Abstract
Eukaryotic DNA-dependent RNA polymerases (Pols I–III) encode two distinct alpha-like heterodimers where one is shared between Pols I and III, and the other is unique to Pol II. Human alpha-like subunit mutations are associated with several diseases including Treacher Collins Syndrome (TCS), 4H leukodystrophy, and primary ovarian sufficiency. Yeast is commonly used to model human disease mutations, yet it remains unclear whether the alpha-like subunit interactions are functionally similar between yeast and human homologs. To examine this, we mutated several regions of the yeast and human small alpha-like subunits and used biochemical and genetic assays to establish the regions and residues required for heterodimerization with their corresponding large alpha-like subunits. Here we show that different regions of the small alpha-like subunits serve differential roles in heterodimerization, in a polymerase- and species-specific manner. We found that the small human alpha-like subunits are more sensitive to mutations, including a “humanized” yeast that we used to characterize the molecular consequence of the TCS-causingPOLR1D G52E mutation. These findings help explain why some alpha subunit associated disease mutations have little to no effect when made in their yeast orthologs and offer a better yeast model to assess the molecular basis of POLR1D associated disease mutations.
Keywords: RNA polymerase, alpha subunits, Rpb11, AC19, POLR2J2, POLR1D, homologs
INTRODUCTION
DNA-dependent RNA polymerases (Pols) are responsible for synthesizing RNA from DNA in the process of transcription. In eukaryotes, there are at least three nuclear Pols (Pols I–III). Pol I synthesizes three of the four ribosomal RNAs (rRNAs), while Pol II synthesizes messenger RNAs (mRNAs) and many noncoding RNAs. Finally, Pol III synthesizes transfer RNAs (tRNAs), small nuclear RNAs (snRNAs), and the 5S rRNA. Although each Pol has a distinct number of subunits, the general structure of each complex is conserved, including a heterodimer of alpha-like subunits.1,2
The alpha-like subunits are evolutionarily related to the bacterial Pol alpha subunit, which forms a homodimer.3–6 The archaeal and eukaryotic alpha-like heterodimers are composed of both a large and a small subunit that share a conserved dimerization structural fold resembling their bacterial counterpart.2,7–9 The alpha-like heterodimers share common structural features, including a highly conserved alpha-motif (⍺M) and a long C-terminal helix (CTH). Together, the ⍺M and the CTH form a heterodimerization domain which uses a leucine-zipper mechanism to dimerize.10,11 In eukaryotes, there are two alpha-like heterodimers: one in Pol II and one that is shared between Pol I and III. Specifically, the Pol II alpha-like heterodimer is composed of POLR2C and POLR2J in humans and their yeast orthologs, Rpb3 and Rpb1112 (Fig. 1A; Table S1). Pols I/III alpha-like heterodimer is composed of POLR1C and POLR1D in humans, and their orthologs AC40 and AC19 in yeast2 (Fig. 1A; Table S1). The alpha-like heterodimers, like their bacterial counterpart, aid in Pol assembly and interact with other transcription factors.2–4,12–14
FIG 1.
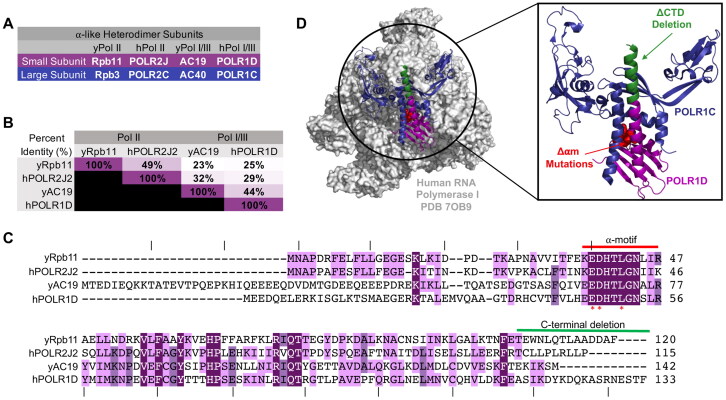
Conservation of the alpha-like subunits in yeast and human. (A) Common names for small and large ⍺-like heterodimer subunits. y, yeast; h, humans. (B) Percent identity between small alpha-like subunit protein homologs. (C) Alignment of yeast and human small alpha-like subunit homologs protein sequences. Residue conservation between homologs is indicated with purple highlighting. Light purple is conserved between two homologs and it ranges up to dark purple where the residue is conserved between all four homologs. Red bar indicates the boundaries of the alpha-motif. An asterisk denotes residues mutated to alanine in the alpha-motif mutant (Δ⍺m), and the green bar denotes residues deleted in the C-terminal helix deletion mutant (ΔCTD). Dashes above the protein sequence indicate every 10 residues. (D) Surface representation of the human RNA polymerase I structure (PDB 7OB9). The highlighted box is a zoomed in ribbon structure of the alpha–like heterodimer. POLR1C and POLR1D are colored in blue and purple, respectively. Residues mutated in the Δ⍺m mutant are colored in red, and the residues deleted in the ΔCTD mutant are colored in green.
The alpha-like heterodimers are essential and clinically significant as several diseases are associated with mutations in each heterodimer. For example, mutations in human alpha-like subunits POLR1D and POLR1C, shared between Pols I and III, cause Treacher Collins Syndrome (TCS), a craniofacial disorder that is characterized by underdeveloped facial structures.15–22 TCS results from decreased ribosome biogenesis in neural crest cells that differentiate into the different facial bones and tissues. During prenatal development neural crest cells fail to migrate properly and apoptoses, leading to craniofacial malformations.23–27
Mutations in several Pol III subunits, including POLR1C, are associated with the neurological disorder 4H (hypomyelination, hypodontia and hypogonadotropic hypogonadism) and leukodystrophy, also known as POLR3-related leukodystrophy.28–36 Current molecular pathogenesis models suggest that Pol III subunit mutations cause complex assembly impairments and localization defects, leading to diminished tRNA synthesis and protein yield. Lack of protein synthesis causes defects in the synthesis or maintenance of the myelin sheath, which results in a variety of neurological and systemic defects throughout the body.37–40 Lastly, a truncation mutation in the Pol II subunit POLR2C, is associated with primary ovarian insufficiency in women and a missense mutation has been found to cause hearing loss and infertility in males.41,42 Currently, the molecular pathogenesis of these Pol II diseases is unclear.
Although there are many disease mutations found within the alpha-like subunits, few have been fully characterized at the molecular and biochemical level. This is in part due to model systems failing to recapitulate a disease phenotype. For instance, in yeast, only one TCS point mutation introduced into AC19, the yeast ortholog of human POLR1D, causes a discernable phenotype.43 Likewise, in a yeast model of 4H leukodystrophy, there is not a discernable phenotype from single point mutations in the largest Pol III subunit (Rpc160). However, when two point mutations in Rpc160 are combined a disease phenotype is seen.44 This parallels a mouse model of 4H leukodystrophy, wherein a point mutation in POLR3A fails to recapitulate any of the defects seen in humans with the same point mutation in the orthologous protein.45 Together, these findings suggest a level of species-specificity in how the subunits function, highlighting the importance of investigating the molecular and biochemical differences between the alpha-like subunit orthologs.
Previous studies using yeast-two hybrid interaction assays to examine the regions necessary for yeast and human Pol II alpha-like heterodimer formation found that different residues within the heterodimerization domain of each protein contribute to heterodimerization to varying degrees.46 Therefore, it is possible that homologous heterodimers have different residue requirements for heterodimerization and would respond differently to disease mutations. Understanding the differences in how the orthologous and paralogous alpha-like subunits form their respective heterodimers will help us improve the use of model systems to characterize pathogenic disease mutations.
In this study, we investigated which domains and amino acids of the yeast and human small alpha-like subunits are required for interaction with the large alpha-like subunits, polymerase assembly, and for cell growth. We used several biochemical assays to show that each small alpha-like homolog requires different residues for function and heterodimerization. We found that the human alpha-like subunits and a humanized version of yeast AC19 were more sensitive to mutations than their yeast orthologs, including a mutation associated with TCS. Together, our results show that the alpha-like subunits use distinct interaction mechanisms for their heterodimerization in both a species- and polymerase-specific manner, and this helps explain why some disease mutations may not reveal disease phenotypes in model organisms.
