Abstract
The biological risks of the deep space environment must be elucidated to enable a new era of human exploration and scientific discovery beyond low earth orbit (LEO). There is a paucity of deep space biological missions that will inform us of the deleterious biological effects of prolonged exposure to the deep space environment. To safely undertake long-term missions to Mars and space habitation beyond LEO, we must first prove and optimize autonomous biosensors to query the deep space radiation environment. Such biosensors must contain organisms that can survive for extended periods with minimal life support technology and must function reliably with intermittent communication with Earth. NASA's BioSentinel mission, a nanosatellite containing the budding yeast Saccharomyces cerevisiae, is such a biosensor and one of the first biological missions beyond LEO in nearly half a century. It will help fill critical gaps in knowledge about the effects of uniquely composed, chronic, low-flux deep space radiation on biological systems and in particular will provide valuable insight into the DNA damage response to highly ionizing particles. Due to yeast's robustness and desiccation tolerance, it can survive for periods analogous to that of a human Mars mission. In this study, we discuss our optimization of conditions for long-term reagent storage and yeast survival under desiccation in preparation for the BioSentinel mission. We show that long-term yeast cell viability is maximized when cells are air-dried in trehalose solution and stored in a low-relative humidity and low-temperature environment and that dried yeast is sensitive to low doses of deep space-relevant ionizing radiation under these conditions. Our findings will inform the design and development of improved future long-term biological missions into deep space.
Key Words: Saccharomyces cerevisiae, Desiccation tolerance, Anhydrobiosis, Biosensor, Deep space, CubeSat
1. Introduction
Robotic missions to Mars and science conducted aboard the International Space Station (ISS) currently facilitate the development and demonstration of the technology, life support, and communication systems necessary for deep space exploration and increase our understanding of the effects of space flight on astronaut health and performance. However, ISS studies of space radiation effects on biological processes cannot be directly extrapolated to extended space flight missions beyond low earth orbit (LEO). Astronauts on the ISS are exposed to a complex radiation environment and elevated doses compared to people on Earth's surface, but the orbiting laboratory is shielded from the full spectrum of deep space radiation by Earth's magnetic field.
Astronauts on long-term deep space missions will be directly exposed to galactic cosmic rays (GCRs)—highly ionizing radiation composed of high-energy protons and heavy ions—and solar particle events (SPEs)—composed primarily of energetic protons (Cucinotta, 2014). Without the appropriate shielding and radiation resistance countermeasures, these sources of ionizing radiation can induce DNA damage, as well as damage to proteins and other components of the cell, and increase astronauts' risk of cancer, circulatory disease, cataracts, hypothyroidism, exposure induced death, and other genetic effects (Daly, 2012; Cucinotta, 2014; Elgazzar and Kazem, 2015). Due to the near-impossibility of simulating prolonged exposure to the diverse, full-spectrum, low-flux cocktail of deep space radiation in terrestrial facilities, long-term space flight missions are needed to characterize the radiobiological hazards of this environment.
NASA's BioSentinel mission is part of an emergent effort to query the deep space environment with biological systems (Straume et al., 2017). The BioSentinel nanosatellite, a 6U deep space CubeSat housing microorganisms, will launch no earlier than October 2020 as the sole biological mission among the 13 CubeSat experiments planned to fly as secondary payloads on Artemis-1. Artemis-1 is a foundational mission in a series of increasingly complex endeavors that will enable human exploration beyond LEO. This test will be the first integrated demonstration of NASA's deep space exploration systems, including the uncrewed Orion spacecraft, the Space Launch System (SLS) rocket, and the ground systems at Kennedy Space Center (KSC) in Cape Canaveral, FL (NASA, 2018). After deployment from the SLS rocket, the BioSentinel spacecraft will undergo a cis-lunar flyby trajectory and enter a heliocentric orbit. It is expected to travel over 40 million kilometers from Earth during the 6- to 12-month mission. A copy of the BioSentinel payload is also planned to be flown on the ISS in 2020.
To characterize the deep space radiation environment, BioSentinel contains two strains of the budding yeast Saccharomyces cerevisiae: a wild type strain that serves as a control for yeast health, and a rad51 deletion mutant (rad51Δ) that is defective for recombinational repair of double stranded breaks (DSBs) in DNA, and is therefore sensitive to ionizing radiation (Bärtsch et al., 2000; Bennett et al., 2001). Previous space experiments in LEO used similar yeast strains to investigate the effects of microgravity and ionizing radiation on DNA repair processes and mutagenesis (Kiefer and Pross, 1999; Pross and Kiefer, 1999; Pross et al., 2000; Takahashi et al., 2001). In these short-term experiments, yeast cells were exposed to high doses of ionizing radiation before flight or during flight using external sources. These studies found that DNA repair, particularly DSB repair, is not affected by microgravity, thus providing evidence that this DNA repair pathway is ideal for assessing the effects of space radiation on biological systems.
Yeast will serve as a robust proxy for human cells in this study. Although yeast and humans diverged from a common ancestor ∼1 billion years ago, nearly half of essential yeast genes are replaceable with the human ortholog (Kachroo et al., 2015). Furthermore, RAD51 constitutes part of an evolutionarily conserved eukaryotic pathway for homologous recombination and recombinational repair (Baumann and West, 1998). Due to the translational power granted by this close genetic relationship and the long duration of the BioSentinel mission, the DNA damage response observed in yeast cells will be directly relevant to that of astronauts.
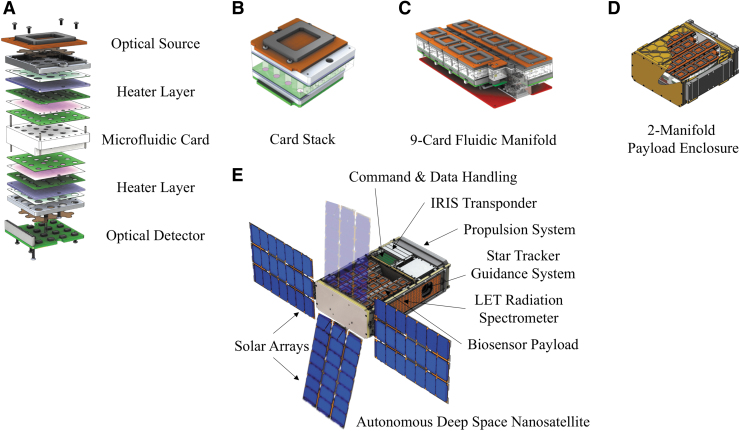
To measure the effects of ionizing radiation on yeast, cells are desiccated then rehydrated at specific time points over the course of the mission with a mixture of synthetic complete (SC) growth medium and alamarBlue (aB), a redox dye that serves as a metabolism and cell health indicator. Approximately 1 × 105 cells are dried inside 100-μL wells in polycarbonate microfluidic cards, two of which will be rehydrated at a time. These cards are then assembled into stacks which include filters, a thermal control system, and a three-color LED optical source and detector that takes optical measurements of the wells. The optical system has the capability to independently record changes to turbidity, as well as the colorimetric changes of aB, enabling both specific and unified interpretations of growth and metabolic data. Card stacks are placed on nine-card fluidic manifolds, two of which are integrated into the 4U biosensor payload enclosure along with the corresponding reagent bags, tubing, pumps, valves, and optical calibration cells (Fig. 1A–D). Attached to the 4U biosensor enclosure, a radiation sensor—a TimePix-based spectrometer—will measure the linear energy transfer (LET) of the different ion species that traverse the sensor.
FIG. 1.
BioSentinel payload architecture. (A, B) Yeast cells are loaded into the wells of microfluidic cards and air-dried in a trehalose solution. Fluidic cards are assembled into stacks, which include filters, a thermal control system, and a three-LED optical source and detector. (C) Card stacks are placed on a nine-card fluidic manifold. Each card is a distinct experiment that can be run autonomously. (D) Two 9-card manifolds and the corresponding reagent bags, fluidic pumps, tubing, and electronics are hermetically sealed in the 4U aluminum payload enclosure (shown here with cutaway displaying interior), with a radiation sensor attached to the side. One such payload will be flown on the International Space Station. (E) For the autonomous deep space nanosatellite, the biosensor payload is integrated with a solar-powered spacecraft bus, which contains propulsion and guidance systems, and a data transponder.
The BioSentinel CubeSat also contains a 2U spacecraft bus, which is solar powered, and houses propulsion, a guidance system, data storage, and a data transponder (Fig. 1E). The data gathered from the deep space nanosatellite will be telemetered back to Earth and compared to both synchronous ground controls and the ISS payload, which will serve as a microgravity control in addition to a probe for this distinct radiation environment. For each payload, yeast growth and metabolism rates will be correlated with the onboard physical radiation dosimetry and spectroscopy.
