Abstract
Cytidine to uridine editing of apolipoprotein B (apoB) mRNA requires the cytidine deaminase APOBEC-1 as well as a tripartite sequence motif flanking a target cytidine in apoB mRNA and an undefined number of auxiliary proteins that mediate RNA recognition and determine site-specific editing. Yeast engineered to express APOBEC-1 and apoB mRNA supported editing under conditions of late log phase growth and stationary phase. The cis-acting sequence requirements and the intracellular distribution of APOBEC-1 in yeast were similar to those described in mammalian cells. These findings suggest that auxiliary protein functions necessary for the assembly of editing complexes, or ‘editosomes’, are expressed in yeast and that the distribution of editing activity is to the cell nucleus.
INTRODUCTION
Cytidine 6666 (C6666) of the mammalian apolipoprotein B (apoB) mRNA is edited to uridine by deamination (1) producing a codon change from glutamine to STOP (2). Unedited and edited mRNAs are translated into full length apoB100 and a truncated variant (apoB48) respectively (2–5), the latter of which is associated with a reduced risk of atherogenic diseases. ApoB mRNA editing requires a 26 nt tripartite motif consisting of the mooring sequence and spacer element downstream of C6666, and a regulatory element upstream of C6666 (6–9).
APOBEC-1 is the zinc-dependent cytidine deaminase required for apoB mRNA editing (10–13). The enzyme cannot edit apoB mRNA in the absence of other proteins (collectively referred to as auxiliary proteins), and alone has only weak binding affinity for AU-rich RNA sequence (10,14,15). Biochemical purification of editing activity from tissues or cultured mammalian cells identified a multi-protein complex ranging in size from 11S to 60S depending on the tissue studied (16–20).
Candidate auxiliary proteins such as hnRNP C and an hnRNP A/B homolog ABBP-1 (21,22), mooring-sequence selective RNA binding proteins of 100, 66 and 55 kDa (21–25) and general RNA binding proteins 40–44 kDa (23,26) have been identified through their affinity for APOBEC-1 or apoB RNA. A complex of proteins (referred to as AUX240 for the 240 kDa antigenic protein it contains) identified with monoclonal antibodies raised against in vitro assembled 27S editing complexes (editosomes) (27) has also been proposed to contain auxiliary proteins. HnRNP A/B, C, AUX240 and RNA binding protein p66 have been shown to have effects on in vitro editing activity (21–27). The specific requirement and function of these candidate auxiliary proteins in apoB mRNA editing remains to be demonstrated in tissues.
Transfection of cells with apobec-1 cDNA or addition of recombinant APOBEC-1 to cell extracts induced editing and thereby revealed that auxiliary proteins are broadly expressed, independent of whether tissues and cells naturally express apoB mRNA or editing activity (10,28–30). This suggests that in the absence of APOBEC-1, the auxiliary proteins may have roles in other cellular processes. Given the advantages of yeast as an experimental organism for characterizing macromolecular interactions and function, the budding yeast Saccharomyces cerevisiae was evaluated for the expression of proteins that could complement APOBEC-1 in C-to-U mRNA editing. The data presented here show that yeast express activities that allow APOBEC-1 to edit apoB RNA in a sequence dependent fashion.
MATERIALS AND METHODS
Plasmids
Rat APOBEC-1 modified with a hexa-histidine sequence, a nine amino acid hemagglutinin (HA) epitope tag and with or without an eight amino acid SV40 NLS motif (23,30) was cloned into pYES2 (Invitrogen) to create plasmids pGD307 and pGD309 respectively. This allowed galactose inducible expression of APOBEC-1 from the GAL1 promoter. 449 bp of wild-type human apoB, or the mutants Δ3′TL and MI (6,7), were amplified by PCR using primers Rsa13KpnI5′ and Rsa13BamHI3′ which incorporated terminal BamHI sites, and cloned into the BglII site in pPE282 (31) to create plasmids pGD308, pPE282Δ3′TL and pPE282MI respectively. This allowed constitutive expression from the PGK promoter and 3′ end formation using the PGK termination and polyadenylation signals.
Yeast strains, growth and transformation
The yeast strain used was CL51 (32). All yeast strains harboring plasmids were grown in synthetic dropout medium lacking uracil and/or leucine to select for maintenance of the plasmids (33). APOBEC-1 was expressed by growth in the presence of 2% galactose. Transformation was by standard methods (34).
Immunofluorescence microscopy
Yeast cells were fixed, permeabilized and immunostained as described (35). Confocal microscopy was performed with a Leica confocal microscope using an SP scan head, with a 100× objective and zoom set to 4×. Images were processed with Leica TCS NT and Adobe Photoshop software.
