Abstract
The fission yeast (Schizosaccharomyces pombe) taz1 gene encodes a telomere-associated protein. It contains a single copy of a Myb-like motif termed the telobox that is also found in the human telomere binding proteins TRF1 and TRF2, and Tbf1p, a protein that binds to sequences found within the sub-telomeric regions of budding yeast (Saccharomyces cerevisiae) chromosomes. Taz1p was synthesised in vitro and shown to bind to a fission yeast telomeric DNA fragment in a sequence specific manner that required the telobox motif. Like the mammalian TRF proteins, Taz1p bound to DNA as a preformed homodimer. The isolated Myb-like domain was also capable of sequence specific DNA binding, although with less specificity than the full-length dimer. Surprisingly, a protein extract produced from a taz1–fission yeast strain still contained the major telomere binding activity (complex I) we have characterised previously, suggesting that there could be other abundant telomere binding proteins in fission yeast. One candidate, SpX, was also synthesised in vitro, but despite the presence of two telobox domains, no sequence specific binding to telomeric DNA was detected.
INTRODUCTION
Telomeres are specialised nucleoprotein structures at the physical ends of eukaryotic chromosomes. They are involved in ensuring the complete replication of chromosomes, preventing chromosome fusions and in protecting chromosomes from nucleolytic degradation (1–3). They have also been implicated in mediating chromosomal localisation within the nucleus and in repressing expression of telomere adjacent genes (4–7). In most eukaryotes, the DNA component of telomeres consists of tandem repeats of TG-rich sequences that are in a defined orientation and are essential for telomere function (8). The TG-rich sequence forms a 3′ single stranded overhang at the chromosome termini (9,10). Both the double stranded and single stranded regions are involved in specific interactions with proteins that play important roles in telomere function. The best characterised double stranded telomere binding protein is budding yeast Rap1p (11). This is an essential multifunctional protein that is involved in activating and repressing transcription, as well as performing important functions at telomeres (12). These functions include helping to protect the ends of the chromosomes and playing a central role in the mechanism by which telomere length is controlled (12). Rap1p interacts with DNA as a monomer via a large central DNA binding domain, the structure of which has been solved by X-ray crystallography (13). It contains two sub-domains, each of which shows structural similarity to a repeat motif present in the DNA binding region of the vertebrate c-Myb oncogene product. These Myb-like sub-domains each consist of three alpha helices, the second and third of which comprise a helix–turn–helix motif (13).
In mammalian cells, two double stranded telomere binding proteins have been identified, TRF1 and TRF2 (14–16). These are very different to Rap1p in both their amino acid sequences and their overall organisation, although they appear to perform some functions in common with this protein. Each binds to DNA as a homodimer via a single Myb-like domain located at the C-terminus (16,17). Detailed analysis of TRF1 has revealed that in vitro the isolated Myb-like domain can also interact with DNA as a monomer that recognises the sequence 5′-AGGGTTA-3′ (18). One common feature between Rap1p and TRF1/TRF2 is the presence of a Myb-like DNA binding domain. This motif is also present within Taz1p, a telomere binding protein in fission yeast (Fig. 1A). This yeast is distantly related to both budding yeast and mammalian cells. It therefore provides a useful model organism in which to compare the organisation and properties of telomeres with these well characterised systems. Previous analysis using gel retardation assays and a probe containing the double stranded repeat region of a cloned fission yeast telomere (19) identified four specific DNA–protein complexes in total protein extracts (I, I′, IIa and IIb) (20). At least two distinct factors are responsible for these complexes, as determined by differing binding site specificities. The taz1 gene was subsequently identified in a one-hybrid screen for genes encoding double stranded telomere binding factors (21). It was found to be a non-essential gene that is not a sequence homologue of RAP1. Taz1p is involved in telomere length regulation, repression of telomere adjacent genes and the interactions between telomeres and the spindle pole body during meiotic prophase (22,23). In terms of overall organisation, Taz1p broadly resembles TRF1 and TRF2. It has a single Myb-like domain at the C-terminus. However, direct DNA binding by Taz1p has not yet been demonstrated and the role, if any, that the Myb-like domain plays has not been investigated. In this study we have synthesised Taz1p in vitro and studied the interaction between this protein and telomeric DNA. We have also used a taz1– yeast strain to investigate the relationship between Taz1p and the telomeric binding proteins identified previously using gel retardation assays. These experiments have shown significant similarities between Taz1p and TRF1/TRF2, reinforcing the idea that fission yeast is an ideal model organism for genetic studies of telomere biology. They have also shown that the major DNA:protein complex detected using a telomere probe in gel retardation assays does not require Taz1p.
Figure 1.


