Abstract
Simple Summary
A variety of MET aberrations that lead to the dysregulation of the MET oncogene and thus the activation of various signaling pathways have been described. These include MET overexpression, the activation of MET mutations comprising exon 14 skipping mutations, MET gene amplifications, and MET fusions. Patients with such aberrations can be treated using a targeted inhibitor such as crizotinib, cabozantinib, tepotinib, and capmatinib. Therefore, the implementation of high-quality and sensitive methods for the detection of the various MET aberrations is essential.
Abstract
MET tyrosine kinase receptor pathway activation has become an important actionable target in solid tumors. Aberrations in the MET proto-oncogene, including MET overexpression, the activation of MET mutations, MET mutations that lead to MET exon 14 skipping, MET gene amplifications, and MET fusions, are known to be primary and secondary oncogenic drivers in cancer; these aberrations have evolved as predictive biomarkers in clinical diagnostics. Thus, the detection of all known MET aberrations in daily clinical care is essential. In this review, current molecular technologies for the detection of the different MET aberrations are highlighted, including the benefits and drawbacks. In the future, another focus will be on the standardization of detection technologies for the delivery of reliable, quick, and affordable tests in clinical molecular diagnostics.
Keywords: MET, NSCLC, MET exon 14 skipping mutation, MET gene amplification, MET fusion
1. Introduction
The MET gene (MET proto-oncogene, receptor tyrosine kinase), which consists of 21 exons separated by 20 introns, is located on chromosome 7q21-31 and encodes the MET receptor tyrosine kinase (190 kDa). Together with its ligand, hepatocyte growth factor (HGF), MET plays an important role in tumor proliferation, angiogenesis, and migration [1,2]. A variety of MET aberrations that lead to the dysregulation of the MET oncogene and thus the activation of various signaling pathways such as MAPK, PI3K-AKT, and JAK-STAT have already been described. These include MET overexpression, the activation of MET mutations comprising MET exon 14 skipping mutations, MET gene amplifications, and MET fusions [3,4]. Patients with these types of aberrations can be treated by using an inhibitor targeting either MET, such as capmatinib and tepotinib, or by using multikinase inhibitors, such as crizotinib and cabozantinib [5]. Capmatinib (Tabrecta®, Novartis Pharma GmbH, Basel, Switzerland) and tepotinib (Tepmetko®, Merck KGaA, Darmstadt, Germany) have been approved for the treatment of patients with advanced non-small cell lung carcinoma (NSCLC) and MET exon 14 skipping mutation who are undergoing systemic therapy after platinum-based chemotherapy or require treatment using immunotherapy. Thus, the implementation of high-quality and sensitive detection methods is essential for the identification of the various MET aberrations.
2. MET Receptor
The MET receptor functions as a disulfide-linked heterodimer tyrosine kinase receptor and is composed of an extracellular, transmembrane, and intracellular domain. The extracellular domain is the binding site of its ligand, HGF, and consists of the semaphorin (SEMA), plexin semaphorin integrin (PSI), and immunoglobulin plexin transcription factor (IPT) domains. The intracellular domain consists of the juxtamembrane (JM) domain with the E3 ubiquitin ligase casitas B-lineage lymphoma (c-CBL) binding site, tyrosine kinase (TK) domain, and C-terminal multifunctional docking site (Figure 1) [2,6,7,8]. Binding of the ligand HGF to the SEMA domain on the extracellular portion of the MET receptor induces MET homodimerization and the subsequent autophosphorylation of the tyrosine residues at codon Y1234 and Y1235 (NM_000245 (Y1252 and Y1253 NM_0001127500)) in the intracellular TK domain, thus leading to the activation of the kinase domain. This is followed by the phosphorylation of Y1349 and Y1356 (NM_000245 (Y1367 and Y1374 NM_0001127500)) in the multifunctional docking site, which opens and forms a docking site for intracellular adaptors that recruit SRC (SRC proto-oncogene, non-receptor tyrosine kinase) adapter protein; additionally, the subsequent activation of several downstream pathways occurs, such as the PI3K/AKT pathway, RAS mitogen activated protein kinase (MAPK) cascade, signal transducer and activator of transcription (STAT), and NF-κB pathway [4]. These signaling pathways play an important role in proliferation, organogenesis, liver regeneration, embryogenesis, wound healing, and cell motility [6,7,9].
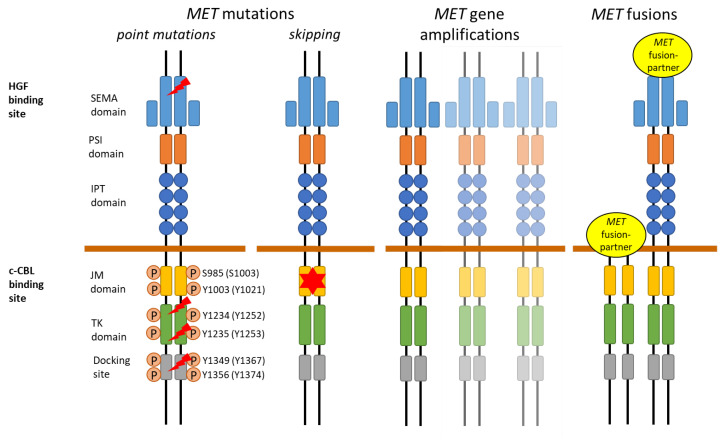
Figure 1.
Schematic representation of MET aberrations in the MET receptor.
A variety of MET aberrations have been described, including MET mutations, MET gene amplifications, and MET fusions. MET point mutations (lightning) can occur in the tyrosine kinase (TK) domain, in the Sema domain, and at the multifunctional docking site.
The main amino acid residues (transcript NM_000245 and NM_0001127500) involved in MET regulation through phosphorylation (P) are depicted. MET exon 14 skipping mutations (star) are located in the juxtamembrane (JM) domain, which contains the E3 ubiquitin ligase casitas B-lineage lymphoma (c-CBL) binding site. MET fusions are very heterogeneous and can result in different fusion proteins.
3. MET Aberrations in Lung Cancer
3.1. MET Overexpression
MET overexpression was discovered to be one of the first mechanisms of dysregulation of MET; since its discovery, it has been detected in a variety of cancers [10,11,12]. MET overexpression increases ligand-independent phosphorylation and activation of signaling pathways, and it has been linked to metastases, enhanced tumor invasion, and poor survival [13]. In non-small cell lung cancers (NSCLC), MET overexpression has been found with a varying frequency of 35–72% through immunohistochemistry (IHC) [14,15,16].
Several MET IHC antibodies have been developed, including monoclonal and polyclonal antibodies and antibodies against phosphorylated MET [17,18,19,20]. Most commonly, the anti-c-MET (SP44) rabbit monoclonal primary antibody (Ventana Medical Systems Inc.) is used; however, comparative studies of the different antibodies are still missing. IHC slides are evaluated by pathologists, and MET protein expression is semi-quantitatively measured (Table 1). At present, a variety of MET IHC scoring systems and cutoff values have been published. Mostly, staining intensities are classified as negative (0), weak (1+), moderate (2+), and strong (3+), and a staining intensity of 2+ in at least 50% of tumor cells is classified as MET overexpression [15,17].
Studies that use MET protein expression as a biomarker for MET-targeted therapies with monoclonal antibodies and MET tyrosine kinase inhibitors have not been successful up to this point. This poses the question of whether the cutoff values for patient enrollment have been chosen sufficiently [17,21,22] or whether MET overexpression determined by IHC may have a low predictive value for MET activation and ET tumor dependency [23] in contrast to, for example, ALK overexpression in ALK fusion-positive NSCLCs [20]. This is supported by growing evidence that MET IHC cannot be used as a screening tool for MET activation by MET exon 14 skipping mutations or MET gene amplifications in NSCLC [15]. One study revealed that only 3% of MET IHC positive samples had MET exon 14 skipping mutations, and only 1% showed a MET gene amplification [15]. Another study showed an only 16.1% concordance between IHC and MET DNA alterations (MET exon 14 skipping mutations and MET gene amplifications) [24]. Thus, MET overexpression as a primary biomarker and oncogenic driver remains unclear and thus has not reached clinical use.
