Abstract
In this study, a series of novel 3-(5-fluoropyridine-3-yl)-2-oxazolidinone derivatives were designed and synthesized based on compounds previously reported, and their antibacterial activity was investigated. Then their antibacterial activity was investigated for the first time. Preliminary screening results showed that all these compounds exhibited antibacterial activity against gram-positive bacteria, including 7 drug-sensitive strains and 4 drug-resistant strains, among which compound 7j exhibited an 8-fold stronger inhibitory effect than linezolid, with a minimum inhibitory concentration (MIC) value of 0.25 µg/mL. Further molecular docking studies predicted the possible binding mode between active compound 7j and the target. Interestingly, these compounds could not only hamper the formation of biofilms, but also have better safety, as confirmed by cytotoxicity experiments. All these results indicate that these 3-(5-fluoropyridine-3-yl)-2-oxazolidinone derivatives have the potential to be developed into novel candidates for the treatment of gram-positive bacterial infections.
Keywords: 3-(5-fluoropyridine-3-yl)-2-oxazolidinone derivatives, antibacterial activity, molecular docking, antibiofilm activity, drug resistance
1. Introduction
Bacterial infections can lead to skin suppuration, bacteremia, local and systemic inflammation, and other serious diseases [1]. With the discovery and development of antibiotics, revolutionary changes have taken place in the treatment of bacterial infections, effectively reducing infection rates and mortality. However, the irregular use of antimicrobial drugs has led to the emergence of various drug-resistant strains, which are considered dangerous and stubborn clinical pathogens that cause difficult-to-treat, life-threatening illnesses [2]. Furthermore, it has been found in clinical studies that many pathogenic bacteria can adhere to the surfaces of objects, secrete metabolites, and generate extracellular polymeric substances (EPS), thus forming biofilms with a “cell population-metabolite” structure, which can effectively protect strains and produce drug resistance, leading to a high incidence of nosocomial infection and a high treatment cost [3,4]. Although great efforts have been made to create novel and effective antimicrobial methods, the pace of development is too slow to meet clinical needs [5,6,7,8].
At present, many pharmaceutical chemists are trying to develop new antibacterial drugs with novel structures, unique mechanisms of action, and long-term effectiveness [9,10,11]. Oxazolidinones are a class of chemosynthetic antibacterial drugs with a brand-new chemical structure, similar to sulfonamides and quinolones, which are used to treat skin and tissue infections, pneumonia, untreatable bacterial infections, and other infectious diseases caused by gram-positive bacteria [12]. Oxazolidinones have been widely used and studied because of their unique antibacterial mechanism, that is, inhibition of protein synthesis at the initial stage and no cross-resistance with other antibacterial drugs [13,14,15,16]. Moreover, some oxazolidinone analogues have been marketed previously [17,18,19]. Although the initial effect is satisfactory, drug resistance appears after long-term use, accompanied by thrombocytopenia and other adverse reactions [20]. This has prompted pharmaceutical chemists to continue to improve oxazolidinone antimicrobials with higher antimicrobial activity and sustained sensitivity [21,22,23,24,25,26,27,28,29].
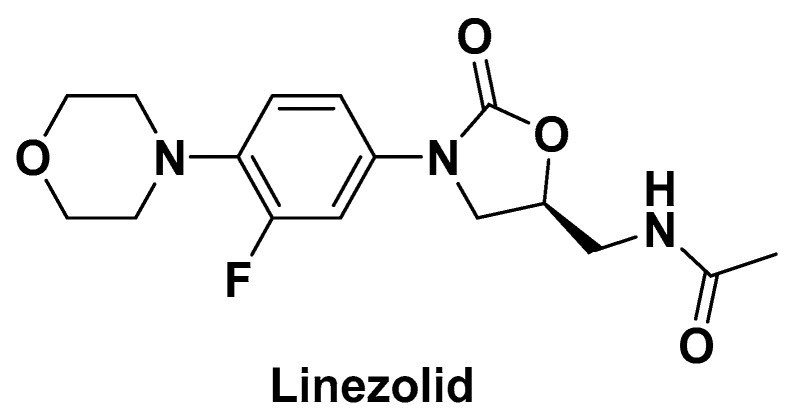
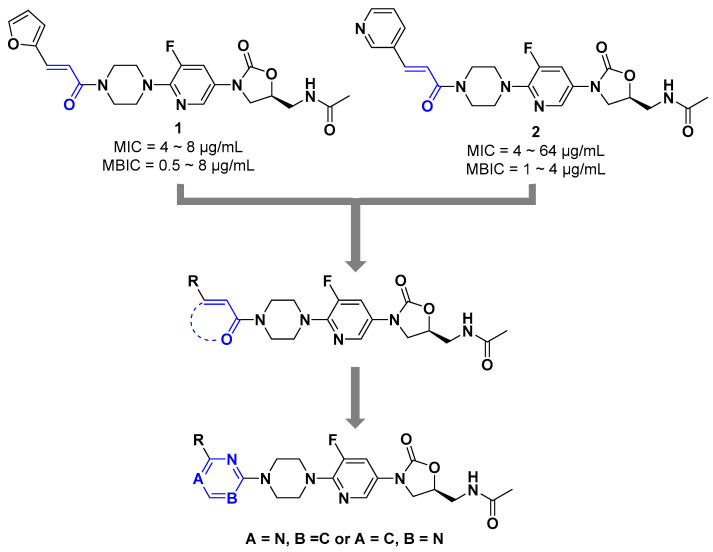
Our research group previously modified the structure of linezolid (Figure 1) [30,31,32,33,34], found that compounds 1 and 2 have efficient antibacterial activity for S. aureus, Streptococcus pneumoniae, Enterococcus faecalis, etc. (MICs = 4~64 µg/mL) and exciting antibiofilm activity (MBIC = 0.5~8 µg/mL). The vinyl structure in these molecules is retained and cyclized to form a new aromatic heterocyclic ring. Considering the unique electronegativity of pyrimidines and their ability to form hydrogen bonds [35], pyrimidine aromatic rings were introduced into the structure [36]. Accordingly, a series of 3-(5-fluoropyridine-3-yl)-2-oxazolidinone derivatives were synthesized and tested for antibacterial activity (Figure 2).
Figure 1.
The structure of linezolid.
Figure 2.
Optimization strategy of novel oxazolidinone derivatives.
2. Results and Discussion
2.1. Chemistry
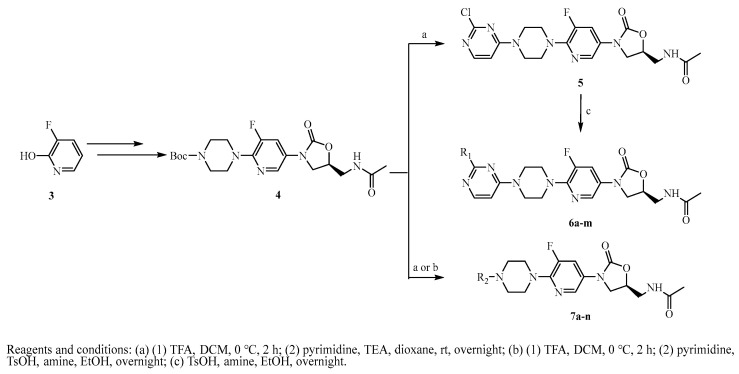
According to the steps described in the literature [37], intermediate 4 was produced through a multi-step reaction by commercially purchased compound 3. After removing the Boc protection group, compound 4 was coupled with 2,4-dichloropyrimidine to produce intermediate 5. The intermediate 5 was linked with different amines to generate the final products 6a-m, as displayed in Scheme 1. Furthermore, the final products 7a-n were obtained by a similar method.
Scheme 1.
Synthesis of target compounds 5, 6a-m and 7a-n.
2.2. Antibacterial Activity Assay
2.2.1. Minimum Inhibitory Concentration against Standard Strains
Intermediate 5 and further derivatives 6a-m were synthesized and tested against a panel of gram-positive bacteria using the double dilution method. As shown in Table 1, all these compounds had moderate antibacterial activity against all six tested gram-positive bacteria but no activity against gram-negative bacteria (E. coli). The MICs of compounds 6a-m were 2~32 µg/mL against gram-positive bacteria, while the MIC was 1~2 µg/mL when R1 = Cl (5), which had better antibacterial activity. It was speculated that the cavity of the target near the R1 side chain was not large enough for these substituent groups.
Table 1.
The MICs (µg/mL) of compounds 5, 6a-m against 7 standard strains. (MIC: minimal inhibit concentration).
