Abstract
Objective
To propose Deep-GA-Net, a 3-dimensional (3D) deep learning network with 3D attention layer, for the detection of geographic atrophy (GA) on spectral domain OCT (SD-OCT) scans, explain its decision making, and compare it with existing methods.
Design
Deep learning model development.
Participants
Three hundred eleven participants from the Age-Related Eye Disease Study 2 Ancillary SD-OCT Study.
Methods
A dataset of 1284 SD-OCT scans from 311 participants was used to develop Deep-GA-Net. Cross-validation was used to evaluate Deep-GA-Net, where each testing set contained no participant from the corresponding training set. En face heatmaps and important regions at the B-scan level were used to visualize the outputs of Deep-GA-Net, and 3 ophthalmologists graded the presence or absence of GA in them to assess the explainability (i.e., understandability and interpretability) of its detections.
Main Outcome Measures
Accuracy, area under receiver operating characteristic curve (AUC), area under precision-recall curve (APR).
Results
Compared with other networks, Deep-GA-Net achieved the best metrics, with accuracy of 0.93, AUC of 0.94, and APR of 0.91, and received the best gradings of 0.98 and 0.68 on the en face heatmap and B-scan grading tasks, respectively.
Conclusions
Deep-GA-Net was able to detect GA accurately from SD-OCT scans. The visualizations of Deep-GA-Net were more explainable, as suggested by 3 ophthalmologists. The code and pretrained models are publicly available at https://github.com/ncbi/Deep-GA-Net.
Financial Disclosure(s)
The author(s) have no proprietary or commercial interest in any materials discussed in this article.
Keywords: Deep Learning, Detection, Explainability, GA, Geographic atrophy, OCT
Age-related macular degeneration (AMD), the leading cause of vision impairment and blindness in adults in industrialized countries, is a degenerative disease of the retina predominantly affecting the macula.1 Geographic atrophy (GA) is the defining lesion of the atrophic form of late AMD.2, 3, 4 It is predicted that, by 2040, atrophic AMD will affect > 5 million people worldwide.5 The detection of GA has important implications in both clinical practice and research. Most cases of GA do not affect the central macula at the onset; however, GA lesions enlarge progressively with time. The median time to central involvement, when visual acuity becomes severely affected, is estimated at 3 years.2 The United States Food and Drug Administration has recently approved the first therapy to slow the rate of GA enlargement.6 Therefore, early detection of GA, before the involvement of the central macula, might provide an optimum therapeutic window when treatments such as this could be initiated.7 Thus, rapid accurate identification of eyes with GA is an important task.
Spectral domain OCT (SD-OCT)8 has become an important imaging modality in ophthalmology, because it can provide in vivo noninvasive high-resolution volumetric (3-dimensional [3D]) images of the eye. Each SD-OCT volumetric scan (or “cube”) consists of a series of 2-dimensional (2D) grayscale images—the B-scans. Detection of GA from volumetric SD-OCT scans is considered superior than that from 2D imaging modalities such as color fundus photography because SD-OCT scans can provide detailed characterization of the inner and outer retinal layers, with the potential for quantitative assessment.3,9 Spectral domain OCT has been proposed as the reference standard to diagnose GA, in the form of complete retinal pigment epithelium (RPE) and outer retinal atrophy.3 However, the detection of GA on OCT scans is time-consuming because of the large volume of OCT data (i.e., many B-scans). It is also challenging because the ophthalmologist or grader must ensure that each of several criteria for GA is met (e.g., a region of hypertransmission of ≥ 250 μm in diameter in any lateral dimension and a zone of attenuation or disruption of the RPE of ≥ 250 μm in diameter, together with overlying photoreceptor degeneration).10 Automated methods may be useful to improve the detection of GA on SD-OCT scans.11, 12, 13, 14, 15
Deep learning has become the state-of-the-art method for computer vision as well as medical image processing.16,17 Many deep learning methods for GA detection have been proposed.15,18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30 Most of these methods require extensive annotations, which is a very time-consuming and subjective task. Of these methods, the Med-XAI-Net28 network uses weak labeling of the SD-OCT scans, where only binary labels have been provided (i.e., presence or absence of GA at the macular cube level). This method uses 2D convolutional neural networks (CNNs) to process the 3D SD-OCT scans; features are extracted from each B-scan individually and are then combined using attention weights. This leads to longer processing time at testing. Also, the 2D convolutions extract only 2D spatial features from individual B-scans and cannot extract 3D spatial features, so 2D convolutions are not robust and cannot capture the order and adjacency of B-scans.
In this study, we propose the first 3D classification network, named Deep-GA-Net, to detect GA on OCT scans. Deep-GA-Net consists of a 3D backbone residual CNN with a 3D loss-based attention layer. The purpose of the 3D attention layer is to capture the context within B-scans (i.e., B-scan attention) as well as the context within each B-scan (i.e., region attention). The difference between 2D networks and 3D networks is demonstrated in Figure 1. Thus, it helps to interpret the contribution of each voxel in determining GA presence or absence in the OCT volume scan. We compare Deep-GA-Net to the best performing network to date, Med-XAI-Net, on the same dataset as described by Shi et al.28 Also, as an ablation study, we compare Deep-GA-Net to (1) its backbone CNN without the attention layer and (2) its backbone CNN with a multiple-instance-learning attention layer. We used a dataset of 1284 OCT scans from 311 participants from the Age-Related Eye Disease Study 2 (AREDS2) Ancillary SD-OCT Study. We used 10-fold cross-validation with overlapping folds where the test fold did not contain any participant from the training set and the micro-average metrics were reported.