Sequence specific binding of Taz1p to telomeric DNA. (A) Relative positions of Myb-like domains in Taz1p (21) and TRF1 (14), with percentage amino acid identity indicated. The region of Taz1p that corresponds to the ‘isolated Myb domain’ is shown (Isolated Region). (B) Gel retardation assay showing in vitro translated (IVT) Taz1p binding to the 140 bp telomeric fragment. Lanes 1 and 3, no added protein; lane 2, fission yeast total protein extract; lane 4, control lysate; lane 5, IVT Taz1p; lane 6, IVT Taz1p + unlabelled probe fragment as competitor; lane 7, IVT Taz1p + unlabelled AC1/2 oligonucleotide competitor; lane 8, IVT Taz1p + unlabelled AC7/8 oligonucleotide competitor. T indicates the position of the major Taz1p complex; ΔT indicates faster migrating complexes produced by non-full length Taz1p. I indicates the position of complex I formed using the total protein extract. F indicates the position of the unbound probe. (C) Gel retardation assay comparing complexes formed using total protein extracts (TPE) of taz1+ and taz1– strains with the 140 bp telomeric fragment. Lane 1, fragment alone; lane 2, taz1– TPE; lane 3, taz1+ TPE. I, I′, IIa and IIb indicate the positions of DNA:protein complexes described previously (20). F indicates the position of the unbound probe.
MATERIALS AND METHODS
Strains and media
Plasmid manipulations were carried out using Escherichia coli MC1061 [F– araD139Δ(ara-leu)7696 Δ(lac)174 galU galK hsdR strA (strr]. The haploid Schizosaccharomyces pombe wild type strain 972h– was compared to the taz1– strain J28 (h– ade6-M210 leu1-32 ura4-D18 taz1::ura4+) (21). Saccharomyces cerevisiae strain HF7c (Clontech) (a ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3 112 gal4-542 gal80-538 LYS2::GAL-HIS3 URA::(GAL4 17-mers)3 -CYC1-LacZ) was used for two- hybrid analysis. Strains were routinely grown using YEPD medium on plates and in liquid culture, and transformants selected on SC minimal media supplemented with the appropriate amino acids.
Cloning and expression of full length Taz1p, GFP-tagged Taz1p, and the isolated Myb domain derivative
The taz1 sequence was retrieved from the EMBL database (accession number YO9406). Primers were designed to the flanking regions of the gene (+ strand: –24 to –4, – strand: +1997 to 2016) incorporating novel EcoRI (position –13) and BamHI (position +2007) sites for ease of cloning. The gene was amplified from S.pombe genomic DNA prepared using the ‘10 minute’ method (24). The 2 kb PCR product was cut with EcoRI and BamHI and cloned into the corresponding sites in plasmid pPE722, downstream of an SP6 promoter. pPE722 is a derivative of pSP56 in which the polylinker PstI site has been destroyed (25).
To construct the GFP–taz1 fusion, the taz1 DNA was isolated from pPE722-taz1, by digestion with EcoRI, end-filling using dNTPs and Klenow polymerase, then digestion with BamHI to excise the 2 kb gene fragment. This was ligated into the GFP plasmid pKS406 (26), that had been cut with NotI, end-filled, then cut with BamHI. The GFP–taz1 fragment was excised from pKS406 by digesting with XhoI, end-filling, then cutting with BamHI. This fragment was ligated into pSP46 (25) that had been cut with BamHI and SmaI.
The taz1 Myb domain was isolated from pPE722-taz1 and ligated into pSP56 (25), downstream of an SP6 promoter. A 477 bp fragment was excised from pPE722-taz1 using XmnI and BamHI. This fragment was cloned into pSP56 cut with SmaI and BamHI (positions 1529–1992 of the taz1 gene). Within this fragment was an in frame ATG codon at position 1590, thus it encoded a protein of 134 amino acids (residues 530–664 of Taz1p). This included the entire Myb-like domain of Taz1p (Fig. 1A). This 134 amino acid protein, as well as the full length and GFP tagged Taz1p were produced by coupled in vitro transcription/translation using the ‘TNT’ reagents according to the manufacturer’s instructions (Promega).
Cloning and expression of SpX
The spx coding region (SWISS-PROT Q10274) (27) was isolated by PCR from a commercial S.pombe cDNA library (Clontech Matchmaker library). The primers used corresponded to the + strand (+1 to +21) and to the – strand (+1294 to +1315). The PCR product was first cloned into the vector pNoTA/T7 (5 prime–3 prime Inc. ), then isolated by BamHI digestion and cloned into the BamHI site of pSP46 (25). SpX was produced by coupled in vitro transcription/translation using the ‘TNT’ reagents according to the manufacturer’s instructions (Promega).