Table 1.
Detection techniques for MET aberrations.
| MET Aberration | Detection Technique | Tested Material | Evaluation Criteria | Advantages | Disadvantages |
|---|---|---|---|---|---|
| MET overexpression [14,15,16,17,18,19,20,21,22,23] |
IHC antibodies | FFPE slide | Semi-quantitative score 0–3+ | Technique widely used and available, fast and cheap | Observer-dependent, tissue sectioning artefacts, new FFPE slide for every analysis, no consensus on scoring system and cutoff |
|
MET exon 14 skipping [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39] |
RNA NGS (amplicon-, AMP-, or hybridization-based) | RNA from FFPE or fresh frozen material | Mutation, coverage, MAF, fusion product of exon 13 and 15 | Sensitive, reliable, direct detection of alternative splicing, multiplexing | RNA degradation, underlying mutation cannot be determined |
| RT-PCR | RNA from FFPE or fresh frozen material | Fusion product of exon 13 and 15 | Sensitive, reliable, direct detection of alternative splicing, fast turnaround time, widely used and available | RNA degradation, underlying mutation cannot be determined, targeted mutations only | |
|
MET exon 14 skipping mutations and point mutations [33,35,37,40,41,42,43,44] |
DNA NGS (amplicon- or hybridization-based) | DNA from FFPE, fresh frozen material, or liquid biopsy | Mutation, coverage, VAF | Sensitive, reliable, detection of exact mutation, multiplexing | No assessment of splicing effect |
| Sanger sequencing | DNA from FFPE, fresh frozen material, or liquid biopsy | Mutation, VAF | Detection of exact mutation, fast turnaround time, widely used and available | Sensitivity, single assay for each target, no assessment of splicing effect | |
|
MET amplifications [5,41,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61] |
FISH | FFPE slides |
MET GCN, MET/CEN7 ratio |
Technique widely used and available, detection of focal amplification, polysomy, and chromosome duplications | Observer-dependent, tissue sectioning artefacts, new FFPE slide for every analysis, no consensus on scoring system and cutoff |
| DNA NGS (amplicon- or hybridization-based) | DNA from FFPE, fresh frozen material, or liquid biopsy | Mutation, coverage, VAF | Sensitive, DNA from FFPE easily accessible, multiplexing | High number of false negatives, no standardized cutoff or bioinformatics, no morphological correlation | |
| Other DNA-based technologies (ddPCR, NanoString nCounter technology) | DNA from FFPE, fresh frozen material, or liquid biopsy | Expression, GCN | DNA from FFPE easily accessible | High number of false negatives, no morphological correlation, no standardized cutoff, large amounts of DNA needed | |
|
MET fusions [35,39,62,63,64,65,66,67,68,69,70,71] |
RNA NGS (AMP- or hybridization-based) | RNA from FFPE or fresh frozen material | Fusionreads, 3′-5′ imbalance | Sensitive, reliable detection of known and novel fusion partners, multiplexing | RNA degradation |
| DNA NGS (Hybridization-based) | DNA from FFPE | Fusionreads, 3′-5′ imbalance, coverage, | DNA from FFPE easily accessible, detection of known and novel fusion partners if region is covered, multiplexing | False negative results, novel fusions are problematic due to location of fusion break point | |
| FISH | FFPE slides | n.a. break-apart events | Technique widely used and available | No standardized assay available, observer-dependent, tissue sectioning artefacts, new FFPE slide for every analysis | |
| RT-PCR | RNA from FFPE | Fusion product | Technique widely used and available | No standardized assay available, only for known fusion partners, RNA degradation |
IHC: immunohistochemistry; FISH: fluorescence in situ hybridization; FFPE: formalin-fixed paraffin-embedded; RT-PCR: quantitative real-time polymerase chain reaction; ddPCR: digital droplet PCR; n.a.: not available; GCN: gene copy number; CEN7: centromere of chromosome 7; AMP: anchored multiplex polymerase chain reaction; NGS: next-generation sequencing; VAF: variant allele frequency.
3.2. MET Mutations
3.2.1. MET Exon 14 Skipping
MET exon 14 skipping mutations occur in 3–4% of NSCLC and are very heterogeneous [3]. MET exon 14 skipping mutations gained importance when Frampton et al. and Paik et al. first reported large studies that featured patients with stage IV lung adenocarcinomas harboring a variety of MET exon 14 splice variants, which resulted in MET exon 14 skipping and showed clinical sensitivity towards MET inhibitors [25,27]. To date, over 100 different mutations that can lead to MET exon 14 skipping have been described [26]. Mutations leading to MET exon 14 skipping interfere with the normal regulation of MET transcription. These include numerous point mutations, insertions, and deletions disrupting the splice acceptor at the branch point or the polypyrimidine tract site in exon 14, the 5′ end of exon 14, or the splice donor site at the 3′ end [28]. Furthermore, silent mutations in the splice sites can lead to MET exon 14 skipping. As a consequence, the spliceosome skips transcribing exon 14, leading to the loss of the entire exon and thus the JM domain encompassing 141 base pairs with the c-CBL binding site (Y1003 (NM_000245); Y1021 (NM_0001127500)). Without the c-CBL binding site, ubiquitination by CBL and the subsequent lysosomal degradation of MET is impaired; thus, its downstream signaling pathway is constitutively activated [72]. Additionally, the JM domain contains a second phosphorylation site at codon S985 NM_000245 (S1003 NM_0001127500). Phosphorylation of S985 negatively regulates kinase activity [7].
MET skipping mutations are mutually exclusive with mutations in EGFR, KRAS, and ERBB2 and fusions in ALK, RET, and ROS [25,27,28,73,74]. Some of the previously reported MET mutations seem to represent SNPs; thus, their clinical importance is highly questionable [74]. This problem especially arises when two different mRNA transcripts are frequently used in the literature and when the mutations are reported without the corresponding transcript (transcript NM_000245 and transcript NM_0001127500). For example, in transcript NM_0001127500, a rare polymorphism at codon 1010 (T1010I) is reported; however, in transcript NM_000245, the splice site of exon 14 is located at codon 1010, which can lead to a misconception of the detected mutation in the clinic. The same has been reported with two different MET resistance mutations, which are identical variants reported on different transcripts [75]. Thus, mutations should always be reported with the utilized mRNA transcript number [76].
There are several methods available for the detection of MET exon 14 skipping mutations (Table 1). They can be either detected on the DNA or RNA level. Increasingly, for the detection of MET exon 14 skipping mutations, amplicon- or hybridization-based next-generation sequencing (NGS) methods are used. Although single gene analyzes such as Sanger sequencing or quantitative real-time polymerase chain reaction (RT-PCR) can detect these aberrations, they are used less often and are considered impractical due to the large number of biomarkers that have to be tested simultaneously, especially in lung cancer, and at the same time the low availability of material. The following paragraphs highlight the different methods available.
Parallel Sequencing (NGS) Multigene Assays
DNA-based NGS assays analyze the DNA variant that underlies the skipping of exon 14 on the RNA level. For this, MET exon 14 as well as the intronic regions up- and downstream of exon 14 have to be covered by NGS assays sufficiently, as MET exon 14 skipping alterations are very heterogenous and deletions can reach far into the intronic sites of exon 14 [25]. At the DNA level, the exact mutation can be specified according to the human genome variation society (HGVS) nomenclature, which is not possible on RNA level. However, the splicing effect cannot be directly assessed at the DNA level. To verify the splicing effect, a confirmatory RNA-based assay is necessary or the effect should be substantiated by the literature [77].