Figure 1.
The difference between the 2D and 3D approaches. With the 2D approach, each B-scan is processed individually using a shared 2D convolutional network (CNN), region attention is performed at the 2D pixel level, and B-scan attention is performed at the axial location (i.e., point) level. With the 3D approach, the OCT scan is processed in one go, and attention is performed at the 3D voxel level.
Methods
Datasets
We used data from the AREDS2 Ancillary SD-OCT study, as described by Shi el. al.28 The details of the AREDS2 design and protocol have been previously described.8, 31 In brief, the AREDS2 was a multicenter, phase 3, randomized controlled clinical trial designed to study the effects of nutritional supplements in people at moderate to high risk of progression to late AMD. At baseline, the participants, aged between 50 and 85 years, had either bilateral large drusen or large drusen in 1 eye and advanced AMD in the fellow eye. At each study visit (baseline and yearly), the participants received comprehensive eye examinations and ophthalmic imaging performed by certified personnel using standardized protocols.
The AREDS2 SD-OCT Study enrolled AREDS2 participants from the Devers Eye Institute, Emory Eye Center, Duke Eye Center, and the National Eye Institute. The study was approved by the institutional review boards of the study sites and was registered at ClinicalTrials.gov (identifier NCT00734487). The study adhered to the tenets of the Declaration of Helsinki and complied with the Health Insurance Portability and Accountability Act. Written informed consent was obtained from all participants. The participants underwent imaging using the Bioptigen Tabletop SD-OCT system at each annual study visit.8 Spectral domain OCT scans with a volume of 6.7 × 6.7 mm were captured (with 1000 A-scans per B-scan and 67-μm spacing between each B-scan). The ground truth grading of the SD-OCT scans for the presence or absence of GA was described previously.3,32 In brief, the OCT scans were viewed using the Duke OCT Retinal Analysis Program and were graded independently by 2 human experts, with any disagreement adjudicated by another human expert. These grades (for the presence or absence of GA at the level of each volume scan) provided the ground truth labels used for training and testing purposes in this study.
The dataset consisted of 1284 SD-OCT scans from 311 participants, such that participants contributed multiple OCT scans from multiple study visits. This comprised 321 volume scans with GA and 963 without GA. Similar to the approach described by Shi et al28 to train Med-XAI-Net, we used 10-fold cross-validation with overlapping folds, such that the test fold did not contain any participant from the training set.
Image Preprocessing
We resized the OCT images in the AREDS2 dataset31 from 1024 × 1000 pixels to 128 × 128 pixels for graphical processing unit memory considerations and interpolated the channel dimension from 100 to 128, i.e., to be at a power of 2 to have proper pooling. Thus, the final resized OCT scan size was set to 128 × 128 × 128. The whole scan values were normalized between –1 and 1. We augmented the scans during the training using random rotation, flipping, and erasing for augmentation.
Deep-GA-Net
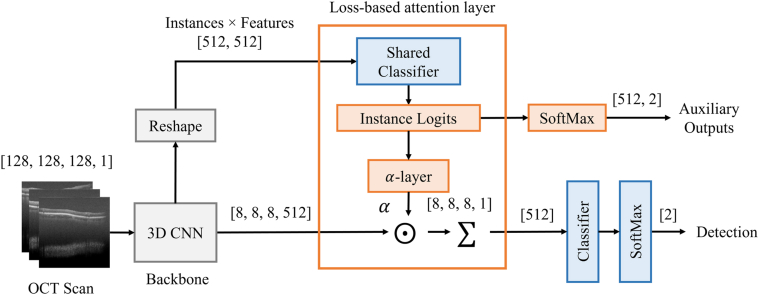
A high-level block diagram of the architecture of the proposed Deep-GA-Net is shown in Figure 2. The proposed network comprises a 3D input layer, a 3D backbone CNN, a 3D loss-based attention layer, and a classifier. The details of Deep-GA-Net architecture are summarized in Table S1 (available at www.ophthalmologyscience.org). In short, the backbone CNN contains an initial block of 2 3D convolutional layers with 64 filters followed by 3 blocks; each contains 2 3D residual convolutional layers with filters 128, 256, and 512, respectively, for each block. All convolution filters are of size 3 × 3 × 3. Each block is followed by a 3D maximum pooling operation of size 2 × 2 × 2. The classifier contains a fully connected layer of length 256 with Rectified Linear Unit activation function and preceded by a dropout layer with rate of 0.5. The fully connected layer is followed by another dropout layer, then a SoftMax layer of length 2 (i.e., binary classification). For the attention layer, we extended the loss-based attention method in the previously published paper by Shi et al33 to 3D settings. Instance logits were obtained by sharing the classifier and applying it to the instance-based convolutional features. The auxiliary outputs were obtained by applying SoftMax layer to the instance logits. The attention weights were derived from the instance logits using a custom layer named -Layer (see Appendix 1 for more details). The loss functions are described in Appendix 2.