Editing assay
Total RNA was prepared from 40 ml of yeast cultures grown to late log phase, using TRIReagent (MRC, Inc.) according to the manufacturer’s instructions. RNAs were digested with RQ DNaseI (Promega) and an appropriate restriction enzyme for which there is a site between the PCR primer annealing sites to ensure the removal of contaminating DNA. Reverse transcript PCR (RT–PCR) was performed using 500 ng of total RNA and oligo-dT for first-strand cDNA synthesis. Oligonucleotides MS2 and MS3 were used for PCR amplification of apoB cDNAs. Poisoned primer extension analysis was performed using 50 ng of RT–PCR product and deoxyoligonucleotide primer DD3 as previously described (36). The no edit controls are from poisoned primer extension on PCR products derived from the plasmids encoding the test RNAs. The reaction products were separated by electrophoresis on a 10% polyacrylamide 7 M urea gel, and visualized by autoradiography. Quantification was by phosphorimage scanning densitometry (Molecular Dynamics) and was calculated as the sum of CAA, UAA and promiscuous editing sites divided into UAA plus promiscuous editing sites times 100. Note that the calculation for editing efficiency is internally controlled and therefore reflects the proportion of total apoB RNAs in the yeast cells that were edited.
Primers
Rsa13KpnI5′ (5′-CTCGGTACCGGATCCCCACAGAGAAAC), Rsa13BamHI3′ (5′-CGCGGATCCACTACTTCCACTTTTG-TTA), MS2 (5′-GTACTTCCACTTTTGTTAAAATC), MS3 (5′-GAAAATACAGAGCAGCCCTG), DD3 (5′-AATCATGTAAATCATAACTATCTTTAATATACTGA).
Western blot analysis
1.8 ml of cells were lysed by disruption with glass beads and treated as described (37). Lysates from 0.75 A600 units of cells were resolved on 10.5% SDS–PAGE, transferred to nitrocellulose, and reacted with the anti-HA monoclonal antibody (BAbCo) under blotting conditions described previously (30). Detection was by enhanced chemiluminescence (ECL, Amersham) according to the manufacturer’s instructions.
RESULTS
ApoB RNA editing in vivo
A standard laboratory yeast strain was transformed with plasmids that enabled the expression of 449 bp of human apoB RNA, encompassing the editing site at C6666, with and without expression of rat His6-HA-tagged APOBEC-1 (henceforth referred to as APOBEC). Expression of APOBEC had no adverse effect on cell growth as judged by colony morphology and cell doubling time in liquid culture. These strains were grown to late log phase in media containing 2% galactose to induce expression of APOBEC, total RNA was isolated and editing of polyadenylated apoB RNA determined by the poisoned primer extension assay. Yeast had little endogenous capacity to edit apoB RNA in the absence of APOBEC, with only background levels (0.3%) of C-to-U conversion detected (Fig. 1, lane 1). Expression of APOBEC stimulated editing of apoB RNA at C6666 to 5.7% (lane 2). As APOBEC alone cannot edit apoB RNA in vitro or in mammalian cell lines that do not express auxiliary proteins (10,30), the current data suggest that yeast express proteins that complement APOBEC in editing activity. Primer extension beyond C6666 (additional stops in lane 2) indicated that C6655 was also edited. ‘Promiscuous’ editing of C6655 and other cytidines has also been observed in mammalian cells (36,38,39) and transgenic animals (40,41) following experimental overexpression of APOBEC-1.
Figure 1.
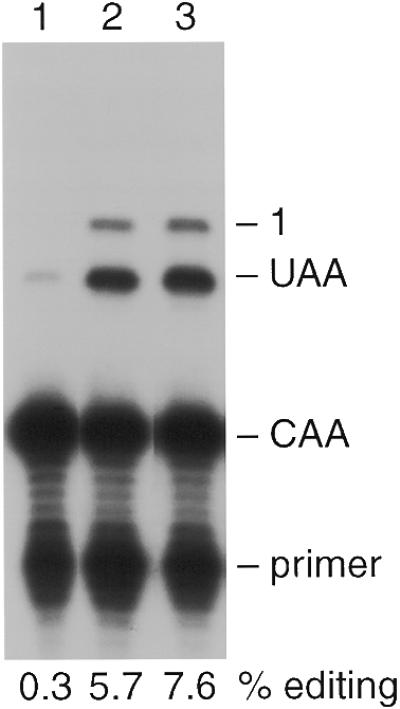
ApoB RNA editing in vivo. Yeast expressing APOBEC-1 and/or apoB RNA were grown to late log phase (A600 = >1.8). RNA was extracted from 40 ml of cells, and editing was assayed by primer extension. CAA and UAA denote the primer extension products from unedited and edited apoB PCR products respectively. Promiscuous editing of an additional 5′ cytidine is denoted as ‘1’. The percentage of edited apoB RNAs (editing efficiency) is indicated below each lane. Lanes 1–3 show primer extension products from strains expressing apoB (pGD308), His6-HA–APOBEC-1 and apoB (pGD309 and pGD308), and His6-HA–NLS–APOBEC-1 and apoB (pGD307 and pGD308) respectively.