Telomeric DNA fragments and competitor oligonucleotides
The standard 140 bp telomere probe used in gel retardation assays was isolated by digestion of plasmid pAJ34 with BamHI and PstI (20). The oligonucleotides for use in competition experiments consisted of the following sequences:
AC1: 5′-GATCTCAGCT GG TTACAGG TTACAGG TT G-3′
AC2: 5′-GATCC AA CCTGTAA CCTGTAA CC AGCTGA-3′
AC3: 5′-GG TTACAGGGGGG TT-3′
AC4: 5′-AA CCCCCCTGTAA CC-3′
AC5: 5′-TTACAGG TTACAGG-3′
AC6: 5′-CCTGTAA CCTGTAA-3′
AC7: 5′-GATCTCAGCT GG TGACAGG TGACAGG TT G-3′
AC8: 5′-GATCC AA CCTGTCA CCTGTCA CC AGCTGA-3′
AS1: 5′-GATCCTAAATATAAAAA-3′
AS2: 5′-GATCTTTTTATATTTAG-3′
These oligonucleotides were annealed in pairs to generate double stranded fragments for use as competitor DNA. In the telomeric repeat oligonucleotide pairs AC1/2, AC3/4, AC5/6 and AC7/8, the telomeric sequence is indicated in bold and the repeats are underlined. AS1/2 is a control oligonucleotide used to test for non-specific competition. For competition analysis, 2 µg of annealed competitor was added to the binding reactions for gel retardation assays.
The minimal telomeric probe used in gel retardation assays contained the telomeric sequence of the AC1/2 oligonucleotide pair. The AC1 and AC2 oligonucleotides were annealed to form double stranded AC1/2 fragment with 5′ and 3′ overhangs producing BamHI ends. This fragment was cloned into the BamHI site of vector pSP46 (25) so that a 160 bp fragment containing AC1/2 sequence + plasmid sequence could be excised with EcoRI and RsaI to be used as a probe in gel retardation assays.
Protein extracts and gel retardation analysis
Schizosaccharomyces pombe cultures were grown to mid-log phase in 50 ml YEPD medium and protein extracts prepared as described previously (20). The protein concentration of each extract was determined using the Bradford assay (28). Typically, 5 µg of this total protein extract or 2 µl of in vitro translated product was used in each gel retardation reaction. Binding reactions were performed at room temperature in a total reaction volume of 20 µl. The labelled DNA fragment for use in the assay was generated by end-labelling ~120 ng of isolated DNA fragment using [γ-32P]ATP (>185 MBq/mmol; Amersham International plc) and T4 polynucleotide kinase (Life Technologies Inc.). The protein sample was incubated with 4 µg poly(dI:dC) and 2 ng of the labelled DNA as described previously (20). For the mixing experiment using in vitro produced and endogenous S.pombe proteins, equal amounts of either 35S-labelled Taz1p or SpX were added to 5 µg of total protein extract in the presence of binding buffer and poly(dI:dC), but in the absence of telomere probe fragment. This mix was incubated at 30°C for 30 min to allow heterodimers to form, before the addition of 2 ng of unlabelled probe fragment. After incubation for 30 min at room temperature, DNA–protein complexes were resolved by electrophoresis at 180 V for 2 h using 16 cm-long 5% polyacrylamide gels containing 0.5× TBE. Gels were dried and visualised either by autoradiography or by using a Molecular Dynamics Phosphorimager system and ImageQuant software. Band intensities were measured and corrected for background.
Two-hybrid analysis and β-galactosidase quantification
The two-hybrid method was based on the protocol by Clontech. Two vectors were used, one containing the Gal4p DNA binding domain (pAS2-1), the other containing the Gal4p activation domain (pACT-2). The taz1 gene was cloned into each of these vectors so that in-frame fusions were generated. Both DBD:taz1 and AD:taz1 fusion constructs were created as follows. Plasmids pACT2 and pAS2-1 were prepared for cloning by digestion with NcoI, end-filling, then digestion with BamHI. The taz1 gene was excised from plasmid pPE722-taz1 by digestion with EcoRI, end-filling, then digestion with BamHI. This fragment was ligated with the prepared pACT-2 and pAS2-1 DNA to generate the fusion plasmids. Positive and negative controls were generated using plasmids pVA3-1, pTD1-1 and pLAM5′-1 (Clontech).
These plasmids were co-transformed into HF7c and activation of the HIS3 reporter gene tested by growing on His– plates supplemented with 5 mM 3-AT. Liquid β-galactosidase assays were carried out using ONPG as a substrate as previously described (29).
