Abstract
Selenocysteine (Sec) incorporation requires the TGA opal codon and a downstream Sec insertion sequence (SECIS), which can be partially randomized and cloned into M13 pIII fusion constructs for phage display. This combinatorial approach provides a convenient non-radioactive assay that couples phage production to opal suppression. Two SECIS libraries were prepared, with the immediate downstream nucleotide either randomized (TGAN) or fixed as thymidine (TGAT). The TGAN library resulted in a majority of clones with a downstream purine and selenium-independent phage production, implicating the endogenous tryptophan-inserting opal suppression pathway. Although the addition of sodium selenite to the growth medium did not affect phage production, it did increase the level of Sec insertion, as shown by the chemical reactivity of the resulting phage. The TGAT phage library yielded clones with strictly selenium-dependent phage production and reactivity consistent with the presence of Sec. These clones were prone to spontaneous mutation upon further propagation, however, resulting in loss of the selenium-dependent phenotype. We conclude that the immediate downstream nucleotide determines whether the endogenous opal suppression pathway competes with co-translational Sec insertion.
INTRODUCTION
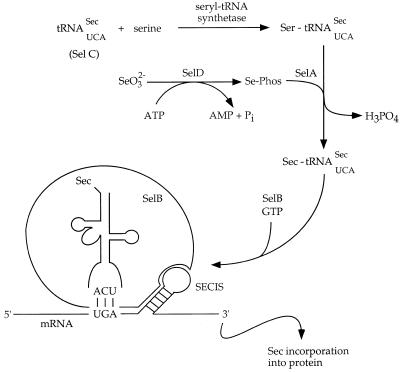
Prokaryotic selenocysteine (Sec) incorporation, as depicted in Figure 1, has been well characterized and requires the constitutively expressed selA, selB, selC and selD gene products (1,2). Sec is encoded by the TGA opal stop codon (3), which is suppressed in the presence of a specific downstream hairpin structure termed the selenocysteine insertion sequence (SECIS). Sec is incorporated via a unique tRNA species, the selC gene product, which is initially aminoacylated with serine by seryl-tRNA synthetase. The loaded serine is converted to selenocysteine by the selA gene product, using a selenium phosphate donor synthesized by the selD gene product. Translation by the resulting Sec-tRNASec is mediated by the selB product, an analog of EF-Tu which simultaneously recognizes Sec-tRNASec and the SECIS.
Figure 1.
Biosynthetic pathway for co-translational selenocysteine incorporation. SelC, the opal codon-specific Sec tRNA, is first charged with serine. ATP-dependent SelD catalysis transforms environmental selenite to an activated Se-phosphate species. This species is utilized by SelA to displace the serine hydroxyl with a selenol moiety, forming a Sec-charged tRNA that recognizes the UGA opal codon. In the presence of GTP, the SelB elongation factor effects Sec translation by binding both the Sec-tRNA and the mRNA SECIS.
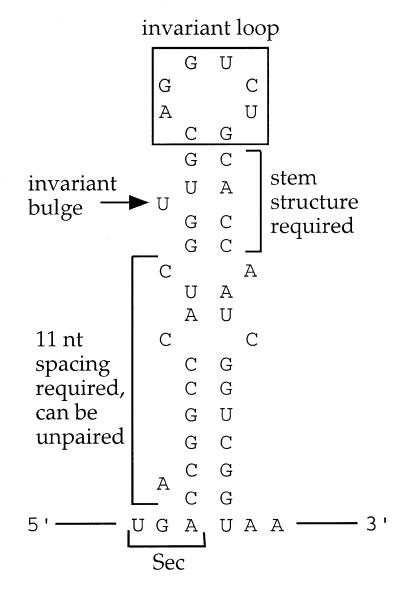
The mRNA requirements for Escherichia coli Sec incorporation, summarized in Figure 2, have been determined by cloning the E.coli formate dehydrogenase gene (fdh) with non-native SECIS variants upstream of a β-galactosidase reporter gene and then measuring either 75Se incorporation by SDS–PAGE or β-galactosidase expression by a colorimetric assay (4–7). Although reporter gene expression is the more quantitative of the two approaches, it is a measure of TGA suppression but not necessarily Sec incorporation. Many E.coli strains possess endogenous opal suppression activity resulting in tryptophan (Trp) incorporation (8), suggesting that a portion of the reporter gene expression could have been independent of Sec incorporation.
Figure 2.
The E.coli formate dehydrogenase SECIS, with permissible mutations as reported in Heider et al. (5), Liu et al. (7) and Klug et al. (12).
