Abstract
A group of highly efficient Zn(II)-dependent RNA-cleaving deoxyribozymes has been obtained through in vitro selection. They share a common motif with the ‘8–17’ deoxyribozyme isolated under different conditions, including different design of the random pool and metal ion cofactor. We found that this commonly selected motif can efficiently cleave both RNA and DNA/RNA chimeric substrates. It can cleave any substrate containing rNG (where rN is any ribonucleotide base and G can be either ribo- or deoxyribo-G). The pH profile and reaction products of this deoxyribozyme are similar to those reported for hammerhead ribozyme. This deoxyribozyme has higher activity in the presence of transition metal ions compared to alkaline earth metal ions. At saturating concentrations of Zn2+, the cleavage rate is 1.35 min–1 at pH 6.0; based on pH profile this rate is estimated to be at least ~30 times faster at pH 7.5, where most assays of Mg2+-dependent DNA and RNA enzymes are carried out. This work represents a comprehensive characterization of a nucleic acid-based endonuclease that prefers transition metal ions to alkaline earth metal ions. The results demonstrate that nucleic acid enzymes are capable of binding transition metal ions such as Zn2+ with high affinity, and the resulting enzymes are more efficient at RNA cleavage than most Mg2+-dependent nucleic acid enzymes under similar conditions.
INTRODUCTION
Deoxyribozymes, or DNA enzymes, have been obtained through in vitro selection (1–5). Although so far no deoxyribozymes have been found in nature, a variety of deoxyribozymes have been isolated in vitro that can catalyze different chemical reactions, including DNA or RNA cleavage (6–12), ligation (13), phosphorylation (14), cleavage of phosphoramidate bonds (15) and porphyrin metallation (16). Of particular interest are deoxyribozymes with RNA-cleaving activity. In addition to their low cost and high stability against chemical and nuclease degradations, the combination of catalytic activity and substrate recognition ability makes RNA-cleaving deoxyribozymes attractive candidates for biochemical and pharmaceutical applications. The catalytic efficiency of 109 M–1min–1 observed for the ‘10–23’ deoxyribozyme by Santoro and Joyce (12) rivals protein ribonuclease. It has been shown to efficiently destroy the hepatitis B viral RNA (17), abnormal BCR-ABL fusion mRNA (18,19) and c-myc RNA in vitro (20,21).
Most nucleic acid enzymes require metal ions for catalytic functions, either through facilitation of the folding of DNA and RNA into stable tertiary structures, or through direct participation in the chemical reaction (22–25). Compared to protein enzymes, nucleic acid enzymes utilize a limited number of metal ions. Protein enzymes recruit metal ions from almost all groups of metals, including even the second and third period transition metal ions such as Mo and W (26–28). Most naturally occurring nucleic acid enzymes are only active in the presence of alkaline earth metal ions and, in some cases, Mn2+ (22–25). One exception is the hammerhead ribozyme, which is also active in the presence of the transition metal ions Co2+, Zn2+ and Cd2+, although spermine is required for Zn2+- and Cd2+-catalyzed reactions (29).
We are interested in isolating nucleic acid enzymes that specifically use transition metal ions. At least two benefits can be envisioned from this endeavor. First, recruiting transition metal ions may broaden the scope and increase the efficiency of nucleic acid enzyme catalysis. Divalent metal ions in RNA-cleaving nucleic acid enzymes have been proposed to serve either as Lewis acids in the form of M2+ or as general bases in the form of M(OH)+ (22). Since transition metal ions are generally better Lewis acids and their hydroxides are usually more basic than alkali and alkaline earth metal hydroxides, transition metal ions such as Zn2+ may be superior metal cofactors for DNA or RNA enzymes. Indeed, many hydrolytic protein enzymes, such as carboxypeptidase, phosphotriesterase and alkaline phosphatase, use Zn2+, Fe2+ and Mn2+ for catalysis (30–32). Second, transition metal ions are more amenable to spectroscopic studies than alkaline earth metal ions, and as a result, will be able to provide more information about the structure of metal-binding sites. A clear understanding of the structure of metal-binding sites and their changes during catalysis is necessary for designing better nucleic acid enzymes. As an initial target, we searched for highly efficient Zn(II)-dependent RNA-cleaving deoxyribozymes. As has been shown for zinc proteins, structural information about spectroscopically silent Zn(II)-binding sites can be gained through metal substitution studies (33,34).
In vitro selection has been employed successfully to search for nucleic acid enzymes that require a specific metal ion. For example, Pan and Uhlenbeck have obtained a Pb2+-dependent ribozyme through in vitro selection (35). It uses Pb2+ as the only metal-cofactor (36). The metal-binding specificity of the group I intron (37) and RNase P ribozymes (38) has been changed through in vitro selection, resulting in Ca(II)- rather than Mg(II)-dependent enzymes. Cu(II)-dependent DNA enzymes have been selected that can perform either ligase (13) or transesterase (9,39) activity. Similarly, small Zn2+ binding motifs have been selected using Zn2+-affinity column (40,41). Finally, Breaker and Joyce have carried out in vitro selection of RNA-cleaving DNA enzymes in the presence of 1 mM of Zn2+ (7). However, no sequence or detailed characterizations were reported. In vitro selection can also be used to improve metal-binding affinity. High metal-binding affinity is desirable because it can improve the activity as well as facilitate spectroscopic studies of the metal-binding sites. Hammerhead ribozyme variants with a lowered concentration requirement for Mg2+ have been isolated through decreasing metal ion concentrations during in vitro selection (42,43).
Here we present the results of an in vitro selection search for a highly efficient DNA enzyme with low level of requirement for Zn2+. Selection stringency was increased through decreasing both the reaction time and concentration of Zn2+. After 12 rounds of selective amplification and 6 rounds of re-selection, we obtained a group of Zn(II)-dependent DNA enzymes (17E) that are highly active and require 10–100-fold less metal ion than most ribozymes. Surprisingly, the catalytic core sequence of this deoxyribozyme was also found to be similar to the 8–17 motif selected under different conditions including different designs of the random pool and different metal ion cofactors (12). Previous studies have focused mainly on the Mg(II)-dependent 10–23 deoxyribozyme obtained during the same selection (44). No detailed mechanistic characterization of the 8–17 motif has been reported. Because this commonly selected motif is highly efficient in RNA cleavage, it has great potential in biochemical and therapeutic applications. In addition, since 17E deoxyribozyme is specific for transition metal ions and has low metal concentration requirement, it enables spectroscopic characterizations to address the role of metal ions in nucleic acid enzyme catalysis. Therefore, we carried out a detailed study of this commonly selected motif, including its sequence requirement, kinetic properties, metal ion preference and mechanism of RNA cleavage.