There are two main types of NGS assays, amplicon-based and hybridization-based NGS [29,33,35,37,43]. Either custom panels or commercially available panels can be used. However, commercially available targeted as well as whole exome sequencing panels sometimes lack the intronic regions around MET exon 14 and are unable to detect all relevant MET exon 14 skipping mutations. Amplicon-based assays use a defined set of primers for the enrichment of the target regions via multiplex PCR followed by library preparation and sequencing. The advantage of an amplicon-based approach is the faster turn-around time, demand of smaller DNA amounts, and ability to use even chemically modified and fragmented DNA derived from formalin-fixed and paraffin-embedded (FFPE) tissue [34]. The disadvantages are the limitation of the total target size, possible allele dropout and thus false-negative results if either the mutation is localized within the primer binding site, or the occurrence of primer mismatches, especially in repetitive sequences [77]. Studies have shown that MET exon 14 skipping alterations can be missed by amplicon-based sequencing if the assay is not optimized for this purpose [31,36]. When using hybridization-based panels, DNA is initially sheared. During library preparation, target-specific biotinylated capture probes are hybridized to target regions, and the probes are enriched by streptavidin beads before sequencing [29,33,37]. The advantages of this method are that it circumvents allelic dropout and that duplicate sequences can be removed. The disadvantages are that larger amounts of DNA are needed and that the data analysis is highly complex [29,31,33,37]. Furthermore, some assays have poor intronic coverage and off-target sequencing reads reduce the sequencing coverage. In general, hybrid-capture assays often fail to detect larger deletions if the bioinformatic analysis does not enable their detection [77].
In contrast to DNA-based assays, RNA-based assays for the detection of MET exon 14 skipping mutations permit the direct detection of alternative splicing of MET exon 14, resulting in a fusion of exons 13 and 15. This method’s limitation is that the underlying mutation cannot be determined. However, only the splicing effect is clinically relevant and qualifies for targeted therapy [3,31]. For RNA-based assays, amplicon-based and hybridization-based panels can also be used. Additionally, an anchored multiplex polymerase chain reaction (AMP) approach can be utilized, which has shown promising results [39]. This technology uses a single-primer extension approach without predefined amplicon sizes. With this technology, fusions and splice variants can be detected without knowledge of the fusion partner, as only one target primer is included [4]. RNA-based assays, however, are highly dependent on the RNA integrity. RNA quality should be closely monitored, especially when using FFPE material. Ideally both DNA- and RNA-based approaches should be simultaneously used. However, as many laboratories perform the DNA extraction first followed by the DNA-based NGS analysis, often no material is left for RNA-based sequencing, especially when using lung cancer biopsies [31,77]. Thus, ideally a combined DNA and RNA extraction from the same tissue slides should be performed.
Single Gene Analyzes
Sanger sequencing can also be used for the detection of MET exon 14 skipping mutations. It is material-consuming, as a single DNA fragment is sequenced at the time and shows low sensitivity because of it only being able to detect mutations with an allele frequency above 20%. Thus, a higher tumor cell content is needed than for other sequencing technologies. On the other hand, it is easy to implement and allows for the detection of previously unknown alterations if the region is covered. At the RNA level, the RT-PCR technique is widely used in laboratories for the detection and confirmation of MET exon 14 skipping variants, as this method is a cost-effective and fast approach to test FFPE material with high sensitivity [30,38]. However, these assays fail to detect additional mutations and should only be used as a pre-screening or confirmatory tool. Newly developed multigene RT-PCR assays from Biocartis NV (Mechelen, Belgium), Diatech Pharmacogenetics S.R.L. (Jesi AN, Italy), or AmoyDx (Xiamen, China) overcome this issue and allow for the detection of a variety of gene fusions and splice variants at the same time in an easy-to-use and time-sensitive manner [32].
3.2.2. Other MET Mutations
In addition to MET exon 14 skipping mutations, activating point mutations in the TK, JM, and extracellular domains have been reported in cancer, leading to ligand-independent receptor phosphorylation and signaling (Figure 1 and Table 1) [78,79,80]. A variety of these mutations, including H1094Y/R/L (NM_000245; H1112, NM_0001127500) and D1228H/N (NM_000245; D1246, NM_0001127500), were first described in hereditary papillary renal cell carcinoma (HPRCC) [44] and were later also found in sporadic papillary renal cell carcinoma (PRCC) with up to a 15% frequency [40,42].
In NSCLC, MET mutations in the TK domain are rare and mainly emerge as an acquired resistance mechanism to MET tyrosine kinase inhibitors or as a resistance mechanism to combinational therapy with EGFR and MET TKI in MET exon 14 skipping positive patients or EGFR-mutant and MET gene amplification positive patients. Mutations such as Y1230C (NM_000245; Y1248C, NM_0001127500), Y1230H (NM_000245; Y1248H, NM_0001127500), D1228H (NM_000245; D1246H, NM_0001127500), and D1228N (NM_000245; D1246N, NM_0001127500) were found to mediate resistance by disrupting the drug binding site of crizotinib [3,81].
MET mutations in the SEMA domain and extracellular compartment have also been reported to possibly affect ligand binding. However, the functional significance and relevance of these mutations is still unknown [13]. Thus, it remains important to evaluate the functional consequences of other MET mutations and their clinical implications.
3.3. MET Gene Amplification and Gene Copy Number Alterations
MET gene amplifications and copy number alterations have been reported in 1–6% of NSCLC [61]. Therapy approaches for NSCLC with MET gene amplifications, which both occur as the primary driver aberration and as the resistance mechanism to other kinase inhibitors, have currently been evaluated in numerous studies. Studies have shown that MET inhibitors are particularly effective in highly amplified (high level) tumors with a gene copy number [GCN] ≥ 10, were MET gene amplification acts as an oncogenic driver [5,55,61]. Thus, patient selection and the exact analysis of the MET gene amplification status is crucial.
MET gene amplifications and copy number alterations arise from the focal or regional amplification of the MET genomic region or from polysomy (Figure 1). In cases with focal amplification, MET GCN gains occur without chromosome 7 duplication, whereas MET GCN gains due to polysomy arise from the duplication of parts or the entire chromosome 7; thus, multiple parts of chromosome 7 are present [3,47].
MET gene amplifications can be assessed using a variety of methodologies, which determine either the average MET GCN and/or the ratio to the centromeric region of chromosome 7 (Table 1). However, the cutoff point for setting MET positivity is still very variable. Different clinical studies have used a variety of thresholds for the definition of amplifications, from amplification positive only to defining an exact MET GCN [22,46,49,56,61]. Depending on the method used, thresholds are set at different levels, which causes problems in the interpretation of the potential of MET GCN as true biomarker [47].
Additionally, true gene amplifications without chromosome 7 duplication are more likely to lead to oncogene addiction [3,61].
3.3.1. Fluorescence In Situ Hybridization (FISH)
MET gene amplifications and MET GCN can be detected using various methods. The gold standard for the detection of MET gene amplifications and the most accurate detection method is still fluorescence in situ hybridization (FISH), which is currently still superior to DNA-based methods and mainly used in clinical trials. Bicolor FISH probes label both the MET gene and the centromere of chromosome 7 (CEN7). The number of signals identified in a nucleus represent the number of copies present. The signals in a predefined number of cell nuclei are counted and scored based on evaluation criteria such as the MET gene/CEN7 ratio and/or the average MET GCN. According to the resulting score, cases are divided into different groups [48,50,53,54,58]. Various evaluation scores have been published so far. Thus far, publications have defined MET gene amplification by GCN only, either as 5 or more copies per cell [48], or as MET GCN ≥ 6, ≥10, or 15 [54,55]. Other studies have also included the number of chromosomes present by calculating MET/CEN7 ratio; thus, true amplification can be distinguished from polysomy. A MET/CEN7 ratio of ≥2.0 is commonly defined as MET gene amplification [50,53,58]. Other studies have categorized the degree of amplification into low, intermediate, high-level, and top-level amplified cases. A top-level amplification was classified as an average MET GCN per cell of ≥10. a high-level amplification was defined in tumors with a MET/CEN7 ratio ≥2.0 or an average MET GCN per cell of ≥6. An intermediate level of GCN gain means that ≥50% of cells contain ≥5 MET signals. A low level of GCN gain was defined as ≥40% of tumor cells showing ≥4 MET signals [55,58].