Subcellular localization of APOBEC-1
Based on the occurrence of edited unspliced apoB pre-mRNA (42) and the ability of introns to interfere with editing activity (36), mammalian apoB mRNA editing has been proposed to be a nuclear event. APOBEC-1 has both nuclear and cytoplasmic distributions in editing competent mammalian cells. It has been suggested that this is due to the influence of the strong C-terminal nuclear export/cytoplasmic retention signal, NES/CRS over a weaker N-terminal nuclear localization signal, NLS (30). Importantly, the NES/CRS was completely dominant in cells which did not express complementing auxiliary proteins as experimentally expressed APOBEC was only detectable in the cytoplasm of these cells.
To evaluate whether editing in yeast could be improved by overcoming the influence of APOBEC-1’s NES/CRS, the SV40 NLS was fused to the N-terminus of APOBEC (NLS–APOBEC) and expressed. Editing was increased by 33% (from 5.7 to 7.6%, Fig. 1, lane 3) in the presence of NLS–APOBEC, suggesting that the nuclear distribution of the enzyme was important for yeast editing. Confocal immunofluorescence microscopy demonstrated a diffuse staining throughout the cell with the exception of the unstained central vacuole (Fig. 2A). Quantification of the fluorescence intensity due to NLS–APOBEC within the optical plane of the nucleus (DAPI staining, Fig. 2B) demonstrated a higher enzyme concentration in the nucleus relative to the cytoplasm. The localization of NLS–APOBEC in both the nucleus and the cytoplasm despite the presence of the SV40 NLS was also observed in mammalian cells (30), suggesting that the mechanism determining the intracellular distribution of APOBEC-1 and editing activity is similar in yeast and mammalian cells.
Figure 2.
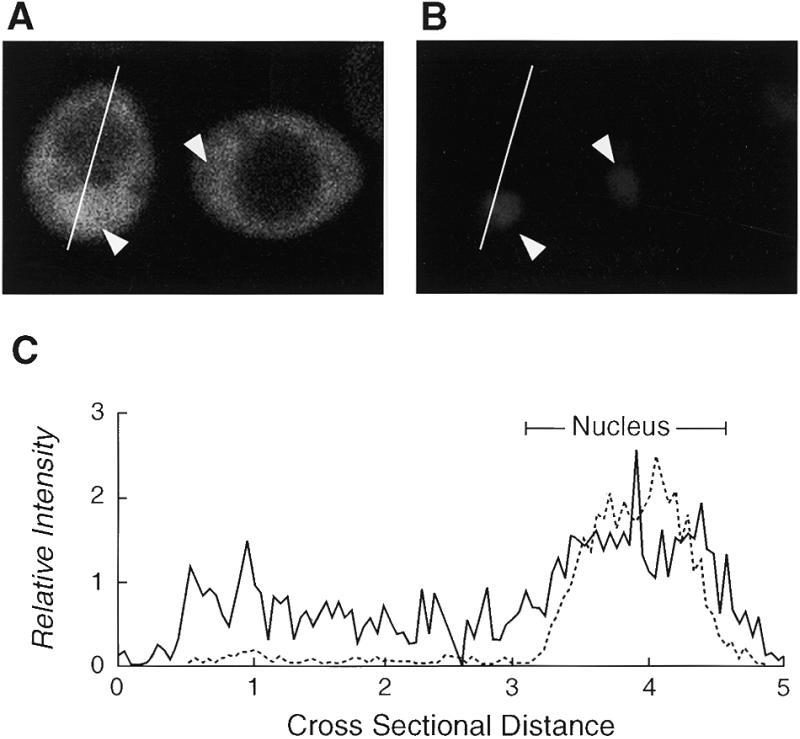
Subcellular localization of APOBEC-1. Yeast expressing NLS–APOBEC were visualized by immunofluorescence confocal microscopy. A representative field is shown for anti-HA antibody staining (A) and DAPI staining (B). Only the cell on the left is in the optical plane of the cell nucleus. Arrowheads indicate the positions of nuclei. The line through the cell on the left (A) indicates from top to bottom the path of the fluorescence quantification scan shown left to right in (C). The relative subcellular distribution of fluorescence was characterized by quantification of pixel intensities for the DAPI (broken line) and FITC (solid line).
Editing is growth-phase dependent
To further evaluate the optimal conditions for editing, NLS–APOBEC/apoB expressing yeast cells were analyzed at various times during growth. RNA was isolated from aliquots of a 750 ml culture at the indicated times and apoB RNA editing determined. ApoB RNA was edited with low efficiency (1–2%) until the cells reached late log phase (A600 = 1.6) at which point editing activity increased markedly (Fig. 3A and C). Moreover, promiscuous editing (C6655, C6651 and C6648) became more apparent as the cells entered stationary phase (Fig. 3C and 4B). In contrast, control cells expressing only apoB RNA showed only background levels of C6666 editing throughout the time course (Fig. 3A). RNA editing assays were quantified as the ratio of primer extension products from unedited and edited mRNAs (36,38,39). Each assay is therefore internally controlled and is not subject to sample to sample variation due to differences in mRNA abundance (36). Taken together, the editing efficiencies determined during the growth curve demonstrate that upon cessation of proliferation yeast exhibit more apoB RNA editing activity.