Figure 2.
The architecture of the proposed Deep-GA-Net. CNN denotes the backbone 3D convolutional network, denotes attention weights, and denotes pointwise multiplication. The size of output tensor is shown near each block. The tensor sizes are enclosed between square brackets.
An ablation study was performed to evaluate the effectiveness of using attention layers. For the ablation study, we developed another 2 networks: (1) a 3D network with a backbone CNN and classifier similar to that of Deep-GA-Net, but with the attention layer replaced by global average pooling (GAP3D); (2) a 3D network with a backbone CNN and classifier similar to that of Deep-GA-Net, but with a 3D attention layer (MIL3D) derived from the 2D multiple-instance-learning approach by Ilse et. al.34 The attention layers were used to highlight the B-scans and B-scan regions that contributed most to the GA classification for the volume scan.
We also trained 2D networks (i.e., with similar architectures to that of the 3D networks but with 3D layers replaced by 2D layers as summarized in Table S2; available at www.ophthalmologyscience.org) on the en face projections of the SD-OCT volumes to explore the effectiveness of using 3D networks. They were named en face global average pooling 2D network (enGAP), en face multiple-instance learning 2D network (enMIL), and en face loss-based attention 2D network (enLBA).
For all networks, the SoftMax activation function was used to generate the class likelihood probabilities; the binary predictions were made using the class with the highest probability. To train networks, we used Adam optimizer35 with learning rate of 0.0001, batch size of 16, and maximum epoch number of 100 to avoid overfitting (see Figure S3, available at www.ophthalmologyscience.org). We used class weights to handle the imbalance in class size in our dataset. All experiments were performed using python 3.8 and a TensorFlow 2.3 deep learning library running on a server with 48 Intel Xeon CPUs with 754 Gb RAM and an NVIDIA GeForce GTX 1080 Ti 32-Gb graphical processing unit. To train the networks, we used categorical cross-entropy loss function, as given by equation 1 in Appendix 2. For Deep-GA-Net, we added a term for the attention loss,33 as given by equation 2 in Appendix 2.
Performance Evaluation and Comparison
We used accuracy, precision, recall, F1 score, kappa (κ),36 area under receiver operating characteristic curve (AUC) and area under precision-recall curve (APR) to evaluate and compare all networks. The 95% confidence intervals (CI) were computed for all metrics. We evaluated the performance metrics on the 7 models: (1) the 2D network with loss-based attention layer (Med-XAI-Net) that was proposed by the study by Shi et al28; (2) the 2D CNN with global average pooling (enGAP); (3) the 2D CNN with an attention-based multiple-instance-learning layer (enMIL); (4) the 2D CNN with loss-based attention layer (enLBA); (5) the 3D CNN with global average pooling (GAP3D); (6) the 3D CNN with an attention-based multiple-instance-learning layer (MIL3D); and (7) the proposed 3D CNN with loss-based attention layer (Deep-GA-Net). We used pretrained Med-XAI-Net networks from Shi et al,28 which were trained using images with size of 224 × 224 pixels. Our proposed networks (5–7) were trained on images with size of 128 × 128 pixels because of memory considerations. All 2D networks were trained on images with 224 × 224 pixels. We compared the networks’ outputs using the 2 independent samples test (ttest_ind) from the Scipy Python package.
Visualization of Deep-GA-Net
Gradient-weighted Class Activation Mappings37 were used to visualize the features learned by GAP3D, MIL3D, and Deep-GA-Net. The details are described in Appendix 3. In short, the weighted features obtained from the Gradient-weighted Class Activation Mappings were 3D. Therefore, to visualize and interpret these 3D learned features, we converted them into 2D fundus heatmaps by projecting the weighted features in the axial dimension. To determine the most relevant B-scans, we projected the weighted features along the axial and transverse dimensions and selected the location with maximum weight. To determine the most relevant regions within the selected B-scans, we used the weights corresponding to those selected B-scans. For Med-XAI-Net, we used the attention weights to visualize the network.
Visualization Grading by Ophthalmologists
In addition, a masked test was conducted independently by 3 ophthalmologists (T.K., A.T., and S.B.) to grade the explainability of all networks’ visualizations. A subset of 50 OCT scans labeled with GA was selected randomly from the test set where all networks had the best performance. The ophthalmologists were instructed to perform 2 tasks: to grade the heatmaps generated by each network and to grade their detections at the B-scan level. In the first task, the heatmaps were graded by the ophthalmologists as positive if the heatmap corresponded well to the actual GA location(s) on the en face image. In the second task, the B-scans were graded as positive if the bounding box (with size of 32 × 32 pixels) corresponded well to the actual GA location(s) on the B-scan. For each task, the final score for each network was computed as the average of the individual grades.
