Abstract
Summary
This manuscript describes an open-source program, DrawGlycan-SNFG (version 2), that accepts IUPAC (International Union of Pure and Applied Chemist)-condensed inputs to render Symbol Nomenclature For Glycans (SNFG) drawings. A wide range of local and global options enable display of various glycan/peptide modifications including bond breakages, adducts, repeat structures, ambiguous identifications etc. These facilities make DrawGlycan-SNFG ideal for integration into various glycoinformatics software, including glycomics and glycoproteomics mass spectrometry (MS) applications. As a demonstration of such usage, we incorporated DrawGlycan-SNFG into gpAnnotate, a standalone application to score and annotate individual MS/MS glycopeptide spectrum in different fragmentation modes.
Availability and implementation
DrawGlycan-SNFG and gpAnnotate are platform independent. While originally coded using MATLAB, compiled packages are also provided to enable DrawGlycan-SNFG implementation in Python and Java. All programs are available from https://virtualglycome.org/drawglycan; https://virtualglycome.org/gpAnnotate.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Glycans are among the most important and complex post-translational modifications (PTMs) identified to date. The Symbol Nomenclature For Glycans (SNFG) has been developed as a community standard to streamline the study of this PTM. This system simplifies the depiction of complex carbohydrate structures using various colored geometric shapes (Neelamegham et al., 2019; Varki et al., 2015). This has been adopted by several software programs and international databases (reviewed in Neelamegham et al., 2019). For wider adoption, versatile, easy to use, and robust open-source programs are needed. These codes should be easily integrated into software that focus on mass spectrometry (MS; glycomics and glycoproteomics), lectin/glycan array databases and additional glycoinformatics applications.
To address the above needs, the current manuscript presents DrawGlycan-SNFG (version 2). Unique features of this version include: (i) a variety of local and global options that vastly enhance the usage of the original code (Cheng et al., 2017), particularly for MS spectrum annotation; (ii) an enhanced web interface with intuitive, user-friendly features; and (iii) bindings so that the compiled MATLAB code can be part of Python and Java packages. As an application of DrawGlycan-SNFG usage, this manuscript also presents gpAnnotate. This lightweight program enables the annotation of individual MS/MS spectrum in order to optimize fragmentation rules suitable for different MS fragmentation modes, and to scrutinize alternate glycopeptide assignments that may explain a given experimental result. This is the first example where SNFG sketches have been integrated into MS data analysis software.
2 Software description and usage
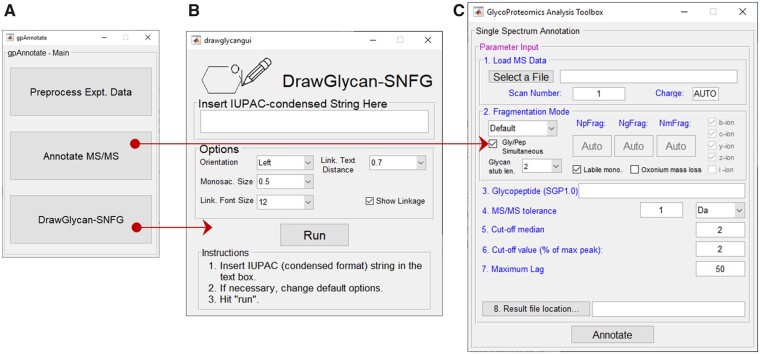
Both DrawGlycan-SNFG and gpAnnotate are written in MATLAB R2018b, and they have exhaustive usage instructions (see Supplementary Material and www.VirtualGlycome.org). Among these, DrawGlycan-SNFG is available as an open-source application with a web interface and standalone GUI (Fig. 1). gpAnnotate is only provided as a standalone application, currently.
Fig. 1.
GUI interfaces. (A) Main interface for gpAnnotate, and pop-up interfaces for (B) DrawGlycan-SNFG and (C) annotate MS/MS
DrawGlycan-SNFG (version 2) contains a vast range of global and local options to fully accommodate the new updates to the SNFG (Neelamegham et al., 2019). At the MATLAB command prompt, SNFG figures can be generated using a simple generic input:
≫drawglycan(‘IUPAC STRING with local options’, ‘Global Option name’, ‘Global Option parameter’);
Additionally, efforts have been undertaken to package the software so that it can be incorporated into JAVA and Python code. New features are also included in this version to depict bond fragmentation, text annotations including characters within SNFG symbols and text to replace symbols at arbitrary locations with the figure, repeating glycan structures, ambiguous assignments, fuzzy brackets, anomeric groups, adduct ions, different line styles to depict different types of glycosydic linkages and methods to modify specific bond orientations. A user manual provides additional details including exhaustive examples.