The biggest scientific risk to the success of the BioSentinel mission is long-term cell survival. Compared to human cells and many other model organisms, yeast is ideally suited for prolonged biosensor mission durations due to its ability to use anhydrobiosis. This rare and remarkable strategy, also found in organisms such as tardigrades (Boothby et al., 2017), nematodes (Madin and Crowe, 1975), insects (Sakurai et al., 2008), and prokaryotes (Potts, 1994), allows organisms to survive extreme desiccation and reversibly halt metabolic activity under these conditions (Dupont et al., 2014).
Due to its colonization of terrestrial interfacial niches, including the surface of soil, rocks, plants, and animals, S. cerevisiae is often subjected to severe fluctuations in hydration. As yeast cells undergo desiccation, they face mechanical, structural, and oxidative challenges, including intracellular crowding, plasma membrane lysis and permeabilization, structural modifications to proteins, protein aggregation, lipid phase transitions, and oxidative stress. To survive these insults, yeast has evolved a variety of endogenous protective mechanisms, including a uniquely elastic cell wall, the accumulation of intracellular glycerol and trehalose, and the induction of stress proteins and biochemical antioxidative systems. Rehydration can also cause significant stress to cells, and yeast uses many of the same antioxidative systems and stress proteins to abrogate the resulting damage (Dupont et al., 2014).
Although yeast is highly adapted to climatic instability and cellular and molecular stress, the viability of dry cell populations can decline under unfavorable environmental conditions, especially over prolonged periods (Dupont et al., 2014). Due to the possibility of launch slippage, BioSentinel's prelaunch period may last over 12 months. The experimental space flight phase of the mission will be ∼6 months, with a potential extension to 12 months. Such a long mission duration poses an acute viability challenge for a biological space flight experiment with autonomous and finite life support. To ensure adequate survival of S. cerevisiae for the entire BioSentinel mission duration, yeast preservation conditions were modulated to enhance endogenous protective mechanisms and to maximize viability. The described findings and methodology will inform the design of improved future biological missions into deep space.
2. Materials and Methods
2.1. Yeast strains
Standard genetic cross and tetrad dissection procedures were used to generate the wild type (YBS21-A) and rad51Δ (YBS29-1) strains used in this study. All strains are diploid prototrophic derivatives of the W303 background (MATa/MATα ADE2 CAN1 HIS3 LEU2 TRP1 URA3).
2.2. Growth and predrying preparation
Wild type and rad51Δ diploid strains from −80°C glycerol stocks were plated on yeast extract - peptone - dextrose (YPD) agar (Sigma-Aldrich; Y1500) and incubated at 30°C for 2–3 days. Samples from freshly grown patches were then used to inoculate 5-mL YPD liquid cultures (Sigma-Aldrich; Y1375), which were grown on a rotating mixer at ambient room temperature (∼23°C) for 7 days. Liquid cultures were pelleted and washed with 1 mL sterile deionized water and then resuspended in 1 mL sterile deionized water. Cell density was determined by hemocytometer counting, and suspensions were diluted to a final density of 1 × 107 cells/mL in 10% (w/v) trehalose (Sigma-Aldrich; T0167). Ten microliters of trehalose suspensions (containing ∼105 cells) were gently dispensed into the bottom edge of the wells of 96-well Stripwell™ microplates (Costar®; 9102). Undesiccated controls were plated on YPD agar (Sigma-Aldrich; Y1500) for colony counting.
2.3. Desiccation methods
2.3.1. Air-drying
Stripwell microplates containing 10% trehalose yeast cell preparations were covered with a Breathe-Easy® gas permeable sealing membrane (Sigma-Aldrich; Z380059) and air-dried without the microplate lid for 3 days in an incubator at 23°C and 20–30% relative humidity (R.H.). After air-drying, microplates were covered with lids (which continued to allow sterile gas exchange through the Breathe-Easy membrane) for long-term storage at 23°C and 0% R.H.
2.3.2. Vacuum drying
Stripwell microplates containing 10% trehalose yeast cell preparations were covered with the microplate lid and placed under vacuum at a final pressure of 100 mTorr for 24 h in an Advantage XL freeze dryer (SP Scientific; AD20XL). After vacuum drying, microplates were covered with a Breathe-Easy membrane and the microplate lid for long-term storage at 23°C and 0% R.H.
2.3.3. Freeze-drying
Stripwell microplates containing 10% trehalose yeast cell preparations were covered with the microplate lid and placed in an Advantage XL Freeze Dryer. Freeze-drying program parameters (Połomska et al., 2012) were as follows: −38°C, 760 Torr, 180 min; −38°C, 120 mTorr, 270 min; −20°C, 770 mTorr, 900 min; −10°C, 930 mTorr, 270 min; and 23°C, 120 mTorr, 55 min. After freeze-drying, microplates were covered with a Breathe-Easy membrane and the microplate lid for long-term storage at 23°C and 0% R.H.
2.4. Storage condition optimization
2.4.1. Relative humidity
Ten percent trehalose yeast cell preparations air-dried in Stripwell microplates and sealed with a Breathe-Easy membrane were stored at 23°C in various R.H. conditions for up to 1 year. Cells maintained at 0% R.H. were placed in a nonvacuum glass desiccator (Thermo Fisher Scientific; 08-615A) containing Drierite™ (W.A. Hammond Drierite™ Drying Desiccant; 21001). Cells maintained at 40–50% R.H. were placed in a glass desiccator containing a layer of saturated potassium carbonate salt solution at the bottom. Cells maintained at 100% R.H. were placed in a desiccator with a layer of Milli-Q water at the bottom. Each desiccator was equipped with an Onset HOBO UX100-011 Temp/RH Logger and sealed with parafilm. Viability over this period was determined using survival assay (see section 2.7).
2.4.2. Temperature
Ten percent trehalose yeast cell preparations air-dried in Stripwell microplates and sealed with a Breathe-Easy membrane were stored at 0% R.H. in a glass desiccator containing Drierite at 23°C for 7 months to mimic the “pre-launch” period at KSC. After 7 months, microwell plates were placed in sealed mylar bags containing Drierite (0% R.H.) and stored in incubators at 4°C, 9°C, 15°C, or 23°C for 6 months to mimic potential thermal space flight conditions. Viability following this period was determined using survival assay (see section 2.7).
2.5. SC medium preparation
SC medium was prepared with deionized water, Yeast Nitrogen Base without Amino Acids (Sigma-Aldrich; Y0626), d-(+)-glucose (Sigma-Aldrich; G5767), Yeast Synthetic Dropout (−) Leu (Sigma-Aldrich; Y1376), and 2% l-leucine (Sigma-Aldrich; L8000). A 1 M 3-(N-Morpholino)propanesulfonic acid (MOPS) (Sigma-Aldrich; M1254) solution, adjusted to pH 7.00 with 10 M NaOH (Sigma-Aldrich; 72068), was added to the medium for a final concentration of 50 mM MOPS. The pH of the medium was adjusted to pH 7.00 with 10 M NaOH (Sigma-Aldrich; 72068). The SC was then filter sterilized with a 0.22 μm filtration system (Corning®; 431096) and stored at 23°C. After 3 days, the medium was moved to 4°C overnight. The SC was then refiltered into sterile glass bottles (Pyrex) with a 0.22 μm filtration system (Corning; 431096) to remove precipitated crystals. Some bottles were frozen overnight at −20°C, thawed at room temperature, and filter sterilized again to remove further precipitation, and others were stored at 23°C as unfrozen controls.
2.6. Long-term reagent storage conditions and assay
Fluorinated ethylene propylene (FEP) bags (Saint-Gobain Life Sciences VueLife “C” Series) and glass bottles (Pyrex) were filled with SC medium or 10 × aB metabolic indicator dye (Invitrogen; DAL1100). FEP bags contained frozen and filtered medium, and glass bottles contained either frozen and filtered medium or unfrozen control medium. Glass bottles were tightly sealed and kept at 23°C in the dark. FEP bags were placed in a nonvacuum glass desiccator (Thermo Fisher Scientific; 08-615A) with a 350 mL layer of distilled water at the bottom (100% R.H.) to simulate vapor–liquid equilibrium within the reagent box in the payload. The glass desiccator was sealed with parafilm and stored at 23°C in the dark. Glass bottles and FEP bags were stored for 19 months.