RESULTS
Conservation of the small alpha-like subunits
The eukaryotic alpha-like subunits are well conserved in terms of their sequence and structure. For example, pairwise alignment of yeast Rpb11 and its human ortholog POLR2J2, reveals a 49% protein identity47 (Fig. 1B and C). To lesser degree, but still significant, the pairwise comparison of yeast Rbp11 to its paralogs yeast AC19 and human POLR1D have a 23–32% protein identity, respectively (Fig. 1B and C). The alpha-like subunit heterodimers also occupy a conserved peripheral position within their respective Pols14,48–51 (Fig. 1D). Given their conservation in sequence, structure, and position, we expected that the alpha-like heterodimers would function similarly and likely heterodimerize by a similar mechanism, but as we demonstrate below the heterodimers seem to dimerize by distinct mechanisms.
The Pol II alpha-like complex heterodimerization
Previous studies showed that the Pol II small alpha-like subunits, yeast Rpb11 and its human ortholog POLR2J2, require distinct and species-specific heterodimerization interfaces for interaction with yeast Rpb3 and human POLR2C, respectively.46 More specifically in a yeast two hybrid assay, a cluster mutation in the highly conserved alpha-motif (Δ⍺M) of Rpb11 (E38A, D39A, L42A) has relatively no effect on its genetic interaction with Rpb3, while deletion of the Rpb11 C-terminal (ΔCTD) end residues 106–120 abolished genetic interaction with Rpb3 (Fig. 1C and D). In the human ortholog, the corresponding ⍺M mutation and CTD in POLR2J2 reduced but did not abolish genetic interaction with POLR2C. To complement these findings in a more direct manner, we employed a combination of protein-protein interaction assays and yeast growth assays to examine the importance of the ⍺M and CTD for yeast Rpb11 biological function and heterodimerization.
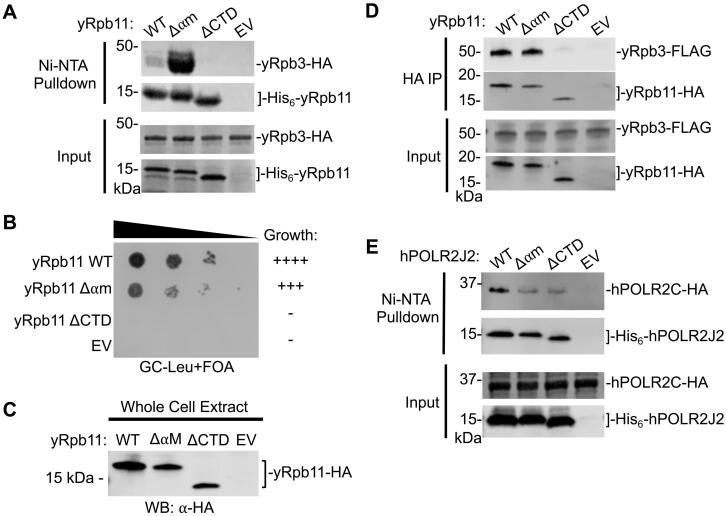
We first developed an in vitro bacterial heterodimer co-expression system for both the yeast and human Pol II alpha-like subunit complexes to investigate the direct physical interactions between the subunits. Yeast Rpb11 and its human POLR2J2 ortholog contain a His6 tag fused to the N-terminus to allow for Ni-NTA affinity-based chromatography. Using this system, we tested whether the Δ⍺M mutation and the ΔCTD affected the formation of the yeast Pol II heterodimer. We found that the ΔCTD mutation in Rpb11 completely abolished interaction with Rpb3, whereas the Δ⍺M mutation retained and enhanced binding to Rpb3 (Fig. 2A). These in vitro results are consistent with the previous genetic interaction results described above for yeast Rpb11 and human POLR2J2.
FIG 2.
Effect of alpha-motif and C-terminal helix mutations on yeast Rpb11 and its human ortholog POLR2J2. (A) In vitro Ni-NTA pulldown of yeast His6-tagged Rpb11 variants and HA-tagged Rpb3 co-expressed in bacteria. SDS-PAGE analysis and Coomassie stain of Ni-NTA purified proteins and inputs are shown. (B) Yeast plasmid shuffle assay of Rpb11 variants grown on glucose media containing 5-FOA. (C) Western blot analysis assessing protein expression levels of yeast Rpb11 variants. (D) Western blot analysis of immunopurified HA-tagged Rpb11 variants in a FLAG-tagged Rpb3 yeast strain. Input of the immunopurified proteins is shown. (E) In vitro Ni-NTA pulldown of human His6-tagged POLR2J2 variants and HA-tagged POLR2C co-expressed in bacteria. Western blot analysis of Ni-NTA purified proteins and SDS-PAGE analysis and Coomassie staining of Ni-NTA pulldown inputs are shown.
We next examined how the Rpb11 mutations impact yeast growth and Rpb3 interaction in vivo. Using a yeast plasmid shuffle assay, we found that the ΔCTD mutation caused lethality, while the Rpb11 Δ⍺M mutant grew similar to wild-type, indicating that the three mutated residues are not required for function of Rpb1152,53 (Fig. 2B). Further, both Rpb11 mutants stably expressed in yeast cell compared to wild-type Rpb11, indicating the lack of cell growth observed for the ΔCTD was not due to lack of protein expression (Fig. 2C). Finally, immunoprecipitation of yeast Rpb11 and its mutant variants showed that the ΔCTD mutation completely abolished Rpb3 interaction, while the Δ⍺M mutation retained wild-type levels of interaction with Rpb3 (Fig. 2D). Taken together, these findings confirm previous findings using both in vitro and in vivo approaches that the C-terminus of yeast Rpb11 is critical for Rpb3 interaction.
To extend this further, we also examined the interaction of human POLR2J2 with POLR2C in our in vitro co-expression system. Unlike the Rpb11Δ⍺M mutation, which did not affect the interaction between Rpb11 and Rpb3, we found that the Δ⍺M mutation in human POLR2J2 markedly reduced, but did not completely abolish, the interaction with POLR2C. And, unlike the Rpb11 ΔCTD mutation, which completely abolished the interaction between Rpb11 and Rpb3, we found that the ΔCTD mutation in human POLR2J2 retained some interaction with POLR2C (Fig. 2E). These results are also consistent with previous yeast-two hybrid interaction assays showing that both the ⍺M and CTD of POLR2J2 are required for heterodimerization in humans. Likewise, our results utilizing direct interaction assays confirm that yeast Rpb11 and human POL2J2, although evolutionarily conserved in terms of their structure and function, rely on different protein-protein surfaces to different degrees in heterodimer formation.
The Pol I/III alpha-like complex heterodimerization
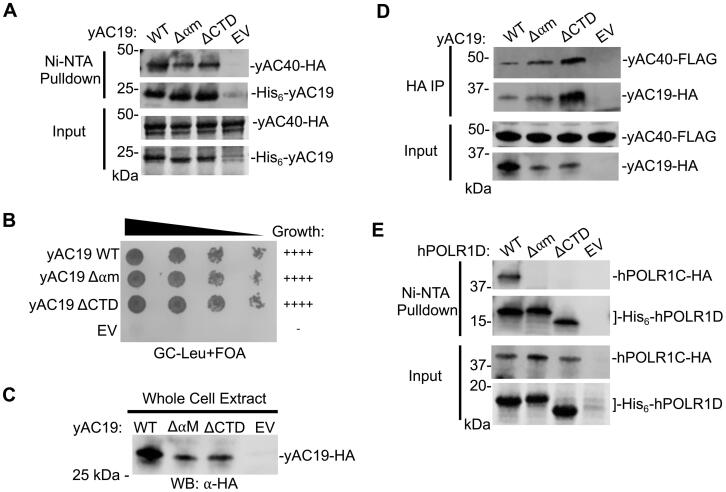
To investigate the paralog-specificity of the alpha-like subunit interactions, we used our co-expression system to examine the interaction between the yeast Pol I/III heterodimer subunits AC40 and AC19, which are paralogs of Rpb3 and Rpb11, respectively. Interestingly, mutation of neither the ⍺M or CTD of AC19 had any effect on the interaction with AC40 in vitro (Fig. 3A). Likewise in vivo, neither mutation in AC19 had an effect on yeast cell growth and little impact on AC19 protein expression levels (Fig. 3B and C). Consistent with these findings, both mutations immunoprecipitated near wild-type levels of AC40 in vivo (Fig. 3D). Together, these findings suggest neither the ⍺M or CTD alone is required for AC40 interaction, which is distinctly different than the interactions between the Pol II heterodimers, which required at least one or both regions being required for the yeast and human paralogs.
FIG 3.