GpAnnotate consists of two parts for: (i) pre-processing of Expt data and (ii) annotating MS/MS spectrum. The program also includes the standalone version of DrawGlycan-SNFG for the convenience of users who wish to install both programs simultaneously. Among these, the preprocessing module accepts selected proprietary and open-source MS input file types: .raw, .mgf, .mzXML and .mzML. Although ProteoWizard installation is necessary in order to handle .raw files (Chambers et al., 2012), gpAnnotate has built-in functions to handle other file formats. Data pre-processing is performed once for each experimental data file in order to extract various data fields from the input files to generate a final .mat file that is used in subsequent steps.
The Annotate MS/MS module is the main code in gpAnnotate. It accepts the .mat file from the preprocessing step as ‘MS Data’. Given a scan number and a candidate glycopeptide, Annotate MS/MS compares the theoretical spectrum for the candidate glycopeptide with the experimental spectrum. To achieve this, gpAnnotate generates the theoretical MS/MS fragmentation spectrum for the candidate glycopeptide depending on the selected fragmentation mode (details provided in user manual): CID, Collision Induced Dissociation; HCD, beam-type CID or higher-energy collision dissociation; ETD, electron-transfer dissociation; ETciD, electron-transfer-CID, and EThcD, electron-transfer-HCD. Additional facilities are also available to define custom fragmentation rules by varying the maximum number of cleavages on the peptide, glycan or non-glycan PTMs, ion types, enabling co-fragmentation of glycan and peptide backbone, and including additional fragments generated due to neutral loss. The ability of the theoretical spectrum to match the experimental data is then quantitatively evaluated by estimating an ‘Ensemble Score (ES)’, a statistical measure that incorporates metrics for the cross-correlation analysis score (XCorr), probability based P-values, ability to match the Top10 most intense peaks, and the % of the theoretical glycan and peptide fragments that are explained by the experimental data (Liu et al., 2017).
Analysis results are presented in a figure with three tabs: (i) ‘Summary’, which presents the candidate glycopeptide using SNFG rendering, with all experimentally detected cleavages marked using DrawGlycan-SNFG, a window with statistical scores including the Ensemble Score and additional parameters, and the fully annotated experimental spectrum with each of the identified peaks marked; (ii) ‘Quantification’, which contains the parent ion isotope distribution, the elution curve of the candidate ion with label-free area under the curve quantitation, and the total ion current of the run; and (iii) ‘Detailed Annotation’, which contains a fully annotated glycopeptide structure in SNFG format and a table showing the identity of each of the identified peaks in the spectrum. Output generated by gpAnnorate can be stored in graphical format as image, or as text exported into spreadsheets.
Overall, DrawGlycan-SNFG is designed to aid the study of glycans using the SNFG. Integration of this standard into gpAnnotate enables a handy tool that can be used for scoring and re-scoring MS/MS spectra.
Supplementary Material
Acknowledgements
We thank Edward J. Sobczak for systems administration of VirtualGlycome.org.
Funding
This work was supported by National Institutes of Health [grants HL103411 and GM126537].
Conflict of Interest: none declared.
Contributor Information
Kai Cheng, Department of Chemical and Biological Engineering, Clinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14260, USA.
Gabrielle Pawlowski, Department of Chemical and Biological Engineering, Clinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14260, USA.
Xinheng Yu, Department of Chemical and Biological Engineering, Clinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14260, USA.
Yusen Zhou, Department of Chemical and Biological Engineering, Clinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14260, USA.
Sriram Neelamegham, Department of Chemical and Biological Engineering, Clinical and Translational Research Center, University at Buffalo, The State University of New York, Buffalo, NY 14260, USA.
References
- Chambers M.C. et al. (2012) A cross-platform toolkit for mass spectrometry and proteomics. Nat. Biotechnol., 30, 918–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K. et al. (2017) DrawGlycan-SNFG: a robust tool to render glycans and glycopeptides with fragmentation information. Glycobiology, 27, 200–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G. et al. (2017) A comprehensive, open-source platform for mass spectrometry-based glycoproteomics data analysis. Mol. Cell Proteomics, 16, 2032–2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neelamegham S. et al. (2019) Updates to the symbol nomenclature for glycans guidelines. Glycobiology, 29, 620–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varki A. et al. (2015) Symbol nomenclature for graphical representations of glycans. Glycobiology, 25, 1323–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.