MATERIALS AND METHODS
Oligonucleotides
DNA oligonucleotides were purchased from Integrated DNA Technologies Inc. Sequences of the random DNA template and the primers (P1, P2 and P3) used in PCR amplifications are shown in Figure 1a. Primer P1b and P3b are the 5′-biotinylated version of primers P1 and P3, respectively. Primer P1a and P3a were prepared by 5′-labeling P1 and P3 with [γ-32P]ATP (Amersham) and T4 polynucleotide kinase (Gibco). The DNA/RNA chimeric substrate (17DS) for trans-cleavage assays has the sequence 5′-ACTCACTATrAGGAAGAGATG-3′, where rA denotes a single ribonucleotide. The all-RNA substrate (17RS) with the same sequence was synthesized by Dharmacon Research. All oligonucleotides were purified using denaturing polyacrylamide gel electrophoresis.
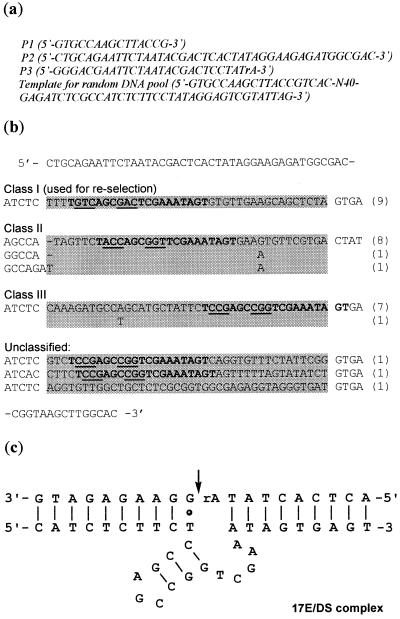
Figure 1.
(a) Sequence of the primers and template for in vitro selection. (b) Sequence alignment of individual clones of cis-cleaving DNA enzymes after 12 rounds of selection. Sequences corresponding to the 40 initially randomized nucleotides are highlighted. The highly conserved region of 20 nt is shown in bold. The nucleotides that can form base pairs within each sequence are underlined. The numbers in the parentheses are the numbers of clones having the indicated sequence. (c) The proposed secondary structure of the engineered trans-cleaving deoxyribozyme–substrate complex 17E/DS. The arrow indicates the cleavage site.
Preparation of random DNA pool
The initial pool for DNA selection was prepared by template-directed extension followed by PCR amplification. The extension was carried out with 200 pmol of DNA template containing a 40-nt random sequence region and 400 pmol of primer P3b in 20× 100 µl reaction mixtures for four thermal-cycles (1 min at 92°C, 1 min at 52°C and 1 min at 72°C). Reaction buffer also included 0.05 U/µl Taq polymerase (Gibco), 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris–HCl (pH 8.3 at 25°C), 0.01% gelatin and 0.2 mM of each dNTP. Subsequently, 1 nmol each of P1 and P3b were added to the extension product to allow four more cycles of PCR amplification. The products were precipitated with ethanol and dissolved in 0.5 ml of 50 mM HEPES (pH 7.0) containing 500 mM NaCl and 20 µM EDTA (buffer A).
In vitro selection
In vitro selection was carried out using the protocol designed by Breaker and Joyce (7). The random DNA pool was immobilized on a NeutrAvidin column (Pierce) by incubating with the column materials for 30 min. The unbound DNA strands were washed off the column with at least 5× 100 µl of buffer A. The non-biotinylated strands of immobilized DNA were washed off with 5× 100 µl of freshly prepared 0.2 N NaOH. The column was then neutralized with 5× 100 µl of 500 mM NaCl in 50 mM HEPES (pH 7.0). The cleavage reaction was carried out by incubating the immobilized single-stranded DNA containing rA with 3× 20 µl of reaction buffer containing 1 mM ZnCl2, 500 mM NaCl and 50 mM HEPES (pH 7.0 at 25°C) over 1 h. The eluted DNA strands were pooled and precipitated with ethanol. A fraction of the selected DNA was amplified in 100 µl PCR reaction mixtures containing 40 pmol each of primers P1 and P2 over 10–20 thermal cycles. One- tenth of the PCR product was further amplified for six cycles with 50 pmol of primers P1 and P3b. The final PCR product was ethanol precipitated and used to initiate the next round of selection. The concentration of ZnCl2 in the reaction buffer for the following rounds of selection was kept constant at 100 µM. Reaction time was gradually decreased from 1 h to 30 s within 12 rounds of selection.
The twelfth generation of selectively amplified DNA was cloned using TA-TOPO Cloning Kit (Invitrogen) and sequenced with T7 Sequenase 2.0 Quick-denatured Plasmid Sequencing Kit (Amersham). Based on the sequence of class I DNA (Fig. 1b), a partially degenerate DNA pool was synthesized (Integrated DNA Technology Inc.) with 20% degeneracy at the N40 region. In other words, during the oligonucleotide synthesis of the N40 region, the wild type sequence was introduced at a probability of 80% at each position, while the other three nucleotides occurred at a probability of 6.67% (45). There were approximately eight mutations per sequence on average. Re-selection was initiated with 100 pmol of the partially randomized pool. The reaction time was decreased from 20 to 1 min over six generations, and the concentration of ZnCl2 was decreased from 20 to 5 µM.
Kinetic analysis
Kinetic studies of intermolecular cleavage were carried out in the presence of excess enzyme unless specified otherwise. Deoxyribozyme and radioisotope-labeled substrate were annealed in 50 mM HEPES (pH 7.0) or MES (pH 6.0) buffer by heating at 95°C for 2 min and then cooling to room temperature over 15 min. The reaction was initiated by adding divalent metal ion to the desired concentrations shown in the plots. The final concentrations were 5 µM deoxyribozyme and 0.5 nM substrate unless otherwise noted. The reaction was quenched at various time points with stop-buffer containing 8 M urea, 50 mM EDTA, 0.05% xylene cyanol and 0.05% bromophenol blue. The products and the un-cleaved substrate were separated on a 20% denaturing polyacrylamide gel and analyzed with a PhosphorImager (Molecular Dynamics). The percentage of product was plotted against time. Since the reaction was carried out with the deoxyribozyme in large excess over the substrate, it followed the rate-law of pseudo-first order kinetics. The observed rate constant kobs was obtained by curve fitting using SigmaPlot based on the equation y = y0 + a(1 – e–kt), where y is the percentage of product at time t, y0 is the background cleavage at t = 0 and is typically <1%, a is the fraction reacted at t = ∞, and k is the observed rate constant. Increasing the enzyme or substrate concentration 5-fold had little effect on the cleavage rate. The kinetic parameters obtained from duplicate assays performed on different days varied by <20%.