Effect of alpha-motif and C-terminal helix mutations on the yeast AC19 and its human ortholog POLR1D. (A) In vitro Ni-NTA pulldown of yeast His6-tagged AC19 variants and HA-tagged AC40 co-expressed in bacteria. SDS-PAGE analysis and Coomassie stain of Ni-NTA purified proteins and inputs are shown. (B) Yeast plasmid shuffle assay of AC19 variants grown on glucose media containing 5-FOA. (C) Protein expression levels of AC19 variants in yeast analyzed by Western blot. (D) Western blot analysis of immunopurified HA-tagged AC19 variants in a FLAG-tagged AC40 yeast strain. Input of the immunopurified proteins is shown. (E) In vitro Ni-NTA pulldown of human His6-tagged POLR1D variants and HA-tagged POLR1C co-expressed in bacteria. Western blot analysis of Ni-NTA purified proteins and SDS-PAGE analysis and Coomassie staining of Ni-NTA pulldown inputs are shown.
Finally, we used our in vitro co-expression system to investigate the human Pol I/III POLR1C and POLR1D heterodimer interactions. In striking contrast to AC19, the ⍺M and CTD mutations in POLR1D completely abolished interaction with POLR1C (Fig. 3E), indicating that the human Pol I/III is very sensitive to mutations in either of these regions. This is similar to the human Pol II heterodimer, in that both regions are required for heterodimerization, yet to a greater degree. More broadly, these findings suggest that each alpha-like heterodimer in eukaryotes rely on the ⍺M and CTD to differing degrees and reveals both species and paralog specific requirements for their heterodimerization.
Paralog-specific residues of yeast Rpb11 and AC19 are required for cell growth
Given the different responses to regional mutations between the eukaryotic alpha-like heterodimers, we hypothesized that they utilize distinct residues for their interaction even though many of the residues within the dimerization domains are conserved in terms of the biochemical nature and atomic position.48,54 To test this, we first analyzed the structures of yeast Rbp11 and AC19 both visually and computationally using the BIPSPI program to predict residues within the dimerization interfaces of each protein, that might contribute to their interactions with Rpb3 and AC40, respectively.55 Nearly all the Rpb11 residues within the dimerization domain were also predicted to be important for AC19, further showing the conservation of the dimerization interface structure.
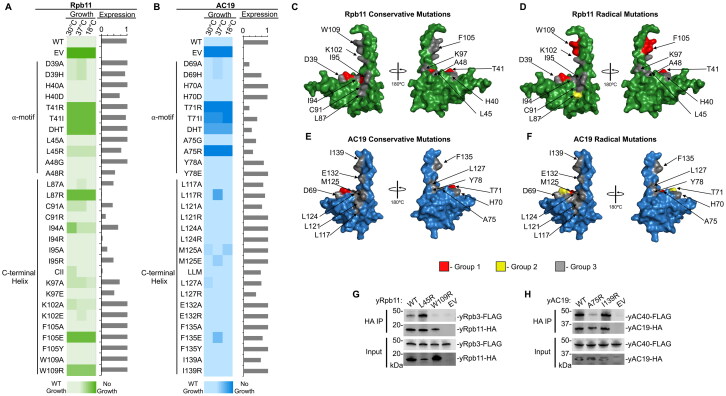
We designed mutations in Rpb11 and AC19 where selected residue positions were mutated to either a conservative alanine mutation or a radical mutation changing either the charge or bulkiness of the residue. For each protein, we created a total of 29 single amino acid mutations in 13 residues: 10-point mutations in the ⍺M at five residue positions and 17-point mutations in the C-terminal helix (CTH) at eight residue positions. We also included two alanine cluster mutations: one in the ⍺M in Rpb11 residues D39, H40, and T41 and AC19 residues D69, H70, and T71, and the second in the CTH, where we mutated Rpb11 residues C91, I94, and I95, AC19 residues L121, L124, and M125. Next, we examined their effect on yeast cell growth at three different temperatures (30 °C, 37 °C, and 18 °C) (Fig. 4A and B). We were interested in mutations that displayed a severe or lethal growth phenotypes yet had minimal impact on protein expression.
FIG 4.
Different residues are required for yeast Rpb11 and AC19 biological function and heterodimerization. (A and B) Heat map summary of yeast plasmid shuffle growth assays of various Rpb11 (Panel A) and AC19 (Panel B) variants grown at the indicated temperatures. “WT,” wild-type; “EV,” empty vector. Boxes with light coloring indicates wild-type-like growth while dark coloring indicates a lethal mutation. The bar graph to the right of the heat map shows the protein expression relative to respective wild-type proteins. (C to F) Surface representation of Rpb11 (PDB 1WCM) (Panels C and D) and AC19 (PDB 4C2M) (Panels E and F). Residues classified as Group 1 (growth defect and stable protein) are colored in red. Group 2 residues (growth defect and unstable protein) are colored in yellow, and Group 3 residues (no growth defect) are colored in gray. The classification of conservative mutations are depicted in Panels C and E, and radical mutations are depicted in Panels D and F. (G) Western blot analysis of immunopurified HA-tagged Rpb11 variants in a FLAG-tagged Rpb3 yeast strain and inputs are shown. (H) Western blot analysis of immunopurified HA-tagged AC19 variants in a FLAG-tagged AC40 yeast strain and inputs are shown.
We categorized these mutations into three distinct groups based on their growth phenotype and protein expression levels (Fig. 4C to F and Fig. S1 and Fig. S2). Group 1 displayed lethal to near-lethal growth phenotypes and retained at or near wild-type protein expression levels. Group 2 also displayed a sick to lethal growth phenotype, but protein expression was markedly reduced, indicating its importance in protein stability. Finally, group 3 were mutations that retained at or near wild-type growth under all temperature conditions, indicating a lack of importance for biological function under the conditions tested.
Overall, we found Rpb11 is more sensitive to mutations within the heterodimer interaction interface compared to its paralog AC19 under all growth conditions (Fig. 4C to F). For instance, nearly twice as many Rpb11 mutations (seven in total) cause severe growth phenotypes, while this is only observed in four AC19 mutants. Among the seven Rpb11 mutants displaying severely sick or lethal growth phenotypes at all temperature conditions, three of the mutants in the ⍺M (T41R, T41I, DHT) and three of the mutants in the CTH (I94A, F105E, W109R) expressed proteins at or near wild-type levels (Fig. 4A and Fig. S2A). The remaining Rpb11 mutant (L87R) lacked any detectable protein expression which is likely the reason for the severe growth defect. Of the AC19 mutations that caused a severe growth defect, only two of the mutations (T71I and A75R) expressed protein, while the remaining two mutations (T71R, DHT) lessened protein expression (Fig. 4B and Fig. S2B).
None of the AC19 CTH mutations caused a severe growth defect, although the mutations L117R and F135E caused a slight growth defect at 37 °C, indicating that the mutations potentially cause thermal instability. In contrast, four Rpb11 mutations (L87R, I94A, F105E, W109R) displayed severe or lethal growth defects at all growth temperatures. These findings agree well with our results (Fig. 2A to D and Fig. S2A) that show the CTH is necessary in Rpb11 for yeast cell growth and heterodimerization, while it is dispensable in AC19 (Fig. 3A to D), further emphasizing that there is a clear difference in how the C-terminal helix is utilized by AC19 versus Rpb11.
Interestingly, the Rpb11 L45 residue in the ⍺M and its corresponding AC19 A75 residue position displayed vastly different growth defects. In AC19, the A75R mutation caused a lethal phenotype at 30 °C and while not affecting AC19 protein expression (Fig. 4B and Fig. S2B), while Rpb11 L45R grew similar to wild type (Fig. 4A and Fig. S2A). The remaining mutations in Rpb11 and AC19 displayed largely similar growth phenotypes. Mutations of the Rpb11 T41 residue and the corresponding AC19 T71 residue, and the ⍺M cluster mutation (DHT) caused moderate to severe growth defects at all temperatures tested in both paralogs (Fig. 4A and B). The remaining ⍺M mutations did not cause a severe growth defect in either paralog (Fig. 4A and B). In terms of yeast cell growth, these results further demonstrate that AC19 and Rpb11 rely on distinct residues for their biological functions and likely for heterodimerization.
Finally, another key difference between AC19 and Rpb11 are the residues required for protein stability and/or expression. Five CTH mutations in Rpb11 caused protein instability (L87R, C91R, I94R, I95A, CII), while three AC19 ⍺M mutations (D69A, T71R, DHT) caused protein instability (Fig. 4A and B and Fig. S2). Together with the growth assays, the differences in residues required for protein stability suggest that AC19 is more resistant to mutations in general compared to Rpb11, and the C-terminus of Rpb11 is highly sensitive to mutations.