Results
Performance of Deep-GA-Net in Detecting GA
The performance metrics of Deep-GA-Net and the other 3 networks in correctly detecting GA from OCT scans are summarized in Table 3. Deep-GA-Net achieved the best overall performance scores over all the cross-validation splits, where it achieved macro average performance metrics of 0.93 (95% CI, 0.92–0.94), 0.90 (95% CI, 0.89–0.92), 0.90 (95% CI, 0.89–0.91), 0.80 (95% CI, 0.77–0.83), 0.94 (95% CI, 0.93–0.95), and 0.91 (95% CI, 0.90–0.93) for accuracy, recall, F1 score, κ, AUC, and APR, respectively. For precision, it came second, with a score of 0.90 (95% CI, 0.88–0.91). The receiver operating characteristic and precision-recall curves are shown in Figure 4 for all networks. The scores of Deep-GA-Net (i.e., except precision) were significantly different from all other networks (P < 0.01). It had insignificant APR compared with that of GAP3D (P = 0.1048).
Table 3.
The Micro-Average Performance Metrics Along with 95% Confidence Intervals for Deep-GA-Net and Other Methods on the Testing Dataset
| Network | Accuracy | Precision | Recall | F1 Score | κ | AUC | APR |
|---|---|---|---|---|---|---|---|
| Med-XAI-Net | 0.92 (0.91, 0.93) | 0.89 (0.88, 0.91) | 0.88 (0.87, 0.90) | 0.89 (0.87, 0.90) | 0.78 (0.75, 0.81) | 0.94 (0.93, 0.95) | 0.91 (0.90, 0.93) |
| enGAP | 0.87 (0.86, 0.88) | 0.84 (0.82, 0.86) | 0.79 (0.77, 0.81) | 0.81 (0.79, 0.83) | 0.62 (0.58, 0.66) | 0.87 (0.85, 0.88) | 0.83 (0.81, 0.86) |
| enMIL | 0.87 (0.85, 0.88) | 0.84 (0.82, 0.86) | 0.77 (0.75, 0.79) | 0.80 (0.78, 0.81) | 0.59 (0.56, 0.63) | 0.85 (0.83, 0.87) | 0.82 (0.79, 0.84) |
| enLBA | 0.87 (0.85, 0.88) | 0.83 (0.81, 0.85) | 0.79 (0.77, 0.81) | 0.80 (0.78, 0.82) | 0.61 (0.57, 0.65) | 0.85 (0.83, 0.87) | 0.81 (0.79, 0.83) |
| GAP3D | 0.93 (0.92, 0.94) | 0.91 (0.89, 0.92)∗ | 0.89 (0.87, 0.90) | 0.90 (0.88, 0.91) | 0.79 (0.76, 0.82) | 0.93 (0.92, 0.95) | 0.91 (0.90, 0.93)† |
| MIL3D | 0.92 (0.91, 0.93) | 0.89 (0.87, 0.90) | 0.89 (0.88, 0.91) | 0.89 (0.88, 0.90) | 0.78 (0.75, 0.81) | 0.93 (0.92, 0.95) | 0.91 (0.89, 0.93) |
| Deep-GA-Net | 0.93 (0.92, 0.94)∗ | 0.90 (0.88, 0.91) | 0.90 (0.89, 0.92)∗ | 0.90 (0.89, 0.91)∗ | 0.80 (0.77, 0.83)∗ | 0.94 (0.93, 0.95)∗ | 0.91 (0.90, 0.93)† |
AUC = area under receiver operating characteristic curve; APR = area under precision-recall curve; enGAP = en face global average pooling; enLBA = en face loss-based attention network; enMIL = en face multiple-instance learning network; GAP3D = global average pooling; MIL3D = multiple-instance-learning layer.
Bold font denotes highest metric values (i.e., before rounding).
Score was significantly different from other scores in the same column (P < 0.01).
Score was significantly different from other scores in the same column (P < 0.01) and insignificant to each other (P = 0.1048).
Figure 4.
The micro-average ROC (A) and precision-recall curves (B) for all networks over all cross-validation sets. APR = area under precision-recall curve; AUC = area under receiver operating characteristic curve; enGAP = en face global average pooling; enLBA = en face loss-based attention network; enMIL = en face multiple-instance learning network; GAP3D = global average pooling; MIL3D = multiple-instance-learning layer; ROC = receiver operating characteristic.
Explainability of the Visualizations of Deep-GA-Net
The grading of the 3 ophthalmologists for the 2 tasks performed on the 50 OCT scans labeled with GA is summarized in Table 4. Deep-GA-Net achieved the best performance in the 2 tasks, as suggested independently by all 3 ophthalmologists.
Table 4.
The Grading of Ophthalmologists on the 50 OCT Scans
| Task |
Heatmaps |
Region |
||||
|---|---|---|---|---|---|---|
| Method | Ophth. #1 | Ophth. #2 | Ophth. #3 | Ophth. #1 | Ophth. #2 | Ophth. #3 |
| Med-XAI-Net | 0.18 | 0.60 | 0.34 | 0.26 | 0.11 | 0.09 |
| GAP3D | 0.76 | 0.70 | 0.62 | 0.59 | 0.33 | 0.30 |
| MIL3D | 0.70 | 0.74 | 0.56 | 0.50 | 0.42 | 0.33 |
| Deep-GA-Net | 0.98 | 0.84 | 0.72 | 0.68 | 0.55 | 0.41 |
GAP3D = global average pooling; MIL3D = multiple-instance-learning layer. Bold text denotes the highest scores.