Product identification with MALDI-TOF mass spectrometry
The products of the reaction catalyzed by the deoxyribozyme were generated in a mixture (150 µl) containing 0.2 nmol each of the deoxyribozyme and the substrate (17DS or 17RS), 0.5 mM ZnCl2 and 50 mM HEPES (pH 7.0, 25°C). The reaction was stopped by adding 5 µl of 100 mM EDTA after 20 min, at which time the substrate was cleaved to >95% as analyzed with 32P-labeled substrate under similar conditions. The reaction mixture was desalted with Sep-Pak C18 cartridge (Waters), lyophilized, and dissolved with 50% CH3CN/H2O. After being diluted to 10 µM with the matrix containing 2,4,6-trihydroxy acetophenone and diammonium citrate, 1 µl of the sample was spotted on a gold-coated plate. The spectrum was acquired with a PerSeptive Biosystems Voyager DE-STR MALDI-TOF workstation in the linear-extraction, positive-ion mode. The acceleration voltage was 20 000 V and the delay was 150 ns.
RESULTS
In vitro selection
The DNA molecules capable of RNA-cleaving in the presence of Zn2+ were obtained through in vitro selection. The protocol designed by Breaker and Joyce for the in vitro selection of RNA-cleaving deoxyribozyme (7) was adopted in this study. The initial selection pool contained ~1014 out of the possible 1024 DNA sequences. These molecules contained a random sequence domain of 40 nt flanked by two conserved primer-binding regions (Fig. 1a). A ribonucleic adenosine was embedded in the 5′-conserved sequence region and was intended to be the cleavage site due to the relative lability of the RNA bond toward hydrolytic cleavage. The DNA pool was immobilized on a NeutrAvidin column through the biotin moiety labeled on the 5′ of the DNA. The sequences that underwent cleavage at the internal RNA bond in the presence of Zn2+ were eluted off the column, amplified and used to seed the following round of selection. The activity of the selected DNA gradually increased until the twelfth generation, of which the DNA was cloned and sequenced.
The sequences of 30 individual clones were divided into three major classes based on sequence similarities. Differences among members of each class were limited to a few point mutations (Fig. 1b). A highly conserved sequence of 20 nt, 5′-TX3AGCY3TCGAAATAGT-3′, was observed in all but one sequence albeit at different locations. X3 and Y3 are complimentary and covariant, indicating that they may form base pairs. Representative members from each sequence class were tested for their catalytic activity. The observed rate constants of cis-cleaving DNA enzymes from class I and class II were 0.1–0.2 min–1 in the selection buffer containing 100 µM Zn2+, while class III was less active with kobs around 0.02 min–1. The cleavage rate of the initial pool under the same conditions was 2 × 10–7 min–1. Therefore, a 106-fold increase in cleavage rate was achieved after 12 rounds of in vitro selection. In order to further improve the activity and decrease the metal ion requirement, random mutations were introduced into the sequence of class I DNA (see Materials and Methods section). Six rounds of re-selection with incrementally decreasing reaction time and Zn2+ concentration were carried out. Sequence analysis revealed that variations of the re-selected DNA sequences were mostly outside of the 5′-TX3AGCY3TCGAAATAGT-3′ region.
Secondary structure analysis and sequence requirement investigation
Sequence analysis of the Zn(II)-dependent DNA enzymes from both primary selection and re-selection revealed a common structural motif, which was then engineered into a trans-cleaving complex by separating the enzyme and substrate domains. It consists of two double helices flanking the cleavage site and a catalytic core of 14 nt with the sequence 5′-X3AGCY3TCGAA-3′ (Fig. 1c). This motif is similar to that of the 8–17 deoxyribozymes selected by Santoro and Joyce for the cleavage of an all-RNA strand in the presence of Mg2+ (12). The unpaired region in the 8–17 deoxyribozymes has the sequence 5′-WCGR-3′ or 5′-WCGAA-3′ (W = A or T; R = A or G). Among the Zn(II)-dependent DNA enzymes obtained after re-selection, 85% of their sequences fell within the 5′-WCGAA-3′ regime (W = A or T). The sequence of the two double helices flanking the catalytic core is totally different between the 8–17 deoxyribozymes and the Zn(II)-dependent deoxyribozymes presented here.
As observed with the 8–17 deoxyribozymes, the sequence of the 3-bp stem was variable and contained at least two G-C base pairs. Immediately downstream of the scissile phosphodiester bond is an invariant G-T wobble pair. It was reported with the 8–17 deoxyribozymes that substitution of a Watson–Crick pair at this position eliminated catalytic activity (12). We wondered if the order of G-T pair was also important, therefore a pair of enzyme–substrate complexes were synthesized with the G-T wobble pair switched to T-G. No cleavage activity was observed. In the sequence of the class I DNA there are two G-T pairs near the cleavage site, while all other sequence classes contain only one G-T pair and are still active. An enzyme/substrate complex (17E/DS) was constructed (Fig. 1c) consisting of one G-T pair immediately downstream of the cleavage site and three G-C base pairs in the stem of the short hairpin. 17E/DS showed improved kcat and Km when compared to the original sequence derived from class I DNA. The single-turnover kcat and Km were 0.24 min–1 and 654 nM for 17E/DS (Table 1), and 0.07 min–1 and 1.6 µM for class I DNA respectively in the presence of 100 µM Zn2+ at pH 6.0.
Table 1. Kinetic parameters of 17E deoxyribozyme in the presence of Zn2+ at pH 6.0.
| Substrate | kcat (min–1) | Km (µM) | kmax (min–1) | Kd,Zn (mM) |
|---|---|---|---|---|
| All-RNA (17RS) | 0.064 | 1.1 | 0.64 | 0.88 |
| DNA/RNA chimera (17DS) | 0.24 | 0.65 | 1.35 | 0.97 |
The kcat and Km were obtained with 10 µM Zn2+.
The Zn(II)-dependent 17E deoxyribozymes and the 8–17 deoxyribozymes cleave after an unpaired ribonucleotide A. In order to investigate the specificity of this base, four substrates were assayed simultaneously. Their sequences differed only at the target site by being rA, rC, rG or rU. Under single turnover condition with 500 µM Zn2+ at pH 6.0, the observed rate constants were 0.49, 0.066, 0.57, 0.029 min–1 for 17DS-rA, rC, rG, rU respectively.