Wild-type yeast from −80°C glycerol stocks was plated on YPD agar (Sigma-Aldrich; Y1500) and incubated at 23°C for 3 days. Single colonies were used to inoculate 5 mL fresh SC cultures, which were grown overnight on a rotating mixer at ambient room temperature (∼23°C). The OD600 was calculated, and the overnight culture was used to inoculate 12 mL of fresh SC at a starting OD600 of 0.2 (0.25 × 107 cells/mL). Cells were grown in an Erlenmeyer flask on an orbital shaker at 23°C until log phase (OD600 0.5–0.7). Cells were pelleted and resuspended in 1 mL sterile deionized water to prevent further growth. Cell density was determined by hemocytometer counting, and suspensions were diluted in sterile deionized water to a final density of 5 × 107 cells/mL.
SC and aB were withdrawn from fresh stocks, FEP bags, and glass bottles with a pipette or sterile syringe. 1 × aB + SC solutions were made for each storage condition. One hundred microliter of each 1 × aB + SC solution was inoculated with ∼105 cells and loaded into a clear 96-well nontreated polystyrene flat-bottom microplate (Falcon®; 351172) in triplicate. Plate readings were taken once per hour on a SpectraMax 250/340 plate reader at 690 nm to measure cell growth. A full spectrum plate reading of 100 μL of each solution (no cells) was also taken at 350–750 nm with a 5 nm increment to assess whether the reagents had undergone optical changes due to age or storage condition.
2.7. Survival assay
To determine cell viability at each time point, yeast cells desiccated in microplates were rehydrated with 100 μL of SC medium and incubated for 30 min. Cell suspensions were then pipette mixed and serially diluted in sterile deionized water. Appropriate dilutions were plated on YPD agar to count colony forming units.
2.8. High-energy heavy ion irradiation at NASA Space Radiation Laboratory
Wild-type and rad51Δ cells dried in 10% trehalose in Stripwell microplates, sealed with a Breathe-Easy membrane, and stored for 1 month before irradiation inside a 23°C incubator, 20–30% R.H., were placed in the beam line at NSRL and exposed to 0, 0.1, or 2.5 Gy (or 0, 10, 250 cGy, respectively) of 1 GeV protons (H-1). After sample release, three wells of dried cells per condition were rehydrated with 100 μL 1 × aB + SC, incubated for 30 min, then gently pipette mixed. Cell suspensions were then transferred to clear 96-well nontreated polystyrene flat-bottom microplates (Falcon; 351172) for plate reading.
2.9. aB assay
Following irradiation at NSRL, optical measurements of cells in 1 × aB + SC were taken once per hour on a SpectraMax 250/340 plate reader at 570, 600, and 690 nm. Percent reduction of aB was calculated for each experimental well using the following formula, which is modified from the manufacturer's manual to account for optical differences related to cell density (captured by absorbance readings at 690 nm):
where:
O1 = molar extinction coefficient (E) of oxidized aB (Blue) at 570 nm = 80,586 cm−1 M−1
O2 = E of oxidized aB at 600 nm = 117,216 cm−1 M−1
R1 = E of reduced aB (Red) at 570 nm = 155,677 cm−1 M−1
R2 = E of reduced aB at 600 nm = 14,652 cm−1 M−1
A1 = absorbance of test wells at 570 nm—absorbance of test wells at 690 nm
A2 = absorbance of test wells at 600 nm—absorbance of test wells at 690 nm
N1 = absorbance of negative control well (1 × aB + SC but no cells) at 570 nm—absorbance of negative control well at 690 nm
N2 = absorbance of negative control well (1 × aB + SC but no cells) at 600 nm—absorbance of negative control well at 690 nm
2.10. Statistical analyses
2.10.1. Survival assay
Percentages of viable cells were determined by normalizing colony counts at each time point to: undesiccated controls to analyze the effects of drying method; viability immediately following the conclusion of the desiccation process to analyze the effects of R.H. and optimized flight-like conditions; or viability following prolonged storage in the desiccated state (to simulate the prelaunch period plus a variable experimental period) to analyze the effect of stasis temperature. Plotted values represent the mean survival of three replicates, and bars represent one standard deviation. p-values were calculated by Student's t test. Differences between conditions were considered significant at p-values ≤0.05 (*p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001).
2.10.2. aB and cell growth assays
To assess cell growth characteristics, the average absorbance of the triplicates at 690 nm was plotted over time with standard deviation error bars. To observe whether medium preparation method or storage conditions had an impact on yeast growth kinetics, the slope between an OD690 of 0.1 and 0.5 was determined and slope values were compared between each medium condition by Student's t test. A p-value of <0.05 indicates a significant difference in growth rate between medium conditions. R2 values for the linear fit were >0.98.
To assess the metabolic effects of radiation, calculated triplicate values for percent reduction of aB in each dose condition were averaged and plotted over time with standard deviation error bars. For each experimental well, the slope between 7% and 52% reduction was determined, and slope values were compared between each radiation dose condition by Student's t test. A p-value of <0.05 denotes radiation sensitivity in comparisons with the 0 cGy dose and a significant dose-dependent difference in metabolic response to radiation between irradiation conditions. R2 values for the linear fit were >0.96.
To observe whether radiation exposure had an impact on yeast cell survival and, consequently, on growth kinetics, the average absorbance of the triplicates at 690 nm was plotted over time, with standard deviation error bars. For each experimental well, the slope between an OD690 of 0.1 and 0.4 was determined up to 20 h of growth, and slope values were compared between each radiation dose condition by Student's t test. A p-value of <0.05 indicates that radiation exposure resulted in a significant difference in growth rate, likely due to changes in the viability of the starting cell population. R2 values for the linear fit were >0.97.
3. Results
3.1. Among tested desiccation methods, air-drying is optimal for long-term yeast preservation
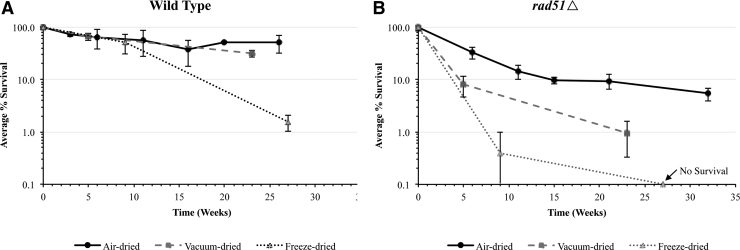
To mitigate viability declines in the wild-type and sensitized rad51Δ yeast strains during the prelaunch period at KSC and the 6- to 12-month experimental phase of the BioSentinel mission, a variety of drying methods were tested to assess effects on long-term survival (Fig. 2). As expected, both wild type and rad51Δ underwent a steep decline in survival by the first time point due to stress during the desiccation process. The subsequent time points illustrate viability loss due to rehydration stress and prolonged stasis in a dry state. Wild type had significantly better postdesiccation survival than rad51Δ—as a sensitized strain, rad51Δ is expected to accumulate unrepaired DNA damage and to be less robust than the wild-type strain in the face of deleterious conditions as a result.
FIG. 2.
Long-term survival of yeast following desiccation by three methods. (A) Wild type and (B) rad51Δ cells in 10% trehalose were air-dried, vacuum-dried, or freeze-dried in microwell plates and stored at 23°C, 0% R.H. Survival was measured after cell rehydration, plating, and incubation and normalized to undesiccated controls. Freeze-dried rad51Δ cells exhibited no survival at t = 27 weeks. R.H., relative humidity.
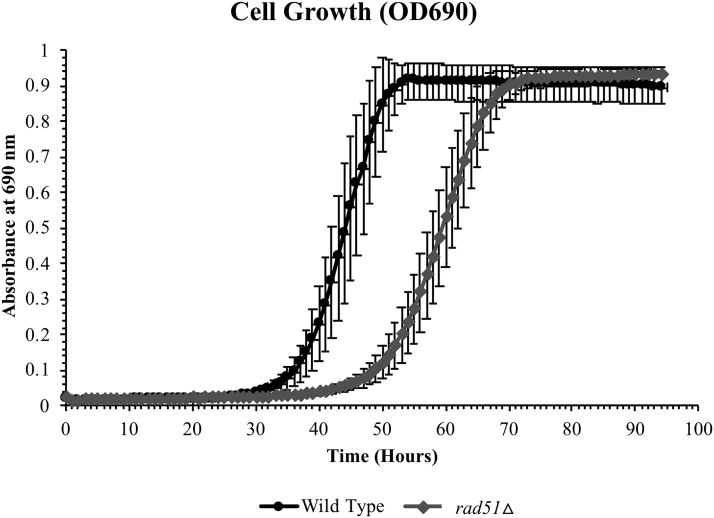
For both strains, air-drying resulted in the highest postdesiccation survival, followed by vacuum-drying and freeze-drying, although differences between the desiccation methods were less pronounced for wild-type cells. Following several months of storage in the dry state, air-dried cells continued to display the best viability for both strains, while freeze-dried cells had the lowest survival (Fig. 2). As a result of these findings, air-drying in 10% trehalose was selected as the desiccation method to prepare yeast cells for space flight. With this methodology, in conjunction with the subsequent optimization of storage conditions, the BioSentinel experiment will be able to withstand long periods of storage. Each of the wells in the fluidic cards contains ∼105 cells. Thus, even when viability is reduced to a fraction of the starting population over prolonged stasis, a sufficient number of viable cells will remain to successfully assay cell growth (Fig. 3).
