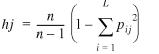Abstract
1.33 Mb of sequence from the human Y chromosome was searched for tri- to hexanucleotide microsatellites. Twenty loci containing a stretch of eight or more repeat units with complete repeat sequence homogeneity were found, 18 of which were novel. Six loci (one tri-, four tetra- and one pentanucleotide) were assembled into a single multiplex reaction and their degree of polymorphism was investigated in a sample of 278 males from Pakistan. Diversities of the individual loci ranged from 0.064 to 0.727 in Pakistan, while the haplotype diversity was 0.971. One population, the Hazara, showed particularly low diversity, with predominantly two haplotypes. As the sequence builds up in the databases, direct methods such as this will replace more biased and technically demanding indirect methods for the isolation of microsatellites.
INTRODUCTION
Microsatellites are tandemly repeated arrays of two to six base-pair units and are consequently also known as simple tandem repeats (STRs). They are dispersed ubiquitously around the genomes of many species, including humans. Their abundance, high degree of polymorphism and ease of scoring have resulted in their being very widely used in diverse fields including genetic mapping, forensic investigations and evolutionary studies (1). Human Y-specific DNA polymorphisms have an important role in several of these areas, and Y-specific microsatellites (2,3) have been used for forensic (4), genealogical (5) and evolutionary (6–8) purposes. There is a need to characterise novel Y-specific microsatellites in order to increase discrimination in forensic applications and to provide a choice of loci with simple or complex structure and high or low variability for application to evolutionary questions on different timescales. Microsatellites have traditionally been identified by cloning fragments containing a predetermined repeat motif. The extensive human sequence data that are accumulating in publicly available databases now allow a simpler, more direct and less biased ascertainment of microsatellites. We have therefore investigated how readily useful Y markers can be derived from available sequence information, and report six novel polymorphic microsatellites showing a range of diversities. They have been assembled into a single convenient multiplex reaction which extends the MS1 and MS2 kits currently available (9) and now allows 16 loci to be scored in three reactions.
MATERIALS AND METHODS
Blood samples in ACD vacutainers were collected from unrelated volunteers from nine different ethnic groups of Pakistan and lymphoblastoid cell lines were established (10). DNA was isolated from these cell lines using the standard organic extraction method (11). A total of 278 samples were analysed in the present study consisting of 12 Kashmiri, 17 Makrani, 18 Pathan, 12 Sindhi, 36 Syed, 88 Parsi, 46 Burusho, 23 Hazara and 26 Kalash.
Y-chromosomal DNA sequence data were obtained from the Whitehead Institute/MIT Genome Sequencing Project’s database (http://www-seq.wi.mit.edu/public_release/humanY.shtml ). The clones screened were: AC005820, AC004474, AC002531, AC004810, AC004617, AC002992, AC004772, AC002509, AC000021, AC000022, AC006565 and AC005942. Sequences were downloaded in FASTA format and used as input for the program Tandem Repeats Finder (12) (http://c3.biomath.mssm. edu/trf.html ). Microsatellites were chosen from the output of this program according to the criteria: (i) unit size 3–6 bp, >90% matches between units and array of eight or more copies, or (ii) unit size 3–6 bp, >80% matches between units and array of 25 or more copies. Primers were designed using Primer3 software (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi ). Unlabelled primers were synthesised on an ABI392 DNA/RNA synthesiser using phosphoramidite chemistry, and labelled primers (Table 1) were supplied by MWG Oligo.