Examples of the generated heatmaps from all networks are shown in Figures 5 and 6. In Figure 5, the 3D networks generated superior heatmaps to those of the Med-XAI-Net (i.e., more meaningful to human ophthalmologists or graders). In Figure 6, in the examples shown, Deep-GA-Net demonstrated heatmaps that were superior to those of GAP3D and MIL3D; on qualitative inspection, its areas of high signal tended to correspond more closely with the regions of GA. The 2D networks highlighted smaller regions compared with the 3D networks because they were trained with the same network depth on larger image size, so their receptive field (i.e., focus region) was smaller. Examples of the detections at the B-scan level obtained from Deep-GA-Net are shown in Figure 7 for the OCT scan with the fundus image shown in the first row in Figure 5.
Figure 5.
Visualization of all networks on SD-OCT scans labeled with GA presence, which shows that all 3D networks performed better than Med-XAI-Net. enGAP = en face global average pooling; enLBA = en face loss-based attention network; enMIL = en face multiple-instance learning network; GA = geographic atrophy; GAP3D = global average pooling; MIL3D = multiple-instance-learning layer.
Figure 6.
Visualization of all networks on SD-OCT scans labeled with GA presence, which shows that Deep-GA-Net performed better than other networks. GA = geographic atrophy; SD = spectral density; enGAP = en face global average pooling; enLBA = en face loss-based attention network; enMIL = en face multiple-instance learning network; GA = geographic atrophy; GAP3D = global average pooling; MIL3D = multiple-instance-learning layer.
Figure 7.
Examples of the highlighted B-scans by Deep-GA-Net for the OCT scans with en face image shown in the first row in Figure 3. For clarity, we mapped a 2 × 2 pixel window of maximum score from the final 8 × 8 heatmap to a bounding box with size of 56 × 56 pixels on a B-scan image with size of 224 × 224 pixels.
Discussion
Main Findings and Interpretation
Deep-GA-Net achieved superior accuracy, AUC, and APR compared with those of the other networks (see Table 3 and Figure 4) in the detection of GA from OCT scans. Also, Deep-GA-Net achieved the highest F1 score, which suggests that it has the best balance between precision and recall. In addition, the 3D networks achieved superior performance compared with the 2D networks, which suggests the utility of 3D networks in processing 3D volumetric OCT scans.
On the subjective test, the manual grading by the ophthalmologists of the heatmaps and bounding boxes at the B-scan level suggested that Deep-GA-Net was more explainable, i.e., the visualizations that accompanied its grading corresponded more closely to areas of actual GA, as determined by ophthalmologists. Overall, Deep-GA-Net achieved superior performance compared with the other networks in the 2 tasks (see Table 4). This also suggests that Deep-GA-Net can be of great utility in detecting GA areas from OCT scans.
Clinical Importance
Geographic atrophy is the defining lesion of the atrophic form of late AMD,2, 3, 4 which is predicted to affect > 5 million people worldwide by 2040.5 Geographic atrophy enlarges progressively with time; when central involvement is not present at incidence, relentless progression toward central involvement is expected within several years, with severe effects on visual acuity.2 Because the first treatment to slow GA enlargement has now been approved,6 and more are likely to follow in the future, rapid, accurate detection of GA is very important to identify individuals who may benefit. In this way, automated approaches to GA detection could assist ophthalmologists in clinical diagnosis and decision making. They could also assist with clinical trials, both in identifying individuals with GA for potential recruitment and in helping detect GA as an outcome measure.
Comparison with Literature
Many automated methods for the detection of GA have been proposed using fundus autofluorescence images,20 color fundus photos,15 and SD-OCT scans.18,19,21, 22, 23, 24, 25, 26, 27, 28, 29, 30 Most of these methods are segmentation-based and require extensive annotations in 2D or 3D, which are laborious and time-consuming tasks. The feasibility of annotation depends on many factors, including the level of annotation (i.e., image level, en face level, or scan level), the number of examples in the dataset, the image quality, which affects the boundaries of GA area, and the agreement on how GA appears on the imaging modality (e.g., GA is difficult to highlight on en face projections). In this study, we focus on deep learning work performed on SD-OCT scans. Hu et al,25 Chen et al,26 and Niu et al27 segmented the choroidal region to create an enhanced fundus image for GA detection. Xu et al23 trained a deep neural network (DNN) to detect GA at the A-scan level, then used an autoencoder to generate a GA map based on the detected A-scans. This approach required manual segmentation of GA area on the fundus projection of the SD-OCT scans to label each A-scan based on the segmentation. Ji et al24 trained many DNNs to detect GA at the A-scan level from the input SD-OCT scan. The output of each DNN was a binary map that showed the GA area. Binary maps from all DNNs were combined using a voting scheme. This approach required manual delineation of the GA boundaries at the B-scan level for some OCT scans. Also, it required manual segmentation of GA area on fundus autofluorescence images, which were then registered to the fundus projection of the OCT scans to label the A-scans. Zhang et al29 used a 2D segmentation U-Net to detect the horizontal extent of RPE loss, overlying photoreceptor degeneration, and hypertransmission from the input B-scan. Then, a binary map of GA area was obtained based on these segmented features. This approach required the demarcation of the horizontal extent of RPE loss, overlying photoreceptor degeneration, and hypertransmission. Xu et al21 used a 3D segmentation CNN to segment GA in 3D where this approach required creating 3D segmentation data. Shi et al28 used a 2D shared backbone CNN to extract features from one B-scan at a time. Then, the extracted features were combined using 2 attention layers: B-scan attention layer and region attention layer. This approach achieved superior performance compared with other approaches, using weak labels for the whole SD-OCT scan (i.e., 1 binary label). However, this approach requires a large time at testing because of processing each B-scan individually. Also, it does account for the relationship between the B-scans.