To further investigate the E.coli SECIS, we designed a phage display assay for Sec incorporation. The native M13 proteins do not contain Sec (9,10) and M13 phage infectivity requires expression of the coat protein pIII (11). The fusion of putative selenopeptides to the N-terminus of pIII therefore should couple phage plaque formation to opal suppression. Because of the relatively high level of protein synthesis required for plaque formation, we also anticipated that selenium-supplemented media would be required for selenopeptide phage display. We thus expected to identify selenopeptide–pIII fusions based on the selenium dependence of plaque formation and to use this in vivo method to identify critical SECIS elements from phage libraries. The utility of in vitro combinatorial methods was previously shown when RNA aptamer libraries were screened for SelB binding (12). The SECIS U17 bulge, noted in Figure 2, was found to be required for SelB binding, which in turn is necessary for Sec incorporation. Whereas the aptamer method only revealed in vitro binding events, the phage display method provides a direct readout of prokaryotic Sec incorporation requirements in vivo.
In addition to selenium dependency of phage formation, Sec incorporation can also be assessed by modifying phage samples with readily detectable reagents. The pKa of Sec is 5.2, compared to 8.1 for cysteine (Cys), so that at pH 6–7, nucleophilic substitution reactions can specifically alkylate a Sec residue without modifying neighboring Cys residues (13). We developed a method to assay phage for selenopeptide display by treatment with an electrophilic iodoacetamido-biotin reagent, followed by detection of biotinylated phage with an anti-biotin antibody. Because the reactivity of Sec is unique from that of any other naturally occurring amino acid side chain, chemical reactivity is a more specific indicator of Sec than opal suppression.
MATERIALS AND METHODS
All enzymes, western detection reagents, dNTPs and bacterial strains were from New England Biolabs. Sodium selenite was from Aldrich. Iodoacetamido-LC-biotin (I-Bt) was from Pierce.
Vectors
The sequence of the phage cloning vector M13KE (New England Biolabs) is available at http://www.neb.com/neb/tech/ nucleotide_seq_maps/sequences/m13ke.seq . The pMal–pIII protein expression vector (GenBank accession no. AF031088) is available from the authors upon request.
Oligonucleotide inserts
Oligodeoxyribonucleotides were synthesized by the phosphoramidite method by the Organic Synthesis Division of New England Biolabs. The inserts were as follows, with Acc65I and EagI restriction sites in bold: M = A or C; N = A, C, T or G.
Duplex extension primer (NEB product no. 8101), d(CATG-CCCGGGTACCTTTCTATTCTC); TGAN library insert, d(CATGTTTCGGCCGATTGGTGCAGACCTGCAACCG-AMNNMNNMNNTCAMNNMNNMNNMNNAGAGTGAG-AATAGAAAGGTACCCGGG); TGAT library insert, d(CA-TGTTTCGGCCGATTGGTGCAGACCTGCAACCGAMN-NMNNMNATCAMNNMNNMNNMNNAGAGTGAGAATAGAAAGGTACCCGGG).
Phage library construction
To construct phage display libraries, the oligonucleotide inserts listed above were synthesized, gel purified and annealed to the duplex extension primer. The duplex was extended with dNTPs and Klenow fragment, digested with Acc65I and EagI, gel purified and ligated into Acc65I/EagI-digested phage cloning vector M13KE. The ligation products were electroporated into either E.coli ER2537 [F′ lacIq Δ(lacZ)M15 proA+B+/fhuA2 supE thi Δ(lac-proAB)Δ(hsdMS-mcrB)5] or ER2738 [F′ lacIq Δ(lacZ)M15 zzf::Tn10 (TetR)/fhuA2 supE thi Δ(lac-proAB)Δ(hsdMS-mcrB)5] and plated with 100 µl of a late log phase culture in 3 ml of agarose top on LB agar plates with 210 µM IPTG and 98 µM X-gal. The agarose top contained 10 g/l tryptone (Difco), 5 g/l yeast extract (Difco), 86 mM sodium chloride, 5 mM magnesium chloride and 7 g/l agarose (American Bioanalytical) supplemented with 2 µM sodium selenite. After incubation for 16 h at 37°C, blue plaques were selected and the individual clones were amplified in early log phase cultures supplemented with 2 µM sodium selenite. Sequencing templates were prepared by ethanol precipitation of phage DNA from 4 M sodium iodide (14). Phage clones were stored at 4°C in 150 mM Tris–HCl pH 7.4, 50 mM sodium chloride, 100 µM DTT containing 0.02% (w/v) sodium azide. Automated DNA sequencing was performed on a PE-ABD 377 or 373 instrument using Dye-Deoxy™ terminator chemistry (PE Applied Biosystems) with the –96 gIII sequencing primer [NEB product no.1259, d(CCCTCATAGTTAGCGTAACG)].