Table 1. Primer sequences.
| Primer | Primer sequence | Repeat | Size | Dye | Final concentration |
|---|---|---|---|---|---|
| name | unita | (bp)a | label | (µM) | |
| DYS434L | CAC TCC CTG AGT GCT GGA TT | CTAT | 114 | TET | 0.2 |
| DYS434R | GGA GAT GAA TGA ATG GAT GGA | 0.2 | |||
| DYS437L | GAC TAT GGG CGT GAG TGC AT | TCTA | 192 | HEX | 0.1 |
| DYS437R | AGA CCC TGT CAT TCA CAG ATG A | 0.1 | |||
| DYS435L | AGC ATC TCC ACA CAG CAC AC | TGGA | 210 | TET | 0.05 |
| DYS435R | TTC TCT CTC CCC CTC CTC TC | 0.05 | |||
| DYS438L | TGG GGA ATA GTT GAA CGG TAA | TTTTC | 221 | HEX | 0.2 |
| DYS438R | GTG GCA GAC GCC TAT AAT CC | 0.2 | |||
| DYS436L | CCA GGA GAG CAC ACA CAA AA | GTT | 133 | FAM | 0.025 |
| DYS436R | GCA ATC CAA CTT CAG CCA AT | 0.025 | |||
| DYS439L | TCC TGA ATG GTA CTT CCT AGG TTT | GATA | 252 | TET | 0.2 |
| DYS439R | GCC TGG CTT GGA ATT CTT TT | 0.2 |
aIn the allele sequenced (GenBank).
The multiplex PCR was performed in a 10 µl final volume containing 1X Super Taq Buffer [10 mM Tris–HCl, pH 9.0, 1.5 mM MgCl2, 50 mM KCl, 0.1% Triton X-100, 0.01% (w/v) stabiliser], additional MgCl2 to give a final concentration of 2.2 mM, 300 µM dNTPs, 30 ng DNA, 0.13 U Super TAQ (HT Biotechnology Ltd) and 0.357 µg TaqStart Antibody (Clontech). Primer sequences and concentrations were as in Table 1. The Super TAQ enzyme was incubated with the TaqStart Antibody in the presence of TaqStart Antibody dilution buffer for 5–7 min at room temperature and was then added to the master mix. In the TouchDown PCR protocol the DNA was initially denatured at 94°C for 2 min. This was followed by eight cycles starting with 94°C for 1 min, 60°C for 1 min and 72°C for 1 min. The annealing temperature was decreased by 0.5°C in each cycle. These eight cycles were then followed by 30 cycles of 94°C for 1 min, 56°C for 1 min and 72°C for 1 min. After a final extension step at 72°C for 5 min the samples were kept at 4°C until electrophoresis. A portion of the sample (0.3 µl) was mixed with TAMRA350 internal lane size standard and the samples were electrophoresed on a 5% polyacrylamide gel using an ABI377 DNA sequencer according to the manufacturer’s instructions. Data were collected using the ABI collection software where the fragment sizes were estimated using GeneScan software (v2.1) and the alleles were called using Genotyper software (v2.0).
Gene frequencies of alleles were obtained for each locus or haplotype by simple gene/haplotype counting, and gene/haplotype diversity values were calculated according to the equation (13)
where n is the number of individuals, hj is the diversity of a given locus or haplotype with L alleles and pij is the gene frequency of allele i in population j.
Standard errors were calculated according to the following equation (13):
Fifteen alleles (at least two from each locus) were sequenced (http://www.bioch.ox.ac.uk/~dnaseq/ ) to determine the correspondence between the number of repeat units and the allele size in base pairs. These values were then used to calibrate the number of units in all the samples based upon their size. A set of DNA samples was used as external standards in each gel, in order to correct for gel-to-gel variations. However, no such variations were observed.