Figure 3.
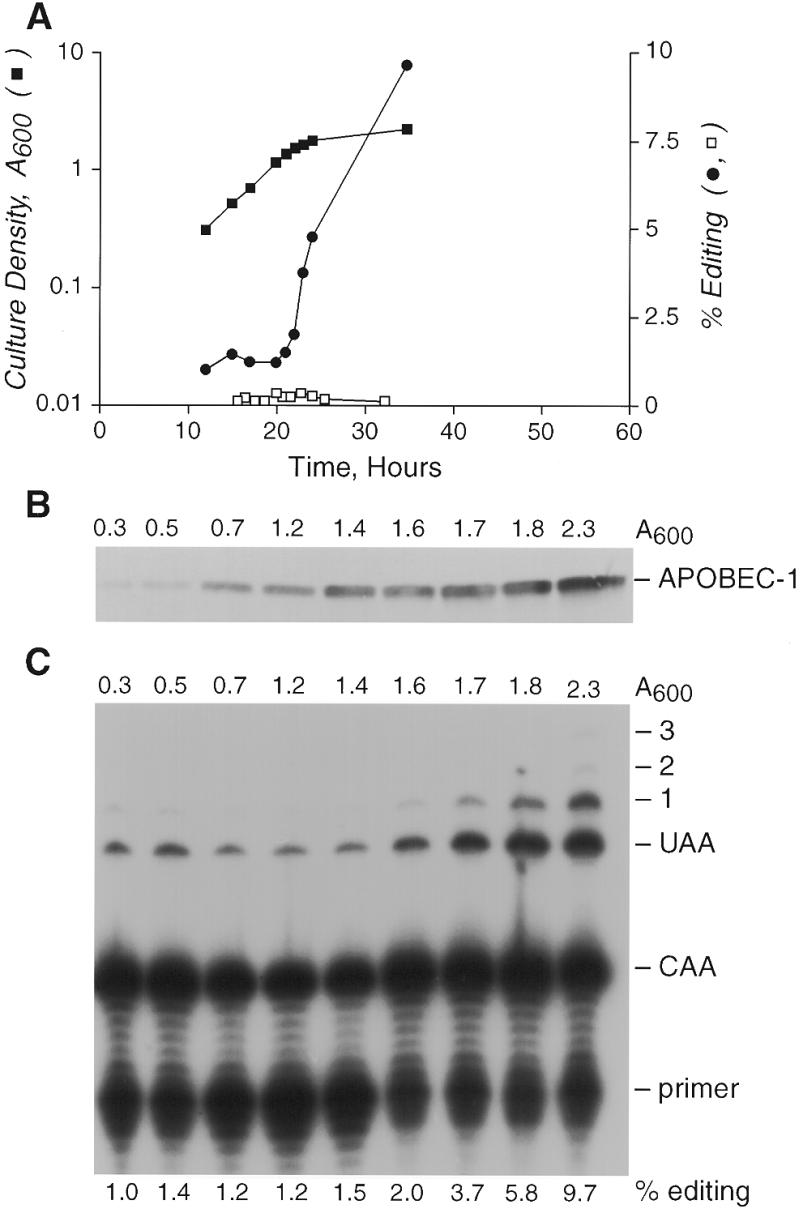
Editing shows a growth phase dependence. Aliquots (35 ml) of yeast expressing NLS–APOBEC and/or apoB RNA were removed from a 750 ml culture for RNA isolation during the time course. Aliquots (1.8 ml) were also removed at these times for western analysis. Editing was assayed by RT–PCR and primer extension. (A) Semi-log plot (left y-axis) of the culture A600 against time (closed square) and percentage of apoB RNA edited at C6666 (right y-axis) either in the absence of APOBEC-1 (open squares), or in the presence of APOBEC-1 (closed circles). (B) Western blot showing expression of NLS–APOBEC during the course of the experiment. (C) Primer extension gel showing editing in the presence of NLS–APOBEC during the course of the experiment. The percentage editing at C6666 is shown below each lane.
Figure 4.