Selenium dependence studies
Individual phage clones were diluted in LB and combined with 200 µl of a late log phase ER2537 or ER2738 culture. After a 5 min incubation at room temperature, the bacteria and phage were combined with 3 ml of agarose top, with or without supplemental 2 µM sodium selenite, and plated on LB agar plates with IPTG and X-gal. After a 16 h 37°C incubation the plates were inspected for the presence of blue plaques.
pMal–pIII fusion protein expression
The library inserts from individual phage clones were amplified by PCR from ~100 ng of sequencing template DNA using the duplex extension primer and the –96 gIII sequencing primer. The amplified products were gel purified and digested with Acc65I and EagI. The digested insert was gel purified and ligated into Acc65I/EagI-digested pMal–pIII protein expression vector. The heat-killed (65°C, 15 min) ligation products were electroporated into ER2537 and plated on LB containing 100 µg/ml ampicillin and individual clones were analyzed by restriction mapping and automated DNA sequencing. The pMal–pIII fusion proteins were expressed in ER2537 and purified as previously described (15). For N-terminal protein sequencing, proteins were subjected to electrophoresis and electroblotted according to the procedure of Matsudaira (16), with modifications as previously described (17,18). The membrane was stained with Coomassie blue R-250 and the protein band of ~46 kDa was excised and subjected to sequential degradation on a PE-Biosystems 494A Protein/Peptide Sequencer using standard gas phase cycles (18).
Chemical modification of phage
Phage (1010 p.f.u.) in 150 mM sodium chloride, 50 mM glycine–HCl (pH 2.5) were combined with 50 µM iodoacetyl-LC-biotin in dimethylformamide (5% v/v) and incubated in the dark at room temperature for 10 min. The reactions were quenched by the addition of SDS gel loading buffer with 42 mM DTT, and samples were promptly denatured at 100°C for 5 min and loaded on a 10–20% SDS–polyacrylamide gel. Immunoblotting was performed according to standard procedures and the blots were probed with HRP-conjugated anti-biotin antibody (1:1000 dilution) or a mouse monoclonal anti-pIII antibody (Bio 101, 1:500 dilution) followed by an HRP-conjugated anti-mouse antibody. The blots were developed using a Phototope Chemiluminescence Kit (NEB).
RESULTS
TGAN and TGAT library clones
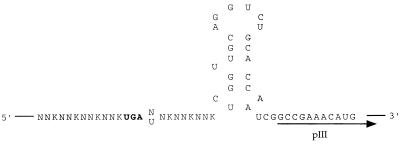
Based on the reported (7) minimal SECIS requirements (Fig. 2), a library consisting of the SECIS element with four upstream and three downstream randomized codons and a minimal mRNA SECIS (Library TGAN, Fig. 3) was prepared. After duplex extension and restriction digestion, the resulting insert was ligated into the M13KE vector, an M13mp19 derivative designed with Acc65I and EagI sites for pentavalent N-terminal pIII expression (15). This vector also carries the lacZα fragment, resulting in characteristic blue plaques when plated with an α-complementing strain on X-gal medium. The sequences of selected clones are shown in Table 1. Although the immediate downstream nucleotide was fully randomized in the TGAN library insert, the majority (74%) of the resulting phage clones possessed a downstream purine. One-third of the displayed peptides in the TGAN library possessed an unpaired Cys residue, which corresponds to 4.8% of the total randomized amino acids. Based on random codon usage, the calculated expected frequency for Cys is 3.1%, whereas the typically observed Cys frequency using this phage display system is <0.5% (C.J.Noren and K.A.Noren, unpublished results). Because M13 proteins are exported to the periplasm, the pairing of the eight Cys residues in M13 pIII into four disulfide bonds (11) could be disrupted by a single unpaired Cys within the displayed peptide. This phenomenon would likely not be observed with a cytoplasmically expressed peptide library.
Figure 3.
The randomized library inserts expressed as pIII fusions in this report. N = A, G, C or U; K = G or U.
Table 1. Selected TGAN library clones.
