Abstract
The Drosophila melanogaster ovo locus codes for several tissue- and stage-specific proteins that all possess a common C-terminal array of four C2H2 zinc fingers. Three fingers conform to the motif framework and are evolutionarily conserved; the fourth diverges considerably. The ovo genetic function affects germ cell viability, sex identity and oogenesis, while the overlapping svb function is a key selector for epidermal structures under the control of wnt and EGF receptor signaling. We isolated synthetic DNA oligomers bound by the OVO zinc finger array from a high complexity starting population and derived a statistically significant 9 bp long DNA consensus sequence, which is nearly identical to a consensus derived from several Drosophila genes known or suspected of being regulated by the ovo function in vivo. The DNA consensus recognized by Drosophila OVO protein is atypical for zinc finger proteins in that it does not conform to many of the ‘rules’ for the interaction of amino acid contact residues and DNA bases. Additionally, our results suggest that only three of the OVO zinc fingers contribute to DNA-binding specificity.
INTRODUCTION
The C2H2 zinc finger, perhaps the most versatile nucleic acid-binding motif known, is composed of 22–26 amino acids that form a two strand β-sheet region and an α-helical region (reviewed in 1). This compact ββα structure is stabilized by two cysteine residues and two histidine residues that coordinate a divalent zinc ion and by three additional conserved residues that contribute hydrophobic interactions. While the C2H2 class of zinc fingers can bind both RNA and DNA, most zinc finger-containing proteins are known for their roles in regulating transcription. Three amino acids in each zinc finger, located at positions –1, +3 and +6 relative to the α-helical region, are oriented so that their side chains make the principal specific contacts with the edges of three DNA nucleotides exposed in the major groove; successive fingers in an array may make specific contacts with successive DNA triplets (2,3). An additional residue at position +2 sometimes makes auxiliary contacts with the DNA minor groove (4–6). The α-helix is oriented in the major groove such that the protein–DNA contacts are ‘anti-parallel’, i.e. the N→C polarity of the peptide backbone is aligned with the 3′→5′ polarity of the DNA strand whose bases face the major groove. Many zinc finger proteins contain tandem arrays of the motif, which can number up to several dozen. Thus both amino acid sequence diversity within C2H2 zinc fingers and the combinatorial possibilities arising from tandem arrays explain the versatility and ubiquity of zinc fingers in nucleic acid recognition.
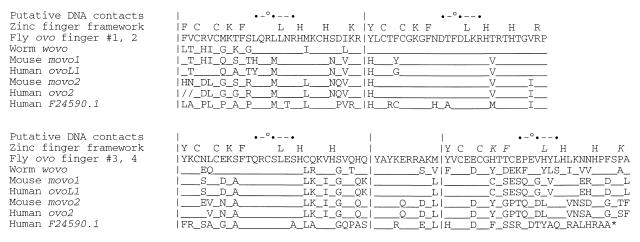
Among the genes encoding C2H2 zinc fingers is the family defined by the Drosophila melanogaster ovo locus [also known as the shavenbaby (svb)–ovo gene region]. Drosophila melanogaster ovo is a complex locus that uses at least three transcription start sites, alternative RNA processing and alternative polyadenylation (7–9). The two 5′-ends used in germline cells are regulated by the germline X:A ratio (10) and are responsible for the ovo genetic function: transcripts originating from these promoters are translated to form two polypeptides, denoted OVO-B and OVO-A. The former is 1028 amino acids long and the latter differs from it by an N-terminal extension of 372 amino acids (8,9,11). Flies carrying distinct ovo mutations show female-specific germline defects that include cell death, aberrant cell proliferation, partial transformation of sex identity and abnormal egg chamber differentiation (12–16). The third promoter, as yet incompletely characterized, is apparently responsible for the svb genetic function and is apparently regulated by the wingless and EGF receptor signal transduction pathways (17). Animals mutated for svb exhibit epidermal differentiation defects that, under most conditions, are organism lethal (18). Complementation between some svb mutations and some ovo mutations is complete, although a sizable class of mutations exists that inactivate both functions (13). This genetic complexity is due to the extensive overlap of exons utilized in the germline ovo mRNAs and in the somatic epidermal svb mRNAs (7–9,19). All known svb–ovo gene region mRNAs code for proteins that have identical 268 amino acid long C-terminal sequences that contain four C2H2 zinc fingers. As shown in Figure 1, the canonical features of this motif are present in all four Drosophila OVO zinc fingers, most notably in the first three copies, which are in close order tandem array and separated from the final copy by a 10 amino acid long ‘linker’ homologous to nuclear localization signals. When the four Drosophila OVO zinc fingers are compared to zinc finger proteins whose three-dimensional structures are known, the Drosophila array can be described as possessing DNA-contacting amino acid quartets LRLR-DFDR-QCSS-EEVL.
Figure 1.
The D.melanogaster svb–ovo gene region codes for four C2H2 zinc fingers that are evolutionarily conserved. Amino acid residues are given in standard single letter code. Potential major groove-contacting amino acids (•) and potential minor groove-contacting amino acids (°) are indicated, as are features of the canonical C2H2 zinc finger motif (italicized framework residues above Drosophila finger 4 are generally absent from the sequence collection). Alignment of the D.melanogaster svb–ovo zinc finger array with arrays coded by putative cognate genes from other species is also shown (underscoring represents residues identical to the fly protein). The C.elegans wovo sequence (GenBank accession no. AF134806) is based on GeneFinder analysis of cosmid F34D10 (20) and on cDNA clones isolated by Schonbaum et al. (21). The mouse movo1 (23) sequence is from GenBank accession no. AF134804 (21). movo1 is syntenic to (23), and likely the ortholog of, the human ovo-like-1 gene located at chromosome 11q13 (GenBank accession no. AF016045; 24). The mouse movo2 sequence is based on cDNA and genomic DNA clones retrieved by reduced stringency hybridization (22) and on mouse EST clones with accession nos W29566, AA591249 and AA757523. The human ortholog of movo2 is defined by human ESTs with accession nos AL079276 and AI151181 (slashes indicate data end points). A third human ovo cognate gene is predicted by XGRAIL analysis of the human chromosome 19 cosmid F24590 (accession no. AD001527, which is overlapped by EST accession no. AI206718). The F24590.1 ORF has a termination codon (*) truncating the rapidly evolving finger 4. Note the absolute invariance of DNA-contacting residues in finger 2, the near invariance in fingers 3 and 1 and the diversity of potential DNA-contacting residues in finger 4.