Specific residues of yeast Rpb11 and AC19 paralogs are required for heterodimerization
Many of the mutations in the heterodimer interaction interface that caused severe growth defects did not impact protein stability, therefore we hypothesized that these mutations could disrupt the heterodimerization between the small and large alpha-like subunits. To examine this further, we selected two positions in Rpb11 and AC19 positions that showed a differential growth response between the paralogs. These mutants include the Rpb11 mutant L45R in the ⍺M and Rpb11 W109R in the CTH and their paralogous AC19 A75R and AC19 I139R mutations, respectively (Fig. 4C to F). We transformed HA-tagged versions of wild-type and mutant variants of Rpb11 and AC19 in yeast strains with the large alpha-like subunit Rpb3 and AC40 genomically FLAG-tagged for heterodimer immunoprecipitation interaction assays.
We found that the Rpb11 W109R and AC19 A75R mutations that cause severe growth phenotypes, significantly reduced interaction between Rpb11 and Rpb3, and AC19 and AC40. (Fig. 4G and H). In contrast, their paralogous mutants, AC19 I139R and Rpb11 L45R, grew at or near wild-type levels and retained near wild-type levels of interaction with AC40 and Rpb3 (Fig. 4G and H). These differences highlight the polymerase-specificity of the alpha-like subunits that we previously established using the regional mutations and further show that the small alpha-like paralogs utilize distinct residues for their interaction, even though many of the residues within the dimerization domains are conserved in terms of the biochemical nature and atomic position.
Yeast-human AC19 chimera is more sensitive to mutations than AC19
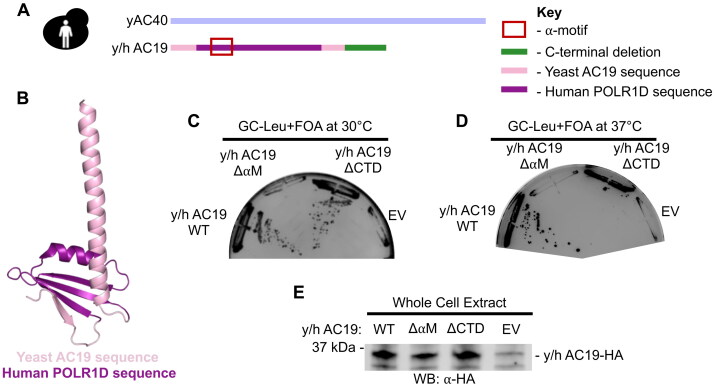
Although the eukaryotic Pol alpha-like heterodimers are highly evolutionary conserved, we have shown that there is species-specificity and polymerase-specificity in how the alpha-like subunits heterodimerize. These findings may help explain why many TCS point mutations in yeast AC19 have no observable phenotype. The highly conserved ⍺M, which is a hotspot for TCS mutations, can be exchanged with human POLR1D to complement yeast cell growth in a yeast-human AC19 chimera (y/h AC19) (Fig. 5A and B)43. If y/h AC19 is in fact humanized, we anticipate that it is sensitive to mutations like human POLR1D.
FIG 5.
Yeast-human AC19 chimera is more sensitive to regional mutations than yeast AC19. (A) Cartoon schematic of humanized AC19 chimera. (B) Ribbon structure of yeast AC19 (PDB 4C2M) made in PyMOL. The AC19 sequence is colored in pink and the region replaced with the human POLR1D sequence is colored in purple. (C and D) Yeast plasmid shuffle assay of AC19 chimera variants grown on glucose media containing 5-FOA at 30 °C (Panel C) and at 37 °C (Panel D). (E) Protein expression levels of AC19 chimera variants in yeast analyzed by Western blot.
We first tested the impact of regional mutations in y/h AC19. The ⍺M mutation in y/h AC19 caused a lethal growth phenotype, unlike the same mutation in yeast AC19 that had no impact on yeast cell growth (Fig. 5C). Likewise, the CTD mutation caused a conditional temperature sensitive phenotype (Fig. 5D), indicating that y/h AC19 is more sensitive to regional mutations like human POLR1D. Each of the mutations were expressed at or near wild-type protein levels (Fig. 5E).
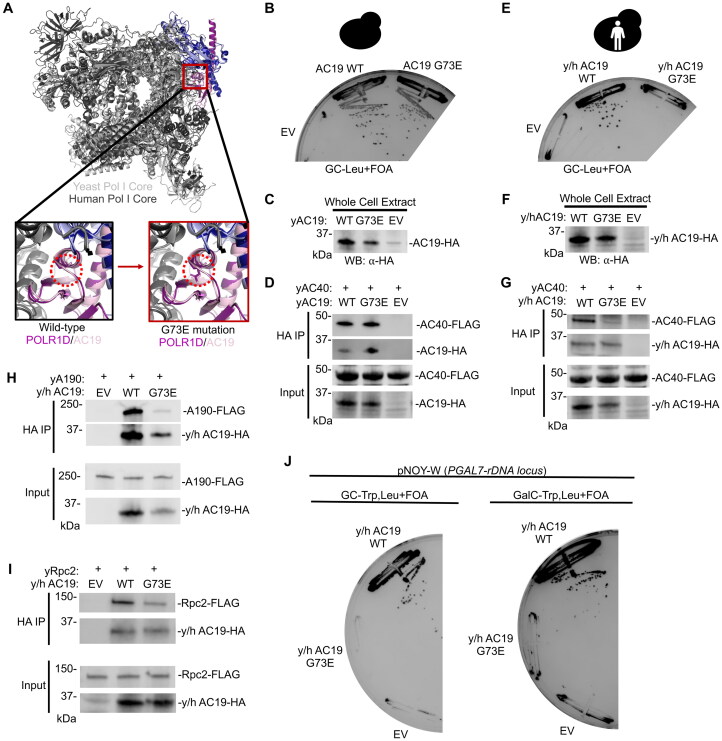
To investigate the y/h AC19 residue requirements further, a clinically relevant and well-characterized mutation (G73E) was made in yeast AC19 and y/h AC19 (Fig. 6A).15,56 Previous in vitro studies showed the orthologous mutation in the context of human and fly POLR1D reduced heterodimer formation.57 Using HHpred to align yeast AC19 and y/h AC19, the yeast G73E mutation is equivalent to the TCS-causing G52E mutation in human POLR1D.47 Given the glycine position in the heterodimer interaction interface within the Pol I structure, it is theorized that replacing glycine with a large negatively charged glutamic acid would sterically disrupt AC40 interaction (Fig. 6A).51
FIG 6.
Yeast-human AC19 chimera is sensitive to the clinically relevant G73E mutation associated with TCS. (A) Structural alignment of Yeast Pol I (PDB 4C2M) and Human Pol I (PDB 7OB9) ribbon structures. Human Pol I in dark gray, yeast Pol I in light gray, human POLR1C in dark blue, yeast AC40 in light blue, human POLR1D in purple, and yeast AC19 in light pink. Boxes below zoom in on the wild-type POLR1D G52 and AC19 G73 position (left box) and their mutated G52E and G73E forms (right box). Red dotted circle indicates the location of the G52 and G73 side chains. (B) Yeast plasmid shuffle assay of indicated AC19 variants grown on glucose media containing 5-FOA. (C) Protein expression levels of indicated AC19 variants in yeast analyzed by Western blot. (D) Western blot analysis of immunopurified HA-tagged AC19 variants in a FLAG-tagged AC40 yeast strain and inputs are shown. (E) Yeast plasmid shuffle assay of indicated y/h AC19 chimera variants grown on glucose media containing 5-FOA. (F) Protein expression levels of indicated y/h AC19 chimera variants in yeast analyzed by Western blot. (G) Western blot analysis of immunopurified HA-tagged y/h AC19 chimera variants in a FLAG-tagged AC40 yeast strain and inputs are shown. (H and I) Western blot analysis of immunopurified HA-tagged y/h AC19 chimera variants in a FLAG-tagged A190 (Panel H) and Rpc2 (Panel I) yeast strain and inputs are shown. (J) Yeast plasmid shuffle assay of indicated y/h AC19 chimera variants grown on glucose or galactose media containing 5-FOA.
Using a plasmid shuffle assay, we found that the G73E mutation did not cause a growth defect in yeast AC19 (Fig. 6B), but the same mutation did cause a severe growth phenotype in y/h AC19 (Fig. 6E). Each of the G73E mutants in the yAC19 and y/h AC19 chimera expressed at or near wild-type protein levels (Fig. 6C and F). Therefore, we tested whether the G73E mutation in y/h AC19 also impacted heterodimerization with AC40. We expressed HA-tagged yeast AC19 and the y/h AC19 chimera in their wild-type and mutant forms in a yeast strain harboring genomically FLAG-tagged AC40. We found that the G73E mutation in y/h AC19 significantly reduced interaction with AC40 (Fig. 6G), whereas the G73E mutation in yAC19 retained interaction with AC40 (Fig. 6D). Together these results show that humanization of yeast AC19 make it more sensitive to mutations, similar to human POLR1D, and may serve as a better yeast model to assess human TCS disease mutations.