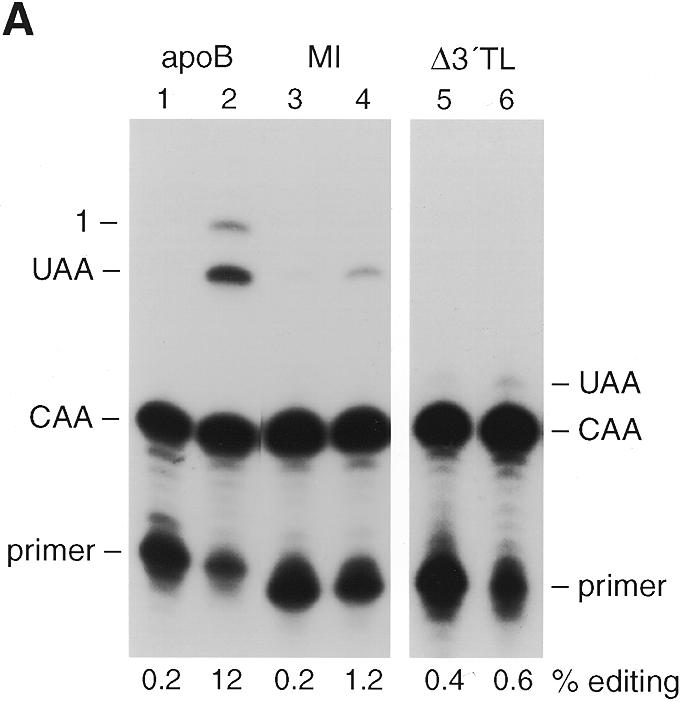
Editing is sequence dependent. (A) Lanes 2, 4 and 6 show editing assays for strains expressing NLS–APOBEC and either apoB RNA (from plasmid pGD308), MI RNA (pPE282MI) or Δ3′TL RNA (pPE282Δ3′TL) respectively. Lanes 1, 3 and 5 are no edit controls. Primer extension products CAA and UAA correspond to unedited and edited RNA respectively. ‘1’ indicates the primer extension product on mRNAs edited at C6666 and promiscuously edited at C6655. The percentage editing of the expressed mRNAs at C6666 is given below each lane. (B) Nucleotide sequence flanking the editing site at C6666 of rat apoB and the mutant substrates. Sequences are aligned and numbered such that the edited C of apoB is C6666, the edited C in the translocation construct, Δ3′TL is C6434. Sequence mismatches with the wild-type sequence flanking C6666 are shown in lower case letters. The location of the enhancer, spacer and mooring sequence components of the tripartite motif are underlined on the wild-type apoB RNA sequence.
Western blotting analysis of total cellular proteins from a constant number of cells revealed a gradual increase in NLS–APOBEC levels during the growth phase that continued into stationary phase (Fig. 3C). Our interpretation of these data is that the conditional nature of apoB editing activity arose from the expression of an appropriate complement of auxiliary proteins during late-log and stationary phase or the reduction of an inhibitory factor. We cannot exclude the possibility that the increase in editing activity resulted from a threshold concentration of NLS–APOBEC being achieved in cells at late-log phase growth that enabled interactions with pre-existing auxiliary proteins.
Editing is sequence dependent
Mooring sequence dependence is an essential characteristic of the mechanism of mammalian apoB mRNA editing (6–9,28,29,36). To evaluate this in yeast, two apoB mutant constructs were constitutively expressed with NLS–APOBEC. The MI mutant RNA construct contains a rearranged mooring sequence (Fig. 4B) and did not support editing in rat intestinal extracts (36) nor in wild-type rat liver cell lines, and supported markedly reduced editing in cells over expressing APOBEC (38,39). The MI RNA was inefficiently edited in yeast (Fig. 4, compare lanes 4 and 2) at only 1.2%.
Mutant RNA construct Δ3′TL was created by translocating the wild-type mooring sequence at C6666 downstream of an otherwise unedited cytidine at C6434 (6–8). The spacer element of this construct is 1 nt longer, and the enhancer element weaker, than that at C6666. Δ3′TL RNA was edited weakly in rat intestinal extracts (36) and in wild-type liver cell lines, but was efficiently and promiscuously edited in cells over expressing APOBEC (38,39). In yeast, Δ3′TL editing efficiency was low (0.6%) with no promiscuous editing detectable (Fig. 4, lane 3). The data suggested that APOBEC-dependent apoB RNA editing in yeast has similar cis-acting RNA sequence requirements as that seen in mammalian cells. It is interesting that yeast did not promiscuously edit Δ3′TL even though NLS–APOBEC had been over expressed. These findings suggest that there may be subtle variations in the yeast complementation of APOBEC and/or auxiliary protein interactions with apoB RNA. In this regard, the location and extent of promiscuous editing is known to vary with the cellular source of mammalian auxiliary factors (36,38,39).