A variety of methods have identified other members of the ovo subclass of zinc fingers. These include one gene in the genome of the nematode Caenorhabditis elegans (20,21), at least two non-allelic genes in the mouse Mus musculus (21–23) and potentially three non-allelic genes in the human (24; human genome project data available at http://ncbi.nlm. nih.gov ). RNA expression patterns and mutagenesis experiments suggest that the mouse ovo genes have a conserved function in epidermal differentiation, while the germline functions are male specific (22,23). Figure 1 includes a BLAST alignment (25,26) of these protein sequences. Fingers 2 and 3 of the domain are almost identical among the sequences, finger 1 is somewhat less conserved and finger 4 least conserved. The predicted DNA-recognizing side chains are invariant in fingers 2 and 3, nearly so in finger 1 and most variable in finger 4.
Several studies indicate that the Drosophila OVO zinc finger domain is capable of sequence-specific DNA binding in vitro (27,28; S.Lee and M.D.Garfinkel, submitted) and that the Drosophila OVO proteins (8) and the mouse movo proteins (22,23) are nuclear localized in vivo. Both the ovo germline promoter and the ovarian tumor promoter, when fused to Escherichia coli lacZ, demonstrate ovo-dependent expression in genetically transformed flies (10,11,28), showing that ovo plays an important role in regulating gene activity in vivo. However, the patterns of sequence conservation and divergence raise important questions concerning the nature of the OVO protein–DNA interaction. For example, do all four zinc finger motifs contribute to DNA recognition, in which case a DNA target sequence of 12–13 bp is predicted, or do only the three evolutionarily near invariant zinc fingers contribute to DNA recognition, in which case a smaller, 9–10 bp, consensus site is predicted? Here we report on the isolation of synthetic DNA oligomers based only on their ability to bind a bacterially expressed fusion protein containing the Drosophila OVO zinc finger array and on three independent statistical analyses of their sequences. All the analyses reveal a DNA target motif nine bases long, nearly identical to the target motif identified in the promoters of five Drosophila genes (27,28; S.Lee and M.D.Garfinkel, submitted), including those known to be regulated by the ovo function in vivo (10,11,28), suggesting that only three of the four OVO zinc fingers are engaged in specific DNA contacts.
MATERIALS AND METHODS
General recombinant DNA methods
Standard methods of plasmid preparation, agarose gel electrophoresis, acrylamide gel electrophoresis, restriction mapping, ligation, etc. were performed essentially as described (29–31).
Protein overexpression and purification
A near full-length ovo cDNA isolated by Garfinkel et al. (9) was used as template for PCR amplification, from which a 562 bp cDNA fragment coding for 179 amino acids from the C-terminus of the common portion of the svb–ovo open reading frames, which includes all four zinc finger motifs, was directionally cloned in pUC18. The synthetic oligomer primers 5′-AGTCATGGATCCGCCGAAGCCCTTTG-3′, corresponding to positions 5058–5083 in the canonical ovo genomic sequence (GenBank accession no. X59772), and 5′-TAACTAAAATCCGTCGACTAATTGTGGACTGGCAT-3′, corresponding to the reverse complement of positions 5681–5715 in GenBank accession no. X59772, with two substitutions creating a SalI site, were obtained from Integrated DNA Technologies (Coralville, IA). Thirty cycles of PCR were conducted as follows: denaturation, 95°C for 1 min; annealing, 55°C for 1 min; elongation, 72°C for 1 min. The amplification product was digested with BamHI (corresponding to codon 850 of the OVO-B open reading frame) and SalI (introduced by the oligomer just downstream of the naturally occurring termination codon), gel purified and ligated to pUC18. Escherichia coli TB1 was used as host strain for transformation. Recombinant colonies were detected on X-gal-containing plates, minipreps made and plasmid structures verified by restriction mapping and DNA sequencing. DNA from a representative PCR clone was purified in large scale and the zinc finger-coding fragment transferred into the vector pMAL™-c2 (32,33). For large scale purification of the MalE–OVO179 fusion protein, E.coli were grown in 2 l LB broth supplemented with 2% glucose at 37°C to A600 ≈ 0.50 and then induced with 0.5 mM IPTG. After centrifugation, the cell pellet was resuspended in 50 ml 20 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride (PMSF), frozen at –20°C overnight, thawed in cold water and sonicated in short pulses (≤15 s). Release of protein was monitored spectrophotometrically (34). When protein release had reached its maximum, the cell extract was centrifuged at 9000 g for 30 min. The supernatant was loaded onto a 2.5 × 10 cm column of amylose resin (New England Biolabs) which was washed with 10–12 column vol of column buffer (20 mM Tris–HCl, 200 mM NaCl, 1 mM EDTA, 10 mM β-mercaptoethanol) at a flow rate of 1 ml/min. Fusion protein was eluted with column buffer + 10 mM maltose. Fifty 1.5 ml fractions were collected. The protein-containing fractions were identified (34) and pooled.
SDS–PAGE
Separating gels contained 12% (w/v) acrylamide:bis-acrylamide in a ratio of 37.5:1 and stacking gels contained 4% (w/v) acrylamide:bis-acrylamide. Gels were cast and run using the Mini-Protean II electrophoresis cell (Bio-Rad). Protein samples were combined with 40 mM Tris–HCl pH 6.8, 10% (v/v) glycerol, 2% (w/v) SDS, 5% 2-mercaptoethanol and were heated in boiling water for 2 min, chilled on ice and loaded onto gels. Electrophoresis was performed at 70 V. Gels were stained in 0.125% (w/v) Coomassie Brilliant Blue R-250, 40% (v/v) methanol and 10% (v/v) acetic acid and destained in the same methanol/acetic acid mix.
