predictions of each fold. Red cross is the point estimate of the original data. DL = deep learning; Lasso = least absolute shrinkage and selection operator; min = minimum; RF = random forest; SE = standard error.
Table 2.
Median Absolute Error (Letters) of Models on First Cross-Validation Split
| Fold | Benchmark | Lasso OCT Min | Lasso OCT 1SE | Lasso Min | Lasso 1SE | RF | CatBoost | DL |
|---|---|---|---|---|---|---|---|---|
| 1 | 7.77 | 10.08 | 10.59 | 7.61 | 8.23 | 7.95 | 8.06 | 7.58 |
| 2 | 8.41 | 9.04 | 9.27 | 7.63 | 7.73 | 6.90 | 7.93 | 8.24 |
| 3 | 8.39 | 8.54 | 9.03 | 8.05 | 8.80 | 7.42 | 6.66 | 8.05 |
| 4 | 7.55 | 10.04 | 9.66 | 7.22 | 7.65 | 7.01 | 6.43 | 9.23 |
| 5 | 8.85 | 11.95 | 11.63 | 8.84 | 9.73 | 9.23 | 9.67 | 10.30 |
| Mean | 8.20 | 9.93 | 10.04 | 7.87 | 8.43 | 7.70 | 7.75 | 8.68 |
DL = deep learning; Lasso = least absolute shrinkage and selection operator; Min = minimum; RF = random forest; SE = standard error.
The calibration plots for the first CV (Figs S1 and S2; available at www.ophthalmologyscience.org) show that the DL model was less calibrated on 1 of 5 folds and had a narrower range of predictions, whereas all other models seemed to be well calibrated.
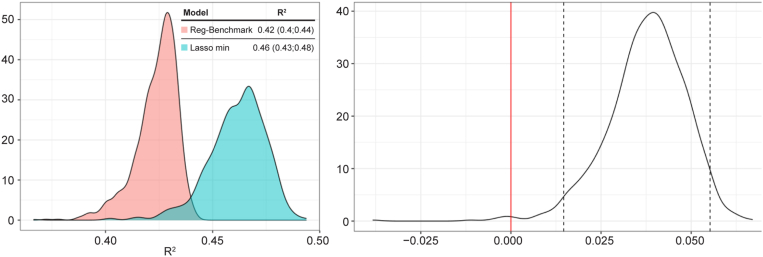
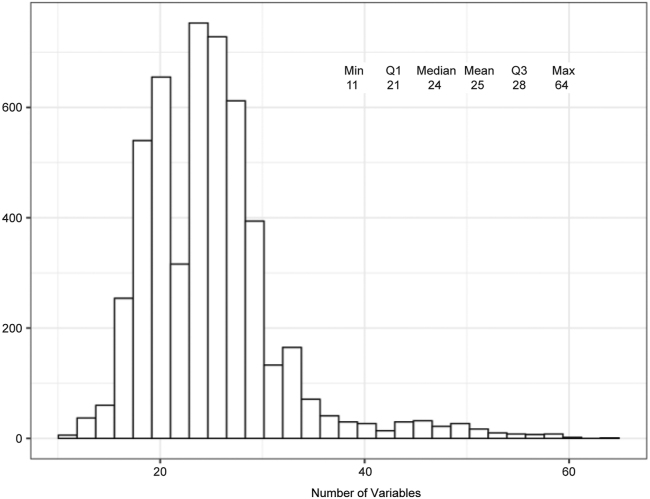
To confirm reproducibility, the experiment was repeated 1000 times for the benchmark model and Lasso min, and the mean R2 of Lasso min and the benchmark model were 0.46 and 0.42, respectively (Fig 5). Of note, the 95% confidence interval for mean difference did not include zero, indicating a statistically significant difference. On average, as shown in Figure 6, the Lasso min model included 25 variables. Table 3 lists the frequency of the variables used in the Lasso min model. Age and BCVA were always included in the model. Because of the highly correlated nature of these variables, no in-depth interpretation of the relative importance of OCT variables was made based on the frequency of the variables.
Figure 5.
Distribution of mean coefficient of determination (R2) and mean R2 difference for 1000 cross-validations. In the distribution of mean R2 difference between the least absolute shrinkage and selection operator (Lasso) minimum (min) and benchmark models, the dotted lines indicate the 2.5 and 97.5 quantiles. Zero does not belong to the 95% confidence interval for mean difference.