In this study, we proposed Deep-GA-Net, a 3D classification CNN, to overcome the limitations in the study by Shi et al.28 Training and evaluation was performed using a large multicenter dataset obtained and curated from the AREDS2 Ancillary SD-OCT study. Deep-GA-Net consists of a 3D backbone residual CNN with a loss-based attention layer. We compared Deep-GA-Net to the Med-XAI-Net28 on the same dataset. Also, we compared Deep-GA-Net to other 3D baseline networks and 2D networks trained on en face images. Comparison with other methods was not feasible because of the lack of annotations. All 3D CNNs outperformed the 2D CNN in Shi et al28 as well as the 2D en face networks. In particular, Deep-GA-Net significantly outperformed all networks. The visualizations of all networks were graded by 3 ophthalmologists, and their gradings suggested that 3D CNNs generated better GA detections than those of the 2D CNN, with Deep-GA-Net performing the best. This suggests that Deep-GA-Net could detect GA with better accuracy and more interpretable visualizations.
Strengths, Limitations, and Future Work
The strengths of this work include the use of a large multicenter dataset curated from the AREDS2, which represents an ideal dataset for training and testing. Also, the strengths include detection of GA from volumetric OCT scans with high accuracy as well as proposing and generating heatmaps that can provide interpretable decisions, as suggested by the subjective test.
The limitations of this work include using SD-OCT scans with relatively small size, because of memory limitations. The GA hyperreflectivity appeared as quite large areas on individual SD-OCT images. Therefore, GA detection was feasible using SD-OCT scans with small size. However, the detection of subtle features might be challenging. Another limitation is that changing the input size would require a new network design, so the input/network relationship needs to be studied further. Moreover, GA detection is explained by the generated heatmaps without precise demarcation or quantification of the GA area, because area measurements are not available in the OCT dataset (i.e., the network was not trained to measure the GA area).
Conclusions
In conclusion, we proposed Deep-GA-Net, a 3D convolutional neural network with an attention layer, for the detection of GA on SD-OCT scans. Deep-GA-Net was able to detect GA from OCT scans with high accuracy, compared with other networks. The visualizations of Deep-GA-Net were more explainable, compared with those of other networks, as suggested by 3 ophthalmologists. The visualizations could show 2D heatmaps of GA areas in the fundus projection as well as detect the hypertransmission areas related to GA at the B-scan level. The code and pretrained models will be publicly available at https://github.com/ncbi/Deep-GA-Net for the transparency and reproducibility of this study work and to provide a benchmark for further studies.
Manuscript no. XOPS-D-22-00241R1.
Footnotes
Supplemental material available atwww.ophthalmologyscience.org.
Emily Chew, Editor-in-Chief of this journal, was recused from the peer-review process of this article and had no access to information regarding its peer-review.
Disclosure(s):
All authors have completed and submitted the ICMJE disclosures form.
The authors have made the following disclosures:
T.D.L.K.: Travel expenses – David R. Hinton Scholarship to the 2023 Conference of the Ryan Initiative for Macular Research; Patent – Co-inventor on a patent application: Methods and Systems for Predicting Rates of Progression of Age-related Macular Degeneration; Advisory Board – Voting member of the Safety Monitoring Committee for the APL2-103 interventional study (Apellis Pharmaceuticals), 2020–2021.
This research is supported by the NIH Intramural Research Program, National Library of Medicine and National Eye Institute, and NIH/NLM 1K99LM014024-01.
HUMAN SUBJECTS: Human subjects were included in this study. The study was approved by the institutional review boards of the study sites and was registered at ClinicalTrials.gov (identifier NCT00734487). The study adhered to the tenets of the Declaration of Helsinki and complied with the Health Insurance Portability and Accountability Act. Written informed consent was obtained from all participants.
No animal subjects were used in this study.
Author Contributions:
Conception and design: Elsawy, Keenan, Chen, Chew, Lu
Analysis and interpretation: Elsawy, Keenan, Chew, Lu
Data collection: Elsawy, Keenan, Chen, Shi, Thavikulwat, Bhandari, Chew, Lu
Obtained funding: Study was performed as part of regular employment duties at National Center for Biotechnology Information, National Library of Medicine, National Institutes of Health, and Division of Epidemiology and Clinical Applications, National Eye Institute, National Institutes of Health.