Binding of MalE–OVO179 fusion protein to high complexity DNA oligomer populations
A 72mer oligonucleotide with 32 completely degenerate central bases was purchased from Integrated DNA Technologies (Coralville, IA). The sequence was 5′-CGCTCGAGGGATCCGAATTC(N32)TCTAGAAAGCTTGTCGACGC-3′ (35). Six micrograms of the 72mer were mixed with 5 µg of SalI–XbaI primer (5′-GCGTCGACAAGCTTTCTAGA-3′; 35), annealed and converted into [α-32P]dCTP-labeled double-stranded DNA as described by Pierrou et al. (35). The 72mer, theoretically 12 µg, was precipitated and resuspended in 100 µl of 1× binding buffer [20 mM HEPES, pH 7.9, 50 mM KCl, 2 mM MgCl2, 0.5 mM EDTA, 10% (v/v) glycerol, 0.1 mg/ml bovine serum albumin, 2 mM dithiothreitol, 0.5 mM PMSF, 10 µg poly(dI·dC)]. Ten nanograms of purified MalE–OVO179 fusion protein were added and the mixture incubated for 15 min at 25°C.
Affinity selection of MalE–OVO179 protein–DNA complexes
After the above incubation, 50 µl of a 1:10 slurry of amylose in 1× binding buffer were added, the tube was flicked gently for 2 min and amylose was collected by a 1 min centrifugation at 5000 r.p.m. The supernatant was aspirated and the pellet washed four times with 1 ml of 1× binding buffer, changing tubes at the first and last washes.
PCR
The washed amylose pellet was resuspended in 100 µl of 1× PCR buffer (Boehringer Mannheim) containing 1 mM each of the SalI–XbaI primer and XhoI–EcoRI primer (5′-CGCTCGAGGGTACCGAATTC-3′; 35) and 0.2 mM dNTPs. Amplification was performed as follows: denaturation at 96°C for 1 min, annealing at 60°C for 30 s and extension at 72°C for 30 s, for a total of 30 cycles. An aliquot was checked on a 4% polyacrylamide gel; the rest of the PCR was precipitated, resuspended in 120 µl H2O and filtered through an Amicon Micropure EZ capsule. The binding reaction for the next round was identical to the first, except that 10 µl of the Micropure EZ filtrate was substituted for the initial 72mer. A total of five rounds of binding and amplification were performed.
Cloning of binding-selected PCR-amplified oligomers
Products from the final amplification were gel purified, digested with EcoRI and SalI and cloned into pUC18. Escherichia coli TB1 was transformed to ampicillin resistance with the ligation mixture and recombinant clones were identified on X-gal-containing plates. Miniprep DNAs were digested with EcoRI and SalI singly and in combination to identify recombinants possessing precisely one insert oligomer. Single insert clones were subjected to gel mobility shift assay as described below.
Gel mobility shift assay
Purified DNAs from clones containing a single insert were digested with a mixture of SalI and EcoRI and end-labeled by incubation with Klenow fragment, [α-32P]dCTP and unlabeled dATP, dGTP and TTP. Binding reactions were carried out in 20 µl of 1× binding buffer containing ~20 ng DNA (36). MalE–OVO179 fusion protein was added and equilibration was achieved by incubation at 25°C for 15 min. Five microliters of loading buffer were added, the samples were loaded onto 4–6% polyacrylamide gels made up in 1× TBE and electrophoresis was carried out at 10–15 V/cm for 90–150 min. Gels were fixed, dried onto 3MM paper and exposed either to X-ray film or to a storage phosphor screen (Bio-Rad Molecular Imager model 363).
DNA sequencing
DNA sequencing was performed using the dideoxynucleotide chain termination method, double-stranded DNA template, [α-35S]dATP (Amersham) and the Sequenase Quick-Denature plasmid sequencing kit (US Biochemicals). Inserts were sequenced in their entirety on both strands. Sequencing reactions were resolved on 40 cm long, 0.3 mm thick, 8% polyacrylamide gels containing 1× TBE and 50% (w/v) urea run at 50 W. Gels were fixed, dried and exposed to X-ray film.
Computer analyses
Gibbs motif sampling (37) was performed using MACAW v.2.0.5 for Macintosh (available from ncbi.nlm.nih.gov ), a web-based interface at bayesweb.wadsworth.org or a SUN workstation running source code obtained from ncbi.nlm.nih.gov modified and compiled by Casey M. Bergman (University of Chicago). Alignments and motif lengths obtained using wconsensus (38) were generated by a web-based interface at bioweb.pasteur.fr . Further details of the results of this analysis are available from the authors’ web site at URL www.iit.edu/~garfinkl/LeeNARsupp.html . χ2 analysis of hand tabulated aligned sequences was performed using the CHITEST function of Microsoft Excel for Macintosh v.4.
RESULTS
Overexpression and purification of MalE–OVO179 fusion protein
We used the pMAL™-c2 expression vector to overexpress and purify a fusion protein containing the OVO zinc finger domain. We chose a protein fragment coded by the ovo cDNA that is 179 amino acids long and has a predicted molecular weight of 21 kDa based on the primary sequence, and appended it to the C-terminus of the amylose-binding domain of the E.coli malE gene product, which is predicted to contribute 42 kDa to an in-frame fusion protein. A gel depicting typical induction and purification of the MalE–OVO179 fusion protein is given in Figure 2. Protein extracts were isolated from uninduced cells and from cells exposed to IPTG for up to 3 h. Cells induced for 3 h were sonicated and centrifuged to separate an insoluble pellet from soluble protein. The soluble fraction was incubated with amylose resin, which was washed to remove unbound proteins and then eluted with maltose. Protein samples were resolved by SDS–PAGE. As can be seen in the Coomassie stained gel, the plasmid construct we designed directs the synthesis of an ~63 kDa protein that is IPTG-induced, soluble and has affinity for amylose. This fusion protein binds with sequence specificity and high affinity to DNA fragments in the germline promoters of several Drosophila genes, including ovo, that are known or suspected of being in vivo targets of transcriptional control by the ovo function (27; S.Lee and M.D.Garfinkel, submitted).