Comparison of the activity of 17E for DNA/RNA chimeric and all-RNA substrates
17E was selected to cleave a DNA/RNA chimeric substrate where a single RNA bond was embedded within a DNA strand. This approach has been successfully utilized to obtain deoxyribozymes that can catalyze RNA bond cleavage (6–8,11). However, the DNA enzymes obtained from previous studies were either inactive or ~1000-fold less active toward all-RNA substrates. Since 17E has a similar catalytic core as the 8–17 motif, which targets at RNA strand, it is expected that 17E should also be able to cleave all-RNA substrate. The cleavage efficiency of 17E was assayed for the DNA/RNA chimeric substrate 17DS and the all-RNA substrate 17RS (Table 1). Both substrates were specifically cleaved at a single position. Under similar conditions, the cleavage rate of 17RS was only two to three times slower than that of 17DS. The apparent Zn2+ binding affinity was comparable for 17DS and 17RS.
Effect of metal ions and pH
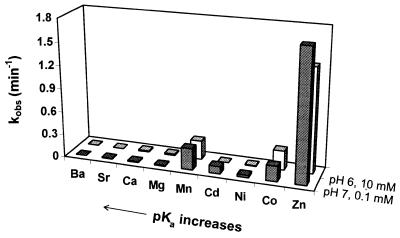
Divalent metal ions were required for the activity of 17E as no more than 1% of the substrate was cleaved after 48 h in the presence of 100 µM EDTA. The rate of 17E-catalyzed RNA cleavage was assayed in the presence of different metal ions. The activity of 17E with each metal ion was measured under two different conditions, either with 100 µM M2+ at pH 7.0 or with 10 mM M2+ at pH 6.0. As shown in Figure 2 and Table 2, the cleavage rate followed the order of Zn2+ >> Mn2+ ~ Co2+ > Cd2+ > Ni2+ > Mg2+ ~ Ca2+ > Sr2+ ~ Ba2+. The activity of 17E was significantly higher with Zn2+ than with other divalent metal ions. In general, transition metal ions were favored over alkaline earth metal ions.
Figure 2.
The rate of 17DS catalyzed by17E deoxyribozyme in the presence of different divalent metal ions. The assays were carried out under enzyme excess conditions with either 100 µM M2+ at pH 7.0 (shaded bars) or 10 mM M2+ at pH 6.0 (non-shaded bars). The values of the rate constants and the pKa of the hydrated metal are listed in Table 1.
Table 2. Activity of 17E/DS under single turnover conditions with 100 µM M2+ at pH 7.0 or with 10 mM M2+ at pH 6.0.
| Cofactor | kobs (min–1) | pKa (59) | |
|---|---|---|---|
| 100 µM, pH 7 | 10 mM, pH 6 | ||
| Ba2+ | – | 0.0003 | 13.82 |
| Sr2+ | 0.0058 | 0.0005 | 13.18 |
| Mg2+ | 0.011 | 0.017 | 11.42 |
| Ca2+ | 0.012 | 0.015 | 12.7 |
| Ni2+ | 0.029 | 0.008 | 9.86 |
| Cd2+ | 0.095 | – | 10.08 |
| Co2+ | 0.20 | 0.25 | 9.65 |
| Mn2+ | 0.28 | 0.24 | 10.6 |
| Zn2+ | 1.7 | 1.35 | 8.96 |
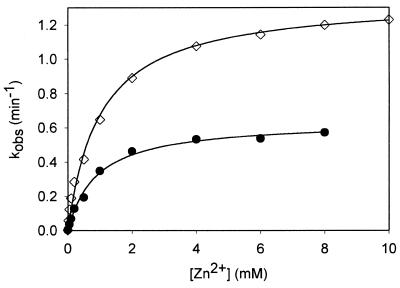
The dependence of the cleavage rate of 17E/DS complex on Zn2+ concentration was investigated. More than 95% of the substrate was cleaved for the Zn2+ concentrations tested, suggesting that the majority of the enzyme–substrate complexes were catalytically active. At pH 7.0 and Zn2+ concentrations >0.3 mM, the high cleavage rates (>1 min–1) precluded accurate determination of the rate constants with common assay method of initiating and terminating the reaction with manual pipette mixing. Thus, the catalytic reactions were performed at pH 6.0, where the cleavage rate should be 10-fold slower (based on the pH-dependent rate profile presented below). The catalytic rate was half-maximal at 1 mM Zn2+, and the apparent maximal rate of cleavage (kmax) was 1.35 min–1 (Fig. 3). Under simulated physiological condition containing 150 mM NaCl, 2 mM Mg2+ and 20 µM Zn2+ at pH 7.5 and 37°C, the kobs for the cleavage of 17DS catalyzed by 17E deoxyribozyme was found to be 0.38 min–1. Therefore 17E is well suited to function under physiological condition.
Figure 3.
Zinc-binding curves of the all-RNA (circle) and DNA/RNA chimeric substrates (diamond) at pH 6.0. The data were fitted with non-linear regression into equation kobs = kmax[Zn2+]/(Kd,Zn + [Zn2+]); kmax is the maximal cleavage rate obtained at saturating concentrations of Zn2+, Kd,Zn is the apparent binding constant for Zn2+.
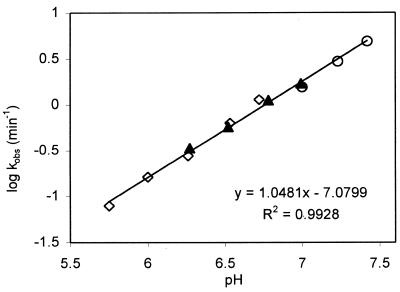
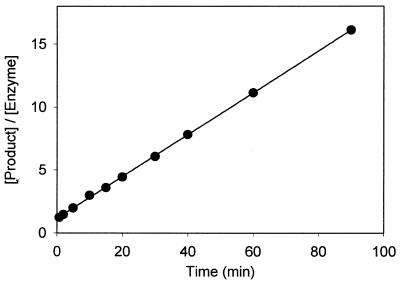
The pH dependence of 17E/DS in the presence of Zn2+ was studied in three different buffers spanning a pH range from 5.7 to 7.5 (Fig. 4). The log value of kobs increased linearly with a slope of 1.05 as pH increased. With this pH profile and a kmax of 1.35 min–1 at pH 6.0 (Table 3), the maximal cleavage rate of 17E was estimated to be ~50 min–1 at pH 7.5. As shown in Figure 5, the 17E deoxyribozyme was also able to catalyze multiple-turnovers of substrate cleavage with rate (k2 = 0.17 min–1) comparable to that obtained under single-turnover conditions (kobs = 0.2 min–1) in the presence of 100 µM Zn2+ at pH 6.0.