The FISH technique is especially useful in cases with low tumor cell content, tumor heterogeneity, and focal amplifications, as FISH is performed on slides and evaluated under the microscope [52,58,60]. However, in situ-based approaches like FISH are also thereby hampered. The evaluation is observer-dependent, and tissue sectioning artefacts can impact the analysis. Furthermore, a new slide of material must be used for each additional parameter that is tested by FISH, which can be problematic when using small biopsies [52,60].
3.3.2. DNA-Based Methods
Another option for the detection of MET gene amplifications are a variety of DNA-based methods that work with extracted nucleic acids, such as digital droplet PCR (ddPCR), next-generation sequencing (NGS), or the NanoString nCounter technology. The detection of MET gene amplifications by GCN changes using DNA-based methods is still under evaluation. These methods allow for an easier quantification of GCN in comparison to FISH but do not allow for morphological correlation. At present, the performance of NGS-based assays for the detection of MET gene amplification have been characterized the best. Data for the other DNA-based methods mentioned above are very limited [52]. Studies that have compared NGS and FISH assays showed low consistency between both methods. Currently, only high-level and top-level amplified samples with GCN ≥ 10 and negative samples determined by FISH can be reliably detected by an NGS analysis [3,52,57,59,60]. Comparative studies have further shown that MET gene amplifications can be missed by NGS assays due to a variety of reasons. On the one hand, the tumor material itself can pose problems in the evaluation due to low tumor purity (inclusion of normal, necrotic, and inflammatory cells), low tumor cell content, the overall amount of material present, the FFPE DNA quality, tumor heterogeneity, or focal amplifications and polysomy [52,57,59,60]. On the other hand, analyzing large genomic alterations can be very challenging and can create computational challenges, such as call accuracy and noise reduction. Additionally, a defined set of normal samples or standardized set of controls and the tumor cell content of the samples have to be used for bioinformatic analyses [41,45,51].
In molecular diagnostics, amplicon-based as well as hybridization-based NGS assays are used for MET GCN detection. Hybridization-based NGS assays can assess MET GCN variations more accurately than amplicon-based NGS assays, as the sequencing bias is reduced, duplicate reads can be filtered out, and the true mean coverage used for GCN determination is less affected by DNA quality, tumor complexity such as tumor purity, and heterogeneity [3,60,78,79,80]. However, to date, there are no methodologically or clinically defined cutoffs for the definition of MET positivity when utilizing NGS assay, nor is there an accepted standard or general consensus regarding the protocols and bioinformatics used, which inevitably leads to discordant results across studies.
3.4. MET Fusions
MET gene fusions are rare oncogenic driver alterations in a variety of cancers, including hepatocellular carcinoma, gastric carcinoma, sarcoma, and NSCLC. Only in glioblastomas, MET fusions are described in 12% of cases [3]. The frequency of MET fusions in NSCLC is < 0.5%, and they are found to be mutually exclusive with other oncogenic drivers [66]. The first MET fusion identified in lung cancer was the TPR-MET fusion [82]. Since then, several fusion partners have been characterized, such as KIF5B, CLIP2, TFG, STARD3NL, ATXN7L1, PTPRZ, and CD74 [63,64,66,67,71]. MET fusions occur through inter- or intra-chromosomal rearrangement and mostly include the kinase domain on exon 15 and downstream, resulting in ligand-independent constitutive MET activation (Figure 1) [65,66,67,68]. The TPR-MET fusion, however, does not include exon 14 of MET and can show the same oncogenic behavior as NSCLCs with MET exon 14 skipping [70]. Fusions such as KIF5B-MET and PTPRZ-MET that include exon 14 appear to be less oncogenic than the TRP-MET fusion [65]. In the PTPRZ-MET fusion protein, the MET gene is present in the full length, including in the dimerization domain in exon 2, resulting in MET overexpression and increased activation [65]. Therefore, the knowledge of the exact fusion break point seems to be important for the success of MET TKIs. Currently, clinical trials are evaluating the efficiency of MET TKIs in MET fusion-positive cancers.
For the detection of MET fusions, an RNA-based NGS approach that uses either AMP- or hybridization-based technologies is the first choice. In this way, both unknown fusion partners and the involved exons can be determined [62]. Alternatively, FISH, RT-PCR, and DNA-based NGS techniques can be used. However, an RNA-based NGS panel analysis is the most sensitive approach for such rare and novel events, as all relevant fusions and MET exon 14 skipping mutations can be detected in just one assay, thus making FISH and RT-PCR inadequate detection tools. Additionally, intrachromosomal rearrangements may lead to false negative FISH results, as the distance between the 5′ and 3′ probes are too short [35,39,62,71]. DNA-based hybrid-capture NGS approaches that can detect both mutations and fusions have proven to be unreliable in the past and often lead to false negative results for fusion detection, especially in cases of novel fusions. This is due to the localization of fusion breakpoints in large intronic regions with repetitive sequences, which are difficult to cover using capture probes [35,54,69].
4. Conclusions
In recent years, the large, growing number of detected MET alterations in NSCLC and other carcinomas as well as the better understanding of the diverse biology driving MET dysregulation in cancer has shown the important role of this kinase for targeted therapy approaches.
Particularly, since the FDA and EMA approval of MET inhibitors for NSCLCs with MET exon 14 skipping mutations, testing for all MET alterations, e.g., MET expression, MET mutations, MET gene amplifications, and MET fusions, should be routine standard of care for patients with NSCLC. However, there is still the need for the further development of quality assured and sensitive molecular detection methods, especially under the new In Vitro Diagnostics Regulation (IVDR) and when it comes to the detection of MET gene amplifications, as these are still widely analyzed reliably by only FISH while the cutoffs for other technologies are lacking. Additionally, inconsistent nomenclature of somatic variants and the different transcripts used in the literature are still of concern, as this can lead to a misconception of the detected mutations in the clinic and thus therapeutic failure. As a final note, quality assured and sensitive molecular detection methods are especially important in Europe, as laboratories are free to choose the diagnostic method used for the detection of the different MET alterations.
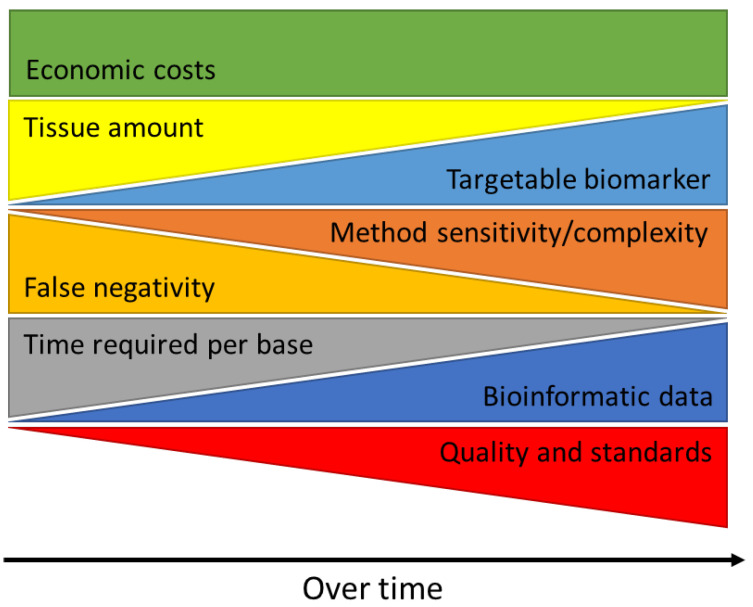
In the future, the number of targetable biomarkers will increase more and more, and the amount of tissue, effort, and time required to complete complex diagnostic tests will become even more limiting. As molecular targets and therapeutic approaches are continuously changing, the ongoing development and implementation of high-quality molecular testing, and the continuous adaption to the latest findings in cancer research will become increasingly important while limiting economic costs at the same time (Figure 2).