Figure 2.
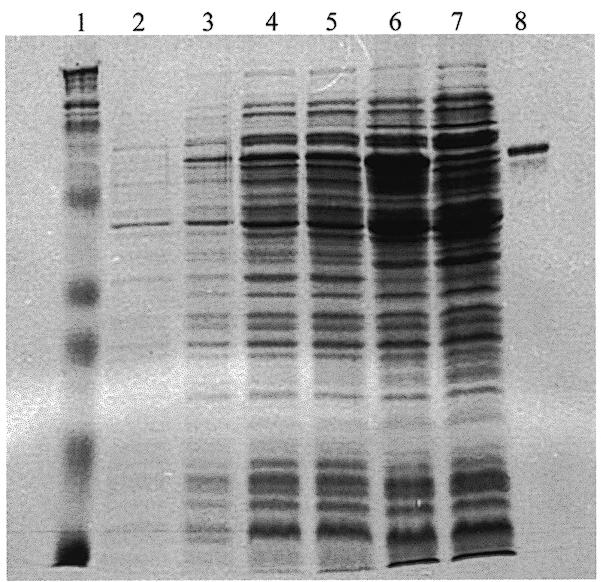
Overexpression and purification of the MalE–OVO179 fusion protein. Escherichia coli TB1 carrying a construct in which the pMAL™-c2 vector was fused in-frame to a 564-bp cDNA fragment of the OVO-B mRNA were grown in LB + 2% glucose medium. When the culture reached A600 ≈ 0.50, IPTG was added. Samples were withdrawn at 0, 1, 2 and 3 h following induction. From the 3 h induced sample, cells were frozen, sonicated and a soluble extract prepared. The soluble material was used in amylose affinity purification. Lane 1, protein size markers; lane 2, uninduced control; lane 3, 1 h induced; lane 4, 2 h induced; lane 5, 3 h induced; lane 6, soluble extract; lane 7, amylose column unbound fraction; lane 8, bound fraction after maltose elution.
Strategy of selection
We wanted to extend the initial characterization of DNA binding specificity by identifying synthetic oligomers based strictly on this property. Accordingly, we adopted an in vitro biochemical selection strategy (35,39) composed of successive iterations of a three-stage process: (i) binding of a DNA oligomer population to the MalE–OVO179 fusion protein; (ii) purification of the DNA–protein complexes by their affinity for amylose; and (iii) primer-directed PCR amplification of the bound DNA oligomers. In the first iteration, the first stage involves a very highly degenerate synthetic oligomer population. In each subsequent iteration, the amplified material from the preceding third stage is used as input for binding in stage one. With each iteration, the complexity of the DNA oligomer population decreases as it becomes increasingly enriched for the subset of oligomers that have sufficiently high affinity for the MalE–OVO179 fusion protein to be retained on amylose resin. Finally, single inserts are purified by cloning in a plasmid vector and checked by gel mobility shift assay and DNA sequencing.
Identification of clones containing single oligomer inserts recognized by MalE–OVO179 fusion protein
After five rounds of binding, affinity purification and amplification, we generated pUC18 plasmid subclones. Our ligation conditions permitted the recovery of clones containing more than one insert; to avoid confusion from clones containing concatamers of binding and non-binding sequences, we tested individual recombinant clones with EcoRI and SalI in both single and double digests. Out of 238 lac– colonies we found 103 clones that possessed single inserts, of which 21 clones were judged by gel mobility shift assay to contain OVO-binding sites; the remainder may have passed the selection due to non-specific binding to amylose resin. An example of our gel mobility shift assay is depicted in Figure 3.
Figure 3.
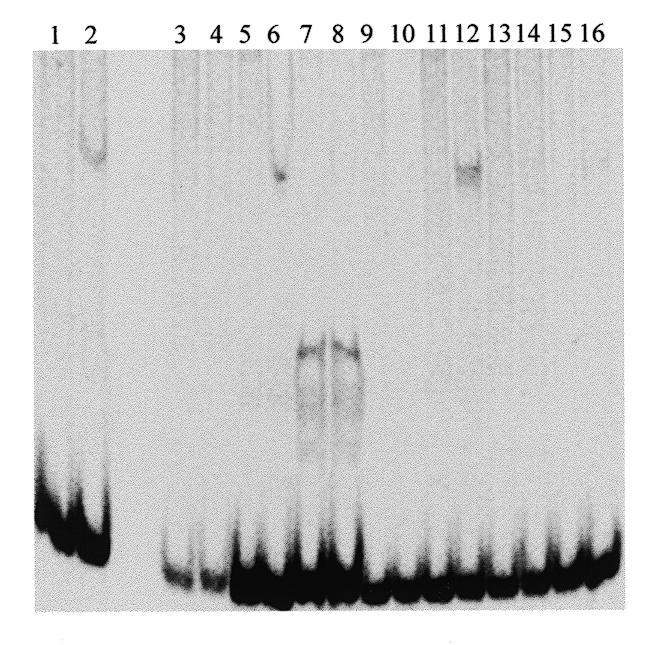
Gel mobility shift assay screen of selected clones. Following five rounds of binding, affinity purification and PCR amplification, the selected 72mer population was digested with EcoRI and SalI and cloned into pUC18. Recombinant clones were subjected to miniprep and restriction analysis to identify those with single inserts. Eight independent single insert-bearing clones were restriction digested, end-labeled and incubated with affinity-purified MalE–OVO179 fusion protein (even numbered lanes) or mock incubated (odd numbered lanes). After non-denaturing polyacrylamide gel electrophoresis, fixation and drying, migration of radioactive DNA was visualized by phosphorimaging.