Figure 4.
The pH dependence of the cleavage rate of substrate 17DS. Reactions were carried out with 100 µM ZnCl2 in 50 mM MES (diamond, pH 5.7–6.7), or PIPES (triangle, pH 6.2–7.0) or HEPES (circle, pH 7.0–7.4) at 25°C. The data were fitted linearly with a slope of 1.05.
Table 3. Effect of monovalent salts on the cleavage rate of 17E/DS in the presence of 100 µM ZnCl2 and 50 mM MES (pH 6.0).
| Type of salt | kobs (min–1) |
|---|---|
| Buffer only | 0.20 |
| LiCl | 0.089 |
| NaCl | 0.12 |
| KCl | 0.10 |
| NH4Cl | 0.097 |
| RbCl | 0.18 |
| NaN3 | 0.065 |
| NaF | 0.082 |
| NaNO3 | 0.11 |
| NaBr | 0.12 |
| NaI | 0.12 |
| NaClO4 | 0.13 |
The concentration of monovalent cation (Na+) in buffer only is <50 mM. The concentration of added monovalent salt is 150 mM.
Figure 5.
Profile of multiple turnover assays with 10 µM 17DS and 0.1 µM 17E in the presence of 100 µM ZnCl2 at pH 6.0. The obtained kcat is 0.17 min–1 based on linear curve fitting.
Since 17E deoxyribozyme was selected in the presence of 500 mM NaCl, the role of NaCl and other monovalent salts in enzyme activity was investigated (Table 3). In fact, NaCl was not required for the intermolecular cleavage activity of 17E. Moreover, monovalent salts in the concentration range tested (50 mM to 2 M) inhibited the substrate cleavage in the presence of low to moderate concentrations of Zn2+. Adding 150 mM of the chloride salt of Li+, Na+, K+ and NH4+ reduced the cleavage rate ~2-fold (Table 3). Stronger inhibition was observed with N3- and F–, probably due to their tighter binding with Zn2+.
Product identification
The cleavage products of both the DNA/RNA chimeric substrate (17DS) and the all-RNA substrate were identified using MALDI mass spectrometry. Two major signals were detected for the cleavage products of 17DS with the mass of 3050 ± 2 Da and 3141 ± 2 Da, corresponding to the 5′-product with 2′,3′-cyclic phosphate terminus (calculated mass = 3048) and the 3′-product with a free 5′-OH group (calculated mass = 3141), respectively. The mass spectrum of the cleavage products of 17RS was also consistent with the formation of 2′,3′-cyclic phosphate (calculated mass = 3150; observed mass = 3152 ± 2 Da) and 5′-hydroxyl termini (calculated mass = 3287, observed mass = 3288 ± 2 Da).
DISCUSSION
Two different in vitro selection schemes resulted in a common structural motif
The sequence of the proposed catalytic core (5′-TX3AGCY3WCGAA-3′, W = A or T, X3 and Y3 form base pairs) of 17E deoxyribozyme was highly conserved. It was surprising that this region is similar to one of the sequences of 8–17 motif selected by Santoro and Joyce (12). The sequence of the unpaired region of the 8–17 deoxyribozymes can be either 5′-WCGR-3′ or 5′-WCGAA-3′ (W = A or T; R = A or G). The selection of 17E was carried out with 106-base oligonucleotides containing a 40-nt random sequence region with a single ribonucleotide as the target cleavage site, while the selection of the 8–17 motif was performed on 99-base oligonucleotides containing 50-base random sequence with the target cleavage site consisting of 12 ribonucleotides. Selections of the 17E and the 8–17 motif not only started with different designs of the initial library, but were also carried out under different selection conditions (17E: ZnCl2, 500 mM NaCl, 50 mM HEPES, pH 7.0, 25°C; 8–17: MgCl2, 1 M NaCl, 50 mM Tris–HCl, pH 7.5, 37°C). A common solution to the same task of phosphodiester cleavage has been obtained from different approaches. It demonstrated a remarkable example of convergent evolution.
As observed for both the 17E and the 8–17 deoxyribozymes, the G-T wobble base pair located immediately downstream of the cleavage site was highly conserved. Substitution of a Watson–Crick base pair at this position in the 8–17 motif (12) eliminated catalytic activity. A G-U wobble base pair was also observed immediately downstream of the cleavage site of the HDV ribozyme and the group I intron (46,47). As revealed by the crystal structure of the hammerhead ribozyme and the leadzyme (48–50), the active sites of ribozymes are usually not properly positioned to allow inline nucleophilic attack in its ground state. An unstable G-U or G-T base pair around the cleavage site may be required for conformational changes toward transition state structures. However, stability may not be the only explanation for the importance of this wobble pair. When we switched the G-T pair in the 17E deoxyribozyme to T-G, the enzyme was also inactivated, indicating that not only the identity, but also the order of this wobble pair is important. The G-T wobble base pair could form an important metal binding site required for chemical reaction. As observed with the group I intron, two out of the three metal binding sites are formed by G-U wobble base pairs (51).
A ribonucleotide adenosine was the cleavage target of 17E deoxyribozyme during the in vitro selection. The results presented here indicate that this adenosine can be substituted with any ribonucleotide. Therefore, the target preference of 17E deoxyribozyme can be defined as 5′-NG-3′ (rN is any ribonucleotide and G can be either ribo- or deoxyribo-G). Although sequences of the form 5′-RG-3′ (R = A or G) have higher cleavage rates, the cleavage efficiency of 17E at the purine residues (C and U) can be improved by optimizing reaction conditions, such as pH and metal ion concentration. The G at the 3′-side of the target nucleotide can be either a DNA or an RNA base. The target choice of the 10–23 deoxyribozyme has the form of 5′-RY-3′ (R = unpaired A or G; Y = paired U or C) (12), while that of the hammerhead ribozyme is 5′-HH-3′ (H = any nucleotide except G) (52). Therefore the 17E deoxyribozyme, the 10–23 motif, and the hammerhead ribozyme can be selectively utilized to target different sequence regions in the inhibition of gene expression.