| Clone | Random insert | Se-dependent? | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TGAN-1 | TCG | TCT | TTT | CCT | TGA | AAG | TCG | CCT | |
| S | S | F | P | opa | K | S | P | – | |
| TGAN-2 | AAG | TGT | ACG | CTT | TGA | TCT | ATG | CTG | |
| K | C | T | L | Sec | S | M | L | + | |
| TGAN-3 | TTG | CTT | TTG | CCT | TGA | AAT | GTT | CTT | |
| L | L | L | P | op | N | V | L | – | |
| TGAN-4 | ATG | ACT | ACG | CAG | TGA | CCT | TCT | CTG | |
| M | T | T | Q | Sec | S | M | L | + | |
| TGAN-5 | CAT | ATT | CCG | CCG | TGA | ACG | AAT | CCT | |
| H | I | P | P | op | T | N | P | – | |
| TGAN-6 | AAG | GCT | CTG | TGT | TGA | CAG | GAT | TCG | |
| K | A | L | C | Sec | Q | D | S | + | |
| TGAN-7 | CTT | CTT | CCG | TGT | TGA | GCT | CAG | CCG | |
| L | L | P | C | Sec | A | Q | P | +b | |
| TGAN-8 | CAT | CAT | CCG | ACT | TGA | GCT | AAG | CAG | |
| H | H | P | T | op | A | K | Q | – | |
| TGAN-9 | ATG | CCT | CCT | ACG | TGA | ATG | GCT | ACG | |
| M | P | P | T | op | M | A | T | – | |
| TGAN-10 | AAT | TGG | TTT | TCT | TGA | CTG | ACT | ACG | |
| N | W | F | S | op | L | T | T | – | |
| TGAN-11 | CTG | CAT | CCG | ACG | TGA | GCT | CGG | CCT | |
| L | H | P | T | op | A | R | P | – | |
| TGAN-12 | GAT | AGG | GGG | CCT | TGA | GCG | AAG | ATT | |
| D | R | G | P | op | A | K | I | – | |
| TGAN-13 | GCG | TCT | TTG | CCT | TGA | AGG | ACG | AGT | |
| A | S | L | P | op | R | T | S | – | |
TGAN-1 was expressed as a pMal–pIII fusion and tryptophan incorporation was verified by N-terminal sequencing.
aop, opal suppression, Sec or W, depending on Se availability.
bTGAN-7 production was Se-enhanced. Se supplementation yielded larger (3–4× diameter) and more (10×) plaques.
Library TGAT (Fig. 3) was nearly identical to Library TGAN, but the immediate downstream nucleotide was fixed as T to investigate the effect of a downstream pyrimidine. Representative DNA sequences from this library are shown in Table 2. Single Cys residues occurred in the TGAT library at a higher than normal frequency of 4.3% of all random amino acids. The TGAT library also had occasional (<10% frequency) mutations within the opal codon, such as the point mutation in clone TGAT-13 (Table 2), which converted the TGA opal codon to the CGA Arg codon.
Table 2. Selected TGAT library clones.
| Clone | Random insert | Se-dependent? | |||||||
|---|---|---|---|---|---|---|---|---|---|
| TGAT-1 | L | P | R | Q | Sec | W | S | P | |
| TTG | CCG | CGT | CAG | TGA | TGG | TCT | CCG | + | |
| TGAT-2 | L | T | G | T | Sec | C | Q | N | |
| TTG | ACT | GGT | ACG | TGA | TGT | CAG | AAT | + | |
| TGAT-3 | E | A | S | R | Sec | C | S | T | |
| GAG | GCG | TCG | CGT | TGA | TGT | TCG | ACT | + | |
| TGAT-4 | K | L | A | R | Sec | S | A | S | |
| AAG | TTG | GCT | CGT | TGA | TCG | GCG | TCG | + | |
| TGAT-5 | N | G | A | Q | Sec | S | R | H | |
| AAT | GGG | GCG | CAG | TGA | TCG | AGG | CAT | + | |
| TGAT-6 | A | S | P | T | Sec | F | K | P | |
| GCG | AGT | CCT | ACT | TGA | TTT | AAG | CCG | + | |
| TGAT-7 | C | A | H | P | Sec | S | T | R | |
| TGT | GCT | CAT | CCG | TGA | TCT | ACT | CGT | + | |
| TGAT-8 | Q | S | T | R | Sec | W | N | D | |
| CAG | TCG | ACG | CGG | TGA | TGG | AAT | GAT | + | |
| TGAT-9 | I | V | E | S | Sec | L | N | P | |
| ATT | GTG | GAG | TCG | TGA | TTG | AAT | CCG | + | |
| TGAT-10 | T | Q | R | M | Sec | L | P | P | |
| ACG | CAG | CGT | ATG | TGA | TTG | CCG | CCC | + | |
| TGAT-11 | V | Q | Y | T | Sec | L | P | K | |
| GTG | CAG | TAT | ACG | TGA | TTG | CCG | AAG | + | |
| TGAT-12 | A | G | Q | S | Sec | S | T | D | |
| GCT | GGG | CAG | TCG | TGA | TCG | ACT | GAT | + | |
| TGAT-13 | L | S | A | S | R | S | Q | F | |
| CTG | TCT | GCG | AGT | CGA | TCG | CAG | TTT | – | |
TGAT-13 carried a T→C point mutation within the opal codon.