DNA sequence analysis
The 21 single insert clones that exhibited MalE–OVO179 fusion protein binding were sequenced (Table 1). Consistent with their origins in a pool of synthetic oligomers of form 5′-TCT-AGA(N32)GAATTC-3′, 17 clones had precisely 32 nt between the flanking XbaI and EcoRI sites. The remaining four clones had 28, 30, 33 and 37 nt each, presumably due to errors in either chemical synthesis of the initial oligomer population or in Taq DNA polymerase-mediated amplification. Their sequences are presented in Table 1; all the inserts possess at least one 5′-AAC-3′ triplet (clone 106 has two 5′-AAC-3′ triplets and clone 26 has three 5′-AAC-3′ triplets), which suggested potential alignments among all the cloned inserts. Further visual inspection revealed that, in addition to the precise identity of the 5′-AAC-3′ triplet, the clone inserts could be aligned so that the position 2 nt downstream of the triplet also yielded an identity for guanine and that the position 2 nt upstream of the triplet yielded a strong preference (14/21) for another guanine residue.
Table 1. Sequences of synthetic DNA oligomers selected by MalE–OVO179 protein binding.
Sequences of 21 single insert clones chosen for affinity to the MalE–OVO179 fusion protein. Upper case letters represent nucleotides from the interior of the insert; lower case letters represent restriction site linkers flanking the insert. Bold type accentuates the 9 nt window containing the binding site consensus. Note that 18 sequences have the XbaI site on the left hand side while three sequences are presented as reverse complements, with the EcoRI site on the left hand side, to facilitate the alignment.
The statistical significance of this ‘core’ seven base long 5′-GN-AACNG-3′ alignment and a measure of the length of the consensus sequence were ascertained in three ways. First, we used the χ2 test to measure the statistical significance of deviations from randomness for a total of 23 positions along the aligned oligomers (Table 2). We excluded from the χ2 calculations any nucleotide that originated in the non-random restriction site linker portion of the synthetic oligomers. Six contiguous nucleotides, positions 8–13 in Table 2, scored as having highly significant deviations from randomness; these residues include all the identities that were observed by human inspection. Position 14 does not deviate from random. Position 15 scored as highly significant. Positions 4 and 16 scored as somewhat significant. Overall, the χ2 test identified a ‘window’ of nine contiguous nucleotides that contains seven highly significant nucleotides and one somewhat significant nucleotide.
Table 2. χ2 analysis of the OVO-binding consensus sequence determined from affinity-selected oligomers.
| Position | G | A | T | C | n | Σχ2 | Significance | mfn |
|---|---|---|---|---|---|---|---|---|
| 1 | 4 | 4 | 4 | 3 | 15 | 0.20 | 0.15 << P | |
| 2 | 4 | 4 | 3 | 5 | 16 | 0.50 | 0.15 << P | |
| 3 | 2 | 5 | 3 | 7 | 17 | 3.47 | 0.15 << P | |
| 4 | 4 | 0 | 8 | 5 | 17 | 7.71 | P ≈ 0.053 | t |
| 5 | 2 | 5 | 7 | 4 | 18 | 2.89 | 0.10 << P | |
| 6 | 3 | 7 | 5 | 4 | 19 | 1.84 | 0.10 << P | |
| 7 | 2 | 5 | 8 | 6 | 21 | 3.57 | 0.10 << P | |
| 8 | 0 | 11 | 6 | 4 | 21 | 11.95 | P << 0.05 | A |
| 9 | 14 | 1 | 2 | 4 | 21 | 20.33 | P << 0.05 | G |
| 10 | 2 | 4 | 13 | 2 | 21 | 15.76 | P << 0.05 | T |
| 11 | 0 | 21 | 0 | 0 | 21 | 63.00 | P << 0.05 | A |
| 12 | 0 | 21 | 0 | 0 | 21 | 63.00 | P << 0.05 | A |
| 13 | 0 | 0 | 0 | 21 | 21 | 63.00 | P << 0.05 | C |
| 14 | 3 | 8 | 5 | 5 | 21 | 2.43 | 0.10 << P | N |
| 15 | 21 | 0 | 0 | 0 | 21 | 63.00 | P << 0.05 | G |
| 16 | 2 | 5 | 9 | 4 | 20 | 5.20 | P ≈ 0.15 | t |
| 17 | 5 | 4 | 3 | 8 | 20 | 2.80 | 0.10 << P | |
| 18 | 3 | 6 | 3 | 7 | 19 | 2.68 | 0.10 << P | |
| 19 | 3 | 3 | 4 | 6 | 16 | 1.50 | 0.10 << P | |
| 20 | 4 | 5 | 2 | 5 | 16 | 1.50 | 0.10 << P | |
| 21 | 3 | 6 | 1 | 6 | 16 | 4.50 | 0.10 << P | |
| 22 | 4 | 3 | 5 | 3 | 15 | 0.73 | 0.10 << P | |
| 23 | 3 | 3 | 4 | 5 | 15 | 0.73 | 0.10 << P |
For each of 23 positions centered on the putative consensus binding site, observed nucleotides were tabulated. Only nucleotides from the originally random insert portions (upper case nucleotides in Table 1) were counted; nucleotides derived from the adjoining linker regions (lower case nucleotides in Table 2) were excluded. Σχ2 was calculated taking n/4 as the expectation for each of the four nucleotides at each of the 23 positions. Probability values were calculated using the CHITEST function of Microsoft Excel v.4 for Macintosh. mfn denotes the most frequent nucleotide at the position.