The 17E deoxyribozymes are efficient in cleaving both chimeric DNA/RNA and all-RNA substrates
Although the DNA enzyme in the present study was selected to cleave a single ribonucleotide bond within a DNA substrate, it can also specifically cleave an all-RNA substrate in a sequence- dependent manner (Table 1). This finding indicates that the 2′-OH groups in the enzyme–substrate recognition regions are not required for 17E-enzyme catalysis. As was also observed with the 10–23 motif (53), the 17E deoxyribozyme has higher Km and lower kcat when the substrate is all-RNA instead of DNA/RNA chimera. Studies with the 10–23 deoxyribozyme attributed this enhancement in rate to a higher level of B-form like helix formed by the DNA/DNA complex than by the DNA/RNA complex (53). Under the conditions of chemical cleavage being the rate-limiting step, enhancements of the cleavage rate of some hammerhead ribozymes were also observed when deoxyribonucleotide substitutions were made in the enzyme/substrate hybridizing arms (54–57). It was proposed that deoxyribonucleotide substitution in the enzyme–substrate-binding region made the structure of the ribozyme–substrate complex closer to the structure of the transition state and therefore entropically favored the reaction (54). Such proposals may also apply to 17E deoxyribozyme for its higher activity with DNA/RNA chimeric substrate than with RNA substrate.
17E prefers transition metals to alkaline earth metal ions
It is interesting that this common motif survived from selections in the presence of either Mg2+ (in the work by Santoro and Joyce) (12) or Zn2+ (in the present study). In the selection of deoxyribozymes using 50 mM histidine and 0.5 mM Mg2+ as cofactors, some lineages do not require histidine and have higher activity with Ca2+ than with Mg2+, although Ca2+ was not present during the selection (8,58). These findings call for the need to test the activity of in vitro selected nucleic acid enzymes with all the common metal ions, in addition to the metal ion used during selection. Among the metal ions we tested, the cleavage efficiency of 17E displayed the following trend: Zn2+ >> Mn2+ ~ Co2+ > Cd2+ > Ni2+ > Mg2+ ~ Ca2+ > Sr2+ ~ Ba2+. This trend is roughly the reverse order of the pKa value of the corresponding metal hydrate (see Fig. 2 and Table 2; 59). However, this correlation does not hold for all metal ions. Similar trend and some exceptions have also been observed in hammerhead ribozymes (60) and in 10–23 deoxyribozyme (12,44).
In general, transition metals are favored over alkaline earth metal ions. The preference of 17E deoxyribozyme for transition metal over alkaline earth metal most likely reflects differences in the metal binding sites, including the binding affinity, ligand-set, ionic radii and geometry. Alkaline earth metal ions tend to maintain their hydration state and bind nucleic acids through outer-sphere coordination with low to moderate binding affinity (61,62). In contrast, transition metal ions like Zn2+, Co2+, Mn2+ and Cd2+ can bind to both the non-bridging phosphate oxygen and the O or N groups on the nucleic acid bases through either inner-sphere or outer-sphere coordination (61,63,64). It is likely that the metal binding sites in 17ES complex contain certain nitrogen ligands that favor transition metal ions. Tighter binding of the transition metal ion Mn2+ than of alkaline earth metal ions (Mg2+ and Ca2+) have also been reported for the 10–23 deoxyribozyme (44), the hammerhead ribozyme (29,60,65,66) and the HDV ribozyme (67).
Various roles of metal ions in DNA and RNA enzyme catalysis have been proposed (22–25). In addition to playing structural roles, divalent metal ions can also directly participate in the chemical step. A coordinated metal hydroxide can act as a general base and deprotonate the 2′-hydroxyl at the cleavage site. Alternatively, the metal ion can serve as a Lewis acid by direct coordination to the oxygen of 2′-hydroxyl, thereby weakening the 2′-O-H bond. Metal ions may also coordinate to the non-bridging phosphodiester oxygen to either make the phosphorus center more susceptible to nucleophilic attack, or help stabilize the developing negative charge of the oxy-anion in the trigonal-bipyramidal transition state. In addition, direct coordination of divalent metal ions to the 5′-oxygen leaving group will stabilize the developing negative charge and accelerate the cleavage of 5′-O-P bond (68,73). The inverse correlation between the pKa value of the metal hydrates and the cleavage efficiency of 17E provides evidence for metal ions playing some or all of these catalytic roles.
The 17E deoxyribozyme is a highly efficient catalyst for RNA transesterification. In the presence of saturating Zn2+ ions at pH 6.0, the apparent maximum rate was 1.35 min–1. While we were unable to directly measure the rate of 17E at pH 7.5, it can be estimated based on the linear increase of log kobs with pH. The estimated maximal rate for Zn2+ should be ~50 min–1 at pH 7.5, which is the physiological pH and the pH used for most ribozyme characterizations. Therefore 17E deoxyribozyme is among the fastest of natural and in vitro selected nucleic acid enzymes under similar conditions. The high cleavage efficiency of 17E is probably due to the appropriately positioned metal-binding sites, and the preference for metal ions with a favorable pKa of metal bound water and/or a better Lewis acidity.
The reaction mechanism of 17E deoxyribozyme is similar to that of hammerhead ribozyme
The cleavage activity of 17E deoxyribozyme was assayed under pre-steady state conditions. The observed rate constants therefore reflect the rate of the chemical step of enzyme catalysis. In the presence of Zn2+, log kobs increased linearly with pH with a slope close to 1 (Fig. 4). This is consistent with a single deprotonation event during the rate-limiting step.
For both the DNA/RNA chimera and the all-RNA substrates, the cleavage catalyzed by the deoxyribozyme 17E in the presence of Zn2+ resulted in a 5′-fragment with a 2′,3′-cyclic phosphate terminus and a 3′-fragment with a free 5′-hydroxyl group. This cleavage pattern is similar to those catalyzed by the hammerhead (69), hairpin (70), Neurospora (71) and HDV ribozymes (72), as well as RNA-cleaving deoxyribozymes obtained through in vitro selection (12,44). The formation of the 2′,3′-cyclic phosphate during cleavage is consistent with an internal transesterification mechanism, in which the 2′-OH group at the cleavage site undergoes in-line attack on the scissile phosphorus, forming a penta-coordinated phosphate intermediate followed by the leaving of 5′-oxygen (68,73).
The above observations, including the pH-rate profile, the correlation of rate with the pKa of the metal hydrate, and the identity of cleavage products, were also known with the hammerhead ribozyme. Therefore, 17E probably undergoes similar chemical mechanism as the hammerhead ribozyme, despite the fact that they have different metal preference.