Overall responsibility: Elsawy, Keenan, Chen, Thavikulwat, Chew, Lu
Contributor Information
Emily Y. Chew, Email: emily.chew@nih.gov.
Zhiyong Lu, Email: luzh@ncbi.nlm.nih.gov.
Supplementary Data
The training and validation loss curves for all networks. We limited the epochs to 100 epochs because the validation losses almost was not improving and the networks were overfitting
References
- 1.GBD 2019 Blindness and Vision Impairment Collaborators, Vision Loss Expert Group of the Global Burden of Disease Study Causes of blindness and vision impairment in 2020 and trends over 30 years, and prevalence of avoidable blindness in relation to VISION 2020: the Right to Sight: an analysis for the Global Burden of Disease Study. Lancet Glob Health. 2021;9:e144–e160. doi: 10.1016/S2214-109X(20)30489-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keenan T.D., Agrón E., Domalpally A., et al. Progression of geographic atrophy in age-related macular degeneration: AREDS2 report number 16. Ophthalmology. 2018;125:1913–1928. doi: 10.1016/j.ophtha.2018.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sadda S.R., Guymer R., Holz F.G., et al. Consensus definition for atrophy associated with age-related macular degeneration on OCT: classification of atrophy report 3. Ophthalmology. 2018;125:537–548. doi: 10.1016/j.ophtha.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fleckenstein M., Keenan T.D.L., Guymer R.H., et al. Age-related macular degeneration. Nat Rev Dis Primers. 2021;7:31. doi: 10.1038/s41572-021-00265-2. [DOI] [PubMed] [Google Scholar]
- 5.Wong W.L., Su X., Li X., et al. Global prevalence of age-related macular degeneration and disease burden projection for 2020 and 2040: a systematic review and meta-analysis. Lancet Glob Health. 2014;2:e106–e116. doi: 10.1016/S2214-109X(13)70145-1. [DOI] [PubMed] [Google Scholar]
- 6.Apellis Pharmaceuticals FDA approves SYFOVRE™ (pegcetacoplan injection) as the first and only treatment for geographic atrophy (GA), a leading cause of blindness. https://investors.apellis.com/news-releases/news-release-details/fda-approves-syfovretm-pegcetacoplan-injection-first-and-only
- 7.Keenan T.D.L. Local complement inhibition for geographic atrophy in age-related macular degeneration: prospects, challenges, and unanswered questions. Ophthalmol Sci. 2021;1 doi: 10.1016/j.xops.2021.100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leuschen J.N., Schuman S.G., Winter K.P., et al. Spectral-domain optical coherence tomography characteristics of intermediate age-related macular degeneration. Ophthalmology. 2013;120:140–150. doi: 10.1016/j.ophtha.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spaide R.F., Jaffe G.J., Sarraf D., et al. Consensus nomenclature for reporting neovascular age-related macular degeneration data: consensus on neovascular age-related macular degeneration nomenclature study group. Ophthalmology. 2020;127:616–636. doi: 10.1016/j.ophtha.2019.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chew E.Y., Clemons T.E., Harrington M., et al. Effectiveness of different monitoring modalities in the detection of neovascular age-related macular degeneration: the home study, report number 3. Retina. 2016;36:1542–1547. doi: 10.1097/IAE.0000000000000940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keenan T.D.L., Chen Q., Peng Y., et al. Deep learning automated detection of reticular pseudodrusen from fundus autofluorescence images or color fundus photographs in AREDS2. Ophthalmology. 2020;127:1674–1687. doi: 10.1016/j.ophtha.2020.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q.A.O., Keenan T.D.L., Allot A., et al. Multimodal, multitask, multiattention (M3) deep learning detection of reticular pseudodrusen: toward automated and accessible classification of age-related macular degeneration. J Am Med Inform Assoc. 2021;28:1135–1148. doi: 10.1093/jamia/ocaa302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen Q., Peng Y., Keenan T., et al. A multi-task deep learning model for the classification of age-related macular degeneration. AMIA Jt Summits Transl Sci Proc. 2019;2019:505–514. [PMC free article] [PubMed] [Google Scholar]
- 14.Peng Y., Dharssi S., Chen Q., et al. DeepSeeNet: A deep learning model for automated classification of patient-based age-related macular degeneration severity from color fundus photographs. Ophthalmology. 2019;126:565–575. doi: 10.1016/j.ophtha.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Keenan T.D., Dharssi S., Peng Y., et al. A deep learning approach for automated detection of geographic atrophy from color fundus photographs. Ophthalmology. 2019;126:1533–1540. doi: 10.1016/j.ophtha.2019.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Suzuki K. Overview of deep learning in medical imaging. Radiol Phys Technol. 2017;10:257–273. doi: 10.1007/s12194-017-0406-5. [DOI] [PubMed] [Google Scholar]