Our second, totally independent, sequence alignment algorithm was the Gibbs motif sampling method described by Lawrence et al. (37). The Gibbs procedure uses iterative sampling to generate local multiple alignments based on ‘informational content’ intrinsic to the sequences analyzed. One important application of this algorithm is to determine the length of a sequence motif by plotting ‘information per parameter’ (IPP), a quantitative measure of the contribution made by each residue position versus window size (37). In the present case, window sizes 5–18 nt in length were used. Windows up to 8 nt gave nearly identical, very high IPP scores (data not shown). A slight reduction in IPP was noted with 9 and 10 nt windows and a steep fall-off in IPP occurs with windows ≥11 nt in length. IPP frequency histograms were also obtained for each window size. For windows up to 10 nt in length, the IPP values obtained were very narrowly distributed around the peak value. IPP peaks became broader as window size increased above 10 nt, indicating that suboptimal alignments became increasingly prevalent (data not shown). The Gibbs sampling procedure yielded a consensus alignment identical to that derived by human inspection and presented in Table 1. The motif identified corresponds to positions 8–16, inclusive, of Table 2.
The third independent method of alignment and motif length determination was the wconsensus algorithm (38). The same alignment as obtained by eye and by Gibbs motif sampling was obtained, which again corresponds to the nine base wide window given in Table 2, positions 8–16 inclusive. The P value of this nine base window was 3 × 10–45. These calculations independently support the conclusion of the χ2 analysis shown in Table 2: the MalE–OVO179 fusion protein recognizes nine contiguous DNA bases.
As shown in Table 2, the consensus DNA target site recognized by Drosophila OVO protein, based on the most frequent nucleotides observed in each position, is 5′-AGTAACNGT-3′. Referring to the sequences presented in Table 1, we see that only one clone, 15, matches the proposed consensus at all eight of the specified positions. Eight clones matched the proposed consensus at seven of the specified positions, nine clones matched at six of the specified positions and three clones matched at five of the eight specified positions. Experiments to determine the relationship between relative binding affinities and number of matches to the derived consensus are in progress in this laboratory.
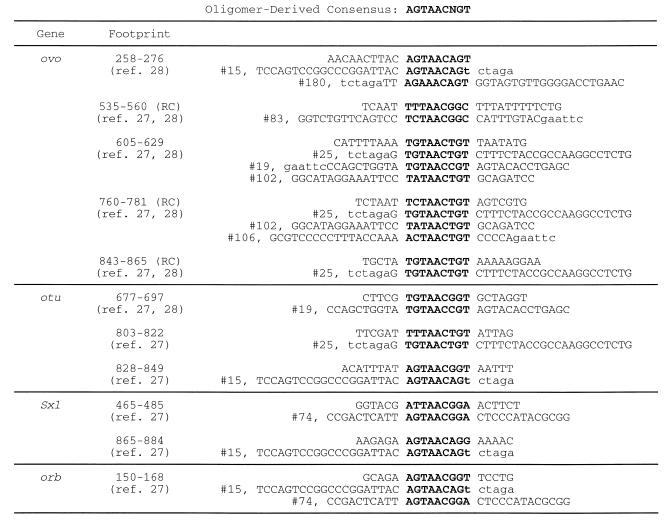
The nine base long consensus given in Table 2 is essentially identical to the consensus we obtained using gel mobility shift assays and DNA footprinting on the promoters of five Drosophila genes involved in germline sex determination and oogenic differentiation (27; S.Lee and M.D.Garfinkel, submitted), four of which interact genetically and biochemically with ovo. Two clones (15 and 25) each matched a single footprint site along the entire nine base consensus and they each matched an additional three footprints at eight positions (Fig. 4). Six more clones matched at least one gene footprint in eight positions (Fig. 4). Eleven more clones matched at least one of the gene footprint sites in seven positions along the nine base consensus (data not shown). Overall, one-third (8/21) of the selected oligomers match at least one footprinted site at eight or nine positions; these footprinted sites correspond to 11 of the 17 reported on Drosophila genes. Thus, the selected oligomers share sequence characteristics with OVO-binding sites defined by DNase I footprinting on Drosophila genes. The consensus sequence is unusual for one recognized by a zinc finger array in its AT-richness. In these and other respects, the consensus sequence deviates from what one would predict from the DNA–protein co-crystal structures of other zinc finger proteins and from other experimental determinations of the relationship between DNA bases and zinc finger amino acid side chains.
Figure 4.
OVO-binding oligomers share the nonamer consensus with OVO-binding sites determined by DNase I footprinting of Drosophila genes. Eleven Drosophila OVO-binding sites are aligned with eight oligomer clones matching at eight or nine positions of the nonamer consensus motif (bold). For oligomer sequences, lower case indicates restriction site linkers. ‘Footprint’ gives nucleotide coordinates of the footprinted site as per the indicated reference (footprints attributed to ref. 27 are also in Lee and Garfinkel, submitted). Certain footprints are shown as reverse complements (RC) of the GenBank sequence entries (ovo, accession no. X59772; otu, accession no. M30825; Sxl, accession no. D84425; orb, accession no. X64412).
DISCUSSION
In this report we have shown that bacterially produced MalE–OVO179 fusion protein, which contains an array of four C2H2 zinc fingers, of form LRLR-DFDR-QCSS-EEVL, binds synthetic DNA oligomers in vitro. Alignment of the 21 resulting oligomers enabled us to derive a consensus recognition site. This consensus is essentially the same as the consensus we derived from binding sites identified in five different natural Drosophila gene promoters (27; S.Lee and M.D.Garfinkel, submitted) and closely similar to one derived by others from a smaller sample of footprinted sites observed on two of the same genes (28). Our statistical analyses robustly demonstrate that the DNA consensus is only 9 nt long, suggesting that in vitro one of the four zinc fingers either binds DNA with no specificity or perhaps does not bind DNA at all. The marked lack of evolutionary conservation of OVO finger 4, as evidenced by its score in PROSITE and PFAM analyses (M.D.Garfinkel, unpublished observations) suggests that it is the non-contributing zinc finger within the array. Experiments to test this hypothesis are in progress in our laboratory.