RESULTS
Taz1p interacts specifically with fission yeast telomeric DNA in vitro
The taz1 gene was identified in a one-hybrid screen for genes encoding proteins that interact with S.pombe telomeric DNA sequence (21). However, direct binding of the Taz1 protein to telomeric DNA was not demonstrated. To address this, a gel retardation assay was performed using in vitro produced Taz1p and a radioactively labeled 140 bp telomeric probe fragment. This fragment contains a total of 13 repeats, the majority of which conform to the consensus 5′-TTACAG1-8-3′. A strong complex (T) between Taz1p and this probe was detected (Fig. 1B, lanes 5 and 8). The complexes (ΔT) migrating faster than the major complex were probably formed by incompletely synthesised or partially degraded Taz1p, because in some preparations of the protein these were not detected (see Fig. 4B lane 2 for example) and because these complexes were always competed with the same kinetics as the major complex. The faint, slower migrating complexes were probably formed by multiple Taz1p molecules binding to the probe. The major Taz1p complex was shown to be specific because it was by competed by an excess of unlabelled 140 bp fragment (lane 6) and an oligonucleotide pair containing two complete telomeric repeats (AC1/2) (lane 7), but there was no significant competition by the oligonucleotide pair AC7/8 which contained two mutated telomeric repeats (lane 8). A truncated version of Taz1p was also produced in vitro. This lacked the region C-terminal to amino acid 551 that contains the Myb-like domain. When an equal amount of this truncated protein was tested in gel retardation assays, no specific complexes were formed (data not shown).
Figure 4.



SpX does not bind to telomeric DNA in vitro. (A) Protein gel showing 35S methionine-labelled IVT proteins. Lane 1, control lysate; lane 2, IVT Taz1p; lane 3, IVT SpX. The positions of molecular weight markers of 66 and 45 kDa are shown. (B) Gel retardation assay using the 140 bp telomere probe fragment with the control lysate (lane 1), IVT Taz1p (lane 2) and IVT SpX (lane 3). T indicates the position of the Taz1p complex; F shows the position of the unbound probe fragment. (C) Gel retardation assays using 35S-labelled IVT Taz1p (lanes 2–4) and 35S-labelled IVT SpX (lanes 5–7). Lanes 2 and 5 contain the 35S protein with no added probe fragment. Lanes 3 and 6 contain the 35S protein with added unlabelled telomere probe. Lanes 4 and 7 contain the 35S protein pre-incubated with a fission yeast total protein extract before the addition of the unlabelled telomere fragment. Lane 1 contains labelled telomere probe fragment plus the same amount of fission yeast protein extract that was used in lanes 4 and 7.
The major complex detected using in vitro produced Taz1p did not migrate to the same position in gel retardation assays as the major complex (I) formed using the same probe and a fission yeast total protein extract (Fig. 1B, lane 2) (20), suggesting that Taz1p may not be responsible for this complex. This is supported by the fact that complex I was found previously to be competed efficiently by both the wild type and the mutant telomeric oligonucleotide pairs (20), whereas the Taz1 complex was only competed efficiently by the wild type oligonucleotide pair (Fig. 1B, lane 7).
In order to confirm this suggestion, gel retardation assays were performed using protein extracts from wild type (972h-) and taz1– (J28) fission yeast strains (Fig. 1C). It is clear from this assay that the protein(s) responsible for complex I is present in the taz1– protein extract (lane 2), although it is not obvious which of the other defined complexes, if any, involve Taz1p binding. It is possible that Taz1p is involved in the formation one or both of complexes IIa and IIb because these complexes are not produced clearly in the taz1- strain and they have similar binding site specificities to Taz1p in competition assays (20). However, the mobility of the major complex formed by in vitro produced Taz1p is not identical to the mobility of these complexes (see Fig. 4C).
Taz1p binds as a dimer to telomeric DNA
By comparison with human TRF1 it would be predicted that Taz1p dimerises, thereby uniting two Myb-like domains for sequence-specific DNA recognition. Two separate approaches have been used to examine the potential for Taz1p to form dimers. First, Taz1p was fused to the 26 kDa green fluorescent protein (GFP) and gel retardation analysis was carried out using in vitro translated Taz1p, GFP–Taz1p and a co-translated mix of GFP–Taz1p + Taz1p. In this experiment the probe contained only two complete telomeric repeat units (minimal telomeric probe) (Fig. 2A). As expected, the major shift caused by binding of the tagged version of Taz1p (lane 3) was greater than the untagged version (lane 1) due to the increase in molecular weight of the protein. Where both Taz1p derivatives were co-translated (lane 2), a new complex (*) that migrated to an intermediate position was observed. This additional complex was dependent on both derivatives being present, implying binding to the telomeric probe by a GFP–Taz1p:Taz1p heterodimer.
Figure 2.