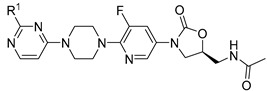
| ||||||||
| Compound | R1 | Sa a | Sp b | Ef c | Bs d | Sx e | Lm f | Ec g |
|---|---|---|---|---|---|---|---|---|
| 5 | -Cl | 1 | 1 | 1 | 1 | 1 | 2 | >128 |
| 6a |
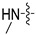
|
8 | 16 | 8 | 8 | 8 | 32 | >128 |
| 6b |
|
16 | 16 | 16 | 16 | 8 | 16 | >128 |
| 6c |
|
8 | 8 | 8 | 8 | 8 | 8 | >128 |
| 6d |
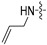
|
8 | 8 | 8 | 4 | 8 | 32 | >128 |
| 6e |
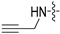
|
8 | 8 | 16 | 8 | 16 | 64 | >128 |
| 6f |
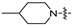
|
16 | 32 | 32 | 32 | 16 | 128 | >128 |
| 6g |
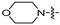
|
16 | 16 | 16 | 16 | 16 | 32 | >128 |
| 6h |
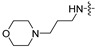
|
8 | 8 | 8 | 8 | 8 | 16 | >128 |
| 6i |
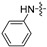
|
8 | 16 | 8 | 8 | 8 | 64 | >128 |
| 6j |
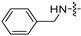
|
8 | 8 | 8 | 8 | 8 | 16 | >128 |
| 6k |
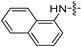
|
32 | 16 | 32 | 32 | 8 | 32 | >128 |
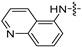
| 6l |
|
8 | 8 | 8 | 8 | 8 | 16 | >128 |
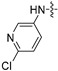
| 6m |
|
2 | 2 | 8 | 4 | 4 | 32 | >128 |
| linezolid | - | 2 | 2 | 2 | 2 | 2 | 2 | >128 |
a Sa, Staphylococcus aureus (ATCC25923); b Sp, Streptococcus pneumoniae (ATCC49619); c Ef, Enterococcus faecalis (ATCC29212); d Bs, Bacillus subtilis (ATCC6633); e Sx, Staphylococcus xylosus (ATCC35924); f Lm, Listeria monocytogenes (ATCC19111); g Ec, Escherichia coli (ATCC25922).
After that, a series of pyrimidine derivatives, 7a-n without a further substituted group, was synthesized, and their antibacterial activity was tested (Table 2). All these compounds, except 7m, had better antibacterial activity than compounds 1 and 2, but these compounds still exhibited no effect against gram-negative bacteria. Among them, compound 7j exhibited the best activity with a MIC of 0.25~1 µg/mL. First, while keeping X1 = N, three substituent groups on the pyrimidine ring (R1 = Cl, NH2, or R1 = R3 = Cl) were examined (5, 7e and 7h). All of them exhibited better activities than the compounds with X1 = C (7b, 7f and 7g). Then, by comparing the activity of chlorine-substituted compounds 5, 7a, 7h, 7j and 7n, it was found that the number of chlorine atoms had no significant influence on activity. Meanwhile, keeping X1, X2, R1, and R3 constant (X1 = N, X2 = C, R1 = Cl, and R3 = H), F, Cl, Br, and methyl substituent groups on R4 were examined. All these derivatives displayed similar biological activities (MICs = 0.25~4 µg/mL), indicating that F, Cl, Br, or methyl were acceptable as substituents.
Table 2.
MICs (µg/mL) of compounds 7a-n against 7 standard bacteria. (MIC: minimal inhibit concentration).
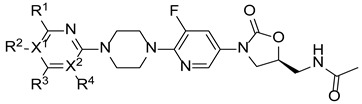
| |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Compound | X1 | X2 | R1 | R2 | R3 | R4 | Sa a | Sp b | Ef c | Bs d | Sx e | Lm f | Ec g |
| 7a | N | C | H | - | Cl | H | 2 | 4 | 2 | 2 | 2 | 2 | >128 |
| 7b | C | N | Cl | H | H | - | 2 | 1 | 2 | 2 | 2 | 8 | >128 |
| 7c | C | N | H | CH3 | H | - | 2 | 2 | 4 | 2 | 1 | 1 | >128 |
| 7d | C | N | H | Br | H | - | 2 | 4 | 2 | 4 | 2 | 2 | >128 |
| 7e | N | C | NH2 | - | H | H | 0.5 | 0.5 | 1 | 1 | 1 | 0.5 | >128 |
| 7f | C | N | NH2 | H | H | - | 2 | 1 | 1 | 1 | 2 | 4 | >128 |
| 7g | C | N | Cl | H | Cl | - | 2 | 4 | 8 | 16 | 2 | 2 | >128 |
| 7h | N | C | Cl | - | Cl | H | 2 | 2 | 1 | 2 | 1 | 2 | >128 |
| 7i | N | C | Cl | - | H | F | 0.5 | 0.5 | 1 | 1 | 1 | 0.5 | >128 |
| 7j | N | C | Cl | - | H | Cl | 1 | 0.25 | 1 | 1 | 1 | 0.25 | >128 |
| 7k | N | C | Cl | - | H | Br | 1 | 0.5 | 0.5 | 1 | 2 | 0.5 | >128 |
| 7l | N | C | Cl | - | H | CH3 | 0.5 | 1 | 4 | 4 | 2 | 0.5 | >128 |
| 7m | N | C | CH3S | - | H | C2H5OCO | 32 | 32 | 32 | 32 | 32 | 32 | >128 |
| 7n | N | C | Cl | - | Cl | Cl | 2 | 4 | 2 | 2 | 2 | 2 | >128 |
| linezolid | - | - | - | - | - | - | 2 | 2 | 2 | 2 | 2 | 2 | >128 |
a Sa, Staphylococcus aureus (ATCC25923); b Sp, Streptococcus pneumoniae (ATCC49619); c Ef, Enterococcus faecalis (ATCC29212); d Bs, Bacillus subtilis (ATCC6633); e Sx, Staphylococcus xylosus (ATCC35924); f Lm, Listeria monocytogenes (ATCC19111); g Ec, Escherichia coli (ATCC25922).
2.2.2. Minimum Inhibitory Concentration against Drug-Resistant Strains
After evaluating the antibacterial potential of these derivatives, they were further tested against clinically isolated resistant bacteria. As shown in Table 3, these MIC results show that compounds 7i-l had significant antibacterial activity against MRSA and VRE but no effect against linezolid-resistant strains.
Table 3.
The MICs (µg/mL) of compounds 7i-l against four drug-resistance bacteria. (MIC: minimal inhibit concentration).
| Compound | MRSA a | VRE b | LRSA c | LRSP d |
|---|---|---|---|---|
| 7i | 1 | 1 | >128 | >128 |
| 7j | 1 | 1 | >128 | >128 |
| 7k | 1 | 1 | >128 | >128 |
| 7l | 1 | 1 | >128 | >128 |
| linezolid | 2 | 2 | >128 | >128 |
a MRSA, Methicillin-resistant Staphylococcus aureus; b VRE, Vancomycin-resistant Enterococcus; c LRSA, Linezolid-resistant Staphylococcus aureus; d LRSP, Linezolid-resistant Streptococcus pneumoniae.
2.3. Molecular Docking Study
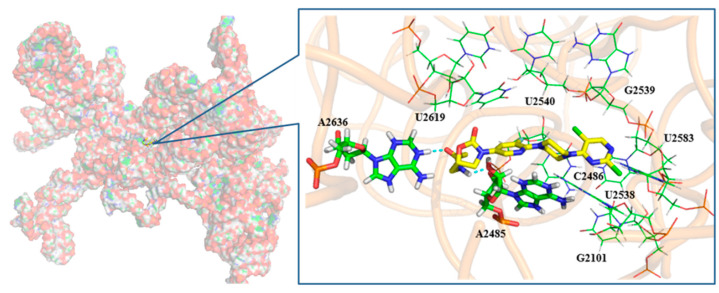
To understand binding site, state, conformation, and interaction, the promising compound 7j was selected for further docking study with the 50S ribosomal subunit from Haloarcula Marismortui (PDB ID: 3CPW) [38,39]. As shown in Figure 2, the compound that expanded linearly bound to the peptidyl transferase center (PTC) of the 50S ribosomal subunit. The potential compound existed in the cavity of PTC, which was composed by U2619, U2540, G2539, U2583, U2538, C2486, G2101, and A2485. Moreover, the H atom and O atom on the 5-side chain amide group of the oxazolidinone ring formed a hydrogen bond with A2485 and A2636, respectively. In addition, the pyrimidine ring of compound 7j and the pyrimidine ring formed π-π conjugations with C2486 and U2538.
As can be seen from Figure 3, the 2-Cl atom on the pyrimidine substituent extended into a shallow pocket, which was too small to accommodate other groups on the pyrimidine. That was probably the reason why compounds 6a-m had worse antibacterial activity. Meanwhile, there was a larger space between the 5-Cl atom on the pyrimidine substituent and the surface of the cavity, which might be the reason why compounds 7i-m had a better antibacterial effect.
Figure 3.
Docking results for 7j with the 50S ribosomal subunit from Haloarcula Marismortui (PDB ID: 3CPW). The O atom (red), the N atom (blue), and the C atom (yellow) are shown. Hydrogen bonds are shown as dashed green lines.
2.4. Inhibition of Biofilm Formation
Using the microtiter dish biofilm formation assay [40], four potent compounds were selected for further evaluation of their effects on bacterial biofilm formation against four drug-resistant strains. As shown in Table 4, the results show that all these compounds significantly inhibited the formation of biofilms, with the minimum biofilm inhibitory concentrations (MBICs) of 0.5 µg/mL against MRSA and VRE and 1~4 µg/mL against LRSA and LRSP. The above results indicate that these compounds can significantly inhibit the growth of biofilm, and it could be speculated that they have stable effects and do not easily develop resistance to bacteria. Meanwhile, the results show that all compounds are more effective than linezolid against four drug-resistant strains, which indicates all compounds have different mechanisms with linezolid.
Table 4.