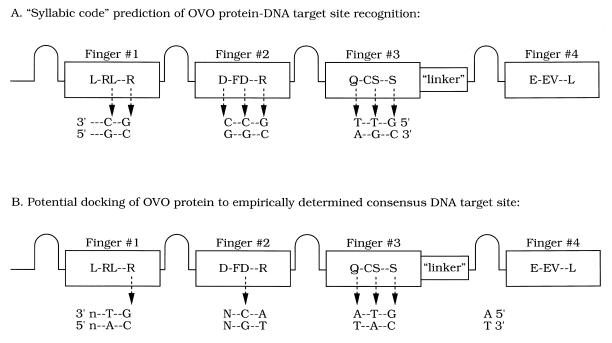
Numerous studies (for example 4–6,40–44) have attempted to establish a correspondence between each of the DNA triplets and the amino acids at each of the principal contact-critical positions in the C2H2 zinc finger. For example, the ‘syllabic code’ proposed by Choo and Klug (44,45), based on phage display of Zif268-derived proteins, makes several predictions for DNA recognition by the Drosophila OVO finger array (Fig. 5A). OVO finger 1, with leucine in the +3 position and arginine in the +6 position, should prefer DNA triplets that have a 5′-position guanine residue and a middle position cytosine. OVO finger 2, which contains the major groove-contacting residues DDR, should prefer the DNA target subsite 5′-GCC-3′ (a preference also noted in 42). OVO finger 3, with major groove-contacting residues QSS, should prefer the DNA target subsite 5′-GT(T or A)-3′, but this requires assumptions concerning side chain–side chain interactions within finger 3 and between finger 3 and finger 4. Assembling these predicted preferences yields the octamer 5′-GT(A or T)GCCGC-3′, a GC-rich sequence that bears little resemblance to either strand of our empirically derived, AT-rich, nonamer consensus. That said, we can ‘dock’ our empirically derived nonamer consensus to the OVO finger array so as to preserve some of the features of the syllabic code predictions (Fig. 5B). However, in doing so, three discrepancies arise. The first is the distinctly different behaviors proposed for the two aspartates in OVO finger 2 and the second is the distinctly different behaviors proposed for the two arginines located in the +6 positions of OVO fingers 1 and 2. How might this arise? It should be emphasized that ‘preferences’ of zinc finger major groove-contacting amino acids for DNA bases appear to be protein sequence context dependent. For example, interaction of arginine residues at the +6 position with guanine residues in the 5′-most position of the DNA target subsite is not an absolute specificity. The assessed zinc finger of a Zif268-derived protein that contained the three major groove-contacting residues DDR exhibited a marked preference for the DNA target subsite 5′-GCC-3′, while the corresponding DAR finger had little specificity at any nucleotide (42). The third discrepancy is that, in Figure 5B, we dislocated the nonamer DNA consensus one position relative to the putative DNA-contacting residues of OVO fingers 1–3. We note that the OVO protein has amino acids in the –1, +2 and +3 positions of each zinc finger that have not been characterized by other studies for their contributions either to protein–DNA interaction or to side chain–side chain interactions, and so they may be expected to have novel characteristics.
Figure 5.
Potential alignment of the Drosophila OVO zinc finger domain contact residues with target DNA. The OVO zinc finger domain is diagrammed as a series of four loops and four boxes, the latter depicting the α-helical portion of each finger. Putative contact amino acid residues are given in single letter code. The 10 amino acid linker separating fingers 3 and 4 is identified. (A) Amino acid/base pair interactions (dotted arrows) based on ‘syllabic code’ rules (44,45). Both strands of a rules-derived DNA binding site are shown, with their 5′- and 3′-ends labeled. (B) Potential alignment of the OVO zinc finger domain to the empirically determined consensus DNA target site. Both strands of DNA corresponding to the experimentally determined binding site are shown, with their 5′- and 3′-ends labeled. Only five of the amino acid residues conform to ‘syllabic code’ rules (see Discussion for more details).
Details of the model depicted in Figure 5B may be refined by computational methods such as threading and homology modeling, and are experimentally testable by biophysical techniques, including nuclear magnetic resonance spectrometry in solution, and by X-ray diffraction of single crystals. That one of the human ovo cognate genes may be associated with clinical disease (24) greatly increases the significance of understanding the structural basis of how this subfamily of C2H2 zinc finger proteins recognizes and regulates the expression of developmental target genes.
Acknowledgments
ACKNOWLEDGEMENTS
We thank our laboratory partners, in particular Gregory J. Sahli, and William W. Mattox, Benjamin C. Stark and Kenneth C. Stagliano for critical comments on the manuscript. Initial Gibbs sampling alignments were performed by Casey M. Bergman (The University of Chicago), to whom we are very grateful and with whom we had valuable conversations. The experimental data presented in this paper were submitted by S.L. in partial fulfillment of the requirements of the doctor of philosophy in biology at the Illinois Institute of Technology. This research was supported by grants awarded to M.D.G. by the National Institutes of Health (1R15 GM52631-01), the American Cancer Society Illinois Division (94-49) and the Educational Research and Initiative Fund of the Illinois Institute of Technology. Additional support was provided by a grant to the Illinois Institute of Technology from the Howard Hughes Medical Institute Program in Undergraduate Science.