FIG. 3.
Cell growth following long-term storage. Air-dried wild-type and rad51Δ cells in 10% trehalose were stored at 23%, 0% R.H., for 7 months to simulate the prelaunch period at KSC. Cells were then moved to 4–9°C, 0% R.H., for an additional 19 months to simulate flight-like conditions. Following this 26-month period, cells were rehydrated with 100 μL fresh 1 × aB + SC. aB, alamarBlue; SC, synthetic complete.
3.2. Desiccated yeast viability is maximized when cells are stored in low-temperature, low-humidity R.H. conditions
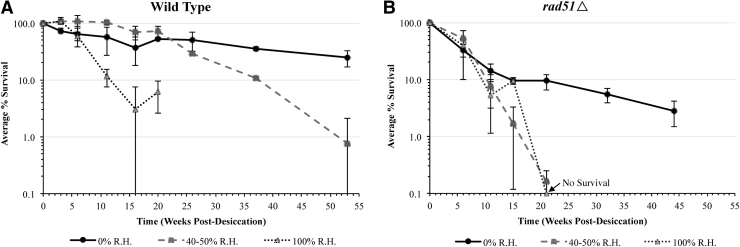
The BioSentinel aluminum payload enclosure will be hermetically sealed to maintain atmospheric pressure in the vacuum of space. This allows for the passive control of the R.H. inside the payload through the use of desiccant, during both the prelaunch storage period and the experimental phase after launch. To determine how R.H. affects the viability of dried yeast cells stored for prolonged periods, air-dried yeast were stored at 23°C at 0%, 40–50%, or 100% R.H (Fig. 4). Yeast cells will remain at 23°C only, while the experiment remains unpowered for the first 6–9 months of the mission before launch. We tested the effects of R.H. under this temperature for longer durations, as it is the highest stasis temperature the payload is likely to encounter. Cells were rehydrated and plated to assess viability following various lengths of storage in each R.H. condition. Viability readings at each time point were normalized to survival immediately following the initial desiccation process.
FIG. 4.
Long-term survival of air-dried yeast stored in varying R.H. conditions. Air-dried (A) wild-type and (B) rad51Δ cells in 10% trehalose were stored at 23°C in 0%, 40–50%, or 100% R.H. conditions for close to a year. Survival was measured after cell rehydration, plating, and incubation and normalized to survival immediately following the desiccation process at the start of the experiment. At t = 21, rad51Δ cells stored at 100% R.H. exhibited no survival, so wild-type survival measurements were also discontinued.
Storage at 0% R.H. resulted in the highest retention of viability, and viability declined with increasing R.H. for both strains after 21 weeks of storage, with 100% R.H. resulting in a complete survival loss for rad51Δ by that time (Fig. 4). Since low-humidity conditions were demonstrated to be ideal for long-term desiccated yeast survival, desiccant chambers in vapor contact with the microfluidic cards were incorporated into the BioSentinel payload design to ensure maintenance of low humidity conditions during space flight and to optimize prolonged yeast survival.
As mentioned previously, the BioSentinel spacecraft is expected to experience temperatures around 23°C before and during integration into the launch vehicle during the 6–9 months at KSC. During this period, the spacecraft will occasionally be taken outdoors to the launch pad and may experience temperatures higher than 23°C. To determine whether these excursions would have a significant impact on yeast viability, air-dried yeast was exposed to either 34°C or 40°C (the highest potential outdoor temperature according to annual averages), for 8 h a day for five consecutive days. These acute temperature insults did not significantly alter viability compared to cells that remained in 23°C storage (data not shown).
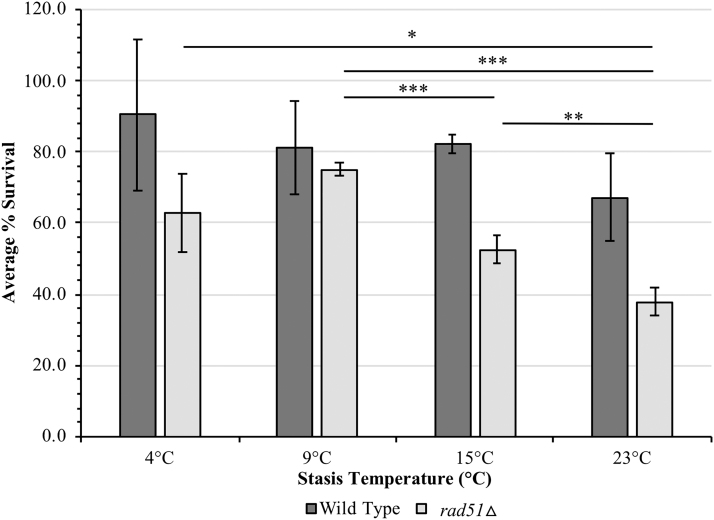
Since space is cold biased, the payload is designed to autonomously maintain a temperature of at least 4°C during the stasis periods of the mission with the use of heaters. However, communication passes, during which data are telemetered to Earth by the transponder, may cause the payload to experience higher average temperatures. Since the payload has no active cooling systems, yeast cell survival was tested at a variety of potential space flight stasis temperatures at 4°C and above. After air-drying in 10% trehalose, cells were stored for 7 months at 23°C, 0% R.H., to mimic the expected prelaunch temperature conditions, then moved to 4°C, 9°C, 15°C, or 23°C at 0% R.H. for an additional 6 months to mimic the experimental phase of the mission (Fig. 5). Following this 13-month space flight-like test, wild-type cells did not exhibit a significant survival difference between the various temperature conditions. The rad51Δ cells were more sensitive to thermal stress and displayed a significant temperature-dependent decrease in viability at 15°C and above (Fig. 5), indicating that the maintenance of cool conditions (4°C–9°C) within the payload is important for maximizing cell survival during stasis periods.
FIG. 5.
Long-term survival of air-dried yeast stored at varying “flight-like” stasis temperatures. Air-dried wild-type and rad51Δ cells in 10% trehalose were stored at 23°C, 0% R.H., for 7 months to mimic the prelaunch period. Data are normalized to the cell survival following this period. The strains were then moved to a variety of potential “flight-like” stasis temperatures for an additional 6 months. Survival following this 13-month period was measured after cell rehydration, plating, and incubation. *p ≤ 0.05; **p ≤ 0.01; ***p ≤ 0.001.
3.3. Desiccated yeast survives for Mars mission-relevant periods in optimized flight-like conditions
To determine how long wild-type and rad51Δ cells could survive under flight-like conditions using the optimal conditions indicated by prior testing, the strains air-dried in 10% trehalose were first stored at 23°C, 0% R.H., for 8 months to simulate a prolonged prelaunch period. The cells were then moved to 4°C, 0% R.H., for another 16 months to simulate an extended space flight mission. Following this period, wild-type cells exhibited an average survival of over 18%. As expected, rad51Δ cells had a greater viability loss, with an average survival of over 1% (Fig. 6). For both strains, survival at ∼2 years postdesiccation is still sufficient to show growth (Fig. 3). Since the experimental phase of the BioSentinel mission has the possibility of being extended from 6 to 12 months, these data indicate that there is little risk of complete viability loss, particularly for rad51Δ, if a relatively low-temperature, low-humidity environment is maintained within the payload.
FIG. 6.
Long-term survival of air-dried yeast stored in optimized flight-like conditions. Air-dried wild-type and rad51Δ cells in 10% trehalose were stored at 23°C, 0% R.H., for 8 months to simulate the prelaunch period at KSC. Cells were then moved to 4°C, 0% R.H., for an additional 16 months to simulate flight-like conditions. Survival was measured after cell rehydration, plating, and incubation and is normalized to survival immediately following desiccation. KSC, Kennedy Space Center.