Figure 2.
Clinical utility of the analysis of tumor material in molecular pathology diagnostics over time.
Author Contributions
Conceptualization, C.H.; investigation, C.H., M.A.I. and S.M.-B., data curation, C.H., M.A.I. and S.M.-B.; writing—original draft preparation, C.H., M.A.I. and S.M.-B.; writing—review and editing, C.H., M.A.I. and S.M.-B.; supervision, C.H. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
Carina Heydt has received honoraria from AstraZeneca, BMS, Illumina, and Molecular Health; Michaela Angelika Ihle has received honoraria from AstraZeneca, BMS, Novartis, and Merck; Sabine Merkelbach-Bruse has received honoraria and travel support from Amgen, AstraZeneca, Bayer, BMS, GSK, Janssen, Merck, MSD, Molecular Health, Novartis, Pfizer, QuIP, Roche, and Targos.
Funding Statement
This research received no external funding.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Fujino T., Suda K., Mitsudomi T. Lung Cancer with MET exon 14 Skipping Mutation: Genetic Feature, Current Treatments, and Future Challenges. Lung Cancer. 2021;12:35–50. doi: 10.2147/LCTT.S269307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gelsomino F., Rossi G., Tiseo M. MET and Small-Cell Lung Cancer. Cancers. 2014;6:2100–2115. doi: 10.3390/cancers6042100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Guo R., Luo J., Chang J., Rekhtman N., Arcila M., Drilon A. MET-dependent solid tumours-molecular diagnosis and targeted therapy. Nat. Rev. Clin. Oncol. 2020;17:569–587. doi: 10.1038/s41571-020-0377-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee M., Jain P., Wang F., Ma P.C., Borczuk A., Halmos B. MET alterations and their impact on the future of non-small cell lung cancer (NSCLC) targeted therapies. Expert Opin. Targets. 2021;25:249–268. doi: 10.1080/14728222.2021.1925648. [DOI] [PubMed] [Google Scholar]
- 5.Jørgensen J.T., Mollerup J. Companion Diagnostics and Predictive Biomarkers for MET-Targeted Therapy in NSCLC. Cancers. 2022;14:2150. doi: 10.3390/cancers14092150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Organ S.L., Tsao M.S. An overview of the c-MET signaling pathway. Ther. Adv. Med. Oncol. 2011;3:S7–S19. doi: 10.1177/1758834011422556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cecchi F., Rabe D.C., Bottaro D.P. Targeting the HGF/Met signaling pathway in cancer therapy. Expert Opin. Targets. 2012;16:553–572. doi: 10.1517/14728222.2012.680957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kawaguchi M., Kataoka H. Mechanisms of Hepatocyte Growth Factor Activation in Cancer Tissues. Cancers. 2014;6:1890–1904. doi: 10.3390/cancers6041890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eder J.P., Vande Woude G.F., Boerner S.A., LoRusso P.M. Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009;15:2207–2214. doi: 10.1158/1078-0432.CCR-08-1306. [DOI] [PubMed] [Google Scholar]
- 10.Catenacci D.V., Ang A., Liao W.L., Shen J., O’Day E., Loberg R.D., Cecchi F., Hembrough T., Ruzzo A., Graziano F. MET tyrosine kinase receptor expression and amplification as prognostic biomarkers of survival in gastroesophageal adenocarcinoma. Cancer. 2017;123:1061–1070. doi: 10.1002/cncr.30437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gayyed M.F., Abd El-Maqsoud N.M., El-Hameed El-Heeny A.A., Mohammed M.F. c-MET expression in colorectal adenomas and primary carcinomas with its corresponding metastases. J. Gastrointest. Oncol. 2015;6:618–627. doi: 10.3978/j.issn.2078-6891.2015.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graveel C.R., Tolbert D., Vande Woude G.F. MET: A critical player in tumorigenesis and therapeutic target. Cold Spring Harb. Perspect. Biol. 2013;5:a009209. doi: 10.1101/cshperspect.a009209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Recondo G., Che J., Janne P.A., Awad M.M. Targeting MET Dysregulation in Cancer. Cancer Discov. 2020;10:922–934. doi: 10.1158/2159-8290.CD-19-1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watermann I., Schmitt B., Stellmacher F., Muller J., Gaber R., Kugler C., Reinmuth N., Huber R.M., Thomas M., Zabel P., et al. Improved diagnostics targeting c-MET in non-small cell lung cancer: Expression, amplification and activation? Diagn. Pathol. 2015;10:130. doi: 10.1186/s13000-015-0362-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guo R., Berry L.D., Aisner D.L., Sheren J., Boyle T., Bunn P.A., Jr., Johnson B.E., Kwiatkowski D.J., Drilon A., Sholl L.M., et al. MET IHC Is a Poor Screen for MET Amplification or MET Exon 14 Mutations in Lung Adenocarcinomas: Data from a Tri-Institutional Cohort of the Lung Cancer Mutation Consortium. J. Thorac. Oncol. 2019;14:1666–1671. doi: 10.1016/j.jtho.2019.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salgia R. MET in Lung Cancer: Biomarker Selection Based on Scientific Rationale. Mol. Cancer Ther. 2017;16:555–565. doi: 10.1158/1535-7163.MCT-16-0472. [DOI] [PubMed] [Google Scholar]
- 17.Spigel D.R., Edelman M.J., O’Byrne K., Paz-Ares L., Mocci S., Phan S., Shames D.S., Smith D., Yu W., Paton V.E., et al. Results From the Phase III Randomized Trial of Onartuzumab Plus Erlotinib Versus Erlotinib in Previously Treated Stage IIIB or IV Non-Small-Cell Lung Cancer: METLung. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017;35:412–420. doi: 10.1200/JCO.2016.69.2160. [DOI] [PubMed] [Google Scholar]
- 18.Ryan C.J., Rosenthal M., Ng S., Alumkal J., Picus J., Gravis G., Fizazi K., Forget F., Machiels J.P., Srinivas S., et al. Targeted MET inhibition in castration-resistant prostate cancer: A randomized phase II study and biomarker analysis with rilotumumab plus mitoxantrone and prednisone. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2013;19:215–224. doi: 10.1158/1078-0432.CCR-12-2605. [DOI] [PubMed] [Google Scholar]
- 19.Iveson T., Donehower R.C., Davidenko I., Tjulandin S., Deptala A., Harrison M., Nirni S., Lakshmaiah K., Thomas A., Jiang Y., et al. Rilotumumab in combination with epirubicin, cisplatin, and capecitabine as first-line treatment for gastric or oesophagogastric junction adenocarcinoma: An open-label, dose de-escalation phase 1b study and a double-blind, randomised phase 2 study. Lancet. Oncol. 2014;15:1007–1018. doi: 10.1016/S1470-2045(14)70023-3. [DOI] [PubMed] [Google Scholar]