REFERENCES
- 1.Tan S. and Richmond,T.J. (1998) Curr. Opin. Struct. Biol., 8, 41–48. [DOI] [PubMed] [Google Scholar]
- 2.Pavletich N.P. and Pabo,C.O. (1991) Science, 252, 809–817. [DOI] [PubMed] [Google Scholar]
- 3.Pavletich N.P. and Pabo,C.O. (1993) Science, 261, 1701–1707. [DOI] [PubMed] [Google Scholar]
- 4.Kim C.A. and Berg,J.M. (1996) Nature Struct. Biol., 3, 940–945. [DOI] [PubMed] [Google Scholar]
- 5.Elrod-Erickson M., Rould,M.A., Nekludova,L. and Pabo,C.O. (1996) Structure, 4, 1171–1180. [DOI] [PubMed] [Google Scholar]
- 6.Elrod-Erickson M., Benson,T.E. and Pabo,C.O. (1998) Structure, 6, 451–464. [DOI] [PubMed] [Google Scholar]
- 7.Mével-Ninio M., Terracol,R. and Kafatos,F.C. (1991) EMBO J., 10, 2259–2266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mével-Ninio M., Terracol,R., Salles,C., Vincent,A. and Payre,F. (1995) Mech. Dev., 49, 83–95. [DOI] [PubMed] [Google Scholar]
- 9.Garfinkel M.D., Wang,J., Liang,Y.-P. and Mahowald,A.P. (1994) Mol. Cell. Biol., 14, 6809–6818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Oliver B., Singer,J., Laget,V., Pennetta,G. and Pauli,D. (1994) Development, 120, 3185–3195. [DOI] [PubMed] [Google Scholar]
- 11.Mével-Ninio M., Fouilloux,E., Guénal,I. and Vincent,A. (1996) Development, 122, 4131–4138. [DOI] [PubMed] [Google Scholar]
- 12.Busson D., Gans,M., Komitopoulou,K. and Masson,M. (1983) Genetics, 105, 309–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Oliver B., Perrimon,N. and Mahowald,A.P. (1987) Genes Dev., 1, 913–923. [DOI] [PubMed] [Google Scholar]
- 14.Oliver B., Pauli,D. and Mahowald,A.P. (1990) Genetics, 125, 535–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rodesch C., Geyer,P.K., Patton,J.S., Bae,E. and Nagoshi,R.N. (1995) Genetics, 141, 191–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Staab S. and Steinmann-Zwicky,M. (1996) Mech. Dev., 54, 205–210. [DOI] [PubMed] [Google Scholar]
- 17.Payre F., Vincent,A. and Carreno,S. (1999) Nature, 400, 271–275. [DOI] [PubMed] [Google Scholar]
- 18.Wieschaus E., Nüsslein-Volhard,C. and Jürgens,G. (1984) Wilhelm Roux’s Arch. Dev. Biol., 193, 296–307. [DOI] [PubMed] [Google Scholar]
- 19.Garfinkel M.D., Lohe,A.R. and Mahowald,A.P. (1992) Genetics, 130, 791–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson R., Ainscough,R., Anderson,K., Baynes,C., Berks,M., Bonfield,J., Burton,J., Connell,M., Copsey,T., Cooper,J. et al. (1994) Nature, 368, 32–38. [DOI] [PubMed] [Google Scholar]
- 21.Schonbaum C.P., Fantes,J. and Mahowald,A.P. (Unpublished) Attribution in GenBank accession nos AF134804, AF134805 and AF134806
- 22.Masu Y., Ikeda,S., Okuda-Ashitaka,E., Sato,E. and Ito,S. (1998) FEBS Lett., 421, 224–228. [DOI] [PubMed] [Google Scholar]
- 23.Dai X., Schonbaum,C., Degenstein,L., Bai,W., Mahowald,A. and Fuchs,E. (1998) Genes Dev., 12, 3452–3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chidambaram A., Allikmets,R., Chandrasekarappa,S.C., Guru,S.C., Modi,W., Gerrard,B. and Dean,M. (1997) Mamm. Genome, 8, 950–951. [DOI] [PubMed] [Google Scholar]
- 25.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 26.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee S. (1998) PhD dissertation, Illinois Institute of Technology, Chicago, IL.
- 28.Lü J., Andrews,J., Pauli,D. and Oliver,B. (1998) Dev. Genes Evol., 208, 213–222. [DOI] [PubMed] [Google Scholar]
- 29.Davis R.W., Botstein,D. and Roth,J.R. (1980) Advanced Bacterial Genetics, A Manual for Genetic Engineering. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 30.Maniatis T., Fritsch,E.F. and Sambrook,J. (1982) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Sambrook J., Fritsch,E.F. and Maniatis,T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 32.Guan C., Li,P., Riggs,P.D. and Inouye,H. (1987) Gene, 67, 21–30. [Google Scholar]
- 33.Maina C.V., Riggs,P.D., Grandea,A.G., Slatko,B.E., Moran,L.S., Tagliamonte,J.A., McReynolds,L.A. and Guan,C.D. (1988) Gene, 74, 365–373. [DOI] [PubMed] [Google Scholar]
- 34.Bradford M.M. (1976) Anal. Biochem., 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 35.Pierrou S., Hellqvist,M., Samuelsson,L., Enerback,S. and Carlson,P. (1994) EMBO J., 13, 5002–5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garner M.M. and Revzin,A. (1981) Nucleic Acids Res., 9, 3047–3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lawrence C.E., Altschul,S.F., Boguski,M.S., Liu,J.S., Neuwald,A.F. and Wootton,J.C. (1993) Science, 262, 208–214. [DOI] [PubMed] [Google Scholar]
- 38.Hertz G.Z. and Stormo,G.D. (1996) Methods Enzymol., 273, 30–42. [DOI] [PubMed] [Google Scholar]
- 39.Oliphant A.R., Brandl,C.J. and Struhl,K. (1989) Mol. Cell. Biol., 9, 2944–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Desjarlais J.R. and Berg,J.M. (1992) Proc. Natl Acad. Sci. USA, 89, 7345–7349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Desjarlais J.R. and Berg,J.M. (1993) Proc. Natl Acad. Sci. USA, 90, 2256–2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Desjarlais J.R. and Berg,J.M. (1994) Proc. Natl Acad. Sci. USA, 91, 11099–11103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Choo Y. and Klug,A. (1994) Proc. Natl Acad. Sci. USA, 91, 11163–11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Choo Y. and Klug,A. (1994) Proc. Natl Acad. Sci. USA, 91, 11168–11172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Choo Y. and Klug,A. (1997) Curr. Opin. Struct. Biol., 7, 117–125. [DOI] [PubMed] [Google Scholar]