3.4. Prolonged reagent storage does not alter optical characteristics or cell growth
Alongside the optimization of cell viability conditions, materials and reagents to be used in the experiment were also tested to ensure that they would withstand the long durations of the BioSentinel mission. To determine whether prolonged storage of 10 × aB or the SC medium in the payload would alter cell growth or the optical characteristics of the metabolic indicator dye and growth medium mixture, each reagent was stored in FEP bags planned for use in flight or in glass bottles for 19 months. The SC medium was either unfrozen or frozen at −20°C and filter sterilized to remove precipitated crystals, to test whether freezing and precipitate filtration could also have undesirable effects on the reagents.
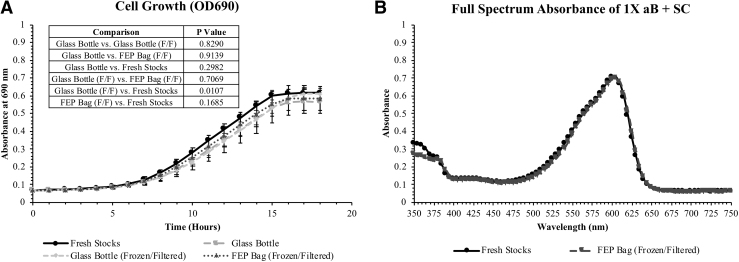
Long-term storage of the reagents in FEP bags did not alter cell growth or result in significant optical differences compared to controls, indicating that FEP bags can be used for prolonged medium storage in the BioSentinel payload (Fig. 7). Similarly, the freezing and filtration of the growth medium did not lead to optical differences or change the growth kinetics of the yeast. Thus, this methodology can be effectively used to prepare reagents for space flight without risk of altering their properties or mission findings (Fig. 7).
FIG. 7.
Optical and growth effects of long-term reagent storage. (A) Cell growth of undesiccated wild-type yeast in 100 μL 1 × aB + SC. Cell growth in fresh 1 × aB + SC (fresh stocks) was compared to growth in reagents stored in glass bottles or FEP bags for 19 months at 23°C. The media in the glass bottles were either frozen and filtered to remove precipitated crystals before long-term storage or were unfrozen. Media stored in FEP bags were frozen and filter sterilized to remove precipitated crystals before storage. (B) Full spectrum absorbance of 1 × aB + SC (no cells). FEP, fluorinated ethylene propylene.
3.5. Desiccated yeast cells are sensitive to deep space-relevant ionizing radiation
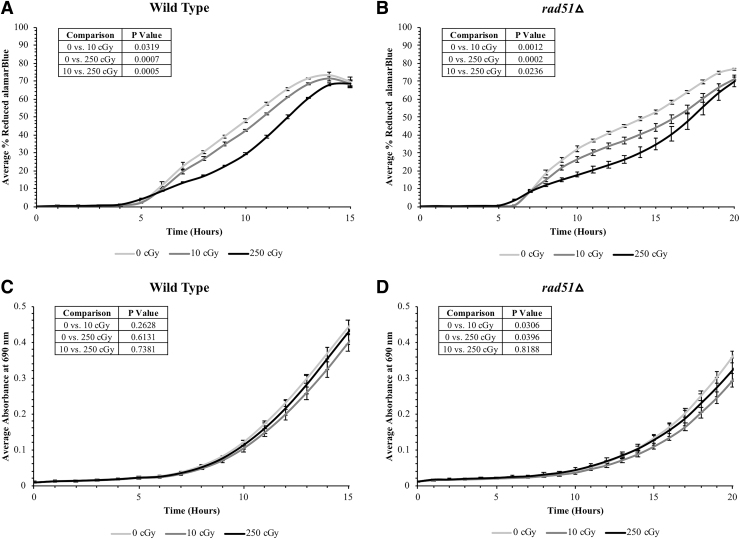
To test whether yeast cells air-dried in trehalose are sensitive to doses and species of ionizing radiation relevant to the deep space environment, wild-type and rad51Δ cells were exposed to 1 GeV protons at the NASA Space Radiation Laboratory (NSRL) in Brookhaven National Laboratory, NY. Cells were irradiated with 0, 10, or 250 cGy doses in a desiccated state and then rehydrated and grown in 1 × aB + SC to assess the growth and metabolic effects of the exposures. Both strains displayed a significant decrease in the rate at which they reduced aB with increasing doses of radiation (Fig. 8A, B). This indicates the presence of dose-dependent physiological changes induced by ionizing radiation at doses as low as 10 cGy. This effect was more pronounced for rad51Δ cells, likely due to its defective DNA damage repair pathway and resultant radiosensitivity (Fig. 8B).
FIG. 8.
Desiccated yeast is sensitive to low doses of deep space relevant ionizing radiation. Air-dried (A, C) wild-type and (B, D) rad51Δ cells in 10% trehalose were irradiated with 0, 10, or 250 cGy of 1 GeV protons at the NASA Space Radiation Laboratory. Cells were then rehydrated and grown in 100 μL 1 × aB in SC medium, then assessed for (A, B) metabolic activity and (C, D) growth kinetics.
Wild-type cells grow and metabolize faster than rad51Δ cells; therefore, both the aB reduction (Fig. 8B) and the OD690 growth responses (Fig. 8D) of rad51Δ cells were monitored for a slightly longer time period. Wild-type cells did not exhibit a significant difference in growth rate between radiation doses (Fig. 8C), indicating that the observed metabolic effects in aB reduction profiles are likely due to changes in the activity of live cells, rather than due to loss of survival in the starting cell population following radiation exposure. rad51Δ cell growth was slightly different between unirradiated and irradiated cells, but not in a dose-dependent manner (Fig. 8D). This indicates that rad51Δ cell populations may have undergone some viability loss following irradiation, which is to be expected for a radiosensitive mutant. However, it appears likely that survivability is not the underlying cause of the dose-responsive sensitivity exhibited in the metabolic assay. Therefore, for both wild type and rad51Δ, the observed shift in metabolic response with increasing radiation dose is likely due to a live-cell response to radiation exposure.
4. Discussion
Budding yeast is an ideal model organism for querying the deep space radiation environment due to its unique ability to undergo anhydrobiosis and survive for prolonged periods in a desiccated state. The dehydration and rehydration of S. cerevisiae induce a variety of adverse effects that cells must mitigate with protective endogenous molecular mechanisms to avoid lysis and cell death. These adaptive mechanisms, particularly the stress-resistance functions of trehalose, informed our optimization approach for long-term yeast preservation and also point to additional avenues for improving yeast desiccation tolerance.
During dehydration, yeast cells are faced with mechanical constraints and osmotic stress, which can result in osmotic shrinkage, permeabilization, and rupture of the plasma membrane and intracellular crowding of macromolecules and organelles, which in turn can affect protein function and intracellular signaling speed (Dupont et al., 2011, 2014; Babazadeh et al., 2013; Miermont et al., 2013). S. cerevisiae resists osmotic stress through the accumulation of intracellular solutes, including trehalose and glycerol, and resists mechanical perturbations with unique cell wall elasticity, the ability to undergo lateral plasma membrane organization, and the high concentration of ergosterol in the plasma membrane, which increases its capacity to deform and recover without vesicle formation and lysis (Schneiter et al., 1999; Tamás et al., 1999; Dupont et al., 2010; Kodedová and Sychrová, 2015).
In addition to the mechanical constraints caused by osmotic stress, yeast cells are faced with structural challenges during dehydration (Despa, 2005; Ball, 2008). The loss of water leads to the denaturation of proteins and lipid phase transitions in membranes (Carpenter and Crowe, 1989; Tokumasu et al., 2003; Veatch and Keller, 2003, 2005), which result in membrane reorganization, permeabilization, and solute leakage during rehydration (Crowe et al., 1989, 1992; Fernández Murga et al., 1999; Hays et al., 2001). To resist these deleterious effects, yeast utilizes the disaccharide trehalose and stress proteins like hydrophilins, which help stabilize membranes, protect enzyme activity, and prevent protein aggregation either through direct interactions or by molecular shielding (Crowe et al., 1992; Goyal et al., 2005; Reyes et al., 2005; Tunnacliffe and Wise, 2007).
S. cerevisiae is also subjected to oxidative stress during dehydration and during rehydration. Following dehydration, yeast cells can exhibit a 10-fold increase in oxidation level, which is attributable to the production of free radical species, including reactive oxygen species (ROS) (de Jesus Pereira et al., 2003). When oxidative damage repair or stress–response mechanisms are overwhelmed by excessive ROS generation, severe damage to proteins, lipids, and DNA can result (Leprince et al., 1994; de Jesus Pereira et al., 2003), and DNA repair mutants like rad51Δ will be particularly ill-equipped to counteract such damage. Yeast cells have evolved antioxidative biochemical systems to defend against the oxidative damage, including glutathione and thioredoxin, protective nonenzymatic scavengers of ROS (Jamieson, 1998), and the enzymes superoxide dismutase (Longo et al., 1996), catalase (França et al., 2005), and peroxidases (Jamieson, 1998), which catalyze ROS degradation. These oxidative stress pathways are also implicated in the physiological response to ionizing radiation (Azzam et al., 2012). We are therefore interested in elucidating the transcriptomic response of yeast cells to deep space-relevant ionizing radiation before attempting to increase desiccation tolerance by perturbing these pathways; a better understanding of live-cell behavior after radiation exposure will enable us to balance radiation sensitivity and desiccation hardiness in future biosensors.