CONCLUSION
By using in vitro selection, we have isolated a class of Zn(II)-dependent deoxyribozymes with a reduced metal ion requirement and increased activity that surpasses many natural and in vitro selected nucleic acid enzymes. This work represents a comprehensive characterization of a nucleic acid-based endonuclease that favors transition metal ions over alkaline earth metal ions. 17E displays a similar pH-dependent activity profile as the hammerhead ribozyme and thus may proceed through the same reaction intermediates as the hammerhead ribozyme. Our work illustrates the power of in vitro selection in searching for nucleic acid enzymes that utilize other transition metal ions in order to increase their catalytic efficiency and diversity. The selected nucleic acid enzymes will be more amenable to spectroscopic characterization and will provide invaluable information about the structure of metal binding sites and the role of metal ions in the folding and chemical mechanism of nucleic acid enzymes.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Gerald F. Joyce at the Scripps Research Institute for the help at the initial phase of the project, and the advice during the project. We also thank Joyce group members, Dr Ronald R. Breaker at Yale University for helpful discussions, and Chia-chen Chu for fellowship support to J.L. This research was supported by the NIH FIRST Award (GM53706), and the Donors of the Petroleum Research Fund, administered by the American Chemical Society. Y.L. is a Sloan Research Fellow of Alfred Sloan Foundation, a Cottrell Scholar of Research Corporation, a Camille Dreyfus Teacher-Scholar of the Camille and Henry Dreyfus Foundation, and a Beckman Young Investigator of the Arnold and Mabel Beckman Foundation.
REFERENCES
- 1.Breaker R.R. (1997) Nature Biotechnol., 15, 427–431. [DOI] [PubMed] [Google Scholar]
- 2.Sen D. and Geyer,C.R. (1998) Curr. Opin. Chem. Biol., 2, 680–687. [DOI] [PubMed] [Google Scholar]
- 3.Breaker R.R. (1997) Chem. Rev., 97, 371–390. [DOI] [PubMed] [Google Scholar]
- 4.Joyce G.F. (1994) Curr. Opin. Struct. Biol., 4, 331–336. [DOI] [PubMed] [Google Scholar]
- 5.Lorsch J.R. and Szostak,J.W. (1996) Acc. Chem. Res., 29, 103–110. [DOI] [PubMed] [Google Scholar]
- 6.Breaker R.R. and Joyce,G.F. (1994) Chem. Biol., 1, 223–229. [DOI] [PubMed] [Google Scholar]
- 7.Breaker R.R. and Joyce,G.F. (1995) Chem. Biol., 2, 655–660. [DOI] [PubMed] [Google Scholar]
- 8.Faulhammer D. and Famulok,M. (1997) Angew. Chem., Int. Ed. Engl., 35, 2837–2841. [Google Scholar]
- 9.Carmi N., Shultz,L.A. and Breaker,R.R. (1996) Chem. Biol., 3, 1039–1046. [DOI] [PubMed] [Google Scholar]
- 10.Geyer C.R. and Sen,D. (1997) Chem. Biol., 4, 579–593. [DOI] [PubMed] [Google Scholar]
- 11.Roth A. and Breaker,R.R. (1998) Proc. Natl Acad. Sci. USA, 95, 6027–6031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Santoro S.W. and Joyce,G.F. (1997) Proc. Natl Acad. Sci. USA, 94, 4262–4266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cuenoud B. and Szostak,J.W. (1995) Nature, 375, 611–614. [DOI] [PubMed] [Google Scholar]
- 14.Li Y. and Breaker,R.R. (1999) Proc. Natl Acad. Sci. USA, 96, 2746–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Burmeister J., von Kiedrowski,G. and Ellington,A.D. (1997) Angew. Chem., Int. Ed. Engl., 36, 1321–1324. [Google Scholar]
- 16.Li Y. and Sen,D. (1996) Nat. Struct. Biol., 3, 743–747. [DOI] [PubMed] [Google Scholar]
- 17.Asahina Y., Ito,Y., Wu,C.H. and Wu,G.Y. (1998) Hepatology, 28, 547–554. [DOI] [PubMed] [Google Scholar]
- 18.Warashina M., Kuwabara,T., Nakamatsu,Y. and Taira,K. (1999) Chem. Biol., 6, 237–250. [DOI] [PubMed] [Google Scholar]
- 19.Kuwabara T., Warashina,M., Tanabe,T., Tani,K., Asano,S. and Taira,K. (1997) Nucleic Acids Res., 25, 3074–3081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun L.-Q., Cairns,M.J., Gerlach,W.L., Witherington,G., Wang,L. and King,A. (1999) J. Biol. Chem., 274, 17236–17241. [DOI] [PubMed] [Google Scholar]
- 21.Cairns M.J., Hopkins,T.M., Witherington,C., Wang,L. and Sun,L.-Q. (1999) Nature Biotechnol., 17, 480–486. [DOI] [PubMed] [Google Scholar]
- 22.Pyle A.M. (1993) Science, 261, 709–714. [DOI] [PubMed] [Google Scholar]
- 23.Yarus M. (1993) FASEB J., 7, 31–39. [DOI] [PubMed] [Google Scholar]
- 24.Pan T., Long,D.M. and Uhlenbeck,O.C. (1993) In Gesteland,R.F., and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 271–302.
- 25.Feig A.L. and Uhlenbeck,O.C. (1999) In Eckstein,F. and Lilley,D.M.J. (eds), The RNA World, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, Vol. 37, pp. 287–319.
- 26.Bertini I., Gray,H.B., Lippard,S.J. and Valentine,J.S. (1994) Bioinorganic Chemistry. University Science Books, Sausalito, CA.
- 27.Lippard S.J. and Berg,J.M. (1994) Principles of Bioinorganic Chemistry. University Science Books, Mill Valley, CA.