MBICs (µg/mL) of compounds 7i-l against 4 drug-resistant bacteria.
| Compound | MRSA a | VRE b | LRSA c | LRSP d |
|---|---|---|---|---|
| 7i | 0.5 | 0.5 | 2 | 4 |
| 7j | 0.5 | 0.5 | 1 | 4 |
| 7k | 0.5 | 0.5 | 1 | 4 |
| 7l | 0.5 | 0.5 | 2 | 4 |
| Linezolid | 64 | 16 | 128 | 128 |
a MRSA, Methicillin-resistant Staphylococcus aureus; b VRE, Vancomycin-resistant Enterococcus; c LRSA, Linezolid-resistant Staphylococcus aureus; d LRSP, Linezolid-resistant Streptococcus pneumoniae.
2.5. Cytotoxicity Determination
When a chemical substance is used to treat infection, it may affect the physiological activity of both cells and bacteria, thereby reducing the cell’s survival rate [41]. Therefore, it was necessary to evaluate the toxicity of active derivatives. The cytotoxicity of compound 7j against the Hela cell line was detected via the MTT colorimetric assay, as shown in Table 5. The result shows that the cytotoxicity of the tested compound increased in a dose-dependent manner, and cell survival at 256 µg/mL and lower concentrations was higher than 85%. Considering that the cytotoxicity only appeared above 256 µg/mL, which was 64~512 times that of its MICs. Consequently, the compound 7j has the potential to be further developed as an antibacterial drug.
Table 5.
Analyzing the toxicity of compound 7j against human cervical-cancer cells (Hela) using the MTT assay.
| Concentration (µg/mL) | 32 | 64 | 128 | 256 | 500 | 1000 |
|---|---|---|---|---|---|---|
| cell viability (%) | 100 | 97.5 | 96.9 | 93.5 | 33.3 | 7.3 |
3. Experimental Section
3.1. Materials and Methods
All the chemicals and solvents used in this study were of analytical grade. All the reagents were purchased from Tianjin Tianli Chemical Reagent Co., Ltd., Tianjin, China. All solvents and chemicals were purified by standard methods. Unless otherwise stated, the synthesis of all compounds was monitored by thin layer chromatography (TLC) and purified by rapid column chromatography. Thin layer chromatography (TLC) was performed on silica gel G plates (Taizhou Luqiao Sijia Biochemical Plastic Products Factory, Taizhou, China). The melting point (m. p.) of all products was measured by the SGW X-4A micro-melting point meter apparatus. All compounds were tested to verify their purity by HPLC (Shimadzu Corporation, Kyoto, Japan). Using the Diamonsil C18 column, the mobile phase was acetonitrile and water of different gradients at a flow rate of 1.0 mL•min−1. The column temperature was set at 35 °C and the injection volume of each sample was 10 μL. Every sample was quantitatively diluted with methanol to 1 mg•mL−1. 1H NMR spectra (600 MHz) and 13C NMR spectra (150 MHz) were recorded on a Bruker Advance spectrometer with tetramethyl-silane (TMS) as the internal standard and DMSO-d6 as the solvent. The used standard strains were purchased from the American Type Culture Collection (ATCC), and the drug-resistant strains were isolated from clinical sources. The Hela cells were donated by Dr. Tao Wang (Qiqihar Medical University).
3.2. Chemistry
3.2.1. Synthesis of (S)-N-((3-(6-(4-(2-chloropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide(5)and(S)-N-((3-(6-(4-(4-chloropyrimidin-2-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7b)
A solution of compound 4 (218 mg, 0.5 mmol) in DCM (5 mL) at 0 °C was dropwise added to TFA (1 mL) and then stirred for 2 h. After the reaction was complete, TEA was added to the solution at 0 °C to adjust pH. The filtrate was concentrated in vacuo. To a solution of the concentrate in ethanol (3 mL) was added TEA (0.14 mL, 1 mmol) and 2, 4-dichloropyrimidine (97 mg, 0.65 mmol), and then stirred at reflux overnight. After the reaction was complete and concentrated, the mixture was extracted with DCM (5 mL × 3). The organic phase was washed with brine and concentrated in vacuo. The residue was purified by silica gel column chromatography (DCM/MeOH/TEA = 50:1:1) to yield compounds 5 and 7b.
Compound 5 was white solid; yield 5.8%. m. p. 171.1–173.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.32 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 8.11 (d, J = 6.0 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 6.88 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.79 − 3.72 (m, 5H), 3.46 − 3.40 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.6, 163.0, 160.0, 158.0, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.6, 133.0, 130.0, 115.4, 102.9, 72.6, 55.4, 47.6, 47.4, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H21ClFN7O3: 449.87; Found: 450.129.
Compound 7b was white solid; yield 40.4%. m. p. 160.0–161.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.35 (d, J = 5.4 Hz, 1H), 8.25 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 6.77 (d, J = 5.4 Hz, 1H), 4.89 − 4.63 (m, 1H), 4.21 − 4.02 (m, 1H), 3.82 − 3.79 (m, 4H), 3.75 − 3.67 (m, 1H), 3.44–3.33 (m, 5H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 161.5, 160.5, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.9, 133.0, 130.0, 128.7, 115.4, 109.7, 72.6, 47.7, 46.1, 43.7, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H21ClFN7O3: 449.87; Found: 450.119.
Raw data for the above products are presented in Supplementary Materials (Figures S1–S3 and S46–S48).
3.2.2. General Procedure for the Synthesis of 6a and 6b
A solution of compound 5 (200 mg, 0.44 mmol) in an amine solution (4 mL) was stirred at room temperature for three days. After the reaction was complete, the filtrate was concentrated in vacuo. The mixture was extracted with DCM (5 mL × 3). The organic phase was washed with brine and concentrated in vacuo. The residue was purified by silica gel column chromatography (DCM/MeOH = 30:1) to yield compounds 6a and 6b.
(S)-N-((3-(5-fluoro-6-(4-(2-(methylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6a).
Compound 6a was a yellow solid; yield 40.5%. m. p. 193.3–195.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.27 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.86 (d, J = 6.6 Hz, 1H), 7.31 − 7.30 (s, 1H), 6.31 (d, J = 6.6 Hz, 1H), 4.78 − 4.73 (m, 1H), 4.13 − 4.10 (m, 1H), 3.84 − 3.78 (s, 4H), 3.75 − 3.73 (m, 1H), 3.43 − 3.42 (m, 6H), 2.82 (d, J = 4.8 Hz, 3H), 1.84 (s, 3H).13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.0, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.7, 145.7, 133.0, 133.0, 130.0, 115.6, 115.5, 72.6, 47.7, 47.6, 43.9, 41.9, 28.1, 22.9. HRMS (ESI) (positive mode) m/z calculated for C20H25FN8O3: 444.47; Found: 445.150.
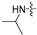
(S)-N-((3-(5-fluoro-6-(4-(2-(isopropylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6b).
Compound 6b was a pink solid; yield 44.4%.m. p. 196.8–199.5 °C.1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 7.82 (d, J = 6.0 Hz, 1H), 6.31 (s, 1H), 6.04 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.12 − 4.10 (m, 1H), 4.04 − 3.95 (m, 1H), 3.74 − 3.72 (m, 1H), 3.68 − 3.66 (m, 4H), 3.49 − 3.36 (m, 5H), 1.84 (s, 3H), 1.12 (d, J = 6.6 Hz, 6H), 1.08 − 0.94 (m, 1H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.7, 161.7, 157.2, 154.8, 149.1 (d, JC-F = 256.3 Hz), 146.0, 133.0, 129.9, 115.5, 115.4, 72.6, 47.8, 47.7, 47.6, 43.5, 42.2, 41.9, 23.1, 22.9. HRMS (ESI) (positive mode) m/z calculated for C22H29FN8O3: 472.53; Found: 473.189.
Raw data for the above products are presented in Supplementary Materials (Figures S4–S9).
3.2.3. General Procedure for the Synthesis of 6c–m
To a solution of compound 5 (150 mg, 0.33 mmol) in dioxane (4 mL), p-toluene sulfonic acid monohydrate (11.4 mg, 0.066 mmol) and amine (1 mmol) were added and stirred at reflux overnight. After the reaction was complete, the filtrate was concentrated in vacuo. The mixture was extracted with DCM (5 mL × 3). The organic phase was washed with brine and concentrated in vacuo. The residue was purified by silica gel column chromatography (DCM/MeOH = 30:1) to yield compounds 6c–m.
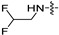
(S)-N-((3-(6-(4-(2-((2,2-difluoroethyl)amino)pyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6c).
Compound 6c was a pink solid; yield 66.7%.m. p. 228.1–231.2 °C.1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.87 (d, J = 6.0 Hz, 1H), 7.02 (s, 1H), 6.20 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.76 − 3.73 (m, 1H), 3.73 − 3.68 (m, 5H), 3.68 − 3.58 (m, 2H), 3.46 − 3.38 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.5, 154.8, 149.1 (d, JC-F = 257.2 Hz), 145.9, 145.8, 133.1, 133.0, 129.9, 115.6, 115.4, 72.6, 47.7, 47.64, 43.9, 43.7, 43.6, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C21H25F3N8O3: 494.48; Found: 495.163.