We have also evaluated whether expression of APOBEC resulted in the editing of yeast endogenous mRNAs. A search for potential editing sites in the yeast ORF database using the 11 nt human mooring sequence as the query sequence identified 40 candidate mRNAs with a match ≥9/11. No yeast mRNAs were identified which contained a tripartite motif identical to apoB mRNA. Five mRNAs representative of the variations in the tripartite motif seen in the candidate group were amplified out of late log phase cells expressing NLS–APOBEC and assayed for editing. [The yeast genes assayed were RAD50 (700), PRP8 (2986), IRA1 (4848), JEM1 (414) and YLL015W (3447), using the nomenclature of S.G.D. (http://genome-www. stanford.edu/Saccharomyces/ ). The numbers in parentheses refer to the nucleotide position of the start of the putative mooring sequence, with respect to the start of the open reading frame.] None of the target cytidines within these mRNAs were edited (data not shown). These data suggest that expression of NLS–APOBEC did not result in random base modification of RNA. We propose that editing of apoB RNA was due to selective interactions of yeast auxiliary proteins with sequence and/or secondary structure specific to apoB RNA.
DISCUSSION
The identification of APOBEC-1 and subsequent publications have shown that, while APOBEC-1 is the cytidine deaminase responsible for editing the apoB mRNA, it requires an unknown complement of auxiliary factors to catalyze this reaction (10,14,15,29). APOBEC-1 has a non-selective RNA binding capacity (14,15), and therefore mooring sequence specificity must be imparted through interactions with a sequence-specific RNA-binding auxiliary factor or factors. These auxiliary proteins are widely expressed in vertebrate tissues, often independent of whether those tissues express or edit apoB mRNA, raising the possibility that the auxiliary proteins may have other roles in the cell (29).
The study described here is the first in which a mammalian mRNA editing enzyme has been expressed in yeast. We have shown that C-to-U editing of polyadenylated mRNA can be studied in yeast when mammalian APOBEC-1 and an apoB RNA substrate were co-expressed. This suggests that yeast can express appropriate and sufficient auxiliary protein activities to form functional editosomes with mammalian APOBEC-1. Recombinant APOBEC-1 has been shown to bind with some specificity to AU-rich RNA, but binds with low affinity as it is readily displaced by competitor RNAs (14,15). It is possible that APOBEC-1 expressed in yeast acquires a previously uncharacterized capacity for autonomous editing site recognition and editing in the absence of auxiliary proteins. We believe it is more likely that mooring sequence specificity is imparted through interactions with a sequence-specific RNA-binding auxiliary factor or factors. We have also shown the interesting characteristic that APOBEC-1 complementation is conditional, in that it depends on yeast cells entering a growth arrested state. This may have resulted from the onset of expression of one or more auxiliary proteins, the accumulation of a threshold amount of auxiliary proteins or the removal of negative regulators of editing. Why these auxiliary protein activities were expressed in yeast remains to be determined. An interesting possibility is that the proteins function in the modification and processing of other yeast RNAs or, consistent with the current hypothesis in the apoB mRNA editing field (28,29), in cellular processes not necessarily related to RNA processing. The low level of editing seen in early and mid log phase, and also in the controls not expressing APOBEC-1, could be a consequence of an endogenous editing activity. In this regard, computational modeling of the deaminase domain has suggested several yeast ORFs (43), of which one or more may be APOBEC-1 homologs. Experiments are in progress to evaluate this possibility.
The subcellular localization data are entirely consistent with APOBEC-1’s distribution in mammalian tissue culture cells (30). APOBEC-1 is known to have a very powerful NES/CRS signal, and this signal appears to be recognized in yeast cells also. The intrinsic weak NLS is also functional in yeast cells, and the presence of the SV40 NLS served to increase the nuclear distribution of the enzyme. However, as has been observed with similar fusions expressed in mammalian cells, the SV40 NLS was unable to substantially retain APOBEC-1 in the nucleus. We have used the NLS–APOBEC fusion throughout in an effort to maximize editing efficiency. It would appear from these data that editing is a nuclear event in yeast. However, we have not yet done experiments that prove conclusively whether editing is nuclear, cytoplasmic or both. The presence of such a strong NES/CRS is at odds with the wealth of data suggesting that editing is a nuclear event (42,44), and has led to the suggestion that the subcellular localization of APOBEC-1 may play a role in the regulation of editing (30). The observation that APOBEC-1 has the same distribution in yeast and mammalian cells suggests that editing in yeast may be regulated in a manner analogous to that in mammalian cells.
Expression of APOBEC-1 in yeast also induced promiscuous editing of apoB mRNA. Promiscuous editing was detected in mammalian cells over expressing APOBEC-1 only when the editing efficiency of C6666 exceeded 30–40% (36,38,39) and then, only at a constant proportion of the total amount of editing activity (45). In this regard the yeast editing system differed from the mammalian system in two ways; (i) promiscuous editing was detected under reduced editing efficiencies at C6666 and (ii) Δ3′TL RNA, a good substrate for promiscuous editing in mammalian cells, was not promiscuously edited in yeast. This suggests that there may be subtle variations in how the auxiliary factors in mammalian and yeast cells interact with either APOBEC-1 or with the tripartite motif in apoB mRNA.