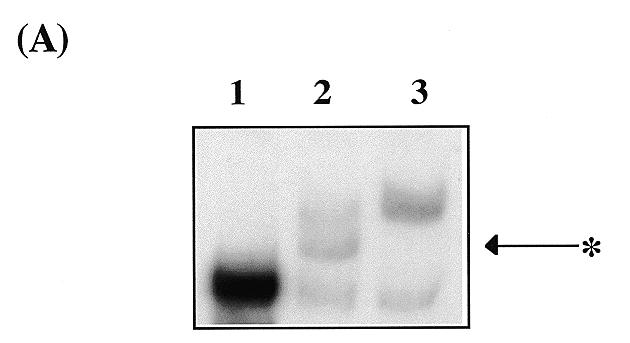

Taz1p binds to DNA as a dimer. (A) Gel retardation assay showing complex formation by Taz1p and GFP–Taz1p. Lane 1, minimal telomere fragment + IVT Taz1p; lane 2, fragment + co-translated IVT Taz1p + IVT GFP–Taz1p; lane 3, fragment + IVT GFP–Taz1p. * indicates the position of the intermediate complex formed only in the co-translated protein mix. (B) Taz1p interacts with itself in the two-hybrid system. Growth of yeast transformants on his– plates. 1, negative control (pAS2-1-Taz1p + pACT-2); 2, negative control (pAS2-1 + pACT-2-Taz1p); 3, negative control (pLAM5′-1 + pACT-2-Taz1p); 4, positive control (pVA3-1 + pTD1-1); 5, pAS2-1-Taz1p + pACT-2-Taz1p. (C) Level of β-galactosidase activity (ΔOD420/min/mg protein) present in the two hybrid transformants. The numbers represent the same combinations as above.
The second strategy to investigate dimerisation of Taz1p was a yeast two-hybrid approach. For this analysis, the taz1 gene was cloned into the two hybrid vectors pAS2-1 and pACT2 to create separate Taz1p fusions with the Gal4p DNA binding domain and activation domain respectively. HF7c yeast transformants expressing both of these fusion proteins were assayed for HIS3 and LacZ reporter gene expression. Growth on his- media is indicated in Figure 2B. Lack of growth for the negative controls indicates that neither Taz1p fusion protein is capable of reporter gene activation alone and that the activation domain fusion protein is not involved in non-specific reactions with LaminC (segments 1–3). Growth on his– media can be seen for the positive control, which indicates the known interaction between p53 and SV40 large T-antigen (segment 4), and for the transformants expressing AD:Taz1p + DBD:Taz1p (segment 5). This is indicative of a Taz1p:Taz1p interaction in the two-hybrid system. The significance of this interaction was clarified when measuring activation of the second reporter gene, LacZ, using liquid β-galactosidase assays. The β-galactosidase activities of the transformants are illustrated in Figure 2C. All three negative controls (columns 1–3) gave very low β-galactosidase levels. In contrast to this very low background activity, transformants containing both taz1 plasmids produced a significant level of β-galactosidase activity (column 5), which was ~40% of the level produced as a result of the strong control interaction (column 4). This indicates that the interaction between Taz1p monomers is relatively strong in this system and that it is not dependent on the Myb-like domains binding to DNA. In agreement with the suggestion that Taz1p binds to DNA as a preformed homodimer, no intermediate complex was observed in the gel retardation experiment where tagged and untagged Taz1p were mixed rather than co-translated (data not shown).
Sequence-specific binding of the isolated Myb domain of Taz1p
The C-terminal Myb-like domain of Taz1p shares 30% amino acid sequence identity with that of TRF1. The conserved regions include basic residues found within the recognition helix of the Myb-like three helical bundle, as well as in the region proposed to form an N-terminal arm in TRF1 (18). The N-terminal arm is involved in direct sequence-specific DNA contacts that are thought to enable the isolated DNA binding domain of TRF1 to bind independently to DNA (18), whereas typical Myb proteins, without this N-terminal arm, require at least two Myb-motifs for sequence-specific DNA binding. To investigate the potential for the Taz1p Myb-like domain to bring about independent DNA binding, this region of Taz1p was produced in vitro and its DNA binding properties examined by gel retardation analysis (Fig. 3A). The C-terminal 134 amino acids of Taz1p that contained the entire Myb domain (Fig. 1A) was expressed in vitro and binding to the 140 bp telomeric probe analysed. This fragment of Taz1p was found to produce a clear complex (Fig. 3A, lane 4) that was competed by the AC1/2 oligonucleotide pair but not by the AS1/2 oligonucleotide pair, in a similar way to full length Taz1p (Fig. 3A, lanes 5, 6 and 13, 14, see also Fig. 3B). This suggested that the isolated Myb domain may bind to DNA strongly and specifically as a monomer. Although it is formally possible that the Myb domain sub-fragment of Taz1p can form a dimer, this seems unlikely because of the relative mobility of the DNA:protein complex and because by analogy with TRF1, it lacks the region most likely to be involved in dimerisation (17). Further cross competition experiments confirmed this view. The oligonucleotide pair AC7/8 contains two complete telomeric repeats with 2 bp of flanking sequence either side, as does the AC1/2 pair. In AC7/8 these two repeats each contain a mutation in a highly conserved T residue. The AC7/8 oligonucleotide pair did not significantly cross compete binding by the dimer of full length Taz1p, but greatly reduced binding by the isolated Myb-like domain (Fig. 3A, lanes 11 and 12, and Fig. 3B) consistent with the idea that the isolated Myb domain is interacting with DNA in a different way to the full length dimer (Discussion).