Figure 6.
Least absolute shrinkage and selection operator minimum (min) model size (1000 cross-validations). Max = maximum; Q = quartile.
Table 3.
Lasso Min Variable Frequency∗
| Frequency | Variable | Ranking |
|---|---|---|
| 100 | Age | 1 |
| 100 | Baseline BCVA | 1 |
| 100 | SRF max width in 3.0 mm radius | 1 |
| 100 | BM_to_IB-RPE min thickness in 1.5 mm radius | 1 |
| 99 | SHRM max width in 3.0 mm radius | 5 |
| 97 | BM_to_IB-RPE mean thickness in 3.0 mm radius | 6 |
| 96 | SRF_volume in 0.5 mm radius | 7 |
| 94 | PED max width in 3.0 mm radius | 8 |
| 94 | IB-RPE_to_ILM min thickness in 1.5 mm radius | 8 |
| 93 | SHRM_volume in 0.5 mm radius | 10 |
| 93 | IB-RPE_to_ILM min thickness in 3.0 mm radius | 10 |
| 86 | IB-RPE_to_OPL-HFL min thickness in 0.5 mm radius | 12 |
| 84 | IB-RPE_to_OPL-HFL max thickness in 3.0 mm radius | 13 |
| 81 | SHRM_volume in 1.5–3.0 mm radius | 14 |
| 77 | SRF max height in 1.5 mm radius | 15 |
| 77 | OPL-HFL_to_ILM max thickness in 3.0 mm radius | 15 |
| 70 | PED max width in 0.5 mm radius | 17 |
| 64 | SHRM_volume in 0.5–1.5 mm radius | 18 |
| 53 | SHRM max height in 3.0 mm radius | 19 |
| 48 | OPL-HFL_to_ILM mean thickness in 3.0 mm radius | 20 |
| 47 | IRF max height in 1.5 mm radius | 21 |
| 46 | IRF max height in 0.5 mm radius | 22 |
| 39 | BM_to_IB-RPE min thickness in 3.0 mm radius | 23 |
| 34 | BM_to_ILM mean thickness in 1.5 mm radius | 24 |
| 32 | IRF_volume in 0.5–1.5 mm radius | 25 |
BCVA = best-corrected visual acuity; BM = Bruch’s membrane; HFL = Henle’s fiber layer; IB_RPE = inner boundary of retinal pigment epithelium; ILM = internal limiting membrane; IRF = intraretinal fluid; Lasso = least absolute shrinkage and selection operator; max = maximum; min = minimum; OPL = outer plexiform layer; PED = pigment epithelial detachment; SE = standard error; SHRM = subretinal hyperreflective material; SRF = subretinal fluid.
Based on 1000 repeated 5-fold cross-validation.
Discussion
Individual responses from patients with nAMD to intravitreal anti-VEGF therapy may be highly variable and can be associated with a significant treatment burden. To address current unmet medical needs in the clinical management of nAMD, new therapeutic options are currently under clinical development or have been recently approved by health authorities.2, 3, 4 Although these new treatment modalities are highly anticipated, the treatment landscape for patients with nAMD is expected to become increasingly complex. In the future, the introduction of new technologies in clinical practice, AI-based tools among them, may help optimize the clinician's toolkit to make more informed treatment decisions.
In this retrospective exploratory analysis of the HARBOR dataset, 7 candidate machine learning or DL models based on OCT and clinical variables were compared with regard to their ability to accurately predict, at baseline, the functional response to ranibizumab at 9 months in patients with nAMD receiving monthly anti-VEGF treatment. Specifically, the aim is not recommending a tool, but rather a technical contribution to the field of OCT-based biomarkers.
For this analysis, data from the monthly ranibizumab 0.5 and 2.0 mg arms were pooled. In the HARBOR study,12 the results of the clinical trial showed that the mean BCVA gain of 10.1 letters (0.5 mg monthly ranibizumab) and 9.2 letters (2.0 mg monthly ranibizumab) was not significantly different between the 2 treatment arms (although the improvements in each arm was clinically meaningful). It is important to emphasize that comparison across candidate models serves to prioritize 1 of them and is not to be taken as an in-depth comparison by itself. We found that the highest-ranking Lasso min model using AI-based OCT segmentation and clinical variables performed statistically significantly better than the benchmark model based on age and BCVA. All 4 models, the Lasso min, Lasso 1SE, Catboost, and RF, performed better than models based on OCT only. The performance of the DL model is consistent with the previous reported results (R2 = 0.33 in predicting BCVA at 12 months in study eyes using baseline OCT).14
Deep learning is capable of automatically extracting and assessing clinically relevant features from OCT, whereas time constraints and human error may challenge manual grading, especially at the point of care. Artificial intelligence-based OCT segmentation can potentially improve OCT image analysis and interpretation, enabling more informed clinical assessment of imaging and clinical data, which is not feasible through currently commercially available standard technologies.