(S)-N-((3-(6-(4-(2-(allylamino)pyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6d)
Compound 6d was a white solid; yield 68.1%.m. p. 182.1–184.6 °C.1H NMR (600 MHz, DMSO-d6) δ 8.25 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.85 (d, J = 6.6 Hz, 1H), 7.51 − 7.08 (m, 1H), 6.28 (d, J = 6.0 Hz, 1H), 5.92 − 5.86 (m, 1H), 5.19 (d, J = 17.4 Hz, 1H), 5.08 (d, J = 10.2 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.13 − 4.10 (m, 1H), 3.93 − 3.91 (m, 2H), 3.78 − 3.75 (m, 4H), 3.74 − 3.72 (m, 1H), 3.43 − 3.39 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.0, 154.8, 149.1 (d, JC-F = 257.5 Hz), 145.8, 138.1, 136.4, 133.0, 130.0, 128.5, 126.0, 115.6, 115.4, 72.6, 47.6, 43.9, 43.4, 41.9, 22.9, 21.2. HRMS (ESI) (positive mode) m/z calculated for C22H27FN8O3: 470.51; Found: 471.133.
(S)-N-((3-(5-fluoro-6-(4-(2-(prop-2-yn-1-ylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6e)
Compound 6e was a white solid; yield 44.9%. m. p. 185.6–188.7 °C.1H NMR (600 MHz, DMSO-d6) δ 8.29 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.88 (d, J = 6.0 Hz, 1H), 7.22 (s, 1H), 6.23 (d, J = 6.0 Hz, 1H), 5.33 (s, 1H), 4.77 − 4.73 (m, 1H), 4.13 − 4.10 (m, 1H), 4.04 − 4.03 (m, 2H), 3.78 − 3.72 (m, 4H), 3.44 − 3.34 (m, 7H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.4, 154.8, 149.1 (d, JC-F = 256.9 Hz), 145.9, 145.8, 133.1, 133.0, 130.0, 115.6, 115.4, 72.6, 63.1, 52.5, 47.7, 43.7, 41.9, 30.6, 22.9, 7.7. HRMS (ESI) (positive mode) m/z calculated for C22H25FN8O3: 468.49; Found: 469.201.
(S)-N-((3-(5-fluoro-6-(4-(2-(4-methylpiperidin-1-yl)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6f).
Compound 6f was a pink solid; yield 64.4%. m. p. 198.1–200.5 °C.1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 7.89 (d, J = 6.0 Hz, 1H), 6.08 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.62 − 4.59 (m, 2H), 4.13 − 4.10 (m, 1H), 3.74 − 3.72 (m, 1H), 3.68 − 3.66 (m, 4H), 3.43 − 3.41 (m, 2H), 3.41 − 3.37 (m, 4H), 2.77 − 2.73 (m, 2H), 1.84 (s, 3H), 1.64 − 1.56 (m, 2H), 1.04 − 0.98 (m, 2H), 0.91 (d, J = 6.6 Hz, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.7, 161.4, 157.1, 154.8, 149.1 (d, JC-F = 256.3 Hz), 146.0, 133.0, 129.9, 115.5, 115.4, 93.2, 72.6, 47.7, 44.0, 43.6, 41.9, 34.1, 31.3, 22.9, 22.4. HRMS (ESI) (positive mode) m/z calculated for C25H33FN8O3: 512.59; Found: 513.208.
(S)-N-((3-(5-fluoro-6-(4-(2-morpholinopyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6g)
Compound 6g was a white solid; yield 46.1%. m. p. 191.3–192.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.96 − 7.90 (m, 2H), 6.17 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.74 − 3.72 (m, 1H), 3.70 − 3.69 (m, 4H), 3.64 − 3.63 (m, 8H), 3.47 − 3.35 (m, 6H), 1.84 (s, 3H).13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.6, 161.6, 157.1, 154.8, 150.0 (d, JC-F = 256.7 Hz), 146.0, 133.0, 129.9, 115.5, 115.4, 94.1, 72.6, 66.6, 47.7, 44.4, 43.6, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C23H29FN8O4: 500.54; Found: 501.176.
(S)-N-((3-(5-fluoro-6-(4-(2-((3-morpholinopropyl)amino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6h)
Compound 6h was brown oil; yield 48.7%. 1H NMR (600 MHz, DMSO-d6) δ 8.28 (t, J = 6.0 Hz, 1H), 8.12 (d, J = 2.4 Hz, 1H), 7.92 (dd, J = 14.4, 2.4 Hz, 1H), 7.81 (d, J = 6.0 Hz, 1H), 6.57 (s, 1H), 6.03 (d, J = 6.0 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.12 − 4.09 (m, 1H), 3.75 − 3.73 (m, 1H), 3.67 − 3.65 (m, 4H), 3.57 − 3.56 (m, 4H), 3.44 − 3.42 (m, 4H), 3.38 − 3.35 (m, 4H), 3.27 − 3.23 (m, 2H), 2.33 − 2.30 (m, 4H), 1.85 (s, 3H), 1.66 − 1.64 (m, 2H). 13C NMR (150 MHz, DMSO-d6) δ 170.6, 162.7, 162.3, 157.2, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.9, 132.9, 129.8, 115.5, 115.3, 72.6, 66.7, 56.7, 53.8, 47.7, 46.1, 43.5, 41.9, 26.5, 22.9, 7.6. HRMS (ESI) (positive mode) m/z calculated for C26H35FN8O4: 557.63; Found: 558.256.
(S)-N-((3-(5-fluoro-6-(4-(2-(phenylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6i)
Compound 6i was a white solid; yield 43%. m. p. 190.6–194.4 °C.1H NMR (600 MHz, DMSO-d6) δ 9.17 (s, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 8.00 (d, J = 6.0 Hz, 1H), 7.95 (dd, J = 14.4, 2.4 Hz, 1H), 7.73 − 7.68 (m, 2H), 7.30 − 7.24 (m, 2H), 6.92 (t, J = 7.2 Hz, 1H), 6.36 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.11 (m, 1H), 3.79 − 3.77 (m, 4H), 3.75 − 3.72 (m, 1H), 3.48 − 3.39 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 165.2, 162.5, 154.8, 149.1 (d, JC-F = 256.2 Hz), 145.7, 141.2, 140.8, 133.0, 129.8, 128.9, 121.8, 119.4, 117.3, 115.6, 95.8, 72.6, 47.7, 43.8, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C25H27FN8O3: 506.54; Found: 507.161.
(S)-N-((3-(6-(4-(2-(benzylamino)pyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6j)
Compound 6j was a white solid; yield 41.5%. m. p. 202.7–204.6 °C.1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 7.82 (d, J = 6.0 Hz, 1H), 7.32 − 7.24 (m, 5H), 7.22 − 7.16 (m, 1H), 6.07 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.10 (m, 1H), 3.74 − 3.72 (m, 1H), 3.74 − 3.64 (m, 4H), 3.47 − 3.38 (m, 2H), 3.36 − 3.34 (m, 4H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.6, 162.3, 154.8, 150.0 (d, JC-F = 256.7 Hz), 146.0, 145.9, 141.7, 133.0, 129.8, 128.5, 127.6, 126.8, 115.5, 115.4, 72.6, 47.8, 44.5, 43.5, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C26H29FN8O3: 520.57; Found: 521.185.
(S)-N-((3-(5-fluoro-6-(4-(2-(naphthalen-1-ylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6k)
Compound 6k was a brown solid; yield 59.3%. m. p. 221.8–223.1 °C.1H NMR (600 MHz, DMSO-d6) δ 8.95 (s, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.14–8.07 (m, 2H), 7.95 − 7.88 (m, 3H), 7.79 (d, J = 7.2 Hz, 1H), 7.67 (d, J = 8.2 Hz, 1H), 7.51–7.44 (m, 3H), 6.29 (d, J = 6.0 Hz, 1H), 4.77–4.72 (m, 1H), 4.15–4.06 (m, 1H), 3.76–3.70 (m, 1H), 3.69–3.64 (m, 4H), 3.43–3.40 (m, 2H), 3.39–3.36(m, 4H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.6, 162.1, 154.8, 152.4, 149.1 (d, JC-F = 257.1 Hz), 146.0, 145.7, 134.9, 134.4, 133.0, 130.0, 128.7, 128.6, 126.4, 126.1, 126.0, 125.1, 123.4, 121.7, 115.6, 115.4, 95.8, 72.6, 63.1, 52.5, 22.9, 7.7. HRMS (ESI) (positive mode) m/z calculated for C29H29FN8O3: 556.60; Found: 557.197.
(S)-N-((3-(5-fluoro-6-(4-(2-(quinolin-5-ylamino)pyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6l)
Compound 6l was a white solid; yield 43%. m. p. 226.2–232.2 °C.1H NMR (600 MHz, DMSO-d6) δ 9.15 (s, 1H), 8.87 (dd, J = 4.2, 1.8 Hz, 1H), 8.53 − 8.48 (m, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.15 − 8.12 (m, 1H), 7.99 − 7.90 (m, 2H), 7.87 (dd, J = 7.2, 1.2 Hz, 1H), 7.77 − 7.71 (m, 2H), 7.489 − 7.47 (m, 1H), 6.33 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.74 − 3.72 (m, 1H), 3.68 − 3.66 (m, 4H), 3.43 − 3.41 (m, 2H), 3.39 − 3.37 (m, 4H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.5, 161.1, 157.3, 154.8, 149.7 (d, JC-F = 259.1 Hz), 148.3, 145.9, 136.9, 133.0, 132.6, 129.9, 129.6, 124.7, 123.6, 120.7, 120.6, 115.6, 115.4, 107.8, 95.9, 72.6, 47.7, 43.5, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C28H28FN9O3: 557.59; Found: 558.207.