The G73E mutation disrupts both Pol I and III complex integrity
Since the G73E mutations disrupt heterodimer formation, we hypothesized that this mutation would also affect Pol I complex integrity by impairing binding to the other Pol I catalytic subunits that include A190, the largest catalytic subunit. Furthermore, the G73 residue is near the largest Pol I subunit A190 (Fig. 6A), so mutation of glycine to a bulky and negatively charged glutamic acid residue could disrupt their interaction. Likewise, disrupting heterodimerization would also impair interaction with A190 since heterodimerization is a prerequisite for the rest of Pol I to assemble.2 Using y/h AC19, we examined the molecular consequence of the G73E mutation on Pol I complex integrity. We transformed a yeast strain containing a genomically FLAG-tagged A190 subunit and immunoprecipitated y/h AC19 and y/h AC19 G73E. We found that the G73E mutation reduced interaction with A190 compared to wild-type y/h AC19, indicating that Pol I complex integrity is impaired (Fig. 6H).
AC19 is a shared subunit of both Pol I and III, therefore we asked whether the G73E mutation is Pol I specific or might also affect Pol III complex integrity. We first performed a rescue experiment with a plasmid containing the entire rDNA locus under the galactose-inducible, Pol II-dependent GAL7 promoter (pNOY-W).58 In the presence of galactose, lethal phenotypes caused by mutations that specifically affect Pol I are rescued by Pol II-dependent expression of rRNA from the pNOY-W plasmid, while lethal phenotypes caused by mutations that would also affect Pol III are not rescued. We cotransformed an AC19 plasmid shuffle strain with our y/h AC19 plasmid variant with the G73E mutation and the pNOY-W rescue plasmid and observed no growth rescue in the presence of galactose (Fig. 6J). This suggest that the G73E mutations is not specific to Pol I and likely impairs Pol III.
To examine Pol III complex integrity, we immunoprecipitated HA-tagged y/h AC19 from a yeast strain containing a FLAG-tagged Rpc2 subunit, the second largest subunit in the Pol III complex.Similar to A190, we found that y/h AC19 G73E reduced interaction with Rpc2 (Fig. 6I). Together, these findings show that in the humanized yeast model the G73E mutation reduces interaction with AC40 and impairs both Pol I and III complex integrity. Overall, our findings demonstrate that the humanized AC19 is more vulnerable to TCS mutations than yeast AC19, therefore, humanized yeast AC19 serves as a better yeast model to examine the molecular consequence of TCS disease mutations. It also provides new information on the molecular and biochemical defects caused by the POLR1D G52E mutation in TCS.
DISCUSSION
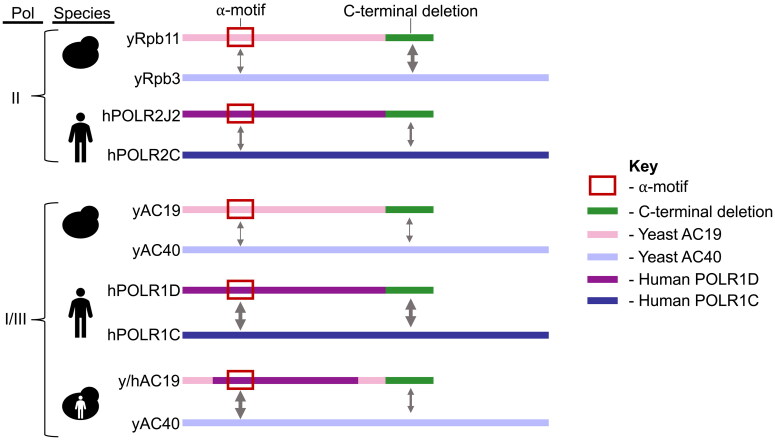
Here we show that distinct regions of the eukaryotic small alpha-like subunits are required for the function and heterodimerization of the Pol alpha-like heterodimer. More specifically, we find that the alpha-like subunits heterodimerize in both a species-specific and polymerase-specific manner (Fig. 7), and the specificity can be altered by “humanizing” a yeast protein. Even though the small alpha-like subunits are evolutionarily conserved throughout all three domains of life in terms of their sequence, structure, and function, the regions and residues required for biological function and heterodimerization are much less conserved between species and their polymerases and may in fact differ greatly between orthologs and paralogs.
FIG 7.
Differential contribution of the small alpha-like subunits alpha-motif and CTD for heterodimerization in yeast and human. Modular schematic of the small and large alpha-like subunit heterodimer pairs in yeast and human depicting the distinct contributions of the alpha-motif (red box) and CTD (green bar) for their interaction. The small yeast and human alpha-like subunits are depicted as pink and purple bars, respectively. The large yeast and human subunits are colored in violet and blue, respectively. Arrow thickness indicates the contribution of the region to the heterodimerization (a greater contribution is shown by thicker arrows). Protein names are labeled to the left of the schematic, along with the corresponding yeast = and human cartoons next to the protein names indicating the species. The yeast-human chimera combines both yeast and human cartoons. The Pol the heterodimer is present and is indicated to the left of the species. “II” indicates Pol II, and “I/III” indicates Pols I and III.
Our studies here help explain why many TCS mutations in yeast AC19 do not result in an observable phenotype. It appears that yeast AC19 is more resistant to mutations in both the ⍺M and CTH, which are both hotspots for TCS mutations in POLR1D.15,56 Likewise, our findings also show that human alpha-like subunits are more sensitive to mutations than their yeast orthologs, which is especially the case for POLR1D. This may explain why many TCS mutations are found in POLR1D as it is more vulnerable to disease mutations. This might also be the case for POLR1C that is also mutated in TCS and 4H leukodystrophy.
This disease vulnerability phenomena may also extend to other Pol subunits and possibly other model systems as some, but not all, clinically relevant mutations cause disease phenotypes. For instance, 4H leukodystrophy mutations examined in yeast POLR3A also lack an observable phenotype unless they are combined with a second mutation (yeast POLR3A G672E) in the pore domain, where nucleotides access the Pol III active site.44,59 In this case, the second mutation further weakens Pol III activity, suggesting again that the yeast proteins are more resistant to mutations than their human orthologs in some fashion. The absence of clinical 4H leukodystrophy phenotypes is also seen in mice models. A homozygous G672E mutation in mouse POLR3A, did not cause neurological abnormalities, and Pol III transcription was unaffected.45 Our work highlights the importance of future studies to understand the biochemical and biophysical properties that make the human, yeast proteins, and other models, more or less disease prone.
Several factors likely influence the susceptibility of the human alpha-like proteins and the resistance of yeast proteins to disease mutations. For instance, changes in the physical properties of the interacting residues between the yeast and human alpha-like subunits might modify their resistance to mutation.60–62 Changes in the hydrophobicity may alter the stability of the protein individually and/or the interaction mechanisms between the large and small subunits.63 In terms of thermal stability of the yeast and human heterodimers, previous temperature shift studies indicate that the human Pol I/III alpha-like heterodimer is slightly more resistant to temperature than its yeast counterpart.43 This suggests that the species-specific differences are possibly at the residue level. It is likely that the yeast proteins can tolerate mutations within the heterodimer as other residues in the small or even large alpha-like subunit compensate better than the corresponding residues in the human counterparts. For instance, the yeast small alpha-like subunits seem more hydrophobic compared to their human counterparts, and we speculate that this may be a reason for the yeast small alpha-like subunits and vice versa. Future work is necessary to determine the precise residues responsible for the species-specific differences.
We used our humanized AC19 yeast model to examine the clinically relevant G52E mutation in human POLR1D associated with TCS. We found that the corresponding G73E mutation in y/h AC19 impaired heterodimer formation, consistent with our previous in vitro findings for the human and Drosophila heterodimers.57 Here, we also extended these findings to show that the G73E mutation in the humanized AC19 disrupted Pol I and III complex integrity. This is another example of where a TCS point mutation disrupts both Pol I and III, and it remains unclear if all POLR1D mutations impact both Pol I and III or if any are Pol I specific. Our studies demonstrate that humanization of yeast AC19 makes it more susceptible to mutations than yeast AC19, including TCS G73E. More specifically our results show that humanizing AC19 and possibly other Pol subunits will more accurately reflect the molecular consequence of disease mutations in humans.