- 20.Srivastava A.K., Hollingshead M.G., Weiner J., Navas T., Evrard Y.A., Khin S.A., Ji J.J., Zhang Y., Borgel S., Pfister T.D., et al. Pharmacodynamic Response of the MET/HGF Receptor to Small-Molecule Tyrosine Kinase Inhibitors Examined with Validated, Fit-for-Clinic Immunoassays. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22:3683–3694. doi: 10.1158/1078-0432.CCR-15-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neal J.W., Dahlberg S.E., Wakelee H.A., Aisner S.C., Bowden M., Huang Y., Carbone D.P., Gerstner G.J., Lerner R.E., Rubin J.L., et al. Erlotinib, cabozantinib, or erlotinib plus cabozantinib as second-line or third-line treatment of patients with EGFR wild-type advanced non-small-cell lung cancer (ECOG-ACRIN 1512): A randomised, controlled, open-label, multicentre, phase 2 trial. Lancet. Oncol. 2016;17:1661–1671. doi: 10.1016/S1470-2045(16)30561-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuler M.H., Berardi R., Lim W.-T., Geel R.V., Jonge M.J.D., Bauer T.M., Azaro A., Gottfried M., Han J.-Y., Lee D.H., et al. Phase (Ph) I study of the safety and efficacy of the cMET inhibitor capmatinib (INC280) in patients (pts) with advanced cMET+ non-small cell lung cancer (NSCLC) J. Clin. Oncol. 2016;34:9067. doi: 10.1200/JCO.2016.34.15_suppl.9067. [DOI] [Google Scholar]
- 23.Finocchiaro G., Toschi L., Gianoncelli L., Baretti M., Santoro A. Prognostic and predictive value of MET deregulation in non-small cell lung cancer. Ann. Transl. Med. 2015;3:83. doi: 10.3978/j.issn.2305-5839.2015.03.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tong J.H., Yeung S.F., Chan A.W., Chung L.Y., Chau S.L., Lung R.W., Tong C.Y., Chow C., Tin E.K., Yu Y.H., et al. MET Amplification and Exon 14 Splice Site Mutation Define Unique Molecular Subgroups of Non-Small Cell Lung Carcinoma with Poor Prognosis. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016;22:3048–3056. doi: 10.1158/1078-0432.CCR-15-2061. [DOI] [PubMed] [Google Scholar]
- 25.Frampton G.M., Ali S.M., Rosenzweig M., Chmielecki J., Lu X., Bauer T.M., Akimov M., Bufill J.A., Lee C., Jentz D., et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–859. doi: 10.1158/2159-8290.CD-15-0285. [DOI] [PubMed] [Google Scholar]
- 26.Kim S.Y., Yin J., Bohlman S., Walker P., Dacic S., Kim C., Khan H., Liu S.V., Ma P.C., Nagasaka M., et al. Characterization of MET Exon 14 Skipping Alterations (in NSCLC) and Identification of Potential Therapeutic Targets Using Whole Transcriptome Sequencing. JTO Clin. Res. Rep. 2022;3:100381. doi: 10.1016/j.jtocrr.2022.100381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paik P.K., Drilon A., Fan P.D., Yu H., Rekhtman N., Ginsberg M.S., Borsu L., Schultz N., Berger M.F., Rudin C.M., et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–849. doi: 10.1158/2159-8290.CD-14-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Onozato R., Kosaka T., Kuwano H., Sekido Y., Yatabe Y., Mitsudomi T. Activation of MET by gene amplification or by splice mutations deleting the juxtamembrane domain in primary resected lung cancers. J. Thorac. Oncol. 2009;4:5–11. doi: 10.1097/JTO.0b013e3181913e0e. [DOI] [PubMed] [Google Scholar]
- 29.Cheng D.T., Mitchell T.N., Zehir A., Shah R.H., Benayed R., Syed A., Chandramohan R., Liu Z.Y., Won H.H., Scott S.N., et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. JMD. 2015;17:251–264. doi: 10.1016/j.jmoldx.2014.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Das R., Jakubowski M.A., Spildener J., Cheng Y.W. Identification of Novel MET Exon 14 Skipping Variants in Non-Small Cell Lung Cancer Patients: A Prototype Workflow Involving in Silico Prediction and RT-PCR. Cancers. 2022;14:4814. doi: 10.3390/cancers14194814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davies K.D., Lomboy A., Lawrence C.A., Yourshaw M., Bocsi G.T., Camidge D.R., Aisner D.L. DNA-Based versus RNA-Based Detection of MET Exon 14 Skipping Events in Lung Cancer. J. Thorac. Oncol. 2019;14:737–741. doi: 10.1016/j.jtho.2018.12.020. [DOI] [PubMed] [Google Scholar]
- 32.Depoilly T., Garinet S., van Kempen L.C., Schuuring E., Clave S., Bellosillo B., Ercolani C., Buglioni S., Siemanowski J., Merkelbach-Bruse S., et al. Multicenter Evaluation of the Idylla GeneFusion in Non-Small-Cell Lung Cancer. J. Mol. Diagn. JMD. 2022;24:1021–1030. doi: 10.1016/j.jmoldx.2022.05.004. [DOI] [PubMed] [Google Scholar]
- 33.Drilon A., Wang L., Arcila M.E., Balasubramanian S., Greenbowe J.R., Ross J.S., Stephens P., Lipson D., Miller V.A., Kris M.G., et al. Broad, Hybrid Capture-Based Next-Generation Sequencing Identifies Actionable Genomic Alterations in Lung Adenocarcinomas Otherwise Negative for Such Alterations by Other Genomic Testing Approaches. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015;21:3631–3639. doi: 10.1158/1078-0432.CCR-14-2683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Heydt C., Fassunke J., Kunstlinger H., Ihle M.A., Konig K., Heukamp L.C., Schildhaus H.U., Odenthal M., Buttner R., Merkelbach-Bruse S. Comparison of pre-analytical FFPE sample preparation methods and their impact on massively parallel sequencing in routine diagnostics. PLoS ONE. 2014;9:e104566. doi: 10.1371/journal.pone.0104566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Heydt C., Wölwer C.B., Velazquez Camacho O., Wagener-Ryczek S., Pappesch R., Siemanowski J., Rehker J., Haller F., Agaimy A., Worm K., et al. Detection of gene fusions using targeted next-generation sequencing: A comparative evaluation. BMC Med. Genom. 2021;14:62. doi: 10.1186/s12920-021-00909-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poirot B., Doucet L., Benhenda S., Champ J., Meignin V., Lehmann-Che J. MET Exon 14 Alterations and New Resistance Mutations to Tyrosine Kinase Inhibitors: Risk of Inadequate Detection with Current Amplicon-Based NGS Panels. J. Thorac. Oncol. 2017;12:1582–1587. doi: 10.1016/j.jtho.2017.07.026. [DOI] [PubMed] [Google Scholar]
- 37.Suh J.H., Johnson A., Albacker L., Wang K., Chmielecki J., Frampton G., Gay L., Elvin J.A., Vergilio J.A., Ali S., et al. Comprehensive Genomic Profiling Facilitates Implementation of the National Comprehensive Cancer Network Guidelines for Lung Cancer Biomarker Testing and Identifies Patients Who May Benefit From Enrollment in Mechanism-Driven Clinical Trials. Oncologist. 2016;21:684–691. doi: 10.1634/theoncologist.2016-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sui J.S.Y., Finn S.P., Gray S.G. Detection of MET Exon 14 Skipping Alterations in Lung Cancer Clinical Samples Using a PCR-Based Approach. Methods Mol. Biol. 2021;2279:145–155. doi: 10.1007/978-1-0716-1278-1_11. [DOI] [PubMed] [Google Scholar]
- 39.Zheng Z., Liebers M., Zhelyazkova B., Cao Y., Panditi D., Lynch K.D., Chen J., Robinson H.E., Shim H.S., Chmielecki J., et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat. Med. 2014;20:1479–1484. doi: 10.1038/nm.3729. [DOI] [PubMed] [Google Scholar]
- 40.Albiges L., Guegan J., Le Formal A., Verkarre V., Rioux-Leclercq N., Sibony M., Bernhard J.C., Camparo P., Merabet Z., Molinie V., et al. MET is a potential target across all papillary renal cell carcinomas: Result from a large molecular study of pRCC with CGH array and matching gene expression array. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014;20:3411–3421. doi: 10.1158/1078-0432.CCR-13-2173. [DOI] [PubMed] [Google Scholar]