In summary, we have presented data which show that yeast express sufficient proteins to form functional editosomes with mammalian APOBEC-1. These editosomes edit cytidines only in the context of the mammalian tripartite motif. A key control mechanism is missing or relaxed such that promiscuous editing occurs, as is also observed in some circumstances in mammalian cells. Finally, the subcellular localization of APOBEC-1 observed in mammalian cells is also seen in yeast. The powerful genetic methods that can be applied to yeast, as well as the availability of the complete genome sequence, should allow for rapid identification of the factors involved in editing in yeast, and by homology, the mammalian auxiliary factors.
Acknowledgments
ACKNOWLEDGEMENTS
The authors wish to thank Chris Guthrie for the gift of strain CL51 and Paul Roberts in the laboratory of David Goldfarb for technical help with the microscopy. The authors are grateful to Jenny M. L. Smith for the preparation of the figures. This work was supported by a Public Health Service Grant DK43738, and grants from the Council for Tobacco Research and the Alcoholic Beverage Medical Research Foundation awarded to H.C.S. and Environmental Health Sciences Center Grant ESO1247 awarded to Thomas Clarkson.
REFERENCES
- 1.Johnson D.F., Poksay,K.S. and Innerarity,T.L. (1993) Biochem. Biophys. Res. Comm., 195, 1204–1210. [DOI] [PubMed] [Google Scholar]
- 2.Chen S.H., Habib,G., Yang,C.Y., Gu,Z.W., Lee,B.R., Weng,S.A., Silberman,S.R., Cai,S.J., Deslypere,J.P., Rosseneu,M. Gotto,A.M., Jr., Li,W.H. and Chen,L. (1987) Science, 238, 363–366. [DOI] [PubMed] [Google Scholar]
- 3.Powell L.M., Wallis,S.C., Pease,R.J., Edwards,Y.H., Knott,T.J. and Scott,J. (1987) Cell, 50, 831–840. [DOI] [PubMed] [Google Scholar]
- 4.Greeve J., Altkemper,I., Dietrich,J.H., Greten,H. and Windler,E. (1993) J. Lipid Res., 34, 1367–1383. [PubMed] [Google Scholar]
- 5.Backus J.W., Eagleton,M.J., Harris,S.G., Sparks,C.E., Sparks,J.D. and Smith,H.C. (1990) Biochem. Biophys. Res. Comm., 170, 513–518. [DOI] [PubMed] [Google Scholar]
- 6.Backus J.W. and Smith,H.C. (1991) Nucleic Acids Res., 19, 6781–6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Backus J.W. and Smith,H.C. (1992) Nucleic Acids Res., 20, 6007–6014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith H.C. (1993) In Stuart,K. (ed.), Seminars in Cell Biology. Saunders Scientific Publishing/Academic Press, UK, Vol. 4, pp. 267–278.
- 9.Shah R.R., Knott,T.J., Legros,J.E., Navaratnam,N., Greeve,J.C. and Scott,J. (1991) J. Biol. Chem., 266, 16301–16304. [PubMed] [Google Scholar]
- 10.Teng B., Burant,C.F. and Davidson,N.O. (1993) Science, 260, 1816–1819. [DOI] [PubMed] [Google Scholar]
- 11.Barnes C. and Smith,H.C. (1993) Biochem. Biophys. Res. Comm., 197, 1410–1414. [DOI] [PubMed] [Google Scholar]
- 12.Navaratnam N., Morrison,J.R., Bhattacharya,S., Patel,D., Funahashi,T., Giannoni,F., Teng,B.B., Davidson,N.O. and Scott,J. (1993) J. Biol. Chem., 268, 20709–20712. [PubMed] [Google Scholar]
- 13.Navaratnam N., Fujino,T., Bayliss,J., Jarmuz,A., How,A., Richardson,N., Somasekaram,A., Bhattacharya,S., Carter,C. and Scott,J. (1998) J. Mol. Biol., 275, 695–714. [DOI] [PubMed] [Google Scholar]
- 14.Anant S., MacGinnitie,A.J. and Davidson,N.O. (1995) J. Biol. Chem., 270, 14762–14767. [PubMed] [Google Scholar]
- 15.Navaratnam N., Bhattacharya,S., Fujino,T., Patel,D., Jarmuz,A.L. and Scott,J. (1995) Cell, 81, 187–195. [DOI] [PubMed] [Google Scholar]
- 16.Smith H.C., Kuo,S.R., Backus,J.W., Harris,S.G., Sparks,C.E. and Sparks,J.D. (1991) Proc. Natl Acad. Sci. USA, 88, 1489–1493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris S.G., Sabio,I., Mayer,E., Backus,J.W., Sparks,J.D., Sparks,C.E. and Smith,H.C. (1993) J. Biol. Chem., 268, 7382–7392. [PubMed] [Google Scholar]
- 18.Smith H.C. (1998) In Chan,L. (ed.), Methods: A Companion to Methods in Enzymology. Academic Press, NY, Vol. 15, pp. 27–39.