Figure 3.
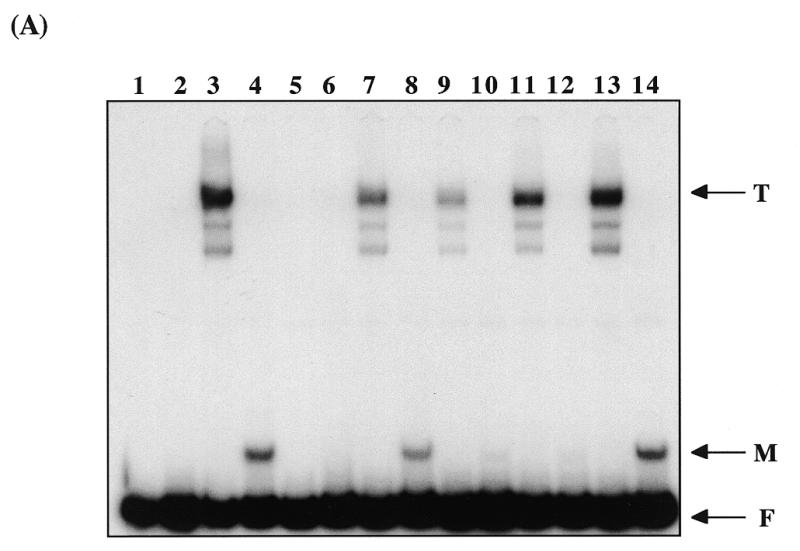
DNA binding by the isolated Myb domain of Taz1p. (A) Gel retardation assay showing binding by in vitro produced Taz1p and Taz1p Myb domain. Lane 1, 140 bp telomere fragment alone; lane 2, control lysate; lane 3, full length (FL) Taz1p; lane 4, Myb domain; lane 5, FL Taz1p + AC1/2; lane 6, Myb + AC1/2; lane 7, FL Taz1p + AC3/4; lane 8, Myb + AC3/4; lane 9, FL Taz1p + AC5/6; lane 10, Myb + AC5/6; lane 11, FL Taz1p + AC7/8; lane 12, Myb + AC7/8; lane 13, FL Taz1p + AS1/2; lane 14, Myb + AS1/2. T, major complex formed by full length Taz1p; M, complex formed by the isolated Myb domain; F, unbound probe fragment. (B) Graphical representation of phosphorimager quantification of the competition assays showing percentage of uncompeted shift for the various competitor oligonucleotides.
SpX does not interact directly with fission yeast telomeric DNA in vitro
SpX is a candidate telomere binding protein in fission yeast (27). It contains two Myb-like domains that are related to the telobox motifs found in Taz1p and TRF1/TRF2. The presence of these motifs suggested that this protein could be responsible for one or more DNA:protein complexes detected in the taz1– protein extract. The spx gene was isolated and used to produce SpX in vitro (Fig. 4A). The in vitro produced protein was then tested in gel retardation assays using the 140 bp telomere probe. Control assays using Taz1p were performed in parallel. The Taz1p bound strongly and specifically to the probe as expected (Fig. 4B, lane 2), but there was no detectable binding by SpX (Fig. 4B, lane 3). Despite performing a number of experiments using different amounts of protein and different buffer conditions, we were not able to detect any specific binding by SpX alone to the telomere probe. To test the possibility that SpX requires an interaction with another protein before it will bind to DNA, we used 35S-labelled in vitro produced SpX in mixing experiments with a fission yeast total protein extract. The in vitro produced protein was mixed with the total protein extract and incubated at 30°C for 30 min to allow interactions with endogenous yeast proteins. Complex formation was detected using an unlabelled telomere probe fragment in gel retardation assays (Fig. 4C). As a control we used 35S-labelled Taz1p in similar mixing experiments. A labelled telomere probe was used with the same batch of total protein extract to show the positions of the previously defined DNA:protein complexes. The Taz1p control generated a major complex (T) both in the absence (lane 3) and presence (lane 4) of the total protein extract. Interestingly, this complex did not align with either complex I or complexes IIa and IIb formed using the labelled probe fragment and the total protein extract (lane 1). In contrast to Taz1p, SpX did not form any detectable complexes either alone (lane 6) or in combination with the total protein extract (lane 7). This confirmed the previous conclusion that SpX does not bind to telomeric DNA alone and suggested that this protein is not involved as part of a heterodimer in any of the telomeric complexes.