The relationship between the model performance and the clinical relevance can be understood as follows: The model needs to be accurate enough to detect a significant treatment effect. In other words, the error of the model prediction needs to be bounded by the treatment effect. Take the MAE as the metric. This means that MAE needs to be smaller than the mean visual acuity gain by a large enough margin such that the model is able to detect the treatment benefit. In our case, those models with a better performance than the benchmark model, that is, Lasso min, RF, and CatBoost, (Table 2) satisfy this condition.
The complex relationship between retinal fluid biomarkers and vision outcomes has been a matter of great clinical interest. Holekamp et al19 assessed visual acuity gains in patients with and without residual retinal fluid in a post hoc analysis of the HARBOR trial. The authors found that residual SRF had a positive effect, whereas residual IRF alone had a negative effect on vision outcomes at month 24. Furthermore, another study revealed that patients who tolerated some SRF achieved vision outcomes at least as good as those when treatment aimed to resolve all SRF.20 Taken together with the results of this current study, this may suggest that a more sophisticated approach, including quantitative volumetric analyses of the retinal fluid biomarkers, may be needed in the assessment of treatment response in nAMD.
In regard to strategies aiming to improve model performance, the present study showed that OCT-segmented features as a whole brought modest improvements to the model performance. In another database study that used machine learning to predict visual acuity at 3 and 12 months after 3 loading doses of intravitreal anti-VEGF therapy in patients with nAMD, Lasso outperformed 4 other machine learning models (AdaBoost.R2, gradient boosting, RF, and extremely randomized trees).21 Rohm et al21 suggested that simple linear models may outperform complex ensemble-based methods. Because numeric measurement data provided limited predictive value to their machine learning models, the authors speculate that future approaches using DL for OCT image recognition may be more valuable.21
In a clinical practice setting, Fu et al11 found that adding quantitative OCT parameters (retinal pigment epithelium, IRF, SRF, pigment epithelium detachment, subretinal hyperreflective material, and hyper-reflective foci) to a DL segmentation algorithm led to a small improvement in performance at 12 months (R2 = 0.38) compared with using baseline BCVA alone (R2 = 0.35). The authors suggested that quantitative OCT biomarkers such as subretinal hyperreflective material and IRF were independently statistically significant and may increase the overall predictive value of a model. Statistical analysis is needed to determine whether the additional OCT data significantly improve model prediction. In this work, we counteracted a potential inflation of R2 values by means of CV and were thus not affected by this problem; specifically, we made predictions on holdout data, as opposed to model introspection/inference. In another analysis of the HARBOR dataset, Schmidt-Erfurth et al22 determined that initial BCVA response at 3 months was the strongest predictive factor of vision outcomes at 12 months, whereas morphologic OCT features, such as SRF or pigment epithelium detachment, demonstrated limited predictive value on top of BCVA. In this study, similar to most models in our current analysis, quantitative features were extracted in a fully automated way from OCT scans and were used in RF models to predict visual acuity, together with baseline BCVA. The feature ranking extracted from these models does not account for multicollinearity (see also below). Five-fold cross validation was applied but only 1 mean R2 value was reported (R2 = 0.21). In contrast, in our analysis, OCT features alone had similar predictive capacity (R2 = 0.20 for Lasso min; Table 1). Further research may reveal other multimodal or functionally relevant OCT features; additional analyses would be of interest to determine whether features hypothesized to more likely reflect anatomical/functional correlation such as photoreceptors’ abnormalities or external limiting membrane disruption, rather than exudative features, may improve algorithm performance.
This study had several limitations. Although BCVA prediction for monthly ranibizumab is a first step toward using AI to potentially enable more informed treatment decisions, this analysis was based on models trained and validated in data from the HARBOR trial, and the model was developed using only the HARBOR monthly regimen dataset. Further work will need to integrate alternative treatment regimens and validate these results in an external dataset.