- 41.Chen Y.-C., Seifuddin F., Nguyen C., Yang Z., Chen W., Yan C., Chen Q., Wang C., Xiao W., Pirooznia M., et al. Comprehensive Assessment of Somatic Copy Number Variation Calling Using Next-Generation Sequencing Data. bioRxiv. 2021 doi: 10.1101/2021.02.18.431906. [DOI] [Google Scholar]
- 42.Pal S.K., Ali S.M., Yakirevich E., Geynisman D.M., Karam J.A., Elvin J.A., Frampton G.M., Huang X., Lin D.I., Rosenzweig M., et al. Characterization of Clinical Cases of Advanced Papillary Renal Cell Carcinoma via Comprehensive Genomic Profiling. Eur. Urol. 2018;73:71–78. doi: 10.1016/j.eururo.2017.05.033. [DOI] [PubMed] [Google Scholar]
- 43.Pfarr N., Stenzinger A., Penzel R., Warth A., Dienemann H., Schirmacher P., Weichert W., Endris V. High-throughput diagnostic profiling of clinically actionable gene fusions in lung cancer. Genes Chromosomes Cancer. 2016;55:30–44. doi: 10.1002/gcc.22297. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt L., Duh F.M., Chen F., Kishida T., Glenn G., Choyke P., Scherer S.W., Zhuang Z., Lubensky I., Dean M., et al. Germline and somatic mutations in the tyrosine kinase domain of the MET proto-oncogene in papillary renal carcinomas. Nat. Genet. 1997;16:68–73. doi: 10.1038/ng0597-68. [DOI] [PubMed] [Google Scholar]
- 45.Budczies J., Pfarr N., Stenzinger A., Treue D., Endris V., Ismaeel F., Bangemann N., Blohmer J.U., Dietel M., Loibl S., et al. Ioncopy: A novel method for calling copy number alterations in amplicon sequencing data including significance assessment. Oncotarget. 2016;7:13236–13247. doi: 10.18632/oncotarget.7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Camidge D.R., Otterson G.A., Clark J.W., Ignatius Ou S.H., Weiss J., Ades S., Shapiro G.I., Socinski M.A., Murphy D.A., Conte U., et al. Crizotinib in Patients With MET-Amplified NSCLC. J. Thorac. Oncol. 2021;16:1017–1029. doi: 10.1016/j.jtho.2021.02.010. [DOI] [PubMed] [Google Scholar]
- 47.Caparica R., Yen C.T., Coudry R., Ou S.I., Varella-Garcia M., Camidge D.R., de Castro G., Jr. Responses to Crizotinib Can Occur in High-Level MET-Amplified Non-Small Cell Lung Cancer Independent of MET Exon 14 Alterations. J. Thorac. Oncol. 2017;12:141–144. doi: 10.1016/j.jtho.2016.09.116. [DOI] [PubMed] [Google Scholar]
- 48.Cappuzzo F., Marchetti A., Skokan M., Rossi E., Gajapathy S., Felicioni L., Del Grammastro M., Sciarrotta M.G., Buttitta F., Incarbone M., et al. Increased MET gene copy number negatively affects survival of surgically resected non-small-cell lung cancer patients. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009;27:1667–1674. doi: 10.1200/JCO.2008.19.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dagogo-Jack I., Moonsamy P., Gainor J.F., Lennerz J.K., Piotrowska Z., Lin J.J., Lennes I.T., Sequist L.V., Shaw A.T., Goodwin K., et al. A Phase 2 Study of Capmatinib in Patients With MET-Altered Lung Cancer Previously Treated With a MET Inhibitor. J. Thorac. Oncol. 2021;16:850–859. doi: 10.1016/j.jtho.2021.01.1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Go H., Jeon Y.K., Park H.J., Sung S.W., Seo J.W., Chung D.H. High MET gene copy number leads to shorter survival in patients with non-small cell lung cancer. J. Thorac. Oncol. 2010;5:305–313. doi: 10.1097/JTO.0b013e3181ce3d1d. [DOI] [PubMed] [Google Scholar]
- 51.Gusnanto A., Wood H.M., Pawitan Y., Rabbitts P., Berri S. Correcting for cancer genome size and tumour cell content enables better estimation of copy number alterations from next-generation sequence data. Bioinformatics. 2011;28:40–47. doi: 10.1093/bioinformatics/btr593. [DOI] [PubMed] [Google Scholar]
- 52.Heydt C., Becher A.K., Wagener-Ryczek S., Ball M., Schultheis A.M., Schallenberg S., Rüsseler V., Büttner R., Merkelbach-Bruse S. Comparison of in situ and extraction-based methods for the detection of MET amplifications in solid tumors. Comput. Struct. Biotechnol. J. 2019;17:1339–1347. doi: 10.1016/j.csbj.2019.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jardim D.L., Tang C., Gagliato Dde M., Falchook G.S., Hess K., Janku F., Fu S., Wheler J.J., Zinner R.G., Naing A., et al. Analysis of 1,115 patients tested for MET amplification and therapy response in the MD Anderson Phase I Clinic. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2014;20:6336–6345. doi: 10.1158/1078-0432.CCR-14-1293. [DOI] [PubMed] [Google Scholar]
- 54.Lee H.E., Kim M.A., Lee H.S., Jung E.J., Yang H.K., Lee B.L., Bang Y.J., Kim W.H. MET in gastric carcinomas: Comparison between protein expression and gene copy number and impact on clinical outcome. Br. J. Cancer. 2012;107:325–333. doi: 10.1038/bjc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Overbeck T.R., Cron D.A., Schmitz K., Rittmeyer A., Korber W., Hugo S., Schnalke J., Lukat L., Hugo T., Hinterthaner M., et al. Top-level MET gene copy number gain defines a subtype of poorly differentiated pulmonary adenocarcinomas with poor prognosis. Transl. Lung Cancer Res. 2020;9:603–616. doi: 10.21037/tlcr-19-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Paik P.K., Felip E., Veillon R., Sakai H., Cortot A.B., Garassino M.C., Mazieres J., Viteri S., Senellart H., Van Meerbeeck J., et al. Tepotinib in Non-Small-Cell Lung Cancer with MET Exon 14 Skipping Mutations. N. Engl. J. Med. 2020;383:931–943. doi: 10.1056/NEJMoa2004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peng L.X., Jie G.L., Li A.N., Liu S.Y., Sun H., Zheng M.M., Zhou J.Y., Zhang J.T., Zhang X.C., Zhou Q., et al. MET amplification identified by next-generation sequencing and its clinical relevance for MET inhibitors. Exp. Hematol. Oncol. 2021;10:52. doi: 10.1186/s40164-021-00245-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schildhaus H.U., Schultheis A.M., Ruschoff J., Binot E., Merkelbach-Bruse S., Fassunke J., Schulte W., Ko Y.D., Schlesinger A., Bos M., et al. MET amplification status in therapy-naive adeno- and squamous cell carcinomas of the lung. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2015;21:907–915. doi: 10.1158/1078-0432.CCR-14-0450. [DOI] [PubMed] [Google Scholar]