DISCUSSION
Taz1p has been shown to interact directly with fission yeast telomeric DNA in gel retardation assays. This sequence specific binding involves dimers of Taz1p, as determined by in vitro binding of a GFP–Taz1p:Taz1p heterodimer. Additionally, Taz1p can dimerise without the need for telomeric DNA binding, as demonstrated by a Taz1p:Taz1p two-hybrid interaction. Dimerisation of Taz1p may increase the DNA binding affinity and specificity of the protein by associating two Myb-like motifs. In this regard Taz1p appears to be similar to TRF1 and TRF2, which also have a single Myb-like motif at the C-terminus and bind to DNA as dimers. The similarity between Taz1p and the human TRF proteins extends beyond the ability of these proteins to bind DNA as homodimers. The C-terminal DNA binding domain of Taz1p shares extensive amino acid identity with the DNA binding domain of TRF1 (Fig. 5). It has been shown previously that the isolated Myb-like domain of TRF1 will bind in a sequence specific manner to telomeric DNA in vitro (18). We have now shown that this is also true for Taz1p. Cross competition experiments demonstrated that the isolated Myb domain can bind to a sequence (AC7/8) that is not recognised by the full length protein. It has been shown previously that the binding site for the TRF1 Myb-like domain corresponds to the sequence 5′-AGGGTTA-3′ (18). In fission yeast telomeric DNA this sequence is not common and the repeat unit is rather variable. In the 140 bp telomere fragment we used as a probe, this sequence occurs only once. However, the truncated sequence GGTTA occurs 12 times, usually preceded by an A or G (eight times). The minimal fragment that was bound by the full length Taz1p dimer (AC1/2) contains two copies of the GGTTA sequence, consistent with the idea that this is the sequence recognised by each Myb-like domain. The AC7/8 oligonucleotide pair cross competed binding by the isolated Myb-like domain, but not the Taz1p dimer. This oligonucleotide contains no exact matches to the GGTTA sequence, but three near matches, TGGTGA, AGGTGA and AGGTTG. It is possible that only one of these near matches can be recognised by the Taz1p Myb domain. This is sufficient for binding by the isolated domain, but not by the dimer, which requires two recognition sites to interact with DNA. It was shown recently that human TRF1 can bind as a homodimer to two recognition sites located at variable distances apart (30). Although we have not tested Taz1p, such a property may be important in allowing this protein to bind along the length of the fission yeast telomere where the basic repeat unit is rather variable in length.
Figure 5.
Alignment of the Myb domains in human TRF1 (TRF1Myb), SpX (SpXMybA, SpXMybB) and Taz1p (Taz1Myb). The boxes indicate residues that conform to the consensus of all four sequences. The position of the recognition helix in the TRF1 Myb domain is underlined.
To date, Taz1p is the only double stranded telomere binding protein to be isolated from fission yeast (21). It was therefore a surprise to find that Taz1p was not required for formation of the major telomere complex produced by a fission yeast total protein extract (complex I). This observation was supported by cross-competition experiments where the Taz1p complex was competed efficiently by the AC1/2 oligonucleotide pair, but not by the AC7/8 pair. In contrast, the protein(s) responsible for complex I was competed efficiently by the AC7/8 sequence (20). Whether Taz1p is involved in the formation of any of the complexes detected in the total protein extract is not clear. The experiments using the taz1– strain indicated that formation of complexes IIa and IIb was affected when Taz1p was absent. However, in vitro produced Taz1p did not form a complex with identical mobility to either complex IIa or IIb in gel retardation assays. It is possible that this reflects differences in post-translational modification between the in vitro produced Taz1p and the endogenous yeast protein.
These observations suggest that the most abundant or high affinity telomere binding protein(s) in fission yeast (responsible for complex I) have yet to be identified. We tested the candidate telomere binding protein SpX for binding to telomeric DNA in vitro. Despite using a range of experimental conditions, no specific binding was observed, either using SpX alone or a mix containing labelled SpX and endogenous fission yeast proteins. Although this does not completely rule out the possibility that SpX does bind to telomeric DNA, it would seem that this is unlikely. SpX contains two Myb-like repeats (Fig. 5), both of which are more similar to TRF1 than to Taz1p, particularly in the recognition helix. It is therefore possible that the sequences recognised by these domains are similar to the AGGGTTA sequence recognised by TRF1, which is not common in fission yeast telomeric DNA.
Overall, the experiments described have established that in many respects fission yeast Taz1p resembles mammalian TRF1/TRF2. This supports the idea that for genetic studies of telomere biology, fission yeast may be a better model for mammalian systems than that currently provided by budding yeast. One important function of TRF1 is the ability of TRF1 multimers to link pairs of telomeric tracts, so helping to mediate intramolecular telomere looping (31,32). It is not known whether yeast telomeres are long enough to allow looping to occur. However, the observation that Taz1p, like TRF1, binds directly to telomeric DNA as a dimer and shares similar sequence-specific DNA binding properties suggests that if looping does occur in fission yeast, Taz1p may play an important role in the process.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Julia Cooper for the generous gift of the taz1– strain J28, Ken Sawin and Paul Nurse for providing the GFP vector pKS406, and Neal Sugawara for telomere subclones. This work was supported by the University of Nottingham Research Opportunities Fund.