(S)-N-((3-(6-(4-(2-((6-chloropyridin-3-yl)amino)pyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (6m)
Compound 6m was a white solid; yield 45.8%. m. p. 134.4–135.6 °C.1H NMR (600 MHz, DMSO-d6) δ 9.46 (s, 1H), 8.74 (d, J = 3.0 Hz, 1H), 8.27 − 8.19 (m, 2H), 8.14 (d, J = 2.4 Hz, 1H), 8.05 (d, J = 6.0 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.40 (d, J = 9.0 Hz, 1H), 6.40 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.11 (m, 1H), 3.78 − 3.71 (m, 5H), 3.47 − 3.42 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.5, 159.5, 157.1, 154.8, 149.1 (d, JC-F = 256.5 Hz), 145.8, 141.3, 140.2, 137.9, 133.0, 129.9, 129.2, 124.2, 115.6, 115.4, 96.6, 72.6, 47.6, 43.7, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C24H25ClFN9O3: 541.97; Found: 542.132.
Raw data for the above products are presented in Supplementary Materials (Figures S10–S42).
3.2.4. General Procedure for the Synthesis of 7a and 7c-n
A solution of compound 4 (130 mg, 0.3 mmol) in DCM (5 mL) at 0 °C was dropwise added to TFA (1 mL) and then stirred for 2 h. After the reaction was complete, TEA was added to the solution at 0 °C to adjust pH. The filtrate was concentrated in vacuo. To a solution of the concentrate in ethanol (3 mL) was added TEA (83 μL, 1 mmol) and pyrimidine derivative (0.4 mmol), and then stirred at reflux overnight. After the reaction was complete and concentrated, the mixture was extracted with DCM (5 mL × 3). The organic phase was washed with brine and concentrated in vacuo. The residue was purified by silica gel column chromatography (DCM/MeOH = 30:1) to yield compounds 7a and 7c-n.
(S)-N-((3-(6-(4-(6-chloropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7a)
Compound 7a was a white solid; yield 45.1%. m. p. 190.0–191.1 °C.1H NMR (600 MHz, DMSO-d6) δ 8.37 (s, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.02 (s, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.10 (m, 1H), 3.81 − 3.75 (m, 4H), 3.74 − 3.71 (m, 1H), 3.43 − 3.42 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.7, 159.7, 158.5, 154.8, 148.2 (d, JC-F = 256.7 Hz), 145.6, 133.0, 129.99, 115.6, 102.3, 72.6, 47.6, 47.5, 43.8, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H21ClFN7O3: 449.87; Found: 450.106.
(S)-N-((3-(5-fluoro-6-(4-(5-methylpyrimidin-2-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7c)
Compound 7c was a white solid; yield 49.4%. m. p. 195.1–197.3 °C.1H NMR (600 MHz, DMSO-d6) δ 8.26 − 8.23 (m, 3H), 8.13 (d, J = 2.4 Hz, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.13 − 4.10 (m, 1H), 3.85 − 3.80 (m, 4H), 3.76 − 3.70 (m, 1H), 3.43 − 3.37 (m, 6H), 2.10 (s, 3H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 160.7, 158.2, 154.8, 149.2 (d, JC-F = 255.3 Hz), 146.1, 133.0, 130.0, 119.1, 115.5, 115.4, 72.6, 47.9, 46.2, 43.9, 22.9, 14.1, 9.1. HRMS (ESI) (positive mode) m/z calculated for C20H24FN7O3: 429.46; Found: 430.181.
(S)-N-((3-(6-(4-(5-bromopyrimidin-2-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7d)
Compound 7d was a white solid; yield 47.6%. m. p. 196.1–196.3 °C.1H NMR (600 MHz, DMSO-d6) δ 8.50 (s, 2H), 8.24 (t, J = 6.0 Hz, 1H), 8.13 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.14 − 4.10 (m, 1H), 3.88 − 3.83 (m, 4H), 3.74 − 3.71 (m, 1H), 3.43 − 3.33 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 166.3, 160.0, 158.5, 154.8, 150.8 (d, JC-F = 253.7 Hz), 133.0, 129.9, 115.3, 106.1, 72.6, 47.6, 43.8, 41.9, 39.0, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H21BrFN7O3: 494.33; Found: 496.095.
(S)-N-((3-(6-(4-(2-aminopyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7e)
Compound 7e was a white solid; yield 60.9%. m. p. 195.6–196.7 °C.1H NMR (600 MHz, DMSO-d6) δ 8.28 − 8.25 (m, 1H), 8.15 (d, J = 2.4 Hz, 1H), 7.97 − 7.88 (m, 3H), 7.89 (dd, J = 7.8, 2.4 Hz, 1H), 6.57 (d, J = 7.8 Hz, 1H), 4.78 − 4.74 (m, 1H), 4.14 − 4.11 (m, 1H), 3.75 − 3.72 (m, 1H), 3.47 − 3.46 (m, 4H), 3.43 − 3.41 (m, 4H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 161.7, 155.1, 154.8, 149.1 (d, JC-F = 256.4 Hz), 145.4, 143.4, 133.0, 130.1, 115.6, 115.5, 95.4, 72.7, 47.6, 47.5, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H23FN8O3: 430.44; Found: 431.144.
(S)-N-((3-(6-(4-(4-aminopyrimidin-2-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7f)
Compound 7f was a white solid; yield 60.9%. m. p. 196.6–197.7 °C.1H NMR (600 MHz, DMSO-d6) δ 8.25 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.77 (d, J = 6.0 Hz, 1H), 7.48 − 7.10 (m, 2H), 5.94 (d, J = 6.0 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.80 − 3.78 (m, 4H), 3.75 − 3.72 (m, 1H), 3.43 − 3.40 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 164.5, 154.8, 149.1 (d, JC-F = 256.7 Hz), 145.9, 138.1, 133.0, 130.0, 128.5, 126.0, 115.6, 72.6, 47.7, 43.9, 41.9, 22.9, 21.3. HRMS (ESI) (positive mode) m/z calculated for C19H23FN8O3: 430.44; Found: 431.141.
(S)-N-((3-(6-(4-(4,6-dichloropyrimidin-2-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide(7g)and (S)-N-((3-(6-(4-(2,6-dichloropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7h)
Compounds 7g and 7h were both white solids, yields were 13.8% and 44.7% respectively, m. p. 191.3–192.5 °C and m. p. 195.2–197.1 °C respectively.
Compound 7g, 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), z7.94 (dd, J = 14.4, 2.4 Hz, 1H), 6.98 (s, 1H), 4.77 − 4.73 (m, 1H), 4.14 − 4.10 (m, 1H), 3.88 − 3.84 (m, 4H), 3.75 − 3.72 (m, 1H), 3.44 − 3.41 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 161.6, 160.5, 154.8, 148.2 (d, J = 256.7 Hz), 145.7, 132.9, 130.0, 115.4, 108.2, 72.6, 47.6, 47.5, 43.9, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H20Cl2FN7O3: 483.10; Found: 484.103.
Compound 7h, 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 7.08 (s, 1H), 4.77 − 4.74 (m, 1H), 4.13 − 4.10 (m, 1H), 3.87 − 3.71 (m, 5H), 3.44 − 3.42 (m, 6H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 163.4, 159.6, 159.0, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.5, 133.0, 130.0, 115.4, 101.4, 72.6, 47.6, 47.4, 43.7, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H20Cl2FN7O3: 483.10; Found: 484.104.
(S)-N-((3-(6-(4-(2-chloro-5-fluoropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7i)
Compound 7i was a white solid; yield 70.1%. m. p. 204.4–205.5 °C.1H NMR (600 MHz, DMSO-d6) δ 8.25 − 8.21 (m, 2H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.10 (m, 1H), 3.90 − 3.86 (m, 4H), 3.75 − 3.71 (m, 1H), 3.48 − 3.46 (m, 4H), 3.43 − 3.41 (m, 2H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 154.8, 153.5, 152.5, 149.1 (d, JC-F = 257.0 Hz), 147.1 (d, JC-F = 256.4 Hz), 145.6, 144.7, 133.0, 130.0, 115.6, 72.6, 47.6, 45.8, 41.9, 40.5, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H20ClF2N7O3: 467.86; Found: 468.113.
(S)-N-((3-(6-(4-(2,5-dichloropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7j)
Compound 7j was a white solid; yield 48.6%. m. p. 193.5–194.2 °C.1H NMR (600 MHz, DMSO-d6) δ 8.35 (s, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.14 − 4.10 (m, 1H), 3.91 − 3.86 (m, 4H), 3.75 − 3.72 (m, 1H), 3.51 − 3.46 (m, 4H), 3.43 − 3.41 (m, 2H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 160.4, 159.1, 157.1, 154.8, 148.3 (d, JC-F = 256.7 Hz), 133.0, 130.0, 115.5, 115.1, 72.6, 47.6, 47.6, 47.0, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H20Cl2FN7O3: 483.10; Found: 484.099.
(S)-N-((3-(6-(4-(5-bromo-2-chloropyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7k)
Compound 7k was a white solid; yield 82.8%. m. p. 196.7–197.6 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.46 (s, 1H), 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 4.77 − 4.73 (m, 1H), 4.14 − 4.10 (m, 1H), 3.87 − 3.83 (m, 4H), 3.75 − 3.72 (m, 1H), 3.50 − 3.46 (m, 4H), 3.43 − 3.41 (m, 2H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 162.7, 161.8, 157.8, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.6, 133.0, 130.0, 115.5, 104.1, 72.6, 47.6, 47.6, 47.3, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H20BrClFN7O3: 528.77; Found: 530.054.