To date, less than half of the yeast genes are functionally interchangeable with their human orthologs.64 Previous yeast growth complementation studies show that human POLR1D cannot replace yeast AC19.65 However, with targeted sequence and/or domain replacement we can expand the number of humanized genes that functionally replace their yeast orthologs, like we accomplished with our humanized yeast AC19. The humanized yeast proteins can be used to explore the molecular genetics and biochemistry of human disease mutations. In summary, our humanized AC19 provides a better yeast model to understand the molecular basis for how POLR1D-associated TCS mutations affect Pol I/III function.
MATERIALS AND METHODS
Protein sequence and structure analysis
Pairwise sequence alignments of the small alpha-like homologs were made using HHPred MPI Bioinformatics Toolkit under default settings.47 The pairwise alignments were used to produce a multiple sequence alignment of all the small alpha-like homologs. Protein sequences were obtained from UniProt and the indicated UniProt ID shown in Table S1. To predict what residues were important for heterodimerization in Rpb11 and AC19, an online software BIPSPI+ (https://bipspi.cnb.csic.es/) was used.55 For Rpb11/Rpb3, the structures for chains K and C were uploaded, and for AC19/AC40, the structures for chains O and G were analyzed by BIPSPI + under default settings. We also visually compared the atomic crystal structures of the two alpha-like heterodimers (Rpb11 PDB 1WCM and AC19 PDB 6RUO) in PyMOL. Rpb11 and AC19 residue side chains within the heterodimerization interface pointing towards or near residues of Rpb3 and AC40, respectively, were predicted to be important for protein-protein interaction. All protein structure images were made using PyMOL. PyMOL mutagenesis wizard was used to make the G73E mutation in the structure.
Heterodimer protein expression and purification
Yeast and human versions of wild-type and mutant Rpb11 homologs and wild-type Rpb3 homologs were recombinantly co-expressed in Rosetta 2 (DE3) pLysS Competent cells (Novagen) by autoinduction in TB5052 media for 18 h at 37 °C. Rpb11 homologs were all inserted into a pET-DUET-1 vector and Rpb3 homologs were all inserted into a pCDF-DUET-1 vector. Cells were harvested by centrifugation, and cell pellets were washed once with Tris-buffered saline (TBS) (50 mM Tris Base pH 7.5, 150 mM sodium chloride). Cell pellets were resuspended in 5 mL/g of lysis buffer (20 mM Tris-HCl pH 7.5, 200 mM KCl, 10% glycerol, 0.1 mM EDTA, 20 mM imidazole, 0.05% tergitol) and lysed by sonication. Lysates were cleared by centrifugation, and cleared lysates were added to Ni-NTA agarose beads and incubated for 18 h at 4 °C. The bound beads were then washed four times with wash buffer (20 mM Tris-HCl pH 7.5, 750 mM KCl, 10% glycerol, 0.1 mM EDTA, 20 mM imidazole, 0.05% tergitol) and eluted with 75 μL elution buffer (20 mM Tris-HCl pH 7.5, 200 mM KCl, 10% glycerol, 0.1 mM EDTA, 250 mM imidazole, 0.05% tergitol) three times. Elutions were pooled and analyzed by SDS-PAGE and Western blots with indicated antibodies.
Yeast strains and cell growth assays
Yeast strains used were made in the BY4705 genetic background (MATα ade2-1 ade2Δ:: hisG his3Δ200 leu2Δ0 lys2Δ0 met15Δ0 trp1Δ63-112 ura3Δ0). Rpb11 and AC19 plasmid shuffle strains contained a chromosomal deletion of Rpb11 and AC19, respectively, using the pAG32 hygromycin B deletion cassette. The deletion strains were then transformed with a plasmid construct pRS316 that included either the Rpb11 or AC19 gene locus with a URA3-selectable marker. The indicated Pol subunits were then C-terminally FLAG-tagged in the shuffle strains. The yeast strains were transformed with indicated plasmids (Table S2). Cells were grown on glucose complete (GC) medium lacking leucine, and then spotted or streaked onto GC-Leu + FOA (5-fluoroorotic acid) plates, incubated at the indicated temperatures for 2–4 days, and then assessed for growth relative to WT.
Yeast immunoprecipitations
HA-tagged proteins were purified from mid-log-phase cultures grown in GC media lacking leucine. For HA-tagged Rpb11 IPs, either WT HA-tagged Rpb11 or mutant HA-tagged Rpb11 constructs were transformed into Rpb11 deleted yeast strains containing chromosomally FLAG-tagged indicated Rpb3 genes. A similar system was used for HA-tagged AC19 IPs. Wild-type HA-tagged AC19 or AC19 variants were transformed into AC19 deleted yeast strains in containing chromosomally FLAG-tagged AC40. Cells were collected by centrifugation and lysed by bead beating in a Bioruptor (info) using zirconia beads in lysis buffer (100 mM Tris pH 7.9, 250 mM ammonium sulfate, 1 mM EDTA, 10% glycerol, supplemented with 0.5 mM DTT, 0.5 mM PMSF, and protease inhibitors). Lysates were cleared by centrifugation and approximately 4 mg of the lysate supernatant was diluted with two volumes of dilution buffer (25 mM HEPES pH 7.5, 50 mM NaCl, 1 mM EDTA) and 10 μL of anti-HA magnetic beads (Bimake). Solutions were then incubated overnight at 4 °C on a rotator. The samples were washed three times in 1 mL of TBS containing 0.1% Tween, and proteins were eluted using 1× Laemmeli sample buffer (BioRad) at 95 °C for 5 min. Samples were run on a 4–20% Tris-glycine polyacrylamide gradient gels (BioRad) in Tris-glycine-SDS running buffer for 35 min at 200 volts and analyzed using Coomassie stain or Western blots. Antibodies used were anti-HA-tag antibody (F-7): sc-7392; rabbit FLAG tag polyclonal antibody (Invitrogen PA1-984B), mouse monoclonal anti-FLAG M2 antibody (Sigma F1804).
Supplementary Material
ACKNOWLEDGEMENTS
We thank Knutson laboratory members for their critical review of the manuscript.
Funding Statement
The work was supported by grants to B.A.K. from the U.S. National Institutes of Health [NIDCR R03-DE027785 and NIGMS R01-GM141033]. H.G.P was partially supported by funds from the Post-Baccalaureate Research Education Program at SUNY-Upstate (PREP-Up).
SUPPLEMENTAL MATERIAL
Supplemental data for this article can be accessed online at https://doi.org/10.1080/10985549.2023.218607.
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. Uncropped protein gel and Western blot images are shown in Supplemental Fig. S3 and can be accessed from Mendeley data repository through accessing the link https://data.mendeley.com/datasets/848yvpr3zj.