- 28.Holm R.H., Kennepohl,P. and Solomon,E.I. (1996) Chem. Rev., 96, 2239–2314. [DOI] [PubMed] [Google Scholar]
- 29.Dahm S.C. and Uhlenbeck,O.C. (1991) Biochemistry, 30, 9464–9469. [DOI] [PubMed] [Google Scholar]
- 30.Coleman J.E. (1992) Annu. Rev. Biophys. Biomol. Struct., 21, 441–483. [DOI] [PubMed] [Google Scholar]
- 31.Rees D.C., Lewis,M. and Lipscomb,W.N. (1983) J. Mol. Biol., 168, 368–387. [Google Scholar]
- 32.Dumas D.P., Caldwell,S.R., Wild,J.R. and Raushel,F.M. (1989) J. Biol. Chem., 264, 19659–19665. [PubMed] [Google Scholar]
- 33.Bertini I. and Luchinat,C. (1983) Metal Ions Biol. Syst., 15, 101–156. [Google Scholar]
- 34.Maret W. and Vallee,B.L. (1993) Methods Enzymol., 226, 52–71. [DOI] [PubMed] [Google Scholar]
- 35.Pan T. and Uhlenbeck,O.C. (1992) Nature, 358, 560–563. [DOI] [PubMed] [Google Scholar]
- 36.Pan T., Dichtl,B. and Uhlenbeck,O.C. (1994) Biochemistry, 33, 9561–9565. [DOI] [PubMed] [Google Scholar]
- 37.Lehman N. and Joyce,G.F. (1993) Nature, 361, 182–185. [DOI] [PubMed] [Google Scholar]
- 38.Frank D.N. and Pace,N.R. (1997) Proc. Natl Acad. Sci. USA, 94, 14355–14360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Carmi N., Balkhi,H.R. and Breaker,R.R. (1998) Proc. Natl Acad. Sci. USA, 95, 2233–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ciesiolka J., Gorski,J. and Yarus,M. (1995) RNA, 1, 538–550. [PMC free article] [PubMed] [Google Scholar]
- 41.Ciesiolka J. and Yarus,M. (1996) RNA, 2, 785–793. [PMC free article] [PubMed] [Google Scholar]
- 42.Conaty J., Hendry,P. and Lockett,T. (1999) Nucleic Acids Res., 27, 2400–2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zillmann M., Limauro,S.E. and Goodchild,J. (1997) RNA, 3, 734–747. [PMC free article] [PubMed] [Google Scholar]
- 44.Santoro S.W. and Joyce,G.F. (1998) Biochemistry, 37, 13330–13342. [DOI] [PubMed] [Google Scholar]
- 45.Breaker R.R. and Joyce,G.F. (1994) Trends Biotechnol., 12, 268–275. [DOI] [PubMed] [Google Scholar]
- 46.Perrotta A.T. and Been,M.D. (1996) Nucleic Acids Res., 24, 1314–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Strobel S.A. and Cech,T.R. (1995) Science, 267, 675–679. [DOI] [PubMed] [Google Scholar]
- 48.Scott W.G., Finch,J.T. and Klug,A. (1995) Cell, 81, 991–1002. [DOI] [PubMed] [Google Scholar]
- 49.Murray J.B., Terwey,D.P., Maloney,L., Karpeisky,A., Usman,N., Beigelman,L. and Scott,W.G. (1998) Cell, 92, 665–673. [DOI] [PubMed] [Google Scholar]
- 50.Wedekind J.E. and McKay,D.B. (1999) Nat. Struct. Biol., 6, 261–268. [DOI] [PubMed] [Google Scholar]
- 51.Cate J.H. and Doudna,J.A. (1996) Structure, 4, 1221–1229. [DOI] [PubMed] [Google Scholar]
- 52.Kore A.R., Vaish,N.K., Kutzke,U. and Eckstein,F. (1998) Nucleic Acids Res., 26, 4116–4120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ota N., Warashina,M., Hirano,K.-i., Hatanaka,K. and Taira,K. (1998) Nucleic Acids Res., 26, 3385–3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warashina M., Takagi,Y., Sawata,S., Zhou,D.-M., Kuwabara,T. and Taira,K. (1997) J. Org. Chem., 62, 9138–9147. [Google Scholar]
- 55.Sawata S., Shimayama,T., Komiyama,M., Kumar,P.K.R., Nishikawa,S. and Taira,K. (1993) Nucleic Acids Res., 21, 5656–5660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimayama T., Nishikawa,S. and Taira,K. (1995) FEBS Lett., 368, 304–306. [DOI] [PubMed] [Google Scholar]
- 57.Shimayama T., Nishikawa,F., Nishikawa,S. and Taira,K. (1993) Nucleic Acids Res., 21, 2605–2611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Faulhammer D. and Famulok,M. (1997) J. Mol. Biol., 269, 188–202. [DOI] [PubMed] [Google Scholar]
- 59.Richens D.T. (1997) The Chemistry of Aqua Ions. John Wiley & Sons, New York, NY.
- 60.Dahm S.C., Derrick,W.B. and Uhlenbeck,O.C. (1993) Biochemistry, 32, 13040–13045. [DOI] [PubMed] [Google Scholar]
- 61.Jack A., Ladner,J.E., Rhodes,D., Brown,R.S. and Klug,A. (1977) J. Mol. Biol., 111, 315–328. [DOI] [PubMed] [Google Scholar]
- 62.Holbrook S.R., Sussman,J.L., Warrant,R.W., Church,G.M. and Kim,S.-H. (1977) Nucleic Acids Res., 4, 2811–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brown R.S., Dewan,J.C. and Klug,A. (1985) Biochemistry, 24, 4785–4801. [DOI] [PubMed] [Google Scholar]
- 64.Rubin J.R., Wang,J. and Sundaralingam,M. (1983) Biochim. Biophys. Acta, 756, 111–118. [DOI] [PubMed] [Google Scholar]
- 65.Horton T.E., Clardy,D.R. and DeRose,V.J. (1998) Biochemistry, 37, 18094–18101. [DOI] [PubMed] [Google Scholar]
- 66.Hansen M.R., Simorre,J.-P., Hanson,P., Mokler,V., Bellon,L., Beigelman,L. and Pardi,A. (1999) RNA, 5, 1099–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Suh Y.A., Kumar,P.K.R., Taira,K. and Nishikawa,S. (1993) Nucleic Acids Res., 21, 3277–3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kuimelis R.G. and McLaughlin,L.W. (1998) Chem. Rev., 98, 1027–1044. [DOI] [PubMed] [Google Scholar]
- 69.Uhlenbeck O.C. (1987) Nature, 328, 596–600. [DOI] [PubMed] [Google Scholar]
- 70.Hampel A. and Tritz,R. (1989) Biochemistry, 28, 4929–4933. [DOI] [PubMed] [Google Scholar]
- 71.Saville B.J. and Collins,R.A. (1990) Cell, 61, 685–696. [DOI] [PubMed] [Google Scholar]
- 72.Thill G., Vasseur,M. and Tanner,N.K. (1993) Biochemistry, 32, 4254–4262. [DOI] [PubMed] [Google Scholar]
- 73.Zhou D.-M. and Taira,K. (1998) Chem. Rev., 98, 991–1026. [DOI] [PubMed] [Google Scholar]