(S)-N-((3-(6-(4-(2-chloro-5-methylpyrimidin-4-yl)piperazin-1-yl)-5-fluoropyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7l)
Compound 7l was a white solid; yield 58.9%. m. p. 198.8–200.8 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 8.06 (s, 1H), 7.93 (dd, J = 14.4, 2.4 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.14 − 4.10 (m, 1H), 3.74 − 3.72 (m, 1H), 3.70 − 3.65 (m, 4H), 3.48 − 3.44 (m, 4H), 3.43 − 3.41 (m, 2H), 2.24 (s, 3H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 165.0, 160.0, 157.2, 154.8, 149.1 (d, JC-F = 257.1 Hz), 145.8, 133.0, 130.0, 116.2, 115.4, 72.6, 47.8, 47.6, 46.9, 41.9, 22.9, 17.3. HRMS (ESI) (positive mode) m/z calculated for C20H23ClFN7O3: 463.90; Found: 464.117.
Ethyl(S)-4-(4-(5-(5-(acetamidomethyl)-2-oxooxazolidin-3-yl)-3-fluoropyridin-2-yl)piperazin-1-yl)-2-(methylthio)pyrimidine-5-carboxylate (7m)
Compound 7m was a white solid; yield 50.6%. m. p. 210.1–213.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.45 (s, 1H), 8.26 (t, J = 6.0 Hz, 1H), 8.11 (d, J = 2.4 Hz, 1H), 7.92 (dd, J = 14.4, 2.4 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.36 − 4.17 (m, 2H), 4.14 − 4.09 (m, 1H), 3.76 − 3.72 (m, 1H), 3.68 − 3.66 (m, 4H), 3.46 − 3.44 (m, 4H), 3.43 − 3.41 (m, 2H), 2.48 (s, 3H), 1.85 (s, 3H), 1.37 − 1.25 (m, 3H). 13C NMR (150 MHz, DMSO-d6) δ 172.6, 170.5, 165.9, 159.8, 159.3, 154.8, 149.0 (d, JC-F = 257.0 Hz), 145.6, 132.9, 129.8, 115.3, 105.7, 72.6, 61.4, 47.6, 47.5, 47.3, 41.9, 22.9, 14.5, 14.1. HRMS (ESI) (positive mode) m/z calculated for C23H28FN7O5S: 533.58; Found: 534.132.
(S)-N-((3-(5-fluoro-6-(4-(2,5,6-trichloropyrimidin-4-yl)piperazin-1-yl)pyridin-3-yl)-2-oxooxazolidin-5-yl)methyl)acetamide (7n)
Compound 7n was a white solid; yield 71.4%. m. p. 210.0–211.1 °C. 1H NMR (600 MHz, DMSO-d6) δ 8.24 (t, J = 6.0 Hz, 1H), 8.14 (d, J = 2.4 Hz, 1H), 7.94 (dd, J = 14.4, 2.4 Hz, 1H), 4.79 − 4.72 (m, 1H), 4.13 − 4.10 (m, 1H), 3.92 − 3.81 (m, 4H), 3.75 − 3.72 (m, 1H), 3.52 − 3.47 (m, 4H), 3.44 − 3.39 (m, 2H), 1.84 (s, 3H). 13C NMR (150 MHz, DMSO-d6) δ 170.5, 161.9, 159.0, 155.1, 154.8, 149.1 (d, JC-F = 255.8 Hz), 145.6, 133.0, 129.8, 115.4, 112.4, 72.6, 47.8, 47.6, 47.5, 41.9, 22.9. HRMS (ESI) (positive mode) m/z calculated for C19H19Cl3FN7O3: 518.76; Found: 520.066.
Raw data for the above products are presented in Supplementary Materials (Figures S43, S45 and S49–S84).
3.3. MIC Determination
The antibacterial activity of the synthesized derivatives was determined by the broth dilution method [42]. The strains, including Staphylococcus aureus (ATCC25923), Streptococcus pneumoniae (ATCC49619), Enterococcus faecalis (ATCC29212), Bacillus subtilis (ATCC6633), Escherichia coli (ATCC25922), Listeria monocytogenes (ATCC19111), Staphylococcus xylosus (ATCC35924), methicillin-resistant Staphylococcus aureus, vancomycin-resistant Enterococcus faecalis, linezolid-resistant Staphylococcus aureus, and linezolid-resistant Streptococcus pneumoniae, were incubated in Mueller-Hinton (MH) medium at 37 °C to mid-logarithm (OD600 = 0.5). The bacteria were diluted to 105 CFU/ mL and added to the 96-well plate, followed by a series of diluted synthesized derivatives (from 128 to 0.25 µg/mL). After incubation at 37 °C for 16–18 h, the minimum inhibitory concentration (MIC) value was the minimum drug concentration without bacterial growth.
3.4. Molecular Docking Studies
The 3D structure of the 50S ribosomal subunit (PDB code: 3CPW) [43] was obtained from the Protein Data Bank and processed by PyMOL 2.5. The original ligand and protein were deleted, and only the required RNA chains were retained and imported into the Auto-Dock for use. The ligand was drawn with ChemOffice2010 Version and imported into the Auto-Dock for later use. The Auto-Dock 4.2.6® software was used for the molecular docking process, and the obtained results were imported into PyMOL 2.5 software in the form of complexes for visual analysis.
3.5. Inhibition of Biofilm Formation Assay
Anti-biofilm activity inhibits biofilm formation and was measured by the crystal violet method [44]. The strains to be tested were placed in a test tube containing 5 mL Tryptic Soy Broth (TSB) and incubated at 37 °C for 24 h. Then the suspension was diluted to 106 CFU/mL and added to a sterile 96-well culture plate, filled with 100 µL per well. All compounds were added to the well according to the selected concentration gradient and incubated at 37 °C for 24 h. After the biofilm was grown, the culture medium was removed from each well, washed twice with sterile PBS, fixed with methanol, and stained with 150 µL 0.1% crystal violet solution at room temperature. Remove the excess solution, wash it twice with water, and add 125 µL 33% acetic acid to each dyeing well for 5 min to dissolve the dye. The microplate reader was used to read at 600 nm to assess the minimum concentration of biofilm inhibition.
3.6. Cytotoxicity Assay
The MTT method was used to detect the effect of typical derivatives on Hela cell viability [45]. Cells (5 × 104 cells/well) were added to a 96-well plate for 24 h in humidified 5% (V/V) CO2/ air at 37 °C. A series of liquid medicines (8, 16, 32, 64, 128, 256, 500, and 1000 µg/mL) were added and incubated for 48 h. Cells treated with equal volumes of DMSO were used as controls. Add 10 µL MTT solution (0.5%) to each well and incubate for 4 h at 37 °C under dark conditions. The culture medium in all wells was discarded. Then 100 µL DMSO was quickly added to each wall and shaken at low speed to dissolve the formed crystals. The absorbance was measured at 570 nm. The cell viability was calculated as follows: cell viability (%) = (treatment sample OD570 − empty OD570)/(control OD570 − empty OD570).
4. Conclusions
In summary, a library of 28 novel 3-(5-fluoropyridine-3-yl)-2-oxazolidinone derivatives was designed, synthesized, and evaluated for their antibacterial properties. The results show that most of the synthesized compounds have potential antibacterial activity against gram-positive bacteria. Amongst them, compounds 7i-l exhibited a better antibacterial effect. The molecular docking results of compounds 7j and PTC were studied to predict the mechanism of action. Further results demonstrated that these compounds have excellent ability to inhibit biofilm formation and meager cytotoxicity. These results provide a basis and reference for the discovery of novel antibacterial compounds and the development of new drugs.
Acknowledgments
We thank J. Ge for expert technical assistance and the National Natural Science Foundation of China (Grant No. 31802227).