RESULTS
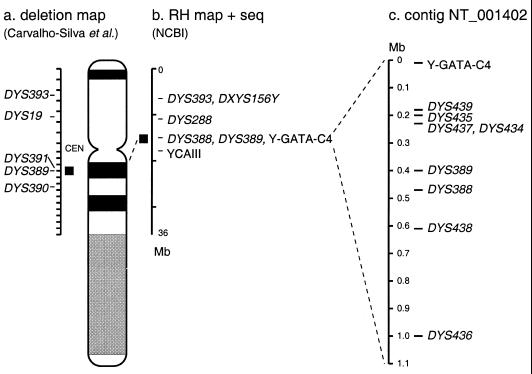
1.33 Mb of Y sequence was screened to identify tandemly repeated sequences. We found 22 loci that matched our first set of criteria and three matching our second set, a total of 25. Two of them corresponded to the previously identified loci DYS388 (AF140633) and Y-GATA-C4 (14). DYS389 is also present in the clones investigated, but was not picked out using our criteria because it was too heterogeneous. The remaining 23 sequences were examined by eye and 18 were found to contain a stretch of eight or more units with complete repeat sequence homogeneity. Two loci from the DAZ cluster were discarded because the region is largely or entirely repeated. Eight of the others (50%) gave a single, male-specific, product; five gave a product in both male and female DNA, and three gave no specific product under the conditions used. Seven of the eight Y-specific loci were tested for polymorphism and all were polymorphic. Six loci could be co-amplified in a single multiplex PCR reaction (Table 1). A map of these loci, in comparison to those previously identified and mapped on the chromosome, is shown in Figure 1.
Figure 1.
Location of the novel microsatellites on the Y chromosome relative to the previously identified microsatellite loci. (a) Known loci mapped on a panel of deleted Y chromosomes by Carvalho-Silva et al. (16). The Y-specific portion of the short arm was divided into seven intervals, shown here as equal in size; the centromere (CEN) lies in interval 8 and the euchromatic portion of the long arm was divided into 13 intervals. (b) Known loci identified in the sequence information available from GenBank and positioned according to the RH mapping information given on http://www.ncbi.nlm.nih.gov/genome/seq/chr.cgi?CHR=Y . In (a) and (b), the location of the new microsatellites is shown by the black square. (c) Positions of known and novel loci in more detail in the NT_001402 contig.
Using this assay, DNA samples from 278 Pakistani males were characterised. Diversities of individual loci in the entire sample ranged from 0.064 to 0.728 (Table 2). Overall haplotype diversity in the Pakistani sample was 0.971. Haplotype diversities in individual populations ranged from 0.656 in the Hazara to 0.949 in the Makrani (Table 3).
Table 2. Characteristics of individual microsatellite loci.
| Locus | Alleles | No. of | Allele | Diversity | |
|---|---|---|---|---|---|
| Units | bpa | Chroms | frequencies | ||
| DYS434 | 0.222 | ||||
| 8 | 110 | 9 | 0.0327 | ||
| 9 | 114 | 242 | 0.8800 | ||
| 10 | 118 | 13 | 0.0473 | ||
| 11 | 122 | 11 | 0.0400 | ||
| DYS437 | 0.664 | ||||
| 8,2,4 | 186 | 109 | 0.3921 | ||
| 9,2,4 | 190 | 75 | 0.2698 | ||
| 10,2,4 | 194 | 93 | 0.3345 | ||
| 11,2,4 | 202 | 1 | 0.0036 | ||
| DYS435 | 0.070 | ||||
| 11 | 220 | 268 | 0.9640 | ||
| 12 | 224 | 8 | 0.0288 | ||
| 13 | 228 | 2 | 0.0072 | ||
| DYS438 | 0.684 | ||||
| 6 | 203 | 2 | 0.0074 | ||
| 9 | 218 | 64 | 0.2353 | ||
| 10 | 223 | 100 | 0.3676 | ||
| 11 | 228 | 97 | 0.3566 | ||
| 12 | 233 | 9 | 0.0331 | ||
| DYS436 | 0.064 | ||||
| 10 | 128 | 1 | 0.0036 | ||
| 11 | 131 | 6 | 0.0218 | ||
| 12 | 134 | 266 | 0.9673 | ||
| 15 | 143 | 2 | 0.0073 | ||
| DYS439 | 0.728 | ||||
| 9 | 238 | 2 | 0.0072 | ||
| 10 | 242 | 59 | 0.2130 | ||
| 11 | 246 | 95 | 0.3430 | ||
| 12 | 250 | 88 | 0.3177 | ||
| 13 | 254 | 29 | 0.1047 | ||
| 14 | 258 | 4 | 0.0144 |
aThe measured size differs slightly from that predicted from the sequence.