- 17.Lee J.G., Jun S., Cho Y.W., et al. Deep learning in medical imaging: general overview. Korean J Radiol. 2017;18:570–584. doi: 10.3348/kjr.2017.18.4.570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Arslan J., Samarasinghe G., Benke K.K., et al. Artificial intelligence algorithms for analysis of geographic atrophy: a review and evaluation. Transl Vis Sci Technol. 2020;9:57. doi: 10.1167/tvst.9.2.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liefers B., Colijn J.M., González-Gonzalo C., et al. A deep learning model for segmentation of geographic atrophy to study its long-term natural history. Ophthalmology. 2020;127:1086–1096. doi: 10.1016/j.ophtha.2020.02.009. [DOI] [PubMed] [Google Scholar]
- 20.Treder M., Lauermann J.L., Eter N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefe Arch Clin Exp Ophthalmol. 2018;256:2053–2060. doi: 10.1007/s00417-018-4098-2. [DOI] [PubMed] [Google Scholar]
- 21.Xu R., Niu S., Gao K., Chen Y. In: Intelligent Computing Theories and Application. Huang D.S., Jo K.H., Zhang X.L., editors. Springer International Publishing; Cham: 2018. Multi-path 3D convolution neural network for automated geographic atrophy segmentation in SD-OCT images; pp. 493–503. [Google Scholar]
- 22.Fang L., Cunefare D., Wang C., et al. Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search. Biomed Opt Express. 2017;8:2732–2744. doi: 10.1364/BOE.8.002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xu R., Niu S., Chen Q., et al. Automated geographic atrophy segmentation for SD-OCT images based on two-stage learning model. Comput Biol Med. 2019;105:102–111. doi: 10.1016/j.compbiomed.2018.12.013. [DOI] [PubMed] [Google Scholar]
- 24.Ji Z., Chen Q., Niu S., et al. Beyond retinal layers: a deep voting model for automated geographic atrophy segmentation in SD-OCT images. Transl Vis Sci Technol. 2018;7:1. doi: 10.1167/tvst.7.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Z., Medioni G.G., Hernandez M., et al. Segmentation of the geographic atrophy in spectral-domain optical coherence tomography and fundus autofluorescence images. Invest Ophthalmol Vis Sci. 2013;54:8375–8383. doi: 10.1167/iovs.13-12552. [DOI] [PubMed] [Google Scholar]
- 26.Chen Q., de Sisternes L., Leng T., et al. Semi-automatic geographic atrophy segmentation for SD-OCT images. Biomed Opt Express. 2013;4:2729–2750. doi: 10.1364/BOE.4.002729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niu S., de Sisternes L., Chen Q., et al. Automated geographic atrophy segmentation for SD-OCT images using region-based C-V model via local similarity factor. Biomed Opt Express. 2016;7:581–600. doi: 10.1364/BOE.7.000581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shi X., Keenan T.D.L., Chen Q., et al. Improving interpretability in machine diagnosis: detection of geographic atrophy in OCT scans. Ophthalmol Sci. 2021;1 doi: 10.1016/j.xops.2021.100038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang G., Fu D.J., Liefers B., et al. Clinically relevant deep learning for detection and quantification of geographic atrophy from optical coherence tomography: a model development and external validation study. Lancet Digit Health. 2021;3:e665–e675. doi: 10.1016/S2589-7500(21)00134-5. [DOI] [PubMed] [Google Scholar]
- 30.Wu M., Cai X., Chen Q., et al. Geographic atrophy segmentation in SD-OCT images using synthesized fundus autofluorescence imaging. Comput Methods Programs Biomed. 2019;182 doi: 10.1016/j.cmpb.2019.105101. [DOI] [PubMed] [Google Scholar]
- 31.Chew E.Y., Clemons T., SanGiovanni J.P., et al. The Age-Related Eye Disease Study 2 (AREDS2): study design and baseline characteristics (AREDS2 report number 1) Ophthalmology. 2012;119:2282–2289. doi: 10.1016/j.ophtha.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Christenbury J.G., Folgar F.A., O’Connell R.V., et al. Progression of intermediate age-related macular degeneration with proliferation and inner retinal migration of hyperreflective foci. Ophthalmology. 2013;120:1038–1045. doi: 10.1016/j.ophtha.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shi X., Xing F., Xu K., et al. Loss-based attention for interpreting image-level prediction of convolutional neural networks. IEEE Trans Image Process. 2021;30:1662–1675. doi: 10.1109/TIP.2020.3046875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ilse M., Tomczak J., Welling M. Attention-based deep multiple instance learning. Proceedings of the 35th International Conference on Machine Learning, PMLR. 2018;80:2127–2136. [Google Scholar]
- 35.Kingma D.P., Ba J. Adam: a method for stochastic optimization. arXiv. 2014;1412:6980. doi: 10.48550/arXiv.1412.6980. [DOI] [Google Scholar]
- 36.McHugh M.L. Interrater reliability: the kappa statistic. Biochem Med (Zagreb) 2012;22:276–282. [PMC free article] [PubMed] [Google Scholar]
- 37.Zhou B., Khosla A., Lapedriza A., et al. Learning deep features for discriminative localization. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) Las Vegas, NV, USA. 2016:2921–2929. doi: 10.1109/CVPR.2016.319. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The training and validation loss curves for all networks. We limited the epochs to 100 epochs because the validation losses almost was not improving and the networks were overfitting