- 59.Schmitt C., Schulz A.A., Winkelmann R., Smith K., Wild P.J., Demes M. Comparison of MET gene amplification analysis by next-generation sequencing and fluorescence in situ hybridization. Oncotarget. 2021;12:2273–2282. doi: 10.18632/oncotarget.28092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Schubart C., Stohr R., Togel L., Fuchs F., Sirbu H., Seitz G., Seggewiss-Bernhardt R., Leistner R., Sterlacci W., Vieth M., et al. MET Amplification in Non-Small Cell Lung Cancer (NSCLC)-A Consecutive Evaluation Using Next-Generation Sequencing (NGS) in a Real-World Setting. Cancers. 2021;13:5023. doi: 10.3390/cancers13195023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wolf J., Seto T., Han J.Y., Reguart N., Garon E.B., Groen H.J.M., Tan D.S.W., Hida T., de Jonge M., Orlov S.V., et al. Capmatinib in MET Exon 14-Mutated or MET-Amplified Non-Small-Cell Lung Cancer. New Engl. J. Med. 2020;383:944–957. doi: 10.1056/NEJMoa2002787. [DOI] [PubMed] [Google Scholar]
- 62.Benayed R., Offin M., Mullaney K., Sukhadia P., Rios K., Desmeules P., Ptashkin R., Won H., Chang J., Halpenny D., et al. High Yield of RNA Sequencing for Targetable Kinase Fusions in Lung Adenocarcinomas with No Mitogenic Driver Alteration Detected by DNA Sequencing and Low Tumor Mutation Burden. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2019;25:4712–4722. doi: 10.1158/1078-0432.CCR-19-0225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Duplaquet L., Kherrouche Z., Baldacci S., Jamme P., Cortot A.B., Copin M.C., Tulasne D. The multiple paths towards MET receptor addiction in cancer. Oncogene. 2018;37:3200–3215. doi: 10.1038/s41388-018-0185-4. [DOI] [PubMed] [Google Scholar]
- 64.Gow C.H., Liu Y.N., Li H.Y., Hsieh M.S., Chang S.H., Luo S.C., Tsai T.H., Chen P.L., Tsai M.F., Shih J.Y. Oncogenic Function of a KIF5B-MET Fusion Variant in Non-Small Cell Lung Cancer. Neoplasia. 2018;20:838–847. doi: 10.1016/j.neo.2018.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.International Cancer Genome Consortium PedBrain Tumor Project Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat. Med. 2016;22:1314–1320. doi: 10.1038/nm.4204. [DOI] [PubMed] [Google Scholar]
- 66.Kim P., Jia P., Zhao Z. Kinase impact assessment in the landscape of fusion genes that retain kinase domains: A pan-cancer study. Brief. Bioinform. 2018;19:450–460. doi: 10.1093/bib/bbw127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Pan Y., Zhang Y., Ye T., Zhao Y., Gao Z., Yuan H., Zheng D., Zheng S., Li H., Li Y., et al. Detection of Novel NRG1, EGFR, and MET Fusions in Lung Adenocarcinomas in the Chinese Population. J. Thorac. Oncol. 2019;14:2003–2008. doi: 10.1016/j.jtho.2019.07.022. [DOI] [PubMed] [Google Scholar]
- 68.Plenker D., Bertrand M., de Langen A.J., Riedel R., Lorenz C., Scheel A.H., Muller J., Bragelmann J., Dassler-Plenker J., Kobe C., et al. Structural Alterations of MET Trigger Response to MET Kinase Inhibition in Lung Adenocarcinoma Patients. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018;24:1337–1343. doi: 10.1158/1078-0432.CCR-17-3001. [DOI] [PubMed] [Google Scholar]
- 69.Siemanowski J., Heydt C., Merkelbach-Bruse S. Predictive molecular pathology of lung cancer in Germany with focus on gene fusion testing: Methods and quality assurance. Cancer Cytopathol. 2020;128:611–621. doi: 10.1002/cncy.22293. [DOI] [PubMed] [Google Scholar]
- 70.Vigna E., Gramaglia D., Longati P., Bardelli A., Comoglio P.M. Loss of the exon encoding the juxtamembrane domain is essential for the oncogenic activation of TPR-MET. Oncogene. 1999;18:4275–4281. doi: 10.1038/sj.onc.1202791. [DOI] [PubMed] [Google Scholar]
- 71.Zhu Y.C., Wang W.X., Xu C.W., Zhang Q.X., Du K.Q., Chen G., Lv T.F., Song Y. Identification of a novel crizotinib-sensitive MET-ATXN7L1 gene fusion variant in lung adenocarcinoma by next generation sequencing. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2018;29:2392–2393. doi: 10.1093/annonc/mdy455. [DOI] [PubMed] [Google Scholar]
- 72.Peschard P., Fournier T.M., Lamorte L., Naujokas M.A., Band H., Langdon W.Y., Park M. Mutation of the c-Cbl TKB domain binding site on the Met receptor tyrosine kinase converts it into a transforming protein. Mol. Cell. 2001;8:995–1004. doi: 10.1016/S1097-2765(01)00378-1. [DOI] [PubMed] [Google Scholar]
- 73.Awad M.M., Oxnard G.R., Jackman D.M., Savukoski D.O., Hall D., Shivdasani P., Heng J.C., Dahlberg S.E., Janne P.A., Verma S., et al. MET Exon 14 Mutations in Non-Small-Cell Lung Cancer Are Associated With Advanced Age and Stage-Dependent MET Genomic Amplification and c-Met Overexpression. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2016;34:721–730. doi: 10.1200/JCO.2015.63.4600. [DOI] [PubMed] [Google Scholar]
- 74.Krishnaswamy S., Kanteti R., Duke-Cohan J.S., Loganathan S., Liu W., Ma P.C., Sattler M., Singleton P.A., Ramnath N., Innocenti F., et al. Ethnic differences and functional analysis of MET mutations in lung cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2009;15:5714–5723. doi: 10.1158/1078-0432.CCR-09-0070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Tsai J.M., Hata A.N., Lennerz J.K. MET D1228N and D1246N are the Same Resistance Mutation in MET Exon 14 Skipping. Oncol. 2021;26:e2297–e2301. doi: 10.1002/onco.13924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li M.M., Datto M., Duncavage E.J., Kulkarni S., Lindeman N.I., Roy S., Tsimberidou A.M., Vnencak-Jones C.L., Wolff D.J., Younes A., et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017;19:4–23. doi: 10.1016/j.jmoldx.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Davies K.D., Ritterhouse L.L., Snow A.N., Sidiropoulos N. MET Exon 14 Skipping Mutations: Essential Considerations for Current Management of Non-Small-Cell Lung Cancer. J. Mol. Diagn. JMD. 2022;24:841–843. doi: 10.1016/j.jmoldx.2022.04.005. [DOI] [PubMed] [Google Scholar]
- 78.Feng Y., Thiagarajan P.S., Ma P.C. MET signaling: Novel targeted inhibition and its clinical development in lung cancer. J. Thorac. Oncol. 2012;7:459–467. doi: 10.1097/JTO.0b013e3182417e44. [DOI] [PubMed] [Google Scholar]
- 79.Ma P.C., Tretiakova M.S., MacKinnon A.C., Ramnath N., Johnson C., Dietrich S., Seiwert T., Christensen J.G., Jagadeeswaran R., Krausz T., et al. Expression and mutational analysis of MET in human solid cancers. Genes Chromosomes Cancer. 2008;47:1025–1037. doi: 10.1002/gcc.20604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jeffers M., Schmidt L., Nakaigawa N., Webb C.P., Weirich G., Kishida T., Zbar B., Vande Woude G.F. Activating mutations for the met tyrosine kinase receptor in human cancer. Proc. Natl. Acad. Sci. USA. 1997;94:11445–11450. doi: 10.1073/pnas.94.21.11445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Heist R.S., Sequist L.V., Borger D., Gainor J.F., Arellano R.S., Le L.P., Dias-Santagata D., Clark J.W., Engelman J.A., Shaw A.T., et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. J. Thorac. Oncol. 2016;11:1242–1245. doi: 10.1016/j.jtho.2016.06.013. [DOI] [PubMed] [Google Scholar]
- 82.Park M., Dean M., Cooper C.S., Schmidt M., O’Brien S.J., Blair D.G., Vande Woude G.F. Mechanism of met oncogene activation. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]