REFERENCES
- 1.Werner F, Grohmann D.. Evolution of multisubunit RNA polymerases in the three domains of life. Nat Rev Microbiol. 2011;9:85–98. doi: 10.1038/nrmicro2507. [DOI] [PubMed] [Google Scholar]
- 2.Wild T, Cramer P.. Biogenesis of multisubunit RNA polymerases. Trends Biochem Sci. 2012;37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
- 3.Ishihama A. Subunit of assembly of Escherichia coli RNA polymerase. Adv Biophys. 1981;14:1–35. [PubMed] [Google Scholar]
- 4.Kimura M, Ishihama A.. Subunit assembly in vivo of Escherichia coli RNA polymerase: role of the amino-terminal assembly domain of alpha subunit. Genes Cells. 1996;1:517–528. doi: 10.1046/j.1365-2443.1996.d01-258.x. [DOI] [PubMed] [Google Scholar]
- 5.Lalo D, Carles C, Sentenac A, Thuriaux P.. Interactions between three common subunits of yeast RNA polymerases I and III. Proc Natl Acad Sci U S A. 1993;90:5524–5528. doi: 10.1073/pnas.90.12.5524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Martindale DW. A conjugation-specific gene (cnjC) from Tetrahymena encodes a protein homologous to yeast RNA polymerase subunits (RPB3, RPC40) and similar to a portion of the prokaryotic RNA polymerase alpha subunit (rpoA). Nucleic Acids Res. 1990;18:2953–2960. doi: 10.1093/nar/18.10.2953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eloranta JJ, Kato A, Teng MS, Weinzierl RO.. In vitro assembly of an archaeal D-L-N RNA polymerase subunit complex reveals a eukaryote-like structural arrangement. Nucleic Acids Res. 1998;26:5562–5567. doi: 10.1093/nar/26.24.5562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mann C, Buhler JM, Treich I, Sentenac A.. RPC40, a unique gene for a subunit shared between yeast RNA polymerases A and C. Cell. 1987;48:627–637. doi: 10.1016/0092-8674(87)90241-8. [DOI] [PubMed] [Google Scholar]
- 9.Vannini A, Cramer P.. Conservation between the RNA polymerase I, II, and III transcription initiation machineries. Mol Cell. 2012;45:439–446. doi: 10.1016/j.molcel.2012.01.023. [DOI] [PubMed] [Google Scholar]
- 10.Azuma Y, Yamagishi M, Ishihama A.. Subunits of the Schizosaccharomyces pombe RNA polymerase II: enzyme purification and structure of the subunit 3 gene. Nucleic Acids Res. 1993;21:3749–3754. doi: 10.1093/nar/21.16.3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruno T, Corbi N, Di Padova M, De Angelis R, Floridi A, Passananti C, Fanciulli M.. The RNA polymerase II core subunit 11 interacts with keratin 19, a component of the intermediate filament proteins. FEBS Lett. 1999;453:273–277. doi: 10.1016/s0014-5793(99)00733-4. [DOI] [PubMed] [Google Scholar]
- 12.Kimura M, Ishiguro A, Ishihama A.. RNA polymerase II subunits 2, 3, and 11 form a core subassembly with DNA binding activity. J Biol Chem. 1997;272:25851–25855. doi: 10.1074/jbc.272.41.25851. [DOI] [PubMed] [Google Scholar]
- 13.Cieśla M, Makała E, Płonka M, Bazan R, Gewartowski K, Dziembowski A, Boguta M.. Rbs1, a new protein implicated in RNA polymerase III biogenesis in yeast Saccharomyces cerevisiae. Mol Cell Biol. 2015;35:1169–1181. doi: 10.1128/MCB.01230-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel C, Plitzko J, Cramer P.. RNA polymerase I-Rrn3 complex at 4.8 A resolution. Nat Commun. 2016;7:12129. doi: 10.1038/ncomms12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dauwerse JG, Dixon J, Seland S, Ruivenkamp CAL, van Haeringen A, Hoefsloot LH, Peters DJM, Boers AC-d, Daumer-Haas C, Maiwald R, et al. Mutations in genes encoding subunits of RNA polymerases I and III cause Treacher Collins syndrome. Nat Genet. 2011;43:20–22. doi: 10.1038/ng.724. [DOI] [PubMed] [Google Scholar]
- 16.Ghesh L, Vincent M, Delemazure AS, Boyer J, Corre P, Perez F, Genevieve D, Laplanche JL, Collet C, Isidor B.. Autosomal recessive Treacher Collins syndrome due to POLR1C mutations: Report of a new family and review of the literature. Am J Med Genet A. 2019;179:1390–1394. doi: 10.1002/ajmg.a.61147. [DOI] [PubMed] [Google Scholar]
- 17.Giampietro PF, Armstrong L, Stoddard A, Blank RD, Livingston J, Raggio CL, Rasmussen K, Pickart M, Lorier R, Turner A, et al. Whole exome sequencing identifies a POLRID mutation segregating in a father and two daughters with findings of Klippel-Feil and Treacher Collins syndromes. Am J Med Genet A. 2015;167A:95–102. doi: 10.1002/ajmg.a.36799. [DOI] [PubMed] [Google Scholar]
- 18.Kolsi N, Boudaya F, Ben Thabet A, Charfi M, Regaieg C, Bouraoui A, Regaieg R, Hentati N, Hamed AB, Gargouri A.. Treacher Collins syndrome: a case report and review of literature. Clin Case Rep. 2022;10:e6782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu M, Yang B, Chen Z, Jiang H, Pan B.. Phenotype analysis and genetic study of Chinese patients with Treacher Collins syndrome. Cleft Palate Craniofac J. 2022;59:1038–1047. doi: 10.1177/10556656211037509. [DOI] [PubMed] [Google Scholar]
- 20.Schaefer E, Collet C, Genevieve D, Vincent M, Lohmann DR, Sanchez E, Bolender C, Eliot MM, Nurnberg G, Passos-Bueno MR, et al. Autosomal recessive POLR1D mutation with decrease of TCOF1 mRNA is responsible for Treacher Collins syndrome. Genet Med. 2014;16:720–724. doi: 10.1038/gim.2014.12. [DOI] [PubMed] [Google Scholar]
- 21.Trainor PA, Andrews BT.. Facial dysostoses: etiology, pathogenesis and management. Am J Med Genet C Semin Med Genet. 2013;163C:283–294. doi: 10.1002/ajmg.c.31375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ulhaq ZS, Nurputra DK, Soraya GV, Kurniawati S, Istifiani LA, Pamungkas SA, Tse WKF.. A systematic review on Treacher Collins syndrome: correlation between molecular genetic findings and clinical severity. Clin Genet. 2023;103:146–155. doi: 10.1111/cge.14243. [DOI] [PubMed] [Google Scholar]
- 23.Lau MC, Kwong EM, Lai KP, Li JW, Ho JC, Chan TF, Wong CK, Jiang YJ, Tse WK.. Pathogenesis of POLR1C-dependent Type 3 Treacher Collins Syndrome revealed by a zebrafish model. Biochim Biophys Acta. 2016;1862:1147–1158. doi: 10.1016/j.bbadis.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 24.Noack Watt KE, Achilleos A, Neben CL, Merrill AE, Trainor PA.. The roles of RNA polymerase I and III subunits Polr1c and Polr1d in craniofacial development and in zebrafish models of Treacher Collins syndrome. PLoS Genet. 2016;12:e1006187. doi: 10.1371/journal.pgen.1006187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ross AP, Zarbalis KS.. The emerging roles of ribosome biogenesis in craniofacial development. Front Physiol. 2014;5:26. doi: 10.3389/fphys.2014.00026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Terrazas K, Dixon J, Trainor PA, Dixon MJ.. Rare syndromes of the head and face: mandibulofacial and acrofacial dysostoses. Wiley Interdiscip Rev Dev Biol. 2017;6: 10.1002/wdev.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Trainor PA. Craniofacial birth defects: the role of neural crest cells in the etiology and pathogenesis of Treacher Collins syndrome and the potential for prevention. Am J Med Genet A. 2010;152A:2984–2994. doi: 10.1002/ajmg.a.33454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashrafi MR, Amanat M, Garshasbi M, Kameli R, Nilipour Y, Heidari M, Rezaei Z, Tavasoli AR.. An update on clinical, pathological, diagnostic, and therapeutic perspectives of childhood leukodystrophies. Expert Rev Neurother. 2020;20:65–84. doi: 10.1080/14737175.2020.1699060. [DOI] [PubMed] [Google Scholar]
- 29.Bernard G, Vanderver A.. 1993. POLR3-related leukodystrophy. In: Adam MP, Everman DB, Mirzaa GM, editors. GeneReviews((R)). Seattle (WA): University of Washington, Seattle. [PubMed] [Google Scholar]
- 30.Gauquelin L, Cayami FK, Sztriha L, Yoon G, Tran LT, Guerrero K, Hocke F, van Spaendonk RML, Fung EL, D'Arrigo S, et al. Clinical spectrum of POLR3-related leukodystrophy caused by biallelic POLR1C pathogenic variants. Neurol Genet. 2019;5:e369. doi: 10.1212/NXG.0000000000000369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kraoua I, Karkar A, Drissi C, Benrhouma H, Klaa H, Samaan S, Renaldo F, Elmaleh M, Ben Hamouda M, Abdelhak S, et al. Novel POLR1C mutation in RNA polymerase III-related leukodystrophy with severe myoclonus and dystonia. Mol Genet Genomic Med. 2019;7:e914. doi: 10.1002/mgg3.914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Potic A, Brais B, Choquet K, Schiffmann R, Bernard G.. 4H syndrome with late-onset growth hormone deficiency caused by POLR3A mutations. Arch Neurol. 2012;69:920–923. doi: 10.1001/archneurol.2011.1963. [DOI] [PubMed] [Google Scholar]