Table 3. Microsatellite haplotype diversities within different populations.
| Population | n | Diversity | Standard error |
|---|---|---|---|
| Parsi | 88 | 0.928 | 0.011 |
| Burusho | 46 | 0.934 | 0.016 |
| Hazara | 23 | 0.656 | 0.053 |
| Kashmiri | 12 | 0.758 | 0.080 |
| Makrani | 17 | 0.949 | 0.014 |
| Pathan | 18 | 0.817 | 0.064 |
| Sindhi | 12 | 0.929 | 0.029 |
| Syed | 36 | 0.948 | 0.011 |
| Kalash | 26 | 0.855 | 0.022 |
DISCUSSION
The method used for identifying new microsatellites was simple and efficient. All of the loci tested after matching our criteria of length and homogeneity proved to be polymorphic. The diversity values for two of them (DYS435 and DYS436) were low, at least in the Pakistani sample surveyed, and it would probably not be useful to analyse shorter or less homogeneous loci. The sequence information available at present from the database is derived from only a single individual, so some microsatellites that are unusually short, or have unusually complex repeat motifs, on this chromosome may have been excluded.
Our findings allow us to compare the Y with other chromosomes and estimate the total number of useful Y microsatellites likely to be available. Since the seven loci tested, and the two previously known loci, were variable, it seems likely that most or all of the 20 microsatellites found in the sequence investigated are polymorphic and could provide useful markers if suitable primers were designed: approximately 1 per 65 kb. Similar analyses of 1.33 Mb of arbitrarily chosen sequence from chromosome 7 (AC006356, AC005154, AC007270, AC005084, AC006318, AC002451, AC002098, AC005371 and AC004854) or chromosome 22 (AC007666, Z82198, AC006285, AC002472, AP000355, AL021937, Z86090, Z82189 and AL022327) revealed 11 and 25 loci, respectively, that matched our search criteria. Thus, despite the lack of recombination on the Y, the frequency of tri- to hexanucleotide microsatellites is similar to that on other chromosomes, as might be expected if microsatellites arise primarily by replication slippage. The Y chromosome is ~60 Mb in length and approximately half of it is euchromatic. Much of the euchromatin contains sequences that are shared with the X chromosome or repeated elsewhere on the Y. If 10 Mb of unique sequences are present, around 150 useful loci are potentially available. In the 1.33 Mb of Y DNA investigated, there were even more (32) dinucleotide microsatellites matching our criteria. Although these markers are less easy to score and compare between laboratories, they may also be useful.
The six loci analysed consisted of one trinucleotide repeat, four tetranucleotide repeats and one pentanucleotide repeat. Four had simple array structures, but two tetranucleotides were part of more complex arrays. Among those with simple structures, the GTT repeat (DYS436; modal unit number = 12) and the TGGA repeat (DYS435; modal number = 11) were the least variable, while the GATA repeat (DYS434; modal number = 9) showed an intermediate level and the TTTTC repeat (DYS438; modal number = 10) was highly variable. The two complex repeats, where the GenBank loci may be represented as (GATA)4(GACA)2(GATA)10 (DYS437) and (GATA)2N4(GA-TA)3N14(GATA)N3(GATA)N7(GATA)13 (DYS439) respectively, were both highly variable. The sequenced alleles showed variation in only the largest block of repeats, but variation in the other blocks may be found in more extensive surveys. Thus the relationship between variability and modal number of repeats is not simple, and may be easier to understand when individual lineages defined by more slowly evolving markers are examined. The TTTTC repeat appears to be derived from the poly(A) tail of an Alu element, as has previously been observed (15). While the three highly variable loci will be the most useful for forensic applications, the four simple loci may be the best for evolutionary work since it will be possible to infer the locus structure from the fragment size.