Although previous studies have performed similar analyses of machine learning models and their ability to predict various aspects of treatment response in patients with nAMD, to our knowledge, the present study is the first systematic comparison of multiple modeling approaches, including machine learning methods and a DL model in these patients, which is a main contribution of this work. One previous study applied a machine learning model to explain the difference in outcomes between clinical practice studies and randomized clinical trials.23 In that study, the main factors found to have the greatest impact on patient outcomes at 1 year were baseline BCVA, baseline SRF, and administration of 3 loading doses of intravitreal anti-VEGF therapy (ranibizumab or aflibercept) within 3 months after start of treatment.23
Consistent with previous studies, our work also identified age and baseline BCVA as features that were most predictive for treatment response. In addition, in our study, we used 105 automatically generated OCT biomarkers; the individual contributions of each of these biomarkers to treatment response and patient outcomes are yet to be explored in future research. To tease out the contributions of each individual OCT biomarker, future work could analyze this high-dimensional data based on clinical knowledge by selecting subsets of variables to investigate and repeating some of the modeling work with these subsets. Such an approach would remove multicollinearity, which commonly hinders model interpretation,24 as well as variables grouped together based on biological and clinical relevance.
In conclusion, this study demonstrated that, in addition to baseline age and BCVA, AI-based OCT-segmented features may improve the performance in predicting visual function response to intravitreal ranibizumab treatment in patients with nAMD to a measurable (statistically significant) extent. Furthermore, the simultaneous assessment of statistical and clinical significance is important. We argue the 3 models (Lasso min, RF, Catboost) that outperform the benchmark model further reduce MAE and increase the signal-to-noise ratio, a precondition to predicting clinically meaningful outcomes. We will conduct further testing on datasets from other studies to confirm or disprove this finding. Of note, the further development and validation of these models would be an important but early step toward the development of AI-based tools that can be deployed in clinical research and clinical practice, aiming to support better patient outcomes through more personalized treatment regimens. The chasm between exploratory research and the deployment of clinical decision support tools at the point of care is still to be overcome in this context.
Acknowledgments
The authors thank Huanxiang Lu for contributions to training models for the dataset described in this manuscript. Funding was provided by Genentech, Inc, a member of the Roche Group, for the study and third-party writing assistance, which was provided by Dinakar Sambandan, PhD, of Envision Pharma Group.
Data Sharing
For up-to-date details on Roche's Global Policy on the Sharing of Clinical Information and how to request access to related clinical study documents, see here: https://go.roche.com/data_sharing. For eligible studies qualified researchers may request access to the clinical data through a data request platform. At the time of writing, this request platform is Vivli. https://vivli.org/ourmember/roche/. For the imaging data underlying this publication, requests can be made by qualified researchers, subject to a detailed, hypothesis-driven proposal and necessary agreements.
Manuscript no. XOPS-D-22-00140.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Disclosures:
All authors have completed and submitted the ICMJE disclosures form.
The authors made the following disclosures: A.M.: Employee – F. Hoffmann-La Roche.
L.B.: Employee – Genentech, Inc; Stock – Roche stock as an employee.
M.G.K.: Employee – Genentech, Inc; Patent – P36847-US, “Treatment Outcome Prediction for Neovascular Age-Related Macular Degeneration Using Baseline Characteristics”; Stock – Roche Restricted stock units and Stock appreciation rights.
J.D.: Employee – Genentech, Inc.
A.Y.L.: Grants or contracts to institution – NIH/NIA, NIH/NEI, Carl Zeiss Meditec, Santen, Regeneron, Microsoft, Research to Prevent Blindness, Lowy Medical Research Institute; Consulting fees – Genentech, Gyroscope, Johnson & Johnson, US FDA.
R.F.S.: Personal compensation – Topcon Medical Systems; Consulting fees – Bayer, Genentech, Heidelberg Engineering, Regeneron, Roche; Payment or honoraria – Topcon Corporation; Support for meeting attendance/travel – Ryan Initiative for Macular Research; Patents – ResMatch; Leadership Role – American Academy of Ophthalmology, Collaborative Communities of Ophthalmology.;Stock or stock options – Index Fund ETF: SPY.