The primary defense of S. cerevisiae against both the structural and oxidative constraints imposed by desiccation is an accumulation of trehalose in the cytosol (Dupont et al., 2014). Trehalose is a nonreducing disaccharide of glucose linked by an alpha-1-1 glycosidic bond that is widespread in the biological world and particularly relevant to anhydrobiotes (Elbein et al., 2003; Tapia and Koshland, 2014). In S. cerevisiae, it has been correlated with tolerance to a variety of adverse environmental conditions in addition to desiccation, including osmotic stress (Hounsa et al., 1998), lethal ethanol stress (Bandara et al., 2009), oxidative stress (Benaroudj et al., 2001), and heat stress (Lee and Goldberg, 1998; Singer and Lindquist, 1998). The mechanistic role of trehalose in desiccation resistance relates to its ability to replace water during dehydration and form hydrogen bonds with macromolecules (Clegg et al., 1982). As a result, trehalose preserves the native folding structure of proteins, prevents protein aggregation, and maintains the structure and fluidity of membranes (Golovina et al., 2009, 2010; Jain and Roy, 2009).
Trehalose also likely confers desiccation and oxidative stress resistance through its ability to form a glass in the cytosol in low-water conditions (Dupont et al., 2014). This vitreous matrix may inhibit the release of free radicals, as well as slow the rate of other deleterious reactions due to its viscosity (Sun and Leopold, 1997). In addition, trehalose itself has been shown to act as a free radical scavenger (Benaroudj et al., 2001; Herdeiro et al., 2006). Due to this pluralistic protective activity, Tapia et al. hypothesized that trehalose alone is sufficient to confer desiccation tolerance in S. cerevisiae in the absence of other stress effectors. They found that increasing intracellular trehalose content by import from growth media can confer high levels of desiccation tolerance (Tapia et al., 2015). Furthermore, intracellular trehalose accumulates in stationary-phase cells and in response to nutrient starvation and results in a significant decrease in sensitivity to drying (Lillie and Pringle, 1980; Hounsa et al., 1998; François and Parrou, 2001).
Given the clear role of trehalose as a stress effector for multiple constraints imposed by desiccation, a prolonged growth period to stationary phase in liquid culture was selected for the BioSentinel mission yeast preparation. Similar strategies have been used previously in stationary-phase yeast cells, either by adding external trehalose in the growth medium or before desiccation (Gadd et al., 1987; Tapia and Koshland, 2014; Tapia et al., 2015). This methodology preconditions cells for desiccation with nitrogen starvation, thereby stimulating an increase in the intracellular content of the disaccharide and improving long-term desiccation tolerance.
After testing a variety of yeast drying buffers, including water, phosphate buffered saline (PBS), microbial freeze-drying buffer (OPS Diagnostics), 10% trehalose, and 10% trehalose in PBS (data not shown), cells air-dried in 10% trehalose buffer had the greatest retention of viability, which is consistent with the benefits of exogenous trehalose for dehydration resistance reported in the literature (Gadd et al., 1987; Tapia and Koshland, 2014; Tapia et al., 2015).
Even though Gadd et al. (1987) observed higher cell survival with higher concentrations of external trehalose (∼19% and above), they did see ∼80% survival with an equivalent concentration to the one used in our studies. It is important to note, however, that they used freeze-drying as their desiccation method and checked for cell survival after a short period of time, while air-drying proved to be far superior for our long-term experiments. Interestingly, Tapia et al. (2015) noticed that higher concentrations of trehalose in growth media (> 5%) did not increase desiccation tolerance; however, they observed significantly lower cell survival after desiccation compared to our data. The differences in cell survival could be attributed to the different strains used and the actual desiccation approach (i.e., external trehalose in growth medium vs. air-drying in 10% trehalose). Consequently, 10% trehalose was selected as the yeast air-drying buffer for the BioSentinel mission to enhance desiccation tolerance.
The natural strategies S. cerevisiae uses to resist desiccation stress point to additional ways in which long-term yeast survival could be improved. However, mutants that are defective in trehalose biosynthesis, hydrophilins, ROS scavenging, and responses to osmotic stress have been shown to retain levels of desiccation tolerance similar to that of wild-type yeast (Calahan et al., 2011). This indicates that desiccation tolerance is likely conferred by a constellation of cooperative stress effectors rather than discrete mechanisms. For future biosensor missions and experiments that require long-term preservation of yeast, it is worth examining the constructive interference and relative impact of each of these endogenous protective molecular mechanisms. Presently, growing yeast to stationary phase and providing extracellular trehalose appear to be the best methods of improving long-term desiccation tolerance.
Air-drying, freeze-drying, and vacuum-drying methods were assessed to elucidate the viability impacts on yeast cells in 10% trehalose. Freeze-drying is often used to prepare S. cerevisiae for commercial and industrial use. A previous study in which yeast cells were used observed almost 100% survival in stationary-phase cells after freeze-drying when provided with external trehalose (Gadd et al., 1987). However, due to the limitations on cell density for space flight and the sensitivity of the rad51Δ strain in our particular mission, it was unclear whether this method would allow for sufficient survival, given that freeze-drying can result in a loss of viability due to thawing and structural and physiological injury to the cells (Mille et al., 2005). Vacuum-drying was also explored due to the lack of oxygen exposure in this method—it could potentially mitigate oxidative stress-induced cell death. Interestingly, for both wild-type and rad51Δ strains, slow convective air-drying led to the highest long-term viability, followed by vacuum-drying and freeze-drying.
Prolonged survival of yeast cells air-dried in 10% trehalose was then examined at a variety of R.H. conditions to simulate potential payload environments during space flight. Both yeast strains exhibited the best survival following storage at 0% R.H. These findings led to the incorporation of desiccant chambers into the payload design to maintain a low-humidity environment throughout the mission. These results are consistent with indications in the literature that low residual moisture is optimal for long-term preservation of dried yeast (Mille et al., 2005; Dupont et al., 2014).
Potential thermal space flight conditions were also tested to determine impacts on long-term survival of yeast cells. As expected, prolonged survival was optimal at 4°C–9°C, the cooler temperatures tested, likely because lower temperature storage serves to slow the rate of protein degradation and other deleterious reactions. These findings led to a 5°C stasis temperature set point for the nonexperimental phases of the mission, when allowed.
Long-term yeast survival was then evaluated under these optimized flight-like drying and storage conditions. Both strains retained sufficient viability following desiccation and long-term storage to support an 8-month prelaunch period and up to 16 additional months in deep space, illustrating the robustness of this anhydrobiotic organism. Furthermore, freezing, filtration, and prolonged storage of reagents used for growing yeast and detecting its metabolic response to radiation did not alter their optical characteristics or the growth of the cells. These results validated yeast survival and reagent integrity for an extended mission duration.
Finally, yeast cells air-dried in 10% trehalose were exposed to high-energy protons at space flight- and human-relevant doses as low as 10 cGy. Both wild-type and rad51Δ strains exhibited a significant dose-dependent response to this radiation exposure, providing proof-of-principle for the ability of the BioSentinel mission to detect the biological response to radiation using this robust model organism. These sensitivity assays are based on cell growth and metabolic activity through continuous optical measurements in liquid cultures. To the best of our knowledge, this is the first time that yeast cells have shown such low sensitivity to ionizing radiation, compared to previous space experiments limited mostly to cell survival and mutagenesis tests after exposure to high radiation levels (Kiefer and Pross, 1999; Pross and Kiefer, 1999; Pross et al., 2000; Takahashi et al., 2001). These short duration studies were performed in LEO, behind Earth's protective magnetosphere, and thus required treatment of yeast cells with very large radiation doses (20 Gy or more) before flight (Kiefer and Pross, 1999; Pross and Kiefer, 1999; Takahashi et al., 2001) or during flight (Pross et al., 2000) using external sources. They also required sample return back to Earth to process the samples, which could have negative effects on the experiment if not handled properly. This is in big contrast to BioSentinel, where our biosensor cells will be exposed to the actual deep space radiation environment for a minimum of 6 months and will allow us to observe the response to cumulative radiation damage at different time points throughout the mission and to compare the biological response with radiation data—both total dose and radiation quality—provided by an onboard LET spectrometer. Furthermore, this system enables us to monitor cell population density and metabolic activity independently, allowing us to parse the effects of ionizing radiation on viability from nonlethal physiological responses. Even when viable cell populations are not significantly impacted by a relatively low radiation dose, we can nevertheless ascertain radiation effects from the metabolic response. This level of sensitivity is promising for deep space biosensors, as a relatively short period of ionizing radiation exposure at low doses may allow us to observe significant effects.