We anticipate that these microsatellites will generally be used in combination with other genetic markers, but even alone the haplotypes found in the different populations provide insights into population structure. For example, the Hazara stand out from the other populations because they have a particularly low diversity (0.656) and the two haplotypes 9,9,11,10,12,12 and 11,8,11,10,12,10 (loci DYS434, DYS437, DYS435, DYS438, DYS436, DYS439), differing by five steps, together account for >80% of the population, suggesting a recent male lineage bottleneck or founder effect.
Acknowledgments
ACKNOWLEDGEMENTS
We thank the DNA donors, Mark Jobling for referring us to Tandem Repeats Finder and for comments on the manuscript and Denise Rejane de Carvalho-Silva for comments on the manuscript. This work was supported by The Wellcome Trust (053584/Z/98/Z).
REFERENCES
- 1.Goldstein D.B. and Pollock,D.D. (1997) J. Hered., 88, 335–342. [DOI] [PubMed] [Google Scholar]
- 2.Roewer L., Arnemann,J., Spurr,N.K., Grzeschik,K.H. and Epplen,J.T. (1992) Hum. Genet., 89, 389–394. [DOI] [PubMed] [Google Scholar]
- 3.Mathias N., Bayés,M. and Tyler-Smith,C. (1994) Hum. Mol. Genet., 3, 115–123. [DOI] [PubMed] [Google Scholar]
- 4.Kayser M., Caglia,A., Corach,D., Fretwell,N., Gehrig,C., Graziosi,G., Heidorn,F., Herrmann,S., Herzog,B., Hidding,M. et al. (1997) Int. J. Legal. Med., 110, 125–133. [DOI] [PubMed] [Google Scholar]
- 5.Foster E.A., Jobling,M.A., Taylor,P.G., Donnelly,P., de Knijff,P., Mieremet,R., Zerjal,T. and Tyler-Smith,C. (1998) Nature, 396, 27–28. [DOI] [PubMed] [Google Scholar]
- 6.Ruiz Linares A., Nayar,K., Goldstein,D.B., Hebert,J.M., Seielstad,M.T., Underhill,P.A., Lin,A.A., Feldman,M.W. and Cavalli Sforza,L.L. (1996) Ann. Hum. Genet., 60, 401–408. [DOI] [PubMed] [Google Scholar]
- 7.Zerjal T., Dashnyam,B., Pandya,A., Kayser,M., Roewer,L., Santos,F.R., Schiefenhovel,W., Fretwell,N., Jobling,M.A., Harihara,S. et al. (1997) Am. J. Hum. Genet., 60, 1174–1183. [PMC free article] [PubMed] [Google Scholar]
- 8.de Knijff P., Kayser,M., Caglia,A., Corach,D., Fretwell,N., Gehrig,C., Graziosi,G., Heidorn,F., Herrmann,S., Herzog,B. et al. (1997) Int. J. Legal. Med., 110, 134–149. [DOI] [PubMed] [Google Scholar]
- 9.Thomas M.G., Bradman,N. and Flinn,H.M. (1999) Hum. Genet., DOI 10.1007/s004399900181. [Google Scholar]
- 10.Walls E.V. and Crawford,D.H. (1987) In Klaus,G.G.B. (ed.), Lymphocytes, A Practical Approach. IRL Press, Oxford, pp. 149–162.
- 11.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 12.Benson G. (1999) Nucleic Acids Res., 27, 573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nei M. (1987) Molecular Evolutionary Genetics. Columbia University Press, New York.
- 14.White P.S., Tatum,O.L., Deaven,L.L. and Longmire,J.L. (1999) Genomics, 57, 433–437. [DOI] [PubMed] [Google Scholar]
- 15.Economou E.P., Bergen,A.W., Warren,A.C. and Antonarakis,S.E. (1990) Proc. Natl Acad. Sci. USA, 87, 2951–2954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Carvalho-Silva D.R., Santos,F.R., Hutz,M.H., Salzano,F.M. and Pena,S.D. (1999) J. Mol. Evol., 49, 204–214. [DOI] [PubMed] [Google Scholar]