J.S.: Was an employee of F. Hoffmann-La Roche at the time of the analysis reported in this manuscript and is currently an employee of Novartis.
D.F.: Employee – Genentech, Inc; Stock – Roche stock/stock options as an employee.
Genentech, Inc., a member of the Roche Group, provided financial support for the study and participated in the study design; conducting the study; data collection, management, analysis, and interpretation; and preparation, review, and approval of the manuscript.
HUMAN SUBJECTS: Human subjects are included in this study.
This article includes analyses of data from a historical study conducted in patients with nAMD (HARBOR clinical trial, ClinicalTrials.gov identifier, NCT00891735); Institutional Review Board approvals were obtained prior to the HARBOR study initiation. Written informed consent was obtained from patients for their data to be used in future medical research and analyses. The trial was conducted in compliance with the principles of the Declaration of Helsinki and the Health Insurance Portability and Accountability Act. No animal subjects were used in this study.
Author Contributions:
Conception and design: Maunz, Barras, Kawczynski, Dai, Ferrara
Data collection: Maunz, Barras, Kawczynski, Dai
Analysis and interpretation: Maunz, Barras, Kawczynski, Dai, Lee, Spaide, Sahni, Ferrara
Obtained funding: N/A
Overall responsibility: Maunz, Barras, Kawczynski, and Dai
Meeting Presentations: Portions of these data were presented at the Association for Research in Vision and Ophthalmology Imaging in the Eye Conference; Virtual meeting; May 1–7, 2021.
Supplementary Data
References
- 1.Ferrara N., Adamis A.P. Ten years of anti-vascular endothelial growth factor therapy. Nat Rev Drug Discov. 2016;15(6):385–403. doi: 10.1038/nrd.2015.17. [DOI] [PubMed] [Google Scholar]
- 2.Arepalli S., Kaiser P.K. Pipeline therapies for neovascular age related macular degeneration. Int J Retina Vitreous. 2021;7:55. doi: 10.1186/s40942-021-00325-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heier J.S., Khanani A.M., Quezada Ruiz C., et al. Efficacy, durability, and safety of intravitreal faricimab up to every 16 weeks for neovascular age-related macular degeneration (TENAYA and LUCERNE): two randomised, double-masked, phase 3, non-inferiority trials. Lancet. 2022;399:729–740. doi: 10.1016/S0140-6736(22)00010-1. [DOI] [PubMed] [Google Scholar]
- 4.Holekamp N.M., Campochiaro P.A., Chang M.A., et al. Archway randomized phase 3 trial of the Port Delivery System with ranibizumab for neovascular age-related macular degeneration. Ophthalmology. 2022;129:295–307. doi: 10.1016/j.ophtha.2021.09.016. [DOI] [PubMed] [Google Scholar]
- 5.Ferrara D., Newton E.M., Lee A.Y. Artificial intelligence-based predictions in neovascular age-related macular degeneration. Curr Opin Ophthalmol. 2021;32:389–396. doi: 10.1097/ICU.0000000000000782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Fauw J., Ledsam J.R., Romera-Paredes B., et al. Clinically applicable deep learning for diagnosis and referral in retinal disease. Nat Med. 2018;24:1342–1350. doi: 10.1038/s41591-018-0107-6. [DOI] [PubMed] [Google Scholar]
- 7.Liefers B., Taylor P., Alsaedi A., et al. Quantification of key retinal features in early and late age-related macular degeneration using deep learning. Am J Ophthalmol. 2021;226:1–12. doi: 10.1016/j.ajo.2020.12.034. [DOI] [PubMed] [Google Scholar]
- 8.Feng D., Chen X., Zhou Z., et al. A preliminary study of predicting effectiveness of anti-VEGF injection using OCT images based on deep learning. Annu Int Conf IEEE Eng Med Biol Soc. 2020;2020:5428–5431. doi: 10.1109/EMBC44109.2020.9176743. [DOI] [PubMed] [Google Scholar]
- 9.Lee H., Kim S., Kim M.A., et al. Post-treatment prediction of optical coherence tomography using a conditional generative adversarial network in age-related macular degeneration. Retina. 2021;41:572–580. doi: 10.1097/IAE.0000000000002898. [DOI] [PubMed] [Google Scholar]