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/molecules28114267/s1. Figure S1: 1H NMR Spectrum of 5; Figure S2: 13C NMR Spectrum of 5; Figure S3: ES-MS for compound 5; Figure S4: 1H NMR Spectrum of 6a; Figure S5: 13C NMR Spectrum of 6a; Figure S6: ES-MS for compound 6a; Figure S7: 1H NMR Spectrum of 6b; Figure S8: 13C NMR Spectrum of 6b; Figure S9: ES-MS for compound 6b; Figure S10: 1H NMR Spectrum of 6c; Figure S11: 13C NMR Spectrum of 6c; Figure S12: ES-MS for compound 6c; Figure S13: 1H NMR Spectrum of 6d; Figure S14: 13C NMR Spectrum of 6d; Figure S15: ES-MS for compound 6d; Figure S16: 1H NMR Spectrum of 6e; Figure S17: 13C NMR Spectrum of 6e; Figure S18: ES-MS for compound 6e; Figure S19 1H NMR Spectrum of 6f; Figure S20: 13C NMR Spectrum of 6f; Figure S21: ES-MS for compound 6f; Figure S22: 1H NMR Spectrum of 6g; Figure S23: 13C NMR Spectrum of 6g; Figure S24: ES-MS for compound 6g; Figure S25: 1H NMR Spectrum of 6h; Figure S26: 13C NMR Spectrum of 6h; Figure S27: ES-MS for compound 6h; Figure S28: 1H NMR Spectrum of 6i; Figure S29: 13C NMR Spectrum of 6i; Figure S30: ES-MS for compound 6i; Figure S31: 1H NMR Spectrum of 6j; Figure S32: 13C NMR Spectrum of 6j; Figure S33: ES-MS for compound 6j; Figure S34: 1H NMR Spectrum of 6k; Figure S35: 13C NMR Spectrum of 6k; Figure S36: ES-MS for compound 6k; Figure S37: 1H NMR Spectrum of 6l; Figure S38: 13C NMR Spectrum of 6l; Figure S39: ES-MS for compound 6l; Figure S40: 1H NMR Spectrum of 6m; Figure S41: 13C NMR Spectrum of 6m; Figure S42: ES-MS for compound 6m; Figure S43: 1H NMR Spectrum of 7a; Figure S44: 13C NMR Spectrum of 7a; Figure S45: ES-MS for compound 7a; Figure S46: 1H NMR Spectrum of 7b; Figure S47: 13C NMR Spectrum of 7b; Figure S48: ES-MS for compound 7b; Figure S49: 1H NMR Spectrum of 7c; Figure S50: 13C NMR Spectrum of 7c; Figure S51: ES-MS for compound 7c; Figure S52: 1H NMR Spectrum of 7d; Figure S53: 13C NMR Spectrum of 7d; Figure S54: ES-MS for compound 7d; Figure S55: 1H NMR Spectrum of 7e; Figure S56: 13C NMR Spectrum of 7e; Figure S57: ES-MS for compound 7e; Figure S58: 1H NMR Spectrum of 7f; Figure S59: 13C NMR Spectrum of 7f; Figure S60: ES-MS for compound 7f; Figure S61: 1H NMR Spectrum of 7g; Figure S62: 13C NMR Spectrum of 7g; Figure S63: ES-MS for compound 7g; Figure S64: 1H NMR Spectrum of 7h; Figure S65: 13C NMR Spectrum of 7h; Figure S66: ES-MS for compound 7h; Figure S67: 1H NMR Spectrum of 7i; Figure S68: 13C NMR Spectrum of 7i; Figure S69: ES-MS for compound 7i; Figure S70: 1H NMR Spectrum of 7j; Figure S71: 13C NMR Spectrum of 7j; Figure S72: ES-MS for compound 7j; Figure S73: 1H NMR Spectrum of 7k; Figure S74: 13C NMR Spectrum of 7k; Figure S75: ES-MS for compound 7k; Figure S76: 1H NMR Spectrum of 7l; Figure S77: 13C NMR Spectrum of 7l; Figure S78: ES-MS for compound 7l; Figure S79: 1H NMR Spectrum of 7m; Figure S80: 13C NMR Spectrum of 7m; Figure S81: ES-MS for compound 7m; Figure S82: 1H NMR Spectrum of 7n; Figure S83: 13C NMR Spectrum of 7n; Figure S84: ES-MS for compound 7n.
Author Contributions
Conceptualization, X.W., B.J. and H.Y.; methodology, X.W., B.J., Y.H., T.W., Z.S., Y.T. and H.Y.; software, Z.S.; validation, X.W., B.J., Y.H., T.W., Z.S., Y.T. and H.Y.; formal analysis, X.W., B.J. and H.Y.; investigation, H.Y.; resources, H.Y.; data curation, X.W., B.J. and H.Y.; writing—original draft preparation, X.W.; writing—review and editing, H.Y.; visualization, Z.S.; supervision, H.Y.; project administration, H.Y.; funding acquisition, H.Y. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Sample Availability
Samples of some compounds may be available from the authors.
Funding Statement
This research was supported by the National Natural Science Foundation of China (Grant No. 31802227).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Yan X., Schouls L.M., Pluister G.N., Tao X., Yu X., Yin J., Song Y., Hu S., Luo F., Hu W., et al. The population structure of Staphylococcus aureus in China and Europe assessed by multiple-locus variable number tandem repeat analysis; clues to geographical origins of emergence and dissemination. Clin. Microbiol. Infect. 2016;22:60.e1–60.e8. doi: 10.1016/j.cmi.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 2.Zaman S.B., Hussain M.A., Nye R., Mehta V., Mamun K.T., Hossain N. A Review on Antibiotic Resistance: Alarm Bells are Ringing. Cureus. 2017;9:e1403. doi: 10.7759/cureus.1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parrino B., Schillaci D., Carnevale I., Giovannetti E., Diana P., Cirrincione G., Cascioferro S. Synthetic small molecules as anti-biofilm agents in the struggle against antibiotic resistance. Eur. J. Med. Chem. 2019;161:154–178. doi: 10.1016/j.ejmech.2018.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Costerton J.W., Stewart P.S., Greenberg E.P. Bacterial Biofilms: A Common Cause of Persistent Infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- 5.Liang J., Sun D., Yang Y., Li M., Li H., Chen L. Discovery of metal-based complexes as promising antimicrobial agents. Eur. J. Med. Chem. 2021;224:113696. doi: 10.1016/j.ejmech.2021.113696. [DOI] [PubMed] [Google Scholar]
- 6.Ling H., Lou X., Luo Q., He Z., Sun M., Sun J. Recent advances in bacteriophage-based therapeutics: Insight into the post-antibiotic era. Acta Pharm. Sin. B. 2022;12:4348–4364. doi: 10.1016/j.apsb.2022.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pisoschi A.M., Pop A., Cimpeanu C., Turcuş V., Predoi G., Iordache F. Nanoencapsulation techniques for compounds and products with antioxidant and antimicrobial activity—A critical view. Eur. J. Med. Chem. 2018;157:1326–1345. doi: 10.1016/j.ejmech.2018.08.076. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y., Lv Q., Chen Y., Xu L., Feng M., Xiong Z., Li J., Ren J., Liu J., Liu B. Bilayer hydrogel dressing with lysozyme-enhanced photothermal therapy for biofilm eradication and accelerated chronic wound repair. Acta Pharm. Sin. B. 2022;13:284–297. doi: 10.1016/j.apsb.2022.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malla T.R., Brewitz L., Muntean D.G., Aslam H., Owen C.D., Salah E., Tumber A., Lukacik P., Strain-Damerell C., Mikolajek H., et al. Penicillin Derivatives Inhibit the SARS-CoV-2 Main Protease by Reaction with Its Nucleophilic Cysteine. J. Med. Chem. 2022;65:7682–7696. doi: 10.1021/acs.jmedchem.1c02214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Naclerio G.A., Abutaleb N.S., Onyedibe K.I., Karanja C., Eldesouky H.E., Liang H.W., Dieterly A., Aryal U.K., Lyle T., Seleem M.N., et al. Mechanistic Studies and In Vivo Efficacy of an Oxadiazole-Containing Antibiotic. J. Med. Chem. 2022;65:6612–6630. doi: 10.1021/acs.jmedchem.1c02034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Y.P., Sangaraiah N., Meng J.P., Zhou C.H. Unique Carbazole-Oxadiazole Derivatives as New Potential Antibiotics for Combating Gram-Positive and -Negative Bacteria. J. Med. Chem. 2022;65:6171–6190. doi: 10.1021/acs.jmedchem.2c00001. [DOI] [PubMed] [Google Scholar]
- 12.Zurenko G.E., Yagi B.H., Schaadt R.D., Allison J.W., Kilburn J.O., Glickman S.E., Hutchinson D.K., Barbachyn M.R., Brickner S.J. In vitro activities of U-100592 and U-100766, novel oxazolidinone antibacterial agents. Antimicrob. Agents Chemother. 1996;40:839–845. doi: 10.1128/AAC.40.4.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lei H., Jiang Y., Wang D., Gong P., Li Y., Dong Y., Dong M. In vitro activity of novel oxazolidinone analogs and 13 conventional antimicrobial agents against clinical isolates of Staphylococcus aureus in Beijing, China. Jpn. J. Infect. Dis. 2014;67:402–404. doi: 10.7883/yoken.67.402. [DOI] [PubMed] [Google Scholar]