Selenium dependency of phage production
To assess the selenium dependency of phage production, phage samples were plated in media with or without 2 µM supplemental sodium selenite. Typical results are illustrated in Figure 4 and summarized in the third column of Tables 1 and 2. Selenium-independent phage clones, such as TGAT-13 and TGAN-8, produced plaques of equal count and size regardless of the media selenium concentration. In contrast, clone TGAT-10 and other Se-dependent phage only produced plaques in Se-supplemented medium. Plaque size and count increased as a function of Se concentration, with half-maximal plaque size attained at ~0.4 µM supplemental selenite (19).
Figure 4.
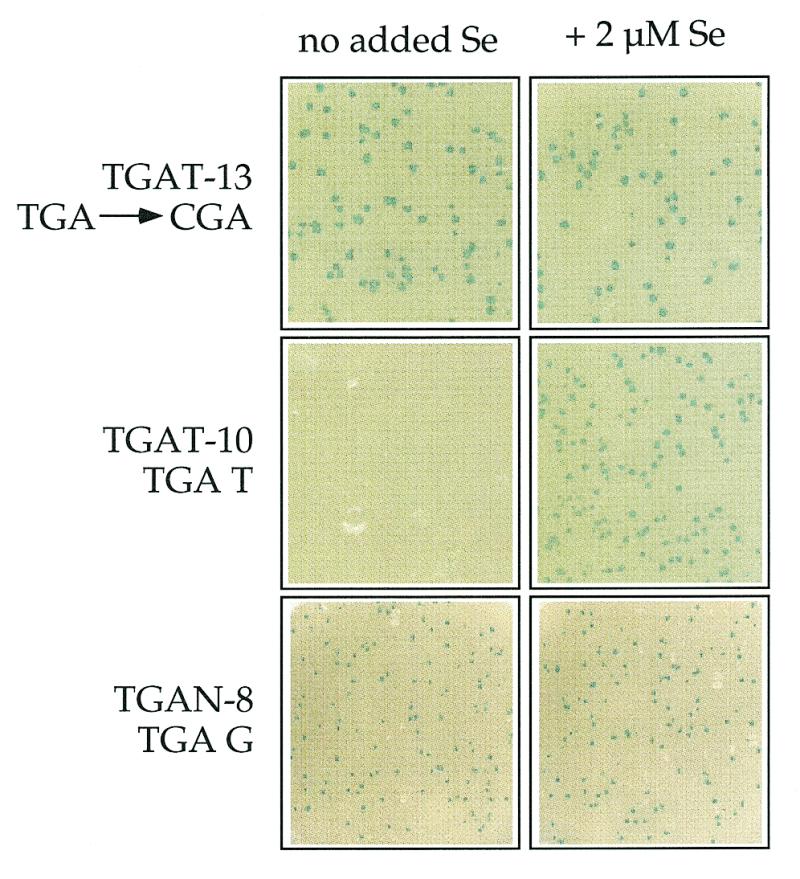
Plating results showing Se-dependent and Se-independent growth of phage clones TGAT-13, TGAT-10 and TGAN-8 (sequences in Tables 1 and 2). Left column, no supplemental Se in plating medium; right column, 2 µM sodium selenite added to plating medium.
All of the clones with an immediate downstream purine grew in a Se-independent manner, with the exception of those containing a single Cys codon within the displayed peptide sequence, e.g. TGAN-7 (Table 1). All of the clones with an opal codon immediately followed by a pyrimidine grew in a Se-dependent manner, with the exception of clone TGAN-10, which had a downstream CTG codon. All of the clones with mutated opal codons, such as TGAT-13, were Se-independent, as was the M13KE phage without insert.
Effect of supplemental selenite on phage amplification
To further explore the effect of sequence context on opal suppression, individual phage clones were amplified in medium with or without supplemental 2 µM selenite. The resulting phage were quantitated by plating diluted samples, and the phage DNA was sequenced. As a test of Sec incorporation in the displayed peptides, phage clones were treated with iodoacetyl-LC-biotin, an electrophilic reagent which should specifically target selenol groups at acidic pH (19). The level of biotinylation was assessed by immunoblotting as described in Materials and Methods. As controls, M13KE phage and clones displaying a single unpaired Cys (Cys-1, displayed peptide SARVLCNH) or Sec (Sec-1, displayed peptide SARVSecHGP, corresponding to the E.coli fdh SECIS) were used.
Clones TGAN-12 and TGAN-8, both of which had a downstream purine, produced equivalent levels of phage in supplemented and unsupplemented media. Growth of both clones in medium with selenite substantially enhanced the reactivity of the resulting phage (Fig. 5A). Production of TGAN-7, which had a downstream purine and a single Cys, was enhanced 50-fold by supplemental Se. Clones with a downstream pyrimidine and a single Cys in the displayed peptide, such as TGAT-7 and TGAT-2, produced 1000-fold less phage without supplemental Se. The phage produced with supplemental Se from these clones had reactivity equal to that of the control Sec-1 phage (Fig. 5B). Clones such as TGAT-1, which possessed a downstream pyrimidine and no Cys in the displayed peptide, either had very low phage production in the absence of supplemental Se or produced phage with opal codon mutations. Figure 5B shows that clone TGAT-1 was reactive when amplified with Se; the TGA→TGG mutant resulting from amplification without Se was unreactive. Occasionally, TGAT clones also developed opal codon point mutations during amplification with supplemental Se.