Our current and future work seeks to quantify the growth, metabolic, and transcriptomic effects caused by the yeast desiccation process and by irradiation with space-relevant particles, not only of single ion species but also of simulated GCRs and SPEs. The results of these studies will be reported elsewhere. These exciting data affirm BioSentinel's promise as a unique and effective biological tool for future Mars and Lunar missions beyond LEO.
5. Conclusions
Deep space has a complex and dangerous radiation environment. Using the well-characterized genetic pathways, the homology to other eukaryotic organisms, and the desiccation tolerance of the model organism S. cerevisiae, BioSentinel is NASA's first biological mission in nearly half a century that will inform with regard to the risks of prolonged deep space travel. By enhancing yeast's ability to defend against the deleterious effects of desiccation, cells can be stored for extensive periods and survive with minimal life support, while retaining sufficient sensitivity to query the biological effects of deep space radiation. BioSentinel's findings will help enable intrepid explorers to go further into our solar system than they have ever ventured before.
Acknowledgments
This work was supported by NASA's Advanced Exploration Systems (AES) Program Office. For critical feedback and technical editing, the authors thank Diana M. Gentry, Michael R. Padgen, and Antonio J. Ricco.
Abbreviations Used
- aB
alamarBlue
- DSB
double stranded break
- FEP
fluorinated ethylene propylene
- GCR
galactic cosmic ray
- ISS
International Space Station
- KSC
Kennedy Space Center
- LEO
low Earth orbit
- LET
linear energy transfer
- NSRL
NASA Space Radiation Laboratory
- PBS
phosphate-buffered saline
- R.H.
relative humidity
- ROS
reactive oxygen species
- SC
synthetic complete
- SLS
space launch system
- SPE
solar particle event
Author Disclosure Statement
No competing financial interests exist.
Funding Information
This work was supported by NASA's Advanced Exploration Systems Division in the Human Exploration and Operations Mission Directorate.
Associate Editor: Petra Rettberg
References
- Azzam, E.I., Jay-Gerin, J.-P., and Pain, D. (2012) Ionizing radiation-induced metabolic oxidative stress and prolonged cell injury. Cancer Lett 327:48–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babazadeh, R., Adiels, C.B., Smedh, M., Petelenz-Kurdziel, E., Goksöro, M., and Hohmann, S. (2013) Osmostress-induced cell volume loss delays yeast Hog1 signaling by limiting diffusion processes and by Hog1-specific effects. PLoS One 8:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball, P. (2008) Water as an active constituent in cell biology. Chem Rev 108:74–108. [DOI] [PubMed] [Google Scholar]
- Bandara, A., Fraser, S., Chambers, P.J., and Stanley, G.A. (2009) Trehalose promotes the survival of Saccharomyces cerevisiae during lethal ethanol stress, but does not influence growth under sublethal ethanol stress. FEMS Yeast Res 9:1208–1216. [DOI] [PubMed] [Google Scholar]
- Bärtsch, S., Kang, L.E., and Symington, L.S. (2000) RAD51 is required for the repair of plasmid double-stranded DNA gaps from either plasmid or chromosomal templates. Mol Cell Biol 20:1194–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann, P. and West, S.C. (1998) Role of the human RAD51 protein in homologous recombination and double-stranded-break repair. Trends Biochem Sci 23:247–251. [DOI] [PubMed] [Google Scholar]
- Benaroudj, N., Lee, D.H., and Goldberg, A.L. (2001) Trehalose accumulation during cellular stress protects cells and cellular proteins from damage by oxygen radicals. J Biol Chem 276:24261–24267. [DOI] [PubMed] [Google Scholar]
- Bennett, C.B., Lewis, L.K., Karthikeyan, G., Lobachev, K.S., Jin, Y.H., Sterling, J.F., and Resnick, M.A. (2001) Genes required for ionizing radiation resistance in yeast. Nat Genet 29:426–434. [DOI] [PubMed] [Google Scholar]
- Boothby, T.C., Tapia, H., Brozena, A.H., Piszkiewicz, S., Smith, A.E., Giovannini, I., and Goldstein, B. (2017) Tardigrades use intrinsically disordered proteins to survive desiccation. Mol Cell 65:975–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calahan, D., Dunham, M., Desevo, C., and Koshland, D.E. (2011) Genetic analysis of desiccation tolerance in Saccharomyces cerevisiae. Genetics 189:507–519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter, J.F. and Crowe, J.H. (1989) An infrared spectroscopic study of the interactions of carbohydrates with dried proteins. Biochemistry 28:3916–3922. [DOI] [PubMed] [Google Scholar]
- Clegg, J.S., Seitz, P., Seitz, W., and Hazlewood, C.F. (1982) Cellular responses to extreme water loss: the water-replacement hypothesis. Cryobiology 19:306–316. [DOI] [PubMed] [Google Scholar]
- Crowe, J.H., Crowe, L.M., and Hoekstra, F.A. (1989) Phase transitions and permeability changes in dry membranes during rehydration. J Bioenerg Biomembr 21:77–91. [DOI] [PubMed] [Google Scholar]
- Crowe, J.H., Hoekstra, F.A., and Crowe, L.M. (1992) Anhydrobiosis. Annu Rev Physiol 54:579–599. [DOI] [PubMed] [Google Scholar]
- Cucinotta, F.A. (2014) Space radiation risks for astronauts on multiple international space station missions. PLoS One 9:16–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daly, M. (2012) Death by protein damage in irradiated cells. DNA Repair 11:12–21. [DOI] [PubMed] [Google Scholar]
- De Jesus Pereira, E., Panek, A.D., and Eleutherio, E.C.A. (2003) Protection against oxidation during dehydration of yeast. Cell Stress Chaperones 8:120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Despa, F. (2005) Biological water: its vital role in macromolecular structure and function. Ann N Y Acad Sci 1066:1–11. [DOI] [PubMed] [Google Scholar]
- Dupont, S., Beney, L., Ritt, J.F., Lherminier, J., and Gervais, P. (2010) Lateral reorganization of plasma membrane is involved in the yeast resistance to severe dehydration. Biochim Biophys Acta Biomembr 1798:975–985. [DOI] [PubMed] [Google Scholar]
- Dupont, S., Beney, L., Ferreira, T., and Gervais, P. (2011) Nature of sterols affects plasma membrane behavior and yeast survival during dehydration. Biochim Biophys Acta Biomembr 1808:1520–1528. [DOI] [PubMed] [Google Scholar]
- Dupont, S., Rapoport, A., Gervais, P., and Beney, L. (2014) Survival kit of Saccharomyces cerevisiae for anhydrobiosis. Appl Microbiol Biotechnol 98:8821–8834. [DOI] [PubMed] [Google Scholar]
- Elbein, A.D., Pan, Y.T., Pastuszak, I., and Carroll, D. (2003) New insights on trehalose: a multifunctional molecule. Glycobiology 13:17–27. [DOI] [PubMed] [Google Scholar]
- Elgazzar, A.H. and Kazem, N. (2015) Biological effects of ionizing radiation. In: Elgazzar, A.H. (eds) The Pathophysiologic Basis of Nuclear Medicine. Springer, Cham, Switzerland, pp 540–548. [Google Scholar]
- Fernández Murga, M.L., Bernik, D., Font De Valdez, G., and Disalvo, A.E. (1999) Permeability and stability properties of membranes formed by lipids extracted from Lactobacillus acidophilus grown at different temperatures. Archiv Biochem Biophys 364:115–121. [DOI] [PubMed] [Google Scholar]
- França, M.B., Panek, A.D., and Araujo Eleutherio, E.C. (2005) The role of cytoplasmic catalase in dehydration tolerance of Saccharomyces cerevisiae. Cell Stress Chaperones 10:167–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- François, J. and Parrou, J.L. (2001) Reserve carbohydrates metabolism in the yeast Saccharomyces cerevisiae. FEMS Microbiol Rev 25:125–145. [DOI] [PubMed] [Google Scholar]
- Gadd, G.M., Chalmers, K., and Reed, R.H. (1987) The role of trehalose in dehydration resistance of Saccharomyces cerevisiae. FEMS Microbiol Lett 48:249–254. [Google Scholar]