- 19.Garcia Z.C., Poksay,K.S., Bostrom,K., Johnson,D.F., Balestra,M.E., Shechter,I. and Innerarity,T.L. (1992) Arterioscl. Thromb., 12, 172–179. [DOI] [PubMed] [Google Scholar]
- 20.Driscoll D.M. and Casanova,E. (1990) J. Biol. Chem., 265, 21401–21403. [PubMed] [Google Scholar]
- 21.Greeve J., Lellek,H., Rautenberg,P. and Greten,H. (1998) J. Biol. Chem., 379, 1063–1073. [DOI] [PubMed] [Google Scholar]
- 22.Lau P.P., Zhu,H.J., Nakamuta,M. and Chan,L. (1997) J. Biol. Chem., 272, 1452–1455. [DOI] [PubMed] [Google Scholar]
- 23.Yang Y., Yang,Y., Kovalski,K. and Smith,H.C. (1997) J. Biol. Chem., 272, 27700–27706. [DOI] [PubMed] [Google Scholar]
- 24.Navaratnam N., Shah,R., Patel,D., Fay,V. and Scott,J. (1993) Proc. Natl Acad. Sci. USA, 90, 222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mehta A. and Driscoll,D.M. (1998) Mol. Cell. Biol., 18, 4426–4432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lau P.P., Chen,S.-H., Wang,J.C. and Chan,L. (1990) Nucleic Acids Res., 18, 5817–5821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schock D., Kuo,S.R., Steinberg,M.F., Bolognino,M., Sparks,J.D., Sparks,C.E. and Smith,H.C. (1996) Proc. Natl Acad. Sci. USA, 93, 1097–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith H.C. and Sowden,M.P. (1996) Trends Genet., 12, 418–424. [DOI] [PubMed] [Google Scholar]
- 29.Smith H.C., Gott,J.M. and Hanson,M.R. (1997) RNA, 3, 1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y., Yang,Y. and Smith,H.C. (1997) Proc. Natl Acad. Sci. USA, 94, 13075–13080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stanway C.A., Chambers,A., Kingsman,A.J. and Kingsman,S.M. (1989) Nucleic Acids Res., 17, 9205–9218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lesser C.F. and Guthrie,C. (1993) Genetics, 133, 851–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sherman F. (1991) In Guthrie,C. and Fink,G.R. (eds), Methods in Enzymology. Academic Press, San Diego, CA, Vol. 194, pp. 3–21.
- 34.Gietz D., St Jean,A., Woods,R.A. and Schiestl,R.H. (1992) Nucleic Acids Res., 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Roberts C.J., Raymond,C.K., Yamashiro,C.T. and Stevens,T.H. (1991) In Guthrie,C. and Fink,G.R. (eds), Methods in Enzymology. Academic Press, San Diego, CA, Vol. 194, pp. 644–661. [DOI] [PubMed]
- 36.Backus J.W., Schock,D. and Smith,H.C. (1994) Biochim. Biophys. Acta, 1219, 1–14. [DOI] [PubMed] [Google Scholar]
- 37.Printen J.A. and Sprague,G.F.,Jr (1994) J. Biol. Chem., 266, 609. [Google Scholar]
- 38.Sowden M.J., Hamm,K. and Smith,H.C. (1996) J. Biol. Chem., 271, 3011–3017. [DOI] [PubMed] [Google Scholar]
- 39.Sowden M.J., Eagleton,M.J. and Smith,H.C. (1998) Nucleic Acids Res., 26, 1644–1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yamanaka S., Balestra,M., Ferrell,L., Fan,J., Arnold,K.S., Taylor,S., Taylor,J.M. and Innerarity,T.L. (1995) Proc. Natl Acad. Sci. USA, 92, 8483–8487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yamanaka S., Poksay,K.S., Arnold,K.S. and Innerarity,T.L. (1997) Genes Dev., 11, 321–333. [DOI] [PubMed] [Google Scholar]
- 42.Lau P.P., Xiong,W.J., Zhu,H.J., Chen,S.H. and Chan,L. (1991) J. Biol. Chem., 266, 20550–20554. [PubMed] [Google Scholar]
- 43.Mian I.S., Moser,M.J., Holley,W.R. and Chatterjee,A.J. (1998) Comput. Biol., 5, 57–72. [DOI] [PubMed] [Google Scholar]
- 44.Sowden M., Hamm,J.K., Spinelli,S. and Smith,H.C. (1996) RNA, 2, 274–288. [PMC free article] [PubMed] [Google Scholar]
- 45.Siddiqui J.F., Van Mater,D., Sowden,M.P. and Smith,H.C. (1999) Exp. Cell. Res., 252, 154–164. [DOI] [PubMed] [Google Scholar]