Figure 5.
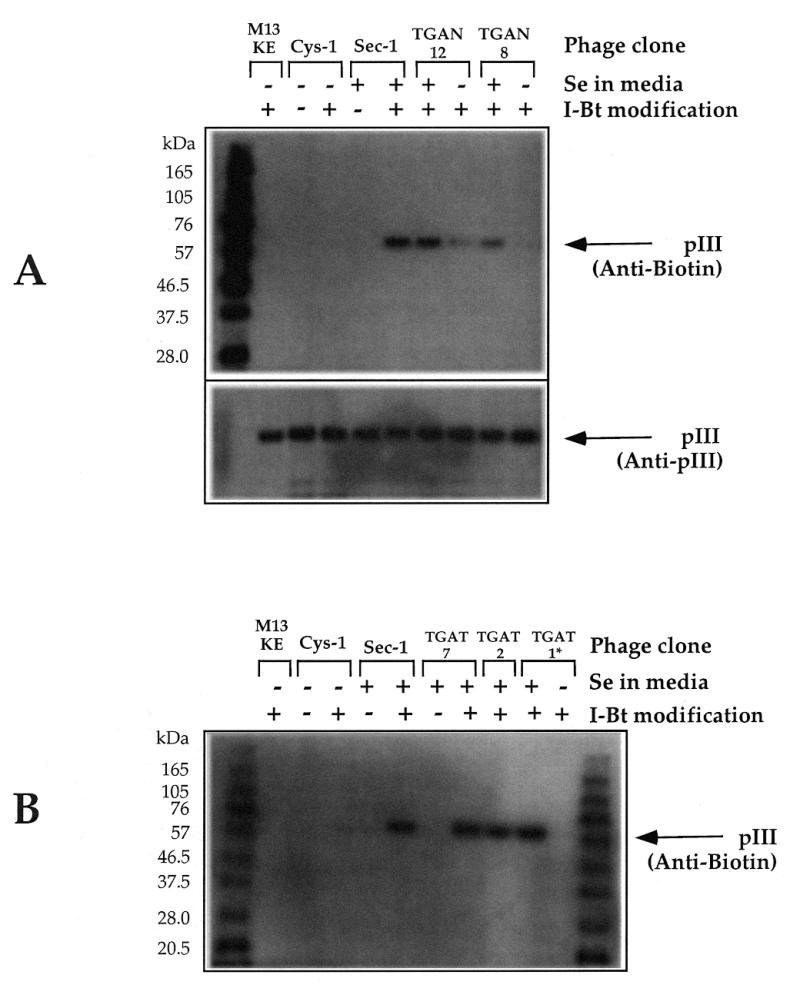
Immunoblots showing specific chemical modification of phage displaying selenopeptides. Individual library clones from the TGAN library (A) or TGAT library (B) were amplified with or without 2 µM supplemental sodium selenite as indicated. Phage were modified as described in Materials and Methods. *Amplification of clone TGAT-1 in unsupplemented medium resulted in a TGA→TGG point mutation.
N-terminal sequencing of a maltose binding protein (MBP) fusion
To further analyze the displayed peptides, the pMal–pIII shuttle vector was employed (15). This vector allows inserts from M13KE to be expressed as fusions to the N-terminus of MBP, with a pIII leader sequence to direct the fusions to the periplasm. The TGAN-1 insert was overexpressed and purified using this system, and N-terminal sequencing revealed mostly (>90%) Trp incorporation at the TGA site. In contrast, the results of sequencing an MBP fusion with the Sec-1 E.coli fdh SECIS insert, in which the opal codon was followed by a cytosine, were consistent with Sec, and not Trp, insertion (19).
DISCUSSION
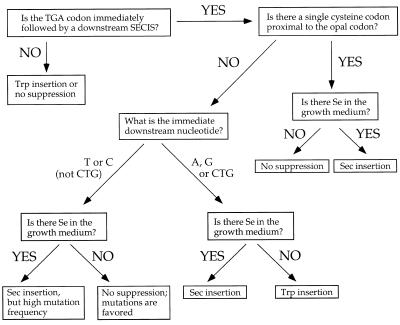
The expression of randomized SECIS elements as N-terminal fusions to M13 pIII couples phage production to opal suppression, providing a combinatorial approach to understanding co-translational Sec insertion. If a sequence fails to produce phage, then it is assumed that there is no opal suppression. If phage is produced in a Se-dependent manner, the opal suppression is presumed to be Sec-inserting. Se-independent phage production can result from Trp insertion or from mutations within the opal codon. The flow chart in Figure 6 summarizes our findings for M13 phage expression in E.coli.