- 33.Han JY, Kim SY, Cheon JE, Choi M, Lee JS, Chae JH.. A familial case of childhood ataxia with leukodystrophy due to novel POLR1C mutations. J Clin Neurol. 2020;16:338–340. doi: 10.3988/jcn.2020.16.2.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lata E, Choquet K, Sagliocco F, Brais B, Bernard G, Teichmann M.. RNA polymerase III subunit mutations in genetic diseases. Front Mol Biosci. 2021;8:696438. doi: 10.3389/fmolb.2021.696438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Naseer MI, Abdulkareem AA, Pushparaj PN, Saharti S, Muthaffar OY.. Next-generation sequencing reveals novel homozygous missense variant c.934T > C in POLR1C gene causing leukodystrophy and hypomyelinating disease. Front Pediatr. 2022;10:862722. doi: 10.3389/fped.2022.862722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yadav N, Saini J, Nagappa M.. Novel mutation in the POLR1C gene causing hypomyelinating leukodystrophy in an adult. Neurol Clin Pract. 2021;11:e367–e369. doi: 10.1212/CPJ.0000000000001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coulombe B, Derksen A, La Piana R, Brais B, Gauthier MS, Bernard G.. POLR3-related leukodystrophy: How do mutations affecting RNA polymerase III subunits cause hypomyelination? Fac Rev. 2021;10:12. doi: 10.12703/r/10-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kashiki H, Li H, Miyamoto S, Ueno H, Tsurusaki Y, Ikeda C, Kurata H, Okada T, Shimazu T, Imamura H, et al. POLR1C variants dysregulate splicing and cause hypomyelinating leukodystrophy. Neurol Genet. 2020;6:e524. doi: 10.1212/NXG.0000000000000524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pelletier F, Perrier S, Cayami FK, Mirchi A, Saikali S, Tran LT, Ulrick N, Guerrero K, Rampakakis E, van Spaendonk RML, et al. Endocrine and growth abnormalities in 4H leukodystrophy caused by variants in POLR3A, POLR3B, and POLR1C. J Clin Endocrinol Metab. 2021;106:e660–e674. doi: 10.1210/clinem/dgaa700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thiffault I, Wolf NI, Forget D, Guerrero K, Tran LT, Choquet K, Lavallee-Adam M, Poitras C, Brais B, Yoon G, et al. Recessive mutations in POLR1C cause a leukodystrophy by impairing biogenesis of RNA polymerase III. Nat Commun. 2015;6:7623. doi: 10.1038/ncomms8623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bitarafan F, Razmara E, Jafarinia E, Almadani N, Garshasbi M.. A biallelic variant in POLR2C is associated with congenital hearing loss and male infertility: case report. Eur J Clin Invest. 2022;53:e13946. [DOI] [PubMed] [Google Scholar]
- 42.Moriwaki M, Moore B, Mosbruger T, Neklason DW, Yandell M, Jorde LB, Welt CK.. POLR2C mutations are associated with primary ovarian insufficiency in women. J Endocr Soc. 2017;1:162–173. doi: 10.1210/js.2016-1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Walker-Kopp N, Jackobel AJ, Pannafino GN, Morocho PA, Xu X, Knutson BA.. Treacher Collins syndrome mutations in Saccharomyces cerevisiae destabilize RNA polymerase I and III complex integrity. Hum Mol Genet. 2017;26:4290–4300. doi: 10.1093/hmg/ddx317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moir RD, Lavados C, Lee J, Willis IM.. Functional characterization of Polr3a hypomyelinating leukodystrophy mutations in the S. cerevisiae homolog, RPC160. Gene. 2021;768:145259. doi: 10.1016/j.gene.2020.145259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choquet K, Yang S, Moir RD, Forget D, Lariviere R, Bouchard A, Poitras C, Sgarioto N, Dicaire MJ, Noohi F, et al. Absence of neurological abnormalities in mice homozygous for the Polr3a G672E hypomyelinating leukodystrophy mutation. Mol Brain. 2017;10:13. doi: 10.1186/s13041-017-0294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benga WJ, Grandemange S, Shpakovski GV, Shematorova EK, Kedinger C, Vigneron M.. Distinct regions of RPB11 are required for heterodimerization with RPB3 in human and yeast RNA polymerase II. Nucleic Acids Res. 2005;33:3582–3590. doi: 10.1093/nar/gki672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zimmermann L, Stephens A, Nam SZ, Rau D, Kubler J, Lozajic M, Gabler F, Soding J, Lupas AN, Alva V.. A completely reimplemented MPI bioinformatics toolkit with a new HHpred server at its core. J Mol Biol. 2018;430:2237–2243. doi: 10.1016/j.jmb.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 48.Armache KJ, Mitterweger S, Meinhart A, Cramer P.. Structures of complete RNA polymerase II and its subcomplex, Rpb4/7. J Biol Chem. 2005;280:7131–7134. doi: 10.1074/jbc.M413038200. [DOI] [PubMed] [Google Scholar]
- 49.Fernandez-Tornero C, Moreno-Morcillo M, Rashid UJ, Taylor NM, Ruiz FM, Gruene T, Legrand P, Steuerwald U, Muller CW.. Crystal structure of the 14-subunit RNA polymerase I. Nature. 2013;502:644–649. doi: 10.1038/nature12636. [DOI] [PubMed] [Google Scholar]
- 50.He Y, Yan C, Fang J, Inouye C, Tjian R, Ivanov I, Nogales E.. Near-atomic resolution visualization of human transcription promoter opening. Nature. 2016;533:359–365. doi: 10.1038/nature17970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Misiaszek AD, Girbig M, Grotsch H, Baudin F, Murciano B, Lafita A, Muller CW.. Cryo-EM structures of human RNA polymerase I. Nat Struct Mol Biol. 2021;28:997–1008. doi: 10.1038/s41594-021-00693-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Boeke JD, Trueheart J, Natsoulis G, Fink GR.. 5-Fluoroorotic acid as a selective agent in yeast molecular genetics. Methods Enzymol. 1987;154:164–175. [DOI] [PubMed] [Google Scholar]
- 53.Goldstein AL, McCusker JH.. Three new dominant drug resistance cassettes for gene disruption in Saccharomyces cerevisiae. Yeast. 1999;15:1541–1553. doi:. [DOI] [PubMed] [Google Scholar]
- 54.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P.. RNA polymerase I structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 55.Sanchez-Garcia R, Sorzano COS, Carazo JM, Segura J.. BIPSPI: a method for the prediction of partner-specific protein-protein interfaces. Bioinformatics. 2019;35:470–477. doi: 10.1093/bioinformatics/bty647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vincent M, Genevieve D, Ostertag A, Marlin S, Lacombe D, Martin-Coignard D, Coubes C, David A, Lyonnet S, Vilain C, et al. Treacher Collins syndrome: a clinical and molecular study based on a large series of patients. Genet Med. 2016;18:49–56. doi: 10.1038/gim.2015.29. [DOI] [PubMed] [Google Scholar]
- 57.Palumbo RJ, Belkevich AE, Pascual HG, Knutson BA.. A clinically-relevant residue of POLR1D is required for Drosophila development. Dev Dyn. 2022;251:1780–1797. doi: 10.1002/dvdy.505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wai HH, Vu L, Oakes M, Nomura M.. Complete deletion of yeast chromosomal rDNA repeats and integration of a new rDNA repeat: use of rDNA deletion strains for functional analysis of rDNA promoter elements in vivo. Nucleic Acids Res. 2000;28:3524–3534. doi: 10.1093/nar/28.18.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Han Y, Yan C, Fishbain S, Ivanov I, He Y.. Structural visualization of RNA polymerase III transcription machineries. Cell Discov. 2018;4:40. doi: 10.1038/s41421-018-0044-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gao M, Zhou H, Skolnick J.. Insights into disease-associated mutations in the human proteome through protein structural analysis. Structure. 2015;23:1362–1369. doi: 10.1016/j.str.2015.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kannan N, Chander P, Ghosh P, Vishveshwara S, Chatterji D.. Stabilizing interactions in the dimer interface of alpha-subunit in Escherichia coli RNA polymerase: a graph spectral and point mutation study. Protein Sci. 2001;10:46–54. doi: 10.1110/ps.26201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Livesey BJ, Marsh JA.. The properties of human disease mutations at protein interfaces. PLoS Comput Biol. 2022;18:e1009858. doi: 10.1371/journal.pcbi.1009858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Redler RL, Das J, Diaz JR, Dokholyan NV.. Protein destabilization as a common factor in diverse inherited disorders. J Mol Evol. 2016;82:11–16. doi: 10.1007/s00239-015-9717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Laurent JM, Young JH, Kachroo AH, Marcotte EM.. Efforts to make and apply humanized yeast. Brief Funct Genomics. 2016;15:155–163. doi: 10.1093/bfgp/elv041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kachroo AH, Laurent JM, Yellman CM, Meyer AG, Wilke CO, Marcotte EM.. Evolution. Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science. 2015;348:921–925. doi: 10.1126/science.aaa0769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials. Uncropped protein gel and Western blot images are shown in Supplemental Fig. S3 and can be accessed from Mendeley data repository through accessing the link https://data.mendeley.com/datasets/848yvpr3zj.