REFERENCES
- 1.Blackburn E.H. (1991) Nature, 350, 569–573. [DOI] [PubMed] [Google Scholar]
- 2.Zakian V.A. (1996) Trends Cell Biol., 6, 29–33. [DOI] [PubMed] [Google Scholar]
- 3.Blackburn E.H. (1994) Cell, 77, 621–623. [DOI] [PubMed] [Google Scholar]
- 4.Chikashige Y., Ding,D., Funabiki,H., Haraguchi,T., Mashiko,S., Yanagida,M. and Hiraoka,Y. (1994) Science, 264, 270–273. [DOI] [PubMed] [Google Scholar]
- 5.Kirk K., Harmon,B., Reichardt,I., Sedat,J. and Blackburn,E. (1997) Science, 275, 1478–1481. [DOI] [PubMed] [Google Scholar]
- 6.Wright J.H., Gottschling,D.E. and Zakian,V.A. (1992) Genes Dev., 6, 197–210. [DOI] [PubMed] [Google Scholar]
- 7.Kyrion G., Liu,K., Liu,C. and Lustig,A.J. (1993) Genes Dev., 7, 1146–1159. [DOI] [PubMed] [Google Scholar]
- 8.Zakian V.A. (1989) Annu. Rev. Genet., 23, 579–604. [DOI] [PubMed] [Google Scholar]
- 9.Pluta A.F., Kaine,B.P. and Spear,B.B. (1982) Nucleic Acids Res., 10, 8145–8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Henderson E.R. and Blackburn,E.H. (1989) Mol. Cell. Biol., 9, 345–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shore D. (1994) Trends Genet., 10, 408–412. [DOI] [PubMed] [Google Scholar]
- 12.Shore D. and Nasmyth,K. (1987) Cell, 51, 721–732. [DOI] [PubMed] [Google Scholar]
- 13.Konig P., Giraldo,R., Chapman,L. and Rhodes,D. (1996) Cell, 85, 125–136. [DOI] [PubMed] [Google Scholar]
- 14.Chong L., van Steensel,B., Broccoli,D., Erdjumant-Bromage,H., Hanish,J., Tempst,P. and de Lange,T. (1995) Science, 270, 1663–1666. [DOI] [PubMed] [Google Scholar]
- 15.Bilaud T., Brun,C., Ancelin,K., Koering,C.E., Laroche,T. and Gilson,E. (1997) Nature Genet., 17, 236–239. [DOI] [PubMed] [Google Scholar]
- 16.Broccoli D., Smogorzewska,A., Chong,L. and de Lange,T. (1997) Nature Genet., 17, 231–235. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi A., Smith,S., Chong,L., Elias,P. and de Lange,T. (1997) EMBO J., 16, 1785–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konig P., Fairall,L. and Rhodes,D. (1998) Nucleic Acids Res., 26, 1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugawara N. (1989) PhD thesis, Harvard University, Cambridge, MA.
- 20.Duffy M. and Chambers,A. (1996) Nucleic Acids Res., 24, 1412–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cooper J.P., Nimmo,E.R., Allshire,R.C. and Cech,T.R. (1997) Nature, 385, 744–747. [DOI] [PubMed] [Google Scholar]
- 22.Cooper J.P., Watanabe,Y. and Nurse,P. (1998) Nature, 392, 828–831. [DOI] [PubMed] [Google Scholar]
- 23.Nimmo E.R., Pidoux,A.L., Perry,P.E. and Allshire,R.C. (1998) Nature, 392, 825–828. [DOI] [PubMed] [Google Scholar]
- 24.Hofmann C.S. and Winston,F. (1987) Gene, 57, 267–272. [DOI] [PubMed] [Google Scholar]
- 25.Ogden J.E., Stanway,C., Kim,S., Mellor,J., Kingsman,A.J. and Kingsman,S.M. (1986) Mol. Cell. Biol., 6, 4335–4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sawin K.E. and Nurse,P. (1996) Proc. Natl Acad. Sci. USA, 94, 15146–15151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brun C., Marcand,S. and Gilson,E. (1997) Trends Cell Biol., 7, 317–324. [DOI] [PubMed] [Google Scholar]
- 28.Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 29.Miller J.H. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Bianchi A., Stansel,R.M., Fairall,L., Griffith,J.D., Rhodes,D. and de Lange,T. (1999) EMBO J., 20, 5735–5744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Griffith J., Bianchi,A. and de Lange,T. (1998) J. Mol. Biol., 278, 79–88. [DOI] [PubMed] [Google Scholar]
- 32.Griffith J.D., Comeau,L., Rosenfield,S., Stansel,R.M., Bianchi,A., Moss,H. and de Lange,T. (1999) Cell, 97, 503–514. [DOI] [PubMed] [Google Scholar]