Figure 6.
Opal suppression flow chart, based on experimental data derived from expressing SECIS variants as N-terminal fusions of M13 pIII.
In addition to the principal requirement for Sec incorporation, the opal codon with a downstream SECIS, our method demonstrates that the presence of a single Cys residue within a peptide displayed on M13 pIII is an important factor in Sec insertion. The occurrence of single Cys residues in selenopeptides was >4%, higher than both the normally observed (<0.5%) and predicted (3.1%) frequencies for similar displayed peptide libraries. Moreover, library clones containing a single Cys residue possessed opal codon mutations with <1% frequency compared to almost 10% for the entire TGAT library. These effects presumably resulted from seleno–sulfide crosslinking, which would stabilize both the Cys and Sec residues in the M13 display system, where the coat protein pIII folds in the periplasm. Because it was possible to obtain and amplify many stable library clones containing an unpaired Sec but not a Cys, it appears that single Sec residues are somewhat more stable than unpaired Cys residues.
Among sequences that did not contain a single Cys residue, the nucleotide immediately downstream was a critical factor in determining whether Sec or Trp insertion occurred. The TGA-purine clones replicated with comparable phage yield and sequence fidelity regardless of the media Se concentration, suggesting that Sec insertion was not the major pathway. Purines in the first downstream position have previously been shown to enhance Trp-inserting opal suppression by endogenous tRNATrp (8). Notably, the TGA CTG sequence present in the Se-independent clone TGAN-10 has also been shown to strongly promote Trp insertion (8). It was recently demonstrated that a downstream SECIS element enhanced opal suppression, presumably by Trp, even in the absence of functional SelB or SelC, possibly by interfering with RF2-dependent termination or by stabilizing the message (20). Although the combination of the immediate downstream purine/CTG and the mRNA SECIS appeared to drive the Trp insertion pathway, it did not prohibit Sec insertion. Amplification of the TGA-purine clones in Se-supplemented medium permitted Sec insertion, with phage reactivity comparable to that of fully Se-dependent clones.
Clones with an immediate downstream pyrimidine, except for CTG (TGAN-10), appeared to utilize primarily the Sec insertion opal suppression pathway; they required supplemental Se in order to produce functional phage, which was reactive with iodoacetyl-LC-biotin. No other opal suppression pathway was implicated, since amplification in unsupplemented media resulted in either very low phage production or opal codon mutations. The occasional tendency of these clones to acquire opal mutations during Se-supplemented amplification was consistent with the recent finding that E.coli Sec insertion is only ~5% efficient (20). A spontaneous mutation in the opal codon would result in more efficient phage production, so that mutants would rapidly dominate the log phase bacterial culture.
It has been shown (21) that the nucleotide immediately downstream of the opal codon influences translational termination efficiency in E.coli, with an overall order of U > G > A > C. It was proposed that the recoding event of Sec insertion is favorable at the UGAC in E.coli fdh because RF2 binding is unfavorable in this context. Our present observation is that any nucleotide in the downstream position is capable of directing Sec insertion and, indeed, UGAU and UGAC are equally proficient. This suggests that the pathway leading to Sec insertion is independent of any effect of the immediate downstream nucleotide on RF2 binding.
All of the factors discussed above would contribute to the observed preference for immediate downstream purines in the TGAN library. The downstream purine, followed by a SECIS, permitted maximal utilization of the endogenous opal suppression pathway without preventing co-translational Sec insertion. This dual pathway strategy effectively maximizes phage production. The fixed downstream pyrimidine (TGAT) library clones strongly favor the Sec insertion pathway, presumably resulting in more homogeneous displayed peptides. The cost of this homogeneity, however, is the likelihood of selection for adventitious mutations. These issues should be considered in cloning strategies for the bacterial expression of Sec-containing peptides and proteins.
CONCLUSIONS
The initial studies of prokaryotic SECIS requirements suggested that certain elements of the mRNA structure were essential, whereas others could be changed without affecting Sec incorporation. Our combinatorial approach provides a useful tool for further study of the SECIS. It is possible to screen thousands of sequences at once and the coupling of plaque formation to Sec incorporation provides a simple, non-radioactive visual readout that specifically indicates Sec incorporation rather than general opal suppression. Using this method, we have already determined that the first nucleotide downstream of TGA strongly influences opal suppression, with purines or CTG promoting a dual pathway approach in which Trp insertion is always possible and Sec insertion occurs if Se is available. TGA-pyrimidine sequences, on the other hand, only permit the inefficient co-translational Sec insertion pathway, thereby occasionally allowing opal codon mutants to dominate a culture. The apparent stabilization of unpaired Cys residues by Sec overrides these rules, so that a phage-displayed peptide with a single Cys will strongly favor the Sec insertion pathway, regardless of the downstream nucleotide.