- 14.Lin A.H., Murray R.W., Vidmar T.J., Marotti K.R. The oxazolidinone eperezolid binds to the 50S ribosomal subunit and competes with binding of chloramphenicol and lincomycin. Antimicrob. Agents Chemother. 1997;41:2127–2131. doi: 10.1128/AAC.41.10.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Swaney S.M., Aoki H., Ganoza M.C., Shinabarger D.L. The oxazolidinone linezolid inhibits initiation of protein synthesis in bacteria. Antimicrob. Agents Chemother. 1998;42:3251–3255. doi: 10.1128/AAC.42.12.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou C.C., Swaney S.M., Shinabarger D.L., Stockman B.J. 1H nuclear magnetic resonance study of oxazolidinone binding to bacterial ribosomes. Antimicrob. Agents Chemother. 2002;46:625–629. doi: 10.1128/AAC.46.3.625-629.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Foti C., Piperno A., Scala A., Giuffrè O. Oxazolidinone Antibiotics: Chemical, Biological and Analytical Aspects. Molecules. 2021;26:4280. doi: 10.3390/molecules26144280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pandit N., Singla R.K., Shrivastava B. Current updates on oxazolidinone and its significance. Int. J. Med. Chem. 2012;2012:159285. doi: 10.1155/2012/159285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yuan S., Shen D.D., Bai Y.R., Zhang M., Zhou T., Sun C., Zhou L., Wang S.Q., Liu H.M. Oxazolidinone: A promising scaffold for the development of antibacterial drugs. Eur. J. Med. Chem. 2023;250:115239. doi: 10.1016/j.ejmech.2023.115239. [DOI] [PubMed] [Google Scholar]
- 20.Vinh D.C., Rubinstein E. Linezolid: A review of safety and tolerability. J. Infect. 2009;59((Suppl. S1)):S59–S74. doi: 10.1016/S0163-4453(09)60009-8. [DOI] [PubMed] [Google Scholar]
- 21.Bai P.-Y., Qin S.-S., Chu W.-C., Yang Y., Cui D.-Y., Hua Y.-G., Yang Q.-Q., Zhang E. Synthesis and antibacterial bioactivities of cationic deacetyl linezolid amphiphiles. Eur. J. Med. Chem. 2018;155:925–945. doi: 10.1016/j.ejmech.2018.06.054. [DOI] [PubMed] [Google Scholar]
- 22.De Rosa M., Zanfardino A., Notomista E., Wichelhaus T.A., Saturnino C., Varcamonti M., Soriente A. Novel promising linezolid analogues: Rational design, synthesis and biological evaluation. Eur. J. Med. Chem. 2013;69:779–785. doi: 10.1016/j.ejmech.2013.09.035. [DOI] [PubMed] [Google Scholar]
- 23.Fortuna C.G., Bonaccorso C., Bulbarelli A., Caltabiano G., Rizzi L., Goracci L., Musumarra G., Pace A., Palumbo Piccionello A., Guarcello A., et al. New linezolid-like 1,2,4-oxadiazoles active against Gram-positive multiresistant pathogens. Eur. J. Med. Chem. 2013;65:533–545. doi: 10.1016/j.ejmech.2013.03.069. [DOI] [PubMed] [Google Scholar]
- 24.Gadekar P.K., Roychowdhury A., Kharkar P.S., Khedkar V.M., Arkile M., Manek H., Sarkar D., Sharma R., Vijayakumar V., Sarveswari S. Design, synthesis and biological evaluation of novel azaspiro analogs of linezolid as antibacterial and antitubercular agents. Eur. J. Med. Chem. 2016;122:475–487. doi: 10.1016/j.ejmech.2016.07.001. [DOI] [PubMed] [Google Scholar]
- 25.Naresh A., Venkateswara Rao M., Kotapalli S.S., Ummanni R., Venkateswara Rao B. Oxazolidinone derivatives: Cytoxazone-linezolid hybrids induces apoptosis and senescence in DU145 prostate cancer cells. Eur. J. Med. Chem. 2014;80:295–307. doi: 10.1016/j.ejmech.2014.04.062. [DOI] [PubMed] [Google Scholar]
- 26.Palumbo Piccionello A., Musumeci R., Cocuzza C., Fortuna C.G., Guarcello A., Pierro P., Pace A. Synthesis and preliminary antibacterial evaluation of Linezolid-like 1,2,4-oxadiazole derivatives. Eur. J. Med. Chem. 2012;50:441–448. doi: 10.1016/j.ejmech.2012.02.002. [DOI] [PubMed] [Google Scholar]
- 27.Wei H., Mao F., Ni S., Chen F., Li B., Qiu X., Hu L., Wang M., Zheng X., Zhu J., et al. Discovery of novel piperonyl derivatives as diapophytoene desaturase inhibitors for the treatment of methicillin-, vancomycin- and linezolid-resistant Staphylococcus aureus infections. Eur. J. Med. Chem. 2018;145:235–251. doi: 10.1016/j.ejmech.2017.12.090. [DOI] [PubMed] [Google Scholar]
- 28.Wu Y., Ding X., Ding L., Zhang Y., Cui L., Sun L., Li W., Wang D., Zhao Y. Synthesis and antibacterial activity evaluation of novel biaryloxazolidinone analogues containing a hydrazone moiety as promising antibacterial agents. Eur. J. Med. Chem. 2018;158:247–258. doi: 10.1016/j.ejmech.2018.09.004. [DOI] [PubMed] [Google Scholar]
- 29.Wu Y., Ding X., Yang Y., Li Y., Qi Y., Hu F., Qin M., Liu Y., Sun L., Zhao Y. Optimization of biaryloxazolidinone as promising antibacterial agents against antibiotic-susceptible and antibiotic-resistant gram-positive bacteria. Eur. J. Med. Chem. 2020;185:111781. doi: 10.1016/j.ejmech.2019.111781. [DOI] [PubMed] [Google Scholar]
- 30.Jin B., Chen J.Y., Sheng Z.L., Sun M.Q., Yang H.L. Synthesis, Antibacterial and Anthelmintic Activity of Novel 3-(3-Pyridyl)-oxazolidinone-5-methyl Ester Derivatives. Molecules. 2022;27:1103. doi: 10.3390/molecules27031103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jin B., Wang T., Chen J.Y., Liu X.Q., Zhang Y.X., Zhang X.Y., Sheng Z.L., Yang H.L. Synthesis and Biological Evaluation of 3-(Pyridine-3-yl)-2-Oxazolidinone Derivatives as Antibacterial Agents. Front. Chem. 2022;10:949813. doi: 10.3389/fchem.2022.949813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang H.-l., Jin B., Chen J.-q., Sheng Z.-l. Synthesis and Biological Activity of Pyridinyl-4,5-2H-isoxazole Heterocyclic Derivatives. Fine Chem. 2019;36:487. [Google Scholar]
- 33.Yang H.-L., Xu G.-X., Bao M.-Y., Zhang D.-P., Li Z.-W., Pei Y.-Z. Design and Synthesis of Pyridinylisoxazoles and Their Anticancer Activities. Chem. J. Chin. Univ. 2014;35:2584. [Google Scholar]
- 34.Yang H.-l., Xu G.-x., Pei Y.-z. Synthesis, preliminary structure-activity relationships and biological evaluation of pyridinyl-4,5-2H-isoxazole derivatives as potent antitumor agents. Chem. Res. Chin. Univ. 2017;33:61–69. doi: 10.1007/s40242-017-6330-8. [DOI] [Google Scholar]
- 35.Elattar K.M., Mert B.D., Monier M., El-Mekabaty A. Advances in the chemical and biological diversity of heterocyclic systems incorporating pyrimido[1,6-a]pyrimidine and pyrimido[1,6-c]pyrimidine scaffolds. RSC Adv. 2020;10:15461–15492. doi: 10.1039/D0RA00411A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albratty M., Alhazmi H.A. Novel pyridine and pyrimidine derivatives as promising anticancer agents: A review. Arab. J. Chem. 2022;15:103846. doi: 10.1016/j.arabjc.2022.103846. [DOI] [Google Scholar]
- 37.Tao Y., Chen J.X., Fu Y., Chen K., Luo Y. Exploratory Process Development and Kilogram-Scale Synthesis of a Novel Oxazolidinone Antibacterial Candidate. Org. Process Res. Dev. 2014;18:511–519. doi: 10.1021/op500030v. [DOI] [Google Scholar]
- 38.Phillips O.A., D’Silva R., Bahta T.O., Sharaf L.H., Udo E.E., Benov L., Eric Walters D. Synthesis and biological evaluation of novel 5-(hydroxamic acid)methyl oxazolidinone derivatives. Eur. J. Med. Chem. 2015;106:120–131. doi: 10.1016/j.ejmech.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 39.Wang Y., Shen J.K., Schroeder S.J. Nucleotide Dynamics at the A-Site Cleft in the Peptidyltransferase Center of H. marismortui 50S Ribosomal Subunits. J. Phys. Chem. Lett. 2012;3:1007–1010. doi: 10.1021/jz3001882. [DOI] [PubMed] [Google Scholar]
- 40.Kotb A., Abutaleb N.S., Seleem M.A., Hagras M., Mohammad H., Bayoumi A., Ghiaty A., Seleem M.N., Mayhoub A.S. Phenylthiazoles with tert-Butyl side chain: Metabolically stable with anti-biofilm activity. Eur. J. Med. Chem. 2018;151:110–120. doi: 10.1016/j.ejmech.2018.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ding R., Wang X., Fu J., Chang Y., Li Y., Liu Y., Liu Y., Ma J., Hu J. Design, synthesis and antibacterial activity of novel pleuromutilin derivatives with thieno[2,3-d]pyrimidine substitution. Eur. J. Med. Chem. 2022;237:114398. doi: 10.1016/j.ejmech.2022.114398. [DOI] [PubMed] [Google Scholar]
- 42.Wiegand I., Hilpert K., Hancock R.E. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nat. Protoc. 2008;3:163–175. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
- 43.Ippolito J.A., Kanyo Z.F., Wang D., Franceschi F.J., Moore P.B., Steitz T.A., Duffy E.M. Crystal Structure of the Oxazolidinone Antibiotic Linezolid Bound to the 50S Ribosomal Subunit. J. Med. Chem. 2008;51:3353–3356. doi: 10.1021/jm800379d. [DOI] [PubMed] [Google Scholar]
- 44.Noolvi M.N., Patel H.M., Kamboj S., Kaur A., Mann V. 2,6-Disubstituted imidazo[2,1-b][1,3,4]thiadiazoles: Search for anticancer agents. Eur. J. Med. Chem. 2012;56:56–69. doi: 10.1016/j.ejmech.2012.08.012. [DOI] [PubMed] [Google Scholar]
- 45.Vaarla K., Kesharwani R.K., Santosh K., Vedula R.R., Kotamraju S., Toopurani M.K. Synthesis, biological activity evaluation and molecular docking studies of novel coumarin substituted thiazolyl-3-aryl-pyrazole-4-carbaldehydes. Bioorg. Med. Chem. Lett. 2015;25:5797–5803. doi: 10.1016/j.bmcl.2015.10.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.