- 10.Nanegrungsunk O., Gu S.Z., Bressler S.B., et al. Correlation of change in central subfield thickness and change in visual acuity in neovascular AMD: post hoc analysis of VIEW 1 and 2. Am J Ophthalmol. 2021;238:97–102. doi: 10.1016/j.ajo.2021.11.020. [DOI] [PubMed] [Google Scholar]
- 11.Fu D.J., Faes L., Wagner S.K., et al. Predicting incremental and future visual change in neovascular age-related macular degeneration using deep learning. Ophthalmol Retina. 2021;5:1074–1084. doi: 10.1016/j.oret.2021.01.009. [DOI] [PubMed] [Google Scholar]
- 12.Busbee B.G., Ho A.C., Brown D.M., et al. Twelve-month efficacy and safety of 0.5 mg or 2.0 mg ranibizumab in patients with subfoveal neovascular age-related macular degeneration. Ophthalmology. 2013;120:1046–1056. doi: 10.1016/j.ophtha.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 13.Ho A.C., Busbee B.G., Regillo C.D., et al. Twenty-four-month efficacy and safety of 0.5 mg or 2.0 mg ranibizumab in patients with subfoveal neovascular age-related macular degeneration. Ophthalmology. 2014;121:2181–2192. doi: 10.1016/j.ophtha.2014.05.009. [DOI] [PubMed] [Google Scholar]
- 14.Kawczynski M.G., Bengtsson T., Dai J., et al. Development of deep learning models to predict best-corrected visual acuity from optical coherence tomography. Transl Vis Sci Technol. 2020;9:51. doi: 10.1167/tvst.9.2.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maunz A., Benmansour F., Li Y., et al. Accuracy of a machine-learning algorithm for detecting and classifying choroidal neovascularization on spectral-domain optical coherence tomography. J Pers Med. 2021;11:524. doi: 10.3390/jpm11060524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ronneberger O., Fischer F., Brox T. In: Medical Image Computing and Computer-Assisted Intervention - MICCAI 2015 Lecture Notes in Computer Science. Navab N., Hornegger J., Wells W., Frangi A., editors. Springer; Cham, Switzerland: 2015. U-Net: convolutional networks for biomedical image segmentation; pp. 234–241. [Google Scholar]
- 17.Friedman J., Hastie T., Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- 18.Regillo C.D., Busbee B.G., Ho A.C., et al. Baseline predictors of 12-month treatment response to ranibizumab in patients with wet age-related macular degeneration. Am J Ophthalmol. 2015;160:1014–1023.e2. doi: 10.1016/j.ajo.2015.07.034. [DOI] [PubMed] [Google Scholar]
- 19.Holekamp N.M., Sadda S., Sarraf D., et al. Effect of residual retinal fluid on visual function in ranibizumab-treated neovascular age-related macular degeneration. Am J Ophthalmol. 2022;233:8–17. doi: 10.1016/j.ajo.2021.06.029. [DOI] [PubMed] [Google Scholar]
- 20.Guymer R.H., Markey C.M., McAllister I.L., et al. Tolerating subretinal fluid in neovascular age-related macular degeneration treated with ranibizumab using a treat-and-extend regimen: FLUID study 24-month results. Ophthalmology. 2019;126:723–734. doi: 10.1016/j.ophtha.2018.11.025. [DOI] [PubMed] [Google Scholar]
- 21.Rohm M., Tresp V., Muller M., et al. Predicting visual acuity by using machine learning in patients treated for neovascular age-related macular degeneration. Ophthalmology. 2018;125:1028–1036. doi: 10.1016/j.ophtha.2017.12.034. [DOI] [PubMed] [Google Scholar]
- 22.Schmidt-Erfurth U., Bogunovic H., Sadeghipour A., et al. Machine learning to analyze the prognostic value of current imaging biomarkers in neovascular age-related macular degeneration. Ophthalmol Retina. 2018;2:24–30. doi: 10.1016/j.oret.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 23.Sagkriotis A., Chakravarthy U., Griner R., et al. Application of machine learning methods to bridge the gap between non-interventional studies and randomized controlled trials in ophthalmic patients with neovascular age-related macular degeneration. Contemp Clin Trials. 2021;104 doi: 10.1016/j.cct.2021.106364. [DOI] [PubMed] [Google Scholar]
- 24.Belsley D.A. Wiley; New York, NY: 1991. Conditioning Diagnostics: Collinearity and Weak Data in Regression. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.