NOTE ADDED IN PROOF
Arnér et al. (J. Mol. Biol., 1999, 292, 1003–1016) have recently reported that co-expression of rat thioredoxin reductase (TrxR) with the selA, selB and selC genes in E.coli did not affect protein yield but did increase the resulting enzyme activity by 6- to 9-fold. The basis for this enhancement was not fully characterized. Noting that the immediate downstream nucleotide in the TrxR gene is guanine, we suggest that the expressed protein was a heterogeneous mixture containing Sec or Trp in the active site, as predicted by Figure 6, and that the sel genes enhanced the Sec insertion pathway.
Acknowledgments
ACKNOWLEDGEMENTS
The authors thank Laurie Mazzola and Jennifer Ware of the NEB DNA sequencing facility for extensive technical support and Jack Benner for protein sequencing. Ira Schildkraut and Devon Byrd are acknowledged for helpful discussions, Karen Noren, Corinna Tuckey, Bill Jack and Tom Evans for critical reading of the manuscript, and Don Comb for support and encouragement.
REFERENCES
- 1.Böck A., Forchhammer,K., Heider,J., Leinfelder,W., Sawers,G., Veprek,B. and Zinoni,F. (1991) Mol. Microbiol., 5, 515–520. [DOI] [PubMed] [Google Scholar]
- 2.Stadtman T.C. (1996) Annu. Rev. Biochem., 65, 83–100. [DOI] [PubMed] [Google Scholar]
- 3.Zinoni F., Birkmann,A., Leinfelder,W. and Böck,A. (1987) Proc. Natl Acad. Sci. USA, 84, 3156–3160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zinoni F., Heider,J. and Böck,A. (1990) Proc. Natl Acad. Sci. USA, 87, 4660–4664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heider J., Baron,C. and Böck,A. (1992) EMBO J., 11, 3759–3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen G.-F.T., Fang,L. and Inouye,M. (1993) J. Biol. Chem., 268, 23128–23131. [PubMed] [Google Scholar]
- 7.Liu Z., Reches,M., Groisman,I. and Engelberg-Kulka,H. (1998) Nucleic Acids Res., 26, 896–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller J.H. and Albertini,A.M. (1983) J. Mol. Biol., 164, 59–71. [DOI] [PubMed] [Google Scholar]
- 9.van Wezenbeek P. and Schoenmakers,J.G. (1979) Nucleic Acids Res., 6, 2799–2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ebright R., Dong,Q. and Messing,J. (1992) Gene, 114, 81–83. [DOI] [PubMed] [Google Scholar]
- 11.Holliger P. and Riechmann,L. (1997) Structure, 5, 265–275. [DOI] [PubMed] [Google Scholar]
- 12.Klug S.J., Hüttenhofer,A., Kromayer,M. and Famulok,M. (1997) Proc. Natl Acad. Sci. USA, 94, 6676–6681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gorlatov S.N. and Stadtman,T.C. (1998) Proc. Natl Acad. Sci. USA, 95, 8520–8525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilson R.K. (1993) Biotechniques, 15, 414–416, 418,–420, 422. [PubMed] [Google Scholar]
- 15.Zwick M.B., Bonnycastle,L.L.C., Noren,K.A., Venturini,S., Leong,E., Barbas,C.F., Noren,C.J. and Scott,J.K. (1998) Anal. Biochem., 264, 87–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsudaira P. (1987) J. Biol. Chem., 262, 10035–10038. [PubMed] [Google Scholar]
- 17.Looney M.C., Moran,L.S., Jack,W.E., Feehery,G.R., Benner,J.S., Slatko,B.E. and Wilson,G.G. (1989) Gene, 80, 193–208. [DOI] [PubMed] [Google Scholar]
- 18.Waite-Rees P.A., Keating,C.J., Moran,L.S., Slatko,B.E., Hornstra,L.J. and Benner,J.S. (1991) J. Bacteriol., 173, 5207–5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sandman K.E., Benner,J.S. and Noren,C.J. (2000) J. Am. Chem. Soc., in press. [Google Scholar]
- 20.Suppmann S., Persson,B.C. and Böck,A. (1999) EMBO J., 18, 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Poole E.S., Brown,C.M. and Tate,W.P. (1995) EMBO J., 14, 151–158. [DOI] [PMC free article] [PubMed] [Google Scholar]