- Golovina, E.A., Golovin, A.V, Hoekstra, F.A., and Faller, R. (2009) Water replacement hypothesis in atomic detail—factors determining the structure of dehydrated bilayer stacks. Biophys J 97:490–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golovina, E.A., Golovin, A., Hoekstra, F.A., and Faller, R. (2010) Water replacement hypothesis in atomic details: effect of trehalose on the structure of single dehydrated POPC bilayers. Langmuir 26:11118–11126. [DOI] [PubMed] [Google Scholar]
- Goyal, K., Walton, L.J., and Tunnacliffe, A. (2005) LEA proteins prevent protein aggregation due to water stress. Biochem J 388:151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hays, L.M., Crowe, J.H., Wolkers, W., and Rudenko, S. (2001) Factors affecting leakage of trapped solutes from phospholipid vesicles during thermotropic phase transitions. Cryobiology 102:88–102. [DOI] [PubMed] [Google Scholar]
- Herdeiro, R.S., Pereira, M.D., Panek, A.D., and Eleutherio, E.C.A. (2006) Trehalose protects Saccharomyces cerevisiae from lipid peroxidation during oxidative stress. Biochim Biophys Acta 1760:340–346. [DOI] [PubMed] [Google Scholar]
- Hounsa, C.-G., Brandt, E.V., Thevelein, J., Hohmann, S., and Prior, B.A. (1998) Role of trehalose in survival of Saccharomyces cerevisiae under osmotic stress. Microbiology 144:671–680. [DOI] [PubMed] [Google Scholar]
- Jain, N.K. and Roy, I. (2009) Effect of trehalose on protein structure. Protein Sci 18:24–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson, D.J. (1998) Oxidative stress responses of the yeast Saccharomyces cerevisiae. Yeast 1527:1511–1527. [DOI] [PubMed] [Google Scholar]
- Kachroo, A.H., Laurent, J.M., Yellman, C.M., Meyer, A.G., Wilke, C.O., and Marcotte, E.M. (2015) Systematic humanization of yeast genes reveals conserved functions and genetic modularity. Science 348:921–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer, J. and Pross, H.D. (1999) Space radiation effects and microgravity. Mutat Res 430:299–305. [DOI] [PubMed] [Google Scholar]
- Kodedová, M. and Sychrová, H. (2015) Changes in the sterol composition of the plasma membrane affect membrane potential, salt tolerance and the activity of multidrug resistance pumps in Saccharomyces cerevisiae. PLoS One 10:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D.H. and Goldberg, A.L. (1998) Proteasome inhibitors cause induction of heat shock proteins and trehalose, which together confer thermotolerance in Saccharomyces cerevisiae. Mol Cell Biol 18:30–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince, O., Atherton, N.M., Deltour, R., and Hendry, G.A.F. (1994) The involvement of respiration in free radical processes during loss of desiccation tolerance in germinating Zea mays L. Plant Physiol 104:1333–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie, S.H. and Pringle, J.R. (1980) Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J Bacteriol 143:1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longo, V.D., Gralla, E.B., and Valentine, J.S. (1996) Superoxide dismutase activity is essential for stationary phase survival in Saccharomyces cerevisiae: mitochondrial production of toxic oxygen species in vivo. J Biol Chem 271:12275–12280. [DOI] [PubMed] [Google Scholar]
- Madin, K.A.C. and Crowe, J.H. (1975) Anhydrobiosis in nematodes: carbohydrate and lipid metabolism during rehydration. J Exp Zool 193:335–342. [Google Scholar]
- Miermont, A., Waharte, F., Hu, S., McClean, M.N., Bottani, S., Leon, S., and Hersen, P. (2013) Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding. Proc Natl Acad Sci U S A 110:5725–5730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mille, Y., Girard, J.P., Beney, L., and Gervais, P. (2005) Air drying optimization of Saccharomyces cerevisiae through its water-glycerol dehydration properties. J Appl Microbiol 99:376–382. [DOI] [PubMed] [Google Scholar]
- NASA. (2018) Moon to Mars: Around the Moon with NASA's First Launch of SLS with Orion. Available online at https://nasa.gov/feature/around-the-moon-with-nasa-s-first-launch-of-sls-with-orion
- Połomska, X., Wojtatowicz, M., Zarowska, B., Szołtysik, M., and Chrzanowska, J. (2012) Freeze-drying preservation of yeast adjunct cultures for cheese production. Pol J Food Nutr Sci 62:143–150. [Google Scholar]
- Potts, M. (1994) Desiccation tolerance of prokaryotes. Microbiol Rev 58:755-805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pross, H.D. and Kiefer, J. (1999) Repair of cellular radiation damage in space under microgravity conditions. Radiat Environ Biophys 38:133–138. [DOI] [PubMed] [Google Scholar]
- Pross, H.D., Casares, A., and Kiefer, J. (2000) Induction and repair of DNA double-strand breaks under irradiation and microgravity. Radiat Res 153:521–525. [DOI] [PubMed] [Google Scholar]
- Reyes, J.L., Rodrigo, M.J., Colmenero-Flores, J.M., Gil, J.V., Garay-Arroyo, A., Campos, F., and Covarrubias, A.A. (2005) Hydrophilins from distant organisms can protect enzymatic activities from water limitation effects in vitro. Plant Cell Environ 28:709–718. [Google Scholar]
- Sakurai, M., Furuki, T., Akao, K., Tanaka, D., Nakahara, Y., Kikawada, T., Watanabe, M., and Okuda, T. (2008) Vitrification is essential for anhydrobiosis in an African chironomid, Polypedilum vanderplanki. Proc Natl Acad Sci U S A 105:5093–5098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneiter, R., Brügger, B., Sandhoff, R., Zellnig, G., Leber, A., Lampl, M., and Kohlwein, S.D. (1999) Electrospray ionization tandem mass spectrometry (ESI-MS/MS) analysis of the lipid molecular species composition of yeast subcellular membranes reveals acyl chain-based sorting/remodeling of distinct molecular species en route to the plasma membrane. J Cell Biol 146:741–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M.A. and Lindquist, S. (1998) Thermotolerance in Saccharomyces cerevisiae: the Yin and Yang of trehalose. Trends Biotechnol 16:460–468. [DOI] [PubMed] [Google Scholar]
- Straume, T., Slaba, T.C., Bhattacharya, S., and Braby, L.A. (2017) Cosmic-ray interaction data for designing biological experiments in space. Life Sci Space Res 13:51–59. [DOI] [PubMed] [Google Scholar]
- Sun, W.Q. and Leopold, A.C. (1997) Cytoplasmic vitrification and survival of anhydrobiotic organisms. Comp Biochem Physiol 117:327–333. [Google Scholar]
- Takahashi, A., Ohnishi, K., Takahashi, S., Masukawa, M., Sekikawa, K., Amano, T., Nakano, T., Nagaoka, S., and Ohnishi, T. (2001) The effects of microgravity on induced mutation in Escherichia coli and Saccharomyces cerevisiae. Adv Space Res 28:555–561. [DOI] [PubMed] [Google Scholar]
- Tamás, M.J., Luyten, K., Sutherland, F.C.W., Hernandez, A., Albertyn, J., Valadi, H., and Hohmann, S. (1999) Fps1p controls the accumulation and release of the compatible solute glycerol in yeast osmoregulation. Mol Microbiol 31:1087–1104. [DOI] [PubMed] [Google Scholar]
- Tapia, H. and Koshland, D.E. (2014) Trehalose is a versatile and long-lived chaperone for desiccation tolerance. Curr Biol 24:2758–2766. [DOI] [PubMed] [Google Scholar]
- Tapia, H., Young, L., Fox, D., Bertozzi, C.R., and Koshland, D. (2015) Increasing intracellular trehalose is sufficient to confer desiccation tolerance to Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 112:6122–6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokumasu, F., Jin, A.J., Feigenson, G.W., and Dvorak, J.A. (2003) Nanoscopic lipid domain dynamics revealed by atomic force microscopy. Biophys J 84:2609–2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe, A. and Wise, M.J. (2007) The continuing conundrum of the LEA proteins. Naturwissenschaften 94:791–812. [DOI] [PubMed] [Google Scholar]
- Veatch, S.L. and Keller, S.L. (2003) Separation of liquid phases in giant vesicles of ternary mixtures of phospholipids and cholesterol. Biophys J 85:3074–3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veatch, S.L. and Keller, S.L. (2005) Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta Mol Cell Res 1746:172–185. [DOI] [PubMed] [Google Scholar]